SHIVs have been extensively used in a nonhuman primate (NHP) model for HIV research. In this study, we investigated viral reservoirs in tissues and immune responses in an NHP model inoculated with newly generated transmitted/founder HIV-1 clade C-based SHIV.C.CH848.
KEYWORDS: SHIV, persistent infection, viral reservoirs, autologous neutralizing Abs and evolution, autologous neutralizing Abs, evolution
ABSTRACT
Chimeric simian/human immunodeficiency viruses (SHIVs) are widely used in nonhuman primate models to recapitulate human immunodeficiency virus (HIV) infection in humans, yet most SHIVs fail to establish persistent viral infection. We investigated immunological and virological events in rhesus macaques infected with the newly developed SHIV.C.CH848 (SHIVC) and treated with combined antiretroviral therapy (cART). Similar to HIV/simian immunodeficiency virus (SIV) infection, SHIV.C.CH848 infection established viral reservoirs in CD4+ T cells and myeloid cells, accompanied by productive infection and depletion of CD4+ T cells in systemic and lymphoid tissues throughout SHIV infection. Despite 6 months of cART-suppressed viral replication, integrated proviral DNA levels remained stable, especially in CD4+ T cells, and the viral rebound was also observed after ART interruption. Autologous neutralizing antibodies to the parental HIV-1 strain CH848 were detected, with limited viral evolution at 5 months postinfection. In comparison, heterogenous neutralizing antibodies in SHIV.C.CH848-infected macaques were not detected except for 1 (1 of 10) animal at 2 years postinfection. These findings suggest that SHIV.C.CH848, a novel class of transmitted/founder SHIVs, can establish sustained viremia and viral reservoirs in rhesus macaques with clinical immunodeficiency consequences, providing a valuable SHIV model for HIV research.
IMPORTANCE SHIVs have been extensively used in a nonhuman primate (NHP) model for HIV research. In this study, we investigated viral reservoirs in tissues and immune responses in an NHP model inoculated with newly generated transmitted/founder HIV-1 clade C-based SHIV.C.CH848. The data show that transmitted founder (T/F) SHIVC infection of macaques more closely recapitulates the virological and clinical features of HIV infection, including persistent viremia and viral rebound once antiretroviral therapy is discontinued. These results suggest this CCR5-tropic, SHIVC strain is valuable for testing responses to HIV vaccines and therapeutics.
INTRODUCTION
Chimeric simian/human immunodeficiency viruses (SHIVs), carrying human immunodeficiency virus (HIV) envelope glycoproteins from transmitted founder (T/F) viruses, are invaluable for testing HIV transmission, pathogenesis, and prevention strategies in nonhuman primate (NHP) models (1–5). However, the majority of current SHIVs have limitations, including differences in coreceptor usage and a lack of sustained viremia or progression to AIDS (5–11). Therefore, the development of functional SHIV clones that better mimic the natural history of HIV infection and that reflect coreceptor usage of globally circulating T/F viruses resulting in establishment of persistent viral reservoirs in animal models is of great significance for testing HIV prevention and cure strategies.
Clade C viruses represent the predominant HIV subtype in the global HIV pandemic, yet most SHIVs to date either have been derived from clade B (SHIVsf262P and SHIVAD8) or utilize different coreceptors from that of HIV T/F strains (SHIV89.6P, SHIV KU, etc.), which exclusively utilize CCR5. The new CCR5-tropic clade C SHIV (SHIV.C.CH848 [SHIVC]) clone (12–14) encodes a clade C Env isolated from an acutely infected Malawian man in 2008 (15) and was developed by a single amino acid substitution at Env residue 375 to increase the affinity of CH848 Env for rhesus CD4 (16, 17). In this study, we investigated immunological and virological events in SHIV.C.CH848-infected animals in acute infection and on antiretroviral therapy, including CD4+ T cells, neutralizing antibody (Ab) responses, and Env viral evolution. SHIV.C.CH848 infection resulted in acute depletion of peripheral CD4+ T cells and persistent viral infection, as indicated by detectable proviral DNA even after 6 months of combined antiretroviral therapy (cART) treatment, and viral rebound after cART interruption. A potent autologous (but not heterogenous) neutralizing antibody response was detected from 5 months to 2 years postinfection (p.i.). These findings suggest that this novel T/F SHIV.C.CH848 is a promising model for HIV latency and cure studies.
RESULTS
Plasma viral load and immunological responses in SHIV.C.CH848-infected rhesus macaques on antiretroviral therapy.
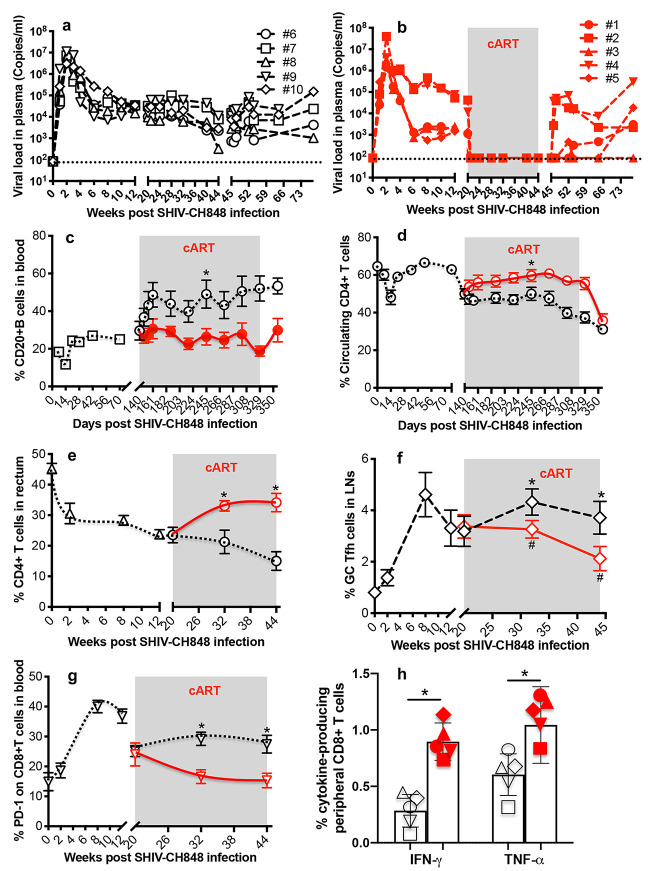
Ten rhesus macaques (RMs) were intravenously inoculated with SHIV.C.CH848, and half of the animals received anti-HIV drugs 5 months postinfection. As indicated in Fig. 1a and b, plasma viral load peaked 14 days post-SHIV infection, followed by a relatively sustained viral set point. Once cART was initiated (5 months postinfection), viremia in treated animals (n = 5) rapidly declined to undetectable levels after 2 weeks, compared with untreated controls (n = 5) that in large part maintained the set point viral load. Notably, viral rebound was detected in 4/5 of the animals after cART interruption, except for 1 animal that remained aviremic up to 6 months after cART cessation. Further, SHIVC infection resulted in significant depletion of peripheral CD4+ T cells by 14 days postinfection and progressive reductions of rectal CD4+ T cells while peripheral CD20+ B cells and GC T follicular help (Tfh) cells (PD-1high CXCR5+ cells gated CD4+ T cells) expanded in untreated animals by 5 months postinfection. In contrast, cART-treated animals significantly recovered CD4+ T cells in blood and rectum while maintaining baseline levels of peripheral CD20+ B cells and GC Tfh cells (Fig. 1c to f). Since PD-1 upregulation reflects CD8+ T cell exhaustion during viral infection (18, 19), we examined PD-1 expression on peripheral CD8+ T cells throughout SHIVC infection. The results showed that SHIVC infection significantly upregulated PD-1 on CD8+ T cells in untreated macaques, whereas reduced frequencies of PD-1+ CD8+ T cells were detected in cART-treated animals (Fig. 1g), consistent with lower simian immunodeficiency virus (SIV) Gag-specific cytotoxic T lymphocyte (CTL) responses in untreated animals at 44 weeks postinfection (Fig. 1h). These data demonstrate that SHIV.C.CH848 recapitulates key virological and immunological characteristics of HIV-1 infection.
FIG 1.
Plasma viral load and immunological events in SHIV.C.CH848-inoculated rhesus macaques on antiretroviral therapy. (a and b) Plasma viral load in SHIV-infected macaques subsequently treated with anti-HIV drugs for 6 months, initiated 5 months post-SHIV infection (n = 5 [a]), compared to untreated controls (n = 5 [b]). (c to f) Changes in peripheral CD20+ B and CD4+ T cells, rectal CD4+ T cells, and T follicular helper cells in untreated and treated animal groups. (g) PD-1 expression on peripheral CD8+ T cells. Note that viral rebound was observed in 4 of 5 animals after cART interruption. (h) SHIV.C.CH848 gag-specific cytokine (TNF‐α and IFN‐γ) responses of peripheral CD8+ T cells in two animal cohorts with (red) or without ART (black) at 44 weeks post-SHIV infection. Error bars indicate SEM. Paired t tests were used to compare ART-treated with untreated groups. *, P < 0.01.
Viral dissemination in systemic and lymphoid compartments of SHIV.C.CH848-infected rhesus macaques on antiretroviral therapy.
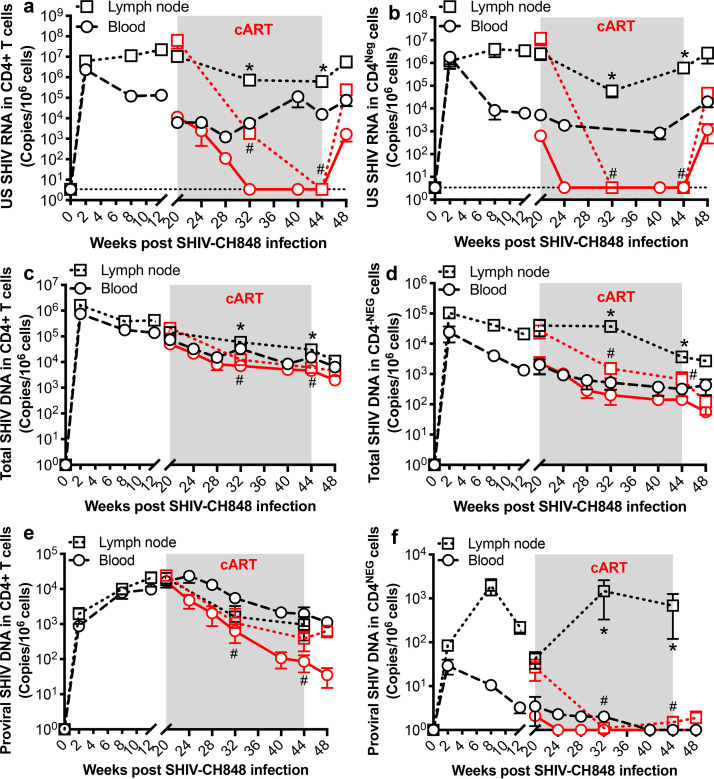
To evaluate the dynamics of cell-associated SHIVC RNA/DNA in systemic and lymphoid tissues after SHIVC infection and their response to cART, unspliced (US) SHIVC RNA, total SHIV DNA, and proviral DNA were longitudinally measured in blood, lymph node (LN), and rectal biopsy specimens from SHIVC-infected macaques. As CD4+ T cells are preferentially targeted in HIV infection, CD4+ T cells were also purified from blood and lymph nodes to further assess cell-associated SHIV RNA/DNA levels, compared with the remaining CD4-negative cell populations. As shown in Fig. 2, both SHIV RNA and DNA were detected in both CD4+ and CD4-negative cell populations in peripheral blood mononuclear cells (PBMCs) and lymph node-derived mononuclear cells. Antiretroviral therapy suppressed SHIVC replication in both cell populations in blood and lymph nodes compared with that in untreated controls. Levels of US SHIVC RNA decreased to undetectable levels in both cell populations by 3 months of cART treatment, although LN-derived CD4+ T cells still had detectable viral RNA (vRNA) at this time point. At 1 month of cART interruption, US SHIVC RNA increased in both cell populations (Fig. 2a and b). Similarly, cART also reduced levels of total SHIVC DNA and proviral DNA in both CD4+ and CD4-negative cell populations within 3 months of treatment (Fig. 2c to f). However, total and proviral DNAs were still detectable in CD4+ T cells throughout SHIVC infection, regardless of treatment duration. Notably, there was no significant reduction of proviral DNA levels in LN-derived CD4+ T cells in treated animals. However, there was no detectable proviral DNA in the corresponding CD4-negative cells after treatment (Fig. 2e and f). These data show that CD4+ T cells are the major reservoir for SHIV.C.CH848 persistence, especially in organized lymphoid tissues (LN), mimicking the hallmark qualities of HIV infection in humans.
FIG 2.
Dynamics of cell-associated SHIV RNA and DNA in CD4+ T cells from PBMCs and lymph nodes of SHIV.C.CH848-infected macaques on antiretroviral therapy, compared with CD4-negative cells. Shown are results of longitudinal analysis of unspliced SHIV RNA (US SHIV RNA) (a and b), total SHIV DNA (c and d), or integrated proviral SHIV DNA (e and f) in CD4+ T cells and CD4-negative cell populations derived from peripheral blood and lymph nodes of SHIV-infected macaques with or without cART. Note that proviral DNA was detectable even under cART, especially in purified CD4+ T cells. Cell-associated SHIV RNA and DNA are expressed as copies per 1 million cells. The dotted line represents the limit of detection (LOD) calculated as described in Materials and Methods. Error bars indicate SEM. *, P < 0.01, determined by two-tailed paired t test in either blood or lymph node tissue of treated and untreated animals; #, P < 0.01, compared between pretreatment (month 5 p.i.) and after treatment in both tissues at different time points.
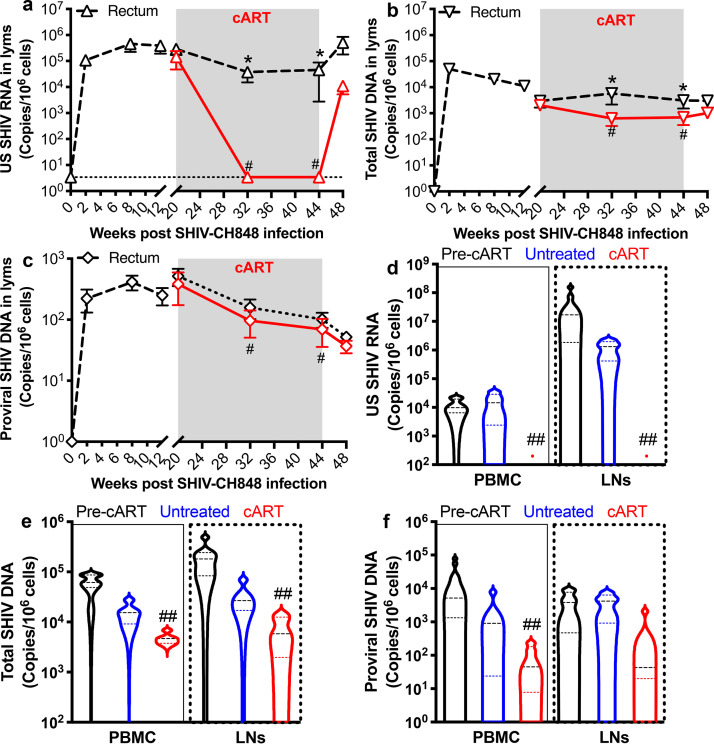
Cell-associated SHIV RNA/DNA was also measured from rectal lymphocytes in SHIV.C.CH848-infected macaques, with or without cART. As shown in Fig. 3a to c, the dynamics of rectal lymphocyte-associated SHIV RNA/DNA were very similar to those in blood and LN-derived CD4+ T cells, and cART did not significantly reduce levels of rectal cell-associated proviral DNA. Cell-associated viral nucleic acids were compared prior to treatment (5 months postinfection) and after 6 months of cART (Fig. 3d to f). Combined, these results showed that changes in SIV RNA/DNA were similar in lymphocytes from the blood, LNs, and rectum and that SHIV.C.CH848 infection results in viral persistence and stable, latent viral reservoirs in both systemic and lymphoid tissue compartments on treatment, which result in viral resurgence after treatment withdrawal.
FIG 3.
Cell-associated SHIV RNA and DNA in total PBMCs and rectal lymphocytes in SHIV.C.CH848-infected rhesus macaques on antiretroviral therapy. (a to c) Changes in levels of US SHIV RNA (a), total SHIV DNA (b), and proviral SHIV DNA (c) in rectal lymphocytes. (d to f) Levels of US SHIV RNA, total SHIV DNA, and proviral SHIV DNA in PBMCs at pretreatment (month 5 p.i.) and in cART-treated and untreated groups at 6 months. Error bars indicate SEM. *, P < 0.01 (paired t test was used to compare groups); #, P < 0.01 (comparison between pretreatment [month 5 p.i.] and after treatment in rectal tissue at different time points); ##, P < 0.05 (comparison with pretreatment or untreated controls in blood).
Plasma neutralizing antibody responses in SHIV.C.CH848-infected macaques.
We next performed TZM-bl neutralization assays to characterize neutralizing antibody responses in these macaques at 5, 15, and 24 months postinfection (Table 1). These animals developed autologous neutralizing antibody against SHIV.C.CH848 with variable potency (50% inhibitory dilution [ID50] titers ranged from less than 1:50 to more than 1:300) at 5 months postinfection, consistent with previous reports that autologous neutralizing antibody responses are common and arise relatively early in viremic macaques in the first few weeks to months postinfection (9, 20–22). At 2 years postinfection, regardless of cART, the plasma neutralization titers against SHIV.C.CH848 increased in 6 of 9 macaques (animal number 9 died before this time point), exhibiting neutralization ID50 titers of ≥1:300.
TABLE 1.
Plasma neutralization ID50 titers of SHIV.C.CH848-infected rhesus macaquesa
| Plasma ID |
ID50 at: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5 mo postinfection |
15 mo postinfection |
2 yrs postinfection |
||||||||
| CH848 (C) | CH848 (C) | CH848 (C) | Du156.12 (C) | ZM109.4 (C) | Q23.17 (A) | BG505 (A) | Yu2 (B) | JR-FL (B) | AD17 (B) | |
| 1 (cART) | <20 | <25 | 27 | ND | ND | ND | ND | ND | ND | ND |
| 2 (cART) | 57 | 97 | 214 | ND | ND | ND | ND | ND | ND | ND |
| 3 (cART) | 96 | 272 | 316 | <25 | <25 | <25 | <25 | <25 | <25 | <25 |
| 4 (cART) | 34 | 508 | 1,350 | <25 | 28 | <25 | <25 | <25 | <25 | <25 |
| 5 (cART) | 57 | 100 | 472 | <25 | <25 | <25 | <25 | <25 | <25 | <25 |
| 6 | 342 | 215 | 113 | <25 | <25 | <25 | <25 | <25 | <25 | <25 |
| 7 | 149 | 554 | 392 | <25 | 51 | 336 | 26 | <25 | 26 | <25 |
| 8 | 21 | 183 | 347 | <25 | <25 | 36 | <25 | <25 | <25 | <25 |
| 9 | 22 | 267 | ND | ND | ND | ND | ND | ND | ND | ND |
| 10 | 48 | 369 | 630 | <25 | <25 | <25 | <25 | <25 | <25 | <25 |
Plasma samples from SHIV.C.CH848-infected RMs at 5 months, 15 months, and 2 years postinfection were tested against a panel of autologous and heterogeneous viruses in TZM-bl cells. Clades are indicated in parentheses. Potency of 50 to 99 is indicated by italics; 100 to 999, underlining; and ≥1,000, underlining and boldface type. ND, not detected.
In contrast, none of the animals developed heterogenous neutralizing antibodies against the seven heterologous HIV-1 strains tested across clades A, B, and C at 15 months postinfection. At 2 years postinfection, there were no appreciable cross-reactive neutralizing antibodies in the 6 animals with CH848 ID50s of ≥1:300, except for animal number 7, in which the plasma cross-neutralized the clade A strain Q23.17 with an ID50 of ∼1:300 and weakly neutralized the clade C strain ZM109.4 with an ID50 of ∼1:50 (Table 1). In summary, 9 out of 10 SHIV.C.CH848-infected macaques mounted potent autologous neutralization antibody responses against the CH848 Env, whereas there were limited heterologous antibodies elicited in 1 infected animal.
Viral diversity in SHIV.C.CH848-infected macaques.
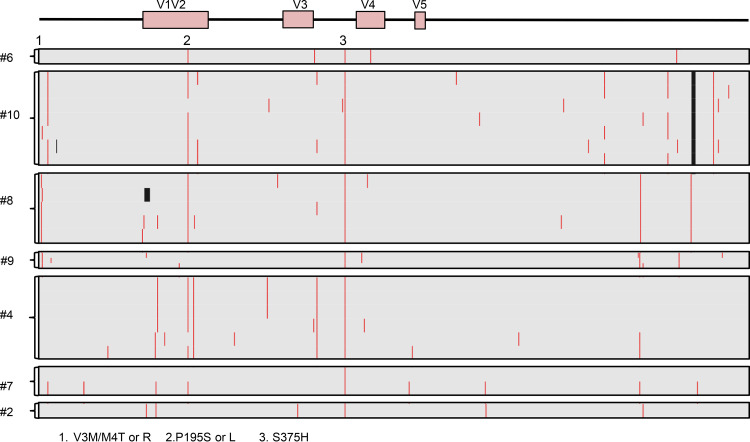
Single-genome sequencing (SGS) of SHIV.C.CH848 gp160 env was used to characterize sequence diversity 5 months postinfection. A total of 31 sequences (median = 5 sequences per animal) were generated from 7 infected rhesus macaques. Sequences were aligned to the human CH848 T/F Env. As indicated in Fig. 4, 2 conserved sites of selection pressure were identified in Env: V3M or M4T/R mutations were detected in 3 of 7 macaques, and the P195S/L mutation was detected in 4 of 7 macaques. Further, while sites of selection pressure across V1V2 were identified in all 7 animals, no mutations were conserved across the entire cohort. These results suggest that at 5 months postinfection, there was minimal sequence evolution in Env in this cohort of SHIV.C.CH848-infected rhesus macaques.
FIG 4.
Viral Env evolution in SHIV.C.CH848-infected macaques. Shown are amino acid highlighter plots showing single-genome env sequences from SHIV.C.CH848-infected RM at 5 months postinfection. Nonsynonymous substitutions compared to the TF sequence are indicated by a red line, and deletions are shown as a black line. Each horizontal line shows one single-genome sequence. Two common sites of selection pressure (Env residues 3/4 and 195) were identified in infected animals.
DISCUSSION
SHIVs have been widely used to explore HIV transmission, pathogenesis, latency, and cure strategies in NHP models of HIV/AIDS for over 20 years. In this study, we characterized the key features of a novel T/F SHIV.C.CH848 infection in rhesus macaques on cART. Our results indicated that SHIV.C.CH848 infection leads to the recapitulation of key immunological and virological characteristics that are hallmarks of HIV-1 infection.
HIV infection establishes a long-lived latent reservoir extremely early after infection (23). However, combined antiretroviral therapy (cART) basically fails to eliminate HIV latency characterized by integrated intact viral genomes, allowing the virus to persist for the lifetime of people living with HIV-1 (24–27). SHIV strains are very useful in NHP models for testing antiviral drugs, HIV vaccines, and functional cure strategies, especially if a small number of replication-competent viral reservoirs/latency are maintained. Although several SHIVs have been used in macaques (7, 28), their low viral persistence and often spontaneous clearance in macaques limit their potential for studying HIV latency in an NHP setting (6, 11). Recently developed CCR5-tropic SHIV.C.CH848 and SHIV.CH505 strains, encoding Env from a transmitted founder HIV-1 subtype C strain, with an increased affinity for rhesus CD4 have shown promise for viral replication kinetics more closely resembling those of HIV infection in humans (16, 17, 29). In this study, we analyzed neutralizing antibody responses and cellular reservoirs in systemic and lymphoid tissues of rhesus macaques infected with T/F SHIV.C.CH848, before and after cART treatment and interruption. These data demonstrate that this novel T/F SHIV.C.CH848 clone is promising as a candidate for testing HIV treatment and cure strategies.
T/F SHIV.C.CH848 infection in rhesus macaques during early and chronic infection closely mirrors HIV infection, as indicated by high peak viremia in primary infection, relatively stable viral set point of 104 to 105 viral RNA copies/ml of plasma, massive gut-associated mucosal CD4+ T cell depletion, stable proviral DNA in systemic and lymphoid tissues even after 6 months of cART, and viral rebound after treatment interruption. Recrudescence of SHIVC is observed within 4 months of treatment interruption, consistent with the viral rebound that occurs in most HIV+ patients after ART interruption, ranging from ∼5 days to 48 days (30, 31). However, viral rebound was not still observed in one animal at 6 months after ART cessation, compared with the stable viremic set point in untreated animals. Unlike SIV-infected macaques, which show rapid viral rebound after analytic treatment interruption, some SHIV.C.CH848-infected animals showed a delayed viral recrudescence, which might be attribute to limited chronic activation and latency reactivation, as indicated by lower levels of plasma inflammatory cytokines and chemokines (e.g., interleukin 8 [IL-8] and MIP-1β) at 3 weeks after treatment discontinuation, compared with those at pretreatment (data not shown). CCR5-tropic SHIV.C.CH848 efficiently infects rhesus macaques, resulting in persistent high levels of viremia, viral reservoir seeding, and depletion of CD4+ T cells, consistent with findings for SHIVAD8-infected macaques (8, 32, 33). However, circulating CD4+ T cells were rapidly restored in SHIV.C.CH848-infected macaques after ART was initiated. Given that SIVmac-infected macaques show higher viremia, and contain higher proportions of intact viral genome (∼84% for SIV versus 11.7% for HIV) on ART (34–36), SHIV.C.CH848, equivalent to HIV, likely shows less pathogenicity than SIVmac. In contrast to the depletion of CD4+ T cells, peripheral CD20+ B cells progressively increased throughout SHIVC infection in untreated animals, yet cART prevented B cell increases, suggesting that persistent SHIVC infection leads to immune activation and B cell hyperactivity if animals are untreated (37). To address the SHIV.C.CH848 latency, chronic activation, and latency reactivation after treatment interruption, more animals are needed for these studies.
In the HIV/SIV life cycle, the virus produces unspliced RNA (∼9 kb), which is responsible for gag/pol translation and packaging of the viral RNA genome, representing bona fide viral replication (38, 39). Further, integrated proviral DNA is the reliable marker of persistent viral reservoirs with clinical relevance, especially in patients when plasma viremia is undetectable (27, 40–45). Since proviral DNA is a fundamental constituent of the latent reservoir, measurement of this in cells is a simple approach to estimate the persistent cellular reservoir that may fuel viral rebound, although this method cannot distinguish defective forms. Considering that CD4+ T cells are primary targets and predominant cell reservoirs in HIV/SIV infection (46–48), purified CD4+ T cells and non-CD4+ cells were longitudinally compared for cell-associated viral RNA/DNA in blood and lymph nodes. Although levels of SHIV RNA/DNA were significantly different between CD4+ and CD4-negative cell populations, the latter including monocytes and other B and T cell lineage-negative cell subsets, the dynamics of viral RNA/DNA in two cell populations were similar, although CD4-negative cell populations showed levels of SHIV RNA/DNA that were at least 1 log lower.
Since non-CD4 cells can serve as reservoirs, including monocytes, dendritic cells, and macrophages (49–54), CD14+ myeloid cells were also purified to compare levels of viral RNA/proviral DNA from chronically SHIV.C.CH848-infected macaques without treatment. As suspected, viral RNA/DNA levels in purified CD4+ T cells were much higher than in peripheral CD4− CD14+ myeloid cells (containing monocytes and macrophages) (viral RNA, 8.9 × 10e5 versus 6.8 × 10e4; proviral DNA, 5.8 × 10e3 versus 39.57), supporting the concept that CD4+ T cells are the major cellular reservoirs in HIV/SIV infection but also that myeloid cells are an additional source of viral persistence (55). CD4+ T cells constitute the predominant reservoir in HIV infection, yet recent advances highlight the existence of HIV reservoirs in tissue-resident myeloid cells (52, 53, 56–58). However, myeloid cells in the blood and colon likely contain HIV transcripts but few proviruses in a large fraction of HIV+ patients, compared with CD4+ T cells with readily detectable proviral DNA (59, 60). Myeloid cells, including monocytes and macrophages, are infected by HIV/SIV, while circulating monocytes might not be considered reservoirs due to their infrequent HIV infection, low levels of proviral DNA, and short life span (60). Macrophages likely represent long-lived myeloid reservoirs for viral persistence, viral rebound, and reestablishment of productive HIV infection when treatment is interrupted (61–67). Interestingly, the decay of both SHIV RNA and DNA in lymph nodes was slower than in blood under cART, consistent with the notion that lymphoid tissues may have suboptimal anti-HIV drug concentrations and thus serve as sanctuary sites, resulting in viral persistence and viral resurgence after treatment is discontinued (68–71). Importantly, cART did not significantly reduce levels of proviral DNA in blood, LNs, and rectum, suggesting that proviruses exist in multiple tissue sites, which may need to be addressed in HIV cure strategies. As our and other studies report that GC Tfh cells are expanded and impaired in chronic SIV/HIV infection (72–74), LN-derived Tfh cells also increased in untreated SHIV.C.CH848-infected animals yet decreased when cART was initiated.
People living with HIV often generate autologous neutralizing antibodies against transmitted/founder viruses within weeks to months postinfection. Similarly, high levels of autologous neutralizing antibodies were observed against SHIV.C.CH848 in 6 of 9 animals postinfection (ID50 titers ≥1:300). Studies indicate that the appearance and persistence of neutralizing antibodies are mostly determined by the duration of HIV/SIV infection (75, 76), yet heterogenous neutralizing antibodies were essentially undetectable after 2 years of SHIV.C.CH848 infection. This is also consistent with previous reports and suggests that the quality of GC Tfh cells and subsequent maturation of antibody responses may be compromised (77, 78). GC Tfh cells represent a subset of CD4+ T cells that mainly reside in the GC of follicles and involve iterative interaction with GC B cells in the GC reactions for neutralizing antibody generation (79). However, GC Tfh cell loss or functional impairment, in spite of accumulation of GC Tfh cell at the chronic stage while serving as a major source of the latent and productive viral reservoirs in persistent HIV/SIV infection, might be associated with defective Ab responses (68, 74, 80, 81). Further, virus evolution in HIV-infected patients occurs concomitantly with the emergence of selective pressures such as host CD8 T cell and antibody responses in the first weeks postinfection (82, 83), as described in the report of the source patient infected with the SHIV.C.CH848 T/F isolate, who developed first autologous and then heterologous V3-targeting antibodies (15). However, conserved sites of selection pressure in V3 were not observed in any animal 5 months postinfection, which may have been too early to generate Env mutations in response to cell or neutralizing Ab responses.
In summary, here we characterize immunological and virological events in rhesus macaques infected with the novel T/F SHIV.C.CH848, in combination with cART and in response to treatment withdrawal. These findings demonstrate that this novel T/F SHIV.C.CH848 clone has promising applications in NHP models to address questions with regard to HIV reservoirs and persistence as well as relevant HIV vaccine and cure strategies.
MATERIALS AND METHODS
Ethics statement.
All animals in this study were housed at the Tulane National Primate Research Center in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International standards. All studies were reviewed and approved by the Tulane University Institutional Animal Care and Use Committee under protocol number P0305R. Animal housing and studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH, AAALAC number 000594) (84) and with the recommendations of the Weatherall report The Use of Nonhuman Primates in Research (85). All clinical procedures were carried out under the direction of a laboratory animal veterinarian. All procedures were performed under anesthesia using ketamine, and all efforts were made to minimize stress, improve housing conditions, and provide enrichment opportunities (e.g., objects to manipulate in cage, varied food supplements, foraging and task-oriented feeding methods, and interaction with caregivers and research staff).
Animals and virus.
A total of 10 adult Indian-origin rhesus macaques (Macaca mulatta; RMs) were intravenously inoculated with 1,000 50% tissue culture infective doses (TCID50) of SHIV.C.CH848 (1:10 diluted stocks containing 4.4 × 106 infectious units as determined by TZM-bl cells), in which SHIV.C.CH848.375H.dCT was constructed and generated as described previously (16). After 20 weeks, 5 animals received combined antiviral drugs (tenofovir [TFV] at 20 mg/kg of body weight/day, emtricitabine [FTC] at 30 mg/kg/day, and dolutegravir [DTG] at 2.5 mg/kg/day) for 6 months. TFV and FTC were kindly provided by Gilead, Inc., and DTG was kindly provided by ViiV Healthcare. Blood, lymph node, and rectal biopsy specimens were collected at the time scheduled, processed into single-cell suspensions, analyzed by flow cytometry, and examined by quantitative cell-associated viral DNA/RNA analysis.
Cells and plasmids.
The TZM-bl cells and the HIV-1 SG3Δenv backbone were obtained from the NIH AIDS Reagent Program (86, 87). The HIV-1 clade A, B, and C reference rev-env expression plasmids were obtained from the NIH AIDS Reagent Program (88–91). HIV-1 Env pseudoviruses were generated by cotransfecting 293T clone 17 (ATCC, Manassas, VA) with Env plasmids, including the parental HIV-1 clade C CH848 rev-env expression plasmid, along with the SG3Δenv backbone.
Tissue collection and phenotyping.
Flow cytometry for surface and intracellular staining was performed using standard protocols (92). Cells were stained with CD3 (SP34), CD4 (OKT4; BioLegend), CD8 (SK1), CD20 (2H7), and the LIVE/DEAD fixable aqua dead cell stain kit (Invitrogen, Grand Island, NY). Isotype-matched controls were included in all experiments. All antibodies and reagents were purchased from BD Biosciences Pharmingen (San Diego, CA) unless otherwise noted. Samples were resuspended in BD stabilizing fixative (BD Biosciences) and acquired on a FACS FORTESSA (Becton, Dickinson). Data were analyzed with FlowJo software (Tree Star, Ashland, OR).
Measurement of gag-specific CD8+ T cells in blood.
SHIV Gag-specific CD8+ T cells were detected as we previously described (93). In brief, PBMCs were stimulated by a pool of 15-mer Gag peptides (5 μg/ml of each peptide), medium (negative control), or phorbol-12-myristate-13-acetate (PMA; 5 ng/ml; Sigma) plus ionomycin (50 μg/ml) (positive control) for 6 h. The cultures also contained brefeldin A (Sigma) and 1 μg/ml of anti-CD49d and anti-CD28 costimulatory molecules (BD Biosciences). Cultured cells were stained with monoclonal antibodies specific for surface molecules (CD3, CD4, CD8, and LIVE/DEAD cell staining kit). After fixation and permeabilization with Cytofix/Cytoperm solution (BD Biosciences), cells were further stained with antibodies specific for gamma interferon (IFN-γ; clone 4S.B3) and TNF-α (clone MAB11) and washed with Perm/Wash buffer (BD Biosciences). Finally, labeled cells were fixed in 1.5% paraformaldehyde and acquired with a FACSA Verse cytometer (Becton Dickinson, San Jose, CA), and data were analyzed using FlowJo software (Tree Star, Ashland, OR). The background level of cytokine staining varied within different samples and different cytokine patterns but was typically <0.05% of total CD8+ T cells (median, 0.01%). Only samples in which the percentage of cytokine-staining cells was at least twice that of background were considered positive.
Purification of CD4+ T cell subsets.
Fresh PBMCs or LN mononuclear cells (LNMCs) were incubated with anti-CD4 microbeads (Miltenyi) for 30 min, washed, and resuspended in magnetically activated cell sorting (MACS) buffer to purify CD4+ T cells following the manufacturer’s instruction.
Genomic DNA and total RNA extraction.
Fresh single-cell suspensions isolated from EDTA-treated venous blood by density gradient centrifugation with lymphocyte separation medium (MP Biomedicals, Santa Ana, CA) and lymph nodes at different time points were processed to extract total genomic DNA and cellular RNA using the AllPrep DNA/RNA minikit (Qiagen) according to the manufacturer’s instructions. Viral RNA in plasma was directly isolated using the QIAamp viral RNA minikit (Qiagen). The extracted cellular DNA and RNA samples were stored at −80°C for use.
Quantification of plasma viral load and cell-associated SHIV RNA.
The extracted RNA was reverse transcribed into cDNA using the SuperScript III first-strand synthesis system (Invitrogen) according to the manufacturer’s protocol. RT reactions were performed in a thermocycler at 25°C for 5 min and 50°C for 60 min, followed by an enzyme inactivation step at 70°C for 15 min. For quantification of targets, all primer/probe sets were synthesized by Integrated DNA Technologies (IDT; Coralville, IA) to target the SIVmac239 gag region (forward primer, GTC TGC GTC ATC TGG TGC ATT C; reverse primer, CAC TAG GTG TCT CTG CAC TAT CTG TTT TG; and probe, 6-carboxyfluorescein [FAM]-CTT CCT CAG TGT GTT TCA CTT TCT CTT CTG CG-BHQ-1). Plasma viral loads were measured by real-time PCR as we previously described (68). cDNA from cell-derived RNA was used to quantify unspliced RNA transcripts by digital droplet PCR (QX100 droplet digital quantitative PCR [qPCR] system; Bio-Rad) as recently described (94). Samples were run in duplicate in a 20-μl volume containing Supermix, 250 nM primers, 900 nM probe, and 2 μl of undiluted cDNA under the following cycling conditions: 10 min at 95°C, 40 cycles of 94°C for 30 s and 63°C for 60 s, and then 98°C for 10 min. Droplets were analyzed by the QuantaSoft software in the absolute quantification mode. Copies of SIV transcripts expressed as copies per 1 million cells were measured and normalized to cellular input, as determined by copies of genomic CCR5 (single-copy rhesus macaque CCR5 DNA per cell) (95–99). The limit of detection (LOD) was based on three or more replicates and calculated using GenEx 5 (MultiD Analyses AB).
Quantification of cell-associated SHIV DNA.
To ensure that quantifications of total SHIV DNA and proviral DNA were comparable, a series of specific standards (plasmids containing SIV U5 DNA or CCR5 DNA fragment) were prepared to perform nested PCR. Since HIV preferentially integrates into regions of the chromosome close to Alu repeats, two Alu primers were used to amplify the segments of integrated proviral DNA (100). Two-step PCR amplification was run in parallel to quantify viral DNA as described previously (94). Briefly, the preamplification reactions were performed using SIV long terminal repeat primer and two outward Alu primers or using primer pairs of U5 (forward primer, AGG CTG GCA GAT TGA GCC CTG GGA GGT TC; reverse primer, CCA GGC GGC GAC TAG GAG AGA TGG GAA CAC; and probe, FAM-TTC CCT GCT AGA CTC TCA CCA GCA CTT GG-BHQ-1) on 7900HT sequence detectors (Life Technologies). The reactions were performed as follows: 25 μl of the reaction mixture, containing 1× PCR buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), 2 mM MgCl2, 0.8 μM each primer, and 0.5 U of Taq DNA polymerase (Invitrogen Life Technologies), was programmed to perform a 5-min hot start at 95°C, followed by 20 cycles of denaturation at 95°C for 30 s, annealing at 63°C for 30 s, and extension at 72°C for 3 min. Volumes of 2.5 μl of these amplicons were further amplified in triplicate with each primer/probe pair by real-time PCR using 40 cycles at 95°C for 15 s and 63°C for 1 min. The highly reproducible calibration curves were generated by plotting quantification cycle (Cq) values against log-transformed concentrations of serial standard. Internal standard curves were also generated using the known copy number of target plasmids (1 to 500 copies) diluted in cellular DNA from SIV-naive RMs. The calibration curves and the internal regression curves were used for interpolating initial copies of each target in unknown samples. A nontemplate control (NTC) and extracted cellular DNA from the HUT78/SIVmac239 cell line (positive control) were included in the qPCRs. As described above, quantification of SHIV RNA/DNA was expressed as copies per 1 million cells, in which cell numbers were determined by copies of genomic CCR5 DNA per cell.
Antibody neutralization assays.
TZM-bl neutralization assays were performed using single-round infection of TZM-bl cells with Env pseudoviruses as described previously (16, 101–103). For each of the 10 SHIV.C.CH848-infected rhesus macaques, plasma neutralization was tested at 3 time points: 5 months, 15 months, and 2 years postinfection. Briefly, TZM-bl cells were seeded at 10,000 per well. After 24 h, plasma was serially diluted 5-fold, starting at a dilution of 1:20, and incubated with 4,000 IU of virus stock as measured via titration on TZM-bl cells. The sham medium was used in place of plasma in specified control wells. The autologous infectious molecular SHIV clone, SHIV.C.CH848, and the pseudotyped HIV-1 CH848 were used to assess the neutralizing antibody titers. Pseudotyped murine leukemia virus (MLV) was used as a negative control. Antibody-virus mixtures were coincubated for 1 h and then added in triplicate to preseeded TZM-bl cells. After 48 h, cells were simultaneously lysed and mixed with luciferase substrate via the addition of Bright-Glo (Promega). Background-corrected luciferase activity for each sample was determined. Neutralization curves were fitted by the 5-parameter nonlinear regression built in Prism 8.0. The 50% inhibitory dilution (ID50) values were determined by the plasma reciprocal dilutions required to inhibit viral infection by 50%.
Viral sequencing.
Single-genome full-length gp160 env sequences were generated as described previously (16). Briefly, up to 20,000 viral RNA copies were extracted from 400 μl of plasma virus from 5 months postinfection using the Qiagen BioRobot EZ1 workstation with EZ1 virus minikit v2.0 (Qiagen). Eluted vRNA was subsequently used as a template for cDNA synthesis and reverse transcribed using the reverse primer SHIV.Env.R1 (5′-TAC CCC TAC CAA GTC ATC A-3′) and SuperScript III reverse transcriptase (Invitrogen Life Technologies). cDNA was serially diluted in a 96-well plate (Applied Biosystems) to identify the dilution at which <30% of wells contained PCR amplicons of the correct size. The SHIV gp160 env genome was amplified via nested PCR with primers as follows: first-round forward primer SHIV.Env.F1 (5′-CGA ATG GCT AAA CAG AAC A-3′), second-round forward primer SHIV.Env.F2 (CTA CCA AGG GAG CTG ATT TTC), first-round reverse primer SHIV.Env.R1 (5′-TAC CCC TAC CAA GTC ATC A-3′), and second-round reverse primer SHIV.Env.R2 (5′-TAT TTT GTT TTC TGT ATGCT-3′). PCR conditions were as follows for the first round of nested PCR: 94°C, 2 min; 37 cycles of 94°C for 20 s, 55°C for 30 s, and 68°C for 3 min 30 s, and then 68°C for 10 min. For the second round of nested PCR, the PCR conditions were as follows: 94°C for 2 min; 42 cycles of 94°C for 20 s, 54°C for 30 s, and 68°C for 3 min 30 s, and then 68°C for 10 min. Amplicons were sequenced via the MiSeq platform (Illumina). Raw reads were aligned to the SHIV.C.CH848 T/F reference using Geneious R9. Sequences that contained mixed bases at a frequency of >25% per nucleotide position were excluded from further analysis. Single-genome sequences were not able to be generated for 3 of 10 rhesus macaques, likely due to a sample storage issue.
Statistics.
Statistical analyses were performed by GraphPad Prism 7.0 software (GraphPad). Nonparametric tests were used for all statistical comparisons within animal tissues under cART. The Mann-Whitney test was used to test for differences in set point viral loads, cell-associated SHIV RNA/DNA, and specific cell subsets in animals before and after cART. Significant statistic differences (P < 0.05) are indicated in figures with asterisks. The data are presented as means and standard errors of the means (SEM).
ACKNOWLEDGMENTS
We thank Meagan Watkins and Eunice Vincent for technical support.
This work was supported by NIH grants R01 DE025432, R01 AI147372, R01 HD099857, R01 AI099795, R01 AI114380, R01 AI122953, the National Center for Research Resources, and the Office of Research Infrastructure Programs (ORIP) of the National Institutes of Health through grant OD011104-51.
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Widade Ziani performed cell-associated virus measurement by qPCR and flow cytometry; Anya Bauer analyzed viral diversity; Hui Li performed the neutralizing Ab assay; Xiaolei Wang, Xueling Wu, Katharine J. Bar, Dongfang Liu, George M. Shaw, and Ronald S. Veazey assisted with experiments, advice, and manuscript revision; and Huanbin Xu designed the study, analyzed the data, and wrote the manuscript.
We declare no competing financial interests.
REFERENCES
- 1.Del Prete GQ, Lifson JD, Keele BF. 2016. Nonhuman primate models for the evaluation of HIV-1 preventive vaccine strategies: model parameter considerations and consequences. Curr Opin HIV AIDS 11:546–554. 10.1097/COH.0000000000000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsons MS, Le Grand R, Kent SJ. 2018. Neutralizing antibody-based prevention of cell-associated HIV-1 infection. Viruses 10:333. 10.3390/v10060333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Z. 2018. Monkey models and HIV vaccine research. Adv Exp Med Biol 1075:97–124. 10.1007/978-981-13-0484-2_5. [DOI] [PubMed] [Google Scholar]
- 4.Naranjo-Gomez M, Pelegrin M. 2019. Vaccinal effect of HIV-1 antibody therapy. Curr Opin HIV AIDS 14:325–333. 10.1097/COH.0000000000000555. [DOI] [PubMed] [Google Scholar]
- 5.Horsburgh BA, Palmer S. 2019. For viral reservoir studies, timing matters. Trends Microbiol 27:809–810. 10.1016/j.tim.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Pahar B, Wang X, Dufour J, Lackner AA, Veazey RS. 2007. Virus-specific T cell responses in macaques acutely infected with SHIV(sf162p3). Virology 363:36–47. 10.1016/j.virol.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller CJ, McChesney MB, Lu X, Dailey PJ, Chutkowski C, Lu D, Brosio P, Roberts B, Lu Y. 1997. Rhesus macaques previously infected with simian/human immunodeficiency virus are protected from vaginal challenge with pathogenic SIVmac239. J Virol 71:1911–1921. 10.1128/JVI.71.3.1911-1921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimura Y, Shingai M, Willey R, Sadjadpour R, Lee WR, Brown CR, Brenchley JM, Buckler-White A, Petros R, Eckhaus M, Hoffman V, Igarashi T, Martin MA. 2010. Generation of the pathogenic R5-tropic simian/human immunodeficiency virus SHIVAD8 by serial passaging in rhesus macaques. J Virol 84:4769–4781. 10.1128/JVI.02279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shingai M, Donau OK, Schmidt SD, Gautam R, Plishka RJ, Buckler-White A, Sadjadpour R, Lee WR, LaBranche CC, Montefiori DC, Mascola JR, Nishimura Y, Martin MA. 2012. Most rhesus macaques infected with the CCR5-tropic SHIV(AD8) generate cross-reactive antibodies that neutralize multiple HIV-1 strains. Proc Natl Acad Sci U S A 109:19769–19774. 10.1073/pnas.1217443109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mavigner M, Watkins B, Lawson B, Lee ST, Chahroudi A, Kean L, Silvestri G. 2014. Persistence of virus reservoirs in ART-treated SHIV-infected rhesus macaques after autologous hematopoietic stem cell transplant. PLoS Pathog 10:e1004406. 10.1371/journal.ppat.1004406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Tellez T, Huot N, Ploquin MJ, Rascle P, Jacquelin B, Muller-Trutwin M. 2016. Non-human primates in HIV research: achievements, limits and alternatives. Infect Genet Evol 46:324–332. 10.1016/j.meegid.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Bbosa N, Kaleebu P, Ssemwanga D. 2019. HIV subtype diversity worldwide. Curr Opin HIV AIDS 14:153–160. 10.1097/COH.0000000000000534. [DOI] [PubMed] [Google Scholar]
- 13.Gartner MJ, Roche M, Churchill MJ, Gorry PR, Flynn JK. 2020. Understanding the mechanisms driving the spread of subtype C HIV-1. EBioMedicine 53:102682. 10.1016/j.ebiom.2020.102682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tartaglia LJ, Gupte S, Pastores KC, Trott S, Abbink P, Mercado NB, Li Z, Liu PT, Borducchi EN, Chandrashekar A, Bondzie EA, Hamza V, Kordana N, Mahrokhian S, Lavine CL, Seaman MS, Li H, Shaw GM, Barouch DH. 2020. Differential outcomes following optimization of simian-human immunodeficiency viruses from clades AE, B, and C. J Virol 94:e01860-19. 10.1128/JVI.01860-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonsignori M, Kreider EF, Fera D, Meyerhoff RR, Bradley T, Wiehe K, Alam SM, Aussedat B, Walkowicz WE, Hwang KK, Saunders KO, Zhang R, Gladden MA, Monroe A, Kumar A, Xia SM, Cooper M, Louder MK, McKee K, Bailer RT, Pier BW, Jette CA, Kelsoe G, Williams WB, Morris L, Kappes J, Wagh K, Kamanga G, Cohen MS, Hraber PT, Montefiori DC, Trama A, Liao HX, Kepler TB, Moody MA, Gao F, Danishefsky SJ, Mascola JR, Shaw GM, Hahn BH, Harrison SC, Korber BT, Haynes BF. 2017. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci Transl Med 9:eaai7514. 10.1126/scitranslmed.aai7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Wang S, Kong R, Ding W, Lee FH, Parker Z, Kim E, Learn GH, Hahn P, Policicchio B, Brocca-Cofano E, Deleage C, Hao X, Chuang GY, Gorman J, Gardner M, Lewis MG, Hatziioannou T, Santra S, Apetrei C, Pandrea I, Alam SM, Liao HX, Shen X, Tomaras GD, Farzan M, Chertova E, Keele BF, Estes JD, Lifson JD, Doms RW, Montefiori DC, Haynes BF, Sodroski JG, Kwong PD, Hahn BH, Shaw GM. 2016. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc Natl Acad Sci U S A 113:E3413–E3422. 10.1073/pnas.1606636113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bauer AM, Ziani W, Lindemuth E, Kuri-Cervantes L, Li H, Lee FH, Watkins M, Ding W, Xu H, Veazey R, Bar KJ. 2020. Novel transmitted/founder simian-human immunodeficiency viruses for human immunodeficiency virus latency and cure research. J Virol 94:e01659-19. 10.1128/JVI.01659-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Wang X, Pahar B, Moroney-Rasmussen T, Alvarez X, Lackner AA, Veazey RS. 2010. Increased B7-H1 expression on dendritic cells correlates with programmed death 1 expression on T cells in simian immunodeficiency virus-infected macaques and may contribute to T cell dysfunction and disease progression. J Immunol 185:7340–7348. 10.4049/jimmunol.1001642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLane LM, Abdel-Hakeem MS, Wherry EJ. 2019. CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol 37:457–495. 10.1146/annurev-immunol-041015-055318. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds T. 1989. Noninvasive hemodynamic assessment of intracardiac pressures and assessment of ventricular function with cardiac Doppler. Crit Care Nurs Clin North Am 1:629–634. 10.1016/S0899-5885(18)30887-6. [DOI] [PubMed] [Google Scholar]
- 21.Gao N, Wang W, Wang C, Gu T, Guo R, Yu B, Kong W, Qin C, Giorgi EE, Chen Z, Townsley S, Hu SL, Yu X, Gao F. 2018. Development of broad neutralization activity in simian/human immunodeficiency virus-infected rhesus macaques after long-term infection. AIDS 32:555–563. 10.1097/QAD.0000000000001724. [DOI] [PubMed] [Google Scholar]
- 22.Nelson AN, Goswami R, Dennis M, Tu J, Mangan RJ, Saha PT, Cain DW, Curtis AD, Shen X, Shaw GM, Bar K, Hudgens M, Pollara J, De Paris K, Van Rompay KKA, Permar SR. 2019. Simian-human immunodeficiency virus SHIV.CH505-infected infant and adult rhesus macaques exhibit similar Env-specific antibody kinetics, despite distinct T-follicular helper and germinal center B cell landscapes. J Virol 93:e00168-19. 10.1128/JVI.00168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, Smith JY, Brinkman AL, Peter LE, Mathew SI, Smith KM, Borducchi EN, Rosenbloom DI, Lewis MG, Hattersley J, Li B, Hesselgesser J, Geleziunas R, Robb ML, Kim JH, Michael NL, Barouch DH. 2014. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 512:74–77. 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, Khoruts A, Estes JD, Anderson J, Callisto SP, Schmidt TE, Thorkelson A, Reilly C, Perkey K, Reimann TG, Utay NS, Nganou Makamdop K, Stevenson M, Douek DC, Haase AT, Schacker TW. 2015. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A 112:E1126–E1134. 10.1073/pnas.1414926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon KJ, Siliciano RF. 2017. HIV persistence: clonal expansion of cells in the latent reservoir. J Clin Invest 127:2536–2538. 10.1172/JCI95329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bachmann N, von Siebenthal C, Vongrad V, Turk T, Neumann K, Beerenwinkel N, Bogojeska J, Fellay J, Roth V, Kok YL, Thorball CW, Borghesi A, Parbhoo S, Wieser M, Boni J, Perreau M, Klimkait T, Yerly S, Battegay M, Rauch A, Hoffmann M, Bernasconi E, Cavassini M, Kouyos RD, Gunthard HF, Metzner KJ, Swiss HIV Cohort Study. 2019. Determinants of HIV-1 reservoir size and long-term dynamics during suppressive ART. Nat Commun 10:3193. 10.1038/s41467-019-10884-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jayaraman P, Mohan D, Polacino P, Kuller L, Sheikh N, Bielefeldt-Ohmann H, Richardson B, Anderson D, Hu SL, Haigwood NL. 2004. Perinatal transmission of SHIV-SF162P3 in Macaca nemestrina. J Med Primatol 33:243–250. 10.1111/j.1600-0684.2004.00079.x. [DOI] [PubMed] [Google Scholar]
- 29.Bar KJ, Coronado E, Hensley-McBain T, O’Connor MA, Osborn JM, Miller C, Gott TM, Wangari S, Iwayama N, Ahrens CY, Smedley J, Moats C, Lynch RM, Haddad EK, Haigwood NL, Fuller DH, Shaw GM, Klatt NR, Manuzak JA. 2019. Simian-human immunodeficiency virus SHIV.CH505 infection of rhesus macaques results in persistent viral replication and induces intestinal immunopathology. J Virol 93:e00372-19. 10.1128/JVI.00372-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colby DJ, Trautmann L, Pinyakorn S, Leyre L, Pagliuzza A, Kroon E, Rolland M, Takata H, Buranapraditkun S, Intasan J, Chomchey N, Muir R, Haddad EK, Tovanabutra S, Ubolyam S, Bolton DL, Fullmer BA, Gorelick RJ, Fox L, Crowell TA, Trichavaroj R, O’Connell R, Chomont N, Kim JH, Michael NL, Robb ML, Phanuphak N, Ananworanich J, RV411 study group. 2018. Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat Med 24:923–926. 10.1038/s41591-018-0026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferris AL, Wells DW, Guo S, Del Prete GQ, Swanstrom AE, Coffin JM, Wu X, Lifson JD, Hughes SH. 2019. Clonal expansion of SIV-infected cells in macaques on antiretroviral therapy is similar to that of HIV-infected cells in humans. PLoS Pathog 15:e1007869. 10.1371/journal.ppat.1007869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gautam R, Nishimura Y, Lee WR, Donau O, Buckler-White A, Shingai M, Sadjadpour R, Schmidt SD, LaBranche CC, Keele BF, Montefiori D, Mascola JR, Martin MA. 2012. Pathogenicity and mucosal transmissibility of the R5-tropic simian/human immunodeficiency virus SHIV(AD8) in rhesus macaques: implications for use in vaccine studies. J Virol 86:8516–8526. 10.1128/JVI.00644-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Navio JM, Fuchs SP, Pantry SN, Lauer WA, Duggan NN, Keele BF, Rakasz EG, Gao G, Lifson JD, Desrosiers RC. 2019. Adeno-associated virus delivery of anti-HIV monoclonal antibodies can drive long-term virologic suppression. Immunity 50:567–575.e5. 10.1016/j.immuni.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandergeeten C, Fromentin R, DaFonseca S, Lawani MB, Sereti I, Lederman MM, Ramgopal M, Routy JP, Sekaly RP, Chomont N. 2013. Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood 121:4321–4329. 10.1182/blood-2012-11-465625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinkevych M, Fennessey CM, Cromer D, Reid C, Trubey CM, Lifson JD, Keele BF, Davenport MP. 2019. Predictors of SIV recrudescence following antiretroviral treatment interruption. Elife 8:e49022. 10.7554/eLife.49022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bender AM, Simonetti FR, Kumar MR, Fray EJ, Bruner KM, Timmons AE, Tai KY, Jenike KM, Antar AAR, Liu PT, Ho YC, Raugi DN, Seydi M, Gottlieb GS, Okoye AA, Del Prete GQ, Picker LJ, Mankowski JL, Lifson JD, Siliciano JD, Laird GM, Barouch DH, Clements JE, Siliciano RF. 2019. The landscape of persistent viral genomes in ART-treated SIV, SHIV, and HIV-2 infections. Cell Host Microbe 26:73–85.e4. 10.1016/j.chom.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moir S, Fauci AS. 2017. B-cell responses to HIV infection. Immunol Rev 275:33–48. 10.1111/imr.12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasternak AO, DeMaster LK, Kootstra NA, Reiss P, O’Doherty U, Berkhout B. 2016. Minor contribution of chimeric host-HIV readthrough transcripts to the level of HIV cell-associated gag RNA. J Virol 90:1148–1151. 10.1128/JVI.02597-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baxter AE, Niessl J, Fromentin R, Richard J, Porichis F, Charlebois R, Massanella M, Brassard N, Alsahafi N, Delgado GG, Routy JP, Walker BD, Finzi A, Chomont N, Kaufmann DE. 2016. Single-cell characterization of viral translation-competent reservoirs in HIV-infected individuals. Cell Host Microbe 20:368–380. 10.1016/j.chom.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yerly S, Gunthard HF, Fagard C, Joos B, Perneger TV, Hirschel B, Perrin L, Swiss HIV Cohort Study. 2004. Proviral HIV-DNA predicts viral rebound and viral setpoint after structured treatment interruptions. AIDS 18:1951–1953. 10.1097/00002030-200409240-00011. [DOI] [PubMed] [Google Scholar]
- 41.Williams JP, Hurst J, Stohr W, Robinson N, Brown H, Fisher M, Kinloch S, Cooper D, Schechter M, Tambussi G, Fidler S, Carrington M, Babiker A, Weber J, Koelsch KK, Kelleher AD, Phillips RE, Frater J, SPARTACTrial Investigators. 2014. HIV-1 DNA predicts disease progression and post-treatment virological control. Elife 3:e03821. 10.7554/eLife.03821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiselinova M, De Spiegelaere W, Buzon MJ, Malatinkova E, Lichterfeld M, Vandekerckhove L. 2016. Integrated and total HIV-1 DNA predict ex vivo viral outgrowth. PLoS Pathog 12:e1005472. 10.1371/journal.ppat.1005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Avettand-Fenoel V, Hocqueloux L, Ghosn J, Cheret A, Frange P, Melard A, Viard JP, Rouzioux C. 2016. Total HIV-1 DNA, a marker of viral reservoir dynamics with clinical implications. Clin Microbiol Rev 29:859–880. 10.1128/CMR.00015-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rouzioux C, Avettand-Fenoel V. 2018. Total HIV DNA: a global marker of HIV persistence. Retrovirology 15:30. 10.1186/s12977-018-0412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cillo AR, Hong F, Tsai A, Irrinki A, Kaur J, Sloan DD, Follen M, Geleziunas R, Cihlar T, Win SS, Murry JP, Mellors JW. 2018. Blood biomarkers of expressed and inducible HIV-1. AIDS 32:699–708. 10.1097/QAD.0000000000001748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee GQ, Lichterfeld M. 2016. Diversity of HIV-1 reservoirs in CD4+ T-cell subpopulations. Curr Opin HIV AIDS 11:383–387. 10.1097/COH.0000000000000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. 2016. HIV reservoirs: what, where and how to target them. Nat Rev Microbiol 14:55–60. 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- 48.Agosto LM, Herring MB, Mothes W, Henderson AJ. 2018. HIV-1-infected CD4+ T cells facilitate latent infection of resting CD4+ T cells through cell-cell contact. Cell Rep 24:2088–2100. 10.1016/j.celrep.2018.07.079. [DOI] [PubMed] [Google Scholar]
- 49.Baxter AE, Russell RA, Duncan CJ, Moore MD, Willberg CB, Pablos JL, Finzi A, Kaufmann DE, Ochsenbauer C, Kappes JC, Groot F, Sattentau QJ. 2014. Macrophage infection via selective capture of HIV-1-infected CD4+ T cells. Cell Host Microbe 16:711–721. 10.1016/j.chom.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kandathil AJ, Sugawara S, Balagopal A. 2016. Are T cells the only HIV-1 reservoir? Retrovirology 13:86. 10.1186/s12977-016-0323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodrigues V, Ruffin N, San-Roman M, Benaroch P. 2017. Myeloid cell interaction with HIV: a complex relationship. Front Immunol 8:1698. 10.3389/fimmu.2017.01698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abreu CM, Veenhuis RT, Avalos CR, Graham S, Parrilla DR, Ferreira EA, Queen SE, Shirk EN, Bullock BT, Li M, Metcalf Pate KA, Beck SE, Mangus LM, Mankowski JL, Mac Gabhann F, O’Connor SL, Gama L, Clements JE. 2019. Myeloid and CD4 T cells comprise the latent reservoir in antiretroviral therapy-suppressed SIVmac251-infected macaques. mBio 10:e01659-19. 10.1128/mBio.01659-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong ME, Jaworowski A, Hearps AC. 2019. The HIV reservoir in monocytes and macrophages. Front Immunol 10:1435. 10.3389/fimmu.2019.01435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kallianpur KJ, Valcour VG, Lerdlum S, Busovaca E, Agsalda M, Sithinamsuwan P, Chalermchai T, Fletcher JL, Tipsuk S, Shikuma CM, Shiramizu BT, Ananworanich J, SEARCH 011 study group. 2014. HIV DNA in CD14+ reservoirs is associated with regional brain atrophy in patients naive to combination antiretroviral therapy. AIDS 28:1619–1624. 10.1097/QAD.0000000000000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitchell BI, Laws EI, Ndhlovu LC. 2019. Impact of myeloid reservoirs in HIV cure trials. Curr HIV/AIDS Rep 16:129–140. 10.1007/s11904-019-00438-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ganor Y, Real F, Sennepin A, Dutertre CA, Prevedel L, Xu L, Tudor D, Charmeteau B, Couedel-Courteille A, Marion S, Zenak AR, Jourdain JP, Zhou Z, Schmitt A, Capron C, Eugenin EA, Cheynier R, Revol M, Cristofari S, Hosmalin A, Bomsel M. 2019. HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat Microbiol 4:633–644. 10.1038/s41564-018-0335-z. [DOI] [PubMed] [Google Scholar]
- 57.DiNapoli SR, Ortiz AM, Wu F, Matsuda K, Twigg HL, III, Hirsch VM, Knox K, Brenchley JM. 2017. Tissue-resident macrophages can contain replication-competent virus in antiretroviral-naive, SIV-infected Asian macaques. JCI Insight 2:e91214. 10.1172/jci.insight.91214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Avalos CR, Price SL, Forsyth ER, Pin JN, Shirk EN, Bullock BT, Queen SE, Li M, Gellerup D, O’Connor SL, Zink MC, Mankowski JL, Gama L, Clements JE. 2016. Quantitation of productively infected monocytes and macrophages of simian immunodeficiency virus-infected macaques. J Virol 90:5643–5656. 10.1128/JVI.00290-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cattin A, Wiche Salinas TR, Gosselin A, Planas D, Shacklett B, Cohen EA, Ghali MP, Routy JP, Ancuta P. 2019. HIV-1 is rarely detected in blood and colon myeloid cells during viral-suppressive antiretroviral therapy. AIDS 33:1293–1306. 10.1097/QAD.0000000000002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Massanella M, Bakeman W, Sithinamsuwan P, Fletcher JLK, Chomchey N, Tipsuk S, Chalermchai T, Routy JP, Ananworanich J, Valcour VG, Chomont N. 2019. Infrequent HIV infection of circulating monocytes during antiretroviral therapy. J Virol 94:e01174-19. 10.1128/JVI.01174-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abreu CM, Veenhuis RT, Avalos CR, Graham S, Queen SE, Shirk EN, Bullock BT, Li M, Metcalf Pate KA, Beck SE, Mangus LM, Mankowski JL, Clements JE, Gama L. 2019. Infectious virus persists in CD4(+) T cells and macrophages in antiretroviral therapy-suppressed simian immunodeficiency virus-infected macaques. J Virol 93:e00065-19. 10.1128/JVI.00065-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jambo KC, Banda DH, Kankwatira AM, Sukumar N, Allain TJ, Heyderman RS, Russell DG, Mwandumba HC. 2014. Small alveolar macrophages are infected preferentially by HIV and exhibit impaired phagocytic function. Mucosal Immunol 7:1116–1126. 10.1038/mi.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Damouche A, Lazure T, Avettand-Fenoel V, Huot N, Dejucq-Rainsford N, Satie AP, Melard A, David L, Gommet C, Ghosn J, Noel N, Pourcher G, Martinez V, Benoist S, Bereziat V, Cosma A, Favier B, Vaslin B, Rouzioux C, Capeau J, Muller-Trutwin M, Dereuddre-Bosquet N, Le Grand R, Lambotte O, Bourgeois C. 2015. Adipose tissue is a neglected viral reservoir and an inflammatory site during chronic HIV and SIV infection. PLoS Pathog 11:e1005153. 10.1371/journal.ppat.1005153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zalar A, Figueroa MI, Ruibal-Ares B, Bare P, Cahn P, de Bracco MM, Belmonte L. 2010. Macrophage HIV-1 infection in duodenal tissue of patients on long term HAART. Antiviral Res 87:269–271. 10.1016/j.antiviral.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 65.Bernard-Stoecklin S, Gommet C, Corneau AB, Guenounou S, Torres C, Dejucq-Rainsford N, Cosma A, Dereuddre-Bosquet N, Le Grand R. 2013. Semen CD4+ T cells and macrophages are productively infected at all stages of SIV infection in macaques. PLoS Pathog 9:e1003810. 10.1371/journal.ppat.1003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cenker JJ, Stultz RD, McDonald D. 2017. Brain microglial cells are highly susceptible to HIV-1 infection and spread. AIDS Res Hum Retroviruses 33:1155–1165. 10.1089/AID.2017.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andrade VM, Mavian C, Babic D, Cordeiro T, Sharkey M, Barrios L, Brander C, Martinez-Picado J, Dalmau J, Llano A, Li JZ, Jacobson J, Lavine CL, Seaman MS, Salemi M, Stevenson M. 2020. A minor population of macrophage-tropic HIV-1 variants is identified in recrudescing viremia following analytic treatment interruption. Proc Natl Acad Sci U S A 117:9981–9990. 10.1073/pnas.1917034117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu H, Wang X, Malam N, Aye PP, Alvarez X, Lackner AA, Veazey RS. 2016. Persistent simian immunodeficiency virus infection drives differentiation, aberrant accumulation, and latent infection of germinal center follicular T helper cells. J Virol 90:1578–1587. 10.1128/JVI.02471-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, Beilman GJ, Khoruts A, Thorkelson A, Schmidt TE, Anderson J, Perkey K, Stevenson M, Perelson AS, Douek DC, Haase AT, Schacker TW. 2014. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A 111:2307–2312. 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, Morcock D, Swanson T, Legasse AW, Axthelm MK, Hesselgesser J, Geleziunas R, Hirsch VM, Edlefsen PT, Piatak M, Jr, Estes JD, Lifson JD, Picker LJ. 2015. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med 21:132–139. 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong JK, Yukl SA. 2016. Tissue reservoirs of HIV. Curr Opin HIV AIDS 11:362–370. 10.1097/COH.0000000000000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cubas R, Perreau M. 2014. The dysfunction of T follicular helper cells. Curr Opin HIV AIDS 10.1097/COH.0000000000000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fahey LM, Wilson EB, Elsaesser H, Fistonich CD, McGavern DB, Brooks DG. 2011. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J Exp Med 208:987–999. 10.1084/jem.20101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu H, Wang X, Malam N, Lackner AA, Veazey RS. 2015. Persistent simian immunodeficiency virus infection causes ultimate depletion of follicular Th cells in AIDS. J Immunol 195:4351–4357. 10.4049/jimmunol.1501273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bonsignori M, Liao HX, Gao F, Williams WB, Alam SM, Montefiori DC, Haynes BF. 2017. Antibody-virus co-evolution in HIV infection: paths for HIV vaccine development. Immunol Rev 275:145–160. 10.1111/imr.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu F, Ourmanov I, Kirmaier A, Leviyang S, LaBranche C, Huang J, Whitted S, Matsuda K, Montefiori D, Hirsch VM. 2020. SIV infection duration largely determines broadening of neutralizing antibody response in macaques. J Clin Invest 130:5413–5424. 10.1172/JCI139123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cubas RA, Mudd JC, Savoye AL, Perreau M, van Grevenynghe J, Metcalf T, Connick E, Meditz A, Freeman GJ, Abesada-Terk G, Jr, Jacobson JM, Brooks AD, Crotty S, Estes JD, Pantaleo G, Lederman MM, Haddad EK. 2013. Inadequate T follicular cell help impairs B cell immunity during HIV infection. Nat Med 19:494–499. 10.1038/nm.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamamoto T, Lynch RM, Gautam R, Matus-Nicodemos R, Schmidt SD, Boswell KL, Darko S, Wong P, Sheng Z, Petrovas C, McDermott AB, Seder RA, Keele BF, Shapiro L, Douek DC, Nishimura Y, Mascola JR, Martin MA, Koup RA. 2015. Quality and quantity of TFH cells are critical for broad antibody development in SHIVAD8 infection. Sci Transl Med 7:298ra120. 10.1126/scitranslmed.aab3964. [DOI] [PubMed] [Google Scholar]
- 79.Xu H, Wang X, Lackner AA, Veazey RS. 2014. PD-1(HIGH) follicular CD4 T helper cell subsets residing in lymph node germinal centers correlate with B cell maturation and IgG production in rhesus macaques. Front Immunol 5:85. 10.3389/fimmu.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. 2013. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 210:143–156. 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X, Xu H. 2018. Potential epigenetic regulation in the germinal center reaction of lymphoid tissues in HIV/SIV infection. Front Immunol 9:159. 10.3389/fimmu.2018.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wargo AR, Kurath G. 2012. Viral fitness: definitions, measurement, and current insights. Curr Opin Virol 2:538–545. 10.1016/j.coviro.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol Mol Biol Rev 76:159–216. 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 85.Weatherall DJ. 2006. The use of non-human primates in research. Academy of Medical Sciences, London, United Kingdom. [Google Scholar]
- 86.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. 10.1128/aac.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 88.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79:10108–10125. 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol 80:11776–11790. 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Long EM, Rainwater SM, Lavreys L, Mandaliya K, Overbaugh J. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res Hum Retroviruses 18:567–576. 10.1089/088922202753747914. [DOI] [PubMed] [Google Scholar]
- 91.Blish CA, Jalalian-Lechak Z, Rainwater S, Nguyen MA, Dogan OC, Overbaugh J. 2009. Cross-subtype neutralization sensitivity despite monoclonal antibody resistance among early subtype A, C, and D envelope variants of human immunodeficiency virus type 1. J Virol 83:7783–7788. 10.1128/JVI.00673-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu H, Wang X, Liu DX, Moroney-Rasmussen T, Lackner AA, Veazey RS. 2012. IL-17-producing innate lymphoid cells are restricted to mucosal tissues and are depleted in SIV-infected macaques. Mucosal Immunol 5:658–669. 10.1038/mi.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu H, Wang X, Lackner AA, Veazey RS. 2013. CD8 down-regulation and functional impairment of SIV-specific cytotoxic T lymphocytes in lymphoid and mucosal tissues during SIV infection. J Leukoc Biol 93:943–950. 10.1189/jlb.1112580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ziani W, Shao J, Wang X, Russell-Lodrigue K, Liu YZ, Montaner LJ, Veazey RS, Xu H. 6 January 2021. Increased proviral DNA in circulating cells correlates with plasma viral rebound in SIV-infected rhesus macaques after antiretroviral therapy interruption. J Virol 10.1128/JVI.02064-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M, Jr, Lifson JD, Picker LJ. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473:523–527. 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Okoye AA, Hansen SG, Vaidya M, Fukazawa Y, Park H, Duell DM, Lum R, Hughes CM, Ventura AB, Ainslie E, Ford JC, Morrow D, Gilbride RM, Legasse AW, Hesselgesser J, Geleziunas R, Li Y, Oswald K, Shoemaker R, Fast R, Bosche WJ, Borate BR, Edlefsen PT, Axthelm MK, Picker LJ, Lifson JD. 2018. Early antiretroviral therapy limits SIV reservoir establishment to delay or prevent post-treatment viral rebound. Nat Med 24:1430–1440. 10.1038/s41591-018-0130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hong F, Aga E, Cillo AR, Yates AL, Besson G, Fyne E, Koontz DL, Jennings C, Zheng L, Mellors JW. 2016. Novel assays for measurement of total cell-associated HIV-1 DNA and RNA. J Clin Microbiol 54:902–911. 10.1128/JCM.02904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kostrikis LG, Touloumi G, Karanicolas R, Pantazis N, Anastassopoulou C, Karafoulidou A, Goedert JJ, Hatzakis A, Multicenter Hemophilia Cohort Study Group. 2002. Quantitation of human immunodeficiency virus type 1 DNA forms with the second template switch in peripheral blood cells predicts disease progression independently of plasma RNA load. J Virol 76:10099–10108. 10.1128/JVI.76.20.10099-10108.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Deleage C, Immonen TT, Fennessey CM, Reynaldi A, Reid C, Newman L, Lipkey L, Schlub TE, Camus C, O’Brien S, Smedley J, Conway JM, Del Prete GQ, Davenport MP, Lifson JD, Estes JD, Keele BF. 2019. Defining early SIV replication and dissemination dynamics following vaginal transmission. Sci Adv 5:eaav7116. 10.1126/sciadv.aav7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cohn LB, Silva IT, Oliveira TY, Rosales RA, Parrish EH, Learn GH, Hahn BH, Czartoski JL, McElrath MJ, Lehmann C, Klein F, Caskey M, Walker BD, Siliciano JD, Siliciano RF, Jankovic M, Nussenzweig MC. 2015. HIV-1 integration landscape during latent and active infection. Cell 160:420–432. 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84:1439–1452. 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu X, Wang C, O’Dell S, Li Y, Keele BF, Yang Z, Imamichi H, Doria-Rose N, Hoxie JA, Connors M, Shaw GM, Wyatt RT, Mascola JR. 2012. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. J Virol 86:5844–5856. 10.1128/JVI.07139-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jia M, Lu H, Markowitz M, Cheng-Mayer C, Wu X. 2016. Development of broadly neutralizing antibodies and their mapping by monomeric gp120 in human immunodeficiency virus type 1-infected humans and simian-human immunodeficiency virus SHIVSF162P3N-infected macaques. J Virol 90:4017–4031. 10.1128/JVI.02898-15. [DOI] [PMC free article] [PubMed] [Google Scholar]