Abstract
Activation of the RIG-I-like receptors, RIG-I and MDA5, establishes an antiviral state by upregulating interferon (IFN)-stimulated genes (ISGs). Among these is ISG15 whose mechanistic roles in innate immunity still remain enigmatic. Here we report that ISG15 conjugation is essential for antiviral IFN responses mediated by the viral RNA sensor MDA5. ISGylation of the caspase activation and recruitment domains (CARD) of MDA5 promotes its oligomerization and thereby triggers activation of innate immunity against a range of viruses including coronaviruses, flaviviruses and picornaviruses. The ISG15-dependent activation of MDA5 is antagonized through direct de-ISGylation mediated by the papain-like protease (PLpro) of SARS-CoV-2, a recently emerged coronavirus that causes the COVID-19 pandemic. Our work demonstrates a crucial role for ISG15 in the MDA5-mediated antiviral response, and also identifies a key immune evasion mechanism of SARS-CoV-2, which may be targeted for the development of new antivirals and vaccines to combat COVID-19.
Introduction
Viral perturbation of host immune homeostasis is monitored by the innate immune system, which relies on receptors that sense pathogen- or danger-associated molecular patterns1, 2, 3. The RIG-I-like receptors (RLRs) RIG-I and MDA5 are pivotal for virus detection by surveying the cytoplasm for viral or host-derived immunostimulatory RNAs4. Binding of RNA to the C-terminal domain (CTD) and helicase of RIG-I and MDA5 leads to their signaling-primed conformation that allows for the recruitment of several enzymes5. These enzymes modify RLRs at multiple domains and sites, and posttranslational modifications (PTMs) are particularly well studied for the caspase activation and recruitment domains (CARDs), the signaling modules. PP1α/γ dephosphorylate the RIG-I and MDA5 CARDs6. In the case of RIG-I, dephosphorylation promotes K63-linked polyubiquitination of the CARDs by TRIM25 and other E3 ligases7, 8, which stabilizes the oligomeric form of RIG-I, thereby enabling MAVS binding. Compared to those of RIG-I, the individual steps of MDA5 activation and critical PTMs involved are less well understood.
RLR activation induces the production of type I and III interferons (IFNs) which, in turn, propagate antiviral signaling by upregulating ISGs9, 10. Among those is ISG15, a ubiquitin-like protein that can be covalently conjugated to lysine (K) residues of target proteins, a PTM process termed ISGylation11. Although ISG15 conjugation has been widely recognized to act antivirally12, mechanisms of host protein ISGylation that could explain the broad antiviral restriction activity of ISG15 are currently unknown.
The causative agent of the ongoing COVID-19 pandemic, severe acute respiratory syndrome coronavirus 2 (SCoV2), belongs to the Coronaviridae family that contains several other human pathogens. Coronaviruses have an exceptional capability to suppress IFN-mediated antiviral responses, and low IFN production in SCoV-2-infected patients correlated with severe disease13. Among the coronaviral IFN antagonists is the papain-like protease (PLpro) which has deubiquitinating and de-ISGylating activities14, 15.
Here we identify an essential role for ISGylation in MDA5 activation. We further show that SCoV2 PLpro interacts with MDA5 and antagonizes ISG15-dependent MDA5 activation via active de-ISGylation, unveiling that SCoV2 has already evolved to escape immune surveillance by MDA5.
Results
MDA5, but not RIG-I, signaling requires ISG15
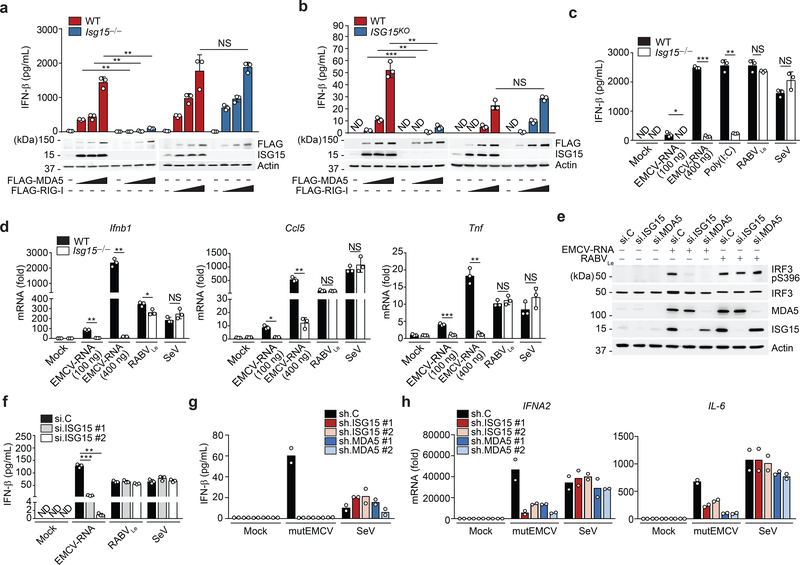
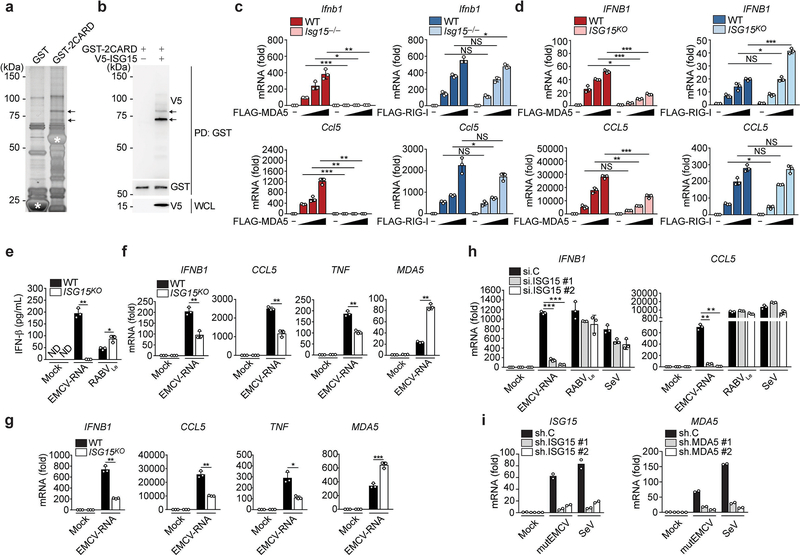
To identify PTMs of the MDA5 CARDs that may regulate MDA5 activation, we subjected affinity-purified MDA5–2CARD fused to glutathione-S-transferase (GST-MDA5–2CARD), or GST alone, to liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) and found that specifically GST-MDA5–2CARD co-purified with ISG15, which appeared as two bands that migrated more slowly (by ~15 and 30 kDa) than unmodified GST-MDA5–2CARD (Extended Data Fig. 1a). Immunoblotting confirmed that GST-MDA5–2CARD is modified by ISG15 (Extended Data Fig. 1b). We next determined the relevance of ISG15 for MDA5-induced signaling. Whereas FLAG-MDA5 expression in wild-type (WT) mouse embryonic fibroblasts (MEFs) induced IFN-β mRNA and protein as well as Ccl5 transcripts in a dose-dependent manner, FLAG-MDA5 expression in Isg15−/− MEFs led to ablated antiviral gene and protein expression (Fig. 1a and Extended Data Fig. 1c). Similarly, antiviral gene induction by FLAG-MDA5 was strongly diminished in ISG15 KO HeLa (human) cells compared to WT control cells (Fig. 1b and Extended Data Fig. 1d), ruling out a species-specific effect. In contrast, FLAG-RIG-I induced comparable amounts of secreted IFN-β protein as well as Ifnb1 and Ccl5 transcripts in Isg15−/− and WT MEFs (Fig. 1a and Extended Data Fig. 1c). IFNB1 transcripts and IFN-β protein production by FLAG-RIG-I were slightly enhanced in ISG15 KO HeLa cells as compared to WT cells (Fig. 1b and Extended Data Fig. 1d), consistent with previous reports that ISGylation negatively impacts RIG-I signaling16, 17.
Fig. 1 |. ISGylation is required for MDA5-, but not RIG-I, signaling.
a, b, ELISA of IFN-β from supernatants of MEFs (WT or Isg15−/−) (a) and HeLa cells (WT or ISG15 KO) (b) transiently transfected with increasing amounts of FLAG-tagged MDA5 or RIG-I for 40 h. Whole cell lysates (WCLs) were probed by immunoblotting (IB) with anti-ISG15, anti-FLAG, and anti-Actin (loading control). c, ELISA of IFN-β from supernatants of WT or Isg15−/− MEFs that were mock-stimulated or transfected with EMCV-RNA (0.1 or 0.4 μg/mL), HMW-poly (I:C) (0.5 μg/mL), or RABVLe (1 pmol/mL), or infected with SeV (10 HAU/mL) for 24 h. d, Quantitative RT-PCR (qRT-PCR) analysis of IFNB1 and CCL5 mRNA in WT and Isg15−/− MEFs stimulated as in (c). e, IRF3 phosphorylation in the WCLs of NHLFs that were transfected with the indicated siRNAs for 30 h and then mock-stimulated or transfected with EMCV-RNA (0.4 μg/mL) or RABVLe (1 pmol/mL) for 6 h, assessed by IB with anti-pS396-IRF3 and anti-IRF3. f, ELISA of IFN-β from supernatants of NHLFs that were transfected with the indicated siRNAs for 30 h and then mock-stimulated or transfected with EMCV-RNA (0.4 μg/mL) or RABVLe (1 pmol/mL), or infected with SeV (10 HAU/mL) for 16 h. g, ELISA of IFN-β from the supernatants of PBMCs that were transduced for 40 h with the indicated shRNAs and then infected with mutEMCV (MOI 10) or SeV (200 HAU/mL) for 8 h. h, qRT-PCR analysis of IFNA2 and IL-6 mRNA in PBMCs that were transduced and infected as in (g). Data are representative of at least two independent experiments with similar results (mean ± s.d. of n = 3 biological replicates in a, b, c, d, f and mean of n = 2 biological replicates in g and h). *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed unpaired t-test). ND, not detected; NS, not significant.
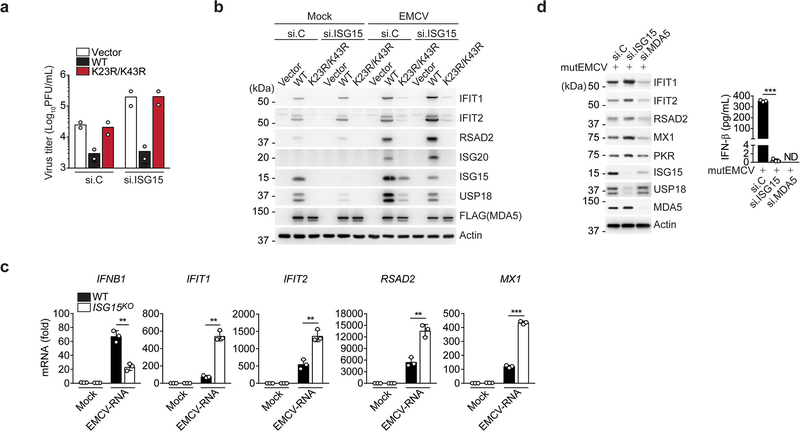
We next tested the effect of ISG15-gene deletion on the activation of endogenous MDA5 and RIG-I by their respective ligands. IFN-β production as well as IFNB1, CCL5, and TNF gene expression induced by transfection of encephalomyocarditis virus (EMCV)-RNA or high-molecular-weight (HMW)-poly(I:C), both of which are predominantly sensed by MDA5, were profoundly attenuated in Isg15−/− MEF, ISG15 KO HeLa, and ISG15 KO HAP-1 cells as compared to their respective control cells (Fig. 1c,d and Extended Data Fig. 1e–g). Importantly, the ablation of antiviral gene induction by EMCV-RNA or HMW-poly(I:C) in ISG15 KO cells was not due to abrogated MDA5 gene expression; on the contrary, MDA5 mRNA expression was enhanced in ISG15 KO cells as compared to WT cells (Extended Data Fig. 1f,g). In contrast to stimulation with MDA5 agonists, stimulation of Isg15−/− MEFs and ISG15 KO HeLa cells by rabies virus leader RNA (RABVLe) transfection or Sendai virus (SeV) infection, which are RIG-I stimuli, led to IFN-β production and antiviral gene expression comparable to WT cells (Fig. 1c,d and Extended Data Fig. 1e). To rule out potential clonal effects that could be associated with ISG15 gene-deleted cells, we performed transient gene silencing experiments in primary normal human lung fibroblasts (NHLFs). ISG15 silencing, similarly to MDA5 knockdown, led to a near-complete loss of phosphorylation of IFN-regulatory factor 3 (IRF3) – a hallmark of RLR-signal activation – upon stimulation with EMCV-RNA but not RABVLe (Fig. 1e). In accord, ISG15 knockdown greatly diminished IFN-β production as well as antiviral transcript expression in NHLFs transfected with EMCV-RNA, but not in cells stimulated with RABVLe or SeV (Fig. 1f and Extended Data Fig. 1h). shRNA-mediated silencing of ISG15 or MDA5 in primary human peripheral blood mononuclear cells (PBMCs) also substantially reduced antiviral protein and transcript expression following infection with a recombinant mutant EMCV (mutEMCV) deficient in MDA5 antagonism18, 19, as compared to infected PBMCs transduced with non-targeting control shRNA (Fig. 1g,h and Extended Data Fig. 1i). By contrast, ISG15 or MDA5 depletion did not affect cytokine responses in PBMCs following SeV infection (Fig. 1g,h and Extended Data Fig. 1i). These results show that ISG15 is essential for immune signaling by MDA5, but not RIG-I.
The MDA5 CARDs are ISGylated at K23 and K43
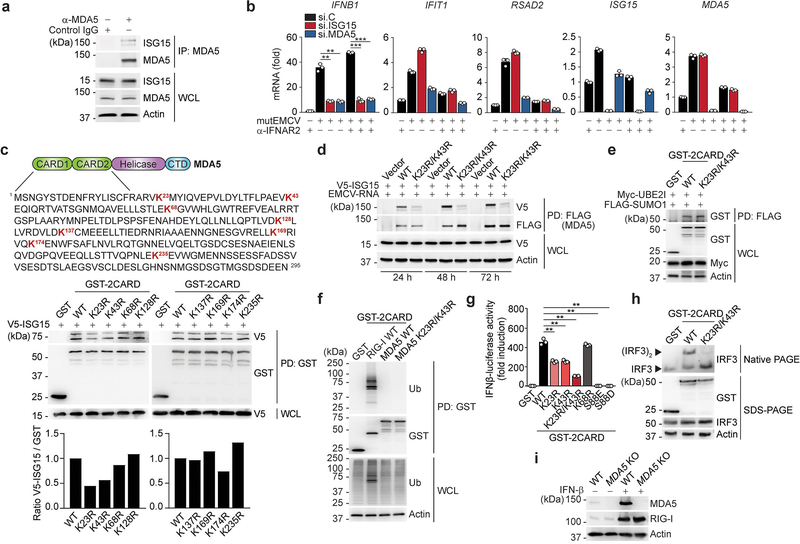
To corroborate our MS analysis that identified MDA5–2CARD ISGylation, we first tested whether also endogenous MDA5 is modified by ISG15. Endogenous MDA5 was robustly ISGylated in cells transfected with HMW-poly(I:C), or infected with dengue (DENV) or Zika (ZIKV) viruses that are sensed by MDA55 (Fig. 2a). Notably, endogenous MDA5 was also ISGylated in uninfected cells, although at very low levels (Extended Data Fig. 2a), which is consistent with previous findings that many host proteins are ISGylated at low levels also in normal (uninfected) conditions20. In cells treated with anti-IFNAR2 to block IFNAR-signaling-mediated ISG upregulation, ISG15 or MDA5 silencing led to a comparable reduction of IFNB1 gene expression following mutEMCV infection (Extended Data Fig. 2b), indicating that ISG15-dependent MDA5 signaling occurs even in the absence of IFNAR signaling.
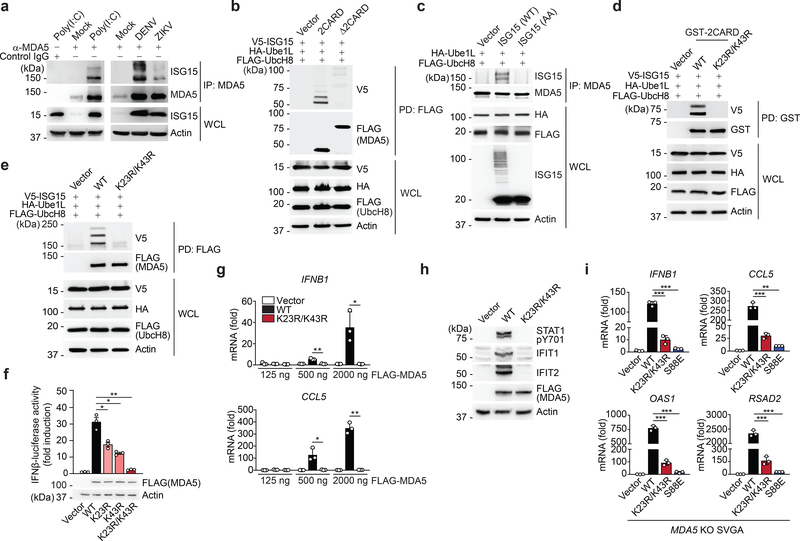
Fig. 2 |. MDA5 activation requires ISGylation at K23 and K43.
a, Endogenous MDA5 ISGylation in NHLFs that were mock-treated, transfected with HMW-poly (I:C) (0.1 μg/mL) for 40 h (left), or infected with DENV or ZIKV (MOI 1 for each) for 48 h (right), determined by immunoprecipitation (IP) with anti-MDA5 (or an IgG isotype control) and IB with anti-ISG15. b, ISGylation of FLAG-tagged MDA5–2CARD and MDA5ΔCARD in transiently transfected HEK293T cells that also expressed V5-ISG15, HA-Ube1L, and FLAG-UbcH8, assessed by FLAG pulldown (PD) and IB with anti-V5 at 40 h post-transfection. c, Endogenous MDA5 ISGylation in ISG15 KO HeLa cells stably reconstituted with vector, WT ISG15 or ISG15-AA and co-transfected with HA-Ube1L and FLAG-UbcH8 after IFN-β treatment (1,000 U/mL) for 24 h, determined by IP with anti-MDA5 and IB with anti-ISG15. d, ISGylation of GST-MDA5–2CARD WT and K23R/K43R in HEK293T cells that were co-transfected with V5-ISG15, HA-Ube1L, and FLAG-UbcH8 for 24 h, determined by GST-PD and IB with anti-V5. e, ISGylation of FLAG-MDA5 WT and K23R/K43R in HEK293T cells that were co-transfected with V5-ISG15, HA-Ube1L, and FLAG-UbcH8, determined by FLAG-PD and IB with anti-V5. f, IFN-β-luciferase reporter activity in HEK293T cells that were transfected for 40 h with vector, or FLAG-MDA5 WT or mutants. Luciferase values are presented as fold induction relative to the values for vector-transfected cells, set to 1. WCLs were probed by IB with anti-FLAG and anti-Actin. g, qRT-PCR analysis of IFNB1 and CCL5 mRNA in HEK293T cells that were transiently transfected with either vector, or increasing amounts of FLAG-MDA5 WT or K23R/K43R. h, STAT1 phosphorylation and ISG (IFIT1 and 2) protein abundance in the WCLs of HEK293T cells that were transiently transfected with vector or FLAG-MDA5 WT or K23R/K43R, determined by IB. i, qRT-PCR analysis of the indicated antiviral genes in MDA5 KO SVGAs that were reconstituted with either empty vector or FLAG-tagged MDA5 WT, K23R/K43R or S88E. Data are representative of at least two independent experiments with similar results (mean ± s.d. of n = 3 biological replicates in f, g, and i). *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed unpaired t-test). NS, not significant.
Biochemical analysis confirmed that the MDA5–2CARD, but not MDA5 Δ2CARD (containing helicase and CTD), is the primary site of MDA5 ISGylation showing two prominent bands for ISGylated 2CARD (Fig. 2b). Reconstitution of ISG15 KO HeLa cells with either WT ISG15, or an unconjugatable ISG15 mutant in which the two glycines needed for conjugation were replaced with alanine (ISG15 AA)21, demonstrated covalent ISG15 conjugation (Fig. 2c).
Mutation of individual K residues in GST-MDA5–2CARD to arginine (R) revealed that single-site mutation of K23 and K43 noticeably reduced ISGylation (Extended Data Fig. 2c), while their combined mutation (K23R/K43R) near-abolished ISGylation (Fig. 2d). Full-length FLAG-MDA5 K23R/K43R also showed markedly diminished ISGylation (Fig. 2e and Extended Data Fig. 2d); the residual ISGylation seen in FLAG-MDA5 K23R/K43R is likely due to additional, minor sites in the CARD and/or Δ2CARD. Of note, the K23R/K43R mutation had no effect on MDA5–2CARD SUMOylation5 (Extended Data Fig. 2e). Furthermore, whereas RIG-I-2CARD was robustly ubiquitinated (which represents K63-linked ubiquitination7), neither MDA5–2CARD WT nor the K23R/K43R mutant showed detectable levels of ubiquitination (Extended Data Fig. 2f). Collectively, these results indicate that the MDA5 CARDs undergo ISGylation at two major sites, K23 and K43.
CARD ISGylation is required for MDA5 activation
When comparing their signal-transducing ability, MDA5–2CARD K23R and K43R single-site mutants showed partially reduced IFN-β promoter activation compared to WT MDA5–2CARD, while the K23R/K43R mutant had a profoundly reduced signaling activity, which was almost as strong as that of the signaling-defective mutants S88E and S88D6 (Fig. 2g and Extended Data Fig. 2g). In contrast, a mutant in which K68, which is the lysine residue that is most proximal to K43 and K23, was substituted with arginine (K68R), showed comparable ISG15 conjugation and signaling competency to WT 2CARD (Extended Data Fig. 2c,g). MDA5–2CARD K23R/K43R, in contrast to WT MDA5–2CARD, also failed to induce IRF3 dimerization (Extended Data Fig. 2h). FLAG-MDA5 K23R, K43R, or K23R/K43R also showed reduced and near-abolished IFN-β promoter-activating abilities, respectively, as compared to FLAG-MDA5 WT (Fig. 2f). MDA5 K23/K43R showed a profound signaling defect even when expressed at high amounts, while WT MDA5 induced antiviral transcripts in a dose-dependent manner (Fig. 2g). In accord, STAT1 phosphorylation, a hallmark of IFNAR signaling, as well as ISG protein expression were highly induced by MDA5 WT, but not K23R/K43R (Fig. 2h). Complementation of MDA5-gene-edited human astrocytes (SVGA) with MDA5 K23R/K43R or S88E led to greatly diminished IFNB1, CCL5, and ISG transcripts compared to cells expressing WT MDA5 (Fig. 2i and Extended Data Fig. 2i). These results demonstrate that ISGylation at K23 and K43 is essential for MDA5-mediated cytokine responses.
Dephosphorylation by PP1 regulates MDA5 ISGylation
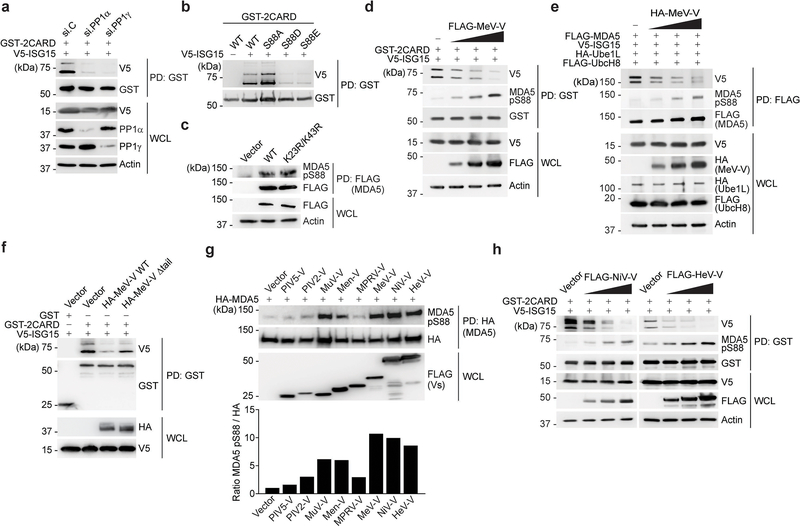
Like RIG-I, MDA5 is phosphorylated within the CARDs in uninfected cells, which prevents auto-activation; dephosphorylation of RIG-I and MDA5 by PP1α/γ is crucial for unleashing RLRs from their signaling-repressed states6, 22, 23, 24. Dephosphorylation of RIG-I allows K63-linked ubiquitination of the CARDs, which promotes RIG-I multimerization and signaling5. The details of how CARD dephosphorylation (at S88) triggers MDA5 activation have remained elusive, and therefore we tested whether dephosphorylation regulates MDA5 ISGylation. Silencing of PP1α or PP1γ strongly diminished MDA5–2CARD ISGylation (Extended Data Fig. 3a). Furthermore, the phosphomimetic S88E and S88D mutants had reduced ISGylation, whereas the ‘phospho-null’ S88A mutant showed stronger ISGylation than WT MDA5 (Extended Data Fig. 3b). Conversely, MDA5 WT and K23R/K43R had comparable S88 phosphorylation (Extended Data Fig. 3c). Together, these data suggested that MDA5 dephosphorylation at S88 precedes CARD ISGylation.
We next made use of the measles virus V protein (MeV-V), which antagonizes MDA5 S88 dephosphorylation through PP1α/γ antagonism25. MeV-V expression enhanced the S88 phosphorylation (indicative of ablated dephosphorylation) of GST-MDA5–2CARD or FLAG-MDA5 in a dose-dependent manner, as previously shown25. Enhanced phosphorylation by MeV-V correlated with a decline in ISGylation (Extended Data Fig. 3d,e). In contrast to WT MeV-V, a mutant MeV-V that has abolished PP1-binding and MDA5-dephosphorylation antagonism (MeV-V Δtail)25, exhibited little effect on MDA5–2CARD ISGylation (Extended Data Fig. 3f), strengthening that the inhibition of ISGylation is primarily due to PP1 inhibition, and not other antagonistic effects, by MeV-V. The V proteins from Nipah and Hendra viruses (NiV-V and HeV-V) also enhanced MDA5 S88 phosphorylation and, correspondingly, dampened MDA5 ISGylation (Extended Data Fig. 3g,h), suggesting that several paramyxoviral V proteins inhibit MDA5 ISGylation through manipulation of S88 phosphorylation, although the precise mechanisms for individual V proteins remain to be determined. Taken together, these data suggest that the MDA5 CARD ISGylation is dependent on dephosphorylation at S88.
ISGylation promotes higher-order MDA5 assemblies
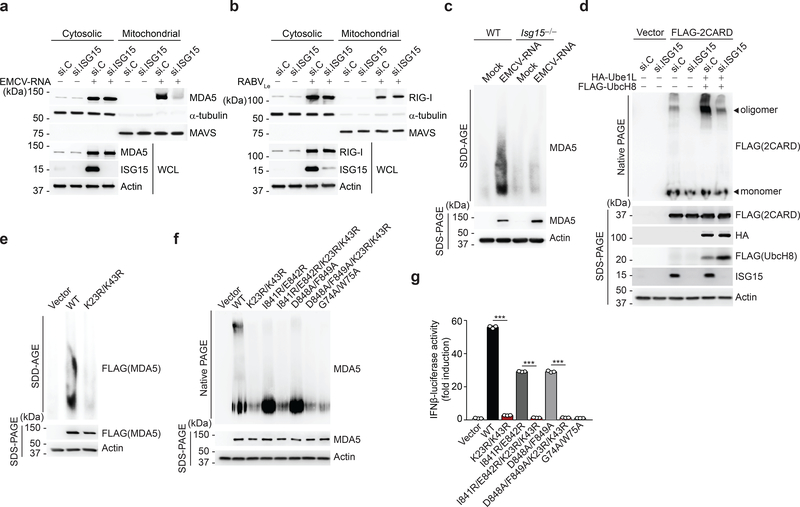
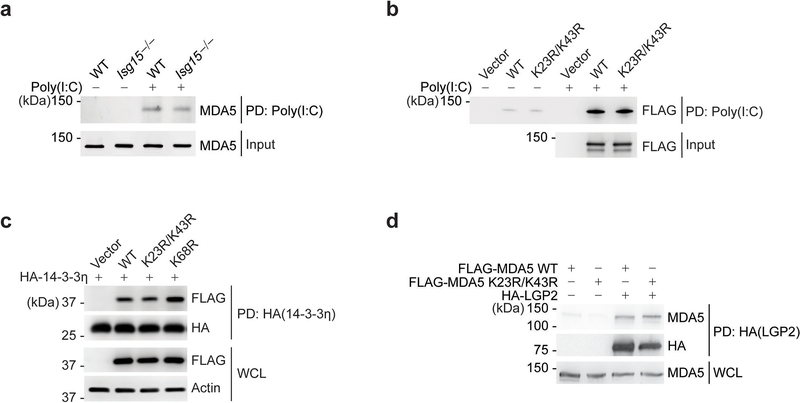
RLR activation requires RNA binding, RLR oligomerization, and their translocation from the cytosol to mitochondria for an interaction with MAVS5. To elucidate the mechanism by which ISGylation impacts MDA5 activity, we first examined whether ISGylation affects RNA binding. Endogenous MDA5 purified from WT or Isg15−/− MEFs interacted equally well with HMW-poly(I:C) in vitro (Extended Data Fig. 4a). MDA5 WT and K23R/K43R showed comparable binding to HMW-poly(I:C), indicating that ISGylation does not affect the RNA-binding ability of MDA5 (Extended Data Fig. 4b). When we monitored the translocation of MDA5 from the cytosol to mitochondria upon EMCV-RNA stimulation we found that ISG15 silencing, but not si.C transfection, abolished MDA5 translocation (Fig. 3a). In contrast, RIG-I translocation following RABVLe transfection was efficient in both ISG15-depleted and si.C-transfected cells (Fig. 3b). These data indicated that ISGylation regulates MDA5 translocation, or a step upstream of it. Since the cytosol-to-mitochondria translocation of MDA5 requires an interaction with 14–3-3η26, we compared 14–3-3η-binding of WT and mutant MDA5. The ability of MDA5 K23R/K43R to bind 14–3-3η was similar to that of WT MDA5 or the K68R mutant (Extended Data Fig. 4c). However, whereas EMCV-RNA stimulation effectively induced MDA5 oligomerization in WT MEFs, the formation of MDA5 oligomers was ablated in ISG15-deficient MEFs (Fig. 3c). ISG15 knockdown in 293T cells also abolished the oligomerization of FLAG-MDA5–2CARD (Fig. 3d). Conversely, co-expression of the ISGylation machinery components, Ube1L and UbcH8, strongly enhanced MDA5–2CARD oligomerization in si.C-transfected cells, but not in ISG15-depleted cells (Fig. 3d), indicating that ISGylation is required for MDA5 oligomer formation. In support of this concept, FLAG-MDA5 K23R/K43R showed near-abolished oligomerization, while WT MDA5 oligomerized efficiently (Fig. 3e). We also compared the effect of the K23R/K43R mutation with that of oligomerization-disruptive mutations which localize either to the interface between MDA5 monomers and impede RNA-binding-mediated MDA5 filamentation (I841R/E842R and D848A/F849A)27, 28, or to the CARDs (G74A/W75A) and disrupt 2CARD oligomerization27. Unlike WT MDA5, the K23R/K43R mutant, similarly to MDA5 G74A/W75A, showed deficient oligomerization and, consistent with this, abolished IFN-β promoter-activating ability (Fig. 3f,g). Introduction of K23R/K43R into the I841R/E842R or D848A/F849A background, either of which by itself decreased MDA5 oligomerization and signaling, also abolished MDA5 oligomer formation and IFN-β induction (Fig. 3f,g). Since LGP2 facilitates MDA5 nucleation on dsRNA and thereby MDA5 oligomerization29, 30, we compared LGP2 binding of MDA5 WT and K23R/K43R. MDA5 K23R/K43R interacted with LGP2 as efficiently as WT MDA5 (Extended Data Fig. 4d), strengthening that CARD ISGylation promotes MDA5 oligomerization independently of RNA-binding-mediated filamentation. Collectively, these results establish that ISGylation facilitates CARD oligomerization and higher-order MDA5 assemblies.
Fig. 3 |. CARD ISGylation is essential for formation of higher-order MDA5 assemblies.
a,b, Cytosol-mitochondria fractionation of WCLs from NHLFs that were transfected for 30 h with non-targeting control siRNA (si.C) or ISG15-specific siRNA (si.ISG15) and then mock-treated or transfected with EMCV-RNA (0.4 μg/mL) (a) or RABVLe (1 pmol/mL) (b) for 16 h. IB was performed with anti-MDA5 (a), anti-RIG-I (b), anti-ISG15 and anti-Actin (a, b). α-Tubulin and MAVS served as purity markers for the cytosolic and mitochondrial fraction, respectively (a, b). c, Endogenous MDA5 oligomerization in WT and Isg15−/− MEFs that were transfected with EMCV-RNA (0.5 μg/mL) for 16 h, assessed by SDD-AGE and IB with anti-MDA5. WCLs were further analyzed by SDS-PAGE and probed by IB with anti-MDA5 and anti-Actin. d, Oligomerization of FLAG-MDA5–2CARD in HEK293T cells that were transfected with the indicated siRNAs together with or without HA-Ube1L and FLAG-UbcH8 for 48 h, determined by native PAGE and IB with anti-FLAG. WCLs were further analyzed by SDS-PAGE and probed by IB with anti-FLAG, anti-HA, anti-ISG15, and anti-Actin. e, Oligomerization of FLAG-MDA5 WT and K23R/K43R in transiently transfected MDA5 KO HEK293 cells, assessed by SDD-AGE and IB with anti-FLAG. WCLs were further analyzed by SDS-PAGE and IB with anti-FLAG and anti-Actin. f, Oligomerization of FLAG-tagged MDA5 WT and mutants in transiently transfected MDA5 KO HEK293 cells, assessed by native PAGE and IB with anti-MDA5. WCLs were further analyzed by SDS-PAGE and probed by IB with anti-MDA5 and anti-Actin. g, IFN-β-luciferase reporter activity in MDA5 KO HEK293 cells that were transfected for 24 h with either empty vector, or FLAG-tagged MDA5 WT or mutants. Luciferase activity is presented as fold induction relative to the values for vector-transfected cells, set to 1. Data are representative of at least two independent experiments with similar results (mean ± s.d. of n = 3 biological replicates in f). ***p < 0.001 (two-tailed unpaired t-test).
ISGylation-dependent MDA5 signaling restricts virus replication
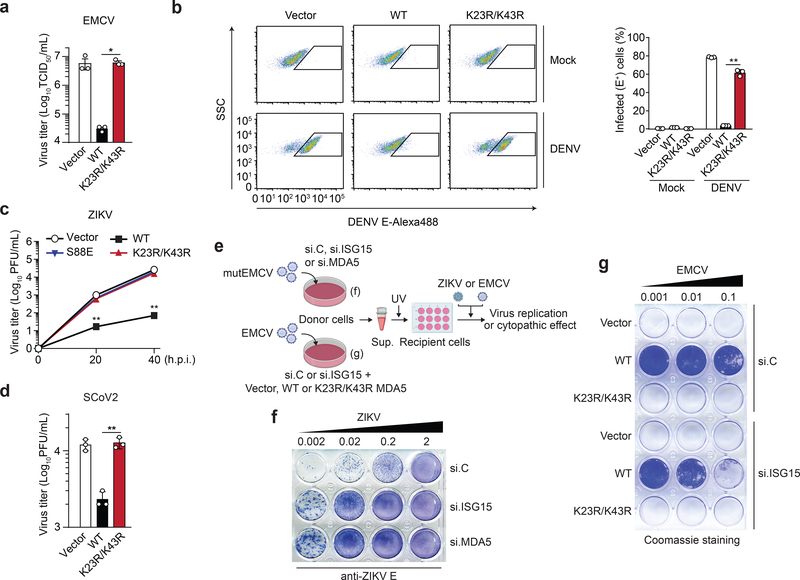
We next assessed whether ISGylation of MDA5 is required for its ability to restrict virus replication. FLAG-MDA5 WT, but not K23R/K43R, potently (by ~2-log) inhibited EMCV replication (Fig. 4a). Similarly, MDA5 KO HEK293 cells reconstituted with WT MDA5, but not cells complemented with the K23R/K43R mutant, effectively restricted DENV replication (Fig. 4b). We also reconstituted MDA5 KO astrocyte SVGAs, a physiologically relevant cell type for ZIKV infection, with either vector, or MDA5 WT or K23R/K43R and then assessed ZIKV replication over a 40-hour time course. ZIKV replication was attenuated by ~100-fold in cells reconstituted with WT MDA5 as compared to vector-expressing cells. In contrast, cells complemented with MDA5 K23R/K43R did not restrict ZIKV, similarly to cells expressing MDA5 S88E (Fig. 4c). WT MDA5, but not K23R/K43R, also restricted SCoV2 replication, although to a lesser extent than that seen for the other viruses tested (Fig. 4d).
Fig. 4 |. ISGylation is required for viral restriction by MDA5.
a, EMCV titers in the supernatant of HEK293T cells that were transfected for 40 h with either vector, or FLAG-MDA5 WT or K23/K43R and then infected with EMCV (MOI 0.001) for 24 h, determined by TCID50 assay. b, Percentage of DENV-infected MDA5 KO HEK293 cells that were transfected for 24 h with either vector or FLAG-MDA5 WT or K23R/K43R and then mock-treated or infected with DENV (MOI 5) for 48 h, assessed by FACS using anti-flavivirus E (4G2). SSC, side scatter. c, ZIKV titers in the supernatant of MDA5 KO SVGAs that were transfected for 30 h with vector or FLAG-tagged MDA5 WT, K23R/K43R, or S88E and then infected with ZIKV (MOI 0.1) for the indicated times, determined by plaque assay. d, SCoV2 titers in the supernatant of HEK293T-hACE2 cells that were transfected for 24 h with either empty vector, or FLAG-MDA5 WT or K23/K43R and then infected with SCoV2 (MOI 0.5) for 24 h, determined by plaque assay. e, Schematic of the experimental approach to ‘decouple’ the role of ISG15 in MDA5-mediated IFN induction from its role in dampening IFNAR signaling. f, NHLF ‘donor’ cells were transfected for 40 h with the indicated siRNAs and then infected with mutEMCV (MOI 0.1) for 16 h. Cell supernatants were UV-inactivated and transferred onto Vero ‘recipient’ cells. 24 h later, cells were infected with ZIKV (MOI 0.002 to 2) for 72 h, and ZIKV-positive cells determined by immunostaining with anti-flavivirus E (4G2) and TrueBlue peroxidase substrate. g, RIG-I KO HEK293 ‘donor’ cells were transfected with si.C or si.ISG15 together with either vector or FLAG- MDA5 WT or K23R/K43R for 24 h, followed by EMCV infection (MOI 0.001) for 16 h. UV-inactivated cell supernatants were transferred onto Vero ‘recipient’ cells for 24 h, followed by infection with EMCV (MOI 0.001 to 0.1) for 40 h. EMCV-induced cytopathic effects were visualized by Coomassie blue staining. Data are representative of at least two independent experiments with similar results (mean ± s.d. of n = 3 biological replicates in a, b, c). *p < 0.05, **p < 0.01 (two-tailed unpaired t-test).
We next determined the effect of ISG15 silencing on MDA5’s ability to inhibit virus replication. While MDA5 K23R/K43R failed to suppress EMCV replication regardless of ISG15 silencing, WT MDA5 effectively restricted EMCV replication in si.C-transfected cells, and unexpectedly, also in ISG15–depleted cells (Extended Data Fig. 5a). In an exploration of the underlying mechanism of these unexpected results, we found that the EMCV-infected cells that expressed WT MDA5 had markedly enhanced levels of ISG protein expression when ISG15 was silenced as compared to infected cells transfected with si.C (Extended Data Fig. 5b). Similarly, elevated ISG transcript and protein expression was observed in ISG15-deficient cells that were transfected with EMCV-RNA or infected with mutEMCV, despite abrogation of IFN-β induction (Extended Data Fig. 5c,d). In contrast, MDA5 knockdown abrogated both IFN-β and ISG protein expression, as expected (Extended Data Fig. 5d). We noticed that the protein abundance of USP18, a deubiquitinating enzyme that negatively regulates IFNAR signaling31, was greatly diminished in ISG15-depleted cells upon EMCV infection as compared to infected cells that were transfected with si.C or MDA5-specific siRNA (Extended Data Fig. 5b,d), which is consistent with the reported role of ISG15 in preventing USP18 degradation32. Together, these data suggested that in experimental settings of ISG15-gene targeting (i.e. silencing or KO) the antiviral effect of MDA5 ISGylation is masked by aberrant ISG upregulation due to the ablation of ISG15’s inhibitory effect on IFNAR signaling.
We next employed a virus protection assay that experimentally decouples MDA5 signaling in virus-infected cells from downstream IFNAR signaling in the same cells (Fig. 4e). Supernatants from mutEMCV-infected ‘donor’ cells that were either si.C-transfected, or depleted of either ISG15 or MDA5, were UV-inactivated and then transferred onto uninfected ‘recipient’ cells. ‘Primed’ recipient cells were then infected with ZIKV to directly monitor the antiviral effect of MDA5-mediated IFN production by donor cells. Whereas the supernatants from si.C-transfected ‘donor’ cells potently inhibited ZIKV replication, the supernatants from ISG15 or MDA5 knockdown cells minimally restricted ZIKV infection (Fig. 4f). Similarly, the culture supernatants from EMCV-infected ‘donor’ cells transfected with WT MDA5 together with si.C led to greater protection of ‘recipient’ cells from viral challenge than that from cells expressing WT MDA5 and depleted of ISG15 (Fig. 4g). Collectively, these data demonstrate that ISGylation is important for MDA5-mediated restriction of a range of RNA viruses.
SCoV2 PLpro targets MDA5 for de-ISGylation
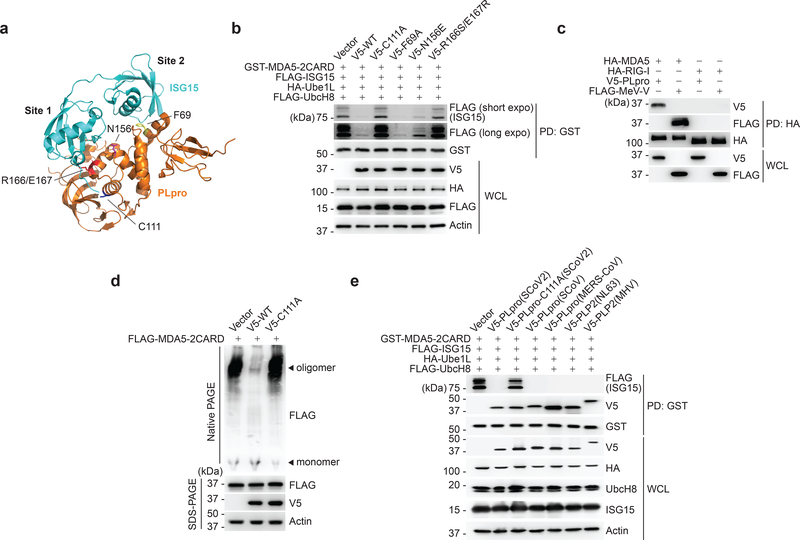
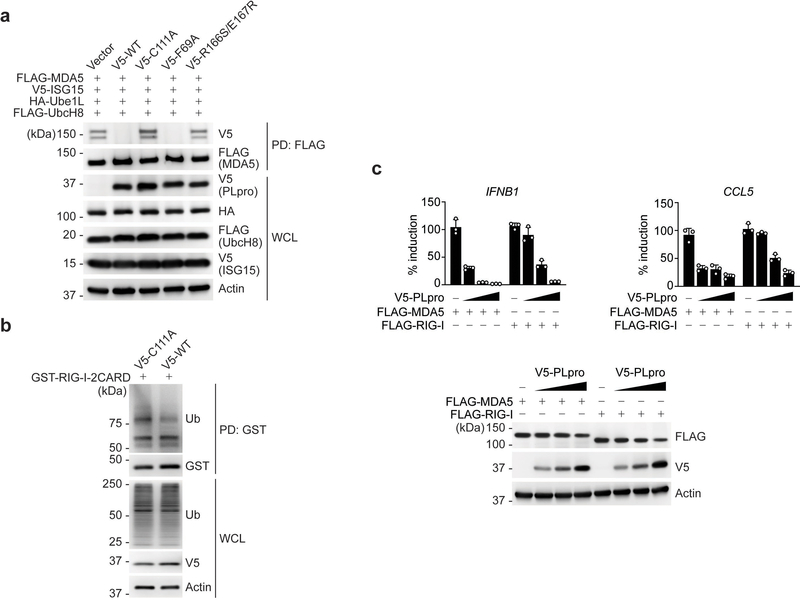
Coronaviruses such as SARS-CoV (SCoV), MERS-CoV, and the recently emerged SCoV2, encode a papain-like protease (PLpro) that mediates viral polyprotein cleavage33. In addition, PLpro has deubiquitinating and de-ISGylating activities. SCoV2 PLpro was recently shown to modulate antiviral responses primarily via its de-ISGylase activity15. Since MDA5 is known to be a major sensor for detecting coronaviruses34, 35, and because our data showed that ISGylation is required for MDA5-mediated virus restriction, we examined whether SCoV2 PLpro enzymatically removes MDA5 ISGylation to antagonize innate immunity. SCoV2 PLpro WT, but not its catalytically-inactive mutant (PLpro-C111A)15, abolished the ISGylation of GST-MDA5–2CARD and FLAG-MDA5 (Fig. 5a,b and Extended Data Fig. 6a). The PLpro N156E and R166S/E167R mutants, which are marginally and severely impaired in ISG15 binding at the ‘site 1’ interface respectively14, 36, did slightly, or not, affect ISGylation. In contrast, PLpro F69A, in which the ‘site 2’ interface that preferentially determines binding to ubiquitin, but not ISG15, is disrupted14, 36, diminished MDA5 ISGylation as potently as WT PLpro (Fig. 5a,b and Extended Data Fig. 6a). SCoV2 PLpro, however, did not suppress RIG-I-2CARD ubiquitination (Extended Data Fig. 6b).
Fig. 5 |. SCoV2 PLpro binds to and de-ISGylates MDA5–2CARD.
a, Ribbon representation of the crystal structure of the SCoV2 PLpro: ISG15 complex (PDB: 6YVA). Key residues that mediate ‘site 1’ interaction (N156 and R166/E167) or ‘site 2’ interaction (F69) in PLpro, as well as its catalytically-active site (C111), are indicated. b, ISGylation of GST-MDA5–2CARD in HEK293T cells that were co-transfected for 20 h with vector or V5-tagged SCoV2 PLpro WT or mutants, along with FLAG-ISG15, HA-Ube1L, and FLAG-UbcH8, determined by GST-PD and IB with anti-FLAG and anti-GST. WCLs were probed by IB with anti-V5, anti-HA, anti-FLAG, and anti-Actin. (c) Binding of HA-tagged MDA5 or RIG-I to V5-SCoV2-PLpro or FLAG-MeV-V (positive control) in transiently transfected HEK293T cells, determined by HA-PD and IB with anti-V5 or anti-FLAG, and anti-HA. WCLs were probed by IB with anti-V5 and anti-FLAG. d, Oligomerization of FLAG-MDA5–2CARD in HEK293T cells that were co-transfected with vector, or V5-SCoV2 PLpro WT or C111A for 24 h, assessed by Native PAGE and IB with anti-FLAG. WCLs were further analyzed by SDS-PAGE and probed by IB with anti-FLAG, anti-V5 and anti-Actin. e, ISGylation of GST-MDA5–2CARD in HEK293T cells that also expressed FLAG-ISG15, HA-Ube1L and FLAG-UbcH8, and were co-transfected for 40 h with vector or the indicated V5-tagged coronaviral PLpro, determined by GST-PD and IB with anti-FLAG, anti-V5, and anti-GST. Data are representative of at least two independent experiments with similar results.
We found that PLpro interacted specifically with MDA5, but not RIG-I, as did MeV-V which binds MDA5 and served as control37 (Fig. 5c). Low amounts of PLpro inhibited signaling by MDA5, but not RIG-I, whereas higher amounts of PLpro suppressed antiviral signaling by both RLRs (Extended Data Fig. 6c). This strengthens that MDA5 is a direct target of PLpro. De-ISGylation of IRF3 likely accounts for the inhibitory effect that higher doses of PLpro have on RLR signaling15, 38.
When examining the effect of PLpro on MDA5–2CARD oligomerization, PLpro WT but not C111A efficiently blocked MDA5–2CARD oligomerization (Fig. 5d), indicating that SCoV2 PLpro inhibits the ISGylation-dependent MDA5 oligomer formation via its enzymatic activity.
The PLpro enzymes of the related beta-coronaviruses, SCoV, MERS-CoV, and murine hepatitis virus (MHV), as well as of the alpha-coronavirus HCoV-NL63 (NL63) also bound to and efficiently reduced MDA5–2CARD ISGylation (Fig. 5e), suggesting that MDA5 antagonism by PLpro may be widely conserved among coronaviruses.
SCoV2 PLpro antagonizes ISG15-dependent MDA5 signaling
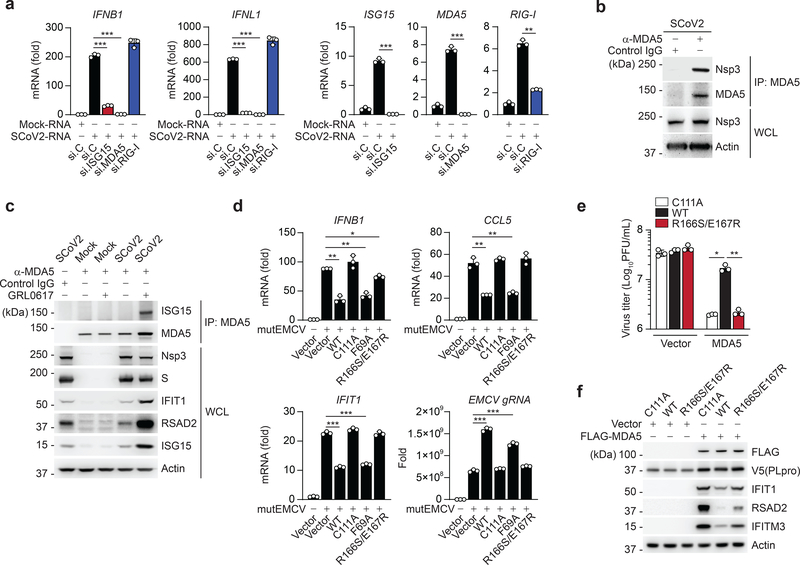
We next determined the relevance of ISG15-dependent MDA5 signaling for antiviral cytokine induction elicited by SCoV2. Since SCoV2 infection is known to minimally induce type I IFNs due to effective viral antagonisms39, we isolated total RNA from SCoV2-infected cells and then re-transfected it into cells to stimulate innate immune signaling. SCoV2-RNA, but not RNA from mock-treated cells, robustly induced IFN transcripts; however, this induction was markedly diminished when ISG15 or MDA5 was silenced (Fig. 6a). RIG-I knockdown did not adversely affect the antiviral gene expression elicited by SCoV2-RNA, indicating that SCoV2 RNA-PAMP(s) are primarily sensed by the ISG15-MDA5 axis (Fig. 6a).
Fig. 6 |. SCoV2 PLpro inhibits ISG15-mediated MDA5 signaling via its deISGylase activity.
a, qRT-PCR analysis of IFNB1, IFNL1, ISG15, MDA5, and RIG-I transcripts in NHLFs that were transfected with the indicated siRNAs for 40 h and then transfected with mock-RNA or SCoV2-RNA (0.4 μg/mL) for 24 h. b, Binding of SCoV2 Nsp3 to endogenous MDA5 in A549-hACE2 cells that were infected with SCoV2 (MOI 0.5) for 24 h, determined by IP with anti-MDA5 (or an IgG isotype control) followed by IB with anti-PLpro and anti-MDA5. WCLs were probed by IB with anti-PLpro (Nsp3) and anti-Actin. c, Endogenous MDA5 ISGylation in A549-hACE2 cells that were mock-infected or infected with SCoV2 (MOI 0.5) for 40 h in the presence of PLpro inhibitor (GRL-0617; 50 μM) or vehicle control (DMSO), determined by IP with anti-MDA5 (or an IgG isotype control) followed by IB with anti-ISG15 and anti-MDA5. Protein abundance of IFIT1, RSAD2, ISG15 and actin in the WCLs were probed by IB. Efficient virus replication was verified by immunoblotting WCLs with anti-PLpro (Nsp3) and anti-Spike (S). d, qRT-PCR analysis of IFNB1, CCL5, IFIT1 transcript, and EMCV genomic RNA (gRNA) in HeLa cells that were transiently transfected for 24 h with vector, or V5-SCoV2 PLpro WT or mutants and then infected with mutEMCV (MOI 0.5) for 12 h. e, EMCV titers in the supernatant of RIG-I KO HEK293 cells that were transiently transfected for 24 h with vector or FLAG-MDA5 along with V5-SCoV2 PLpro WT, C111A, or R166S/E167R and then infected with EMCV (MOI 0.001) for 16 h, determined by plaque assay. f, Protein abundance of the indicated ISGs in the WCLs from the experiment in (e), determined by IB with the indicated antibodies. Data are representative of at least two independent experiments with similar results (mean ± s.d. of n = 3 biological replicates in a, d, e). *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed unpaired t-test).
We found that SCoV2 non-structural protein 3 (Nsp3), within which PLpro lies, readily interacted with endogenous MDA5 during authentic SCoV2 infection (Fig. 6b). Endogenous MDA5 ISGylation was undetectable in SCoV2-infected cells, although the virus triggered ISG15 expression; however, in infected cells treated with a specific PLpro inhibitor15 MDA5 ISGylation and downstream ISG induction were strongly enhanced (Fig. 6c), supporting that PLpro effectively suppresses MDA5 ISGylation and signaling during live SCoV2 infection.
We next examined the effect of WT and mutant PLpro on the activation of endogenous MDA5 during mutEMCV infection. Consistent with their effect on MDA5–2CARD ISGylation (Fig. 5b), SCoV2 PLpro WT and F69A prevented antiviral transcript induction, whereas MDA5 R166S/E167R, similarly to the C111A mutant, did not affect antiviral gene expression (Fig. 6d). In accord, mutEMCV replication was enhanced in cells expressing PLpro WT or F69A, but not in cells expressing PLpro C111A or R166S/E167R (Fig. 6d). Likewise, WT PLpro, but not the R166S/E167R or C111A mutant, blocked EMCV restriction by FLAG-MDA5 (Fig. 6e); the effect on virus replication correlated with induced ISG proteins (Fig. 6f). Collectively, this establishes SCoV2 PLpro as an effective IFN antagonist that actively de-ISGylates MDA5.
Discussion
ISG15 conjugation is known to confer antiviral activity to a multitude of viruses; however, only few bona fide substrates have been identified12. On the other hand, ISG15 in its unconjugated form acts provirally by fortifying USP18-mediated IFNAR-signal inhibition31, 32, 40. This study identifies a key role for ISGylation in MDA5-mediated IFN induction. Our work also stresses the importance of experimental design in which decoupling the role of ISG15 in MDA5 activation from that in IFNAR signaling is essential to reveal ISG15’s potent antiviral activity. In an infected organism, it is likely the sum of multiple ISGylation events (affecting both host and viral proteins) that determine the outcome of infection and pathogenesis, which may be context-dependent12.
Our findings indicate that ISGylation of MDA5 acts analogously to the K63-linked ubiquitination of RIG-I5: both PTMs are 1) regulated by PP1-induced dephosphorylation and 2) promote CARD oligomerization and RLR higher-order assemblies. However, whereas ubiquitin is abundant in both uninfected and infected cells, ISG15 expression is strongly increased by IFN stimulation. Nevertheless, even at basal levels, ISG15 is conjugated to many host proteins20, including MDA5 as our work showed, which may be sufficient for initial MDA5 activation. During viral infections that are sensed by multiple PRRs, MDA5 ISGylation may be a ‘priming’ mechanism whereby ISG15 upregulation by an immediate innate sensor (e.g. RIG-I)41 primes MDA5 to enter a ‘kick-start’ mode41. As ISG15 negatively regulates RIG-I16, 17, ISGylation may trigger ‘sensor switching’ where MDA5 activation is promoted when ISG15 levels increase, while RIG-I activity is being dampened.
We identified that SCoV2 PLpro antagonizes MDA5 ISGylation via its enzymatic activity after binding to the sensor; this strategy is likely conserved among coronaviruses, which warrants further investigation. Cryo-EM analyses revealed that coronaviral Nsp3 is part of a pore complex that spans ER-derived double-membrane vesicles and exports newly-synthesized viral RNA42. Thus, MDA5 may position itself in close proximity to the site of viral RNA export to facilitate PAMP detection; however, the PLpro domain of Nsp3 (which is on the cytoplasmic side) blocks MDA5 signaling through direct de-ISGylation. Some viruses may also inhibit MDA5 ISGylation through dysregulation of MDA5 phosphorylation, as shown for MeV-V.
In summary, our study uncovers a prominent role for ISGylation in activating MDA5-mediated immunity as well as its inhibition by SCoV2, unveiling a potential molecular target for the design of therapeutics against COVID-19.
Methods
Cell culture.
HEK293T (human embryonic kidney cells), Vero (African green monkey kidney epithelial cells), BHK-21 (Baby hamster kidney), and Aedes albopictus clone C6/36 cells were purchased from ATCC. Human peripheral blood monocuclear cells (PBMCs) were isolated from unidentified healthy donor peripheral blood (HemaCare) and purified by Lymphoprep density gradient centrifugation (STEMCELL Technologies). The WT and isogenic Isg15−/− MEFs (mouse embryonic fibroblasts) were kindly provided by Deborah Lenschow (Washington University in St. Louis). SVGAs (human fetal glial astrocytes) were kindly provided by Ellen Cahir-McFarland (Biogen)43. SVGA MDA5 KO cells were generated by CRISPR/Cas9-mediated genome editing using a guide RNA (5’-AACTGCCTGCATGTTCCCGG-3’) targeting the exon 1 of IFIH1/MDA5. The MDA5 KO and RIG-I KO HEK293 cells were a gift from Jan Rehwinkel (University of Oxford)44. The WT and isogenic ISG15 KO HeLa cells were kindly provided by Elmar Schiebel (University of Heidelberg)45. ISG15 KO HeLa cells stably expressing FLAG-ISG15 WT or FLAG-ISG15 AA (GG156/157AA) were generated by lentiviral transduction followed by selection with puromycin (2 μg/mL). HAP-1 WT and isogenic ISG15 KO cells were purchased from Horizon Discovery. HEK293T-hACE2 and Vero-E6-hACE2 were a gift from Jae U. Jung (Cleveland Clinic). A549-hACE2 were kindly provided by Benjamin R. tenOever (Icahn School of Medicine at Mount Sinai)39. HEK293T, HEK293, HeLa, MEFs, NHLFs, Vero, A549-hACE2, and BHK-21 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco), 2 mM GlutaMAX (Gibco), 1 mM sodium pyruvate (Gibco), and 100 U/mL penicillin-streptomycin (Gibco). HEK293T-hACE2 and Vero-E6-hACE2 were maintained in DMEM containing 200 μg/mL hygromycin B and 2 μg/mL puromycin, respectively. SVGA and HAP-1 cells were cultured in Eagle’s Minimum Essential Medium (MEM, Gibco) and Iscove’s Modified Dulbecco’s Medium (IMDM, Gibco), respectively, supplemented with 10% FBS and 100 U/mL penicillin-streptomycin. PBMCs were maintained in RPMI 1640 (Gibco) supplemented with 10% FBS and 100 U/mL penicillin-streptomycin. C6/36 cells were cultured in MEM with 10% FBS and 100 U/mL penicillin-streptomycin. Except for C6/36 cells that were maintained at 28°C, all cell cultures were maintained at 37°C in a humidified 5% CO2 atmosphere.
Commercially obtained cell lines were authenticated by vendors and were not validated further in our laboratory. Cell lines that were obtained and validated by other groups were not further authenticated. KO cell lines were validated by confirming the absence of target protein expression. All cell lines used in this study have been regularly tested for potential mycoplasma contamination by PCR or using the MycoAlert Kit (Lonza).
Viruses.
DENV (serotype 2, strain 16681) and ZIKV (strain BRA/Fortaleza/2015) were propagated in C6/36 and Vero cells, respectively46, 47. Encephalomyocarditis virus (EMCV, EMC strain) was purchased from ATCC and propagated in HEK293T cells6. mutEMCV (EMCV-ZnC19A/C22A), which carries two point mutations in the zinc domain of the L protein18, was kindly provided by Frank J.M. van Kuppeveld (Utrecht University) and was propagated in BHK-21 cells. Sendai virus (strain Cantell) was purchased from Charles River Laboratories. SCoV2 (strain 2019-nCoV/USA_WA½020) was kindly provided by Jae U. Jung (Cleveland Clinic Lerner Research Center) and was propagated in Vero E6-hACE2 cells. All work relating to SCoV2 live virus and SCoV2-RNA was conducted in the BSL-3 facility of the Cleveland Clinic Florida Research and Innovation Center in accordance with institutional biosafety committee (IBC) regulations.
DNA constructs and transfection.
The human MDA5 ORF containing an N-terminal FLAG tag was amplified from the pEF-Bos-FLAG-MDA56 and subcloned into pcDNA3.1/Myc-His B between XhoI and AgeI. Site-directed mutagenesis on pcDNA3.1-FLAG-MDA5 (K23R/K43R, S88A, S88E, I841R/E842R, D848A/F849A, and G74A/W75A) was introduced by overlapping PCR. HA-MDA5 was cloned into pcDNA3.1(+) between KpnI and XhoI. GST-MDA5–2CARD (in pEBG vector) and its S88A, S88D, S88E derivatives were described previously6. The single (K23R, K43R, K68R, K128R, K137R, K169R, K174R, and K235R) and double (K23R/K43R) mutations of MDA5–2CARD (aa 1–295) were introduced by site-directed mutagenesis into GST-MDA5–2CARD. Additionally, MDA5–2CARD and its K23R/K43R mutant were subcloned into pcDNA3.1(−) harboring an N-terminal 3×FLAG tag between NheI and NotI. pCR3-FLAG-MV-V (strain Schwarz) was a gift from Karl-Klaus Conzelmann (LMU, Munich). pEF-Bos-FLAG-NiV-V, pCAGGS-HA-MeV-V, and pCAGGS-HA-MeV-VΔtail were described previously25. Myc-tagged PIV2-V, MenV-V, MPRV-V, and HeV-V constructs were kindly provided by Stephen Goodbourn (University of London). FLAG-tagged PIV5-V, PIV2-V, MenV-V, MPRV-V, and HeV-V were subcloned into pEF-Bos containing an N-terminal FLAG tag between NotI and SalI. pEF-Bos-FLAG-MuV-V was a gift from Curt Horvath (Addgene #4490848). pCAGGS-V5-hISG15 was a gift from Adolfo García-Sastre (Icahn School of Medicine at Mount Sinai)49. pCAGGS-HA-Ube1L and pFLAG-CMV2-UbcH8 were kindly provided by Jae U. Jung (University of Southern California). pcDNA3.1-Myc-UBE2I was cloned by ligating a synthetic UBE2I ORF into pcDNA3.1/Myc-His B between HindIII and NotI. FLAG-SUMO1 was obtained from Florian Full (University of Erlangen-Nuremberg, Germany). V5-tagged SARS-CoV-PLpro, MERS-CoV-PLpro, NL63-PLpro, MHV-PLP2 in pcDNA3.1-V5/His-B were kindly provided by Susan C. Baker (Loyola University of Chicago). The SARS-CoV-2 PLpro ORF (aa. 746–1060) was amplified from pDONR207 SARS-CoV-2 NSP3 (a gift from Fritz Roth; Addgene # 14125750) and subcloned into pcDNA3.1-V5. The C111A, F69A, N156E, R166S/E167R mutations of SARS-CoV-2-PLpro were introduced by site-directed mutagenesis. The correct sequence of all constructs was confirmed by DNA sequencing. Transient DNA transfections were performed using linear polyethylenimine [1 mg/mL solution in 10 mM Tris-HCl (pH 6.8); Polysciences], Lipofectamine 2000 (Invitrogen), Lipofectamine LTX with Plus Reagent (Invitrogen), TransIT-HeLaMONSTER (Mirus), or TransIT-X2 Transfection Reagent (Mirus) as per the manufacturers’ instructions.
Antibodies and other reagents.
Primary antibodies used in this study include anti-GST (1:5,000; Sigma-Aldrich), anti-V5 (1:5,000, R960–25; Novex), anti-FLAG (M2, 1:2,000; Sigma-Aldrich), anti-HA (1:3,000, HA-7; Sigma-Aldrich), anti-Phospho-IRF-3 (Ser396) (1:1,000, D6O1M; CST), anti-IRF3 (1:1,000, D6I4C; CST), anti-Phospho-STAT1 (Tyr701) (1:1,000, 58D6; CST), anti-IFIT1 (1:1,000, PA3–848; Invitrogen and 1:1,000, D2X9Z; CST), anti-IFIT2 (1:1,000; Proteintech), anti-ISG15 (1:500, F-9; Santa Cruz), anti-MAVS (1:1,000; CST), anti-RIG-I (1:2,000, Alme-1; Adipogen), anti-MDA5 (1:1,000, D74E4; CST), anti-Phospho-MDA5 (Ser88)6, anti-PP1α (1:2,000; Bethyl laboratories), anti-PP1γ (1:2,000; Bethyl laboratories), anti-USP18 (1:1000, D4E7; CST), anti-RSAD2 (1:1,000, D5T2X; CST), anti-PKR (1:1,000, D7F7; CST), anti-MX1 (1:1,000, D3W7I; CST), anti-IFITM3 (1:1,000, D8E8G; CST), anti-ISG20 (1:1,000, PA5–30073; Invitrogen), anti-ubiquitin (1:1,000, P4D1; Santa Cruz), anti-NS347, anti-PLpro (Nsp3) (1:1,000, GTX135589; GeneTex), anti-Spike (1:1,000, 1A9; GeneTex), anti-α-tubulin (1:1,000; CST), and anti-β-Actin (1:1,000, C4; Santa Cruz). Monoclonal anti-MDA5 antibody was purified from mouse hybridoma cell lines kindly provided by Jan Rehwinkel (University of Oxford)44. Monoclonal anti-IFNAR2 neutralizing antibody (1:250, MMHAR-2) was obtained from PBL Assay Science. Monoclonal anti-flavivirus E antibody (4G2) was purified from the mouse hybridoma cell line D1–4G2–4-15 (ATCC). Anti-mouse and anti-rabbit HRP-conjugated secondary antibodies (1:2,000) were purchased from CST. Anti-FLAG M2 magnetic beads (Sigma-Aldrich), anti-FLAG agarose beads (Sigma-Aldrich), Glutathione Sepharose 4B resin (GE Healthcare), and Protein G Dynabeads (Invitrogen) were used for protein immunoprecipitation. Protease and phosphatase inhibitors were obtained from Sigma-Aldrich. Poly(I:C) (HMW)/LyoVec and Poly(I:C) (HMW) Biotin were obtained from Invivogen. Human IFN-β was purchased from PBL Biomedical Laboratories. GRL-0617 was purchased from AdooQ Bioscience.
Mass spectrometry.
Large-scale GST-pulldown and mass spectrometry (MS) analysis were performed as previously described7, 22. Briefly, HEK293T cells were transfected with GST or GST-MDA5–2CARD, and the cells were collected at 48 h post-transfection and lysed in Nonidet P-40 (NP-40) buffer [50 mM HEPES (pH 7.4), 150 mM NaCl, 1% (v/v) NP-40, 1 mM EDTA, and 1× protease inhibitor cocktail (Sigma)]. Cell lysates were cleared by centrifugation at 20,000 ×g at 4°C for 20 min, and cleared supernatants were subjected to GST-pulldown using glutathione Sepharose 4B beads (GE Healthcare) at 4°C for 4 h. The beads were extensively washed with NP-40 buffer and proteins eluted by heating in 5× Laemmli SDS sample buffer at 95°C for 5 min. Eluted proteins were resolved on a NuPAGE 4–12% Bis-Tris gel (Invitrogen) and then stained at room temperature using the SilverQuest Silver Staining Kit (Invitrogen). The bands that were specifically present in the GST-MDA5–2CARD sample, but not the GST control sample, were excised and analyzed by LC-MS/MS (Taplin Mass Spectrometry Facility, Harvard University).
Immunoprecipitation and immunoblotting.
Cells were transfected with FLAG-MDA5, GST-MDA5–2CARD, or FLAG-MDA5–2CARD in the absence or presence of ISGylation machinery components (i.e. HA-Ube1L, FLAG-UbcH8, and V5-ISG15) as indicated. Forty-eight hours later, cells were lysed in NP-40 buffer and cleared by centrifugation at 20,000 ×g at 4°C for 20 min. Cell lysates were then subjected to GST or FLAG pulldown using glutathione magnetic agarose beads (Pierce) and anti-FLAG M2 magnetic beads (Millipore) at 4°C for 4 h or 16 h, respectively. The beads were extensively washed with NP-40 buffer and proteins eluted by heating in 1× Laemmli SDS sample buffer at 95°C for 5 min or by competition with FLAG peptide (Millipore) 4°C for 4 h. For endogenous MDA5 immunoprecipitation, NHLFs were stimulated with poly(I:C) (HMW)/LyoVec (0.1 μg/mL) or infected with DENV or ZIKV at the indicated MOI for 40 h. Cell lysates were precleared with Protein G Dynabeads (Invitrogen) at 4°C for 2 h and then incubated with Protein G Dynabeads conjugated with the anti-MDA5 antibody or an IgG1 isotype control (G3A1; CST) at 4°C for 4 h. The beads were washed four times with RIPA buffer [20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% (v/v) NP-40, 1% (w/v) deoxycholic acid, 0.01% (w/v) SDS] and protein eluted in 1× Laemmli SDS sample buffer. Protein samples were resolved on Bis-Tris SDS-PAGE gels, transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad), and visualized using the SuperSignal West Pico PLUS or Femto chemiluminescence reagents (Thermo Scientific) on an ImageQuant LAS 4000 Chemiluminescent Image Analyzer (General Electric) as previously described47.
Enzyme-linked immunosorbent assay (ELISA).
Human or mouse IFN-β in the culture supernatants of NHLFs, HeLa, and MEFs was determined by ELISA using the VeriKine Human Interferon Beta ELISA Kit or VeriKine Mouse Interferon Beta ELISA Kit (PBL Assay Science) as previously described6.
siRNA- and shRNA-mediated knockdown.
Transient knockdown in NHLFs, HeLa, HAP-1, HEK293T, and HEK293 cells was performed using non-targeting or gene-specific siGENOME SMARTpool siRNAs (Dharmacon). These are the Non-Targeting siRNA Pool #2 (D-001206–14), IFIH1 (M-013041–00), DDX58 (M-012511–01), PPP1CA (M-008927–01), PPP1CC (M-006827–00) and ISG15 (D-004235–17 and D-004235–18). Transfection of siRNAs was performed using the Lipofectamine RNAiMAX Transfection Reagent (Invitrogen) as per the manufacturer’s instructions. Scrambled shRNA control lentiviral particles and shRNA lentiviral particles targeting ISG15 (TL319471V) or IFIH1 (TL303992V) were purchased from OriGene. Lentiviral transduction of human PBMCs (1 ×105 cells; MOI 8) was performed in the presence of 8 μg/mL polybrene (Santa Cruz). Knockdown efficiency was determined by qRT-PCR or immunoblotting as indicated.
Quantitative real-time PCR (qRT-PCR).
Total RNA was purified using the E.Z.N.A. HP Total RNA Kit (Omega Bio-tek) as per the manufacturer’s instructions. One-step qRT-PCR was performed using the SuperScript III Platinum One-Step qRT-PCR Kit (Invitrogen) and predesigned PrimeTime qPCR Probe Assays (IDT) on a 7500 Fast Real-Time PCR System (Applied Biosystems). Relative mRNA expression was normalized to the levels of GAPDH and expressed relative to the values for control cells using the ΔΔCt method.
Luciferase reporter assay.
IFN-β reporter assay was performed as previously described51. Briefly, HEK293T or MDA5 KO HEK293 cells were transfected with IFN-β luciferase reporter construct and β-galactosidase (β-gal) expressing pGK-β-gal, along with GST-MDA5–2CARD (WT or mutants) or FLAG-MDA5 (WT or mutants). At the indicated time points after transfection, luciferase and β-gal activities were determined using respectively the Luciferase Assay System (Promega) and β-Galactosidase Enzyme Assay System (Promega) on a Synergy HT microplate reader (BioTek). Luciferase activity was normalized to β-gal values, and fold induction was calculated relative to vector-transfected samples, set to 1.
Cytosol-mitochondria fractionation assay.
The cytosol-mitochondria fractionation assay was performed using a Mitochondria/Cytosol Fractionation Kit (Millipore) as previously described46, 47. Briefly, NHLFs were transfected for 30 h with either non-targeting control siRNA or ISG15-specific siRNA and then transfected with EMCV-RNA or RABVLe for 16 h. Cells were homogenized in an isotonic buffer using a Dounce homogenizer and the lysates were centrifuged at 600 ×g to pellet the nuclei and unbroken cells. The supernatant was further centrifuged at 10,000 ×g at 4°C for 30 min to separate the cytosolic (supernatant) and mitochondrial (pellet) fractions. The protein concentration of both fractions was determined by a bicinchoninic acid (BCA) assay (Pierce), and equal amounts of proteins were analyzed by immunoblotting. Anti-α-tubulin and anti-MAVS immunoblotting served as markers for the cytosolic and mitochondrial fractions, respectively.
In vitro RNA-binding assay.
WT and Isg15−/− MEFs were stimulated with IFN-β (1,000 U/mL) for 24 h. Cells were lysed in a buffer containing 50 mM HEPES (pH 7.4), 200 mM NaCl, 1% (v/v) NP-40, 1 mM EDTA, and 1× protease inhibitor cocktail (Sigma). NeutrAvidin agarose beads (Pierce) were conjugated with the biotinylated HMW-Poly(I:C) at 4°C for 4 h. Cell lysates were incubated with the conjugated beads at 4°C for 16 h. The beads were washed three times with lysis buffer and then boiled at 95°C in 1× Laemmli SDS sample buffer to elute the proteins. Precipitated proteins were resolved on Bis-Tris SDS-PAGE gels and analyzed by IB with anti-MDA5. Equal input MDA5 protein amounts were confirmed by IB with anti-MDA5.
Native PAGE.
Native PAGE for analyzing endogenous IRF3 dimerization was performed as previously described52. For measuring MDA5 oligomerization, HEK293T or HEK293 MDA5 KO cells were transfected with WT or mutant FLAG-MDA5–2CARD or FLAG-MDA5 as indicated. Twenty-four hours later, cells were lysed in 1× NativePAGE sample buffer (Invitrogen) containing 1% (v/v) NP-40 on ice for 30 min and then lysates were cleared by centrifugation at 16,000 ×g at 4°C for 10 min. Cleared lysates were resolved on a 3–12% Bis-Tris NativePAGE gel (Invitrogen) as per the manufacturer’s instructions and analyzed by immunoblotting with the indicated antibodies.
Semi-denaturating detergent agarose gel electrophoresis (SDD-AGE).
MDA5 oligomerization in MEFs transfected with EMCV-RNA, or in HEK293 MDA5 KO cells reconstituted with WT or mutant FLAG-MDA5, was determined by SDD-AGE as previously described with modifications26. Briefly, cells were lysed in a buffer containing 50 mM HEPES (pH 7.4), 150 mM NaCl, 0.5% (v/v) NP-40, 10% (v/v) glycerol, and 1× protease inhibitor cocktail (Sigma) at 4°C for 20 min. Cell lysates were cleared by centrifugation at 16,000 ×g at 4°C for 10 min and then incubated on ice for 1 h. Cell lysates were subsequently incubated in 1× SDD-AGE buffer (0.5× TBE, 10% (v/v) glycerol, and 2% (w/v) SDS) for 15 min at room temperature and resolved on a vertical 1.5% agarose gel containing 1× TBE and 0.1% (w/v) SDS at 80 V for 90 min at 4°C. Proteins were transferred onto a PVDF membrane and analyzed by immunoblotting with the indicated antibodies.
Viral RNA purification.
EMCV-RNA was produced as previously described6. Briefly, Vero cells were infected with EMCV (MOI 0.1) for 16 h, and total RNA was isolated using the Direct-zol RNA extraction kit (Zymo Research) as per the manufacturer’s instructions. Mock-RNA and SCoV2-RNA were produced by isolating total RNA from uninfected or SCoV2-infected (MOI 1 for 24 h) Vero-hACE2 cells. RABVLe was generated by in vitro transcription using the MEGAshortscript T7 Transcription Kit (Invitrogen) as previously described53.
Virus infection and titration.
All viral infections were performed by inoculating cells with the virus inoculum diluted in MEM or DMEM containing 2% FBS. After 1–2 h, the virus inoculum was removed and replaced with the complete growth medium (MEM or DMEM containing 10% FBS) and cells were further incubated for the indicated times. EMCV titration was performed either on Vero cells using the median tissue culture infectious dose (TCID50) methodology as previously described54, or on BHK-21 cells by the standard plaque assay. The titers of ZIKV were determined by plaque assay on Vero cells as previously described47. Titration of SCoV2 was performed on Vero-hACE2 cells by plaque assay.
Flow cytometry.
To quantify the percentage of DENV-infected cells, reconstituted HEK293 MDA5 KO cells were washed with PBS (Gibco) and fixed with 4% (v/v) formaldehyde in PBS at room temperature for 30 min. Cells were subsequently permeabilized with 1× BD Perm/Wash buffer (BD Biosciences) for 15 min and incubated with an anti-flavivirus E antibody (4G2; 1:100 in 1× BD Perm/Wash buffer) at 4°C for 30 min. Cells were further washed three times with 1× BD Perm/Wash buffer and incubated with a goat anti-mouse Alexa Fluor 488-conjugated secondary antibody (#A10667, 1:500 in 1× BD Perm/Wash buffer; Invitrogen) at 4°C for 30 min in the dark. After washing three times with 1× BD Perm/Wash buffer, cells were analyzed on a FACSCalibur flow cytometer (BD Biosciences). Data analysis was performed using the FlowJo software.
Virus protection assay.
The culture supernatants from mutant or WT EMCV-infected NHLFs or RIG-I KO HEK293 cells were UV-inactivated in a biosafety cabinet under a UV-C lamp (30W) at a dose of 5,000 μJ/cm2 for 15 min. Complete inactivation of EMCV was confirmed by plaque assay on BHK-21 cells. The inactivated supernatants were then transferred onto fresh Vero cells for 24 h, and the primed Vero cells were subsequently infected with ZIKV (MOI 0.002 to 2) for 72 h, or with EMCV (MOI 0.001 to 0.1) for 40 h. ZIKV-positive cells were determined by immunostaining with anti-flavivirus E antibody (4G2) and visualized using the KPL TrueBlue peroxidase substrate (SeraCare). EMCV-induced cytopathic effect was visualized by Coomassie blue staining.
Statistical analysis.
Two-tailed unpaired t-test was used to compare differences between two experimental groups in all cases. Significant differences are denoted by *p < 0.05, **p < 0.01, or ***p < 0.001. Pre-specified effect sizes were not assumed, and in general, three biological replicates (n) for each condition were used.
Data Availability
The data that support the findings of this study are available from the corresponding author upon request.
Extended Data
Extended Data Fig. 1. ISG15 is required for MDA5, but not RIG-I, mediated signal transduction.
(a) Silver-stained affinity-purified GST and GST-MDA5–2CARD from transiently transfected HEK293T cells. Asterisks denote the GST and GST-MDA5–2CARD (aa 1–295) proteins. Arrows indicate the bands that identified ISG15 by MS analysis. (b) GST-MDA5–2CARD ISGylation in transiently transfected HEK293T cells with or without co-expressed V5-ISG15, determined by GST pulldown (PD) and immunoblot (IB) with anti-V5 and anti-GST. Whole cell lysates (WCL) were probed by IB with anti-V5. (c, d) qRT-PCR analysis of IFNB1 and CCL5 transcripts in WT and Isg15−/− MEFs (c) or WT and ISG15 KO HeLa cells (d) that were transfected with empty vector or increasing amounts of FLAG-MDA5 or FLAG-RIG-I for 40 h. (e) ELISA of IFN-β in the supernatants of WT or ISG15 KO HeLa cells that were mock-transfected or transfected with EMCV-RNA (0.4 μg/mL) or RABVLe (1 pmol/mL) for 24 h. (f) qRT-PCR analysis of IFNB1, CCL5, TNF, and MDA5 mRNA in WT and ISG15 KO HeLa cells that were mock-transfected or transfected with EMCV-RNA (0.4 μg/mL) for 24 h. (g) qRT-PCR analysis of IFNB1, CCL5, TNF, and MDA5 mRNA in WT and ISG15 KO HAP-1 cells that were stimulated as in (f). (h) qPCR analysis of IFNB1 and CCL5 mRNA in NHLFs that were transfected with the indicated siRNAs for 30 h and then mock-stimulated or transfected with EMCV-RNA (0.4 μg/mL) or RABVLe (1 pmol/mL), or infected with SeV (10 HAU/mL) for 16 h. (i) qRT-PCR analysis of ISG15 and MDA5 mRNA in PBMCs that were transduced for 40 h with the indicated shRNA lentiviral particles and then infected with mutEMCV (MOI 10) or SeV (200 HAU/mL) for 8 h Data are representative of at least two independent experiments (mean ± s.d. of n = 3 biological replicates in c, d, e, f, g, and h; mean of n = 2 biological replicates in i). *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed unpaired t-test). NS, not significant; ND, not detected.
Extended Data Fig. 2. ISGylation at K23 and K43 is essential for MDA5 activation.
(a) ISGylation of endogenous MDA5 in uninfected NHLFs, determined by IP with anti-MDA5 (or IgG isotype control) and IB with anti-ISG15. (b) qRT-PCR analysis of the indicated transcripts in NHLFs that were transfected for 30 h with the indicated siRNAs and then infected with mutEMCV (MOI 0.005) for 16 h in the absence or presence of anti-IFNAR2 (2 μg/mL). (c) Upper panels: Amino acid sequence of the MDA5 CARDs. Lysine (K) residues are highlighted in red. CTD, C-terminal domain. Middle panels: ISGylation of GST-MDA5–2CARD WT or mutants in HEK293T cells that co-expressed V5-ISG15, determined by GST-PD and IB with anti-V5. Lower panels: Densitometric analysis of the ISGylation levels of GST-MDA5–2CARD WT and mutants, normalized to protein levels in GST-PD. (d) ISGylation of FLAG-MDA5 WT and K23R/K43R in transfected MDA5 KO HEK293 cells that co-expressed V5-ISG15 and were stimulated with EMCV-RNA (0.4 μg/mL) for the indicated times, determined by FLAG-PD and IB with anti-V5. (e) SUMOylation of GST-MDA5–2CARD WT and K23R/K43R in transfected HEK293T cells that co-expressed Myc-UBE2I and FLAG-SUMO1, determined by FLAG-PD and IB with anti-GST. (f) Ubiquitination of GST-RIG-I-2CARD WT, or GST-MDA5–2CARD WT and K23R/K43R in HEK293T cells, determined by GST-PD and IB with anti-Ub. (g) IFN-β-luciferase activity in HEK293T cells transfected for 40 h with GST, or GST-MDA5–2CARD (GST-2CARD) WT or mutants. (h) Endogenous IRF3 dimerization in HEK293T cells transfected with GST, or GST-MDA5–2CARD WT or K23R/K43R for 24 h, determined by Native PAGE and IB with anti-IRF3. WCLs were analyzed by SDS-PAGE and IB with anti-GST, anti-IRF3, and anti-Actin. (i) Validation of MDA5 gene editing in SVGAs. MDA5 protein abundance in WT control and MDA5 KO SVGAs treated with IFN-β (1,000 U/mL) for 16 h, or left untreated, assessed by IB with anti-MDA5. Immunoblotting for RIG-I and Actin served as control. Data are representative of at least two independent experiments with similar results (mean ± s.d. of n = 3 biological replicates in b and g). **p < 0.01, ***p < 0.001 (two-tailed unpaired t-test).
Extended Data Fig. 3. Dephosphorylation of MDA5 induces ISGylation.
(a) ISGylation of GST-MDA5–2CARD in HEK293T cells transfected with V5-ISG15 and the indicated siRNAs for 48 h, assessed by GST-PD and IB with anti-V5 and anti-GST. WCLs were probed by IB with anti-V5, anti-PP1α, anti-PP1γ, and anti-Actin. (b) ISGylation of GST-MDA5–2CARD WT or S88A, S88D and S88E mutants in transiently transfected HEK293T cells that also co-expressed V5-ISG15, determined by GST-PD and IB with anti-V5 and anti-GST forty hours after transfection. (c) Phosphorylation of FLAG-MDA5 WT or K23R/K43R mutant in HEK293T cells, determined by FLAG-PD and IB with anti-pS88-MDA5 and anti-FLAG. WCLs were probed by IB with anti-FLAG and anti-Actin. (d) ISGylation and phosphorylation of GST-MDA5–2CARD in HEK293T cells transfected with V5-ISG15 and either empty vector or increasing amounts of FLAG-MeV-V for 24 h, determined by GST-PD and IB with anti-pS88-MDA5, anti-V5, and anti-GST. (e) ISGylation and phosphorylation of FLAG-tagged MDA5 in transfected HEK293T cells that co-expressed V5-ISG15, HA-Ube1L, and FLAG-UbcH8 as well as either empty vector or increasing amounts of HA-MeV-V for 24 h, determined by FLAG-PD and IB with anti-pS88-MDA5, anti-V5, and anti-FLAG. WCLs were probed by IB with the indicated antibodies. (f) ISGylation of GST-MDA5–2CARD, or GST (negative control), in HEK293T cells transiently transfected with V5-ISG15 and either empty vector or HA-tagged MeV-V WT or Δtail for 48 h, determined by GST-PD and IB with anti-V5 and anti-GST. (g) Upper panel: Phosphorylation of HA-MDA5 in transfected HEK293T cells that co-expressed empty vector or the indicated FLAG-tagged paramyxoviral V proteins, assessed by HA-PD and IB with anti-pS88-MDA5 and anti-HA. WCLs were probed by IB with anti-FLAG. Lower panel: Densitometric analysis of the levels of MDA5 S88 phosphorylation, normalized to protein levels in HA-PD. (h) ISGylation and phosphorylation of GST-MDA5–2CARD in transfected HEK293T cells that co-expressed V5-ISG15 and either empty vector or increasing amounts of FLAG-NiV-V or FLAG-HeV-V for 24 h, determined by GST PD and IB with anti-V5, anti-pS88-MDA5, and anti-GST. WCLs were probed by IB with anti-FLAG, anti-V5, and anti-Actin. Data are representative of at least two independent experiments with similar results.
Extended Data Fig. 4. CARD ISGylation does not affect the ability of MDA5 to bind RNA, 14–3-3η, or LGP2.
(a) In vitro RNA-binding ability of endogenous MDA5 from WT or Isg15−/− MEFs that were stimulated with IFN-β (1,000 U/mL) for 24 h, assessed by biotin-HMW-poly(I:C)-PD and IB with anti-MDA5. Equal input MDA5 protein amounts were confirmed by IB with anti-MDA5. (b) In vitro RNA-binding ability of FLAG-MDA5 WT and K23R/K43R from transiently transfected HEK293T cells, assessed by biotin-HMW-poly(I:C)-PD and IB with anti-FLAG. Equal input FLAG-MDA5 protein amounts were confirmed by IB with anti-FLAG. (c) Binding of FLAG-tagged MDA5–2CARD WT or mutants to HA-14–3-3η in transiently transfected HEK293T cells, determined by HA-PD and IB with anti-FLAG and anti-HA. WCLs were probed by IB with anti-FLAG and anti-Actin. (d) Binding of FLAG-tagged MDA5 WT and K23R/K43R to HA-tagged LGP2 in transiently transfected HEK293T cells, determined by HA-PD and IB with anti-MDA5 and anti-HA. WCLs were probed by IB with anti-MDA5. Data are representative of at least two independent experiments with similar results.
Extended Data Fig. 5. Aberrant ISG upregulation in ISG15-deficient cells upon MDA5 stimulation.
(a) EMCV titers in the supernatant of RIG-I KO HEK293 cells that were transfected for 24 h with nontargeting control siRNA (si.C) or ISG15-specific siRNA (si.ISG15) and then transfected with either empty vector or FLAG-tagged MDA5 WT or K23R/K43R for 24 h prior to infection with EMCV (MOI 0.001) for 16 h, determined by plaque assay. (b) Protein abundance of the indicated ISGs and USP18 in mock-infected or EMCV-infected (MOI 0.001 for 16 h) RIG-I KO HEK293 cells that were transfected with the indicated siRNAs, and 24 h later, transfected with empty vector or FLAG-MDA5 WT or K23/K43R for 24 h, determined by IB with the indicated antibodies. WCLs were further immunoblotted with anti-FLAG and anti-Actin. (c) qRT-PCR analysis of IFNB1 and ISG transcripts in WT and ISG15 KO HeLa cells that were mock-treated or transfected with EMCV-RNA (0.4 μg/mL) for 16 h. (d) Left panel: Protein abundance of the indicated ISGs and USP18 in NHLFs that were transfected for 40 h with the indicated siRNAs and then infected with mutEMCV (MOI 0.1) for 16 h, determined by IB with the indicated antibodies. Right panel: ELISA of IFN-β from supernatants of NHLFs from the same experiment (left panel). Data are representative of at least two independent experiments with similar results (mean of n = 2 biological replicates in a; mean ± s.d. of n = 3 biological replicates in c and d). **p < 0.01, ***p < 0.001 (two-tailed unpaired t-test). ND, not detected.
Extended Data Fig. 6. SCoV2 PLpro does not affect RIG-I ubiquitination and preferentially antagonizes the MDA5 pathway.
(a) ISGylation of FLAG-MDA5 in HEK293T cells that were co-transfected for 20 h with vector or V5-tagged SCoV2 PLpro WT or mutants, along with V5-ISG15, HA-Ube1L, and FLAG-UbcH8, determined by FLAG-PD and IB with anti-V5 and anti-FLAG. WCLs were probed by IB with anti-V5, anti-HA, anti-FLAG, and anti-Actin. (b) Ubiquitination of GST-RIG-I-2CARD in HEK293T cells that were transfected with V5-tagged SCoV2 PLpro WT or C111A for 24 h, determined by GST-PD and IB with anti-Ub and anti-GST. WCLs were probed by IB with anti-Ub, anti-V5, and anti-Actin. (c) Upper panels: qPCR analysis of IFNB1 and CCL5 transcripts in HeLa cells that were transfected with FLAG-MDA5 or FLAG-RIG-I together with either empty vector (Vec) or increasing amounts of V5-tagged SCoV2 PLpro (10 ng, 25 ng, and 50 ng) for 24 h. Data are presented as percentage of induction relative to the values for cells transfected with the respective RLR (i.e. FLAG-MDA5 or FLAG-RIG-I) and vector, set to 100%. Lower panels: WCLs from the same experiment were probed by IB with the indicated antibodies. Data are representative of at least two independent experiments with similar results (mean ± s.d. of n = 3 biological replicates in c).
Supplementary Material
Acknowledgements
We greatly thank Deborah Lenschow (Washington University in St. Louis), Elmar Schiebel (University of Heidelberg), Ellen Cahir-McFarland (Biogen), Jan Rehwinkel (University of Oxford), Frank J.M. van Kuppeveld (Utrecht University), Karl-Klaus Conzelmann (LMU, Munich), Stephen Goodbourn (University of London), Susan C. Baker (Loyola University Chicago), Benjamin R. tenOever (Icahn School of Medicine at Mount Sinai), Adolfo García-Sastre (Icahn School of Medicine at Mount Sinai), and Jae U. Jung (Cleveland Clinic Lerner Research Institute) for providing reagents. We are also grateful to Sara Tavakoli and Jessica Poole for their help in the BSL-3 facility at the Cleveland Clinic Florida Research and Innovation Center. This study was supported in part by the US National Institutes of Health grants R01 AI087846 and R01 AI127774 (to M.U.G.).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Liu G & Gack MU Distinct and Orchestrated Functions of RNA Sensors in Innate Immunity. Immunity 53, 26–42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J & Chen ZJ Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32, 461–488 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Chow KT, Gale M Jr. & Loo YM RIG-I and Other RNA Sensors in Antiviral Immunity. Annu Rev Immunol 36, 667–694 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Schlee M Master sensors of pathogenic RNA - RIG-I like receptors. Immunobiology 218, 1322–1335 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rehwinkel J & Gack MU RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol 20, 537–551 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wies E et al. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity 38, 437–449 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gack MU et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Chiang C & Gack MU Post-translational Control of Intracellular Pathogen Sensing Pathways. Trends Immunol 38, 39–52 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazear HM, Schoggins JW & Diamond MS Shared and Distinct Functions of Type I and Type III Interferons. Immunity 50, 907–923 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoggins JW Interferon-Stimulated Genes: What Do They All Do? Annu Rev Virol 6, 567–584 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Zhang D & Zhang DE Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res 31, 119–130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perng YC & Lenschow DJ ISG15 in antiviral immunity and beyond. Nat Rev Microbiol 16, 423–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hadjadj J et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369, 718–724 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klemm T et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J, e106275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin D et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim MJ, Hwang SY, Imaizumi T & Yoo JY Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J Virol 82, 1474–1483 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Y et al. LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J 37, 351–366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hato SV et al. The mengovirus leader protein blocks interferon-alpha/beta gene transcription and inhibits activation of interferon regulatory factor 3. Cell Microbiol 9, 2921–2930 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Deddouche S et al. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. Elife 3, e01535 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durfee LA, Lyon N, Seo K & Huibregtse JM The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol Cell 38, 722–732 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenschow DJ et al. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J Virol 79, 13974–13983 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gack MU, Nistal-Villan E, Inn KS, Garcia-Sastre A & Jung JU Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J Virol 84, 3220–3229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nistal-Villan E et al. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J Biol Chem 285, 20252–20261 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maharaj NP, Wies E, Stoll A & Gack MU Conventional protein kinase C-alpha (PKC-alpha) and PKC-beta negatively regulate RIG-I antiviral signal transduction. J Virol 86, 1358–1371 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis ME et al. Antagonism of the phosphatase PP1 by the measles virus V protein is required for innate immune escape of MDA5. Cell Host Microbe 16, 19–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin JP, Fan YK & Liu HM The 14–3-3eta chaperone protein promotes antiviral innate immunity via facilitating MDA5 oligomerization and intracellular redistribution. PLoS Pathog 15, e1007582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu B et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152, 276–289 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Yu Q, Qu K & Modis Y Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis. Mol Cell 72, 999–1012 e1016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruns AM, Leser GP, Lamb RA & Horvath CM The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell 55, 771–781 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uchikawa E et al. Structural Analysis of dsRNA Binding to Anti-viral Pattern Recognition Receptors LGP2 and MDA5. Mol Cell 62, 586–602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malakhova OA et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J 25, 2358–2367 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X et al. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature 517, 89–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harcourt BH et al. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol 78, 13600–13612 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth-Cross JK, Bender SJ & Weiss SR Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol 82, 9829–9838 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menachery VD et al. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2’-o-methyltransferase activity. J Virol 88, 4251–4264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bekes M et al. Recognition of Lys48-Linked Di-ubiquitin and Deubiquitinating Activities of the SARS Coronavirus Papain-like Protease. Mol Cell 62, 572–585 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Childs K et al. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359, 190–200 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Shi HX et al. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol 30, 2424–2436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanco-Melo D et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181, 1036–1045 e1039 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speer SD et al. ISG15 deficiency and increased viral resistance in humans but not mice. Nat Commun 7, 11496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Errett JS, Suthar MS, McMillan A, Diamond MS & Gale M Jr. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol 87, 11416–11425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolff G et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 369, 1395–1398 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Major EO et al. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc Natl Acad Sci U S A 82, 1257–1261 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hertzog J et al. Infection with a Brazilian isolate of Zika virus generates RIG-I stimulatory RNA and the viral NS5 protein blocks type I IFN induction and signaling. Eur J Immunol 48, 1120–1136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cerikan B et al. Cell-Intrinsic Adaptation Arising from Chronic Ablation of a Key Rho GTPase Regulator. Dev Cell 39, 28–43 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Chan YK & Gack MU A phosphomimetic-based mechanism of dengue virus to antagonize innate immunity. Nat Immunol 17, 523–530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riedl W et al. Zika Virus NS3 Mimics a Cellular 14–3-3-Binding Motif to Antagonize RIG-I- and MDA5-Mediated Innate Immunity. Cell Host Microbe 26, 493–503 e496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ulane CM, Rodriguez JJ, Parisien JP & Horvath CM STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J Virol 77, 6385–6393 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oudshoorn D et al. HERC6 is the main E3 ligase for global ISG15 conjugation in mouse cells. PLoS One 7, e29870 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim DK et al. A Comprehensive, Flexible Collection of SARS-CoV-2 Coding Regions. G3 (Bethesda) 10, 3399–3402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiang C et al. The Human Papillomavirus E6 Oncoprotein Targets USP15 and TRIM25 To Suppress RIG-I-Mediated Innate Immune Signaling. J Virol 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gack MU et al. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci U S A 105, 16743–16748 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiang JJ et al. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat Immunol 19, 53–62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sparrer KMJ et al. TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat Microbiol 2, 1543–1557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.