Methanogens from the order Methanomicrobiales are thought to prefer H2 as an electron donor for growth. They are ubiquitous in anaerobic environments, such as in wastewater treatment facilities, anaerobic digesters, and the rumen, where they catalyze the terminal steps in the breakdown of organic matter.
KEYWORDS: Methanomicrobiales, flavin-based electron bifurcation, formate dehydrogenase, heterodisulfide reductase, methanogenesis
ABSTRACT
Hydrogenotrophic methanogens produce CH4 using H2 as an electron donor to reduce CO2. In the absence of H2, many are able to use formate or alcohols as alternate electron donors. Methanogens from the order Methanomicrobiales are capable of growth with H2, but many lack genes encoding hydrogenases that are typically found in other hydrogenotrophic methanogens. In an effort to better understand electron flow in methanogens from the Methanomicrobiales, we undertook a genetic and biochemical study of heterodisulfide reductase (Hdr) in Methanoculleus thermophilus. Hdr catalyzes an essential reaction by coupling the first and last steps of methanogenesis through flavin-based electron bifurcation. Hdr from M. thermophilus copurified with formate dehydrogenase (Fdh) and only displayed activity when formate was supplied as an electron donor. We found no evidence of an Hdr-associated hydrogenase, and H2 could not function as an electron donor, even with Hdr purified from cells grown on H2. We found that cells catalyze a formate hydrogenlyase activity that is likely essential for generating the formate needed for the Hdr reaction. Together, these results highlight the importance of formate as an electron donor for methanogenesis and suggest the ability to use formate is closely integrated into the methanogenic pathway in organisms from the order Methanomicrobiales.
IMPORTANCE Methanogens from the order Methanomicrobiales are thought to prefer H2 as an electron donor for growth. They are ubiquitous in anaerobic environments, such as in wastewater treatment facilities, anaerobic digesters, and the rumen, where they catalyze the terminal steps in the breakdown of organic matter. However, despite their importance, the metabolism of these organisms remains understudied. Using a genetic and biochemical approach, we show that formate metabolism is closely integrated into methanogenesis in Methanoculleus thermophilus. This is due to a requirement for formate as the electron donor to heterodisulfide reductase (Hdr), an enzyme responsible for catalyzing essential reactions in methanogenesis by linking the initial CO2 fixing step to the exergonic terminal reaction of the pathway. These results suggest that hydrogen is not necessarily the preferred electron donor for all hydrogenotrophic methanogens and provide insight into the metabolism of methanogens from the order Methanomicrobiales.
INTRODUCTION
The methanogenic archaea (methanogens) grow exclusively by making CH4 and are responsible for the majority of global CH4 production. The three major pathways of methanogenesis are classified as hydrogenotrophic, methylotrophic, and acetoclastic based on the major substrate(s) used to generate CH4 (1, 2). Hydrogenotrophic methanogens generally specialize in CO2 reduction to CH4, methylotrophic methanogens can additionally reduce methyl compounds such as methanol, methylamines, and methylsulfides, and acetoclastic methanogens split acetate to CH4 and CO2 (2). H2 is thought to be the preferred electron donor for hydrogenotrophs, but some can also use alternative electron donors such as formate, ethanol, or secondary alcohols for CO2 reduction. Hydrogenotrophic methanogens from the order Methanomicrobiales are some of the most metabolically versatile. In addition to H2, >90% of species are experimentally confirmed to use formate and >50% are confirmed to use alcohols for methanogenesis (3). The Methanomicrobiales are common in anoxic environments such as the rumen, wastewater treatment plants, and anaerobic digesters and are model organisms for the study of syntrophic partnerships with fermentative bacteria (4). However, their metabolism has remained poorly studied compared to that of other groups.
Hydrogenotrophic methanogenesis from CO2 requires four reduction steps (5). This begins with CO2 reduction by reduced ferredoxin (Fdred) to a formyl group attached to the C1 carrier methanofuran to generate formylmethanofuran. This reaction is catalyzed by formylmethanofuran dehydrogenase (Fwd). The formyl group is transferred to a second carrier, tetrahydromethanopterin (H4MPT), and is further reduced to generate methyl-H4MPT. These two reduction steps rely on reduced coenzyme F420 (F420H2), which can be generated from H2 by F420-reducing hydrogenase (Frh) or from formate by formate dehydrogenase (Fdh). A methyltransferase transfers the methyl group of methyl-H4MPT to coenzyme M (CoM-SH), forming methyl-S-CoM. This reaction is exergonic and coupled to the buildup of electrochemical sodium ion potential across the membrane. Finally, methyl-S-CoM is further reduced with coenzyme B (CoB-SH) to generate CH4 and the heterodisulfide CoB-S-S-CoM. To regenerate CoB-SH and CoM-SH, CoB-S-S-CoM is reduced by heterodisulfide reductase (Hdr). The reduction of CoB-S-S-CoM is an exergonic reaction coupled to the endergonic reduction of Fd via flavin-based electron bifurcation (FBEB) (5–7). Fdred is necessary for the next initial reaction of CO2 reduction (6), and the coupling of CoB-S-S-CoM and Fd reductions renders the overall pathway a cycle known as the Wolfe cycle (5). A second pathway to generate Fdred relies on a membrane-bound energy-converting hydrogenase such as Eha or Ech. This Fdred facilitates CO2 fixation when methanogenic intermediates are depleted in anabolic reactions (8). The energy-converting hydrogenase-dependent pathway of Fd reduction requires H2 as an electron donor; therefore, H2 is essential for methanogenesis (8). When methanogens are grown with formate, H2 for the Eha reaction is produced via a formate hydrogenlyase activity that is dependent on formate oxidation by Fdh and H2 production by either F420-reducing hydrogenase or the combined activities of H2- and F420-dependent methylene-H4MPT dehydrogenases (the Hmd-Mtd cycle) (9, 10).
Hdr exists in a multisubunit protein complex that is a central metabolic hub for energy conservation in hydrogenotrophs. All hydrogenotrophic methanogens tested to date are capable of utilizing H2 as an electron donor to Hdr (6, 11–16). Electron pairs likely flow to a bound flavin in HdrA, the site of FBEB, and one electron is transferred to Fd and another is transferred to HdrB, the site of CoB-S-S-CoM reduction (13). When H2 is the electron donor for growth, an F420-nonreducing hydrogenase (Mvh) binds to Hdr to oxidize H2 (12). In addition to H2, Methanococcus maripaludis was shown to incorporate Fdh into the Hdr complex (8, 11) and utilize formate as an electron donor (15). In both cases, the δ subunit of Mvh (MvhD) is required as a binding site for Mvh or Fdh (17). Some members of the order Methanomicrobiales lack genes encoding the large and small subunits of the Mvh hydrogenase (MvhAG) (18) but still possess a gene encoding MvhD. In these organisms, it was proposed that FrhABG can substitute for MvhAG as the Hdr-associated hydrogenase (19), but this hypothesis remains untested.
In an effort to better understand the metabolism of methanogens from the order Methanomicrobiales, we undertook a study to characterize electron donors to Hdr in Methanoculleus thermophilus. We focus on Hdr because of its central role in methanogenesis. M. thermophilus is an excellent model due to its rapid growth under laboratory conditions, its ability to use either H2 or formate as an electron donor for methanogenesis (20), and a recently developed genetic system (21). M. thermophilus also possesses genes encoding Mvh, Frh, and Fdh, the three enzymes that have been proposed as electron donors for Hdr. Through biochemical characterization of Hdr, we show that formate is the electron donor for Hdr activity in M. thermophilus, even when cells are grown with H2 as the electron donor for growth. We found no enzymatic activity with H2, suggesting a central role for formate as an electron donor for methanogenesis.
RESULTS
Identification of hydrogenases and formate dehydrogenases in M. thermophilus.
M. thermophilus utilizes both H2 and formate as electron donors for growth (20). To assess how electrons from H2 and formate are used by M. thermophilus for methanogenesis, we analyzed the draft genome (22) for genes potentially encoding hydrogenases or formate dehydrogenases. M. thermophilus possesses two homologs of membrane-bound hydrogenases, Eha (NCBI accession numbers WP_066954065 to WP_066954673) and Ech (WP_066957700 to WP_066957709), that may participate in Fd reduction. We also found the genes that encode the Frh (WP_066956025 to WP_066956925) and Mvh (WP_066954307 to WP_066954310) hydrogenases (Fig. 1). Methanogens that grow with formate as an electron donor generally possess multiple copies of Fdh. Homology searches for genes encoding Fdh identified three putative operons. Hence, we refer to these as fdh1 (WP_066953711 and WP_066953708), fdh2 (WP_066958300 and WP_066958302), and fdh3 (WP_066958856 and WP_066958857) (Fig. 1).
FIG 1.
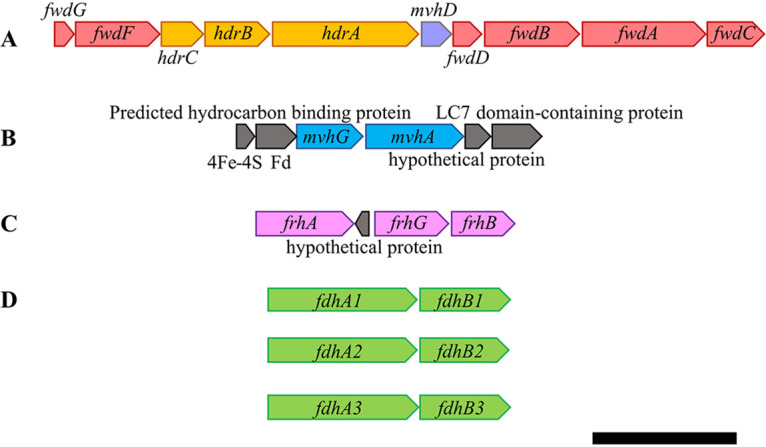
Heterodisulfide reductase, hydrogenase, and formate dehydrogenase genes in M. thermophilus. The genome organizations of genes encoding heterodisulfide reductase (Hdr) (WP_066957203 to WP_066957207) and formylmethanofuran dehydrogenase (Fwd) (WP_066957199 to WP_066957217) (A) are distinct from those of cytoplasmic hydrogenases, F420-nonreducing hydrogenase (Mvh) (WP_066954307 to WP_066954310) (B), F420-reducing hydrogenase (Frh) (WP_066956025 to WP_066956925) (C), and formate dehydrogenases (Fdh) Fdh1 (WP_066953711 to WP_066953708), Fdh2 (WP_066958300 to WP_066958302), and Fdh3 (WP_066958856 to WP_066958857) (D). The scale bar represents 2 kbp.
As F420-reducing enzymes, Frh and Fdh likely provide the reducing equivalents needed for the reduction of methenyl carbon to methyl carbon in methanogenesis: Frh when cells are grown with H2 and Fdh when cells are grown with formate. A protein complex composed of Hdr, an associated hydrogenase or formate dehydrogenase, and, in some methanogens, Fwd catalyzes the other two reductive steps of methanogenesis. While hdrABC and fwdABCDFG are coincident in a putative operon, there are no hydrogenase or fdh genes present in this putative operon that could indicate which compound serves as an electron donor to the complex (Fig. 1). It should be noted that the gene encoding MvhD is present in this putative operon; MvhD is essential for electron transfer from Mvh or Fdh to HdrA (17). The colocalization of genes encoding MvhD, HdrABC, and FwdABCDFG is consistent with their coupled activities (11).
Fdh2 copurifies with Hdr from H2-grown cultures.
Because Hdr catalyzes an essential energy-conserving step in methanogenesis, we sought to determine which substrates act as electron donors. We generated an M. thermophilus strain expressing hdrB with a C-terminal polyhistidine tag (HdrB-His) under the control of the native chromosomal promoter. HdrB was selected because it is the active site of CoB-S-S-CoM reduction and tagging this subunit in M. maripaludis did not affect growth (11). We also generated a strain with a polyhistidine tag on the α-subunit of Mvh (MvhA-His), as this enzyme has copurified with Hdr in every methanogen tested to date (6, 11–16). Cultures of M. thermophilus wild type, HdrB-His, or MvhA-His were grown with H2 as the electron donor, and cells were harvested and lysed under anoxic conditions. Anaerobic immobilized metal affinity chromatography (IMAC) was carried out to purify protein complexes. A Coomassie-stained gel showed that protein was only recovered from the HdrB-His strain, with several bands that match the expected molecular masses (kDa) of HdrA, HdrB, HdrC, and MvhD (Fig. 2). Other bands are also visible, presumably reflecting the presence of Mvh, Frh, Fdh, or Fwd subunits. The lack of visible protein bands from the wild-type (WT) and MvhA-His samples indicated that protein eluted from HdrB-His is not due to nonspecific interactions with the IMAC column.
FIG 2.
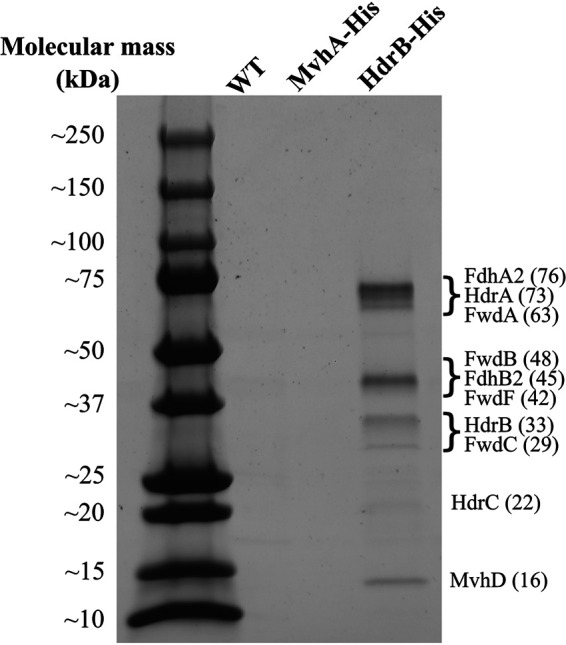
Proteins present in elution fractions after IMAC. The wild-type (WT) strain and strains with chromosomal genes encoding MvhA-His or HdrB-His were grown with H2 as the sole electron donor before lysis and purification. The predicted masses of the protein bands are indicated in parentheses (in kDa).
All samples were analyzed with mass spectrometry analysis in order to determine the composition of the purified proteins. The WT and MvhA-His elution fractions had similar compositions (see Table S1 in the supplemental material). There was not a significant increase in Mvh proteins from the MvhA-His strain; this suggests that either the His-tagged protein was not expressed, Mvh is in low abundance in cells grown on H2, or Mvh proteins could not be detected by mass spectrometry. Furthermore, these preparations did not contain a significant enrichment of Hdr proteins (Table 1). For the HdrB-His samples, we detected a significant increase in several components of the Hdr complex, including HdrA, HdrB, HdrC, MvhD, FwdC, and FwdF. FwdA and FwdB were detected in a subset of samples but were not significantly enriched across all data sets. Copurification of Hdr and Fwd does not always occur in species where this interaction is known to take place, suggesting that this interaction is transient or that these proteins readily dissociate upon purification (17).
TABLE 1.
Peptide CPT of proteins identified from His tag purifications
| Accession no. | Protein | Mass (kDa) | CPTa |
P valueb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WTc |
MvhA-Hisc |
HdrB-His |
||||||||
| Sample 1 | Sample 2 | Sample 1 | Sample 2 | Sample 1 | Sample 2 | Sample 3 | ||||
| Hdr subunits | ||||||||||
| WP_066957207.1 | HdrA | 73 | 4 | 34 | 21 | 13 | 130 | 120 | 113 | 0.00005 |
| WP_066957205.1 | HdrB | 33 | ND | 5 | 9 | ND | 33 | 31 | 28 | 0.00013 |
| WP_066957203.1 | HdrC | 22 | ND | 3 | ND | ND | 49 | 24 | 46 | 0.03625 |
| Mvh subunits | ||||||||||
| WP_066954307.1 | MvhA | 50 | ND | ND | ND | ND | ND | ND | ND | NS |
| WP_066954310.1 | MvhG | 34 | ND | ND | ND | ND | ND | ND | ND | NS |
| WP_066957209.1 | MvhD | 16 | ND | 3 | 9 | 2 | 39 | 33 | 32 | 0.00057 |
| Fdh subunits | ||||||||||
| WP_066953711.1 | FdhA1 | 76 | ND | 2 | 4 | ND | 12 | 22 | 20 | 0.02522 |
| WP_066953708.1 | FdhB1 | 46 | ND | 2 | 8 | 2 | 9 | 8 | 16 | NS |
| WP_066958300.1 | FdhA2 | 76 | 4 | 28 | 38 | 11 | 139 | 101 | 147 | 0.00516 |
| WP_066958302.1 | FdhB2 | 45 | 1 | 16 | 30 | 5 | 94 | 88 | 78 | 0.00025 |
| WP_066958856.1 | FdhA3 | 76 | ND | ND | ND | ND | ND | ND | ND | NS |
| WP_066958857.1 | FdhB3 | 45 | ND | 3 | 11 | 3 | 34 | 16 | 28 | 0.03749 |
| Fwd subunits | ||||||||||
| WP_066957215.1 | FwdA | 64 | 30 | 2 | 13 | 7 | 13 | 62 | 48 | NS |
| WP_066957213.1 | FwdB | 48 | 3 | 2 | 4 | 2 | 5 | 48 | 19 | NS |
| WP_066957217.1 | FwdC | 29 | ND | 12 | 17 | 4 | ND | 46 | 39 | 0.01092 |
| WP_066957211.1 | FwdD | 14 | ND | ND | ND | ND | ND | ND | ND | NS |
| WP_066957201.1 | FwdF | 42 | ND | 11 | 38 | 25 | 65 | 60 | 52 | 0.01117 |
| WP_066957199.1 | FwdG | 10 | ND | ND | ND | ND | ND | ND | ND | NS |
CPT, counts per thousand; ND, not detected.
Comparing HdrB-His sample to all other samples. NS, not significant (P > 0.05).
No significant differences between these two groups for proteins listed.
Interestingly, despite growing cultures with H2 as the electron donor for methanogenesis, we did not detect a hydrogenase associated with Hdr in any of the samples (Table 1). Only Fdh was detected, suggesting that only formate could serve as an electron donor to the complex. In particular, Fdh2 was significantly enriched compared to Fdh1 and Fdh3.
Hdr exhibits activity with formate, but not H2, as the electron donor.
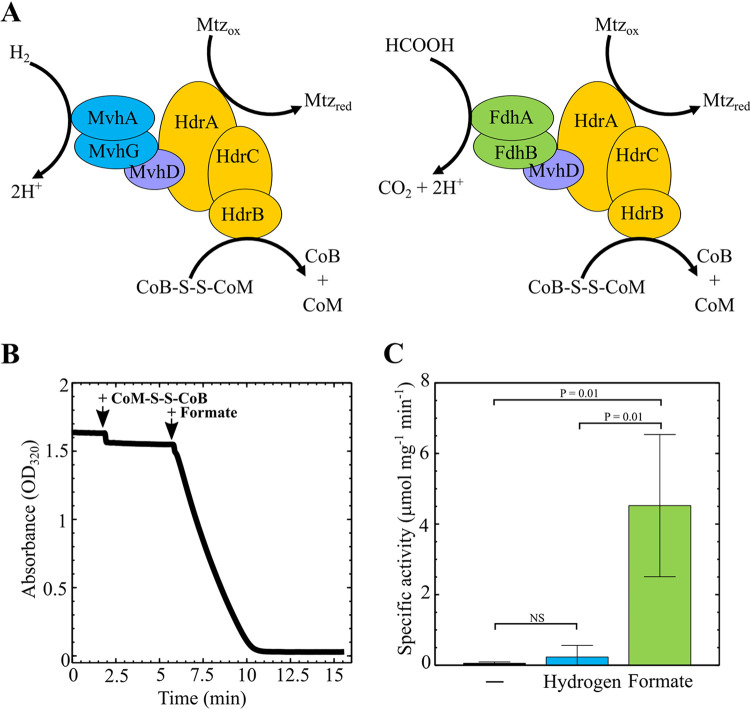
To determine if the purified Hdr complex is functional and to determine whether it is activated by H2 or formate, we carried out an in vitro activity assay with the HdrABC-FdhAB-MvhD complex purified from H2-grown cells. Four independent protein preparations were used in these experiments. Hdr catalyzes an FBEB-based reaction with both CoB-S-S-CoM and Fd required as electron acceptors. For our assay, we substituted metronidazole in place of Fd, as this is a known alternative to Fd in this assay and is easily traced spectrophotometrically (6). Activity was followed by monitoring absorbance at 320 nm to track metronidazole reduction (6). Purified protein complex was suspended in metronidazole-containing buffer under an H2 atmosphere to measure the background rate of metronidazole reduction. Under these reaction conditions, we did not observe a change in absorbance (Fig. 3). Addition of CoB-S-S-CoM to the reaction mixture did not stimulate activity, suggesting that H2 does not serve as an electron donor. This is consistent with the absence of a hydrogenase in the purified protein complex.
FIG 3.
Activity assay for purified Hdr. (A) Proposed enzyme activity with either H2 or formate as the electron donor. (B) Time course of metronidazole reduction with protein purified from strains expressing hdrB-His. The assay was performed at 55°C in 0.7-ml anaerobic cuvettes containing 0.5 ml final assay volume with 80% H2 in the headspace. The final assay mixture consisted of 1.6 M potassium phosphate (pH 7), 180 μM metronidazole, 144 μM CoB-S-S-CoM, 3 μg protein, and 30 mM sodium formate. The H2-dependent assay was started by addition of CoB-S-S-CoM. The formate-dependent assay was started by addition of sodium formate. (C) Average specific activity of metronidazole (Mtz) reduction in the presence of H2 (0.23 μmol · min−1 mg−1) or in the presence of formate (4.52 μmol · min−1 mg−1). –, the background rate of Mtz reduction without CoB-S-S-CoM. Data are averages and standard deviations from four independent experiments.
Upon addition of formate to the reaction mixture, an immediate decrease in absorbance occurred, indicating that formate is an electron donor to Hdr (Fig. 3). Over the course of several minutes, metronidazole reduction proceeded to completion. Based on the extinction coefficient of metronidazole at 320 nm (23) and the concentration of protein in the reaction mixture, the average specific activity of purified Hdr complex was determined to be 4.52 ± 2.01 μmol · min−1 mg−1 protein (Fig. 3); this rate is consistent with the CoB-S-S-CoM- and H2-dependent rate of metronidazole reduction observed with HdrABC-MvhADG protein complexes purified from Methanothermobacter marburgensis (2 to 4 μmol · min−1 mg−1) (6).
FdhA2 is sufficient for growth of M. thermophilus.
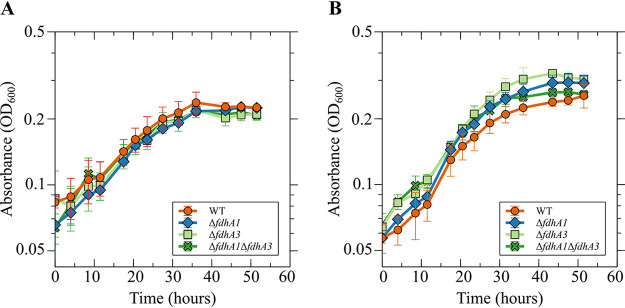
In M. maripaludis, deletion of a single formate dehydrogenase gene abolishes growth on formate (24). M. maripaludis possesses a second formate dehydrogenase that is not essential for growth on formate and may be expressed specifically under conditions when formate concentrations limit growth (25). Our genomic analysis found that M. thermophilus harbors three Fdh paralogs, and mass spectrometry suggested that FdhA2 is primarily responsible for donating electrons for Hdr activity (Fig. 1 and Table 1). Hence, we hypothesized that Fdh2 is primarily responsible for growth. To test this, we attempted to generate mutant strains lacking each fdhA gene using allelic replacement. Assuming that the fitness of a mutant carrying a deletion is similar to that of the wild type, the mutagenesis procedure should generate mutant strains at the same frequency as the wild type (i.e., 50% of recovered colonies should carry the mutant allele) (21). We were unable to isolate a ΔfdhA2 mutant, consistent with the importance of Fdh2 for CoB-S-S-CoM reduction (Table 2). Interestingly, while 50% of colonies screened for a deletion of fdhA3 contained the mutant allele, only ∼15% of colonies screened for a deletion of fdhA1 contained the mutant allele. The reason for a <50% mutant recovery rate during fdhA1 mutagenesis is unknown, but it may be a result of a fitness advantage for the wild type or differing rates of homologous recombination around the genomic locus for fdh1. Mutants lacking fdhA1, fdhA3, or both fdhA1 and fdhA3 did not exhibit a growth defect in medium with either H2 or formate as the electron donor for growth, suggesting that Fdh2 is sufficient to support growth (Fig. 4).
TABLE 2.
Number of colonies screened for fdh deletions
| Strain genotype | No. of colonies tested | No. of mutants detected | Mutant vs WT ratio |
|---|---|---|---|
| ΔfdhA1 | 40 | 5 | 0.13 |
| ΔfdhA2 | 54 | 0 | NA |
| ΔfdhA3 | 14 | 7 | 0.50 |
| ΔfdhA1 ΔfdhA3 | 40 | 6 | 0.15 |
FIG 4.
fdhA1 and fdhA3 are not essential for growth. Growth of M. thermophilus wild-type, ΔfdhA1, ΔfdhA3, and ΔfdhA1 ΔfdhA3 mutant strains with H2 (A) or formate (B) as the sole electron donor. Data are averages and standard deviations from triplicate cultures. There was not a significant difference in the specific growth rates of any of the mutant strains, suggesting that Fdh2 is sufficient for growth on both H2 and formate.
Whole cells of M. thermophilus catalyze formate hydrogenlyase activity.
The requirement of formate for Hdr activity in cultures grown with H2 raises a question of the source of formate in these cultures. Previous work in M. maripaludis identified formate hydrogenlyase activity that can convert formate to H2 (9, 10). This activity is dependent on Frh and Fdh. We hypothesized that when H2 is the only available electron donor for growth, M. thermophilus catalyzes the reverse reaction to generate formate from H2. To test this hypothesis, we suspended whole cells from an H2-grown culture to a final optical density (OD) at 600 nm of ∼0.2 in buffer containing 2-bromoethanesulfonate, a specific inhibitor of methanogenesis (26), and assayed for H2-dependent formate production. Cells were incubated under a headspace of either N2-CO2 or H2-CO2 (80:20 for each). In the presence of H2, formate concentrations increased over time, with up to 3.5 μmoles produced after 2 h (Fig. 5). No formate production was observed in the absence of H2. These data indicate that H2-grown M. thermophilus can produce the formate required for Hdr via formate hydrogenlyase activity. We hypothesize that, in analogy to other methanogens, this activity is dependent on Frh and Fdh. However, we have not been able to test this hypothesis as we were unable to generate a strain lacking all three fdhA genes (Table 2), and efforts to generate a strain lacking frh have been unsuccessful.
FIG 5.
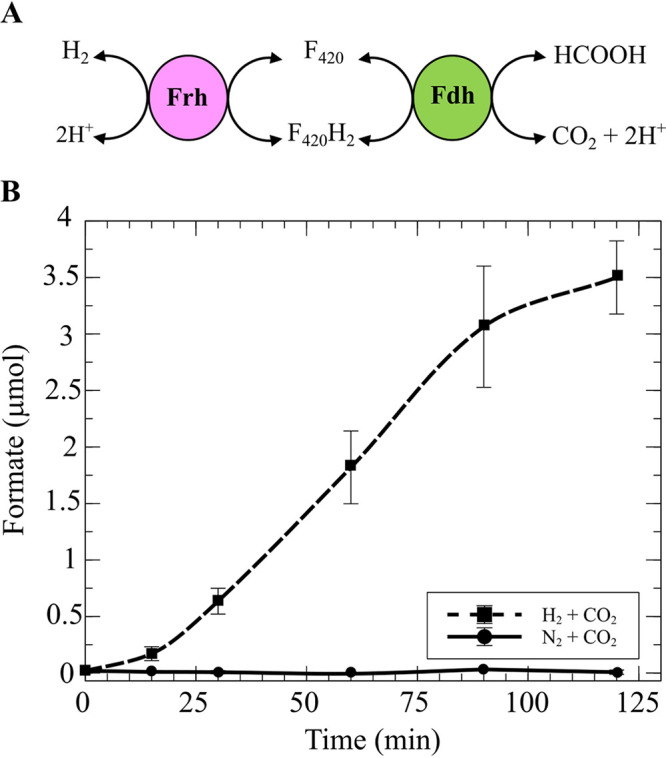
M. thermophilus exhibits formate hydrogenlyase activity. (A) Formate hydrogenlyase in hydrogenotrophic methanogens is dependent on Frh and Fdh with coenzyme F420 as an intermediate. (B) Production of formate by M. thermophilus in the presence of H2. The assay was initiated by addition of 280 kPa H2-CO2 (80:20) or N2-CO2 (80:20). At various time points, supernatant from each culture was analyzed with the EnzymeChrome formate assay kit (BioAssay System) to detect the presence of formate. Data are averages and standard deviations from triplicate cultures.
DISCUSSION
Hydrogenotrophic methanogens are thought to prefer H2 as the electron donor for methanogenesis. However, our results suggest that formate is the preferred electron donor for the essential Hdr-dependent reduction of CoB-S-S-CoM in M. thermophilus (Fig. 6). Only one other organism, M. maripaludis, is known to additionally utilize formate as a direct electron donor to Hdr; in this organism, multiple forms of the Hdr protein complex are present that are capable of utilizing either H2 (via an Mvh homolog, Vhu) or formate (via Fdh) as an electron donor (15, 17). Many members of the order Methanomicrobiales lack mvhAG, and so it has been hypothesized that Frh could substitute as an Hdr-associated hydrogenase (19, 27). While M. thermophilus possesses predicted homologs of both mvhAG and frhABG, we did not detect either hydrogenase associated with Hdr, even when MvhA was histidine tagged and targeted for purification. Additionally, we found no activity when purified Hdr was incubated with H2 as the sole electron donor. It is possible that Mvh is important in another cellular reaction or that it is not expressed. There is precedence for hydrogenase genes not being expressed in methanogens from the Methanomicrobia class. Methanosarcina acetivorans carries genes that encode several hydrogenases that are not expressed under laboratory growth conditions (28, 29).
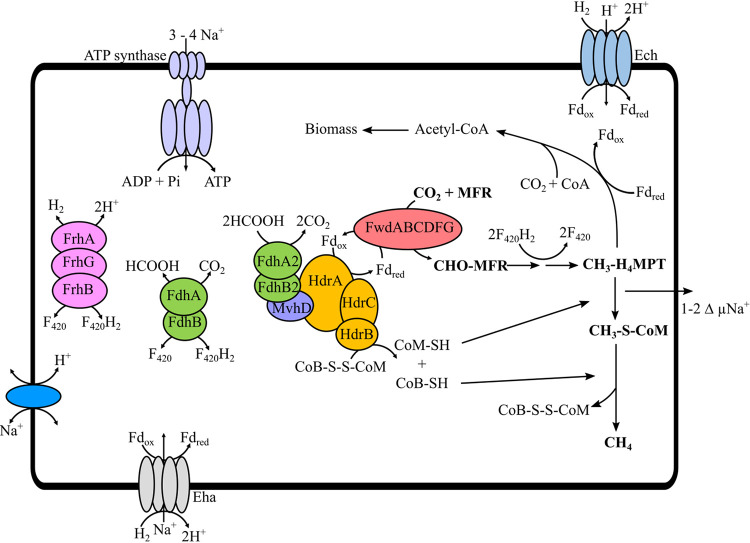
FIG 6.
Overview of the methanogenic pathway in M. thermophilus based on gene content and Hdr activity. H2 or formate can act as an electron donor for growth. The core methanogenic pathway is shown in boldface font. H2 can be generated from formate or formate can be generated from H2 via a reversible formate hydrogenlyase activity. H2 is essential for energy-converting hydrogenase (Eha/Ech)-dependent ferredoxin reduction and formate is essential for heterodisulfide reduction. Either H2 or formate can act as an electron donor for F420 reduction, via Frh or Fdh, respectively. The CH3-H4MPT:CoM methyltransferase translocates Na+ across the cell membrane. The resulting Na+ gradient is used to drive ATP synthesis via ATP synthase. CH3-H4MPT and CO2 are substrates for synthesis of acetyl coenzyme A (acetyl-CoA) with Fdred required as an electron donor.
The observation that H2-grown M. thermophilus relies on formate as an electron donor to Hdr raises a question of the source of formate in the culture medium. Hydrogenotrophic methanogens that utilize formate possess formate hydrogenlyase activity that can generate H2 and CO2 from formate (10). The reverse reaction—formate production from H2—has been observed in bacteria (30) but, to our knowledge, has not been documented in hydrogenotrophic methanogens. Methanogenic formate hydrogenlyase activity primarily requires Fdh and Frh, with coenzyme F420 acting as an intermediate electron carrier (9, 10). In some organisms, an alternative pathway for H2 production exists that is dependent on Hmd (8, 9); however, M. thermophilus lacks genes encoding Hmd. We hypothesize that formate hydrogenlyase activity is essential during growth on either H2 or formate in M. thermophilus. During growth on formate, H2 production is needed for membrane-bound hydrogenases such as Eha or Ehb (8), and during growth on H2, formate production is necessary for Hdr.
Hydrogenotrophic methanogens that utilize formate as an electron donor for growth often possess genes encoding multiple copies of Fdh in their genome, with some not essential or only activated when the correct cofactor is available (10, 24, 31). Our data suggest that Fdh2 complexes with Hdr in M. thermophilus and that Fdh2 can donate electrons for all four reductive steps of methanogenesis. The roles of Fdh1 and Fdh3 are unknown, but they may be useful for growth under nutrient-limiting conditions, as has been suggested for alternative Fdh enzymes in M. maripaludis (25).
Based on the results presented here, we hypothesize that both H2 and formate are essential for the growth of M. thermophilus: formate as an electron donor for Hdr and H2 as an electron donor to membrane-bound energy-converting hydrogenases (Eha/Ech) (Fig. 6). Thus, formate is required for catabolic reactions, and H2 is likely required to reduce ferredoxin for anabolic reactions (32). In addition to CoB-S-S-CoM and ferredoxin reduction, a source of F420H2 is required for the reduction of both methenyl-H4MPT to methylene-H4MPT and methylene-H4MPT to methyl-H4MPT. Frh is likely essential for F420 reduction in cultures grown on H2 and Fdh in cultures grown on formate. While additional work is necessary to determine the role for formate in methanogenesis in other Methanomicrobiales methanogens, many members of this group lack genes encoding Mvh (exceptions being Methanoculleus spp., Methanolacinia spp., and Methanofollis spp.), grow with formate as an electron donor, and harbor several copies of fdh on their genomes. Therefore, we hypothesize that formate plays a central role as an electron donor for methanogenesis in organisms from the order Methanomicrobiales.
MATERIALS AND METHODS
Strains, medium, and growth conditions.
Strains used in this study are listed in Table 3. M. thermophilus DSM 2373 was purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). M. thermophilus DSM 2373 and its mutants were grown anaerobically at 55°C with liquid or solid McTry medium as previously described (21). For 1 liter of liquid McTry medium, 5 g NaHCO3, 14 g NaCl, 0.5 g NH4Cl, 0.34 g KCl, 2.8 g MgCl2·6H2O, 3.5 g MgSO4·6H2O, 0.14 g CaCl2·2H2O, 0.07 g K2HPO4, 0.0095 g FeSO4, 4.8 g NaCH3COO, 2 g Casamino Acids, 4 g peptone, 4 g tryptone, and 0.001 g resazurin were added. Trace mineral and vitamin solutions were added to the medium as previously described (21). The solution was boiled under a stream of N2-CO2 (80:20) before 0.5 g dithiothreitol was added as a reductant. Five milliliters of liquid McTry medium was aliquoted into Balch tubes in a Coy anaerobic chamber (2% to 3% H2, 10% CO2, balance nitrogen in the atmosphere) before the headspace was vacuum exchanged to H2-CO2 (80:20). Then, 0.1 ml of a 0.5% (NH4)2S solution was added to all tubes immediately prior to inoculation. For 1 liter of solid McTry medium, 15 g Noble agar was added and NaHCO3 was reduced to 2 g · liter−1. Plates were poured in the anaerobic chamber after autoclaving. For growth, plates were incubated in an anaerobic incubation vessel with a reservoir containing 15 ml of 25% Na2S as a sulfur source. For growth on formate (McTry-formate), the following modifications were made to the McTry medium: the NaCl concentration was reduced to 2.92 g · liter−1, and 41.8 g · liter−1 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 7) and 13.6 g · liter−1 sodium formate were added. Liquid cultures in McTry-formate medium were pressurized with N2-CO2 (80:20) at 280 kPa followed by incubation at 55°C. For selection of transformed M. thermophilus DSM 2373, neomycin (0.25 mg · ml−1) was used, and for the counterselection against the merodiploid, 6-azauracil (0.2 mg · ml−1) was used.
TABLE 3.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description | Reference, source, or sequence (5′→3′) |
|---|---|---|
| Strains | ||
| KC58 | Wild-type M. thermophilus DSM 2373 | 20 |
| KC59 | DSM 2373 ΔfdhA1 | This study |
| KC60 | DSM 2373 ΔfdhA3 | This study |
| KC61 | DSM 2373 ΔfdhA1 ΔfdhA3 | This study |
| KC62 | DSM 2373 with C-terminal His-tagged mvhA | This study |
| KC63 | DSM 2373 with C-terminal His-tagged hdrB | This study |
| Plasmids | ||
| pCRuptneo | pCRprtneo containing upt gene | 11 |
| pJAL1 | Plasmid containing the codon-optimized high-temp neomycin resistance gene (co-htk) | Gift from Thomas Lie |
| pCRuptneo_co-htk | pCRuptneo containing the codon-optimized high-temp neomycin resistance gene (co-htk) from pJAL1 | This study |
| pCRuptΔfdhA1 | pCRuptneo_co-htk containing in-frame deletion of fdhA1 | This study |
| pCRuptΔfdhA2 | pCRuptneo_co-htk containing in-frame deletion of fdhA2 | This study |
| pCRuptΔfdhA3 | pCRuptneo_co-htk containing in-frame deletion of fdhA3 | This study |
| pCRuptHdrB-His | pCRuptneo_co-htk containing sequence for histidine tag insertion into HdrB C terminus | This study |
| pCRuptMvhA-His | pCRuptneo_co-htk containing sequence for histidine tag insertion into MvhA C terminus | This study |
| Primers | ||
| hdrB_His_upXbaI | Amplifies ∼3.2-kbp fragment upstream of the hdrB stop codon, contains XbaI cut site | CTAGTCTAGACTTGTGGCTTCTGGGAGATCTTACCTGGTCTCAACGCTACTG |
| hdrB_His_up_RV | Amplifies ∼3.2-kbp fragment upstream of the hdrB stop codon, contains 6× His sequence | ATGTGTTTGCACCTCAGTGATGGTGATGGTGATGACGACCTTCGATCAGGATCTTCTTCA |
| hdrB_His_dw_FW | Amplifies ∼3.2-kbp fragment downstream of the hdrB stop codon, contains 6× His sequence | GAAGAAGATCCTGATCGAAGGTCGTCATCACCATCACCATCACTGAGGTGCAAACACATG |
| hdrB_His_dwNotI | Amplifies ∼3.2-kbp fragment downstream of the hdrB stop codon, contains NotI cut site | GGCCGCGGCCGCGCAGACGGTTGCCGTGTTGTCCATGACCGCTCCGA |
| mvhAHis_usF_Xba | Amplifies ∼1.0-kbp fragment upstream of the mvhA stop codon, contains XbaI cut site | CTATAGGGCGAATTGGGCCCTCTAGACGCCCCGGACTTCATCC |
| mvhA_His_usR | Amplifies ∼1.0-kbp fragment upstream of the mvhA stop codon, contains 6× His sequence | CTTTCACCTCAGTGATGGTGATGGTGATGACGACCTTCGATCGTTCGTCTGACGCATTCG |
| mvhA_His_dsF | Amplifies ∼1.0-kbp fragment downstream of the mvhA stop codon, contains 6× His sequence | ATCGAAGGTCGTCATCACCATCACCATCACTGAGGTGAAAGAATGTTAAAACAGATCCTG |
| mvhAHis_dsR_Not | Amplifies ∼1.0-kbp fragment downstream of the mvhA stop codon, contains NotI cut site | GATATCCATCACACTGGCGGCCGCTCAGATTCGCCCGCCC |
| fdhA1_usF_XbaI | Amplifies ∼1.3-kbp fragment upstream of the fdhA1 start codon, contains XbaI cut site | CTATAGGGCGAATTGGGCCCTCTAGACCTCGCCCGATGCTTG |
| fdhA1KO_usR2 | Amplifies ∼1.3-kbp fragment upstream of the fdhA1 start codon | CATATCTCATGGCGCGCCCATTATTCTCCCTCTGGCAGGG |
| fdhA1KO_dsF2 | Amplifies ∼1.3-kbp fragment of fdhA1 3′ end | GAATAATGGGCGCGCCATGAGATATGGCAGGAAAAGGCGATTTAC |
| fdhA1_dsR2_NotI | Amplifies ∼1.3-kbp fragment of fdhA1 3′ end, contains NotI cut site | GATATCCATCACACTGGCGGCCGCTCAGCGAAAGATATCAAAGATCTGG |
| fdhA2_usF_XbaI | Amplifies ∼1.0-kbp fragment upstream of the fdhA2 start codon, contains XbaI cut site | CTATAGGGCGAATTGGGCCCTCTAGACTATTTATACCGCTGATCCCGCTC |
| fdhA2KO_usR | Amplifies ∼1.0-kbp fragment upstream of the fdhA2 start codon | GCCATTTTCATGGCGCGCCCATGGTAAAACCTCGGATACACAAATATG |
| fdhA2KO_dsF | Amplifies ∼0.8-kbp fragment downstream of the fdhA2 stop codon | GTTTTACCATGGGCGCGCCATGAAAATGGCAGCAAAAGGCGAC |
| fdhA2_dsR_NotI | Amplifies ∼0.8-kbp fragment downstream of the fdhA2 stop codon, contains NotI cut site | GATATCCATCACACTGGCGGCCGCCTTTGCCGCGGATCTCGATAC |
| fdhA3_usF_XbaI | Amplifies ∼1.0-kbp fragment upstream of the fdhA3 start codon, contains XbaI cut site | CTATAGGGCGAATTGGGCCCTCTAGATCAATATGCTATTTTTTCGTAATAGGACTG |
| fdhA3KO_usR | Amplifies ∼1.0-kbp fragment upstream of the fdhA3 start codon | CCATCTCATGGCGCGCCCATTCACATTCTCCAAATTGTTGCG |
| fdhA3KO_dsF | Amplifies ∼0.8-kbp fragment downstream of the fdhA3 stop codon | GAATGTGAATGGGCGCGCCATGAGATGGTAGCAAAGGGAGATATGG |
| fdhA3_dsR_NotI | Amplifies ∼0.8-kbp fragment downstream of the fdhA3 stop codon, contains NotI cut site | GATATCCATCACACTGGCGGCCGCGTTCTCGACCTTGCCGC |
For growth rate calculations, triplicate cultures were grown in reduced medium with either H2 or formate as a sole electron donor. Growth was initiated with a 2% (vol/vol) inoculum from an overnight culture. Optical density at 600 nm (OD600) was monitored on a Genesys 30 visible spectrophotometer (Thermo Scientific) blanked with an uninoculated tube of McTry or McTry-formate medium.
Plasmid construction.
Plasmids and oligonucleotides used to construct the recombinant plasmids are listed in Table 3. All PCRs were carried out using Phusion high-fidelity DNA polymerase (New England Biolabs). All recombinant plasmids were generated by restriction cloning or Gibson assembly (33) using enzymes/reagents from New England Biolabs. Recombinant plasmid transformation into Escherichia coli and selection were performed as previously described (21). All constructs were sequence verified by Sanger sequencing at the University of Minnesota Genomics Center (UMGC).
To generate M. thermophilus carrying a chromosomal copy of hdrB with a C-terminal histidine tag, 3,200-bp DNA fragments upstream and downstream of the stop codon of hdrB were PCR amplified with primers encoding the histidine tag. Fragments were fused via fusion PCR, and the resulting 6,400-bp fragment was digested with XbaI and NotI and ligated into XbaI/NotI-digested pCRuptneo_co-htk using T4 DNA ligase. A 6,400-bp construct was used to ensure the resulting plasmid could integrate into the M. thermophilus genome without interrupting expression of essential genes in the hdrB operon (Fig. 1). The ligated plasmid was transferred to E. coli via electroporation and plated on lysogeny broth solidified with 1.5% agar and ampicillin (50 μg · ml−1). Colonies were screed by PCR followed by restriction digestion of the PCR product using HphI to identify mutants. The plasmid was transferred to M. thermophilus via natural transformation. A similar approach was employed to generate an mvhA C-terminal His tag, except the PCR to amplify regions flanking mvhA generated fused fragments of ∼2,000 bp to ensure the resulting plasmid could integrate into the M. thermophilus genome without interrupting expression of genes in the mvhA operon (Fig. 1).
To generate the deletion constructs for fdhA1, fdhA2, and fdhA3, ∼1,000-bp fragments flanking the region of interest were PCR amplified, fused, and cloned into pCRuptneo_co-htk as described above. After transformation into M. thermophilus, potential mutants were screened by PCR. To generate the ΔfdhA1 ΔfdhA3 double deletion mutant, the construct for deletion of fdhA1 was introduced into the ΔfdhA3 background, and mutants were selected.
Transformation of M. thermophilus DSM 2393.
Cultures for transformation were inoculated and transformed using a natural transformation method as described previously (21). Briefly, the cultures were grown to an OD600 of 0.2 to 0.3. At this point, DNA was added anaerobically, tubes were repressurized with H2-CO2, and cultures were incubated overnight to allow for plasmid uptake. After overnight incubation, cultures were subinoculated and selected by growth in 5 ml McTry containing neomycin; this resulted in a merodiploid with the deletion plasmid integrated into the genome. Merodiploids were resolved by growth without selection for 48 h to enable loss of the vector in a subset of the population, 10-fold serially diluted, plated onto solid McTry medium containing 6-azauracil, and incubated for 5 to 7 days. Multiple colonies were PCR screened for the mutation of interest. Mutants were verified to have acquired neomycin sensitivity. For long-term storage, 3 ml of culture was mixed with 2 ml 50% glycerol under an N2 atmosphere and stored at −80°C.
Immobilized metal affinity chromatography.
Cell pellets of M. thermophilus wild type, MvhA-His, and HdrB-His were used for protein purification with all procedures performed anaerobically in a room temperature Coy anaerobic chamber (2 to 3% H2, balance N2 in the atmosphere). Two 200-ml cultures of each strain were grown in 1-liter anaerobic Schott bottles with an H2-CO2 (80:20) headspace pressurized to 138 kPa. Upon reaching an OD600 of ∼0.2, cultures were pooled and cells were anaerobically harvested by centrifugation at 4,000 × g for 10 min. The cell pellet was resuspended in 3 ml binding buffer (containing 100 mM NaCl, 12.5 mM MgCl2, 25 mM HEPES [pH 7.5], 10 mM imidazole, 0.5 mM dithionite, and 20 μM flavin adenine dinucleotide), and then 2 ml 50% glycerol was added. Samples were stored at −80°C for up to 4 weeks before use.
For protein purification, the frozen cell pellet was brought into a Coy anaerobic chamber (2 to 3% H2 atmosphere, balance N2), thawed on ice, and transferred to a 15-ml conical tube. Additional binding buffer was added to the sample until the volume reached 6 ml. The suspension was sonicated using a QSonica model XL-2000 sonicator at a setting of 4 for ∼3 s, followed by a 10-s rest on ice for a total of 10 pulses. Debris was removed by centrifugation at 16,000 × g for 10 min. The supernatant was added to nickel-chelating resin (G-Biosciences) and allowed to bind for 1 h at room temperature with gentle mixing by inverting the tube every 10 min. The cell lysate-resin suspension was poured into a disposable gravity flow column (Thermo Scientific), and the flowthrough was collected. The column was washed three times with 5 ml binding buffer followed by three washes with 5 ml wash buffer (same components as binding buffer except with 30 mM imidazole). Finally, proteins were eluted with 2.5 ml elution buffer (same components as binding buffer except with 300 mM imidazole). The eluate was concentrated using a Vivaspin 500 column (3-kDa molecular weight cutoff) by centrifuging for 20 min at 13,000 rpm.
Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue. Samples were also analyzed by mass spectrometry. The concentrated eluate for MvhA-His and HdrB-His samples was biochemically assayed to determine the specific enzyme activity with either H2 or formate as the electron donor.
Mass spectrometry analysis of WT, MvhA-His, and HdrB-His copurified proteins.
Each concentrated protein sample (10 μl) of two biological replicates for WT and MvhA-His as well as three biological replicates from HdrB-His was mixed with 10 μl of 4× SDS load buffer and heated for 10 min at 95°C. The samples were loaded on a 10% Bio-Rad Criterion Tris-HCl gel and run at 25 mA constant current for 35 min. The gel was stained with Thermo Scientific’s Imperial Protein stain. Stained gel regions for each sample were excised and proteolytically digested with trypsin as previously described (34), except that iodoacetamide was used instead of methyl methanethiosulfonate during the reduction and alkylation step of the digestion. Extracted peptides were dried in vacuo and then cleaned with a C18 stage tip (35).
The dried peptide pellets were resuspended in load solvent (97.99:2:0.01, water/acetonitrile/formic acid), and approximately 0.2 to 0.5 μg of material was loaded on the LTQ Orbitrap Velos mass spectrometer (Thermo Scientific) as previously described (36) with the following revisions: the liquid chromatography (LC) gradient was 2% to 5% B solvent from 0 to 2 min and 5% to 30% B solvent from 2 to 67 min with a flow rate of 330 nl/min; lock mass was not invoked; MS1 survey scan was 380 to 1,800 m/z; dynamic exclusion list size was 200, duration was 45 s, and window was ±15 ppm; the top 12 most intense ions were selected for MS2 fragmentation; and MS1 maximum injection time (IT) was 150 ms while MS2 maximum IT was 200 ms.
The tandem mass spectrometry (MS/MS) data were analyzed using Sequest (37) (version ISE 1.1.0.189, ×64 in Proteome Discoverer 2.4.0.305; Thermo Fisher Scientific, San Jose, CA, USA). Sequest was set up to search the Methanoculleus thermophilus (taxon identifier [ID] 2200) reference sequence protein database downloaded from NCBI on 21 December 2019 after concatenation of the common lab contaminants' protein sequences from https://www.thegpm.org/crap/. The total number of protein sequences was 2,256. The search parameters included trypsin enzyme with full specificity, fragment ion mass tolerance of 0.1 Da, precursor ion tolerance of 20 ppm, carbamidomethyl cysteine as a fixed amino acid modification, acetylation of protein N terminus, and protein N-terminal loss of methionine or acetylated methionine, oxidation of methionine, pyroglutamic acid modification of glutamine, and asparagine deamidation as variable modifications.
Scaffold (version 4.9; Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 91.0% probability to achieve a false-discovery rate (FDR) of less than 1.0% by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 53.0% probability to achieve an FDR of less than 1.0% and contained at least 2 identified peptides. Protein identity probabilities were assigned by the Protein Prophet algorithm (38). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
The resulting MS/MS data from Scaffold were analyzed using R (version 3.6.2) (39). The total spectral counts (SC) corresponding to each protein were normalized by multiplying each protein total SC by its protein identity probability and dividing by the whole total spectral counts of the respective sample, which was then divided by 1,000, resulting in a counts per thousand (CPT) ratio for each protein accession number. Proteins with a maximum CPT value of less than 20 across samples were removed from the analysis. Significant differences of mean CPT values (P < 0.05) were assessed using a two-sample t test comparing HdrB-His to the combined MvhA-His and WT-His samples. Data from mass spectrometry experiments can be found in Table S1 in the supplemental material.
In vitro flavin-based electron bifurcation assay.
The flavin-based electron bifurcation assay was carried out in 0.7-ml UV quartz semi-micro cuvettes with a screw cap (FireflySci). All reagents used were anaerobically prepared. The assay mixture consisted of 1.6 M potassium phosphate (pH 7), 180 μM metronidazole, 144 μM CoB-S-S-CoM, 1 to 3 μg protein from concentrated His tag elution fractions, and 30 mM formate. CoB-S-S-CoM was synthesized by the Good Manufacturing Practices Services Core at the Institute for Therapeutics Discovery and Development at the University of Minnesota. Initially, potassium phosphate buffer (pH 7), metronidazole, and protein sample were added to the cuvette (0.5-ml volume) inside a Coy anaerobic chamber (2% to 3% H2, balance N2 in the atmosphere). The cuvette was sealed with the supplied screw cap fitted with a butyl rubber stopper. The cuvette headspace was flushed with H2 gas mix (0.8 atm) and pressurized to 140 kPa (6, 15). The assay was carried out in a preheated (55°C) Cary 3500 UV-visible (UV-Vis) spectrophotometer (Agilent). The cuvette was allowed to equilibrate for 10 min before the assay was started. The reaction proceeded for ∼2 min to establish a baseline reading; then CoB-S-S-CoM was added using a Hamilton syringe, and data were collected for ∼4 min (H2 as an electron donor). Finally, sodium formate was added using a Hamilton syringe, and data were collected for ∼14 min. Absorbance was measured at 320 nm. The specific activity of metronidazole reduction was calculated from the first ∼10% of the reaction curve as follows. The rate of absorbance change (Abs min−1) was divided by the extinction coefficient of metronidazole at 320 nm (ε = 9,300 M−1 cm−1). The resulting value was multiplied by 0.0005 liters and divided by the amount of protein in the sample (milligrams) to obtain the specific activity in micromoles per minute per milligram.
Formate hydrogenlyase activity assay.
M. thermophilus was grown to an OD600 of 0.1 to 0.2 and anaerobically harvested by centrifugation at 2,500 × g for 10 min. The cell pellet was washed once with 5 ml of assay buffer (50 mM morpholinepropanesulfonic acid [MOPS] [pH 7.0], 250 mM NaCl, 20 mM KCl, 20 mM MgCl2, 1 mM CaCl2, 5 mM dithiothreitol [DTT], and 1 mM bromoethanesulfonate) and resuspended in the same volume of assay buffer. The tubes containing the cell suspension were flushed with N2-CO2 (80:20) for 20 min to remove residual H2 from the headspace. At time zero, a sample was taken from each tube, and the assay was initiated by addition of 140 kPa H2-CO2 (80:20) or N2-CO2 (80:20). Samples were incubated at 55°C with shaking at 250 rpm. At 15, 30, 60, 90, and 120 min, 100 μl of liquid was sampled from each tube and centrifuged to remove the cells. The supernatant was analyzed with the EnzymeChrome formate assay kit (BioAssay System) according to the manufacturer’s instructions.
Data accessibility.
Spectral data for mass spectrometry experiments has been archived and are freely available at the Data Repository for University of Minnesota (DRUM) (https://doi.org/10.13020/g0xp-gf10).
Supplementary Material
ACKNOWLEDGMENTS
We thank Thomas Niehaus for use of the spectrophotometer for enzyme assays and the University of Minnesota Center for Mass Spectrometry and Proteomics for assistance in data analysis.
This work was supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences, under grant number DE-SC0019148.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 2.Costa KC, Leigh JA. 2014. Metabolic versatility in methanogens. Curr Opin Biotechnol 29:70–75. doi: 10.1016/j.copbio.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Oren A. 2014. Other major lineages of bacteria and the archaea, p 195–290. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes, 4th ed. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 4.Maegaard K, Garcia-Robledo E, Kofoed MVW, Agneessens LM, de Jonge N, Nielsen JL, Ottosen LDM, Nielsen LP, Revsbech NP. 2019. Biogas upgrading with hydrogenotrophic methanogenic biofilms. Bioresour Technol 287:121422. doi: 10.1016/j.biortech.2019.121422. [DOI] [PubMed] [Google Scholar]
- 5.Thauer RK. 2012. The Wolfe cycle comes full circle. Proc Natl Acad Sci U S A 109:15084–15085. doi: 10.1073/pnas.1213193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaster A-K, Moll J, Parey K, Thauer RK. 2011. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci U S A 108:2981–2986. doi: 10.1073/pnas.1016761108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckel W, Thauer RK. 2013. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim Biophys Acta 1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Lie TJ, Costa KC, Lupa B, Korpole S, Whitman WB, Leigh JA. 2012. Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc Natl Acad Sci U S A 109:15473–15478. doi: 10.1073/pnas.1208779109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrickson EL, Leigh JA. 2008. Roles of coenzyme F420-reducing hydrogenases and hydrogen- and F420-dependent methylenetetrahydromethanopterin dehydrogenases in reduction of F420 and production of hydrogen during methanogenesis. J Bacteriol 190:4818–4821. doi: 10.1128/JB.00255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupa B, Hendrickson EL, Leigh JA, Whitman WB. 2008. Formate-dependent H2 production by the mesophilic methanogen Methanococcus maripaludis. Appl Environ Microbiol 74:6584–6590. doi: 10.1128/AEM.01455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa KC, Wong PM, Wang T, Lie TJ, Dodsworth JA, Swanson I, Burn JA, Hackett M, Leigh JA. 2010. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc Natl Acad Sci U S A 107:11050–11055. doi: 10.1073/pnas.1003653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stojanowic A, Mander GJ, Duin EC, Hedderich R. 2003. Physiological role of the F420-non-reducing hydrogenase (Mvh) from Methanothermobacter marburgensis. Arch Microbiol 180:194–203. doi: 10.1007/s00203-003-0577-9. [DOI] [PubMed] [Google Scholar]
- 13.Wagner T, Koch J, Ermler U, Shima S. 2017. Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science 357:699–703. doi: 10.1126/science.aan0425. [DOI] [PubMed] [Google Scholar]
- 14.Yan Z, Ferry JG. 2018. Electron bifurcation and confurcation in methanogenesis and reverse methanogenesis. Front Microbiol 9:1322. doi: 10.3389/fmicb.2018.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milton RD, Ruth JC, Deutzmann JS, Spormann AM. 2018. Methanococcus maripaludis employs three functional heterodisulfide reductase complexes for flavin-based electron bifurcation using hydrogen and formate. Biochemistry 57:4848–4857. doi: 10.1021/acs.biochem.8b00662. [DOI] [PubMed] [Google Scholar]
- 16.Setzke E, Hedderich R, Heiden S, Thauer RK. 1994. H2:heterodisulfide oxidoreductase complex from Methanobacterium thermoautotrophicum. Composition and properties. Eur J Biochem 220:139–148. doi: 10.1111/j.1432-1033.1994.tb18608.x. [DOI] [PubMed] [Google Scholar]
- 17.Costa KC, Lie TJ, Xia Q, Leigh JA. 2013. VhuD facilitates electron flow from H2 or formate to heterodisulfide reductase in Methanococcus maripaludis. J Bacteriol 195:5160–5165. doi: 10.1128/JB.00895-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson I, Ulrich LE, Lupa B, Susanti D, Porat I, Hooper SD, Lykidis A, Sieprawska-Lupa M, Dharmarajan L, Goltsman E, Lapidus A, Saunders E, Han C, Land M, Lucas S, Mukhopadhyay B, Whitman WB, Woese C, Bristow J, Kyrpides N. 2009. Genomic characterization of Methanomicrobiales reveals three classes of methanogens. PLoS One 4:e5797. doi: 10.1371/journal.pone.0005797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thauer RK, Kaster A-K, Goenrich M, Schick M, Hiromoto T, Shima S. 2010. Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu Rev Biochem 79:507–536. doi: 10.1146/annurev.biochem.030508.152103. [DOI] [PubMed] [Google Scholar]
- 20.Rivard CJ, Smith PH. 1982. Isolation and characterization of a thermophilic marine methanogenic bacterium, Methanogenium thermophilicum sp. nov. Int J Syst Bacteriol 32:430–436. doi: 10.1099/00207713-32-4-430. [DOI] [Google Scholar]
- 21.Fonseca DR, Halim MFA, Holten MP, Costa KC. 2020. Type IV-like pili facilitate transformation in naturally competent archaea. J Bacteriol 202:e00355-20. doi: 10.1128/JB.00355-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narihiro T, Kusada H, Yoneda Y, Tamaki H. 2016. Draft genome sequences of Methanoculleus horonobensis strain JCM 15517, Methanoculleus thermophilus strain DSM 2373, and Methanofollis ethanolicus strain JCM 15103, hydrogenotrophic methanogens belonging to the family Methanomicrobiaceae. Genome Announc 4:e00199-16. doi: 10.1128/genomeA.00199-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JS, Blanchard DK. 1979. A simple hydrogenase-linked assay for ferredoxin and flavodoxin. Anal Biochem 93:216–222. doi: 10.1016/S0003-2697(79)80140-2. [DOI] [PubMed] [Google Scholar]
- 24.Wood GE, Haydock AK, Leigh JA. 2003. Function and regulation of the formate dehydrogenase genes of the methanogenic archaeon Methanococcus maripaludis. J Bacteriol 185:2548–2554. doi: 10.1128/jb.185.8.2548-2554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa KC, Yoon SH, Pan M, Burn JA, Baliga NS, Leigh JA. 2013. Effects of H2 and formate on growth yield and regulation of methanogenesis in Methanococcus maripaludis. J Bacteriol 195:1456–1462. doi: 10.1128/JB.02141-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunsalus RP, Romesser JA, Wolfe RS. 1978. Preparation of coenzyme M analogues and their activity in the methyl coenzyme M reductase system of Methanobacterium thermoautotrophicum. Biochemistry 17:2374–2377. doi: 10.1021/bi00605a019. [DOI] [PubMed] [Google Scholar]
- 27.Gilmore SP, Henske JK, Sexton JA, Solomon KV, Seppälä S, Yoo JI, Huyett LM, Pressman A, Cogan JZ, Kivenson V, Peng X, Tan Y, Valentine DL, O'Malley MA. 2017. Genomic analysis of methanogenic archaea reveals a shift towards energy conservation. BMC Genomics 18:639. doi: 10.1186/s12864-017-4036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lessner DJ, Li L, Li Q, Rejtar T, Andreev VP, Reichlen M, Hill K, Moran JJ, Karger BL, Ferry JG. 2006. An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics. Proc Natl Acad Sci U S A 103:17921–17926. doi: 10.1073/pnas.0608833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guss AM, Kulkarni G, Metcalf WW. 2009. Differences in hydrogenase gene expression between Methanosarcina acetivorans and Methanosarcina barkeri. J Bacteriol 191:2826–2833. doi: 10.1128/JB.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinske C, Sargent F. 2016. Exploring the directionality of Escherichia coli formate hydrogenlyase: a membrane-bound enzyme capable of fixing carbon dioxide to organic acid. Microbiologyopen 5:721–737. doi: 10.1002/mbo3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones JB, Stadtman TC. 1981. Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J Biol Chem 256:656–663. [PubMed] [Google Scholar]
- 32.Major TA, Liu Y, Whitman WB. 2010. Characterization of energy-conserving hydrogenase B in Methanococcus maripaludis. J Bacteriol 192:4022–4030. doi: 10.1128/JB.01446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang R-Y, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi Z-Q, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison CA, Smith HO, Venter JC. 2010. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 34.Thu YM, Van Riper SK, Higgins L, Zhang T, Becker JR, Markowski TW, Nguyen HD, Griffin TJ, Bielinsky AK. 2016. Slx5/slx8 promotes replication stress tolerance by facilitating mitotic progression. Cell Rep 15:1254–1265. doi: 10.1016/j.celrep.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rappsilber J, Ishihama Y, Mann M. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 36.Lin-Moshier Y, Sebastian PJ, Higgins L, Sampson ND, Hewitt JE, Marchant JS. 2013. Re-evaluation of the role of calcium homeostasis endoplasmic reticulum protein (CHERP) in cellular calcium signaling. J Biol Chem 288:355–367. doi: 10.1074/jbc.M112.405761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eng JK, McCormack AL, Yates JR. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 38.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 39.R Core Development Team. 2019. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Spectral data for mass spectrometry experiments has been archived and are freely available at the Data Repository for University of Minnesota (DRUM) (https://doi.org/10.13020/g0xp-gf10).