A significant barrier to implementing renewable chemical technologies is high production costs relative to those for petroleum-derived products. Existing technologies using engineered organisms have difficulty competing with petroleum-derived chemicals due to the cost of feedstocks (such as glucose), product extraction, and purification.
KEYWORDS: isoprene, methanogen, archaea, isoprenogen, mevalonate, ispS, Methanosarcina acetivorans, Methanosarcina barkeri, methanogenesis, isoprenoids, synthetic biology, metabolic engineering, mevalonate pathway
ABSTRACT
Isoprene is a valuable petrochemical used for a wide variety of consumer goods, such as adhesives and synthetic rubber. We were able to achieve a high yield of renewable isoprene by taking advantage of the naturally high-flux mevalonate lipid synthesis pathway in anaerobic methane-producing archaea (methanogens). Our study illustrates that by genetically manipulating Methanosarcina species methanogens, it is possible to create organisms that grow by producing the hemiterpene isoprene. Mass balance measurements show that engineered methanogens direct up to 4% of total carbon flux to isoprene, demonstrating that methanogens produce higher isoprene yields than engineered yeast, bacteria, or cyanobacteria, and from inexpensive feedstocks. Expression of isoprene synthase resulted in increased biomass and changes in gene expression that indicate that isoprene synthesis depletes membrane precursors and redirects electron flux, enabling isoprene to be a major metabolic product. Our results demonstrate that methanogens are a promising engineering chassis for renewable isoprene synthesis.
IMPORTANCE A significant barrier to implementing renewable chemical technologies is high production costs relative to those for petroleum-derived products. Existing technologies using engineered organisms have difficulty competing with petroleum-derived chemicals due to the cost of feedstocks (such as glucose), product extraction, and purification. The hemiterpene monomer isoprene is one such chemical that cannot currently be produced using cost-competitive renewable biotechnologies. To reduce the cost of renewable isoprene, we have engineered methanogens to synthesize it from inexpensive feedstocks such as methane, methanol, acetate, and carbon dioxide. The “isoprenogen” strains we developed have potential to be used for industrial production of inexpensive renewable isoprene.
INTRODUCTION
Isoprene (2-methyl-1,3-butadiene, C5H8) is a valuable chemical used to synthesize synthetic rubber, styrene-isoprene-styrene (SIS block) copolymer adhesives, flavorings, cosmetics, and pharmaceuticals. Approximately 800,000 tons of isoprene is refined from petroleum annually, in which over 95% of it is used to produce cis-1,4-polyisoprene (synthetic rubber) (1). The global market for isoprene, including natural and synthetic polyisoprene rubber, is estimated at 1.3 metric tons per year, at a value approaching $4.3 billion (2). Cost-effective, high-yield synthesis of renewable isoprene from biomass feedstocks has the potential to supplant the need for petroleum-derived isoprene and would contribute to reducing use of fossil fuels.
In addition to the cis-polyisoprene natural rubber secreted in tree sap, isoprene monomer or isoprenoid lipids can be naturally synthesized by various species of plants and microbes (3–5). It is estimated some plant species channel up to 10% of total fixed carbon into isoprene, which transpires through photosynthetic tissues (6).
To obtain industrial quantities of renewable isoprene, synthetic biologists have introduced the isoprene synthase ispS gene from plants into microbial host organisms such as Escherichia coli (3, 7), Saccharomyces yeast (8, 9), and Synechocystis cyanobacteria (10). Synthesis of isoprene by engineered microbes or algae is advantageous over natural plant isoprene synthesis because microbes can be grown in enclosed bioreactors that facilitate isoprene recovery from the gas phase. Because the isoprene monomer has a low vapor point (35°C) and evaporates rapidly at room temperature, it can be easily captured from the gas phase of a microbial culture. While efforts to engineer isoprene monomer synthesis using microbes have been successful at small scales, there are remaining issues with cost of production and yield optimization (11). Three factors limit industrial-scale renewable isoprene technologies: scale-up development costs, production costs, and metabolic flux of the chassis organism. We hypothesized that using methane-producing archaea as a chassis could simultaneously solve all three of these limiting factors.
Methane-producing archaea (methanogens) are strict anaerobes that use gaseous or liquid C1 substrates or acetate to grow, converting 60 to 99% of C to pure methane gas (12). Technologies for growing methanogens at industrial scale are already well established (13). Methanogens are currently used in large-scale anaerobic digesters worldwide, where waste biomass is used to produce methane-containing biogas that is recovered, upgraded, and compressed to be used to generate electricity and transportation fuel. Production costs for methanogen-based technologies are also very low. When cultivated in anaerobic digesters at large scale, methanogens do not require light or aeration, and substrates for methanogenesis (CO, CO2, methanol, acetate, etc.) are inexpensive and abundant. Methanosarcina species have been coaxed to grow on other substrates (14), to synthesize bioproducts such as lactate in the reverse methanotrophic direction (15), and to increase tolerance to oxygen exposure (16, 17). Hydrogenotrophic Methanococcus strains have also been engineered to produce geraniol, a monoterpene derived from the mevalonate pathway for use as a fragrance (Table 1). Combined with an expanding list of available genetic tools, methanogens are emerging as viable chassis for bioproduct synthesis from inexpensive feedstocks. Due to the low feedstock cost, high yield, and existing anaerobic digestion infrastructure, methanogens have the potential to be an efficient and adaptable platform for renewable isoprene synthesis.
TABLE 1.
Comparison of isoprene and terpenoid yields in engineered bacteria and archaea
| Organism | Yield (μmol mol−1 g−1) | Yield (μmol liter−1) | Condition | Citation |
|---|---|---|---|---|
| Batch culture yieldsa | ||||
| Clostridium ljungdahliib | 0.000147 | 0.21 | MES-fructose, batch | (60) |
| Synechocyctisb | 4.93 | not reported | Light + CO2, batch | (61) |
| Methanococcus maripaludisc | 41.1 | not reported | H2 + CO2, batch (geraniol) | (39) |
| Methanococcus maripaludisc | 67.53 | not reported | Formate, batch (geraniol) | (39) |
| Methanosarcina acetivorans | 886.67 | 855.01 | Methanol, batch | This study |
| Fed-batch yields | ||||
| Escherichia coli | not reported | 352,319.4 | 0.1–2% glucose, fed-batch | (45) |
| Saccharomyces cerevisiaed | not reported | 174,691.7 | 25 g liter−1 initial glucose, fed-batch | (46) |
Batch culture yields were reported in mg isoprene per mol substrate per g dry cell weight (DCW) or mg isoprene per liter.
Strains contain two or more mutations to enhance isoprene production.
Values are for the terpene geraniol.
The rate of feed solution and final concentration of glucose utilized were not reported.
The substrate for the isoprene synthase enzyme, IspS, is dimethylallyl pyrophosphate (DMAPP), an isomer of isopentenyl diphosphate (IPP) (3). IPP/DMAPP is synthesized by one of two known biochemical pathways, the 2-C-methyl-d-erythritol 4-phosphate/1-deoxy-d-xylulose 5-phosphate (MEP/DOXP) pathway or the mevalonate pathway (Fig. 1). The two pathways differ in starting substrates (the branch point from central metabolism), enzyme steps, substrate intermediates, and energetic requirements, particularly due to a variation in the conversion of mevalonate to IPP. The MEP/DOXP pathway is primarily used by bacteria, while the mevalonate pathway is used by eukaryotes and archaea. The MEP/DOXP pathway uses 8 enzymes, 2 NADPH, 2 CTP, ATP, and reduced ferredoxin (2 e−) to produce DMAPP from pyruvate. The mevalonate pathway used by eukaryotes and archaea requires 2 or 3 ATP and 2 NADPH (18–21). Using the MEP/DOXP pathway results in a substrate pool IPP:DMAPP ratio of 5:1, and thus isoprene synthesis using this pathway also necessitates increased activity of isopentenyl diphosphate isomerase idi (22). The mevalonate pathway uses seven or eight enzymes to synthesize DMAPP from acetyl-CoA and results in a more favorable IPP:DMAPP ratio of 3:7. It has been found in previous attempts to produce isoprenoids at an industrial scale that the mevalonate pathway produces superior yields (23). The yeast Saccharomyces cerevisiae has been engineered to synthesize isoprene; unfortunately, isoprene yields were low because eukaryote enzymes are feedback-inhibited (24, 25). Thus, efforts to increase flux through the mevalonate pathway to increase isoprene yields by manipulating intracellular substrate pools are unsuccessful unless archaeal enzymes, which are resistant to feedback inhibition, are used (26–28). Methanogens, in contrast, already grow on inexpensive feedstocks (CO2, CO, C1 compounds, acetate) that are 3 to 20× less expensive than glucose, do not require illumination or aeration, and naturally direct 5% of biomass to isoprenoid lipid synthesis (Table S1 in the supplemental material) (29). We surmised that so long as they are still able to synthesize isoprenoid lipid membranes for growth, methanogens may be able to produce renewable isoprene in high yields due to their inherently high metabolic flux through the archaeal mevalonate pathway (Fig. 1). The purpose of our study was to test whether Methanosarcina can be used to synthesize isoprene from C1 substrates and acetate.
FIG 1.
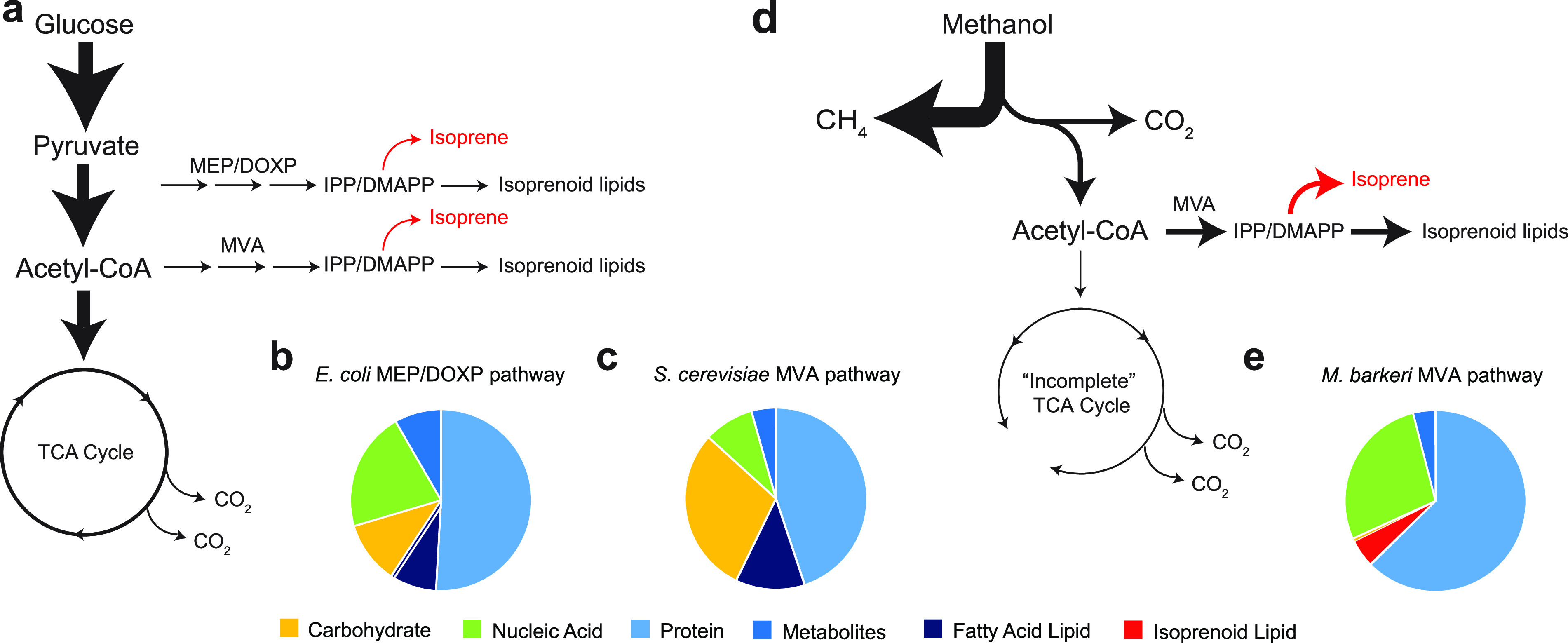
Isoprenoid biosynthesis pathways and macromolecular compositions of representative bacteria, eukarya, and archaea. (a) Isoprene is synthesized from isopentenyl pyrophosphate/dimethylallyl pyrophosphate (IPP/DMAPP) derived from glucose via the methylerythritol phosphate/deoxy xylulose phosphate (MEP/DOXP) pathway in bacteria or mevalonate (MVA) pathway in eukarya. (b and c) Relative amounts of macromolecules in E. coli bacterium (58) and S. cerevisiae yeast (59), respectively. (d) Isoprenoid lipids are synthesized from IPP/DMAPP by the archaeal MVA pathway in methanogens. (e) Isoprenoid lipids in methanogens comprise 5% of biomass dry weight (29). Arrow sizes and line widths depict published carbon fluxes through each pathway. One or more genes are required for most organisms to produce isoprene monomer (red arrows). See Table S1 for macromolecular composition values shown in panels b, c, and e.
RESULTS
Creation and characterization of Methanosarcina acetivorans ispS+ strains.
In plants, the isoprene monomer is synthesized by cleavage of the C-O bond of dimethylallyl pyrophosphate (DMAPP) to produce isoprene monomer and pyrophosphate by the enzyme isoprene synthase (IspS) (3). To generate isoprene-synthesizing methanogens, we cloned the codon-optimized ispS gene from poplar (Populus alba) into the Methanosarcina spp. overexpression suicide vector pNB730 (Figure 2a) (30, 31). Once transfected into cells, the resulting plasmid pJA2 integrates into the chromosome, resulting in constitutive overexpression of synthetic ispS from the methyl coenzyme M reductase (mcr) promoter, PmcrB (Figure 2a) (32). Integration of pJA2 was confirmed by PCR, and transcription of ispS mRNA was validated by reverse transcriptase-PCR (RT-PCR) of an 88-bp fragment of the ispS cDNA transcript (Figure 2b). A vector-only control (att:pNB730) strain was also created by integrating pNB730 onto the M. acetivorans chromosome. These results indicate successful integration of pJA2 onto the chromosome and transcription of ispS in M. acetivorans.
FIG 2.
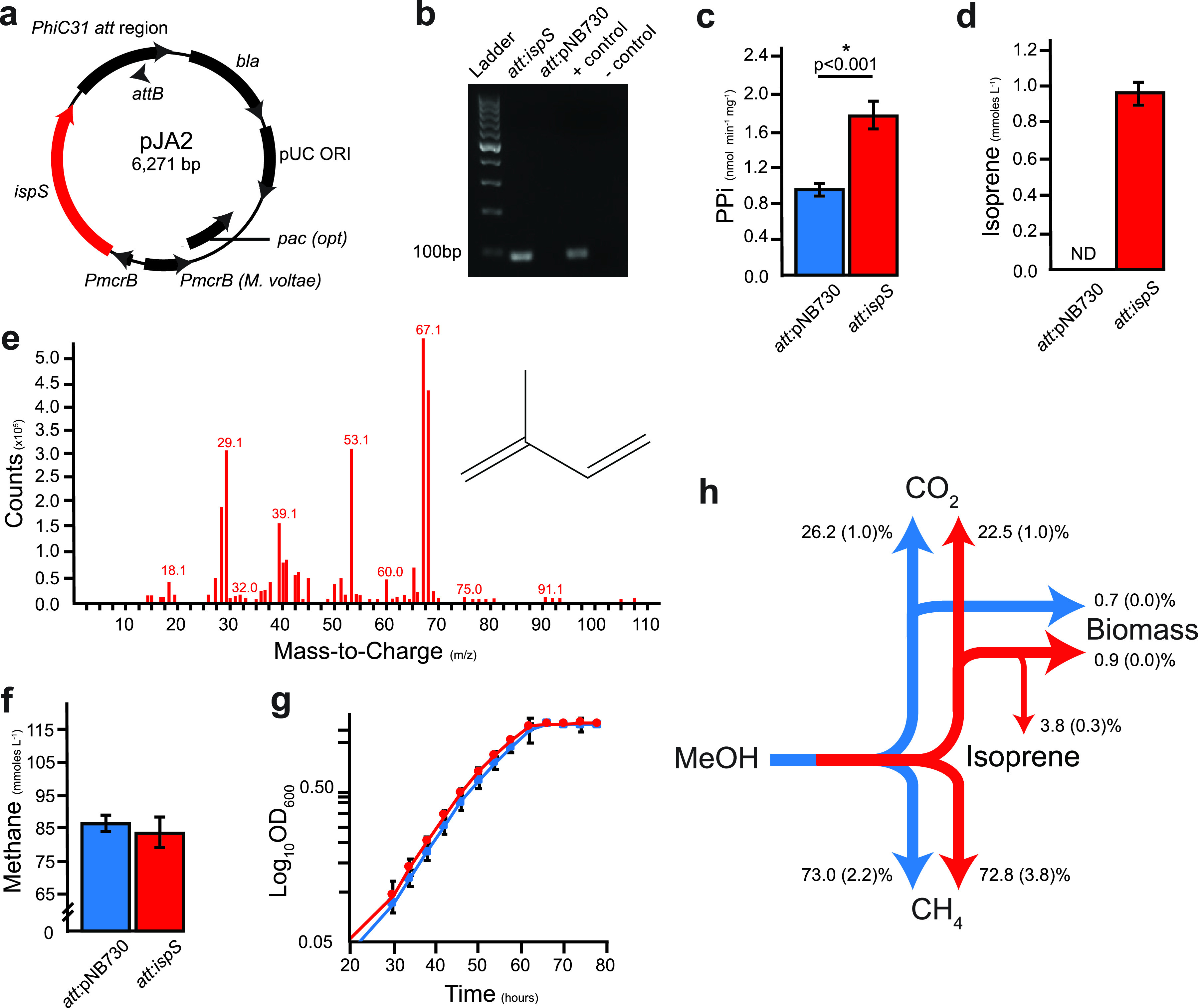
Strain construction and validation of isoprene production from methanol. (a) Plasmid map of pJA2 for constitutive expression of isoprene synthase IspS in Methanosarcina spp. (b) Validation of ispS transcription by RT-qPCR. Plasmid DNA from pJA2 was used as a positive control, while genomic DNA from the parent strain NB34 was used as a negative control. (c) Dimethylallyl pyrophosphate pyrophosphatase activity in cell extracts. (d) Isoprene production measured by gas chromatography. (e) Validation of isoprene production by mass spectrometry. (f) Endpoint methane production. (g) Growth curve of att:pNB730 and att:ispS strains. (h) Mass balance of M. acetivorans att:pNB730 (blue) and att:ispS (red) strains showing percent molar carbon fluxes. Standard deviations are shown in parentheses. Blue bars, att:pNB730 strain; red bars, att:ispS strain. Data presented in panels c, d, f, and h were obtained from quadruplicate biological and triplicate technical replicates (n = 12). Data presented in panel e were from a double-blinded experiment. Data from panel g were from five biological replicates.
Gene integration and transcription alone is not necessarily sufficient to ensure an enzyme will be translated, folded, and maintain stable biochemical activity in a new host cell. P. alba IspS is most enzymatically active at 40°C and has a high Km for DMAPP (30). To determine if the newly designed, synthetic ispS was translated and maintained enzyme activity in methanogen cells grown at 35°C, the cell extract was assayed for DMAPP pyrophosphatase activity (Figure 2c). Cells expressing ispS had a 2-fold higher release of PPi from DMAPP activity versus vector-only control (att:pNB730) cells, suggesting IspS is enzymatically active in methanogen cells. Isoprene synthesis by methanol-grown cells was confirmed by gas chromatography using flame ionization and mass spectrometry (Figure 2d and e). These data suggest the synthetic ispS gene was transcribed, translated, and folded into an active enzyme that could access the intracellular DMAPP pool in growing M. acetivorans cells at 35°C to yield the end product isoprene.
M. acetivorans att:ispS+ strains were cultured and physiologically characterized to determine if ispS expression had an effect on growth of the organism (Table S2). Because methanogens synthesize cell membrane lipids entirely from DMAPP (Figure 1d) (4, 29, 33), it was initially expected that high constitutive expression of ispS from PmcrB could decrease viability or may even be lethal. However, when grown on methanol, there was no detectable difference in methane produced (Figure 2e) or in population doubling time (Figure 2g), thus demonstrating that ispS expression does not result in decreased cell viability.
Some possible explanations for tolerance of high ispS expression by M. acetivorans include substrate channeling (such that DMAPP is preferentially funneled to membrane synthesis and only excess intracellular DMAPP pools are available to IspS), the high Km of IspS (34), or auto-titration of gene copies. Previous work has shown that methanogens vary the number of copies of the entire chromosome determined by growth phase and type of growth substrate, with chromosomal copies ranging from 3 to 18 (35). We hypothesized that this variance could possibly modulate ispS gene copy number and therefore expression levels by homologous recombination at the site of pJA2 integration. To test this hypothesis, the stability of the ispS gene in the culture population was assessed by serial passaging of ispS+ strains with and without puromycin antibiotic selection. If expression of ispS caused a decrease in reproductive fitness, serial passaging in the absence of antibiotic selection should have selected for fewer copies of the ispS gene in the total population, which could be detected using quantitative PCR (qPCR) of ispS versus an unlinked essential housekeeping gene, such as rpoA1. The rpoA1 gene encodes the sole DNA-dependent RNA polymerase gene on the chromosome and can be used as a reference gene in a qPCR assay. By comparing ispS gene copy number to copies of the chromosomal reference gene rpoA1, we could calculate the average number of ispS genes per chromosome and per cell. If expression of ispS was neither beneficial nor detrimental under the culturing conditions, we would have expected no change in the ispS:rpoA1 ratio. With constant antibiotic pressure, M. acetivorans strains transformed with pJA2 were found to have an average of 0.57 copies of ispS per chromosome after 20 generations (Table 2). The ispS:rpoA1 ratio was relatively unchanged at 0.56 after 140 generations. Without antibiotic selection, the ispS:rpoA1 ratios were 0.59 at 20 generations and 0.53 at 140 generations. We next tested whether we could drive an increase in ispS:rpoA1 ratio through homologous recombination by selecting for increased expression of the linked puromycin resistance cassette; however, these efforts were unsuccessful and the ispS:rpoA1 ratio remained unchanged. These data suggest pJA2 is stably integrated onto the chromosome. Additional experiments are needed to further explore the effects of gene dosage and gene expression on isoprene production.
TABLE 2.
Gene copies per cell
| Gene | Puromycina | Generation number (days)b |
P value | |||||
|---|---|---|---|---|---|---|---|---|
| 20 (7) | Std dev | 56 (19) | Std dev | 140 (47) | Std dev | |||
| rpoA1 | − | 16 | 0 | 16 | 0 | 16 | 0 | 1 |
| IspS | − | 9.39 | 0.10 | 8.32 | 0.51 | 8.52 | 0.53 | 0.545 |
| Ratio | − | 0.59 | 0.56 | 0.53 | ||||
| rpoA1 | + | 16 | 0 | 16 | 0 | 16 | 0 | 1 |
| IspS | + | 9.19 | 0.34 | 9.51 | 0.23 | 8.91 | 0.18 | 0.548 |
| Ratio | + | 0.57 | 0.59 | 0.56 | ||||
Without antibiotic selection (−), with constant antibiotic selection (+).
Values were obtained from biological and technical triplicates (n = 9).
Mass balance of isoprene synthesis from methanol.
The primary metabolic products of methylotrophic methanogenesis by M. acetivorans are methane, CO2, and biomass. Mass balance experiments were used to measure the molar carbon partitioning between control and ispS+ strains to determine if isoprene was derived from the methane, CO2, or biomass pools (Table S3). Mass balance experiments showed both control and ispS+ strains consumed 100% of the substrate methanol and produced nearly equivalent amounts of methane (73.0 ± 2.2 and 72.8 ± 3.8, P = 0.0191, respectively, for a 0.2% C flux difference). However, the ispS+ strain shows 3.7% less CO2 flux than the control strain (P = 0.0001) but 6.7% more biomass C according to dry weight (P = 0.0041). The ispS+ strain directed 3.8% of total C to isoprene. The yields of methane were not significantly different for control or ispS+ strains on bicarbonate-buffered medium versus MOPS-buffered medium (Table S3). These results show cells benefit with increased biomass synthesis when carbon is siphoned from the oxidative branch of the methylotrophic methanogenesis pathway to produce isoprene (Figure 2h).
Isoprene synthesis affects transcription of Mcr, mevalonate, and TCA enzymes.
Isoprene synthesis would be expected to reduce the intracellular pool of DMAPP that normally feeds into membrane synthesis. Therefore, as a result of isoprene synthesis, we would expect the ispS+ strain to upregulate expression of mevalonate pathway genes that supply DMAPP/IPP to the lipid synthesis pathway (Fig. S1). In addition, if isoprene synthesis is affecting electron flux through the electron transport system, we would expect to observe downregulation of one or more genes in the tricarboxylic acid (TCA) or methanogenesis cycles that are responsible for maintaining redox balance in the cell (Fig. 1). Compared to the control strain, the M. acetivorans att::ispS+ strain was found to have a slight decrease in 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase transcripts (1.5-fold) compared to the control strain and increased mRNA abundance for genes downstream of HMG-CoA synthase. These changes in mRNA levels suggest the cell is reacting to a depletion of downstream metabolite pools that includes DMAPP (Table 3). We also observed that methyl coenzyme M reductase (mcrB), pyruvate carboxylase, and malate dehydrogenase were upregulated, while fumarate hydratase was significantly downregulated. These results suggest the cell is sensing an imbalance between methanogenesis, biomass synthesis, and redox-dependent reactions and supports the hypothesis that electron flux is decreased through the rate-limiting terminal electron acceptor CoM-CoB heterodisulfide and the membrane electron carrier methanophenazine, which is critical for ATP synthesis (36, 37). Together, these results suggest that isoprene synthesis may relieve known kinetic bottlenecks in CoM-CoB and methanophenazine redox balance, thereby contributing to increased biomass. Future experiments using mutant strains could provide further evidence of this process.
TABLE 3.
Relative transcript abundance between att:ispS and att:pNB730 strains of M. acetivoransa
| Pathway | Gene | Fold change | P value |
|---|---|---|---|
| Mevalonate pathwayb | HMG-CoA synthase | 0.66 | 0.001271 |
| HMG-CoA reductase | 1.49 | 0.045242 | |
| Mevalonate kinase | 1.31 | 0.004854 | |
| Phosphomevalonate dehydratase | NT | NT | |
| Anhydromevalonate phosphate decarboxylase | NT | NT | |
| Isopentenyl phosphate kinase | 1.88 | 0.117332 | |
| Isopentenyl diphosphate delta-isomerase | 2.26 | 0.011509 | |
| TCA pathwayc | Pyruvate synthase | 0.80 | 0.211017 |
| Pyruvate carboxylase | 1.96 | 0.000206 | |
| Malate dehydrogenase | 1.68 | 0.007657 | |
| Fumarate hydratase | 0.21 | 1.53E−05 | |
| Methanogenesis | Methyl coenzyme M reductase B (mcrB) | 1.55 | 0.000121 |
aValues were obtained from triplicate biological replicates and 5 technical replicates (n = 15); NT, not tested.
bThe proposed archaeal mevalonate pathway in M. acetivorans differs from the eukaryotic pathway in the conversion of mevalonate to isopentenyl phosphate (19–21).
cAll annotated TCA cycle enzymes in M. acetivorans. Methanogens have an incomplete TCA cycle.
Isoprene production utilizing other carbon sources.
Isoprene production and ispS+ strain physiology were assessed on additional carbon sources trimethylamine (TMA) and acetate to determine if isoprene yields changed depending on growth substrate (Fig. 3). When growing on methanol or TMA, M. acetivorans uses the methylotrophic methanogenesis pathway, while when growing on acetate uses the acetoclastic pathway. The methylotrophic and acetoclastic methanogenesis pathways differ with respect to intracellular carbon and electron fluxes, which could have an impact on isoprene yields. Cells grown on TMA and acetate had similar DMAPP pyrophosphatase activity and isoprene yields as methanol-grown cells (Fig. 3, Table S2). Endpoint methane production by ispS+ strains was 10% lower than control strains when grown on TMA, despite the fact that methanol and TMA are both metabolized by the methylotrophic methanogenesis pathway (Table S2). Similar to methanol-grown cells, TMA- and acetate-grown ispS+ strains had population doubling times that were the same as control strains (Table S2). These data show engineered M. acetivorans can produce high quantities of isoprene from a variety of inexpensive carbon sources and production is independent of whether the methylotrophic or acetoclastic methanogenesis pathway is used by the cell.
FIG 3.

Characterization of ispS+ strains grown on trimethylamine (TMA) or acetate substrates. (a and e) Dimethylallyl pyrophosphate pyrophosphatase activity in cell extracts. (b and f) Endpoint methane production. (c and g) Isoprene production measured by gas chromatography. (d and h) Growth curves of att:pNB730 and att:ispS strains. Blue bars, att:pNB730 strain; red bars, att:ispS strain. Data presented in panels a to c and e to g were obtained from quadruplicate biological and triplicate technical replicates (n = 12). Data in panels d and h were from five biological replicates.
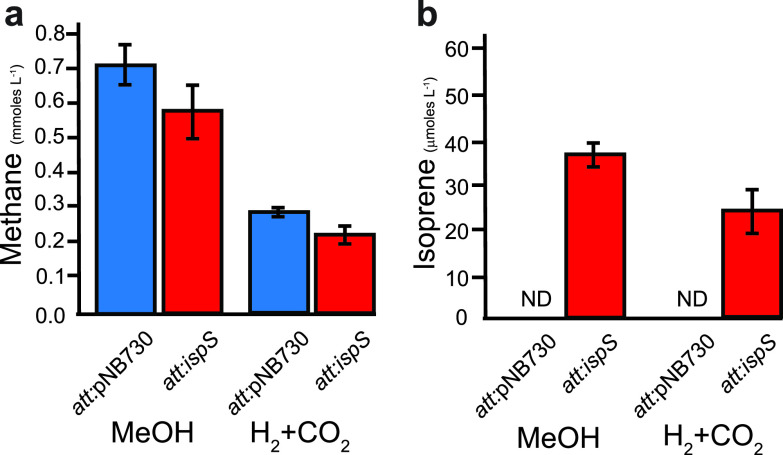
Isoprene synthesis by engineered Methanosarcina barkeri.
Methanosarcina acetivorans is a versatile organism capable of growth on the widest range of methanogenic substrates, including C1 chemicals (carbon monoxide, methanol, methylamines, methylsulfides, etc.) and acetate (38). Methanosarcina barkeri, a related methanogen, can grow on C1 compounds and acetate similar to M. acetivorans, except it has maintained the ability to use H2 as an electron donor via the hydrogenotrophic (reducing carbon dioxide to methane) or methyl respiration (reducing methanol or other C1 compounds to methane) methanogenesis pathways. To expand the possible feedstocks for isoprene synthesis and to determine whether the different electron transport system configuration found in M. barkeri affects isoprene yields, we transformed M. barkeri with the pJA2 plasmid.
After confirmation of successful integration of pJA2 onto the chromosome, biochemical tests were used to confirm isoprene production and its effect on growth of the organism (Table S4). Similar to M. acetivorans, the methanol-grown M. barkeri ispS+ strain had increased DMAPP pyrophosphatase levels (136% ± 0.4%) compared to the parental strain (100% ± 0.2%), indicating the introduced ispS gene was translated into active enzyme in methanogen whole-cell lysate. M. barkeri ispS+ strains had identical growth rates as control strains; however, methane yields were 18% less on methanol and 10% less on H2 + CO2 versus control strains, similar to what was observed with TMA-grown M. acetivorans (Figure 4a). Isoprene yield with M. barkeri was 3.8% of the isoprene produced by M. acetivorans during growth on methanol. Isoprene yield on H2 + CO2 was 2.4% of the M. acetivorans yield on methanol (Figure 4b), and roughly equivalent to the reported yield of geraniol diterpene by Methanococcus maripaludis (Table 1, Table S4) (39).
FIG 4.
Demonstration of isoprene production by Methanosarcina barkeri. (a) Endpoint methane assays for M. barkeri att:pNB730 and att:ispS strains. (b) Isoprene production by M. barkeri att:pNB730 and att:ispS strains as measured by gas chromatography. Blue, att:pNB730 strains; red, att:ispS strains. Data for panels a and b were obtained from quadruplicate biological and triplicate technical replicates (n = 12).
Differences in isoprene yield between these methanogens likely results from expression of hydrogenases in M. barkeri and M. maripaludis. Hydrogenases are essential in the hydrogenotrophic methanogenesis pathway (40) and are necessary for conserving energy through hydrogen cycling in M. barkeri (41). As a result of hydrogenase expression, redox balancing in M. barkeri kinetically favors hydrogen synthesis rather than acetyl-CoA and DMAPP synthesis. M. acetivorans, which does not use hydrogen cycling for energy conservation, is poised to donate electrons to CoM-CoB heterodisulfide reductase or to acetyl-CoA synthesis (42). Previous work has shown that further decreasing flux through the CoM-CoB terminal electron acceptor in M. acetivorans results in increased biomass synthesis and increased metabolic efficiency (37). The degree of similarity in substrate channeling and redox balance mechanisms between M. acetivorans and M. barkeri is an ongoing area of research, but it is clear that isoprene optimization will require tailored metabolic engineering approaches depending on whether methanogens are capable of hydrogenotrophic growth. Our results indicate that while M. acetivorans produces higher yields than hydrogenotrophic methanogens, both M. acetivorans and M. bakeri can be engineered to produce isoprene from various inexpensive feedstocks without significantly sacrificing growth kinetics or biomass yields.
DISCUSSION
Our results confirm the hypothesis that archaea, and Methanosarcina spp. in particular, can be engineered to synthesize high yields of isoprene. Under batch-growth conditions using methanol as a substrate, M. acetivorans was able to produce 6 × 106 times more isoprene than the bacterium Clostridium ljungdahlii, and 179 times more isoprene than the autotrophic cyanobacterium Synechocystis. The high carbon fluxes we measured (4% total C) and the observation of increased biomass (Figure 2h) suggest that in M. acetivorans ispS+ strains, isoprene is an abundant metabolic product that benefits cells. Furthermore, the engineered strains showed no detectable changes in population doubling rate, maximum culture optical density, and methane production compared to a vector-only control strain. Mass balance data indicated a 16% decrease in CO2 production, suggesting that C for isoprene was derived from the oxidative branch of the methylotrophic methanogenesis pathway. The data support the conclusion that M. acetivorans and M. barkeri ispS+ strains have become “isoprenogens,” i.e., methanogens capable of growing by synthesizing mixed products of isoprene and methane. Importantly, the isoprene yields and 4% total C flux we observed were obtained by expression of a single terpene synthase without extensive pathway optimization.
Can we push isoprene yields further? Other investigators using S. cerevisiae and E. coli chassis were challenged by unstable expression and low activity of terpene synthases, low substrate pathway fluxes, and enzyme feedback inhibition that had to be overcome through biochemical and metabolic engineering. Optimization of these factors and use of inducible promoters has the potential to further increase isoprene yield using Methanosarcina spp. The results of genetic selection experiments, shown in Table 2, suggest that 4% flux seems to be an upper limit that still provides enough isoprenoid lipid synthesis for central metabolism and maintaining redox balance at wild-type growth rates. However, by identifying metabolic bottlenecks and addressing these with additional mutations, such as using an engineered IspS enzyme with a lower Km (43), it is possible that yields may be increased.
Methanogens survive on the very edge of thermodynamic favorability, producing approximately 0.3 ATP per mole of carbon substrate utilized (44). This lean metabolism creates a high flux of carbon with an exceedingly small fraction of overall carbon being utilized for biomass, all of which is coupled to the rate-limiting reactions of methanogenesis. As such, the growth of methanogens can be predicted predominantly from the energetics of substrate utilization. In a steady-state culture, methylotrophic methanogenesis can be modeled based on the mass balance equation: 4CH3OH → 1CO2 + 3CH4 + 2H2O (ΔG° = −84.25 kJ Cmol−1). Assuming all mevalonate pathway flux is devoted to isoprene synthesis at the expense of CO2 or membrane lipid synthesis in a nonreplicating culture, up to 75% of C could be directed to isoprene synthesis from methylotrophic substrates according to the mass balance equation: 40CH3OH → 9CH4 + 6C5H8 + 38H2O (ΔG° = −5.2 kJ Cmol−1). Based on current understanding of metabolism in methanogens, up to 85.7% of substrate carbon could be used to synthesize isoprene using hydrogenotrophic methanogens (35CO2 + 104H2 → 5CH4 + 6C5H8 + 70H2O, ΔG° = −9.9 kJ Cmol−1) and up to 71.4% at near equilibrium from acetoclastic methanogens (7CH3CO2H → 2C5H8 + 4CO2 + 6H2O, ΔG° = −16.6 kJ Cmol−1). While growing cells must divert some C flux to lipid synthesis, as long as cells can couple additional isoprene synthesis to generation of a transmembrane ion gradient, they will be able to conserve energy via ATP synthesis. Recent studies have shown that E. coli and S. cerevisiae strains were able to significantly increase isoprene yields utilizing nonreplicating cells in fed-batch fermentation (Table 1) (45, 46). The data reported here for Methanosarcina spp. were obtained from batch-grown cultures to facilitate mass balance measurements and likely represent an underestimate compared to the isoprene yield that could be obtained from larger-scale fed-batch or chemostat bioreactors.
The lack of change in growth rate of isoprene-producing Methanosarcina species strains, as well as the decrease in CO2 production, are consistent with the interpretation that isoprene synthesis does not negatively affect the cell’s ability to conserve energy. In methanogens, the methanogenesis pathway is linked to biosynthesis by the carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) complex, which facilitates growth and ATP generation by the regeneration of the cofactor ferredoxin (Fdx) and the synthesis of acetyl-CoA (36). We speculate that by providing the ability to synthesize an alternative metabolic by-product, isoprene, the cell can overcome the kinetic bottleneck caused by reduced Fdx and high acetyl-CoA pools. Further investigation is needed to test this hypothesis and to clarify how the introduction of isoprene synthase may have altered metabolism in M. acetivorans and M. barkeri.
MATERIALS AND METHODS
Anaerobic techniques.
Anaerobic procedures were performed in a custom B-type Coy anaerobic chamber (Coy Labs, Grass Lake, MI). Internal environment of the chamber is maintained at 5% H2/20% CO2/75% N2 (± 3%) (Matheson Gas, Lincoln, NE). Cells incubated outside the anaerobic chamber are contained in glass Balch tubes secured with butyl rubber stoppers (Belco Glass, Vineland, NJ) and aluminum crimps (Wheaton, Millville, NJ).
Methanogen cell culture.
Cells listed in Table 4 were grown in anaerobic high-salt (HS) medium (200 mM NaCl, 45 mM NaHCO3, 13 mM KCl, 54 mM MgCl2·6H2O, 2 mM CaCl2·2H2O, 2 μM 0.1% resazurin [w v−1], 5 mM KH2PO4, 19 mM NH4Cl, 2.8 mM cysteine·HCl, 0.1 mM Na2S·9H2O, trace elements, vitamin solution) (47) supplemented with a carbon and energy source (methanol, 125 mM; trimethylamine, 50 mM; sodium acetate, 120 mM) and 2 mg liter−1puromycin as needed (48, 49). 3-(N-morpholino)propanesulfonic acid (MOPS) high-salt medium (MHS) was created by substituting 45 mM NaHCO3 with 50 mM MOPs buffer (50). Cells in liquid medium were incubated at 35°C without shaking. For growth on solid medium, 1.4% agar was added to HS medium. All chemicals and reagents were sourced from Millipore Sigma (St. Louis, MO) or Thermo Fisher Scientific (Waltham, MA).
TABLE 4.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description | Purpose | Source |
|---|---|---|---|
| Strains | |||
| Methanosarcina acetivorans C2A | |||
| 394 | Δhpt::φC31 int, att:pJA2 | Isoprene production (att:ispS) | This study |
| 452 | Δhpt::φC31 int, att:pNB730 | Vector-only control (att:pNB730) |
(55) |
| Methanosarcina barkeri | |||
| 396 | Δhpt::φC31 int, att:pJA2 | Isoprene production | This study |
| 459 | Δhpt::φC31 int, att:pNB730 | Vector-only control (att:pNB730) |
(31) |
| Escherichia coli | |||
| 3 | F′proA+B+ lacIq Δ(lacZ)M15 zzf::Tn10 (TetR)/fhuA2Δ(argF-lacZ)U169 phoA glnV44 Φ80Δ(lacZ)M15 gyrA96 recA1 endA1 thi-1 hsdR17 | Cloning and plasmid propagation | New England Biolabs |
| 134 | F′ proA+B+ lacIq Δ(lacZ)M15 zzf::Tn10 (TetR)/fhuA2Δ(argF-lacZ)U169 phoA glnV44 Φ80Δ(lacZ)M15 gyrA96 recA1 endA1 thi-1 hsdR17/ pNB730 | Production of pNB730 plasmid | (31) |
| 453 | F′ proA+B+ lacIq Δ(lacZ)M15 zzf::Tn10 (TetR)/fhuA2Δ(argF-lacZ)U169 phoA glnV44 Φ80Δ(lacZ)M15 gyrA96 recA1 endA1 thi-1 hsdR17/ pJA2 | Expression of ispS in M. acetivorans | (55) |
| Plasmids | |||
| pNB730 | pUC ori bla PmcrB pac(opt) φC31 attB | Methanosarcina spp. integration and expression vector | (31) |
| pJA2 | pNB730 ispSΔ1−13 | Integration of ispS into genome for constitutive expression of isoprene synthase | (55) |
| Primers and DNA strings | |||
| sNB19 | GenBank accession no. MW295460 (see the supplemental material) | Synthetic optimized Populus alba ispS (isoprene synthase) | This study |
| oNB568 | ATTAAGGAGGAAATTCATATGTCCGTTTCCACCGAAAATGT | ispSΔ1−13 DNA string amplification and cloning, fwd | (55) |
| oNB576 | CGAGGGCCCAAGCTTGGATCCTCATCTTTCAAAAGGAAGAATAG | DNA string amplification and cloning, rev | (55) |
| oNB729 | CATATGCCTGACGACCTCATTA | RNA polymerase, rpoA1 qRT fwd housekeeping gene | This study |
| oNB730 | GAATTTGATTTCGAGCTGTTCC | RNA polymerase, rpoA1 qRT rev housekeeping gene | This study |
| oNB733 | GTTTACAAAAGTAGCTGCAAGGGTA | Methyl-coenzyme M reductase, mcrB qRT fwd | This study |
| oNB734 | ATACAAATTCTACAAGGCAAACGAC | Methyl-coenzyme M reductase, mcrB qRT rev | This study |
| oNB735 | GGATTCGATGCAGTTACCAAA | ispS qPCR primer, fwd | This study |
| oNB736 | TGCTTCCTGGCTAACTTCAAA | ispS qPCR primer, rev | This study |
| oNB930 | CCGTGCCTGATGTCGACGAA | HMG-CoA synthase fwd | This study |
| oNB931 | TGGAGGGATCTACGCCGCTT | HMG-CoA synthase rev | This study |
| oNB932 | GCCGGCCTTCTGAAAGTAAACG | HMGR Fwd | This study |
| oNB933 | TCCGCGGTTTACACTGGCAA | HMGR rev | This study |
| oNB934 | CCCGTGTGCGGGTGGAATTA | Mevalonate kinase fwd | This study |
| oNB935 | ACCACTGCGGAGATATAAGGATGT | Mevalonate kinase rev | This study |
| oNB936 | GAGGCAGCGCCATTACCGAT | Isopentenyl phosphate kinase fwd | This study |
| oNB937 | GCCTGAAACTTCCCGCGCAA | Isopentenyl phosphate kinase rev | This study |
| oNB938 | CAGCCAGAGAGCCGCAATCG | Isopentenyl-diphosphate δ-isomerase fwd | This study |
| oNB939 | CCGTAGACAAAGGCGTTCGGA | Isopentenyl-diphosphate δ-isomerase rev | This study |
| oNB940 | GCTCATGCACGAGGTGCTCT | Pyruvate synthase fwd | This study |
| oNB941 | GCACTGACTGCCCTGTTTGC | Pyruvate synthase rev | This study |
| oNB942 | TCATGCGTGCCTGCAGAGAG | Pyruvate carboxylase fwd | This study |
| oNB943 | GCCTCATCGGCATACTTGGCA | Pyruvate carboxylase rev | This study |
| oNB944 | CCGAACTGGAACCTGGCGAA | Malate dehydrogenase fwd | This study |
| oNB945 | TGCCTGCATGAGGTCAAGGG | Malate dehydrogenase rev | This study |
| oNB946 | TCCTCGACCTGCCTATCGGT | Fumarate hydratase fwd | This study |
| oNB947 | GGTCGGCTGGAACCTCAACC | Fumarate hydratase rev | This study |
Cloning and genetic techniques.
Methods for genetic manipulation of M. acetivorans have been described previously (51). All plasmids and primers shown in Table 4 were designed using VectorNTI software (Thermo Fisher Scientific, Waltham MA). PCR primers were synthesized by Integrated DNA Technologies (IDT, Coralville, IA). All plasmids were verified by sequencing (Eurofins, Louisville, KY). Plasmid pNB730 was used as a parent vector (31). Key features of pNB730 include: (i) pUC ori for high-copy replication in E. coli; (ii) φC31 phage recombinase att site for chromosomal insertion of the vector into the host genome; (iii) resistance to ampicillin for selection during amplification in Escherichia coli; and (iv) puromycin resistance for selection in Methanosarcina spp. The cDNA sequence of ispS was obtained from NCBI (locus BAD98243, gi: 63108310) from the isoprene-producing poplar plant, Populus alba (30). The P. alba ispS gene was codon-optimized for translation in archaea and inverted repeats were removed to create sNB19, which was commercially synthesized by Life Technologies Corporation (Grand Island, NY). PCR amplification of synthetic genes designed with predicted chloroplast localization signal intact or truncated (ispS and ispSΔ1-13) was achieved using the primers listed in Table 4 with Phusion Flash PCR Master Mix as a proofreading DNA polymerase (Thermo Fisher Scientific, Waltham, MA). DNA purification was accomplished using Promega Wizard SV Gel and PCR clean-up kits (Madison, WI). Fast Digest restriction enzymes (BamHI and Ndel) were purchased from Thermo Fisher Scientific (Waltham, MA). DNA fragments were assembled using the sequence- and ligation-independent cloning (SLIC) protocol previously described (52). The synthesized ispS genes were expressed from the constitutive methyl-CoM reductase promoter (Pmcr) at the pNB730 multiple cloning site. Electroporated E. coli cells were plated on lysis broth (LB) agarose plates with 100 mg liter−1 ampicillin and colonies were selected after overnight growth at 37°C (53). Plasmids were screened by PCR as described and sequenced (31). Plasmids were transfected into Methanosarcina spp. cells according to established procedures using Roche DOTAP liposomal transfection reagent (Roche Diagnostics Corporation, Indianapolis, IN) (48, 51). Cells transfected with pNB730 lacking ispS were used as a vector-only control.
Quantitative PCR (qPCR) was used to quantify integrated gene copies of ispS in the population relative to the unique rpoA1 gene found on each chromosome. Cells were grown in HS + MeOH medium until late exponential (optical density at 600 nm [OD600] = 0.8) and harvested by vacuum filtration followed by lysis using TRI reagent (Sigma-Aldrich, St. Louis, MO) according to the manufacturer’s instructions. cDNA was synthesized with random hexamers (Promega, Madison, WI) using GoTaq 2-step RT-qPCR system (Promega, Madison, WI) and the ispS transcript was confirmed by qPCR using primers oNB735 and oNB736, listed in Table 4.
Cellular growth measurements.
Cell growth rate was determined by measuring culture optical density at 600 nm using a Spectronic D spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Biomass measurements for each strain were obtained as previously described (37).
Pyrophosphate assay.
M. acetivorans cell extracts were assayed for isopentenyl pyrophosphate pyrophosphatase activity using EnzChek pyrophosphate assay kit (Molecular Probes, Eugene, OR). Briefly, cells were harvested at late exponential phase of growth from a 100-ml culture by centrifugation in a Thermo Fisher Scientific Sorvall Legend XTR centrifuge using a TX-750 swinging bucket rotor with 50-ml conical tube adapters at 4,000 × g for 5 min at room temperature. Cell pellets were washed twice using 1 ml of 0.4 M NaCl to remove spent culture medium. After resuspension, cells were lysed using 9 ml ddH2O and centrifuged at 10,000 × g in a Thermo Fisher Scientific Sorvall Legend Micro21 rotor to pellet cell debris. The resulting supernatant was transferred to a fresh microcentrifuge tube and kept on ice. Cell lysate was used to test for enzymatic activity of IspS by following the protocol described by the manufacturer using dimethylallyl diphosphate purchased from Sigma-Aldrich (St. Louis, MO) as the substrate for the reaction. The reaction was monitored spectrophotometrically at 360 nm on a Jenway 7305 spectrophotometer (Burlington, NJ).
Methane production assay.
Methane in culture headspace was measured by gas chromatography using a flame ionization detector (GC-FID) as previously described (54). Briefly, 10-ml cultures were grown to stationary phase. After growth, 100 μl of headspace was captured using a gastight Hamilton syringe and transferred to an empty crimped 2 ml autosampler serum vial (Wheaton, Millville, NJ). Vial contents were analyzed by flame ionization using a custom Agilent 7890A gas chromatography (GC) system. The GC is equipped with an autosampler for consistent sample injection and utilized a GS CarbonPLOT column (Agilent Technologies) at 145°C for separation of volatile metabolites. Quantification of methane was achieved by comparison to a methane standard curve (Matheson, Lincoln, NE) run in parallel with experimental samples.
Isoprene production assay.
The same GC-FID system as above was deployed to quantify isoprene (55). M. acetivorans strains were grown in 10-ml cultures with a 1 ml paraffin oil overlay. Once grown to stationary phase, the oil was harvested and transferred to a 2 ml stoppered and crimped autosampler vial. The GC-FID method for isoprene quantification was as follows: 160°C for 35 min, ramp to 200°C at 75°C/min for 20 min, ramp to 275°C at 75°C/min for 20 min, 275°C for 5 min, and ramp to 160°C at 75°C/min to equilibrate the system for the next run. Isoprene quantification was achieved using a standard of known volumes of isoprene injected into 1 ml of paraffin oil in a 2 ml autosampler vial.
Mass balance measurements.
M. acetivorans was grown to early stationary phase in 100-ml cultures. Cultures were centrifuged and concentrated to 10 ml in MHS medium. Cells were washed twice with MHS and resuspended in 10 ml MHS. Then, 0.250 ml of resuspended cells was transferred to sterile, anaerobic autosampler vials, after which 0.250 ml of 2× MeOH MHS was added to the autosampler vials, which were then stoppered and crimped. The headspaces of the autosampler vials were flushed with N2 to remove residual CO2. Prepared samples were incubated for 36 h at 35°C. After incubation, remaining methanol in spent medium was analyzed by GC-FID and the methanol peak area was compared to standard curves generated by serial dilutions of HS MeOH medium. CO2 and CH4 in the headspace were quantified using a thermal conductivity detector (GC-TCD). Peak areas of headspace gases were compared to standard curves generated for each gas using methane and CO2 reference standards (Airgas, Randor, PA, and Matheson Gas, Lincoln, NE).
qRT-PCR methods.
Cultures of att:ispS+ and control strains were grown anaerobically in triplicate in HS + MeOH until exponential phase (OD600 of 0.56 to 0.74). Cells were anaerobically concentrated in a clinical centrifuge (5,000 × g) and RNA isolation was performed using TRI reagent (Invitrogen) as per the manufacturer’s protocol. DNase treatment was performed using TURBO DNase (Invitrogen) and DNA digestion was confirmed by lack of PCR amplification after 35 cycles using primers oNB733 and oNB734. The cDNA was synthesized using GoTaq 2-step RT-qPCR system (Promega, Madison, WI) with random hexamers and cDNA integrity was verified via agarose gel. qPCR was performed using the primers in Table 4 and GoTaq qPCR Master Mix with SYBR Green I (Promega, Madison, WI) on a Mastercycler RealPlex (2) thermocycler (Eppendorf). Data were obtained from three biological replicates and five technical replicates each (n = 15). Threshold cycle (CT) values from qPCR were normalized to the expression of rpoA1, the DNA-dependent RNA polymerase found in a single copy in M. acetivorans (38) and transcript abundance of each gene was compared using the 2^(-ΔΔCT) method as described (56, 57).
Data availability.
Plasmids, strains, and growth and assay data that support the findings of this study are available from the corresponding author (N.R.B.) upon reasonable request. The sequence for sNB19 was submitted to GenBank (MW295460) but not released prior to publication; it is expected to be released shortly. This sequence may also be found in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Water Environment & Research Foundation Grant NTRY6R14 and Nebraska Center for Energy Science awards and is described in United States patent application US 20170175145 A1. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the funding agencies.
N.R.B. has disclosed a significant financial interest in RollingCircle Biotech, LLC, and Molecular Trait Evolution, LLC.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kim JH, Wang C, Jang HJ, Cha MS, Park JE, Jo SY, Choi ES, Kim SW. 2016. Isoprene production by Escherichia coli through the exogenous mevalonate pathway with reduced formation of fermentation byproducts. Microb Cell Fact 15:214. doi: 10.1186/s12934-016-0612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Rubber Study Group. Rubber statistical bulletin. 2012. International Rubber Study Group. http://www.rubberstudy.com/reports.
- 3.Miller B, Oschinski C, Zimmer W. 2001. First isolation of an isoprene synthase gene from poplar and successful expression of the gene in Escherichia coli. Planta 213:483–487. doi: 10.1007/s004250100557. [DOI] [PubMed] [Google Scholar]
- 4.Matsumi R, Atomi H, Driessen AJM, van der Oost J. 2011. Isoprenoid biosynthesis in Archaea—biochemical and evolutionary implications. Res Microbiol 162:39–52. doi: 10.1016/j.resmic.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Wagner WP, Helmig D, Fall R. 2000. Isoprene biosynthesis in Bacillus subtilis via the methylerythritol phosphate pathway. J Nat Prod 63:37–40. doi: 10.1021/np990286p. [DOI] [PubMed] [Google Scholar]
- 6.Sharkey TD, Yeh S. 2001. Isoprene emission from plants. Annu Rev Plant Physiol Plant Mol Biol 52:407–436. doi: 10.1146/annurev.arplant.52.1.407. [DOI] [PubMed] [Google Scholar]
- 7.Yang JM, Xian M, Su SZ, Zhao G, Nie QJ, Jiang XL, Zheng YN, Liu W. 2012. Enhancing production of bio-isoprene using hybrid MVA pathway and isoprene synthase in E. coli. PLoS One 7:e33509. doi: 10.1371/journal.pone.0033509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lv XM, Xie WP, Lu WQ, Guo F, Gu JL, Yu HW, Ye LD. 2014. Enhanced isoprene biosynthesis in Saccharomyces cerevisiae by engineering of the native acetyl-CoA and mevalonic acid pathways with a push-pull-restrain strategy. J Biotechnol 186:128–136. doi: 10.1016/j.jbiotec.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 9.Hong SY, Zurbriggen AS, Melis A. 2012. Isoprene hydrocarbons production upon heterologous transformation of Saccharomyces cerevisiae. J Appl Microbiol 113:52–65. doi: 10.1111/j.1365-2672.2012.05319.x. [DOI] [PubMed] [Google Scholar]
- 10.Lindberg P, Park S, Melis A. 2010. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab Eng 12:70–79. doi: 10.1016/j.ymben.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Meadows AL, Hawkins KM, Tsegaye Y, Antipov E, Kim Y, Raetz L, Dahl RH, Tai A, Mahatdejkul-Meadows T, Xu L, Zhao L, Dasika MS, Murarka A, Lenihan J, Eng D, Leng JS, Liu CL, Wenger JW, Jiang H, Chao L, Westfall P, Lai J, Ganesan S, Jackson P, Mans R, Platt D, Reeves CD, Saija PR, Wichmann G, Holmes VF, Benjamin K, Hill PW, Gardner TS, Tsong AE. 2016. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature 537:694–697. doi: 10.1038/nature19769. [DOI] [PubMed] [Google Scholar]
- 12.Ferry JG. 2012. Methanogenesis: ecology, physiology, biochemistry & genetics. Springer Science+Business Media, Berlin, Germany. [Google Scholar]
- 13.Shen YW, Linville JL, Urgun-Demirtas M, Mintz MM, Snyder SW. 2015. An overview of biogas production and utilization at full-scale wastewater treatment plants (WWTPs) in the United States: challenges and opportunities towards energy-neutral WWTPs. Renew Sust Energ Rev 50:346–362. doi: 10.1016/j.rser.2015.04.129. [DOI] [Google Scholar]
- 14.Lessner DJ, Lhu L, Wahal CS, Ferry JG. 2010. An engineered methanogenic pathway derived from the domains Bacteria and Archaea. mBio 1:e00243-10. doi: 10.1128/mBio.00243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McAnulty MJ, Poosarla VG, Li J, Soo VW, Zhu F, Wood TK. 2017. Metabolic engineering of Methanosarcina acetivorans for lactate production from methane. Biotechnol Bioeng 114:852–861. doi: 10.1002/bit.26208. [DOI] [PubMed] [Google Scholar]
- 16.Jennings ME, Schaff CW, Horne AJ, Lessner FH, Lessner DJ. 2014. Expression of a bacterial catalase in a strictly anaerobic methanogen significantly increases tolerance to hydrogen peroxide but not oxygen. Microbiology (Reading) 160:270–278. doi: 10.1099/mic.0.070763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jasso-Chavez R, Santiago-Martinez MG, Lira-Silva E, Pineda E, Zepeda-Rodriguez A, Belmont-Diaz J, Encalada R, Saavedra E, Moreno-Sanchez R. 2015. Air-adapted Methanosarcina acetivorans shows high methane production and develops resistance against oxygen stress. PLoS One 10:e0117331. doi: 10.1371/journal.pone.0117331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstein JL, Brown MS. 1990. Regulation of the mevalonate pathway. Nature 343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 19.Vinokur JM, Korman TP, Cao Z, Bowie JU. 2014. Evidence of a novel mevalonate pathway in archaea. Biochemistry 53:4161–4168. doi: 10.1021/bi500566q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayakawa H, Motoyama K, Sobue F, Ito T, Kawaide H, Yoshimura T, Hemmi H. 2018. Modified mevalonate pathway of the archaeon Aeropyrum pernix proceeds via trans-anhydromevalonate 5-phosphate. Proc Natl Acad Sci U S A 115:10034–10039. doi: 10.1073/pnas.1809154115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida R, Yoshimura T, Hemmi H. 2020. Reconstruction of the “archaeal” mevalonate pathway from the methanogenic archaeon Methanosarcina mazei in Escherichia coli cells. Appl Environ Microbiol 86:e02889-19. doi: 10.1128/AEM.02889-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heuston S, Begley M, Gahan CGM, Hill C. 2012. Isoprenoid biosynthesis in bacterial pathogens. Microbiology (Reading) 158:1389–1401. doi: 10.1099/mic.0.051599-0. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L, Chang W-c, Xiao Y, Liu H-w, Liu P. 2013. Methylerythritol phosphate pathway of isoprenoid biosynthesis. Annu Rev Biochem 82:497–530. doi: 10.1146/annurev-biochem-052010-100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray JC, Kekwick RGO. 1972. Inhibition of plant mevalonate kinase preparations by prenyl pyrophosphates. Biochim Biophys Acta 279:290–296. doi: 10.1016/0304-4165(72)90145-6. [DOI] [PubMed] [Google Scholar]
- 25.Fu Z, Voynova NE, Herdendorf TJ, Miziorko HM, Kim J-JP. 2008. Biochemical and structural basis for feedback inhibition of mevalonate kinase and isoprenoid metabolism. Biochemistry 47:3715–3724. doi: 10.1021/bi7024386. [DOI] [PubMed] [Google Scholar]
- 26.Huang KX, Scott AI, Bennett GN. 1999. Overexpression, purification, and characterization of the thermostable mevalonate kinase from Methanococcus jannaschii. Protein Expr Purif 17:33–40. doi: 10.1006/prep.1999.1106. [DOI] [PubMed] [Google Scholar]
- 27.Kazieva E, Yamamoto Y, Tajima Y, Yokoyama K, Katashkina J, Nishio Y. 2017. Characterization of feedback-resistant mevalonate kinases from the methanogenic archaeons Methanosaeta concilii and Methanocella paludicola. Microbiology (Reading) 163:1283–1291. doi: 10.1099/mic.0.000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu CL, Bi HR, Bai Z, Fan LH, Tan TW. 2019. Engineering and manipulation of a mevalonate pathway in Escherichia coli for isoprene production. Appl Microbiol Biotechnol 103:239–250. doi: 10.1007/s00253-018-9472-9. [DOI] [PubMed] [Google Scholar]
- 29.Feist AM, Scholten JC, Palsson BO, Brockman FJ, Ideker T. 2006. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol Syst Biol 2:2006.0004. doi: 10.1038/msb4100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki K, Ohara K, Yazaki K. 2005. Gene expression and characterization of isoprene synthase from Populus alba. FEBS Lett 579:2514–2518. doi: 10.1016/j.febslet.2005.03.066. [DOI] [PubMed] [Google Scholar]
- 31.Shea MT, Walter ME, Duszenko N, Ducluzeau AL, Aldridge J, King SK, Buan NR. 2016. pNEB193-derived suicide plasmids for gene deletion and protein expression in the methane-producing archaeon, Methanosarcina acetivorans. Plasmid 84–85:27–35. doi: 10.1016/j.plasmid.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guss AM, Rother M, Zhang JK, Kulkarni G, Metcalf WW. 2008. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2:193–203. doi: 10.1155/2008/534081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sprott GD. 1992. Structures of archaebacterial membrane lipids. J Bioenerg Biomembr 24:555–566. doi: 10.1007/BF00762348. [DOI] [PubMed] [Google Scholar]
- 34.Schnitzler JP, Zimmer I, Bachl A, Arend M, Fromm J, Fischbach RJ. 2005. Biochemical properties of isoprene synthase in poplar (Populus x canescens). Planta 222:777–786. doi: 10.1007/s00425-005-0022-1. [DOI] [PubMed] [Google Scholar]
- 35.Hildenbrand C, Stock T, Lange C, Rother M, Soppa J. 2011. Genome copy numbers and gene conversion in methanogenic archaea. J Bacteriol 193:734–743. doi: 10.1128/JB.01016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buan NR, Metcalf WW. 2010. Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol Microbiol 75:843–853. doi: 10.1111/j.1365-2958.2009.06990.x. [DOI] [PubMed] [Google Scholar]
- 37.Catlett JL, Ortiz AM, Buan NR. 2015. Rerouting cellular electron flux to increase the rate of biological methane production. Appl Environ Microbiol 81:6528–6537. doi: 10.1128/AEM.01162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, Brown A, Allen N, Naylor J, Stange-Thomann N, DeArellano K, Johnson R, Linton L, McEwan P, McKernan K, Talamas J, Tirrell A, Ye W, Zimmer A, Barber RD, Cann I, Graham DE, Grahame DA, Guss AM, Hedderich R, Ingram-Smith C, Kuettner HC, Krzycki JA, Leigh JA, Li W, Liu J, Mukhopadhyay B, Reeve JN, Smith K, Springer TA, Umayam LA, White O, White RH, Conway de Macario E, Ferry JG, Jarrell KF, Jing H, Macario AJL, Paulsen I, Pritchett M, Sowers KR, et al. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res 12:532–542., doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyu Z, Jain R, Smith P, Fetchko T, Yan Y, Whitman WB. 2016. Engineering the autotroph Methanococcus maripaludis for geraniol production. ACS Synth Biol 5:577–581. doi: 10.1021/acssynbio.5b00267. [DOI] [PubMed] [Google Scholar]
- 40.Buan NR. 2018. Methanogens: pushing the boundaries of biology. Emerging Topics in Life Sciences 2:629–646. doi: 10.1042/etls20180031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulkarni G, Mand TD, Metcalf WW. 2018. Energy conservation via hydrogen cycling in the methanogenic archaeon Methanosarcina barkeri. mBio 9:e01256-18. doi: 10.1128/mBio.01256-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieber DJ, Catlett J, Madayiputhiya N, Nandakumar R, Lopez MM, Metcalf WW, Buan NR. 2014. A multienzyme complex channels substrates and electrons through acetyl-CoA and methane biosynthesis pathways in Methanosarcina. PLoS One 9:e107563. doi: 10.1371/journal.pone.0107563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bott RR, Cervin MA, Kellis JT, McAuliffe JC, Miasnikov A, Peres CM, Rife CL, Wells DH, Weyler W, Whited GM. 2008. Isoprene synthase variants for improved microbial production of isoprene. US patent US20100003716A1. https://patents.google.com/patent/US20100003716A1/en.
- 44.Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 45.Yang C, Gao X, Jiang Y, Sun B, Gao F, Yang S. 2016. Synergy between methylerythritol phosphate pathway and mevalonate pathway for isoprene production in Escherichia coli. Metab Eng 37:79–91. doi: 10.1016/j.ymben.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 46.Yao Z, Zhou P, Su B, Su S, Ye L, Yu H. 2018. Enhanced isoprene production by reconstruction of metabolic balance between strengthened precursor supply and improved isoprene synthase in Saccharomyces cerevisiae. ACS Synth Biol 7:2308–2316. doi: 10.1021/acssynbio.8b00289. [DOI] [PubMed] [Google Scholar]
- 47.Sowers KR, Boone JE, Gunsalus RP. 1993. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl Environ Microbiol 59:3832–3839. doi: 10.1128/AEM.59.11.3832-3839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metcalf WW, Zhang JK, Apolinario E, Sowers KR, Wolfe RS. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc Natl Acad Sci U S A 94:2626–2631. doi: 10.1073/pnas.94.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang JK, White AK, Kuettner HC, Boccazzi P, Metcalf WW. 2002. Directed mutagenesis and plasmid-based complementation in the methanogenic archaeon Methanosarcina acetivorans C2A demonstrated by genetic analysis of proline biosynthesis. J Bacteriol 184:1449–1454. doi: 10.1128/jb.184.5.1449-1454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welander PV, Metcalf WW. 2008. Mutagenesis of the C1 oxidation pathway in Methanosarcina barkeri: new insights into the Mtr/Mer bypass pathway. J Bacteriol 190:1928–1936. doi: 10.1128/JB.01424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buan N, Kulkarni G, Metcalf W. 2011. Genetic methods for methanosarcina species. Methods Enzymol 494:23–42. doi: 10.1016/B978-0-12-385112-3.00002-0. [DOI] [PubMed] [Google Scholar]
- 52.Li MZ, Elledge SJ. 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 53.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. doi: 10.1128/JB.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aldridge JT, Catlett JL, Smith ML, Buan NR. 2016. Methods for detecting microbial methane production and consumption by gas chromatography. Bio Protoc 6:e1779. doi: 10.21769/bioprotoc.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aldridge J, Carr S, Weber KA, Buan NR. 2018. Production of bioisoprene from wastewater. Report no. NTRY6R14/4822. The Water Research Foundation, Denver, CO. https://www.waterrf.org/research/projects/production-bioisoprene-wastewater. [Google Scholar]
- 56.Bose A, Kulkarni G, Metcalf WW. 2009. Regulation of putative methyl-sulphide methyltransferases in Methanosarcina acetivorans C2A. Mol Microbiol 74:227–238. doi: 10.1111/j.1365-2958.2009.06864.x. [DOI] [PubMed] [Google Scholar]
- 57.Rao X, Huang X, Zhou Z, Lin X. 2013. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath 3:71–85. [PMC free article] [PubMed] [Google Scholar]
- 58.Egan AJ, Vollmer W. 2013. The physiology of bacterial cell division. Ann N Y Acad Sci 1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- 59.Yamada EA, Sgarbieri VC. 2005. Yeast (Saccharomyces cerevisiae) protein concentrate: preparation, chemical composition, and nutritional and functional properties. J Agric Food Chem 53:3931–3936. doi: 10.1021/jf0400821. [DOI] [PubMed] [Google Scholar]
- 60.Diner BA, Fan J, Scotcher MC, Wells DH, Whited GM. 2017. Synthesis of heterologous mevalonic acid pathway enzymes in Clostridium ljungdahlii for the conversion of fructose and of syngas to mevalonate and isoprene. Appl Environ Microbiol 84:e01723-17. doi: 10.1128/AEM.01723-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pade N, Erdmann S, Enke H, Dethloff F, Duhring U, Georg J, Wambutt J, Kopka J, Hess WR, Zimmermann R, Kramer D, Hagemann M. 2016. Insights into isoprene production using the cyanobacterium Synechocystis sp. PCC 6803. Biotechnol Biofuels 9:89. doi: 10.1186/s13068-016-0503-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmids, strains, and growth and assay data that support the findings of this study are available from the corresponding author (N.R.B.) upon reasonable request. The sequence for sNB19 was submitted to GenBank (MW295460) but not released prior to publication; it is expected to be released shortly. This sequence may also be found in the supplemental material.