Abstract
Our understanding of folate metabolism in Leishmania has greatly benefited from studies of resistance to the inhibitor methotrexate (MTX). Folates are reduced in Leishmania by the bifunctional dihydrofolate reductase thymidylate synthase (DHFR-TS) and by pteridine reductase (PTR1). To further our understanding of folate metabolism in Leishmania, a Cos-seq genome-wide gain of function screen was performed against MTX and against the two thymidylate synthase (TS) inhibitors 5-fluorouracil and pemetrexed. The screen revealed DHFR-TS and PTR1 but also the nucleoside transporter NT1 and one hypothetical gene derived from chromosome 31. For MTX, the concentration of folate in the culture medium affected the enrichment pattern for genes retrieved by Cos-seq. We generated a L. infantum DHFR-TS null mutant that was thymidine auxotroph, a phenotype that could be rescued by the addition of thymidine or by transfection of the flavin dependent bacterial TS gene ThyX. In these DHFR-TS null mutants it was impossible to obtain a chromosomal null mutant of PTR1 except if DHFR-TS or PTR1 were provided episomally. The transfection of ThyX however did not allow the elimination of PTR1 in a DHFR-TS null mutant. Leishmania can survive without copies of either DHFR-TS or PTR1 but not without both. Provided that our results observed with the insect stage parasites are also replicated with intracellular parasites, it would suggest that antifolate therapy in Leishmania would only work if both DHFR-TS and PTR1 would be targeted simultaneously.
Author summary
The protozoan parasite Leishmania is auxotroph for folate and unconjugated pterins and salvages both from the mammalian host. Two enzymes of the folate metabolism pathway, namely the bifunctional dihydrofolate reductase-thymidylate synthase (DHFR-TS) and the pteridine reductase 1 (PTR1), are being evaluated for drug discovery and repurposing of existing anti-metabolites. Despite their apparent potential, development of DHFR-TS and PTR1 targeted chemotherapy against Leishmania is still awaiting. Here we revisited folate metabolism at the genomic level and report on the identification of known resistance genes alongside some new ones. Through gene disruption studies we found that L. infantum DHFR-TS null mutants are thymidine auxotroph and that these can be rescued by the bacterial flavin dependent thymidylate synthase ThyX. We also found that PTR1 is essential in the absence of a functional DHFR-TS even in the presence of ThyX or thymidine supplementation, indicating the essential role of reduced pterins or folate beyond thymidine synthesis. This study indicates that simultaneous targeting of DHFR-TS and PTR1 will be required for the development of anti-folate chemotherapy against Leishmania.
Introduction
As early as in the 50’s and the 60’s biopterin and folic acid were found to be required to sustain the growth of Leishmania parasites in vitro [1,2]. This was of interest as the folic acid pathway had been the target of many chemotherapeutic compounds against bacteria (e.g. trimethoprim), parasites (pyrimethamine) or cancer cells (methotrexate). While the main role of reduced folates is well established in Leishmania in thymidylate biosynthesis [3] the exact role of reduced pterins is still unclear [4,5]. They provide protection against oxidative stresses [6,7], are involved in the process of metacyclogenesis [8,9] but other roles as growth promoter are likely. Our understanding of folate/pterin metabolism in Leishmania has greatly benefited from studies on mechanisms of resistance to the model drug methotrexate (MTX). Indeed, upon MTX selection the parasite will amplify part of its genome (reviewed in [10]). Characterization of the gene content of these amplified loci led to the discovery of the dihydrofolate reductase (DHFR) fused to thymidylate synthase (TS) DHFR-TS [11] and of the pteridine reductase PTR1 [12,13]. Many other proteins involved in folate metabolism and transport were described, often related to studies of MTX resistance (reviewed in [4,5]). Intriguingly there are links between the two one-carbon metabolic donors that are reduced folates and S-adenosylmethionine [14], and these two metabolites are transported inside the Leishmania cells by the same family of transporters [15]. The genomic screen Cos-seq, an approach based on functional cloning coupled to next-generation sequencing, have helped to improve our understanding of one carbon metabolism in Leishmania when applied to inhibitors of this metabolic pathway [16,17].
Here we report a series of Cos-seq screens with different inhibitors of the folate metabolism that discovered already known as well as new markers of resistance. The genes PTR1 and DHFR-TS were prevalent in most of our screens and these two genes, as well as their genetic interactions, were further studied by gene disruption experiments in L. infantum.
Methods
Parasite culture
L. infantum MHOM/MA/67/ITMAP-263 parasites and their transfectants were maintained as promastigotes at 25°C in SDM-79 or M199. Depending on requirements, thymidine and folate were supplemented. Cell growth and EC50 against drugs were monitored by measuring the absorbance at 600 nm.
Cos-seq procedures
L. infantum cells transfected with a cosmid library have been described before [17]. The Cos-seq procedure and the cosmid extraction after passages with increased concentration of either MTX, 5-FU or PMX was performed essentially as described by Potvin et al. [18]. Briefly, cosmid DNA extracted from parasites by SDS/alkali lysis and phenol/CHCl3 extraction was quantified with the QuantiFluor dsDNA System staining kit (Promega) and fifty nanograms were used for paired-end library preparation using Nextera DNA Sample preparation kit (Illumina). Sequencing libraries were quantified with the QuantiFluor dsDNA System, their size distribution was checked with Bioanalyzer (Agilent) and these were sequenced using an Illumina HiSeq2500 system at a final concentration of 8 pM. Sequencing reads from each sample were independently aligned to the L. infantum JPCM5 reference genome obtained from TriTrypDB (http://tritrypdb.org/tritrypdb/) using the BWA software [19]. BEDTools was used to convert BAM files to BED files [20] for read depth coverage. The detection of genes enriched with the screens relied on the Trinity software version 2.1.1 [21], which includes all third-party tools required for the analysis. Gene abundance within samples was quantified using the Kallisto software [22]. Clusters of genes significantly enriched by drug selection were retrieved with edgeR [23] using the default parameters (false discovery rate≤0.001). Gene clusters were then plotted according to the median-centered log2 fragment per kilobase per million mapped reads (FPKM) values using R scripts included in the Trinity package. Only genes with a log2-fold change ≥4 at any stage of selection were retained.
DNA constructs, cosmid isolation and transfection
The genes of L. infantum were amplified from genomic DNA using compatible primer pairs and cloned in the Leishmania expression vectors pSP72αBlastα or pSP72αPuroα unless mentioned otherwise. Each plasmid insert was sequenced to confirm the absence of mutation. A total of 20 μg of plasmid DNA for episomal expression were transfected into 5×108 Leishmania promastigotes by electroporation using a BioRad Gene Pulser II Electroporation System with one electrical pulse at 450 Volts with capacitance at 500μF. Transfected parasites were selected and maintained with either puromycin (100 μg/ ml) or blasticidine (100 μg/ ml).
For generating knockout lines, replacement constructs with neomycin, hygromycin, puromycin or zeocin resistance markers with ~600 bp upstream or downstream of target ORFs were prepared by fusion PCR as described earlier [24]. Each of these constructs was cloned in the pGEM-T-Easy vector. After confirming proper fusion by sequencing, replacement construct DNA was prepared by digesting the plasmids with NotI and subsequent purification. 10 μg of DNA was transfected in target cells followed by selection with appropriate antibiotics (zeocin, 400 μg/ml; puromycin, 100 μg/ml; hygromycin, 400 μg/ml and G418, 40 μg/ml). Clones were isolated on agar plates. Genomic DNAs for Southern blot analysis were isolated from clones using the DNAzol reagent (Invitrogen) as recommended by the manufacturer, except for DHFR-TSN/H PTR1P/Z/+ parasites with or without a ThyX episome for which gDNA was extracted from the population of transfectants. The list of all PCR primers from this study can be found in S1 Table.
[H3]-thymidine transport
1×108 mid-log phase promastigotes were washed and resuspended in transport assay buffer (33 mM HEPES, 98 mM NaCl, 4.6 mM KCl, 0.55 mM CaCl2, 0.07 mM MgSO4, 5.8 mM NaH2PO4, 0.3 mM MgCl2, 23 mM NaHCO3 and 14 mM glucose, pH 7.3) supplemented with 250 nM of [3H] thymidine (44.2 Ci mmol−1) (PerkinElmer). Radioactivity accumulation was measured as previously described [25]. Total cellular protein from the same cell population was quantified using Bradford reagent (Biorad). The uptake was normalized to total protein in lysate and the background transport level (t = 0) was removed by subtracting the accumulation values obtained on ice from each of the test readings
Thymidine auxotrophy
To test thymidine auxotrophy, 108 promastigotes grown in thymidine supplemented SDM-79 were transferred to thymidine-free medium and grown for one day. After monitoring for proper cell physiology under the microscope, 2×106 cells were transferred to fresh SDM-79 medium and growth was monitored by measuring OD600 each day after inoculation. To determine minimal thymidine concentration, the SDM-79 was supplemented with various concentrations of thymidine and growth was monitored similarly.
Statistical analysis
For statistical analysis a two-tailed unpaired t-test with GraphPad Prism 5.01 software was performed unless mentioned otherwise.
Results
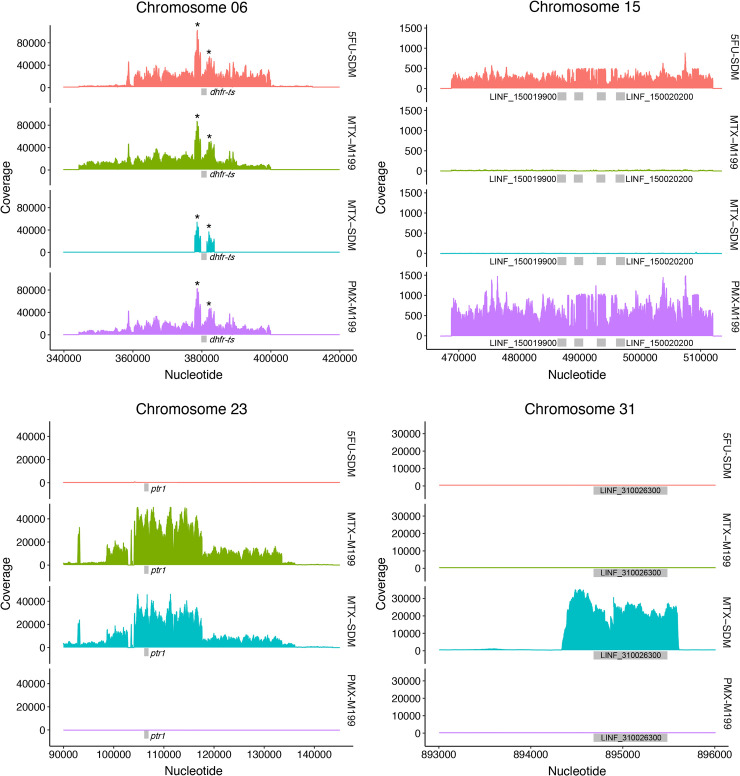
A Cos-seq screen has already been done using MTX selection in M199 medium and this led to the isolation of DHFR-TS and PTR1 [17]. In order to delve further into the one carbon metabolism in Leishmania we carried out additional screens using MTX and the two TS inhibitors 5-fluorouracil (5-FU) and pemetrexed (PMX), as described in the Cos-seq procedures section of the Methods. Folate concentration influences Leishmania response to MTX [26–28] and this was confirmed by contrasting EC50s between cells grown in M199 or SDM-79 medium with close to one thousand fold difference (Table 1). Since our initial Cos-seq MTX screen was carried out in the folate poor M199 medium, we carried out our screen in the folate rich SDM-79 medium in the hope of diversifying gene discovery. This screen led to a cosmid derived from chromosome 23 that encodes a number of genes (S2 Table), including PTR1 (Fig 1). Enriched cosmids, as deduced from an increase in the number of sequence reads, also led to a single gene locus (LINF_310026300) within chromosome 31 (Fig 1). Single genes are unusual for cosmids and this cosmid must have rearranged either in E. coli or Leishmania prior to its isolation. Surprised by the absence of enrichment of the DHFR-TS locus we carried out a new MTX Cos-seq screen in M199 medium and under these conditions we could observe enrichment for both the PTR1 and DHFR-TS loci (Fig 1), indicating a difference in cosmid/gene enrichment with MTX selection depending on folate concentration in the medium. In contrast to MTX, susceptibility to 5-FU is similar in both M199 and SDM-79 medium (Table 1) and the Cos-seq screen was carried out solely in SDM-79 medium. The drug PMX is inactive against Leishmania when grown in SDM-79 but equally active to MTX when parasites are grown in M199 medium (Table 1). The Cos-seq screen with PMX was therefore carried out in M199. The Cos-seq screens with both 5-FU and PMX led to the same two regions enriched in terms of sequence reads, one derived from chromosome 6 and encoding DHFR-TS and another from a locus located on chromosome 15 (Fig 1) and encoding for a number of genes including the nucleoside transporter NT1 (S2 Table). NT1 is known to transport purines and pyrimidines [29,30].
Table 1. Susceptibilities to antimetabolites of Leishmania infantum transfected with genes pinpointed by a Cos-seq screen.
| L. infantum strains1 | EC50 (μM) and fold change2 | ||
|---|---|---|---|
| 5-FU | MTX | PMX | |
| WT | 5.35±0.90 | 0.45±0.04 | 0.25±0.03 |
| cL-Hyg | 8.75±0.21 | 0.32±0.04 | 0.23±0.01 |
| cL-Hyg (SDM) | 14.09±0.37 | 259.56±2.29 | NS2 |
| psp72 α-blast-α | 5.38±0.90 | 0.32±0.59 | 0.24±0.03 |
| DHFR-TS α-blast-α | 15.80±2.84 (3.1±0.9**, n = 3) |
0.95±0.02 (3.0±0.5***, n = 3) |
2.02±0.08 (8.4±0.9**, n = 3) |
| PTR1 α-blast-α | 12.2±0.81 (2.3±0.4**, n = 3) |
1.11±0.14 (4.1±0.8**, n = 3) |
0.71±0.06 (3.0±0.6***, n = 3) |
| psp72 α-puro-α | 7.33±1.04 | 0.34±0.65 | 0.34±0.08 |
| NT1 α-puro-α | 11.94±0.44 (1.7±0.3***, n = 5) |
NA | 0.97±0.26 (2.9±0.6**, n = 4) |
| LINF_310026300 α-puro-α | NA | 0.78±0.28 (2.2±0.4**, n = 6) |
NA |
| DHFR-TS-/+ | 3.88±0.35 (-1.4 ±0.1*, n = 3) |
0.17±0.07 (-2.7±0.3***, n = 3) |
0.18±0.01 (-2.1±0.3**, n = 3) |
| PTR1-/- | 3.62±0.11 (-1.5±0.2*, n = 3) |
0.09±0.02 (-5.0±0.5***, n = 3) |
0.18±0.005 (-1.3±0.1*, n = 3) |
1 The L. infantum PTR1-/- strain has been described in [6].
2 All carried out in M199, except for a wild-type line transfected with cL-Hyg that was also measured in SDM-79 medium.
NS: not susceptible; NA: not applicable
Statistical analyses were performed using unpaired two-tailed t-tests.
***P < 0.001
**P < 0.01
*P < 0.05
Fig 1. Cos-Seq identification of loci upon 5 fluorouracil (5-FU), methotrexate (MTX) and pemetrexed (PMX) selection.
Visualization of the four enriched loci on chromosomes 6, 15, 23, and 31 as delimited by regions of higher read density. For LinJ.06, the asterisk denotes a bias in read counts coming from the DHFR-TS flanking regions originating from the cLHYG backbone [51]. The medium (M199 or SDM-79) in which the screen was done is also indicated. The key genes are shown with boxes and the full list of genes part of the enriched cosmids can be found in S2 Table.
We cloned the four genes PTR1, DHFR-TS, NT1 and LINF_310026300 potentially involved in resistance to either MTX, 5-FU or PMX into Leishmania expression vectors and tested those transfectants for resistance. As reported previously, transfection of the individual DHFR-TS and PTR1 genes conferred resistance to MTX (Table 1), but also to 5-FU and PMX (Table 1). Transfection of NT1 led to parasites resistant to both 5-FU and PMX (Table 1). The NT1 transfectant also had an increased capacity to transport thymidine (Fig 2). Episomal expression of the LINF_310026300 hypothetical gene enriched in the screen with MTX conferred a two-fold increase in resistance to MTX (Table 1). Searches in databases did not reveal any structural features that could help in determining the possible function of this hypothetical protein.
Fig 2. Transport of thymidine in L. infantum transfected with the nucleoside transporter NT1.
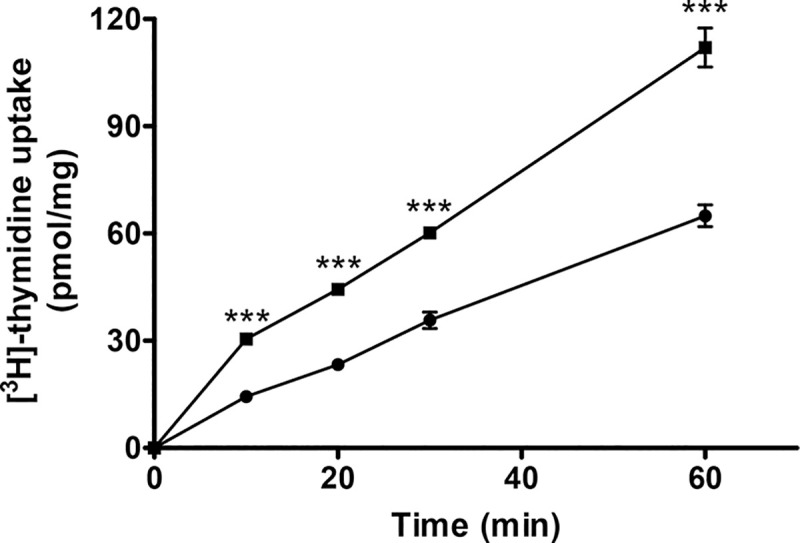
The uptake of thymidine in L. infantum transfected with pSP72αpuroα (●) or with pSP72αpuroα-NT1 (◼). Average of three experiments with standard error of mean (SEM) are presented, ***p <0.005.
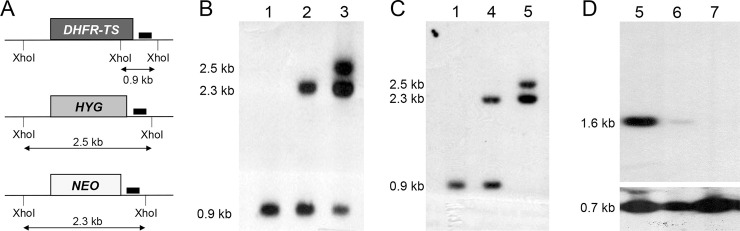
Since the Cos-seq screen did not reveal the anticipated diversity of new genes involved in MTX resistance, we resorted in studying the consequences of gene replacement of both PTR1 and DHFR-TS, the two main markers revealed by our screen. A knockout of DHFR-TS has been described in L. major and these parasites were thymidine auxotroph [31]. A knockout of DHFR-TS has not been reported in other species, including species associated with visceral leishmaniasis. The PTR1 gene has been inactivated in at least three species [6,27,32]. Leishmania is a pterin auxotroph but a PTR1 knockout cell will grow well if sufficient reduced pterins are available [27,32]. While a biopterin transporter 1 (BT1)/PTR1 knock out was achieved in Leishmania [33], there are no published attempts at inactivating both DHFR-TS and PTR1 in the same cell. In order to attempt this double knockout, we first tested whether we can inactivate the DHFR-TS gene in L. infantum by a classical gene knockout strategy using neomycin (NEO) and hygromycin (HYG) selection cassettes (Fig 3A). Transfection of the NEO cassette led to a DHFR-TSNEO/+ single knockout (Fig 3B, lane 2) but transfection of the HYG cassette in the DHFR- TSNEO/+ cells led to a DHFR- TSNEO/HYG/+ parasite (Fig 3B, lane 3) with a remaining DHFR-TS wild-type allele. Five clones were analyzed and all five exhibited similar aneuploidies at the DHFR-TS locus (S1A Fig). This aneuploidy is often encountered in Leishmania when attempting to disrupt genes reputed essential [34,35]. We thus first transfected an episomal copy of DHFR-TS as part of a BLAST vector (psp72αblastα-DHFR-TS) in the DHFR-TSNEO/+ cells (Fig 3C, lane 4) and in these cells, upon transfection of the HYG inactivation cassette, we could obtain a DHFR-TSNEO/HYG chromosomal null mutant (Fig 3C, lane 5). These cells were cloned and harbored the DHFR-TS episome as demonstrated by the 1.6 kb band hybridizing to a probe internal to the DHFR-TS gene (Figs 3D, lane 5 and see S2A). At 20 passages in the absence of blasticidin, the copy number of the episome decreased (Fig 3D, lane 6) and after 25 passages it was cured in 4 independent clones (Figs 3D, lane 7 and S2B) provided that cells were supplemented with thymidine. We tried to decrease thymidine supplementation but this failed, indicating the thymidine auxotrophy of the L. infantum DHFR-TSNEO/HYG cells (Fig 4). Episomal expression of PTR1 in these cells did not revert thymidine auxotrophy (Fig 4). However, episomal expression of the bacterial flavin dependent TS ThyX [36] derived from Mycobacterium tuberculosis did allow growth of the Leishmania DHFR-TSNEO/HYG cells without the addition of thymidine (Fig 4).
Fig 3. Deletion of the DHFR-TS gene in Leishmania infantum.
(A) Schematic representation of the DHFR-TS locus and of the integration of the hygromycin phosphotransferase B (HYG) and neomycin phosphotransferase (NEO) deletion cassettes. Also shown are the expected sizes after digestion with XhoI and the location of probe 1 (small black boxes) used for the hybridization of Southern blots. (B) Southern blot analysis of genomic DNAs digested with XhoI from L. infantum wild-type (1), the single allele knock out DHFR-TSNEO/+ (2), or the aneuploid cell DHFR-TSNEO/HYG/+ (3) hybridized to probe 1. (C) Southern blot analysis of genomic DNAs digested with XhoI from L. infantum wild-type (1), the single allele knockout clone DHFR-TSNEO/+ transfected with psp72αblastα-DHFR-TS (4), or the DHFR-TSNEO/HYG null mutant harboring psp72αblastα-DHFR-TS (5) with probe 1. (D) Southern blot analysis of total DNAs digested with XbaI and HindIII (see S2A Fig) from a clone of the DHFR- TSNEO/HYG chromosomal null mutant harboring psp72αblastα-DHFR-TS (5) or of these cells grown for 20 (lane 6) and 25 (lane 7) passages without blasticidin but supplemented with 50 μg/ml of thymidine. This blot was hybridized to a DHFR-TS intragenic probe (see S2 Fig). The blot was re-hybridized with a PTR1 probe to monitor the amount of DNA analyzed (lower panel).
Fig 4. Metabolic and genetic complementation of the DHFR-TS null mutant.
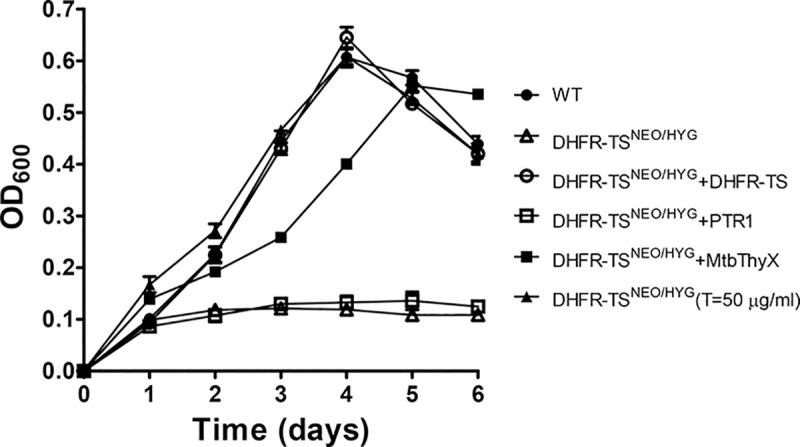
Cells were grown in SDM-79 medium and their growth was monitored by measuring their OD at 600 nm. L. infantum wild-type cells (●); or L. infantum DHFR-TSNEO/HYG complemented with either psp72αblastα-DHFR-TS (○), psp72αblastα-ThyX (■); psp72αblastα-PTR1 (☐); or supplemented with 50 μg/ml thymidine (▲); or no supplementation at all (ρ). Average of three experiments with standard error of mean (SEM) are presented.
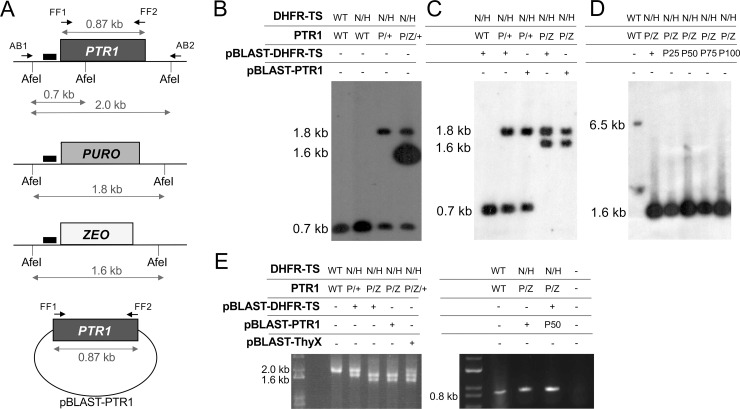
We attempted to generate a PTR1 null mutant in the DHFR-TSNEO/HYG cells using puromycin (PURO) and zeocin (ZEO) inactivation cassettes (Fig 5A). Transfection of the PURO cassette in DHFR-TSNEO/HYG cloned cells led to PTR1PURO/+ parasites (Fig 5B, lane 3). Transfection of the ZEO cassette in these parasites led to the correct integration at the PTR1 locus but with a remaining intact copy of PTR1 in the population of cells analyzed (Fig 5B, lane 4). It is unlikely that genuine PTR1 null mutants occurred in this population and that these were outgrown by cells retaining a wild-type PTR1 allele. First, the same aneuploidy at the PTR1 locus was repeated when the ZEO replacement cassette was introduced in an independent DHFR-TSNEO/HYG clone (S1B Fig). Also, genuine double null mutants are addicted to episomal expression of either DHFR-TS or PTR1 (see below). In light of the fact that inactivation of PTR1 was easily achieved in the same L. infantum wild-type strain [6], the PTR1PURO/ZEO/+ genotype is suggesting that it is not possible to obtain a viable Leishmania cell without a functional copy of either DHFR-TS or PTR1. To further investigate this putative essentiality, we transfected DHFR-TSNEO/HYG PTR1PURO/+ cells with an episomal copy (as part of a BLAST vector) of either DHFR-TS (Fig 5C lane 2) or PTR1 (Fig 5C lane 3). In both cases, we could obtain a chromosomal PTR1 null mutant upon transfection of the ZEO PTR1-inactivation cassette (Fig 5C lanes 4 and 5). We attempted to grow the double chromosomal DHFR-TS-PTR1 null mutant overexpressing DHFR-TS or PTR1 in the absence of blasticidin but in the presence of thymidine metabolic complementation. We grew those cells for up to 100 passages in the absence of blasticidin but failed to lose the DHFR-TS episome, as shown by Southern blot hybridization (Fig 5D, lanes 3–6), or the PTR1 episome, as shown by PCR amplification (Fig 5E, right panel). For the latter, the PCR band is genuinely derived from the episome as the gene is absent from the chromosome (Fig 5E, left panel). This suggests that L. infantum cells need a functional copy of either DHFR-TS or PTR1 to thrive even in thymidine rich medium. This was further supported by our attempt to delete PTR1 in a DHFR-TS null mutant in the presence of ThyX, a gene product reversing thymidine auxotrophy (Fig 4). For this, we first complemented DHFR-TSNEO/HYG PTR1PURO/+ cells with episomally-expressed ThyX (S3A and S3B Fig, lane 3). These cells were able to grow in the absence of thymidine supplementation. Upon the transfection of the ZEO PTR1-inactivation cassette we obtained integration at the right locus but the hybridization signals were consistent with the population of parasites having the PTR1PURO/ZEO/+ genotype (S3B Fig, lane 4). This PTR1PURO/ZEO/+ genotype was also obtained in an independent PTR1 gene knock out experiment where ThyX was episomally expressed (Fig 5E, left panel lane 5). When the genetic complementation was done with DHFR-TS instead of ThyX, the transfection of the ZEO PTR1-inactivation led to a PTR1 chromosomal null mutant (S3B Fig lane 5), as already shown in Fig 5C (lane 4).
Fig 5. Disruption of PTR1 in chromosomal DHFR-TS null mutants.
(A) Schematic representation of the PTR1 locus and of the integration of the puromycin N-acetyl transferase (PURO) and zeocin resistance gene (ZEO) deletion cassettes. The expected sizes after digestion with AfeI when hybridized to probe 2 (small black boxes) are shown. Also shown are the expected size of PCR fragments generated with primers FF75 and FF76 amplifying the coding sequence of PTR1, or with primers AB1 and AB2 located in its 5’ and 3’UTRs, respectively. The pSP72αBlastα-PTR1 episome is shown at the bottom. All cells from this Fig were grown in the presence of 50 μg/ml thymidine. (B and C) Southern blot analysis of genomic DNAs digested with AfeI hybridized with probe 2. The genotype of the strains for each lane is indicated on top of the blots. In panel B, the hybridized band with the ZEO integration is more intense than expected. This is likely due to tandem integration of the marker during selection. WT and +, wildtype; N, NEO; H, HYG; P, PURO; Z, ZEO; pBLAST, pSP72αBlastα. (D) Southern blot analysis of total DNAs (genomic and episomal) digested with XbaI and HindIII and hybridized to an intragenic DHFR-TS probe (see also S2 Fig). The genotype of the strains for each lane is indicated on top of the blot. WT and +, wildtype; N, NEO; H, HYG; P, PURO; Z, ZEO; pBLAST, pSP72αBlastα. P25, P50, P75 and P100 mean that parasites harboring the pSP72αBlastα-DHFR-TS episome were grown for 25, 50, 75 and 100 passages in absence of blasticidin in the presence of excess thymidine. (E) Left panel. Amplification of the PTR1 locus with primers AB1 and AB2 shows the replacement of PTR1 alleles by PURO and ZEO markers in the chromosome of DHFR-TSNEO/HYG PTR1PURO/ZEO cells harboring episomes coding for DHFR-TS or PTR1. Right panel. Amplification of the PTR1 coding sequence with primers FF75 and FF76 confirms that the psp72αblastα-PTR1 episome is not lost from DHFR-TSNEO/HYG PTR1PURO/ZEO cells in the absence of blasticidine. The genotype of the strains for each lane is indicated on top of the gels. WT and +, wildtype; N, NEO; H, HYG; P, PURO; Z, ZEO; pBLAST. P50, parasites harboring the pSP72αBlastα-PTR1 episome grown for 50 passages in the absence of blasticidin in the presence of excess thymidine.
Discussion
Our understanding of folate metabolism in Leishmania has benefited from studies of mechanisms of resistance to the model drug MTX. Here we used a Cos-seq screen [17] while selecting for MTX resistance in medium with different concentrations of folate and with two inhibitors, 5-FU and PMX, targeting primarily TS. Inhibitory action of PMX was detectable only in low folate medium (Table 1), a finding consistent to what observed in mammalian cells, where PMX activity is greatly affected by the cellular folate concentration [37]. For both 5-FU and PMX, the Cos-seq screen led to two loci; one encoding the target DHFR-TS (Fig 1) and the other encoding the nucleoside transporter NT1 (Fig 1). Amplification of the DHFR-TS locus, but not of NT1, has been observed in Leishmania cells selected for resistance to 5-FU [25]. NT1 is known to transport pyrimidine [29,38] and here we confirmed its ability of transporting thymidine (Fig 2). This increase in uptake of thymidine likely renders the TS function dispensable hence rendering parasites less susceptible to TS inhibitors. PTR1 is not a target of 5-FU or PMX and a Cos-seq screen selected with these two drugs did not lead to PTR1 (Fig 1). Yet PMX was shown to bind to the T. brucei PTR1 orthologue [39] and overexpression of PTR1 leads to 5-FU and PMX cross resistance (Table 1) while its inactivation modestly increased sensitivity to the two drugs (Table 1). Possibly the ability of PTR1 to reduce folates [32, 40,41], an essential co-factor for TS activity, may contribute to the observed phenotypes of PTR1 overexpression on 5-FU and PMX cross-resistance by competing with the binding sites of the drugs.
This laboratory has carried out numerous screens with MTX and when carried out in SDM-79 medium we never observed DHFR-TS amplification except if PTR1 is deleted [42]. When screens are carried out in M199 medium, we, as others [43,44] can observe DHFR-TS amplification [45]. Our Cos-seq data is consistent with these observations where DHFR-TS selection was seen only when the screen was carried out in M199 but not in SDM-79 (Fig 1). Folate concentration in the medium (and likely inside the parasite with its full complement of folate transporters [46–48]) have thus a huge impact on genes that can be selected for resistance. For example, we isolated the biopterin transporter BT1 by virtue that it confers MTX resistance [49] but this is only observed in SDM-79 medium with its high folate levels. Thus, medium with its variation in metabolite concentration must be considered when measuring the impact of a gene in drug resistance. This may apply also to field isolates if they need to be grown prior to resistance analysis. Cosmid based screens using MTX as the selective force [17,50] have led primarily to DHFR-TS and PTR1. Cos-seq highlighted those two genes as well as additional ones but they are associated with low level of resistance only ([17], Table 1). Possibly other type of screens such as Mut-seq [24] or loss of function screens could help in widening the discovery of genes involved in folate metabolism and MTX resistance.
DHFR-TS has been knocked out in L. major and was found to be a thymidine auxotroph [31]. We now show that this is also true for L. infantum, a species responsible for visceral leishmaniasis. This auxotrophy was proven by both genetic and metabolic means (Fig 4). Interestingly we could rescue this thymidine auxotrophy by expressing the bacterial TS ThyX gene, demonstrating that the DHFR portion is dispensable (Fig 4) provided that PTR1 is present (see below). DHFR-TS is also dispensable if cells are supplemented with thymidine as long as PTR1 is present.
DHFR-TS and PTR1 are the main resistance mechanisms towards MTX in Leishmania and both were again isolated in this Cos-seq screen (Fig 1). Both DHFR-TS and PTR1 can be deleted independently provided that thymidine or reduced pterins are in sufficient amounts in the medium. One genetic analysis never reported was the attempt of inactivating both genes in the same strain. Theoretically this should work with the appropriate metabolites’ complementation. This, however, proved not to be the case. Indeed, we could obtain chromosomal null mutant for both DHFR-TS and PTR1 in the same cell but only when either of those two genes were included as part of an episome (Fig 5). Addition of thymidine or expression of ThyX did not remove the need for an episomal rescue with either DHFR-TS or PTR1. Growth in the absence of the selective drug blasticidin for serial passages did not lead to a loss of the DHFR-TS or PTR1 containing episome. DHFR and to a lesser extent PTR1 (PTR1 was estimated to provide up to 10% of reduced folates in Leishmania [40]), reduce folate into tetrahydrofolate. The latter is a key intermediate that enters the one carbon metabolic pathways and is converted, by the action of many enzymes, into a number of key metabolites required in Leishmania for the synthesis of thymidine, methionine, or the mitochondrial methionyl tRNA formylation, but in contrast to many cells, not for purines [4,5]. Possibly some of these reduced folate intermediates are needed in small amounts and PTR1 can lead to sufficient levels of intermediates even when DHFR-TS is absent. However, those levels cannot be reached when both DHFR-TS and PTR1 are absent. Thus, Leishmania cells need at least one copy of either DHFR-TS or PTR1 to thrive. One could attempt metabolic rescue with some of these intermediates although this is complicated by their rapid oxidation. It has been commented [5] that antifolate therapy in Leishmania would only work if both DHFR-TS and PTR1 would be targeted, and this study is further supporting this claim. Further studies testing this hypothesis would also require work with intracellular amastigotes, the stage of the parasite ultimately targeted by drugs.
Supporting information
(A) Southern blot analysis of genomic DNAs digested with XhoI from L. infantum wild-type (1), from the single allele knock out DHFR-TSNEO/+ (2), or from five aneuploid DHFR-TSNEO/HYG/+ clones (3–7) hybridized to probe 1 derived from the 3’UTR of DHFR-TS (see Fig 3A). Lane 3 is the same DHFR-TSNEO/HYG/+ clone as in Fig 3B. (B) Southern blot analysis of genomic DNAs digested with AfeI from L. infantum wild-type (1), DHFR-TSNEO/HYG PTR1PURO/+ cells (8), DHFR-TSNEO/HYG PTR1PURO/ZEO cells with (9) or without (10) psp72αblastα-DHFR-TS hybridized with probe 2 derived from the 5’UTR of PTR1 (see Fig 5A). Lane 10 is an attempt at deleting the second PTR1 allele in DHFR-TSNEO/HYG PTR1PURO/+ cells in the absence of DHFR-TS or PTR1 episomes that is distinct from the one shown in Fig 5B. The vertical white line in the panel comes from the cropping of the blot to remove irrelevant lanes in its middle section.
(TIF)
(A) Schematic representation of the DHFR-TS locus and of the psp72αblastα plasmid into which DHFR-TS was cloned. HindIII and XbaI sites are shown and the size of bands when hybridized to probe 3 (represented by small black boxes) within the DHFR-TS coding sequence. (B) A DHFR-TSNEO/HYG null mutant was obtained provided that an episomal DHFR-TS construct was present (lane 1). In four DHFR-TSNEO/HYG clones we lost the episome if cells were supplemented with 50 μg/ml of thymidine (lanes 2–5). Lane 6 is L. infantum wild-type cells and its intact DHFR-TS chromosomal copy. Lower panel shown the same blot hybridized to a PTR1 probe for monitoring the amount of DNA analyzed in each lane.
(TIF)
(A) Schematic map of the PTR1 locus (see also Fig 5A) and the psp72αblastα plasmid into which the Mycobacterium tuberculosis ThyX gene was cloned. The location of the probe used for the hybridization of the Southern blot in B is indicated by small black boxes. (B) Southern blot of L. infantum WT (1) and of DHFR-TSNEO/HYG PTR1PURO/+ parasites transfected with psp72αblastα-ThyX (2) or psp72αblastα-DHFR-TS (3). These parasites were transfected with a ZEO PTR1-inactivation cassette to lead to DHFR-TSNEO/HYG PTR1PURO/ZEO/+ (4) or DHFR-TSNEO/HYG PTR1PURO/ZEO (5) parasites harboring the episomal psp72αblastα-ThyX and psp72αblastα-DHFR-TS vectors, respectively.
(TIF)
(DOCX)
This table provides the list of genes located on cosmids enriched by MTX, 5-FU or PMX shown in Fig 1.
(DOCX)
Acknowledgments
We thank Fereshteh Fani for help with S1 Fig.
Data Availability
The NGS dataset is available in the Sequencing Read Archive (https://www.ncbi.nlm.nih.gov/sra) repository under BioProject accession PRJNA682394, biosample accessions SAMN16988551 to SAMN16988582.
Funding Statement
This work was supported by a Canadian Institutes of Health Research Foundation Grant (FDN 167289) to MO. M.O. holds a Canada Research Chair in Antimicrobial Resistance. This research was enabled in part by computing infrastructure provided by Calcul Québec (www.calculquebec.ca) and Compute Canada (www.computecanada.ca). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kidder GW, Dutta BN. The growth and nutrition of Crithidia fasciculata. J Gen Microbiol. 1958;18(3):621–38. 10.1099/00221287-18-3-621 [DOI] [PubMed] [Google Scholar]
- 2.Trager W. Pteridine requirement of the hemoflagellate Leishmania tarentolae. J Protozool. 1969;16(2):372–5. 10.1111/j.1550-7408.1969.tb02284.x [DOI] [PubMed] [Google Scholar]
- 3.Cruz A, Beverley SM. Gene replacement in parasitic protozoa. Nature. 1990;348(6297):171–3. 10.1038/348171a0 [DOI] [PubMed] [Google Scholar]
- 4.Ouellette M, Drummelsmith J, El-Fadili A, Kundig C, Richard D, Roy G. Pterin transport and metabolism in Leishmania and related trypanosomatid parasites. Int J Parasitol. 2002;32(4):385–98. 10.1016/s0020-7519(01)00346-0 [DOI] [PubMed] [Google Scholar]
- 5.Vickers TJ, Beverley SM. Folate metabolic pathways in Leishmania. Essays Biochem. 2011;51:63–80. 10.1042/bse0510063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreira W, Leblanc E, Ouellette M. The role of reduced pterins in resistance to reactive oxygen and nitrogen intermediates in the protozoan parasite Leishmania. Free Radic Biol Med. 2009;46(3):367–75. 10.1016/j.freeradbiomed.2008.10.034 [DOI] [PubMed] [Google Scholar]
- 7.Nare B, Garraway LA, Vickers TJ, Beverley SM. PTR1-dependent synthesis of tetrahydrobiopterin contributes to oxidant susceptibility in the trypanosomatid protozoan parasite Leishmania major. Curr Genet. 2009;55(3):287–99. 10.1007/s00294-009-0244-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham ML, Titus RG, Turco SJ, Beverley SM. Regulation of differentiation to the infective stage of the protozoan parasite Leishmania major by tetrahydrobiopterin. Science. 2001;292(5515):285–7. 10.1126/science.1057740 [DOI] [PubMed] [Google Scholar]
- 9.Papadopoulou B, Roy G, Breton M, Kundig C, Dumas C, Fillion I, et al. Reduced infectivity of a Leishmania donovani biopterin transporter genetic mutant and its use as an attenuated strain for vaccination. Infect Immun. 2002;70(1):62–8. 10.1128/iai.70.1.62-68.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laffitte MN, Leprohon P, Papadopoulou B, Ouellette M. Plasticity of the Leishmania genome leading to gene copy number variations and drug resistance. F1000Res. 2016;5:2350. 10.12688/f1000research.9218.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coderre JA, Beverley SM, Schimke RT, Santi DV. Overproduction of a bifunctional thymidylate synthetase-dihydrofolate reductase and DNA amplification in methotrexate-resistant Leishmania tropica. Proc Natl Acad Sci U S A. 1983;80(8):2132–6. 10.1073/pnas.80.8.2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callahan HL, Beverley SM. A member of the aldoketo reductase family confers methotrexate resistance in Leishmania. J Biol Chem. 1992;267(34):24165–8. [PubMed] [Google Scholar]
- 13.Papadopoulou B, Roy G, Ouellette M. A novel antifolate resistance gene on the amplified H circle of Leishmania. EMBO J. 1992;11(10):3601–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummelsmith J, Girard I, Trudel N, Ouellette M. Differential protein expression analysis of Leishmania major reveals novel roles for methionine adenosyltransferase and S-adenosylmethionine in methotrexate resistance. J Biol Chem. 2004;279(32):33273–80. 10.1074/jbc.M405183200 [DOI] [PubMed] [Google Scholar]
- 15.Dridi L, Ahmed Ouameur A, Ouellette M. High affinity S-Adenosylmethionine plasma membrane transporter of Leishmania is a member of the folate biopterin transporter (FBT) family. J Biol Chem. 2010;285(26):19767–75. 10.1074/jbc.M110.114520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharya A, Sharma M, Pakkinathan C, Rosen BP, Leprohon P, Ouellette M. Genomewide Analysis of Mode of Action of the S-Adenosylmethionine Analogue Sinefungin in Leishmania infantum. mSystems. 2019;4(5). 10.1128/mSystems.00416-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gazanion E, Fernandez-Prada C, Papadopoulou B, Leprohon P, Ouellette M. Cos-Seq for high-throughput identification of drug target and resistance mechanisms in the protozoan parasite Leishmania. Proc Natl Acad Sci U S A. 2016;113(21):E3012–21. 10.1073/pnas.1520693113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Potvin JE, Leprohon P, Gazanion E, Sharma M, Fernandez-Prada C, Ouellette M. Cos-Seq: A High-Throughput Gain-of-Function Screen for Drug Resistance Studies in Leishmania. Methods Mol Biol. 2019;1971:141–67. 10.1007/978-1-4939-9210-2_7 [DOI] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34(5):525–7. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- 23.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhattacharya A, Leprohon P, Bigot S, Padmanabhan PK, Mukherjee A, Roy G, et al. Coupling chemical mutagenesis to next generation sequencing for the identification of drug resistance mutations in Leishmania. Nat Commun. 2019;10(1):5627. 10.1038/s41467-019-13344-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritt JF, Raymond F, Leprohon P, Legare D, Corbeil J, Ouellette M. Gene amplification and point mutations in pyrimidine metabolic genes in 5-fluorouracil resistant Leishmania infantum. PLoS Negl Trop Dis. 2013;7(11):e2564. 10.1371/journal.pntd.0002564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaur K, Coons T, Emmett K, Ullman B. Methotrexate-resistant Leishmania donovani genetically deficient in the folate-methotrexate transporter. J Biol Chem. 1988;263(15):7020–8. [PubMed] [Google Scholar]
- 27.Papadopoulou B, Roy G, Mourad W, Leblanc E, Ouellette M. Changes in folate and pterin metabolism after disruption of the Leishmania H locus short chain dehydrogenase gene. J Biol Chem. 1994;269(10):7310–5. [PubMed] [Google Scholar]
- 28.Petrillo-Peixoto ML, Beverley SM. Amplified DNAs in laboratory stocks of Leishmania tarentolae: extrachromosomal circles structurally and functionally similar to the inverted-H-region amplification of methotrexate-resistant Leishmania major. Mol Cell Biol. 1988;8(12):5188–99. 10.1128/mcb.8.12.5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alzahrani KJH, Ali JAM, Eze AA, Looi WL, Tagoe DNA, Creek DJ, et al. Functional and genetic evidence that nucleoside transport is highly conserved in Leishmania species: Implications for pyrimidine-based chemotherapy. Int J Parasitol Drugs Drug Resist. 2017;7(2):206–26. 10.1016/j.ijpddr.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter NS, Landfear SM, Ullman B. Nucleoside transporters of parasitic protozoa. Trends Parasitol. 2001;17(3):142–5. 10.1016/s1471-4922(00)01806-7 [DOI] [PubMed] [Google Scholar]
- 31.Cruz A, Coburn CM, Beverley SM. Double targeted gene replacement for creating null mutants. Proc Natl Acad Sci U S A. 1991;88(16):7170–4. 10.1073/pnas.88.16.7170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bello AR, Nare B, Freedman D, Hardy L, Beverley SM. PTR1: a reductase mediating salvage of oxidized pteridines and methotrexate resistance in the protozoan parasite Leishmania major. Proc Natl Acad Sci U S A. 1994;91(24):11442–6. 10.1073/pnas.91.24.11442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El Fadili A, Kundig C, Roy G, Ouellette M. Inactivation of the Leishmania tarentolae pterin transporter (BT1) and reductase (PTR1) genes leads to viable parasites with changes in folate metabolism and hypersensitivity to the antifolate methotrexate. J Biol Chem. 2004;279(18):18575–82. 10.1074/jbc.M400652200 [DOI] [PubMed] [Google Scholar]
- 34.Cruz AK, Titus R, Beverley SM. Plasticity in chromosome number and testing of essential genes in Leishmania by targeting. Proc Natl Acad Sci U S A. 1993;90(4):1599–603. 10.1073/pnas.90.4.1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee A, Roy G, Guimond C, Ouellette M. The gamma-glutamylcysteine synthetase gene of Leishmania is essential and involved in response to oxidants. Mol Microbiol. 2009;74(4):914–27. 10.1111/j.1365-2958.2009.06907.x [DOI] [PubMed] [Google Scholar]
- 36.Myllykallio H, Lipowski G, Leduc D, Filee J, Forterre P, Liebl U. An alternative flavin-dependent mechanism for thymidylate synthesis. Science. 2002;297(5578):105–7. 10.1126/science.1072113 [DOI] [PubMed] [Google Scholar]
- 37.Chattopadhyay S, Tamari R, Min SH, Zhao R, Tsai E, Goldman ID. Commentary: a case for minimizing folate supplementation in clinical regimens with pemetrexed based on the marked sensitivity of the drug to folate availability. Oncologist. 2007;12(7):808–15. 10.1634/theoncologist.12-7-808 [DOI] [PubMed] [Google Scholar]
- 38.Vasudevan G, Carter NS, Drew ME, Beverley SM, Sanchez MA, Seyfang A, et al. Cloning of Leishmania nucleoside transporter genes by rescue of a transport-deficient mutant. Proc Natl Acad Sci U S A. 1998;95(17):9873–8. 10.1073/pnas.95.17.9873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dawson A, Tulloch LB, Barrack KL, Hunter WN. High-resolution structures of Trypanosoma brucei pteridine reductase ligand complexes inform on the placement of new molecular entities in the active site of a potential drug target. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 12):1334–40. 10.1107/S0907444910040886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nare B, Hardy LW, Beverley SM. The roles of pteridine reductase 1 and dihydrofolate reductase-thymidylate synthase in pteridine metabolism in the protozoan parasite Leishmania major. J Biol Chem. 1997;272(21):13883–91. 10.1074/jbc.272.21.13883 [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Leblanc E, Chang CF, Papadopoulou B, Bray T, Whiteley JM, et al. Pterin and folate reduction by the Leishmania tarentolae H locus short-chain dehydrogenase/reductase PTR1. Arch Biochem Biophys. 1997;342(2):197–202. 10.1006/abbi.1997.0126 [DOI] [PubMed] [Google Scholar]
- 42.Kundig C, Leblanc E, Papadopoulou B, Ouellette M. Role of the locus and of the resistance gene on gene amplification frequency in methotrexate resistant Leishmania tarentolae. Nucleic Acids Res. 1999;27(18):3653–9. 10.1093/nar/27.18.3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beverley SM, Coderre JA, Santi DV, Schimke RT. Unstable DNA amplifications in methotrexate-resistant Leishmania consist of extrachromosomal circles which relocalize during stabilization. Cell. 1984;38(2):431–9. 10.1016/0092-8674(84)90498-7 [DOI] [PubMed] [Google Scholar]
- 44.Hightower RC, Wong ML, Ruiz-Perez L, Santi DV. Electron microscopy of amplified DNA forms in antifolate-resistant Leishmania. J Biol Chem. 1987;262(30):14618–24. [PubMed] [Google Scholar]
- 45.Guimond C, Trudel N, Brochu C, Marquis N, El Fadili A, Peytavi R, et al. Modulation of gene expression in Leishmania drug resistant mutants as determined by targeted DNA microarrays. Nucleic Acids Res. 2003;31(20):5886–96. 10.1093/nar/gkg806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cunningham ML, Beverley SM. Pteridine salvage throughout the Leishmania infectious cycle: implications for antifolate chemotherapy. Mol Biochem Parasitol. 2001;113(2):199–213. 10.1016/s0166-6851(01)00213-4 [DOI] [PubMed] [Google Scholar]
- 47.Richard D, Kundig C, Ouellette M. A new type of high affinity folic acid transporter in the protozoan parasite Leishmania and deletion of its gene in methotrexate-resistant cells. J Biol Chem. 2002;277(33):29460–7. 10.1074/jbc.M204796200 [DOI] [PubMed] [Google Scholar]
- 48.Richard D, Leprohon P, Drummelsmith J, Ouellette M. Growth phase regulation of the main folate transporter of Leishmania infantum and its role in methotrexate resistance. J Biol Chem. 2004;279(52):54494–501. 10.1074/jbc.M409264200 [DOI] [PubMed] [Google Scholar]
- 49.Kundig C, Haimeur A, Legare D, Papadopoulou B, Ouellette M. Increased transport of pteridines compensates for mutations in the high affinity folate transporter and contributes to methotrexate resistance in the protozoan parasite Leishmania tarentolae. EMBO J. 1999;18(9):2342–51. 10.1093/emboj/18.9.2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cotrim PC, Garrity LK, Beverley SM. Isolation of genes mediating resistance to inhibitors of nucleoside and ergosterol metabolism in Leishmania by overexpression/selection. J Biol Chem. 1999;274(53):37723–30. 10.1074/jbc.274.53.37723 [DOI] [PubMed] [Google Scholar]
- 51.Ryan KA, Dasgupta S, Beverley SM. Shuttle cosmid vectors for the trypanosomatid parasite Leishmania. Gene. 1993;131(1):145–50. 10.1016/0378-1119(93)90684-u [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Southern blot analysis of genomic DNAs digested with XhoI from L. infantum wild-type (1), from the single allele knock out DHFR-TSNEO/+ (2), or from five aneuploid DHFR-TSNEO/HYG/+ clones (3–7) hybridized to probe 1 derived from the 3’UTR of DHFR-TS (see Fig 3A). Lane 3 is the same DHFR-TSNEO/HYG/+ clone as in Fig 3B. (B) Southern blot analysis of genomic DNAs digested with AfeI from L. infantum wild-type (1), DHFR-TSNEO/HYG PTR1PURO/+ cells (8), DHFR-TSNEO/HYG PTR1PURO/ZEO cells with (9) or without (10) psp72αblastα-DHFR-TS hybridized with probe 2 derived from the 5’UTR of PTR1 (see Fig 5A). Lane 10 is an attempt at deleting the second PTR1 allele in DHFR-TSNEO/HYG PTR1PURO/+ cells in the absence of DHFR-TS or PTR1 episomes that is distinct from the one shown in Fig 5B. The vertical white line in the panel comes from the cropping of the blot to remove irrelevant lanes in its middle section.
(TIF)
(A) Schematic representation of the DHFR-TS locus and of the psp72αblastα plasmid into which DHFR-TS was cloned. HindIII and XbaI sites are shown and the size of bands when hybridized to probe 3 (represented by small black boxes) within the DHFR-TS coding sequence. (B) A DHFR-TSNEO/HYG null mutant was obtained provided that an episomal DHFR-TS construct was present (lane 1). In four DHFR-TSNEO/HYG clones we lost the episome if cells were supplemented with 50 μg/ml of thymidine (lanes 2–5). Lane 6 is L. infantum wild-type cells and its intact DHFR-TS chromosomal copy. Lower panel shown the same blot hybridized to a PTR1 probe for monitoring the amount of DNA analyzed in each lane.
(TIF)
(A) Schematic map of the PTR1 locus (see also Fig 5A) and the psp72αblastα plasmid into which the Mycobacterium tuberculosis ThyX gene was cloned. The location of the probe used for the hybridization of the Southern blot in B is indicated by small black boxes. (B) Southern blot of L. infantum WT (1) and of DHFR-TSNEO/HYG PTR1PURO/+ parasites transfected with psp72αblastα-ThyX (2) or psp72αblastα-DHFR-TS (3). These parasites were transfected with a ZEO PTR1-inactivation cassette to lead to DHFR-TSNEO/HYG PTR1PURO/ZEO/+ (4) or DHFR-TSNEO/HYG PTR1PURO/ZEO (5) parasites harboring the episomal psp72αblastα-ThyX and psp72αblastα-DHFR-TS vectors, respectively.
(TIF)
(DOCX)
This table provides the list of genes located on cosmids enriched by MTX, 5-FU or PMX shown in Fig 1.
(DOCX)
Data Availability Statement
The NGS dataset is available in the Sequencing Read Archive (https://www.ncbi.nlm.nih.gov/sra) repository under BioProject accession PRJNA682394, biosample accessions SAMN16988551 to SAMN16988582.