A fungal effector interferes with Ca2+ cross-linking in plant cell walls to loosen their structure in aid of infections.
Abstract
Effectors are small, secreted proteins that promote pathogen virulence. Although key to microbial infections, unlocking the intrinsic function of effectors remains a challenge. We have previously shown that members of the fungal Avr4 effector family use a carbohydrate-binding module of family 14 (CBM14) to bind chitin in fungal cell walls and protect them from host chitinases during infection. Here, we show that gene duplication in the Avr4 family produced an Avr4-2 paralog with a previously unknown effector function. Specifically, we functionally characterize PfAvr4-2, a paralog of PfAvr4 in the tomato pathogen Pseudocercospora fuligena, and show that although it contains a CBM14 domain, it does not bind chitin or protect fungi against chitinases. Instead, PfAvr4-2 interacts with highly de-esterified pectin in the plant’s middle lamellae or primary cell walls and interferes with Ca2+-mediated cross-linking at cell-cell junction zones, thus loosening the plant cell wall structure and synergizing the activity of pathogen secreted endo-polygalacturonases.
INTRODUCTION
Plant immunity generally consists of passive and inducible immune responses (1). The plant cell wall has a major part in passive defenses, both due to its function as a structural barrier to infections and because changes in its physicochemical composition affect plant development and immunity (2, 3).
Plant cell walls are mainly composed of a dynamic and intricate matrix of cellulose and hemicellulose microfibrils embedded in pectins, a family of complex polysaccharides consisting primarily of homogalacturonan and rhamnogalacturonans (4). Homogalacturonan is a linear homopolymer of α-1,4–linked d-galacturonic acid units (GalA) that is deposited in plant cell walls in a highly methyl-esterified form but is subsequently de-esterified by wall-based pectin methylesterases (PMEs) that remove methyl-ester groups (5). De-esterified homogalacturonan can then be readily cross-linked by Ca2+, leading to the formation of Ca2+ pectate gels known as “egg boxes” that promote cell-cell adhesion (6) and further affect the rheology, stiffening, porosity, and permeability of the plant cell wall to enzymes (7). Egg-box formation is particularly evident in the middle lamella, the thin pectin layer deposited at the interface between the primary cell walls of adjacent plant cells that bonds them together and prevents loosening of the plant tissue (8). Thus, the pattern and degree of esterification of homogalacturonan have profound effects on the cell wall properties and, consequently, on plant tissue plasticity and susceptibility to microbial attacks (2, 3, 7, 9).
Owing to its key role in defense and development, plants have developed elaborate mechanisms to regulate the methyl-esterification status of homogalacturonan during growth (2, 3, 7, 9). Conversely, microbes also secrete an array of pectic enzymes such as PMEs, pectate lyases, pectin lyases (PNLs), and polygalacturonases (PGs) to degrade de-esterified homogalacturonan and loosen the plant cell wall structure in aid of the infections (9). However, the downside of this enzymatic activity is the release from the damaged cell walls of pectic fragments that are perceived by the host as “danger signals,” thus activating innate immune responses (10). In general, innate immunity in plants is triggered by the perception from cell-surface pattern recognition receptors of danger signals, including broadly conserved microbial components known as “pathogen-associated molecular patterns” and endogenous “damage-associated molecular patterns” (DAMPs) molecules that emanate from cellular damage as a result of microbial activity or trauma (11). Oligogalacturonides (OGs), for example, with a degree of polymerization between 10 and 15 (OG10-15) that are released from the hydrolysis of de-esterified homogalacturonan by microbial pectin-degrading enzymes, are well-known elicitors of DAMP-triggered immunity in plants (12). Their perception is mediated by the wall-associated kinase 1 receptor and is contingent on their arrangement in an egg-box configuration (10, 13). Notably, a recent study revealed that OGs released from damaged plant cell walls during infections consist of a complex mixture of acetyl- and methyl-esterified OGs that are mainly spurred by pathogen-secreted PNLs (14). This contrasts the current model that unsterified OG10-15s derived from the enzymatic activity of PGs are the major DAMPs that accumulate during microbial infections and further highlights the importance of pectin degrading enzymes in plant-microbe interactions.
To suppress or evade innate immunity, in response, pathogens secrete an array of effector proteins that promote parasitic infections. In resistant plants, however, effectors are recognized by cognate resistance proteins, thus eliciting effector-triggered immunity that typically manifests in the hypersensitive response (HR) (15). To date, a great deal of effort has been placed on elucidating the function of effectors secreted by plant pathogens (16). An example is CfAvr4, an effector from the hemibiotrophic extracellular pathogen of tomato Cladosporium fulvum, which uses a family 14 carbohydrate-binding module (CBM14) to bind chitin in fungal cell walls and protect them from chitinases (17). Although functional orthologs of CfAvr4 exist in many fungi, microbes generally exhibit diverse effector repertoires that differ even among phylogenetically closely related species (18).
Here, we show that gene duplication of Avr4 in the tomato pathogen Pseudocercospora fuligena (19), a close relative of C. fulvum with a similar infection biology, led to the emergence of a candidate effector with a diversified function. Specifically, we show that PfAvr4-2 binds to homogalacturonan with a low degree of esterification in the middle lamella, thus interfering with Ca2+-mediated cross-linking of neighboring homogalacturonan chains and the integrity of the plant cell wall.
RESULTS
PfAvr4-2 is a paralog of PfAvr4
Query of the P. fuligena genome (JABCIY000000000) with CfAvr4 revealed the presence of PfAvr4 (19) and an additional 101–amino acid–long PfAvr4-2 protein that shares 33% amino acid identity with PfAvr4. PfAvr4-2 (MK410883) consists of a predicted 16-residue N-terminal signal peptide and an 85-residue mature protein that contains a predicted CBM14 chitin-binding domain (Pfam01607) with 10 cysteine (Cys) residues instead of the 8 defining the CBM14 domain of Avr4 proteins (fig. S1A). The PfAvr4 and PfAvr4-2 genes are located on different scaffolds of the P. fuligena assembly, and no repetitive DNA was identified immediately adjacent to them (fig. S1B), suggesting that they are not embedded within repeat-rich regions of the genome, as is the case with other fungal effectors (16). Further examination of the allelic variation present in PfAvr4-2 in 30 strains of P. fuligena collected from Taiwan (n = 1) and Japan (n = 29) indicated the absence of polymorphisms, suggesting the lack of selection pressure from cognate resistance proteins. A blast search against the National Center for Biotechnology Information nr and the Joint Genome Institute MycoCosm databases identified two homologs of PfAvr4-2 in the phylogenetically related species Pseudocercospora musae (KXS93729.1; 74.3% identity) and Pseudocercospora eumasae (KXS98644.1; 69.3% identity). However, PfAvr4-2 was absent from Pseudocercospora fijiensis that, together with P. eumasae and P. musae, constitute the sigatoka disease complex in banana (20). A phylogenetic tree constructed based on an amino acid alignment of the Avr4 and/or Avr4-2 proteins present in P. fuligena, P. musae, P. eumusae, and C. fulvum showed that Avr4-2 proteins form a separate orthologous group to the Avr4 ones (fig. S1C), suggesting a gene duplication in an ancestral species that generated two distinct lineages within the Avr4 family. The alignment also showed that amino acids that are critical to chitohexaose [(GlcNAc)6] binding in CfAvr4 (17) are not fully conserved in Avr4-2 proteins, suggesting alterations in substrate binding affinity or specificity (fig. S1A).
Further modeling of PfAvr4-2 on the previously determined CfAvr4 and PfAvr4 tertiary structures (17, 19) predicts that the proteins adopt an overall similar architecture of a distorted β-sandwich fold formed by a central β-sheet with three antiparallel β-strands (A1, A2, and A3) and a smaller C-terminal β-sheet with two antiparallel β-strands (B1 and B2). The short N-terminal (H1) and C-terminal (H2) α-helixes as well as the four disulfide bonds that are formed between the eight cysteines present in Avr4 are also predicted to be conserved in PfAvr4-2 (fig. S2, A and B). However, despite these similarities, some notable discrepancies were observed as well. Specifically, the two extra cysteine residues present in PfAvr4-2 are predicted to form an additional disulfide bond (Cys57-Cys62) in the β-turn that is connecting β-strands A2 and A3, and which likely contributes toward stabilizing the turn and constraining the three-dimensional (3D) conformation of the protein. Moreover, the β-turn connecting β-strands B1 and B2 in the C-terminus of Avr4 is predicted to be replaced in PfAvr4-2 by an α-helix (H2). This helix would create an outward bulge that would widen PfAvr4-2’s structure at its C-terminus and would further change its electrostatic surface potential by creating a pocket of positive charge that could contribute to the biding of negatively charged molecules (fig. S2, A and B). Notably, the aromatic residues Trp100 and Tyr103 in CfAvr4 that are critical for (GlcNAc)6 binding are predicted to be replaced in PfAvr4-2 by the positively charged Arg84 and the generally unreactive Gly87, respectively, whereas the placement of the small glycine (Gly) and isoleucine (Ile) residues at positions 87 and 88, results in another positively charged residue (Lys90) that is predicted to be protruding toward the substrate (fig. S2C). Overall, the lack of conservation in amino acids that are critical to (GlcNAc)6 binding and the change in the electrostatic surface potential in the C-terminus of Avr4 and PfAvr4-2 suggest that they could be interacting with different substrates.
PfAvr4-2 has functionally diversified from PfAvr4
To search for evidence of substrate diversification, we used an in vitro affinity precipitation assay to test the binding of PfAvr4-2 to chitin and various other insoluble polysaccharides of fungal and plant origin (19). Binding specificity was assessed at pH 5.5, pH 7.0, and pH 8.5, as the pH environment may influence the ligand-binding characteristics of carbohydrate-binding modules. At all three pHs, PfAvr4-2 was present only in the unbound fraction, indicating the absence of binding to the chitinous substrates and to the other polysaccharides used in the assay (Fig. 1, A and B). Localization studies on germlings of Trichoderma viride (19) using PfAvr4 and PfAvr4-2 conjugated to the Alexa Fluor 488 green fluorescent dye further showed that unlike PfAvr4488, PfAvr4-2488 does not localize onto fungal cell walls (Fig. 1C). Similar results were obtained when examining the localization of PfAvr4RR and PfAvr4-2RR conjugated to the red fluorescent dye Rhodamine Red (RR) on mycelia of P. fuligena after their treatment with β-1,3-glucans to expose the underlying chitin layer (Fig. 1D). Last, in contrast to PfAvr4, external application of 10 μg of PfAvr4-2 on germlings of T. viride failed to protect them from the chitinolytic activity of whole tomato leaf extracts (Fig. 1E) and from bacterial chitinases supplemented with β-1,3-glucanases (Fig. 1F). Together, these results indicate that PfAvr4-2 has functionally diversified from PfAvr4 and other Avr4 family members.
Fig. 1. PfAvr4-2 does not bind chitin or protects fungal hyphae against chitinases.
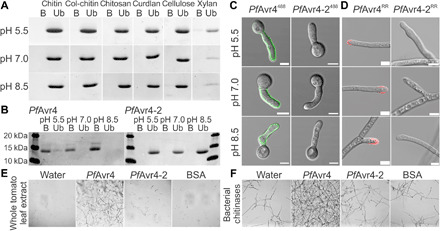
(A and B) In vitro polysaccharide precipitation assay testing binding of PfAvr4-2 to insoluble polysaccharides of fungal (chitin, colloidal chitin, chitosan, and curdlan) and plant (cellulose and xylan) origin (A) or binding of PfAvr4 (control) and PfAvr4-2 to shrimp shell chitin (B). Binding was tested at pH 5.5, pH 7.0, and pH 8.5. B, bound fraction; Ub, unbound fraction. (C and D) Localization of PfAvr4 and PfAvr4-2 conjugated to Alexa Fluor 488 (488) or Rhodamine Red (RR) on germlings of T. viride (C) and mycelia of P. fuligena (D), following their treatment with β-1,3-glucanases to remove the overlaying β-glucan layer in the fungal cell wall. White bars correspond to 5 μm. (E and F) In vitro assays testing whether PfAvr4 and PfAvr4-2 protect germlings of T. viride against plant-derived (E) and bacterial-derived (F) chitinases. Bovine serum albumin (BSA) is used as control.
PfAvr4-2 binds to pectin with a low degree of esterification
To explore the carbohydrate-binding specificity of PfAvr4-2, we initially used a carbohydrate microarray spotted with plant and fungal-related polysaccharides, including pectins with various degrees of esterification (fig. S3A). On the basis of the mean intensity of the spots on the microarray, PfAvr4-2 bound primarily to pectins with a low degree of esterification, whereas a negative correlation was observed between the degree of esterification and the intensity of PfAvr4-2 binding. Specifically, binding was observed on lime pectins that were de-esterified either by fungal (F series) and plant (P series) PMEs to a degree of esterification of ≤11 in case of fungal and to a degree of esterification of ≤32 in case of plant PMEs. Binding to polygalacturonic acid and sugar-beet pectin with a degree of esterification of 1% was also observed. In contrast, PfAvr4-2 did not interact with other plant or fungal-derived polysaccharides on the array, including short chitin oligosaccharides, β-glucan, xyloglucan, mannan, arabinose, and cellulose (fig. S3A).
To further explore the interaction between PfAvr4-2 and homogalacturonan with a low degree of esterification, we used isothermal titration calorimetry (ITC) to determine the thermodynamic binding parameters between PfAvr4-2 and OG10-15 (fig. S3B). Control titrations of 2 mM OG10-15 mixture into 0.5 mM PfAvr4-2 resulted in a mild heat exchange, indicating weak binding. Although a saturation point could not be reached within a reasonable range of ligand and protein concentrations, the thermodynamic values obtained from fitting a one-site binding model to the isotherm curve showed that PfAvr4-2 bound to the OG10-15 mixture with a dissociation constant (Kd) of 159.9 ± 32.01 μM, a binding enthalpy (ΔH) of −20.48 ± 1.76 kJ/mol, and a stoichiometric ratio (n) of 0.84 ± 0.03. In contrast, control titrations of OG10-15 into the buffer or of the buffer titrated into PfAvr4-2 resulted in no measurable heat change (fig. S3B). Collectively, despite the weak affinity of PfAvr4-2 to OG10-15, the ITC data are suggestive of PfAvr4-2’s interaction with galacturonan structures.
PfAvr4-2 interacts with de-esterified pectin in the middle lamella or the primary cell wall
Since pectin is abundantly present in plant cell walls, we examined the localization pattern of PfAvr4-2488 on stem cross sections of Arabidopsis plants. Labeling was performed at pH 7.0, pH 8.5, and pH 5.5, with the latter reflecting the pH of the tomato apoplast and of the plant cell walls (21).
In contrast to PfAvr4488, PfAvr4-2488 localized on most stem tissue types, thus corroborating that the two proteins have functionally diversified (Fig. 2A). The localization of PfAvr4-2488 on stems was seen only at pH 5.5 (fig. S4A), whereas labeling of the plant stems with PfAvr4-2RR produced the same results, indicating that fluorophore selection did not alter the localization pattern (fig. S4B). PfAvr4-2RR localized in both the dermal and ground tissues of the stem including the epidermis, cortex, and pith parenchyma but only partially in vascular tissue, as the protein bound in the phloem region but not in the xylem (Fig. 2B). Observations at higher magnification showed that PfAvr4-2RR fills the tricellular junctions in the interfascicular fibers (fig. S4C) and in the young pith parenchyma (fig. S4D) but occupies the corners of intercellular spaces at tricellular junctions in the older pith parenchyma toward the stem’s center, where cell junctions are expanded and form air-filled intercellular spaces (Fig. 2B and fig. S4D). Staining with the cellulose-binding dye calcofluor white (CW) further showed that PfAvr4-2RR occupies mainly the outer cell wall layers facing the intercellular air spaces, whereas CW binds to the inner cell wall components adjacent to the plasma membrane (Fig. 2B). Similar results were also obtained when examining the localization pattern of PfAvr4-2RR in cross sections of tomato leaves infected with P. fuligena, as PfAvr4-2RR was detected proximally to the CW but as a discrete thin layer lining the outer cell wall regions of epidermal cells (Fig. 2C).
Fig. 2. PfAvr4-2 localizes to pectin-rich regions of the plant cell wall.
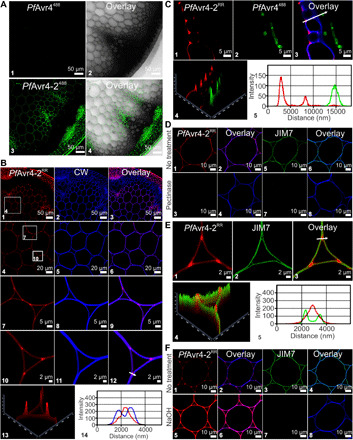
(A) Binding of PfAvr4488 and PfAvr4-2488 conjugated to Alexa Fluor 488 onto Arabidopsis stem cross section; only a quarter of the cross section is shown. (B) Dual labeling of Arabidopsis stem cross sections with PfAvr4-2RR conjugated to Rhodamine Red (RR) (red signal) and the cellulose-binding protein calcofluor white (CW) (blue signal). White boxes in (B1) and (B4) indicate the sections shown at higher magnification in succeeding panels. Panel (B13) is a 2.5D representation of (B10), whereas the fluorescence intensities of PfAvr4-2RR and CW at the region marked by an arrow in (B12) are shown in (B14). (C) Labeling with PfAvr4-2RR, PfAvr4488, and CW of cross sections of tomato leaves infected with P. fuligena and sampled at 9 days after inoculation. Panel (C4) is a 2.5D representation of (C3), whereas the fluorescence intensities of PfAvr4-2RR and PfAvr4488 at the region marked by an arrow in (C3) are shown in (C5). (D) Localization of PfAvr4-2RR, CW, and the JIM7 monoclonal antibody (mAb) on Arabidopsis stem cross sections before and after pectinase treatment. A secondary mAb conjugated to Alexa Fluor 488 is used for detecting JIM7. (E) Labeling of Arabidopsis stem cross sections with PfAvr4-2RR and JIM7. Cells are pith parenchyma and labeling was performed sequentially with the two probes. Panel (E10) is a 2.5D representation of (E9), whereas the fluorescence intensities of PfAvr4-2RR and JIM7 at the region marked by an arrow in (E9) are shown in (E11). (F) Labeling of the Arabidopsis stem cross sections with PfAvr4-2RR, CW, and JIM7 after pretreatment of the stem with NaOH to chemically de-esterify plant cell walls.
The above observations are consistent with PfAvr4-2 localizing at the corners of intercellular spaces at tricellular junctions, the cell wall region facing the intercellular air spaces, and the thin flat sheet lining the outer cell wall regions of neighboring cells, spaces typically occupied by the plant’s middle lamella (8). Since pectin is the predominant polysaccharide in the middle lamella (22), the Arabidopsis stem cross sections were treated with commercially available fungal pectinases and were subsequently labeled with PfAvr4-2RR, CW, and JIM7, a monoclonal antibody that specifically recognizes methyl-esterified homogalacturonan (23). The pectinase treatment fully abolished the localization of PfAvr4-2RR and JIM7, whereas it had no discernable effect on the labeling intensity and localization pattern of CW (Fig. 2D), thereby suggesting that PfAvr4-2 associates with pectin. Moreover, labeling of Arabidopsis stem cross sections with PfAvr4-2RR and JIM7 showed that the two localize to discrete regions of the cell walls in pith parenchyma cells (Fig. 2E), implying that PfAvr4-2 does not interact with the methyl-esterified homogalacturonan epitope that is recognized by JIM7. Instead, it is plausible that PfAvr4-2 binds pectin in plant cell walls in its fully or highly de-esterified form, as suggested by the carbohydrate microarray data.
To assess this hypothesis, the Arabidopsis stem sections were treated with NaOH to induce a random chemical de-esterification of pectin, and labeling was repeated with PfAvr4-2RR and the JIM7 monoclonal antibody. The alkaline pretreatment did not alter their localization pattern on the stem but nearly doubled the labeling intensity of PfAvr4-2RR and almost fully abolished the fluorescence signal of JIM7 (Fig. 2F). This points to a negative correlation between the degree of esterification of pectin in either the middle lamella or the primary cell wall and its relevance as a binding substrate for PfAvr4-2. This association was further corroborated by examining the localization of PfAvr4-2RR on stem cross sections obtained from two tomato PME-silenced lines (PME RNAi_1 and PME RNAi_2) (24). The silencing of PMEs in tomato reduces the de-esterification of homogalacturonan during tissue maturation, thus lowering the amount of potential binding sites. Labeling with PfAvr4-2RR showed that the intensity of the fluorescence signal was nearly abolished (PME RNAi_1) or almost halved (PME RNAi_2) in the stem cross sections of the two PME-silenced lines as compared to the intensity of the fluorescence signal on the wild-type plants (fig. S4, E and F). Furthermore, the difference in fluorescence intensity between the two PME-silenced lines correlated with the silencing levels of PMEs in these plants (24). Collectively, these results allude that PfAvr4-2 specifically interacts with homogalacturonan pectin in its highly de-esterified form in either the plant’s middle lamella or primary cell wall.
PfAvr4-2 interferes with Ca2+-mediated cross-linking at tricellular junctions
Under native conditions, nonmethylated GalA residues in adjacent homogalacturonan chains can be ionically cross-linked with Ca2+ to form egg-box structures that affect the rheological properties of the plant cell wall (7). The interaction of PfAvr4-2 with homogalacturonan of low degree of esterification suggests that it is competing with Ca2+ for binding to the deprotonated carboxyl groups (COO−) of nonmethylated GalA residues or, alternatively, that PfAvr4-2 binds to Ca2+-mediated cross-linked homogalacturonan chains. To investigate these hypotheses, we compared the localization pattern of PfAvr4-2RR on Arabidopsis stem cross sections with that of the 2F4 monoclonal antibody that specifically binds to Ca2+-mediated cross-linked homogalacturonan (egg boxes) (13), and with the OG7-13488 probe, a mixture of OG oligomers with a degree of polymerization of 7 to 13 that does not bind to existing egg boxes but forms de novo associations with nonmethylated GalA residues in homogalacturonan chains in the presence of Ca2+ (25). PfAvr4-2RR and OG7-13488 accumulated mostly at the corners and the cell wall region facing the intercellular air spaces at tricellular junctions as well as the thin flat sheet between adjacent cells (Fig. 3A). This suggests the presence in these regions of abundant nonmethylated GalA residues with which the OG7-13488 probe can form new egg boxes in the presence of Ca2+ ions. The 2F4 monoclonal antibody mainly localized at the cell wall region facing the intercellular air spaces but was less abundant at the corners of tricellular junctions, indicating the limited presence of Ca2+-mediated cross-linked homogalacturonan chains in these regions (Fig. 3B). When a strong Ca2+ chelating agent such as EGTA (50 mM) was added to the labeling buffer, the fluorescence intensity of PfAvr4-2RR remained the same, whereas that of the 2F4 monoclonal antibody was nearly abolished, suggesting that the binding of PfAvr4-2 to homogalacturonan with a low degree of esterification does not require Ca2+ (Fig. 3C). In contrast, labeling of the Arabidopsis stem sections in the presence of increasing calcium concentrations ([Ca2+]) showed that the fluorescence intensities of PfAvr4-2RR and of 2F4 were negatively and positively correlated with [Ca2+] in the labeling buffer, respectively (Fig. 3D). Collectively, the above suggest that PfAvr4-2 competes with Ca2+ for binding to free COO− groups of nonmethylated GalA residues along the homogalacturonan chains.
Fig. 3. PfAvr4-2 competes with Ca2+ for binding to de-esterified homogalacturonan at junction zones.
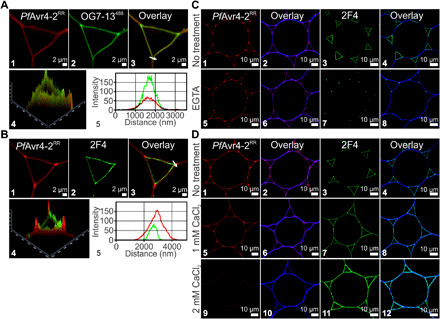
(A) Labeling of cross sections of Arabidopsis stems with PfAvr4-2RR and the OG7-13488 probe. PfAvr4-2RR is conjugated to the red fluorochrome Rhodamine Red (RR), whereas the OG7-13488 probe is conjugated to the green fluorophore Alexa Fluor 488. Cells are pith parenchyma, and labeling was performed sequentially with the two probes. Panel (A4) is a 2.5D representation of (A3), whereas the fluorescence intensities of PfAvr4-2RR and the OG7-13488 probe at the corner of the tricellular junction marked by an arrow in (A3) are shown in (A5). (B) Labeling of cross sections of Arabidopsis stems with PfAvr4-2RR and the 2F4 mAb. A secondary mAb conjugated to the green fluorophore Alexa Fluor 488 was used for detecting the 2F4 mAb. Cells are pith parenchyma, and labeling was performed sequentially with the two probes. Panel (B4) is a 2.5D representation of (B3), whereas the fluorescence intensities of PfAvr4-2RR and the 2F4 mAb at the corner of the tricellular junction marked by an arrow in (B3) are shown in (B5). (C) Labeling of cross sections of Arabidopsis stems with PfAvr4-2RR, the 2F4 mAb, or CW (blue signal) after pretreatment of the stems with the Ca2+ chelating agent EGTA. Cells are pith parenchyma. (D) Labeling of Arabidopsis stem cross sections with PfAvr4-2RR, the 2F4 mAb, or CW in increasing calcium concentrations ([Ca2+]).
PfAvr4-2 can partly disrupt egg boxes, thus loosening the cell wall structure
To further assess whether binding of PfAvr4-2 to nonmethylated GalA residues in intermittently Ca2+-mediated cross-linked homogalacturonan chains has any effect on egg boxes, the 2F4 monoclonal antibody and the OG7-13488 probe were used to monitor their integrity and formation in the presence of PfAvr4-2. For this purpose, Arabidopsis sections were pretreated with PfAvr4-2, and after washing the protein off the tissue, the stem sections were labeled with the OG7-13448 probe, the 2F4, or the JIM7 monoclonal antibodies. As controls, the stems were also pretreated with PfAvr4 and wheat germ agglutinin (WGA), as these proteins do not bind pectin or other polysaccharides in plant cell walls.
Labeling with the 2F4 monoclonal antibody of stems pretreated with PfAvr4-2 produced a patchier and less intense fluorescence signal as compared to stems pretreated with PfAvr4 and WGA, whereas the reverse observations were made with the OG7-13488 probe (Fig. 4A). This suggest that some of the Ca2+-mediated cross-links in the tricellular junction zones were disrupted by PfAvr4-2, thus generating after its removal more sites for the de novo formation of egg boxes between the OG7-13488 probe and nonmethylated GalA residues along the homogalacturonan chains. Moreover, pretreatment with PfAvr4-2 produced no changes in the fluorescence intensity of the JIM7 monoclonal antibody, suggesting that the above observations were not due to PfAvr4-2 modification of the state of pectin methyl-esterification (Fig. 4A).
Fig. 4. PfAvr4-2 can disrupt existing Ca2+-mediated cross-links formed between adjacent homogalacturonan chains.
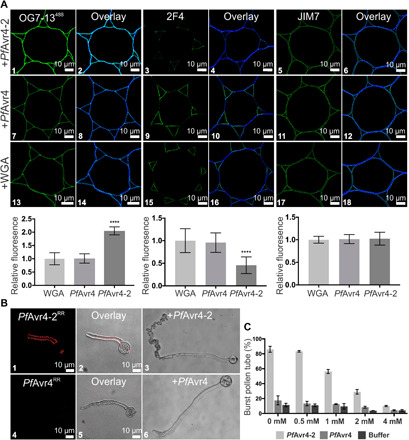
(A) Labeling of cross sections of Arabidopsis stems with the OG7-13488 probe and the 2F4 and JIM7 monoclonal antibodies (mAbs) after pretreatment of the stems with 10 μM PfAvr4-2, PfAvr4, or WGA. The OG7-13488 probe is conjugated to the green fluorophore Alexa Fluor 488, whereas a secondary mAb also conjugated to Alexa Fluor 488 was used for detecting the 2F4 and JIM7 mAbs. Cells are pith parenchyma. Labeling with the OG7-13488 probe necessitates the addition of 1 mM Ca2+ in the labeling buffer. Bottom graphs show the fluorescence intensity of the OG7-13488 probe, the 2F4, and JIM7 mAbs, relative to WGA (signal set to 1.0). Error bars indicate SD (OG7-13488, n = 15; 2F4488, n = 20; JIM7, n = 12). Statistical differences between PfAvr4-2 and other treatments were determined by ANOVA using Tukey’s test. ****P < 0.0001. (B) Microscope images of germinating pollen tubes of tomato incubated at increasing calcium concentrations ([Ca2+]) with PfAvr4-2RR or PfAvr4RR conjugated to the red fluorochrome Rhodamine Red (RR). Shown are the images where 1 mM CaCl2·2H2O was used in the labeling buffer. (C) Effect of PfAvr4-2, PfAvr4, and a buffer control under increasing [Ca2+] on bursting of tomato pollen tubes. The experiment was repeated in triplicate, and 100 pollen tubes were counted in each of the replicates. Error bars indicate SD from three replicates.
To further support the notion that PfAvr4-2 might be interfering with Ca2+-mediated cross-linking of homogalacturonan chains, we exposed germinating pollen tubes of tomato to PfAvr4-2RR and examined the effect on pollen tube growth and integrity. Pollen tubes are known for the distinctive spatial distribution of homogalacturonan with a high and low degree of esterification in the tube apex and the tube flanks, respectively. Moreover, Ca2+-mediated cross-linking of homogalacturonan with low degree of esterification in the tube wall is crucial to pollen viability, as it stiffens the tube until cellulose and callose are deposited at the tube flanks to further strengthen the wall and maintain a unidirectional polarized growth (26). Labeling of the pollen tubes with PfAvr4-2RR showed that the protein localizes along the shank of the pollen tube walls but not at the growing tip, thus alluding to the specific binding of PfAvr4-2 to fully or partially de-esterified pectin. In contrast, labeling of the tubes with PfAvr4RR did not show any binding of the protein along the pollen tube (Fig. 4B). PfAvr4-2RR induced bursting of the pollen tubes around the apical area in a [Ca2+]-dependent manner, shortly after their exposure to the protein. Specifically, in the absence of Ca2+, 86% of the pollen tubes burst within 2 hours of their exposure to the protein as compared to 18% of the pollen tubes bursting when exposed to PfAvr4 or the pollen germination media alone. The addition of increasing amounts of CaCl2·2H2O to the pollen germination media resulted in an analogous gradual reduction in the percentage of pollen tubes bursting in the presence of PfAvr4-2, whereas this effect was not seen in the presence of PfAvr4 or the pollen germination media alone (Fig. 4B). Collectively, these results suggest that PfAvr4-2 weakens the structural integrity of cell walls in pollen tubes in a [Ca2+]-dependent manner, possibly by interfering with the Ca2+-mediated cross-linking and formation of the egg-box structures in the pollen tubes.
Stable expression of PfAvr4-2 in tomato alters the plant’s growth and textural properties
To examine whether PfAvr4-2 could be interfering with Ca2+-mediated cross-linking under in planta conditions, we generated two transgenic tomato lines of cv. Moneymaker that constitutively express under the control of the cauliflower mosaic virus 35S promoter (CaMV 35S), the mature form of PfAvr4-2, fused to the PR1a secretion signal of Nicotiana tabacum for secretion into the apoplast. Primary transgenic plants were selected on kanamycin and two homozygous T2 lines (i.e., 35S::PfAvr4-2_1 and 35S::PfAvr4-2_2) obtained by segregation analysis of T1 kanamycin-resistant plants were lastly selected for further analysis.
Quantification of transgene expression using quantitative polymerase chain reaction (qPCR) showed that PfAvr4-2 expression levels were 5.92 times higher in the 35S::PfAvr4-2_1 line as compared to the 35S::PfAvr4-2_2 line (Fig. 5A). Notably, immunolabeling with the 2F4 monoclonal antibody of pectins extracted from leaves showed a substantially weaker and inversely correlated with the levels of PfAvr4-2 expression, labeling of the pectin fraction obtained from the two 35S::PfAvr4-2 transgenic lines than from the control Moneymaker line, suggesting reduced Ca2+-mediated cross-linking in their plant cell walls. In contrast, labeling with the JIM5 monoclonal antibody showed no differences in signal intensity, indicating that pectin levels were similar among the three lines (Fig. 5B). To further investigate whether the stable expression of PfAvr4-2 in tomato plants instigated any changes in the composition of the cell wall, leaf material from the two transgenic lines and the control Moneymaker line was subjected to comprehensive microarray polymer profiling (CoMPP) of major plant cell wall components using 21 unique anti-glycan monoclonal antibodies (fig. S5A) (27). The CoMPP analysis indicated that there were no substantial changes among the three lines in the signal intensities of the anti-homogalacturonan monoclonal antibodies JIM5 and JIM7. Signal intensities from antibodies that recognize the backbone of rhamnogalacturonan I (INRA-RU1) and its side chains (LM5 and LM6), xyloglucan (LM15 and LM25), and arabinogalactan (JIM13) were slightly decreased in the transgenic lines as compared to the control Moneymaker line, whereas the opposite was observed in signal intensities from antibodies recognizing epitopes of the hemicelluloses xylan (LM10) and arabinoxylan (INRA-AX1). However, with the exception of the change observed for arabinoxylan, all others were not significant at α = 0.05 and were mostly observed in the NaOH fraction. Moreover, further monosaccharide composition analysis using high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) indicated that the changes in the CoMPP profiles were not supported by any substantial changes in monosaccharide profile (fig. S5B), suggesting that they are either too small to yield any changes in monosaccharide content or/and that the results reflect changes in organization rather than amounts of cell wall polymers. Collectively, the above results suggest that the stable expression of PfAvr4-2 in tomato plants does not lead to any major changes in the composition of the plant cell wall other than reducing their Ca2+-mediated cross-linking.
Fig. 5. The 35S::PfAvr4-2 transgenic tomato plants have altered anatomical characteristics.
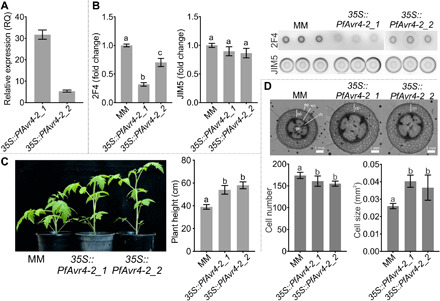
(A) PfAvr4-2 expression levels in the two 35S::PfAvr4-2 tomato lines (35S::PfAvr4-2_1 and 35S::PfAvr4-2_2), relative to the tomato RLP2 gene (set to 1.0 RQ). Error bars indicate SD from six samples. (B) immunodot blot analysis of Ca2+-mediated cross-linked homogalacturonan and of pectin extracted from the cell walls of 6-week-old plants. An equal amount of 33 μg of Ca2+-mediated cross-linked homogalacturonan or of pectin was spotted on each dot and probed with the 2F4 or JIM5 monoclonal antibodies, respectively. Bars indicate signal intensity, error bars are SD from minimum nine spots per line, and letters are significant differences based on a Tukey’s test (α = 0.05). (C) Height of 6-week-old 35S::PfAvr4-2 and control cv. Moneymaker (MM) plants. Bars represent the average height of seven plants per line, error bars are SD, and letters are significant differences based on a Tukey’s test (α = 0.05). (D) Cell size and cell number comparisons between the 35S::PfAvr4-2 and control Moneymaker plants in stem cross sections made at 1 cm from the crown base. Main anatomical features are indicated; ep, epidermis; co, cortex; en, endodermis; vb, vascular bundle (xylem-cambium-phloem); pi, pith. Bars represent the average cell number or size in the cortex of 2-week-old plants per line. Cell numbers were quantified by measuring the number of the cells in the cortex of seven plants, and cell size was quantified by measuring the area of the cortex and dividing it by the number of cells. Error bars indicate SD and letters are significant differences based on a Tukey’s test (α = 0.05). Photo credit: Li-Hung Chen, University of California, Davis.
Further phenotypic analysis of the transgenic plants indicated that the reduction in Ca2+-mediated cross-linking is accompanied by changes in their morphological, anatomical, and textural properties that are associated with relaxation in plant cell wall stresses. Transgenic plants had elongated stems and were taller as compared to the control Moneymaker ones (Fig. 5C), whereas fewer but larger in size cortical cells were observed in their stems that likely contributed to their increase in length (Fig. 5D). As Ca2+-mediated cross-linking of homogalacturonan in plant cell walls confers mechanical strength to the plant tissue (7), characterization of the plants’ textural properties via an indentation tested on the plants’ petioles using a texture analyzer (Fig. 6A) showed that the 35S::PfAvr4-2–expressing plants were softer as compared to the control Moneymaker plants (Fig. 6B). Specifically, compared to the control Moneymaker plants, the 35S::PfAvr4-2–expressing plants offered less resistance to indentation, as the maximum force on the probe achieved during the test was significantly lower for the transgenic lines. Stiffness of the petiole tissue, defined as the maximum force achieved during the test divided by the total distance traveled by the probe, was also lower for the transgenic lines than for the Moneymaker line. Last, the distance at fracture, quantified as the displacement of the probe when the first significant fracture (breakage) of the tissue occurred, was again greater for the transgenic lines than for the Moneymaker line (Fig. 6B). This parameter showed that the petiole tissue from the transgenic plants was both softer and more compliant than the Moneymaker plants, as it could be deformed to a greater extent before fracturing.
Fig. 6. The 35S::PfAvr4-2 transgenic tomato plants have altered textural characteristics.
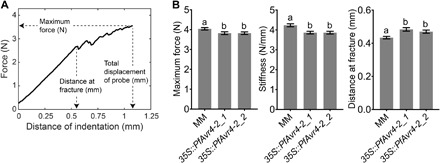
(A) Exemplary texture curve showing the parameters that were defined using texture analysis. The textural properties of the petioles were tested by an indentation test. Parameters measured were “maximum force (in newton),” which refers to the maximum normal force exerted by the petiole on the probe during the test; “distance at fracture (in millimeter),” which refers to the displacement of the probe when the first significant fracture (breakage) of the tissue occurred; and “stiffness (in newton per millimeter),” which refers to the maximum force (in newton) achieved during the test divided by the total distance traveled by the probe (in millimeter) (achieved at 33% strain). (B) Textural properties of the two 35S::PfAvr4-2–expressing lines (35S::PfAvr4-2_1 and 35S::PfAvr4-2_2) and the control cv. Moneymaker (MM) tomato line, as measured with the indentation test. Bars show that maximum force and stiffness, which positively correlate with tissue firmness, and the distance at fracture, which indicates the extent to which the tissue can be deformed before fracture occurs. Error bars indicate SD and were obtained from performing indentation tests at four equally distanced sites per petiole. At least 29 petioles were tested for each line (resulting in a minimum of 115 total compression tests per line). Letters show significant differences based on a Tukey’s test (α = 0.05).
PfAvr4-2 contributes to virulence by enhancing the activity of a fungal endo-PG
Since PfAvr4-2 interferes with the integrity of the plant cell walls, we next examined whether this activity contributes to fungal virulence. Virulence assays performed with the wild-type P. fuligena on the two the two 35S::PfAvr4-2–expressing lines and the control Moneymaker line indicated that the fungus induced considerably more necrosis (Fig. 7, A and B) and accumulated more biomass (Fig. 7C) in inoculated leaves of the two transgenic lines as compared to the Moneymaker line. Similarly, spot inoculations with the necrotrophic pathogen Botrytis cinerea on leaves of the two transgenic lines and the control Moneymaker line also showed that the diameter of the infection lesions was significantly larger in the former (Fig. 7, D and E). To further confirm that PfAvr4-2 is a virulence factor, we examined whether it is expressed under in planta conditions and whether its deletion from the genome of P. fuligena would have an effect on fungal infectivity. Analysis of the in planta expression of PfAvr4-2, showed that it exhibits a transient expression pattern in which transcription is steadily increased during the initial biotrophic stage of the infection (1 to 6 days post inoculation), reaching a maximum at the time of transitioning from biotrophy to necrotrophy (8 to 9 days after inoculation) and gradually decreasing thereafter (Fig. 7F). Virulence assays with two PfAvr4-2 deletion mutants (∆PfAvr4-2_1 and ∆PfAvr4-2_2) that were generated showed that relative to the wild-type P. fuligena strain, the two ∆PfAvr4-2 mutants induced less symptoms and produced less biomass in leaves of infected Moneymaker plants (Fig. 7, G and H). Collectively, these results support that PfAvr4-2 is a virulence factor.
Fig. 7. PfAvr4-2 is a virulence factor.

(A) Disease symptoms at 15 days post inoculation (dpi) induced by P. fuligena on leaves of the 35S::PfAvr4-2 tomato lines (35S::PfAvr4-2_1 and 35S::PfAvr4-2_2) and the control cv. Moneymaker (MM) line. (B) Magnification of the sections marked by white boxes in (A). (C) Quantification (9 dpi) of P. fuligena biomass in leaves of the 35S::PfAvr4-2 plants relative to its biomass in leaves of Moneymaker (set to 1.0). Error bars indicate SEM from three assays and letters significant differences based on a Tukey’s test (α = 0.05). (D) Infection lesions (3 dpi) of B. cinerea on leaves of Moneymaker and 35S::PfAvr4-2 plants. (E) Mean diameter of the infection lesions produced on the 35S::PfAvr4-2 plants relative to the Moneymaker line (set to 1.0). Error bars indicate SD in lesion diameters from three assays and ten leaves per line. Letters show significant differences based on a Tukey’s test (α = 0.05). (F) In vitro and in planta PfAvr4-2 expression levels, relative to the P. fuligena GAPDH gene (set to 1.0 RQ). Shown are average RQ values from two inoculation assays on the Moneymaker line. Error bars indicate SE from four qPCRs. (G) Virulence (15 dpi) of a wild-type (WT) P. fuligena and of two PfAvr4-2 deletion mutants (∆PfAvr4-2_1 and ∆PfAvr4-2_2) on Moneymaker plants. (H) Quantification of fungal biomass in tomato leaves infected with the ∆PfAvr4-2 mutants, relative to the biomass of the wild-type (WT) strain (set to 1.0 RQ). Error bars indicate SEs from three virulence assays. Letters show significant differences based on a Tukey’s test (α = 0.05). Photo credit: Li-Hung Chen, University of California, Davis.
We next sought to investigate how PfAvr-4-2 could be contributing to fungal virulence. Ca2+-mediated cross-linking of homogalacturonan chains increases cell-cell adhesion, which, in turn, may further limit hydrolyzing enzymes from accessing the plant cell wall (7, 28). We thus used the PECTOPLATE assay (29) to examine whether by interfering with Ca2+-mediated cross-linking in plant cell walls, PfAvr4-2 could be affecting the enzymatic activity of various homogalacturonan-modifying enzymes, including PMEs from B. cinerea, an endo-PG from Aspergillus niger, an exo-PG from Yersinia enterocolitica (exo-PG), and a pectate lyase from Aspergillus sp. Mixing of PfAvr4-2 with the endo-PG resulted in an ~46% increase relative to controls in the diameter of the cleared halo formed around the wells containing the mixture, indicating that PfAvr4-2 enhanced the activity of this enzyme (Fig. 8A). The increase in the diameter of the halo was caused by the endo-PG, as PfAvr4-2 did not show any pectin-related enzymatic activity (Fig. 8B). The positive impact of PfAvr4-2 on the activity of the endo-PG was further confirmed by a spectrophotometric enzyme activity assay that quantified the OGs released from degradation of homogalacturonan in the presence/absence of PfAvr4-2 (Fig. 8C). In contrast to endo-PG activity, there were no changes relative to controls in the pattern or diameters of the fuchsia-colored haloes formed around wells containing mixtures of PfAvr4-2 with the PME, exo-PG, or pectate lyase (fig. S6A). In contrast, PfAvr4 and WGA had no impact on the activity of the endo-PG, indicating that the effect seen with PfAvr4-2 was specific (fig. S6B).
Fig. 8. PfAvr4-2 enhances the activity of fungal endo-PGs.
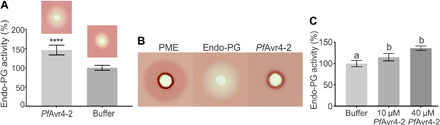
(A) Relative quantification of fungal endo-PG activity in the presence of PfAvr4-2 using the PECTOPLATE. The bar graph shows relative quantification based on the intensity of Ruthenium Red staining around the inoculation wells of endo-PG activity when the enzyme is applied alone or in a mixture with PfAvr4-2. Experiments were done in triplicate, and the intensity of the fuchsia-colored halo produced by the activity of the endo-PG is set to 100%. Error bars indicate SD from the three experiments. Student’s t test was used to evaluate statistical significance of the difference between the two treatments. ****P < 0.001. The images above the bars provide a visual representation of the results. (B) Visual representation of the PECTOPLATE radial diffusion assay used to compare the pectin-related enzymatic activity of PME, endo-PG, and PfAvr4-2. The enzymatic activity of the three pectin-related proteins is assessed on the basis of the size of the fuchsia-stained haloes in the PECTOPLATE. (C) Relative quantification of endo-PG activity in the presence of increasing amounts of PfAvr4-2. The enzymatic activity of endo-PG was monitored by measuring using 2-cyanoacetamide, the amount of oligogalacturonides (OGs) released in the buffer. The activity of endo-PG alone was set to 100%, and the activity of the enzyme in mixtures with different amounts of PfAvr4-2 was expressed relative to this activity. Experiments were done in triplicate, and error bars indicate SD. Treatments with the same letter were not significantly different by a Tukey’s test (α = 0.05).
PfAvr4-2 does not suppress OG-induced immunity in tomato and is not recognized by Cf-4
As OGs can be perceived as DAMPs, thereby eliciting immune responses (12), we examined whether PfAvr4-2 has the ability to suppress OG-triggered immunity, as demonstrated for other carbohydrate-binding effectors (30). However, treatment of the two 35S::PfAvr4-2–expressing lines and the control Moneymaker line with 100 μM OG10-15 solution did not result in statistically significant differences among the three lines in the accumulation of reactive oxygen species (ROS), indicating that PfAvr4-2 is unable to inhibit the OG-induced ROS burst in tomato (fig. S7A). Last, since effectors may mediate avirulence in the presence of cognate resistance genes, we examined whether PfAvr4-2 is perceived by the Cf-4 immune receptor from tomato that mediates HR upon recognition of Avr4 proteins, including of PfAvr4 (19). Transient coexpression of PfAvr4 or PfAvr4-2 with Cf-4 in N. tabacum and Nicotiana benthamiana using an Agrobacterium tumefaciens transient transformation assay showed that only PfAvr4 induced HR in the presence of the Cf-4 receptor, thus suggesting the lack of PfAvr4-2 recognition by Cf-4 (fig. S7B). However, lack of an anti–PfAvr4-2 antibody impeded us from testing whether the absence of a HR was due to the lack of protein accumulation in the infiltrated tissue.
DISCUSSION
Here, we show that a candidate effector from the tomato pathogen P. fuligena assists infections by binding to homogalacturonan with a low degree of esterification in the plant’s middle lamella or the primary cell wall and interfering with the Ca2+-mediated cross-linking of homogalacturonan chains. This, in turn, loosens the plant cell wall and synergizes the activity of fungal endo-PGs (fig. S8).
Carbohydrate-binding modules are generally known to synergize the activity of appending CAZymes by promoting the interaction of the enzyme with the target substrate and/or through the noncatalytic disruption of the substrate (31). It is shown, for example, that cellulose-binding domains enhance the activity of cellulases by sliding like a wedge and thus disrupting the hydrogen bonding between two adjacent cellulose chains, thereby enabling the enzyme to penetrate more deeply into crystalline cellulose (32). Through binding to nonmethylated GalA residues in intermittently Ca2+-mediated cross-linked homogalacturonan chains, PfAvr4-2 could likewise part adjacent homogalacturonan chains and disrupt their Ca2+-mediated cross-linking, thus providing endo-PGs better access to unesterified regions of homogalacturonan and creating more cleavage sites along the homogalacturonan backbone (fig. S8). Notably, endo-PGs are most active against homogalacturonan with a low degree of esterification, and therefore, they often function in cohort with pathogen secreted PMEs (33, 34). As primary cell walls consist of a matrix of cellulose microfibrils associated with pectins and hemicelluloses (35), the enhanced destruction of the pectin network could subsequently indirectly aid the enzymatic degradation of other plant cell wall–based polysaccharides, thus further accelerating the decomposition of the plant tissue. This may also explain why PfAvr4-2 exhibits higher expression levels during the switch to the necrotrophic phase rather than the initial biotrophic phase, as is the case with many effectors (16). Plants, on the other hand, are also known to tightly regulate the degree and pattern of pectin methyl-esterification during host-microbe interactions through the precise and timely activation of endogenous PMEs and PME inhibitors. Overexpression of PME inhibitors, for example, is shown in various plant species to increase resistance against necrotrophic and hemibiotrophic pathogens, owing to the reduced susceptibility of methyl-esterified pectin to pectic enzymes secreted by pathogens. As the status of pectin methyl-esterification is also crucial to PfAvr4-2’s function, it is plausible that PME inhibitors further promote resistance by indirectly interfering with the protein’s activity. Decreased binding of PfAvr4-2 to plant cell walls was observed in the tomato PME-silencing lines, suggesting that PME inhibitors would negatively affect the virulence function of PfAvr4-2. Thus, by reducing the levels of pectin methyl-esterification, PME inhibitors lower both pectin’s susceptibility to enzymatic digestion and its amenability by PfAvr4-2.
The disruption by PfAvr4-2 of the Ca2+-mediated cross-links in plant cell walls could also loosen their structure, a function that is somewhat reminiscent of the function of expansins. Expansins are a large superfamily of mainly plant proteins that mediate the noncatalytic loosening of the cell wall by disrupting hydrogen bonds linking adjacent wall polysaccharides and thus reducing their adhesion (36). Despite the lack of sequence similarity between PfAvr4-2 and known expansins of plant or microbial origin, there are interesting similarities between the function of the two groups. For instance, they both act on noncovalent bonds and are most active at acidic pH. They also lack lytic activity on polysaccharide substrates while mediating the reversible disruption of cross-links between adjacent polysaccharide chains, thus reducing their adhesion and loosening the cell wall. However, PfAvr4-2 does not interact with cellulose, a nearly universally property in plant and microbial expansins and has a CBM14 domain instead of the typical CBM63 cellulose-binding domain and family 45 glycosyl hydrolase (GH45) domain found in canonical expansins (36).
Although chitin is the only known binding substrate of CBM14 modules, PfAvr4-2 is an exception as it interacts with pectin in plant cell walls. This finding expands the substrate specificity of the CBM14 family and further broadens the role of fungal apoplastic effectors in structural modification of plant cell walls. Other examples of effectors that fulfill their role in virulence through carbohydrate binding include the chitin-binding Avr4 (17, 19) and Ecp6 (30) core effector proteins that protect fungal chitin against chitinases or inhibit chitin-induced innate immunity by sequestering chito-oligosaccharides from the apoplastic space and the FGB1 effector from Piriformospora indica that suppresses β-glucan–triggered innate immunity by binding to β-glucans released from the fungal cell wall (37). These examples show that microbial lectins can serve multiple virulence roles as effectors during infections, including protection or noncatalytic disruption of polysaccharide substrates, enhancement of the hydrolytic activity of cell wall–degrading enzymes, and subversion of plant defense responses. Such functions may be particularly important for extracellular pathogens, as the carbohydrate-rich plant and fungal cell walls form the main interface of interaction between plants and pathogens in the apoplast and are consequently reciprocally targeted by each side seeking to gain an advantage.
MATERIALS AND METHODS
Plant materials and growth conditions
Tomato plants were grown in the growth chamber with 16-hour light and 70% relative humidity at 25°C for 6 weeks. Arabidopsis thaliana plants were grown in a controlled environment chamber with a 10-hour photoperiod at 23°C and 75% relative humidity.
Allelic variation
Allelic variation was examined in 30 strains of P. fuligena originating from Taiwan (n = 1) and nine prefectures of Japan, i.e., Aichi (n = 1), Chiba (n = 2), Fukuoka (n = 1), Gifu (n = 6), Mie (n = 14), Shizuoka (n = 1), Tokushima (n = 2), Tottori (n = 1), and Wakayama (n = 1). Genomic DNA was extracted from the strains using the DNeasy Plant Mini Kit (QIAGEN), and the PfAvr4-2 gene was amplified with primers PfAvr4-2_F1/PfAvr4-2_1 (table S1) using the REDTaq DNA Polymerase (Sigma-Aldrich). PCR reaction cycle consisted of an initial 95°C for 10 min, followed by 15 s at 95°C, 30 s at 58°C, and 30 s at 72°C for 40 cycles. PCR products were sequenced with primer PfAvr4-2_R2 (table S1), and obtained electropherograms were aligned using the MEGA software package (v6.0) (38).
Protein structure modeling
The 3D model of PfAvr4-2 was generated by I-TASSER (Iterative Threading ASSEmbly Refinement) with the default settings (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) (39) and using the PfAvr4 [Protein Data Bank (PDB): 4Z4A] and CfAvr4 (PDB: 6BN0) structures as threading templates. The highest C-score (=0.14) model was used for further analysis. Protein structure alignments and coloring of the electrostatic surfaces and of o the side chains of Avr4 proteins were conducted using UCSF Chimera (v1.14) (40).
Protein purification from culture filtrates of Pichia pastoris
Recombinant PfAvr4-2 protein was produced using the methylotrophic yeast P. pastoris GS115 as described by Kohler et al. (19). The produced by PfAvr4-2 protein was purified through a NiNTA column (Thermo Fisher Scientific). The protein was eluted in 50 mM tris-HCl (pH 7.0) and 50 mM NaCl with 250 mM imidazole and concentrated in a Amicon Ultra-15 centrifugal filter unit (3-kDa cutoff; Millipore). The concentrated protein was further purified by passing through a Superdex 75 column (GE Healthcare) with 25 mM tris-HCl (pH 7.0) and 50 mM NaCl on an AKTA FPLC system (GE Healthcare; software UNICORN 5.2). The eluted protein was subsequently analyzed by a MiniDAWN TREOS MALS detector (Wyatt Technology) with a laser source at 658.8 nm, followed by an Optilab T-rEX refractometer (Wyatt Technology) with a 658.0-nm light source. The final protein was verified by SDS–polyacrylamide gel electrophoresis, concentrated, and stored at −80°C.
In vitro polysaccharide precipitation assays and fungal protection assays against chitinases
Binding assays of PfAvr4-2 or PfAvr4 with various insoluble polysaccharides that are common constituents of fungal and plant cell walls—such as shrimp-shell chitin (Sigma-Aldrich), colloidal chitin (Sigma-Aldrich), chitosan (Sigma-Aldrich), curdlan AL (InvivoGen), cellulose (Sigma-Aldrich), and xylan (Sigma-Aldrich)—were performed at three pHs (5.5, 7.0, and 8.5) as described previously (17, 19), with some modifications (Supplementary Materials and Methods). To test whether PfAvr4-2 can protect fungal hyphae against chitinases of tomato or bacterial origin, we followed the procedure described before (17, 19) with some modifications (Supplementary Materials and Methods).
Defined carbohydrate microarray
The defined carbohydrate microarray was prepared as described before (25). Briefly, the carbohydrates were printed on a nitrocellulose membrane using a microarray printer (ArrayJet, Roslin, UK). PfAvr4-2 was conjugated with fluorescein isothiocyanate (FITC; Thermo Fisher Scientific) following the manufacturer’s protocol. The array was blocked with 5% bovine serum albumin (BSA) dissolved in a NaOAc/NaCl buffer [25 mM NaOAc and 50 mM NaCl (pH 5.5)] and incubated with PfAvr4-2FITC at a 1:100 dilution for 1 hour at 4°C. The array was then washed twice with the NaOAc/NaCl buffer and scanned by a GenePix scanner using a 488-nm laser. The intensity of the fluorescence signals on the array was quantified using ProScanArray Express-MicroArray Analysis System software (v4.0.0.0004, PerkinElmer), and values were normalized from 0 to 100. Highest value was set as 100, and the cutoff value was 5. The experiment was performed once.
Isothermal titration calorimetry
For the ITC assays, PfAvr4-2 was dialyzed against the NaOAc/NaCl buffer at 4°C for 2 days. The OG10-15 mixture (Carbosynth, catalog no. OG59704) was dissolved in the same buffer to a final concentration of 2 mM. ITC experiments were conducted on a NanoITC (TA Instruments). The heat change was quantified by titrating 2.5 μl of 2 mM OG10-15 into the cell containing 0.5 mM PfAvr4-2 in a total volume of 170 μl with a 1.5-min equilibration time and a stir rate of 250 rpm for 20 titrations. Data were analyzed using the NanoITC analysis software (v3.7.5, TA Instruments) and fit with an independent binding model provided by the software. The first titration was done with 1 μl of 2 mM OG10-15, and the value of this point was used for fitting the model. ITC experiments were repeated twice with reproducible results.
Localization studies on fungal and plant cell walls
Localization of PfAvr4 and PfAvr4-2, labeled with Alexa Fluor 488 succinimidyl ester (488) (Life Technologies) or RR-X, succinimidyl ester (RR) (Life Technologies), respectively, on cell walls of T. viride was examined as described before (19). The experiment was repeated three times, and protein localization on fungal cell walls was examined on at least 30 germlings of T. viride with reproducible results. Localization of PfAvr4 and PfAvr4-2 on cell walls of P. fuligena was performed in a similar way (19), with some modifications. Specifically, mycelia of P. fuligena were first treated at 25°C for 1 hour with 5 U of Zymolyase (Zymo Research, catalog no. E1004) to remove the β-glucan layer in the fungal cell wall and expose the underlying chitin layer. The mycelia were then incubated with 3 μM PfAvr4RR or PfAvr4-2RR in the NaOAc/NaCl buffer at room temperature for 40 min. After washing three times with the same buffer, the fluorescent signal was detected with a Zeiss LSM710 confocal microscope using the 40× water objective. The experiment was repeated three times with reproducible results.
Localization studies of PfAvr4488, PfAvr4-2488, and PfAvr4-2RR; the OG7-13488 probe; and the JIM7 and 2F4 (PlantProbes, http://plantprobes.net) antibodies on Arabidopsis stem cross sections were done according to Mravec et al. (25), with modifications. All localization experiments on Arabidopsis stem cross sections were repeated at least three times with reproducible results. In each experiment, a minimum of two stem cross sections were labeled with the individual proteins or antibodies, and at least five pictures were taken per stem cross section. The Arabidopsis stem samples were collected at 2 cm above the leaf of 1-month-old Arabidopsis plants and fixed in 4% formaldehyde for 1 hour at room temperature. After washing with water, the stem samples were embedded in 5% agarose and sectioned into 150-μm-thick sections using a Vibratome-1000. The stem cross sections were then washed three times with the NaOAc/NaCl buffer and 2 μM of the assayed proteins was incubated with the sections in 500 μl of the same buffer for 40 min. CW (50 μg/ml) (Sigma-Aldrich) was used for labeling cellulose in plant cell walls on the stem cross sections for 10 min at room temperature. After washing three times with the NaOAc/NaCl buffer, the fluorescence signal was detected with a Zeiss LSM710 confocal microscope using the 40× water or 63× oil objectives. Images were analyzed using the Zen imaging software (v2.3, blue edition, ZEISS).
For immunostaining of the Arabidopsis stem cross sections with the JIM7 and 2F4 monoclonal antibodies, the antibodies were diluted 1:50 in the NaOAc/NaCl buffer and incubated with the stem samples for 1 hour at room temperature. After washing with the NaOAc/NaCl buffer three times, anti-rat immunoglobulin G (IgG; H + L) secondary monoclonal antibody with Alexa Flour 488 (A-11006, Invitrogen) was used for detecting the JIM7 monoclonal antibody, whereas anti-mouse IgG (H + L) secondary monoclonal antibody with Alexa Flour 488 (A28175, Invitrogen) was used for detecting the 2F4 monoclonal antibody. The secondary antibodies were diluted 250× in the NaOAc/NaCl buffer and mixed with the samples for 1 hour at room temperature. For labeling of the stem samples with the OG7-13488 probe, 2 μM probe was incubated with the Arabidopsis stem cross sections in the NaOAc/NaCl buffer that also contained 1 mM CaCl2. The fluorescence intensity of images was analyzed and quantified using the Zen imaging software (v2.3, blue edition, ZEISS).
To localize PfAvr4-2RR and PfAvr4488 on tomato leaves infected with P. fuligena, the leaves of 6-week-old Moneymaker tomato plants were inoculated with the mycelial fragments of the wild-type P. fuligena strain as described before (19). The infected tomato leaves were collected at 9 days after inoculation and washed with 100% ethanol for 2 days to remove all chlorophyll. The samples were subsequently washed with the NaOAc/NaCl buffer and labeled with PfAvr4-2RR and PfAvr4488, as described above. Images were taken with a Zeiss LSM710 microscope using 63× oil objective and analyzed with the Zen imaging software (v2.3, blue edition, ZEISS). The experiment was repeated three times with reproducible results, and at least ten images were taken from two infected leaves in each experiment.
The PME-silenced tomato lines (cv. Rutgers) were a gift from E. Mitcham in the Department of Plant Sciences, UC Davis. Tomato stems (n = 2) from 5-week-old tomato plants were embedded, sectioned, and labeled with PfAvr4-2RR as described above for Arabidopsis stems. Images (n = 5 per stem) of tomato stems stained with PfAvr4-2RR were taken with a Zeiss LSM710 microscope using 20× and 40× water objectives and analyzed with the Zen imaging software (v2.3, blue edition, ZEISS). The experiment was repeated three times with reproducible results.
Enzymatic and chemical treatment of Arabidopsis stem cross sections
To investigate how pH and calcium concentration ([Ca2+]) influence the localization of PfAvr4-2 on plant cell walls, Arabidopsis stem cross sections (n = 6 per treatment) were incubated for 40 min at room temperature with 2 μM PfAvr4-2RR in a NaOAc/NaCl buffer and a tris-HCl/NaCl solution (25 mM tris-HCl and 50 mM NaCl) buffered at pH 7.0 and pH 8.5 as well as the NaOAc/NaCl buffer supplemented with 1 and 2 mM CaCl2. After washing the samples with the same buffers, images (n = 5 per stem cross section per treatment) of the stem cross sections were taken with a Zeiss LSM710 confocal microscope and analyzed using the Zen imaging software (v2.3, blue edition, ZEISS). For the pectinase, NaOH, and EGTA treatments, Arabidopsis stem cross sections (n = 6 per treatment) were pretreated with 8 mU of a fungal pectinase mix consisting of pectin transeliminase, PG, and pectin esterase (Sigma-Aldrich, catalog no. P2736). Treatments were done at room temperature in 100 mM NaOH for 2 hours or 50 mM EGTA for 15 min in a total volume of 500 μl of the NaOAc/NaCl buffer. After treatment, the samples were washed three times with the same buffer and then probed with PfAvr4-2RR, as described above. Images (n = 5 per stem cross section per treatment) were taken with the Zeiss LSM710 microscope using 20× and 40× water objectives and were analyzed with the Zen imaging software (v2.3, blue edition, ZEISS). The experiment was repeated three times with reproducible results.
Monitoring the state of Ca2+ cross-links (i.e., egg boxes) in junction zones
Stem segments of 1-month-old Arabidopsis plants were embedded in 5% agarose and thin sectioned using a Vibratome-1000. The thin cross sections were then incubated for 2 hours with 10 μM PfAvr4, PfAvr4-2, or WGA in 500 μl of the NaOAc/NaCl buffer. To label them with the OG7-13488 probe, the stem cross sections (n = 3 per pretreatment) were washed three times with the NaOAc/NaCl buffer, and then 2 μM OG7-13488 probe dissolved in the same buffer containing 1 mM CaCl2 was added to the samples. To label the stem cross sections with the 2F4 monoclonal antibody, the sections (n = 3 per pretreatment) were first washed with the NaOAc/NaCl buffer for 1 hour to remove PfAvr4-2, PfAvr4, or WGA and then immunostained in the same buffer with the 2F4 monoclonal antibody and the anti-mouse IgG (H + L) secondary monoclonal antibody conjugated to the Alexa Fluor 488. After washing three times with the NaOAc/NaCl buffer, images (n = 5 per stem cross section per treatment) were taken with a Zeiss LSM710 confocal microscope using a 40× water lens or 63× oil lens and analyzed with the Zen imaging software (v2.3, blue edition, ZEISS). Quantification of relative fluorescence was done by measuring the fluorescence signal at the corners of cell junction from more than 12 images using the Zen software (Zeiss). The fluorescence intensity of 2F4 or OG7-13488 labeling after WGA treatment was set to 1.0 for calculating the relative fluorescence of other treatments. Experiments were performed three times with reproducible results. Statistical analysis was performed with the GraphPad Prism software (v8.3.1., Graphpad software Inc.). Student’s t test was conducted to evaluate the significance of difference among treatments.
Pollen tube assays
The germination of pollen tubes in pollen germination media was examined as described before (41). Briefly, fully open flowers of young Moneymaker tomato plants were collected into 2-ml tubes containing 1 ml of pollen germination media buffer [24% (w/v) polyethylene glycol 4000, 0.01% (w/v) H3BO3, 2% (w/v) sucrose, 20 mM Hepes buffer (pH 6.0), 0.02% MgSO4, and 1 mM KNO3]. The tubes were then gently vortexed for 15 s to release the pollen grains from flowers. Pollen grains (500 grains/ml) were pregerminated in pollen germination media buffer amended with CaCl2 at different concentrations (0, 0.5, 1, 2, and 4 mM) in a 48-well plate for 1 hour at room temperature. A 20 μM PfAvr4 or PfAvr4-2 was then added into each well in a total volume of 500 μl and incubated for 2 hours. Images of germinated pollen grains and the formed pollen tubes were taken with a Nikon Diaphot inverted microscope. The percentage of burst pollen tubes was calculated by the amount of burst pollen tubes from 100 counted pollen grains per treatment. Experiments were repeated more than three times. To further image PfAvr4-2RR on pollen tubes, tomato pollen grains were pregerminated in the pollen germination media buffer amended with 0.5 mM CaCl2 for 2 hours at room temperature. The germinated pollen grains (n = 1000) were incubated with 2 μM PfAvr4-2RR in 500 μl of pollen germination media buffer for 40 min at room temperature. The pollen grains were collected via centrifugation at 2000 rpm for 3 min and washed with the pollen germination media buffer three times. Images of at least 10 pollen tubes were taken with a Leica LAS-AF SPE confocal microscope (Leica Camera) at ×40 magnification.
Stable expression of PfAvr4-2 in tomato plants
To express PfAvr4-2 in Moneymaker plants, A. tumefaciens GV3101 that was transformed with a binary pMOG800 vector in which the mature PfAvr4-2 was cloned was submitted to the Plant Transformation Facility of the UC Davis (https://ptf.ucdavis.edu/) for generating transgenic plants according to an established protocol used by the facility (Supplementary Materials and Methods). The PfAvr4-2 transgene expressed in Moneymaker consisted of the mature PfAvr4-2 protein, fused to the N. tabacum PR1A signal sequence for secretion into the apoplast, and placed in-between the cauliflower mosaic virus 35S promoter (CaMV 35S) and the Agrobacterium nopaline synthase terminator. The presence of the PfAvr4-2 transgene in T1 plants was confirmed by PCR using primers PR1A_ClaI_F/PfAvr4-2_NotI_R (table S1). At least five T1 plants with the PfAvr-2 transgene were selected and self-fertilized. The derived T2 lines were germinated on Murashige and Skoog minimal organics (MSO) medium amended with kanamycin sulfate (50 mg/liter), and the presence of the PfAvr4-2 transgene was reconfirmed by PCR using the above primers. T2 plants were then self-fertilized, and 11 seeds were tested for segregation by placing them on MSO medium amended with kanamycin sulfate (50 mg/liter). Of the 11 seeds, seven produced seedlings that survived the selection and continued to grow, indicating that they are homozygous for PfAvr4-2. Two T2 lines (35S::PfAvr4-2_1 and 35S::PfAvr4-2_2) were lastly selected, and the expression levels of the PfAvr4-2 transgene in each line were examined by qPCR relative to the tomato RPL2 gene using the primers PfAvr4-2_qPCR-F/PfAvr4-2_qPCR-R and TmRRL2-F/TmRPL2-R (table S1).
Extraction of pectin from tomato leaves and immunodot blot analysis
Extraction of pectin and of Ca2+-mediated cross-linked homogalacturonan from plant cell walls of the two 35S::PfAvr4-2 transgenic lines and the control Moneymaker line was performed as described before (42), with some modifications. Briefly, 1 g of tomato leaves collected from 6-week-old plants was ground to fine powder in liquid nitrogen, washed with 5 ml of 70% ethanol, incubated at room temperature with rotation for 10 min, and pelleted by centrifugation at 5000 rpm for 10 min. The washing step was repeated five times until all chlorophyll was removed. The pellet was then resuspended in 5 ml of 100% acetone, incubated for 10 min at room temperature with rotation, and centrifuged at 5000 rpm for 10 min. The supernatant was decanted, and the pellet that contained the alcohol-insoluble residue (AIR) was air-dried overnight. Pectin was extracted from 10 mg of AIR by adding 300 μl of 50 μM cyclohexane diamine tetraacetic acid (CDTA; pH 7.0), vortexing the sample at room temperature for 2 hour, and centrifuging at 12,000 rpm for 10 min. The supernatant was then stored at 4°C until immunodot blot analysis. Since CDTA removes Ca2+ from pectin, to extract Ca2+–cross-linked homogalacturonan, 10 mg of AIR was mixed with 300 μl of ddH2O and then vortexed at room temperature for 2 hours. After centrifugation, the supernatant was stored at 4°C.
Immunodot blot analysis was performed as described before (43), with some modifications. Briefly, to determine the amount of Ca2+-mediated cross-linking in the above samples, they were diluted 10×, 20×, and 40× in ddH2O, and 1 μl of each diluted sample was applied to a nitrocellulose membrane (Sigma-Aldrich). The membrane was then dried at 37°C for 1 hour and blocked with 1× phosphate-buffered saline with 3% milk (PBS/M) [137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 (pH 7.4)] for 1 hour. Next, the 2F4 antibody was diluted 1:250 in PBS/M buffer and incubated with the membrane overnight. The next day, the membrane was washed three times with the PBS buffer and incubated with the goat anti-mouse IgG (H + L) secondary monoclonal antibody conjugated to the horseradish peroxidase (HRP) enzyme (Cell Signaling Technology) in PBS/M buffer in a 1:2500 dilution for 1 hour. The membrane was then washed with PBS buffer three times, and the signal was developed using the TMB substrate Kit (Vector Laboratories), following the manufacturer’s protocol. To quantify the amount of pectin extracted with CDTA from the AIR samples, we followed the same procedure but using the JIM5 antibody and anti-rat IgG (H + L) secondary monoclonal antibody conjugated with HRP (Cell Signaling Technology). The fluorescent signal from the 2F4 or JIM5 antibodies was detected using ChemiDoc MP (Bio-Rad). All immunodot blot results were analyzed and quantified using the Image Lab software (v6.0.0 build 26, Bio-Rad). The experiments were repeated five times with reproducible results. Statistical analysis was performed using GraphPad Prism software (v8.3.1, GraphPad software Inc.). One-way analysis of variance (ANOVA) with a Tukey’s test (α = 0.05) was performed to evaluate the difference between different treatments.
CoMPP and monosaccharide composition analysis
Freeze-dried leaves of the two 35S::PfAvr4-2 transgenic lines and the control Moneymaker line were homogenized into fine powder, and the material was washed five times for 10 min and two times for 60 min in 70% ethanol under rotation at room temperature. The material was pelleted between washes by centrifugation for 10 min at 4500 rpm, and the supernatant was decanted. The pellet was subsequently washed in CHCl3:CH3OH 1:1 (v/v) for 2 hours and lastly in 100% acetone for 2 hours. Residual acetone was evaporated overnight in a fume hood, and the resulting cell wall–enriched fractions of AIR were used for CoMPP and monosaccharide composition analysis.
CoMPP was carried out as described before (27), with some modifications. Briefly, 10 mg of AIR was sequentially treated with 400 μl of 50 mM CDTA (pH 7.5) to primarily extract pectins and 4 M NaOH with 0.1% (w/v) NaBH4 to primarily extract hemicelluloses. Each extraction was carried out for 2 hours under shaking in a Tissue lyser (QIAGEN) at 7 s−1 at room temperature. Samples were pelleted in between extractions by centrifugation at 4500 rpm for 15 min. The extracts were printed on nitrocellulose membrane in two technical replicates and four dilutions using an Arrayjet Marathon (Arrayjet, Roslin, UK). The resulting microarrays were probed with different monoclonal antibodies and quantified as described by Moller et al. (44). The details on antiglycan antibodies and respective epitopes can be found in Rydahl et al. (45).
For monosaccharide composition analysis, AIR material was destarched as follows: 40 mg of AIR was transferred to 2-ml tubes, together with two glass beads and 450 μl of preheated (85°C) MOPS buffer [50 mM 4-morpholinepropanesulfonic acid sodium salt, 5 mM CaCl2, and 0.02% NaN3 (pH 7.0)]. The mixture was heated to 85°C for 5 min to allow for starch gelatinization, and 4 μl of termamyl ultrapure amylase (activity, 179 KNU/g; Novozymes) was added to it and incubated at 85°C for 1.5 hours in a heating block at 500 rpm. The mixture was then cooled to 50°C, and the pH was adjusted to 4.5 with CH₃COOH before adding 1.8 U of amyloglucosidase (A-3042, Sigma-Aldrich) and 0.5 U of isoamylase (E-ISAMY, Megazyme) and incubating at 50°C for 1 hour in a heating block at 500 rpm. Last, ethanol was added to 70% and the sample was allowed to precipitate at 4°C overnight. The following day, the samples were centrifuged at 14,000 rpm for 15 min, washed three times in 70% ethanol, and dried overnight under vacuum. For the analysis, 10 mg of destarched AIR was hydrolyzed in 2 M trifluoroacetic acid (TFA) at 120°C for 1 hour. TFA was removed by drying under vacuum. The hydrolysates were dissolved in ddH2O, and monosaccharide composition was determined by high HPAEC-PAD using a PA20 column (Dionex) at a flow rate of 0.45 ml/min. Monosaccharide standards were l-Fuc, l-Rha, l-Ara, d-Gal, d-Glc, d-Xyl, d-Man, d-GalUA, and d-GlcA (Sigma-Aldrich). Statistical analysis was performed using RStudio (v4.0.0, RStudio Core Team). One-way ANOVA with a Tukey’s test (α = 0.05) was performed to evaluate the difference between different lines.
Characterization of the morphological, anatomical, and textural properties of the 35S::PfAvr4-2–expressing tomato plants
Transgenic tomato plants were grown as described above. The height of 14 6-week-old plants per line was determined by measuring stem length from the cotyledons till the shoot tip. To determine the size and number of cells in the cortex, 2-week-old seedlings were cut at 1 cm from the base of the crown and sectioned into 150-μm-thick sections using a Vibratome-1000, as described above. Images of the stem cross sections were taken with a Leica SP8 confocal microscope (Leica Camera) at ×40 magnification, and they were combined and constructed using the navigation tool feature of the Leica LAS X software (v2.0.0.144332). The average cell size in the cortex was determined by calculating the total cortex area in the stem using the drawing tool in the Fiji software (v2.0.0-rc-62/1.51s) and dividing it by the number of cells in the cortex. All experiments were repeated three times with reproducible results.
The texture of petiole tissue from transgenic (35S::PfAvr4-2_1 and 35S::PfAvr4-2_2) and control Moneymaker plants was assessed using an indentation test (46). Briefly, petioles were carefully removed with a razor from the main stem, placed on their side on a metal platform, and immobilized with tape. Petioles were then compressed in the direction perpendicular to their long axis using a texture analyzer (TA-XT Plus, Texture Technologies Corp.) equipped with a 1-mm-diameter cylindrical probe. The test was conducted at 0.1 mm/s and was stopped when 33% strain was reached. The test was conducted at room temperature, and data were acquired at 250 Hz. All indentation tests on a given petiole were carried out within 10 min of removal from the plant stem. The parameters that were obtained from texture analysis were maximum force (in newton), defined as the highest normal on the probe achieved during the test, stiffness (in newton per millimeter), defined as the maximum force achieved during the test divided by the total displacement of the probe, and distance at fracture (in millimeter), quantified as the displacement of the probe when the first significant fracture (breakage) of the petiole tissue occurred (47). The first significant fracture of the petiole tissue was defined as the first peak in the texture curve (force versus distance) with prominence >0.015 N, which was used to ensure that a consistent criterion was applied to all samples. Parameters obtained from texture analysis are presented using an exemplary texture curve in Fig. 6A. The experiment was repeated three times and at least 29 petioles were tested per line, resulting in a minimum of 115 total compression tests per line. Statistical analysis was performed using GraphPad Prism software (v8.3.1, GraphPad software Inc.). One-way ANOVA with a Tukey’s test (α = 0.05) was conducted to compare different treatments.
Expression analysis and virulence assays
Inoculation of the Moneymaker tomato line or of the 35S::PfAvr4-2 transgenic plants with P. fuligena wild-type (WT) strain or the two derivative ΔPAfr4-2 knockout strains (ΔPfAvr4-2_1 and ΔPfAvr4-2_2), analysis of PfAvr4 and PfAvr4-2 expression, and quantification of the fungal biomass at 9 days after inoculation were performed as described before (19). The PfAvr4-2 deletion mutants were generated by replacing the PfAvr4-2 gene with the geneticin resistance cassette as described before (19), with some modifications (Supplementary Materials and Methods). For each assayed strain, nine inoculated leaves from three tomato plants were collected at 1, 3, 6, 9, and 12 days after inoculation. Total RNA was extracted using TRIzol (Life Technologies), and complementary DNA (cDNA) was produced using the iScript cDNA Synthesis Kit (Bio-Rad). The biomass of P. fuligena during fungal infection was quantified by qPCR using fungal actin and the tomato RPL2 gene for calibration, with the primers listed in table S1. qPCR was conducted in a CFX96 Touch Real-Time PCR detection system (Bio-Rad) equipped with the CFX Maestro Software. The reaction cycles consisted of an initial 95°C for 10 min, followed by 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C for 40 cycles. The expression of PfGAPDH was used as an endogenous control that was set to 1.0 for calculating the relative expression of PfAvr4-2. For the virulence assays, the relative biomass of P. fuligena during infection was based on the expression of PfActin as compared to TmRPL2 transcripts. Virulence assays and qPCRs were performed in triplicate. Statistical analysis was performed using GraphPad Prism software (v8.3.1, GraphPad software Inc.). One-way ANOVA with a Tukey’s test (α = 0.05) was conducted to compare different treatments. Expression analysis of PfAvr4-2 was performed twice.
To inoculate leaves of the 35S::PfAvr4-2 transgenic plants with B. cinerea strain B05.10, detached leaves from 6-week-old tomato plants were placed on 1% water phytoagar (Thomas Scientific) in plastic trays covered with a lid to retain humidity. Conidia of B. cinerea were collected from a 2-week-old culture grown on potato dextrose agar and resuspended to 5000 spores/ml in 50% organic grape juice. A single 4-μl droplet of the conidial suspension was then placed on the abaxial side of the tomato leaves. Images of five to six inoculated leaves were taken at 4 days after inoculation, and the diameters (i.e., sizes) of infection lesions from 10 inoculation sites were measured with the Fiji software (48). The size of the infection lesions on leaves of the two 35S::PfAvr4-2 transgenic lines was calculated relative to their size on the control Moneymaker plants, which was set to 1.0. Infection assays with B. cinerea were repeated three times with reproducible results. Statistical analysis was performed using GraphPad Prism software (v8.3.1, GraphPad software Inc.). One-way ANOVA with a Tukey’s test (α = 0.05) was conducted to compare different treatments.
Quantification of PME, PG, and pectate lyase activity
To quantify the enzymatic activity of PMEs, PGs, and pectate lyases, we used an agarose diffusion assay as described before (29), with some modifications. Briefly, 100 μg of PfAvr4-2 was mixed with 10 ml of SDS buffer (62.5 mM tris, 715 mM β-mercaptoethanol, and 346.78 mM NaC12H25SO4) and boiled for 10 min. The boiled protein was precipitated with 20% (w/v) trichloroacetic acid overnight at 4°C, pelleted by centrifugation at 14,000 rpm for 30 min at 4°C, and washed with cold 100% acetone. After centrifuging the samples at 14,000 rpm for 20 min at 4°C, the acetone was removed by aspiration, and the pelleted protein was dried at room temperature for 30 min. Last, the protein was resuspended in the NaOAc/NaCl buffer. Fungal PMEs were extracted from the culture media of 4-day-old B. cinerea cultures in Gamborg B5 medium in which 0.5% apple pectin (w/v; Sigma-Aldrich) was used as the sole carbon source. The culture filtrates were first passed through a Whatman GFA filter paper (Sigma-Aldrich) and then through a bottle-top vacuum filter with a 0.45-μm polyethersulfone membrane (Olympus Plastics). The total protein was concentrated to a small volume using an Amicon Ultra-15 centrifugal filter unit (3-kDa cutoff, Millipore) and stored at −80°C. PfAvr4-2 was next incubated with a fungal PME, fungal endo-PG (Sigma-Aldrich, catalog no. 17389), exo-PG (Megazyme, catalog no. E-EXPGA), or pectate lyase (Megazyme, catalog no. E-PCLYAN) in wells (n = 2 per pectoplate per treatment) of the pectoplates (n = 3 per treatment) as follows: The pectoplate was prepared with 0.1% (w/v) apple pectin, 1% agarose, 25 mM NaOAc (pH 5.5), and 50 mM NaCl for measuring PME and pectate lyase activities, whereas instead of apple pectin, 0.1% (w/v) polygalacturonic acid (Sigma-Aldrich, catalog no. 81325) was used for detecting the enzymatic activities of the endo- and exo-PG. Once the gel is polymerized in 120-mm petri dish (50 ml per plate), the mixture of enzymes and proteins (2 mU of enzyme with 40 μM PfAvr4-2 or boiled PfAvr4-2) was loaded into 4-mm wells cut into the pectoplate with a cork borer and incubated at 30°C for 16 hours. The plates were next stained with 0.05% (w/v) Ruthenium Red (Sigma-Aldrich) for 30 min and washed with water until the fuchsia-colored haloes become noticeable on the gel. The size of the fuchsia-stained haloes (n = 9) was measured using the Fiji software (v2.0.0-rc-62/1.51s). The PECTOPLATE assays were repeated three times, with two replicates for each treatment included in each experiment. Results were reproducible. Statistical analysis was performed using GraphPad Prism software (v8.3.1, GraphPad software Inc.). Student’s t test or one-way ANOVA with a Tukey’s test (α = 0.05) was conducted to compare different treatments.
To further confirm the impact of PfAvr4-2 on the enzymatic activity of the endo-PGs, we used a spectrophotometric approach to measure the enzymatic degradation of pectin using 2-cyanoacetamide as described before (49). Briefly, 0.5 mg of polygalacturonic acid, 5 mU of the fungal endo-PG, and 10 μM of protein samples (PfAvr4, PfAvr4-2, or WGA) were mixed in the NaOAc/NaCl buffer to a final volume of 500 μl at 37°C for 30 min. A total of 1.2 ml of TBC reagent (100 mM Na2B4O7, 100 mM H3BO3, and 0.1% 2-cyanoacetamide) was added to the reaction mixture and boiled for 10 min to stop the reaction. After cooling, the absorbance of the mixture was determined spectrophotometrically at 276 nm using a SpectraMax Plus 384 microplate reader (Molecular Devices). The experiment was repeated three times with at least three replicates per experiment and reproducible results. Statistical analysis was performed using GraphPad Prism software (v8.3.1, GraphPad software Inc.). Student’s t test or one-way ANOVA with a Tukey’s test (α = 0.05) was conducted to evaluate the statistical significant differences between treatments.
Recognition of the PfAvr4 and PfAvr4-2 by the tomato Cf-4 receptor.
A. tumefaciens–based transient transformation assays were performed as described before (19). The pMOG800 plasmid containing the mature PfAvr4-2 protein was prepared as described above, whereas pMOG800 plasmid containing the mature PfAvr4 or the tomato Cf-4–resistant gene was available by the study of Kohler et al. (19). All plasmids were transformed into A. tumefaciens strain GV3101 via electroporation. Transformed agrobacterium with the pMOG800/Cf-4 plasmid was grown to an optical density at 600 nm (OD600) of 0.5, whereas agrobacterium transformed with the pMOG800/PfAvr4 or PfAvr4-2 plasmid was grown to an OD600 of 1.0. Each mixture of agrobacterium cultures was infiltrated at least three times, on three leaves of three N. benthamiana or N. tabacum plants, and infiltrated plants were kept in a growth chamber with 16-hour light and 70% humidity at 25°C for 6 days. The experiment was repeated twice with reproducible results.
Monitoring of ROS burst.
To monitoring ROS-burst following treatment of tomato leaves with an OG10-15 solution, leaf discs of 3.8 mm in diameter from 4- to 5-week-old tomato plants of the two 35S::PfAvr4-2 transgenic lines and the control Moneymaker line were collected and placed overnight at room temperature in a well (one leaf disk per well) of a white 96-well plate (Corning Inc.) with their abaxial side phasing up and in 200 μl of water. After removing the water from the wells, 100 μl of assay solution containing 1.25 μM chemiluminescent probe L-012 (Wako Chemicals, catalog no. 075-05111), peroxidase (20 μg/ml; Sigma-Aldrich, catalog no. P6782), and 10 μM OG10-15 solution (Carbosynth, catalog no. OG59704) was added to the bottom of each well without damaging and submerging the leaf discs. Mock treatments consisted of the same assay solution without the OG10-15 solution. The production of chemiluminescence from each well was measured immediately using a TriStar LB 941 microplate reader (Berthold Technologies). Experiments were repeated three times with reproducible results and with four leaves monitored per line per assay.
Statistical analysis
Student’s t test or one-way ANOVA with a Tukey’s test (α = 0.05) was conducted to evaluate the statistical significance among treatments, as described in individual experiments.
Acknowledgments
We are thankful to A. Kohler and A. Salvucci for the assistance during the initial phases of this work. We thank M. G. Rydahl for the assistance with the carbohydrate microarrays. We are grateful to B. Mattei and G. de Lorenzo for the critical comments and assisting with some of the early assays. We are thankful to G. Bornhorst and C. Swackhamer for the critical contributions to equipment and for assistance with the texture analyzer experiments. We thank E. Mitcham for providing the PME-silenced tomato lines and C. Nakashima for providing the strains of P. fuligena. L. Epstein and R. Bostock are acknowledged for the critical discussions. This paper is dedicated to the memory of J.L. who passed away on 24 September 2019 while this manuscript was in preparation. Funding: I.S. was partially supported by the USDA-NIFA Hatch project CA-D-PPA-2185-H. S.K.K., J.M., and B.J. were funded by the Villum Foundation projects PLANET (00009283). This work was partially supported by the Research Investments in Science and Engineering (RISE) program of the UC Davis (Award RI-091). Author contributions: Conceptualization: L.-H.C., S.K.K., J.M., and I.S.. Primary data collection and analysis: L.-H.C. Secondary data collection and analysis: L.-H.C., S.K.K., K.S.N., and J.M. Funding acquisition: I.S. and B.J. Methodology: L.-H.C., S.K.K., J.M., J.L., B.J., and I.S. Materials: S.K.K., J.M., and B.J. Project administration: I.S. Writing (original draft): L.-H.C. and I.S. Writing (review and editing): L.-H.C., S.K.K., K.S.N., J.M., B.J., J.L., and I.S. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/19/eabe0809/DC1
REFERENCES AND NOTES
- 1.Jones J. D. G., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Bacete L., Mélida H., Miedes E., Molina A., Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 93, 614–636 (2018). [DOI] [PubMed] [Google Scholar]
- 3.De Lorenzo G., Ferrari S., Giovannoni M., Mattei B., Cervone F., Cell wall traits that influence plant development, immunity, and bioconversion. Plant J. 97, 134–147 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Caffall K. H., Mohnen D., The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344, 1879–1900 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Sénéchal F., Wattier C., Rustérucci C., Pelloux J., Homogalacturonan-modifying enzymes: Structure, expression, and roles in plants. J. Exp. Bot. 65, 5125–5160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngouémazong D. E., Jolie R. P., Cardinaels R., Fraeye I., Van Loey A., Moldenaers P., Hendrickx M., Stiffness of Ca2+−pectin gels: Combined effects of degree and pattern of methylesterification for various Ca2+ concentrations. Carbohydr. Res. 348, 69–76 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Wolf S., Mouille G., Pelloux J., Homogalacturonan methyl-esterification and plant development. Mol. Plant 2, 851–860 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Zamil M. S., Geitmann A., The middle lamella-more than a glue. Phys. Biol. 14, 015004 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Lionetti V., Cervone F., Bellincampi D., Methyl esterification of pectin plays a role during plant-pathogen interactions and affects plant resistance to diseases. J. Plant Physiol. 169, 1623–1630 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Brutus A., Sicilia F., Macone A., Cervone F., De Lorenzo G., A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107, 9452–9457 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi H. W., Klessig D. F., DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol. 16, 232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari S., Savatin D. V., Sicilia F., Gramegna G., Cervone F., De Lorenzo G., Oligogalacturonides: Plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 4, 49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabrera J. C., Boland A., Messiaen J., Cambier P., Van Cutsem P., Egg box conformation of oligogalacturonides: The time-dependent stabilization of the elicitor-active conformation increases its biological activity. Glycobiology 18, 473–482 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Voxeur A., Habrylo O., Guénin S., Miart F., Soulié M.-C., Rihouey C., Pau-Roblot C., Domon J.-M., Gutierrez L., Pelloux J., Mouille G., Fagard M., Höfte H., Vernhettes S., Oligogalacturonide production upon Arabidopsis thaliana–Botrytis cinerea interaction. Proc. Natl. Acad. Sci. U.S.A. 116, 19743–19752 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui H., Tsuda K., Parker J. E., Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Toruño T. Y., Stergiopoulos I., Coaker G., Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54, 419–441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurlburt N. K., Chen L.-H., Stergiopoulos I., Fisher A. J., Structure of the Cladosporium fulvum Avr4 effector in complex with (GlcNAc)6 reveals the ligand-binding mechanism and uncouples its intrinsic function from recognition by the Cf-4 resistance protein. PLOS Pathog. 14, e1007263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stergiopoulos I., van den Burg H. A., Ökmen B., Beenen H. G., van Liere S., Kema G. H. J., de Wit P. J. G. M., Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. U.S.A. 107, 7610–7615 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohler A. C., Chen L.-H., Hurlburt N., Salvucci A., Schwessinger B., Fisher A. J., Stergiopoulos I., Structural analysis of an Avr4 effector ortholog offers insight into chitin binding and recognition by the Cf-4 receptor. Plant Cell 28, 1945–1965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang T.-C., Salvucci A., Crous P. W., Stergiopoulos I., Comparative genomics of the Sigatoka disease complex on banana suggests a link between parallel evolutionary changes in Pseudocercospora fijiensis and Pseudocercospora eumusae and increased virulence on the banana host. PLOS Genet. 12, e1005904 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.C. Brett, K. Waldron, Physiology and Biochemistry of Plant Cell Walls (Unwin Hyman, London, Boston, 1990), Topics in plant physiology, pp. xiv, 194 p. [Google Scholar]
- 22.Mohnen D., Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11, 266–277 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Clausen M. H., Willats W. G. T., Knox J. P., Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr. Res. 338, 1797–1800 (2003). [DOI] [PubMed] [Google Scholar]
- 24.de Freitas S. T., Handa A. K., Wu Q., Park S., Mitcham E. J., Role of pectin methylesterases in cellular calcium distribution and blossom-end rot development in tomato fruit. Plant J. 71, 824–835 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Mravec J., Kračun S. K., Rydahl M. G., Westereng B., Pontiggia D., De Lorenzo G., Domozych D. S., Willats W. G. T., An oligogalacturonide-derived molecular probe demonstrates the dynamics of calcium-mediated pectin complexation in cell walls of tip-growing structures. Plant J. 91, 534–546 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Parre E., Geitmann A., More than a leak sealant. The mechanical properties of callose in pollen tubes. Plant Physiol. 137, 274–286 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.S. K. Kračun, J. U. Fangel, M. G. Rydahl, H. L. Pedersen, S. Vidal-Melgosa, W. G. T. Willats, Carbohydrate microarray technology applied to high-throughput mapping of plant cell wall glycans using comprehensive microarray polymer profiling (CoMPP), in High-Throughput Glycomics and Glycoproteomics (Springer, 2017), pp. 147–165. [DOI] [PubMed] [Google Scholar]
- 28.Ferguson I. B., Calcium in plant senescence and fruit ripening. Plant Cell Environ. 7, 477–489 (1984). [Google Scholar]
- 29.Lionetti V., PECTOPLATE: The simultaneous phenotyping of pectin methylesterases, pectinases, and oligogalacturonides in plants during biotic stresses. Front. Plant Sci. 6, 331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Jonge R., van Esse H. P., Kombrink A., Shinya T., Desaki Y., Bours R., van der Krol S., Shibuya N., Joosten M. H. A. J., Thomma B. P. H. J., Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329, 953–955 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J., Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reyes-Ortiz V., Heins R. A., Cheng G., Kim E. Y., Vernon B. C., Elandt R. B., Adams P. D., Sale K. L., Hadi M. Z., Simmons B. A., Kent M. S., Tullman-Ercek D., Addition of a carbohydrate-binding module enhances cellulase penetration into cellulose substrates. Biotechnol. Biofuels 6, 93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sella L., Castiglioni C., Paccanaro M. C., Janni M., Schäfer W., D’Ovidio R., Favaron F., Involvement of fungal pectin methylesterase activity in the interaction between Fusarium graminearum and wheat. Mol. Plant Microbe Interact. 29, 258–267 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Valette-Collet O., Cimerman A., Reignault P., Levis C., Boccara M., Disruption of Botrytis cinerea pectin methylesterase gene Bcpme1 reduces virulence on several host plants. Mol. Plant Microbe Interact. 16, 360–367 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Cosgrove D. J., Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Cosgrove D. J., Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 25, 162–172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wawra S., Fesel P., Widmer H., Timm M., Seibel J., Leson L., Kesseler L., Nostadt R., Hilbert M., Langen G., Zuccaro A., The fungal-specific β-glucan-binding lectin FGB1 alters cell-wall composition and suppresses glucan-triggered immunity in plants. Nat. Commun. 7, 13188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y., The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Covey P. A., Subbaiah C. C., Parsons R. L., Pearce G., Lay F. T., Anderson M. A., Ryan C. A., Bedinger P. A., A pollen-specific RALF from tomato that regulates pollen tube elongation. Plant Physiol. 153, 703–715 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moller I. E., Pettolino F. A., Hart C., Lampugnani E. R., Willats W. G. T., Bacic A., Glycan profiling of plant cell wall polymers using microarrays. J. Vis. Exp., e4238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.C. Hervé, S. E. Marcus, J. P. Knox, Monoclonal antibodies, carbohydrate-binding modules, and the detection of polysaccharides in plant cell walls, in The Plant Cell Wall (Springer, 2011), pp. 103–113. [DOI] [PubMed] [Google Scholar]
- 44.Moller I., Sørensen I., Bernal A. J., Blaukopf C., Lee K., Øbro J., Pettolino F., Roberts A., Mikkelsen J. D., Knox J. P., Bacic A., Willats W. G. T., High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. 50, 1118–1128 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Rydahl M. G., Hansen A. R., Kračun S. K., Mravec J., Report on the current inventory of the toolbox for plant cell wall analysis: Proteinaceous and small molecular probes. Front. Plant Sci. 9, 581 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uluisik S., Chapman N. H., Smith R., Poole M., Adams G., Gillis R. B., Besong T. M. D., Sheldon J., Stiegelmeyer S. M., Pérez L., Samsulrizal N., Wang D., Fisk I. D., Yang N., Baxter C., Rickett D., Fray R., Blanco-Ulate B., Powell A., Harding S., Craigon J., Rose J., Fich E. A., Sun L., Domozych D., Fraser P., Tucker G., Grierson D., Seymour G., Genetic improvement of tomato by targeted control of fruit softening. Nat. Biotechnol. 34, 950–952 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Skamniotis C., Elliott M., Charalambides M., Computer simulations of food oral processing to engineer teeth cleaning. Nat. Commun. 10, 3571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Honda S., Nishimura Y., Takahashi M., Chiba H., Kakehi K., A manual method for the spectrophotometric determination of reducing carbohydrates with 2-cyanoacetamide. Anal. Biochem. 119, 194–199 (1982). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/19/eabe0809/DC1


