Abstract
Background:
There are large knowledge gaps regarding how transmission of 2019 novel coronavirus disease (COVID-19) occurred in different settings across the world. This study aims to summarize basic reproduction number (R0) data and provide clues for designing prevention and control measures.
Methods:
Several databases and preprint platforms were retrieved for literature reporting R0 values of COVID-19. The analysis was stratified by the prespecified modeling method to make the R0 values comparable, and by country/region to explore whether R0 estimates differed across the world. The average R0 values were pooled using a random-effects model.
Results:
We identified 185 unique articles, yielding 43 articles for analysis. The selected studies covered 5 countries from Asia, 5 countries from Europe, 12 countries from Africa, and 1 from North America, South America, and Australia each. Exponential growth rate model was most favored by researchers. The pooled global R0 was 4.08 (95% CI, 3.09–5.39). The R0 estimates for new and shifting epicenters were comparable or even higher than that for the original epicenter Wuhan, China.
Conclusions:
The high R0 values suggest that an extraordinary combination of control measures is needed for halting COVID-19.
Keywords: 2019 novel coronavirus disease (COVID-19), basic reproduction number (R0), meta-analysis
1. Introduction
In January 2020, the general public became aware of an outbreak of a novel coronavirus strain, now termed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which had been affecting Wuhan city, China. The disease has been spreading rapidly worldwide, leading the World Health Organization (WHO) to declare a pandemic on March 11, 2020. While majority of cases have been relatively mild outside of Wuhan,[1] a lot of uncertainties remain about the severity of the 2019 novel coronavirus disease (COVID-19) on a per-case basis. Furthermore, higher-than-normal spread rates expected from this virus may result in population-level severe morbidity and mortality even if the case-fatality ratio remains low.
There exist large knowledge gaps regarding how transmission of SARS-CoV-2 occurred in different settings across the world. The basic reproduction number (R0) is one of the fundamental and most often used metrics that describes the contagiousness or transmissibility of the infectious agent at the beginning of an epidemic. The proportion of the population needed to be vaccinated for the elimination of an infection can be based on R0 values.[2] Estimating R0 is a requisite for designing prevention and control measures for infectious diseases such as COVID-19.
Although many researchers estimated the reproductive number of COVID-19, their results, as well as stages of infection, measurement methods, and applied preventive interventions differ substantially across the studies. Considering the variability of the reproductive numbers among the countries, we attempt to summarize available R0 estimates of COVID-19 at the global level. The pooled statistical findings might help to characterize the spread of the disease and inform public health policy.
2. Methods
2.1. Search strategy
Our study conforms to the Preferred Reporting Items for Systematic Reviews and Meta-analysis guidelines.[3] We retrieved literature from the PubMed, EmBase, China National Knowledge Infrastructure (Chinese), WanFang (Chinese) database, and BioRxiv, MedRxiv, arXiv preprint platforms in August, 2020. The search terms included: (“novel coronavirus” or “SARS-CoV-2” or “2019 novel coronavirus disease” or “COVID-19”) and (“basic reproduction number” or “R0” or “transmission” or “epidemic dynamics”). To identify additional studies, we reviewed and hand searched the references of important articles.
2.2. Study selection
Two independent authors screened the titles and abstracts for relevance. Articles were evaluated for inclusion according to the following criteria: described the early epidemic dynamics of COVID-19 in the country or region; reported basic reproduction number based on daily data of COVID-19 case counts; and presented in English or Chinese language.
Nonhuman and laboratory studies were excluded, as well as studies merely reporting time-dependent reproductive number. No exclusions were made for modeling methods used for R0 estimation.
2.3. Data extraction
The name of the first author, country or region, estimation period, measurement method, the estimated R0 value (with certain confidence interval [CI]) and digital object identifier were extracted from the articles. All studies that estimated R0 for COVID-19 were used for systemic review, while only those with 95% CI were entered into the meta-analysis.
2.4. Statistical analysis
We stratified the analysis by prespecified modeling method to make the R0 values comparable, and by country/region to examine if R0 estimates differed across the world. The average R0 values were pooled using a random-effects model. The I2 values were used for heterogeneity evaluation. A threshold of I2 ≥ 50% indicated high heterogeneity. Publication bias was assessed with Begg and Egger tests, as well as the funnel plots visually. Moreover, sensitivity analysis was performed using a 1-study-removed analysis.
All statistical analyses were conducted using Stata software (version 15.0, StataCorp, TX, USA). Statistical significance was defined as a P value < .05. The map was performed using ArcGIS software (version 10.6, Esri, USA).
3. Result
3.1. Study selection and characteristics
Our database search led to 413 studies, 228 of which were duplicates, and 72 of which were deemed irrelevant based on review of their titles and abstracts. Of the remaining 113 studies, 32 did not report the reproduction number, 21 reported effective reproduction number (Re) or real-time reproduction number (Rt) rather than R0, 9 lacked data source and 8 lacked methodology for estimation. Finally, a total of 43 studies were included for the analysis (Fig. 1).[4–46]
Figure 1.
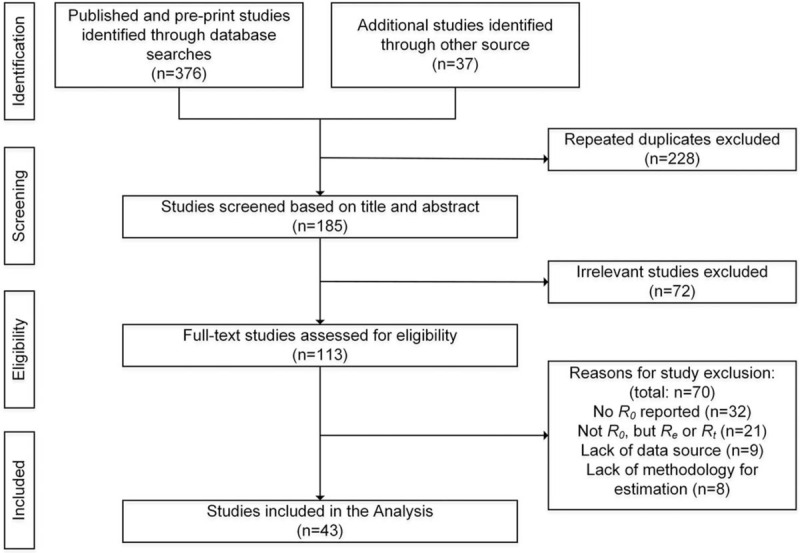
Flow diagram for study selection.
From the 43 studies (Tables 1 and 2), 61 estimates of R0 values were exacted, majority (39) of which were for China (including for Wuhan city specifically). The remaining estimates were presented for Korea (6), United States (3), Italy (2), Iran (2), followed by Japan (1), India (1), The United Kingdom (1), France (1), Germany (1), Spain (1), Brazil (1), Australia (1), and top 12 countries in Africa (1).
Table 1.
R0 estimates for cities and provinces within China.
| Author | Location | Estimation period | Methods | R0 | 95% CI |
| Li JH[7] | Wuhan | January 10–23, 2020 | EGR | 5.54 | 5.07–6.06 |
| Liu T[8] | Wuhan | By February 7, 2020 | EGR | 4.40 | 4.30–4.60 |
| Liu T[8] | China | By February 7, 2020 | EGR | 4.50 | 4.40–4.60 |
| Sanche S[9] | Wuhan | January 15–30, 2020 | EGR | 5.70 | 3.80–8.90 |
| Song QQ[10] | China | January 15–31, 2020 | EGR | 3.74 | 3.63–3.87 |
| Wang Y[5] | China | Jan 17 to February 8, 2020 | EGR | 3.49 | 3.42–3.58 |
| Zhao QY[11] | Wuhan | By January 23, 2020 | EGR | 5.70 | 3.40–9.20 |
| Zhao S[12] | China | January 10–24, 2020 | EGR | 2.24 | 1.96–2.55 |
| Zhao S[12] | China | January 10–24, 2020 | EGR | 3.58 | 2.89–4.39 |
| Zhao S[13] | Wuhan | January 1–15, 2020 | EGR | 2.56 | 2.49–2.63 |
| Li JH 7 | Wuhan | January 10–23, 2020 | SEIR | 3.55 | 2.97–4.21 |
| Read J[14] | China | January 1–22, 2020 | SEIR | 3.11 | 2.39–4.13 |
| Shen MW[15] | China | December 12, 2019 to January 22, 2020 | SEIR | 4.71 | 4.50–4.92 |
| Song QQ[10] | China | January 15–31, 2020 | SEIR | 3.91 | 3.71–4.11 |
| Tang B[16] | China | By January 22, 2020 | SEIR | 6.47 | 5.71–7.23 |
| Zhou T[17] | China | By 25 January, 2020 | SEIR | 2.80–3.30 (3.05) | / |
| Zhou WK[18] | China | By January 10, 2020 | SEIR | 5.32 | / |
| Cao ZD[19] | China | By January 23, 2020 | SEIRDC | 4.08 | |
| Chen TM[20] | Wuhan | December 7, 2019 to January 1, 2020 | SEIAR | 3.58 | / |
| LI Y[21] | China | By January 23, 2020 | SEIQR | 5.60 | / |
| Li JH[7] | Wuhan | January 10–23, 2020 | MLE | 2.65 | 2.64–2.67 |
| Song QQ[10] | China | January 15–31, 2020 | MLE | 3.16 | 2.90–3.43 |
| Tang B[22] | China | January 1–23, 2020 | MLE | 3.80 | 3.50–4.20 |
| Tang B[22] | Guangdong | January 19–31, 2020 | MLE | 3.00 | 2.60–3.30 |
| Wang Y[5] | China | January 17 to February 8, 2020 | MLE | 2.99 | 2.93–3.06 |
| Jung SM[23] | China | By January 24, 2020 | Epidemic growth model | 2.10 | 2.00–2.20 |
| Jung SM[23] | China | By January 24, 2020 | Epidemic growth model | 3.20 | 2.70–3.70 |
| Li JH[7] | Wuhan | January 10–23, 2020 | Sequential Bayesian method | 1.68 | 1.09–2.33 |
| Wang Y[5] | China | January 17 to February 8, 2020 | Sequential Bayesian method | 2.80 | 2.42–3.15 |
| Li JH[7] | Wuhan | January 10–23, 2020 | Time dependent reproduction number | 5.95 | 4.96–7.03 |
| Wang Y[5] | China | January 17 to February 8, 2020 | Time dependent reproduction number | 4.48 | 4.26–4.71 |
| Cao ZD[24] | Wuhan | By January 23, 2020 | Geo-stratified debiasing estimation framework | 3.24 | / |
| Chinazzi M[25] | China | By January 23, 2020 | GLEAM and SLIR | 2.57 | 2.37–2.78 (90% CI) |
| Li Q[26] | China | By January 22, 2020 | Fitted transmission model with zoonotic infection | 2.20 | 1.40–3.90 |
| Du ZW[27] | Wuhan | By January 22, 2020 | Hierarchical model | 1.90 | 1.47–2.59 |
| Imai N[28] | China | By January 18, 2020 | Mathematical model | 1.50–3.50 | / |
| Majumder M[29] | Wuhan | December 8, 2019 to January 26, 2020 | Incidence Decay and Exponential Adjustment (IDEA) model | 2.00–3.10 (2.50) | / |
| Wu J[30] | Wuhan | December 31, 2019 to January 28, 2020 | Markov Chain Monte Carlo methods | 2.68 | 2.47–2.86 |
| Riou J[31] | China | By January 18, 2020 | Stochastic simulations of early outbreak trajectories | 2.20 | 1.40–3.80 (90% HDI) |
CI = confidence interval, EGR = exponential growth rate, GLEAM = global epidemic and mobility model, HDI = high-density interval, MLE = maximum likelihood estimation, SEIAR = susceptible, exposed symptomatic, infectious asymptomatic, infectious removed, SEIQR = susceptible, exposed, infected but not hospitalized, infectious and isolated recovered, SEIR = susceptible-exposed-infected-removed, SEIRDC = SEIR with death cumulative, SLIR = susceptible latent infectious recovered.
Table 2.
Country-level R0 estimates across the world except China.
| Author | Location | Estimation period | Methods | R0 | 95% CI |
| de Souza W[32] | Brazil | February 25 to March 19, 2020 | EGR | 3.10 | 2.40–5.50 |
| Dwivedi L[33] | India | March 14 to April 3, 2020 | EGR | 2.56 | / |
| Ki M[34] | Korea | January 20 to February 10, 2020 | EGR | 0.48 | 0.25–0.84 |
| Musa SS[4] | Africa | March 1–19, 2020 | EGR | 2.37 | 2.22–2.51 |
| Yuan J[35] | France | February 23 to March 9, 2020 | EGR | 6.32 | 5.72–6.99 |
| Yuan J[35] | Germany | February 21 to March 9, 2020 | EGR | 6.07 | 5.51–6.69 |
| Yuan J[35] | Italy | February 23 to March 9, 2020 | EGR | 3.27 | 3.17–3.38 |
| Yuan J[35] | Spain | February 19 to March 9, 2020 | EGR | 5.08 | 4.51–5.74 |
| Choi S[36] | Korea | January 20 to February 17, 2020 | SEIR | 0.56 | 0.51–0.60 |
| Dropkin G[37] | United Kingdom | January 30 to March 31, 2020 | SEIR | 6.94 | 6.52–7.39 |
| Kuniya T[38] | Japan | January 15 to February 29, 2020 | SEIR | 2.60 | 2.40–2.80 |
| D’Arienzo M[39] | Italy | January 25 to March 12, 2020 | SIR | 2.43–3.10 | / |
| Khosravi A[40] | Iran | February 20 to March 5, 2020 | MLE | 2.74 | 2.10–3.40 |
| Tang B[22] | Korea | January 23 to March 2, 2020 | MLE | 2.60 | 2.50–2.70 |
| Muniz-Rodriguez K[41] | Iran | February 19 to March 1, 2020 | Generalized growth model | 4.40 | 3.90–4.90 |
| Shim E[6] | Korea | January 20 to February 26, 2020 | Generalized growth model | 1.50 | 1.40–1.60 |
| Fellows IE[42] | United States | January 22 to March 14, 2020 | Sequential Bayesian method | 2.37 | 2.22–2.52 |
| Gunzler D[43] | USA | By March 17, 2020 | Sequential Bayesian method | 4.02 | 3.69–5.15 |
| Zhuang Z[44] | Korea | January 31 to March 1, 2020 | Stochastic model | 2.60 | 2.30–2.90 |
| Zhuang Z[44] | Korea | February 5 to March 1, 2020 | Stochastic model | 3.20 | 2.90–3.50 |
| Rockett R[45] | Australia | January 21 to March 28, 2020 | Agent-based model | 2.27 | / |
| Ives AR[46] | New York state, USA | February 26 to April 20, 2020 | Time-varying autoregressive state-space model | 6.40 | 4.30–9.00 |
CI = confidence interval, EGR = exponential growth rate, MLE = maximum likelihood estimation, SEIR = susceptible-exposed-infected-removed, SIR = susceptible- infected-removed.
3.2. Methodological approaches used by the selected studies
As shown in Table 1, the evaluation techniques or models for R0 estimation were diverse. Exponential growth rate (EGR) model, susceptible-exposed-infected-removed (SEIR) model or other susceptible-infected-removed-based models, maximum likelihood estimation (MLE) model were the top 3 methods favored by researchers. Other methods include generalized growth model, sequential Bayesian method, stochastic model, etc. (Tables 1 and 2).
3.3. Overview of R0 estimates across the world
As shown in Figure 2, the R0 estimates were conducted for 6 continents, Asia (5 countries), Europe (5 countries), Africa (12 countries), Australia (1 country), North America (1 country), and South America (1 country). The map represented individual R0 estimate for 12 countries, and a single R0 for Africa covering 12 countries, which was a study at a regional scale.[4] The highest country-level R0 estimates are for France (R0, 6.32; 95% CI, 5.72–6.99), following Germany (R0, 6.07; 95% CI, 5.51–6.69) and Spain (R0, 5.08; 95% CI, 4.51–5.74). The R0 for Wuhan city, China and New York state, USA were 4.47 (95% CI 3.10–6.44) and 6.40 (95% CI, 4.30–9.00) respectively.
Figure 2.
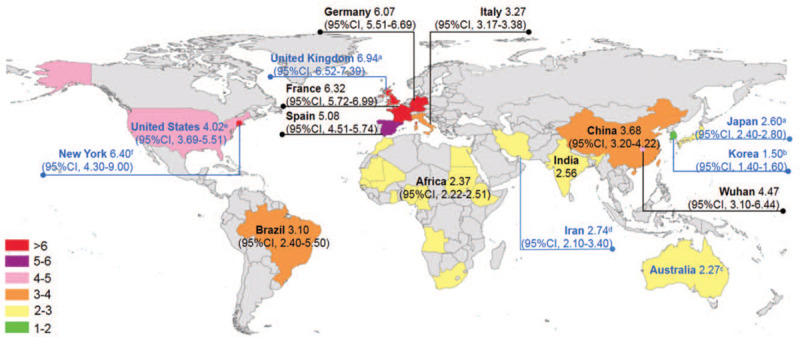
Distribution map of R0 estimates (EGR model-based if not specified) aSEIR model; bGeneralized growth model; cAgent-based model; dMLE model; eSequential Bayesian method; fTime-varying autoregressive state-space model. EGR = exponential growth rate, MLE = maximum likelihood estimation, SEIR = susceptible-exposed-infected-removed.
After pooling the 5 R0 estimates based on EGR models, we got the average R0 as 4.47 (95% CI, 3.10–6.44, I2 = 99.5%) for Wuhan. The pooled R0 for China was estimated as 3.83 (95% CI, 3.30–4.44; I2 = 98.8%). Moreover, combining the pooled result for China and EGR model-based estimates for other 7 countries or regions, we obtained the pooled global R0 for COVID-19 was 3.42 (95% CI, 2.59–4.51; I2 = 98.6%). If the R0 estimate for Korea based on data collected before its exponential growth is excluded, the pooled global R0 may be 4.08 (95% CI 3.09–5.39) (Fig. 3).
Figure 3.
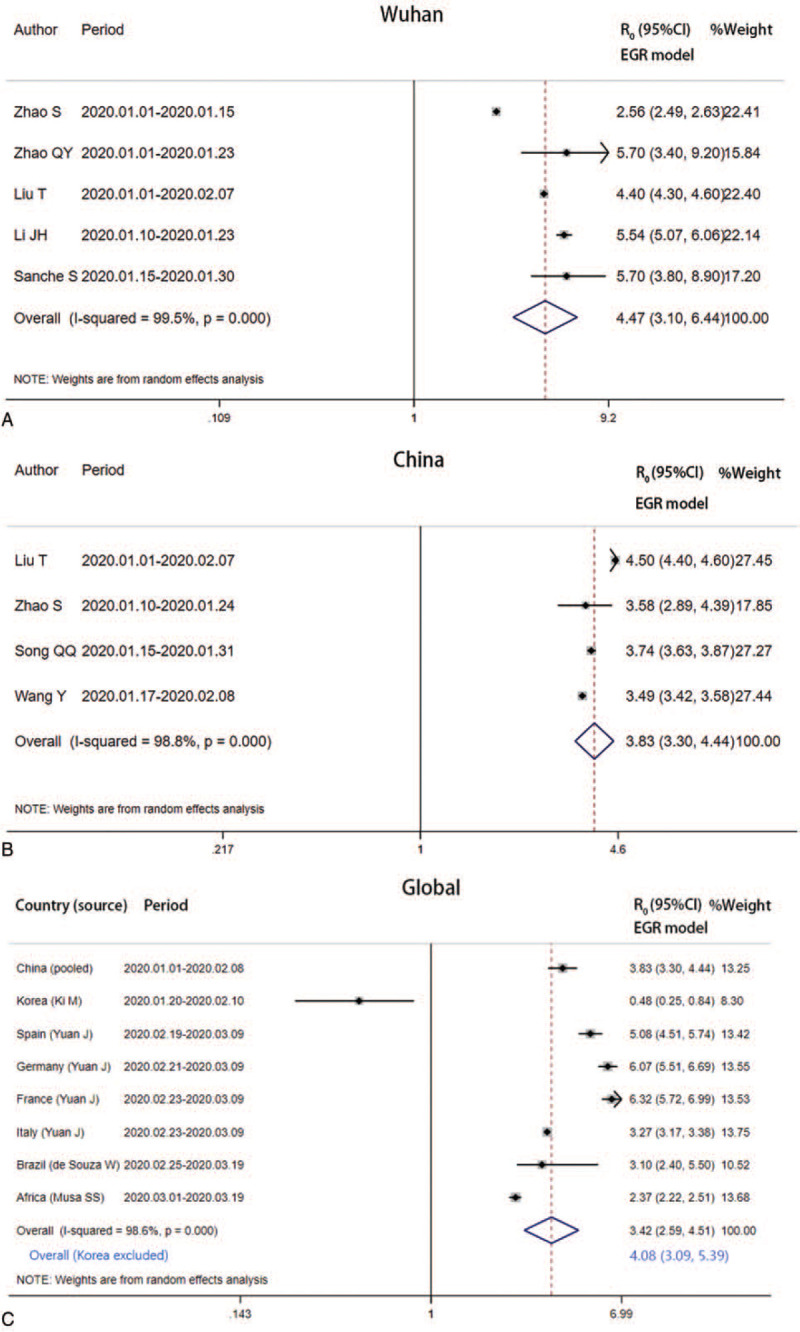
Forest plot of the pooled EGR model-based R0 estimates. EGR = exponential growth rate.
SEIR models have been used for R0 estimates for China, Japan, and Korea. The pooled R0 for China was estimated as 4.50 (95% CI, 3.71–5.46; I2 = 96.1%). The pooled Asian R0 was 3.50 (95% CI, 1.64–5.36; I2 = 99.8%) from the meta-analysis (see Figure s1, Supplemental Content, which illustrates the forest plot of the pooled SEIR model-based R0 estimates). Meanwhile, MLE models have also been applied to estimate R0 for China, Korea, and Iran. The pooled R0 for China was estimated as 3.28 (95% CI, 2.87–3.76, I2 = 92.3%). The pooled Asian R0 estimate was 2.85 (95% CI, 2.41–3.37; I2 = 81.1%) (see Figure s2, Supplemental Content, which illustrates the forest plot of the pooled MLE model-based R0 estimates).
3.4. Sensitivity analysis and publication bias
The sensitivity analysis showed that no individual study significantly affected the summarized results of R0 (see Figures 3, which illustrates the sensitivity analysis plot of meta-analysis for EGR model-based R0 estimates). The publication bias was P > .05 in both Begg and Egger tests (data not shown). Figures 4 (Supplemental Content, which illustrates the funnel plot of meta-analysis for EGR model-based R0 estimates) showed the Begg funnel plots.
4. Discussion
Knowledge of the basic reproductive number is critical for understanding the dynamics of the novel coronavirus disease. As the pandemic progresses in space and time, this needs to be re-evaluated. To the best of our knowledge, this study for the first time summarized the R0 values at the global level. This study might help to characterize the spread scale of the disease, and convey a clear message to public health decision makers.
R0 is affected by lots of biosocial factors and is estimated by various complex mathematical models. Therefore, the R0 values are usually dependent on model structures and assumptions. It is recommended not to compare values based on different models.[2] Based on this review, the EGR model was the most common method addressed by the available studies. It has been more than 3 decades since the development of EGR model which embraces both the infection cycle and the change in number of new case counts within the Lotka–Euler framework.[47] By comparing 4 methods, Wang et al[5] found that EGR model fitted the Chinese COVID-19 data best. Our study calculated pooled R0 values with the same method respectively, avoiding the limitation of different modeling. Through this approach we got more comparable estimates than previous meta-analyses.[48]
Using the assumption of exponential growth (EGR models), we found R0 values of 4.54 (95% CI 3.18–5.90) and 3.69 (95% CI 3.17–4.21) for Wuhan and China respectively. Compared with severe acute respiratory syndrome (SARS, R0 = 3, range 2.2–3.6) and Middle East respiratory syndrome (R0 range 0.8–1.3),[49,50] COVID-19 has a higher R0, suggesting the novel coronavirus be more contagious and stringent public health strategies be necessary. One modeling study indicated that if R0 were above 3.5, even near perfect case isolation and contact tracing would not be sufficient to control COVID-19 outbreaks.[51] Lessons from influenza pandemics also tell us that timely implementation of nonpharmaceutical interventions (NPIs), including infection control, social distancing, small area lockdown, and travel restrictions, is warranted.[52]
It is on January 30, 2020 that the WHO declared the COVID-19 a public health emergency of international concern, 1 week after Chinese government launched the unprecedented lockdown in Wuhan epicenter and other 12 cities in Hubei province. Significant decrease in the growth rate of COVID-19 cases within China was reported by us and other researchers.[53,54] In contrast, new epicenters were developing and shifting across the world during the same period. Some epidemiological parameters in countries except China could be different based on control strategies. However, it is clear that the R0 values for United States, France, Germany, Italy, and Spain from February to March were comparable or even higher than that of Wuhan. Africa, the last continent to be hit by the COVID-19 pandemic, is expected to be the most vulnerable continent. Interestingly, most of the identified COVID-19 cases in Africa had been imported from Europe and the United States, rather than from the original COVID-19 epicenter China.[55] Maybe, this situation would have been different if the alert call from WHO and the NPI example from China were taken into consideration on time by these authorities.[56]
It is worth noting that some estimates of R0 were significantly lower for Korea than those for most other countries. The previous 2 studies reporting subexponential growth dynamic in Korea used data collected before February 17, 2020. However, it was on February 19, 2020 when the number of confirmed COVID-19 cases started to increase rapidly. The increased spread of COVID-19 in Korea may be attributed to one superspreading event that had resulted in more than 3900 secondary cases stemming from church services in the city of Daegu.[6]
Admittedly, there are several limitations in this study. First, the study covered a small number of world countries and was not enough to cover the geographical dimensions of the continents. Second, the study did not assess meteorological characteristics of a location and demographic characteristics of that location's population which might influence the disease transmission pattern. Last, some articles included for analysis are preprints that might affect the overall quality of the review to some extent.
In conclusion, the relatively high value for R0 suggests that an extraordinary combination of control measures is needed for halting COVID-19. Indeed, at the expected transmissibility of the pandemic pathogen, timely NPIs should be implemented before a highly efficacious vaccine could become available. Such efforts will be the key to quell local outbreaks and reduce the risk of further global dissemination.
Acknowledgments
The authors thank Prof Lei Zhang from Xi’an Jiaotong University Health Science Center for his valuable scientific comments and advices.
Author contributions
GQ designed the study protocol. CJY, ZXW, and YX did the literature search. The titles, abstracts, and full texts were screened and selected by CJY and MXH. The data were extracted and analysed by CJY, MXH and KC. CJY, ZXW, and YX drafted the manuscript. GQ edited the draft. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: COVID-19 = 2019 novel coronavirus disease, EGR = exponential growth rate, MLE = maximum likelihood estimation, NPI = nonpharmaceutical interventions, R0 = basic reproduction number, SARS = severe acute respiratory syndrome, SARS-CoV-2 = severe acute respiratory syndrome corona virus 2, SEIR = susceptible-exposed-infected-removed, WHO = World Health Organization.
How to cite this article: Yu CJ, Wang ZX, Xu Y, Hu MX, Chen K, Qin G. Assessment of basic reproductive number for COVID-19 at global level: a meta-analysis. Medicine. 2021;100:18(e25837).
C-JY, Z-XW, and YX contributed equally to this work.
Summary: The pooled global R0 estimate for COVID-19 is 4.08 (95% CI, 3.09–5.39). New and shifting epicenters have been developed across the world. The high R0 estimates suggest that an extraordinary combination of control measures is needed for halting the disease.
This work was supported in part by grants from Jiangsu Provincial Health Committee (QNRC2016402 and LGY2017039) and from Nantong Municipal Bureau of Science and Technology (HS2016002 and XG202009-3). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
The key information and data generated and/or analyzed during this study were included in this article and/or its supplementary information files.
GQ is an Academic Editor of this journal.
The authors have no conflicts of interest to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Supplemental digital content is available for this article.
References
- [1].Zhang Y. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi 2020;41:145–51. [DOI] [PubMed] [Google Scholar]
- [2].Delamater P, Street E, Leslie T, et al. Complexity of the Basic Reproduction Number (R0). Emerg Infect Dis 2019;25:01–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Moher D, Shamseer L, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev 2015;4:01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Musa SS, Zhao S, Wang MH, et al. Estimation of exponential growth rate and basic reproduction number of the coronavirus disease 2019 (COVID-19) in Africa. Infect Dis Poverty 2020;9:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang Y, You X, Wang Y, et al. Estimating the basic reproduction number of COVID-19 in Wuhan, China. Zhonghua Liu Xing Bing Xue Za Zhi 2020;41:476–9. [DOI] [PubMed] [Google Scholar]
- [6].Shim E, Tariq A, Choi W, et al. Transmission potential and severity of COVID-19 in South Korea. Int J Infect Dis 2020;93:339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li J, Wang Y, Gilmour S, et al. Estimation of the epidemic properties of the 2019 novel coronavirus: a mathematical modeling study. medRxiv 2020;preprint. doi: 10.1101/2020.02.18.20024315. [Google Scholar]
- [8].Liu T, Hu J, Xiao J, et al. Time-varying transmission dynamics of Novel Coronavirus Pneumonia in China. bioRxiv 2020;preprint. doi: 10.1101/2020.01.25.919787. [Google Scholar]
- [9].Sanche S, Lin YT, Xu C, et al. High contagiousness and rapid spread of severe acute respiratory syndrome Coronavirus 2. Emerg Infect Dis 2020;26:1470–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Song Q, Zhao H, Fang L-Q, et al. Study on assessing early epidemiological parameters of coronavirus disease epidemic in China. Zhonghua Liu Xing Bing Xue Za Zhi 2020;41:461–5. [DOI] [PubMed] [Google Scholar]
- [11].Zhao Q, Chen Y, Small DS. Analysis of the epidemic growth of the early 2019-nCoV outbreak using internationally confirmed cases. medRxiv 2020;preprint. doi: 10.1101/2020.02.06.20020941. [Google Scholar]
- [12].Zhao S, Lin Q, Ran J, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int J Infect Dis 2020;92:214–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhao S, Musa SS, Lin Q, et al. Estimating the Unreported Number of Novel Coronavirus (2019-nCoV) cases in China in the First Half of January 2020: a data-driven modelling analysis of the early outbreak. J Clin Med 2020;9:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Read JM, Bridgen JR, Cummings DA, et al. Novel coronavirus 2019-nCoV: early estimation of epidemiological parameters and epidemic predictions. medRxiv 2020;preprint. doi: 10.1101/2020.01.23.20018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shen M, Peng Z, Xiao Y, et al. Modelling the epidemic trend of the 2019 novel coronavirus outbreak in China. bioRxiv 2020;doi: 10.1016/j.xinn.2020.100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tang S, Wang Z, Li Y-S, et al. Estimation of the transmission risk of the 2019-nCoV and its implication for public health interventions. J Clin Med 2020;9:462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhou T, Liu Q, Yang Z, et al. Preliminary prediction of the basic reproduction number of the Wuhan novel coronavirus 2019-nCoV. J Evid Based Med 2020;13:03–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhou W, Wang A, Xia F, et al. Effects of media reporting on mitigating spread of COVID-19 in the early phase of the outbreak. Math Biosci Eng 2020;17:2693–707. [DOI] [PubMed] [Google Scholar]
- [19].Cao Z, Zhang Q, Lu X, et al. Estimating the effective reproduction number of the 2019-nCoV in China. medRxiv 2020;preprint. doi: 10.1101/2020.01.27.20018952. [Google Scholar]
- [20].Chen T-M, Rui J, Wang Q-P, et al. A mathematical model for simulating the phase-based transmissibility of a novel coronavirus. Infect Dis Poverty 2020;9:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li Y, Wang LW, Peng ZH, et al. Basic reproduction number and predicted trends of coronavirus disease 2019 epidemic in the mainland of China. Infect Dis Poverty 2020;9:article 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tang B, Xia F, Bragazzi N, et al. Lessons drawn from China and South Korea for managing COVID-19 epidemic: insights from a comparative modeling study. medRxiv 2020;preprint doi: 10.1101/2020.03.09.20033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jung S-M, Akhmetzhanov AR, Hayashi K, et al. Real time estimation of the risk of death from novel coronavirus (2019-nCoV) infection: Inference using exported cases. medRxiv preprint doi:10.3390/jcm9020523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cao Z, Zhang Q, Lu X, et al. Incorporating human movement data to improve epidemiological estimates for 2019-nCoV. medRxiv 2020;preprint. doi: 10.1101/2020.02.07.20021071. [Google Scholar]
- [25].Chinazzi M, Davis JT, Ajelli M, et al. The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science 2020;368:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of Novel coronavirus-infected pneumonia. N Engl J Med 2020;382:1199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Du Z, Wang L, Cauchemez S, et al. Risk for transportation of 2019 Novel Coronavirus (COVID-19) from Wuhan to Cities in China. medRxiv 2020;preprint. doi: 10.1101/2020.01.28.20019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Imai N. <Report 1 Estimating the potential total number of novel Coronavirus cases in Wuhan City, China.>. Imperial College London COVID-19 Response Team 2020. [Google Scholar]
- [29].Majumder M, Mandl K. Early transmissibility assessment of a novel coronavirus in Wuhan, China. SSRN Electron J 2020;preprint. doi: 10.2139/ssrn.3524675. [Google Scholar]
- [30].Wu J, Leung K, Leung G. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet 2020;395:689–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Riou J, Althaus C. Pattern of early human-to-human transmission of Wuhan 2019-nCoV. bioRxiv 2020;preprint doi: 10.1101/2020.01.23.917351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].de Souza WM, Buss LF, Candido DDS, et al. Epidemiological and clinical characteristics of the COVID-19 epidemic in Brazil. Nat Hum Behav 2020;4:856–65. [DOI] [PubMed] [Google Scholar]
- [33].Dwivedi L, Rai B, Shukla A, et al. Assessing the impact of complete lockdown on COVID-19 Infections in India and its burden on public health facilities. Demography India 2020;49:37–50. [Google Scholar]
- [34].Ki M. Epidemiologic characteristics of early cases with 2019 novel coronavirus (2019-nCoV) disease in Republic of Korea. Epidemiol Health 2020;42:e2020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yuan J, Li M, Lv G, et al. Monitoring transmissibility and mortality of COVID-19 in Europe. Int J Infect Dis 2020;95:311–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Choi S, Ki M. Estimating the reproductive number and the outbreak size of COVID-19 in Korea. Epidemiol Health 2020;42:e2020011–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dropkin G. COVID-19 UK Lockdown Forecasts and R0. 2020. Preprint. doi: 10.3389/fpubh.2020.00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kuniya T. Prediction of the epidemic peak of coronavirus disease in Japan, 2020. J Clin Med 2020;9:789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].D’Arienzo M, Coniglio A. Assessment of the SARS-CoV-2 basic reproduction number, R0, based on the early phase of COVID-19 outbreak in Italy. Biosafety Health 2020;2:57–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Khosravi A, Chaman R, Rohani-Rasaf M, et al. The basic reproduction number and prediction of the epidemic size of the novel coronavirus (COVID-19) in Shahroud, Iran. medRxiv 2020;preprint. doi:10.1017/S0950268820001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Muniz-Rodriguez K, Fung IC-H, Ferdosi SR, et al. Transmission potential of COVID-19 in Iran. medRxiv 2020;preprint doi: 10.1101/2020.03.08.20030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fellows IE, Slayton RB, Hakim2 AJ. The COVID-19 Pandemic, Community Mobility and the Effectiveness of Non-pharmaceutical interventions: The United States of America, February to May 2020. arXiv 2020;https://arxiv.org/ftp/arxiv/papers/2007/2007.12644.pdf [Google Scholar]
- [43].Gunzler D, Sehgal AR. Time-Varying COVID-19 Reproduction Number in the United States. medRxiv 2020;preprint. doi: 10.1101/2020.04.10.20060863. [Google Scholar]
- [44].Zhuang Z, Zhao S, Lin Q, et al. Preliminary estimating the reproduction number of the coronavirus disease (COVID-19) outbreak in Republic of Korea from 31 January to 1 March 2020. medRxiv 2020;preprint. doi: 10.1101/2020.03.02.20030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rockett RJ, Arnott A, Lam C, et al. Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat Med 2020;26:1398–404. [DOI] [PubMed] [Google Scholar]
- [46].Ives AR, Bozzuto C. State-by-state estimates of R0 at the start of COVID-19 outbreaks in the USA. medRxiv 2020; preprint doi: 10.1101/2020.05.17.20104653 [Google Scholar]
- [47].Wallinga J, Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc Biol Sci 2007;274:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].He W, Yi GY, Zhu Y. Estimation of the basic reproduction number, average incubation time, asymptomatic infection rate, and case fatality rate for COVID-19: meta-analysis and sensitivity analysis. J Med Virol 2020;92:2543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cauchemez S, Fraser C, Van Kerkhove MD, et al. Middle East respiratory syndrome coronavirus: quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect Dis 2014;14:50–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lipsitch M, Cohen T, Cooper B, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science 2003;300:1966–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hellewell J, Abbott S, Gimma A, et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob Health 2020;8:e488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Halloran ME, Ferguson NM, Eubank S, et al. Modeling targeted layered containment of an influenza pandemic in the United States. Proc Natl Acad Sci U S A 2008;105:4639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ji T, Chen HL, Xu J, et al. Lockdown contained the spread of 2019 novel coronavirus disease in Huangshi City, China: early epidemiological findings. Clin Infect Dis 2020;71:1454–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lau H, Khosrawipour V, Kocbach P, et al. The positive impact of lockdown in Wuhan on containing the COVID-19 outbreak in China. J Travel Med 2020;27:taaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ruth M. Africa braces for coronavirus, but slowly. The New York Times 20202020. [Google Scholar]
- [56].Institute of Labor Economics (IZA), Amuedo-Dorantes C, Kaushal N, Muchow AN. Is the Cure Worse than the Disease? County-Level Evidence from the COVID-19 Pandemic in the United States. 2020;http://ftp.iza.org/dp13695.pdf [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


