Spikes in rickettsioses occur as deforestation, urbanization, and homelessness increase human exposure to blood-feeding arthropods. Still, effective Rickettsia vaccines remain elusive.
KEYWORDS: Rickettsia, rickettsioses, spotted fever group, transitional group, typhus group, lipid A, lipopolysaccharide, pathogenesis
ABSTRACT
Species of Rickettsia (Alphaproteobacteria: Rickettsiales) are obligate intracellular parasites of a wide range of eukaryotes, with recognized arthropod-borne human pathogens belonging to the transitional group (TRG), typhus group (TG), and spotted fever group (SFG) rickettsiae. Growing in the host cytosol, rickettsiae pilfer numerous metabolites to make a typical Gram-negative bacterial cell envelope. The O-antigen of rickettsial lipopolysaccharide (LPS) is immunogenic and has been shown to tether the S-layer to the rickettsial surface; however, little is known about the structure and immunogenicity of the Rickettsia lipid A moiety. The structure of lipid A, the membrane anchor of LPS, affects the ability of this molecule to interact with components of the host innate immune system, specifically the MD-2/TLR4 receptor complex. To dissect the host responses that can occur during Rickettsia in vitro and in vivo infection, structural analysis of Rickettsia lipid A is needed. Lipid A was extracted from four Rickettsia species and structurally analyzed. R. akari (TRG), R. typhi (TG), and R. montanensis (SFG) produced a similar structure, whereas R. rickettsii (SFG) altered the length of a secondary acyl group. While all structures have longer acyl chains than known highly inflammatory hexa-acylated lipid A structures, the R. rickettsii modification should differentially alter interactions with the hydrophobic internal pocket in MD2. The significance of these characteristics toward inflammatory potential as well as membrane dynamics between arthropod and vertebrate cellular environments warrants further investigation. Our work adds lipid A to the secretome and O-antigen as variable factors possibly correlating with phenotypically diverse rickettsioses.
IMPORTANCE Spikes in rickettsioses occur as deforestation, urbanization, and homelessness increase human exposure to blood-feeding arthropods. Still, effective Rickettsia vaccines remain elusive. Recent studies have determined that Rickettsia lipopolysaccharide anchors the protective S-layer to the bacterial surface and elicits bactericidal antibodies. Furthermore, growing immunological evidence suggests vertebrate sensors (MD-2/TLR4 and noncanonical inflammasome) typically triggered by the lipid A portion of lipopolysaccharide are activated during Rickettsia infection. However, the immunopotency of Rickettsia lipid A is unknown due to poor appreciation for its structure. We determined lipid A structures for four distinct rickettsiae, revealing longer acyl chains relative to highly inflammatory bacterial lipid A. Surprisingly, lipid A of the Rocky Mountain spotted fever agent deviates in structure from other rickettsiae. Thus, lipid A divergence may contribute to variable disease phenotypes, sounding an alarm for determining its immunopotency and possible utility (i.e., as an adjuvant or anti-inflammatory) for development of more prudent rickettsiacidal therapies.
OBSERVATION
Lipopolysaccharide (LPS), an amphipathic molecule comprising the majority of the outer leaflet of the Gram-negative bacterial outer membrane, is composed of extracellular polysaccharide chains (O-antigen) linked to a membrane phosphoglycolipid (lipid A) by a core oligosaccharide. Depending on its structure, lipid A can be a potent activator of the mammalian immune system through detection by MD-2/TLR4 (1, 2) and the noncanonical inflammasome (3, 4). However, not all lipid A structures are equal in their ability to activate these mammalian cellular receptors (5); for instance, some bacterial pathogens employ lipid A modification as a mechanism of immune evasion when infecting a mammalian host (6, 7). Given the importance of lipid A for membrane integrity, resistance to antibiotics, and use as a vaccine adjuvant, it is crucial to understand the structure and immunostimulatory potential of lipid A from Gram-negative pathogens.
Species of Rickettsia, Gram-negative obligate intracellular Alphaproteobacteria, are metabolic parasites of a wide range of eukaryotic hosts (8). Across the Rickettsia tree, agents of human disease from the transitional group (TRG), typhus group (TG), and spotted fever group (SFG) rickettsiae are interspersed with numerous invertebrate and protist endosymbionts, most with unknown pathogenicity. All described rickettsioses are vector-borne diseases that differ in their severity of illness and clinical manifestations (9), facts undoubtedly linked to variability in the Rickettsia secretome (10) and O-antigen epitopes (11, 12). Rickettsia O-antigen contains the sugar quinovosamine (11–13), and transposon-mediated disruption of an epimerase involved in its production abrogates S-layer formation, dampens pathogenicity, and abolishes recognition by bactericidal antibodies (14). While the immunopotency of the lipid A moiety of Rickettsia LPS remains unknown, it could be proinflammatory given MD-2/TLR4 and noncanonical inflammasome activation during infection (15–20). However, the lone published Rickettsia lipid A structure (an unreported strain of Rickettsia typhi, the agent of murine typhus) revealed a bisphosphorylated hexa-acylated structure with acyl chains ranging from C14 to C18 in length (21), much longer than the C12-C14 chains of the highly inflammatory hexa-acylated lipid A of Escherichia coli (Fig. 1A). These collective observations warrant determining Rickettsia lipid A structures for phenotypically diverse species to assess if any structural variability correlates with disease severity.
FIG 1.

Variable acyl chain lengths in Rickettsia lipid A. (A) The structure of the highly inflammatory lipid A of E. coli. Asterisks depict acyl chains that diverge in length in Rickettsia lipid A. (B) Structure of lipid A isolated from R. akari strain Hartford, R. typhi strain Wilmington, and R. montanensis strain M5/6 during Vero 76 cell infection. (C) Structure of lipid A isolated from R. rickettsii strains Sheila Smith and Iowa during Vero 76 cell infection (for full spectra, see Fig. S1 in the supplemental material). (D) Schematic representation of analytical methods used to determine fatty acid compositions, with the R. rickettsii lipid A shown as an example. Lipid A from R. rickettsii strain Sheila Smith was subjected to sequential release of fatty acids and analyzed at each step with MALDI-TOF analysis (for full spectra, see Fig. S2). m/z, mass-to-charge ratio of lipid A ions identified during MALDI-TOF analysis.
MALDI-TOF analysis of Rickettsia lipid A. Lipid A microextraction from rickettsial cultures was performed as previously described (A. El Hamidi, A. Tirsoaga, A. Novikov, A. Hussein, et al., J Lipid Res 46:1773–1778, 2005; A. J. Scott, B. Flinders, J. Cappell, T. Liang, et al., Pathog Dis 74, 2016; A. Tirsoaga, A. El Hamidi, M. B. Perry, M. Caroff, et al., J Lipid Res 48:2419–2427, 2007). See the text for additional details. (A) R. typhi strain Wilmington, (B) R. montanensis strain M5/6, (C) R. akari strain Hartford, and (D) R. rickettsii strain Sheila Smith. Insets identify the vector host and relative virulence to humans. Peak labels are masses of singly charged ions. Major peaks are indicated by colored labels (A to C, gray; D, red), with minor peaks (likely representing heterogeneity in fatty acid incorporation) noted by dashed lines. Download FIG S1, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2021 Guillotte et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fatty acid analysis of Rickettsia lipid A. (A, D, G, J, and M) Lipid A of R. akari strain Hartford, R. typhi strain Wilmington, R. montanensis strain M5/6, R. rickettsii strain Sheila Smith, and R. rickettsii strain Iowa was subjected to sequential fatty acid release prior to MALDI-TOF analysis of daughter molecules (A. El Hamidi, A. Tirsoaga, A. Novikov, A. Hussein, et al., J Lipid Res 46:1773–1778, 2005; A. Tirsoaga, A. El Hamidi, M. B. Perry, M. Caroff, et al., J Lipid Res 48:2419–2427, 2007). (B, E, H, K, and N) Primary ester-linked fatty acids (3′ position, green; 3 position, blue), which are more readily released than secondary ester-linked fatty acids, were liberated using mild alkali treatment with ammonium hydroxide, yielding predominate ions of either m/z ∼1,247/1,273 (gray for palmitate/stearate in R. akari, R. typhi, and R. montanensis) or m/z 1,191 (red for laurate in R. rickettsii) that are tri-acyl daughter molecules. (C, F, I, L, and O) The remaining secondary ester-linked fatty acid attached to the hydroxypalmitate at the 2′ position was liberated using a second alkaline reaction with methylamine, yielding the di-acyl daughter ion at m/z 1009 for all analyzed strains. Structures (insets) depict the major lipid A ion in each spectrum, with minor peaks (dashed lines) likely representing heterogeneity in fatty acid incorporation. Download FIG S2, PDF file, 2.5 MB (2.5MB, pdf) .
Copyright © 2021 Guillotte et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To investigate possible lipid A diversity between rickettsiae, we isolated and analyzed lipid A from three human pathogens (R. akari, R. typhi, and R. rickettsii) and a nonpathogen (R. montanensis), all grown in Vero cell cultures. Lipid A extracted directly from infected-Vero cells for R. akari, R. typhi, and R. montanensis produced major ions at m/z 1,936 that are largely consistent with the published R. typhi lipid A structure (21) and likely represent bisphosphorylated hexa-acyl structures (Fig. 1B). However, there is considerable variance between species in the number and intensity of minor lipid A ions (m/z 1,963, 1,909, and 1,882) that likely represents heterogeneity in fatty acid (FA) incorporation (see Fig. S1 in the supplemental material) known to occur in some bacteria (22). In contrast, the Rocky Mountain spotted fever agent, R. rickettsii, produces a lower-molecular-weight lipid A molecule (major ion, m/z 1,882) (Fig. 1C), corresponding to a loss of four carbons from one or more FA chains (Fig. 1D). Sequential FA release by alkaline treatment indicates a laurate (C12) on 2′-hydroxypalmitate (C16-OH) as opposed to palmitate/stearate (C16/C18) in the other Rickettsia structures (Fig. S2).
The underlying mechanisms for acyl chain heterogeneity within species and divergence between R. rickettsii and other rickettsiae are not readily apparent. The enzymes for lipid A biosynthesis are highly conserved across rickettsiae (Fig. S3), including the motifs that function as a hydrocarbon ruler in late acyltransferase LpxJ (23). However, inspection of analogous motifs in late acyltransferase LpxL revealed an active-site substitution (Leu-Ile) conserved in SFG rickettsiae that diverge after R. gravesii (Fig. 2, Fig. S4). The potential for this modified LpxL active site to incorporate shorter 2′ acyl chains into Rickettsia lipid A awaits experimentation. Despite conserved FA synthesis machinery, malonyl-ACP is synthesized in rickettsiae using host-derived precursors and cofactors (24), making FA availability and incorporation contingent on host metabolic burden during infection. Variance in FA acyl chain length may also reflect engrained flexibility for adapting to divergent cellular environments encountered across invertebrate and vertebrate hosts, making it a priority to assess lipid A structures directly from arthropod and mammalian infections.
FIG 2.
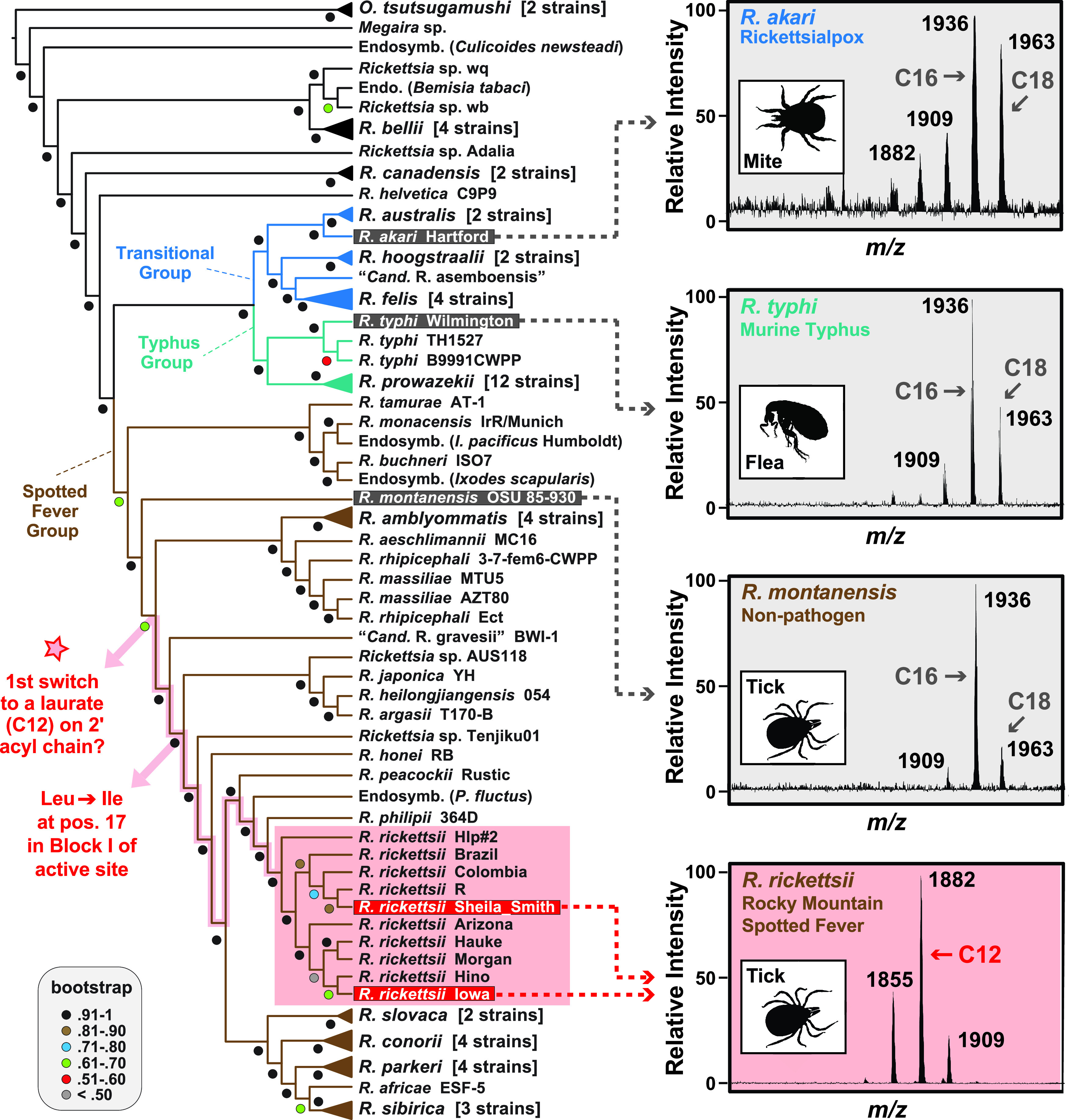
Evolution of structural variability in Rickettsia lipid A. Genome-based phylogeny on the left was estimated as previously described (25). Mass spectra on the right depict MALDI-TOF analyses of lipid A extracted from R. akari strain Hartford, R. typhi strain Wilmington, R. montanensis strain M5/6, and R. rickettsii strains Sheila Smith and Iowa (for full spectra, see Fig. S1 in the supplemental material). Peak labels are masses of singly charged ions; arrows denote major peaks. Insets show typical arthropod vectors. The star on the phylogeny indicates the earliest point in SFG rickettsia evolution where a switch from palmitate/stearate (C16/18) to laurate (C12) on lipid A 2′ hydroxypalmitate could have occurred. The node reflecting a switch to a conserved Ile in position 17 of block I of the LpxL active site is also noted (see Fig. S4 for more details).
Raetz pathway for biosynthesis of lipid A in rickettsiae. Rickettsiae lack enzymes for amino sugar metabolism and synthesis of pentose phosphates, necessitating the acquisition of host N-acetylglucosamine-1-P and ribose-5-P to initiate biosynthesis of lipid IV(A) and 3-deoxy-d-manno-octulosonate (Kdo), respectively (T. P. Driscoll, V. I. Verhoeve, M. L. Guillotte, S. S. Lehman, et al., mBio 8:e00859-17, 2017). Using these two pilfered metabolites, 14 enzymes (conserved in Rickettsia genomes, inset at top) and three cofactors (ATP, UTP, and phosphoenolpyruvate) are required for Kdo2-lipid A biosynthesis. Kdo2 is not shown on the final structure, as our lipid A microextraction protocol removed Kdo residues prior to lipid A analysis (the structure of Kdo2 and the inner and outer core oligosaccharide have not been determined for Rickettsiae to date). Enzymes contain locus tags for R. typhi strain Wilmington enzymes that were used in blastp searches against the NCBI Rickettsia database (taxid 780) to confirm the strict conservation of all 14 CDS in the five strains analyzed in this study. Searches were performed with composition-based statistics, with no filter used. Default matrix parameters (BLOSUM62) and gap costs (existence, 11; extension, 1) were implemented, with an inclusion threshold of 0.005. Download FIG S3, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2021 Guillotte et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparative analysis of Rickettsia LpxL enzymes. Lipid A biosynthesis in E. coli utilizes the late acyltransferase LpxL to add a laurate (C12) to the 2′ hydroxymyristate (D. A. Six, S. M. Carty, Z. Guan, and C. R. H. Raetz, Biochemistry 47:8623–8637, 2008). Our work on Rickettsia lipid A indicates that LpxL enzymes add either a palmitate or stearate on the 2′-hydroxypalmitate (R. akari, R. typhi, and R. montanensis) or a laurate (C12) on the 2′-hydroxypalmitate (R. rickettsii strains Sheila Smith and Iowa). (A) Multiple-sequence alignment of LpxL homologs using MUSCLE (default parameters) (R. C. Edgar, Nucleic Acids Res 32:1792–1797, 2004) indicates strong conservation across the entirety of the proteins (the proteins from R. rickettsii strains Sheila Smith and Iowa are identical). Amino acid similarity (% identity) between homologs is shown below the alignment. Structural modeling of the R. typhi LpxL protein to Acinetobacter baumannii LpxM (PDB entry 5KNK) using Phyre2 (L. A. Kelley and M. J. E. Sternberg, Nat Protoc 4:363–371, 2009) indicates strong conservation within the three blocks recognized within GPAT, LPAAT, DHAPAT, and LPEAT acyltransferases (J. Yao and C. O. Rock, Biochim Biophys Acta 1831:495–502, 2013; T. M. Lewin, P. Wang, and R. A. Colema, Biochemistry 38:5764–5771, 1999). Active-site residues within blocks are colored according to charge. Asp193 is proposed to participate in the active-site charge relay system with His122 (R. J. Heath, C. O. Rock, J Bacteriol 180:1425–30, 1998; A. F. Neuwald, Curr Biol 7:R465-R466, 1997). There is strict conservation within the regions analogous to the deep hydrophobic cleft of LpxM predicted to function as a hydrocarbon ruler (D. Dovala, C. M. Rath, Q. Hu, W. S. Sawyer, et al., Proc Natl Acad Sci U S A 113:E6064–E6071, 2016). (B) Mapping of character states for the five unique R. rickettsii LpxL residues over the Rickettsia phylogeny from Fig. 1. Shared residues between R. rickettsii strains and other rickettsiae are highlighted yellow. The strict conservation of Ile130 within block I of the LpxL active site is illustrated at left. All other information follows the description for Fig. 1. Download FIG S4, PDF file, 0.7 MB (692.2KB, pdf) .
Copyright © 2021 Guillotte et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The membrane or immunological function of shorter 2′ secondary acyl chains in derived SFG rickettsiae remains to be determined. If other human pathogens in this lineage diverging after R. montanensis (Fig. 2, red star) have acyl chain lengths similar to those of R. rickettsii, this lipid A structure may serve as a candidate drug target or vaccine adjuvant for treatment of these particular SFG rickettsioses. Furthermore, determining lipid A structures for other species throughout the rickettsial tree stands to illuminate additional structural diversity that bolsters joining lipid A with the secretome and O-antigen as variable factors defining phenotypically diverse rickettsioses.
Rickettsiae are rare among obligate intracellular bacteria in that they lyse the phagosome and colonize the host cytosol, risking inordinate metabolic thievery in the face of host surveillance systems (24). The pilfering of host metabolites for cell envelope synthesis entangles rickettsial growth and virulence. Further characterization of rickettsial lipid A, particularly its immunopotency, is imperative for advancing knowledge on host-pathogen interactions and a highly unique mode of obligate intracellular parasitism.
Bacterial strains and cell culture.
Vero 76 cells (African green monkey kidney; ATCC CRL-1587) were maintained in Dulbecco’s modification of Eagle’s medium (DMEM with 4.5 g/liter glucose and 480 l-glutamine) supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37°C with 5% CO2. Rickettsiae were propagated in Vero 76 cells grown in DMEM (supplemented with 5% FBS at 34°C with 5% CO2) for 48 to 72 h until confluence before harvesting. Rickettsial cultures were partially purified by mild sonication (one 10-s pulse; power output 6) followed by 5.0-μm filtration and collected by gentle centrifugation (2,000 × g; 15 min). Pellets were washed once in ultrapure water prior to lipid A extraction.
Lipid A extraction.
Lipid A microextraction from rickettsial cultures initiated by placing pellets from 1 to 4 confluent T75 cm2 culture flasks (maintained as described above) in 400 μl of solution (5 parts isobutyric acid, 3 parts 1 M NH4OH) and heated at 100°C for 1 h followed by a 15-min incubation on ice and centrifugation at 2,000 × g for 15 min. The lipid A-containing bottom layer of supernatant was collected, mixed in equal parts with H2O, frozen, and then lyophilized. Contaminants were washed from the dried material by two rounds of methanol (MeOH) washes (1 ml MeOH with sonicating and pelleting at 10,000 × g for 5 min). The final product was reconstituted in 50 μl chloroform-MeOH-H2O (2:1:0.25) along with 4 to 8 grains of Dowex ion exchange resin (ThermoFisher), incubated at room temperature with vortexing (5 min). Solubilized lipid A (1 to 2 μl) was spotted onto a stainless steel target plate with 1 μl of Norharmane matrix (10 mg/ml in 2:1 chloroform:methanol) for matrix-assisted laser desorption ionization (MALDI) analysis on a Bruker microflex MALDI-time of flight mass spectrometry (TOF MS) instrument in negative-ion mode calibrated with Agilent tuning mix (Santa Clara, CA), and data were processed in flexAnalysis (Bruker Daltonics). All microextraction chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
Sequential fatty acid release by alkaline treatment.
To liberate primary ester-linked fatty acids, lipid A samples were suspended in 100 μl of 28% NH4OH and incubated at 50°C for 5 h with occasional vortexing. To liberate secondary fatty acids, lipid A or treated derivatives were suspended in 40% methylamine and incubated at 50°C for 3 h. Ultrapure H2O (100 μl) was added before samples were frozen and lyophilized. Final products were reconstituted in chloroform-MeOH-H2O (2:1:0.25) and analyzed by MALDI-TOF. The entire workflow described above was performed in triplicate for all five strains.
ACKNOWLEDGMENTS
We acknowledge Francesca Gardner, Kelsey Gregg, Erin Harberts, Belita Opene, and Alison Scott for their support.
This work was supported with funds from the National Institutes of Health/National Institute of Allergy and Infectious Diseases grants (R01AI017828 and R01AI126853 to A.F.A., R21AI26108 and R21AI146773 to J.J.G. and M.S.R., and R01AI123820 and R01AI147314 to R.K.E.). M.L.G. was supported in part by the NIH/NIAID grant T32AI095190 (Signaling Pathways in Innate Immunity).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 2.Miller SI, Ernst RK, Bader MW. 2005. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol 3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 3.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. 2013. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aachoui Y, Leaf IA, Hagar JA, Fontana MF, Campos CG, Zak DE, Tan MH, Cotter PA, Vance RE, Aderem A, Miao EA. 2013. Caspase-11 protects against bacteria that escape the vacuole. Science 339:975–978. doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Needham BD, Trent MS. 2013. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol 11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoyagi KL, Brooks BD, Bearden SW, Montenieri JA, Gage KL, Fisher MA. 2015. LPS modification promotes maintenance of Yersinia pestis in fleas. Microbiology 161:628–638. doi: 10.1099/mic.0.000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanistanon D, Powell DA, Hajjar AM, Pelletier MR, Cohen IE, Way SS, Skerrett SJ, Wang X, Raetz CRH, Ernst RK. 2012. Role of francisella lipid A phosphate modification in virulence and long-term protective immune responses. Infect Immun 80:943–951. doi: 10.1128/IAI.06109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillespie JJ, Nordberg EK, Azad AA, Sobral BW. 2012. Phylogeny and comparative genomics: the shifting landscape in the genomics era, p 84–141. In Azad AF, Palmer GH (ed), Intracellular pathogens II: rickettsiales. ASM Press, Washington, DC. [Google Scholar]
- 9.Sahni A, Fang R, Sahni SK, Walker DH. 2019. Pathogenesis of rickettsial diseases: pathogenic and immune mechanisms of an endotheliotropic infection. Annu Rev Pathol Mech Dis 14:421058251. doi: 10.1146/annurev-pathmechdis-012418-012800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillespie JJ, Kaur SJ, Sayeedur Rahman M, Rennoll-Bankert K, Sears KT, Beier-Sexton M, Azad AF. 2014. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev 39:47–80. doi: 10.1111/1574-6976.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amano K, Fujita M, Suto T. 1993. Chemical properties of lipopolysaccharides from spotted fever group rickettsiae and their common antigenicity with lipopolysaccharides from Proteus species. Infect Immun 61:4350–4355. doi: 10.1128/IAI.61.10.4350-4355.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amano KI, Williams JC, Dasch GA. 1998. Structural properties of lipopolysaccharides from Rickettsia typhi and Rickettsia prowazekii and their chemical similarity to the lipopolysaccharide from Proteus vulgaris OX19 used in the Weil-Felix test. Infect Immun 66:923–926. doi: 10.1128/IAI.66.3.923-926.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peturova M, Vitiazeva V, Toman R. 2015. Structural features of the O-antigen of Rickettsia typhi, the etiological agent of endemic typhus. Acta Virol 59:228–233. doi: 10.4149/av_2015_03_228. [DOI] [PubMed] [Google Scholar]
- 14.Kim HK, Premaratna R, Missiakas DM, Schneewind O. 2019. Rickettsia conorii O antigen is the target of bactericidal Weil-Felix antibodies. Proc Natl Acad Sci U S A 116:19659–19664. doi: 10.1073/pnas.1911922116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quevedo-Diaz MA, Song C, Xiong Y, Chen H, Wahl LM, Radulovic S, Medvedev AE. 2010. Involvement of TLR2 and TLR4 in cell responses to Rickettsia akari. J Leukoc Biol 88:675–685. doi: 10.1189/jlb.1009674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan JM, Woods ME, Olano J, Walker DH. 2008. The absence of toll-like receptor 4 signaling in C3H/HeJ mice predisposes them to overwhelming rickettsial infection and decreased protective Th1 responses. Infect Immun 76:3717–3724. doi: 10.1128/IAI.00311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bechelli J, Smalley C, Zhao X, Judy B, Valdes P, Walker DH, Fang R. 2016. MyD88 mediates instructive signaling in dendritic cells and protective inflammatory response during rickettsial infection. Infect Immun 84:883–893. doi: 10.1128/IAI.01361-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordan JM, Woods ME, Soong L, Walker DH. 2009. Rickettsiae stimulate dendritic cells through toll-like receptor 4, leading to enhanced NK cell activation in vivo. J Infect Dis 199:236–242. doi: 10.1086/595833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smalley C, Bechelli J, Rockx-Brouwer D, Saito T, Azar SR, Ismail N, Walker DH, Fang R. 2016. Rickettsia australis activates inflammasome in human and murine macrophages. PLoS One 11:e0157231. doi: 10.1371/journal.pone.0157231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burke TP, Engström P, Chavez RA, Fonbuena JA, Vance RE, Welch MD. 2020. Inflammasome-mediated antagonism of type I interferon enhances Rickettsia pathogenesis. Nat Microbiol 5:688–696. doi: 10.1038/s41564-020-0673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fodorová M, Vadovič P, Toman R. 2011. Structural features of lipid A of Rickettsia typhi. Acta Virol 55:31–44. doi: 10.4149/av_2011_01_31. [DOI] [PubMed] [Google Scholar]
- 22.Bainbridge BW, Karimi-Naser L, Reife R, Blethen F, Ernst RK, Darveau RP. 2008. Acyl chain specificity of the acyltransferases LpxA and LpxD and substrate availability contribute to lipid A fatty acid heterogeneity in Porphyromonas gingivalis. J Bacteriol 190:4549–4558. doi: 10.1128/JB.00234-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillotte ML, Gillespie JJ, Chandler CE, Rahman MS, Ernst RK, Azad AF. 2018. Rickettsia lipid A biosynthesis utilizes the late acyltransferase LpxJ for secondary fatty acid addition. J Bacteriol 200:e00334-18. doi: 10.1128/JB.00334-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Driscoll TP, Verhoeve VI, Guillotte ML, Lehman SS, Rennoll SA, Beier-Sexton M, Rahman MS, Azad AF, Gillespie JJ. 2017. Wholly Rickettsia ! Reconstructed metabolic profile of the quintessential bacterial parasite of eukaryotic cells. mBio 8:e00859-17. doi: 10.1128/mBio.00859-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagen R, Verhoeve VI, Gillespie JJ, Driscoll TP. 2018. Conjugative transposons and their cargo genes vary across natural populations of Rickettsia buchneri infecting the tick Ixodes scapularis. Genome Biol Evol 10:3218–3229. doi: 10.1093/gbe/evy247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MALDI-TOF analysis of Rickettsia lipid A. Lipid A microextraction from rickettsial cultures was performed as previously described (A. El Hamidi, A. Tirsoaga, A. Novikov, A. Hussein, et al., J Lipid Res 46:1773–1778, 2005; A. J. Scott, B. Flinders, J. Cappell, T. Liang, et al., Pathog Dis 74, 2016; A. Tirsoaga, A. El Hamidi, M. B. Perry, M. Caroff, et al., J Lipid Res 48:2419–2427, 2007). See the text for additional details. (A) R. typhi strain Wilmington, (B) R. montanensis strain M5/6, (C) R. akari strain Hartford, and (D) R. rickettsii strain Sheila Smith. Insets identify the vector host and relative virulence to humans. Peak labels are masses of singly charged ions. Major peaks are indicated by colored labels (A to C, gray; D, red), with minor peaks (likely representing heterogeneity in fatty acid incorporation) noted by dashed lines. Download FIG S1, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2021 Guillotte et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fatty acid analysis of Rickettsia lipid A. (A, D, G, J, and M) Lipid A of R. akari strain Hartford, R. typhi strain Wilmington, R. montanensis strain M5/6, R. rickettsii strain Sheila Smith, and R. rickettsii strain Iowa was subjected to sequential fatty acid release prior to MALDI-TOF analysis of daughter molecules (A. El Hamidi, A. Tirsoaga, A. Novikov, A. Hussein, et al., J Lipid Res 46:1773–1778, 2005; A. Tirsoaga, A. El Hamidi, M. B. Perry, M. Caroff, et al., J Lipid Res 48:2419–2427, 2007). (B, E, H, K, and N) Primary ester-linked fatty acids (3′ position, green; 3 position, blue), which are more readily released than secondary ester-linked fatty acids, were liberated using mild alkali treatment with ammonium hydroxide, yielding predominate ions of either m/z ∼1,247/1,273 (gray for palmitate/stearate in R. akari, R. typhi, and R. montanensis) or m/z 1,191 (red for laurate in R. rickettsii) that are tri-acyl daughter molecules. (C, F, I, L, and O) The remaining secondary ester-linked fatty acid attached to the hydroxypalmitate at the 2′ position was liberated using a second alkaline reaction with methylamine, yielding the di-acyl daughter ion at m/z 1009 for all analyzed strains. Structures (insets) depict the major lipid A ion in each spectrum, with minor peaks (dashed lines) likely representing heterogeneity in fatty acid incorporation. Download FIG S2, PDF file, 2.5 MB (2.5MB, pdf) .
Copyright © 2021 Guillotte et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Raetz pathway for biosynthesis of lipid A in rickettsiae. Rickettsiae lack enzymes for amino sugar metabolism and synthesis of pentose phosphates, necessitating the acquisition of host N-acetylglucosamine-1-P and ribose-5-P to initiate biosynthesis of lipid IV(A) and 3-deoxy-d-manno-octulosonate (Kdo), respectively (T. P. Driscoll, V. I. Verhoeve, M. L. Guillotte, S. S. Lehman, et al., mBio 8:e00859-17, 2017). Using these two pilfered metabolites, 14 enzymes (conserved in Rickettsia genomes, inset at top) and three cofactors (ATP, UTP, and phosphoenolpyruvate) are required for Kdo2-lipid A biosynthesis. Kdo2 is not shown on the final structure, as our lipid A microextraction protocol removed Kdo residues prior to lipid A analysis (the structure of Kdo2 and the inner and outer core oligosaccharide have not been determined for Rickettsiae to date). Enzymes contain locus tags for R. typhi strain Wilmington enzymes that were used in blastp searches against the NCBI Rickettsia database (taxid 780) to confirm the strict conservation of all 14 CDS in the five strains analyzed in this study. Searches were performed with composition-based statistics, with no filter used. Default matrix parameters (BLOSUM62) and gap costs (existence, 11; extension, 1) were implemented, with an inclusion threshold of 0.005. Download FIG S3, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2021 Guillotte et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparative analysis of Rickettsia LpxL enzymes. Lipid A biosynthesis in E. coli utilizes the late acyltransferase LpxL to add a laurate (C12) to the 2′ hydroxymyristate (D. A. Six, S. M. Carty, Z. Guan, and C. R. H. Raetz, Biochemistry 47:8623–8637, 2008). Our work on Rickettsia lipid A indicates that LpxL enzymes add either a palmitate or stearate on the 2′-hydroxypalmitate (R. akari, R. typhi, and R. montanensis) or a laurate (C12) on the 2′-hydroxypalmitate (R. rickettsii strains Sheila Smith and Iowa). (A) Multiple-sequence alignment of LpxL homologs using MUSCLE (default parameters) (R. C. Edgar, Nucleic Acids Res 32:1792–1797, 2004) indicates strong conservation across the entirety of the proteins (the proteins from R. rickettsii strains Sheila Smith and Iowa are identical). Amino acid similarity (% identity) between homologs is shown below the alignment. Structural modeling of the R. typhi LpxL protein to Acinetobacter baumannii LpxM (PDB entry 5KNK) using Phyre2 (L. A. Kelley and M. J. E. Sternberg, Nat Protoc 4:363–371, 2009) indicates strong conservation within the three blocks recognized within GPAT, LPAAT, DHAPAT, and LPEAT acyltransferases (J. Yao and C. O. Rock, Biochim Biophys Acta 1831:495–502, 2013; T. M. Lewin, P. Wang, and R. A. Colema, Biochemistry 38:5764–5771, 1999). Active-site residues within blocks are colored according to charge. Asp193 is proposed to participate in the active-site charge relay system with His122 (R. J. Heath, C. O. Rock, J Bacteriol 180:1425–30, 1998; A. F. Neuwald, Curr Biol 7:R465-R466, 1997). There is strict conservation within the regions analogous to the deep hydrophobic cleft of LpxM predicted to function as a hydrocarbon ruler (D. Dovala, C. M. Rath, Q. Hu, W. S. Sawyer, et al., Proc Natl Acad Sci U S A 113:E6064–E6071, 2016). (B) Mapping of character states for the five unique R. rickettsii LpxL residues over the Rickettsia phylogeny from Fig. 1. Shared residues between R. rickettsii strains and other rickettsiae are highlighted yellow. The strict conservation of Ile130 within block I of the LpxL active site is illustrated at left. All other information follows the description for Fig. 1. Download FIG S4, PDF file, 0.7 MB (692.2KB, pdf) .
Copyright © 2021 Guillotte et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.


