Abstract
Backgrounds:
Due to difficulty in early diagnosis of chronic pancreatitis (CP), it is urgent to find novel biomarkers to detect CP. Exosomal microRNAs (Exo-miRNAs) located in the serum may be potential diagnostic and therapeutic targets for CP.
Objective:
To identify differentially expressed Exo-miRNAs (DE-Exo-miRNAs) in the serum of CP patients, we performed a bioinformatics analysis.
Methods:
The dataset GSE128508 was downloaded from the Gene Expression Omnibus (GEO) database. The analysis was carried out using BRB-ArrayTools and significance analysis of microarrays (SAM). The target genes of DE-S-Exo-miRNAs were predicted by miRWalk databases. Further gene ontology (GO) term and Kyoto Encyclopedia of Genomes (KEGG) pathway analyses were performed with plug-in ClueGO in Cytoscape software 3.7.0. Subsequently, the interaction regulatory network between encoded proteins of target genes was performed with the Search Tool for the Retrieval of Interacting Genes (STRING) database and analyzed using plug-in Molecular Complex Detection (MCODE) and cytoHubba in Cytoscape software 3.7.0.
Results:
We identified 227 DE-Exo-miRNAs in the serum. Further analysis using the miRWalk database identified 5164 target genes of these miRNAs. The protein–protein interaction (PPI) regulatory network of 1912 potential target genes for hub 10 up-regulated miRNAs with high degrees and one down-regulated miRNAs were constructed using the STRING database and Cytoscape software. The functional analysis using Cytoscape software tool highlighted that target genes involved in pancreatic cancer. Acinar-ductal metaplasia (ADM) in the inflammatory environment of CP is a precursor of pancreatic cancer. Subsequently, we constructed a network of target genes associated with ADM and their miRNAs.
Conclusions:
Exo-miRNAs in the serum as well as their target genes may be promising targets for the early diagnosis and treatment of CP. In addition, we identified potential Exo-miRNAs involved in ADM that is a precursor of pancreatic cancer associated with CP.
Keywords: acinar-ductal metaplasia, bioinformatics analysis, chronic pancreatitis, exosome, miRNA
1. Introduction
Chronic pancreatitis (CP) is a form of continuing fibro-inflammatory disease associated with pathologically morphological changes of the pancreas, typically causing endocrine and exocrine dysfunction, especially in persons carrying genetic risk factors.[1] The diagnosis of CP can be difficulty in early phases of disease. Here, pancreatic functions are preserved and tests of laboratory and imaging may only display minimally abnormal results.[2] Thus, a comprehensive understanding about the molecular mechanism underlying CP and the identification of novel targets to diagnose CP in early phase are necessary.
MicroRNAs (miRNAs) are associated with acute or chronic pancreatitis.[3–7] miRNAs are single-stranded RNAs ∼21–23-nucleotides in length which have important functions in almost every aspect of biology.[8] miRNAs play crucial gene regulatory roles by directing post-transcriptional repression of protein-coding genes.[9] The roles of miRNAs in many human diseases have made them promising targets for diagnostic and therapeutic strategies.[10–12] miRNAs have been discovered in body fluids, such as serum and plasma.[13,14] Additionally, researchers have also extracted miRNAs from exosomes.[15]
Exosomes are formed by the fusion of multivesicular bodies with the plasma membrane and are then released from the cell into the microenvironment.[16] Exosomes regulate many pathophysiological processes, such as inflammation, infection, immune responses, and tumor growth.[17] Exosomes contain proteins, lipid, and miRNAs.[18,19] As miRNAs in exosomes are prevented from RNase degradation, they are easier to detect compared with freely circulating miRNAs.[20] In addition, the miRNA expression profile in exosomes is different compared with the corresponding intracellular miRNAs.[21] Acute pancreatitis plasma exosomes contained more pro-inflammatory miRNA than exosomes from plasma control.[22] Therefore, exosomal microRNAs (Exo-miRNAs) in the serum could be potential biomarkers for diagnosing CP.
In this study, we analyzed the dataset GSE128508 from the public Gene Expression Omnibus (GEO) database. The samples displayed in the dataset are from the serum exosomes from CP patients, autoimmune pancreatitis (AIP) patients, and healthy control (HC) participants. Previous study identified 165 differentially expressed Exo-miRNAs (DE-Exo-miRNAs) when researchers focused on AIP and HC. However, they only analyzed 10 DE-Exo-miRNAs.[23] There, we analyzed more interesting DE-Exo-miRNAs in the serum by only comparing CP patients and HC participants. Subsequently, functional analysis of target genes for DE-Exo-miRNAs from the miRWalk databases was performed using Cytoscape software tool. Then the target genes of 10 up-regulated miRNAs with high degrees and one down-regulated miRNAs selected from miRNAs-mRNA networks were used to construct a protein–protein interaction (PPI) regulatory network. As functionally grouped network of Kyoto Encyclopedia of Genomes (KEGG) pathways show that pancreatic cancer is significant and acinar-ductal metaplasia (ADM) in the inflammatory environment of CP is a precursor of pancreatic cancer,[24] the regulatory network of target genes associated with ADM and their DE-Exo-miRNAs in the serum were also constructed.
2. Materials and methods
2.1. Data acquisition
In this study, we retrieve datasets from public GEO database. The search keyword was “pancreatitis” and the selection was restricted to series. Finally, the miRNA dataset GSE128508 using the GPL21263 platform, which included results from 10 CP and 10 healthy adults’ samples, were downloaded for further analyses. The characteristics of patients and healthy adults were shown in Table 1. Both miRNA profiles data and clinical characteristics of the healthy adults and CP patients are publicly available and open-access. Therefore, approval by a local ethics committee and informed consent were not needed.
Table 1.
The characteristics of patients and healthy participants.
| Variable | Healthy adults (10) | CP patients (10) |
| Gender, male: female | 6:4 | 7:3 |
| Age, median (range), y | 61.5 (49–82) | 62 (50–78) |
CP = chronic pancreatitis.
2.2. Data processing and identifying serous DE-Exo-miRNAs
The data from GSE128508 were analyzed by BRB-ArrayTools 4.6.0, R 4.0.1, and Significance Analysis of Microarrays (SAM) 5.0 to identify serous DE-Exo-miRNAs between CP and normal samples. We set Delta = 2.45 as threshold. For more information about statistic method, please visit the url https://brb.nci.nih.gov/BRB-ArrayTools/download.html and http://www-stat.stanford.edu/∼tibs/SAM.
2.3. Identification of serous DE-Exo-miRNA target genes
The miRWalk database is a large collection of predicted and experimentally verified binding sites information for miRNAs-targets.[25] The information for DE-Exo-miRNA target genes was obtained from the miRWalk databases. We selected genes from the intersection between the Targetscan and miRDB databases, as well as experimentally verified miRTarBase database to increase the accuracy of the predictions.
2.4. Function analysis of target genes
Potential functions of target genes in biological process were analyzed using gene ontology (GO) term. KEGG is a database for the systematic analysis of gene functions and utilities in biological system from molecular-level information. We obtained information from both the GO term and KEGG pathway using the plug-in ClueGO (2.5.6) in Cytoscape software 3.7.0.[26]
2.5. Construction of miRNA-mRNA networks and miRNAs screening
Many gene targets of DE-Exo-miRNA in the serum may be associated with CP. The miRNA-mRNA networks were visualized with Cytoscape software 3.7.0. We identified hub miRNAs in networks according to their intra-network connectivity and correlation with their target genes. The10 up-regulated miRNAs with high degrees and 1 down-regulated miRNAs were identified from the networks.
2.6. Construction of PPI networks and significant submodule screening
The functional interactions between encoded proteins of target genes for 11 dysregulated miRNAs were explored using the Search Tool for the Retrieval of Interacting Genes (STRING) database.[27] We downloaded the interaction information from the STRING database and use Cytoscape software 3.7.0 to construct the PPI regulatory network. We applied the Molecular Complex Detection (MCODE) tool of Cytoscape software 3.7.0 to identify the significant submodule from the PPI network.[28] Furthermore, the hub genes were identified using cytoHubba plugin of Cytoscape software 3.7.0 from the significant submodule.[29] We set degree cutoff = 2, node score cutoff = 0.2, K-core = 2, and maximum depth = 100 as the thresholds.
2.7. Construction of the regulatory network associated with ADM
KEGG pathways analysis involved in pancreatic cancer and pathways in cancer. Pancreas acinar cells show high plasticity and can undergo ADM after injury. Cells undergoing ADM are precursors for pancreatic intraepithelial neoplasia lesions that can further progress to pancreatic ductal adenocarcinoma (PDAC).[30] Among target genes of DE-Exo-miRNAs in the serum, signal transducer and activator of transcription 3 (STAT3), pyruvate dehydrogenase kinase 1 (PDK1), kirsten rat sarcoma 2 viral oncogene homolog (KRAS), Rac family small GTPase 1 (RAC1), and ras homolog family member A (RHOA) are key factors involving in ADM.[31–34] Therefore, we construct the regulatory network of these target genes and their DE-Exo-miRNAs.
3. Results
3.1. DE-Exo-miRNAs in CP serum
A total of 227 DE-Exo-miRNAs in the serum were identified from the dataset GSE128508. Among these DE-Exo-miRNAs, 226 were significantly up-regulated and 1 were down-regulated. One down-regulated DE-Exo-miRNAs and the top 50 up-regulated DE-Exo-miRNAs in the serum can be found summarized in Table 2. In supplementary materials, all 227 DE-Exo-miRNAs in the serum are listed.
Table 2.
List of partial DE-Exo-miRNAs in the serum∗.
| e | Numerator | Denominator | Fold change | |
| Down-regulated microRNAs | ||||
| hsa-miR-5100 | –4.20 | –1.32 | 0.31 | 0.82 |
| Up-regulated microRNAs (Top 50) | ||||
| hsa-miR-5006-5p | 13.60 | 1.65 | 0.12 | 1.50 |
| hsa-miR-6748-5p | 13.56 | 2.11 | 0.16 | 1.63 |
| hsa-miR-4485-5p | 12.34 | 1.74 | 0.14 | 1.52 |
| hsa-miR-5192 | 11.75 | 2.34 | 0.20 | 1.70 |
| hsa-miR-4514 | 11.73 | 2.22 | 0.19 | 1.67 |
| hsa-miR-6760-5p | 11.36 | 2.83 | 0.25 | 1.77 |
| hsa-miR-6165 | 10.97 | 2.44 | 0.22 | 1.63 |
| hsa-miR-4298 | 10.95 | 2.44 | 0.22 | 1.63 |
| hsa-miR-6846-5p | 10.92 | 2.19 | 0.20 | 1.62 |
| hsa-miR-6887-5p | 10.91 | 3.26 | 0.30 | 1.78 |
| hsa-miR-6830-5p | 10.83 | 1.71 | 0.16 | 1.51 |
| hsa-miR-6828-5p | 10.82 | 1.97 | 0.18 | 1.59 |
| hsa-miR-370-3p | 10.68 | 2.50 | 0.23 | 1.69 |
| hsa-miR-6766-5p | 10.65 | 2.31 | 0.22 | 1.61 |
| hsa-miR-3151-5p | 10.64 | 1.95 | 0.18 | 1.57 |
| hsa-miR-6795-5p | 10.60 | 2.20 | 0.21 | 1.54 |
| hsa-miR-3622b-5p | 10.50 | 1.66 | 0.16 | 1.50 |
| hsa-miR-4419b | 10.49 | 2.51 | 0.24 | 1.61 |
| hsa-miR-513a-5p | 10.41 | 2.01 | 0.19 | 1.61 |
| hsa-miR-3154 | 10.41 | 2.11 | 0.20 | 1.55 |
| hsa-miR-6826-5p | 10.22 | 2.01 | 0.20 | 1.55 |
| hsa-miR-4728-5p | 10.08 | 2.81 | 0.28 | 1.66 |
| hsa-miR-4727-3p | 9.93 | 1.76 | 0.18 | 1.51 |
| hsa-miR-3619-3p | 9.87 | 2.74 | 0.28 | 1.49 |
| hsa-miR-6849-5p | 9.86 | 2.34 | 0.24 | 1.56 |
| hsa-miR-6717-5p | 9.82 | 2.74 | 0.28 | 1.60 |
| hsa-miR-5010-5p | 9.81 | 1.68 | 0.17 | 1.50 |
| hsa-miR-491-5p | 9.77 | 1.65 | 0.17 | 1.50 |
| hsa-miR-6746-5p | 9.69 | 2.29 | 0.24 | 1.51 |
| hsa-miR-6890-5p | 9.66 | 1.87 | 0.19 | 1.54 |
| hsa-miR-6824-5p | 9.55 | 2.13 | 0.22 | 1.53 |
| hsa-miR-6757-5p | 9.54 | 2.51 | 0.26 | 1.54 |
| hsa-miR-2110 | 9.53 | 1.80 | 0.19 | 1.50 |
| hsa-miR-4257 | 9.51 | 2.13 | 0.22 | 1.48 |
| hsa-miR-6731-5p | 9.44 | 1.86 | 0.20 | 1.52 |
| hsa-miR-3714 | 9.40 | 1.93 | 0.21 | 1.54 |
| hsa-miR-4462 | 9.29 | 1.95 | 0.21 | 1.50 |
| hsa-miR-920 | 9.28 | 2.58 | 0.28 | 1.53 |
| hsa-miR-7846-3p | 9.10 | 2.19 | 0.24 | 1.55 |
| hsa-miR-7847-3p | 9.08 | 2.43 | 0.27 | 1.55 |
| hsa-miR-6812-5p | 9.00 | 2.04 | 0.23 | 1.53 |
| hsa-miR-3934-5p | 8.84 | 1.90 | 0.21 | 1.49 |
| hsa-miR-583 | 8.83 | 1.45 | 0.16 | 1.44 |
| hsa-miR-5189-5p | 8.83 | 1.83 | 0.21 | 1.51 |
| hsa-miR-6777-5p | 8.71 | 2.28 | 0.26 | 1.51 |
| hsa-miR-6511b-5p | 8.68 | 1.79 | 0.21 | 1.47 |
| hsa-miR-8059 | 8.66 | 2.29 | 0.26 | 1.46 |
| hsa-miR-551b-5p | 8.61 | 1.83 | 0.21 | 1.51 |
| hsa-miR-6769b-5p | 8.61 | 1.79 | 0.21 | 1.43 |
| hsa-miR-6756-5p | 8.59 | 2.03 | 0.24 | 1.36 |
All 227 DE-Exo-miRNAs in the serum are listed in supplementary materials. DE-Exo-miRNAs = differentially expressed exosomal miRNAs, Denominator = The denominator of the T-statistic, Numerator = The numerator of the T-statistic, Score = The T-statistic value.
3.2. Target genes for serous DE-Exo-miRNAs
We utilized the miRWalk database to screen for target genes for the 227 DE-Exo-miRNAs in the serum to explore the potential functions of these DE-Exo-miRNAs. To increase the accuracy of the search, the intersection between the TargetScan and miRDB databases, as well as experimentally verified genes in the miRTarBase database, were selected as target genes. Finally, we obtained 5164 target genes for the 227 serous DE-Exo-miRNAs. Among the target genes, many of them are associated with CP, such as SMAD family member 3 (SMAD3), transient receptor potential cation channel subfamily V member 1 (TRPV1), MORC family CW-type zinc finger 4 (MORC4), and so on.[35,36] The DE-Exo-miRNAs (hsa-miR-491–5p, hsa-miR-6807–5p, and hsa-miR-6731-5p, etc) may be valuable for the diagnosis and treatment of CP by their target genes.
3.3. Function analysis of target genes for serous DE-Exo-miRNAs
The function analysis indicated that the enriched GO terms of 5164 target genes primarily included those involved in the cell cycle process, cellular response to signal transduction by p53 class mediator, lymphocyte mediated immunity, regulation of cell death, regulation of cell differentiation, regulation of gene expression, regulation of cell communication, regulation of cellular metabolic process, and regulation of B cell activation, amongst others. The typical 20 GO terms are summarized in Fig. 1.
Figure 1.
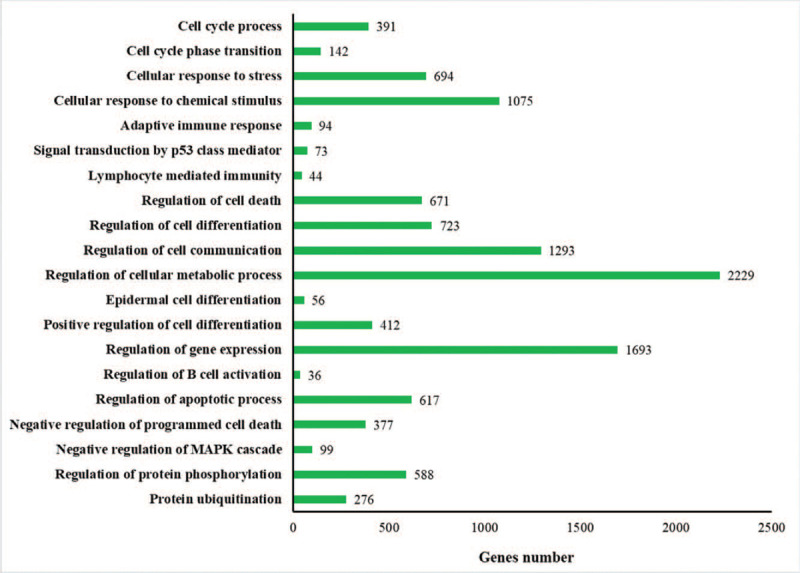
GO function annotation of miRNA target genes. Levels of GO terms increase from the upper to the lower. GO = gene ontology, miRNAs = microRNAs.
The KEGG pathways analysis revealed that target genes were involved in numerous different pathways, including pancreatic cancer, pathways in cancer, p53 signaling pathway, mitogen-activated protein kinase (MAPK) signaling pathway, cellular senescence, and signaling pathways regulating pluripotency of stem cells, amongst others. These pathways are shown in Fig. 2.
Figure 2.
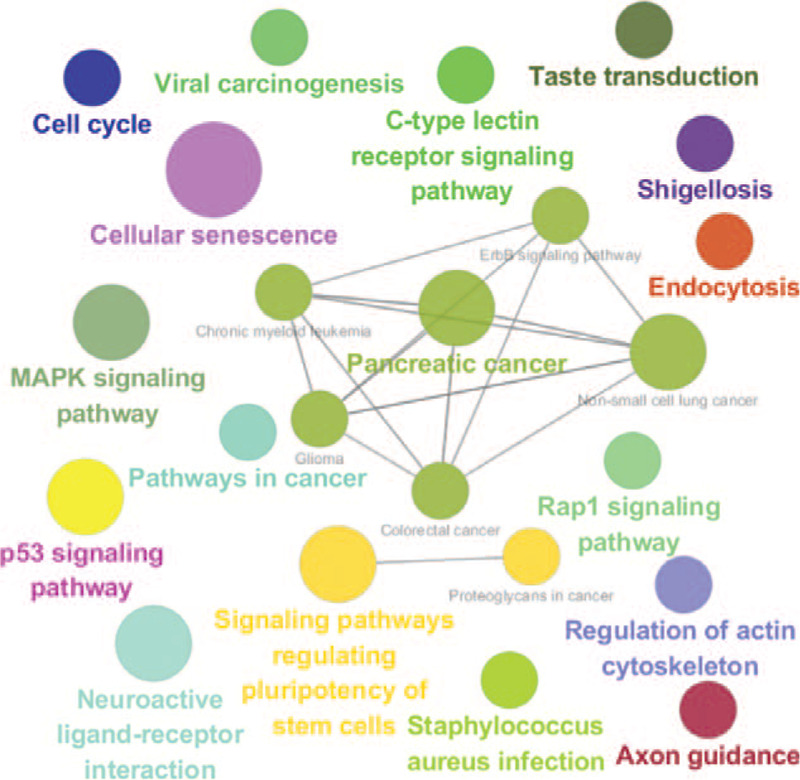
Functionally grouped network of KEGG pathways of serous DE-Exo-miRNA target genes. The node size stands for the pathway significance. The node size increases when the P-value of pathways decreases. KEGG = Kyoto Encyclopedia of Genes and Genomes.
3.4. The serous DE-Exo-miRNAs with high degrees
We constructed the miRNAs-mRNA networks between the 227 DE-Exo-miRNAs and 5164 target genes using Cytoscape software 3.7.0. The hub 10 up-regulated miRNAs with high degrees identified from the miRNAs-mRNA networks are summarized in Table 3.
Table 3.
The top 10 miRNAs with high degrees from miRNAs-mRNA networks.
| Rank | miRNA | Degree | Number of target genes | Feature |
| 1 | hsa-miR-24-3p | 516 | 516 | Up |
| 2 | hsa-miR-149-3p | 449 | 449 | Up |
| 3 | hsa-miR-6785-5p | 448 | 448 | Up |
| 4 | hsa-miR-4728-5p | 446 | 446 | Up |
| 5 | hsa-miR-6808-5p | 402 | 402 | Up |
| 6 | hsa-miR-6779-5p | 388 | 388 | Up |
| 7 | hsa-miR-6799-5p | 384 | 384 | Up |
| 8 | hsa-miR-6086 | 343 | 343 | Up |
| 9 | hsa-miR-4722-5p | 286 | 286 | Up |
| 10 | hsa-miR-4433a-3p | 245 | 245 | Up |
Degree stands for miRNA intra-network connectivity and correlation with their target genes. The miRNA with most high-degree are the most hub node in the network.
miRNAs = microRNAs.
3.5. The significant submodule from the PPI regulatory network
The 10 up-regulated miRNAs, one down-regulated miRNAs (hsa-miR-5100), and their 1912 target genes were explored further. Using the interaction information of the target genes obtained from the STRING database, the PPI regulatory network of 1912 target genes for 11 key serous DE-Exo-miRNAs were constructed. We selected a significant submodule with the highest scores of 31.226 from the PPI regulatory network (Fig. 3). Furthermore, hub genes were identified from the significant submodule, including von Hippel-Lindau tumor suppressor (VHL), NEDD4 like E3 ubiquitin protein ligase (NEDD4L), ubiquitin-conjugating enzyme E2 N (UBE2N), ubiquitin-conjugating enzyme E2 B (UBE2B), beta-transducin repeat containing E3 ubiquitin protein ligase (BTRC), ubiquitin conjugating enzyme E2 D3 (UBE2D3), ubiquitin-conjugating enzyme E2 K (UBE2K), ring finger protein 111 (RNF111), cyclin-F (CCNF), ubiquitin-conjugating enzyme E2 S (UBE2S).
Figure 3.
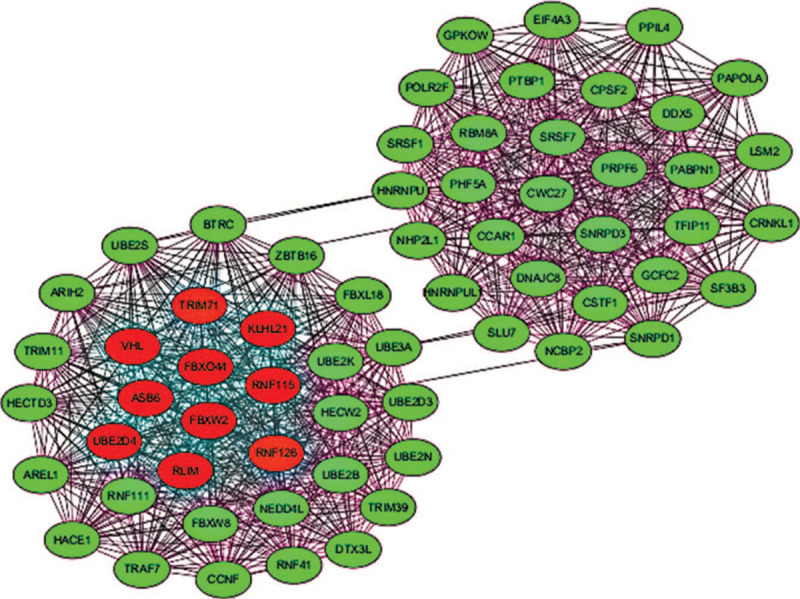
The submodule from the PPI network. The hub genes (nodes) are shown with red color. PPI = protein–protein interaction.
3.6. Regulatory network of target genes associated with ADM and their miRNAs
Among the target genes of DE-Exo-miRNAs in the serum, STAT3, PKD1, KRAS, RAC1, and RHOA are key factors participating in ADM. miRNAs play crucial gene regulatory roles by directing post-transcriptional repression of protein-coding genes. The expression of these target genes may be repressed by the DE-S-Exo-miRNAs in the serum (Fig. 4).
Figure 4.
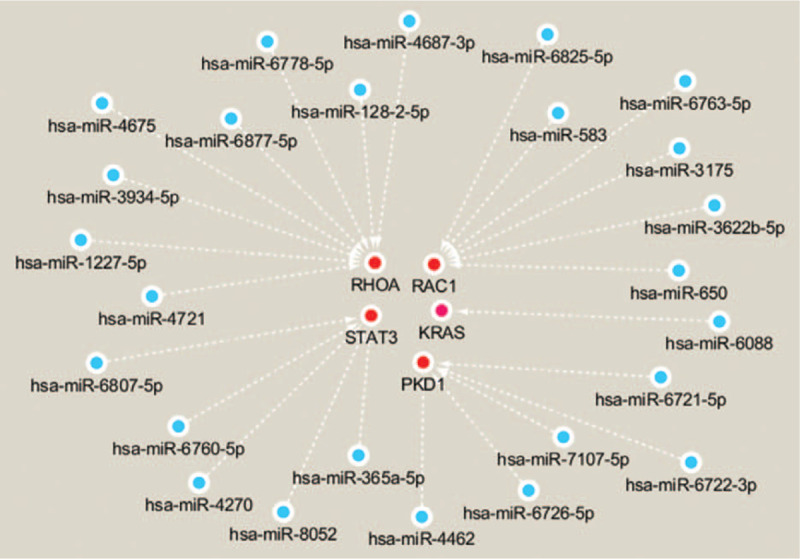
The Regulatory network of target genes associated with ADM and their miRNAs. The target genes (nodes) are shown with red color. The expression of these genes may be repressed by DE-S-Exo-miRNAs in the serum. ADM = acinar-ductal metaplasia, miRNAs = microRNAs.
4. Discussion
The molecular mechanisms of CP are complex and diagnosis can be difficulty in early phases of the disease.[2] Therefore, it is of great importance to understand the molecular mechanism of CP, as well as identify new targets to diagnose CP. Exo-miRNAs in the serum could be effective biomarkers for CP diagnosis, as they are stable and informative. Here, we downloaded the publicly accessible CP patient dataset GSE128508 and performed bioinformatic analysis. Previous study focused on AIP and only identified 10 differentially expressed Exo-miRNAs (DE-Exo-miRNAs).[23] When only comparing CP and normal samples, we identified 227 interesting DE-Exo-miRNAs in the serum. Moreover, we identified 5164 target genes of the 227 dysregulated Exo-miRNAs in CP serum. GO term and KEGG pathway analysis of these 5164 target genes indicated that they involved in the regulation of numerous biological processes and signaling pathways such as pancreatic cancer, amongst many others. ADM in the inflammatory environment of pancreatitis is a precursor of pancreatic cancer. The expression of the target genes associated with ADM (STAT3, PKD1, KRAS, RAC1, and RHOA) may be repressed by DE-Exo-miRNAs in the serum, which may contribute to blocking ADM initiation.
GO term analysis indicated that these target genes were enriched in the cell cycle process, cellular response to signal transduction by p53 class mediator, lymphocyte mediated immunity, regulation of cell death, regulation of cell differentiation, regulation of gene expression, regulation of cell communication, regulation of cellular metabolic process, and regulation of B cell activation, amongst others. These biological processes are associated with CP. Cyclin D1, an important regulator of the G1 phase of the cell cycle, increases in the pancreas of patients with CP, which may contribute to a hyperproliferative state in CP.[37] Inhibition of p53 expression decreased levels of inflammatory factors and suppressed progression of CP in the mouse model.[38] The chronic inflammatory process in CP markedly affects immune function in CP patients.[39] Pancreatitis also leads cell death, such as apoptosis and necrosis.[40] Pancreatic acinar cells undergoing ADM contribute to the regeneration of the pancreas in response to injury.[41] The target genes of miRNA in serum exosomes may promote ADM by regulation of cell differentiation and gene expression. Cell communication of pancreatic acinar cells is associated with basal release of amylase.[42] The disruption of the metabolism at a molecular level occurs in the pancreas during pancreatitis.[43] Finally, the pancreatic B cell can secrete insulin.[44] The target genes of miRNA in serum exosomes may remedy or exacerbate diabetes mellitus (DM) secondary to chronic pancreatitis by regulation of B cell activation.
The information regarding KEGG pathway analysis indicated that the target genes involve in pancreatic cancer, pathways in cancer, p53 signaling pathway, MAPK signaling pathway, cellular senescence, and signaling pathways regulating pluripotency of stem cells, amongst others. Chronic pancreatitis increases the risk of pancreatic cancer.[45] p53 is one of the most important tumor suppressor genes that is frequently mutated in human cancers.[46] p53 mutations can be detected in some cases of CP.[47] The target genes associated with pancreatic cancer, pathways in cancer, and p53 signaling pathway may contribute to pancreatic cancer associated with CP. As the major effector cells, activation of pancreatic stellate cells (PSCs) contributes to fibrosis of PC.[48] PSCs can be activated by many cellular signals, such as MAPK pathway.[49] The number of senescent PSCs are associated with the severity of inflammation and the extension of fibrosis of CP.[50] Pancreatic acinar cells show plasticity and dedifferentiate to an embryonic progenitor phenotype in experimental chronic pancreatitis.[51] Signaling pathways regulating pluripotency of stem cells may involve in this process. Therefore, a comprehensive understanding of the KEGG pathway analysis could be helpful in exploring complicated mechanisms underlying CP.
We selected the top 10 up-regulated miRNAs with high degrees and 1 up-regulated miRNAs to constructed a PPI regulatory network and selected a significant submodule with the highest scores from the PPI regulatory network. Furthermore, hub genes were identified from the significant submodule. Our current understanding for the functions of hue genes in CP are limited. As tumor suppressor gene, VHL inhibits the proliferation, migration, and invasion of pancreatic cancer cells.[52] Moreover, UBE2S regulates pancreatic cancer cell metastasis and invasion by epithelial-mesenchymal transition.[53] VHL and UBE2S may involve in pancreatic cancer associated with CP. The roles of other hub genes in CP need to be researched further in future studies.
Functionally grouped network of KEGG pathways show that pancreatic cancer and pathways in cancer are significant. ADM in the inflammatory environment of CP is a precursor of pancreatic cancer. Among the target genes of DE-Exo-miRNAs in the serum, STAT3, PKD1, KRAS, RAC1, and RHOA are key factors participating in ADM. STAT3 and PDK1 are critical effectors of oncogenic KRAS in the pancreas, mediating ADM formation.[31,32] Activation of Rac1 is required for transdifferentiation of acinar cells into ADM.[33] Moreover, activation of RhoA/ROCK signaling pathway promotes the formation of ADM.[34] We construct the regulatory network of these target genes and their DE-Exo-miRNAs in the serum. Nevertheless, the relationship between DE-Exo-miRNAs and target genes associated with ADM require future experimental verification and could provide interesting therapeutic leads of preventing pancreatic cancer associated with CP.
As a class of non-coding RNAs, one major function of miRNAs is the post-transcriptional repression of their target genes and they may be potential therapeutic targets for CP by interfering with the relative genetic factors.[54] miRNA-21 and connective tissue growth factor (CCN2) in PSC-derived exosomes are newly identified participants in PSC fibrogenic signaling during CP.[55] The exosomes may easily transport miRNAs to their target cells by encapsulating and protecting them. Therefore, studying the functions of DE-Exo-miRNAs in the serum and target genes is a promising avenue to identifying novel therapeutics.
In our work, we use bioinformatics analysis to identify serous DE-Exo-miRNAs between CP and normal samples. We then obtained their target genes from the miRWalk database. Functional analysis of these target genes as well as interactions between them provided insights into the molecular mechanism of CP. We constructed a network of target genes associated with ADM and their DE-Exo-miRNAs in the serum, as the interactions between these could likely underlie ADM in CP that is a precursor of pancreatic cancer. Nevertheless, further experimental validation is required for these predictions, which was a limitation for our study. Additionally, the fold changes of 227 DE-Exo-miRNAs are lower than 1.78 and detection of the difference is very difficult. That is another limitation.
In summary, Exo-miRNAs in the serum and their target genes may be potential tools for the early diagnosis and treatment of CP. In addition, we identified potential Exo-miRNAs (hsa-miR-4270, hsa-miR-4462, hsa-miR-3622b-5p, hsa-miR-6088, and hsa-miR-3934-5p, etc.) involved in ADM that is a precursor of pancreatic cancer associated with CP and their target genes (STAT3, PKD1, RAC1, KRAS, and RHOA).
5. Authors’ contributions
Conceptualization: Li-Ping Sheng.
Data curation: Li-Ping Sheng.
Methodology: Li-Ping Sheng.
Project administration: Zhen Ding.
Supervision: Xuan-Ji Li, Xin-Ru Xie, Rong Lin.
Software: Li-Ping Sheng, Chi Nie, Tao Xu, Kun Zhang, Xuan-Ji Li.
Validation: Li-Ping Sheng, Chi Nie, Tao Xu, Kun Zhang, Xuan-Ji Li, Xin-Ru Xie.
Visualization: Li-Ping Sheng, Tao Xu, Kun Zhang, Xin-Ru Xie.
Writing – original draft: Zhen Ding.
Writing – review & editing: Zhen Ding.
Footnotes
Abbreviations: ADM = acinar-ductal metaplasia, AIP = autoimmune pancreatitis, BTRC = beta-transducin repeat containing E3 ubiquitin protein ligase, CCNF = cyclin-F, CP = chronic pancreatitis, DE-Exo-miRNAs = differentially expressed Exo-miRNAs, DM = diabetes mellitus, Exo-miRNAs = Exosomal microRNAs, GEO = Gene Expression Omnibus, GO = gene ontology, HC = healthy control, KEGG = Kyoto Encyclopedia of Genes and Genomes, KRAS = Kirsten rat sarcoma 2 viral oncogene homolog, MAPK = mitogen-activated protein kinase, MCODE = molecular complex detection, miRNAs = microRNAs, MORC4 = MORC family CW-type zinc finger 4, NEDD4L = NEDD4 like E3 ubiquitin protein ligase, PDAC = pancreatic ductal adenocarcinoma, PDK1 = pyruvate dehydrogenase kinase 1, PPI = protein–protein interaction, PSCs = pancreatic stellate cells, RAC1 = Rac family small GTPase 1, RHOA = ras homolog family member A, RNF111 = ring finger protein 111, SAM = significance analysis of microarrays, SMAD3 = SMAD family member 3, STAT3 = signal transducer and activator of transcription 3, STRING = Search Tool for the Retrieval of Interacting Genes, TRPV1 = transient receptor potential cation channel subfamily V member 1, UBE2B = ubiquitin-conjugating enzyme E2 B, UBE2D3 = ubiquitin conjugating enzyme E2 D3, UBE2K = ubiquitin-conjugating enzyme E2 K, UBE2N = ubiquitin-conjugating enzyme E2 N, UBE2S = ubiquitin-conjugating enzyme E2 S, VHL = von Hippel-Lindau tumor suppressor.
How to cite this article: Sheng LP, Han CQ, Nie C, Xu T, Zhang K, Li XJ, Xie XR, Lin R, Ding Z. Identification of potential serum exosomal microRNAs involved in acinar-ductal metaplasia that is a precursor of pancreatic cancer associated with chronic pancreatitis. Medicine. 2021;100:18(e25753).
This study was supported in part by the National Natural Science Foundation of China (81770637).
The authors have no conflicts of interest to disclose.
Data available statement: Data comes from GEO database, which is a public open platform.
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Whitcomb DC, Frulloni L, Garg P, et al. Chronic pancreatitis: an international draft consensus proposal for a new mechanistic definition. Pancreatology 2016;16:218–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gupte A, Goede D, Tuite R, et al. Chronic pancreatitis. BMJ 2018;361:k2126. [DOI] [PubMed] [Google Scholar]
- [3].Yu P, Liu K, Gao X, et al. Transforming growth factor-β and bone morphogenetic protein 2 regulation of MicroRNA-200 family in chronic pancreatitis. Pancreas 2018;47:252–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang Z, Li D, Jin J, et al. Association between microRNA polymorphisms and chronic pancreatitis. Pancreatology 2016;16:244–8. [DOI] [PubMed] [Google Scholar]
- [5].Xin L, Gao J, Wang D, et al. Novel blood-based microRNA biomarker panel for early diagnosis of chronic pancreatitis. Sci Rep 2017;7:40019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang D, Xin L, Lin J-H, et al. Identifying miRNA-mRNA regulation network of chronic pancreatitis based on the significant functional expression. Medicine (Baltimore) 2017;96:e6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fan L, Hui X, Mao Y, et al. Identification of acute pancreatitis-related genes and pathways by integrated bioinformatics analysis. Dig Dis Sci 2020;65:1720–32. [DOI] [PubMed] [Google Scholar]
- [8].Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov 2014;13:622–38. [DOI] [PubMed] [Google Scholar]
- [9].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203–22. [DOI] [PubMed] [Google Scholar]
- [11].Párrizas M, Novials A. Circulating microRNAs as biomarkers for metabolic disease. Best Pract Res Clin Endocrinol Metab 2016;30:591–601. [DOI] [PubMed] [Google Scholar]
- [12].Wang J, Liew OW, Richards AM, et al. Overview of MicroRNAs in cardiac hypertrophy, fibrosis, and apoptosis. Int J Mol Sci 2016;17:749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ortiz-Quintero B. Cell-free microRNAs in blood and other body fluids, as cancer biomarkers. Cell Prolif 2016;49:281–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Arner P, Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol 2015;11:276–88. [DOI] [PubMed] [Google Scholar]
- [15].Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–9. [DOI] [PubMed] [Google Scholar]
- [16].Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 2010;73:1907–20. [DOI] [PubMed] [Google Scholar]
- [17].Barile L, Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther 2017;174:63–78. [DOI] [PubMed] [Google Scholar]
- [18].Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569–79. [DOI] [PubMed] [Google Scholar]
- [19].Ferguson SW, Nguyen J. Exosomes as therapeutics: the implications of molecular composition and exosomal heterogeneity. J Control Release 2016;228:179–90. [DOI] [PubMed] [Google Scholar]
- [20].Gallo A, Tandon M, Alevizos I, et al. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 2012;7:e30679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Squadrito ML, Baer C, Burdet F, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep 2014;8:1432–46. [DOI] [PubMed] [Google Scholar]
- [22].Jiménez-Alesanco A, Marcuello M, Pastor-Jiménez M, et al. Acute pancreatitis promotes the generation of two different exosome populations. Sci Rep 2019;9:19887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nakamaru K, Tomiyama T, Kobayashi S, et al. Extracellular vesicles microRNA analysis in type 1 autoimmune pancreatitis: increased expression of microRNA-21. Pancreatology 2020;20:318–24. [DOI] [PubMed] [Google Scholar]
- [24].Pinho AV, Chantrill L, Rooman I. Chronic pancreatitis: a path to pancreatic cancer. Cancer Lett 2014;345:203–9. [DOI] [PubMed] [Google Scholar]
- [25].Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 2015;12:697. [DOI] [PubMed] [Google Scholar]
- [26].Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bader GD, Hogue CWV. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chin C-H, Chen S-H, Wu H-H, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8: suppl: S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol 2017;14:296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Corcoran RB, Contino G, Deshpande V, et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res 2011;71:5020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Eser S, Reiff N, Messer M, et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell 2013;23:406–20. [DOI] [PubMed] [Google Scholar]
- [33].Saponara E, Visentin M, Baschieri F, et al. Serotonin uptake is required for Rac1 activation in Kras-induced acinar-to-ductal metaplasia in the pancreas. J Pathol 2018;246:352–65. [DOI] [PubMed] [Google Scholar]
- [34].Tao X, Chen Q, Li N, et al. Serotonin-RhoA/ROCK axis promotes acinar-to-ductal metaplasia in caerulein-induced chronic pancreatitis. Biomed Pharmacother 2020;125:109999. [DOI] [PubMed] [Google Scholar]
- [35].Liu L, Zhu Y, Noë M, et al. Neuronal transforming growth factor beta signaling via SMAD3 contributes to pain in animal models of chronic pancreatitis. Gastroenterology 2018;154:2252.e2–65.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhu Y, Colak T, Shenoy M, et al. Nerve growth factor modulates TRPV1 expression and function and mediates pain in chronic pancreatitis. Gastroenterology 2011;141:370–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kornmann M, Ishiwata T, Arber N, et al. Increased cyclin D1 expression in chronic pancreatitis. Pancreas 1998;17:158–62. [DOI] [PubMed] [Google Scholar]
- [38].Zhou L, Tan J-H, Cao R-C, et al. ATF6 regulates the development of chronic pancreatitis by inducing p53-mediated apoptosis. Cell Death Dis 2019;10:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gansauge F, Gansauge S, Eh M, et al. Distributional and functional alterations of immunocompetent peripheral blood lymphocytes in patients with chronic pancreatitis. Ann Surg 2001;233:365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gukovskaya AS, Pandol SJ. Cell death pathways in pancreatitis and pancreatic cancer. Pancreatology 2004;4:567–86. [DOI] [PubMed] [Google Scholar]
- [41].Puri S, Folias AE, Hebrok M. Plasticity and dedifferentiation within the pancreas: development, homeostasis, and disease. Cell Stem Cell 2015;16:18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Meda P, Bruzzone R, Knodel S, et al. Blockage of cell-to-cell communication within pancreatic acini is associated with increased basal release of amylase. J Cell Biol 1986;103:475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ma C, Tian B, Wang J, et al. Metabolic characteristics of acute necrotizing pancreatitis and chronic pancreatitis. Mol Med Rep 2012;6:57–62. [DOI] [PubMed] [Google Scholar]
- [44].Petit P, Loubatières-Mariani MM. Potassium channels of the insulin-secreting B cell. Fundam Clin Pharmacol 1992;6:123–34. [DOI] [PubMed] [Google Scholar]
- [45].Kirkegård J, Mortensen FV, Cronin-Fenton D. Chronic pancreatitis and pancreatic cancer risk: a systematic review and meta-analysis. Am J Gastroenterol 2017;112:1366–72. [DOI] [PubMed] [Google Scholar]
- [46].Hong B, van den Heuvel APJ, Prabhu VV, et al. Targeting tumor suppressor p53 for cancer therapy: strategies, challenges and opportunities. Curr Drug Targets 2014;15:80–9. [DOI] [PubMed] [Google Scholar]
- [47].Gansauge S, Schmid RM, Muller J, et al. Genetic alterations in chronic pancreatitis: evidence for early occurrence of p53 but not K-ras mutations. Br J Surg 1998;85:337–40. [DOI] [PubMed] [Google Scholar]
- [48].Apte MV, Haber PS, Darby SJ, et al. Pancreatic stellate cells are activated by proinflammatory cytokines: implications for pancreatic fibrogenesis. Gut 1999;44:534–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jin G, Hong W, Guo Y, et al. Molecular mechanism of pancreatic stellate cells activation in chronic pancreatitis and pancreatic cancer. J Cancer 2020;11:1505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Fitzner B, Müller S, Walther M, et al. Senescence determines the fate of activated rat pancreatic stellate cells. J Cell Mol Med 2012;16:2620–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pinho AV, Rooman I, Reichert M, et al. Adult pancreatic acinar cells dedifferentiate to an embryonic progenitor phenotype with concomitant activation of a senescence programme that is present in chronic pancreatitis. Gut 2011;60:958–66. [DOI] [PubMed] [Google Scholar]
- [52].Sun J, Jiang Z, Li Y, et al. Downregulation of miR-21 inhibits the malignant phenotype of pancreatic cancer cells by targeting VHL. Onco Targets Ther 2019;12:7215–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wang L, Liang Y, Li P, et al. Oncogenic activities of UBE2S mediated by VHL/HIF-1α/STAT3 signal via the ubiquitin-proteasome system in PDAC. Onco Targets Ther 2019;12:9767–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hu L-H, Ji J-T, Li Z-S. Potential application of miRNAs as diagnostic and therapeutic tools in chronic pancreatitis. J Cell Mol Med 2015;19:2049–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Charrier A, Chen R, Chen L, et al. Connective tissue growth factor (CCN2) and microRNA-21 are components of a positive feedback loop in pancreatic stellate cells (PSC) during chronic pancreatitis and are exported in PSC-derived exosomes. J Cell Commun Signal 2014;8:147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]


