The nonsegmented negative-sense (NNS) RNA viruses share many common replication and gene expression strategies. There are no vaccines or antiviral drugs for many of these viruses.
KEYWORDS: N6-methyladenosine, innate immunity, negative-strand RNA virus
ABSTRACT
N6-Methyladenosine (m6A) is the most abundant internal RNA modification catalyzed by host RNA methyltransferases. As obligate intracellular parasites, many viruses acquire m6A methylation in their RNAs. However, the biological functions of viral m6A methylation are poorly understood. Here, we found that viral m6A methylation serves as a molecular marker for host innate immunity to discriminate self from nonself RNA and that this novel biological function of viral m6A methylation is universally conserved in several families in nonsegmented negative-sense (NNS) RNA viruses. Using m6A methyltransferase (METTL3) knockout cells, we produced m6A-deficient virion RNAs from the representative members of the families Pneumoviridae, Paramyxoviridae, and Rhabdoviridae and found that these m6A-deficient viral RNAs triggered significantly higher levels of type I interferon compared to the m6A-sufficient viral RNAs, in a RIG-I-dependent manner. Reconstitution of the RIG-I pathway revealed that m6A-deficient virion RNA induced higher expression of RIG-I, bound to RIG-I more efficiently, enhanced RIG-I ubiquitination, and facilitated RIG-I conformational rearrangement and oligomerization. Furthermore, the m6A binding protein YTHDF2 is essential for suppression of the type I interferon signaling pathway, including by virion RNA. Collectively, our results suggest that several families in NNS RNA viruses acquire m6A in viral RNA as a common strategy to evade host innate immunity.
IMPORTANCE The nonsegmented negative-sense (NNS) RNA viruses share many common replication and gene expression strategies. There are no vaccines or antiviral drugs for many of these viruses. We found that representative members of the families Pneumoviridae, Paramyxoviridae, and Rhabdoviridae among the NNS RNA viruses acquire m6A methylation in their genome and antigenome as a means to escape recognition by host innate immunity via a RIG-I-dependent signaling pathway. Viral RNA lacking m6A methylation induces a significantly higher type I interferon response than m6A-sufficient viral RNA. In addition to uncovering m6A methylation as a common mechanism for many NNS RNA viruses to evade host innate immunity, this study discovered a novel strategy to enhance type I interferon responses, which may have important applications in vaccine development, as robust innate immunity will likely promote the subsequent adaptive immunity.
INTRODUCTION
The nonsegmented negative-strand (NNS) RNA viruses of the order Mononegavirales include a wide range of human, animal, and plant pathogens. The NNS RNA viruses are classified into five families: Pneumoviridae, exemplified by human respiratory syncytial virus (RSV) and human metapneumovirus (hMPV); Paramyxoviridae, exemplified by human parainfluenza virus type 3, measles virus (MeV), mumps virus, Sendai virus (SeV), Nipah virus, and Hendra virus; Rhabdoviridae, exemplified by vesicular stomatitis virus (VSV) and rabies virus; Filoviridae, exemplified by Ebola and Marburg viruses; and Bornaviridae, exemplified by Borna disease virus. For many of these viruses, there are no effective vaccines or antiviral therapeutic agents. These viruses share many common strategies in viral replication, gene expression, and innate immune evasion (1, 2). The active template for replication and transcription of NNS RNA viruses is the nucleocapsid (N) protein-encapsidated genome complex (N-RNA). During replication, the viral RNA-dependent RNA polymerase recognizes the N-RNA template and copies the negative-sense genomic RNA to produce an exact, positive-sense full-length cRNA antigenome, which is encapsidated by N protein and serves as the template for synthesis of progeny genome RNA (1). During transcription of mRNAs from the genomes, the mRNAs are capped, the cap is guanine N-7 (G-N-7) and ribose 2′-O methylated, and the 3′ end is polyadenylated (3, 4).
The host immune system has developed mechanisms to detect incoming microbes, triggering signaling pathways that result in the production of type I/III interferon (IFN), which provides an antiviral state that restricts viral infection. Both the genome and antigenome of NNS RNA viruses bear 5′-terminal triphosphate that is recognized by RIG-I (5, 6). Replication of some NNS RNA viruses also yields long double-stranded RNA (dsRNA), which can be detected by MDA5 (7, 8). In turn, NNS RNA viruses encode multifunctional proteins (such as NS1 and NS2 of RSV, G protein of hMPV, and V/C protein of paramyxoviruses) to impair IFN signaling and/or to counteract cellular antiviral factors (2, 9, 10). In addition to using virus-encoded proteins to interfere with the IFN pathway, many viruses have developed other extraordinarily diverse strategies, including modifying their nucleotides, to escape detection by the sensor molecules of the innate immune system. In coronaviruses, flaviviruses, and poxviruses, ribose 2′-O methylation of the viral 5′ mRNA cap prevents the recognition of viral mRNA by host innate immunity via MDA5 and IFN-induced proteins with a tetratricopeptide repeat (IFIT)-dependent mechanism (11, 12). Short synthetic RNA-containing modified nucleosides (m5C, m6A, m5U, s2U, or pseudouridine) ablate Toll-like receptor 3 (TLR3), TLR7, and TLR8 signaling pathways in dendritic cells (13). Similarly, short synthetic poly(U/UC) RNAs derived from hepatitis C virus containing common RNA modifications, such as m6A, Ψ, N-1-methylpseudouridine (m1Ψ), m5C, 5-hydroxymethylcytidine (hm5C), 5-methoxycytidine (5moC), and 2′ fluoro-deoxyribose (2′ fluoro-deoxyuridine [2FdU] and 2′ fluoro-deoxycytidine [2FdC]), suppress RIG-I responses (14). The presence of m6A RNA modification on human circular RNAs inhibits innate immunity (15). Nucleotide modifications (such as Ψ and m5C) decrease the innate immune response induced by synthetic analogs of small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) (16). Although these studies showed that RNA modifications on short synthetic RNA or circular RNA can inhibit innate immunity, whether this indeed occurs in the context of virus infection is not clear.
Among over 160 RNA modifications, m6A is the most prevalent and abundant posttranscriptional RNA modification on eukaryotic mRNA (17, 18). RNA m6A methylation is installed by host m6A methyltransferase (m6A writer), a catalytic subunit (METTL3), and a key accessary subunit (METTL14) (19) and can be reversibly removed by host demethylases (m6A erasers), such as FTO and ALKBH5 (20, 21). The m6A functions are mediated by m6A binding proteins (m6A readers) YTHDF1, YTHDF2, and YTHDF3 (22). Recent studies have found that m6A methylation affects many fundamental functions, such as pre-mRNA processing, nuclear transport, RNA stability, translation, and microRNA biogenesis, and is associated with numerous human diseases (23–25). It was also suggested that m6A machinery alters host m6A-modified antiviral transcripts and modulates innate immunity (26, 27). Interestingly, depletion of METTL3 in cells directly decreases the m6A methylation of IFNB mRNA, leading to elevated levels of type I IFN (28, 29).
Viruses are obligate intracellular parasites; they must utilize host machinery to synthesize their own genomes and proteins. The presence of m6A in their genome RNA and/or mRNA has been reported for many RNA viruses and DNA viruses (30, 31). Unexpectedly, viral m6A RNA can play either antiviral or proviral roles in the virus life cycle via poorly understood mechanisms (30, 31). Similar to host mRNA, m6A in viral mRNA (such as some positive-sense RNA viruses and DNA viruses) has been shown to be important for RNA stability and translation (31, 32). The genome and antigenome (replicative intermediate) of NNS RNA viruses are triphosphorylated and cannot be directly translated into viral proteins. However, why the genomes and antigenomes of NNS RNA viruses are m6A methylated is mysterious.
Recently, we provided the first evidence that m6A methylation in the hMPV genome and antigenome prevents innate immune detection in a RIG-I-dependent manner (33). Here, we demonstrate that this novel function of viral m6A in innate immunity is universally conserved in the representative members of the Pneumoviridae (hMPV), Paramyxoviridae (MeV and SeV), and Rhabdoviridae (VSV) in the order Mononegavirales. Specifically, m6A-deficient virion RNA purified from viruses grown in METTL3 knockout (KO) U2OS cells induced significantly larger amounts of type I IFN than m6A-sufficient virion RNA grown in wild-type (WT) U2OS cells in a RIG-I-dependent manner. Reconstitution of the RIG-I signaling pathway revealed that m6A-deficient virion RNA enhances RIG-I expression, binding affinity to RIG-I, RIG-I ubiquitination, and RIG-I conformational changes. Furthermore, our results suggest that YTHDF2, the m6A binding protein, sequesters m6A-sufficient viral RNA, which is essential for suppression of type I IFN signaling. Thus, this novel function of m6A methylation of viral RNA in innate immunity appears universally conserved in the Pneumoviridae, Paramyxoviridae, and Rhabdoviridae among NNS RNA virus families.
RESULTS
RNA purified from virions grown in METTL3 knockout cells is defective in m6A methylation.
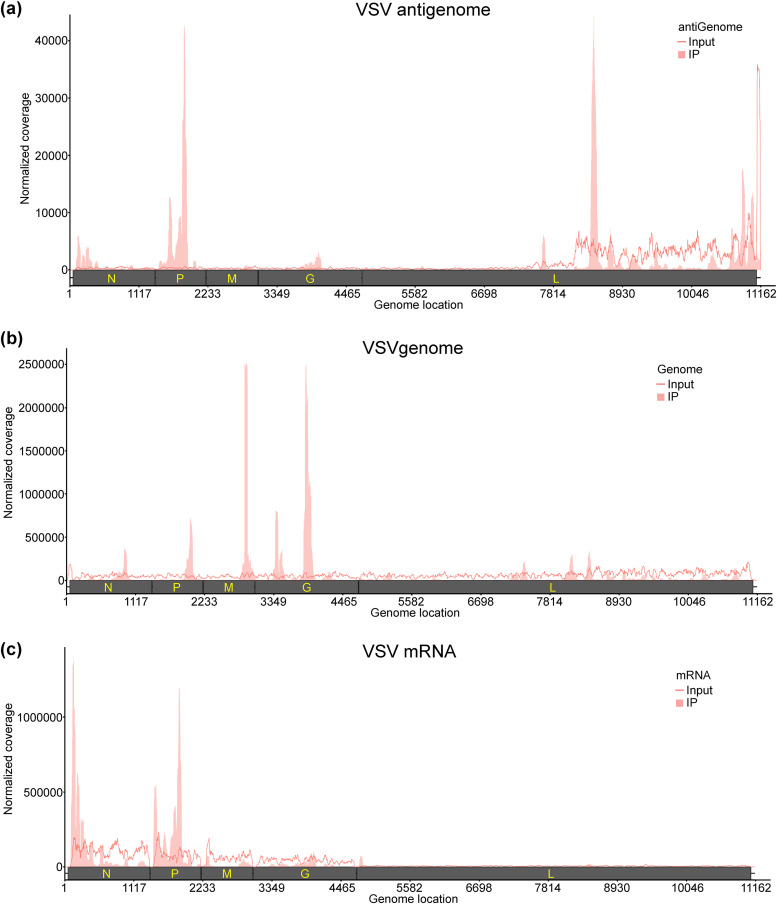
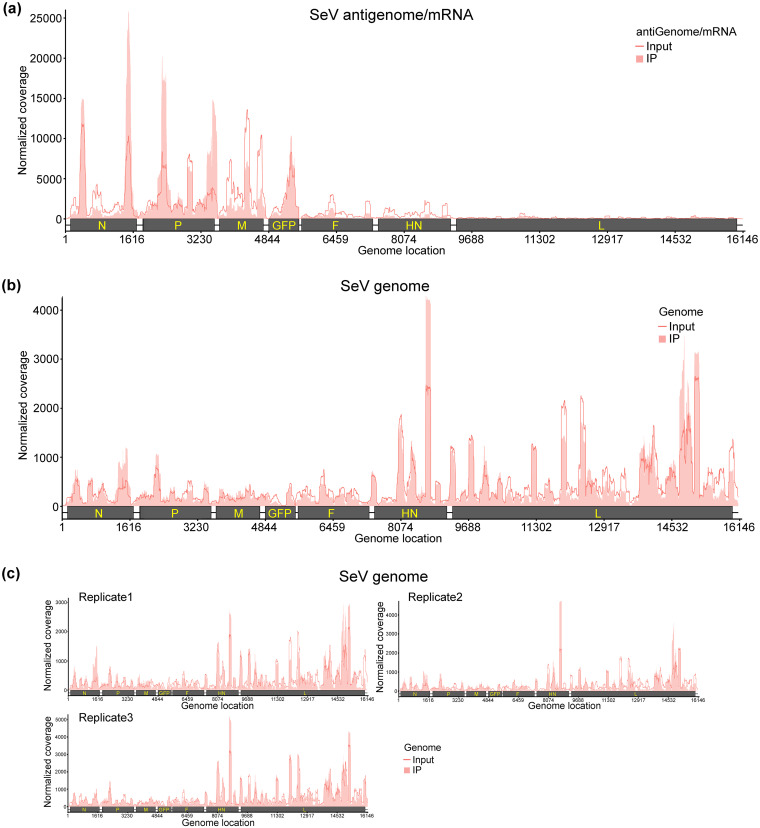
We previously showed that the genome, antigenome, and mRNAs of RSV and hMPV, two members of the family Pneumoviridae, are m6A methylated (33, 34). Here, we performed high-throughput sequencing of m6A RNA (m6A-seq) of VSV and SeV, representative members of the families Rhabdoviridae and Paramyxoviridae, respectively. For VSV, virion RNA (n = 2) extracted from highly purified virions and polyadenylated mRNAs (n = 2) isolated from VSV-infected A549 cells were subjected to m6A-seq. Similar to RSV and hMPV, we found that VSV genome, antigenome, and mRNAs contain m6A. Several m6A peaks were identified on both strands of the viral RNA (Fig. 1a and b). The VSV antigenome RNA contained 21 major m6A peaks in the regions complementary to the N, P, M, G, and L genes (Fig. 1a). In the genome RNA, six m6A peaks were detected in the N, P, M, G, and L genes (Fig. 1b). The P and L gene regions from the antigenome have the strongest m6A enrichment, while the most enriched m6A peaks are located at M and G gene regions from the genome (Table 1). We also mapped m6A peaks in mRNAs purified from VSV-infected cells and identified six m6A peaks from VSV N, P, and L mRNAs, with peak regions largely overlapping those of the antigenome (Fig. 1c and Table 1). For SeV m6A-seq, an SeV strain expressing GFP (rSeV-GFP) was used. Briefly, total RNA (n = 3) isolated from rSeV-GFP-infected or mock-infected A549 cells was subjected to m6A-seq. We identified 6 m6A peaks (consistent across all replicates) in SeV positive-sense RNAs, which include both antigenome and mRNAs. Among them, 2 peaks, 3 peaks, and 1 m6A peak are located in the N, P, and GFP genes, respectively (Fig. 2a and Table 2). In addition, several m6A peaks are observed in negative-sense RNA (genome) (Fig. 2b). However, these peaks are not consistently observed across three replicates (Fig. 2c).
FIG 1.
The VSV RNAs are m6A methylated. (a and b), Distribution of m6A peaks in the VSV antigenome (a) and genome (b). A schematic diagram of the VSV antigenome containing all genes (N, P, M, G, and L) is shown. Total RNAs were extracted from purified VSV virions grown in A549 cells and were analyzed by m6A immunoprecipitation (IP) followed by m6A-seq. Shaded areas show the distribution of m6A immunoprecipitation reads mapped to the VSV antigenome (a) or genome (b). The baseline signal from input samples is shown as a line. (c) Distribution of m6A peaks in the VSV mRNAs. Polyadenylated mRNAs were isolated from VSV-infected A549 cells and subjected to m6A-seq. Shaded pink areas show the distribution of m6A immunoprecipitation reads mapped to the VSV mRNAs. The baseline signal from input samples is shown as a line. Data are presented as the averages from two independent virus-infected A549 cell samples (n = 2).
TABLE 1.
m6A peaks in VSV RNAs
| VSV RNAa | Peak no. | Peak range (nt)b | Gene locationc | Peak size (nt) | Fold enrichmentd | Putative m6A site(s)e |
|---|---|---|---|---|---|---|
| Genome | 1 | 898–1047 | N | 149 | 2.57 | AAACC |
| 2 | 1944–2143 | P | 199 | 5.26 | GAACA, AGACA | |
| 3 | 2891–3040 | M | 149 | 20.7 | GAACA, GAACA | |
| 4 | 3389–3588 | G | 199 | 6.16 | GAACA, GAACA, AAACA, GAACT | |
| 5 | 3838–4036 | G | 198 | 22.56 | AGACC, AAACC | |
| 6 | 7426–7475 | L | 49 | 2.10 | NA | |
| Antigenome | 1 | 1–449 | N | 448 | 11.99 | AAACT, AGACA, AAACT, GAACT, AGACA |
| 2 | 499–548 | N | 49 | 3.51 | NA | |
| 3 | 599–698 | N | 99 | 2.39 | GAACA, GAACA | |
| 4 | 898–997 | N | 99 | 2.84 | AGACT, AGACA | |
| 5 | 1147–1246 | N | 99 | 2.55 | AAACC | |
| 6 | 1396–1894 | P | 498 | 43.8 | AAACC, GAACA, GGACT, GGACT, GAACT | |
| 7 | 1944–1993 | P | 49 | 6.79 | AGACC, AAACA | |
| 8 | 2742–2890 | M | 148 | 4.50 | AAACA, AGACC | |
| 9 | 3389–3488 | G | 99 | 3.64 | NA | |
| 10 | 3688–4036 | G | 348 | 10.26 | AGACC, AGACT, GAACC, AGACA, GAACC | |
| 11 | 4685–4784 | L | 99 | 3.62 | GAACT, GGACT | |
| 12 | 4884–4933 | L | 49 | 2.57 | NA | |
| 13 | 5582–5681 | L | 99 | 3.13 | GGACT | |
| 14 | 6678–6727 | L | 49 | 2.46 | GAACA | |
| 15 | 7575–7624 | L | 49 | 7.38 | GGACT | |
| 16 | 8323–8471 | L | 148 | 11.02 | GAACC, AAACC, AGACC, GAACC | |
| 17 | 8771–8820 | L | 49 | 2.02 | NA | |
| 18 | 8970–9119 | L | 149 | 2.08 | AAACC, AGACT | |
| 19 | 10615–10664 | L | 49 | 4.80 | AAACT | |
| 20 | 10764–10863 | L | 99 | 10.25 | GGACC | |
| 21 | 10914–11062 | L | 148 | 2.71 | AGACC | |
| mRNAs | 1 | 51–249 | N | 198 | 5.18 | AGACA, AAACT, GAACT, AGACA |
| 2 | 1346–1445 | gs, ge, and ig | 99 | 5.73 | GGACT, GAACT | |
| 3 | 1496–1595 | P | 99 | 2.01 | GAACA, GGACT | |
| 4 | 1645–1844 | P | 199 | 6.62 | AAACC | |
| 5 | 4685–4784 | G, gs, ge, ig, L | 99 | 12.64 | GAACT, GGACT | |
| 6 | 8372–8471 | L | 99 | 2.06 | GAACC, AAACC, AGACC, GAACC | |
Genome and antigenome were extracted from highly purified virions grown in A549 cells. VSV mRNAs were extracted from VSV-infected A549 cells and purified by poly(A) bead.
Nucleotide sequence position with reference to the VSV Indiana strain (GenBank accession no. J02428). Nucleotide ranges are indicated.
The VSV genes and regulatory elements are covered by m6A peaks. These regions may contain m6A sites. VSV gene start, gene end, and intergenic sequence are indicated by gs, ge, and ig, respectively.
Log2 enrichment of the m6A peaks identified in the VSV genome, antigenome, and mRNAs.
Putative m6A sites are identified based on searching for the presence of the m6A motif Pu [G > A]m6AC[A/C/U] (Pu represents purine) in the peak ranges. NA indicates no known m6A motifs are found in this region.
FIG 2.
The SeV RNAs are m6A methylated. (a and b), Distribution of m6A peaks in the SeV antigenome/mRNAs (a) and genome (b). A schematic diagram of the rSeV-GFP antigenome containing all genes (N, P, M, GFP, F, HN, and L) is shown. Total RNA was extracted from rSeV-GFP or mock-infected A549 cells and subjected to sonication. RNA containing m6A methylation was pulled down by m6A antibody, followed by m6A-seq. The shaded pink areas show the distribution of m6A immunoprecipitation (IP) reads mapped to the SeV antigenome/mRNAs (a) or genome (b). The baseline signal from input samples is shown as a line. Data presented in panels a and b are the averages from three independent virus-infected A549 cell samples (n = 3). (c) Distribution of m6A peaks in the SeV genome in individual replicates.
TABLE 2.
m6A peaks in Sendai virus RNAs
| SeV RNA peak no.a | Peak range (nt)b | Gene locationc | Peak size (nt) | Fold enrichmentd | Putative m6A site(s)e |
|---|---|---|---|---|---|
| 1 | 351–450 | N | 99 | 1.34 | AGACC |
| 2 | 1401–1750 | N | 349 | 2.63 | AAACA, GGACC, AAACC, AGACA, AGACT, AAACT |
| 3 | 2101–2500 | P | 399 | 2.37 | GAACA, AAACC, AAACA, GAACT, AAACT, AGACT, AAACA, AGACC, GGACT, GAACT |
| 4 | 2651–2750 | P | 99 | 1.56 | GGACA, AGACC, AAACC, GGACC |
| 5 | 3251–3600 | P | 349 | 3.35 | AAACC, GGACC, GGACA, GAACC, AGACA, AGACC, GAACT, GGACA |
| 6 | 5301–5400 | GFP | 99 | 1.39 | GAACT |
Total RNA was extracted from rSeV-GFP or mock-infected A549 cells and subjected to sonication. RNA containing m6A methylation was pulled down by m6A antibody. Three replicates (n = 3) of RNA from virus-infected cells were m6A sequenced.
Nucleotide sequence position with reference to the Sendai virus Z strain expressing GFP (GenBank accession no. AB855655). Nucleotide ranges are indicated.
The SeV genes and GFP gene are covered by m6A peaks. These regions may contain m6A sites.
Log2 enrichment of the m6A peaks identified in the SeV antigenome and mRNAs.
Putative m6A sites are identified based on searching for the presence of the m6A motif Pu [G > A]m6AC[A/C/U] (where Pu represents purine) in the peak ranges.
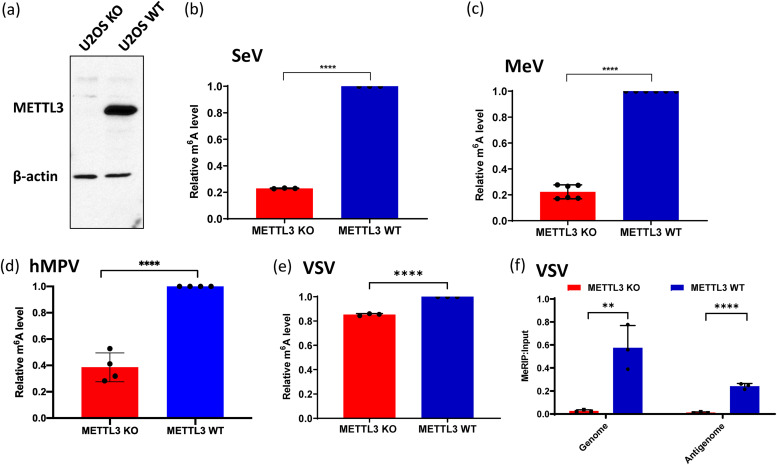
In order to determine the role of viral m6A methylation in innate immunity, we first developed an approach to generate viral RNA that was naturally defective in m6A methylation. To do this, we took advantage of METTL3 knockout (KO) U2OS cells (Fig. 3a), which are susceptible to infection by many NNS RNA viruses. RNA purified from virions grown in METTL3 KO U2OS cells would likely be defective in m6A because METTL3 is the catalytic subunit of m6A methyltransferase (19). Selected NNS RNA viruses, including hMPV, MeV, SeV, and VSV, were grown in wild-type (WT) U2OS or METTL3 KO U2OS cells. To avoid any contamination of defective interfering (DI) particles, all viruses were grown at a relatively low multiplicity of infection (MOI), and virus particles were purified by linear sucrose density gradient ultracentrifugation. Virion RNA was extracted from these virus particles. For many NNS RNA viruses, such as SeV (35, 36), hMPV (33), and Newcastle disease virus (NDV) (35), it has been reported that both genome and antigenome (replicative intermediate) are packaged into virions. To confirm this, both genome and antigenome virion RNAs of hMPV, MeV, SeV, and VSV were quantified by real-time reverse transcription-PCR (RT-PCR). Both genome and antigenome were found in virions of all these viruses. However, the amount of antigenome incorporated into virions varies. The ratio between antigenome and genome in hMPV and MeV virions was approximately 1:1 from both WT and KO U2OS cells. The majority of the RNA from SeV virions was genome RNA, with a ratio between antigenome and genome RNAs of approximately 1:100 in both cells. However, the ratios between antigenome and genome of VSV virions grown in WT and KO U2OS cells were different, depending on the cell source, with approximately 1:5 and 1:10 from WT compared to KO U2OS cells, respectively. Therefore, we extracted total RNA from VSV-infected WT and KO U2OS cells and quantified VSV genome and antigenome and found that the ratios of VSV genome and antigenome in virus-infected cells were similar (1:1) in both cell lines. Next, the m6A content of virion RNA was quantified using a commercial m6A RNA methylation assay. As expected, m6A content in virion RNAs of virus particles purified from METTL3 KO U2OS cells had reduced levels of m6A compared to virion RNAs from WT U2OS cells (P < 0.05) (Fig. 3b to e). For VSV RNA extracted from virus-infected cells, a methylated RNA immunoprecipitation (MeRIP) assay was used for quantification of m6A levels. Both VSV genome and antigenome from METTL3 KO U2OS cells were defective in m6A methylation compared to those from WT U2OS cells (Fig. 3f). However, in all cases, m6A content in virion RNAs from METTL3 KO U2OS cells was not completely defective in m6A, consistent with the previous observation that METTL3 is the major but not the only host RNA m6A methyltransferase. Unlike m6A-deficient virion RNA generated by mutagenesis to disrupt m6A addition sites, these virion RNAs were naturally defective in m6A methylation due to METTL3 KO, but their nucleotide sequences had not been altered.
FIG 3.
The viral RNA of virus grown in METTL3 knockout U2OS cells is defective in m6A methylation. (a) Western blot showing METTL3 expression in METTL3-knockout U2OS cells and wild-type U2OS cells. (b to d) Quantification of m6A level in virion RNA. Shown are the relative m6A levels in virion RNAs from SeV (b), MeV (c), hMPV (d), and VSV (e) grown on METTL3 KO/WT U2OS cells. Each virus was purified through 30 to 50% linear sucrose gradient ultracentrifugation. Virion RNA was extracted, and the total m6A level of each virion RNA was quantified by m6A RNA methylation assay. (f) Total viral RNA from VSV-infected METTL3 KO U2OS cells is defective in binding to m6A antibody by MeRIP assay. An MeRIP assay was carried out to determine the binding of RNA to m6A antibody using the Magna MeRIPTM m6A kit. Anti-m6A antibody was first conjugated to magnetic beads. Total RNA (15 μg) was extracted from VSV-infected METTL3 KO/WT U2OS cells and incubated with m6A antibody-associated beads at 4°C for 2 h with rotation. The RNA-associated magnetic beads were then washed for 3 times. Total RNA was extracted from beads by TRIzol reagent and quantified by real-time RT-PCR using primers annealing to VSV antigenome and genome. Data shown are the mean ± standard deviation (SD) from n = 3 (b, e, and f), n = 6 (c), or n = 4 (d) biologically independent experiments. Statistical significance was determined by two-sided Student's t test: **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
m6A-deficient viral RNA induces significantly higher type I interferon.
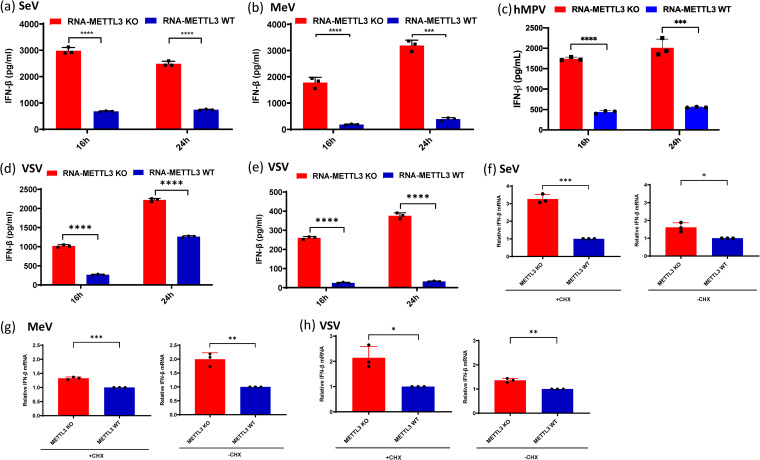
To assess the impact of viral RNA m6A methylation on innate immunity, equal amounts of m6A-deficient or m6A-sufficient virion RNA of each virus were transfected into A549 cells, and their ability to induce IFN-β was measured. Under these conditions, there was no viral replication, thus avoiding the inhibitory effects of viral proteins on the IFN response. As expected, m6A-deficient virion RNA from SeV, MeV, and hMPV induced significantly higher IFN-β than m6A-sufficient virion RNA (Fig. 4a, b, and c). Since the ratios between VSV antigenome and genome from WT and KO U2OS cells were different, we extracted the total RNA from VSV-infected METTL3 KO U2OS cells and WT U2OS cells and found that the ratios between antigenome and genome in VSV-infected cells were similar. Therefore, we transfected A549 cells with equal amounts of total RNA from VSV-infected cells and tested the IFN response. Similarly, m6A-deficient VSV viral RNA induced significantly stronger IFN responses at two different doses (108 and 107 genome RNA copies) (Fig. 4d and e). This phenotype was similar to our previous finding that virion RNA from hMPV grown in A549 cells overexpressing ALKBH5 (an m6A eraser protein) induced more IFN-β (33).
FIG 4.
m6A-deficient viruses and their viral RNAs induce higher type I IFN production. (a to c) Comparison of IFN production triggered by virion RNAs of SeV (a), MeV (b), and hMPV (c) produced from METTL3 KO or WT U2OS cells. A549 cells were transfected with 105 RNA copies of SeV (a), 5 × 106 RNA copies of MeV (b), and 107 RNA copies of hMPV (c), with each virus grown on METTL3 KO or WT U2OS cells. IFN-β production was measured by an ELISA kit at indicated time points. (d to e) IFN-β response in A549 cells transfected with total RNA from VSV-infected cells. Total RNA was extracted from VSV-infected METTL3 KO or WT U2OS cells, and the antigenome/genome was quantified by real-time RT-PCR. A549 cells were transfected with 108 (d) and 107 (e) copies of viral RNA. IFN-β was measured by ELISA at indicated time points. (f to h) IFN-β mRNA level in A549 after viral infection with or without cycloheximide (CHX) treatment. A549 cells were treated for 1 h with 0 or 50 μg/ml CHX and then infected with either SeV (f), MeV (g), or VSV (h) grown in METTL3 KO U2OS or WT U2OS cells. Total RNA was extracted from virus-infected cells, and IFN-β mRNA was quantified by real-time RT-PCR. The relative mRNA level between METTL3 KO- and WT U2OS cell-derived viruses was calculated. The data shown are the mean ± SD from n = 3 biologically independent experiments. Statistical significance was determined by two-sided Student's t test: *, P < 0.5; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Although it is clear that direct transfection of m6A-deficient virion RNA induced more type I IFN, proving that this phenomenon occurs during virus infection would be complicated by the production of new copies of viral genome and antigenome that would be m6A methylated by host m6A machinery. Furthermore, many viral proteins are known to suppress IFN induction. Thus, we quantified IFN mRNA expression in A549 cells infected by m6A-deficient or -sufficient SeV, MeV, and VSV in the presence of cycloheximide (CHX), a drug that inhibits protein synthesis. In infected cells, CHX also inhibits primary transcription and genome replication of many NNS RNA viruses, presumably by limiting the level of viral protein production and therefore their contributions to transcription and replication (37). This strategy should minimize the impact of viral proteins and newly synthesized viral RNA on IFN mRNA expression. As shown in Fig. 4f to h, infection of cells with m6A-deficient SeV, MeV, and VSV induced significantly higher IFN mRNA than did m6A-sufficient viruses in the presence of CHX, as well as in its absence. Thus, infection of A549 cells with the m6A-deficient virion does induce more IFN. This finding is similar to our previous observation that infection of cells with hMPV mutants whose m6A sites had been removed by mutagenesis induced significantly higher IFN compared to WT hMPV (33).
Taken together, these results indicate that the ability of m6A-deficient virion RNA to induce IFN is conserved among NNS RNA viruses, and conversely, m6A modification of virion RNA prevents it from inducing IFN.
m6A-deficient viral RNA activates type I interferon in an RIG-I-dependent manner.
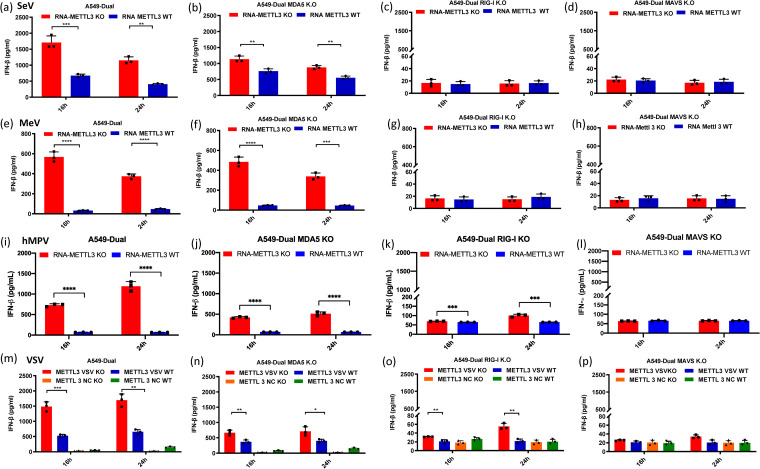
RIG-I and MDA5 are the two major RNA sensors that recognize viral RNA, leading to the induction of IFN. RIG-I detects single-stranded RNA (ssRNA) with 5′-pp or 5′-ppp and short dsRNA with blunted 5′-pp or 5′-ppp, whereas MDA5 recognizes long dsRNA (38). The genome and antigenome of NNS RNA viruses are ssRNAs bearing 5′ triphosphate and thus are recognized by RIG-I. To identify which RNA sensor detects m6A-deficient RNA, equal amounts of m6A-sufficient virion RNA or m6A-deficient virion RNA were transfected into A549-Dual cells lacking RIG-I, MDA5, or their downstream adapter protein, MAVS, and the amount of IFN-β released was measured. As expected, m6A-deficient SeV virion RNAs triggered more IFN-β production than m6A-sufficient SeV virion RNAs (P < 0.05, Student's t test) (Fig. 5a). IFN-β production was totally abrogated in A549-Dual cells lacking RIG-I (Fig. 5c) and MAVS (Fig. 5d). However, a considerable amount of IFN-β was detected in MDA5 KO cells (Fig. 5b), although the amount was significantly reduced compared to that in the parental A549-Dual cells (Fig. 5a). Similar results were observed for m6A-sufficient virion RNA of MeV (Fig. 5e to h), hMPV (Fig. 5i to l), and VSV (Fig. 5m to p). Thus, RIG-I is the main RNA sensor involved in detecting m6A-deficient viral RNA of NNS RNA viruses, although MDA5 also plays a minor role.
FIG 5.
IFN response in A549 knockout cells transfected with m6A-deficient viral RNA. (a to l) IFN production after transfection of virion RNA. Confluent wild-type (a, e, i), MDA5 knockout (b, f, j), RIG-I knockout (c, g, k), or MAVS knockout (d, h, l) A549-Dual cells were transfected with the same amounts of SeV (a to d, 105 RNA copies), MeV (e to h, 2 × 105 RNA copies), and hMPV (i to l, 107 RNA copies) virion RNAs from either METTL3 KO or WT cells. (m to p) IFN production after transfection of total RNA from VSV-infected cells. Confluent wild-type (m), MDA5 knockout (n), RIG-I knockout (o), or MAVS knockout (p) A549-Dual cells were transfected with the same amounts of total RNA from VSV-infected METTL3 KO cells or METTL3 WT cells containing 5 × 106 copies of viral RNA. Cell culture supernatants were harvested at 16 and 24 h after inoculation, IFN-β in the supernatant at indicated time points was measured by a commercial ELISA kit. Data shown are the mean ± SD from n = 3 biologically independent experiments. Statistical significance was determined by two-sided Student's t test: *, P < 0.5; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
m6A-deficient viral RNA enhances multiple steps in the RIG-I signaling pathway.
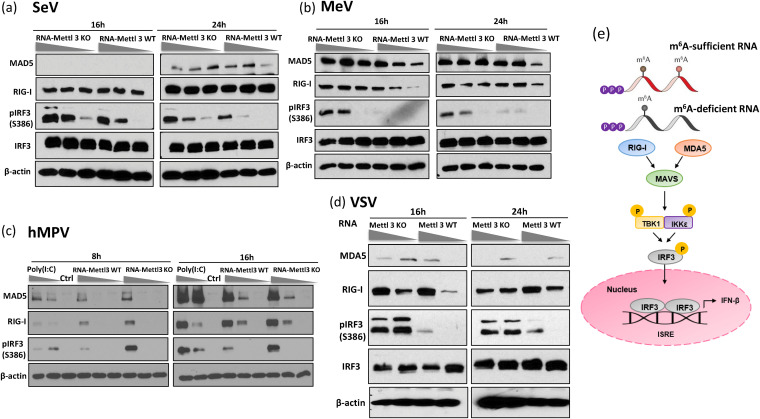
Having demonstrated that RIG-I is the major RNA sensor recognizing m6A-deficient viral RNA, we next investigated the activation of the IFN pathway after viral RNA stimulation (Fig. 6). Various doses of m6A-sufficient or m6A-deficient viral RNA were transfected into A549 cells, and MDA and RIG-I expression and phosphorylation of interferon regulatory factor 3 (IRF3) at S386 were detected by Western blotting. Except for SeV, RIG-I expression significantly increased in cells transfected with m6A-deficient viral RNA compared to cells transfected with m6A-sufficient viral RNA. However, except for MeV, there was no significant increase in MDA5 expression. Importantly, m6A-deficient viral RNA stimulated a significantly larger amount of IRF3 phosphorylation than m6A-sufficient viral RNA for all viruses tested in a dose-dependent manner (Fig. 6), which is consistent with the observation that they induced higher expression of type I IFN (Fig. 4).
FIG 6.
m6A-deficient viral RNA enhances expression of molecules involved in type I IFN signaling pathway. (a to c), m6A-deficient virion RNA increases expression of RIG-I and MDA5 and induces higher IRF3 phosphorylation. A549 cells were transfected with virion RNA of SeV (a, at doses of 2 × 105, 1 × 105, and 5 × 104 RNA copies), MeV (b, at doses of 1 × 106, 5 × 105, and 2 × 105 RNA copies), and hMPV (c, at doses of 107, 106, and 105 RNA copies) grown on METTL3 KO or WT U2OS cells. (d) m6A-deficient total RNA from VSV-infected cells increases expression of RIG-I and MDA5 and induces higher IRF3 phosphorylation. A549 cells were transfected with total RNA from VSV-infected cells at doses of 1 × 108 and 1 × 107 RNA copies grown on METTL3 KO or WT U2OS cells. At indicated times, cell lysates were analyzed by Western blotting using antibodies specific to RIG-I, MDA5, IRF3, IRF3 phosphorylated at S386, or β-actin. Western blots are representative of n = 3 biologically independent experiments. (e) A cartoon model for type I IFN signaling pathway. m6A-sufficient or -deficient viral RNA is detected by either RIG-I or MDA5, engaging the adaptor protein MAVS, which leads to the phosphorylation of IRF‐3 by TBK1/inducible IκB kinase (IKK‐i), the formation of IRF‐3 homodimers and/or heterodimers, and translocation into the nucleus, resulting in the expression of type I IFNs.
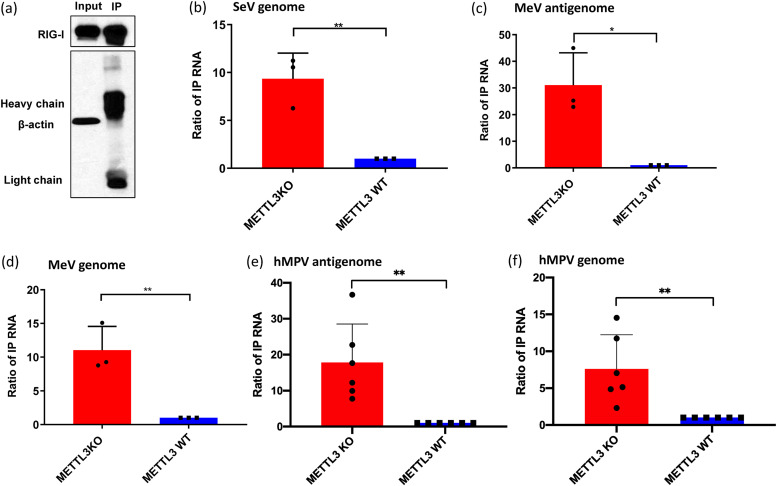
Next, we determined whether virion RNA deficient in m6A enhances the binding to RIG-I. Briefly, cells expressing Flag-tagged RIG-I were lysed and divided into 7 aliquots, mixed with each of the virion RNAs, and pulled down with Flag antibody-conjugated magnetic beads (Fig. 7a), and the RNA in the complex was quantified by quantitative reverse transcription-PCR (RT-qPCR). For SeV virion RNA, a 10-fold increase in RIG-I binding was observed for m6A-deficient virion RNA compared to m6A-sufficient virion RNA (Fig. 7b). For MeV virion RNA, RIG-I binding to m6A-deficient antigenome and genome RNAs was increased 30- and 10-fold, respectively, compared to those of m6A-sufficent virion RNA (Fig. 7c and d). For hMPV virion RNA (Fig. 7e and f), 18- and 8-fold increases in RIG-I binding were observed for the antigenome and genome of m6A-deficient RNA, respectively, compared to those of m6A-sufficent RNA. Thus, m6A deficiency enhances binding to RIG-I of SeV, MeV, and hMPV virion RNA.
FIG 7.
m6A-deficient virion RNA increases RIG-I binding. Shown are the results from an affinity binding assay of RIG-I with each virion RNA. RIG-I-conjugated magnetic beads were incubated with virion RNA purified from METTL3 KO or WT U2OS cells. (a) One aliquot of beads was analyzed by Western blotting. (b to e) Antigenome or genome RNA bound to magnetic beads was quantified by real-time RT-PCR. Results were normalized as the ratio between immunoprecipitated RNA from METTL3 KO cells and that from METTL3 WT cells. The data shown are the mean ± SD from n = 3 (b to d) or n = 6 (e and f) biologically independent experiments. Statistical significance was determined by two-sided Student's t test: *, P < 0.5, **, P < 0.01.
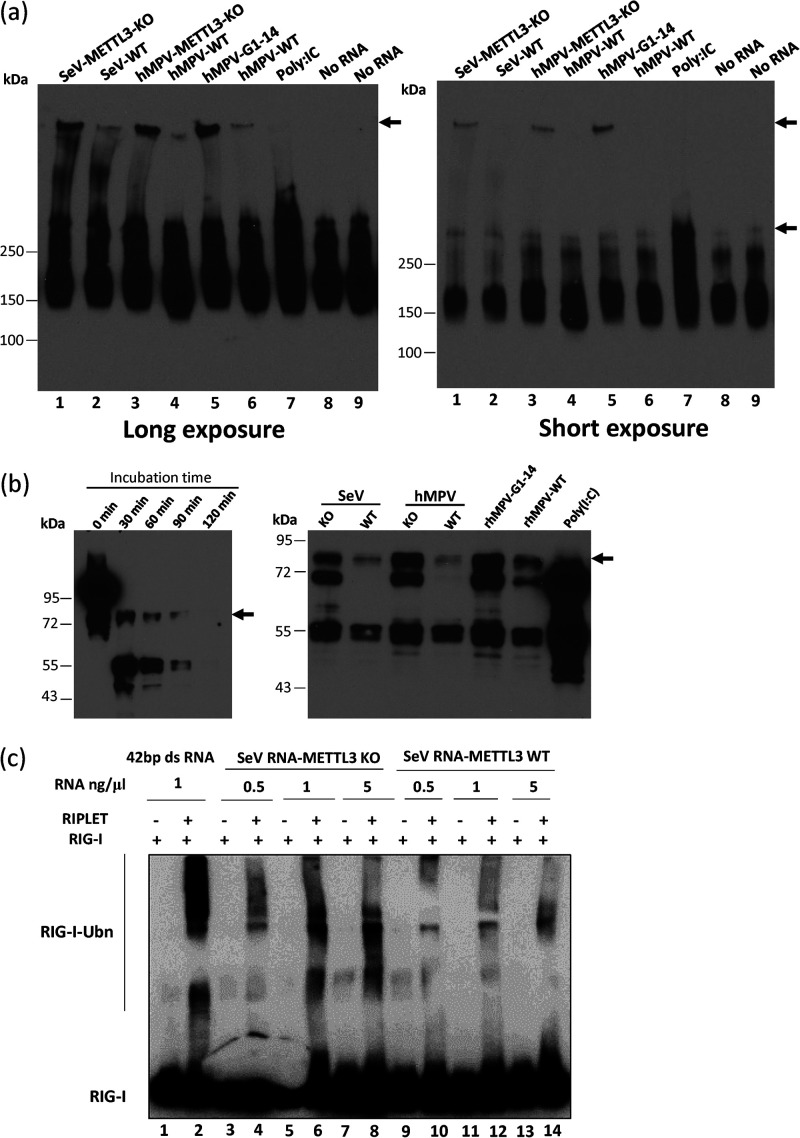
We next determined whether m6A-deficient RNA can stimulate the formation of RIG-I:RNA complex and RIG-I oligomerization. Purified RIG-I was incubated with m6A-sufficient RNA or m6A-deficient RNA in the presence of ATP. Formation of RIG-I:RNA complexes was resolved by native PAGE gel and detected by Western blotting. In the absence of RNA, monomeric RIG-I with a molecular weight of approximately 170 kDa on the native gel was detected (Fig. 8a, lanes 8 and 9). Incubation of RIG-I with poly(I·C) leads to the formation of RIG-I:RNA complexes with a molecular weight of approximately 350 kDa (Fig. 8a, lane 7), suggesting that RIG-I oligomerization occurs in the presence of poly(I·C). Interestingly, incubation of RIG-I with SeV and hMPV RNA formed much larger RIG-I:RNA complexes than the complex of RIG-I with poly(I·C) (Fig. 8a), which is probably due to the difference in the sizes of the RNAs [14 kb for hMPV, 15 kb for SeV, and 1.0 to 1.5 kb for poly(I·C)] and the difference in protein-RNA interaction. Importantly, incubation of RIG-I with m6A-deficient virion RNA from SeV, hMPV, and rhMPV-G1-14 formed more RIG-I:RNA complexes compared to the m6A-sufficient RNA (Fig. 8a, compare lanes 1 and 2, 3 and 4, and 5 and 6). These results suggests that m6A-deficient RNA can stimulate the formation of RIG-I:RNA complex and RIG-I oligomerization.
FIG 8.
m6A-deficient virion RNA facilitates RIG-I:RNA conformation change and increases the ubiquitination of RIG-I. (a) Analysis of RIG-I:RNA complex in native PAGE gel. Purified RIG-I was incubated with poly(I·C) or 107 copies of virion RNA in MOPS buffer in the presence of RNase inhibitor and AMP-PNP (2 mM). The reaction mixtures were incubated at 37°C for 30 min to enable RIG-I:RNA complex formation. Ten microliters of the mixture was mixed with an equal volume of native PAGE buffer, and RIG-I:RNA complex was separated in native PAGE gel, followed by Western blotting with anti-RIG-I helicase antibody to detect the RIG-I:RNA complex. Gels with short and long exposures are shown. The Western blots shown are the representatives of three independent experiments. (b) Analysis of RIG-I:RNA conformation by limited trypsin digestion in denaturing SDS-PAGE. The RIG-I:RNA complex was formed as described for panel a. Limited trypsin digestion of RIG-I protein in the absence of RNA ligand for 0 to 2 h (left panel) or in the presence of poly(I·C) or viral RNA (right panel) for 2 h is shown. The Western blots shown are the representatives of three independent experiments. (c) In vitro ubiquitination analysis of RIG-I. A 1.0 μM concentration of purified RIG-I was incubated with 1 ng/μg of 42-bp dsRNA and different doses of SeV virion RNA from METTL3 KO or WT U2OS cells. Ubiquitination of RIG-I was analyzed by anti-RIG-I blotting. The blots shown are the representatives of three independent experiments.
Upon binding of an RNA ligand, RIG-I oligomerization leads to a significant conformational rearrangement that can be detected by limited trypsin proteolysis of RIG-I:RNA complexes in denaturing SDS-PAGE (14, 39). In the absence of ligand RNA, trypsin treatment of the RIG-I protein yielded a 55-kDa RIG-I fragment (helicase domain), representing the trypsin-sensitive autorepressed RNA-free RIG-I conformation. Trypsin digestion of poly(I·C)-bound RIG-I yielded an 80-kDa fragment, the caspase activation and recruitment domain (CARD)-helicase domain, which is recognized by a monoclonal anti-helicase domain antibody (Fig. 8b, left panel). This band represents the trypsin resistance of the RNA-bound RIG-I (14, 33). Natural m6A-deficient RNA from SeV and hMPV virions grown in METTL3 KO cells yielded more 80-kDa protein than m6A-sufficient virion RNA (Fig. 8b, right panel). Similar to our previous observation, virion RNA of rhMPV-G1-14, which contains a total of 14 m6A mutations, yielded more trypsin-resistant 80-kDa protein than wild-type hMPV virion RNA did (Fig. 8b, right panel). Thus, m6A-deficient viral RNA facilitates the conformational change of RIG-I.
Upon conformational change, RIG-I exposes its two CARD domains to K63-linked polyubiquitination and binding of K63-linked ubiquitin chains, which activate the RIG-I conformer, engaging the adaptor protein MAVS for activation of transcription factors (40, 41). Recent studies have found that RIG-I ubiquitination and activation are dependent on RIPLET, a ubiquitin E3 ligase (42). Thus, we tested the abilities of m6A-deficient RNA in promoting ubiquitination of RIG-I (Fig. 8c). As a positive control, incubation of purified RIG-I with 42-bp dsRNA triggered strong ubiquitination of RIG-I only in the presence of RIPLET (Fig. 8c, lane 2). SeV RNA isolated from virions grown in METTL3 KO cells promoted RIG-I ubiquitination in a dose-dependent manner (Fig. 8c, compare lanes 4, 6, and 8). In comparison, SeV RNAs isolated from virions grown in WT cells were significantly less efficient in stimulating RIG-I ubiquitination (Fig. 8c, compare lanes 4 and 10, 6 and 12, and 8 and 14). These data demonstrate that RIG-I is intrinsically capable of sensing m6A in viral RNAs and that SeV RNA lacking m6A has a greater ability to stimulate RIG-I, providing a mechanistic insight into its higher activity in triggering IFN production.
YTHDF2, the m6A binding protein, is essential for suppression of type I IFN expression.
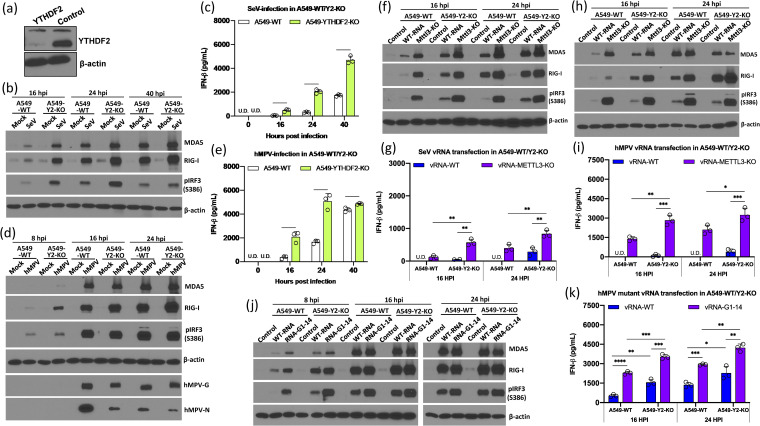
The functionality of m6A can be also mediated by m6A binding proteins (m6A readers) (18). We hypothesized that m6A reader proteins bind and protect m6A-methylated viral RNA, thereby preventing recognition by RNA sensors and suppressing type I IFN signaling. To test this hypothesis, we generated YTHDF2 KO A549 cells (Fig. 9a) using CRISPR-Cas9 technology and compared the activation of type I signaling in WT A549 cells and YTHDF2 KO A549 cells upon virus infection or viral RNA transfection. Briefly, WT A549 cells and YTHDF2 KO A549 cells were infected by SeV or hMPV, and IFN expression and the IFN signaling pathway were examined. Both SeV infection (Fig. 9c) and hMPV infection (Fig. 9e) induced significantly higher type I IFN production in YTHDF2 KO A549 cells than in WT A549 cells. Consistent with this, MDA5 and RIG-I expression and IRF3 phosphorylation increased in YTHDF2 KO A549 cells upon SeV (Fig. 9b) and hMPV (Fig. 9d) infection, suggesting that YTHDF2 suppresses the type I IFN signaling pathway.
FIG 9.
YTHDF2 is essential for suppression of the IFN signaling pathway. (a) Western blot analysis of YTHDF2 in YTHDF2 knockout A549 cells and control sgRNA-treated A549 cells. (b) SeV infection induces a higher IFN signaling pathway in YTHDF2 knockout A549 (A549-Y2 KO) cells compared to WT A549 cells. Confluent cells were infected by SeV at an MOI of 1.0, and cell lysates were analyzed by Western blotting at the indicated time points. (c) IFN production in WT and YTHDF2 knockout A549 cells upon SeV infection at an MOI of 1.0. (d) hMPV infection induces a higher IFN signaling pathway in A549-Y2 KO cells compared to A549 WT cells. Cells were infected by hMPV at an MOI of 1.0. (e) IFN production in WT and YTHDF2 knockout A549 cells upon hMPV infection at an MOI of 1.0. (f) Comparison of IFN signaling pathways in A549-Y2 KO cells and A549 WT cells upon transfection of equal amounts (106 RNA copies) of WT SeV RNA and m6A-deficient SeV RNA. (g) IFN production in WT and YTHDF2 knockout A549 cells upon SeV RNA transfection (106 RNA copies). (h) Comparison of IFN signaling pathway in A549-Y2 KO cells and A549 WT cells upon transfection of equal amounts (106 RNA copies) of WT hMPV RNA and m6A-deficient hMPV RNA. (i) IFN production in WT and YTHDF2 knockout A549 cells upon hMPV RNA transfection (106 RNA copies). (j) Comparison of IFN signaling pathways in A549-Y2 KO cells and A549 WT cells upon transfection of equal amounts (106 RNA copies) of WT hMPV RNA and hMPV G1-14 RNA. (k) IFN production in WT and YTHDF2 knockout A549 cells upon WT hMPV RNA and hMPV G1-14 RNA transfection (106 RNA copies). The Western blots shown are the representatives of three independent experiments. The interferon data shown are the mean ± SD from n = 3 biologically independent experiments. Statistical significance was determined by two-sided Student's t test: *, P < 0.5; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Next, we transfected WT viral RNA or m6A-deficient viral RNA (from METTL3 KO U2OS cells) into WT A549 cells and YTHDF2 KO A549 cells and examined type I IFN signaling. Similar to SeV infection, transfection of WT SeV RNA induced significantly higher RIG-I expression, IRF3 phosphorylation (Fig. 9f), and IFN production (Fig. 9g) in YTHDF2 KO A549 cells than in WT A549 cells. Thus, YTHDF2 suppresses the type I IFN signaling pathway upon RNA transfection.
Consistent with previous observations, m6A-deficient SeV RNA induced significantly higher expression of MDA5, RIG-I, and IRF3 phosphorylation (Fig. 9f) and IFN production (Fig. 9g) than WT SeV RNA in WT A549 cells. Since YTHDF2 was not available to bind to viral RNA in YTHDF2 KO A549 cells, the difference in RIG-I, MDA5, and IRF3 phosphorylation (Fig. 9f) and IFN-β production (Fig. 9g) between WT and m6A-deficient RNA was diminished in YTHDF2 KO A549 cells compared to WT A549 cells. However, m6A-deficient RNA induced significantly more IFN (Fig. 9g) and related proteins (Fig. 9f) in YTHDF2 KO A549 cells compared to those in WT A549 cells, which may due to the fact that these viral RNAs still retained some m6A methylation, although they had significantly less (Fig. 3). Similar results were observed when m6A-deficient hMPV RNA (derived from METTL3 KO cells) was transfected into YTHDF2 KO A549 cells and WT A549 cells (Fig. 9h and i). To confirm these results, we compared the IFN induction by WT rhMPV and rhMPV-G1-14 RNA, whose m6A sites in its G gene have been removed by mutagenesis. The results were similar to those observed for m6A-deficient SeV RNA or m6A-deficient hMPV RNA derived from METTL3 KO cells (Fig. 9j and k). Collectively, these results indicate that YTHDF2 may sequester m6A-sufficient virion RNA, thereby suppressing host innate immunity.
DISCUSSION
Internal RNA m6A methylation was discovered in the early 1970s (43, 44). Studies have shown that m6A plays many important roles in mRNA metabolism, translation, and splicing (22, 45). However, the biological functions of m6A in viral RNA remain poorly understood. In this study, we found that m6A on the genomes and antigenomes of several families in NNS RNA viruses enables viral RNA to escape recognition by RIG-I. This novel role of m6A in innate immunity is universally conserved in the representative members of the families Pneumoviridae (hMPV), Paramyxoviridae (SeV and MeV), and Rhabdoviridae (VSV). Abrogation of m6A on virion RNA enhances its recognition by RIG-I and facilitates the RIG-I conformational change, oligomerization, and ubiquitination, which activates the RIG-I-mediated IFN signaling pathway. In addition, the m6A binding protein, YTHDF2, is essential for suppression of type I IFN responses. The data presented here suggest that three families in NNS RNA viruses have evolved a common mechanism, which is to mask their genome and replication intermediate antigenome with m6A, to mimic host RNA and evade host innate immunity that is dependent on RNA sensor RIG-I.
Our finding suggests that m6A serves as a molecular marker for host innate immunity to discriminate self from nonself RNA. All NNS RNA virus genomes and antigenomes have triphosphate at their 5′ termini. RIG-I plays a dominant role in the recognition of NNS viral genome and antigenome RNAs, consistent with the fact that RIG-I detects 5′-triphosphorylated single-strand RNA. The addition of m6A to NNS genomes and antigenomes appears to reduce RIG-I detection of the triphosphorylated 5′ terminus of these RNAs. The fact that IFN production was significantly reduced but not completely inhibited in RNA-transfected MDA5 knockout cells suggest that MDA5 might play a secondary role in recognizing m6A-deficient RNA of NNS RNA viruses. However, we also cannot rule out the possibility of other cytosolic RNA sensors (such as protein kinase R [PKR] and Toll-like receptors [TLRs]) in detection of m6A-deficient RNA. To date, many positive-sense RNA viruses (such as HIV, HCV, Zika virus, and EV71) and segmented negative-sense RNA viruses (such as influenza virus) have also been shown to contain m6A methylation in their viral genomes and mRNAs (32, 46, 47). The 5′ structures of their RNA genome, such as ppp, cap0, cap1, cap2, untranscribed region (UTR), or internal ribosome entry site (IRES), the nature of the RNA genome (ssRNA or dsRNA), and the replication strategies of these RNA viruses are highly diverse: different pattern recognition receptors (PRRs) may be involved in the detection of their m6A-sufficient and -deficient genomes (48). For example, m6A-modified poly(U/UC) RNA sequence derived from hepatitis C virus (HCV) prevented its recognition by RIG-I and therefore the conformational changes of RIG-I (14). It was shown that unmodified circular RNA, but not m6A-modified circular RNA, directly activates the RIG-I-mediated IFN signaling pathway (15). Similarly, it was found that m6A modifications of hepatitis B virus (HBV) and HCV RNAs were less effective in RIG-I signaling and that single-nucleotide mutations in the m6A motif in viral RNAs enhanced RIG-I sensing activity (49). Recently, it was found that loss of METTL3 and m6A activates an aberrant innate immune response during hematopoietic development, mediated by the formation of endogenous double-strand RNAs (dsRNAs) (50). This leads to activation of the 2′,5′-oligoadenylate synthetase (OAS)-RNase L and PKR-eIF2α pathways and upregulation of the dsRNA sensors MDA5 and RIG-I, which signal through MAVS (50).
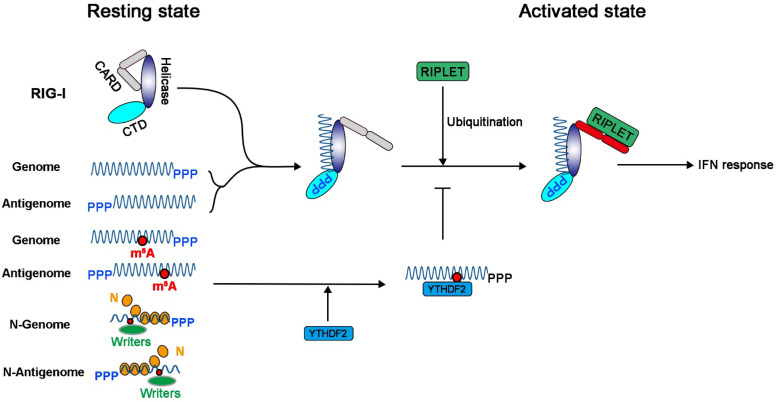
It is not known how RIG-I accesses or recognizes the m6A-deficient genome and antigenome of NNS RNA viruses in the context of virus infection, as both are encapsidated by N protein (1). Studies have demonstrated that RIG-I mainly recognizes the full-length genome RNA bearing 5′-triphosphates for influenza virus and SeV (51). However, others have shown that RIG-I preferentially associates with shorter 5′-triphosphate-containing viral RNA segments (such as defective interfering [DI] particles) in cells infected with influenza virus and SeV (52, 53). It is poorly understood how RIG-I gains access to these encapsidated RNAs to induce IFN signaling, regardless of whether they are full-length or DI genomes and antigenomes. Nevertheless, the fact that both the genome and antigenome of NNS RNA viruses are m6A methylated suggests that m6A writer proteins must gain access and methylate genome and antigenome prior to their encapsidation, or m6A methylation occurs between the elongation of the genome or antigenome and its encapsidation occurs concurrently. One possibility is that some of the genomes and antigenomes that are synthesized are not encapsidated (Fig. 10). This would seem most likely to occur in the early stage of the virus replication cycle. Some of these genomes and antigenomes may not be encapsidated because of the low concentration of the N protein, allowing RIG-I access to both the 5′-ppp and RNA downstream from it. Our data suggest that m6A methylation of the viral genome and antigenome inhibits recognition by RIG-I, preventing RIG-I conformational change, oligomerization, and ubiquitination, thereby suppressing type I IFN and its protective effects (Fig. 10).
FIG 10.
Model for how m6A methylation invades host innate immunity in NNS RNA viruses. The newly synthesized genome and antigenome of NNS RNA viruses are first methylated by m6A writer proteins, which are encapsidated by N protein to serve as a template for replication and transcription. Some of the genome and antigenome may not be encapsidated at an early stage in the virus replication cycle, particularly when the concentration of N protein is low. These unmodified antigenomes or genomes would be recognized by RIG-I to induce the IFN signaling pathway. m6A methylation in genome and antigenome prevents RIG-I binding, conformational change, oligomerization, and K63-linked polyubiquitination, which result in lower activation of MAVS and subsequent phosphorylation of IRF3, thereby inhibiting type I IFN production. In addition, YTHDF2 binds to the m6A-methylated genome and antigenome, which suppresses type I IFN signaling.
YTHDF2 is one of the major m6A binding proteins in mRNA, functioning as a double-edged sword, either promoting RNA decay or promoting translation (23–25). Our results suggest that YTHDF2 has another novel function, which is to suppress innate immunity during virus infection. IFN expression was significantly enhanced in YTHDF2 KO A549 cells upon WT virus infection or WT viral RNA transfection. However, the difference in IFN production between m6A-deficient and WT virion RNA was diminished in YTHDF2 knockout A549 cells, compared to those in WT A549 cells (Fig. 9). Although we have not demonstrated the direct interaction of YTHDF2 and viral RNA, it is likely that the binding of YTHDF2 to m6A-methylated viral RNA sequesters these viral RNAs and prevents the RIG-I activation. We previously found that YTHDF2 was partially colocalized with N proteins of RSV and hMPV (33, 34). It is possible that other effector functions of YTHDF2 (such as interaction with viral N protein and enhancement of N protein-mediated viral RNA encapsidation) may play a role in preventing viral RNA from recognition by RNA sensors. Our results are consistent with a recent study that showed that YTHDF2 was essential for suppression of circular RNA-induced immunity (15). Specifically, YTHDF2 sequestered m6A-modified circular RNAs and blocked endogenous circular RNAs from activating the RIG-I antiviral pathway (15). Similarly, it was shown that YTHDF2 inhibited RIG-I signaling by occupying m6A-modified RNAs and inhibiting RIG-I recognition in HBV and HCV (49). Recent work also suggests that RNA methylation (such as internal 2′-O methylation) in short dsRNA serves as a “brake” or “throttle” to prevent RIG-I translocation, oligomerization, and downstream signaling (54). Thus, the binding of YTHDF2 to m6A sites will likely be an additional barrier, preventing RIG-I conformational changes and thereby suppressing type I IFN responses (Fig. 10).
Our work highlights the need to elucidate the molecular determinants of regulation and functions of other epitranscriptomic modifications in viral RNA. To date, more than 160 RNA modifications have been reported. Several RNA modifications, including internal 2′-O methylation and m5C, have been detected in viral RNAs (55–57). For example, West Nile virus (57) and HIV (55) RNAs were reported to contain internal 2′-O methylation. Interestingly, host FTSJ3, a 2′-O-methyltransferase (2′O-MTase), was shown to catalyze the internal 2′-O methylation of HIV RNA (55). Knockdown of FTSJ3 in cells led to a reduction in internal 2′-O methylation in viral RNA and triggered a higher expression of type I IFNs in human dendritic cells through the RNA sensor MDA5 (55). Since viral RNA modification modulates many biological functions (such as RNA decay, translation, and innate immunity), it is possible that they may contribute to the pathogenesis and virulence of different virus strains.
In conclusion, NNS RNA viruses acquire m6A modification in their genome and antigenome, as do other cellular RNAs, enabling them to escape innate immune recognition by the host cytosolic RIG-I. The m6A methylation on viral RNA reduces its binding affinity to RIG-I, preventing RIG-I conformational change, oligomerization, and K63-linked polyubiquitination, which result in lower activation of MAVS and subsequent phosphorylation of IRF3, thereby inhibiting type I IFN production.
MATERIALS AND METHODS
Cell lines.
A549 cells (ATCC CCL-185) were purchased from the American Type Culture Collection. A549-Dual, A549-Dual KO RIG-I, A549-Dual KO MDA5, and A549-Dual KO MAVS knockout cells were purchased from InvivoGen (San Diego, CA) and were supplemented with Normocin (100 μg ml−1), blasticidin (10 μg ml−1), and Zeocin (100 μg ml−1). The wild-type U2OS and METTL3 knockout U2OS cell lines were a gift from Yang Shi (Harvard Medical School, Boston, MA). All cell lines were kept in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and were confirmed free from mycoplasma by the LookOut Mycoplasma PCR detection kit (Sigma, St. Louis, MO).
Generation of YTHDF2 knockout A4549 cell line.
CRISPR-Cas9 was used to knock out the YTHDF2 gene in A549 cells. Three single guide RNAs (sgRNAs) specific for exon 4 of the YTHDF2 gene (sgRNA1, 5′-GCAGTATATGCATTATTCTG-3′; sgRNA2, 5′-GGGTAAGTAGGAATCTGACA-3′; and sgRNA3, 5′-GAATAGGAGAAGCCAATGGA-3′) and one control sgRNA (5′-GTACGTCGGTATAACTCCTC-3′) were designed and cloned into the lentiviral vector plentiCRISPR v2. 293T cells (2.5 × 106) were transfected with the lentiviral vector expressing both the sgRNA and Cas9 (5 μg), the packaging plasmid (5 μg), and the VSV-G envelope expression plasmid (2.5 μg) to generate lentiviral particles that were used to transduce A549 cells (2 × 105). After puromycin (2 μg/ml) selection, the transduced cells were single cell cloned. The knockout of YTHDF2 expression in A549 cells was confirmed by Western blotting. Genome editing of the targeted region was also confirmed by PCR of exon 4 of YTHDF2 from genomic DNA followed by sequencing.
Virus production and purification.
Ten T150 flasks of wild-type U2OS cells and METTL3 knockout U2OS cells were infected with measles virus (MeV) at an MOI of 0.01, Sendai virus (SeV) at an MOI of 1, or human metapneumovirus (hMPV) at an MOI of 0.5. When extensive cytopathic effects (CPE) were observed, cell culture supernatants were harvested and clarified by centrifugation at 10,000 × g for 30 min. Virus was concentrated through a 10% (wt/vol) sucrose cushion by centrifugation at 30,000 × g for 2 h at 4°C in a type 50.2 Ti rotor (Beckman, Brea, CA). The pellet was resuspended in NTE buffer (0.05 M Tris-HCl, 0.15 M NaCl, 15 mM CaCl2, pH 6.5). Virus was further purified through 30 to 50% linear sucrose gradient ultracentrifugation. The bands containing virus particles were collected and centrifuged at 25,000 × g for 2 h at 4°C. The final pellets were resuspended in NTE buffer with 10% trehalose.
Isolation of total viral RNA and virion RNA.
Confluent METTL3 KO U2OS cells or WT cells in 60-mm-diameter dishes were mock infected or infected with VSV at an MOI of 0.1. At 24 h after infection, total RNA was isolated from virus-infected cells using the TRIzol reagent (Life Technologies) and dissolved in RNase-free water. Virion RNA was extracted from sucrose gradient-purified virions of each virus.
Real-time RT–PCR.
Viral RNA was isolated from purified virion or infected cells by using TRIzol reagent (Life Technologies, Carlsbad, CA) following the manufacturer’s instruction. Reverse transcription (RT) real-time PCR was conducted by using primers annealing to the leader and N sequence of each virus in TB-Green premix Ex Taq (TaKaRa, Kusatsu, Shiga Prefecture, Japan).
Colorimetric quantification of viral m6A methylation.
Virion RNA was extracted from viruses purified by sucrose cushion ultracentrifugation as described above. The total m6A modification level of virion RNA was quantified by an m6A RNA methylation assay kit (Abcam; ab185912) based on the manufacturer’s instruction. Briefly, each amount of virion RNA was bound to strip wells using an RNA high-binding solution, and m6A was detected using a specific capture anti-m6A antibody and then quantified colorimetrically by reading the absorbance in a microplate spectrophotometer at a wavelength of 450 nm. A standard curve was generated using known m6A-methylated RNA (range from 0.02 to 1 ng of m6A) as a positive control. The m6A content was calculated from each RNA sample based on its optical density at 450 nm (OD450) values. The percentage of change was calculated by dividing m6A levels in virion RNA from U2OS KO cells by those from the WT U2OS cells.
m6A-seq.
High-throughput sequencing of the VSV was carried out using m6A-seq (MeRIP-seq) as described previously. Briefly, for m6A-seq of the VSV genome and antigenome, RNAs were extracted from purified VSV virions and purified with the RiboMinus Eukaryote System v2 kit (Thermo Fisher). For m6A-seq of VSV mRNA, total RNAs were extracted from mock-infected or VSV-infected A549 cells, and polyadenylated RNAs were isolated using the Dynabeads mRNA DIRECT Purification kit (Thermo Fisher). Purified RNAs were sonicated with Bioruptor Pico (Diagenode) with 30 s on and 30 s off for 30 cycles, mixed with 1 μl of affinity-purified anti-m6A monoclonal antibody (Cell Signaling Tech) in IPP buffer (150 mM NaCl, 0.1% NP-40, 10 mM Tris-HCl, pH 7.4), and incubated for 2 h at 4°C. Enriched and input mRNA fragments were purified with RNA Clean & Concentrator kit (Zymo) and used for library generation with the TruSeq Stranded mRNA Library Prep kit (Illumina). Sequencing was carried out on an Illumina HiSeq 4000 according to the manufacturer’s instructions. Two replicates of RNA samples from virions, virus-infected cells, and mock-infected cells were subjected to m6A-seq. To prepare m6A-seq of SeV, total RNAs of rSeV-GFP/mock-infected A549 cells were subjected to sonication and m6A pulldown as described above. Enriched and input RNA fragments were further processed by SMARTer Stranded Total RNA-Seq kit v2—Pico Input Mammalian (TaKaRa). Sequencing was carried out on Illumina Nova-seq according to the manufacturer’s instructions. Three replicates of virus-infected cells were m6A sequenced. For data analysis, the reads were mapped to the VSV and SeV genome and antigenome by using Hisat2 (58) followed by peak calling using R package MeRIP tools (59). We reported peaks that are consistent across all replicates.
Determination of IFN-β by ELISA.
For virus infection, A549 cells or YTHDF2 knockout A549 cells were infected by SeV, hMPV, VSV, or MeV at an MOI of 1.0, cell supernatants were harvested at 16, 24, and 40 h after infection, and the IFN-β concentrations were determined by commercial enzyme-linked immunosorbent assays (ELISA) according to the manufacturer’s instructions (PBL). Known concentrations of human IFN-β were used to generate the standard curve. For RNA transfection, A549 cells or YTHDF2 knockout A549 cells in 24-well plates were transfected with equal amounts of virion RNA of SeV, hMPV, VSV, or MeV by using Lipofectamine 3000. At 16 and 24 h after transfection, culture medium was harvested for IFN-β quantification by ELISA.
Determination of IFN-β mRNA by RT-qPCR.
For viral infection, A549 cells were first treated or untreated with cycloheximide (CHX) at a concentration of 50 μg/ml 1 h before infection and then infected with SeV grown from METTL3 KO U2OS or WT U2OS cells at an MOI of 5. A 50-μg/ml concentration of CHX was maintained in the culture medium after inoculation. At 24 h postinfection, cell lysates were collected, and total RNA was extracted by TRIzol reagent following the manufacturer’s instructions. IFN mRNA was quantified by real-time RT-PCR and normalized with mRNA of β-actin.
Antibodies and Western blotting.
The antibodies used in this study were anti-METTL3 (Proteintech; 15073-1-AP), anti-RIG-I (Abcam; ab180675), anti-MDA5 (Abcam; ab79055), anti-IRF3(phospho-S386) (Abcam; ab76493), anti-IRF3 (Abcam; ab25950), anti-YTHDF2 (Abcam; ab220163), and antiactin (Proteintech; 66009). For viral RNA transfection, the same copies of viral RNA at different deses were transfected into regular A549 or A549-Dual KO cells using Lipofectamine 3000 (ThermoFisher Scientific, Waltham, MA). Cells were harvested and lysed in 1× radioimmunoprecipitation assay (RIPA) buffer (Abcam; ab156034). Protein expression was measured by Western blotting using antibody against RIG-I, MDA5, IRF3 and IRF3(phospho-S386). β-Actin was used as the loading control.
RIG-I RNA binding assay.
To assess the binding efficiency of viral RNA to RIG-I, an immunoprecipitation assay of RIG-I and viral RNA was conducted as described previously. Briefly, confluent 60-mm dishes of A549 cells were transfected with 2 μg of plasmid pEF-BOS-RIG-I-Flag (provided by J. Yount, The Ohio State University College of Medicine). At 24 h after transfection, cells were lysed in lysis buffer (Abcam; ab152163). Cell lysates were harvested after centrifugation at 13,000 × g for 10 min and incubated with anti-Flag M2 magnetic beads (Sigma-Aldrich; M8823) at room temperature for 60 min. The mixture was then divided into 7 aliquots (150 μl per tube). Four aliquots were incubated with 106 copies of SeV virion RNA, 108 copies of MeV virion RNA, or 108 copies of hMPV virion RNA at 37°C for 1 h, respectively. Beads with RNA:protein complex were washed in lysis buffer three times, and total RNA was extracted from beads using TRIzol reagent and quantified by real-time RT-PCR. The 7th aliquot was washed and analyzed by Western blotting.
Analysis of RIG-I:RNA complex in native SDS-PAGE.
Recombinant human RIG-I protein was purified from HEK-293T cells transfected with a plasmid encoding Flag-tagged RIG-I (pEF-BOS-RIG-I-Flag) and was incubated with 107 copies of virion RNA of virus grown from METTL3-KO U2OS cells or WT cells in 30 μl MOPS (morpholinepropanesulfonic acid)-buffered reaction system (10 mM MOPS, pH 7.4, 1 mM dithiothreitol [DTT], 1 mM MgCl2, 0.002% Tween 20) in the presence of RNase inhibitor and AMP-PNP (adenylyl-imidodiphosphate; 2 mM). The reaction mixtures were incubated at 37°C for 30 min to enable RIG-I:RNA complex formation. Ten microliters of the mixture was transferred into a new tube containing 10 μl of 2× native sample buffer (Bio-Rad; 1610738) and then loaded on 4∼15% Mini-Protean Tgx gel (Bio-Rad; 4561086) in glycine buffer (Bio-Rad; 1610734) for native PAGE, followed by Western blotting with anti-RIG-I helicase antibody to test the presence of RIG-I oligomer. Poly(I·C) (5 μg) was used as a positive control.
Limited proteolysis of RIG-I:RNA complexes using trypsin.
Limited trypsin digestion of RIG-I:RNA complex was carried out to examine the conformational change of RIG-I after it bound with viral RNA (14, 33). RIG-I:RNA complex was formed as described above. The complex was mixed with 8 μl of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (2.5 ng μl−1) and incubated at room temperature. At indicated time points (0 to 120 min), 10 μl was removed, mixed with 5× SDS-PAGE loading dye, separated by SDS-PAGE, and analyzed by Western blotting probed with an anti-RIG-I helicase antibody. Poly(I·C) (5 μg) was used as a positive control.
Determination of ubiquitination of RIG-I.
Ubiquitination plays essential roles in RIG-I activation, as well as the type I IFN signaling pathway. Here, we measured the activation of RIG-I by virion RNA using a ubiquitination assay as previously described. Specifically, purified RIG-I (1.0 mM) was first incubated with purified SeV virion RNA (1, 2, or 10 ng/μl) in buffer A (20 mM HEPES, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 2 mM ATP and 5 mM DTT) at room temperature for 15 min. The RIG-I:RNA complex (to the final RIG-I concentration of 0.5 mM) was then further incubated with 20 mM ubiquitin, 1 mM mE1, 5 mM ubiquitin-conjugating enzyme E2 13 (Ubc13), 2.5 mM ubiquitin-conjugating enzyme E2 variant 1A (Uev1A), and 0.25 mM E3 RIPLET in buffer B (50 mM Tris, pH 7.5, 10 mM MgCl2, 5 mM ATP, and 0.6 mM DTT) at 37°C for 30 min. Reactions were quenched with SDS loading buffer, and then the mixtures were boiled at 96°C for 5 min and analyzed on SDS-PAGE by anti-RIG-I or antiubiquitin Western blotting.
Statistical analyses.
All data were analyzed with GraphPad Prism v.8. The error bars represent standard deviations in all figures, and P values were determined using two-tailed Student’s t tests: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ***, P < 0.0001.
Data availability.
The raw sequencing data obtained from the MeRIP-seq for VSV and SeV have been deposited in the Gene Expression Omnibus under accession no. GSE162339 for VSV and GSE164882 for SeV.
ACKNOWLEDGMENTS
This study was supported by grants from the National Institutes of Health (no. R01AI090060 to J.L., no. P01 AI112524 to M.E.P. and J.L., no. R01 HG008688 and RM1 HG008935 to C.H., and no. R01 AI111784 to S.H. and S.A.). C.H. is an investigator of the Howard Hughes Medical Institute. H.W. is supported by a Roche fellowship.
We thank Yang Shi (Harvard Medical School) for the METTL3 knockout U2OS and WT U2OS cells, J. Yount (The Ohio State University) for the RIG-I plasmid, and members of the J. Li laboratory for critical reading of the manuscript.
C.H. is a scientific founder of Accent Therapeutics, Inc. J.L., C.H., and M.E.P. have filed a provisional patent (application no. 62/748,175).
REFERENCES
- 1.Whelan SP, Barr JN, Wertz GW. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol 283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- 2.Chatterjee S, Basler CF, Amarasinghe GK, Leung DW. 2016. Molecular mechanisms of innate immune inhibition by non-segmented negative-sense RNA viruses. J Mol Biol 428:3467–3482. doi: 10.1016/j.jmb.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li JR, Wang JT, Whelan SPJ. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc Natl Acad Sci U S A 103:8493–8498. doi: 10.1073/pnas.0509821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Rahmeh A, Morelli M, Whelan SPJ. 2008. A conserved motif in region V of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J Virol 82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 2006. 5'-triphosphate RNA is the ligand for RIG-I. Science 314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 6.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Sousa CRE. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science 314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 7.Ostertag D, Hoblitzell-Ostertag TA, Perrault J. 2007. Overproduction of double-stranded RNA in vesicular stomatitis virus-infected cells activates a constitutive cell-type-specific antiviral response. J Virol 81:503–513. doi: 10.1128/JVI.01218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Son KN, Liang ZG, Lipton HL. 2015. Double-stranded RNA is detected by immunofluorescence analysis in RNA and DNA virus infections, including those by negative-stranded RNA viruses. J Virol 89:9383–9392. doi: 10.1128/JVI.01299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee S, Luthra P, Esaulova E, Agapov E, Yen BC, Borek DM, Edwards MR, Mittal A, Jordan DS, Ramanan P, Moore ML, Pappu RV, Holtzman MJ, Artyomov MN, Basler CF, Amarasinghe GK, Leung DW. 2017. Structural basis for human respiratory syncytial virus NS1-mediated modulation of host responses. Nat Microbiol 2:17101. doi: 10.1038/nmicrobiol.2017.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao X, Liu T, Shan Y, Li K, Garofalo RP, Casola A. 2008. Human metapneumovirus glycoprotein G inhibits innate immune responses. PLoS Pathog 4:e1000077. doi: 10.1371/journal.ppat.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2'-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daffis S, Szretter KJ, Schriewer J, Li JQ, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong HP, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Shi PY, Diamond MS. 2010. 2'-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kariko K, Buckstein M, Ni H, Weissman D. 2005. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Durbin AF, Wang C, Marcotrigiano J, Gehrke L. 2016. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio 7:e00833-16. doi: 10.1128/mBio.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, Hur S, Chang HY. 2019. N6-Methyladenosine modification controls circular RNA immunity. Mol Cell 76:96–109.e9. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stepanov G, Zhuravlev E, Shender V, Nushtaeva A, Balakhonova E, Mozhaeva E, Kasakin M, Koval V, Lomzov A, Pavlyukov M, Malyants I, Zhorov M, Kabilova T, Chernolovskaya E, Govorun V, Kuligina E, Semenov D, Richter V. 2018. Nucleotide modifications decrease innate immune response induced by synthetic analogs of snRNAs and snoRNAs. Genes (Basel) 9:531. doi: 10.3390/genes9110531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue Y, Liu J, He C. 2015. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev 29:1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, Doxtader KA, Nam Y. 2016. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell 63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia GF, Fu Y, Zhao X, Dai Q, Zheng GQ, Yang Y, Yi CQ, Lindahl T, Pan T, Yang YG, He C. 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng GQ, Dahl JA, Niu YM, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, Lu ZK, Bosmans RPG, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia GF, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y, Dominissini D, Rechavi G, He C. 2014. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet 15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. 2015. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 2015. 5' UTR m(6)A promotes cap-independent translation. Cell 163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Q, Hou J, Zhou Y, Li Z, Cao X. 2017. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat Immunol 18:1094–1103. doi: 10.1038/ni.3830. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, Zhou Q, Cao X. 2019. Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat Commun 10:1898. doi: 10.1038/s41467-019-09903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, Nachshon A, Tai-Schmiedel J, Friedman N, Le-Trilling VTK, Trilling M, Mandelboim M, Hanna JH, Schwartz S, Stern-Ginossar N. 2019. m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol 20:173–182. doi: 10.1038/s41590-018-0275-z. [DOI] [PubMed] [Google Scholar]
- 29.Rubio RM, Depledge DP, Bianco C, Thompson L, Mohr I. 2018. RNA m(6)A modification enzymes shape innate responses to DNA by regulating interferon beta. Genes Dev 32:1472–1484. doi: 10.1101/gad.319475.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai K, Cullen BR. 2020. Epigenetic and epitranscriptomic regulation of viral replication. Nat Rev Microbiol 18:559–570. doi: 10.1038/s41579-020-0382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams GD, Gokhale NS, Horner SM. 2019. Regulation of viral infection by the RNA modification N6-methyladenosine. Annu Rev Virol 6:235–253. doi: 10.1146/annurev-virology-092818-015559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. 2016. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 1:16011. doi: 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu M, Zhang Z, Xue M, Zhao BS, Harder O, Li A, Liang X, Gao TZ, Xu Y, Zhou J, Feng Z, Niewiesk S, Peeples ME, He C, Li J. 2020. N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat Microbiol 5:584–598. doi: 10.1038/s41564-019-0653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue M, Zhao BS, Zhang Z, Lu M, Harder O, Chen P, Lu Z, Li A, Ma Y, Xu Y, Liang X, Zhou J, Niewiesk S, Peeples ME, He C, Li J. 2019. Viral N(6)-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nat Commun 10:4595. doi: 10.1038/s41467-019-12504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolakofsky D, Bruschi A. 1975. Antigenomes in Sendai virions and Sendai virus-infected cells. Virology 66:185–191. doi: 10.1016/0042-6822(75)90189-0. [DOI] [PubMed] [Google Scholar]
- 36.Mottet G, Roux L. 1989. Budding efficiency of Sendai virus nucleocapsids: influence of size and ends of the RNA. Virus Res 14:175–187. doi: 10.1016/0168-1702(89)90037-3. [DOI] [PubMed] [Google Scholar]
- 37.Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S, Jacob R, Devignot S, Kochs G, Garcia-Sastre A, Weber F. 2013. Incoming RNA virus nucleocapsids containing a 5'-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe 13:336–346. doi: 10.1016/j.chom.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr, Patel SS, Marcotrigiano J. 2011. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peisley A, Wu B, Yao H, Walz T, Hur S. 2013. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell 51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 41.Cadena C, Hur S. 2019. Filament-like assemblies of intracellular nucleic acid sensors: commonalities and differences. Mol Cell 76:243–254. doi: 10.1016/j.molcel.2019.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cadena C, Ahmad S, Xavier A, Willemsen J, Park S, Park JW, Oh SW, Fujita T, Hou F, Binder M, Hur S. 2019. Ubiquitin-dependent and -independent roles of E3 ligase RIPLET in innate immunity. Cell 177:1187–1200.e16. doi: 10.1016/j.cell.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei CM, Gershowitz A, Moss B. 1975. Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell 4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 44.Perry RP, Kelley DE, Friderici K, Rottman F. 1975. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5' terminus. Cell 4:387–394. doi: 10.1016/0092-8674(75)90159-2. [DOI] [PubMed] [Google Scholar]
- 45.Shi HL, Wei JB, He C. 2019. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell 74:640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, Ilkayeva OR, Law BA, Holley CL, Garcia-Blanco MA, Evans MJ, Suthar MS, Bradrick SS, Mason CE, Horner SM. 2016. N6-Methyladenosine in Flaviviridae viral RNA genomes regulates Infection. Cell Host Microbe 20:654–665. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hao H, Hao S, Chen H, Chen Z, Zhang Y, Wang J, Wang H, Zhang B, Qiu J, Deng F, Guan W. 2019. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res 47:362–374. doi: 10.1093/nar/gky1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiel V. 2020. Viral RNA in an m(6)A disguise. Nat Microbiol 5:531–532. doi: 10.1038/s41564-020-0689-x. [DOI] [PubMed] [Google Scholar]
- 49.Kim GW, Imam H, Khan M, Siddiqui A. 2020. N(6)-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J Biol Chem 295:13123–13133. doi: 10.1074/jbc.RA120.014260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao Y, Vasic R, Song Y, Teng R, Liu C, Gbyli R, Biancon G, Nelakanti R, Lobben K, Kudo E, Liu W, Ardasheva A, Fu X, Wang X, Joshi P, Lee V, Dura B, Viero G, Iwasaki A, Fan R, Xiao A, Flavell RA, Li HB, Tebaldi T, Halene S. 2020. m(6)A modification prevents formation of endogenous double-stranded RNAs and deleterious innate immune responses during hematopoietic development. Immunity 52:1007–1021.e8. doi: 10.1016/j.immuni.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 52.Baum A, Sachidanandam R, Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A 107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baum A, Garcia-Sastre A. 2011. Differential recognition of viral RNA by RIG-I. Virulence 2:166–169. doi: 10.4161/viru.2.2.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devarkar SC, Schweibenz B, Wang C, Marcotrigiano J, Patel SS. 2018. RIG-I uses an ATPase-powered translocation-throttling mechanism for kinetic proofreading of RNAs and oligomerization. Mol Cell 72:355–368.e4. doi: 10.1016/j.molcel.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ringeard M, Marchand V, Decroly E, Motorin Y, Bennasser Y. 2019. FTSJ3 is an RNA 2'-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565:500–504. doi: 10.1038/s41586-018-0841-4. [DOI] [PubMed] [Google Scholar]
- 56.Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR. 2019. Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26:217–227.e6. doi: 10.1016/j.chom.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong H, Chang DC, Hua MH, Lim SP, Chionh YH, Hia F, Lee YH, Kukkaro P, Lok SM, Dedon PC, Shi PY. 2012. 2'-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog 8:e1002642. doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z, Luo K, Zou Z, Qiu M, Tian J, Sieh L, Shi H, Zou Y, Wang G, Morrison J, Zhu AC, Qiao M, Li Z, Stephens M, He X, He C. 2020. Genetic analyses support the contribution of mRNA N(6)-methyladenosine (m(6)A) modification to human disease heritability. Nat Genet 52:939–949. doi: 10.1038/s41588-020-0644-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequencing data obtained from the MeRIP-seq for VSV and SeV have been deposited in the Gene Expression Omnibus under accession no. GSE162339 for VSV and GSE164882 for SeV.