Abstract
Purpose
This work was initiated to offer solid evidence regarding the expression and roles of long noncoding RNA (lncRNA) CCDC144NL-AS1 in hepatocellular carcinoma (HCC).
Patients and Methods
Cell Counting Kit-8 assay, flow cytometric analysis, and invasion assays were used to explore the malignant biological characteristics of cells. Immunohistochemistry (IHC), Western blotting analysis, and real-time quantitative PCR (RT-qPCR) were used to analyze the expression level of related proteins and nucleic acids. Bl6/Rag2/GammaC double knockout mice were used for HCC modeling to address the therapeutic value of CCDC144NL-AS1.
Results
CCDC144NL-AS1 was significantly upregulated in HCC tissue and had a marked relationship with the 5-year prognosis. In vitro study revealed that CCDC144NL-AS1 was highly expressed in HCC cell line MHCC97H but lowly expressed in normal hepatic cell line L02. Overexpression of CCDC144NL-AS1 in L02 enhanced the invasion and proliferation abilities of cells but inhibited the apoptosis rate. Knockdown of CCDC144NL-AS1 in MHCC97H weakened the invasion and proliferation abilities of cells but increased the apoptosis rate. CCDC144NL-AS1 was found to sponge miR-940 to induce the expression of WD repeat domain 5 (WDR5). ChIP-seq analysis identified that matrix metalloproteinase (MMP) 2, MMP9, and cyclin-dependent kinase (CDK) 1, CDK2, and CDK4 were all targets of WDR5. The recruitment of WDR5 to the promoter of these target genes upregulated the histone H3 lysine 4 trimethylation (H3K4me3) level in these regions and further induced the transcription of MMP2, MMP9, CDK1, CDK2, and CDK4. In vivo study revealed that compared to the normal liver tissue, CCDC144NL-AS1, WDR5, MMP2, MMP9, CDK1, CDK2, and CDK4 were all significantly upregulated in HCC tissue from the same mouse, while miR-940 was decreased. Besides, knockdown of CCDC144NL-AS1 or WDR5 or overexpression of miR-940 could all inhibit tumor growth.
Conclusion
CCDC144NL-AS1 drives HCC development by inducing MMP2/MMP9 and CDK1/CDK2/CDK4 expressions through miR-940/WDR5-regulated epigenetic pathway.
Keywords: long non-coding RNA CCDC144NL-AS1, WD repeat domain 5, matrix metalloproteinase, cyclin-dependent kinase, prognosis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies all over the world. It is also the leading cause of cancer death worldwide, ranking 2nd and 6th cause of cancer death for men and women respectively.1 The most used treatment for HCC patients in early-stage remains complete surgical resection, but the curative effect is not sufficient yet, especially for patients with huge tumor size and metastatic disease that is not apparent at the time of surgery.2 The prognosis of HCC is considered to be the poorest one of all solid cancers, with the 5-year survival rate of advanced HCC at ~1%.3 Controlling tumor growth and metastasis will provide more opportunities for the treatment of HCC, and will also greatly improve the prognosis of patients.
Aberrant epigenetic processes were demonstrated to play key roles in carcinogenesis. As for HCC, previous in vitro and in vivo studies had observed the dysregulation of histone deacetylases.4,5 Histone methylation, serving as a major epigenetic modification, might also be involved in the pathogenesis of HCC.6 The aberrant histone methylation pattern contributes to the initiation, maintenance, and development of HCC, and is related to the poor prognosis of patients.7,8
Long non-coding RNA (lncRNA) CCDC144NL-AS1 was first reported to be involved in carcinogenesis in 2020.9 Aberrantly expressed CCDC144NL-AS1 was observed in gastric cancer,9 however, its roles in HCC development and the potential mechanism have not been reported yet. In this study, we investigated the expression pattern of CCDC144NL-AS1 using human HCC tissues, and explored the underlying mechanism from the epigenetic pathway. We also estimated the therapy value of CCDC144NL-AS1 using HCC mouse model. Our study may provide novel therapeutic targets for HCC treatment to improve the prognosis of HCC.
Patients and Methods
Tissue Collection
From April 2014 to December 2015, 135 pairs of HCC tissues and the matched adjacent normal tissues were collected from the Department of Infectious Diseases of the First Affiliated Hospital of Zhengzhou University. Patients who received treatments before or complicated with distant metastasis or other systemic diseases such as diabetes, cardiovascular, or cerebrovascular diseases were excluded from this study.
This study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University, and was conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from every patient before sample collection.
Cell Culture
HCC cell lines Huh-7, HepG2, Hep3B, SMMC-7721, MHCC97H, SNU-368, HCCLM3, and normal hepatic cell line L02 were purchased from the American Type Culture Collection (USA) and were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Thermo Fisher Scientific, USA) supplemented with 10% (v/v) fetal bovine serum (FBS) (Clark, USA). Cells were placed in an incubator at 37 °C with 5% CO2. The culture medium was replaced every other day.
Transfections and Transductions
Small interfering RNA (siRNA) for CCDC144NL-AS1, miR-940, and WDR5 were purchased (Hippobio, China) and were used to knock down the target transcription. miRNA mimics were also used to overexpress miR-940. The transfection was conducted using Lipofectamine 3000® Transfection Reagent (Invitrogen, USA) according to the manufacturers’ instructions. Briefly, after starvation for 24 hours (h), cells were incubated with Opti-MEM™ medium (Gibco, USA) containing Lipofectamine™ 3000 Reagent, P3000™ Reagent, and siRNAs (Opti-MEM™ medium: Lipofectamine™ 3000 Reagent: P3000™ Reagent: siRNAs = 250 ul: 3.75 ul: 5 ul: 2.5 ug) for 4 h. Then the medium was replaced with fresh DMEM containing 10% FBS. The knockdown efficiency was analyzed 48 h after transfection using real-time quantitative PCR (RT-qPCR). The sequence of siRNAs and primers used in RT-qPCR was provided in Table 1.
Table 1.
Sequence of Primers and siRNAs
| Name | Forward/Sense (from 5ʹ to 3ʹ) | Reverse/Antisense (from 5ʹ to 3ʹ) | |
|---|---|---|---|
| Primers used for ChIP-qPCR | MMP2 | TCCTTTCTCACTAGGTGGC | TCATACCTGACGCTGTGCTA |
| MMP9 | TGCTGGCTTTTTGGACACCCACT | CCGCCGCCACCTGTTGAGGA | |
| CDK1 | GAGACUUGAAUUAAUAAGUGAdTdT | UCACUUAUUAAUUCAAGUCUCdTdT | |
| CDK2 | GCUCAUGAUCAAACGCUCUAAdTdT | UUAGAGCGUUUGAUCAUGAGCdTdT | |
| CDK4 | GAUUCAGAUUCUCUAGCGUGAdTdT | CUUCAAGCAGUUCAUCAUUCUCdTdT | |
| Primers used for RT-qPCR | CCDC144NL-AS1 | AACGGTTTCAGCTGCCTCTA | TTTGGGTCACCTGAACTTCC |
| WDR5 | ATCCCCTTCCTCCTCCACTC | GAAGGAGTGGCGAACACTGA | |
| miR-940 | TCGTUTCCAGCGGCATGGC | TGCGCGCTAGCGTACGTAGC | |
| siRNAs | siNC | UUCUCCGAACGUGUCACGUdTdT | ACGUGACACGUUCGGAGAAdTdT |
| WDR5 | AATGGTCCGCGTATCGCGTdTdT | GCGGTTCCAAGCGTCGATCdCdG | |
| siCCDC144NL-AS1 | CGTGCGGCACTCCGGACAATCdTdT | AGCCGGTGGGACTGCATTCGdTdT | |
| simiR-940 | AGGGAGAAUGUUGAAACACAAdTdT | UUGUGUUUCAACAUUCUCCCUdTdT |
Abbreviations: CDK, cyclin-dependent kinase; MMP, matrix metalloproteinase; WDR, WD repeat domain.
pPLK-shRNA (pPLK) vectors for CCDC144NL-AS1 and WD repeat domain 5 (WDR5) were constructed by Public Protein/Plasmid Library (Jiangsu, China) to permanently knockdown CCDC144NL-AS1 and WDR5 respectively. pLenti-CMV (pLV) vector was used to permanently overexpress miR-940. The empty vector was used as the negative control (NC). All these vectors were packaged with lentivirus. Viral supernatants were used to transduce cells. Stable transductants were selected by growth in media containing Ampicillin.
Induction of HCC in Mouse
Six-week male Bl6/Rag2/GammaC double knockout nude mice were purchased (Cyagen, China) and adaptively fed for one week. Unlike NOD-SCID mice, these mice lack T-, B-, and NK-cells but show no spontaneous tumor formation and have a normal hematopoietic function. Mice were housed in a facility with a 12 h light/dark cycle maintained at 25 ± 0.5°C and 50% to 60% humidity. All animal treatments were performed in accordance with the 3R principle of experimental animals. The Center for Animal Experiments of The First Affiliated Hospital of Zhengzhou University approved the in vivo study.
The xenograft HCC model was established as described before.10 Briefly, MHCC97H cells were prepared as a single-cell suspension, and 1×106 prepared cells in 50 ul matrigel were injected subcutaneously into the right axillary fossa. After the injection, mice were continued fed for 30 days. The tumor size was measured every week using a vernier caliper.
Luciferase Assays
The target miRNA of CCDC144NL-AS1 and the potential binding site were identified using DIANA Tools. The target messenger RNA (mRNA) of miR-940 and the potential binding site were predicted by the TargetScan algorithm. CCDC144NL-AS1 and the 3ʹ-UTR fragments of WDR5 mRNA were amplified and cloned into the pGL3-basic vectors (Genomeditech, China). The binding sites for miR-940 were mutated with QuikChange® Multi Site-Directed Mutagenesis Kit (TIANGEN, China). Plasmids were co-transfected into cells with NC mimic, miR-940 mimic, siNC, and simiR-940, respectively, using Lipofectamine 3000® Transfection Reagent (Invitrogen, USA). Twenty-four hours after transfection, cells were lysed and luciferase activity was measured.
Invasion Assay
Invasion assay was performed using invasion chambers with 6.4 mm diameters and 8 mm pore size (Corning, USA) coated with 100 ul matrigel (BD, USA). Briefly, cells resuspended in DMEM culture medium with no FBS were seeded into the upper chamber. DMEM medium containing 20% FBS was filled into the lower chamber as the chemoattractant. After 48 h of incubation, the non-invasive cells were removed from the upper side of the chamber, and the invasive cells were stained using crystal violet. The chamber was then observed and photographed under a light microscope to count invasive cells.
Cell Counting Kit-8 (CCK-8) Assay
After 24 h of transfection, cells were seeded into the 96-well plate at a density of 1×103 per well. Cells were then cultured for 0, 24, 48, and 72 h at 37°C. To analyze cell proliferation, each well was loaded with 10 µL CCK-8 solution (Beyotime, China) and was incubated at 37°C for another 2 h. A microplate reader (Tecan Group Ltd., Switzerland) was used to monitor the absorbance at 450 nm wavelength.
RNA Pull-Down Assay
Plasmids used in RNA pull-down assay were constructed by Public Protein/Plasmid Library (Jiangsu, China) and were transfected to cells using Lipofectamine 3000® Transfection Reagent (Invitrogen, USA) to express exogenous miR-940. These exogenous RNAs were labeled using the Biotin RNA Labeling Mix (Roche, USA). Total RNA extracts from MHCC97H and L02 cells were mixed with biotin-labeled-miR-940, incubated with Dynabeads™ M-280 Streptavidin (Invitrogen, USA) at 4 °C for 3 h. Finally, the nucleic acids of CCDC144NL-AS1 and WDR5 contained in the pulled-down complexes were analyzed using RT-qPCR.
Flow Cytometric Analysis
Cell apoptosis was analyzed with an Annexin V-FITC apoptosis detection kit (KeyGen Biotech Co., Ltd., China). Approximately 1×106 cells were rinsed with phosphate-buffered saline (PBS). The collected cells were resuspended in 500 μL binding buffer. Later, 5 μL Annexin V-APC and 5 μL propidium iodide were introduced into the cell suspension, and additional cultivation was performed at room temperature for 15 min without light. Finally, the late and early apoptotic rates were determined using flow cytometry (Becton Dickinson, USA).
For cell cycle analysis, approximately 1×106 cells were isolated and resuspended in cold PBS. PI was introduced into the cell suspension at the final concentration of 50 ug/mL. Incubation was performed at 4°C for 30 min without light. After being filtered by 80 μm pore size mesh, cells were analyzed using flow cytometry (Becton Dickinson, USA).
Immunofluorescence
Cells were fixed with 4% formaldehyde and incubated overnight at 4°C with primary antibody for the detection of Ki67 (ab16667, Abcam, USA). Cells were then incubated with the secondary antibody (BA1031, BOSTER, China) at 37°C for another 30 minutes. 4ʹ,6-diamidino-2-phenylindole (DAPI) (Beyotime, China) was added to stain the nucleus. Cells were observed by an inverted fluorescence microscope (Olympus, Japan) using a ×20 objective.
Immunohistochemistry (IHC)
IHC was performed in paraffin-embedded sections using IHC kit (SV0002, BOSTER, China). Briefly, after being deparaffinized using xylene, and rehydrated using an ethanol gradient, the sections were incubated with 5% bovine serum albumin for 30 min at 37°C. Sections were probed with WDR5 (ab245479, Abcam, USA), CDK1 (ab201008, Abcam, USA), CDK2 (ab32147, Abcam, USA), CDK4 (ab68266, Abcam, USA), MMP2 (ab86607, Abcam, USA), and MMP9 (ab237782, Abcam, USA) overnight at 4°C, followed by incubation with the secondary antibody provided in the kits for another 30 min. The section was then stained with 3,3ʹ- diaminobenzidine, and was photographed by an Olympus inverted microscope using a ×20 objective.
Western Blotting Analysis
Total proteins (30 ug) from cell lysates of each group were separated using 10% SDS-polyacrylamide gel and were transferred to a polyvinyl difluoride (PVDF) membrane. After being blocked using blocking buffer (Beyotime, China), membrane was incubated with antibodies anti-WDR5 (ab178410, Abcam, USA), MMP2 (ab92536, Abcam, USA), MMP9 (ab76003, Abcam, USA), CDK1 (ab133327, Abcam, USA), CDK2 (ab101682, Abcam, USA), CDK4 (ab108357, Abcam, USA), N-cadherin (ab76011, Abcam, USA), E-cadherin (ab40772, Abcam, USA), β-catenin (ab32572, Abcam, USA), vimentin (ab92547, Abcam, USA), caspase-3 (ab32351, Abcam, USA), Bcl-2 (ab32124, Abcam, USA), Bax (ab32503, Abcam, USA), and tubulin (ab6046, Abcam, USA) separately overnight at 4°C. The membrane was then incubated with the second antibodies (SA00001-1 or SA00001-2, Proteintech Group, USA) for another 0.5 h. The blots were visualized using BeyoECL Plus kit (Beyotime, China), and the band was quantified by Image J (National Institutes of Health, USA).
RT-qPCR
Total RNAs were extracted using Invitrogen™ TRIzol™ Reagent (Thermo Fisher Scientific, USA) and were reversely transcribed into cDNA using PrimeScriptTM RT Master Mix (Takara, Japan). RT-qPCR was performed on the Bio-Rad CFX96 (Bio-Rad Laboratories, USA) using SYBR Premix Ex TaqTM (Takara, Japan). The sequence of primers is provided in Table 1. The reaction process includes hold (95°C for 30s, 1 cycle), PCR (95°C for 5s, followed by 60°C for 30s, 40 cycles), and dissociation (95°C for 15s, then 60°C for 30s, followed by 95°C for 15s, 1 cycle). The expression level of each transcript was calculated using the comparative threshold cycle (Ct), and the relative expression of the target gene was normalized by GAPDH.
Chromatin Immunoprecipitation (ChIP) and ChIP Followed by Deep Sequencing (ChIP-Seq)
ChIP assay was performed according to the previous report.11 Briefly, cells were cross-linked using 4% methanol for 10 minutes and sheared by sonication to produce DNA fragments of appropriate length (~500bp). 1% of the chromatin fragments were separated as the input. Chromatin fragments were immunoprecipitated by antibodies against WDR5 (13105S, Cell Signaling Technology, USA) and histone H3 lysine 4 trimethylation (H3K4me3) (9751S, Cell Signaling Technology) respectively. An equal amount of normal rabbit IgG was added as the negative control. Input and ChIP-seq analysis were performed using the ChIP-IT High Sensitivity Kit (ab185908, USA). The model-based analysis of ChIP-seq (MACS) peak-finding algorithm was used to normalize ChIP against the input control. The sequence of the primers used for ChIP-qPCR was provided in Table 1.
Statistical Analysis
Statistical analyses were all performed using GraphPad Prism 6.0 (GraphPad Software, USA). The difference between the two groups was evaluated by Student’s t-test, and one-way ANOVA was used for the comparison among multiple groups. Chi-square test was used to analyze the categorical variables. The survival curve was analyzed by the Kaplan-Meier method. In this study, results were obtained from at least three independent experiments. P<0.05 was considered to be statistically significant.
Results
CCDC144NL-AS1 Was Upregulated in Human HCC Tissue and is Associated with the Severity of HCC
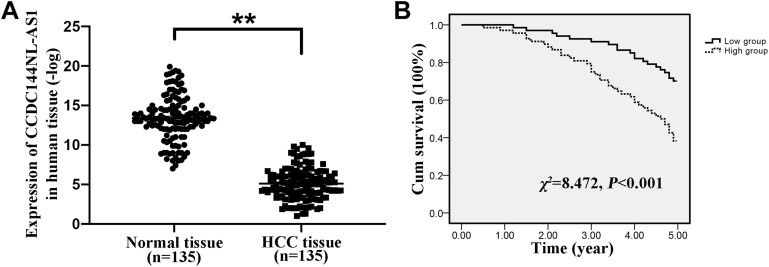
Firstly, we analyzed the expression pattern of CCDC144NL-AS1 in human HCC tissue and the paired adjacent normal tissue. Results showed that CCDC144NL-AS1 was upregulated markedly in HCC tissue compared to that in normal tissue (Figure 1A). Besides, we found that the expression level of CCDC144NL-AS1 in HCC tissue was significantly correlated with the infection of hepatitis B virus (HBV) and hepatitis C virus (HCV), the cirrhosis state, the differentiation state, the T stage, and the N stage of patients (Table 2), and was also negatively correlated with the 5-year prognosis of patients (χ2=8.472, P<0.001, Figure 1B).
Figure 1.
Expression pattern of CCDC144NL-AS1 in human HCC tissue. (A) CCDC144NL-AS1 was significantly upregulated in human HCC tissue compared to that in the matched normal tissue. (B) CCDC144NL-AS1 expression in HCC tissue was negatively correlated with the 5-year prognosis of patients. Data were expressed as mean ± SEM. **P<0.001.
Table 2.
The Relationships Between CCDC144NL-AS1 Expression in HCC Tissue and Patients’ Clinical Characteristics
| Characteristics | Case (n=135) | CCDC144NL-AS1 (-log) | P |
|---|---|---|---|
| Gender | 0.556 | ||
| Male | 83 | 5.13 ± 1.12 | |
| Female | 52 | 5.03 ± 1.55 | |
| Age | 0.455 | ||
| < 50 | 56 | 5.17± 0.96 | |
| ≥50 | 79 | 5.04 ± 1.08 | |
| BMI | 0.773 | ||
| < 18.5 | 47 | 5.05 ± 1.16 | |
| 18.5~24.9 | 67 | 5.08 ± 1.09 | |
| ≥25 | 21 | 5.22 ± 1.21 | |
| Smoking | 0.671 | ||
| No | 69 | 5.15 ± 0.88 | |
| Yes | 66 | 5.03 ± 1.31 | |
| Alcohol using | 0.542 | ||
| No | 44 | 5.14 ± 1.18 | |
| Yes | 91 | 5.07 ± 1.05 | |
| Etiological classification | < 0.001 | ||
| HBV | 81 | 3.32 ± 0.63 | |
| HCV | 23 | 3.73 ± 0.59 | |
| Others | 31 | 10.73 ± 1.32 | |
| With/without cirrhosis | < 0.001 | ||
| Cirrhosis | 97 | 3.80 ± 0.69 | |
| Non-cirrhosis | 38 | 8.39 ± 1.27 | |
| Histological classification/differentiation | 0.001 | ||
| Well/moderate | 81 | 6.42 ± 1.29 | |
| Poor | 54 | 3.10 ± 0.78 | |
| T stage | < 0.001 | ||
| Tis-2 | 21 | 9.15 ± 1.22 | |
| T3–4 | 114 | 4.34 ± 1.03 | |
| N stage | < 0.001 | ||
| N0–1 | 37 | 8.23 ± 1.31 | |
| N2–3 | 98 | 3.91 ± 0.93 |
Note: Statistical significance (P < 0.05) is shown in bold.
Abbreviation: BMI, body mass index.
CCDC144NL-AS1 Promotes HCC Cells Invasion and Proliferation
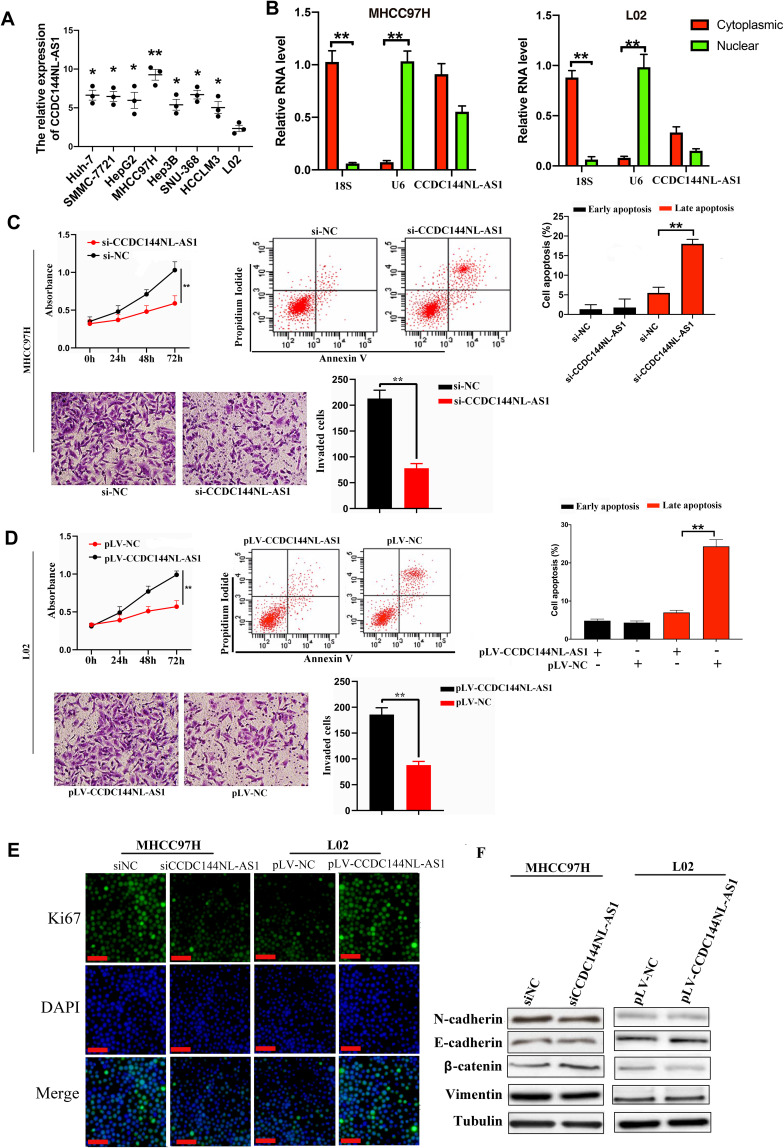
We explored the expression pattern of CCDC144NL-AS1 in different kinds of cell lines, and found that CCDC144NL-AS1 was significantly upregulated in HCC cell lines compared to normal hepatic cells L02, and it was upregulated most significantly in MHCC97H (Figure 2A). MHCC97H and L02 were used in the further study. Sub-cellular location study identified that CCDC144NL-AS1 was expressed in both cytoplasm and nucleus (Figure 2B). Knockdown of CCDC144NL-AS1 markedly weakened the invasion and proliferation abilities, but increased the late apoptosis rate of MHCC97H cells. Overexpressing CCDC144NL-AS1 enhanced the invasion and proliferation abilities, but decreased the level of late apoptosis in L02 cells (Figure 2C and D). Similarly, we observed that Ki67 expression was also regulated by CCDC144NL-AS1. Knockdown of CCDC144NL-AS1 significantly decreased Ki67 expression in MHCC97H, while overexpressing CCDC144NL-AS1 increased Ki67 expression (Figure 2E). Analysis of migration-related proteins showed that N-cadherin, E-cadherin, β-catenin, and vimentin were not involved in CCDC144NL-AS1-regulated pathways (Figure 2F). These results indicate that CCDC144NL-AS1 is crucial for the regulation of malignant biological behaviors such as invasion, proliferation, and apoptosis of HCC cells.
Figure 2.
CCDC144NL-AS1 promotes invasion and proliferation, while inhibited apoptosis of HCC cells. (A) CCDC144NL-AS1 expression was highest in HCC cell line MHCC97H compared to other HCC cell lines and normal hepatic cell line L02. (B) The expression of CCDC144NL-AS1 is located in both cytoplasm and nucleus of cells. (C and D) Knockdown of CCDC144NL-AS1 markedly inhibited the invasion and proliferation abilities, and enhanced the level of late apoptosis in MHCC97H cells, while overexpressing CCDC144NL-AS1 enhanced the invasion and proliferation abilities, and inhibited the level of late apoptosis in L02 cells. (E) Knockdown of CCDC144NL-AS1 led to decreased Ki67 expression in MHCC97H, while overexpressing CCDC144NL-AS1 increased Ki67 expression in L02. (F) Detection of migration-related proteins showed that there’s no significant change in their expression levels. The scale bar is 50 um. Data were expressed as mean ± SEM. *P<0.05, **P<0.001.
miR-940 is Involved in CCDC144NL-AS1 Regulating Cell Invasion and Proliferation
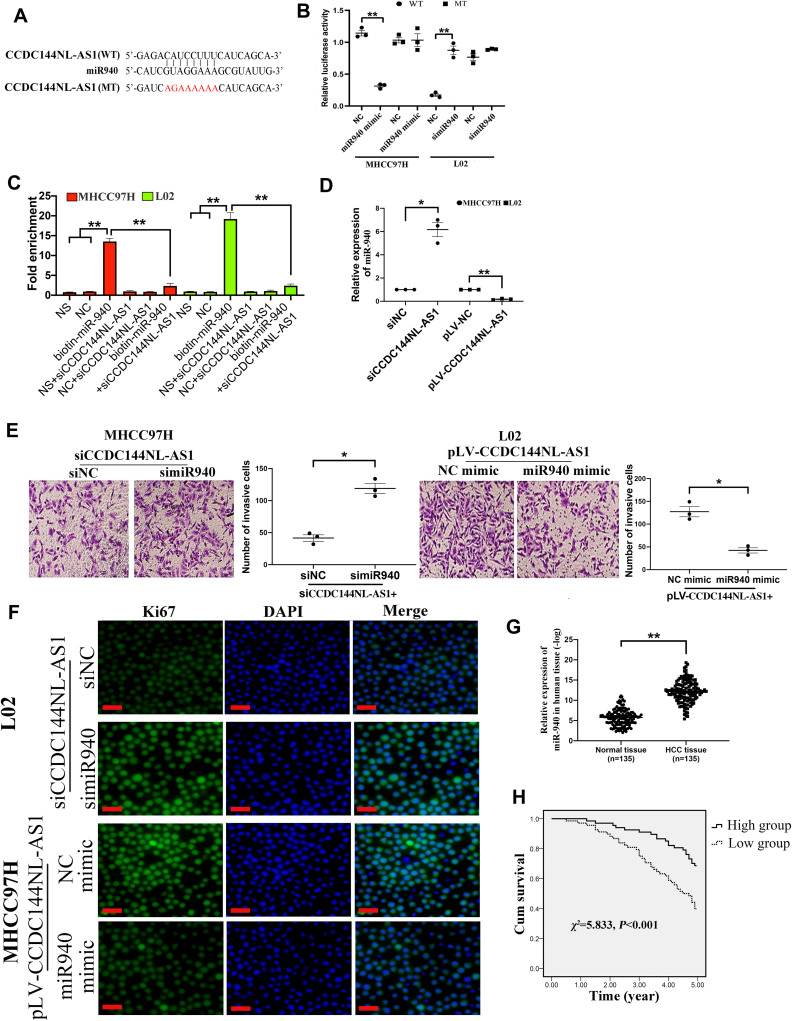
To further clarify the function of CCDC144NL-AS1, we analyzed its possible miRNA partner using DIANA TOOLS. Results showed that miR-940 could be targeted by CCDC144NL-AS1 (Figure 3A), and this was confirmed by luciferase assay and RNA pull-down assay (Figure 3B and C). Besides, knockdown of CCDC144NL-AS1 in MHCC97H increased miR-940 level, while its overexpression in L02 led to the decrease of miR-940 (Figure 3D). Knockdown of miR-940 in MHCC97H significantly blocked the effects of siCCDC144NL-AS1 on cell invasion and Ki67 expression, and overexpressing miR-940 in L02 could also reverse the effects of pLV-CCDC144NL-AS1 (Figure 3E and F). We next analyzed the expression level of miR-940 in human HCC tissue and its relationship with patients’ clinical characteristics. Results showed that miR-940 was markedly downregulated in HCC tissue compared to that in the normal tissue (Figure 3G), and was significantly correlated with the infection of HBV and HCV, the cirrhosis state, the differentiation state, the T stage, and the N stage of patients (Table 3), and positively correlated with patients’ prognosis in 5 years (χ2=5.833, P<0.001, Figure 3H).
Figure 3.
miR-940 is involved in CCDC144NL-AS1 regulating cell invasion and proliferation. (A) miR-940 was predicted to be targeted by CCDC144NL-AS1. (B) Luciferase assays, (C) RNA pull-down assay, and (D) RT-qPCR confirmed the interaction between CCDC144NL-AS1 and miR-940. (E and F) Knockdown of miR-940 in MHCC97H significantly blocked the effects of siCCDC144NL-AS1 on cell invasion and Ki67 expression, and overexpressing miR-940 in L02 could also reverse the effects of pLV-CCDC144NL-AS1 in L02. (G) Expression of miR-940 was significantly downregulated in human HCC tissue compared to that in the matched normal tissue. (H) Expression of miR-940 in HCC tissue was positively correlated with the 5-year prognosis of patients. The scale bar is 50 um. Data were expressed as mean ± SEM. *P<0.05, **P<0.001.
Table 3.
The Relationships Between miR-940 Expression in HCC Tissue and Patients’ Clinical Characteristics
| Characteristics | Case (n=135) | miR-940 (-log) | P |
|---|---|---|---|
| Gender | 0.267 | ||
| Male | 83 | 12.12 ± 1.91 | |
| Female | 52 | 12.28 ± 2.13 | |
| Age | 0.698 | ||
| < 50 | 56 | 12.15 ± 1.79 | |
| ≥50 | 79 | 12.20 ± 1.98 | |
| BMI | 0.423 | ||
| < 18.5 | 47 | 12.18 ± 1.62 | |
| 18.5~24.9 | 67 | 12.19 ± 2.07 | |
| ≥25 | 21 | 12.16 ± 2.55 | |
| Smoking | 0.556 | ||
| No | 69 | 12.17 ± 2.28 | |
| Yes | 66 | 12.19 ± 2.64 | |
| Alcohol using | 0.874 | ||
| No | 44 | 12.16 ± 2.13 | |
| Yes | 91 | 12.19 ± 2.37 | |
| Etiological classification | < 0.001 | ||
| HBV | 81 | 15.04 ± 1.87 | |
| HCV | 23 | 14.88 ± 2.22 | |
| Others | 31 | 2.71 ± 1.02 | |
| With/without cirrhosis | < 0.001 | ||
| Cirrhosis | 97 | 14.08 ± 1.55 | |
| Non-cirrhosis | 38 | 7.34 ± 1.34 | |
| Histological classification/differentiation | 0.011 | ||
| Well/moderate | 81 | 10.23 ± 1.17 | |
| Poor | 54 | 15.11 ± 1.82 | |
| T stage | < 0.001 | ||
| Tis-2 | 21 | 6.67 ± 2.12 | |
| T3–4 | 114 | 13.20 ± 3.08 | |
| N stage | < 0.001 | ||
| N0–1 | 37 | 5.86 ± 1.76 | |
| N2–3 | 98 | 14.57 ± 2.14 |
Note: Statistical significance (P < 0.05) is shown in bold.
Abbreviation: BMI, body mass index.
WDR5 is Involved in CCDC144NL-AS1 Regulating Cell Invasion and Proliferation
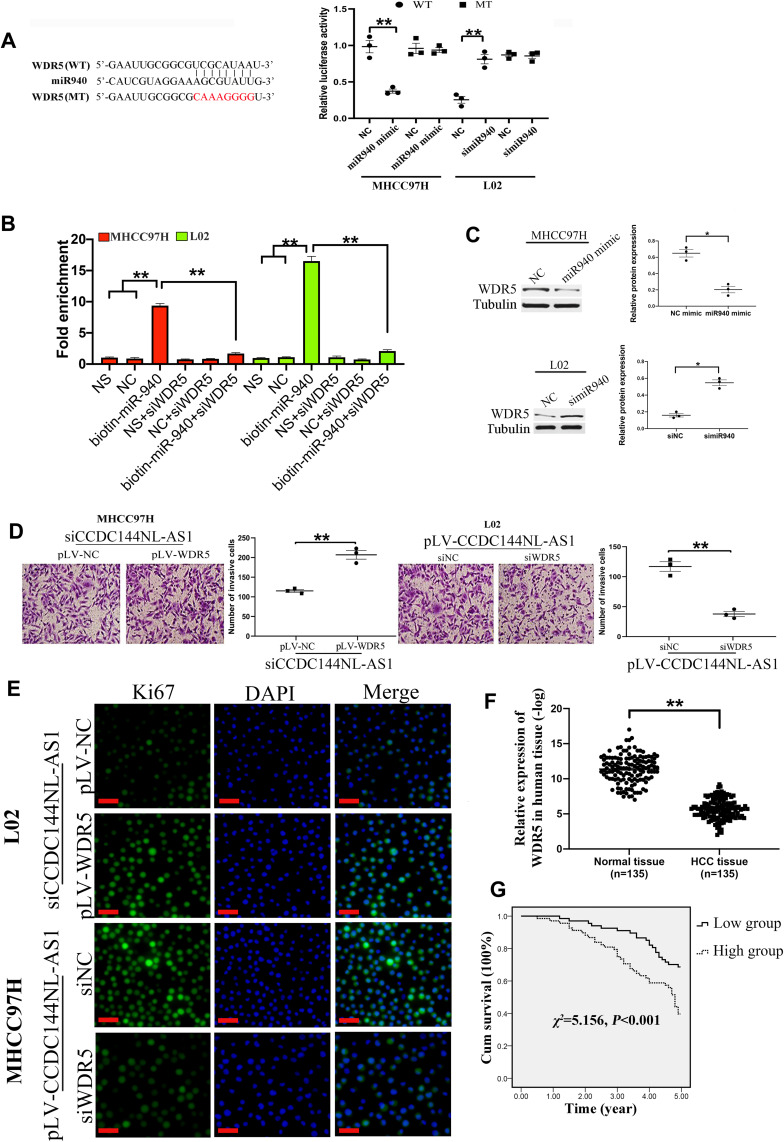
We next analyzed the target mRNA of miR-940 using the TargetScan algorithm and found that miR-940 could bind to the 3ʹ-UTR regions of WDR5 mRNA (Figure 4A). The relationship between miR-940 and WDR5 was confirmed by luciferase assays (Figure 4A) and RNA pull-down assay (Figure 4B). Besides, overexpression of miR-940 in MHCC97H significantly inhibited the expressions of WDR5, while knockdown of miR-940 in L02 led to the increased WDR5 expression (Figure 4C). The function of WDR5 in regulating cell invasion and proliferation was further analyzed. Results showed that overexpression of WDR5 in MHCC97H could significantly block the effects of siCCDC144NL-AS1, and siWDR5 could also reverse the effects of pLV-CCDC144NL-AS1 (Figure 4D and E). We next analyzed the expression level of WDR5 in human HCC tissue and its relationship with patients’ clinical characteristics. Results showed that WDR5 was markedly upregulated in HCC tissue compared to that in the normal tissue (Figure 4F), and was significantly correlated with the infection of HBV and HCV, the cirrhosis state, the differentiation state, the T stage, and the N stage of patients (Table 4), and negatively correlated with patients’ prognosis in 5 years (χ2=5.156, P<0.001, Figure 4G).
Figure 4.
WDR5 is involved in CCDC144NL-AS1 regulating cell invasion and proliferation. (A) Luciferase assays, (B) RNA pull-down assay, and (C) Western blotting analysis confirmed the interaction between WDR5 mRNA and miR-940. (D and E) Overexpression of WDR5 in MHCC97H significantly blocked the effects of siCCDC144NL-AS1 on cell invasion and Ki67 expression, and knockdown of WDR5 also reversed the effects of pLV-CCDC144NL-AS1. (F) Expression of WDR5 was significantly upregulated in human HCC tissue compared to that in the matched normal tissue. (G) Expression of WDR5 in HCC tissue was negatively correlated with the 5-year prognosis of patients. The scale bar is 50 um. Data were expressed as mean ± SEM. *P<0.05, **P<0.001.
Table 4.
The Relationships Between WDR5 Expression in HCC Tissue and Patients’ Clinical Characteristics
| Characteristics | Case (n=135) | WDR5 (-log) | P |
|---|---|---|---|
| Gender | 0.549 | ||
| Male | 83 | 5.62 ± 1.32 | |
| Female | 52 | 5.58 ± 1.17 | |
| Age | 0.707 | ||
| < 50 | 56 | 5.55 ± 0.89 | |
| ≥50 | 79 | 5.64 ± 1.08 | |
| BMI | 0.338 | ||
| < 18.5 | 47 | 5.58 ± 1.42 | |
| 18.5~24.9 | 67 | 5.65 ± 1.33 | |
| ≥25 | 21 | 5.51 ± 1.11 | |
| Smoking | 0.668 | ||
| No | 69 | 5.57 ± 0.97 | |
| Yes | 66 | 5.64 ± 1.31 | |
| Alcohol using | 0.633 | ||
| No | 44 | 5.66 ± 1.48 | |
| Yes | 91 | 5.58 ± 1.31 | |
| Etiological classification | < 0.001 | ||
| HBV | 81 | 4.23 ± 0.83 | |
| HCV | 23 | 4.28 ± 1.11 | |
| Others | 31 | 10.18 ± 1.42 | |
| With/without cirrhosis | < 0.001 | ||
| Cirrhosis | 97 | 4.25 ± 0.94 | |
| Non-cirrhosis | 38 | 9.06 ± 1.53 | |
| Histological classification/differentiation | 0.003 | ||
| Well/moderate | 81 | 6.24 ± 0.86 | |
| Poor | 54 | 4.65 ± 1.21 | |
| T stage | < 0.001 | ||
| Tis-2 | 21 | 9.13 ± 2.12 | |
| T3–4 | 114 | 4.96 ± 1.18 | |
| N stage | < 0.001 | ||
| N0–1 | 37 | 8.56 ± 2.45 | |
| N2–3 | 98 | 4.49 ± 0.93 |
Note: Statistical significance (P < 0.05) is shown in bold.
Abbreviation: BMI, body mass index.
CCDC144NL-AS1 Promotes the Recruitment of WDR5 to the Promoter Region of MMPs and CDKs
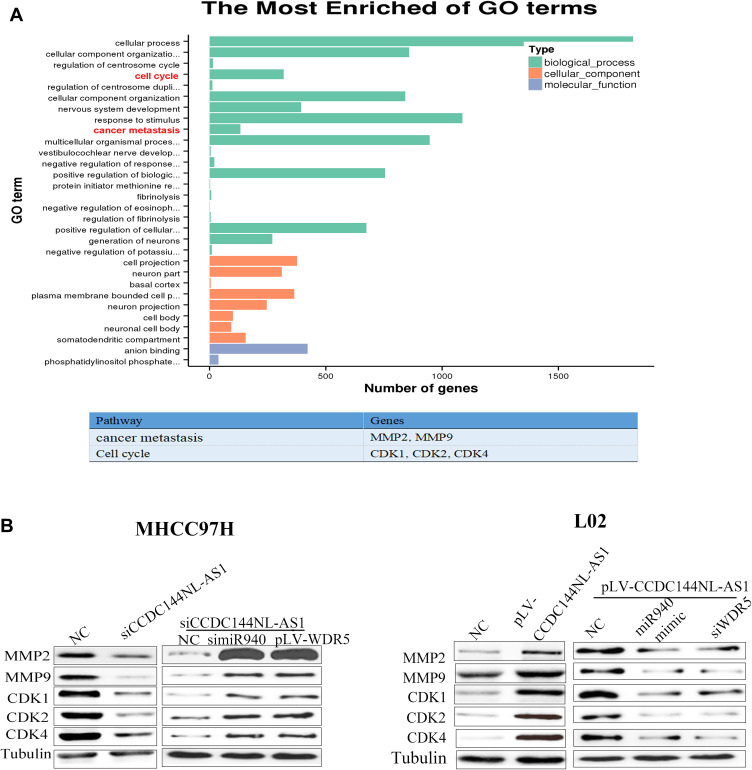
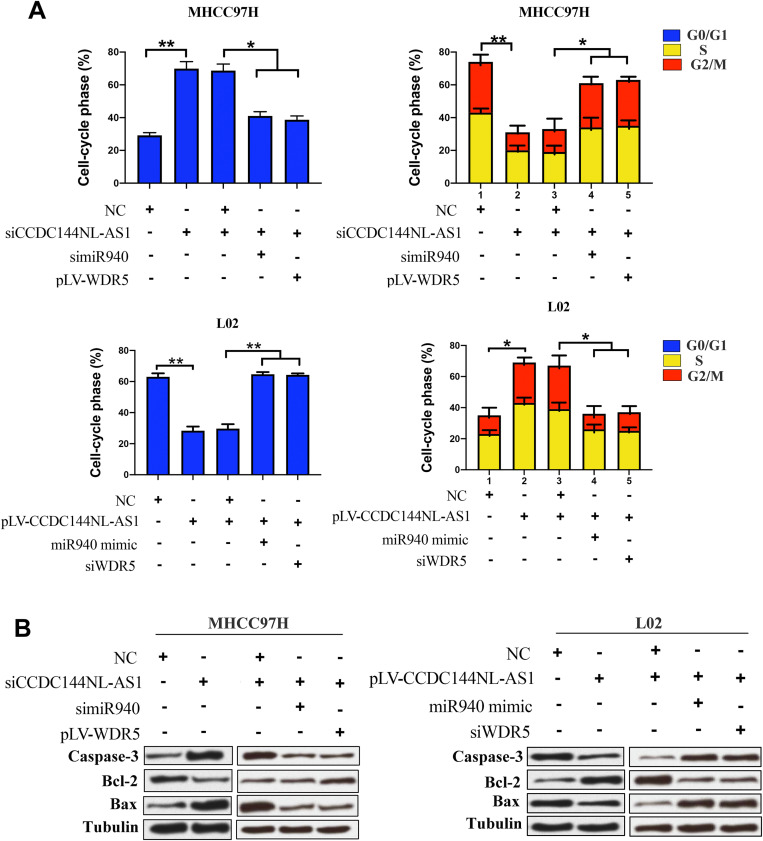
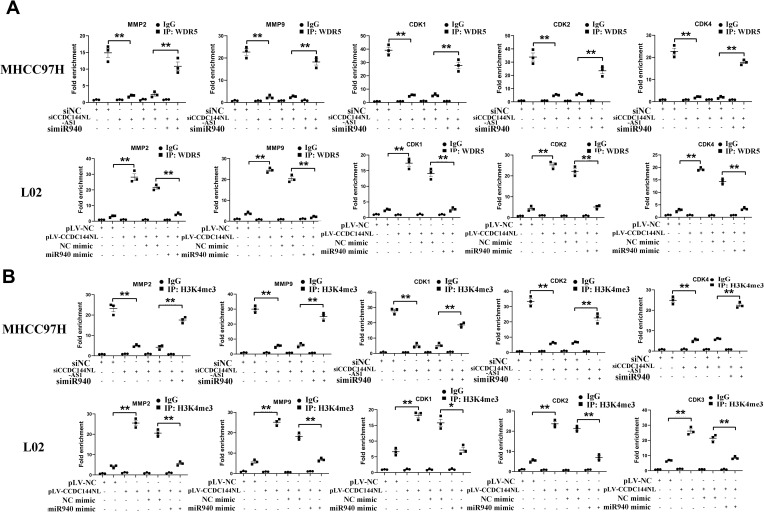
As a component of histone methyltransferases, WDR5 is essential for the catalysis of H3K4me3.12 We next performed ChIP-seq and pathway analysis to find the potential target of WDR5. Results showed the involvement of cancer metastasis and the cell cycle pathways (Figure 5A). GeneGo analysis revealed that a large number of DNA fragments of MMPs and CDKs were enriched by anti-WDR5 antibody in cells overexpressing CCDC144NL-AS1compared to normal cells (Figure 5A). Further results verified that MMP2, MMP9, CDK1, CDK2, and CDK4 could all be regulated by CCDC144NL-AS1/miR-940/WDR5 axis (Figure 5B). Cell-cycle analysis revealed that CCDC144NL-AS1 and WDR5 promoted cell transition from G0/G1 to G2/M phase, while miR-940 showed the opposite effect (Figure 6A). Analysis of apoptosis-related proteins showed that CCDC144NL-AS1 and WDR5 inhibited the expression of caspase-3 and Bax, and enhanced the expression of Bcl-2, while miR-940 showed the opposite effect (Figure 6B). We next investigated the recruitment of WDR5 and the H3K4me3 level in the promoter region of the above five genes, and results showed that CCDC144NL-AS1 promoted the recruitment of WDR5 to the promoter region of MMP2, MMP9, CDK1, CDK2, and CDK4, and resulted in the increased H3K4me3 level in these regions, while miR-940 played the opposite role to reverse the effects of CCDC144NL-AS1 (Figure 7).
Figure 5.
MMPs and CDKs are the targets of WDR5. (A) Results of ChIP-seq and bioinformatics analysis on L02 with or without CCDC144NL-AS1 overexpression showed the involvement of cancer metastasis (MMP2 and MMP9) and the cell cycle (CDK1, CDK2, and CDK4) pathways in CCDC144NL-AS1-overexpressed cells. (B) Results of Western blotting showed that the expression of MMPs and CDKs are regulated by CCDC144NL-AS1/miR-940/WDR axis.
Figure 6.
CCDC144NL-AS1 regulates cell cycle by miR-940/WDR5 pathway. (A) Analysis of flow cytometry showed that CCDC144NL-AS1 significantly upregulated the proportion of cells in S/G2/M through miR-940/WDR5 pathway. (B) Apoptosis-related proteins such as caspase-3, Bcl-2, and Bax were also involved in the pathway regulated by CCDC144NL-AS1/miR-940/WDR5. *P<0.05, **P<0.001.
Figure 7.
CCDC144NL-AS1 promotes WDR5 to be recruited to the promoter region of MMPs and CDKs. ChIP assay showed that (A) the recruitment of WDR5 and (B) the H3K4me3 level in the promoter region of MMP2, MMP9, CDK1, CDK2, and CDK4 was regulated by CCDC144NL-AS1/miR-940 axis. Data were expressed as mean ± SEM. *P<0.05, **P<0.001.
CCDC144NL-AS1, miR-940, WDR5, MMPs, and CDKs are All Aberrantly Expressed in HCC Tissue in Mice Model
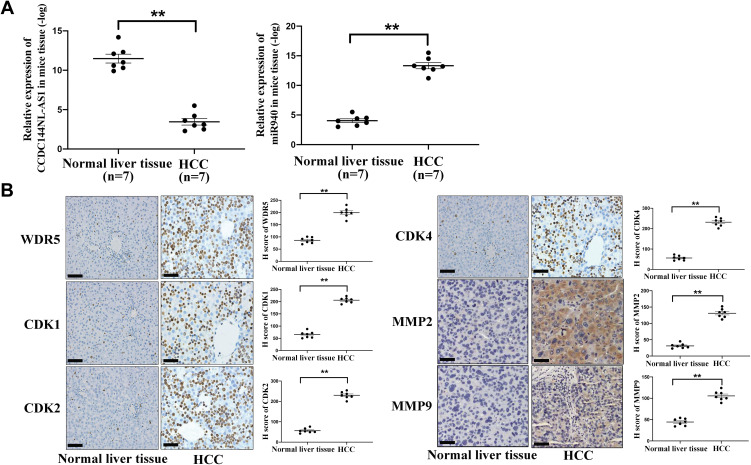
We next explored the expression pattern of CCDC144NL-AS1, miR-940, WDR5, MMPs, and CDKs in mouse model using IHC assay and RT-qPCR. As we expected, expressions of CCDC144NL-AS1, WDR5, MMP2, MMP9, CDK1, CDK2, and CDK4 all increased significantly in mouse HCC tissue compared to those in paired normal liver tissue, while the expression of miR-940 decreased markedly in mouse HCC tissue (Figure 8).
Figure 8.
Expression pattern of CCDC144NL-AS1/miR-940/WDR5, MMPs, and CDKs in mice HCC model. (A) RT-qPCR showed that the expression of CCDC144NL-AS1 increased markedly in mouse HCC tissue compared to that in the normal liver tissue, while the expression of miR-940 was significantly decreased. (B) IHC assay showed that the expression of WDR5, CDK1, CDK2, CDK4, MMP2, and MMP9 were all increased significantly in mouse HCC tissue compared to those in normal tissue. The scale bar is 50 um. Data were expressed as mean ± SEM. **P<0.001.
Targeting CCDC144NL-AS1/WDR5 or Overexpressing miR-940 Significantly Suppresses Tumor Growth and Improves the Prognosis of HCC in Mice Model
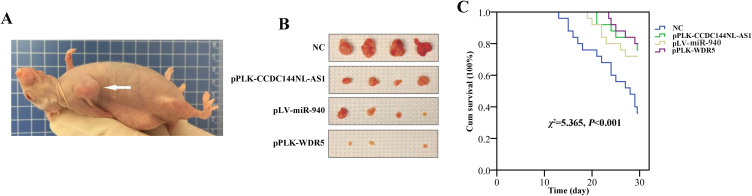
We next used the mice model to explore the therapeutic value of CCDC144NL-AS1/miR-940/WDR5. Mouse burdening xenograft HCC is shown in Figure 9A. Targeting CCDC144NL-AS1 and WDR5 could both suppress tumor growth. Similarly, overexpressing miR-940 could also suppress tumor growth (Figure 9B). Results of survival analysis indicated that targeting CCDC144NL-AS1/WDR5 or overexpressing miR-940 could all improve the prognosis of HCC mice significantly (χ2=5.365, P<0.001, Figure 9C).
Figure 9.
Targeting CCDC144NL-AS1/WDR5 or overexpressing miR-940 significantly suppresses tumor growth and improves the prognosis of HCC in mice model. (A) Mouse burdening xenograft HCC. (B) Targeting CCDC144NL-AS1 or WDR5 suppressed tumor growth significantly, and similar results were observed in miR-940-overexpressed group. (C) Targeting CCDC144NL-AS1/WDR5 or overexpressing miR-940 could all improve the prognosis of HCC mice significantly.
Discussion
According to the recent work of the Encyclopedia of DNA elements (ENCODE), only 56 (0.1%) of the 41,204 genes showed mass spectrum evidence consistent with protein expression, indicating that most RNA transcripts are non-coding.13 The production of such a huge amount of non-coding RNA (ncRNA) transcripts suggests that RNA plays a more diverse role in biological processes than we originally expected. ncRNAs can be divided into different types according to their size. lncRNAs include transcripts of around 200 nucleotides or more.14 Although having been extensively studied in cancer biology, the function and the mechanism of action of lncRNAs remain unclear. Before our study, CCDC144NL-AS1 was reported to be involved in the carcinogenesis of gastric cancer and osteosarcoma.9,15 As for HCC, no study has investigated the role of CCDC144NL-AS1 in HCC development. In the current study, for the first time, we explored the function of CCDC144NL-AS1 in HCC development through the epigenetic pathway and found that CCDC144NL-AS1 could enhance the invasion and proliferation abilities of HCC cells by inducing WDR5 expression via sponging miR-940.
The dysregulation of epigenetic modifiers of histones and their role in hepatocarcinogenesis are being actively investigated, and have made great progress. For instance, many works have found that the overexpression of histone deacetylases (HDACs), like HDAC1 and HDAC2 in HCC tissues, is associated with increased mortality.16 Other HDACs, such as HDAC4, HDAC5, and mitochondrial deacetylase sirtuins (SIRTs), have also been reported to be upregulated in HCC tissues, and are correlated with tumor progression.17–19 The mechanisms underlying the aberrant histone modifications are not fully understood yet, but the role of specific lncRNAs and miRNAs is being elucidated.4 Besides to histone deacetylation, histone methylation is another important type of histone modification. H3K4me3, which usually marks the activation of gene transcription, is much less understood in HCC compared to histone deacetylation.12 WDR5 is an important component of the histone methyltransferase family, and it is essential for the catalysis of H3K4me3 in gene promoter.12 In this study, we found that WDR5 was regulated by CCDC144NL-AS1/miR-940 pathway, and its overexpression resulted in the high H3K4me3 level in the promoter region of MMP2, MMP9, CDK1, CDK2, and CDK4, leading to the high expression level of these genes, and further enhanced the invasion and proliferation of HCC cells.
Before our study, many documents reported the epigenetic therapy for HCC. For example, HDAC inhibitor (HDACi) is an attractive choice for the treatment of HCC that addresses different molecular mechanisms compared to chemotherapy or other targeted therapies to inhibit tumor cell growth and promote cell death.20 Targeting the histone-lysine-N-methyltransferase enhancer of zeste homolog 2 (EZH2) reactivated transcriptionally repressed chemokines genes and augmented T cell trafficking to the tumor.21 Besides to epigenetic therapy, non-coding RNAs also provide us novel targets for HCC treatment. Upregulated ncRNAs in HCC are thought to have an oncogenic function, whereas a few ncRNAs exhibiting downregulated expression in HCC may act as tumor suppressors.22,23 Notably, using the mice model, we observed that targeting CCDC144NL-AS1/WDR5 or overexpressing miR-940 could all suppress HCC growth and improve the prognosis of HCC mice, suggesting that CCDC144NL-AS1/miR-940/WDR5 pathway could serve as therapeutic targets for HCC.
Conclusion
In conclusion, in this study, we found that CCDC144NL-AS1 promotes the development of HCC by inducing WDR5 expression via sponging miR-940. The highly expressed WDR5 is recruited to the promoter of MMP2, MMP9, CDK1, CDK2, and CDK4, and further enhances the invasion and proliferation of HCC cells.
Acknowledgment
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi: 10.1016/S0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 2.Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34(2):153–159. doi: 10.1053/j.semdp.2016.12.011 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 4.Zhao J, Gray SG, Greene CM, et al. Unmasking the pathological and therapeutic potential of histone deacetylases for liver cancer. Expert Rev Gastroenterol Hepatol. 2019;13(3):247–256. doi: 10.1080/17474124.2019.1568870 [DOI] [PubMed] [Google Scholar]
- 5.Quint K, Agaimy A, Di Fazio P, et al. Clinical significance of histone deacetylases 1, 2, 3, and 7: HDAC2 is an independent predictor of survival in HCC. Virchows Arch. 2011;459(2):129–139. doi: 10.1007/s00428-011-1103-0 [DOI] [PubMed] [Google Scholar]
- 6.Feng B, Zhu Y, Su Z, et al. Basil polysaccharide attenuates hepatocellular carcinoma metastasis in rat by suppressing H3K9me2 histone methylation under hepatic artery ligation-induced hypoxia. Int J Biol Macromol. 2018;107:2171–2179. doi: 10.1016/j.ijbiomac.2017.10.088 [DOI] [PubMed] [Google Scholar]
- 7.Wahid B, Ali A, Rafique S, et al. New insights into the epigenetics of hepatocellular carcinoma. Biomed Res Int. 2017;2017:1609575. doi: 10.1155/2017/1609575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu W, Gao L, Huang C, et al. Mechanisms and importance of histone modification enzymes in targeted therapy for hepatobiliary cancers. Discov Med. 2019;28(151):17–28. [PubMed] [Google Scholar]
- 9.Fan H, Ge Y, Ma X, et al. Long non-coding RNA CCDC144NL-AS1 sponges miR-143-3p and regulates MAP3K7 by acting as a competing endogenous RNA in gastric cancer. Cell Death Dis. 2020;11(7):521. doi: 10.1038/s41419-020-02740-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y, Chen E, Tang Y, et al. miR-223 overexpression inhibits doxorubicin-induced autophagy by targeting FOXO3a and reverses chemoresistance in hepatocellular carcinoma cells. Cell Death Dis. 2019;10(11):843. doi: 10.1038/s41419-019-2053-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim TH, Yoo JY, Choi KC, et al. Loss of HDAC3 results in nonreceptive endometrium and female infertility. Sci Transl Med. 2019;11(474):7533. doi: 10.1126/scitranslmed.aaf7533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue W, Yao X, Ting G, et al. BPA modulates the WDR5/TET2 complex to regulate ERβ expression in eutopic endometrium and drives the development of endometriosis. Environ Pollut. 2021;268:115748. doi: 10.1016/j.envpol.2020.115748 [DOI] [PubMed] [Google Scholar]
- 13.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- 15.He J, Guan J, Liao S, et al. Long noncoding RNA CCDC144NL-AS1 promotes the oncogenicity of osteosarcoma by acting as a molecular sponge for microRNA-490-3p and thereby increasing HMGA2 expression. Onco Targets Ther. 2021;14:1–13. doi: 10.2147/OTT.S280912 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Freese K, Seitz T, Dietrich P, et al. Histone deacetylase expressions in hepatocellular carcinoma and functional effects of histone deacetylase inhibitors on liver cancer cells in vitro. Cancers (Basel). 2019;11(10):1587. doi: 10.3390/cancers11101587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Wang H, Shen P, et al. Expression of HDAC4 in stage B hepatocellular carcinoma and its influence on survival. Ann Clin Lab Sci. 2019;49(2):189–192. [PubMed] [Google Scholar]
- 18.Ye M, Fang Z, Gu H, et al. Histone deacetylase 5 promotes the migration and invasion of hepatocellular carcinoma via increasing the transcription of hypoxia-inducible factor-1α under hypoxia condition. Tumour Biol. 2017;39(6):1010428317705034. doi: 10.1177/1010428317705034 [DOI] [PubMed] [Google Scholar]
- 19.Zeng X, Wang N, Zhai H, et al. SIRT3 functions as a tumor suppressor in hepatocellular carcinoma. Tumour Biol. 2017;39(3):1010428317691178. doi: 10.1177/1010428317691178 [DOI] [PubMed] [Google Scholar]
- 20.He B, Dai L, Zhang X, et al. The HDAC inhibitor quisinostat (JNJ-26481585) supresses hepatocellular carcinoma alone and synergistically in combination with sorafenib by G0/G1 phase arrest and apoptosis induction. Int J Biol Sci. 2018;14(13):1845–1858. doi: 10.7150/ijbs.27661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong YK, Li Y, Pandit H, et al. Epigenetic modulation enhances immunotherapy for hepatocellular carcinoma. Cell Immunol. 2019;336:66–74. doi: 10.1016/j.cellimm.2018.12.010 [DOI] [PubMed] [Google Scholar]
- 22.Niu ZS, Niu XJ, Wang WH. Long non-coding RNAs in hepatocellular carcinoma: potential roles and clinical implications. World J Gastroenterol. 2017;23(32):5860–5874. doi: 10.3748/wjg.v23.i32.5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D, Zhang J, Li J. Role of miRNA sponges in hepatocellular carcinoma. Clin Chim Acta. 2020;500:10–19. doi: 10.1016/j.cca.2019.09.013 [DOI] [PubMed] [Google Scholar]