Though an increasing number of studies have indicated the existence of an interaction between the virome and human health or disease, the specific role of these plasma viral components remains largely unsolved. We provide evidence here that an altered plasma virome profile is associated with different immune statuses of HIV-1 infection.
KEYWORDS: MSM; plasma virome; HIV-1 infection; viral metagenome; pegivirus; anellovirus,
ABSTRACT
Altered gut virome and expanded abundance of certain viruses were found in HIV-1-infected individuals. It remains largely unknown how plasma virus composition changes during HIV-1 infection and antiretroviral therapy (ART). We performed viral metagenomic analysis on viral particles enriched from human plasma from 101 men who have sex with men (MSM) with or without HIV-1 infection and whether or not on ART and compared the differences in the plasma virome. An increased plasma viral abundance of main eukaryotic viruses was observed during HIV-1 infection in MSM, especially in AIDS patients (CD4+ T cell counts of <200). Anellovirus, pegivirus and hepatitis B virus (HBV) were the most abundant blood-borne viruses detected among MSM and HIV-1-infected individuals, and anellovirus and pegivirus were closely related to HIV-1 infection. High diversity of anelloviruses was found mostly in HIV-1-infected MSM, and their abundance was positively correlated with the HIV-1 viral load, but negatively correlated with both CD4+ T cell counts and CD4+/CD8+ ratio; in contrast, the abundance of pegivirus showed opposite correlations. ART usage could restore the plasma virome toward that of HIV-1-negative individuals. These data showed an expansion in abundance of certain viruses during HIV-1 infection, indicating the higher risk of shedding some blood-borne viruses in these individuals. These investigations indicate that both anellovirus and pegivirus may play certain roles in HIV disease progression.
IMPORTANCE Though an increasing number of studies have indicated the existence of an interaction between the virome and human health or disease, the specific role of these plasma viral components remains largely unsolved. We provide evidence here that an altered plasma virome profile is associated with different immune status of HIV-1 infection. Specific resident viruses, such as anellovirus and pegivirus, may directly or indirectly participate in the disease progression of HIV-1 infection. These results can help to determine their clinical relevance and design potential therapies.
INTRODUCTION
Viruses are the most abundant and variable components of the microbiome in human bodies (1, 2), and they could affect human health by direct interaction with host immunity or indirect interaction with other components of human microbiome. Previous studies showed that the human virome may change under different pathological conditions and immune status (3, 4). For example, altered viral composition of gut has been observed in cancer, diabetes, inflammatory bowel disease (IBD), and AIDS (5–8), and changes of a specific virome profile may be predictive of disease development (9).
An impaired immune system after HIV-1 infection can lead to substantial changes of the host residential microbial composition, including an increased diversity or abundance of gut viral and bacterial communities. For example, during HIV-1 infection, an expansion of adenoviruses in the gut has been demonstrated in association with a decrease in CD4+ T cell counts (10). Similarly, increased abundance of enteroviruses, including adenoviruses, was found to be associated with AIDS symptoms in simian immunodeficiency virus (SIV)-infected macaques (11). These data indicated a close relationship of particular viruses in immunodeficiency and AIDS-associated enteropathy. Though relatively less diverse than gut microbial communities, human plasma is major carrier of pathogenic viruses, as well as many commensal viruses (12). Previous studies found that anelloviruses, adenoviruses, human endogenous retrovirus (HERV), herpesviruses, papillomaviruses, polyomaviruses, and pegivirus could be present in the general population without specific disease status, indicating the complex sources and still undefined roles of most viruses in certain individuals (1). Early studies showed an altered plasma virome after HIV-1 infection, and an increased abundance of anellovirus and HERV was seen in AIDS patients (8, 13). Besides, specific plasma viral components were also associated with positive outcomes: for example, coinfection with pegivirus could inhibit HIV-1 replication and had a beneficial impact on patients’ survival (14, 15).
The population of men who have sex with men (MSM) has higher risk of infection by HIV-1 and other infectious viruses. The rate of new HIV-1 infection among this population has still been increasing in most countries over the past 20 years (16, 17). An estimated 1.7 million people were newly infected by HIV-1 in 2018, and 17% of them were MSM. Recent HIV-1 prevalence surveys in different MSM communities showed prevalence ranging from 4% to 15% (18). Using viral metagenomics, we characterized the plasma virome during HIV-1 infection of MSM, trying to determine a comprehensive profile of the circulating virome of these individuals, as well as how the plasma virome changes in response to HIV-1 infection, antiretroviral therapy (ART) usage, and immune status. This will help to understand the role of plasma virome in HIV-1 progression and identify potential viral biomarkers.
RESULTS
Participants.
A total of 101 MSM with or without HIV-1 infection and 20 HIV-1-negative non-MSM as a control were recruited. All individuals were between 23 and 42 years old. All MSM samples were divided into four groups: HIV-1-negative MSM (n = 22), HIV-1-infected ART-naive MSM with CD4 > 200 (n = 30) or CD4 < 200 (n = 20), and HIV-1-infected ART-treated MSM (n = 29). ART usage in the HIV-1-infected ART-treated MSM group lasted for at least 8 months (median = 38.7; interquartile range [IQR], 24.7 to 65.3), plasma HIV-1 viral loads were below the limit of detection (<20 copies/ml), and CD4+ T cell counts were above 200 cells/μl. HIV-1 viral load in the ART-naive groups ranged from 0.56 × 105 to 11.35 × 105 copies/ml and was negatively correlated with CD4+ T cell counts (Spearman’s r = −0.41, P = 0.0032) or CD4+/CD8+ ratio (Spearman’s r = −0.44, P = 0.0017) (Table 1; see Fig. S1A in the supplemental material).
TABLE 1.
Cohort characteristics
| Patient characteristic | Result for groupa |
||||
|---|---|---|---|---|---|
| Non-MSM | HIV-1− MSM | HIV-1+ ART-naive MSM |
ART-treated MSM (CD4 > 200) | ||
| CD4 > 200 | CD4 < 200 | ||||
| No. of samples | 20 | 22 | 30 | 20 | 29 |
| Age (yr) | 28 (24–35) | 29.5 (25–38) | 26 (23–30) | 32 (27–36) | 37 (30–42) |
| HIV-1 viral load (copies/ml) | − | <20b | 1.03 ×105 (0.56 × 105–2.86 × 105) | 4.8 ×105 (2.02 × 105–11.35 ×105) | <20b |
| T cell count (cells/μl) | |||||
| CD4+ | − | − | 325.5 (277–386) | 84.5 (35.5–161) | 527 (433–621) |
| CD8+ | − | − | 1,014 (681–1,272) | 658 (595.5–829) | 682 (521–935) |
| CD4+/CD8+ ratio | − | − | 0.35 (0.28–0.46) | 0.13 (0.06–0.20) | 0.77 (0.59–1.04) |
| No. of mo on ART | − | − | − | − | 38.7 (24.7–65.3) |
Data are shown as medians with interquartile ranges in parentheses. A minus sign (−) indicates not tested.
The detection limit of the HIV-1 quantification kit used is 20 copies/ml.
Correlation analysis of HIV-1 viral load with CD4+ T cell counts, CD8+ T cell counts, and CD4+/CD8+ ratio. (A) Correlation between peripheral circulating CD4+ T cell counts, CD8+ T cell counts, CD4+/CD8+ ratio, and HIV-1 viral load in HIV-1-infected ART-naive MSM. (B) Spearman's correlation between HIV-1 viral load and HIV-1 read count (normalized by RPM). Download FIG S1, TIF file, 0.5 MB (507.7KB, tif) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Viral composition in human plasma of MSM.
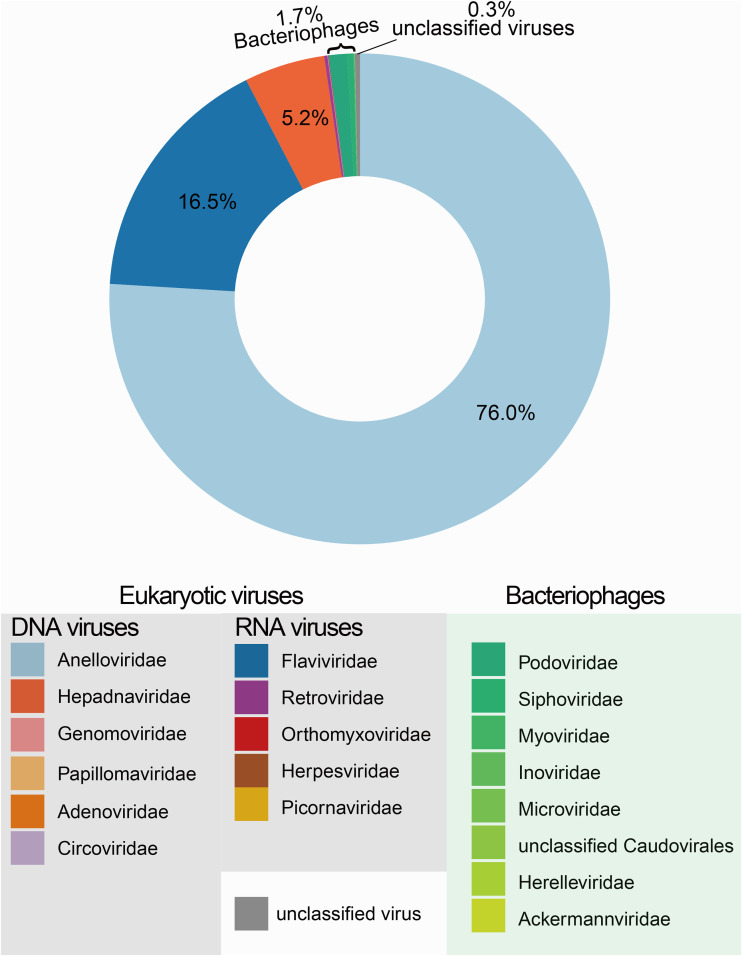
All plasma samples were first processed through virus-like particle (VLP) enrichment and then subjected to the Illumina NovaSeq platform. On average, 67 million reads were obtained from each sample, and after low-quality and human genome reads were removed, 17 million reads from each sample were obtained for subsequent analyses. The majority of cleaned reads were unclassified (∼57%); viral reads accounted for ∼10% of cleaned reads (see Fig. S2A in the supplemental material). In general, the plasma virome of these participants were dominated by eukaryotic viruses (98.0% of total viral reads), followed by bacteriophages (1.7%) and unclassified viruses (0.3%) (see Table S1 in the supplemental material). Here, we focused on the main pathogenic and commensal human plasma viruses: in total, 11 eukaryotic viral families were detected, including DNA viruses Anelloviridae, Hepadnaviridae, Genomoviridae, Papillomaviridae, Adenoviridae, Circoviridae, and Herpesviridae and RNA viruses Flaviviridae (e.g., pegivirus and hepatitis C virus [HCV]), Retroviridae, Orthomyxoviridae, and Picornaviridae (Fig. 1; Table S1).
FIG 1.
Viral metagenome composition from all individuals of this study. The donut chart shows the viral composition of the plasma virome from all individuals of this study. Eukaryotic viruses and bacteriophages are labeled in different colors.
Taxonomic composition of NGS results and correlation analysis of anellovirus and pegivirus with CD4+ T cell counts, CD8+ T cell counts, and CD4+/CD8+ ratio. (A) Taxonomic composition of different plasma samples as determined by NGS. Abundance is shown as reads per million (RPM); other terms represent unclassified eukaryotic reads or unknown reads. (B) Spearman’s correlation between anellovirus and CD8+ T cell counts, CD4+ T cell counts, and the CD4+/CD8+ ratio in ART-treated or ART-naive groups. (C) Spearman’s correlation between pegivirus and CD4+ T cell counts, CD8+ T cell counts, and CD4+/CD8+ ratio in ART-treated or ART-naive groups. Download FIG S2, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
All the main viral families detected and number of sequencing reads. Download Table S1, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
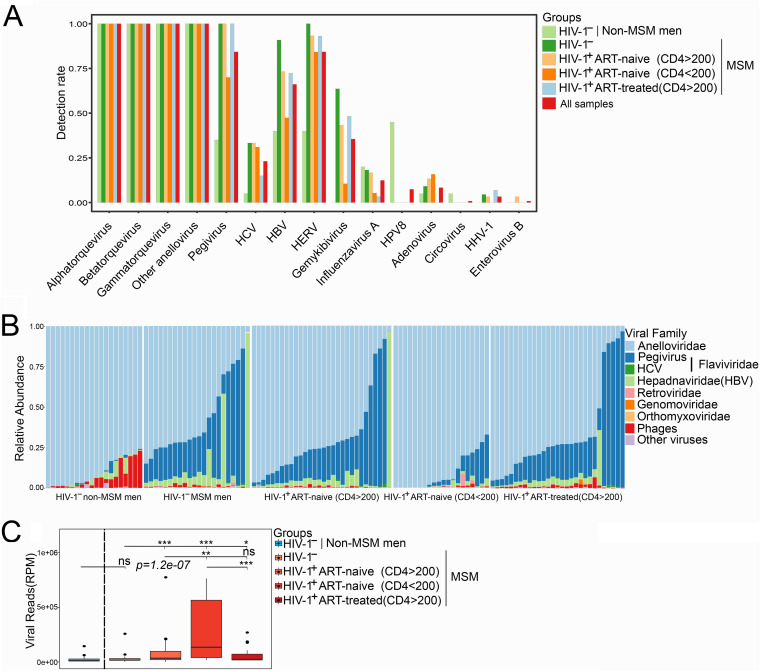
HIV-1 reads were identified in 65 out of 79 HIV-1-positive samples and positively correlated with HIV-1 viral load (Spearman’s r = 0.55, P = 5.7e−05) (Fig. S1B), reflecting consistent results from both next-generation sequencing (NGS) and quantitative reverse-transcription PCR (RT-qPCR) for HIV-1 detection. Besides HIV-1, the most frequently detected viruses from these individuals included anellovirus, pegivirus, hepatitis B virus (HBV), HCV, and HERV, whose detection rates were 100% (121/121), 84.3% (102/121), 43.8% (53/121), 23.1% (28/121), and 84.3% (102/121), respectively. Other viruses included papillomavirus (9/121 [7.4%]), influenza virus (15/121 [12.4%]), alphaherpesvirus (human herpesvirus 1 [HHV-1]) (4/121 [3.3%]), adenovirus (10/121 [8.3%]), and gemykibivirus (43/121 [35.5%]). Circovirus or enterovirus B was only detected in a single individual (Fig. 2A; see Table S2 in the supplemental material).
FIG 2.
Distribution of the main plasma viruses in different groups. (A) Positive rate for main viruses from all individuals. (B) Family-level taxonomic composition of the plasma virome; the relative abundance of the top 6 virus families is shown in the bar plot. (C) Comparison of viral abundance between different groups, shown as reads per million (RPM).
Detection rate of plasma viruses in different groups. Download Table S2, DOCX file, 0.01 MB (15.7KB, docx) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
HIV-1 infection reshaped the plasma virome of MSM.
Compared to non-MSM individuals, MSM did not show obvious changes in overall plasma viral reads; however, HIV-1 infection significantly increased the abundance of plasma viral reads. AIDS patients (CD4 < 200) had the highest abundance of viral reads (excluding HIV-1 reads). Even though the viral abundance of individuals receiving ART (CD4 > 200) was higher than that of HIV-1-negative MSM, ART successfully controlled the shedding of different viruses into plasma (Fig. 2C).
When specific viral components were examined, HIV-1 infection led to a decrease in phage abundance and increase in eukaryotic virus abundance in MSM (especially pegivirus), compared to non-MSM (Fig. 2B). MSM and HIV-1-infected individuals displayed similar plasma viral profiles, with anellovirus being dominant in almost all individuals, followed by pegivirus and HBV. ART usage could restore the virome profile toward that of HIV-1-negative individuals. There was no difference in detection rate of anellovirus among different groups, while HIV-1-infected MSM with CD4 < 200 had relatively lower detection rate for several viruses, including pegivirus, HBV, HCV, and HERV compared to other MSM (Fig. 2A).
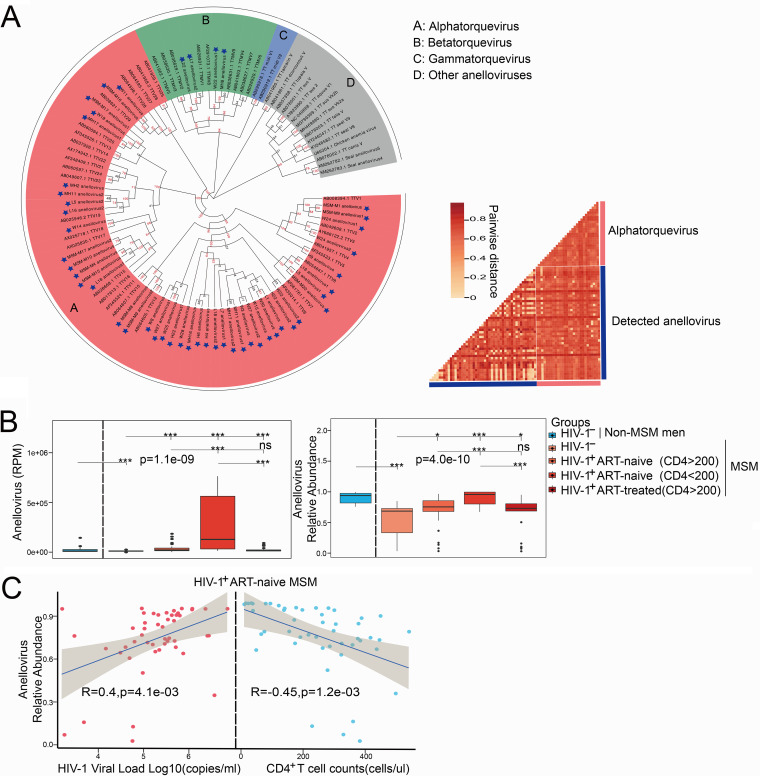
Association of anellovirus with the progress of HIV-1 infection.
As anellovirus dominated the plasma virome, we further analyzed its changes and diversity in different groups. Compared to non-MSM, MSM had a lower abundance of anellovirus. HIV-1 infection increased the abundance of anellovirus, with AIDS patients (CD4 < 200) showing the highest abundance. In addition, ART could decrease the abundance of anellovirus (Fig. 3B). Importantly, anellovirus had a significantly negative correlation with CD4+ T cell counts (Spearman’s r = 0.4, P = 0.0041) and CD4+/CD8+ ratio (Spearman’s r = −0.44, P = 1.4e−03), and a positive correlation was observed between anellovirus and HIV-1 viral load. After ART, those correlations disappeared (Fig. 3C; and Fig. S2B in the supplemental material).
FIG 3.
Associations of anellovirus with HIV-1 infection and CD4+ T cell counts. (A, left) Phylogenetic tree of anellovirus; newly discovered anelloviruses are labeled with blue stars. The sample source of each anellovirus is indicated as follows: MSM-M, HIV-1-negative non-MSM; MH, HIV-1-negative MSM; W, HIV-1-infected ART-naive MSM (CD4 > 200); L, AIDS patients (CD4 < 200); H, HIV-1-infected ART-treated MSM (CD4 > 200). The number at the end of the name represents the number of full-length ORF1 genes obtained. (Right) The heat map represents the pairwise distance of anelloviruses. (B, left) Read count (normalized by RPM) of pegivirus in samples from different groups. (Right) Relative abundance of pegivirus in samples from different groups. (C) Spearman's correlation between anellovirus and HIV-1 viral load (left) and CD4+ T cell counts (right) in HIV-1-infected ART-naive MSM.
All three main genera (alpha, beta, and gamma) of anelloviruses could be annotated using reads and partial contigs. In total, we obtained 44 alphatorqueviruses and 4 betatorqueviruses with full-length open reading frame 1 (ORF1) sequences. Phylogenetic analysis based on these 48 new anelloviruses and other references showed that these anelloviruses were highly divergent from each other, with a mean pairwise distance of about 0.69 (Fig. 3A).
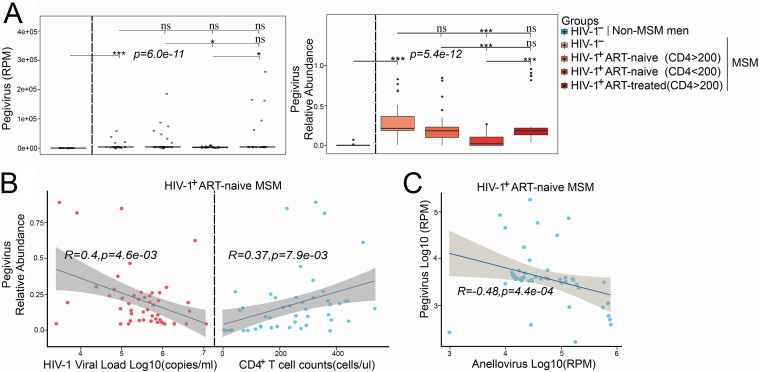
Association of pegivirus with the progress of HIV-1 infection.
Pegivirus C was the only species from Pegivirus genus identified from these participants. MSM had a higher abundance of pegivirus than non-MSM, and HIV-1 infection slightly decreased the abundance of pegivirus. The lowest abundance of pegivirus was observed in AIDS patients (CD4 < 200), even though they had the highest overall plasma viral abundance (Fig. 4A). To explore whether pegivirus changes along the HIV-1 disease progression, we compared the relative abundances of pegivirus among HIV-1-infected MSM with different clinical indexes. In HIV-1-infected ART-naive MSM, pegivirus abundance showed a positive correlation with CD4+ T cell counts (Spearman’s r = 0.37, P = 0.0079) and CD4+/CD8+ ratio (Spearman’s r = 0.41, P = 0.038) and a negative correlation with HIV-1 viral load (Spearman’s r = 0.4, P = 0.0046) (Fig. 4B; Fig. S2C). No significant correlation between pegivirus and any clinical indexes (CD4+ T cell counts, CD8+ T cell counts, and CD4+/CD8+ ratio) was found in MSM receiving ART (Fig. S2C). A negative correlation was observed between the abundances of pegivirus and anellovirus (Fig. 4C).
FIG 4.
Associations of pegivirus with HIV-1 infection and CD4+ T cell counts. (A, left) Read count (normalized by RPM) of pegivirus in samples from different groups. (Right) Relative abundance of pegivirus in samples from different groups. (B) Spearman's correlation between pegivirus and HIV-1 viral load (left) and CD4+ T cell counts (right) in HIV-1-infected ART-naive MSM. (C) Spearman's correlation between anelloviridae and pegivirus read counts in HIV-1-infected ART-naive MSM.
DISCUSSION
In this study, we reported the total plasma virome in MSM and its change during HIV-1 infection. Overall, same as in previous reports, the most frequently found viruses in these populations are blood-borne viruses, such as anellovirus, pegivirus, HBV, HCV, and HERV. Except for anellovirus, HIV-1 infection in MSM led to higher positive rate for pegivirus, HBV, HCV, as well as HERV, indicating a higher risk of infection by other viruses in this Chinese population, due possibly to unsafe sex or impaired immune status. An increased plasma viral load was observed after HIV-1 infection, which was associated with lower CD4+ T cell counts. Besides HIV-1 infection, ART could also reduce the shedding of several other viruses into the plasma.
Anelloviruses constitute a diverse group of species and are highly prevalent in the healthy population. Increased anellovirus abundance was observed after immunosuppressive therapy and organ transplantation (9, 19), and it is considered a potential indicator for immune status or disease progression (20). As the most abundant viruses in the blood, a recent study showed anellovirus was almost ubiquitous in the general population (21). Even though the prevalence of anellovirus was also the same here in MSM and after HIV-1 infection, its abundance greatly increased after HIV-1 infection and was negatively correlated with CD4+ T cell counts, which could be restored by ART. Our data again highlighted the potential use of anellovirus as a predictor of disease pathophysiology or immune status. However, decreased anellovirus abundance was observed in MSM compared to the non-MSM group, probably due to a bloom in pegivirus. Pegivirus is an RNA virus that belongs to the Flaviviridae family and commonly causes persistent infection (22, 23). The global prevalence of pegivirus in healthy blood donors ranges from 1% to 9% among different regions (24), but in HIV-1-infected populations, the prevalence is higher—up to more than 80% (25, 26). Recent studies in China showed that the prevalences of pegivirus in intravenous drug users (IDUs) and MSM were ∼30% and 18.3%, respectively (25, 27), indicating a higher risk of transmission through blood. The increased abundance of plasma pegivirus in MSM here simply reflected the higher infection risk for them and raised concerns that unsafe blood sharing or transfusion may further increase the transmission of pegivirus among the MSM population. Unlike anellovirus, AIDS patients had lowest abundance of pegivirus, and the shedding of pegivirus in AIDS patients was inversely associated with the host’s immune status. Previous studies reported that pegivirus inhibited HIV-1 replication, and coinfection with pegivirus in HIV-1-infected individuals could promote survival (14, 15, 28). Anellovirus and pegivirus are the main components of the plasma virome with the highest abundance: their inverse correlation indicates that both viruses’ persistent infections may have different modulation effects on the immune system, and they could be potential indicators of the disease progression of HIV-1 infection. However, the causal relationship of both viruses and disease pathophysiology or immune status remains to be elucidated.
Other viruses were also detected in the plasma, reflecting their complex sources. For example, all participants were sampled without clear signs of respiratory symptoms: the detection of influenza virus A in 15 individuals could be the results of asymptomatic infections of these people, since influenza virus infections with no clear symptoms are quite common (29). Gemykibivirus belongs to a divergent group of single-stranded DNA viruses, and it was detected in some MSM with or without HIV-1 infection. Several studies showed its presence/enrichment in HIV-1-infected people and also patients with other diseases (30–33), which suggested this virus could be associated with human disease (34). Even though more evidence is still needed, the detection of gemykibivirus here raises question about its pathogenic role in human. That 9 of 10 adenovirus cases and all human herpesvirus 1 (HHV-1 [4/4]) and human enterovirus B (HEV-B [1/1]) cases were detected in MSM and HIV-1-infected individuals indicates the increased risk of infections of different transmissible viruses in these populations. Papillomaviruses can persist in peripheral blood mononuclear cells (PBMCs), and healthy blood donors from a previous study showed 8 to 15% positive rate for the HPV genome (35), so the positivity for HPV in the healthy group could represent a general distribution of this virus in these investigated individuals.
Even though this study compared the plasma virome changes in different groups, this is a cross-sectional study, and further research on dynamics of the virome, especially the abundance changes of anellovirus and pegivirus along with disease progression, is needed to validate the clinical implications of them. Another limitation is that we used random genome amplification to enrich virus genomes, which may magnify the real abundance of some viruses such as anellovirus.
In summary, we systematically compared plasma virome in MSM with or without HIV-1 infection and compared the virome changes at different CD4+ T cell counts. Altered plasma viral composition was observed in MSM and those with HIV-1 infection. Overall, anellovirus shedding was proportional to CD4+ T cell counts and positively correlated with HIV-1 viral load; opposite correlations were observed between the clinical parameters of HIV infection and pegivirus abundance, suggesting these two types of viruses may have different roles in the regulation of immune status. Further functional studies are needed to investigate the specific functions of both viruses in MSM and patients with HIV-1 infection.
MATERIALS AND METHODS
Ethics statement.
This study was approved by the Ethics Committees of Shenzhen Center for Disease Control and Prevention (Shenzhen CDC) and Institut Pasteur of Shanghai and complied with all relevant ethical regulations. Oral or written informed consent was obtained from volunteers before sample collection.
Participants, sampling and viral metagenomics processing.
All participants were recruited from Shenzhen CDC, China (Table 1). At least 2 ml blood was collected from each participant, and plasma was separated within 24 h at Shenzhen CDC, China. All plasma samples were stored at –80°C until use. Quantification of HIV-1 viral load was done by quantitative reverse transcription-PCR (RT-qPCR; diagnostic kit from Da An Gene Co., Ltd., of Sun Yat-Sen University). Virus-like particle (VLP) enrichment and nucleic acid preamplification were performed as previously described (36). Plasma samples were centrifuged at 12,000 rpm at 4°C for 10 min, and filtrates were passed through sterile 0.45-μm-pore filters (Corning, USA) to reduce the background of human and bacterial cells. Filtered samples were incubated with 20 U Benzonase (Merck, Germany), 15 U Turbo DNase (Life Technologies, USA), and 20 U RNase I (Promega, USA) to digest free nucleic acids (NAs) at 37°C for 1.5 h, 20 mM EDTA (Sigma-Aldrich, Germany) was added, and the mixture was incubated at 65°C for 10 min to terminate the digestion. Viral nucleic acids were extracted with the QIAamp miniElute virus kit (Qiagen, Germany). Total RNA extracts were reverse transcribed using the SuperScript III kit with a mixture of random hexamer primers and oligo(dT) primers. The cDNA was treated with RNase H (New England BioLabs, Inc., USA) prior to whole-genome amplification using a multiple annealing and looping-based amplification cycle (MALBAC) single-cell DNA Quick-Amp kit (37) (Yikon Genomics, China). Amplified DNA was sheared to an average fragment length of 250 bp using Covaris E210 (Covaris, USA). Sheared DNA was purified and used for Illumina library construction using NEBNext Ultra II DNA (New England BioLabs, Inc., USA). The sequencing libraries were quantified by Qubit3.0 (Invitrogen, USA). Sequencing was done on the Illumina Nova-seq platform (Illumina, USA) with 2 × 150-bp paired reads, yielding an average of 67 million reads per library.
Bioinformatic analysis.
Sequencing data were demultiplexed using Illumina software to generate FastQ files for each sample. The raw data of NGS were first cleaned by Cutadapt v.1.18 and Trimmomatic v.0.38 (38) by removing Illumina sequencing adaptor and low-quality sequences (SLIDINGWINDOW:4:20 MINLEN:50). Human- and bacterium-derived sequences were filtered out by Bowtie2 v.2.3.4.3 (39). The remaining high-quality reads were de novo assembled by Megahit v.1.1.3 (40). Assembled contigs were dereplicated and clustered at 95% identity using Vsearch v.2.10.4 (41). After low-complexity contigs were removed, high-quality contigs with a length of more than 500 nucleotides (nt) were further filtered for human and bacterial sequences. BLASTX from DIAMOND v.0.9.24 (42) was used to search against the NCBI nonredundant protein sequence (nr) database to annotate viral contigs; the remaining contigs were mapped to the NCBI nucleotide sequence (nt) database by BLASTN (43, 44). Furthermore, cleaned reads were mapped to viral contigs by BLASTN. Viral reads were first normalized by reads per million (RPM), and then abundance was calculated.
Phylogenetic tree construction.
The amino acid sequences of the anellovirus ORF1 gene were extracted from all anellovirus contigs using NCBI’s orfFinder tool. Reference ORF1 sequences of anelloviruses were downloaded from the NCBI’s GenBank database. All ORF1 sequences were aligned using MUSCLE (45), and the resulting alignment was used to construct a neighbor-joining phylogenetic tree with a p-distance model and 1,000 bootstraps by MEGA X (46). The tree was visualized and modified with evolview (47).
Statistics.
Comparative statistical analyses between two groups were performed using the Mann-Whitney-Wilcoxon test. Comparative statistical analyses of three or more groups were performed using the Kruskal-Wallis H test. As shown on the figures, differences were considered significant at P < 0.001 (***), P < 0.01 (**), or P < 0.05 (*), and “ns” indicates no statistically significant differences (P > 0.05). For all studies, data are presented as representative of one independent experiment. All statistical analyses were performed by RStudio v.3.8.
Data availability.
Sequencing data have been uploaded to the Sequence Read Archive (SRA) database under accession no. PRJNA634526.
ACKNOWLEDGMENTS
This study was supported by the Shanghai Science & Technology Innovation Action Program (20MC1920100), the National Natural Science Foundation of China (81672033, U1302224), the Guangdong Basic and Applied Basic Research Foundation (2019B1515120003), the Shenzhen Health System Research Project (SZGW201810003), Shenzhen Key Medical Discipline Construction Fund (SZXK064), and the San-Ming Project of Medicine in Shenzhen (SZSM201811071).
C.Z. conceived the study idea. C.Z., J.Z., and P.H. supervised the study. K.L., R.X., J.Z., Y.Z., H.L., Z.Y., and Z.W. performed the experiments. K.L. and Y.L. interpreted the results and wrote the manuscript. X.J. provided thoughtful suggestions on the interpretation of results and contributed to critical revision of the manuscript.
The authors have declared they have no conflicts of interest.
REFERENCES
- 1.Moustafa A, Xie C, Kirkness E, Biggs W, Wong E, Turpaz Y, Bloom K, Delwart E, Nelson KE, Venter JC, Telenti A. 2017. The blood DNA virome in 8,000 humans. PLoS Pathog 13:e1006292. doi: 10.1371/journal.ppat.1006292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Virgin HW. 2014. The virome in mammalian physiology and disease. Cell 157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shkoporov AN, Clooney AG, Sutton TDS, Ryan FJ, Daly KM, Nolan JA, McDonnell SA, Khokhlova EV, Draper LA, Forde A, Guerin E, Velayudhan V, Ross RP, Hill C. 2019. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 26:527–541.e5. doi: 10.1016/j.chom.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Rascovan N, Duraisamy R, Desnues C. 2016. Metagenomics and the human virome in asymptomatic individuals. Annu Rev Microbiol 70:125–141. doi: 10.1146/annurev-micro-102215-095431. [DOI] [PubMed] [Google Scholar]
- 5.Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, Li X, Szeto CH, Sugimura N, Lam TY, Yu AC, Wang X, Chen Z, Wong MC, Ng SC, Chan MTV, Chan PKS, Chan FKL, Sung JJ, Yu J. 2018. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology 155:529–541.e5. doi: 10.1053/j.gastro.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 6.Vehik K, Lynch KF, Wong MC, Tian X, Ross MC, Gibbs RA, Ajami NJ, Petrosino JF, Rewers M, Toppari J, Ziegler AG, She JX, Lernmark A, Akolkar B, Hagopian WA, Schatz DA, Krischer JP, Hyoty H, Lloyd RE, TEDDY Study Group. 2019. Prospective virome analyses in young children at increased genetic risk for type 1 diabetes. Nat Med 25:1865–1872. doi: 10.1038/s41591-019-0667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, Stappenbeck TS, McGovern DP, Keshavarzian A, Mutlu EA, Sauk J, Gevers D, Xavier RJ, Wang D, Parkes M, Virgin HW. 2015. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Deng X, Linsuwanon P, Bangsberg D, Bwana MB, Hunt P, Martin JN, Deeks SG, Delwart E. 2013. AIDS alters the commensal plasma virome. J Virol 87:10912–10915. doi: 10.1128/JVI.01839-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thijssen M, Tacke F, Beller L, Deboutte W, Yinda KC, Nevens F, Laleman W, Van Ranst M, Pourkarim MR. 2020. Clinical relevance of plasma virome dynamics in liver transplant recipients. EBioMedicine 60:103009. doi: 10.1016/j.ebiom.2020.103009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, Lankowski A, Baldridge MT, Wilen CB, Flagg M, Norman JM, Keller BC, Luevano JM, Wang D, Boum Y, Martin JN, Hunt PW, Bangsberg DR, Siedner MJ, Kwon DS, Virgin HW. 2016. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 19:311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Handley SA, Desai C, Zhao G, Droit L, Monaco CL, Schroeder AC, Nkolola JP, Norman ME, Miller AD, Wang D, Barouch DH, Virgin HW. 2016. SIV infection-mediated changes in gastrointestinal bacterial microbiome and virome are associated with immunodeficiency and prevented by vaccination. Cell Host Microbe 19:323–335. doi: 10.1016/j.chom.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu GJ, Kula T, Xu Q, Li MZ, Vernon SD, Ndung'u T, Ruxrungtham K, Sanchez J, Brander C, Chung RT, O'Connor KC, Walker B, Larman HB, Elledge SJ. 2015. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science 348:aaa0698. doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li SK, Leung RK, Guo HX, Wei JF, Wang JH, Kwong KT, Lee SS, Zhang C, Tsui SK. 2012. Detection and identification of plasma bacterial and viral elements in HIV/AIDS patients in comparison to healthy adults. Clin Microbiol Infect 18:1126–1133. doi: 10.1111/j.1469-0691.2011.03690.x. [DOI] [PubMed] [Google Scholar]
- 14.Xiang J, Wünschmann S, Diekema DJ, Klinzman D, Patrick KD, George SL, Stapleton JT. 2001. Effect of coinfection with GB virus C on survival among patients with HIV infection. N Engl J Med 345:707–714. doi: 10.1056/NEJMoa003364. [DOI] [PubMed] [Google Scholar]
- 15.Williams CF, Klinzman D, Yamashita TE, Xiang J, Polgreen PM, Rinaldo C, Liu C, Phair J, Margolick JB, Zdunek D, Hess G, Stapleton JT. 2004. Persistent GB virus C infection and survival in HIV-infected men. N Engl J Med 350:981–990. doi: 10.1056/NEJMoa030107. [DOI] [PubMed] [Google Scholar]
- 16.Beyrer C, Baral SD, van Griensven F, Goodreau SM, Chariyalertsak S, Wirtz AL, Brookmeyer R. 2012. Global epidemiology of HIV infection in men who have sex with men. Lancet 380:367–377. doi: 10.1016/S0140-6736(12)60821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver Ratmann AS, Bezemer D, Gavryushkina A, Jurriaans S, Wensing A, de Wolf F, Reiss P, Fraser C, ATHENA observational cohort. 2016. Sources of HIV infection among men having sex with men and implications for prevention. Sci Transl Med 8:320–320ra2. doi: 10.1126/scitranslmed.aad1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.UNAIDS. 2019. UNAIDS data 2019. https://www.unaids.org/sites/default/files/media_asset/2019-UNAIDS-data_en.pdf. Accessed 3 August 2020.
- 19.De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, Okamoto J, Snyder TM, Cornfield DN, Nicolls MR, Weill D, Bernstein D, Valantine HA, Quake SR. 2013. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 155:1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E, Haas AR, Abbas A, Frye L, Christie JD, Bushman FD, Collman RG. 2015. Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. Am J Transplant 15:200–209. doi: 10.1111/ajt.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Focosi D, Spezia PG, Macera L, Salvadori S, Navarro D, Lanza M, Antonelli G, Pistello M, Maggi F. 2020. Assessment of prevalence and load of torquetenovirus viraemia in a large cohort of healthy blood donors. Clin Microbiol Infect 26:1406–1410. doi: 10.1016/j.cmi.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Smith DB, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff AS, Pletnev A, Rico-Hesse R, Stapleton JT, Simmonds P. 2016. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J Gen Virol 97:2894–2907. doi: 10.1099/jgv.0.000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. 2011. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol 92:233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang N, Dai R, Zhang X. 2020. Global prevalence of human pegivirus-1 in healthy volunteer blood donors: a systematic review and meta-analysis. Vox Sang 115:107–119. doi: 10.1111/vox.12876. [DOI] [PubMed] [Google Scholar]
- 25.Taklual W, Tang S, Yue W. 2019. Effect of human pegivirus route of transmission on the genetic distribution of the virus: an institution based cross-sectional study. Virol J 16:50. doi: 10.1186/s12985-019-1161-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anggorowati N, Yano Y, Subronto YW, Utsumi T, Heriyanto DS, Mulya DP, Rinonce HT, Widasari DI, Lusida MI, Soetjipto, Hayashi Y. 2013. GB virus C infection in Indonesian HIV-positive patients. Microbiol Immunol 57:298–308. doi: 10.1111/1348-0421.12033. [DOI] [PubMed] [Google Scholar]
- 27.Miao Z, Gao L, Song Y, Yang M, Zhang M, Lou J, Zhao Y, Wang X, Feng Y, Dong X, Xia X. 2017. Prevalence and clinical impact of human pegivirus-1 infection in HIV-1-infected individuals in Yunnan, China. Viruses 9:28. doi: 10.3390/v9020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tillmann HL, Heiken H, Knapik-Botor A, Heringlake S, Ockenga J, Wilber JC, Goergen B, Detmer J, McMorrow M, Stoll M, Schmidt RE, Manns MP. 2001. Infection with GB virus C and reduced mortality among HIV-infected patients. N Engl J Med 345:715–724. doi: 10.1056/NEJMoa010398. [DOI] [PubMed] [Google Scholar]
- 29.Leung NH, Xu C, Ip DK, Cowling BJ. 2015. The fraction of influenza virus infections that are asymptomatic: a systematic review and meta-analysis. Epidemiology 26:862–872. doi: 10.1097/EDE.0000000000000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uch R, Fournier PE, Robert C, Blanc-Tailleur C, Galicher V, Barre R, Jordier F, de Micco P, Raoult D, Biagini P. 2015. Divergent gemycircularvirus in HIV-positive blood, France. Emerg Infect Dis 21:2096–2098. doi: 10.3201/eid2111.150486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bezerra RS, Bitencourt HT, Covas DT, Kashima S, Slavov SN. 2020. Metagenomic identification of human Gemykibivirus-2 (HuGkV-2) in parenterally infected blood donors from the Brazilian Amazon. Int J Infect Dis 98:249–251. doi: 10.1016/j.ijid.2020.06.096. [DOI] [PubMed] [Google Scholar]
- 32.Zhou C, Zhang S, Gong Q, Hao A. 2015. A novel gemycircularvirus in an unexplained case of child encephalitis. Virol J 12:197. doi: 10.1186/s12985-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamberto I, Gunst K, Müller H, Zur Hausen H, de Villiers EM. 2014. Mycovirus-like DNA virus sequences from cattle serum and human brain and serum samples from multiple sclerosis patients. Genome Announc 2:e00848-14. doi: 10.1128/genomeA.00848-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Li Y, He X, Ma J, Hong W, Hu F, Zhao L, Li Q, Zhang J, Zhang C, Zhang F. 2019. Gemykibivirus genome in lower respiratory tract of elderly woman with unexplained acute respiratory distress syndrome. Clin Infect Dis 69:861–864. doi: 10.1093/cid/ciz072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen ACH, Keleher A, Kedda MA, Spurdle AB, McMillan NA, Antonsson A. 2009. Human papillomavirus DNA detected in peripheral blood samples from healthy Australian male blood donors. J Med Virol 81:1792–1796. doi: 10.1002/jmv.21592. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Fu X, Ma J, Zhang J, Hu Y, Dong W, Wan Z, Li Q, Kuang YQ, Lan K, Jin X, Wang JH, Zhang C. 2019. Altered respiratory virome and serum cytokine profile associated with recurrent respiratory tract infections in children. Nat Commun 10:2288. doi: 10.1038/s41467-019-10294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zong C, Lu S, Chapman AR, Xie XS. 2012. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 41.Rognes T, Flouri T, Nichols B, Quince C, Mahe F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchfink B, Xie C, Huson DH. 2015. fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–63. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 43.NCBI Resource Coordinators. 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 45.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, Gao S, Lercher MJ, Hu S, Chen WH. 2012. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res 40:W569–W572. doi: 10.1093/nar/gks576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation analysis of HIV-1 viral load with CD4+ T cell counts, CD8+ T cell counts, and CD4+/CD8+ ratio. (A) Correlation between peripheral circulating CD4+ T cell counts, CD8+ T cell counts, CD4+/CD8+ ratio, and HIV-1 viral load in HIV-1-infected ART-naive MSM. (B) Spearman's correlation between HIV-1 viral load and HIV-1 read count (normalized by RPM). Download FIG S1, TIF file, 0.5 MB (507.7KB, tif) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Taxonomic composition of NGS results and correlation analysis of anellovirus and pegivirus with CD4+ T cell counts, CD8+ T cell counts, and CD4+/CD8+ ratio. (A) Taxonomic composition of different plasma samples as determined by NGS. Abundance is shown as reads per million (RPM); other terms represent unclassified eukaryotic reads or unknown reads. (B) Spearman’s correlation between anellovirus and CD8+ T cell counts, CD4+ T cell counts, and the CD4+/CD8+ ratio in ART-treated or ART-naive groups. (C) Spearman’s correlation between pegivirus and CD4+ T cell counts, CD8+ T cell counts, and CD4+/CD8+ ratio in ART-treated or ART-naive groups. Download FIG S2, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
All the main viral families detected and number of sequencing reads. Download Table S1, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detection rate of plasma viruses in different groups. Download Table S2, DOCX file, 0.01 MB (15.7KB, docx) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Sequencing data have been uploaded to the Sequence Read Archive (SRA) database under accession no. PRJNA634526.