Aerobic bacteria are frequent primocolonizers of the human naive intestine. Their generally accepted role is to eliminate oxygen, which would allow colonization by anaerobes that subsequently dominate bacterial gut populations.
KEYWORDS: Bacteroides, Clostridium scindens, Escherichia coli, germfree mice, intestine, oxygen, primocolonization
ABSTRACT
Aerobic bacteria are frequent primocolonizers of the human naive intestine. Their generally accepted role is to eliminate oxygen, which would allow colonization by anaerobes that subsequently dominate bacterial gut populations. In this hypothesis-based study, we revisited this dogma experimentally in a germfree mouse model as a mimic of the germfree newborn. We varied conditions leading to the establishment of the dominant intestinal anaerobe Bacteroides thetaiotaomicron. Two variables were introduced: Bacteroides inoculum size and preestablishment by bacteria capable or not of consuming oxygen. High Bacteroides inoculum size enabled its primocolonization. At low inocula, we show that bacterial preestablishment was decisive for subsequent Bacteroides colonization. However, even non-oxygen-respiring bacteria, a hemA Escherichia coli mutant and the intestinal obligate anaerobe Clostridium scindens, facilitated Bacteroides establishment. These findings, which are supported by recent reports, revise the long-held assumption that oxygen scavenging is the main role for aerobic primocolonizing bacteria. Instead, we suggest that better survival of aerobic bacteria ex vivo during vectorization between hosts could be a reason for their frequent primocolonization.
OPINION/HYPOTHESIS
Initial microbial colonization of the naive intestine may have lasting consequences on the host (1, 2), yet the factors that influence this crucial step are mainly unknown (3). The temporal sequence of microbial establishment varies greatly among individual human newborns (4–6). The concentration and composition of the microbial bolus encountered by neonates and the uniqueness of each individual are likely crucial to the colonization of the naive intestine, making the identification of factors governing colonization a major challenge.
Bacteroides species are dominant heme auxotrophs and obligate anaerobes of human and animal intestinal microbiota (7–10), which coexist in symbiosis with the healthy host. These bacteria are proposed to contribute to host well-being, e.g., by (i) providing membrane-permeable nutrients such as short-chain fatty acids, (ii) occupying the intestinal mucosal space and thus preventing access to pathogens (this role relies on a large repertoire of Bacteroides enzymes that catabolize complex sugars lining the intestinal mucosal wall), and (iii) producing antimicrobial molecules that may limit the outgrowth of bacterial competitors, including pathogens (1, 11–13).
Oxygen depletion in the intestine by precolonizing bacteria is considered the sine qua non for Bacteroides thetaiotaomicron colonization. Aerobic bacteria such as Escherichia coli, which are often among the primocolonizers, are proposed to be responsible for consuming toxic oxygen, thus enabling subsequent B. thetaiotaomicron establishment (4, 14). However, to our knowledge, this dogma remains unproven. Moreover, microbial footprints of neonate feces indicate that aerobes are not systematically the first to colonize the intestines (5). In this work, we therefore revisit this hypothesis by giving evidence in a germfree mouse model that primocolonizing bacteria promote B. thetaiotaomicron establishment regardless of their capacity to consume oxygen.
B. thetaiotaomicron primocolonization of the mouse intestine is inoculum dependent.
Most colonization studies involving B. thetaiotaomicron use 106 to 108 CFU for implantation (15). We reasoned that under natural conditions, B. thetaiotaomicron concentrations that reach the intestines might be far lower. Even if higher concentrations are ingested at childbirth, contact with gastric products during passage through the intestine could decrease microbial survival (16, 17). All methodologies are described in Text S1 in the supplemental material. Accordingly, 103 and 104 CFU of B. thetaiotaomicron, determined by first establishing the correlation with optical density at 600 nm (OD600) readings, were orally administered at time zero (T0) to two mouse cohorts (n = 6). B. thetaiotaomicron colonization was assessed by CFU determinations in feces, sampled at 4-h intervals for 28 h, starting at T0. The capacity to colonize was found to be inoculum dependent (Fig. 1). Administration of 104 CFU led to colonization at 8 h postinoculation (p.i.), whereas the 10-fold-lower concentration did not promote B. thetaiotaomicron establishment even at 28 h p.i. These findings suggest that inoculum size is a contributing factor for B. thetaiotaomicron primocolonization.
FIG 1.
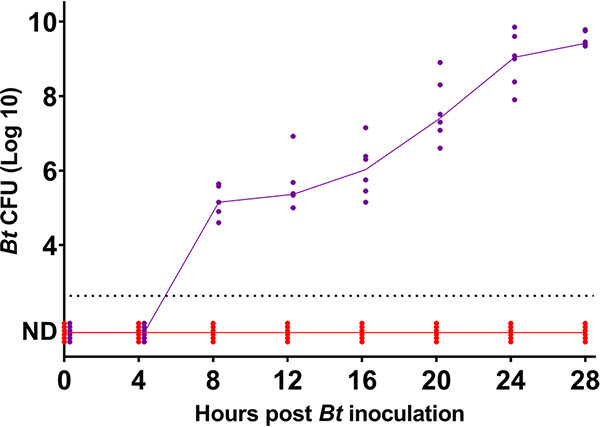
B. thetaiotaomicron (Bt) implantation in the intestine as a function of inoculum size. B. thetaiotaomicron was administered orally to germfree BALB/c mice at two concentrations by gastric probe. Fecal samples were taken at the time of implantation and every 4 h for 28 h. Oral administration was with 103 CFU B. thetaiotaomicron (red) or 104 CFU B. thetaiotaomicron (purple). Individual values are shown for each time point; the intersection of lines with values indicates the median CFU per gram in fecal samples of mice for each cohort. The detection threshold was 5 × 102 CFU/g feces. ND, not detected. Data for mice where no CFU were detected are expanded at the baseline to distinguish the number of mice tested.
Word file describing the materials and methods used for experimentation: strains, constructions, growth conditions, oxygen consumption measurements, colonization of germfree mice, determination of colonization efficiency, and microscopy. Download Text S1, PDF file, 0.2 MB (201.7KB, pdf) .
Copyright © 2021 Halpern et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
E. coli enables B. thetaiotaomicron colonization in a germfree mouse intestinal model.
Although Escherichia coli is a minor constituent of the adult microbiota, it is frequently among the first species to transiently dominate the naive newborn intestinal microbiota (4, 5, 18). E. coli is unique among the major intestinal bacteria to be fully equipped for aerobic respiration and to thereby eliminate oxygen (19, 20). We examined the capacity of the “low” B. thetaiotaomicron inoculum (103 CFU) to colonize intestines of mice that were preimplanted (16 h prior to the B. thetaiotaomicron inoculum [T−16]) or not with E. coli strain MG1655 (108 CFU) (Fig. 2). As mentioned above, no B. thetaiotaomicron bacteria were detected in feces of monocolonized mice when sampled up to 72 h p.i. In marked contrast, mice preimplanted with E. coli were colonized by B. thetaiotaomicron at 109 to 1010 CFU calculated per g of feces at 24 h p.i. This range is comparable to the CFU reported after mouse colonization with a high B. thetaiotaomicron inoculum (2 × 1010 CFU) (15).
FIG 2.
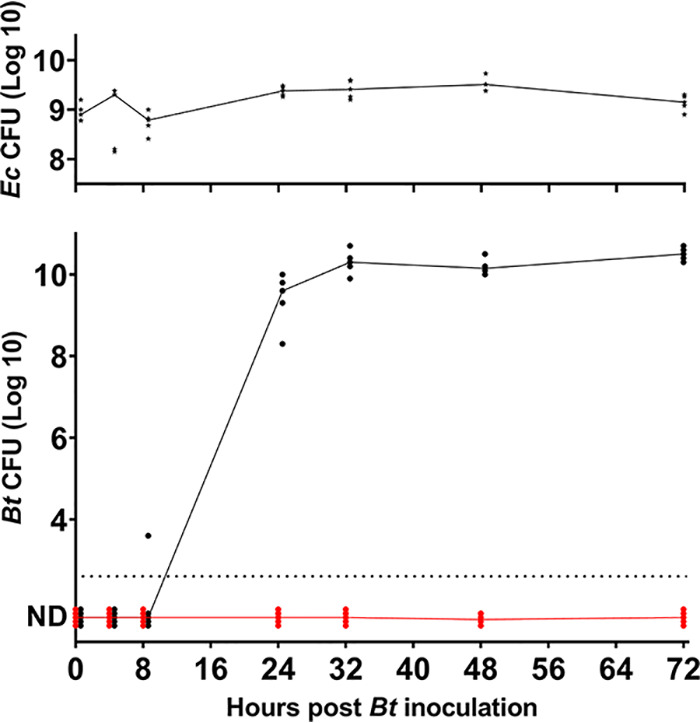
E. coli facilitates B. thetaiotaomicron (Bt) establishment in germfree animals. E. coli MG1655 (WT) was established in germfree BALB/c mice by oral administration. Sixteen hours later (T0), 2 × 103 CFU of B. thetaiotaomicron were administered to the group precolonized by E. coli and to a second naive group. All mouse groups received the B. thetaiotaomicron doses at the same time and from the same bacterial preparation. Fecal samples were taken at the indicated times over a 72-h period for CFU determinations. Dilutions were spotted on Bacteroides bile esculin agar with amikacin (BBE) medium incubated anaerobically for B. thetaiotaomicron and on LB medium incubated aerobically for E. coli. (Top) E. coli (Ec) CFU per gram; (bottom) B. thetaiotaomicron CFU per gram of feces. Red, B. thetaiotaomicron administered alone; black, B. thetaiotaomicron administered after E. coli precolonization. Individual values are shown for each time point; the intersection of lines with values indicates the median CFU per gram in fecal samples of the mice for each cohort. The detection threshold was 5 × 102 CFU/g feces. ND, not detected. Data for mice where no CFU were detected are expanded at the baseline to distinguish the number of mice tested.
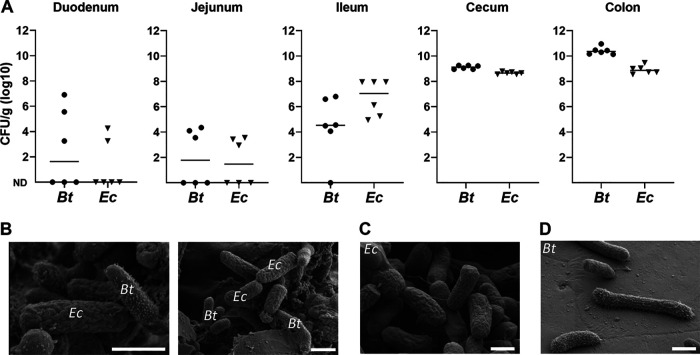
The marked impact of E. coli on B. thetaiotaomicron colonization at low inocula (Fig. 2) might suggest the proximity of the two species in the gut. Bacterial loads in cocolonized mice were determined from the different intestinal compartments (Fig. 3A). For each given compartment, E. coli and B. thetaiotaomicron showed comparable CFU, ranging from about 102 to 103 CFU/g in the duodenum and jejunum to 109 to 1011 CFU/g in the cecum and colon. Scanning microscopy of feces of cocolonized mice (Fig. 3B) revealed two discrete bacterial forms, which were distinguishable as E. coli and B. thetaiotaomicron, as identified in monocultures (Fig. 3C and D). B. thetaiotaomicron and E. coli contact and metabolic exchanges were suggested and shown to occur in dysbiosis and infection (21, 22). The proximity of these bacteria as observed here suggests that similar exchanges are possible in the healthy host in early stages of colonization.
FIG 3.
E. coli and B. thetaiotaomicron (Bt) colocalize in the mouse intestinal tract. (A) Bacterial loads in intestinal compartments. Intestinal samples were recovered from E. coli WT (Ec)- and B. thetaiotaomicron-cocolonized mice used in the experiment shown in Fig. 2, 72 h after the start of experiments. Intestinal contents were recovered from the five indicated locations of dissected mice, and CFU were determined. Bars represent the median values of CFU obtained from individual samples. ND, below the detection level. (B) Visualization by field emission scanning electron microscopy of feces from cocolonized mice. E. coli and B. thetaiotaomicron are identified by their distinct morphologies. Small particles may correspond to food particles or shed mucus. (C and D) Purified cultures were used for identification. White bars, 1 μM.
E. coli facilitates B. thetaiotaomicron colonization independently of a role as an oxygen scavenger.
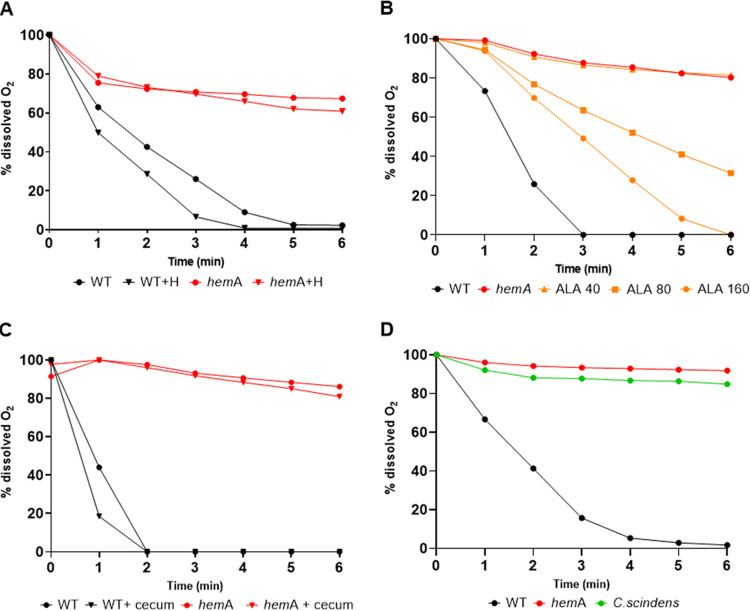
B. thetaiotaomicron growth is inhibited by oxygen, which led to the simple and generally accepted hypothesis that respirative aerobic bacteria consume intestinal oxygen, thus facilitating the subsequent implantation of anaerobes such as B. thetaiotaomicron (14). We tested this hypothesis by assessing B. thetaiotaomicron establishment in germfree mice precolonized by an E. coli strain that does not consume oxygen, compared to a wild-type (WT) E. coli strain. We chose a hemA mutant, which does not synthesize heme and thus cannot carry out aerobic respiration, the main pathway for oxygen reduction to water (19). Unlike other respiration-related genes, which are mostly redundant in E. coli, the hemA mutation disables respiration and oxygen-consuming functions (19). It also disables anaerobic respiration by nitrate, which is reportedly used in the gut upon inflammation (23). This choice allowed us to inactivate a single rather than multiple genes without compromising fermentation growth. We first validated the differences in oxygen consumption of the MG1655 WT and hemA mutant strains. As expected, only the WT strain consumed oxygen (Fig. 4). It was possible that intestinal heme (24) or δ-aminolevulinic acid (ALA) (the HemA product) (25) could alter the capacity of the hemA strain to consume oxygen. However, heme addition did not affect hemA mutant oxygen consumption, which is consistent with observations that MG1655 does not assimilate exogenous heme (26, 27) (Fig. 4A). In contrast, while ALA has not, to our knowledge, been reported in intestinal contents, it was detected in blood plasma at trace levels (<0.1 μM in healthy humans [28]) and in urine (up to ∼20 μM in healthy individuals [29]). The MG1655 hemA mutant consumed oxygen in the presence of 80 μM to 160 μM ALA but not at 40 μM ALA (Fig. 4B). To determine whether intestinal contents might stimulate hemA oxygen consumption, WT and hemA strains were grown in a pooled murine cecal sample, and oxygen consumption was measured (Fig. 4C). Cecum addition had no effect on WT strain oxygen consumption and had no stimulatory effect on oxygen consumption by the hemA strain. We therefore considered that hemA would not consume oxygen during gut passage.
FIG 4.
Bacterial oxygen consumption. (A) E. coli MG1655 and hemA strains were grown in LB supplemented or not with 5 μM heme (H). (B) E. coli WT and hemA strains were grown in LB. The hemA strain was also grown in LB supplemented with the indicated concentrations of δ-aminolevulinic acid (ALA) (micromolar). (C) WT and hemA E. coli strains were grown in LB or in 90% murine cecum containing 10% of a 10×-concentrated LB medium. (D) C. scindens and E. coli WT and hemA control strains were compared for their capacity to consume oxygen. See Text S1 in the supplemental material for protocols. Dissolved oxygen (milligrams per liter) is normalized to 100% for all samples at T0.
The capacity of the hemA mutant to enable B. thetaiotaomicron colonization was tested in the germfree mouse model as described above. Mice were precolonized (T−16) with either the MG1655 WT or the hemA strain. A third group of germfree mice was not precolonized. At T0, all groups were administered 2 × 103 CFU of B. thetaiotaomicron. Fecal samples were collected at 4-h intervals over a 28-h period for E. coli and B. thetaiotaomicron CFU determinations (Fig. 5A). As described above, B. thetaiotaomicron only colonized mice that were precolonized with E. coli. In mice precolonized with the hemA mutant, compared to the WT E. coli strain, B. thetaiotaomicron establishment was delayed by about 4 h. The hemA strain phenotypes (kanamycin resistance and no growth on aerobically incubated solid medium) were confirmed in bacteria recovered from feces at the 28-h time point, indicating that the strain did not revert to the WT in the gut. The E. coli hemA strain thus had nearly the same stimulatory effect on B. thetaiotaomicron establishment as did WT E. coli. These findings suggest a marginal, if any, role for E. coli as an oxygen scavenger in promoting B. thetaiotaomicron establishment. These in vivo findings argue against the currently accepted hypothesis that respiratory aerobic bacteria eliminate toxic oxygen from the intestine to facilitate Bacteroides establishment.
FIG 5.
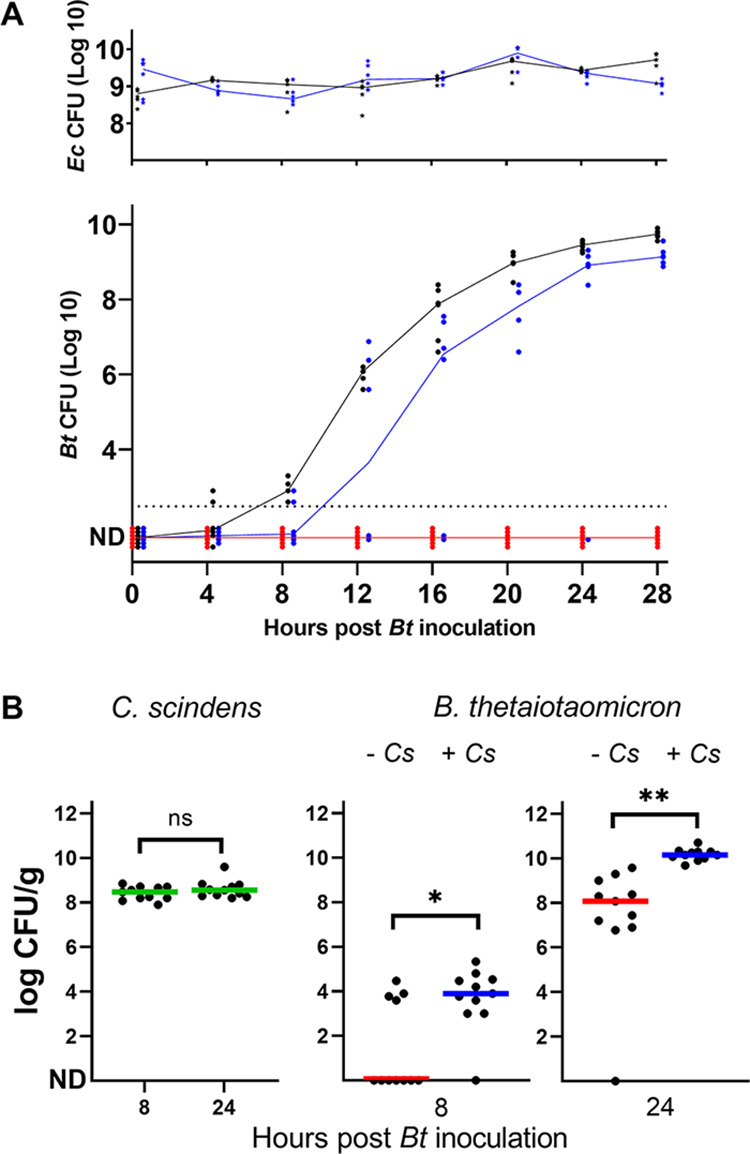
Precolonizing aerobic and anaerobic bacteria facilitate B. thetaiotaomicron (Bt) establishment in germfree mice. (A) E. coli WT and hemA mutant strains (108 CFU) were each orally administered to germfree BALB/c mice. Sixteen hours later (T0), 2 × 103 CFU of B. thetaiotaomicron were administered to the two groups of animals precolonized with E. coli and to a group of naive mice. All mouse groups received the B. thetaiotaomicron doses at the same time and from the same bacterial preparation. Fecal samples were taken at 4-h intervals for 28 h for CFU determinations. E. coli hemA CFU were determined on LB plates containing δ-aminolevulinic acid (200 μM) incubated aerobically. (Top) E. coli (Ec) (CFU per gram feces); (bottom) B. thetaiotaomicron (CFU per gram of feces). Red, mice colonized with B. thetaiotaomicron alone; black, mice colonized with E. coli WT and then B. thetaiotaomicron; blue, mice colonized with E. coli hemA and then B. thetaiotaomicron. Individual CFU values are shown for each time point; the intersecting line represents the median of CFU for each cohort. The detection threshold was 5 × 102 CFU/g feces. ND, not detected. Data for mice where no CFU were detected are expanded at the baseline to distinguish the number of mice tested. (B) Anaerobic C. scindens (Cs) was administered as described above for panel A for the administration of E. coli to germfree animals, while a second group of mice received no C. scindens as a control. Sixteen hours later, both groups of mice received 103 CFU B. thetaiotaomicron by oral administration. CFU determinations were performed at 8 h and 24 h p.i. (Left) C. scindens CFU at 8 h and 24 h in fecal samples of mice precolonized with this bacterium. (Right) B. thetaiotaomicron CFU at 8 h and 24 h in feces samples of mice with (+ Cs) or without (− Cs) C. scindens precolonization. Results of two independent experiments were pooled. Bars indicate median CFU per gram for the mice in each group. The detection threshold was 5 × 102 CFU/g feces. ND, below the detection level. *, P = 0.5; **, P = 0.05; ns, not significant.
The role of accessory bacteria in enabling B. thetaiotaomicron establishment was then investigated using Clostridium scindens, an obligate anaerobe and common isolate of the healthy human intestine (30), in place of E. coli as a primocolonizer. As expected, the tested C. scindens strain ATCC 35704 did not consume oxygen (Fig. 4D). The capacity of B. thetaiotaomicron to colonize mouse intestines was tested as described above, in the absence or presence of C. scindens. In these experiments, which were performed twice independently, B. thetaiotaomicron CFU appeared even in the absence of precolonizing bacteria. This observed shift might be related to a change in germfree BALB/c mouse suppliers and/or to subtle changes in animal housing conditions that occur over time (e.g., water or food supply). Nevertheless, precolonization with C. scindens significantly improved B. thetaiotaomicron establishment (Fig. 5B). Altogether, these findings rule out species specificity and demonstrate that oxygen consumption by aerobic bacteria is not a sine qua non for B. thetaiotaomicron establishment.
Limitations of the primocolonization germfree model.
To our knowledge, this is the first description of a germfree model that tests intestinal primocolonization with low bacterial doses. In developing this approach, we confronted two notable technical issues. The first concerns the use of low inocula: while great care was taken to ensure reproducible conditions, the use of low inocula increases the risk of variation during inoculation and amplifies differences between individuals within a cohort. The second concerns the handling of anaerobic bacteria, which are oxygen sensitive. After anaerobic growth, B. thetaiotaomicron bacteria are briefly exposed to oxygen during inoculum preparation for oral administration. These steps need careful coordination to ensure repeatability and minimize the period of oxygen exposure. The combination of these limitations was considered when choosing the minimal B. thetaiotaomicron colonization dose (1 × 103 to 2 × 103 CFU per mouse) and by simultaneously administering doses from a single bacterial stock. We recommend that these technical steps be carefully prepared and timed in experimentations involving low-dose bacterial administrations, particularly when dealing with anaerobic bacteria.
Anaerobic bacteria may encode functions involved in oxygen management.
Properties of B. thetaiotaomicron itself might suggest why bacterially mediated oxygen removal is not needed for its establishment: (i) B. thetaiotaomicron encodes an aerobic respiration system involving quinol oxidase, which allows it to withstand nanomolar concentrations of oxygen (shown for the closely related species Bacteroides fragilis [31]); (ii) B. thetaiotaomicron and B. fragilis encode a catalase and other peroxide-scavenging enzymes, which may eliminate toxic oxygen radicals (32, 33); and (iii) frequently arising mutations in oxe (BF638R_0963), a B. fragilis flavoprotein, reportedly led to greater oxygen resistance and are common in clinical isolates (B. thetaiotaomicron carries an oxe homolog [BT_4126] sharing 92% identity [34]). Moreover, B. thetaiotaomicron colonizes germfree rats when the oxidoreduction potential is high, in keeping with its tolerance to an oxidative environment (15). Importantly, C. scindens is itself anaerobic and was directly established in the mouse intestine albeit at a high inoculum (Fig. 5B), further supporting the proposal that oxygen removal is not the main role of primocolonizing bacteria.
Further studies point to alternative roles of primocolonizing bacteria, without direct oxygen consumption.
The above-described results revise the accepted main role of primocolonizing bacteria and raise questions on their roles in enabling B. thetaiotaomicron establishment without involving respiratory oxygen consumption (Fig. 1). This function is not E. coli specific and can be fulfilled by an anaerobic bacterium, as shown here with C. scindens. Colonization is associated with rapid changes in intestinal volume and cell histology (35, 36), some within hours of colonization, as well as changes in mucus glycan composition and the production of metabolites (11, 24, 36–38). Evidence for an indirect modulation of intestinal oxygen homeostasis by bacteria is suggested from recent studies. Interestingly, bacterial pathogens, but also the normal microbiota, may trigger an anoxic response, depleting oxygen in their surrounding tissues. The bacterial metabolite butyrate, which is produced by anaerobic bacteria, was proposed to stimulate oxygen elimination via β-oxidation in host cells (see reference 39 and references therein; 40, 41). More generally, lipid β-oxidation triggered by the microbiota was suggested as a means of removing oxygen (42), further supporting an alternative role for primocolonizing bacteria in modulating intestinal oxygen. Interestingly, previous studies also give evidence that no notable differences in oxygen status exist between germfree and conventional intestines, further questioning the need for oxygen consumption by aerobic bacteria (42, 43). These and our conclusions are also consistent with an exhaustive study of primocolonizing bacteria in human neonates, where in some babies, the dominant primocolonizing bacteria were members of Bacteroidetes genera (5). In a simpler hypothesis that reconciles our and previous findings, we suggest that aerobic bacteria have a better chance of survival ex vivo, during transmission between donor and recipient. This is consistent with (i) recent studies indicating that intestinal E. coli bacteria develop essentially by anaerobic growth (44) and (ii) observations of a greater abundance of aerobic bacteria in babies born by Caesarean than in babies born by vaginal delivery (45).
Importance of oxygen consumption in infection conditions?
While our findings rule out the need for aerobic respiring bacteria during primocolonization, this property may be important in other situations. For example, intestinal dysbiosis due to infection, postantibiotic treatment, or inflammation might lead to high E. coli populations (46–48). The proximity of E. coli to B. thetaiotaomicron in the dysbiotic host could increase the availability of metabolites (e.g., bacterial growth-promoting heme and quinones [24, 49]) and may also protect anaerobes in the stressed host by respiring oxygen. Oxygen elimination by aerobic bacteria might thus be relevant to Bacteroides survival during polymicrobial intra-abdominal infection (22, 50).
ACKNOWLEDGMENTS
We are grateful to A. Foussier, F. Joly, and C. Maudet (INRAE, Micalis, Anaxem) for technical help in germfree animal studies and the INRAE IERP team for use of the Hach electrode. M. H. Malamy (Tufts University, Boston, MA) and C. Wandersman (Institut Pasteur, France) kindly provided strains. We thank C. Poyart (Hôpital Cochin, Paris, France), J. M. Ghigo (Institut Pasteur, Paris, France), P. Bouloc (I2BC, Orsay, France), T. Rochat (INRAE-VIM), and laboratory colleagues D. Lechardeur, M. De Paepe, P. Serror, E. Borezée-Durant, and P. Gaudu for valuable comments and technical advice.
This work received support from the French National Research Agency ANR-11-IDEX-0003-02, ALIAS project.
We state that there is no conflict of interest concerning this work.
REFERENCES
- 1.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Thompson-Chagoyan OC, Maldonado J, Gil A. 2007. Colonization and impact of disease and other factors on intestinal microbiota. Dig Dis Sci 52:2069–2077. doi: 10.1007/s10620-006-9285-z. [DOI] [PubMed] [Google Scholar]
- 3.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-17. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orrhage K, Nord CE. 1999. Factors controlling the bacterial colonization of the intestine in breastfed infants. Acta Paediatr Suppl 88:47–57. doi: 10.1111/j.1651-2227.1999.tb01300.x. [DOI] [PubMed] [Google Scholar]
- 5.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. 2007. Development of the human infant intestinal microbiota. PLoS Biol 5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka M, Nakayama J. 2017. Development of the gut microbiota in infancy and its impact on health in later life. Allergol Int 66:515–522. doi: 10.1016/j.alit.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 8.McKnite AM, Perez-Munoz ME, Lu L, Williams EG, Brewer S, Andreux PA, Bastiaansen JW, Wang X, Kachman SD, Auwerx J, Williams RW, Benson AK, Peterson DA, Ciobanu DC. 2012. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One 7:e39191. doi: 10.1371/journal.pone.0039191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozutsumi Y, Hayashi H, Sakamoto M, Itabashi H, Benno Y. 2005. Culture-independent analysis of fecal microbiota in cattle. Biosci Biotechnol Biochem 69:1793–1797. doi: 10.1271/bbb.69.1793. [DOI] [PubMed] [Google Scholar]
- 10.Shanson DC, Singh J. 1981. Effect of adding cysteine to brain-heart infusion broth on the isolation of Bacteroides fragilis from experimental blood cultures. J Clin Pathol 34:221–223. doi: 10.1136/jcp.34.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Bayona L, Comstock LE. 2018. Bacterial antagonism in host-associated microbial communities. Science 361:eaat2456. doi: 10.1126/science.aat2456. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 14.Adlerberth I, Wold AE. 2009. Establishment of the gut microbiota in Western infants. Acta Paediatr 98:229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 15.Wrzosek L, Miquel S, Noordine ML, Bouet S, Chevalier-Curt MJ, Robert V, Philippe C, Bridonneau C, Cherbuy C, Robbe-Masselot C, Langella P, Thomas M. 2013. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol 11:61. doi: 10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conway PL, Gorbach SL, Goldin BR. 1987. Survival of lactic acid bacteria in the human stomach and adhesion to intestinal cells. J Dairy Sci 70:1–12. doi: 10.3168/jds.S0022-0302(87)79974-3. [DOI] [PubMed] [Google Scholar]
- 17.Drasar BS, Shiner M, McLeod GM. 1969. Studies on the intestinal flora. I. The bacterial flora of the gastrointestinal tract in healthy and achlorhydric persons. Gastroenterology 56:71–79. doi: 10.1016/S0016-5085(69)80067-3. [DOI] [PubMed] [Google Scholar]
- 18.Fanaro S, Chierici R, Guerrini P, Vigi V. 2003. Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl 91:48–55. doi: 10.1111/j.1651-2227.2003.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 19.Richardson DJ. 2000. Bacterial respiration: a flexible process for a changing environment. Microbiology 146(Part 3):551–571. doi: 10.1099/00221287-146-3-551. [DOI] [PubMed] [Google Scholar]
- 20.Gruss A, Borezee-Durant E, Lechardeur D. 2012. Environmental heme utilization by heme-auxotrophic bacteria. Adv Microb Physiol 61:69–124. doi: 10.1016/B978-0-12-394423-8.00003-2. [DOI] [PubMed] [Google Scholar]
- 21.Cameron EA, Curtis MM, Kumar A, Dunny GM, Sperandio V. 2018. Microbiota and pathogen proteases modulate type III secretion activity in enterohemorrhagic Escherichia coli. mBio 9:e02204-18. doi: 10.1128/mBio.02204-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otto BR, van Dooren SJ, Dozois CM, Luirink J, Oudega B. 2002. Escherichia coli hemoglobin protease autotransporter contributes to synergistic abscess formation and heme-dependent growth of Bacteroides fragilis. Infect Immun 70:5–10. doi: 10.1128/IAI.70.1.5-10.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, Tsolis RM, Baumler AJ. 2013. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio 4:e00430-13. doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halpern D, Gruss A. 2015. A sensitive bacterial-growth-based test reveals how intestinal Bacteroides meet their porphyrin requirement. BMC Microbiol 15:282. doi: 10.1186/s12866-015-0616-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haddock BA, Schairer HU. 1973. Electron-transport chains of Escherichia coli. Reconstitution of respiration in a 5-aminolaevulinic acid-requiring mutant. Eur J Biochem 35:34–45. doi: 10.1111/j.1432-1033.1973.tb02806.x. [DOI] [PubMed] [Google Scholar]
- 26.Letoffe S, Delepelaire P, Wandersman C. 2006. The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc Natl Acad Sci U S A 103:12891–12896. doi: 10.1073/pnas.0605440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mills M, Payne SM. 1995. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J Bacteriol 177:3004–3009. doi: 10.1128/JB.177.11.3004-3009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai T, Morita Y. 1996. δ-Aminolevulinic acid in plasma or whole blood as a sensitive indicator of lead effects, and its relation to the other heme-related parameters. Int Arch Occup Environ Health 68:126–132. doi: 10.1007/BF00381245. [DOI] [PubMed] [Google Scholar]
- 29.Davis JR, Andelman SL. 1967. Urinary delta-aminolevulinic acid (ALA) levels in lead poisoning. I. A modified method for the rapid determination of urinary delta-aminolevulinic acid using disposable ion-exchange chromatography columns. Arch Environ Health 15:53–59. doi: 10.1080/00039896.1967.10664873. [DOI] [PubMed] [Google Scholar]
- 30.Morris G, Winter J, Cato E, Ritchie A, Bokkenheuser VD. 1985. Clostridium scindens sp. nov., a human intestinal bacterium with desmolytic activity on corticoids. Int J Syst Bacteriol 35:478–481. doi: 10.1099/00207713-35-4-478. [DOI] [Google Scholar]
- 31.Baughn AD, Malamy MH. 2004. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427:441–444. doi: 10.1038/nature02285. [DOI] [PubMed] [Google Scholar]
- 32.Mishra S, Imlay JA. 2013. An anaerobic bacterium, Bacteroides thetaiotaomicron, uses a consortium of enzymes to scavenge hydrogen peroxide. Mol Microbiol 90:1356–1371. doi: 10.1111/mmi.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rocha ER, Smith CJ. 1995. Biochemical and genetic analyses of a catalase from the anaerobic bacterium Bacteroides fragilis. J Bacteriol 177:3111–3119. doi: 10.1128/JB.177.11.3111-3119.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meehan BM, Baughn AD, Gallegos R, Malamy MH. 2012. Inactivation of a single gene enables microaerobic growth of the obligate anaerobe Bacteroides fragilis. Proc Natl Acad Sci U S A 109:12153–12158. doi: 10.1073/pnas.1203796109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson GR, Trexler PC. 1971. Gastrointestinal structure and function in germ-free or gnotobiotic animals. Gut 12:230–235. doi: 10.1136/gut.12.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomas J, Wrzosek L, Bouznad N, Bouet S, Mayeur C, Noordine M-L, Honvo-Houeto E, Langella P, Thomas M, Cherbuy C. 2013. Primocolonization is associated with colonic epithelial maturation during conventionalization. FASEB J 27:645–655. doi: 10.1096/fj.12-216861. [DOI] [PubMed] [Google Scholar]
- 37.Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E, Koga Y, Benno Y. 2012. Impact of intestinal microbiota on intestinal luminal metabolome. Sci Rep 2:233. doi: 10.1038/srep00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arena ET, Tinevez JY, Nigro G, Sansonetti PJ, Marteyn BS. 2017. The infectious hypoxia: occurrence and causes during Shigella infection. Microbes Infect 19:157–165. doi: 10.1016/j.micinf.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Van Immerseel F, Ducatelle R, De Vos M, Boon N, Van De Wiele T, Verbeke K, Rutgeerts P, Sas B, Louis P, Flint HJ. 2010. Butyric acid-producing anaerobic bacteria as a novel probiotic treatment approach for inflammatory bowel disease. J Med Microbiol 59:141–143. doi: 10.1099/jmm.0.017541-0. [DOI] [PubMed] [Google Scholar]
- 41.Litvak Y, Byndloss MX, Baumler AJ. 2018. Colonocyte metabolism shapes the gut microbiota. Science 362:eaat9076. doi: 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman ES, Bittinger K, Esipova TV, Hou L, Chau L, Jiang J, Mesaros C, Lund PJ, Liang X, FitzGerald GA, Goulian M, Lee D, Garcia BA, Blair IA, Vinogradov SA, Wu GD. 2018. Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc Natl Acad Sci U S A 115:4170–4175. doi: 10.1073/pnas.1718635115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bornside GH, Donovan WE, Myers MB. 1976. Intracolonic tensions of oxygen and carbon dioxide in germfree, conventional, and gnotobiotic rats. Proc Soc Exp Biol Med 151:437–441. doi: 10.3181/00379727-151-39229. [DOI] [PubMed] [Google Scholar]
- 44.Bittinger K, Zhao C, Li Y, Ford E, Friedman ES, Ni J, Kulkarni CV, Cai J, Tian Y, Liu Q, Patterson AD, Sarkar D, Chan SHJ, Maranas C, Saha-Shah A, Lund P, Garcia BA, Mattei LM, Gerber JS, Elovitz MA, Kelly A, DeRusso P, Kim D, Hofstaedter CE, Goulian M, Li H, Bushman FD, Zemel BS, Wu GD. 2020. Bacterial colonization reprograms the neonatal gut metabolome. Nat Microbiol 5:838–847. doi: 10.1038/s41564-020-0694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McClay R, Mileski M, Naiman JL. 2019. Neonatal bacterial colonization of the intestine—implications for the practitioner. J Ideas Health 2:102–107. doi: 10.47108/jidhealth.Vol2.Iss2.36. [DOI] [Google Scholar]
- 46.Barc MC, Bourlioux F, Rigottier-Gois L, Charrin-Sarnel C, Janoir C, Boureau H, Dore J, Collignon A. 2004. Effect of amoxicillin-clavulanic acid on human fecal flora in a gnotobiotic mouse model assessed with fluorescence hybridization using group-specific 16S rRNA probes in combination with flow cytometry. Antimicrob Agents Chemother 48:1365–1368. doi: 10.1128/AAC.48.4.1365-1368.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barc MC, Charrin-Sarnel C, Rochet V, Bourlioux F, Sandre C, Boureau H, Dore J, Collignon A. 2008. Molecular analysis of the digestive microbiota in a gnotobiotic mouse model during antibiotic treatment: influence of Saccharomyces boulardii. Anaerobe 14:229–233. doi: 10.1016/j.anaerobe.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 48.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 49.Franza T, Delavenne E, Derre-Bobillot A, Juillard V, Boulay M, Demey E, Vinh J, Lamberet G, Gaudu P. 2016. A partial metabolic pathway enables group B streptococcus to overcome quinone deficiency in a host bacterial community. Mol Microbiol 102:81–91. doi: 10.1111/mmi.13447. [DOI] [PubMed] [Google Scholar]
- 50.Rotstein OD, Pruett TL, Simmons RL. 1985. Lethal microbial synergism in intra-abdominal infections. Escherichia coli and Bacteroides fragilis. Arch Surg 120:146–151. doi: 10.1001/archsurg.1985.01390260016003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Word file describing the materials and methods used for experimentation: strains, constructions, growth conditions, oxygen consumption measurements, colonization of germfree mice, determination of colonization efficiency, and microscopy. Download Text S1, PDF file, 0.2 MB (201.7KB, pdf) .
Copyright © 2021 Halpern et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.