Abstract
Mast cells (MCs) play a pivotal role in the hypersensitivity reaction by regulating the innate and adaptive immune responses. Humans have two types of MCs. The first type, termed MCTC, is found in the skin and other connective tissues and expresses both tryptase and chymase, while the second, termed MCT, which only expresses tryptase, is found primarily in the mucosa. MCs induced from human adult-type CD34+ cells are reported to be of the MCT type, but the development of MCs during embryonic/fetal stages is largely unknown. Using an efficient coculture system, we identified that a CD34+c-kit+ cell population, which appeared prior to the emergence of CD34+CD45+ hematopoietic stem and progenitor cells (HSPCs), stimulated robust production of pure Tryptase+Chymase+ MCs (MCTCs). Single-cell analysis revealed dual development directions of CD34+c-kit+ progenitors, with one lineage developing into erythro-myeloid progenitors (EMP) and the other lineage developing into HSPC. Interestingly, MCTCs derived from early CD34+c-kit+ cells exhibited strong histamine release and immune response functions. Particularly, robust release of IL-17 suggested that these early developing tissue-type MCTCs could play a central role in tumor immunity. These findings could help elucidate the mechanisms controlling early development of MCTCs and have significant therapeutic implications.
Keywords: mast cells, human pluripotent stem cells (hPSCs), development, tryptase, chymase
Introduction
Mast cells (MCs) are blood components and are classically known to originate from multipotential hematopoietic progenitor cells in the bone marrow (BM) (Kitamura et al., 1977; Kitamura and Ito, 2005; Kalesnikoff and Galli, 2008). Unlike most hematopoietic BM cells, MCs complete terminal maturation in peripheral tissues under the influence of the local microenvironment (Kitamura, 1989). Mature MCs can be distinguished from immature MCs by surface expression of the high-affinity IgE receptor (FcεRI) and by the presence of high c-Kit levels and characteristic secretory granules. MCs are multifunctional effector cells that play an essential role in innate immunity, host defense, host hypersensitivity, and the pathogenesis of allergic disease. MCs predominantly reside in tissues and are characterized by large cytoplasmic granules containing proteases, which are released upon activation, such as during an allergic reaction (Gilfillan et al., 2009; Da Silva et al., 2014). MC activation induces a wide variety of symptoms in complex diseases that are difficult, often impossible, to diagnose. Recently, MCs have been implicated in pain and neuroinflammation in endometriosis (De Leo et al., 2017). However, the exact physiological and pathogenic functions of MCs remain controversial due to insufficient understanding of human MC types and their development during embryonic/fetal stages.
In mice, MCs are subclassified into two main subsets based on their location, staining characteristics, protease expression patterns, and histamine content. One MC subtype is connective tissue-type (CT-MCs) and the other subtype is mucosal-type (M-MCs) (Gurish and Austen, 2012; Dwyer et al., 2016). M-MCs are found in the normal alveolar wall and small intestinal mucosa. CT-MCs are similar to tissue-resident macrophages; are found in the skin, submucosa, and other connective tissues; and have radio-resistant and self-renewal properties. Further, M-MCs are inducible and transient with a lifespan of only 2 weeks, while CT-MCs are constitutive and long-lasting ( Gurish and Austen, 2012; Dwyer et al., 2016). A recent report demonstrated that MCs and macrophages exhibit similar overall development kinetics with dual hematopoietic origins (Li et al., 2018), indicating diverse MCs in developmental sources, tissue distributions, and functions.
In humans, three types of MCs have been classified on the basis of protease content: (i) Tryptase+Chymase+ MCs (MCTCs), which contain tryptase, chymase, carboxypeptidase, and cathepsin G in their secretory granules, are predominantly located in normal skin and intestinal submucosa and correspond to rodent CT-MCs; (ii) Tryptase+Chymase− MCs (MCTs), which contain tryptase but lack other proteases, are the main MC type in the normal alveolar wall and small intestinal mucosa and correspond to rodent M-MCs; and (iii) Tryptase−Chymase+ MCs (MCCs), which are a rare MC subtype that only contain chymase and are found in endometrial tissue (Irani et al., 1986; Kaminer et al., 1995; Welle, 1997; Kalesnikoff and Galli, 2008; Moon et al., 2009). In patients with T cell deficiency, a marked and selective lack of MCTs in the intestinal mucosa is observed, but these patients have normal numbers of MCTCs in the adjacent submucosa (Irani et al., 1987), suggesting that the development of MCTs is different from that of MCTCs in humans.
Unlike the hematopoietic stem cell (HSC)-dependent hematopoiesis originating in the adult BM, early mammalian embryos generate HSC-independent hematopoietic lineage cells sharing both primitive and definitive properties. This HSC-independent wave predominately produces erythrocytes, megakaryocytes, and other myeloid lineage cells derived from erythro-myeloid progenitors (EMP) and other tissue-resident cell progenitors, such as macrophages (Dzierzak and Bigas, 2018). In humans, many reports exploring the onset of human hematopoiesis that use the in vitro differentiation of hPSCs accumulated data similar to what has been found in mouse ontogeny.
Our group recently reported a unique pathway for early definitive erythropoiesis from hPSCs, which is phenotypically different from more mature adult hematopoietic stem and progenitor cell (HSPC)-derived definitive erythropoiesis (Mao et al., 2016). Moreover, in our previous study, we demonstrated that cynomolgus non-human primate embryonic stem cell (ESC)-derived MCs (ESC-MCs) have similar characteristics to MCTCs with functional maturity in short-term cultures, suggesting a unique embryonic/fetal pathway in primates for early development of MCTCs, which may be from a different pathway other than that of MCTs (Ma et al., 2008). On the other hand, although human cord blood (CB) CD34+ progenitors could give rise to MCTCs in long-term cultures, generally after >10 weeks in vitro, these cells still cannot fully mimic the functions of MCTCs in human skin, such as substance P-induced degranulation (Kinoshita et al., 1999). At present, it is still unclear whether MCs develop via HSC-independent pathways, and the functional properties of these potential lineages remain unknown.
In the present study, we applied a three-stage culture method to generate mature and functional MCTCs from hPSCs. We found that coculture of CD34+c-kit+ cells for 8 days gave rise mainly to a pure MC population phenotypically and functionally similar to human MCTCs as reported.
Single-cell RNA sequencing analysis revealed two distinct pathways of MC development from CD34+c-kit+ cells in the coculture system. One pathway was via EMP-containing erythroids, macrophages, and MC progenitors. The other pathway was via late multilineage HSPC defined by expression of CD34 and CD45, as we previously reported (Zhou et al., 2019). Functional comparison revealed dramatic differences between MCs generated from coculture day 8 (D8) CD34+c-kit+, coculture D14 CD34+CD45+, and CB CD34+ cells, with the earlier coculture D8 CD34+c-kit+ cell-derived MCs (CD34+c-kit+-MCs) exhibiting the strongest histamine release and other innate immune responses. We established a new in vitro model to trace the early development of hPSC-MCs (most MCTCs), which provides a new approach for the development of novel drugs targeting MCTC-mediated diseases.
Results
hPSC-generated MCs share MCTC protease phenotypes and functional characteristics
To generate MCs from human pluripotent stem cells (hPSCs), we applied a three-stage culture method (Figure 1A). The first stage involved producing hematopoietic progenitors by hPSC/AGM-S3 coculture. Generally, a 35-mm diameter culture dish (equal to a single well of a 6-well plate) generated 1.48 × 106 ± 0.48 × 106 total cells (H1 line, n = 9) after 14 days of coculture. We then harvested D14 cocultured total cells and transferred them to nonadhesive culture plates to expand hematopoietic progenitors by adding cytokines SCF, IL-3, IL-6, thrombopoietin (TPO), and flt3 ligand (FL). This second culture allowed stable production of hematopoietic cells with no interference from adherent/stromal cell growth.
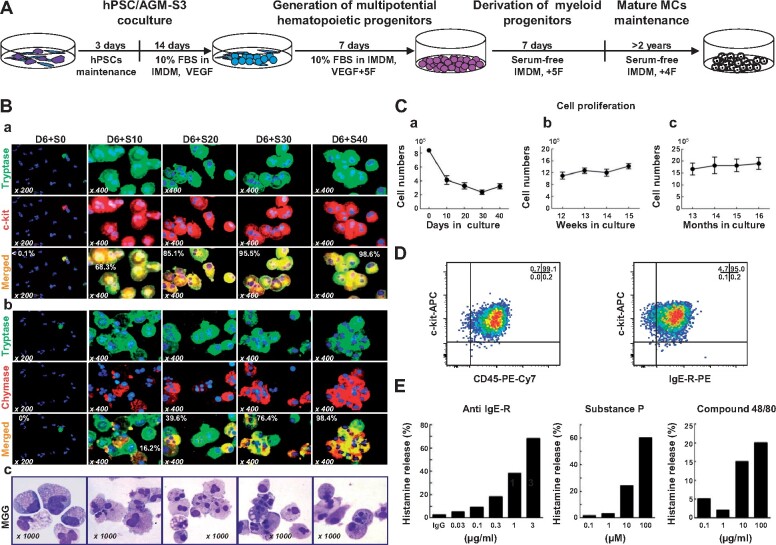
Figure 1.
Early hPSC-MCs share skin MC protease phenotypes and functions. (A) Schematic illustrating the strategy for the generation of MCs from D14 hPSC/AGM-S3 coculture. (B) Immunostaining and MGG staining of early developed hPSC-MCs over the time course of SF-MC cultivation. (C) Persistence of cell proliferation of H1/AGM-S3 coculture D14 (a), short-term (up to 15 weeks, b), and long-term (up to 16 months, c) in SF-MC cultures. (D) Mature hPSC-MCs (20 weeks in SF-MC culture) co-expressing CD45, c-kit, and a high-affinity IgE-R. (E) Histamine release by mature hPSC-MCs following stimulation with an antibody against the human IgE-R (CRA1), substance P, and compound 48/80, respectively.
In the second culture, one single 6-well plate generated 1.75 × 106 ± 0.6 × 106 (n = 5) cells after 7 days, which was 29‒117-fold higher than the initial number of undifferentiated hPSCs, while CD34+ hematopoietic progenitors expanded 9.1-fold. To induce MC differentiation, we used serum-deprived medium containing SCF, IL-6, TPO, and FL, but not IL-3 (SF-MC culture). MCs derived from hPSCs expressing both c-kit and tryptase appeared very early during cultivation and robustly proliferated, resulting in a rapid increase in purity (68.3% at D10 and 98.6% at D40, Figure 1Ba). MCs then gradually increased in number and represented >99% of the cells thereafter.
Interestingly, hPSC-MCs co-expressed a high level of chymase even at early culture times (39.6% at D20 and 98.4% at D40, Figure 1Bb). May-Grunwald Giemsa (MGG) staining of early developed hPSC-MCs over the time course of SF-MC cultivation is shown in Figure 1Bc. In SF-MC cultures, total cell proliferation gradually decreased, while the proportion of MCs increased (Figure 1Ca), as observed previously during in vitro MC differentiation from human CD34+ CB cells. The hPSC-MCs gradually became smaller in size with single nuclei and grew as aggregates as cultivation progressed (Supplementary Figure S1A). We calculated that an initial input of 1× 104‒3 × 104 undifferentiated hPSCs generated 1.1 × 106 mature MCTCs after 12 weeks (37‒110-fold) and 2.64 × 106 mature MCTCs after 15 months (88‒264-fold) of SF-MC culture.
Mature hPSC-MCs exhibited basophilic staining, as revealed by MGG staining (Supplementary Figure S1B). In this protocol, the hPSC-MCs chromatically acquired a purple color upon toluidine blue staining (Supplementary Figure S1C) and were stained blue by Alcian blue solution (Supplementary Figure S1D). hPSC-MCs were positive for carboxypeptase A and cathepsin G (Supplementary Figure S1E and F). The maturity of hPSC-MCs after 10 weeks in SF-MC culture was further confirmed by co-expression of c-kit with tryptase and chymase in all of the cells (Supplementary Figure S1G‒L), confirming the MCTCs protease phenotype. Transmission electron microscopy (TEM) revealed that the hPSC-MCs contained abundant granules of various densities with well-developed mitochondria. Scrolling shaped granules, which are typical of human MCs, were also present (Supplementary Figure S1M). These features were identical to those of mature human MCs. Mature hPSC-MCs underwent slow but constant proliferation throughout cultivation and survived over a period of 16 months (Figure 1Cb and Cc). When further matured (>20 weeks in SF-MC culture), most of the hPSC-MCs expressed high-affinity FcεRI, CD45, and c-kit (Figure 1D), suggesting that they had reached functional maturity.
We then investigated whether the hPSC-MCs had normal functions. hPSC-MCs displayed degranulation and released histamine upon stimulation with CRA1, a monoclonal antibody against the human IgE receptor (anti-IgE-R), in a dose-dependent manner (Figure 1Ea). Consistent with our previous report of non-human primate ESC-MCs, hPSC-MCs also efficiently underwent dose-dependent degranulation in response to substance P and compound 48/80, which resulted in histamine release (Figure 1Eb and Ec). Thus, hPSC-MCs were functionally mature MCTCs, not only because they expressed characteristic proteases but also because stimulation with specific pharmacological agents induced degranulation.
Coculture D8 CD34+c-kit+ cells rapidly develop into mature MCTCs
Our findings suggested the existence of a transient specific cell fraction CD34+c-kit+, which arose from early H1/AGM-S3 coculture on D6, became abundant by D10, peaked on D12, and then gradually decreased (Figure 2A). However, a late multilineage HSPCs (CD34+CD45+) first appeared on D10 and then reached peak value at D14 (Figure 2B), suggesting that CD34+c-kit+ cells were derived from another pathway.
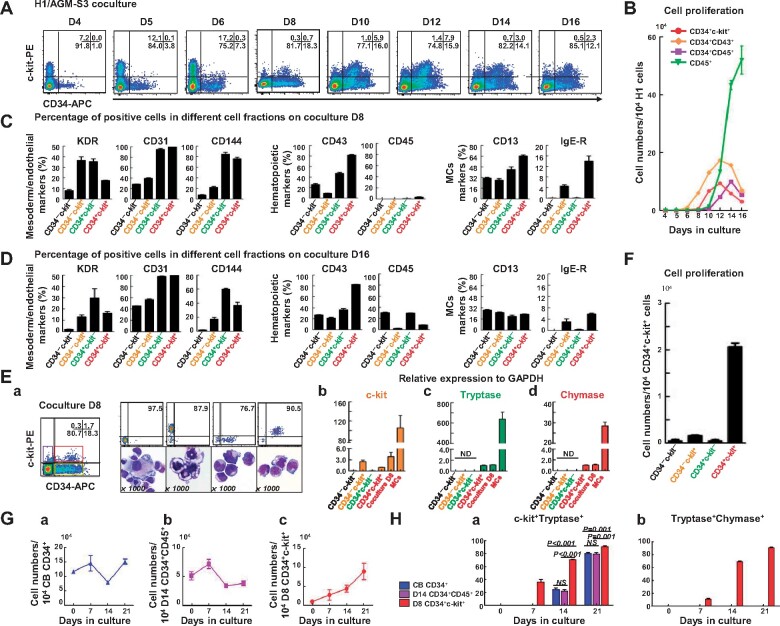
Figure 2.
Rapid maturation of MCTCs generated from a CD34+c-kit+ cell population. (A) Flow cytometric profiles showing expression of CD34+c-kit+ cells derived from H1/AGM-S3 coculture. (B) Proliferation of CD34+CD45+, CD34+CD43+, CD34+c-kit+, and CD45+ cell fractions on D4‒D16 derived from 1 × 104 H1 cells (independent experiments, n = 3; mean ± SD). (C and D) Flow cytometric analysis showing representative phenotypic expression of mesoderm (KDR), endothelial (CD31 and CD144), hematopoietic (CD43 and CD45), and MC (CD13 and IgE-R) markers in the four cell fractions at D8 (C) and D16 (D) during a single culture. (E) Four cell fractions defined by expression of CD34 and c-kit were sorted by FACS from a D8 H1/AGM-S3 coculture. MGG analysis showing typical morphologies of the sorted CD34+c-kit+, CD34+c-kit−, CD34−c-kit+, and CD34−c-kit− cell fractions (a) and qPCR analysis showing mRNA expression of c-kit (b), tryptase (c), and chymase (d) in each cell fraction, total coculture D8 cells, and a pure population of MCs. Independent experiments, n = 3; mean ± SD. ND, not detected. Scale bar, 10 µm. (F) Total cell proliferation of the four cell fractions sorted from D8. (G) Total cell proliferation of CB CD34+ cells (a), coculture D14 CD34+CD45+ cells (b), and coculture D8 CD34+c-kit+ cells (c) in SF-MC culture. (H) Compared with CB CD34+ and coculture D14 CD34+CD45+ progenitor cells, a high proportion (36.3% ± 3.4%) of coculture D8 CD34+c-kit+ cell-derived c-kit+Tryptase+ MCs could be observed early on D7 in SF-MC culture and reached a purity of 90.3% ± 1.4% after 21 days in culture (a) that was associated with a rapid increase in co-expression of chymase (90.2% ± 1.1% at D21, b). NS, non-significant.
To clarify the features of CD34+c-kit+ cells, we detected several important surface markers in four different cell populations, classified according to CD34 and c-kit expression at coculture D8 and D16. As illustrated in Figure 2C and D, compared with other cell fractions, the CD34+c-kit+ cell population at coculture D8 exhibited higher expression of CD144, CD13, and IgE-R and almost no expression of CD45, strongly indicating that before the emergence of the CD34+CD45+ cell population, a subset of cells expressing CD13 and IgE-R was present in the CD34+c-kit+ cell population.
Cells from coculture D8 were divided into four fractions by fluorescence-assisted cell sorting (FACS) on the basis of c-kit and CD34 expression. MGG analysis revealed typical morphology of cells derived from the four cell populations (Figure 2Ea). Quantitative polymerase chain reaction (qPCR) analysis revealed that only the CD34+c-kit+ cell population expressed mRNA encoding tryptase and chymase (Figure 2Eb‒Ed). However, immunofluorescence (IF) staining analysis did not reveal tryptase or chymase protein expression (data not shown). When we placed the four cell fractions in SF-MC culture medium for 7 days (Figure 2F), only the CD34+c-kit+ cell population expanded significantly. These data clearly demonstrated that the CD34+c-kit+ cells could be new MC progenitor cells.
To identify the progenitors of MCTCs, we grew coculture D8 CD34+c-kit+ cells, coculture D14 CD34+CD45+ cells (HSPCs), and CB CD34+ cells (adult HSPCs) in serum-free medium. Total cell proliferation of D14 CD34+CD45+ cells and CB CD34+ cells gradually decreased, while the proportion of MCs increased and the CD34+c-kit+ fraction increased progressively (Figure 2Ga‒Gc). After 3 weeks of culture, c-kit+Tryptase+ cells were present in all three types of MCs, but Tryptase+Chymase+ cells were only present in CD34+c-kit+-MCs (Supplementary Figure S2). In the CD34+c-kit+-MC culture, Tryptase+Chymase+ cells appeared 7 days after initiating differentiation, and by D21, >90% of the cells expressed both chymase and tryptase (Figure 2Ha and Hb). This finding suggested that fast-maturing MCTCs derived from hPSCs were initially generated from the CD34+c-kit+ cell population.
Dual development potential of MCs before and after appearance of hematopoietic stem-like cells
To clarify the origin and characteristics of CD34+c-kit+ cells, we sorted cells at coculture D5 based on expression of mesodermal/endothelium (KDR) and endothelial (CD34) surface markers, as well as an important MC marker c-kit, before the CD34+c-kit+ cells arose (Supplementary Figure S3A). KDR−CD34−c-kit−, KDR+CD34−c-kit−, KDR−CD34+c-kit−, KDR+CD34+c-kit−, and KDR−CD34−c-kit+ cells were sorted and re-cultured on stromal cells for another 7 days. Then the total cells were collected, and expression of CD34+c-kit+ cells were detected by FACS analysis. As showed in Supplementary Figure S3A, the results suggested that CD34+c-kit+ cells were primarily generated from KDR+CD34+c-kit− hematopoietic and endothelial (HE) precursor cells.
To obtain a gene develop chart of the development of CD34+c-kit+ cells, we isolated and sequenced 8901 cells from two successive developmental stages, including KDR+CD34+ cells at coculture D5 and cells generated from KDR+CD34+ cells after another 5 days culture (replacement of AGM-S3 with 10 ng/ml VEGF) (D5_5). Sequencing quality metrics were similar across samples, reflecting minimal technical variation between samples (Supplementary Table S1).
A graph-based, unsupervised clustering analysis was applied to the pooled time points, identifying 13 cell clusters that represented distinct cell types or cell subpopulations (Figure 3Aa). Subsequent barcode deconvolution of single cells revealed the appearance of the population, effectively capturing the dynamics of KDR+CD34+ cell progression (Supplementary Figure S3B). Considering the positive expression of c-kit and CD34, clusters 0, 1, 4, 5, and 7 were selected as ‘target cells’ for reanalysis. Dimension reduction was performed by principal component analysis (PCA) (Supplementary Figure S3C). Of note, cells in cluster 0 could be further divided into two clusters (Supplementary Figure S3Ca). Finally, six clusters were identified and annotated as ‘mesoderm (M)’, ‘hematopoietic and endothelial (HE)’, ‘erythro-myeloid progenitors (EMP)’, ‘mast progenitors (MP)’, ‘hematopoietic stem/progenitor cells (HSPC)’, and ‘myeloid progenitor cells (MPC)’ based on the cell transcriptional profiles (Figure 3A and B). Moreover, PCA also revealed that the EMP cluster was positioned next to the MP cluster and that the HSPC and MPC clusters closely correlated (Supplementary Figure S3Cb).
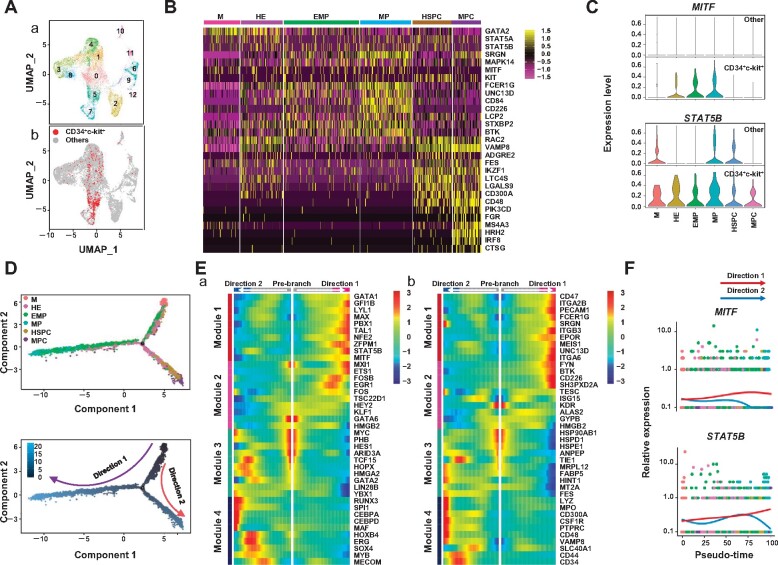
Figure 3.
Dual development potential of MCs before and after HSPCs appeared. (A) Identification of unsupervised clustering for 2 days combined with UMPA. (a) Each dot represents one cell, and colors represent cell clusters as indicated. (b) Expression of CD34+c-kit+ cells in different clusters. (B) Heatmap showing scaled expression of MC-related genes in the six clusters. (C) Violin plots showing expression of featured genes in each cell cluster between CD34+c-kit+ cells and other cells. (D) Trajectory analysis by Monocle 2 combining six clusters from D5 with 13 clusters from D5_5 indicates the development of CD34+c-kit+ cells. (E) Four distinct gene expression patterns along the pseudotime axis were inferred by Monocle 2. Prominent transcription factors (a) and other genes (b) are listed. (F) Expression of representative genes of pattern 1 along pseudotime axis inferred by Monocle 2.
We next sought to confirm the MC clusters by examining the transcriptional profiles of the intermediate subpopulations identified above. Gene expression patterns of mesoderm markers, endothelial markers, hematopoietic markers, megakaryocyte and erythrocyte progenitor cell (MEP) markers, granulocyte and monocyte progenitor cell (GMP) markers, and MC-related genes were compared among the six clusters (Figure 3B;Supplementary Figure S3D). Compared with the other clusters, the MP cluster exhibited MC-like development features, such as high expression of STAT5A/STAT5B, SRGN, MAPK14, and MITF (Shelburne et al., 2003; Abrink et al., 2004; Morii et al., 2004; Hu et al., 2012). Furthermore, several genes related to MC activation, including FCER1G, UNC13D, CD84, CD226, LCP2, STXBP2, and BTK, were highly expressed in this cluster (Figure 3B; Bachelet et al., 2006; Álvarez-Errico et al., 2011; Oksaharju et al., 2011; Bin et al., 2013; Rodarte et al., 2018; Simonowski et al., 2020; Wang et al., 2020). However, in this cluster, the endothelial markers PECAM1 and ITGB3 were highly expressed, while the hematopoietic markers SPN and PTPRC were weakly expressed and absent, respectively (Supplementary Figure S3D). Compared with other cell types, CD34+c-kit+ cells exhibited MCTCs-like developmental features, such as increased expression of MITF and STAT5B (Figure 3C). Moreover, MC activity was also presumably located within the HSPC cluster, which exhibited high expression of MC-related genes. However, this cluster also exhibited granulocyte and monocyte-like features, such as high expression of IL3RA, CSF1R, CSF3R, LYZ, and MPO, as well as the hematopoietic genes SPN and PTPRC (Supplementary Figure S3D; Zhou et al., 2019).
Monocle analysis revealed the developmental trajectory of the dual hematopoietic origin of MCs, clearly illustrating one path from the M cluster via HE, then from EMP to MP, and another path from the M cluster via HE, then from HSPC to MPC (Figure 3D). Genes that were significantly differentially expressed along the pseudotime axis were subjected to k-means clustering analysis, resulting in four distinct gene expression modules (Figure 3E). Genes grouped in module 1 were highly expressed only in MP, including transcription factors such as STAT5B and MITF, and were related to the regulation of MCTC development (Figure 3Ea and F). In addition to the MEP markers CD47 and ITGA2B and endothelial marker PECAM1, both SRGN and FCER1G, which are critical to MC development and function, were placed in module 1 (Figure 3Eb). Genes in module 4 were those upregulated in MCPs, including the genes already highly expressed in HSPCs as compared with the M and HE clusters, such as MYB, CD44, and CD34, and also those specifically expressed in MCPs, such as LYZ, MPO, PTPRC, and CD48 (Figure 3E).
MCTCs from CD34+c-kit+ cells robustly release histamine and cytokines
To understand the characteristics of MCs derived from coculture D8 CD34+c-kit+ cells, we collected coculture D14 CD34+CD45+ HSPCs and CB CD34+ adult HSPCs for functional comparison. We detected a qualitative difference in chymase and tryptase expression between MCs differentiated from the three culture methods. The average CD34+c-kit+-MC yield of chymase/tryptase staining was 258.4 × 106 ± 32.7 × 106 per 1 × 106 human ESCs (hESCs) (n = 4), 157.9 × 106 ± 3.3 × 106 positively stained cells were obtained from 1 × 106 hESCs differentiated from CD34+CD45+ cells (n = 3), and 111.3 × 106 ± 5.96 × 106 per 1 × 106 CD34+ progenitors were present (n = 3) (Table 1).
Table 1.
MC production from hESCs.
| Type | MCs generated from one hESC |
Reference | |
|---|---|---|---|
| c-kit+Tryptase+ | Tryptase+Chymase+ | ||
| Coculture D8 CD34+c-kit+a | 258.35 ± 32.65 (100%) | 258.35 ± 32.65 (100%) | This study |
| Coculture D14 CD34+CD45+b | 163.33 ± 10.36 (100%) | 157.94 ± 3.27 (96.7% ± 2%) | |
| CB CD34+c | 595.28 ± 95.62 (100%) | 111.32 ± 5.96 (18.7% ± 1%) | |
| Coculture D12 CD34+d | NA | 1.5 ± 0.3 (>90%) | Kovarova et al. (2010) |
| Embryoid body (EB) e | NA | 9.7 ± 4.7 (>90%) | |
MCs generated from hESC/AGM-S3 coculture D8 CD34+c-kit+ cells after 12 weeks.
MCs generated from hESC/AGM-S3 coculture D14 CD34+CD45+ cells after 12 weeks.
MCs generated from CB CD34+ cells after 12 weeks.
MCs generated from hESC/OP9 coculture D12 CD34+ cells after 12 weeks.
MCs generated from hESC-derived EB cells after 12 weeks.
We measured the intracellular histamine concentration of cell lysates using a histamine ELISA assay kit. As summarized in Table 2, CB CD34+-MCs contained 40% less histamine than coculture D8 CD34+c-kit+-MCs. Coculture D8 CD34+c-kit+-MCs contained 10.8 ± 1.4 pg/cell (n = 3), coculture D14 CD34+CD45+-MCs contained 8.5 ± 1.5 pg/cell (n = 3), and CB CD34+-MCs contained 6.2 ± 2.1 pg/cell (n = 3). Significant differences were found between CD34+c-kit+-MCs and CB CD34+-MCs (P = 0.014), but there were no differences between CD34+c-kit+-MCs and CD34+CD45+-MCs (P = 0.137) or between CD34+CD45+-MCs and CB CD34+-MCs (P = 0.195).
Table 2.
Intracellular histamine content of human MCs.
| Type | Histamine content (pg/cell) |
|---|---|
| CB CD34+-MCs | 6.15 ± 2.10 |
| Coculture D14 CD34+CD45+-MCs | 8.46 ± 1.49 |
| Coculture D8 CD34+c-kit+-MCs | 10.81 ± 1.35* |
A total of 1 × 105 hESC-MCs (coculture D14 CD34+CD45+-MCs and coculture D8 CD34+c-kit+-MCs) or CB CD34+-MCs cultured for 12 weeks were lysed, and the cell supernatant was analyzed by ELISA (n = 3).
P < 0.05 relative to CB CD34+-MCs.
A series of MC activators were used to stimulate these MCs, and histamine release was measured by ELISA. As shown in Figure 4A, with activation of CRA1 (anti-FcεRI antibody) via FcεRI, IgE-sensitized CB CD34+-MCs released relatively small proportions of histamine (maximum 38% histamine release), whereas the other two types of MCs exhibited stronger and highly significant histamine release, with a maximum of 60% for coculture D14 CD34+CD45+-MCs and 76% for coculture D8 CD34+c-kit+-MCs. When the active concentration of CRA1 was 1 µg/ml, significant differences were detected between not only CD34+c-kit+-MCs and CB CD34+-MCs but also CD34+c-kit+-MCs and CD34+CD45+-MCs (P = 0.137), as well as CD34+CD45+-MCs and CB CD34+-MCs.
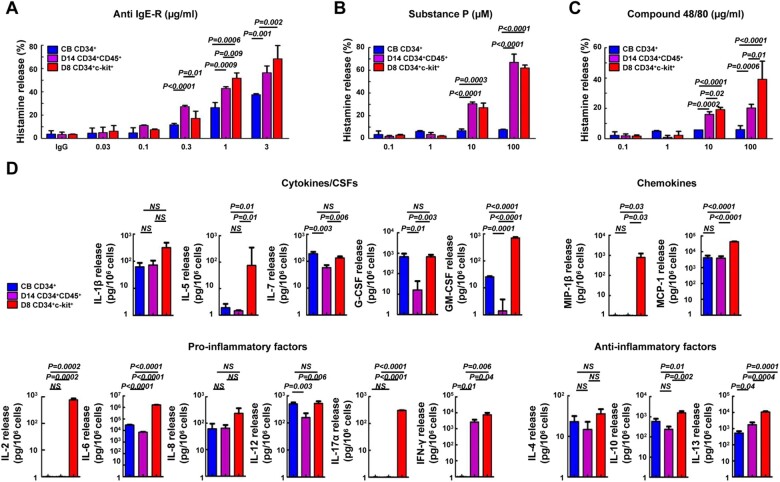
Figure 4.
Histamine and cytokine release by mature MCs derived from three different protocols. (A‒C) IgE-sensitized MCs were stimulated for 30 min with anti-IgE-R (CRA1), Substance P, and compound 48/80. Data are expressed as mean ± SD of at least three sample replicates. (D) Heterogenous cytokine release after anti-IgE-R stimulation. Cytokine levels are expressed as pg/106 cells. Significant differences among MC culture protocols are indicated. Data are expressed as mean ± SD of at least three sample replicates. Blue, CB CD34+-MCs; purple, coculture D14 CD34+CD45+-MCs; red, coculture D8 CD34+c-kit+-MCs. NS, non-significant.
Substance P and compound 48/80 were also used to stimulate the three MC types. Both substance P and compound 48/80 induced a histamine release <10% in CB CD34+-MCs but induced a significant histamine release from the other two MC types (Figure 4B, substance P release: 67.0% ± 5.5% for coculture D14 CD34+CD45+-MCs and 62.1% ± 1.8% for coculture D8 CD34+c-kit+-MCs; Figure 4C, compound 48/80 release: 20.2% ± 1.7% for coculture D14 CD34+CD45+-MCs and 39.2% ± 8.4% for coculture D8 CD34+c-kit+-MCs). Measurement of histamine release revealed that CB CD34+-MCs were poor histamine releasers and were only activated by CRA1. By contrast, coculture D14 CD34+CD45+ and coculture D8 CD34+c-kit+-MCs responded robustly to all compounds tested.
To investigate cytokine production by activated MCs, we stimulated the three different MC types with IgE followed by CRA1. Seventeen human cytokines were measured. As shown in Figure 4D, cytokine release after IgE and CRA1 stimulation was highly heterogenous among the three MC types. None of the MC types released TNF-α in response to the stimuli. Compared with the other two MC types, coculture D8 CD34+c-kit+-MCs released high concentrations of the other 16 cytokines, but the differences in IL-1β, IL-4, and IL-8 were not statistically significantly. The cytokines IL-5 and GM-CSF, which are important for eosinophil development and proliferation, were highly expressed in coculture D8 CD34+c-kit+-MCs stimulated with IgE/CRA1, suggesting that this MC type could activate eosinophils/basophils. Interestingly, IL-17a, which is produced primarily by Th17 cells, was robustly released by early MCTCs derived from coculture D8 CD34+c-kit+ cells, suggesting that MCTCs could also play a central role in tumor immunity.
To better understand the different activation states of the three MC types, we validated the expression of MC surface markers and the presence of MC morphological characteristics. All three MC types highly expressed CD45, c-kit, CD13, and CD81 but did not express CD34 or HLA-DR (Supplementary Figure S4A). Compared with the other two types of MCs, coculture D8 CD34+c-kit+-MCs highly expressed CD203C, CD88, and a high-affinity IgE receptor (FcεRI) (Supplementary Figure S4A). MCs derived from coculture D8 CD34+c-kit+ cells and coculture D14 CD34+CD45+ cells highly expressed whereas MCs derived from CB CD34+ moderately expressed CD31 (Supplementary Figure S4A). Moreover, MGG analysis revealed a typical morphology of CB CD34+-MCs (Supplementary Figure S4Ba), coculture D14 CD34+CD45+-MCs (Supplementary Figure S4Bb), and coculture D8 CD34+c-kit+-MCs (Supplementary Figure S4Bc) after 12 weeks of culture. MCs derived from coculture D8 CD34+c-kit+ cells gradually became smaller in size with single nuclei and grew as aggregates as cultivation progressed. Expression of c-kit, tryptase, and chymase in mature MCs was detected by IF staining, revealing that all of the mature MCs expressed these markers. However, the relative abundance of chymase varied between the three different cell types (Supplementary Figure S4C). Our findings demonstrate that MCs quickly derived from coculture D8 CD34+c-kit+ cells might resemble skin MCs and thus belong to the MCTC lineage.
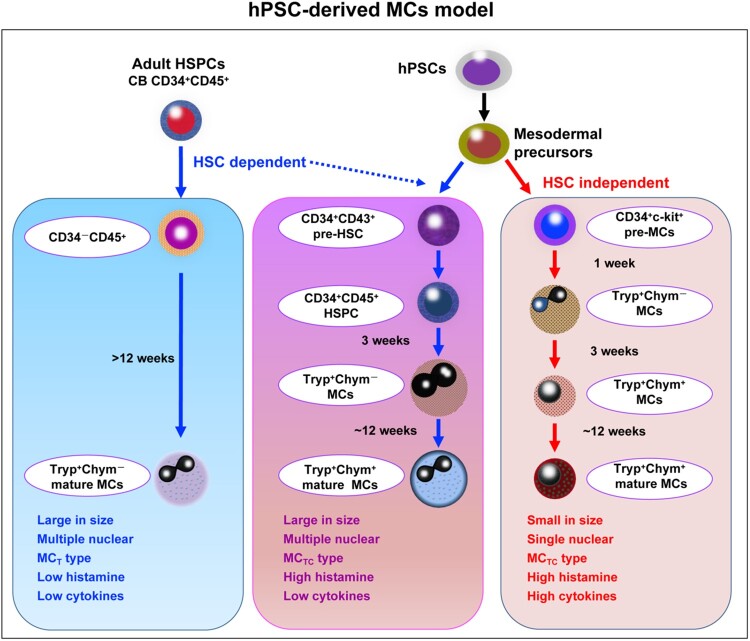
In the present study, we found that hPSC-MCs shared two developmental pathways (Figure 5). MCTCs quickly derived from coculture D8 CD34+c-kit+ progenitor cells, with upregulation of tryptase and chymase. MCs generated from these progenitors were small in size and mononuclear, with functional similarities to MCTCs, including high histamine and cytokine release stimulated by different activators, such as CRA1, substance P, and compound 48/80. MCs derived from coculture D14 CD34+CD45+-MCs were phenotypically similar to CB CD34+-MCs, with a large cell size and multiple nuclei. However, the functions of the two types of MCs were very different. MCs from coculture D14 CD34+CD45+ cells were biased toward the MCTC type, while MCs from CB CD34+ cells were partial to the MCT type. MCs from coculture D14 CD34+CD45+ cells strongly released histamine in response to CRA1, substance P, and compound 48/80, while MCs from CB CD34+ cells did not release histamine in response to substance P or compound 48/80. Both of the two types of MCs exhibited weak release of cytokines in response to IgE and CRA1 stimulation.
Figure 5.
Model depicting the development pathway for hPSC-MCs. There is a specific development stage during MCTCs from hPSC/AGM-S3 coculture. Early MCTCs are derived from CD34+c-kit+ cells, independent of HSCs. Compared with the MCs derived from HSC-dependent pathway, these cells exhibit special morphology and function.
Discussion
Recently, several groups found that MCs have different hematopoietic origins in mouse models, similar to macrophages (Gentek et al., 2018; Li et al., 2018). The potential for differential MC origins during human development remains unclear. Because experimental manipulations cannot be conducted using human embryos, the factors and mechanisms involved in directing differentiation of immature MCs into either functional MCCTs or MCTs remain unknown. Using a sequential coculture system with unbiased displaying for natural progression of human hematopoiesis in vitro, we efficiently produced scalable quantities of functionally mature MCTCs from hPSCs even in short-term cultures (Figure 1; Supplementary Figure S1). Quantitative evaluations revealed that one undifferentiated hESC could generate 258.4 ± 32.7 mature MCTCs at 12 weeks after isolation of coculture D8 CD34+c-kit+ MC progenitors and 157.9 ± 3.3 mature MCTCs from coculture D14 CD34+CD45+ HSPCs, much higher than what had been reported previously, i.e. ∼5.0–14.4 MCs per undifferentiated hESC (Table 1; Kovarova et al., 2010). Due to the lack of a cell model for screening drugs that target human MCTCs, the efficiency of pure MCTC production in our culture system provided an easy and applicable method for investigation of molecular targets and drug screening for MCTC-related conditions on a large scale.
MC progenitors have been reported to develop in a variety of hematopoietic sites and peripheral tissues in embryos and adults (Dahlin et al., 2015). Classically, human MCs are known to arise from CD34+c-kit+ progenitor cells from the BM, peripheral blood, or CB HSPCs that highly co-express CD45 and CD13, which also generate other myeloid cells such as neutrophils, eosinophils, basophils, and monocytes (Kirshenbaum et al., 1991, 1994, 1999; Durand et al., 1994; Rottem et al., 1994; Saito et al., 1996).
Previously, the generation of functional MCs from hESCs has been reported (Kovarova et al., 2010). However, in this study, MC production was very low, and the resultant MCs held smaller granules, less heparin, and lower levels of proteolytic enzymes, similar to CB CD34+ cell-MCs. Because the MC induction culture protocol used comparatively mature hESC-derived HSPCs, this approach could bypass early development pathways for hESC-MCs. A more recent report established an early rapid and robust production of MCs from mPSCs and hPSCs (1 week from mouse cultures and several weeks from human cultures) by genetic manipulation (Kauts et al., 2018). However, hPSC-MCs in this study did not exhibit mature MC functions, and their origins were unknown. Contrastingly, in our coculture system, CD34+c-kit+ cells that differentiated as early as D8 in coculture already expressed tryptase and chymase mRNA (Figure 2Eb). Particularly, the early coculture D8 CD34+c-kit+ MC precursors were CD45−, exhibiting a lack of HSPCs or myeloid properties by their source. In consequential culture, MCTCs derived from coculture D8 CD34+c-kit+ precursors exclusively exhibited MCTC characteristics, typically mimicking mature human skin tissue-type MCs. Phenotypically, these early MCTCs were 100% double-positive for tryptase and chymase, small, and single-nucleated and contained scroll-shaped granules similar to human skin tissue-type MCs. Importantly, these hPSC-derived early MCTCs already exhibited characteristics of mature MCs, including robust histamine release upon substance P stimulation (Figures 2 and 4), a function characteristic of human skin tissue-type MCTCs that is seldom exhibited by CB CD34+ cell-derived M-MCTs. Taken together with our previous findings in non-human primate ESC-derived CT-MCs (Ma et al., 2008), our data point to a common embryonic/fetal pathway for early development of MCTCs independent of HSPC-MCTCs.
The present study was suggestive of two waves of human MC development. In the first wave, a mesodermal HE precursor directly develops into a CD34+c-kit+CD45− MC progenitor (D6–D8 coculture-derived MC precursors in our system) that rapidly differentiated exclusively into MCTCs with functional maturity mimicking human skin tissue-type MCTCs. The other wave of human MC development, as widely reported, is derived from adult-type HSCs and predominately generates mature MCTs functionally expressing themselves as M-MCs (CB CD34+-MCs).
Using a subtle cell sorting assay, we found that early coculture D8 CD34+c-kit+ MC progenitors primarily developed from CD34+KDR+ HE precursors along with a definitive hematopoiesis pathway that could be traced back to KDR+CD34− mesoderm cells. Further analysis by single-cell RNA sequencing confirmed the existence of two different pathways for the development of early CD34+c-kit+ MC progenitors (Figure 3D). One pathway developed with EMP development, and the other pathway developed with a late multi-lineage HSPC. Although MC potential could be detected in both pathways, the MC progenitor cells in the EMP pathway highly expressed MITF and STAT5B, which are both important transcription factors for tissue-type MC development (Figure 3C and E; Shelburne et al., 2003; Morii et al., 2004). These findings were suggestive of a unique pathway for early and rapid development of functional mature MCTCs in a first wave independent of HSPC-MCs.
The MCTCs developed from the early wave (typically as D8 coculture-derived MCTCs) exhibited stronger function response to release various bio-regulators of IgE stimulation. Robust IL-5 and GM-CSF secretion in response to these stimuli suggested a potent and pivotal role for early MCTCs as eosinophil/basophil activators in innate immunity. On the other hand, robust release of IL-17 from early MCTCs suggested that this cell population could also play a central role in regulation of tumor immunity. A recent report of esophageal squamous cell carcinoma suggested that in situ resident IL-17-expressing cells are MCs but not T lymphocytes or macrophages (Wang et al., 2013). These tissue-resident IL-17-expressing MCs suppress tumor growth and are predictive of a favorable prognosis, thus suggesting that MCs could contribute to tumor immunity by releasing IL-17. This cell population is therefore a potential novel prognostic marker and therapeutic target.
Interestingly, our findings suggested that MCTCs from the early wave exhibited the highest response to stimuli, as detected by release of various bio-regulators such as cytokines, chemokines, and pro- and anti-inflammatory factors. This finding again reflected that MCTCs developed in this early wave could complete functional maturation along a rapid and unique pathway, rather than via myeloid development classically recognized as an HSC-dependent pathway.
In conclusion, the present study describes the development of a novel method to uncover the developmental pathway of early functionally mature MCTCs. Subsequent studies in our group will identify the molecular and cellular regulatory mechanisms underlying the early development and maturation of these unique MCTCs, which cannot be recapitulated by animal models. In addition to the theoretical and experimental importance of our findings, this study also provides a new approach for the development of novel drugs targeting MCTC-mediated allergic diseases and tumors.
Materials and methods
MCs generation
MCs generation from coculture D14 CD34+CD45+ hematopoietic progenitor cells, coculture D8 CD34+c-kit+ cells, and human CB CD34+ HSPCs are given in Supplementary material.
MC characterization
Detailed methods of MC characterization, including flow cytometry, mRNA expression, histochemistry, and immunofluorescence staining, are described in Supplementary material.
Functional assays of MCs
Activation of MCs and the histamine and cytokine assay are illustrated in Supplementary material.
Processing of single-cell RNA sequencing and quality control
Cell Ranger (version 3.0.2) software from 10× Genomics (http://www.10xgenomics.com) was used to align and quantify the raw sequencing data with default parameters. Low-quality cells were then filtered out before further analysis. Genes expressed in >3 cells were retained, and cells that expressed <500 genes were removed from the dataset. Cells were also discarded if their mitochondrial gene percentages were >10%.
Dimensionality reduction and cell clustering
After completion of quality control, the Seurat package (version 3.0.2) (Butler et al., 2018) implemented in R (version 3.6) was used to perform downstream analysis and visualization. Specifically, raw count normalizations (NormalizeData, LogNormalize, and the scale factor was set to 10000), high variable gene (HVG) detection (FindVariableFeatures), scaling and centering of data (ScaleData), PCA analysis (RunPCA, with HVGs), and cell clustering (FindClusters, based on a Shared Nearest Neighbor (SNN) graph) were performed using Seurat. Finally, 13 transcriptionally similar clusters (‘resolution’ = 0.4) were identified and used for subsequent analysis, and cells were visualized using UMAP or PCA with default parameters.
Trajectory analysis
To further investigate the differentiation trajectories of ‘target cells’, Monocle2 package (v2.12.0) (Qiu et al., 2017) was used to reconstruct pseudotime analysis. Ordering genes were selected based on PCA loading. The Discriminative Dimensionality Reduction with Trees (DDRTree) method was then used to perform dimensionality reduction and visualized through plot_cell_trajectory. To compare the different molecular characteristics of the two developmental pathways, we used Branched Expression Analysis Modeling (BEAM) to detect different gene expression patterns along the two different pathways. Genes with q value <10−7 were considered statistically significant.
Transcription factors and surface markers
Lists of surface markers and transcription factors were obtained from HumanTFDB3.0 (http://bioinfo.life.hust.edu.cn/HumanTFDB/) and Cell Surface Protein Atlas (http://wlab.ethz.ch/surfaceome/), respectively.
Gene functional annotation and pathway analysis
Geno Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed based on significant differentially expressed genes with R package cluster Profiler (version 3.10.1) (Yu et al., 2012).
Statistical analysis
Prism software (GraphPad) was used for statistical analysis, as indicated in the respective figure legends.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Supplementary Material
Acknowledgements
We thank Professor Tao Cheng at the State Key Laboratory of Experimental Hematology, CAMS & PUMC for generously providing the H1 hESC line.
Funding
This work was supported by the National Basic Research Program (973 Program; 2015CB964902), the National Natural Science Foundation of China (H81170466 and H81370597), and the CAMS Initiatives for Innovative Medicine (2016-I2M-1-018) awarded to F.M.; the CAMS Initiatives for Innovative Medicine (2017-12M-2005); the Union Youth Fund of Chinese Academy of Medical Sciences (81572089) to G.B.; and the National Nature Science Foundation of China Youth Fund (81700107) to B.M.
Conflict of interest: none declared.
References
- Abrink M., Grujic M., Pejler G. (2004). Serglycin is essential for maturation of mast cell secretory granule. J. Biol. Chem. 279, 40897–40905. [DOI] [PubMed] [Google Scholar]
- Álvarez-Errico D., Oliver-Vila I., Ainsua-Enrich E., et al. (2011). CD84 negatively regulates IgE high-affinity receptor signaling in human mast cells. J. Immunol. 187, 5577–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachelet I., Munitz A., Mankutad D., et al. (2006). Mast cell costimulation by CD226/CD112 (DNAM-1/Nectin-2): a novel interface in the allergic process. J. Biol. Chem. 281, 27190–27196. [DOI] [PubMed] [Google Scholar]
- Bin N.R., Jung C.H., Piggott C., et al. (2013). Crucial role of the hydrophobic pocket region of Munc18 protein in mast cell degranulation. Proc. Natl Acad. Sci. USA 110, 4610–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., et al. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva E.Z.M., Jamur M.C., Oliver C. (2014). Mast cell function: a new vision of an old cell. J. Histochem. Cytochem. 62, 698–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin J.S., Ding Z., Hallgren J. (2015). Distinguishing mast cell progenitors from mature mast cells in mice. Stem Cells Dev. 24, 1703–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leo, B.Esnal-Zufiaurre A., Collins F., et al. (2017). Immunoprofiling of human uterine mast cells identifies three phenotypes and expression of ERβ and glucocorticoid receptor. F1000Res. 6, 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B., Migliaccio G., Yee N.S., et al. (1994). Long-term generation of human mast cells in serum-free cultures of CD34+ cord blood cells stimulated with stem cell factor and interleukin-3. Blood 84, 3667–3674. [PubMed] [Google Scholar]
- Dwyer D.F., Barrett N.A., Austen K.F. (2016). Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat. Immunol. 17, 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E., Bigas A. (2018). Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell 22, 639–651. [DOI] [PubMed] [Google Scholar]
- Gentek R., Ghigo C., Hoeffel G., et al. (2018). Hemogenic endothelial fate mapping reveals dual developmental origin of mast cells. Immunity 48, 1160–1171. [DOI] [PubMed] [Google Scholar]
- Gilfillan A.M., Peavy R.D., Metcalfe D.D. (2009). Amplification mechanisms for the enhancement of antigen-mediated mast cell activation. Immunol. Res. 43, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurish M.F., Austen K.F. (2012). Developmental origin and functional specialization of mast cell subsets. Immunity 37, 25–33. [DOI] [PubMed] [Google Scholar]
- Hu P., Carlesso N., Xu M., et al. (2012). Genetic evidence for critical roles of P38α protein in regulating mast cell differentiation and chemotaxis through distinct mechanisms. J. Biol. Chem. 287, 20258–20269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani A.A., Schechter N.M., Craig S.S., et al. (1986). Two types of human mast cells that have distinct neutral protease compositions. Proc. Natl Acad. Sci. USA 83, 4464–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani A.M., Craig S.S., DeBlois G., et al. (1987). Deficiency of the tryptase-positive, chymase-negative mast cell type in gastrointestinal mucosa of patients with defective T lymphocyte function. J. Immunol. 138, 4381–4386. [PubMed] [Google Scholar]
- Kalesnikoff J., Galli S.J. (2008). New developments in mast cell biology. Nat. Immunol. 9, 1215–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminer M.S., Murphy G.F., Zweiman B., et al. (1995). Connective tissue mast cells exhibit time-dependent degranulation heterogeneity. Clin. Diagn. Lab. Immunol. 2, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauts M.L., De Leo B., Rodríguez-Seoane C., et al. (2018). Rapid mast cell generation from Gata2 reporter pluripotent stem cells. Stem Cell Rep. 11, 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Sawai N., Hidaka E., et al. (1999). Interleukin-6 directly modulates stem cell factor-dependent development of human mast cells derived from CD34+ cord blood cells. Blood 94, 496–508. [PubMed] [Google Scholar]
- Kirshenbaum A.S., Kessler S.W., Goff J.P., et al. (1991). Demonstration of the origin of human mast cells from CD34+ bone marrow progenitor cells. J. Immunol. 146, 1410–1415. [PubMed] [Google Scholar]
- Kirshenbaum A.S., Goff J.P., Albert J.P., et al. (1994). Fibroblasts determine the fate of FcεRI+ cell populations in vitro by selectively supporting the viability of mast cells while internalizing and degrading basophils. Int. Arch. Allergy Immunol. 105, 374–380. [DOI] [PubMed] [Google Scholar]
- Kirshenbaum A.S., Goff J.P., Semere T., et al. (1999). Demonstration that human mast cells arise from a progenitor cell population that is CD34+, c-kit+, and expresses aminopeptidase N (CD13). Blood 94, 2333–2342. [PubMed] [Google Scholar]
- Kitamura Y. (1989). Heterogeneity of mast cells and phenotypic change between subpopulations. Annu. Rev. Immunol. 7, 59–76. [DOI] [PubMed] [Google Scholar]
- Kitamura Y., Ito A. (2005). Mast cell-committed progenitors. Proc. Natl Acad. Sci. USA 102, 11129–11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura Y., Shimada M., Hatanaka K., et al. (1977). Development of mast cells from grafted bone marrow cells in irradiated mice. Nature 268, 442–443. [DOI] [PubMed] [Google Scholar]
- Kovarova M., Latour A.M., Chason K.D., et al. (2010). Human embryonic stem cells: a source of mast cells for the study of allergic and inflammatory diseases. Blood 115, 3695–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.Q., Liu S.X., Xu J.F., et al. (2018). Adult connective tissue-resident mast cells originate from late erythro-myeloid progenitors. Immunity 49, 640–653. [DOI] [PubMed] [Google Scholar]
- Ma F., Kambe N., Wang D., et al. (2008). Direct development of functionally mature tryptase/chymase double-positive connective tissue-type mast cells from primate embryonic stem cells. Stem Cells 26, 706–714. [DOI] [PubMed] [Google Scholar]
- Mao B., Huang S., Lu X.L., et al. (2016). Early development of definitive erythroblasts from human pluripotent stem cells defined by expression of glycophorin A/CD235a, CD34, and CD36. Stem Cell Rep. 7, 869–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon T.C. St, Laurent C.D., Morris K.E., et al. (2009). Advances in mast cell biology: new understanding of heterogeneity and function. Mucosal Immunol. 3, 111–128. [DOI] [PubMed] [Google Scholar]
- Morii E., Oboki K., Ishihara K., et al. (2004). Roles of MITF for development of mast cells in mice: effects on both precursors and tissue environments. Blood 104, 1656–1661. [DOI] [PubMed] [Google Scholar]
- Ng E.S., Azzola L., Bruveris F.F., et al. (2016). Differentiation of human embryonic stem cells to HOXA+ hemogenic vasculature that resembles the aorta-gonadmesonephros. Nat. Biotechnol. 34, 1168–1179. [DOI] [PubMed] [Google Scholar]
- Oksaharju A., Kankainen M., Kekkonen R.A., et al. (2011). Probiotic Lactobacillus rhamnosus downregulates FCER1 and HRH4 expression in human mast cells. World J. Gastroenterol. 17, 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., et al. (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodarte E.M., Ramos M.A., Davalos A.J., et al. (2018). Munc13 proteins control regulated exocytosis in mast cells. J. Biol. Chem. 293, 345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottem M., Okada T., Goff J.P., et al. (1994). Mast cells cultured from the peripheral blood of normal donors and patients with mastocytosis originate from a CD34+/FcεRI− cell population. Blood 84, 2489–2496. [PubMed] [Google Scholar]
- Saito H., Ebisawa M., Tachimoto H., et al. (1996). Selective growth of human mast cells induced by Steel factor, IL-6, and prostaglandin E2 from cord blood mononuclear cells. J. Immunol. 157, 343–350. [PubMed] [Google Scholar]
- Shelburne C.P., McCoy M.E., Piekorz R., et al. (2003). Stat5 expression is critical for mast cell development and survival. Blood 102, 1290–1297. [DOI] [PubMed] [Google Scholar]
- Simonowski A., Wilhelm T., Habib P., et al. (2020). Differential use of BTK and PLC in FcεRI− and KIT-mediated mast cell activation: a marginal role of BTK upon KIT activation. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118622. [DOI] [PubMed] [Google Scholar]
- Slukvin I.I. (2016). Generating human hematopoietic stem cells in vitro-exploring endothelial to hematopoietic transition as a portal for stemness acquisition. FEBS Lett. 590, 4126–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen J.G., Michele G., Mindy T. (2008). Immunomodulatory mast cells: negative, as well as positive, regulators of immunity. Nat. Rev. Immunol. 8, 478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz Eldor J., Shapiro S.S., et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- Wang B., Li L., Liao Y., et al. (2013). Mast cells expressing interleukin 17 in the muscularis propria predict a favorable prognosis in esophageal squamous cell carcinoma. Cancer Immunol. Immunother. 62, 1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang Y., Peng M., et al. (2020). UBASH3B is a novel prognostic biomarker and correlated with immune infiltrates in prostate cancer. Front. Oncol. 9, 1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welle M. (1997). Development, significance, and heterogeneity of mast cells with particular regard to the mast cell-specific proteases chymase and tryptase. J. Leukoc. Biol. 61, 13. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., et al. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang Y.G., Chen B., et al. (2019). Overexpression of GATA2 enhances development and maintenance of human embryonic stem cell-derived hematopoietic stem cell-like progenitors. Stem Cell Rep. 13, 31–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.