Human immunodeficiency virus type 1 (HIV-1) remains a treatable, but incurable, viral infection. The establishment of viral reservoirs containing latently infected cells remains the main obstacle in the search for a cure.
KEYWORDS: HIV, interferon, MG1, macrophage, oncolytic virus
ABSTRACT
The use of unique cell surface markers to target and eradicate HIV-infected cells has been a longstanding objective of HIV-1 cure research. This approach, however, overlooks the possibility that intracellular changes present within HIV-infected cells may serve as valuable therapeutic targets. For example, the identification of dysregulated antiviral signaling in cancer has led to the characterization of oncolytic viruses capable of preferentially killing cancer cells. Since impairment of cellular antiviral machinery has been proposed as a mechanism by which HIV-1 evades immune clearance, we hypothesized that HIV-infected macrophages (an important viral reservoir in vivo) would be preferentially killed by the interferon-sensitive oncolytic Maraba virus MG1. We first showed that HIV-infected monocyte-derived macrophages (MDM) were more susceptible to MG1 infection and killing than HIV-uninfected cells. As MG1 is highly sensitive to type I interferons (IFN-I), we then investigated whether we could identify IFN-I signaling differences between HIV-infected and uninfected MDM and found evidence of impaired IFN-α responsiveness within HIV-infected cells. Finally, to assess whether MG1 could target a relevant, primary cell reservoir of HIV-1, we investigated its effects in alveolar macrophages (AM) obtained from effectively treated individuals living with HIV-1. As observed with in vitro-infected MDM, we found that HIV-infected AM were preferentially eliminated by MG1. In summary, the oncolytic rhabdovirus MG1 appears to preferentially target and kill HIV-infected cells via impairment of antiviral signaling pathways and may therefore provide a novel approach to an HIV-1 cure.
IMPORTANCE Human immunodeficiency virus type 1 (HIV-1) remains a treatable, but incurable, viral infection. The establishment of viral reservoirs containing latently infected cells remains the main obstacle in the search for a cure. Cure research has also focused on only one cellular target of HIV-1 (the CD4+ T cell) while largely overlooking others (such as macrophages) that contribute to HIV-1 persistence. In this study, we address these challenges by describing a potential strategy for the eradication of HIV-infected macrophages. Specifically, we show that an engineered rhabdovirus—initially developed as a cancer therapy—is capable of preferential infection and killing of HIV-infected macrophages, possibly via the same altered antiviral signaling seen in cancer cells. As this rhabdovirus is currently being explored in phase I/II clinical trials, there is potential for this approach to be readily adapted for use within the HIV-1 cure field.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) cure research has largely concentrated on the eradication of latently infected CD4+ T cells, while overlooking other cellular constituents of the viral reservoir. Tissue-resident macrophages, for example, contribute to HIV-1 persistence but remain poorly characterized (1–5). Although strategies by which to target these cells have been proposed (6–8), the development of a cure cannot proceed without a better understanding of the macrophage HIV-1 reservoir.
It has been well established that the ability of HIV-1 to infect and replicate within susceptible target cells is due, in part, to its ability to shut down antiviral cellular signaling pathways. As evidence of this, the degradation of key RNA-sensing proteins (9), downregulation of key antiviral interferon (IFN)-stimulated genes (ISG) (10, 11), and impaired release of type I IFN (IFN-I) (12–14), have all been observed within HIV-infected macrophages. In addition to its antiviral effects, impairment of the IFN-I response permits rapid cellular proliferation and survival (15) and is therefore a common feature of malignant cells. This observation has led to the development of therapeutic oncolytic viruses (OV) that are genetically engineered to be highly sensitive to IFN-I and can therefore be used to preferentially eradicate cancer cells while sparing healthy, nonmalignant cells (16, 17).
Given the parallels between malignant and HIV-infected cells, we questioned whether one such OV, the recombinant Maraba virus MG1, could be employed in the context of HIV-1 infection. We have previously shown that both HIV-infected cell lines and primary CD4+ T cells were preferentially infected and killed by MG1 (18, 19), suggesting that MG1 has therapeutic potential for the preferential eradication of the HIV-1 reservoir. We therefore hypothesized that HIV-infected, primary monocyte-derived macrophages (MDM) would also be preferentially infected and killed by MG1.
We first demonstrated the susceptibility of HIV-infected MDM to MG1-mediated killing by measuring markers of cell death and proviral HIV-1 DNA following OV infection. Cell death was not observed after exposure to UV-inactivated MG1, or to filtered supernatants from MG1-infected MDM, confirming that replication-competent MG1 was responsible for the preferential killing of HIV-infected cells. Because MG1 is highly sensitive to IFN-I, we questioned whether HIV-infected MDM possessed changes in IFN-I signaling that were not present in HIV-uninfected cells. Basal levels of two representative ISG were higher in HIV-infected MDM, but IFN-α-induced ISG expression in these cells was impaired, suggesting that alterations in antiviral signaling may be responsible for increased MG1-mediated cytopathogenicity. Finally, we demonstrated the ability of MG1 to eliminate HIV-infected alveolar macrophages (AM) from combination antiretroviral therapy (cART)-treated people living with HIV-1 (PLWHIV), which represent the only readily accessible source of tissue-resident myeloid cells implicated in HIV-1 persistence (2, 20–22).
RESULTS
HIV-infected monocyte-derived macrophages are preferentially infected by MG1.
We have previously shown that the oncolytic Maraba virus MG1 preferentially kills latently HIV-infected primary CD4+ T cells (19). Therefore, we questioned whether HIV-infected MDM could also be preferentially infected by MG1.
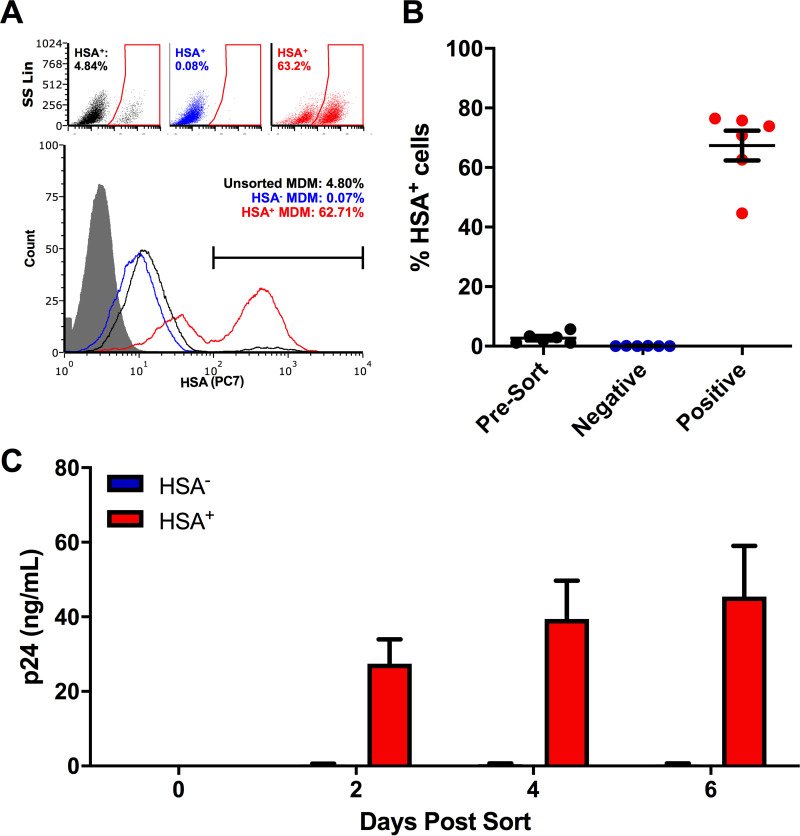
First, we established a model of HIV-1 infection in human MDM that allowed us to reliably differentiate between HIV-infected and HIV-uninfected bystander cells. Studies investigating HIV-1 infection of MDM cultures typically obtain results using bulk cell analysis, overlooking the fact that rates of infection in these cells are typically quite low (23, 24). To avoid this, we used the CCR5-tropic reporter virus HIV NL4.3 BAL-IRES-HSA. This virus has been engineered to express murine heat-stable antigen (HSA) (24), which localizes to the cell surface during productive HIV-1 infection and can be used to differentiate infected from bystander (uninfected) MDM. By 6 days postinfection (dpi), distinct HSA-positive (HSA+) and -negative (HSA−) populations could be detected by flow cytometry (Fig. 1A). Similar to others (25), we found that HSA+ MDM enriched by magnetic bead isolation continued to release HIV-1 p24 antigen into culture supernatants, while HSA− cells did not (Fig. 1B and C). Thus, HSA+ MDM were defined as HIV infected and HSA− MDM were defined as bystander populations.
FIG 1.
Enrichment of HSA+ MDM. (A) Representative dot plots and histogram showing the frequency of HSA+ MDM presort (black), as well as within the HSA-negative (blue) and HSA-enriched (red) cell fractions. The HSA isotype control is shown in gray. Histogram peak counts (y axis) were normalized to that of the isotype control for visualization purposes. SS Lin, side scatter, linear scale. (B) Purity of HSA-enriched MDM fraction postsort (n = 6). (C) HIV-1 p24 antigen release by enriched HSA+ and HSA− MDM (n = 4). Data represent mean ± standard error of the mean (SEM); n values represent separate biological replicates.
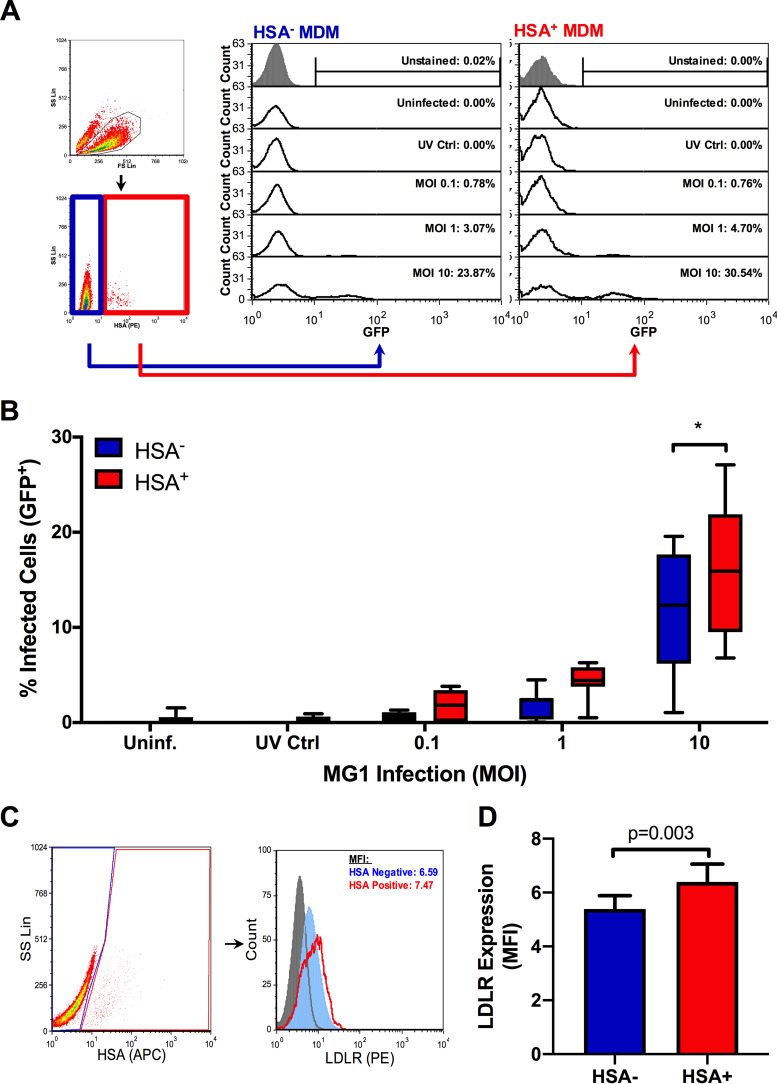
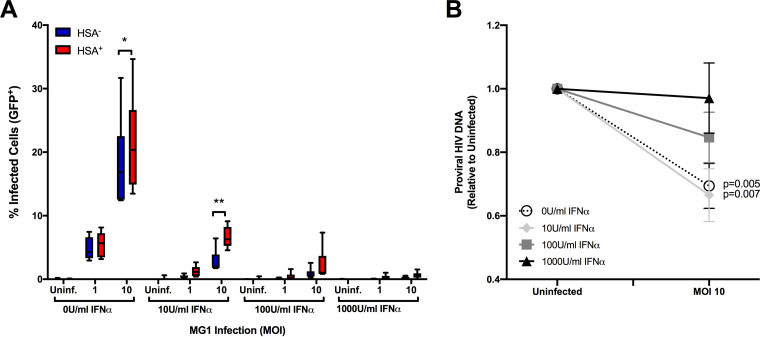
Using this model, bulk MDM cultures consisting of both HSA+ and HSA− cells were exposed to the green fluorescent protein (GFP)-expressing MG1 virus. After 48 h, the proportion of GFP-positive cells was assessed by flow cytometry (Fig. 2A, representative histograms). As evidence for the preferential infection of HIV-infected MDM by MG1, a significantly greater percentage of GFP-positive cells was seen in the HSA+ MDM population than in the HSA− cells (Fig. 2B). Since this preferential infection could be explained, in part, by elevated surface expression of the MG1 receptor on HIV-infected cells, we measured the expression of the low-density lipoprotein receptor (LDLR) (26–28) and found that its expression was slightly higher on HSA+ cells (Fig. 2C and D). Differences in MG1 infection may therefore be partially receptor mediated, although the biological relevance in this small difference in surface expression remains undetermined.
FIG 2.
MG1 preferentially infects HSA+ MDM. At 6 days post-HIV-1 infection, MDM cultures were infected with MG1 or left uninfected. UV-inactivated MG1 was included as an additional experimental condition (UV Ctrl.). UV-inactivated viral particles were added to MDM cultures at a ratio of 10:1, as calculated from pre-UV inactivation virus titers. At 48 h post-OV infection, frequencies of HSA+ and GFP+ MDM were measured by flow cytometry. (A) Example of gating strategy employed during data analysis. Intact cells were analyzed (black gate), after which HSA− (blue gate) and HSA+ (red gate) MDM were gated upon. The percentages of GFP+ cells were then measured within HSA+ and HSA− populations, as shown in representative histograms. Histogram peak counts (y axis) for HSA− and HSA+ populations were normalized to that of the uninfected control for visualization purposes. (B) Frequencies of GFP+ cells within HSA+ and HSA− MDM populations at 48 h post-MG1 infection (n = 7; P < 0.0001 by 2-way repeated-measures ANOVA; *, P < 0.001 by Bonferroni posttest). (C) Flow cytometry gating strategy and representative histograms depicting LDLR expression on HSA− (blue) and HSA+ (red) MDM. Intact cells were gated (black), after which HSA− (blue) and HSA+ (red) MDM were defined. The PE FMO control is shown as a filled, gray peak. Histogram peak counts (y axis) for HSA− and HSA+ populations were normalized to that of the PE FMO control for visualization purposes. (D) LDLR expression on HSA− (blue) and HSA+ (red) MDM (n = 7; P = 0.003 by paired, two-tailed t test). Data represent mean ± SEM; n values represent separate biological replicates.
MG1 preferentially kills HIV-infected monocyte-derived macrophages.
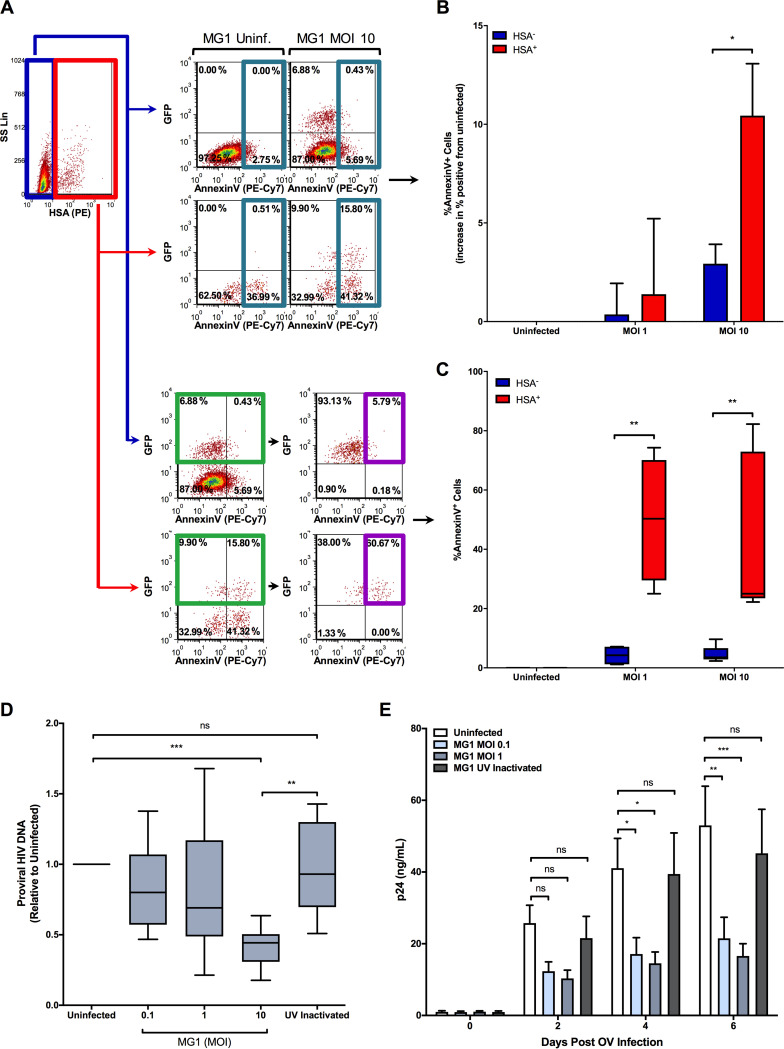
Oncolytic rhabdoviruses, including MG1, have been found to trigger cell death pathways in cancerous cell lines and solid tumors (29–31). To investigate whether preferential infection by MG1 would lead to increased killing of HIV-infected cells, we chose to perform annexin V staining to measure surface levels of phosphatidylserine, an early indicator of apoptosis, on MDM following MG1 infection. First, we assessed whether or not MG1 infection would increase the frequency of annexin V+ cells within HSA+ and HSA− MDM populations as a whole at 24 h post-MG1 infection. Not only did the frequency of annexin V+ cells increase in a multiplicity of infection (MOI)-dependent manner, but this frequency was higher within HSA+ MDM than HSA− MDM (Fig. 3A and B). In order to understand whether productive MG1 infection resulted in a difference in annexin V expression between HSA− and HSA+ MDM, we next measured annexin V staining on the GFP+ cells found within the HSA+ and HSA− cell gates and found that GFP+/HSA+ MDM contained a significantly higher percentage of annexin V+ cells than GFP+/HSA− MDM (Fig. 3C).
FIG 3.
MG1-infected, HSA+ MDM preferentially killed by MG1. (A) Representative dot plots depicting the gating strategy used to identify HSA−/GFP+/annexin V+ and HSA+/GFP+/annexin V+ MDM, as shown in panels B and C. HSA− (blue) and HSA+ (red) MDM were first defined by flow cytometry. To demonstrate that the surface expression of phosphatidylserine was increased following MG1 infection, the frequency of annexin V+ cells within the HSA− or HSA+ gates was measured at 24 h post-MG1 infection (top scatterplots; teal gates). To measure the amount of annexin V+ cells within the MG1-infected population, GFP+ cells were gated upon (bottom; green gate), and the frequency of annexin V+ MDM within HSA+/GFP+ and HSA−/GFP+ cell gates was measured (bottom; purple gate). (B) Cumulative data showing the difference in frequency of annexin V+ cells between MG1-infected and uninfected MDM cultures (n = 4; P = 0.013 by 2-way repeated-measures ANOVA; *, P < 0.05 by Bonferroni posttest). (C) Cumulative data showing the frequency of annexin V+ cells within HSA+/GFP+ and HSA−/GFP+ MDM gates. This was shown as the percentage of HSA−/GFP+/annexin V+ MDM and HSA+/GFP+/annexin V+ MDM at 24 h post-MG1 infection (uninfected and at an MOI of 10, n = 5; at an MOI of 1, n = 4; P = 0.004 by 2-way repeated-measures ANOVA; **, P < 0.01 by Bonferroni posttest). (D) Proviral HIV-1 DNA, measured relative to the MG1-uninfected control, at 48 h post-MG1 infection (n = 10; P = 0.0004 by 1-way repeated-measures ANOVA; **, P < 0.01; ***, P < 0.001 by Bonferroni posttest). (E) Quantification of HIV-1 p24 antigen in culture supernatants following MG1 infection (uninfected, MOI 0.1, and MOI 1, n = 6; UV inactivated, n = 4; P < 0.0001 by 2-way ANOVA; *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Bonferroni posttest; ns, not significant). Data represent mean ± SEM; n values represent separate biological replicates.
As has been previously demonstrated (19, 32–34), the killing of HIV-infected cells was then confirmed by measuring integrated (proviral) HIV-1 DNA at 2 days post-MG1 infection. Consistent with the annexin V staining, an MOI-dependent decrease in proviral HIV-1 DNA was observed in HIV-infected MDM that had been exposed to infectious MG1 (Fig. 3D). The release of HIV-1 p24 antigen into cell supernatants was also inhibited by MG1 infection (Fig. 3E). Importantly, UV-inactivated MG1 had no effect on proviral HIV-1 DNA or p24 release, indicating that replication-competent oncolytic virus was required for the killing of HIV-infected MDM. Together, these data indicate that MG1 infection results in the preferential death of HIV-infected MDM.
MG1-mediated killing of HIV-infected monocyte-derived macrophages is not mediated by soluble factors.
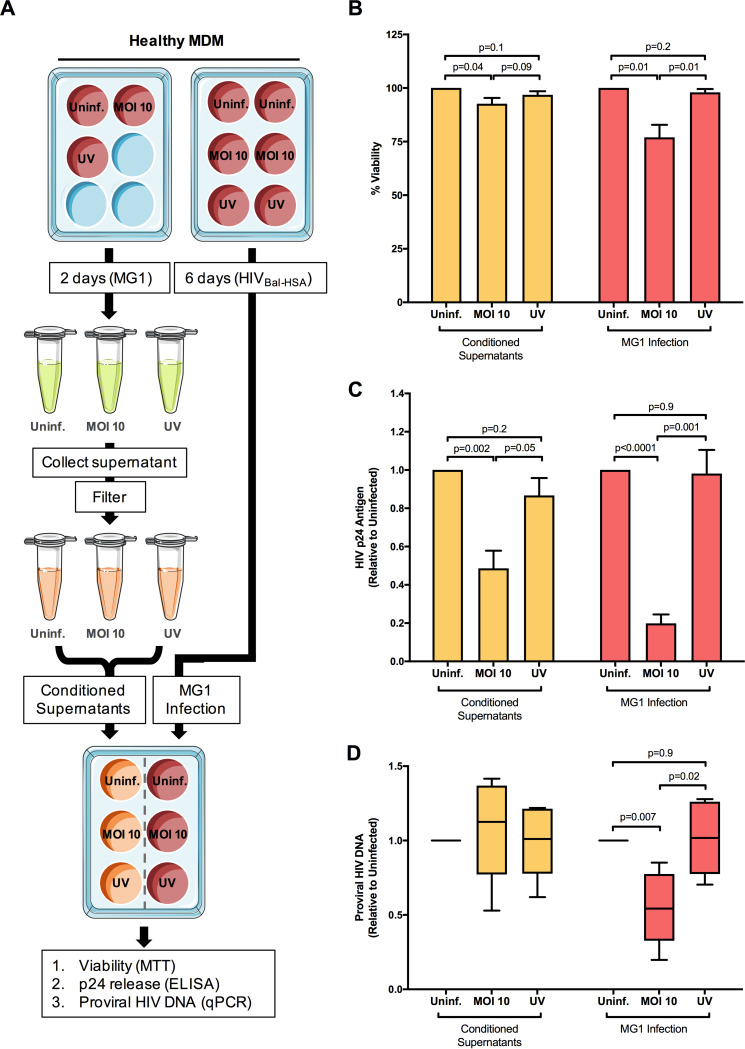
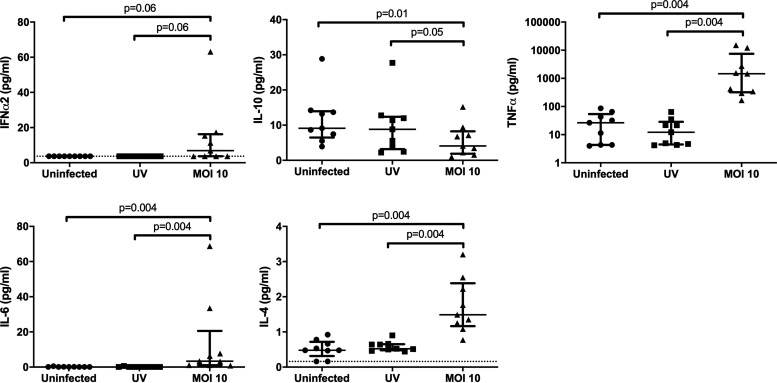
Because cell death was observed in both GFP+/HSA+ and GFP−/HSA+ cells, we considered the possibility that MG1 infection was also causing the indirect, cytokine-mediated killing of HIV-infected cells. To address this, we performed supernatant transfer experiments, as depicted in Fig. 4A. Cells that received conditioned supernatants from MG1-infected MDM showed a small but significant drop in viability as measured by the MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay (Fig. 4B). Both MG1 infection and conditioned supernatants also prevented the accumulation of HIV-1 p24 antigen in culture medium (Fig. 4C), but only MG1 infection resulted in a decrease in proviral HIV-1 DNA (Fig. 4D). Additionally, no effect on HIV-1 p24 release or proviral HIV-1 DNA was seen following exposure to either UV-inactivated MG1 or supernatants from MDM exposed to UV-inactivated MG1. Measurement of cytokines in filtered supernatants showed that concentrations of IFN-α2, tumor necrosis factor alpha (TNF-α), interleukin 4 (IL-4), and IL-6 were elevated at 48 h post-MG1 infection (Fig. 5). IFN-γ was also measured but was undetectable in all samples analyzed. In summary, the eradication of HIV-infected MDM is not mediated by soluble factors and requires the presence of infectious MG1.
FIG 4.
Conditioned supernatants from MG1-infected, HIV-1-negative MDM block p24 release without the preferential killing of HIV-infected MDM. (A) Supernatant transfer experimental workflow. MDM from healthy donors were plated in duplicate and either infected with MG1 (MOI of 10, UV-inactivated MG1 [UV], or uninfected [Uninf.]) or infected with HIV NL4.3 BAL-IRES-HSA. UV-inactivated viral particles were added to MDM cultures at a ratio of 10:1. Supernatants from MG1-infected MDM were collected at 2 dpi, filtered with Amicon ultra centrifugal filter units (MWCO, 100 kDa), and stored at −80°C. At 6 dpi, HIV-infected MDM cultures were infected with MG1 at an MOI of 10, left uninfected, or treated with UV-inactivated MG1. In duplicate, HIV-infected MDM were treated with filtered supernatants collected from autologous HIV-1-uninfected MDM. At 48 h post-MG1 infection, cell viability was assessed by the MTT assay, and proviral HIV-1 DNA was measured by qPCR. In certain experiments, cell supernatants were collected at 0, 2, 4, and 6 days post-MG1 infection for the measurement of HIV-1 p24 release by ELISA. (B) MDM viability, as measured by the MTT assay relative to the respective uninfected control, at 48 h post-MG1 infection or supernatant transfer. (C) HIV-1 p24 antigen in culture supernatants, relative to MG1-uninfected control, at 6 days post-MG1 infection or supernatant transfer. (D) Proviral HIV-1 DNA, measured relative to the respective uninfected control, at 48 h post-MG1 infection or supernatant transfer. n = 6; P values were calculated by a paired, two-tailed t test. Data represent mean ± SEM; n values represent separate biological replicates.
FIG 5.
MG1 infection induces cytokine secretion by MDM. Supernatants were collected at 48 hpi from MG1-infected, HIV-uninfected MDM, and concentrations of IFN-α2, IL-10, TNF-α, IL-6, and IL-4 were measured via Luminex technology. n = 9; P values were calculated by Wilcoxon matched-pairs signed rank test (nonparametric), Data represent median with interquartile range; n values represent separate biological replicates.
ISG induction is impaired within HIV-infected monocyte-derived macrophages.
As MG1 is highly sensitive to IFN-I (17), we next wanted to determine if IFN-I signaling influenced OV-mediated cytopathogenicity. We began by assessing MG1 infection and killing in IFN-α-pretreated MDM cultures and found that stimulation with IFN-α blocked both MG1 infection (Fig. 6A) and the reduction of proviral HIV-1 DNA in a dose-dependent manner (Fig. 6B). Interestingly, preferential infection and the corresponding reduction in proviral HIV-1 DNA were maintained after treatment with low doses of IFN-α, leading us to investigate whether or not differences in ISG expression existed between HIV-infected and bystander cells.
FIG 6.
IFN-α protects HIV-infected and bystander MDM from MG1 infection and killing in a dose-dependent manner. (A) To assess the effect of IFN-I on MG1 infection of HSA− and HSA+ MDM, cells were pretreated with increasing doses of IFN-α 24 h prior to MG1 infection. Frequencies of GFP+ MDM within HSA− and HSA+ cell populations at 48h post-MG1 infection were then measured by flow cytometry (n = 6; P < 0.0001 by 2-way repeated-measures ANOVA; *, P < 0.05, **, P < 0.01 by Bonferroni posttest.). (B) Proviral HIV-1 DNA, measured relative to MG1-uninfected control, at 48 h post-MG1 infection. MDM were pretreated with IFN-α 24 h prior to MG1 infection (n = 7; P values were calculated by paired, two-tailed t test). Data represent mean ± SEM; n values represent separate biological replicates.
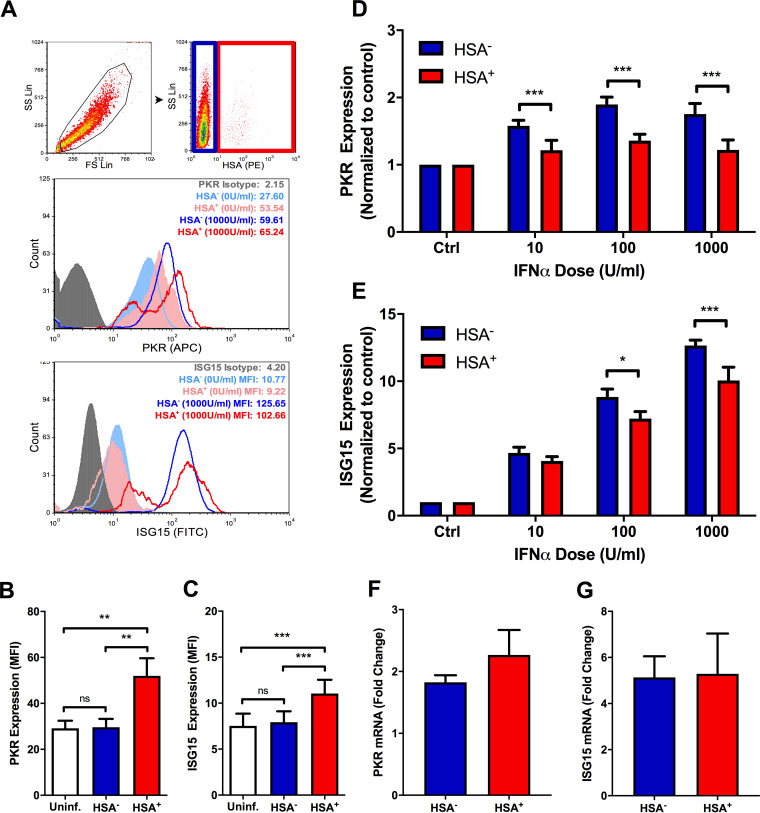
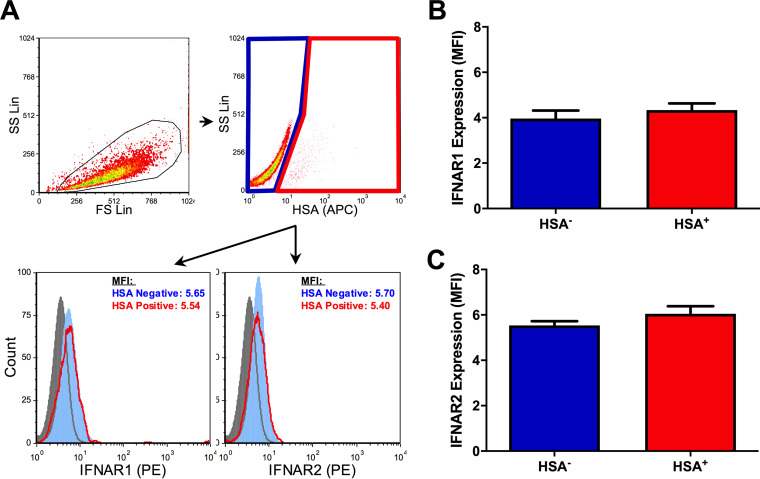
To do this, HIV-1-infected MDM cultures were stimulated with IFN-α, and levels of two representative ISG products, PKR and ISG15, were measured by flow cytometry (Fig. 7A). Basal expression of both proteins was higher in HSA+ MDM than in both HSA− MDM and HIV-1 naive MDM (Fig. 7B and C). Conversely, the relative IFN-α-induced expression of PKR and ISG15 was lower in HSA+ MDM (Fig. 7D and E), indicating impaired IFN-α responsiveness in HIV-infected cells. This was not associated with differences in ISG15 or PKR mRNA levels in sorted HSA+ and HSA− MDM (Fig. 7F and G). Additionally, surface expression of the IFN-α/β receptor (subunits 1 and 2; IFNAR1/2) did not differ between HSA+ and HSA− cells (Fig. 8), suggesting that differences in ISG induction in these cell populations was not due to receptor downregulation.
FIG 7.
Differences in basal and IFN-α-induced ISG expression exist between HIV-infected and bystander MDM. (A) Flow cytometry gating strategy. Intact cells were gated (black), after which HSA− (blue) and HSA+ (red) MDM were defined. Representative histograms show PKR (top) and ISG15 (bottom) induction in IFN-α-stimulated MDM. Respective isotype controls are shown in gray; HSA− MDM are shown in blue (filled, unstimulated; open, stimulated); HSA+ MDM are shown in red (filled, unstimulated; open, stimulated). Histogram peak counts (y axis) for HSA− and HSA+ populations were normalized to that of the isotype control for visualization purposes. SS Lin, side scatter, linear scale; FS Lin, forward scatter, linear scale. (B) Basal PKR expression in uninfected (white), HSA− (blue), and HSA+ (red) MDM (n = 6; P = 0.001 by 1-way repeated measures ANOVA; **, P < 0.01 by Bonferroni posttest). (C) Basal ISG15 expression in uninfected (white), HSA− (blue), and HSA+ (red) MDM (n = 7; P = 0.0001 by 1-way repeated measures ANOVA; ***, P < 0.001 by Bonferroni posttest). (D) Relative PKR induction following 24 h of IFN-α stimulation, normalized to respective unstimulated controls (n = 6; P = 0.023 by 2-way repeated measures ANOVA; ***, P < 0.001 by Bonferroni posttest.). (E) Relative ISG15 induction following 24 h of IFN-α stimulation, normalized to respective unstimulated controls (n = 7; P < 0.0001 by 2-way repeated measures ANOVA, *, P < 0.05, ***, P < 0.001 by Bonferroni posttest; ns, not significant). (F) PKR mRNA measured in HSA+ and HSA− MDM at 16 h poststimulation with 1,000 U/ml IFN-α. (G) ISG15 mRNA measured in HSA+ and HSA− MDM at 16 h poststimulation with 1,000 U/ml IFN-α. ΔΔCTs used to determine fold change were calculated by normalizing threshold cycle (CT) values to those of the respective untreated control and GAPDH (n = 4). Data represent mean ± SEM; n values represent separate biological replicates.
FIG 8.
Surface expression of IFNAR1/2 on HSA+ and HSA− MDM. (A) Flow cytometry gating strategy and representative histograms (filled gray, PE FMO control; filled light blue, HSA− MDM; empty red, HSA+ MDM) depicting IFNAR1 and IFNAR2 expression on HSA− and HSA+ MDM. Intact cells were gated (black), after which HSA− (blue) and HSA+ (red) MDM were defined. Histogram peak counts (y axis) for HSA− and HSA+ populations were normalized to that of the PE FMO control for visualization purposes. (B) IFNAR1 expression on HSA− (blue) and HSA+ (red) MDM, as measured by mean fluorescent intensity (n = 7). (C) IFNAR2 expression on HSA− and HSA+ MDM, as measured by mean fluorescent intensity (n = 7). Data represent mean ± SEM; n values represent separate biological replicates.
MG1 eliminates HIV-infected alveolar macrophages ex vivo.
Finally, to begin to address the potential clinical implications of our findings, we investigated whether MG1-mediated killing of HIV-infected myeloid cells could be replicated in primary alveolar macrophages. As one of the only accessible sources of tissue-resident myeloid cells, AM are known to harbor replication-competent HIV-1 and thus act as a viral reservoir (2, 20–22, 35). To assess whether HIV-infected AM were susceptible to MG1-mediated killing, cells were collected from PLWHIV (on suppressive ART for ≥3 years at the time of collection) via bronchoalveolar lavage, isolated by plate adherence, and infected with MG1.
Following MG1 infection of adherent AM, proviral HIV-1 DNA was measured by digital droplet PCR (ddPCR). As has been previously demonstrated using cells collected from the lungs of PLWHIV (35), the frequency of HIV-infected AM was highly variable. Despite a relatively small number of participants, a statistically significant decrease in proviral HIV-1 DNA was observed at 48 h post-MG1 infection (Fig. 9), emphasizing the consistency of this effect. Additionally, MG1 infection did not appreciably reduce CD3 DNA copy number (as quantified by ddPCR) in comparison to MG1-uninfected AM, suggesting that HIV-uninfected AM were not killed by MG1. As primary, tissue-resident macrophages have been historically overlooked in HIV-1 cure research (reviewed in references 36), these data provide important insights into the future application of MG1 as an HIV-1 cure strategy.
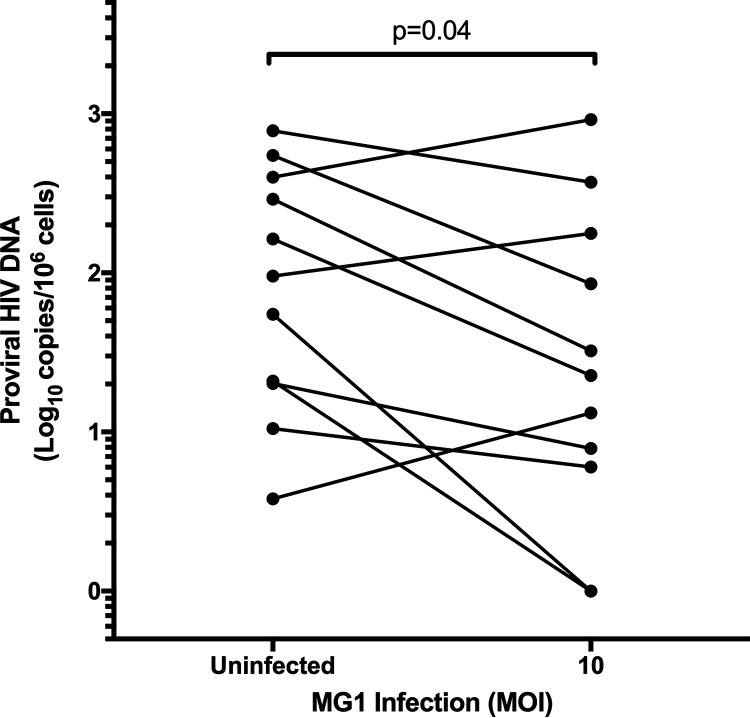
FIG 9.
MG1 infection reduces proviral HIV-1 DNA in alveolar macrophages from PLWHIV. Alveolar macrophages were allowed to adhere for 2 h at 37°C following collection by bronchoalveolar lavage. Subsequently, nonadherent cells were removed by washing, and adherent alveolar macrophages were infected with MG1 at an MOI of 10. Cell pellets were collected at 48 h postinfection, and proviral HIV DNA was measured by ddPCR. n = 11 (representative of separate biological replicates); P value calculated by paired, two-tailed t test.
DISCUSSION
The development of an HIV-1 cure has focused largely on the identification, characterization, and eradication of latently infected CD4+ T cells. Unfortunately, many of these strategies remain untested in other cellular reservoirs of HIV-1. We therefore sought to identify whether MG1, an established oncolytic rhabdovirus capable of targeting latently HIV-infected cell lines and primary CD4+ T cells (19), could also infect and kill HIV-infected macrophages.
In agreement with our previous findings in CD4+ T cells, we observed a higher frequency of MG1 infection in HSA+ (HIV-infected) MDM than in HSA− (bystander) MDM. This coincided with the preferential killing of HIV-infected MDM, as measured by an increase in annexin V+ cells and a reduction in proviral HIV-1 DNA. Next, we investigated whether MG1-mediated killing of HIV-infected MDM could be achieved via indirect means. UV-inactivated viral particles, for instance, have been found to induce the cytolytic killing of acute myeloid leukemia cells in vitro and in a murine model of leukemic blast crisis (37). OV-induced release of proinflammatory cytokines may also facilitate the indirect killing of tumor cells (38). In the context of HIV-1 infection, however, UV-inactivated MG1 did not kill HIV-infected MDM (Fig. 3D) or stimulate the release of proinflammatory cytokines such as TNF-α or IL-6 (Fig. 5).
Similarly, treatment of HIV-infected cells with conditioned supernatants from MG1-infected MDM did not eliminate HIV-infected cells but resulted in a significant reduction in HIV-1 p24 antigen release (Fig. 4C and D). IFN-α2, TNF-α, IL-4, and IL-6 were elevated in supernatants from MG1-infected cells, which is consistent with previous literature describing cytokine release by cells infected with MG1 or with the closely related rhabdovirus, vesicular stomatitis virus (31, 39, 40). Cytokines including TNF-α, IL-6, and IFN-γ have also been shown to block HIV replication in human macrophages (41–44), suggesting that these soluble factors are responsible for the reduction in p24 release following supernatant transfer. Given that the eradication of HIV-infected MDM requires the presence of replication-competent MG1, avoiding off-target effects by enhancing the selectivity of MG1 for HIV-infected cells remains an important objective that is under investigation by our group. Nonetheless, healthy cynomolgus macaques were found to tolerate MG1 doses of 1,011 PFU, providing promise for the ongoing investigation of OV as an HIV-1 cure strategy (45).
Next, we investigated the role of IFN-I signaling in MG1 infection. The MG1 virus is highly sensitive to IFN-I signaling due to two genetically engineered amino acid substitution mutations within the G (Q242R) and M (L123W) proteins (17). It was therefore no surprise that IFN-α pretreatment protected both HSA+ and HSA− MDM from MG1 infection in a dose-dependent manner (Fig. 6).
To investigate whether we could identify global IFN-I signaling differences between HSA+ and HSA− MDM, we measured the basal and IFN-α-induced expression of two representative ISG. The observed baseline elevation of PKR and ISG15 in HSA+ MDM was as expected, given that ISG induction has been shown to occur within the first 24 h of HIV-1 infection in MDM (46). Nonetheless, it was interesting to see that higher levels of ISG were not sufficient to block preferential MG1-mediated cytopathogenicity, as elevated ISG expression in malignant cells has been found to be protective against OV infection and killing (47). One possible explanation is the functional impairment of ISG in HIV-infected MDM. The interaction of PKR with a number of cellular and viral proteins in HIV-1-infected cells has already been shown to block this ISG from fulfilling its role as a key RNA sensing protein needed for the activation of cellular antiviral response pathways (48–51). Additionally, given that pretreatment with low levels of IFN-α did not prevent MG1-mediated killing of HIV-infected cells (Fig. 6B), and that HIV-infected MDM are impaired in their ability to produce IFN-I (14), it is possible that additional defects in antiviral signaling that sensitize these cells to OV infection exist.
As evidence of altered IFN-I signaling in HIV-infected MDM, we also showed that the IFN-α-mediated induction of ISG15 and PKR was impaired in HSA+ MDM. This is consistent with findings previously reported by Ranganath et al. in latently HIV-infected, pro-myeloid OM10.1 cells (18). IFN-α/β receptor downregulation (52, 53) and impaired STAT1/2 activation (10, 54, 55) have been found to prevent IFN-I-induced ISG mRNA expression in the context of HIV-1 infection. No differences in IFNAR1/2 surface expression or PKR and ISG15 mRNA were observed between HSA+ and HSA− MDM, however, suggesting a posttranscriptional mechanism of impairment. In support of this hypothesis, HIV-1-mediated inhibition of cellular translational machinery has been previously reported (56–59). The small number of HSA+ MDM obtained via magnetic bead separation, as well as the fragility of these cells postisolation, was an experimental limitation that unfortunately prevented the further study of translational defects in HIV-infected cells. Nonetheless, these data remain unique in their use of a reporter virus model to study and compare IFN-I signaling in HIV-infected and bystander MDM populations.
Finally, we demonstrated that MG1 could eliminate HIV-infected macrophages ex vivo, using AM from ART-treated PLWHIV. AM form a unique source of accessible, tissue-resident myeloid cells that form a known HIV-1 reservoir (60). The ability to eradicate these cells using an IFN-I-sensitive OV therefore has important implications for the development of an HIV-1 cure. Low cell numbers, and the relative fragility of cells obtained by bronchoalveolar lavage, unfortunately prevented further investigation of markers of MG1 infection, cell death, and IFN-I signaling, as was possible with in vitro HIV-infected MDM. This, combined with the relatively small number of donors recruited for bronchoalveolar lavage and AM collection, represents a current limitation of this study objective. Still, the decision to assess MG1-mediated elimination of HIV-infected cells using primary AM from PLWHIV represents an important step forward in the field of HIV-1 cure research. Primary, tissue-resident macrophages have consistently been overlooked as a source of replication-competent HIV-1 and, consequently, are a necessary target for novel HIV-1 cure strategies. Future work assessing the efficacy of latency reversal agents, allogeneic stem cell transplant, or other gene therapy approaches on HIV-1 reservoir size must therefore consider the myeloid HIV-1 reservoir as part of the experimental design (36).
In summary, these results demonstrate the preferential infection and killing of HIV-infected MDM by the IFN-I-sensitive oncolytic virus MG1. In doing so, we have fulfilled a current objective in HIV-1 cure research, which is to consider and account for myeloid HIV-1 reservoirs when testing new therapeutic strategies that have already been investigated in CD4+ T cells (36). Going forward, the ability of MG1 to reduce the size of the HIV-1 reservoir will need to be further assessed using a relevant in vivo model of HIV-1 infection. Should ongoing phase I/II clinical trials of MG1 in cancer (ClinicalTrials registration numbers NCT02285816, NCT02879760, and NCT03618953) identify this oncolytic virus as safe, a proof-of-concept clinical trial to investigate the safety and impact on HIV reservoir size in PLWHIV may be feasible.
MATERIALS AND METHODS
Reagents.
Gibco RPMI 1640, Gibco Dulbecco’s modified Eagle medium (DMEM), and Gibco phosphate-buffered saline (PBS), pH 7.4, were purchased from Life Technologies (Carlsbad, CA). Gibco Opti-MEM reduced serum medium was purchased from ThermoFisher Scientific. Additional reagents include heat-inactivated fetal bovine serum (FBS), normal goat serum (NGS), penicillin (100 U/ml) and streptomycin (100 µg/ml) (PenStrep), and l-glutamine (all from Life Technologies), heat-inactivated human AB serum (Valley Biomedical, Winchester, VA), universal type I alpha interferon (IFN-α) (PBL Assay Science, Piscataway, NJ), recombinant human macrophage colony-stimulating factor (M-CSF; carrier-free) (BioLegend catalog no. 574802), maraviroc (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH; catalog no. 11580), and Lipofectamine 2000 transfection reagent (Invitrogen, Burlington, ON, Canada).
Ethics statement.
Experiments requiring healthy volunteers were approved by the Ottawa Health Science Network Research Ethics Board (protocol no. 2005388-01H), and all participants provided written informed consent. The collection and use of alveolar macrophages from PLWHIV were approved by the Institutional Review Boards of the MUHC (no. 15-031) and the Université du Québec à Montréal (no. 602). All study participants provided written informed consent.
Cell culture.
Vero (CCL-81) and HEK293T cells (CRL-3216) were obtained from the American Type Culture Collection (Manassas, VA) and cultured in DMEM with 10% fetal calf serum (FCS) and PenStrep. ACH-2 cells were obtained via the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from Thomas Folks (61, 62) and cultured in RPMI 1640 supplemented with 10% FCS, PenStrep, and l-glutamine (2 mM). Cells were maintained at 0.2 × 105 to 1 × 106 cells/ml by passaging every 2 to 3 days.
Monocytes were separated from healthy donor peripheral blood mononuclear cells (PBMC) by plate adherence. Following isolation by density gradient centrifugation, PBMC were resuspended at 6.25 × 106/ml in warm, serum-free RMPI 1640 with PenStrep. A total of 1.25 × 108 PBMC were then plated in 150-cm2 polystyrene tissue culture dishes (Sarstedt, Nümbrecht, Germany) and left to adhere for 2 h at 37°C. Plates were washed 3 times with endotoxin-free PBS (pH 7.4; Gibco) to remove nonadherent lymphocytes, and 20 ml of warmed RPMI 1640, supplemented with PenStrep and 10% heat-inactivated human AB serum (Mϕ medium) and M-CSF (25 U/ml), was added to the plate. Adherent cells were incubated at 37°C with 5% CO2 for 7 days. At 3 days postplating, cells were washed twice with warmed endotoxin-free PBS, and 20 ml of Mϕ medium was added to the plate. On day 8, adherent MDM were washed twice with endotoxin-free PBS, detached using Accutase (Millipore-Sigma) and gentle scraping with a Sarstedt cell scraper, and counted by trypan blue exclusion. MDM were then pelleted by centrifugation (300 × g for 10 min), resuspended at 2.5 × 105 cells/ml in Mϕ medium, and plated in either 6-well (5 × 105 cells/well) or 12-well (2.5 × 105 cells/well) plates for further experiments.
AM were isolated from bronchoalveolar lavage fluid by plate adherence, as described previously (35, 63, 64). Briefly, participants recruited at the McGill University Health Centre (MUHC, Montreal, Canada) were cART-treated PLWHIV, with suppressed plasma viral load for ≥3 years and without respiratory symptoms or active illness. A total of 50 to 100 ml of lavage fluid was collected during bronchoscopy. Cells were pelleted and washed at 180 × g for 10 min and then counted by trypan blue exclusion. Cells were then resuspended in serum-free RPMI 1640 at 5 × 105 cells/ml and plated in 24-well plates at 2.5 × 105 cells/well for 2 h at 37°C. Nonadherent cells were removed by rinsing the wells with endotoxin-free PBS, and adherent cell were covered with 500 µl of Mϕ medium. Prior to collection, adherent AM were rinsed an additional 3 times using endotoxin-free PBS, in order to remove cellular debris and nonadherent lymphocytic cells. AM were detached at 37°C for 30 min using CellStripper dissociation reagent (Corning, Fisher Scientific), followed by gentle pipetting.
Production of virus stocks.
The HIV NL4.3 BAL-IRES-HSA plasmid was obtained from Michel J. Tremblay at Université Laval. HEK293T cells were seeded at 2 × 106 cells/T75 flask and transfected with 20 µg of purified plasmid using Lipofectamine 2000 and Opti-MEM I reduced serum medium, in accordance with the manufacturer’s instructions. Supernatants were collected at 48 h posttransfection and filtered through 0.45-µm and 0.22-µm polyvinylidene fluoride (PVDF) filters (UltiDent Scientific, St. Laurent, QC, Canada). HIV-1 p24 concentration was quantified by ELISA.
The GFP-expressing recombinant OV MG1 (obtained from John Bell and David Stojdl) was propagated and titered on Vero cells, as described previously (17, 65). UV inactivation of MG1 stocks was performed as described previously (19).
In vitro HIV-1 infection and enrichment of HSA+ MDM.
MDM were infected with HIV NL4.3 BAL-IRES-HSA for 6 days at 37°C with 5% CO2. HIV-1 infection was confirmed by quantitative PCR (qPCR) (66), p24 ELISA, and surface expression of virus-encoded murine heat-stable antigen (HSA) by flow cytometry (24). HSA-expressing MDM were isolated by positive selection using Miltenyi LS columns in combination with the MidiMACS magnet (Miltenyi Biotech). The HSA sorting protocol was optimized by the laboratory of Michel Tremblay, as described previously (24, 25). Purity of the HSA+ and HSA− fractions was assessed by flow cytometry.
In vitro MG1 infection and supernatant transfer experiments.
MDM and AM were infected with MG1 in RPMI 1640, supplemented with PenStrep and 10% heat-inactivated human AB serum (Mϕ medium), with 10 µM maraviroc. Cells pellets were collected at 48 hpi and stored at −80°C for quantification of HIV-1 DNA. MG1 infection, frequency of HSA-expressing cells, and cell viability were assessed in MDM cultures by flow cytometry and MTT assay.
For supernatant transfer experiments, MDM from healthy donors were infected with either MG1 or HIV NL4.3 BAL-IRES-HSA. At 48 hpi, supernatants from MG1-infected cells were collected, filtered with Amicon Ultra centrifugal filter units (molecular weight cutoff [MWCO], 100 kDa; Millipore Sigma) and stored at −80°C. Removal of infectious MG1 was confirmed on Vero cells by flow cytometry. At 6 dpi, HIV-infected MDM cultures were infected with MG1 at an MOI of 10, left uninfected, or treated with UV-inactivated MG1. In duplicate, HIV-infected MDM were treated with filtered supernatants, at a 1:1 ratio with fresh Mϕ medium. At 48 h post-MG1 infection, cell viability was assessed by MTT assay and proviral HIV-1 DNA was measured by qPCR. Cell-free supernatants were collected every 2 days for quantification of HIV-1 p24 antigen by ELISA. MDM were pelleted at 6 dpi and stored at −80°C for quantification of proviral DNA.
Measurement of supernatant cytokines.
IFN-α2, IFN-γ, TNF-α, IL-4, IL-6, and IL-10 were measured in filtered supernatants collected at 2 dpi from MG1-infected, HIV-uninfected MDM using the Milliplex human cytokine/chemokine/growth factor panel A kit (catalog no. HCYTA-60K-06; Millipore). The panel was read using Luminex technology on a MAGPIX system (Luminex xPONENT for MAGPIX version 4.2; Luminex Corporation, Austin, TX), Analysis was performed via Milliplex Analyst version 5.1.0.0 software (Vigenetech Inc., Carlisle, MA).
IFN-α stimulation of MDM.
MDM were stimulated with IFN-α at 6 days post-HIV-1 infection. IFN-α was diluted in Mϕ medium and added directly to the wells, after which MDM were incubated at 37°C for 16 h for mRNA isolation or 24 h for flow cytometry analysis.
Flow cytometry.
Analysis was performed using the FC500 Beckman Coulter flow cytometer (Beckman Coulter) and FCS Express Research edition 4.0 (De Novo Software, Los Angeles, CA). In preparation for staining, MDM were detached with Accutase and gentle pipetting, aliquoted into polypropylene tubes (1 × 105 cells/tube), and washed with PBS–1% BSA (460 × g for 5 min).
For HSA staining, cells were resuspended in cold PBS plus 10% human AB serum and 20% NGS and incubated at 4°C for 20 min. The anti-HSA antibody (clone M1/69; BioLegend) or HSA isotype control antibody (rat IgG2b κ isotype clone RTK4530; BioLegend) was then added at 3 µg/ml, and cells were incubated at 4°C for 15 min. Cells were then washed twice and prepared for either further staining or for sorting.
Expression of the IFN-α/β receptor subunit 1 (R&D Systems; clone no. 85228), the IFN-α/β receptor subunit 2 (Miltenyi; clone REA124), and the LDL receptor (LDLR) (R&D Systems; clone no. 472413) was measured by blocking MDM at 4°C for 15 min in PBS–1% BSA with 10 µl of FcR blocking reagent (Miltenyi Biotec). Cells were then stained at 4°C for 30 min in 100 µl of PBS–1% BSA plus 10 µl of the antibody of interest. Prior to analysis, cells were washed and fixed in 1% paraformaldehyde (PFA). Fluorescence minus one (FMO) controls, in which cells were stained only with the allophycocyanin (APC)-conjugated anti-HSA antibody, were used as the negative control.
Annexin V staining was performed using the eBioscience phycoerythrin (PE)/Cy7 annexin V detection kit (ThermoFisher Scientific), by following the manufacturer’s instructions. Cells were stained with 5 µl of annexin V for 15 min at 4°C, washed twice with 1× annexin V binding buffer (ThermoFisher Scientific), and then fixed, as described above.
ISG15 staining was performed on PFA-fixed MDM using 5 μl of the Alexa Fluor 488-conjugated anti-human ISG15 (R&D Systems; clone no. 851701) or 5 μl of the isotype-matched control (R&D Systems; clone no. 54447). PKR staining was performed on PFA-fixed MDM using 1 μl unconjugated mouse anti-human PKR antibody (Abcam; clone 6H3A10), followed by staining with 4 μl of the APC-conjugated goat anti-mouse IgG (Abcam) or 2 μl isotype-matched control (1 mg/ml) (Abcam; clone 1F8). All incubations were performed at 4°C for 30 min, in 100 μl of 0.5% saponin with 10% NGS. Cells were washed and resuspended in PBS–1% BSA for analysis.
MTT assay.
Cell viability was assessed by using a Vybrant MTT cell proliferation assay kit (Invitrogen), in accordance with the manufacturer’s instructions. At 2 days post-MG1 infection, the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] stock solution was diluted 1:15 with phenol red-free RPMI 1640 (supplemented with PenStrep and 10% FCS) and added to MDM. Cells were incubated for 2 h at 37°C, at which point viral particles were lysed for 1 h at 37°C using a 10% sodium dodecyl sulfate (SDS; Fisher Scientific) solution containing 0.01 M HCl (Fisher Scientific). Absorbance was read at 570 nm using a Multiskan Ascent 96 plate reader (MTX Lab Systems Inc., Bradenton, FL).
p24 ELISA.
The HIV-1 p24 antigen capture kit (Frederick National Laboratory for Cancer Research, Frederick, MD; NIH AIDS Reagent Program) was used to quantify HIV-1 p24 antigen in cell-free supernatants, as described previously (19).
DNA and RNA extraction and quantification.
Genomic DNA extraction and quantification of proviral HIV-1 DNA were performed using a previously described protocol and primer-probe set (66). Detection of proviral HIV-1 DNA in AM was performed using digital droplet PCR (ddPCR), as described previously (19), and absolute quantification of proviral HIV-1 DNA and CD3 DNA used the QX200 droplet reader (Bio-Rad) and QuantaSoft software version 1.7 (Bio-Rad).
Cell-associated RNA was extracted using the Illustra RNAspin minikit (GE Healthcare Life Sciences, Mississauga, ON, Canada) in accordance with the manufacturer’s instructions. RNA integrity was monitored by agarose gel electrophoresis, and concentrations were measured using an ND-1000 spectrophotometer (NanoDrop, Wilmington, DE). RNA was converted to cDNA using iScript reverse transcription supermix for reverse transcription (RT)-qPCR (Bio-Rad). ISG15, PKR, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) cDNA was then detected using Bio-Rad’s PrimePCR SYBR green assay (ISG15 assay no. qHsaCED0001967; PKR assay no. qHsaCED0042156; GAPDH assay no. qHsaCED0038674), using the CFX Connect real-time PCR detection system (Bio-Rad). Amplicon purity was confirmed using a 1.5% agarose gel. RNA and cDNA were stored at –80°C in single-use aliquots.
Artwork.
Figure 4A was created using Smart Servier Medical Art image sets. Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License.
Statistics.
Statistics were performed using GraphPad Prism 5.0 software (San Diego, CA), and P values of ≤0.05 were considered significant. As determined a priori, the following statistical analyses were used. PKR and ISG15 expression between untreated and IFN-α-stimulated MDM, cell viability via MTT assay, HIV-1 p24 antigen release, proviral HIV-1 DNA, HIV-1 proviral DNA in MG1-infected AM, and surface expression of LDL-R were analyzed by a paired, two-tailed t test. IFN-α-induced PKR and ISG15 expression, MG1 infection in MDM at 48 hpi, MG1-induced cell death, and HIV-1 p24 antigen release were analyzed by 2-way repeated-measures analysis of variance (ANOVA) with the Bonferroni posttest. Proviral HIV-1 DNA at 48 hpi MG1 infection and basal ISG expression between uninfected, HSA−, and HSA+ MDM was analyzed by 1-way repeated-measures ANOVA with the Bonferroni posttest. MG1-induced cytokine secretion was analyzed by the Wilcoxon matched-pairs signed rank test (nonparametric).
ACKNOWLEDGMENTS
We thank the blood donors and patients, without whom this research would not be possible, the U.S. National Institutes of Health AIDS Reagent Program, Division of AIDS, NIAID, NIH, for reagents, Michel Tremblay for the contribution of the HIV NL4.3 BAL-IRES-HSA plasmid, and John Bell and Dave Stojdl for their contribution of the MG1 virus.
This study was supported by a Canadian HIV Cure Enterprise Team grant (HIG-133050 to J.B.A., C.T.C., and M.-A.J.) from the Canadian Institutes of Health Research (CIHR) in partnership with the Canadian Foundation for AIDS Research and the International AIDS Society, by CIHR project grant PJT-153082 to C.T.C. and M.-A.J., and by the Department of Medicine, University of Ottawa. T.S.S. is a recipient of the CIHR Doctoral Research Award. N.R. is a recipient of the Vanier Canada Graduate Scholarship. M.-A.J. holds the CIHR Canada Research Chair Tier 2 in Immunovirology. C.T.C. holds a Fonds de Recherche du Québec-Santé (FRQ-S) Junior 1 research salary award. J.B.A. is supported by a University of Ottawa Tier 1 Clinical Research Chair in HIV Cure Research.
T.S.S. validated the methodologies, designed and performed the experiments, analyzed the data, and drafted the final manuscript. N.R. assisted in optimizing the MG1 infection experiments, as well as the MTT assay and HIV-1 p24 ELISA utilized in the study. S.C.B.S. assisted in the collection of healthy donor blood, the measurement of supernatant cytokines by Luminex (Fig. 5), and the overall editing of the final manuscript. S.S., O.M., C.T.C., and M.-A.J. assisted in patient recruitment, coordination of patients undergoing bronchoalveolar lavage, and collection of alveolar macrophages. S.C.C. and J.B.A. participated in the overall study design and assisted with data analysis and interpretation, as well as the drafting and editing of the manuscript. All authors read and approved the final manuscript prior to submission.
We have no competing interests to declare.
REFERENCES
- 1.Micci L, Alvarez X, Iriele RI, Ortiz AM, Ryan ES, McGary CS, Deleage C, McAtee BB, He T, Apetrei C, Easley K, Pahwa S, Collman RG, Derdeyn CA, Davenport MP, Estes JD, Silvestri G, Lackner AA, Paiardini M. 2014. CD4 depletion in SIV-infected macaques results in macrophage and microglia infection with rapid turnover of infected cells. PLoS Pathog 10:e1004467. 10.1371/journal.ppat.1004467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honeycutt JB, Thayer WO, Baker CE, Ribeiro RM, Lada SM, Cao Y, Cleary RA, Hudgens MG, Richman DD, Garcia JV. 2017. HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat Med 23:638–643. 10.1038/nm.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abreu CM, Veenhuis RT, Avalos CR, Graham S, Queen SE, Shirk EN, Bullock BT, Li M, Metcalf Pate KA, Beck SE, Mangus LM, Mankowski JL, Clements JE, Gama L. 2019. Infectious virus persists in CD4+ T cells and macrophages in antiretroviral therapy-suppressed simian immunodeficiency virus-infected macaques. J Virol 93:e00065-19. 10.1128/JVI.00065-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abreu CM, Veenhuis RT, Avalos CR, Graham S, Parrilla DR, Ferreira EA, Queen SE, Shirk EN, Bullock BT, Li M, Metcalf Pate KA, Beck SE, Mangus LM, Mankowski JL, Mac Gabhann F, O’Connor SL, Gama L, Clements JE. 2019. Myeloid and CD4 T cells comprise the latent reservoir in antiretroviral therapy-suppressed SIVmac251-infected macaques. mBio 10:e01659-19. 10.1128/mBio.01659-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganor Y, Real F, Sennepin A, Dutertre C-A, Prevedel L, Xu L, Tudor D, Charmeteau B, Couëdel-Courteille A, Marion S, Zenak A-R, Jourdain J-P, Zhou Z, Schmitt A, Capron C, Eugenin EA, Cheynier R, Revol M, Cristofari S, Hosmalin A, Bomsel M. 2019. HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat Microbiol 4:633–644. 10.1038/s41564-018-0335-z. [DOI] [PubMed] [Google Scholar]
- 6.Xue J, Fu C, Cong Z, Peng L, Peng Z, Chen T, Wang W, Jiang H, Wei Q, Qin C. 2017. Galectin-3 promotes caspase-independent cell death of HIV-1-infected macrophages. FEBS J 284:97–113. 10.1111/febs.13955. [DOI] [PubMed] [Google Scholar]
- 7.Boliar S, Gludish DW, Jambo KC, Kamng'ona R, Mvaya L, Mwandumba HC, Russell DG. 2019. Inhibition of the lncRNA SAF drives activation of apoptotic effector caspases in HIV-1-infected human macrophages. Proc Natl Acad Sci U S A 116:7431–7438. 10.1073/pnas.1818662116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao R, Li Y, Gorantla S, Poluektova LY, Lin H, Gao F, Wang H, Zhao J, Zheng JC, Huang Y. 2019. Small molecule ONC201 inhibits HIV-1 replication in macrophages via FOXO3a and TRAIL. Antiviral Res 168:134–145. 10.1016/j.antiviral.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solis M, Nakhaei P, Jalalirad M, Lacoste J, Douville R, Arguello M, Zhao T, Laughrea M, Wainberg MA, Hiscott J. 2011. RIG-I-mediated antiviral signaling is inhibited in HIV-1 infection by a protease-mediated sequestration of RIG-I. J Virol 85:1224–1236. 10.1128/JVI.01635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akhtar LN, Qin H, Muldowney MT, Yanagisawa LL, Kutsch O, Clements JE, Benveniste EN. 2010. Suppressor of cytokine signaling 3 inhibits antiviral IFN-beta signaling to enhance HIV-1 replication in macrophages. J Immunol 185:2393–2404. 10.4049/jimmunol.0903563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wie S-H, Du P, Luong TQ, Rought SE, Beliakova-Bethell N, Lozach J, Corbeil J, Kornbluth RS, Richman DD, Woelk CH. 2013. HIV downregulates interferon-stimulated genes in primary macrophages. J Interferon Cytokine Res 33:90–95. 10.1089/jir.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pathak S, De Souza GA, Salte T, Wiker HG, Åsjö B. 2009. HIV induces both a down-regulation of IRAK-4 that impairs TLR signalling and an up-regulation of the antibiotic peptide dermcidin in monocytic cells. Scand J Immunol 70:264–276. 10.1111/j.1365-3083.2009.02299.x. [DOI] [PubMed] [Google Scholar]
- 13.Sirois M, Robitaille L, Allary R, Shah M, Woelk CH, Estaquier J, Corbeil J. 2011. TRAF6 and IRF7 control HIV replication in macrophages. PLoS One 6:e28125. 10.1371/journal.pone.0028125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harman AN, Nasr N, Feetham A, Galoyan A, Alshehri AA, Rambukwelle D, Botting RA, Hiener BM, Diefenbach E, Diefenbach RJ, Kim M, Mansell A, Cunningham AL. 2015. HIV blocks interferon induction in human dendritic cells and macrophages by dysregulation of TBK1. J Virol 89:6575–6584. 10.1128/JVI.00889-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colamonici OR, Domanski P, Platanias LC, Diaz MO. 1992. Correlation between interferon (IFN) alpha resistance and deletion of the IFN alpha/beta genes in acute leukemia cell lines suggests selection against the IFN system. Blood 80:744–749. 10.1182/blood.V80.3.744.744. [DOI] [PubMed] [Google Scholar]
- 16.Stojdl DF, Lichty B, Knowles S, Marius R, Atkins H, Sonenberg N, Bell JC. 2000. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat Med 6:821–825. 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- 17.Brun J, McManus D, Lefebvre C, Hu K, Falls T, Atkins H, Bell JC, McCart JA, Mahoney D, Stojdl DF. 2010. Identification of genetically modified Maraba virus as an oncolytic rhabdovirus. Mol Ther 18:1440–1449. 10.1038/mt.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ranganath N, Sandstrom TS, Fadel S, Côté SC, Angel JB. 2016. Type I interferon responses are impaired in latently HIV infected cells. Retrovirology 13:66. 10.1186/s12977-016-0302-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranganath N, Sandstrom TS, Burke Schinkel SC, Côté SC, Angel JB. 2018. The oncolytic virus MG1 targets and eliminates cells latently infected with HIV-1: implications for an HIV cure. J Infect Dis 217:721–730. 10.1093/infdis/jix639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landay AL, Schade SZ, Takefman DM, Kuhns MC, McNamara AL, Rosen RL, Kessler HA, Spear GT. 1993. Detection of HIV-1 provirus in bronchoalveolar lavage cells by polymerase chain reaction. J Acquir Immune Defic Syndr (1988) 6:171–175. [PubMed] [Google Scholar]
- 21.Clarke JR, Gates AJ, Coker RJ, Douglass JA, Williamson JD, Mitchell DM. 2008. HIV-1 proviral DNA copy number in peripheral blood leucocytes and bronchoalveolar lavage cells of AIDS patients. Clin Exp Immunol 96:182–186. 10.1111/j.1365-2249.1994.tb06539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cribbs SK, Lennox J, Caliendo AM, Brown LA, Guidot DM. 2015. Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res Hum Retroviruses 31:64–70. 10.1089/aid.2014.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown A, Zhang H, Lopez P, Pardo CA, Gartner S. 2006. In vitro modeling of the HIV-macrophage reservoir. J Leukoc Biol 80:1127–1135. 10.1189/jlb.0206126. [DOI] [PubMed] [Google Scholar]
- 24.Imbeault M, Lodge R, Ouellet M, Tremblay MJ. 2009. Efficient magnetic bead-based separation of HIV-1-infected cells using an improved reporter virus system reveals that p53 up-regulation occurs exclusively in the virus-expressing cell population. Virology 393:160–167. 10.1016/j.virol.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Deshiere A, Joly-Beauparlant C, Breton Y, Ouellet M, Raymond F, Lodge R, Barat C, Roy M-A, Corbeil J, Tremblay MJ. 2017. Global mapping of the macrophage-HIV-1 transcriptome reveals that productive infection induces remodeling of host cell DNA and chromatin. Sci Rep 7:5238. 10.1038/s41598-017-05566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finkelshtein D, Werman A, Novick D, Barak S, Rubinstein M. 2013. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc Natl Acad Sci U S A 110:7306–7311. 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong JG, Valdes YR, Barrett JW, Bell JC, Stojdl D, McFadden G, McCart JA, DiMattia GE, Shepherd TG. 2015. Evidence for differential viral oncolytic efficacy in an in vitro model of epithelial ovarian cancer metastasis. Mol Ther Oncolytics 2:15013. 10.1038/mto.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong JG, Valdes YR, Sivapragasam M, Barrett JW, Bell JC, Stojdl D, DiMattia GE, Shepherd TG. 2017. Spatial and temporal epithelial ovarian cancer cell heterogeneity impacts Maraba virus oncolytic potential. BMC Cancer 17:594. 10.1186/s12885-017-3600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaddy DF, Lyles DS. 2005. Vesicular stomatitis viruses expressing wild-type or mutant M proteins activate apoptosis through distinct pathways. J Virol 79:4170–4179. 10.1128/JVI.79.7.4170-4179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felt SA, Moerdyk-Schauwecker MJ, Grdzelishvili VZ. 2015. Induction of apoptosis in pancreatic cancer cells by vesicular stomatitis virus. Virology 474:163–173. 10.1016/j.virol.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourgeois-Daigneault M-C, St-Germain LE, Roy DG, Pelin A, Aitken AS, Arulanandam R, Falls T, Garcia V, Diallo J-S, Bell JC. 2016. Combination of paclitaxel and MG1 oncolytic virus as a successful strategy for breast cancer treatment. Breast Cancer Res 18:83. 10.1186/s13058-016-0744-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, Winckelmann A, Palmer S, Dinarello C, Buzon M, Lichterfeld M, Lewin SR, Ostergaard L, Søgaard OS. 2014. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 1:e13-21. 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- 33.Azzoni L, Foulkes AS, Papasavvas E, Mexas AM, Lynn KM, Mounzer K, Tebas P, Jacobson JM, Frank I, Busch MP, Deeks SG, Carrington M, O'Doherty U, Kostman J, Montaner LJ. 2013. Pegylated interferon alfa-2a monotherapy results in suppression of HIV type 1 replication and decreased cell-associated HIV DNA integration. J Infect Dis 207:213–222. 10.1093/infdis/jis663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petravic J, Rasmussen TA, Lewin SR, Kent SJ, Davenport MP. 2017. Relationship between measures of HIV reactivation and decline of the latent reservoir under latency-reversing agents. J Virol 91:e02092-16. 10.1128/JVI.02092-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costiniuk CT, Salahuddin S, Farnos O, Olivenstein R, Pagliuzza A, Orlova M, Schurr E, De Castro C, Bourbeau J, Routy J-P, Ancuta P, Chomont N, Jenabian M-A. 2018. HIV persistence in mucosal CD4+ T cells within the lungs of adults receiving long-term suppressive antiretroviral therapy. AIDS 32:2279–2289. 10.1097/QAD.0000000000001962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell BI, Laws EI, Ndhlovu LC. 2019. Impact of myeloid reservoirs in HIV cure trials. Curr HIV/AIDS Rep 16:129–140. 10.1007/s11904-019-00438-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batenchuk C, Le Boeuf F, Stubbert L, Falls T, Atkins HL, Bell JC, Conrad DP. 2013. Non-replicating rhabdovirus-derived particles (NRRPs) eradicate acute leukemia by direct cytolysis and induction of antitumor immunity. Blood Cancer J 3:e123. 10.1038/bcj.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breitbach CJ, Paterson JM, Lemay CG, Falls TJ, McGuire A, Parato KA, Stojdl DF, Daneshmand M, Speth K, Kirn D, McCart JA, Atkins H, Bell JC. 2007. Targeted inflammation during oncolytic virus therapy severely compromises tumor blood flow. Mol Ther 15:1686–1693. 10.1038/sj.mt.6300215. [DOI] [PubMed] [Google Scholar]
- 39.Orzechowska B, Antoszków Z, Siemieniec I, Lorenc M, Jatczak B, Błach-Olszewska Z. 2007. Cytokine production by human leukocytes with different expressions of natural antiviral immunity and the effect of antibodies against interferons and TNF-alpha. Arch Immunol Ther Exp (Warsz) 55:111–117. 10.1007/s00005-007-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chauhan VS, Furr SR, Sterka DG, Nelson DA, Moerdyk-Schauwecker M, Marriott I, Grdzelishvili VZ. 2010. Vesicular stomatitis virus infects resident cells of the central nervous system and induces replication-dependent inflammatory responses. Virology 400:187–196. 10.1016/j.virol.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 41.Cassol E, Cassetta L, Rizzi C, Alfano M, Poli G. 2009. M1 and M2a polarization of human monocyte-derived macrophages inhibits HIV-1 replication by distinct mechanisms. J Immunol 182:6237–6246. 10.4049/jimmunol.0803447. [DOI] [PubMed] [Google Scholar]
- 42.Cassol E, Cassetta L, Alfano M, Poli G. 2010. Macrophage polarization and HIV-1 infection. J Leukoc Biol 87:599–608. 10.1189/jlb.1009673. [DOI] [PubMed] [Google Scholar]
- 43.Cobos Jiménez V, Booiman T, de Taeye SW, van Dort KA, Rits MAN, Hamann J, Kootstra NA. 2012. Differential expression of HIV-1 interfering factors in monocyte-derived macrophages stimulated with polarizing cytokines or interferons. Sci Rep 2:763. 10.1038/srep00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cassetta L, Kajaste-Rudnitski A, Coradin T, Saba E, Chiara Della G, Barbagallo M, Graziano F, Alfano M, Cassol E, Vicenzi E, Poli G. 2013. M1 polarization of human monocyte-derived macrophages restricts pre and postintegration steps of HIV-1 replication. AIDS 27:1847–1856. 10.1097/QAD.0b013e328361d059. [DOI] [PubMed] [Google Scholar]
- 45.Pol JG, Acuna SA, Yadollahi B, Tang N, Stephenson KB, Atherton MJ, Hanwell D, El-Warrak A, Goldstein A, Moloo B, Turner PV, Lopez R, LaFrance S, Evelegh C, Denisova G, Parsons R, Millar J, Stoll G, Martin CG, Pomoransky J, Breitbach CJ, Bramson JL, Bell JC, Wan Y, Stojdl DF, Lichty BD, McCart JA. 2019. Preclinical evaluation of a MAGE-A3 vaccination utilizing the oncolytic Maraba virus currently in first-in-human trials. Oncoimmunology 8:e1512329. 10.1080/2162402X.2018.1512329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nasr N, Alshehri AA, Wright TK, Shahid M, Heiner BM, Harman AN, Botting RA, Helbig KJ, Beard MR, Suzuki K, Kelleher AD, Hertzog P, Cunningham AL. 2017. Mechanism of interferon-stimulated gene induction in HIV-1-infected macrophages. J Virol 91:e00744-17. 10.1128/JVI.00744-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paglino JC, van den Pol AN. 2011. Vesicular stomatitis virus has extensive oncolytic activity against human sarcomas: rare resistance is overcome by blocking interferon pathways. J Virol 85:9346–9358. 10.1128/JVI.00723-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McMillan NA, Chun RF, Siderovski DP, Galabru J, Toone WM, Samuel CE, Mak TW, Hovanessian AG, Jeang KT, Williams BR. 1995. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology 213:413–424. [7491766]. 10.1006/viro.1995.0014. [DOI] [PubMed] [Google Scholar]
- 49.Cai R, Carpick B, Chun RF, Jeang KT, Williams BR. 2000. HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch Biochem Biophys 373:361–367. 10.1006/abbi.1999.1583. [DOI] [PubMed] [Google Scholar]
- 50.Clerzius G, Gelinas JF, Daher A, Bonnet M, Meurs EF, Gatignol A. 2009. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J Virol 83:10119–10128. 10.1128/JVI.02457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clerzius G, Shaw E, Daher A, Burugu S, Gélinas J-F, Ear T, Sinck L, Routy J-P, Mouland AJ, Patel RC, Gatignol A. 2013. The PKR activator, PACT, becomes a PKR inhibitor during HIV-1 replication. Retrovirology 10:96. 10.1186/1742-4690-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lau AS, Read SE, Williams BR. 1988. Downregulation of interferon alpha but not gamma receptor expression in vivo in the acquired immunodeficiency syndrome. J Clin Invest 82:1415–1421. 10.1172/JCI113746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hardy GAD, Sieg SF, Rodriguez B, Jiang W, Asaad R, Lederman MM, Harding CV. 2009. Desensitization to type I interferon in HIV-1 infection correlates with markers of immune activation and disease progression. Blood 113:5497–5505. 10.1182/blood-2008-11-190231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song MM, Shuai K. 1998. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J Biol Chem 273:35056–35062. 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- 55.Yadav A, Fitzgerald P, Sajadi MM, Gilliam B, Lafferty MK, Redfield R, Reid W. 2009. Increased expression of suppressor of cytokine signaling-1 (SOCS-1): a mechanism for dysregulated T helper-1 responses in HIV-1 disease. Virology 385:126–133. 10.1016/j.virol.2008.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ventoso I, Blanco R, Perales C, Carrasco L. 2001. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc Natl Acad Sci U S A 98:12966–12971. 10.1073/pnas.231343498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castelló A, Franco D, Moral-López P, Berlanga JJ, Alvarez E, Wimmer E, Carrasco L. 2009. HIV- 1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One 4:e7997. 10.1371/journal.pone.0007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K. 2012. Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog 8:e1002612. 10.1371/journal.ppat.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kleinman CL, Doria M, Orecchini E, Giuliani E, Galardi S, De Jay N, Michienzi A. 2014. HIV-1 infection causes a down-regulation of genes involved in ribosome biogenesis. PLoS One 9:e113908. 10.1371/journal.pone.0113908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewin SR, Kirihara J, Sonza S, Irving L, Mills J, Crowe SM. 1998. HIV-1 DNA and mRNA concentrations are similar in peripheral blood monocytes and alveolar macrophages in HIV-1-infected individuals. AIDS 12:719–727. 10.1097/00002030-199807000-00008. [DOI] [PubMed] [Google Scholar]
- 61.Clouse KA, Powell D, Washington I, Poli G, Strebel K, Farrar W, Barstad P, Kovacs J, Fauci AS, Folks TM. 1989. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J Immunol 142:431–438. [PubMed] [Google Scholar]
- 62.Folks TM, Clouse KA, Justement J, Rabson A, Duh E, Kehrl JH, Fauci AS. 1989. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc Natl Acad Sci U S A 86:2365–2368. [2784570]. 10.1073/pnas.86.7.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cohen AB, Cline MJ. 1971. The human alveolar macrophage: isolation, cultivation in vitro, and studies of morphologic and functional characteristics. J Clin Invest 50:1390–1398. 10.1172/JCI106622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tomlinson GS, Booth H, Petit SJ, Potton E, Towers GJ, Miller RF, Chain BM, Noursadeghi M. 2012. Adherent human alveolar macrophages exhibit a transient pro-inflammatory profile that confounds responses to innate immune stimulation. PLoS One 7:e40348. 10.1371/journal.pone.0040348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, Brown EG, Durbin RK, Durbin JE, Hiscott J, Bell JC. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275. 10.1016/S1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 66.Vandergeeten C, Fromentin R, Merlini E, Lawani MB, DaFonseca S, Bakeman W, McNulty A, Ramgopal M, Michael N, Kim JH, Ananworanich J, Chomont N. 2014. Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J Virol 88:12385–12396. 10.1128/JVI.00609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]