The potential markets for seaweed-derived products and seaweed processing technologies are growing, yet commercial enzyme cocktails for complete conversion of seaweed to fermentable sugars are not available. Such an enzyme cocktail would require the catalytic properties of a variety of different enzymes, where fucoidanases, laminarinases, and cellulases together with endo- and exo-acting alginate lyases would be the key enzymes.
KEYWORDS: DEH, DHF, alginate lyase, exo-activity, biorefining, Saccharina latissima, thermal stability, NMR, nuclear magnetic resonance
ABSTRACT
Enzymatic depolymerization of seaweed polysaccharides is gaining interest for the production of functional oligosaccharides and fermentable sugars. Herein, we describe a thermostable alginate lyase that belongs to polysaccharide lyase family 17 (PL17) and was derived from an Arctic Mid-Ocean Ridge (AMOR) metagenomics data set. This enzyme, AMOR_PL17A, is a thermostable exolytic oligoalginate lyase (EC 4.2.2.26), which can degrade alginate, poly-β-d-mannuronate, and poly-α-l-guluronate within a broad range of pHs, temperatures, and salinity conditions. Site-directed mutagenesis showed that tyrosine Y251, previously suggested to act as a catalytic acid, indeed is essential for catalysis, whereas mutation of tyrosine Y446, previously proposed to act as a catalytic base, did not affect enzyme activity. The observed reaction products are protonated and deprotonated forms of the 4,5-unsaturated uronic acid monomer, Δ, two hydrates of DEH (4-deoxy-l-erythro-5-hexulosuronate), which are formed after ring opening, and, finally, two epimers of a 5-member hemiketal called 4-deoxy-d-manno-hexulofuranosidonate (DHF), formed through intramolecular cyclization of hydrated DEH. The detection and nuclear magnetic resonance (NMR) assignment of these hemiketals refine our current understanding of alginate degradation.
IMPORTANCE The potential markets for seaweed-derived products and seaweed processing technologies are growing, yet commercial enzyme cocktails for complete conversion of seaweed to fermentable sugars are not available. Such an enzyme cocktail would require the catalytic properties of a variety of different enzymes, where fucoidanases, laminarinases, and cellulases together with endo- and exo-acting alginate lyases would be the key enzymes. Here, we present an exo-acting alginate lyase that efficiently produces monomeric sugars from alginate. Since it is only the second characterized exo-acting alginate lyase capable of degrading alginate at a high industrially relevant temperature (≥60°C), this enzyme may be of great biotechnological and industrial interest. In addition, in-depth NMR-based structural elucidation revealed previously undescribed rearrangement products of the unsaturated monomeric sugars generated from exo-acting lyases. The insight provided by the NMR assignment of these products facilitates future assessment of product formation by alginate lyases.
INTRODUCTION
Environmental challenges and decreasing amounts of fossil resources call for a transition that implies the use of more sustainable resources for production of energy, fuels, and commodity chemicals. Marine macroalgae such as brown algae (seaweed) are currently receiving a lot of attention as a source of renewable biomass. The global seaweed cultivation market has been estimated to be $16.7 billion (U.S. dollars [USD]) in 2020 and is projected to reach $30.2 billion (USD) by 2025, due to inclusion in cosmetics and personal care products, as well as consumer adoption of plant-based food products (data from MarketsandMarkets [1]). These algae are among the fastest-growing organisms in nature, and their growth may lead to production of large amounts of biomass without the use of freshwater, agricultural land, fertilizer, or pesticides (2). Macroalgal biomass therefore represents an attractive and important alternative source for the production of biofuels (3, 4), chemicals (5), or single-cell proteins (6).
Brown seaweeds are the dominating seaweed found along the Norwegian coastline and are rich in carbohydrates like alginate, cellulose, laminarin, and mannitol. Mannitol can be fermented directly (7), and cellulose and laminarin can be hydrolyzed to glucose by using currently available commercial enzyme cocktails (8). The additional degradation of the alginate component would strengthen the potential of seaweed as a source of fermentable sugars. Alginate is the main structural component of the cell walls of brown macroalgae and has been shown to comprise up to 40% of the dry mass of species like Laminaria hyperborea (9). Alginate is a linear copolymer of the uronic acid β-d-mannuronate (M) and its C-5 epimer α-l-guluronate (G), arranged as homopolymeric blocks of 1,4-linked consecutive M residues (polyM) or G residues (polyG) or as blocks of alternating M- and G residues (polyMG) (10).
Alginate lyases are enzymes capable of depolymerizing alginate and have been identified in a range of organisms, including bacteria, marine fungi, animals, algae, and viruses (11). They are categorized as polysaccharide lyases (PLs) in the Carbohydrate-Active enZYme (CAZy) database (12). The majority of alginate lyases are classified into PL families 5 to 7, 14 to 15, and 17 to 18 (13), and there are currently three types: mannuronate-specific lyases that cleave M-M and M-G bonds (EC 4.2.2.3), guluronate-specific lyases that cleave G-G and G-M bonds (EC 4.2.2.11), and oligo-alginate lyases that can exolytically degrade oligomers produced by endolytic alginate lyases (EC 4.2.2.26). Most endolytic alginate lyases are found in families PL5 and PL7, while exolytic alginate lyases are found in families PL6 and -7, PL14 and -15, and PL17 (14).
To achieve full saccharification of alginate, both endo- and exolytic alginate lyases are necessary. Depending on their properties, endolytic lyases convert alginate substrates to di-, tri-, tetra-, or pentamers (15), while exolytic lyases generate monomers from alginate oligomers. Saccharification can also be achieved by thermochemical means, but enzymatic conversion is generally considered more sustainable (16). Saccharification of seaweed biomass, as opposed to just alginate, would require a multitude of enzymes that are capable of degrading the variety of biopolymer structures found within seaweeds, including fucoidanases (17), laminarinases (18), and cellulases (19). However, due to the dominating structural role of alginate, endo- and exo-acting alginate lyases are key enzymes. The viscosity-reducing effect of endo-acting alginate lyases is important for efficient enzymatic degradation of other seaweed macromolecules (20, 21).
Most identified alginate lyases today are endolytic, but at least 13 exolytic alginate lyases have been described in the literature (based on the occurrence of EC 4.2.2.26 in CAZy [summarized in Table 1]) from a range of organisms, including Sphingomonas sp. (22–24), Agrobacterium tumefaciens (25), Saccharophagus degradans (26), Zobellia galactanivorans (27), Vibrio splendidus (4, 28), Shewanella sp. (29, 30), Stenotrophomonas maltophilia (31), Thalassotalea crassostreae (32), and Haliotis discus subsp. hannai (33). Structural data are only available for two of these exolytic alginate lyases so far: Alg17C (34) in the PL17 family and Atu3025 (35) in the PL15 family.
TABLE 1.
Previously characterized exolytic alginate lyasesa
| Name | Organism | CAZy no. | Optimal enzyme condition |
Reference(s) | |
|---|---|---|---|---|---|
| pH | Temp (°C) | ||||
| A1-IV | Sphingomonas sp. strain A1 | PL15 | 7.5–8.5 | 37 | 22 |
| Atu3025 | Agrobacterium tumefaciens C58 | PL15 | 7.3 | 30 | 25 |
| AlgL | Sphingomonas sp. strain MJ-3 | PL17 | 6.5 | 50 | 23 |
| Alg17C | Saccharophagus degradans 2-40 | PL17 | 6.0 | 40 | 26 |
| AlyA5 | Zobellia galactanivorans | PL7 | 7.0 | NRb | 27 |
| OalA | Vibrio splendidus | PL15 | 6.5 | 16 | 4, 28 |
| OalB | Vibrio splendidus | PL17 | 7.0 | 30 | 4, 28 |
| OalC | Vibrio splendidus | PL17 | 7.5 | 35 | 4, 28 |
| OalS17 | Shewanella sp. strain Kz7 | PL17 | 6.2 | 50 | 29 |
| OAL | Stenotrophomonas maltophilia KJ-2 | PL17 | 7.5 | 37 | 31 |
| ZH0-IV | Sphingomonas sp. strain ZH0 | PL15 | 7.0 | 37 | 24 |
| TcAlg1 | Thalassotalea crassostreae | PL17 | 7.0 | 40 | 32 |
| OalS6 | Shewanella sp. strain Kz7 | PL6 | 7.2 | 40 | 30 |
| HdAlex | Haliotis discus subsp. hannai | PL14 | 7.1 | 42 | 33 |
| AlyRm4 | Rhodothermus marinus strain 378 | PL17 | 6.5 | 81 | 43 |
The table shows optimal enzyme conditions reported for previously characterized exolytic alginate lyases (EC 4.2.2.26) in the CAZy database (62). All have been reported to produce unsaturated monomers (i.e., Δ), except for HdAlex, which was reported to produce unsaturated dimers. The site of initial attack has only been reported for two enzymes: AlyA5, which degrades from the nonreducing end, and HdAlex, which degrades from the reducing end.
NR, not reported.
In this study, we describe the cloning, purification, and characterization of a novel, thermostable exolytic alginate lyase (AMOR_PL17A), whose gene was retrieved from a metagenomic data set collected from the Arctic Mid-Ocean Ridge (AMOR). We show that AMOR_PL17A converts alginate to unsaturated monomers referred to as Δ (4-deoxy-l-erythro-4-hexenopyranouronate), which, in an aqueous solution, are spontaneously converted to hydrated 4-deoxy-l-erythro-5-hexulosuronate (DEH). Using in-depth nuclear magnetic resonance (NMR) analyses, we show that intramolecular cyclization of the DEH aldo-hydrate results in two 5-member hemiketal ring epimers [4-deoxy-d-manno-(5S)-hexulofuranosidonate hydrate and 4-deoxy-d-manno-(5R)-hexulofuranosidonate hydrate], referred to here as (5S)- and (5R)-DHF, respectively. Under the conditions examined here, these DHF hydrates are the major products formed, and they have not previously been associated with alginate lyase activity.
RESULTS
Sequence analysis.
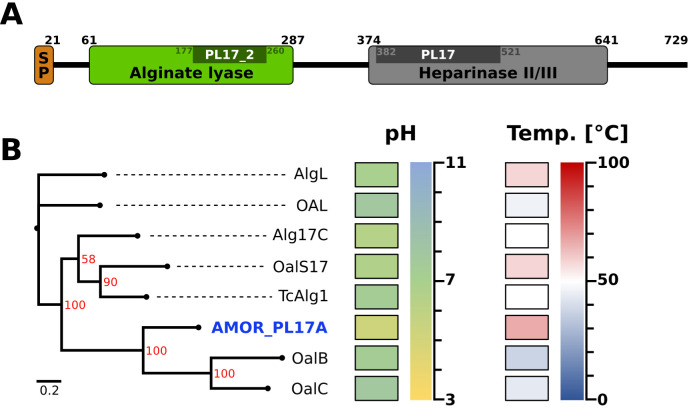
An analysis of the AMOR_PL17 amino acid sequence using Pfam revealed two domains typical of PL17 enzymes: one domain called “alginate lyase domain” (PF05426) and the other called the “heparinase II/III domain” (PF07940) (Fig. 1A). A similar domain structure also occurs in PL15 family exo-acting enzymes. Using SignalP, we detected a signal peptide with a predicted cleavage site between residues 21 and 22. Phylogenetic analysis of all exolytic PL17s described in the literature (Fig. 1B) showed that the two closest relatives of AMOR_PL17A are OalB and OalC from Vibrio splendidus. All residues thought to be essential for catalysis and/or to contribute to substrate binding, based on structural studies of S. degradans Alg17C (SdAlg17C [26]), are conserved in AMOR_PL17A (see Fig. S1 in the supplemental material), including Tyr251 and Tyr446, which are discussed below. AMOR_PL17A was produced without its signal peptide (amino acids 18 to 729) and with a C-terminal His tag (AHHHHHH) and was purified to high purity (see Fig. S2 in the supplemental material).
FIG 1.
Sequence analysis reveals two domains in AMOR_PL17A. (A) Pfam analysis of the AMOR_PL17A amino acid sequence revealed the presence of two domains: an alginate lyase domain (PF05426) and a heparinase II/III domain (PF07940). Darker regions indicate catalytic domains detected by dbCAN. Residues 1 to 21 are predicted to be a signal peptide. AMOR_PL17A was produced without its signal peptide and with a C-terminal His tag (AHHHHHH). (B) Phylogenetic tree of AMOR_PL17A and the PL17 exolytic alginate lyases listed in Table 1 (except AlyRm4, for which the sequence is not available). Differences between the amino acid sequences are indicated by distance in the horizontal lines; the scale bar indicates the number of amino acid substitutions per site. Optimal reaction conditions for pH and temperature are indicated as heat maps (see Table 1 for details). The tree was generated from a multiple-sequence alignment using Clustal Omega and IQ-Tree and visualized with FigTree. Bootstrap support values (red) were generated from 1,000 iterations.
Characterization of AMOR_PL17A. (i) AMOR_PL17A is active on alginate, polyM, and polyG.
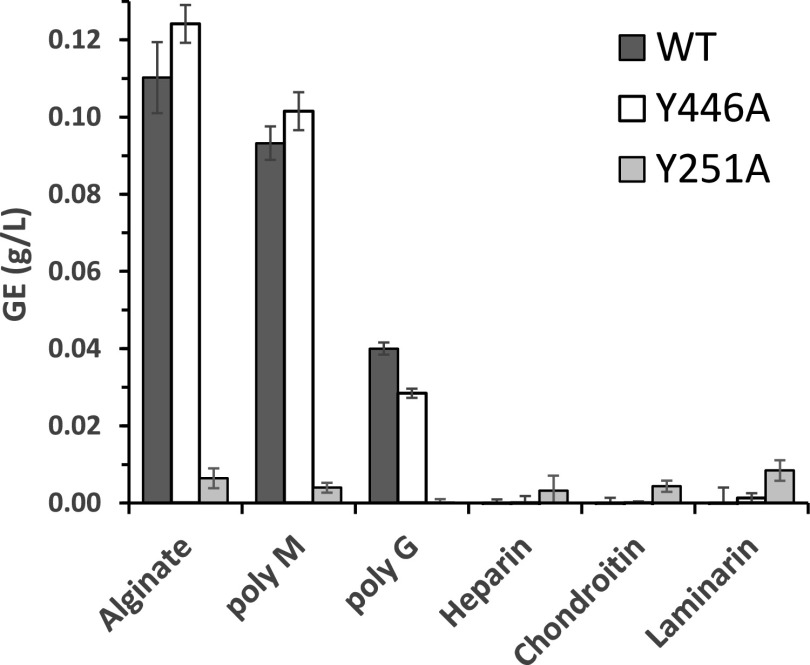
AMOR_PL17A was tested for activity on alginate, polyG, polyM, heparin, chondroitin, and laminarin (Fig. 2). Additionally, two mutant enzymes containing two different Tyr→Ala point mutations of the predicted catalytic acid and base, Y251A and Y446A (34), were tested for activity. Wild-type AMOR_PL17A was active on alginate, polyM, and, to a lesser extent, polyG. This activity was maintained in the Y446A mutant but abolished in the Y251A mutant. Neither the wild-type enzyme nor any of the mutants showed detectable activity on heparin, chondroitin, or laminarin under the conditions tested, when analyzed with the reducing-ends assay (Fig. 2) or by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) for product analysis (data not shown).
FIG 2.
AMOR_PL17A is active on alginate, polyM, and polyG. AMOR_PL17A and the two active site mutants Y251A and Y446A were tested for activity on four different brown algae-derived substrates plus chondroitin and heparin. Product formation was measured as reducing sugars and quantified as glucose equivalents (GE). The reaction conditions were 1 μM enzyme, 0.1% (wt/vol) substrate, 25 mM NaOAc (pH 5.6), and 300 mM NaCl, and the reaction mixture was incubated for 20 h at 60°C. Standard deviations from three independent experiments are indicated. HPAEC-PAD analysis confirmed that no products were generated from heparin and chondroitin.
(ii) AMOR_PL17A is thermostable and active within a broad range of pHs, temperatures, and salinities.
The activity of AMOR_PL17A was screened to find optimal reaction conditions with respect to pH, salinity, temperature, and thermal stability. Initial measurements of progress curves under standard conditions showed that product formation was linear over time for at least 10 min (data not shown). For the enzyme characterization, reactions were run for only 5 min to minimize stability effects on the observed enzyme activity. The enzyme showed maximum activity at a salt concentration of 200 to 300 mM NaCl and was close to inactive in the absence of salt (Fig. 3A). Five-minute reactions with mixtures incubated at 40 to 90°C showed that temperature had a large effect on enzyme activity, with product yields being 18-fold higher at 90°C than at 40°C (Fig. 3B). These results do not account for thermal stability though and cannot be used to make assumptions regarding the enzyme’s response to these temperatures during extended incubations. Assessment of the effect of pH on activity showed that the enzyme has a broad pH optimum in the region of pH 5.0 to 6.0 (Fig. 3C). At acidic pH, there was a dramatic shift from no activity at pH 4.3 to almost maximal activity at pH 4.8. Thermal stability was assessed by preincubating AMOR_PL17A at 60 and 65°C for 0 to 24 h followed by a standard 5-min activity assay at 50°C. This experiment revealed that AMOR_PL17A was stable for 24 h at 60°C, while its activity declined during preincubation at 65°C (Fig. 3D).
FIG 3.
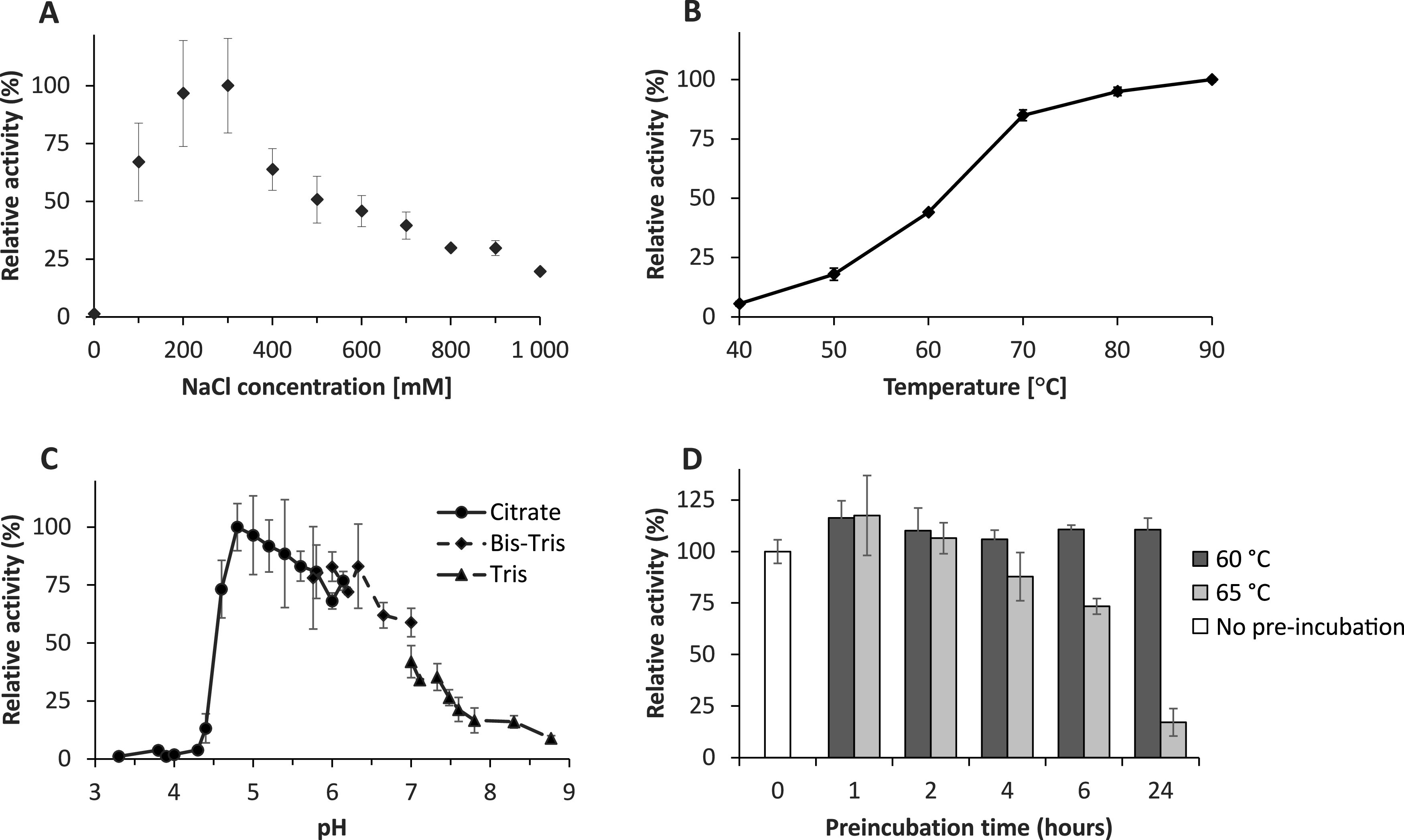
Optimal reaction conditions for AMOR_PL17A when degrading sodium alginate. (A) Activity at various NaCl concentrations. A reaction with 2,000 mM NaCl gave only 7% relative activity (not shown in the figure). (B) Activity at various temperatures. (C) Activity at various pHs, tested using three different buffers (30 to 45 mM), as indicated in the figure (citrate-citric acid, Bis-Tris-HCl, and Tris-HCl). (D) Thermal stability test where the enzyme (pH 5.6, 50 mM NaOAc, 2 μM enzyme, 600 mM NaCl) was preincubated for 0 to 24 h at 60 or 65°C before dilution with a substrate solution to achieve standard reaction conditions. Unless otherwise stated, all reactions were performed in triplicate with 1 μM enzyme, 0.5% (wt/vol) alginate, 25 mM NaOAc (pH 5.6), and 300 mM NaCl (150 mM for the pH screening) at 50°C. Reactions were stopped after 5 min only, to maximize the chance of having initial rate conditions, and enzyme activity was measured as release of reducing sugars quantified as glucose equivalents. The graphs show standard deviations derived from three independent experiments.
(iii) AMOR_PL17A is exolytic and cleaves from the nonreducing end.
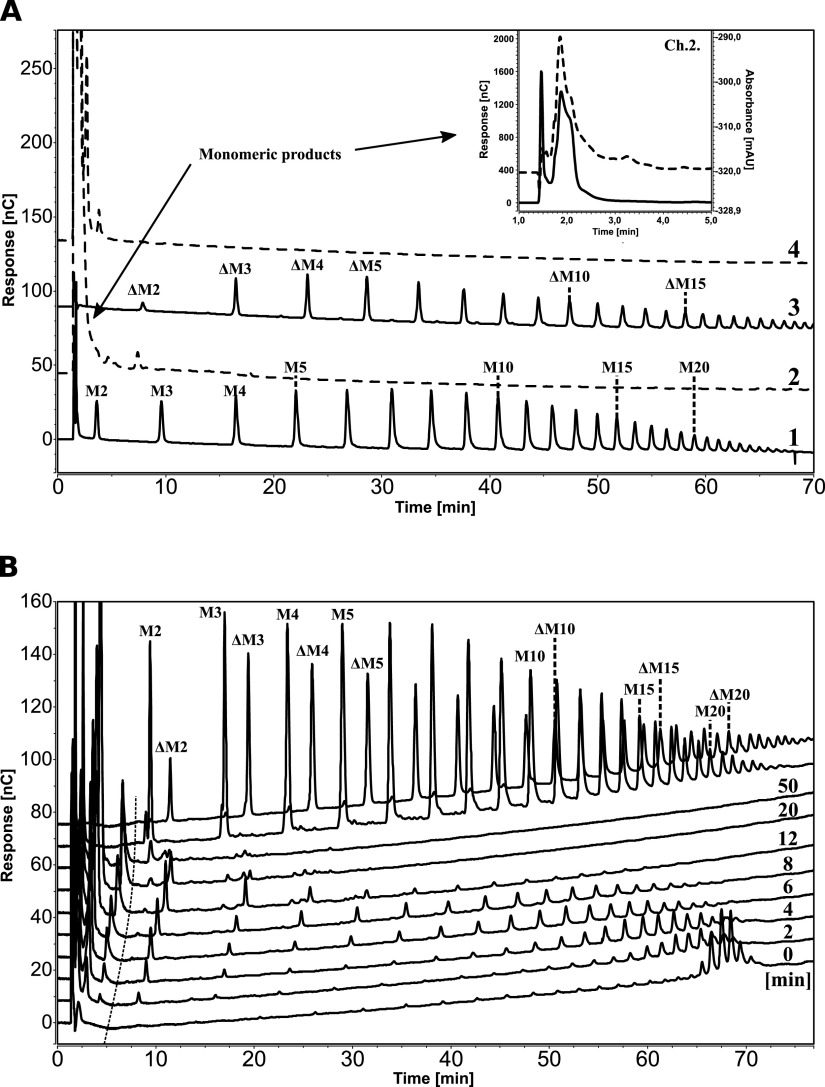
High-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) was used to analyze product formation by AMOR_PL17A when degrading polyM- and polyG-based model substrates. PolyM and polyG are large, saturated polymers reminiscent of the M and G blocks found in natural alginate. One-hour reactions with polyM and polyG yielded only one product peak in the chromatograms, eluting with about a 2-min retention time, indicative of a monomeric product based on the short retention time (see Fig. S3 in the supplemental material). Similarly, two different polyM-derived substrates, an acid hydrolysate (i.e., a mixture of fully saturated M oligomers of various degrees of polymerization [DPs]) and an endo-lyase-treated polyM (i.e., a mixture of M oligomers of various DPs with a 4,5-unsaturated uronic acid residue, Δ, at the nonreducing end), revealed that AMOR_PL17A was able to degrade all mannuronate oligomers irrespective of the nature of the nonreducing end (Fig. 4A) and yielded one large product peak with a retention time of about 2 min (Fig. 4A, inset).
FIG 4.
HPAEC-PAD analysis of products generated by AMOR_PL17A indicates exolytic activity. (A) Conversion of acid-hydrolyzed polyM (a mixture of saturated M oligomers of various DPs) and endo-lyase-treated polyM (a mixture of unsaturated M oligomers of various DPs with a Δ at the nonreducing end). Chromatograms 1 and 3 show the respective substrates, with peak IDs based on previous identifications (20), whereas chromatograms 2 and 4 show products generated from the saturated and unsaturated substrate, respectively, after incubation for 1 h with AMOR_PL17A. The products have retention times of 1 to 5 min. The inset shows an enlargement of the monomeric region for chromatogram 2 with overlay of the UV trace (235 nm [dotted line]). (B) Degradation of purified M24 (polyM with a DP of 24 [61]) over time. The chromatograms are labeled with the time points (0 to 50 min); the two uppermost chromatograms show standards generated by treating polyM with an endo-lyase (unsaturated products) or by acid (saturated products). The dotted almost vertical line indicates a nonlinear drift in the HPAEC-PAD-system causing a time-dependent shift in retention time. Note that all oligomers generated from M24 by AMOR_PL17A were unsaturated.
Alginate lyases introduce a double bond in the sugar that is on the reducing end side of the scissile glycosidic bond, which implies that the product emerging at this side will have an unsaturated sugar at its reducing end. As an example, the action of an exolytic alginate lyase acting on one of the ends of a saturated M pentamer could have the following outcomes: MMMMM → M + ΔMMM or MMMM + Δ. To determine the mode of cleavage (exo/endo) for AMOR_PL17A, we studied the degradation of saturated M24 (i.e., polyM with a DP of 24) over time (Fig. 4B). We observed a time-dependent shift of oligomer lengths from relatively long oligomers in the beginning of the reaction to gradually shorter oligomers after longer incubation times. All intermediate products were unsaturated, which, considering the nature of the lyase reaction, means that they contained a Δ residue at their nonreducing ends. Both endolytic activity and exolytic activity would yield progressively shorter oligomer lengths. However, endolytic catalysis would yield large variation of oligomer lengths early in the reaction as the M24 oligomer would be cleaved at multiple internal sites, resulting in an even distribution of different oligomer lengths. Furthermore, since the starting material is a saturated M24, endolytic activity would initially yield a mixture of saturated and unsaturated oligomeric products, and over time the unsaturated oligomers would dominate. The product profile depicted in Fig. 4B thus clearly shows that AMOR_PL17A is exo-acting: the product length gradually decreases, and in accordance with exo-activity, the peak eluting at about 2 min increases as the reaction proceeds. For example, at an 8-min reaction time, we observe ΔM oligomers with DPs of 2 to 19 and a product peak at about 2 min. The fact that all intermediate products are unsaturated shows that AMOR_PL17A acts from the nonreducing end of the substrate. Thus, AMOR_PL17A degrades M24 by removing one monomer at the time from the nonreducing end of the substrate, generating gradually shorter intermediate oligomers with unsaturated nonreducing ends and accumulating a monomeric product (the large majority of which is unsaturated [see below for further details]). Importantly, the combined data depicted in Fig. 4 and Fig. S3 show that AMOR_PL17A can cleave off M, G, and Δ residues from the nonreducing end.
(iv) The product generated by AMOR_PL17A is monomeric and reacts with water to produce six hydrated structures.
Two preparations of polyM treated with AMOR_PL17A were incubated at two different temperatures and analyzed with NMR spectroscopy to elucidate the structures of the reaction products. The first reaction was completed in deuterated water (D2O) and incubated in tube within the NMR magnet at 25°C for ∼17 h. This low temperature was selected to slow the process in order to allow structural elucidation of the unsaturated monomer. The second reaction was completed in H2O and incubated in a benchtop thermocycler at 50°C for ∼48 h. This was to allow full conversion of the substrate and was used to characterize the major products of the reaction mixture. One final preparation was used for real-time monitoring of product formation and interconversion by time-resolved NMR. This in-tube reaction was completed in D2O, the mixture was incubated within the NMR magnet at 50°C for ∼4 h, and constantly monitored with 1H NMR.
The NMR analyses supported the above HPAEC data—i.e., that AMOR_PL17A lyase is exo-acting and converts polyM substrate into monomers from the nonreducing end. Exolytic activity is confirmed by both the absence of oligomeric signals (Fig. 5A; see Fig. S5 to S8 in the supplemental material) and the presence of monomeric products (Fig. 5 and as described below). Two 1H signals at δH 4.89 and 5.20 that represent the anomeric signal of a saturated monomer, M, were observed. These signals remain constant throughout the reaction, which shows that AMOR_PL17A cleaves from the nonreducing end, initially producing one saturated M monomer followed by production of multiple unsaturated M monomers as the substrate is further degraded.
FIG 5.
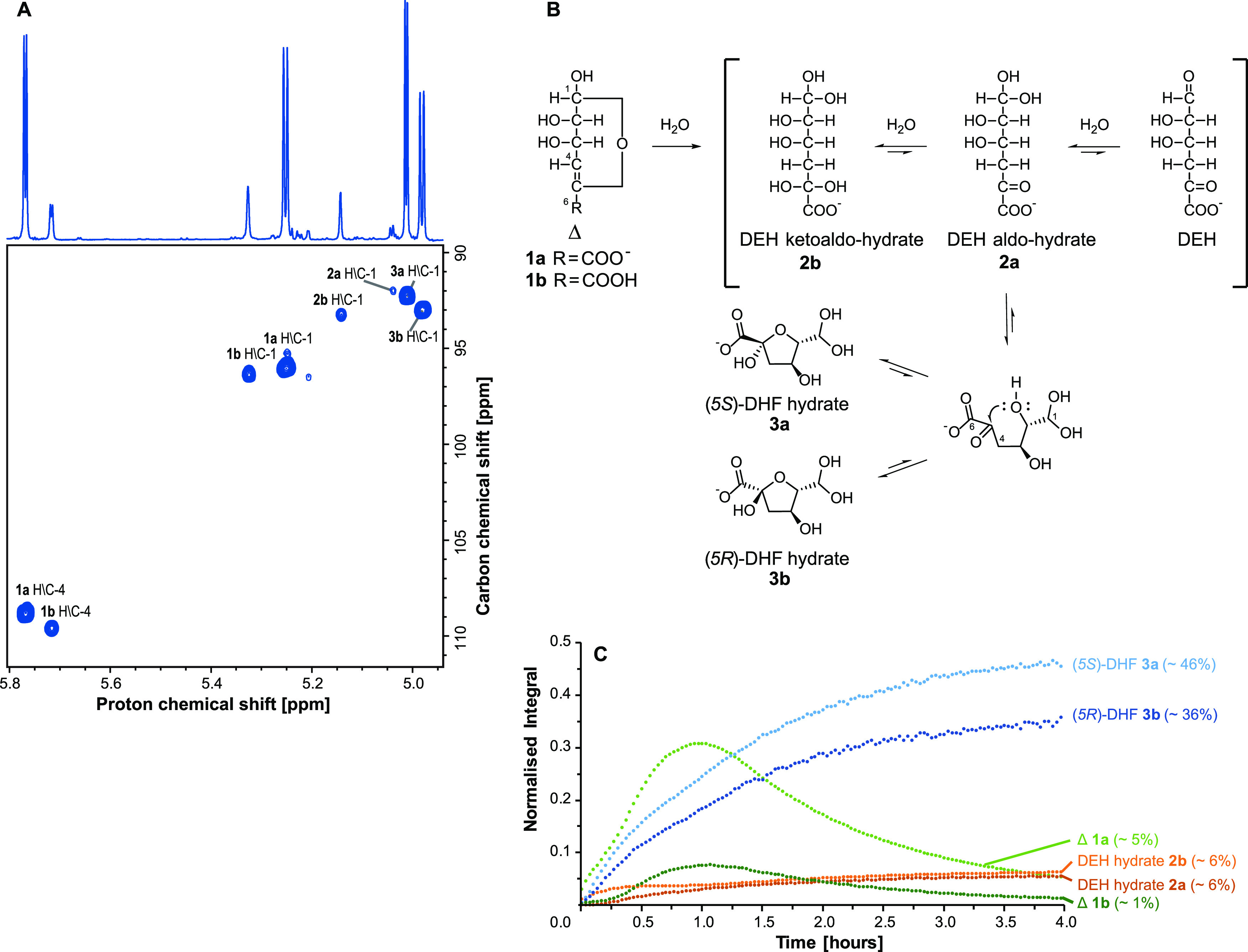
NMR analysis reveals six reaction products when degrading polyM with AMOR_PL17A. (A) Annotated 1H-13C HSQC spectrum showing the region of interest for reaction products obtained after a 16-h 40-min incubation at 25°C (1% [wt/vol] polyM substrate, 2 μM AMOR_PL17A, 10 mM NaOAc, 200 mM NaCl [pH 5.6] in 99.9% D2O). The spectrum shows the assignment of proton and carbon chemical shifts associated with the doubly oxygenated methines for all observed products (C/H-1 products 1a, 1b, 2a, 2b, 3a, and 3b), as well as olefin signals for Δ (C/H-4 products 1a and 1b). The 1D proton spectrum is shown along the top. (B) Scheme detailing the ring opening of Δ (4-deoxy-l-erythro-4-hexenopyranuronate 1a and 4-deoxy-l-erythro-4-hexenopyranuronic acid 1b), formation of DEH (4-deoxy-l-erythro-5-hexulosuronate) and DEH hydrates (2a and 2b), and formation of two DHF epimers (4-deoxy-d-manno-(5S)-hexulofuranosidonate hydrate 3a and 4-deoxy-d-manno-(5R)-hexulofuranosidonate hydrate 3b). Ring opening of Δ leads to the formation of DEH. Structures 2a and 2b were observed by NMR and can be rationalized by the hydration of ketone and aldehyde groups of DEH. Intramolecular cyclization of structure 2a through nucleophilic attack of the C-2 hydroxyl on the C-5 keto group yields two hemiketal C-5 epimers, 3a and 3b. (C) Integrated and sum-normalized H-1 signals of the six products 1a, 1b, 2a, 2b, 3a, and 3b plotted through time. Integrated signals are from 1H NMR spectra acquired while incubating a reaction mixture at 50°C for ∼4 h (1% [wt/vol] polyM substrate, 2 μM AMOR_PL17A, 10 mM NaOAc, 200 mM NaCl [pH 5.6] in 99.9% D2O). The integrals were normalized to the sum of the H-1 signals of all six products in the final spectrum (at time 4 h, the sum of integrals = 1). Relative abundance of each component in the final spectrum (at time 4 h) is represented as a percentage. At the beginning of the incubation (∼1 h), the signals associated with Δ (1a and 1b H-1) and the DHF hydrates (3a and 3b H-1) all increase. As the reaction proceeds, signals associated with the DHF hydrates continue to increase, while those associated with Δ (1a and 1b) decrease as it undergoes ring opening.
The NMR analyses allow the identification of a total of six structures (in the supplemental material, see Table S1 for chemical shift assignments, Fig. S5 to S8 for two-dimensional [2D] NMR spectra, and Text S1 for detailed description of structural assignments). The first two structures are assigned as cyclic 4,5-unsaturated uronate and uronic acid monomers, referred to as Δ. Under the examined reaction conditions (pH 5.6), the anionic carboxylate (structure 1a, 4-deoxy-l-erythro-4-hexenopyranuronate) dominates with a minor amount of the protonated species (structure 1b, 4-deoxy-l-erythro-4-hexenopyranuronic acid). The second two structures are two DEH (4-deoxy-l-erythro-5-hexulosuronate) hydrates that are formed after ring opening of Δ. One is the aldo-hydrate that retains a keto group in position 5 (structure 2a), and the other is the ketoaldo-hydrate (structure 2b). Finally, there are two cyclic structures formed after intramolecular cyclization of DEH aldo-hydrate 2a. These are cyclic hemiketals that are epimers at C-5 and called 4-deoxy-d-manno-(5S)-hexulofuranosidonate hydrate [structure 3a, referred to here as (5S)-DHF] and 4-deoxy-d-manno-(5R)-hexulofuranosidonate hydrate [structure 3b, referred to here as (5R)-DHF].
Interpretation of 2D NMR spectra allowed the structural elucidation of the deprotonated and protonated species of Δ (structures 1a and 1b). These structures are characterized by anomeric methines at position 1 (e.g., in structure 1a, C-1 δC 96.1, H-1 δH 5.25) together with methine signals of the newly formed double bonds at position 4 (e.g., in structure 1a, C-4 δC 108.9, H-4 δH 5.77), as observed in the heteronuclear single quantum coherence (HSQC) spectrum (Fig. 5A). In addition to this, in structure 1a, heteronuclear multiple-bond correlations (HMBCs) (Fig. S6) from H-4 further confirm the presence of a double bond (δC 148.2) and carboxylic acid (δC 172.0) in positions C-5 and C-6, respectively. The observed three-bond HMBC between H-1 and the C-5 quaternary carbon is consistent with a 6-member hemiacetal ring. Additionally, these assignments for protonated and deprotonated Δ are consistent with literature values reported for the unsaturated uronic acid formed from a glucuronic acid residue (a C-2 epimer of mannuronic acid) (36, 37). It is widely accepted that Δ will undergo spontaneous ring opening and produce linear products (3, 30, 36). Cleavage of the C-O bond can result in the formation of an enolate at C-4 and C-5 that can rearrange to afford DEH and various hydrated forms when in the presence of water (e.g., products 2a and 2b in Fig. 5B). Hydration can occur at either or both C-1 and C-5, which can exist either as fully hydrated geminal diols or as aldehyde and keto groups, respectively (38). Under the conditions examined here, two DEH hydrates were characterized. These include DEH aldo-hydrate (structure 2a), which has a geminal diol at C-1 and ketone group at C-5, and a ketoaldo-hydrate (structure 2b) with geminal diols at both C-1 and C-5.
The ring opening and conversion of Δ (structures 1a and 1b) to linear structures was also monitored by UV as the double bond between C-4 and C-5 in Δ can absorb UV light at 235 nm (39). Examination of the UV-activity of AMOR_PL17A’s products over time showed a decline over the first 24 h (see Fig. S4 in the supplemental material). Interestingly, both the initial UV absorption and the rate of decline were dependent on the method used for stopping the enzymatic reaction (heat, HCl, or NaOH [Fig. S4]). Noteworthy, UV absorption of samples containing NaOH was 2.6 times higher than those of samples of heat-inactivated or acid-inactivated reactions. Although details are lacking, already in 1962, it was reported that UV absorption of unsaturated uronic acids depends on pH (40), which could explain these observations. The decline of UV absorption over time can be explained by the ring opening and conversion of Δ to DEH, which leads to loss of the conjugated π-system that is responsible for the UV absorbance.
Partial structures for both the DEH hydrates 2a and 2b were assigned from 2D NMR spectra. In both structures, the 13C and 1H chemical shifts at position 1 are consistent with geminal diols (e.g., in structure 2a, C-1 δC 92.0, H-1 δH 5.04), as observed in the HSQC spectrum (Fig. 5A). Methylene groups are observed in both structures at position 4 (e.g., in structure 2a, C-4 δC 45.0, H-4 δH 3.10, H-4′ δH 3.10 [Fig. S7]), which are expected to result from rearrangement of the enol moiety in Δ after ring opening. Finally, HMBC correlations (Fig. S8) between H-4 and C-5 quaternary carbons revealed a carbonyl present in structure 2a (δC 206.7) and a doubly oxygenated carbon in structure 2b (δC 98.5) consistent with a geminal diol. The final structures characterized in the product mixture are two 5-member hemiketal stereoisomers (DHF hydrates 3a and 3b in Fig. 5B). These structures can be rationalized by intramolecular cyclization of the DEH aldo-hydrate 2a, as a result of nucleophilic attack of the hydroxyl group of C-2 on the carbonyl group at C-5 (shown in the scheme in Fig. 5B).
Position 1, in both of the DHF hydrates, is characterized by 13C and 1H chemical shifts that are again consistent with geminal diols (e.g., in structure 3a, C-1 δC 92.3, H-1 δH 5.01), and those at position 4 are consistent with methylene groups (e.g., in structure 3a, C-4 δC 46.6, H-4 δH 2.55, H-4′ δH 2.06). Terminal carbonyl moieties were again observed at position 6 (e.g., in structure 3a, C-6 δC 179.8) through HMBCs (Fig. S8) from H-4 and H-4′. Several pieces of evidence indicate the structures comprise 5-member rings. The HMBC correlations between H-2 and C-5 (Fig. S8) can occur either as a three-bond correlation through an ether linkage or as a four-bond correlation through C-2, -3, and -4. However, the 13C chemical shifts of both C-2 (e.g., in structure 3a, δC 91.6) and C-5 (e.g., in structure 3a, δC 107.2) are consistent with an ether linkage between C-2 and C-5. Importantly, if the structures were present as 6-member rings, the 13C chemical shift of C-2 should be closer to the range typically observed for a primary alcohol in a carbohydrate residue (e.g., ∼70 ppm), and there should be a HMBC correlation observed between H-1 and C-5, which was completely absent. Intramolecular cyclization of structure 2a can yield two possible stereoisomers at C-5. Structure 3a was observed as the dominant structure (Fig. 5C and as described below) and is therefore assigned as the S-stereoisomer. The S-isomer is expected to be favored as steric hindrance offered by the geminal diol at C-1 likely hinders formation of the R-stereoisomer.
After structural characterization of the products, one further in-tube preparation (∼4 h of incubation at 50°C) was used to monitor the formation of these products using time-resolved NMR, providing additional insight into the formation of the final product mixture. During time-resolved NMR, a proton spectrum was collected every 2 min in order to monitor the product formation and visualize the relative abundance of each product as a function of time. The H-1 signals associated with the six characterized products (1a, 1b, 2a, 2b, 3a, and 3b) were integrated and normalized across all acquired spectra, and the normalized integral (i.e., the relative abundance) was plotted through time (Fig. 5C).
It can be observed that H-1 signals associated with Δ (structures 1a and 1b) increased at the beginning of the reaction, accompanied by an increase in the H-1 signals associated with DHF hydrates 3a and 3b. Initial increase in the DEH hydrates (2a and 2b) toward the beginning of the reaction is also observed. After approximately 1 h, the H-1 signal associated with Δ (structures 1a and 1b) began to reduce due to conversion of this unsaturated monomer, which shows that, as expected, Δ (structures 1a and 1b) is unstable. After 4 h, the relative ratio between products in the reaction shows that the DHF hydrates are the major components comprising ∼46% (structure 3a) and ∼36% (structure 3b) each (Fig. 5C). The remaining four structures (1a, 1b, 2a, and 2b) are the minor products of the reaction, accounting for ∼1, 5, 6, and 6%, respectively. Importantly, the observed products are different hydrated forms, and they are in equilibrium with each other, which means that the distribution of these products is highly dependent on the conditions.
DISCUSSION
It is well known that proteins from the Arctic Mid-Ocean Ridge (AMOR) hot vents are thermostable (20, 41, 42), and here we set out to find a stable exotype alginate lyase for possible use in enzymatic processing of alginate. The new enzyme, AMOR_PL17A, is active on polyG, polyM, and alginate, whereas no activity was observed toward heparin, chondroitin, and laminarin. The high activity on alginate could indicate that the enzyme prefers sequences with alternating M and G, as has been reported for exolytic alginate lyases Atu3025 (25) and OalB (28), but more experiments would be required to validate this. The enzyme is active at high temperatures (>50°C) and works optimally between pHs 5.0 and 6.0 and in the presence of 300 mM NaCl. Interestingly, the thermal vent from which AMOR_PL17A was isolated has a pH of 5.7 (measured at 25°C [E. P. Reeves, personal communication]), which corresponds well with the measured pH optimum of the enzyme. AMOR_PL17A retained 100% activity after a 24-h incubation at 60°C in the absence of substrate, which means that AMOR_PL17A is among the most thermostable exolytic alginate lyases that have been discovered and characterized to date (22–29, 31, 32, 43) (Table 1). Only one other thermostable exolytic alginate lyase has been characterized, namely, a PL17 enzyme called AlyRm4 from Rhodothermus marinus strain 378, which has optimal activity at 81°C and is stable at 70°C for at least 16 h (43).
Although several authors have, through the years, proposed possible links between the amino acid composition of a protein and its thermal stability, such links are not well supported by experimental data. It is well known that protein stability may be drastically increased upon introduction of only a few mutations that cannot be predicted by sequence analysis (44, 45). Nevertheless, it is worth noting that sequence statistics, depicted in Fig. S1B and S1C, show that AMOR_PL17A has more prolines, arginines, phenylalanines, and glutamines and more overall positive charge than its more mesophilic counterpart, Alg17C. These features could relate to both the enzyme’s high stability and its halophilicity. Interestingly, the presence of high numbers of prolines and arginines has been associated with high stability since the early days of protein stability studies (46, 47).
The activity assays with various substrates showed that AMOR_PL17A converts all kinds of oligomeric and polymeric versions of polyM and polyG substrates with saturated or unsaturated nonreducing ends to monomeric products in an exolytic manner, working from the nonreducing end. The primary product, Δ, is UV active, as one would expect for an enzyme using the canonical β-elimination mechanism employed by polysaccharide lyases. Based on these properties, AMOR_PL17A may be classified as an oligo-alginate lyase (EC 4.2.2.26).
In their study of the crystal structure of SdAlg17C, Park et al. (34) identified residues interacting with a ΔMG trisaccharide occupying subsites −1, +1, and +2. Based on their observations, Park et al. suggested Tyr450 to act as a general base that abstracts the proton at C-5, while Tyr258 was proposed to act as general acid. Furthermore, Asn201 and His202 were suggested to stabilize the charge of the carboxylate (residue numbers refer to the Alg17C sequence). All of these residues are conserved in AMOR_PL17A (Fig. S1A), including the proposed general acid and base, which in AMOR_PL17A are Tyr251 and Tyr446, respectively. Mutation of these two residues to alanine in AMOR_PL17A confirmed the importance of Tyr251 but showed that Tyr446 is not essential for catalysis. Although β-elimination using two tyrosines was reported for Alg17C (34), it is atypical for a tyrosine to function as a catalytic base (48, 49). Interestingly, Tyr/Tyr β-elimination has also been reported for a family PL5 alginate lyase, where the same tyrosine is thought to function as both the catalytic acid and base (50). On the other hand, studies of the exolytic PL15 enzyme called Atu3025 led to the conclusion that a histidine and a tyrosine function as the catalytic base and acid, respectively (35). While the PL17 enzymes do contain conserved tyrosines, they also contain conserved histidines (Fig. S1A), which perhaps are more likely to exert a base function during catalysis.
It is widely accepted that Δ resulting from lyase activity can undergo spontaneous ring opening and hydration to yield other reaction products (3, 30, 51, 52). Specifically, it has been proposed that the ring opening of Δ is followed by the formation of cyclic hydrated DEH structures (3, 30, 51). These structures are proposed to form 6-member pyranose-like rings, similar to those that are expected from the intramolecular cyclization of hexose sugars. This would require nucleophilic attack of the C-5 hydroxyl group onto the C-1 aldehyde to form hemiacetals, which, in fact, is unlikely since DEH has a carbonyl at C-5. Such intramolecular cyclization would lead to four possible structures with chiral centers at C-1 and C-5. Yet, only two have been detected by NMR and reported in the literature (3, 30, 51). The NMR data reported here (1H and 13C chemical shifts, and 3JHH coupling constants) match closely to those reported previously for the products of alginate lyases, but our 2D NMR spectra have been used to further resolve the structures. The NMR data reported here do not support the formation of DEH-derived pyranose structures, but rather support formation of two 5-member hemiketal structures with an ether connection between C-2 and C-5 and a geminal diol at C-1 that are epimers at C-5. Of note, 6-member hydrated products have also been reported as the result of enzymatic processing of unsaturated uronic acid products formed from galacturonic (51) or glucuronic (52) substrates. In these reports, 1H chemical shifts and 3JHH coupling constants display some differences from the data recorded here. Without key 13C chemical shifts (for example C-2 and C-5), it is difficult to determine whether the products reported in these previous studies are 5- or 6-member rings.
Efficient utilization of brown seaweed as a carbohydrate feedstock for bioconversion into biofuels, commodity chemicals, or single-cell protein requires microorganisms that can metabolize alginate. Although not straightforward, engineered microbial platforms for ethanol production from seaweed have been reported, based on both Escherichia coli (4) and Saccharomyces cerevisiae (3), where the microbes contain (engineered) systems for degradation (alginate lyases), uptake (transporters), and metabolism (reductases and kinases). Uptake of DEH has been studied in detail, whereas nothing is known about possible uptake of DHF. The six structures described in this study are in equilibrium, and whether only DEH is taken up by microorganisms or if DHF transporters exists remains unknown. When taken up, DEH is reduced to 2-keto-3-deoxy-d-gluconate (KDG) by a DEH reductase (DehR) and further converted to the glycolytic intermediates pyruvate and glyceraldehyde-3-phosphate by KDG kinase (KdgK) and KDG-6-phosphate aldolase (KdgpA) (4). Preliminary BLAST searches showed that many natural yeasts have genes encoding homologues of the DEH transporter from Asteromyces cruciatus, which was used to engineer S. cerevisiae (3) for alginate conversion. Based on the results of this study, further studies on uptake mechanisms, including a quest for possible DHF transporters, are warranted.
MATERIALS AND METHODS
Sampling, DNA extraction, sequencing, and gene identification.
A sample of unbleached Norway spruce (Picea abies) was incubated for 1 year in ∼70°C hot sediments at Mohn’s Ridge situated at 71°N in the Arctic Mid-Ocean Ridge (AMOR), 570 m below sea level (53). As described previously (20), after this incubation, DNA was extracted from the sample using a FastDNA spin kit (MP Biomedicals, Santa Ana, CA), and metagenomics sequencing was done using Illumina MiSeq 300 paired-end chemistry (20, 42). Reads were filtered with the CLC genomics workbench (v.10; Qiagen) using default parameters. Assembly was done using default parameters with an automatic k-mer and bubble size. Minimum contig length was set to 1,000 bases with scaffolding enabled. Open reading frame (ORF) prediction was performed using the –p meta option in Prodigal (54). The metagenomics data set was mined for polysaccharide lyases belonging to family PL17 using dbCAN (55), which resulted in the identification of a putative PL17 alginate lyase gene, amor_pl17a.
Sequence analysis.
The predicted amino acid sequence of AMOR_PL17A was analyzed for the presence of a signal peptide using SignalP version 4.0 (56) and with Pfam for the detection of functional domains. Clustal Omega was used for multiple sequence alignment of AMOR_PL17A with other exolytic alginate lyases, and IQ-Tree (57) was used for maximum likelihood phylogeny.
Cloning, expression, and purification.
The amor_pl17a gene (codon optimized for E. coli expression) was synthesized by GeneScript (Piscataway, NJ), and nucleotides 52 to 2187 were amplified by PCR using Q5 DNA polymerase (New England Biolabs, Ipswich, MA) and forward primer 5′-TTAAGAAGGAGATATACTATGCTGCCGGCGGGT-3′ and reverse primer 5′-AATGGTGGTGATGATGGTGCGCCTTGATTTTCTCCACCTTGTAATT-3′, where the underlined nucleotides overlap the pNIC-CH vector (AddGene, Cambridge, MA). The PCR product was verified by gel electrophoresis before being inserted into the pNIC-CH vector by ligation-independent cloning (LIC). As a result of this cloning strategy, the C terminus of the protein is extended with a seven-residue His tag (AHHHHHH). LIC was followed by heat shock transformation into chemically competent E. coli TOP10 cells (Invitrogen, Carlsbad, CA) according to the supplier’s protocol. The lysogenic broth (LB) agar plates used in the protocol were supplemented with 5% sucrose and 50 μg/ml kanamycin. Cells from a colony were inoculated into liquid LB medium containing 50 μg/ml kanamycin and incubated overnight at 37°C with shaking at 200 rpm. A Nucleospin plasmid kit (Macherey-Nagel) was used to isolate plasmids from the culture, and the cloned DNA fragment containing the amor_pl17a gene was verified by Sanger sequencing (GATC, Constance, Germany). These plasmids were subsequently used to transform E. coli BL-21 Star(DE3) cells (Invitrogen), as outlined by the supplier.
For protein production, a transformant colony was inoculated and grown overnight at 37°C in a preculture of LB medium containing 50 μg/ml kanamycin. This preculture (1.5 ml) was added to 500 ml Terrific Broth (TB) containing 50 μg/ml kanamycin, followed by overnight incubation at 22°C in a Lex-48 bioreactor (Harbinger Biotechnology & Engineering, Markham, Canada). Protein expression was induced by addition of IPTG (isopropyl-β-d-thiogalactopyranoside [0.2 mM, final concentration]), followed by continued incubation for 24 h. Then cells were harvested by centrifugation at 5,000 × g for 15 min at 4°C and kept at −80°C for at least 1 h to promote cell lysis. The cells were subsequently thawed and resuspended in 25 ml 50 mM Tris-HCl (pH 8.0) supplemented with 500 mM NaCl and 5 mM imidazole, followed by sonication on ice using a Vibracell sonicator (Sonics & Materials, Inc., Newtown, CT) with 5-s on/off pulses for 3 min at 30% amplitude. Cell debris was removed by centrifugation at 15,000 × g for 15 min at 4°C, and the supernatant containing the soluble protein was filtered using a sterile 0.45-μm-pore filter (Sarstedt, Nümbrecht, Germany).
The recombinant His-tagged protein was purified by immobilized metal affinity chromatography using an Äkta Pure chromatography system (GE HealthCare, Chicago, IL) equipped with a Ni2+ affinity HisTrap HP 5-ml column (GE HealthCare). The His-tagged protein was eluted using a linear gradient of 5 to 500 mM imidazole in 50 mM Tris-HCl (pH 8.0) supplemented with 500 mM NaCl. Eluted fractions containing the target protein, as indicated by SDS-PAGE, were pooled and concentrated using a 10,000-molecular-weight-cutoff (MWCO) Vivaspin ultrafiltration tube (Sartorius, Göttingen, Germany), with concomitant buffer exchange to 20 mM NaOAc (pH 5.6) and 300 mM NaCl. The purified His-tagged protein, AMOR_PL17A, was stored at 4°C and was used “as is” in subsequent experiments.
The protein concentration was calculated from UV absorbance at 280 nm using the theoretical extinction coefficient (https://web.expasy.org/protparam/).
Two variants of amor_pl17a with single point mutations, Y251A and Y446A, were generated by splicing by overlap extension-PCR (SOE-PCR). The upstream and downstream fragments for each mutant were amplified by PCR before being spliced by overlap extension in subsequent rounds of PCR. Fragments and the complete mutated genes were evaluated by gel electrophoresis using 1% agarose gels. Together with the respective primers, all PCR mixtures contained Q5 DNA polymerase (New England Biolabs, Ipswich, MA) and recombinant amor_pl17a as the template. For the Y251A mutant, the respective forward and reverse primers for the upstream fragment were 5′-TTAAGAAGGAGATATACTATGCTGCCGGCGGGT-3′ and 5′-AGCGTACGGACCTTCCATATAGTAGCC-3′, whereas the downstream fragment was generated using 5′-CTACTATATGGAAGGTCCGTACGCTATCCGTTATGCGCTGCGT-3′ and 5′-AATGGTGGTGATGATGGTGCGCCTTGATTTTCTCCACCTTGTAATT-3′. For the Y446A mutant, the respective forward and reverse primers for the upstream fragment were 5′-TTAAGAAGGAGATATACTATGCTGCCGGCGGGT-3′ and 5′-GGCACGGCCACCAAACTTCGGC-3′, and those for the downstream fragment were 5′-GGAGCCGAAGTTTGGTGGCCGTGCCCTGCCGGAAAACACCACCT-3′ and 5′-AATGGTGGTGATGATGGTGCGCCTTGATTTTCTCCACCTTGTAATT-3′. The mutant enzymes were produced and purified as described above for the wild-type enzyme.
Determination of substrate specificity and reaction optima.
To determine the optimal temperature for AMOR_PL17A activity, reactions were performed at 40 to 90°C in 25 mM NaOAc supplemented with 300 mM NaCl (pH 5.6). Note that the pHs of NaOAc buffers vary little with temperature (d[pKa]/dT = 0.0002). For determining the optimal pH, we used one of three buffers: citric acid-citrate (pH 3.3 to 6.1), Bis-Tris-HCl (pH 5.8 to 7.0), or Tris-HCl (pH 7.0 to 8.8) at 30 to 45 mM and supplemented with 150 mM NaCl. The effect of NaCl on the enzyme activity was investigated by running reactions in 25 mM NaOAc at pH 5.6 and 50°C, supplemented with various concentrations of NaCl (0 to 2.0 M). In all the reactions described above, the enzyme concentration was 1 μM and the concentration of the substrate, sodium alginate (Sigma-Aldrich; monad frequency FG = 0.44, Mw = 107.9 ± 2.7 kDa), was 0.5% (wt/vol).
To investigate the thermal stability of the enzyme, we used 2 μM enzyme in 50 mM NaOAc (pH 5.6) with 600 mM NaCl and preincubated the mixture at 60 or 65°C for 0 to 24 h. Reactions were initiated by adding preheated (50°C) 1% (wt/vol) sodium alginate in a 1:1 ratio, yielding final concentrations comparable to those used in the standard activity assays described above.
To test substrate specificity, we used sodium alginate, isolated polyG (average degree of polymerization [DP] of ∼20, FG = 0.95 [from Laminara hyperborea, as described in reference 58]) and polyM (Mw = 275 kDa, FG = 0.0; obtained from an epimerase-negative AlgG mutant of Pseudomonas fluorescens [59]). In addition, we used heparin, chondroitin, and laminarin—all from Sigma-Aldrich.
All reaction mixtures were incubated in a Thermomixer (Eppendorf) for 5 min at 600 rpm, and the reactions were stopped by boiling for 15 min, unless indicated otherwise. The lyase activity was determined by measuring the release of reducing sugars using the 3,5-dinitrosalicylic acid (DNS) reagent (60), with glucose as the standard.
Product analysis using HPAEC.
Reaction products were analyzed by HPAEC using a Dionex ICS-5000 system (Thermo Scientific, San Jose, CA), equipped with both a UV (235 nm) and a pulsed amperometric detector (PAD) with a disposable electrochemical gold electrode. For analysis of alginate oligomers, a 4- by 250-mm IonPac AS4A column (Dionex, Thermo Scientific) connected to an IonPac AG4A (4- by 50-mm) guard column was used and operated at 30°C. The mobile phases were 0.1 M NaOH (A) and 1 M NaOAc in 0.1 M NaOH (B), and a linear gradient was developed from 1% B to 88.5% B over 100 min: i.e., 8.75 mM NaOAc/min, at a flow rate of 1 ml/min. The PAD detector was set to use an AAA waveform for optimal signal-to-noise detection and data acquisition, and data analysis was done using Chromeleon 7.2 (Thermo Scientific, San Jose, CA).
Characterization of products and monitoring of product formation with UV-Vis and NMR spectroscopy.
UV absorption at 235 nm upon AMOR_PL17A-catalyzed degradation of alginate was monitored over 24 h using a Synergy H4 microplate reader (BioTek, Winooski, VT). The enzymatic degradation reactions were stopped either by boiling, as described above, or by adding NaOH or HCl to a final concentration of 0.1 M, prior to analysis of product stability.
For NMR analyses, all homo- and heteronuclear experiments were acquired on a Bruker AV-IIIHD 800-MHz spectrometer (Bruker BioSpin AG, Fälladen, Switzerland) equipped with a 5-mm cryogenic CP-TCI z-gradient probe. The polyM used for all NMR analyses was subjected to a mild hydrolysis resulting in a substrate with the following characteristics: FG = 0.0, DP of ∼50, Mw of ∼10 kDa. For NMR analyses, the prepared reactions were dissolved in deuterated water (D2O; 99.9% [Sigma-Aldrich]) and placed in a 3-mm LabScape Stream NMR tube (Bruker LabScape).
For structural elucidation, two reaction mixtures were prepared and incubated at two different temperatures (25 and 50°C). The 25°C incubation was completed inside an NMR tube and incubated within the NMR magnet. This reaction mixture contained a solution of 1% (wt/vol) polyM in sodium acetate buffer (10 mM NaOAc, 200 mM NaCl [pH 5.6]) in 99.9% D2O. A volume of 160 μl of this solution was added to a 3-mm NMR tube, and the experiment was started by adding 15 μl of enzyme stock solution to give a final enzyme concentration of 2 μM. After approximately 17 h of incubation, 1- and 2D NMR experiments were acquired for structural elucidation. The 50°C incubation mixture contained 0.4% (wt/vol) polyM in sodium acetate buffer (5 mM NaOAc, 100 mM NaCl [pH 5.6]) in H2O and 0.8 μM enzyme. The reaction mixture was incubated at 50°C in a benchtop Thermomixer (Eppendorf) for ∼48 h at 600 rpm to achieve complete conversion of the substrate. Water was removed by freeze-drying overnight, and the sample was resuspended in 180 μl of 99.9% D2O and transferred to a 3-mm NMR tube. One- and two-dimensional NMR experiments were collected for structural elucidation.
For chemical shift assignment, data from the following 1- and 2D NMR experiments were collected at a temperature of 25°C for both samples: 1D proton, 1H-13C heteronuclear single quantum coherence (HSQC) with multiplicity editing, and 1H-13C heteronuclear multiple bond coherence (HMBC) with suppression of one-bond correlations. For the sample incubated at 25°C, a series of 1D selective correlation spectroscopy (COSY) spectra were also acquired for H-1 to H-4 of structures 1a and 1b. In addition, the following experiments were also collected for the sample incubated at 50°C: 1H-1H double quantum filtered correlation spectroscopy (DQF-COSY) and 1H-13C heteronuclear two-bond correlation (H2BC) spectroscopy.
After structural elucidation, a reaction mixture was prepared and incubated within an NMR tube inside the magnet at a temperature of 50°C to observe the formation and interconversion of products. The reaction mixture contained 160 μl of a solution of 1% (wt/vol) polyM in sodium acetate buffer (10 mM NaOAc, 200 mM NaCl [pH 5.6]) in 99.9% D2O. The reaction was started by adding 15 μl of enzyme stock solution (2 μM final enzyme concentration) and was monitored with time-resolved 1H NMR. The time-resolved experiment consisted of a series of 1D proton spectra recorded over time. A proton spectrum was recorded every 2 min, with a total of 120 recorded spectra over approximately 4 h. Following the time-resolved experiment, the following spectra were also collected: 1D proton, 1H-13C heteronuclear single quantum coherence (HSQC) with multiplicity editing. To plot the change in products through time, the H-1 signals for products 1a, 1b, 2a, 2b, 3a, and 3b were integrated in each of the 120 recorded spectra. The integrals were normalized to the sum of the H-1 signals of all six products in the final spectrum (at time 4 h, the sum of the integrals = 1).
For all experiments, the 1H chemical shift was internally referenced to the residual water signal (4.75 ppm for experiments acquired at 25°C and 4.50 ppm at 50°C), while the 13C chemical shift was referenced indirectly to water, based on the absolute frequency ratios (61). The spectra were recorded, processed, and analyzed using the TopSpin 3.5 or 4.0.1 software (Bruker BioSpin AG). Full acquisition parameters are included in the supplemental material.
Data availability.
The nucleotide sequence of amor_pl17a has been submitted to GenBank under accession no. MT444120.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kiira S. Vuoristo and Line D. Hansen for valuable help during the initial testing of this alginate lyase, as well as Eoghan P. Reeves for sampling and geochemical analysis of hydrothermal vent samples.
This research was supported by the Research Council of Norway through grants 294946 (The Norwegian Seaweed Biorefinery Platform), 229003 (BIOFEED—Novel salmon feed by integrated bioprocessing of nonfood biomass), 237841 (Foods of Norway), and 221568 (NorZymeD). Infrastructure was supported in part by NorBioLab grants 226247 and 270038 and by a grant to the Norwegian NMR Platform (226244), all provided by the Research Council of Norway. The research cruise to the Arctic Mid-Ocean Ridge vent fields was funded by the Research Council of Norway through project no. 179560.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.MarketsandMarkets. 2020. Seaweed cultivation market. https://www.marketsandmarkets.com/Market-Reports/commercial-seaweed-market-152763701.html. Accessed 15 December 2020.
- 2.Song M, Duc Pham H, Seon J, Chul Woo H. 2015. Marine brown algae: a conundrum answer for sustainable biofuels production. Renewable Sustainable Energy Rev 50:782–792. doi: 10.1016/j.rser.2015.05.021. [DOI] [Google Scholar]
- 3.Enquist-Newman M, Faust AM, Bravo DD, Santos CN, Raisner RM, Hanel A, Sarvabhowman P, Le C, Regitsky DD, Cooper SR, Peereboom L, Clark A, Martinez Y, Goldsmith J, Cho MY, Donohoue PD, Luo L, Lamberson B, Tamrakar P, Kim EJ, Villari JL, Gill A, Tripathi SA, Karamchedu P, Paredes CJ, Rajgarhia V, Kotlar HK, Bailey RB, Miller DJ, Ohler NL, Swimmer C, Yoshikuni Y. 2014. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature 505:239–243. doi: 10.1038/nature12771. [DOI] [PubMed] [Google Scholar]
- 4.Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CN, Kim PB, Cooper SR, Raisner RM, Herman A, Sivitz AB, Lakshmanaswamy A, Kashiyama Y, Baker D, Yoshikuni Y. 2012. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 335:308–313. doi: 10.1126/science.1214547. [DOI] [PubMed] [Google Scholar]
- 5.Marinho GS, Alvarado-Morales M, Angelidaki I. 2016. Valorization of macroalga Saccharina latissima as novel feedstock for fermentation-based succinic acid production in a biorefinery approach and economic aspects. Algal Res 16:102–109. doi: 10.1016/j.algal.2016.02.023. [DOI] [Google Scholar]
- 6.Sharma S, Hansen LD, Hansen JO, Mydland LT, Horn SJ, Overland M, Eijsink VGH, Vuoristo KS. 2018. Microbial protein produced from brown seaweed and spruce wood as a feed ingredient. J Agric Food Chem 66:8328–8335. doi: 10.1021/acs.jafc.8b01835. [DOI] [PubMed] [Google Scholar]
- 7.Horn SJ, Aasen IM, Østgaard K. 2000. Production of ethanol from mannitol by Zymobacter palmae. J Ind Microbiol Biotechnol 24:51–57. doi: 10.1038/sj.jim.2900771. [DOI] [Google Scholar]
- 8.Sharma S, Horn SJ. 2016. Enzymatic saccharification of brown seaweed for production of fermentable sugars. Bioresour Technol 213:155–161. doi: 10.1016/j.biortech.2016.02.090. [DOI] [PubMed] [Google Scholar]
- 9.Horn SJ, Moen E, Østgaard K. 1999. Direct determination of alginate content in brown algae by near infra-red (NIR) spectroscopy. J Appl Phycol 11:9–13. doi: 10.1023/A:1008024009954. [DOI] [Google Scholar]
- 10.Haug A, Larsen B, Smidsrød O, Smidsrød O, Eriksson G, Blinc R, Paušak S, Ehrenberg L, Dumanović J. 1967. Studies on the sequence of uronic acid residues in alginic acid. Acta Chem Scand 21:691–704. doi: 10.3891/acta.chem.scand.21-0691. [DOI] [Google Scholar]
- 11.Wong TY, Preston LA, Schiller NL. 2000. Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu Rev Microbiol 54:289–340. doi: 10.1146/annurev.micro.54.1.289. [DOI] [PubMed] [Google Scholar]
- 12.Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, Henrissat B. 2010. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J 432:437–444. doi: 10.1042/BJ20101185. [DOI] [PubMed] [Google Scholar]
- 13.Peng C, Wang Q, Lu D, Han W, Li F. 2018. A novel bifunctional endolytic alginate lyase with variable alginate-degrading modes and versatile monosaccharide-producing properties. Front Microbiol 9:167. doi: 10.3389/fmicb.2018.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu B, Yin H. 2015. Alginate lyase: review of major sources and classification, properties, structure-function analysis and applications. Bioengineered 6:125–131. doi: 10.1080/21655979.2015.1030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ertesvåg H. 2015. Alginate-modifying enzymes: biological roles and biotechnological uses. Front Microbiol 6:523. doi: 10.3389/fmicb.2015.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VG. 2012. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels 5:45. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusaykin MI, Silchenko AS, Zakharenko AM, Zvyagintseva TN. 2016. Fucoidanases. Glycobiology 26:3–12. doi: 10.1093/glycob/cwv072. [DOI] [PubMed] [Google Scholar]
- 18.Adams JM, Ross AB, Anastasakis K, Hodgson EM, Gallagher JA, Jones JM, Donnison IS. 2011. Seasonal variation in the chemical composition of the bioenergy feedstock Laminaria digitata for thermochemical conversion. Bioresour Technol 102:226–234. doi: 10.1016/j.biortech.2010.06.152. [DOI] [PubMed] [Google Scholar]
- 19.Scullin C, Stavila V, Skarstad A, Keasling JD, Simmons BA, Singh S. 2015. Optimization of renewable pinene production from the conversion of macroalgae Saccharina latissima. Bioresour Technol 184:415–420. doi: 10.1016/j.biortech.2014.09.105. [DOI] [PubMed] [Google Scholar]
- 20.Vuoristo KS, Fredriksen L, Oftebro M, Arntzen MO, Aarstad OA, Stokke R, Steen IH, Hansen LD, Schuller RB, Aachmann FL, Horn SJ, Eijsink VGH. 2019. Production, characterization, and application of an alginate lyase, AMOR_PL7A, from hot vents in the Arctic Mid-Ocean Ridge. J Agric Food Chem 67:2936–2945. doi: 10.1021/acs.jafc.8b07190. [DOI] [PubMed] [Google Scholar]
- 21.Ravanal MC, Sharma S, Gimpel J, Reveco-Urzua FE, Øverland M, Horn SJ, Lienqueo ME. 2017. The role of alginate lyases in the enzymatic saccharification of brown macroalgae, Macrocystis pyrifera and Saccharina latissima. Algal Res 26:287–293. doi: 10.1016/j.algal.2017.08.012. [DOI] [Google Scholar]
- 22.Miyake O, Hashimoto W, Murata K. 2003. An exotype alginate lyase in Sphingomonas sp. A1: overexpression in Escherichia coli, purification, and characterization of alginate lyase IV (A1-IV). Protein Expr Purif 29:33–41. doi: 10.1016/s1046-5928(03)00018-4. [DOI] [PubMed] [Google Scholar]
- 23.Park HH, Kam N, Lee EY, Kim HS. 2012. Cloning and characterization of a novel oligoalginate lyase from a newly isolated bacterium Sphingomonas sp. MJ-3. Mar Biotechnol (NY) 14:189–202. doi: 10.1007/s10126-011-9402-7. [DOI] [PubMed] [Google Scholar]
- 24.He M, Guo M, Zhang X, Chen K, Yan J, Irbis C. 2018. Purification and characterization of alginate lyase from Sphingomonas sp. ZH0. J Biosci Bioeng 126:310–316. doi: 10.1016/j.jbiosc.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 25.Ochiai A, Hashimoto W, Murata K. 2006. A biosystem for alginate metabolism in Agrobacterium tumefaciens strain C58: molecular identification of Atu3025 as an exotype family PL-15 alginate lyase. Res Microbiol 157:642–649. doi: 10.1016/j.resmic.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Kim HT, Chung JH, Wang D, Lee J, Woo HC, Choi I-G, Kim KH. 2012. Depolymerization of alginate into a monomeric sugar acid using Alg17C, an exo-oligoalginate lyase cloned from Saccharophagus degradans 2-40. Appl Microbiol Biotechnol 93:2233–2239. doi: 10.1007/s00253-012-3882-x. [DOI] [PubMed] [Google Scholar]
- 27.Thomas F, Lundqvist LCE, Jam M, Jeudy A, Barbeyron T, Sandström C, Michel G, Czjzek M. 2013. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J Biol Chem 288:23021–23037. doi: 10.1074/jbc.M113.467217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jagtap SS, Hehemann JH, Polz MF, Lee JK, Zhao H. 2014. Comparative biochemical characterization of three exolytic oligoalginate lyases from Vibrio splendidus reveals complementary substrate scope, temperature, and pH adaptations. Appl Environ Microbiol 80:4207–4214. doi: 10.1128/AEM.01285-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Li S, Yu W, Gong Q. 2015. Cloning, overexpression and characterization of a new oligoalginate lyase from a marine bacterium, Shewanella sp. Biotechnol Lett 37:665–671. doi: 10.1007/s10529-014-1706-z. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Wang L, Han F, Gong Q, Yu W. 2016. Cloning and characterization of the first polysaccharide lyase family 6 oligoalginate lyase from marine Shewanella sp. Kz7. J Biochem 159:77–86. doi: 10.1093/jb/mvv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin JW, Lee OK, Park HH, Kim HS, Lee EY. 2015. Molecular characterization of a novel oligoalginate lyase consisting of AlgL- and heparinase II/III-like domains from Stenotrophomonas maltophilia KJ-2 and its application to alginate saccharification. Korean J Chem Eng 32:917–924. doi: 10.1007/s11814-014-0282-1. [DOI] [Google Scholar]
- 32.Wang D, Aarstad OA, Li J, McKee LS, Sætrom GI, Vyas A, Srivastava V, Aachmann FL, Bulone V, Hsieh YS. 2018. Preparation of 4-deoxy-l-erythro-5-hexoseulose uronic acid (DEH) and guluronic acid rich alginate using a unique exo-alginate lyase from Thalassotalea crassostreae. J Agric Food Chem 66:1435–1443. doi: 10.1021/acs.jafc.7b05751. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki H, Suzuki K-i, Inoue A, Ojima T. 2006. A novel oligoalginate lyase from abalone, Haliotis discus hannai, that releases disaccharide from alginate polymer in an exolytic manner. Carbohydr Res 341:1809–1819. doi: 10.1016/j.carres.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 34.Park D, Jagtap S, Nair SK. 2014. Structure of a PL17 family alginate lyase demonstrates functional similarities among exotype depolymerases. J Biol Chem 289:8645–8655. doi: 10.1074/jbc.M113.531111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochiai A, Yamasaki M, Mikami B, Hashimoto W, Murata K. 2010. Crystal structure of exotype alginate lyase Atu3025 from Agrobacterium tumefaciens. J Biol Chem 285:24519–24528. doi: 10.1074/jbc.M110.125450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenau T, Potthast A, Zwirchmayr NS, Hosoya T, Hettegger H, Bacher M, Krainz K, Yoneda Y, Dietz T. 2017. Chromophores from hexeneuronic acids (HexA): synthesis of model compounds and primary degradation intermediates. Cellulose 24:3703–3723. doi: 10.1007/s10570-017-1396-5. [DOI] [Google Scholar]
- 37.Heyraud A, Gey C, Leonard C, Rochas C, Girond S, Kloareg B. 1996. NMR spectroscopy analysis of oligoguluronates and oligomannuronates prepared by acid or enzymatic hydrolysis of homopolymeric blocks of alginic acid. Application to the determination of the substrate specificity of Haliotis tuberculata alginate lyase. Carbohydr Res 289:11–23. doi: 10.1016/0008-6215(96)00060-2. [DOI] [PubMed] [Google Scholar]
- 38.Bell RP. 1966. The reversible hydration of carbonyl compounds, p 1–29. In Gold V (ed), Advances in physical organic chemistry, vol 4. Academic Press, Waltham, MA. [Google Scholar]
- 39.Shevchik VE, Condemine G, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. 1999. The exopolygalacturonate lyase PelW and the oligogalacturonate lyase Ogl, two cytoplasmic enzymes of pectin catabolism in Erwinia chrysanthemi 3937. J Bacteriol 181:3912–3919. doi: 10.1128/JB.181.13.3912-3919.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preiss J, Ashwell G. 1962. Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosac-charides and 4-deoxy-l-erythro-5-hexoseulose uronic acid. J Biol Chem 237:309–316. [PubMed] [Google Scholar]
- 41.Stepnov AA, Fredriksen L, Steen IH, Stokke R, Eijsink VGH. 2019. Identification and characterization of a hyperthermophilic GH9 cellulase from the Arctic Mid-Ocean Ridge vent field. PLoS One 14:e0222216. doi: 10.1371/journal.pone.0222216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fredriksen L, Stokke R, Jensen MS, Westereng B, Jameson JK, Steen IH, Eijsink VGH. 2019. Discovery of a thermostable GH10 xylanase with broad substrate specificity from the Arctic Mid-Ocean Ridge vent system. Appl Environ Microbiol 85:e02970-18. doi: 10.1128/AEM.02970-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hreggvidsson GO, Jonsson Wheat JO, Bjornsdottir B, Fridjonsson OH, Altenbuchner J, Watzlawick H, Dobruchowska JM, Kamerling JP. July 2015. Thermostable alginate degrading enzymes and their methods of use. WO patent WO2015104723A1.
- 44.van den Burg B, Eijsink VG. 2002. Selection of mutations for increased protein stability. Curr Opin Biotechnol 13:333–337. doi: 10.1016/s0958-1669(02)00325-7. [DOI] [PubMed] [Google Scholar]
- 45.Eijsink VG, Bjork A, Gaseidnes S, Sirevag R, Synstad B, van den Burg B, Vriend G. 2004. Rational engineering of enzyme stability. J Biotechnol 113:105–120. doi: 10.1016/j.jbiotec.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe K, Masuda T, Ohashi H, Mihara H, Suzuki Y. 1994. Multiple proline substitutions cumulatively thermostabilize Bacillus cereus ATCC7064 oligo-1,6-glucosidase. Irrefragable proof supporting the proline rule. Eur J Biochem 226:277–283. doi: 10.1111/j.1432-1033.1994.tb20051.x. [DOI] [PubMed] [Google Scholar]
- 47.Mrabet NT, Van den Broeck A, Van den Brande I, Stanssens P, Laroche Y, Lambeir AM, Matthijssens G, Jenkins J, Chiadmi M, van Tilbeurgh H. 1992. Arginine residues as stabilizing elements in proteins. Biochemistry 31:2239–2253. doi: 10.1021/bi00123a005. [DOI] [PubMed] [Google Scholar]
- 48.Xu F, Wang P, Zhang YZ, Chen XL. 2017. Diversity of three-dimensional structures and catalytic mechanisms of alginate lyases. Appl Environ Microbiol 84:e02040-17. doi: 10.1128/AEM.02040-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garron ML, Cygler M. 2010. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 20:1547–1573. doi: 10.1093/glycob/cwq122. [DOI] [PubMed] [Google Scholar]
- 50.Mikami B, Ban M, Suzuki S, Yoon HJ, Miyake O, Yamasaki M, Ogura K, Maruyama Y, Hashimoto W, Murata K. 2012. Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III. Acta Crystallogr D Biol Crystallogr 68:1207–1216. doi: 10.1107/S090744491202495X. [DOI] [PubMed] [Google Scholar]
- 51.Hobbs JK, Lee SM, Robb M, Hof F, Barr C, Abe KT, Hehemann JH, McLean R, Abbott DW, Boraston AB. 2016. KdgF, the missing link in the microbial metabolism of uronate sugars from pectin and alginate. Proc Natl Acad Sci U S A 113:6188–6193. doi: 10.1073/pnas.1524214113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jongkees SA, Withers SG. 2011. Glycoside cleavage by a new mechanism in unsaturated glucuronyl hydrolases. J Am Chem Soc 133:19334–19337. doi: 10.1021/ja209067v. [DOI] [PubMed] [Google Scholar]
- 53.Stokke R, Reeves EP, Dahle H, Fedoy AE, Viflot T, Lie Onstad S, Vulcano F, Pedersen RB, Eijsink VGH, Steen IH. 2020. Tailoring hydrothermal vent biodiversity toward improved biodiscovery using a novel in situ enrichment strategy. Front Microbiol 11:249. doi: 10.3389/fmicb.2020.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 57.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aarstad OA, Tøndervik A, Sletta H, Skjåk-Bræk G. 2012. Alginate sequencing: an analysis of block distribution in alginates using specific alginate degrading enzymes. Biomacromolecules 13:106–116. doi: 10.1021/bm2013026. [DOI] [PubMed] [Google Scholar]
- 59.Gimmestad M, Sletta H, Ertesvag H, Bakkevig K, Jain S, Suh SJ, Skjak-Braek G, Ellingsen TE, Ohman DE, Valla S. 2003. The Pseudomonas fluorescens AlgG protein, but not its mannuronan C-5-epimerase activity, is needed for alginate polymer formation. J Bacteriol 185:3515–3523. doi: 10.1128/jb.185.12.3515-3523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 61.Zhang H, Neal S, Wishart DS. 2003. RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR 25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 62.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The Carbohydrate-Active enZYmes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequence of amor_pl17a has been submitted to GenBank under accession no. MT444120.