High prevalence of avian influenza viruses (AIVs) was detected in a small, low-density, isolated population of ruddy turnstones in Australia. Analysis of these viruses revealed relatively recent introductions of viral gene segments from both Eurasia and North America, as well as long-term persistence of introduced gene segments in Australian wild birds.
KEYWORDS: avian influenza virus, Charadriiformes, East Asian-Australasian Flyway, reassortant viruses, shorebirds, whole-genome sequencing, wild bird surveillance
ABSTRACT
Australian lineages of avian influenza A viruses (AIVs) are thought to be phylogenetically distinct from those circulating in Eurasia and the Americas, suggesting the circulation of endemic viruses seeded by occasional introductions from other regions. However, processes underlying the introduction, evolution and maintenance of AIVs in Australia remain poorly understood. Waders (order Charadriiformes, family Scolopacidae) may play a unique role in the ecology and evolution of AIVs, particularly in Australia, where ducks, geese, and swans (order Anseriformes, family Anatidae) rarely undertake intercontinental migrations. Across a 5-year surveillance period (2011 to 2015), ruddy turnstones (Arenaria interpres) that “overwinter” during the Austral summer in southeastern Australia showed generally low levels of AIV prevalence (0 to 2%). However, in March 2014, we detected AIVs in 32% (95% confidence interval [CI], 25 to 39%) of individuals in a small, low-density, island population 90 km from the Australian mainland. This epizootic comprised three distinct AIV genotypes, each of which represent a unique reassortment of Australian-, recently introduced Eurasian-, and recently introduced American-lineage gene segments. Strikingly, the Australian-lineage gene segments showed high similarity to those of H10N7 viruses isolated in 2010 and 2012 from poultry outbreaks 900 to 1,500 km to the north. Together with the diverse geographic origins of the American and Eurasian gene segments, these findings suggest extensive circulation and reassortment of AIVs within Australian wild birds over vast geographic distances. Our findings indicate that long-term surveillance in waders may yield unique insights into AIV gene flow, especially in geographic regions like Oceania, where Anatidae species do not display regular inter- or intracontinental migration.
IMPORTANCE High prevalence of avian influenza viruses (AIVs) was detected in a small, low-density, isolated population of ruddy turnstones in Australia. Analysis of these viruses revealed relatively recent introductions of viral gene segments from both Eurasia and North America, as well as long-term persistence of introduced gene segments in Australian wild birds. These data demonstrate that the flow of viruses into Australia may be more common than initially thought and that, once introduced, these AIVs have the potential to be maintained within the continent. These findings add to a growing body of evidence suggesting that Australian wild birds are unlikely to be ecologically isolated from the highly pathogenic H5Nx viruses circulating among wild birds throughout the Northern Hemisphere.
INTRODUCTION
Influenza A viruses circulating in wild birds represent an omnipresent source of genomic diversity for viral evolution and transmission to livestock and humans (1–3). Global patterns of avian influenza virus (AIV) gene flow in wild birds are therefore important to understanding potential evolutionary pathways of AIVs and the health and economic threats they pose (4). Genomic diversity among AIVs is generated through high mutation rates and segmental reassortment. Influenza A is a negative-sense RNA virus consisting of eight gene segments, two encoding surface glycoproteins (hemagglutinin [HA] and neuraminidase [NA]) and six encoding internal proteins (polymerase basic 2 [PB2], polymerase basic 1 [PB1], polymerase acidic [PA], nucleoprotein [NP], matrix [M], and nonstructural [NS]) (2, 5). Each segment comprises multiple functionally equivalent lineages (6–8) that can be exchanged when host cells are simultaneously coinfected with more than one AIV (9). Reassortment, together with genetic drift, results in transient genome constellations (viral “genotypes”) in which the evolutionary histories of each gene segment are partially independent (7, 10).
Dramatic shifts in the AIV genome, through reassortment, appear crucial to the maintenance of AIVs in the wild bird reservoir and to cross-species transmission (11). In particular, bulk changes in the genome often facilitate the emergence of novel AIVs in domestic hosts (12, 13), and humans (14). Shifts in AIV genotype and phenotype are often more pronounced when gene segments from different lineages are combined (15–17), which typically occurs at times and locations characterized by high rates of interhemispheric mixing of wild birds and their viruses (18). To date, the majority of reassortant AIVs have been reported from locations where distinct communities of long-distance migratory hosts and their viruses regularly converge, such as intercontinental margins (e.g., 19–22), premigratory staging areas (10, 23) and stopover sites (24, 25). Within these locations, rates of AIV reassortment appear to be shaped by prevalence, host identity, and interspecies interactions (11, 21). For instance, although hosts of the family Anatidae show high prevalence of AIVs and high rates of reassortment at intercontinental margins (11, 26, 27), novel genome constellations are disproportionately detected in gulls and terns (order Charadriiformes, family Laridae) (28–30) and waders (order Charadriiformes, family Scolopacidae) (7, 8, 31, 32), despite overall prevalence estimates of 1.4% and 0.8%, respectively (33). Hosts of the order Charadriiformes may therefore play a pivotal role in the redistribution of gene segments over large distances and the emergence of novel genome constellations (18, 34).
Partial separation of avian communities along migratory flyways has shaped the evolution of AIVs, with clear genetic differences between Eurasian and North American lineages for each virus segment (7, 8, 35). However, frequent reassortment and intercontinental gene flow occurs, with a growing body of studies demonstrating American-lineage gene segments in Western Europe (19, 36, 37), and Eurasian-lineage gene segments in North America (e.g., 20, 22, 27–30, 32, 38). Striking evidence for such intercontinental gene flow comes from Iceland during autumn migration, where viruses that were entirely “American,” entirely “Eurasian,” and composed of both American- and Eurasian-lineage gene segments were reported from a single location (21). Occasionally, such introductions result in selective sweeps of locally circulating AIV lineages, as illustrated by the circulation of Eurasian-origin PB1, PA, and H6 HA segments in North American wild birds since the early 2000s (8).
Although Australian AIVs are broadly part of the Eurasian clade, they have formed distinct clades in each gene segment (39, 40). It has therefore been suggested that AIVs within Australia show endemic circulation and partial evolutionary isolation, seeded by occasional introductions from elsewhere (39, 40). These outsider events predominantly originate from Eurasia; however, gene segments from North American lineages have been detected in Australian wild birds (17, 41). One of these gene segments (H10 HA) was detected in wild birds prior to two outbreaks of low-pathogenic H10N7 in biosecure intensive commercial poultry facilities in New South Wales (NSW) and Queensland (QLD) (17, 42). Infections in the NSW poultry facility went on to infect abattoir workers processing clinically normal birds from the farm, indicating the potential for AIVs circulating in Australian wild birds to be incorporated into zoonotic strains (17, 42). Critically, when, where, and in which species such outsider AIVs arrive in Australia, and how they are incorporated into circulating AIVs, remains unknown (17).
The low prevalence of AIVs in Australian wild birds, and the apparent evolutionary isolation of these viruses, has been attributed to the fact that it is extremely rare for Australian members of Anatidae to undertake regular movements within or between continents (39, 43–45). There are, however, millions of birds that migrate to Australia along the East Asian-Australasian Flyway (46). In particular, an estimated 8.5 million waders (order Charadriiformes, family Scolopacidae) from 37 species migrate to Australia to “overwinter” during the Austral summer (47, 48), linking Australia to avian communities in the Russian and Alaskan Arctic, Japan, Indonesia, Taiwan, and the east coast of China. Yet, AIV in waders has received significantly less attention both in Australia and globally, largely because its prevalence tends to be substantially lower than that in Anatidae (33, 34, 49, 50). One notable exception is Delaware Bay, USA, during spring migration, where AIV prevalence in waders averages 4% to 18% in a given year (51–53), approximately 50 times the combined rate of isolation at all other surveillance sites worldwide (54, 55). The vast majority of AIV infections in waders have been detected in just one species—the ruddy turnstone (Arenaria interpres)—with sympatric waders generally showing markedly lower prevalence (51, 56). Genomic evidence also suggests that ruddy turnstones introduce AIVs to Delaware Bay from the southeastern United States (57) and from the Northern Hemisphere to their overwintering sites in South America (58). Ruddy turnstones may therefore play a unique role in AIV gene flow (34), particularly in geographic regions where intercontinental migration of Anatidae is limited.
Several studies have called for long-term studies in waders outside Delaware Bay, to better understand their role in AIV gene flow, reassortment, and maintenance (10, 18, 34, 49). In response, we present 5 years of biannual surveillance for AIV in two populations of ruddy turnstones in southeastern Australia, at the terminus of the East Asian-Australasian Flyway. Using whole-genome sequencing, we illustrate patterns of gene flow and reassortment from the global AIV gene pool, providing unique insights into the evolutionary dynamics of AIVs in Australia.
RESULTS
Infection prevalence.
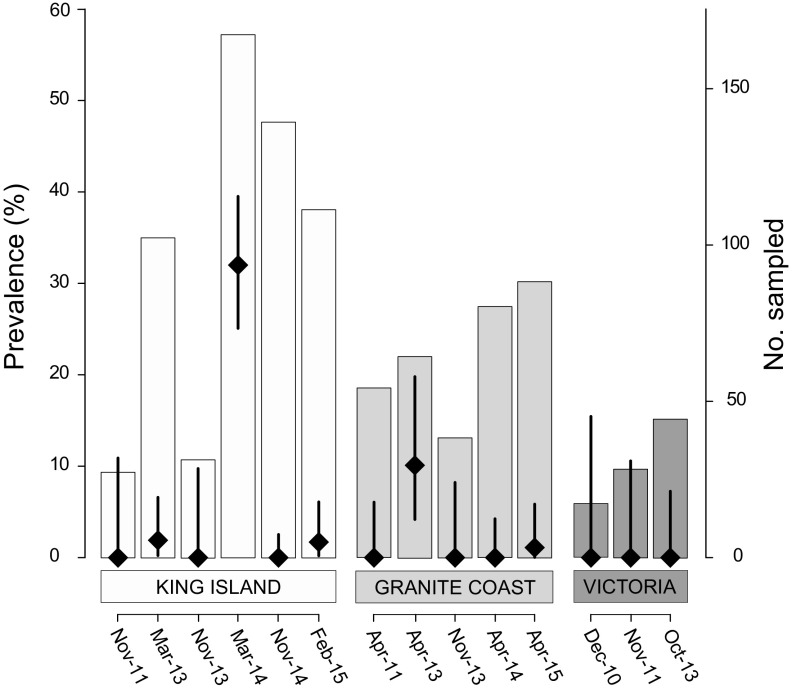
Over the 5-year study period (2010 to 2015), the prevalence of AIVs across all ruddy turnstone populations was 6.3% (n = 1,060; 95% confidence interval [CI] = 4.9 to 8.0%). Prevalence remained at 0 to 2% for the majority of nonbreeding locations across southeastern Australia throughout this period (Fig. 1). However, prior to prebreeding migration in April 2013, 10.1% (7/69; 95% CI = 4.2 to 19.8%) of birds were infected in the South Australian population, and in March 2014, strikingly, 32% (55/172; 95% CI = 25 to 39%) of the ruddy turnstones sampled on King Island, Tasmania, were infected (Fig. 1).
FIG 1.
Surveillance for avian influenza virus in ruddy turnstones in southeastern Australia, 2010 to 2015. Number of birds sampled at each time point at a given location indicated in bars (right axis); prevalence of avian influenza virus and 95% confidence interval (CI) are indicated by diamonds and error bars, respectively (left axis). Where no infection was detected, the upper 95% CI represents the maximum possible prevalence that could remain undetected with 95% confidence.
During the March 2014 epizootic, infections were only detected in four of the six catch events across four sites (Table 1 and Fig. 2A), with one site (Central Manuka) showing no infection in the first catch (95% CI = 0 to 21%; n = 13) but 46% of individuals infected 4 days later (95% CI = 28 to 66%; n = 28; Table 1). Among catches with infection, there was no significant difference in prevalence (χ2 = 4.33, P = 0.23). Juveniles were 3.3 times more likely to be infected than adults (95% CI = 1.3 to 8.8 times more likely; P = 0.15). A catch was also more likely to contain infected individuals when there was a higher proportion of juveniles (χ2 = 7.64, P = 0.006), but was not related to the abundance of turnstones at the site (χ2 = 0.22, P = 0.64). There was no difference in infection between the sexes (P = 0.423). During this epizootic, cycle threshold (CT) values ranged from 18.7 to 36.4 (mean ± standard deviation [SD] = 28.1 ± 4.8), with no difference between adults and juveniles (P = 0.75) or between males and females (P = 0.40).
TABLE 1.
Infection with influenza A virusa and seroprevalence of the nucleoprotein gene segment of influenza A virus in ruddy turnstones on King Island across the six catches in March 2014b
| Date | Site (beach) | PCR positive (n) | Infection prevalence (% [95% CI]) | Seroprevalence (% [95% CI]) | Subtype (n) |
|---|---|---|---|---|---|
| 18 March 2014 | Central Manuka | 0 | 0 (0–21) | 23 (5–54) | |
| 20 March 2014 | Stokes Point | 0 | 0 (0–8) | 6 (1–20) | |
| 22 March 2014 | Central Manuka | 13 | 46 (28–66) | 39 (22–59) | H6N8 (4) |
| 23 March 2014 | Whalebone Bay | 9 | 29 (14–48) | 35 (19–55) | H3N5 (3); H6N8 (2) |
| 24 March 2014 | Surprise Bay | 13 | 48 (29–68) | 44 (24–65) | H10N8 (10); H6/H10N8 (1) |
| 24 March 2014 | Burges Bay | 20 | 53 (36–69) | 34 (19–52) | H6N8 (3) |
| Total | 55 | 32 (25–39) | 30 (23–38) |
Detected by reverse transcriptase PCR (RT-PCR).
See Fig. 2 for spatial reference.
FIG 2.

Ruddy turnstones on King Island, Tasmania—prevalence and number sampled at each capture location in March 2014 (A), abundance in March 2014 (B), and relative number of individuals moving between locations, inferred from capture-recapture data from 2011 to 2014 (C), where colors correspond to locations indicated in the small map inset on the left, numbers indicate the number of birds first banded at each capture location, and width of each arc indicates the proportion of all birds that were banded at one location (set back from the outer ring) and recaptured at the same or another location (closer to the outer ring).
Origins of the March 2014 epizootic.
Influenza A virus was isolated from 23 of the PCR-positive samples from the March 2014 epizootic, with HA titers ranging from 32 to 512. All gene segments were successfully sequenced from the majority of these isolates, including 24 HA sequences (with one isolate coinfected with two HA types), 22 NA, 16 PB2, 22 PB1, 17 PA, 20 NP, 23 MP, and 22 NS gene sequences (Table 2). BLAST analysis of the surface proteins (HA and NA) showed that these isolates represented three influenza subtypes—three H3N5, nine H6N8, 10 H10N8, and one coinfected H6/H10N8 sample (Fig. 3 and Table 2). The H6N8 viruses were detected from central sites, including Whalebone Bay (n = 2), Manuka (n = 4), and Burges Bay (n = 3), while the H3N5 viruses were restricted to Whalebone Bay; the H10/H6N8 mixed infection and all 10 H10N8 viruses were detected at the southern end of the island (Surprise Bay), indicating potential geographical structuring by subtype within sampling locations (Table 2). The genomes of each of the subtypes, including H6N8 viruses sampled across four sites, showed high similarity across each gene segment. Given the high degree of site fidelity shown in recapture records (Fig. 2C), the high genomic similarity suggests recent exposure to this virus genotype across a large part of the island (Fig. 3 and 4 and Tables 1 and 2).
TABLE 2.
Phylogenetic designation for influenza A viruses isolated from ruddy turnstones on King Island in March 2014c
Spatial location of sites is depicted in Fig. 2.
Codes refer to origins of clades, as follows: EA, Eurasia; NAm, North America. H10N7 refers to the Australian 2010 to 2012 poultry outbreak isolates. Blanks indicate that the gene segment was not sequenced.
Shading colors indicate viral genotypes, designated H3N5 (green), H6N8 (blue), and H10N8 (orange).
FIG 3.
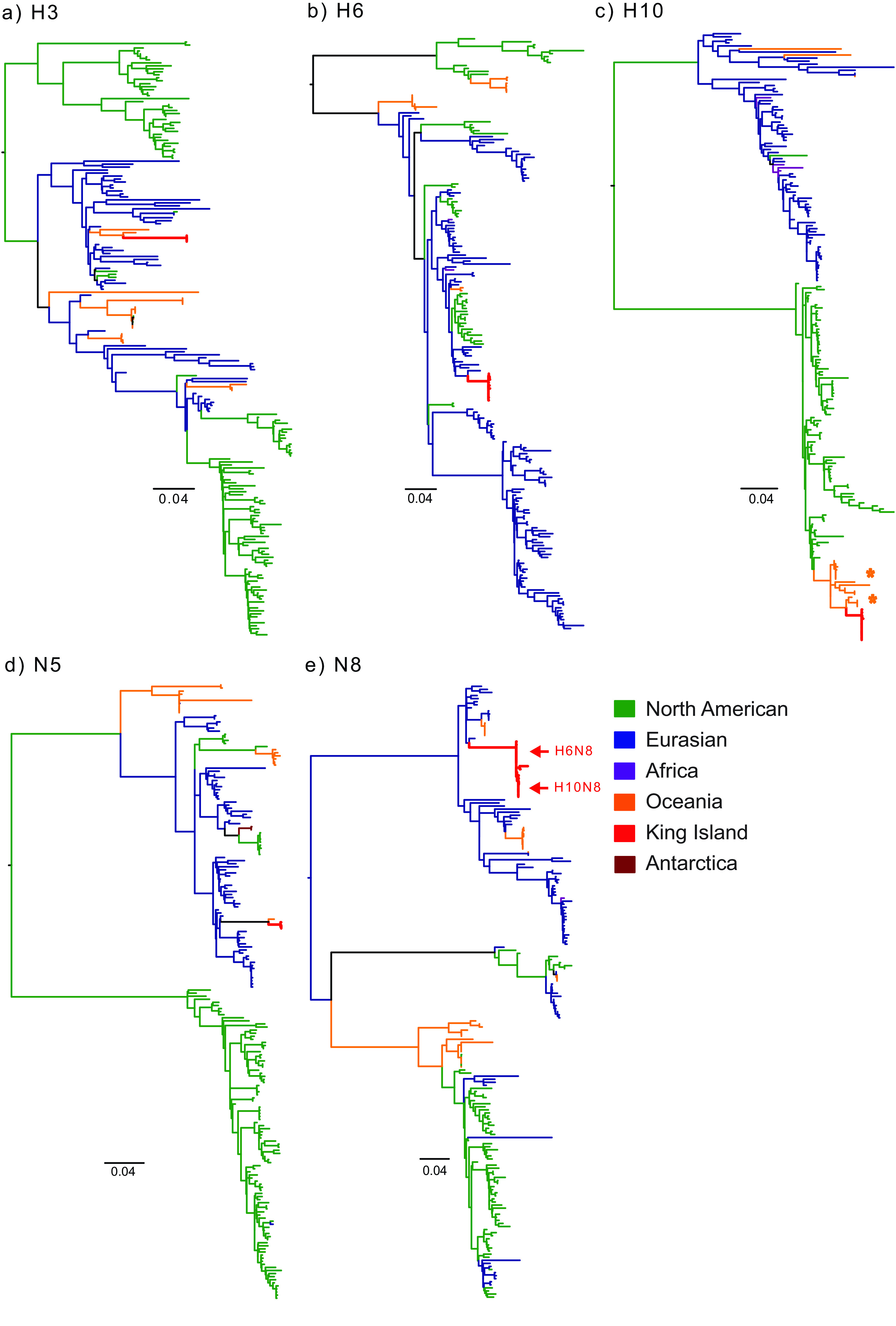
Phylogenetic relationships of the hemagglutinins H3 (a), H6 (b), H10 (c), and neuraminidases N5 (d), and N8 (e) of AIV isolates from ruddy turnstones on King Island in March 2014 (shown in red). Orange asterisks indicate the strains involved in the H10N7 outbreaks in Australian poultry in 2010 and 2012. Bars represent the number of substitutions per site. Trees with full strain names and showing bootstrap support on each node are provided in Fig. S1A to E in the supplemental material.
FIG 4.

Phylogenetic relationships of PB2 (a), PB1 (b), PA (c), NP (d), M (e), and NS (f) of AIV isolates from ruddy turnstones on King Island in March 2014 (shown in red). Orange asterisks indicate the strains involved in the H10N7 outbreaks in Australian poultry in 2010 and 2012. Bars represent number of substitutions per site. Trees with full strain names and showing bootstrap support on each node are provided in Fig. S1F to K.
Phylogenies revealed a wide range of origins for their gene segments. The HA and NA genes of H3N5 viruses were most closely related to viruses previously detected in Australia and New Zealand (A/chestnut-teal/Victoria/1/2004 (H3N2) and A/duck/MW04/Vic/2012 (H12N5), respectively; Fig. 3). This “Oceanic” H3 lineage was estimated to have diverged from the Eurasian avian gene pool 40 years prior to this epizootic (mean time of most recent common ancestor [TMRCA], 1973.1; 95% CI, 1969.3 to 1976.8) (see Fig. S2A in the supplemental material), while the N5-NA diverged from AIV in East Asia during late 1990s (mean TMRCA, 1997; 95% CI, 1994.8 to 1999.2; Fig. S2D). The H6 HA genes showed a more recent divergence from the Eurasian AIV gene pool during 2000 to 2005, predominantly clustering with AIVs detected throughout Asia between 2003 to 2014 (represented by A/duck/Guangxi/113/2012) (Fig. 3B; see also Fig. S1B and Fig. S2B in the supplemental material). The N8-NA genes of H10N8 and H6N8 clustered among the Eurasian avian gene pool but showed the earliest divergence of all external gene segments (mean TMRCA, 1964.7; 95% CI, 1957.5 to 1971) (Fig. S1E and Fig. S2E). The HA gene of H10N8 viruses were derived from the North American H10 lineage that has been detected in Australia since 2009. Our results illustrate that this North American lineage has become endemic in Oceania since 2009 (Fig. 3C, Table 2, and Fig. S1C). Our results also suggest long-term endemicity of N8-NA and H3-HA, and more recent introductions of the H6-HA and N5-NA lineages in Oceania.
The internal gene segments of all subtypes formed either two distinct sublineages with independent evolutionary origins (PB2, PB1, PA, and MP genes) (Fig. 4, Fig. S2F to H and Fig. S2J), or were all derived from a single lineage (NP and NS) (Fig. 4, Table 2, and Fig. S1F to K). All three King Island AIV subtypes contained two to four internal gene segments that shared ancestry with the lineage that gave rise to the Australian H10N7 poultry outbreak genotype (Fig. 4 and 5, Fig. S1F to K). The common NS gene segment of Eurasian/Australian origin that showed 97.37 to 98.32% nucleotide (nt) and 97.04 to 98.54% amino acid (aa) similarity to the 2010/2012 H10N7 outbreak genotype, estimated to have diverged in 2006.1 (95% highest posterior density [HPD], 2003.4 to 2008.4) (Fig. S2K), whereas the NP gene segment was derived independently from that in North America, with the closest available sequences collected from Michigan, Delaware, and Ontario and their divergence estimated during the 1990s (mean TMRCA, 1993.6; 95% CI, 1991 to 1996) (Fig. S2I). Notably, no genes were derived from clades known to circulate predominantly in gulls (Fig. S1).
FIG 5.

Schematic representation of the reassortment of influenza A virus lineages leading to the three AIV genotypes isolated from ruddy turnstones on King Island in March 2014.
AIV genotypes and recent reassortment.
Phylogenetic analysis showed that the epizootic detected in ruddy turnstones on King Island in 2014 involved three distinct AIV genotypes across three subtypes (Table 2). While the NP and NS genes were shared by all three genotypes, the H3N5 and H6N8 viruses shared a common PB1 and MP genes, the H3N5 and H10N8 viruses shared a common PB2 gene, and the H6N8 and H10N8 viruses shared the NA and PA genes. These genomic structures suggest a recent, common introduction into the turnstone population following extensive reassortment among the subtypes. Furthermore, all three genotypes identified on King Island represent intercontinental reassortants, with gene segments derived from the Eurasian lineage, North American lineage, and those endemic to Oceania (Table 2). The AIV gene pool in ruddy turnstones on King Island was dominated by Eurasian-lineage gene segments (20 of a possible 24). In particular, four of the five external gene segments (H3, H6, N5, and N8) appear to have been introduced directly from the Eurasian clade (Fig. 3), with few or no similar sequences identified from the Australian continent (Fig. S1A to E).
All three genotypes showed extensive reassortment with ancestors of the H10N7 viruses collected from outbreaks in commercial poultry in NSW and QLD in 2010 and 2012, as well as from wild birds since 2009 (Fig. 5). The PB2 segment of the H6N8 viruses clustered with previously isolated Oceanic viruses of various subtypes detected in Australia in 2001 to 2012 and were most closely related to H10N7 viruses from poultry outbreaks in NSW and QLD. The viruses comprising the second PB2 cluster, including the H10/H6N8 coinfection, a single H3N5 virus, and all H10N8 viruses were closely related to an H12N5 virus detected in 2012 in a duck in Victoria [A/duck/MW04/Vic/2012 (H12N5)] (Table 2 and Fig. S1F). In contrast, although the PB1 genes of all H10N8 viruses clustered distinctly and were most closely related to those of A/duck/MW04/Vic/2012 (H12N5) and the H10N7 poultry outbreak viruses from QLD and NSW, the PB1 genes of H3N5 and H6N8 viruses clustered together and were closely related to those of viruses isolated from ducks in China and South Korea between 2003 and 2004 (Fig. S1G). The PA genes of H3N5 viruses clustered distinctly and were most closely related to those of contemporary H1N3 and H5N2 viruses from China and South Korea, while the H6N8 and H10N8 viruses formed two closely related clusters within a diverse Oceania sublineage that was most closely related to the H10N7 poultry outbreak strains from 2010 and 2012 (Fig. S1H). Similarly to the PB1 gene, the MP gene of H10N8 viruses formed a monophyletic group, but it clustered with an Australian H10N5 virus (A/duck/Victoria/0305-2/2012) and a H10N7 poultry outbreak virus from QLD (A/Chicken/Queensland/1/2012), as well as with contemporary strains of various subtypes from China and Japan (Fig. S1J). The MP genes of the H3N5 and H6N8 viruses formed two closely related, subtype-specific clusters that were closely related to those of the H10N7 poultry outbreak virus from NSW (A/Chicken/NSW-Sydney/D809/2010) and AIV of various subtypes from China and New Zealand circulating in 2003 and 2004 (Fig. S1J). The NP and NS genes of H10N8, H3N5, and H6N8 viruses clustered together with minor discrete clustering patterns observed based on subtype. The NP genes were most closely related to those of the contemporaneous H12N5 Oceanic virus A/duck/MW04/Vic/2012 and clustered within a North American lineage, while the H10N7 poultry outbreak viruses clustered in the Eurasian lineage (Fig. S1I). The NS genes clustered within the Eurasian lineage and were most closely related to those of the H12N5 Oceanic virus A/duck/MW04/Vic/2012 and H10N7 poultry outbreak viruses from QLD and NSW and contemporary strains from China (Fig. S1K).
Serology.
During the epizootic in March 2014, anti-NP antibodies were found in 30% of turnstones on King Island (95% CI = 23 to 38%; Table 1). Seroprevalence was significantly higher in catches where AIVs were detected (38%; 95% CI = 29 to 47%) compared to catches where AIVs were not detected (10%; 95% CI = 4 to 23%; P = 0.0006), including Central Manuka, where no infection was detected on 18 March 2014, but 46% of individuals were infected just 4 days later. Of the 65 serum samples positive for anti-NP, only one sample (from an adult female that was not infected at the time of capture) showed a titer of 1:40 to H6 A/Turnstone/King Island/7019CP/2014. This virus was isolated from an individual captured at the same site on the same day as the hemagglutination inhibition (HI)-positive individual. All other HI titers were <1:40, with low serum volumes necessitating a high starting dilution. None of the anti-NP positive samples from two subsequent sampling visits (November 2014 and February 2015; n = 83) showed detectable responses to the three viral strains found circulating in March 2014, with HI titers of <1:10. Of the 17 individuals that were PCR positive for AIV in March 2014 and were subsequently sampled in November 2014 and/or February 2015, 10 (59%) had no detectable antibodies in their follow-up samples.
DISCUSSION
Infection prevalence.
Prior to this study, no AIVs had been detected in ruddy turnstones in Australia (59), despite the majority of AIV infections in waders across the globe having been detected in this species (51, 52, 54, 60). The generally low prevalence of AIV (0 to 2%) in ruddy turnstones in southeastern Australia during 2010 to 2015 is in line with estimates for waders from Europe, Asia, the Afrotropics, South America, and the vast majority of sites in North America (e.g., 49, 52, 58, 61–67). However, there were 2 years in which prevalence was markedly higher prior to prebreeding migration (March/April), similar to prevalence seen in ducks globally (ca. 10% globally, seasonal peaks of 20 to 60%; 33, 50) and ruddy turnstones in Delaware Bay during prebreeding migration (ca. 4 to 18%; 51–53). Similar, sporadic detections of higher AIV prevalence have been reported in waders at a few locations outside Delaware Bay (39, 49, 68). However, the epizootic detected in ruddy turnstones on King Island in March 2014 represents one of the highest prevalence levels reported for this species, with 32% of individuals found infected.
Increased prevalence of AIVs in wild birds is thought to be driven by a suite of biological and environmental factors, including large aggregations of host individuals, the pulsed influx of naive hatch-year birds, active migration, successive waves of migrants, confluence of migratory flyways, and the use of aquatic habitats (2, 3, 10, 69–74). In Anatidae, prevalence is consistently higher in hatch-year individuals lacking antibodies to AIV (3, 16, 69, 70). Similarly, we found that hatch-year ruddy turnstones were more likely to be infected and less likely to have detectable antibodies to AIV, suggesting recent introductions into a broadly naive population. Indeed, antibodies to the NP gene segment were only detected in 30% of ruddy turnstones on King Island in March 2014, in contrast to estimates of 75% in Iceland (63) and an increase from 40 to 95% during stopover in Delaware Bay (53). Seroprevalence also increased over the sampling period, averaging 10% at locations on King Island where infection was not detected (sampled 18 and 20 March 2014) and 38% at locations where AIVs were detected (sampled 22 to 24 March 2014). However, in line with experimental observation of low rates of seroconversion (75), we found little evidence of seroconversion following the epizootic. Over half of the infected birds that were resampled 8 to 11 months later lacked detectable antibodies to NP, and only one NP-positive individual displayed a weak HI titer to any of the circulating strains. This individual was captured during the epizootic, with none of the 132 NP-positive individuals from November 2014 or February 2015 displaying detectable antibodies to the epizootic strains. Collectively, these results suggest that naivety to AIV may have been an important factor promoting the epizootic reported here. They also suggest that the epizootic on King Island may represent the local amplification of AIVs recently introduced into the population from an as-yet-undefined reservoir, in line with the epidemiology of AIV in waders reported elsewhere (10, 49, 51, 55, 56).
Many of the other biological or environmental divers do not appear to have been necessary for the epizootic seen in ruddy turnstones on King Island in March. Globally, high prevalences of AIV tend to be reported from migratory stopover sites, particularly at the confluence of major migratory flyways (18, 33) or where migrants from disparate breeding locations pass through in successive waves (72, 73, 76). In contrast, the sites in southeastern Australia reported here represent the terminus of one branch of the East Asian-Australasian Flyway. Moreover, the epizootic on King Island in March 2014 occurred approximately 5 months after ruddy turnstones had arrived on migration. Similar end-of-the-flyway sites in North America report consistently low levels of AIV (11, 57, 77). Notably, the sites included in our study also lack the abundance and diversity of Anatidae seen in many other “overwintering” areas, presumably further limiting the potential for AIV transmission (49, 71). The abiotic conditions of our sites also appear unlikely to support high rates of AIV transmission, with ruddy turnstones foraging along the high tide line and roosting on off-shore rocks—habitats that present unfavorable conditions for AIV persistence (78). Large, high-density aggregations are thought to be a key factor driving the anomalously high prevalence of AIV in ruddy turnstones in Delaware Bay in Spring, with more than one million waders foraging in densities of up to 210 birds/m2 at this time (54, 55), in contrast to low prevalence during breeding, autumn migration, and winter, when ruddy turnstones are seen in small flocks (<50) (53). Yet, populations of ruddy turnstones “overwintering” in southeastern Australia are small and dispersed. For instance, in March 2014, an estimated 577 ruddy turnstones inhabited King Island in flocks of <50 individuals dispersed along the entire western coastline of the island (Fig. 2B), and the estimated population has never exceeded 900 (79). Our findings suggest that large, high-density aggregations may not be necessary for sporadic high-prevalence outbreaks of AIV in waders.
Reassortment and geographic origins.
Phylogenetic analysis revealed that all three genotypes isolated from ruddy turnstones on King Island in 2014 represent extensive intercontinental reassortants, with gene segments derived from Eurasian lineages, North American lineages, and those endemic to Oceania. Previous analysis had suggested that Australian AIVs formed a relatively distinct gene pool within the broader Eurasian lineage, whereby monophyletic clade(s) within each gene segment were thought to arise from circulation among hosts that remain within the continent (39, 40). A number of the gene segment variants detected here appear to fit this description, with the NA, PB2, NS, and MP (all genotypes), PA (H10 and H6), PB1 (H10), and H3 HA gene segments seen to be closely related to other AIVs detected in Australia that display distant similarity to AIVs from the Eurasian gene pool. However, three gene segments (H6 HA, PB1 [H3 and H6] and PA [H3]) represented novel detections in Oceania with high nucleotide similarity to recent isolates from across Eurasia (see Fig. S2B, G, and H in the supplemental material). This diversity of ancestral sources, together with the genomic similarity of multiple gene segments across multiple capture locations, suggests that extensive reassortment had taken place prior to the AIVs being detected in ruddy turnstones on King Island.
In addition to novel outsider events from Eurasia, two gene segments of North American origin were detected. The NP gene segment present in all three genotypes appears to have arrived in Australia during the mid-1990s, suggesting it circulated for up to 20 years before detection in unspecified duck species in Victoria in 2012 and in ruddy turnstones on King Island in 2014. We also detected the H10 HA detected in unspecified duck species in 2009, and subsequently in Australian poultry and poultry abattoir workers in 2010 and 2012 (17). Together with the MP detected in an unspecified species of tern in 2004 (41), our NP and H10 HA findings suggest that AIVs from the North American lineage are likely to be introduced into Australian wild birds more often than previously thought. Our data also provide the first example of reassortment of the North American-derived H10 in wild birds since this HA appeared, supporting previous suggestions that this North American-lineage H10 HA has replaced the Eurasian-origin H10 HAs that were previously circulating in Australian wild birds (Fig. S2C). Previous estimates, based on limited genomic data, suggested initial transmission of this HA gene segment into Australia occurred during 2007 and 2008, and that it was derived from viruses isolated from aquatic birds in the western United States (17). The additional 11 H10 gene segments sequenced from King Island suggest that this gene segment was introduced from viruses circulating through the midwestern United States in the early 2000s, significantly earlier than previous estimates. A group of highly related viruses therefore appears to have circulated in Australian wild birds for roughly a decade before the outbreaks in commercial poultry.
Several other gene segments detected on King Island indicate ongoing circulation of both the Eurasian and North American outsider events. Each of the AIV genotypes detected on King Island contained two or four gene segments, in addition to the H10 HA, derived from the AIVs infecting Australian poultry and poultry abattoir workers in 2010 and 2012 (Fig. 5). Similarly, the North American-lineage NP shows high nucleotide identity to AIVs isolated from ducks on the Australian mainland, approximately 150 km north of King Island in 2012 and 2018 (80). Taken together, these results substantiate earlier suggestions that gene segments introduced into Australia are incorporated into the broader AIV gene pool, continuing to reassort with other introduced and endemic viruses in wild birds to generate new genome constellations for decades after their introduction. In contrast, outsider events are not generally seen to persist in the North American lineage, where Eurasian-lineage gene segments are rarely detected in wintering areas (11, 81), despite being detected in up to 70% of AIVs in wild ducks in Alaska (26, 32), and appear to be lost within a decade of initial detection in waders and gulls (82). Given that the poultry outbreaks occurred 900 to 1,500 km to the north of King Island, our findings suggest that extensive circulation and reassortment of AIVs within Australian wild birds occurs over vast geographic distances, despite the lack of migratory behavior in Australian ducks, geese, and swans (45). Reassortment may therefore represent a critical strategy for the perpetuation of AIVs in regions with low AIV prevalence and limited intercontinental migration, as has been proposed during the winter nadir of infection in regions with strong seasonality (11).
Conclusions.
Our findings indicate that long-term surveillance in waders may yield unique insights into AIV gene flow in Oceania. In particular, our findings pose questions regarding how outsider AIVs arrive in Australia, where (and in which species) reassortment takes place, and how AIVs are introduced into isolated populations of ruddy turnstones. Given that Australian Anatidae rarely undertake intercontinental migrations, migratory waders may be particularly important for the introduction of outsider events into Australia. In support of this suggestion, AIVs have been detected in recently arrived sharp-tailed sandpipers (Calidris acuminata) and red-necked stints (Calidris ruficollis) (68), with the latter also showing serological evidence of AIV infection during migration through East Asia (83). Collectively, these species directly link Australia to both Asia and North America. For instance, adult sharp-tailed sandpipers migrate via East Asia, whereas juveniles migrate from hatching areas in Siberia to staging areas in Alaska, where they are thought to join other species in making direct, nonstop, 10,000-km flights across the Pacific Ocean from Alaska to New Zealand and eastern Australia (84, 85). Interactions between migratory waders and resident wild birds, and movements that introduce AIVs to King Island several months after the arrival of the ruddy turnstone population, are yet to be investigated. Data on movements and AIV infections in Australian gull species in particular may yield novel insights into the introduction of outsider events, given the gull breeding colonies on King Island and the high rates of outsider events detected in gull species in the Northern Hemisphere (20, 21, 29, 30, 86, 87). Regular, long-term surveillance for AIVs in waders along their migratory flyways, in combination with genomic data on AIVs circulating in resident birds, is needed to assess whether epizootics in waders are isolated or recurrent events, and to establish more comprehensive understanding of gene flow, reassortment, and maintenance of AIVs in geographic regions where Anatidae do not display regular inter- or intracontinental migration.
MATERIALS AND METHODS
Capture and sampling.
During the Austral summers of 2010 to 2015, 1,060 samples were collected from “overwintering” (i.e., nonbreeding) populations of ruddy turnstones at several locations across southeastern Australia, including King Island, Tasmania; the “Granite coast” of South Australia; and Flinders (2010), Stoney Point (2011), and Killarney (2013) in Victoria. These locations represent the southern end of the migratory route for this population, which breeds on the New Siberian Islands in northeastern Siberia (88) and migrates via locations in Indonesia, Taiwan, and the east coasts of China and Russia (89). Sampling took place after arrival from postbreeding migration (November/December) and again immediately prior to prebreeding migration (March/April; Fig. 1). Each sampling event comprised either a single catch or multiple separate catches over a period of 7 to 10 days at the same location. Cannon nets were used to target a single flock of 15 to 50 turnstones along the high tide line. Individuals were banded (or recapture recorded), weighed, and measured, and then sterile rayon swabs were used to collect cloacal and oropharyngeal samples. Both swabs from each individual were placed in a single vial of viral transport media, stored on ice throughout the day of sampling, and then maintained at −80°C until analysis. Approximately 200 μl of whole blood was collected from the brachial vein, allowed to clot for ∼8 h, then centrifuged and serum maintained at −20°C until analysis. Birds were aged as either hatch-year or adult on the basis of plumage and sexed using molecular methods. Birds were held in freshly laundered individual cotton bags for the duration of sampling, after which they were released at the shoreline. All efforts were made to minimize any suffering throughout the study.
During the sampling events on King Island, Tasmania, members of the Victorian Wader Study Group systematically scanned all suitable ruddy turnstone habitat and counted individual ruddy turnstones to estimate abundance. All records of captures and recaptures of ruddy turnstones on King Island between 2011 and 2014 were used to quantify the movement of individuals between beaches (sites) within and between seasons. This study was conducted under approval of the Deakin University Animal Ethics Committee (permit numbers A113‐2010 and B37‐2013), the Australian Bird Banding Scheme (permit number 8001), the Department of Environment, Land, Water and Planning, Victoria (permit numbers 10006663 and 10005726), the Department of Environment, Water and Natural Resources, South Australia (permit numbers M25919‐1, -2, -3, -4, and -5), and the Department of Primary Industries, Parks, Water and Environment, Tasmania (permit numbers FA11255, TFA13032, TFA14065, TFA14110, TFA15269).
RT-PCR and virus isolation.
Current infection with AIV was tested using a generic real-time reverse transcriptase PCR (RT-PCR) assay targeting the matrix gene on RNA isolated from the swab samples following the standard diagnostic procedure for the Australian wild bird surveillance program (44), using a SensiFast probe Lo-ROX one-step kit (Bioline, Australia) on an Applied Biosystems 7500 fast real-time PCR system (Life Technologies, Australia). Samples with a cycle threshold (CT) value of <40 were considered positive for the presence of influenza A virus. Factors predicting infection in an individual, including age, sex, and sampling location, were tested using generalized linear models (GLM) in R version 3.6.2 (90). Virus isolation was attempted from all RT-PCR-positive samples, whereby swab specimens diluted 1:1 with phosphate-buffered saline (PBS) containing 1% neomycin-polymyxin solution (bioCSL, Australia) were injected into the allantoic cavity of two 11-day-old embryonated chicken eggs. Allantoic fluid was harvested after 3 days, and influenza virus was detected by hemagglutination with turkey erythrocytes. If, after the first passage, hemagglutination titers were detectable but were too low to store or conduct further assays, then allantoic fluid was repassaged in two more eggs to increase titers.
Whole-genome sequencing.
Viral RNA was extracted from isolates using QIAxtractor (Qiagen, Australia), and each of the eight gene segments was amplified using the multiplex RT-PCR method (91). Amplified RT-PCR products were sequenced by next-generation sequencing (NGS) using the IonTorrent PGM instrument as previously described (92). De novo analysis of the NGS data was first performed using CLC Genomic Benchtop 7 (Qiagen). All of the resulting contigs were then subjected to BLAST searches to find influenza virus-related sequences. The closest-matched complete influenza virus segment sequences were chosen as reference sequences for a second round of analysis to map all the reads to generate final influenza virus segment sequences.
Phylogenetic analysis.
The AIV gene sequences determined in this study were analyzed in conjunction with sequences obtained from the Global Initiative on Sharing All Influenza Data (GISAID) database (93), as well as sequences from the Oceania region available in GenBank but not in the GISAID database. We retained all samples from Oceania and Antarctica, approximately 30 to 50 for HA and NA genes and approximately 100 for internal genes. The remaining data from Eurasia/Africa and the Americas were downsampled by first removing identical sequences using CD-HIT (94), and then retaining a random set of ∼100 sequences each for Eurasia/Africa and Americas, resulting in a final data set of ∼250 sequences for the internal protein genes and ∼300 sequences for the surface protein genes. Multiple-sequence alignments were performed for each segment using MUSCLE v3.5 (95) followed by manual optimization. Maximum-likelihood (ML) phylogenetic trees were estimated utilizing RAxML v8.0 (96), applying the general time-reversible nucleotide substitution model with a gamma distribution of among-site rate variation (GTR + Γ). Support for individual nodes was estimated with 1,000 rapid bootstrap replicates. Nucleotide and amino acid similarities were calculated using the proportional (p) distance algorithm.
We performed exploratory linear regression analysis of the root-to-tip genetic distances using the ML tree and sampling dates in TempEst v1.4 (97). Time-measured evolutionary histories were reconstructed using Bayesian phylogenetic inference in BEAST v1.8.2 (98), using the Hasegawa-Kishino-Yano (HKY) substitution model under a strict clock and a constant coalescent size tree prior (99, 100). Three independent Markov chain Monte Carlo chains were run for 100 million steps and sampled every 10,000th generation, with the first 10% discarded as burn-in. Convergence and mixing of the chains were inspected using Tracer v1.5 (http://tree.bio.ed.ac.uk/software/tracer/); all continuous parameters yielded effective sample sizes greater than 200. A maximum clade credibility (MCC) tree was summarized using TreeAnnotator v1.8.2 (98). Trees were visualized and annotated using FigTree v1.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Serology.
The presence of antibodies to AIV nucleoprotein (NP) in serum was tested using a commercially available blocking enzyme-linked immunosorbent assay (bELISA; MultiS-Screen avian influenza virus antibody test kit). Absorbance was measured at 620 nm using a FLUOstar Omega plate reader. Assays were carried out in duplicate, in combination with supplied positive and negative controls. A sample signal-to-noise ratio of >0.5 was considered negative for the presence of antibodies to AIV. All samples positive for antibodies to NP from March 2014, November 2014, and February 2015 were further tested for specific HA antibodies using a hemagglutination inhibition (HI) assay against three circulating AIVs isolated from March 2014 [A/Turnstone/King Island/7049CP/2014 (H3N5), A/Turnstone/King Island/7019CP/2014 (H6N8), and A/Turnstone/King Island/7104CP/2014 (H10N8)], following WHO standards (101).
Data availability.
The sequences determined in this study are available under GenBank accession numbers MW243404 to MW243569.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Victorian Wader Study Group, including Rob Patrick, Robin Atkinson, Margaret Bennett, Maureen Christie, Penny Johns, and the late Clive Minton, as well as David Roshier, Yaara Rotman, Alice Risley, Justin Eastwood, Jayome Hutchinson, and Jutta Leyrer, for assistance with field logistics and sampling. We also thank Patrick Mileto, Kim O’Riley, Chantal Baas, Heidi Peck, and Malet Aban for laboratory assistance.
This study was supported by the Australian Research Council (grant DP130101935 to M.K.), the National Avian Influenza Wild Bird Program (B.J.H., M.K., and S.W.), the Deakin University Central Research Grant Scheme (B.J.H.), and Department of Job, Precincts & Regions surveillance funding (S.W.). The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health. C.M.D. is an Australian NHMRC Early Career Research Fellow (1113269), and B.J.H. is supported by an ARC DECRA Fellowship (DE200100884). D.V. is supported by contract HHSN272201400006C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Department of Health and Human Services, USA.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Easterday BC, Trainer DO, Tůmová B, Pereira HG. 1968. Evidence of infection with influenza viruses in migratory waterfowl. Nature 219:523–524. 10.1038/219523a0. [DOI] [PubMed] [Google Scholar]
- 2.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. 10.1128/MR.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinshaw VS, Webster RG, Turner B. 1980. The perpetuation of orthomyxoviruses and paramyxoviruses in Canadian waterfowl. Can J Microbiol 26:622–629. 10.1139/m80-108. [DOI] [PubMed] [Google Scholar]
- 4.Brown VL, Drake JM, Barton HD, Stallknecht DE, Brown JD, Rohani P. 2014. Neutrality, cross-immunity and subtype dominance in avian influenza viruses. PLoS One 9:e88817. 10.1371/journal.pone.0088817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. 2010. Genome packaging in influenza A virus. J Gen Virol 91:313–328. 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 6.Chen R, Holmes EC. 2009. Frequent inter-species transmission and geographic subdivision in avian influenza viruses from wild birds. Virology 383:156–161. 10.1016/j.virol.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dugan VG, Chen R, Spiro DJ, Sengamalay N, Zaborsky J, Ghedin E, Nolting J, Swayne DE, Runstadler JA, Happ GM, Senne DA, Wang R, Slemons RD, Holmes EC, Taubenberger JK. 2008. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog 4:e1000076. 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahl J, Vijaykrishna D, Holmes EC, Smith GJD, Guan Y. 2009. Gene flow and competitive exclusion of avian influenza A virus in natural reservoir hosts. Virology 390:289–297. 10.1016/j.virol.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholtissek C. 1995. Molecular evolution of influenza viruses. Virus Genes 11:209–215. 10.1007/BF01728660. [DOI] [PubMed] [Google Scholar]
- 10.Bahl J, Krauss S, Kühnert D, Fourment M, Raven G, Pryor SP, Niles LJ, Danner A, Walker D, Mendenhall IH, Su YCF, Dugan VG, Halpin RA, Stockwell TB, Webby RJ, Wentworth DE, Drummond AJ, Smith GJD, Webster RG. 2013. Influenza A virus migration and persistence in North American wild birds. PLoS Pathog 9:e1003570. 10.1371/journal.ppat.1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill NJ, Ma EJ, Meixell BW, Lindberg MS, Boyce WM, Runstadler JA. 2016. Transmission of influenza reflects seasonality of wild birds across the annual cycle. Ecol Lett 19:915–925. 10.1111/ele.12629. [DOI] [PubMed] [Google Scholar]
- 12.Hill NJ, Hussein IT, Davis KR, Ma EJ, Spivey TJ, Ramey AM, Puryear WB, Das SR, Halpin RA, Lin X, Fedorova NB, Suarez DL, Boyce WM, Runstadler JA. 2017. Reassortment of influenza A viruses in wild birds in Alaska before H5 clade 2.3.4.4 outbreaks. Emerg Infect Dis 23:654–657. 10.3201/eid2304.161668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steel J, Lowen AC. 2014. Influenza A virus reassortment. Curr Top Microbiol Immunol 385:377–401. 10.1007/82_2014_395. [DOI] [PubMed] [Google Scholar]
- 14.Smith GJD, Bahl J, Vijaykrishna D, Zhang J, Poon LLM, Chen H, Webster RG, Peiris JSM, Guan Y. 2009. Dating the emergence of pandemic influenza viruses. Proc Natl Acad Sci U S A 106:11709–11712. 10.1073/pnas.0904991106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615–619. 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkatesh D, Poen MJ, Bestebroer TM, Scheuer RD, Vuong O, Chkhaidze M, Machablishvili A, Mamuchadze J, Ninua L, Fedorova NB, Halpin RA, Lin X, Ransier A, Stockwell TB, Wentworth DE, Kriti D, Dutta J, van Bakel H, Puranik A, Slomka MJ, Essen S, Brown IH, Fouchier RAM, Lewis NS. 2018. Avian Influenza viruses in wild birds: virus evolution in a multihost ecosystem. J Virol 92:e00433-18. 10.1128/JVI.00433-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vijaykrishna D, Deng Y-M, Su YCF, Fourment M, Iannello P, Arzey GG, Hansbro PM, Arzey KE, Kirkland PD, Warner S, O'Riley K, Barr IG, Smith GJD, Hurt AC. 2013. The recent establishment of North American H10 lineage influenza viruses in Australian wild waterfowl and the evolution of Australian avian influenza viruses. J Virol 87:10182–10189. 10.1128/JVI.03437-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahl J, Pham TT, Hill NJ, Hussein ITM, Ma EJ, Easterday BC, Halpin RA, Stockwell TB, Wentworth DE, Kayali G, Krauss S, Schultz-Cherry S, Webster RG, Webby RJ, Swartz MD, Smith GJD, Runstadler JA. 2016. Ecosystem interactions underlie the spread of avian influenza A viruses with pandemic potential. PLoS Pathog 12:e1005620. 10.1371/journal.ppat.1005620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Borm S, Rosseel T, Vangeluwe D, Vandenbussche F, van den Berg T, Lambrecht B. 2012. Phylogeographic analysis of avian influenza viruses isolated from Charadriiformes in Belgium confirms intercontinental reassortment in gulls. Arch Virol 157:1509–1522. 10.1007/s00705-012-1323-x. [DOI] [PubMed] [Google Scholar]
- 20.Hall JS, TeSlaa JL, Nashold SW, Halpin RA, Stockwell T, Wentworth DE, Dugan V, Ip HS. 2013. Evolution of a reassortant North American gull influenza virus lineage: drift, shift and stability. Virol J 10:179. 10.1186/1743-422X-10-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dusek RJ, Hallgrimsson GT, Ip HS, Jónsson JE, Sreevatsan S, Nashold SW, TeSlaa JL, Enomoto S, Halpin RA, Lin X, Fedorova N, Stockwell TB, Dugan VG, Wentworth DE, Hall JS. 2014. North Atlantic migratory bird flyways provide routes for intercontinental movement of avian influenza viruses. PLoS One 9:e92075. 10.1371/journal.pone.0092075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Robertson GJ, Ojkic D, Whitney H, Lang AS. 2014. Diverse inter-continental and host lineage reassortant avian influenza A viruses in pelagic seabirds. Infect Genet Evol 22:103–111. 10.1016/j.meegid.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Sharp GB, Kawaoka Y, Jones DJ, Bean WJ, Pryor SP, Hinshaw V, Webster RG. 1997. Coinfection of wild ducks by influenza a viruses: distribution patterns and biological significance. J Virol 71:6128–6135. 10.1128/JVI.71.8.6128-6135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barton HD, Rohani P, Stallknecht DE, Brown J, Drake JM. 2014. Subtype diversity and reassortment potential for co-circulating avian influenza viruses at a diversity hot spot. J Anim Ecol 83:566–575. 10.1111/1365-2656.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wille M, Tolf C, Avril A, Latorre-Margalef N, Wallerström S, Olsen B, Waldenström J. 2013. Frequency and patterns of reassortment in natural influenza A virus infection in a reservoir host. Virology 443:150–160. 10.1016/j.virol.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Koehler AV, Pearce JM, Flint PL, Franson JC, Ip HS. 2008. Genetic evidence of intercontinental movement of avian influenza in a migratory bird: the northern pintail (Anas acuta). Mol Ecol 17:4754–4762. 10.1111/j.1365-294X.2008.03953.x. [DOI] [PubMed] [Google Scholar]
- 27.Ramey AM, Pearce JM, Flint PL, Ip HS, Derksen DV, Franson JC, Petrula MJ, Scotton BD, Sowl KM, Wege ML, Trust KA. 2010. Intercontinental reassortment and genomic variation of low pathogenic avian influenza viruses isolated from northern pintails (Anas acuta) in Alaska: examining the evidence through space and time. Virology 401:179–189. 10.1016/j.virol.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Wille M, Benkaroun J, Munro H, Bond AL, Fifield DA, Robertson GJ, Ojkic D, Whitney H, Lang AS. 2014. Perpetuation and reassortment of gull influenza A viruses in Atlantic North America. Virology 456-457:353–363. 10.1016/j.virol.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 29.Wille M, Robertson GJ, Whitney H, Ojkic D, Lang AS. 2011. Reassortment of American and Eurasian genes in an influenza A virus isolated from a great black-backed gull (Larus marinus), a species demonstrated to move between these regions. Arch Virol 156:107–115. 10.1007/s00705-010-0839-1. [DOI] [PubMed] [Google Scholar]
- 30.Wille M, Robertson GJ, Whitney H, Bishop MA, Runstadler JA, Lang AS. 2011. Extensive geographic mosaicism in avian influenza viruses from gulls in the Northern Hemisphere. PLoS One 6:e20664. 10.1371/journal.pone.0020664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krauss S, Obert CA, Franks J, Walker D, Jones K, Seiler P, Niles L, Pryor SP, Obenauer JC, Naeve CW, Widjaja L, Webby RJ, Webster RG. 2007. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathog 3:e167. 10.1371/journal.ppat.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramey AM, Pearce JM, Ely CR, Sheffield Guy LM, Irons DB, Derksen DV, Ip HS. 2010. Transmission and reassortment of avian influenza viruses at the Asian-North American interface. Virology 406:352–359. 10.1016/j.virol.2010.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus A, Fouchier R. 2006. Global patterns of influenza A virus in wild birds. Science 312:384–388. 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 34.Runstadler J, Hill N, Hussein ITM, Puryear W, Keogh M. 2013. Connecting the study of wild influenza with the potential for pandemic disease. Infect Genet Evol 17:162–187. 10.1016/j.meegid.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi W, Lei F, Zhu C, Sievers F, Higgins DG. 2010. A complete analysis of HA and NA genes of influenza A viruses. PLoS One 5:e14454. 10.1371/journal.pone.0014454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallensten A, Munster VJ, Elmberg J, Osterhaus A, Fouchier RAM, Olsen B. 2005. Multiple gene segment reassortment between Eurasian and American lineages of influenza A virus (H6N2) in guillemot (Uria aalge). Arch Virol 150:1685–1692. 10.1007/s00705-005-0543-8. [DOI] [PubMed] [Google Scholar]
- 37.Lebarbenchon C, Chang C-M, Gauthier-Clerc M, Thomas F, Renaud F, van der Werf S. 2009. H9N2 avian influenza virus in a Mediterranean gull. J Mol Genet Med 03:121–123. 10.4172/1747-0862.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramey AM, Reeves AB, Sonsthagen SA, TeSlaa JL, Nashold S, Donnelly T, Casler B, Hall JS. 2015. Dispersal of H9N2 influenza A viruses between East Asia and North America by wild birds. Virology 482:79–83. 10.1016/j.virol.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 39.Hansbro PM, Warner S, Tracey JP, Arzey KE, Selleck P, O’Riley K, Beckett EL, Bunn C, Kirkland PD, Vijaykrishna D, Olsen B, Hurt AC. 2010. Surveillance and analysis of avian influenza viruses, Australia. Emerg Infect Dis 16:1896–1904. 10.3201/eid1612.100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulach D, Halpin R, Spiro D, Pomeroy L, Janies D, Boyle DB. 2010. Molecular analysis of H7 avian influenza viruses from Australia and New Zealand: genetic diversity and relationships from 1976 to 2007. J Virol 84:9957–9966. 10.1128/JVI.00930-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kishida N, Sakoda Y, Shiromoto M, Bai G-R, Isoda N, Takada A, Laver G, Kida H. 2008. H2N5 influenza virus isolates from terns in Australia: genetic reassortants between those of the Eurasian and American lineages. Virus Genes 37:16–21. 10.1007/s11262-008-0235-z. [DOI] [PubMed] [Google Scholar]
- 42.Arzey GG, Kirkland PD, Arzey KE, Frost M, Maywood P, Conaty S, Hurt AC, Deng Y-M, Iannello P, Barr I, Dwyer DE, Ratnamohan M, McPhie K, Selleck P. 2012. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis 18:814–816. 10.3201/eid1805.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haynes L, Arzey E, Bell C, Buchanan N, Burgess G, Cronan V, Dickason C, Field H, Gibbs S, Hansbro P, Hollingsworth T, Hurt A, Kirkland P, McCracken H, O’Connor J, Tracey J, Wallner J, Warner S, Woods R, Bunn C. 2009. Australian surveillance for avian influenza viruses in wild birds between July 2005 and June 2007. Aust Vet J 87:266–272. 10.1111/j.1751-0813.2009.00446.x. [DOI] [PubMed] [Google Scholar]
- 44.Grillo V, Arzey K, Hansbro P, Hurt A, Warner S, Bergfeld J, Burgess G, Cookson B, Dickason C, Ferenczi M, Hollingsworth T, Hoque M, Jackson R, Klaassen M, Kirkland P, Kung N, Lisovski S, O'Dea M, O'Riley K, Roshier D, Skerratt L, Tracey J, Wang X, Woods R, Post L. 2015. Avian influenza in Australia: a summary of 5 years of wild bird surveillance. Aust Vet J 93:387–393. 10.1111/avj.12379. [DOI] [PubMed] [Google Scholar]
- 45.Tracey JP, Woods R, Roshier D, West P, Saunders GR. 2004. The role of wild birds in the transmission of avian influenza for Australia: an ecological perspective. Emu 104:109–124. 10.1071/MU04017. [DOI] [Google Scholar]
- 46.Dingle H. 2008. Bird migration in the southern hemisphere: a review comparing continents. Emu 108:341–359. 10.1071/MU08010. [DOI] [Google Scholar]
- 47.Hansen BD, Fuller RA, Watkins D, Rogers DI, Clemens RS, Newman M, Woehler EJ, Weller DR. 2016. Revision of the East Asian-Australasian Flyway population estimates for 37 listed migratory shorebird species. BirdLife Australia, Melbourne, Australia. [Google Scholar]
- 48.Bamford M, Watkins D, Bancroft W, Tischler G, Wahl J. 2008. Migratory shorebirds of the East Asian-Australasian Flyway: population estimates and internationally important sites. Wetlands International–Oceania, Canberra, Australia. [Google Scholar]
- 49.Gaidet N, El Mamy ABO, Cappelle J, Caron A, Cumming GS, Grosbois V, Gil P, Hammoumi S, de Almeida RS, Fereidouni SR, Cattoli G, Abolnik C, Mundava J, Fofana B, Ndlovu M, Diawara Y, Hurtado R, Newman SH, Dodman T, Balanca G. 2012. Investigating avian influenza infection hotspots in old-world shorebirds. PLoS One 7:e46049. 10.1371/journal.pone.0046049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Munster VJ, Baas C, Lexmond P, Waldenström J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WEP, Schutten M, Olsen B, Osterhaus ADME, Fouchier RAM. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog 3:e61. 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanson BA, Luttrell MP, Goekjian VH, Niles L, Swayne DE, Senne DA, Stallknecht DE. 2008. Is the occurrence of avian influenza virus in Charadriformes species and location dependent? J Wildl Dis 44:351–361. 10.7589/0090-3558-44.2.351. [DOI] [PubMed] [Google Scholar]
- 52.Stallknecht DE, Luttrell MP, Poulson R, Goekjian V, Niles L, Dey A, Krauss S, Webster RG. 2012. Detection of avian influenza viruses from shorebirds: evaluation of surveillance and testing approaches. J Wildl Dis 48:382–393. 10.7589/0090-3558-48.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maxted AM, Luttrell MP, Goekjian VH, Brown JD, Niles LJ, Dey AD, Kalasz KS, Swayne DE, Stallknecht DE. 2012. Avian influenza virus infection dynamics in shorebird hosts. J Wildl Dis 48:322–334. 10.7589/0090-3558-48.2.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krauss S, Stallknecht DE, Negovetich NJ, Niles LJ, Webby RJ, Webster RG. 2010. Coincident ruddy turnstone migration and horseshoe crab spawning creates an ecological ‘hot spot’ for influenza viruses. Proc R Soc B 277:3373–3379. 10.1098/rspb.2010.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown VL, Drake JM, Stallknecht DE, Brown JD, Pedersen K, Rohani P. 2013. Dissecting a wildlife disease hotspot: the impact of multiple host species, environmental transmission and seasonality in migration, breeding and mortality. J R Soc Interface 10:20120804. 10.1098/rsif.2012.0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maxted AM, Sitters HP, Luttrell MP, Dey AD, Kalasz KS, Niles LJ, Stallknecht DE. 2016. Spring migration stopover ecology of avian influenza virus shorebird hosts at Delaware Bay. Avian Dis 60:394–405. 10.1637/11079-040515-Reg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poulson R, Carter D, Beville S, Niles L, Dey A, Minton C, McKenzie P, Krauss S, Webby R, Webster R, Stallknecht DE. 2020. Influenza A viruses in ruddy turnstones (Arenaria interpres); connecting wintering and migratory sites with an ecological hotspot at Delaware Bay. Viruses 12:1205. 10.3390/v12111205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Araujo J, de Azevedo SM, Gaidet N, Hurtado RF, Walker D, Thomazelli LM, Ometto T, Seixas MMM, Rodrigues R, Galindo DB, da Silva ACS, Rodrigues AMM, Bomfim LL, Mota MA, Larrazábal ME, Branco JO, Serafini P, Neto IS, Franks J, Webby RJ, Webster RG, Durigon EL. 2014. Avian influenza virus (H11N9) in migratory shorebirds wintering in the Amazon Region, Brazil. PLoS One 9:e110141. 10.1371/journal.pone.0110141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Curran JM, Ellis TM, Robertson ID. 2014. Surveillance of Charadriiformes in Northern Australia shows species variations in exposure to avian influenza virus and suggests negligible virus prevalence. Avian Dis 58:199–204. 10.1637/10634-080913. [DOI] [PubMed] [Google Scholar]
- 60.Kawaoka Y, Chambers T, Sladen W, Webster RG. 1988. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology 163:247–250. 10.1016/0042-6822(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 61.Breed AC, Harris K, Hesterberg U, Gould G, Londt BZ, Brown IH, Cook AJC. 2010. Surveillance for avian influenza in wild birds in the European Union in 2007. Avian Dis 54:399–404. 10.1637/8950-053109-Reg.1. [DOI] [PubMed] [Google Scholar]
- 62.Cheng MC, Lee MS, Ho YH, Chyi WL, Wang CH. 2010. Avian influenza monitoring in migrating birds in Taiwan during 1998–2007. Avian Dis 54:109–114. 10.1637/8960-061709-Reg.1. [DOI] [PubMed] [Google Scholar]
- 63.Hall JS, Hallgrimsson GT, Suwannanarn K, Sreevatsen S, Ip HS, Magnusdottir E, TeSlaa JL, Nashold SW, Dusek RJ. 2014. Avian influenza virus ecology in Iceland shorebirds: intercontinental reassortment and movement. Infect Genet Evol 28:130–136. 10.1016/j.meegid.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 64.Iverson SA, Takekawa JY, Schwarzbach S, Cardona CJ, Warnock N, Bishop MA, Schirato GA, Paroulek S, Ackerman JT, Ip H, Boyce WM. 2008. Low prevalence of avian influenza virus in shorebirds on the Pacific coast of North America. Waterbirds 31:602–610. 10.1675/1524-4695-31.4.602. [DOI] [Google Scholar]
- 65.D’Amico VL, Bertellotti M, Baker AJ, Diaz LA. 2007. Exposure of red knots (Calidris canutus rufa) to select avian pathogens; Patagonia, Argentina. J Wildl Dis 43:794–797. 10.7589/0090-3558-43.4.794. [DOI] [PubMed] [Google Scholar]
- 66.Escudero G, Munster VJ, Bertellotti M, Edelaar P. 2008. Perpetuation of avian influenza in the Americas: examining the role of shorebirds in Patagonia. Auk 125:494–495. 10.1525/auk.2008.2408.2. [DOI] [Google Scholar]
- 67.Ghersi BM, Blazes D, Icochea E, Gonzalez RI, Kochel T, Tinoco Y, Sovero M, Lindstrom S, Shu B, Klimov A, Gonzalez AE, Montgomery JM. 2009. Avian influenza in wild birds from the central coast of Peru. Emerg Infect Dis 15:935–938. 10.3201/eid1506.080981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hurt AC, Hansbro PM, Selleck P, Olsen B, Minton C, Hampson AW, Barr IG. 2006. Isolation of avian influenza viruses from two different transhemispheric migratory shorebird species in Australia. Arch Virol 151:2301–2309. 10.1007/s00705-006-0784-1. [DOI] [PubMed] [Google Scholar]
- 69.van Dijk JGB, Hoye BJ, Verhagen JH, Nolet BA, Fouchier RAM, Klaassen M. 2014. Juveniles and migrants as drivers for seasonal epizootics of avian influenza virus. J Anim Ecol 83:266–275. 10.1111/1365-2656.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoye BJ, Fouchier RAM, Klaassen M. 2012. Host behaviour and physiology underpin individual variation in avian influenza virus infection in migratory Bewick’s swans. Proc Biol Sci 279:529–534. 10.1098/rspb.2011.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gaidet N, Caron A, Cappelle J, Cumming GS, Balança G, Hammoumi S, Cattoli G, Abolnik C, Servan de Almeida R, Gil P, Fereidouni SR, Grosbois V, Tran A, Mundava J, Fofana B, Ould El Mamy AB, Ndlovu M, Mondain-Monval JY, Triplet P, Hagemeijer W, Karesh WB, Newman SH, Dodman T. 2012. Understanding the ecological drivers of avian influenza virus infection in wildfowl: a continental-scale study across Africa. Proc Biol Sci 279:1131–1141. 10.1098/rspb.2011.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gunnarsson G, Latorre-Margalef N, Hobson KA, Van Wilgenburg SL, Elmberg J, Olsen B, Fouchier RAM, Waldenström J. 2012. Disease dynamics and bird migration—linking mallards Anas platyrhynchos and subtype diversity of the influenza A virus in time and space. PLoS One 7:e35679. 10.1371/journal.pone.0035679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Latorre-Margalef N, Tolf C, Grosbois V, Avril A, Bengtsson D, Wille M, Osterhaus ADME, Fouchier RAM, Olsen B, Waldenström J. 2014. Long-term variation in influenza A virus prevalence and subtype diversity in migratory mallards in northern Europe. Proc Biol Sci 281:20140098. 10.1098/rspb.2014.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hill NJ, Takekawa JY, Ackerman JT, Hobson KA, Herring G, Cardona CJ, Runstadler JA, Boyce WM. 2012. Migration strategy affects avian influenza dynamics in mallards (Anas platyrhynchos). Mol Ecol 21:5986–5999. 10.1111/j.1365-294X.2012.05735.x. [DOI] [PubMed] [Google Scholar]
- 75.Hall JS, Krauss S, Franson JC, TeSlaa JL, Nashold SW, Stallknecht DE, Webby RJ, Webster RG. 2013. Avian influenza in shorebirds: experimental infection of ruddy turnstones (Arenaria interpres) with avian influenza virus. Influenza Other Respir Viruses 7:85–92. 10.1111/j.1750-2659.2012.00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Verhagen JH, van Dijk JGB, Vuong O, Bestebroer T, Lexmond P, Klaassen M, Fouchier RAM. 2014. Migratory birds reinforce local circulation of avian influenza viruses. PLoS One 9:e112366. 10.1371/journal.pone.0112366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stallknecht DE, Shane SM, Zwank PJ, Senne DA, Kearney MT. 1990. Avian influenza viruses from migratory and resident ducks of coastal Louisiana. Avian Dis 34:398–405. 10.2307/1591427. [DOI] [PubMed] [Google Scholar]
- 78.Brown JD, Goekjian G, Poulson R, Valeika S, Stallknecht DE. 2009. Avian influenza virus in water: infectivity is dependent on pH, salinity and temperature. Vet Microbiol 136:20–26. 10.1016/j.vetmic.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 79.Minton C, Atkinson R, Leung K, Patrick R. 2019. VWSG King Island visit report 22–31 March 2019. Victorian Wader Study Group Bull 42:56–69. [Google Scholar]
- 80.Bhatta TR, Chamings A, Vibin J, Klaassen M, Alexandersen S. 2020. Detection of a reassortant H9N2 avian influenza virus with intercontinental gene segments in a resident Australian chestnut teal. Viruses 12:88. 10.3390/v12010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pearce JM, Ramey AM, Flint PL, Koehler AV, Fleskes JP, Franson JC, Hall JS, Derksen DV, Ip HS. 2009. Avian influenza at both ends of a migratory flyway: characterizing viral genomic diversity to optimize surveillance plans for North America. Evol Appl 2:457–468. 10.1111/j.1752-4571.2009.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pearce JM, Ramey AM, Ip HS, Gill RE. 2010. Limited evidence of trans-hemispheric movement of avian influenza viruses among contemporary North American shorebird isolates. Virus Res 148:44–50. 10.1016/j.virusres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 83.Wille M, Lisovski S, Risely A, Ferenczi M, Roshier D, Wong FYK, Breed AC, Klaassen M, Hurt AC. 2019. Serologic evidence of exposure to highly pathogenic avian influenza H5 viruses in migratory shorebirds, Australia. Emerg Infect Dis 25:1903–1910. 10.3201/eid2510.190699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gill RE, Tibbitts TL, Douglas DC, Handel CM, Mulcahy DM, Gottschalck JC, Warnock N, McCaffery BJ, Battley PF, Piersma T. 2009. Extreme endurance flights by landbirds crossing the Pacific Ocean: ecological corridor rather than barrier? Proc Biol Sci 276:447–457. 10.1098/rspb.2008.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Handel CM, Gill RE. 2010. Wayward youth: trans-Beringian movement and differential southward migration by juvenile sharp-tailed sandpipers. Arctic 63:273–288. 10.14430/arctic1492. [DOI] [Google Scholar]
- 86.Lewis NS, Verhagen JH, Javakhishvili Z, Russell CA, Lexmond P, Westgeest KB, Bestebroer TM, Halpin RA, Lin X, Ransier A, Fedorova NB, Stockwell TB, Latorre-Margalef N, Olsen B, Smith G, Bahl J, Wentworth DE, Waldenström J, Fouchier RAM, de Graaf M. 2015. Influenza A virus evolution and spatio-temporal dynamics in Eurasian wild birds: a phylogenetic and phylogeographical study of whole-genome sequence data. J Gen Virol 96:2050–2060. 10.1099/vir.0.000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mathieu C, Moreno V, Pedersen J, Jeria J, Agredo M, Gutiérrez C, García A, Vásquez M, Avalos P, Retamal P. 2015. Avian influenza in wild birds from Chile, 2007–2009. Virus Res 199:42–45. 10.1016/j.virusres.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 88.Gosbell K, Minton C, Fox J. 2012. Geolocators reveal incubation and re-nesting characteristics of ruddy turnstones Arenaria interpres and eastern curlews Numenius madagascariensis. Wader Study Group Bull 119:160–171. [Google Scholar]
- 89.Minton C, Gosbell K, Johns P, Christie M, Klaassen M, Hassell C, Boyle A, Jessop R, Fox J. 2011. Geolocator studies on ruddy turnstones Arenaria interpres and greater sandplovers Charadrius leschenaultii in the East Asian-Australasia Flyway reveal widely different migration strategies. Wader Study Group Bull 118:87–96. [Google Scholar]
- 90.R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 91.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza A viruses. J Virol 83:10309–10313. 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hurt AC, Su YCF, Aban M, Peck H, Lau H, Baas C, Deng Y-M, Spirason N, Ellström P, Hernandez J, Olsen B, Barr IG, Vijaykrishna D, Gonzalez-Acuna D. 2016. Evidence for the introduction, reassortment, and persistence of diverse influenza A viruses in Antarctica. J Virol 90:9674–9682. 10.1128/JVI.01404-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shu Y, McCauley J. 2017. GISAID: global initiative on sharing all influenza data—from vision to reality. Euro Surveill 22(13):30494. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 95.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:vew007. 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang J, Xie D, Nie Z, Xu B, Drummond AJ. 2019. Inferring host roles in Bayesian phylodynamics of global avian influenza A virus H9N2. Virology 538:86–96. 10.1016/j.virol.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 100.Escalera-Zamudio M, Golden M, Gutiérrez B, Thézé J, Keown JR, Carrique L, Bowden TA, Pybus OG. 2020. Parallel evolution in the emergence of highly pathogenic avian influenza A viruses. Nat Commun 11:5511. 10.1038/s41467-020-19364-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.World Health Organization. 2011. Manual for the laboratory diagnosis and virological surveillance of influenza. World Health Organization, Geneva, Switzerland. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences determined in this study are available under GenBank accession numbers MW243404 to MW243569.