Antibacterial resistance poses a significant threat to human and animal health and global food security. Surveillance for resistance on farms is important for many reasons, including tracking impacts of interventions aimed at reducing the prevalence of resistance.
KEYWORDS: antibiotic resistance, food-borne pathogens, mathematical modeling, surveillance studies
ABSTRACT
Little is known about the drivers of critically important antibacterial resistance in species with zoonotic potential present on farms (e.g., CTX-M β-lactamase-positive Escherichia coli). We collected samples monthly between January 2017 and December 2018 on 53 dairy farms in South West England, along with data for 610 variables concerning antibacterial usage, management practices, and meteorological factors. We detected E. coli resistant to amoxicillin, ciprofloxacin, streptomycin, and tetracycline in 2,754/4,145 (66%), 263/4,145 (6%), 1,475/4,145 (36%), and 2,874/4,145 (69%), respectively, of samples from fecally contaminated on-farm and near-farm sites. E. coli positive for blaCTX-M were detected in 224/4,145 (5.4%) of samples. Multilevel, multivariable logistic regression showed antibacterial dry cow therapeutic choice (including use of cefquinome or framycetin) to be associated with higher odds of blaCTX-M positivity. Low average monthly ambient temperature was associated with lower odds of blaCTX-M E. coli positivity in samples and with lower odds of finding E. coli resistant to each of the four test antibacterials. This was in addition to the effect of temperature on total E. coli density. Furthermore, samples collected close to calves had higher odds of having E. coli resistant to each antibacterial, as well as E. coli positive for blaCTX-M. Samples collected on pastureland had lower odds of having E. coli resistant to amoxicillin or tetracycline, as well as lower odds of being positive for blaCTX-M.
IMPORTANCE Antibacterial resistance poses a significant threat to human and animal health and global food security. Surveillance for resistance on farms is important for many reasons, including tracking impacts of interventions aimed at reducing the prevalence of resistance. In this longitudinal survey of dairy farm antibacterial resistance, we showed that local temperature—as it changes over the course of a year—was associated with the prevalence of antibacterial-resistant E. coli. We also showed that prevalence of resistant E. coli was lower on pastureland and higher in environments inhabited by young animals. These findings have profound implications for routine surveillance and for surveys carried out for research. They provide important evidence that sampling at a single time point and/or single location on a farm is unlikely to be adequate to accurately determine the status of the farm regarding the presence of samples containing resistant E. coli.
INTRODUCTION
Antimicrobial resistance, and particularly antibacterial resistance (ABR), is a significant global challenge. Many countries are implementing plans to reduce the use of antibacterial drugs (ABs) in food-producing animals. For example, the most recent UK 5-year National Action Plan includes a target to reduce AB use (ABU) in the treatment of food-producing animals by 25% (1). In Europe, AB sales for food-producing animals fell by 20% from 2011 to 2016 (2). In the UK dairy industry, overall ABU dropped from 24 mg/kg in 2015 to 17 mg/kg in 2018 (3, 4). In 2018, additional industry-led policies were enforced in the UK that aimed to almost eliminate the use of highest-priority critically important antimicrobials (HP-CIAs), such as third- and fourth-generation cephalosporins (3GCs and 4GCs), as well as fluoroquinolones, on dairy farms. One reason for reducing ABU in farming is to reduce the prevalence of ABR bacteria carried by farm animals. However, there is a need for better data on drivers of ABR in farming. More granularity of understanding is required concerning the risks of using individual ABs and other management practices. This is especially important in terms of drivers of HP-CIA resistance. A focus within HP-CIAs is on 3GC and fluoroquinolone resistance in Escherichia coli, a species commonly found in animal feces and considered one of the most significant potential zoonotic ABR threats to humans (5).
Resistance to 3GCs is increasingly prevalent in E. coli causing infections in humans (6) and is also found in farmed and domestic animals around the world (7). The production of CTX-M (an extended-spectrum β-lactamase) is the most common mechanism of 3GC resistance in E. coli in humans in the UK; for example, in a recent study of urinary E. coli from humans in South West England, 82.2% of 3GC-resistant isolates carried blaCTX-M (8).
The objective of this study was to describe the prevalence of 3GC-resistant E. coli carrying blaCTX-M, as well as E. coli resistant to amoxicillin, tetracycline, streptomycin, and the fluoroquinolone ciprofloxacin, found in fecally contaminated environments of dairy cattle in a geographically restricted population of dairy farms in South West England. These are, or represent, ABs widely used on dairy farms in the UK (3, 4). Furthermore, this study investigated environmental, ABU, and management practice risk factors for the presence of such E. coli.
RESULTS
Prevalence and PCR characterization of 3GC-resistant E. coli from dairy farms.
A total of 4,581 samples were collected from fecally contaminated sites on 53 dairy farms. Samples were collected on each farm monthly between January 2017 and December 2018. On nonselective agar, 4,145 samples were positive for growth of E. coli. Of these, 384/4,145 (9.3%) samples representing 47/53 (88.7%) of farms were positive for growth of E. coli on agar containing the 3GC cefotaxime. From these, 1,226 3GC-resistant isolates were taken forward for PCR testing for possible cephalosporinase genes of interest (GOIs): blaCTX-M (groups 1, 2, 8, 9, and 25), blaCMY, blaDHA, and blaSHV. Over half (648/1,226; 52.7%) of all isolates tested were found to harbor blaCTX-M genes. Of these, 547/648 (84.4%) were of group 1, 99/648 (15.3%) were of group 9, and, in one case, both gene groups were identified. Twelve isolates harbored a blaCMY gene (one alongside the blaCTX-M group 1 gene) and one isolate was blaDHA-1-positive. No isolates were positive for blaSHV and the remaining 566/1,226 (46.2%) isolates were PCR-negative for all GOIs. These isolates were hypothesized to hyper-produce the chromosomally encoded AmpC β-lactamase; some of these isolates have been characterized in detail in a separate study (9).
Farm- and sample-level risk factors for blaCTX-M E. coli positivity.
Based on PCR, carriage of blaCTX-M was the most common mechanism of 3GC resistance in E. coli from dairy farms in this study. Identifying management practice- and antimicrobial use (AMU)-associated risk factors for blaCTX-M E. coli positivity was therefore considered an important objective. Overall, 5.4% (224/4,145) of samples representing 42/53 (79.2%) of farms contained 3GC-resistant E. coli confirmed to carry blaCTX-M using PCR. Positivity for blaCTX-M E. coli was three times higher in samples collected from the environments of calves (calf samples; 98/631 [15.5%] of samples) than overall (Table 1).
TABLE 1.
Prevalence of E. coli carrying blaCTX-M at farm and sample levels
| Sample type | Farm level | Sample level |
|---|---|---|
| Overall | ||
| Total sample size | 53 | 4,145 |
| Total (%) positive for CTX-M-carrying E. coli | 42 (79.2%) | 224 (5.4%) |
| Adult | ||
| Total sample size | 52 | 1,835 |
| Total (%) positive for CTX-M-carrying E. coli | 25 (48.1%) | 76 (4.1%) |
| Dry Cow | ||
| Total sample size | 46 | 282 |
| Total (%) positive for CTX-M-carrying E. coli | 7 (15.2%) | 8 (2.8%) |
| Calf | ||
| Total sample size | 51 | 631 |
| Total (%) positive for CTX-M-carrying E. coli | 33 (64.7%) | 98 (15.5%) |
| Heifer | ||
| Total sample size | 41 | 1,235 |
| Total (%) positive for CTX-M-carrying E. coli | 18 (44%) | 40 (3.2%) |
| Pastureland | ||
| Total sample size | 47 | 630 |
| Total (%) positive for CTX-M-carrying E. coli | 8 (17%) | 12 (1.9%) |
| Publicly accessible pastureland (footpaths) | ||
| Total sample size | 41 | 395 |
| Total (%) positive for CTX-M-carrying E. coli | 8 (20.0%) | 11 (2.8%) |
Given the high positivity rate for blaCTX-M E. coli in calf samples, a separate risk factor analysis using only calf data was performed. One farm-level fixed effect and three sample-level fixed effects were retained in the final multilevel, multivariable logistic regression model (Table S1 in the supplemental material, Table 2). The use of cefquinome or framycetin dry cow therapies were both associated with higher odds of blaCTX-M E. coli positivity, as was higher average monthly temperature. Plotting sample-level positivity for E. coli carrying blaCTX-M versus average monthly temperature revealed that the relationship between positivity and temperature was primarily driven by low blaCTX-M E. coli positivity rates in months where the average temperature was below 10°C (Fig. 1A).
TABLE 2.
Fixed effects from the multilevel, multivariable logistic regression model predicting blaCTX-M E. coli positivity performed on calf samples
| Risk factor | Odds ratio (95% confidence interval) | P value |
|---|---|---|
| Use of cefquinome dry cow therapy in the last six months | 4.18 (2.11, 8.25) | 0.00003 |
| Daily water trough cleaning | 0.44 (0.29, 0.69) | 0.0002 |
| Avg monthly temperature | 1.57 (1.20, 2.06) | 0.0008 |
| Use of framycetin dry cow therapy in the last six months | 1.91 (1.01, 3.61) | 0.04 |
FIG 1.
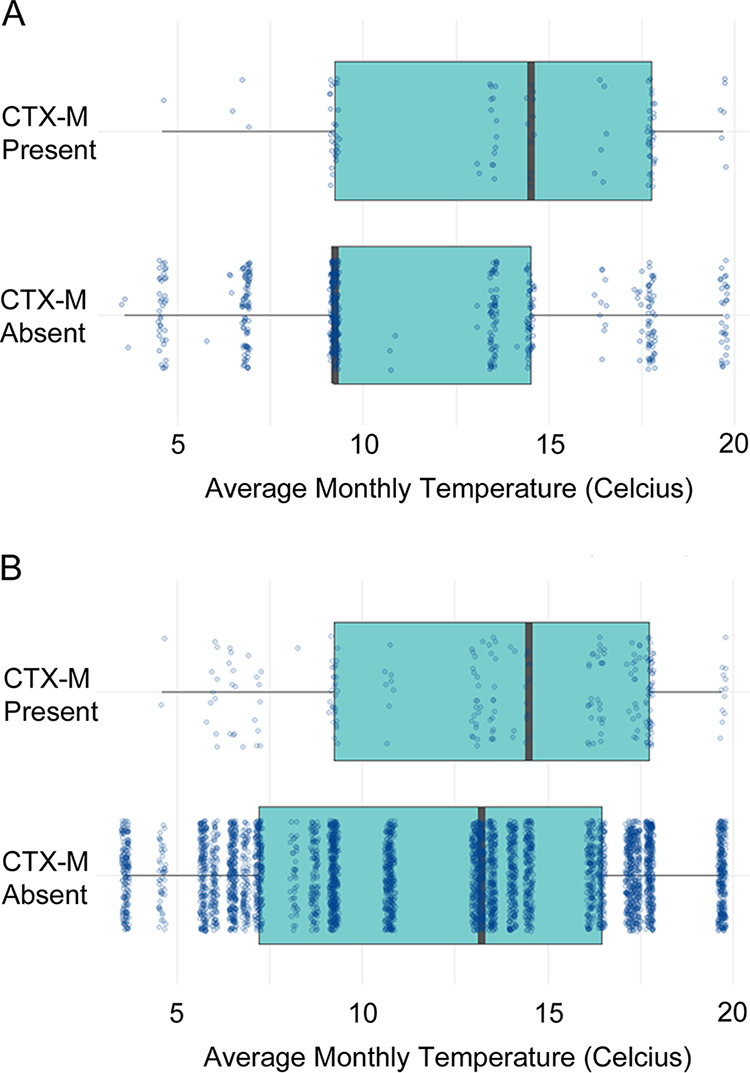
Average monthly temperature versus presence or absence of E. coli positive for blaCTX-M in samples from preweaned calves (A) and all fecally contaminated dairy farm environments (B). Each sample is represented by a dot. A multilevel, multivariable logistic regression model revealed a positive association with increased temperature in both cases (P ≤ 0.0001).
Risk factor analysis was also performed for the full data set. One farm-level fixed effect and three sample-level fixed effects were retained in the final model (Table S2, Table 3). Interestingly, this model revealed that blaCTX-M E. coli was less likely to be found in samples obtained from pastureland, which included publicly accessible farmland (footpaths) compared with other sample types. Analysis of the full data set confirmed what was seen with the calf data set, i.e., higher average monthly temperature was associated with higher odds of blaCTX-M E. coli positivity. Again, visualization of the data confirmed this was primarily driven by a reduction in blaCTX-M E. coli positivity rate in months with an average temperature below 10°C (Fig. 1B). Strikingly, farm-level positivity for blaCTX-M E. coli at the sequential monthly sampling visits was higher in warmer months and lower in the coldest month (Fig. 2A).
TABLE 3.
Fixed effects from the multilevel, multivariable logistic regression model predicting blaCTX-M E. coli positivity performed on the full data set
| Risk factor | Odds ratio (95% confidence interval) | P value |
|---|---|---|
| Sample taken from the environment of preweaned heifers | 4.52 (3.25, 6.27) | <0.00000001 |
| Avg monthly temperature | 1.61 (1.36, 1.90) | 0.00000001 |
| Sample taken from pastureland | 0.32 (0.17,0.61) | 0.0004 |
| Feeding of maize silage | 3.28 (1.50, 7.18) | 0.002 |
FIG 2.
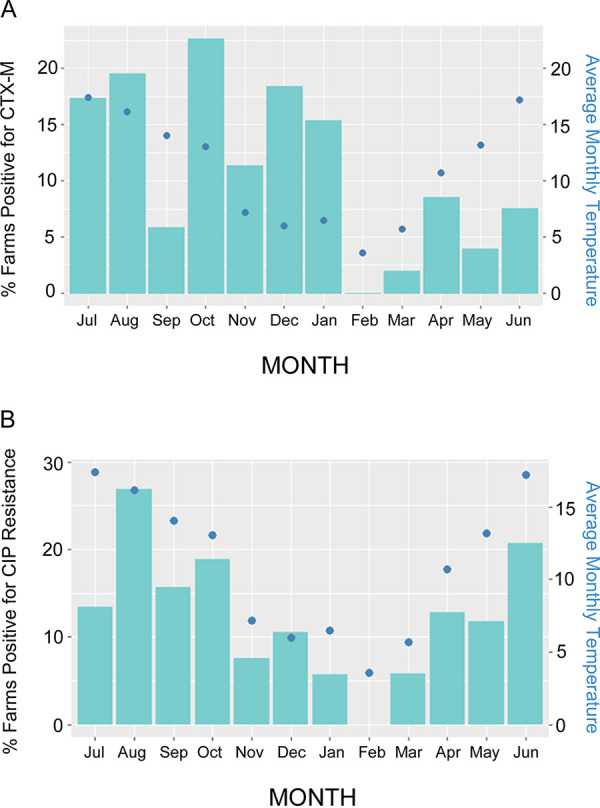
Percentage of farms with E. coli positive for blaCTX-M in samples (A) or ciprofloxacin-resistant E. coli (B). Data are presented by month (bars) and overlaid by a graph of average monthly temperature (dots) representing a year during the middle period of this study. Samples from calves have been excluded.
A Bayesian logistic regression model was also constructed, in which the effect of total farm ABU, and specifically total 3GC and 4GC use, were tested as predictors for blaCTX-M E. coli positivity in the total data set, with 102 potential predictors included. The impact of temperature (odds ratio 1.71 [1.42, 2.05]) on blaCTX-M E. coli positivity was also retained in this alternative model (Table S3).
Defining sample-level positivity for blaCTX-M E. coli is dependent upon finding blaCTX-M using PCR in E. coli colonies that have grown on agar containing cefotaxime. If blaCTX-M E. coli in a sample exists at such low density that it is not detected using selective agar, the sample will be falsely identified as negative for blaCTX-M E. coli. This impact of bacterial density on assay sensitivity is an important consideration in the context of the finding that blaCTX-M positivity is low at low temperatures. To account for this, the logistic link function was adjusted (see Materials and Methods). This only modestly altered the effect sizes and the P values for the risk factors (Fig. S1), confirming that the effect of low temperature on blaCTX-M E. coli positivity was additional to its effect on E. coli prevalence. All values in Tables 2 and 3 come from models with this adjusted logistic link function applied.
Prevalence and risk factor analysis for E. coli resistant to other antibacterial classes.
All 4,145 samples positive for growth of E. coli on nonselective agar were also tested for detectable numbers of E. coli resistant to four noncephalosporins: amoxicillin, tetracycline, streptomycin, and ciprofloxacin, the last being representative of the HP-CIA class, the fluoroquinolones. Resistance to amoxicillin and tetracycline were the most prevalent types of resistance found, with ciprofloxacin resistance being the least commonly detected (Table 4).
TABLE 4.
Farm- and sample-level prevalence of resistance to noncephalosporins
| Antibacterial drug | Farm-level resistance | Sample-level resistance |
|---|---|---|
| Amoxicillin | 53/53 (100%) | 2,754/4,145 (66%) |
| Ciprofloxacin | 49/53 (92%) | 263/4,145 (6%) |
| Streptomycin | 53/53 (100%) | 1,475/4,145 (36%) |
| Tetracycline | 53/53 (100%) | 2,874/4,145 (69%) |
Using a Bayesian logistic regression method, factors associated with the risk of a sample being positive for E. coli resistant to each of the test ABs were identified from the total data set. As seen for blaCTX-M E. coli, the variables of where and when the samples were collected were more consistently associated with the odds of finding resistant E. coli in a sample than farm-level management practices or ABU, with all four models showing a positive association between average monthly temperature and the odds of finding resistant E. coli in a sample (Table 5). Also consistent across all models was the significance of sampling different areas of the farm. Again, as with blaCTX-M E. coli, samples from the environments of calves were more likely to harbor E. coli resistant to all four ABs than samples collected elsewhere on the farm. Samples collected from pastureland were, like blaCTX-M E. coli, negatively associated with the presence of amoxicillin and tetracycline resistance (Table 5).
TABLE 5.
Fixed effects from a Bayesian model predicting resistant E. coli positivity performed on the full data set
| Risk factor | Odds ratio (95% credible interval) |
|---|---|
| Amoxicillin resistance | |
| Avg monthly temperature | 1.91 (1.57, 2.35) |
| Sample taken from the environment of preweaned heifers | 1.99 (1.29, 2.98) |
| Sample taken from pastureland | 0.27 (0.20, 0.37) |
| Ciprofloxacin resistance | |
| Avg monthly temperature | 2.14 (1.63, 2.87) |
| Sample taken from the environment of preweaned heifers | 4.13 (2.79, 6.46) |
| Streptomycin resistance | |
| Avg monthly temperature | 1.53 (1.32, 1.77) |
| Sample taken from the environment of preweaned heifers | 1.95 (1.46, 2.51) |
| Tetracycline resistance | |
| Avg monthly temperature | 1.98 (1.55, 2.55) |
| Sample taken from the environment of preweaned heifers | 3.39 (2.00, 5.82) |
| Sample taken from pastureland | 0.24 (0.15, 0.35) |
| Sample taken during the calving season | 2.02 (1.10, 3.29) |
| Avg monthly rainfall | 1.26 (1.08, 1.47) |
| Total farm streptomycin use in 2017 measured in mg/PCU | 0.76 (0.60, 0.97) |
| Total farm ABU in 2017 measured in mg/PCU | 1.47 (1.01, 2.13) |
Full results for all variables tested can be found in Table S4. Rerunning the models with skeptical priors did not affect the results; only very small differences in the model coefficients were observed (Table S5).
DISCUSSION
Prevalence of blaCTX-M-positive E. coli.
This study is unique in its scale; extensive management practice and ABU data, along with multiple samples from multiple farms, were collected monthly over a 2-year period. Overall, 224/4,145 (5.4%) of samples were positive for E. coli carrying blaCTX-M. This is similar to previously calculated blaCTX-M E. coli carriage of approximately 7% in Danish slaughter pigs (10) and 3.6% in UK broiler chickens and turkeys (11). Various studies have identified much higher prevalence in chicken meat, but this could be due to cross-contamination at slaughter and in the food chain (12, 13).
Studies examining the prevalence of blaCTX-M E. coli in human populations have shown mixed results. A prevalence of 65.7% was found among commensal isolates in Thailand (14). In the UK, a study across four regions reported commensal fecal carriage of blaCTX-M E. coli to be approximately 7% (15). A recent analysis of human urinary samples from the same region as the farms surveyed in this study gave a sample-level prevalence of blaCTX-M E. coli of approximately 5% (8). It should be noted that all farm samples in the present study were from fecally contaminated sites, not individual animals, and so it is possible that the number of animals carrying blaCTX-M E. coli was much lower than the reported sample-level prevalence. Direct comparison with human and other farm animal carriage studies should therefore be made with caution.
Impact of temperature on the odds of finding resistant and blaCTX-M-positive E. coli.
This study found 42/53 (79%) of farms to be positive for blaCTX-M E. coli, based on phenotypic analysis and PCR. This was higher than seen in other studies using similar methodology; for instance, 17/48 (35%) of randomly selected UK dairy farms (16) and 5/25 (20%) of farms in Ohio (17) have been previously shown to be positive. In the present study, samples were collected each month over 2 years, hence the chances of finding a positive sample on each farm may have been greater than in these earlier point-prevalence studies (16, 17). When farm-level positivity for blaCTX-M E. coli was plotted on a month-by-month basis (Fig. 2A), the highest prevalence for a single monthly survey was 22.5%, which fits more closely with these other studies.
Sample-level prevalence of blaCTX-M E. coli was low overall (5.4%). This contrasts with >90% (18) or 50% (19) of blaCTX-M E. coli in samples taken from bovine fecal pats. This difference could be because these earlier studies used enrichment culture prior to testing for resistance at sample level, which increases the chances of finding resistance at sample level. Another possible explanation for the difference is the large number of samples collected in the present study, particularly over winter, given that low temperature was associated with low blaCTX-M E. coli positivity (Fig. 1A and B). Indeed, the observation that average monthly temperature had a significant effect on blaCTX-M E. coli positivity (Table 3), as well as positivity for resistance to amoxicillin, ciprofloxacin, streptomycin, and tetracycline (Table 5), highlights problems with studies where a single time point or sampling season is used. Figure 2 shows the stark impact of this in real terms: blaCTX-M E. coli positivity and positivity for ciprofloxacin-resistant E. coli at farm level was zero in February, the coldest month of the year (based on average temperature) (Fig. 2A and B).
While average annual temperature found at locations across an entire continent has previously been shown to impact average ABR levels at those locations (20), the finding that periods of low temperatures were associated with lower prevalence of a dominant cause of ABR— and particularly HP-CIA resistance at a given location during the course of a year—is particularly important. This observation also leads to concern about the impact of climate change, and especially increasing temperatures, on attempts to reduce ABR. While temperature was associated with the total number of E. coli found in each sample, this was accounted for using a measurement error method incorporated into the model; as such, the effect of temperature on ABR or blaCTX-M-positive E. coli, while in part driven by the effect on total E. coli number, also had an independent association suggestive of a temperature-dependent fitness burden for resistance.
High levels of ABR and blaCTX-M-positive E. coli in farm locations dominated by young animals and low levels on pastureland.
There were clear differences in the risk of encountering blaCTX-M E. coli at different sites on a farm (e.g., 15.5% in calf samples compared to 4.1% in adult samples). The calf environment was also much more likely to have E. coli resistant to amoxicillin, tetracycline, streptomycin, and ciprofloxacin, so this seems to be a universal effect. Other studies have also generally found high levels of resistance in samples collected from or in the environment of younger calves (21–24). There may also be an association with temperature here, since calves are generally kept in warmer environments than are adult cows, but may also be due to some physiological change with age.
This study also identified lower odds of detecting blaCTX-M E. coli in samples collected on pastureland compared with those collected elsewhere on the farm. This relationship also held for E. coli resistant to amoxicillin and tetracycline. Because pastureland may be more affected by the elements, this finding may be partly linked with the association between temperature and ABR.
AB contamination of colostrum as a possible driver of blaCTX-M E. coli positivity in dairy calves—evidence of direct and coselection.
Our analysis identified a small number of specific risk factors. There was an association between calf water trough cleaning and lower odds of calf samples having E. coli with blaCTX-M (Table 2). While there are many reasons for providing the cleanest possible drinking water, this was not seen for other ABR phenotypes and it is unclear why this association was identified. Furthermore, feeding maize silage was associated with higher odds of finding blaCTX-M-positive E. coli across the whole data set (Table 3). It would be interesting to take samples of silage to test if resistant E. coli survive better in this type of medium, but again this association was not seen for other resistance phenotypes. There were also associations between the odds of finding tetracycline-resistant E. coli and ABU, calving, and rainfall (Table 5).
The most interesting AB-related association found was specifically for calf samples. It has been shown experimentally that feeding waste (AB-contaminated) milk to calves increases fecal excretion of ABR bacteria (25). This practice is reducing on UK dairy farms and, in the analysis presented here, waste milk feeding was not associated with an increased risk of finding ABR or blaCTX-M positive E. coli. In contrast, the choice of dry cow therapy (an AB preparation inserted into a cow’s udder between lactations to help treat or prevent mastitis) was associated with blaCTX-M E. coli positivity in calf samples (Table 2). It has previously been shown that colostrum from cows given cefquinome dry cow therapy is heavily contaminated with cefquinome (26), and colostrum management is both a hugely important part of early life for most farmed mammals and universally encouraged in dairy farming. In this study, cefquinome (a 4GC) dry cow therapy was most significantly associated with blaCTX-M E. coli in calf samples (Table 2). This can be explained by direct selection because production of CTX-M confers 4GC resistance in E. coli (27). There was also a clear positive association between the usage of framycetin as part of a dry cow therapy combination and the odds of finding blaCTX-M E. coli in calf samples (Table 2). While no work has been published on the contamination of colostrum with framycetin, its use as a mastitis therapy for milking cows leads to identifiable residues in milk (28), so it is highly likely to also contaminate colostrum. It is possible, therefore, that feeding of colostrum—which can be contaminated with AB used for dry cow therapy—is a driver of blaCTX-M E. coli in calves. An alternative (or indeed an additional) explanation for this observed association is that E. coli (a species known to be found in the udders of dairy cows [29]) that carry blaCTX-M are selected within the udder during AB dry cow therapy and contaminate colostrum alongside the AB used. Others (16) have also identified overall use of 3/4GCs as a risk factor for blaCTX-M E. coli presence on dairy farms but have not made a link between the usage of framycetin and the prevalence of blaCTX-M E. coli. However, it is not always clear whether other studies have separated out different dry cow therapies, since they have tended to focus on systemic AB use. Clearly, an aminoglycoside like framycetin cannot directly select for blaCTX-M E. coli, but aminoglycoside resistance genes are common on plasmids (30), as is blaCTX-M (27). Hence, this may be an example of coselection, where selection of resistance to one antibacterial class increases resistance to another.
Overall, we provide important evidence that sampling at single time points and/or limited locations on a farm are unlikely to be adequate to accurately determine the status of the farm concerning the prevalence of ABR E. coli. This makes comparisons between surveillance studies on farms—designed for research or for regulatory purposes—extremely difficult, and we urge for a standardized, multisample framework accounting for the differential risk factors identified here to be used in the design of future studies.
MATERIALS AND METHODS
Farm recruitment and ethical approval.
A convenience sample of 53 dairy farms was recruited through personal contacts, local veterinary practices, and milk processors. These represented a variety of dairy management systems, ranging from seasonal calving with extensively managed herds to zero-grazed intensive systems. Recruited dairy farms were comparable to farms throughout the UK, with a median herd size of 193 compared to a UK median of 178, a median 305-day milk yield of 7,488 liters compared to a UK median of 8,967 liters, and a median somatic cell count of 167,000 cells/ml of milk compared to a UK median of 178,000 cells/ml of milk. Antibiotic purchasing in 2016 was 26 mg per population corrected unit (mg/PCU) for the UK dairy industry (3) and 21 mg/PCU for the recruited farms.
Of the 53 farms recruited, 43 study farms were in a 50 × 50 km area defined based on the locations of 146 general practices that referred routine urine samples from human patients to the microbiology reference lab at Severn Pathology, Southmead Hospital (8). A further 10 study farms were clustered in a separate region of South West England.
All farmers gave fully informed consent to participate in the study. Ethical approval was obtained from the University of Bristol’s Faculty of Health Sciences Research Ethics Committee (ref 41562).
Farm sampling, sample characteristics, and sample processing.
Farms were visited monthly between January 2017 and December 2018. Samples were collected using sterile overshoes (over-boot socks) to traverse farm areas. Where access was restricted (e.g., for pens containing single or pairs of calves), samples were collected directly from the ground using gloved hands.
Samples were collected of the following six types. (i) “Adult” samples, the fecally contaminated environment of the milking cow population, either the collecting yard, housing shed, or field, collected every month from 52 farms (one farm was a youngstock-only unit). (ii) “Dry cow” samples, the fecally contaminated environment of the dry cow population, collected from 46 farms. (iii and iv) “Heifer” and “calf” samples, the fecally contaminated environment of replacement dairy heifers. These categories were not mutually exclusive and reflected the animals present when the samples were collected. In addition, 209 (5%) samples were collected where no relevant animals were present. On 41 farms, approximately 10 heifers were followed per farm from birth until 18 months of age, with samples collected monthly from their environment, whether housed in a shed or out at pasture. Furthermore, 10 farms had samples collected from additional groups of preweaned replacement heifer calves (calves still being fed milk) to increase the number of samples available of this type. For analysis, because of the differing management practices at different life stages, replacement heifer samples were divided into those associated with preweaned calves (calf samples) and postweaned heifers (heifer samples). (v) “Pasture” samples from the fecally contaminated environments around the above animals while grazing on pastureland on 47 farms were also separated from a subset of (vi) “footpath” samples, which were taken from public footpaths or other rights of way that crossed the farm, where relevant, on 41 farms.
Characteristics for each sample were recorded on a datasheet at the time of collection. Different information was recorded depending on the sample type.
For all samples the following was recorded: sample number; description of sample location; whether animals were housed or in a field; and Ordinance Survey reference for outdoor samples. For calf samples the following was recorded: sample number; ear tag number; description of the sample location; total number of animals in the group; presence or absence of beef calves in the group; date of birth for each calf; and the dry cow therapy used on the dam of each calf in the dry period before the birth. For footpath samples, the presence of livestock at time of sampling was recorded.
Samples were refrigerated (4 to 8°C) from collection to processing, were transferred into individual labeled sterile stomacher bags, and suspended in 10 ml · g−1 of phosphate-buffered saline (PBS Dulbecco A; Oxoid, Basingstoke, UK). Samples were then mixed for 1 min in a stomacher (Stomacher 400, Seward, Worthing, UK). Samples were mixed 50:50 with sterile glycerol and aliquots stored at −80°C.
Microbiology and PCR analysis.
Twenty microliters of sample (diluted 1:10) were spread onto tryptone bile X-glucuronide agar (TBX; Scientific Laboratory Supplies), then 20 μl of undiluted sample were spread onto TBX agar containing 16 mg · liter−1 tetracycline, 8 mg · liter−1 amoxicillin, 0.5 mg · liter−1 ciprofloxacin, 16 mg · liter−1 streptomycin, or 16 mg · liter−1 cephalexin. Plates were incubated at 37°C and the number of blue colonies (E. coli) was counted. Samples yielding no E. coli colonies on antibiotic-free agar were excluded from further analysis. Up to five E. coli isolates from each cephalexin TBX agar plate were transferred onto cefotaxime (CTX, 2 mg · liter−1) TBX agar. All AB concentrations were chosen as those which define clinically relevant resistance in humans according to EUCAST (31). Two multiplex PCRs were performed to screen for β-lactamase genes in CTX-R E. coli. The first was to detect blaCTX-M groups, as previously described (32), and the second was to detect the following additional β-lactamase genes: blaCMY, blaDHA, blaSHV, blaTEM, and blaOXA-1 (8).
Obtaining farm management information.
Four management practice questionnaires were developed. Questionnaire 1 was completed by the researcher in the presence of the farmer at the time of consent and the first farm visit. Questionnaire 2 was completed by the researcher in the presence of the farmer using Epicollect5 approximately six to 9 months into the project. Questionnaire 3 was completed by the researcher in the presence of the farmer using Epicollect5 approximately 12 to 16 months into the project. Questionnaire 4 was completed by the researcher during a telephone call with the farmer within 2 months of the last visit to the farm. All questionnaires are presented in Table S6.
In total, there were 610 variables derived from the four questionnaires used in the analyses. Questions giving rise to these variables were validated and processed in the following way. Questions with a single response or zero variance were removed. Questions which had either been shown in the literature to be important risk factors or which were judged by veterinary experts to be of potential importance were selected. Categorical levels were collapsed to avoid small response counts. Some questions were combined, as individually they provided detail irrelevant to this study. Repeat questions of demographic variables were averaged. Variables with missing values were removed; all variables removed for this reason were also considered likely to be of low importance. As a result, all the categorical variables were dichotomous.
Monitoring antibacterial usage.
All dairy farmer participants gave permission for researchers to contact their veterinary practices and request AB prescription/sales data for a period of at least one year before the beginning of the project through the end of the project.
All practices except one supplied records. This practice serviced two farms and on-farm records were used instead for these two farms.
Data were assessed by a veterinary researcher to ensure consistent naming of products and quantification between practices, given the wide range of variation in product names and the quantity denominators used.
Usage metrics were produced using mg/PCU for the first 12 months of the project, from the date the farm enrolled on the project and had the first samples collected (range January 2017 to July 2017) until 12 months after this date (range January 2018 to July 2018).
Risk factor analysis.
The risk factors examined fell into four categories: farm management, ABU, sample characteristics, and meteorological. The first three are described above; for the last, local meteorological data were extracted from publicly available UK Met Office data (https://www.metoffice.gov.uk/pub/data/weather/uk/climate/stationdata/yeoviltondata.txt).
Sample processing and data analysis workflows are illustrated in Fig. S2. All data analysis was performed using R (https://www.r-project.org/). Two modeling approaches were used: (i) variable selection via univariable screening and stepwise model selection with a multilevel, multivariable logistic regression model and (ii) a regularized Bayesian model. Both were used to analyze risk factors associated with blaCTX-M E. coli positivity; the second was also used to analyze risk factors associated with positivity for E. coli resistant to amoxicillin, ciprofloxacin, streptomycin, and tetracycline. Sensitivity analyses were performed to test for measurement bias, and to account for the fact that resistant E. coli were more likely to be found in a sample if there was a higher density of bacterial CFU. Further details of variable selection and development of the models and model checking are presented below. All code can be found at https://github.com/HannahSchubert1/OH-STAR-modelling-code. Details of all model checking can be found in Fig. S3.
Modeling risk of blaCTX-M β-lactamase gene carriage.
Risk factor analysis was performed separately on calf data and on the full data set (all samples combined). Thirty-seven variables were selected for the calf risk factor analysis and 110 for the full risk factor analysis, as described below. Exploratory data analysis revealed the main source of clustering of observations was at the farm level. There were no noticeable longitudinal patterns or clustering due to the location within each farm. Random intercept logistic regression models with farm as a random effect were used throughout the analysis.
Two approaches to risk factor analysis were performed: (i) a frequentist variable selection method (using univariable screening followed by step-down model selection and maximum likelihood estimation; method 1) and (ii) a Bayesian method with regularization (method 2, on the full data set only).
Method 1: variable selection.
Variable selection was performed by first screening the variables for association with E. coli carrying blaCTX-M using univariable, multilevel logistic regression (33) with each variable entered as a fixed effect and with random intercepts for each farm. Continuous variables were first checked for linearity with the log odds of the outcome to ensure they did not violate model assumptions. For the analysis of the full data set, it was not possible to converge such a model for 27 of the variables (most likely due to the low number of samples that were positive for E. coli carrying blaCTX-M), so 83 variables were examined. See Tables S1 and S2 for the full univariable results.
Variables with associations where the P value was <0.25 after controlling for false discovery rate using the Benjamini-Hochberg procedure (chosen to be the most appropriate method for the exploratory nature of the project) were entered into a multivariable, multilevel logistic regression model with random intercepts for each farm. A backward stepwise procedure was used to further refine the model, selecting only those variables where the regression coefficient was maintained at P < 0.05. Variables which survived this analysis were checked for multicollinearity by removing each variable in turn and checking that the confidence intervals for the estimate for each variable still overlapped. The predictive accuracy was checked using area under the receiver operating characteristic curve (0.84 for the full data set; 0.80 for the calf data).
Method 2: Bayesian model for predicting presence of blaCTX-M-positive E. coli.
A farm-level random intercept model was fit using the R package BRMS (34) on the full data set. All variables were used as predictors of blaCTX-M-positive E. coli but were split into two groups. The first group comprised farm-level (i) total antibiotic use, (ii) total third- and fourth-generation cephalosporin use, and (iii) total first-generation cephalosporin use. The second group contained the remaining meteorological and farm management variables. For the first group, uninformative priors (normal distribution with a mean 0, standard deviation 5) were used; for the second group, a regularizing prior (horseshoe prior with a single degree of freedom [35]) was used. The mean intercept was also given a diffuse prior (normal distribution with mean 0, standard deviation 5), while the standard deviation of the random effects was given a half-Student-T distribution with three degrees of freedom and a scale factor of 10 (the default in BRMS). Four independent Markov chains were sampled with 1,000 warmup iterations and 1,000 sampling iterations. The target acceptance criterion was increased from the default of 0.8 to 0.95 to decrease the chance of sampling divergencies (although two remained, these were not deemed significant). The sampling was assumed to be well converged, as the Gelman-Rubin statistic for each variable was 1.00. Results are shown in Table S3.
Accounting for measurement error.
To account for measurement error of the blaCTX-M-positive E. coli (whereby if more E. coli were found in a sample, the sensitivity of the test for finding blaCTX-M-positive E. coli was higher), the logistic link function was altered to include the sensitivity and specificity of each sample, following the work by Coutinho et al. (36). The altered logistic link function was derived by equating the conditional probability of the true blaCTX-M status to the conditional probability of the observed blaCTX-M status, for a given sensitivity (s) and specificity (e).
The specificity of the blaCTX-M test was assumed to be 100% across samples, whereas the sensitivity (s) was estimated separately for each observation using the number of E. coli colonies grown on antibiotic-free agar (k) and a minimum detectable prevalence of blaCTX-M-positive E. coli (q) through the following equation:
| (1) |
The distribution s across the whole data set using method 1 for different values of q is shown in Fig. S4. While the value of q did change the sensitivity associated with each observation, this change had only a negligible effect on the coefficient estimates (Fig. S1A) and the area under the curve (AUC) (Fig. S1B) over the range of 0.01 to 1.00. The reported values used a q of 0.01. The R-code for this link function is provided in the supplemental material (Fig. S5).
Model for predicting the presence of E. coli resistant to noncephalosporins.
For each resistance phenotype (amoxicillin, ciprofloxacin, streptomycin, and tetracycline), a random intercept model with farm as the random effect was fitted using the R package BRMS on the full data set (https://cran.r-project.org/web/packages/brms/index.html). All variables (see above) were used as predictors of resistant E. coli in a sample but were split into two groups. The first group (the “main” variables) comprised farm-level ABU and average monthly temperature at the time of sample collection, as these were the main variables hypothesized to influence resistance. For all models, total ABU, streptomycin usage, tetracycline usage, amoxicillin usage, fluoroquinolone usage, and cefalexin usage were included as predictors. For each model, the usage of additional antibacterial drugs was tested if they were hypothesized to be important in selecting for resistance to the relevant model: (i) for the amoxicillin model, first-generation cephalosporins, penicillins, and potentiated amoxicillin; (ii) for the ciprofloxacin model, novobiocin and third- and fourth-generation cephalosporins; (iii) for the streptomycin and tetracycline models, no additional ABU variables were tested. The second group of variables (the “regularized” variables) contained the remaining farm management variables.
For the first group (the “main” variables), uninformative priors (normal distribution with a mean 0, standard deviation 5) were used; for the second group (the “regularized” variables), a regularizing prior (horseshoe prior with a single degree of freedom) was used. The mean intercept was also given a diffuse prior (normal distribution with mean 0, standard deviation 5), while the standard deviation of the random effects was given a half-Student-T distribution with three degrees of freedom and a scale factor of 10 (the default in BRMS). Four independent Markov chains were sampled with 1,000 warmup iterations and 10,000 sampling iterations. The target acceptance criterion was increased from the default of 0.8 to 0.98 to decrease the chance of sampling divergencies in all models. Measurement error based on E. coli density was accounted for as described above. All reported results used a q value of 0.01.
In addition, the measurement error of temperature was considered because only an average monthly temperature was available. Data where daily temperature was recorded for a (different) 12-month period were used to calculate an estimated average monthly standard deviation over the 12 months (2.89). The “me” function in the package “BRMS” (https://rdrr.io/cran/brms/man/me.html) was used, whereby the monthly temperature variable was assumed to vary with a standard deviation of 2.89 (0.61 when scaled as the temperature was scaled to mean 0 and standard deviation 1 before entering the model; the actual standard deviation of the temperature was 4.7).
To further test the robustness of the model outputs, all models were rerun using skeptical priors, whereby the prior was set to the opposite of what would be expected. So, for all main variables, if there was an association, a positive association would be expected given prior knowledge. To test the models with skeptical priors, a prior for the main variables was given as a normal distribution with mean −0.5 and a narrow standard deviation 2 (i.e., assuming a negative correlation with low variation; the opposite of expected).
Bayesian model checking.
Convergence is assumed to be good with a Gelman-Rubin statistic (Rhat) of 1.00 for all variables across all models. Figure S6 shows trace plots for the associated variables, also providing evidence of good convergence. There were no divergence issues reported for any of the models. Table S4 shows the full results from all models, including odds ratios, 95% credible intervals, and effective sample sizes. Figure S7 shows the posterior distributions of the associated variables. Rerunning the models with skeptical priors did not alter the model conclusions (Table S5).
Supplementary Material
ACKNOWLEDGMENTS
We thank all the farmers and veterinary surgeons who participated in this study. In addition, we thank the milk processing companies who supported our recruitment process and the veterinary students who helped with data processing and sample collection.
This work was funded by grant NE/N01961X/1 to M.B.A., K.K.R., K.M.T., T.A.C., and D.C.B. from the Antimicrobial Resistance Cross Council Initiative supported by the seven United Kingdom research councils. R.A. was supported by the Jean Golding Institute for data-intensive research at the University of Bristol. G.M.R. was funded by the Langford Trust.
D.C.B. was president of the British Cattle Veterinary Association in 2018 to 2019. Otherwise, we declare no competing interests. Farming and veterinary businesses that contributed data and permitted access for sample collection were not involved in the design of this study or in data analysis, and were not involved in drafting the manuscript for publication.
Conceived the study: D.C.B., K.K.R., and M.B.A. Collection of data: H.S., J.F., K.M., E.F.P., O.M., and V.C.G., supervised by T.A.C., K.K.R., G.M.R., and M.B.A. Cleaning and analysis of data: H.S., J.F., R.A., V.C.G., E.F.P., L.V., and M.E., supervised by K.M.T., K.K.R., and M.B.A. Initial drafting of manuscript: H.S., K.K.R., and M.B.A. Corrected and approved manuscript: all authors.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Department for Environment, Food & Rural Affairs, Department of Health and Social Care, Public Health England, and Veterinary Medicines Directorate. 2019. Collection: antimicrobial resistance (AMR). Government of the United Kingdom, London, UK. https://www.gov.uk/government/collections/antimicrobial-resistance-amr-information-and-resources.
- 2.ESVAC. 2020. European Surveillance of Veterinary Antimicrobial Consumption. European Medicines Agency, Amsterdam, Netherlands. https://www.ema.europa.eu/en/veterinary-regulatory/overview/antimicrobial-resistance/european-surveillance-veterinary-antimicrobial-consumption-esvac.
- 3.Veterinary Medicines Directorate & Animal and Plant Health Agency. 2017. UK Veterinary Antibiotic Resistance and Sales Surveillance. Veterinary Medicines Directorate, Surrey, UK. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/915743/_1691664-v1-VARSS_2017_Watermark_FINALx-accessible.pdf.
- 4.Veterinary Medicines Directorate & Animal and Plant Health Agency. 2019. UK Veterinary Antibiotic Resistance and Sales Surveillance. Veterinary Medicines Directorate, Surrey, UK. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/942772/FINAL_MASTER_VARSS_2019_Report__updated_Dec_2020_.pdf.
- 5.Lazarus B, Paterson DL, Mollinger JL, Rogers BA. 2015. Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin Infect Dis 60:439–452. doi: 10.1093/cid/ciu785. [DOI] [PubMed] [Google Scholar]
- 6.Bevan ER, Jones AM, Hawkey PM. 2017. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 72:2145–2155. doi: 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 7.Madec J-Y, Haenni M. 2018. Antimicrobial resistance plasmid reservoir in food and food-producing animals. Plasmid 99:72–81. doi: 10.1016/j.plasmid.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Findlay J, Gould VC, North P, Bowker KE, Williams OM, MacGowan AP, Avison MB. 2020. Characterization of cefotaxime-resistant urinary Escherichia coli from primary care in South-West England 2017–18. J Antimicrob Chemother 75:65–71. doi: 10.1093/jac/dkz397. [DOI] [PubMed] [Google Scholar]
- 9.Alzayn M, Findlay J, Schubert H, Mounsey O, Gould VC, Heesom KJ, Turner KM, Barrett DC, Reyher KK, Avison MB. 2020. Characterization of AmpC-hyperproducing Escherichia coli from humans and dairy farms collected in parallel in the same geographical region. J Antimicrob Chemother 75:2471–2479. doi: 10.1093/jac/dkaa207. [DOI] [PubMed] [Google Scholar]
- 10.Agersø Y, Aarestrup FM, Pedersen K, Seyfarth AM, Struve T, Hasman H. 2012. Prevalence of extended-spectrum cephalosporinase (ESC)-producing Escherichia coli in Danish slaughter pigs and retail meat identified by selective enrichment and association with cephalosporin usage. J Antimicrob Chemother 67:582–588. doi: 10.1093/jac/dkr507. [DOI] [PubMed] [Google Scholar]
- 11.Randall LP, Clouting C, Horton RA, Coldham NG, Wu G, Clifton-Hadley FA, Davies RH, Teale CJ. 2011. Prevalence of Escherichia coli carrying extended-spectrum β-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J Antimicrob Chemother 66:86–95. doi: 10.1093/jac/dkq396. [DOI] [PubMed] [Google Scholar]
- 12.Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K, Huijsdens X, Kluytmans J. 2011. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg Infect Dis 17:1216–1222. doi: 10.3201/eid1707.110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kola A, Kohler C, Pfeifer Y, Schwab F, Kühn K, Schulz K, Balau V, Breitbach K, Bast A, Witte W, Gastmeier P, Steinmetz I. 2012. High prevalence of extended-spectrum-β-lactamase-producing Enterobacteriaceae in organic and conventional retail chicken meat, Germany. J Antimicrob Chemother 67:2631–2634. doi: 10.1093/jac/dks295. [DOI] [PubMed] [Google Scholar]
- 14.Luvsansharav U-O, Hirai I, Nakata A, Imura K, Yamauchi K, Niki M, Komalamisra C, Kusolsuk T, Yamamoto Y. 2012. Prevalence of and risk factors associated with faecal carriage of CTX-M β-lactamase-producing Enterobacteriaceae in rural Thai communities. J Antimicrob Chemother 67:1769–1774. doi: 10.1093/jac/dks118. [DOI] [PubMed] [Google Scholar]
- 15.McNulty CAM, Lecky DM, Xu-McCrae L, Nakiboneka-Ssenabulya D, Chung K-T, Nichols T, Thomas HL, Thomas M, Alvarez-Buylla A, Turner K, Shabir S, Manzoor S, Smith S, Crocker L, Hawkey PM. 2018. CTX-M ESBL-producing Enterobacteriaceae: estimated prevalence in adults in England in 2014. J Antimicrob Chemother 73:1368–1388. doi: 10.1093/jac/dky007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snow LC, Warner RG, Cheney T, Wearing H, Stokes M, Harris K, Teale CJ, Coldham NG. 2012. Risk factors associated with extended spectrum beta-lactamase Escherichia coli (CTX-M) on dairy farms in North West England and North Wales. Prev Vet Med 106:225–234. doi: 10.1016/j.prevetmed.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Mollenkopf DF, Weeman MF, Daniels JB, Abley MJ, Mathews JL, Gebreyes WA, Wittum TE. 2012. Variable within- and between-herd diversity of CTX-M cephalosporinase-bearing Escherichia coli isolates from dairy cattle. Appl Environ Microbiol 78:4552–4560. doi: 10.1128/AEM.00373-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton RA, Duncan D, Randall LP, Chappell S, Brunton LA, Warner R, Coldham NG, Teale CJ. 2016. Longitudinal study of CTX-M ESBL-producing E. coli strains on a UK dairy farm. Res Vet Sci 109:107–113. doi: 10.1016/j.rvsc.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Brunton LA, Reeves HE, Snow LC, Jones JR. 2014. A longitudinal field trial assesing the impact of feeding waste milk containing antibiotic residues on the prevalence of ESBL-producing Escherichia coli in calves. Prev Vet Med 117:403–412. doi: 10.1016/j.prevetmed.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 20.MacFadden DR, McGough SF, Fisman D, Santillana M, Brownstein JS. 2018. Antibiotic resistance increases with local temperature. Nat Clim Chang 8:510–514. doi: 10.1038/s41558-018-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson E, Jeckel S, Snow L, Stubbs R, Teale C, Wearing H, Horton R, Toszeghy M, Tearne O, Ellis-Iversen J, Coldham N. 2012. Epidemiology of extended spectrum beta-lactamase E. coli (CTX-M-15) on a commercial dairy farm. Vet Microbiol 154:339–346. doi: 10.1016/j.vetmic.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 22.Duse A, Persson Waller K, Emanuelson U, Ericsson Unnerstad H, Persson Y, Bengtsson B. 2016. Occurrence and spread of quinolone-resistant Escherichia coli on dairy farms. Appl Environ Microbiol 82:3765–3773. doi: 10.1128/AEM.03061-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereira RV, Siler JD, Ng JC, Davis MA, Grohn YT, Warnick LD. 2014. Effect of on-farm use of antimicrobial drugs on resistance in fecal Escherichia coli of preweaned dairy calves. J Dairy Sci 97:7644–7654. doi: 10.3168/jds.2014-8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Afema JA, Davis MA, Sischo WM. 2019. Antimicrobial use policy change in pre-weaned dairy calves and its impact on antimicrobial resistance in commensal Escherichia coli: a cross sectional and ecological study. BMC Microbiol 19:217–217. doi: 10.1186/s12866-019-1576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Awosile BB, Smith BA. 2017. Risk assessment modelling of fecal shedding caused by extended-spectrum cephalosporin-resistant Escherichia coli transmitted through waste milk fed to dairy pre-weaned calves. J Dairy Sci 100:9667–9673. doi: 10.3168/jds.2017-13196. [DOI] [PubMed] [Google Scholar]
- 26.Bachmann J, Helmschrodt C, Richter A, Heuwieser W, Bertulat S. 2018. Residue concentration of cefquinome after intramammary dry cow therapy and short dry periods. J Dairy Sci 101:7540–7550. doi: 10.3168/jds.2017-13826. [DOI] [PubMed] [Google Scholar]
- 27.Peirano G, Pitout JDD. 2019. Extended-spectrum β-lactamase-producing Enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs 79:1529–1541. doi: 10.1007/s40265-019-01180-3. [DOI] [PubMed] [Google Scholar]
- 28.Luo PJ, Zhang JB, Wang HL, Chen X, Wu N, Zhao YF, Wang XM, Zhang H, Zhang JY, Zhu L, Jiang WX. 2016. Rapid and sensitive chemiluminescent enzyme immunoassay for the determination of neomycin residues in milk. Biomed Environ Sci 29:374–378. doi: 10.3967/bes2016.048. [DOI] [PubMed] [Google Scholar]
- 29.Bradley AJ, Green MJ. 2001. Adaptation of Escherichia coli to the bovine mammary gland. J Clin Microbiol 39:1845–1849. doi: 10.1128/JCM.39.5.1845-1849.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultsz C, Geerlings S. 2012. Plasmid-mediated resistance in Enterobacteriaceae: changing landscape and implications for therapy. Drugs 72:1–16. doi: 10.2165/11597960-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 31.European Committee on Antimicrobial Susceptibility Testing. 2020. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf. Last accessed May 2020.
- 32.Woodford N, Fagan EJ, Ellington MJ. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother 57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 33.Dohoo I, Martin W, Stryhn H. 2011. Veterinary epidemiologic research, 2nd ed. VER Inc., Charlottetown, Prince Edward Island, Canada. [Google Scholar]
- 34.Buerkner PC. 2017. BRMS: an R package for Bayesian multilevel models using Stan. J Stat Softw 80:1–28. doi: 10.18637/jss.v080.i01. [DOI] [Google Scholar]
- 35.Carvalho CM, Polson NG, Scott JG. 2010. The horseshoe estimator for sparse signals. Biometrika 97:465–480. doi: 10.1093/biomet/asq017. [DOI] [Google Scholar]
- 36.Coutinho C, Bastos LS, Corrêa da Mota J, Toledo L, Costa K, Bertoni N, Bastos FI. 2019. The risks of HCV infection among Brazilian crack cocaine users: incorporating diagnostic test uncertainty. Sci Rep 9:443. doi: 10.1038/s41598-018-35657-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


