The ability of bacteria to take up DNA (competence) and incorporate foreign DNA into their genomes (transformation) permits them to rapidly evolve and gain new traits and/or acquire antibiotic resistances. It also facilitates laboratory-based investigations into mechanisms of specific phenotypes, such as those involved in host colonization. Vibrio fischeri has long been a model for symbiotic bacterium-host interactions as well as for other aspects of its physiology, such as bioluminescence and biofilm formation.
KEYWORDS: competence, Vibrio fischeri, LitR, HapR, quorum sensing, luminescence, Euprymna scolopes, transformation, Vibrio cholerae, symbiosis
ABSTRACT
Vibrio species, including the squid symbiont Vibrio fischeri, become competent to take up DNA under specific conditions. For example, V. fischeri becomes competent when grown in the presence of chitin oligosaccharides or upon overproduction of the competence regulatory factor TfoX. While little is known about the regulatory pathway(s) that controls V. fischeri competence, this microbe encodes homologs of factors that control competence in the well-studied V. cholerae. To further develop V. fischeri as a genetically tractable organism, we evaluated the roles of some of these competence homologs. Using TfoX-overproducing cells, we found that competence depends upon LitR, the homolog of V. cholerae master quorum-sensing and competence regulator HapR, and upon homologs of putative pilus genes that in V. cholerae facilitate DNA uptake. Disruption of genes for negative regulators upstream of LitR, namely, the LuxO protein and the small RNA (sRNA) Qrr1, resulted in increased transformation frequencies. Unlike LitR-controlled light production, however, competence did not vary with cell density under tfoX overexpression conditions. Analogous to the case with V. cholerae, the requirement for LitR could be suppressed by loss of the Dns nuclease. We also found a role for the putative competence regulator CytR. Finally, we determined that transformation frequencies varied depending on the TfoX-encoding plasmid, and we developed a new dual tfoX and litR overexpression construct that substantially increased the transformation frequency of a less genetically tractable strain. By advancing the ease of genetic manipulation of V. fischeri, these findings will facilitate the rapid discovery of genes involved in physiologically relevant processes, such as biofilm formation and host colonization.
IMPORTANCE The ability of bacteria to take up DNA (competence) and incorporate foreign DNA into their genomes (transformation) permits them to rapidly evolve and gain new traits and/or acquire antibiotic resistances. It also facilitates laboratory-based investigations into mechanisms of specific phenotypes, such as those involved in host colonization. Vibrio fischeri has long been a model for symbiotic bacterium-host interactions as well as for other aspects of its physiology, such as bioluminescence and biofilm formation. Competence of V. fischeri can be readily induced upon overexpression of the competence factor TfoX. Relatively little is known about the V. fischeri competence pathway, although homologs of factors known to be important in V. cholerae competence exist. By probing the importance of putative competence factors that control transformation of V. fischeri, this work deepens our understanding of the competence process and advances our ability to genetically manipulate this important model organism.
INTRODUCTION
The ability of bacteria to take up and incorporate foreign DNA into their genomes plays a vital role in bacterial adaptation. For example, bacteria can acquire genetic material that permits them to become resistant to antibiotics or to survive/thrive in a specific environment. The acquisition of extracellular DNA, or competence, can be induced under particular environmental conditions. For example, the pathogen Vibrio cholerae, found in brackish water and associated with chitin-containing zooplankton such as copepods, induces its competence pathway upon exposure to chitin (1, 2).
Once the cells have taken up exogenous DNA, they can recombine it into their genomes in a process known as transformation. Researchers have exploited these phenomena to rationally generate specific bacterial mutants. For example, exposing cells to DNA containing sequences up- and downstream of a gene to be disrupted along with a centrally located antibiotic resistance cassette, followed by selection for the resistance marker, results in growth of the desired mutant. In V. cholerae, competence and transformation have been optimized such that strains carrying multiple unmarked mutations can be engineered in a single step: exposing competent cells to multiple fragments of mutant DNA, followed by selection for a single selectable marker, can result in uptake and recombination of all of the introduced DNA fragments to produce unmarked deletions (3). This technology, termed MuGent (multiplex genome editing by natural transformation), has been applied to the study of genes that are present in multiple copies in the genome, such as those involved in carbohydrate transport (4).
The genes for competence and transformation in V. cholerae have been well studied (5–8). V. cholerae uses a type IV competence pilus to acquire environmental DNA and bring it into the periplasm. A second step results in transport of this transforming DNA (tDNA) across the inner membrane, delivering it as single-stranded DNA into the cytoplasm. Finally, the tDNA is bound by and protected by proteins, including RecA, that facilitate its recombination into the chromosome. At least 19 structural proteins contribute to these different steps of DNA uptake and recombination (7).
Competence in V. cholerae is positively controlled by the transcription factor HapR, which, in turn, is controlled by quorum sensing; HapR activity and competence are low at low cell densities and increase with exposure to autoinducers and/or at high cell density (Fig. 1) (2, 6, 9, 10). In V. cholerae, HapR activates transcription of positive competence regulators, including comEA, which encodes a DNA binding protein required for DNA uptake and thus competence (2, 5, 9, 11). HapR also inhibits expression of the secreted nuclease Dns, which degrades extracellular DNA, thereby preventing DNA uptake (6). A mutant defective for production of Dns exhibits increased transformation, while one lacking the gene for the competence secretin protein PilQ fails to take up DNA (6, 11). Competence is also stimulated by the chitin-induced regulator TfoX, which contributes to induction of comEA and other competence factors (2, 6). A complex regulatory scheme controls the production of TfoX, including chitin-sensing proteins and a small regulatory RNA, TfoR (12). Overexpression of TfoX can bypass the need for chitin (2). Finally, HapR and TfoX control downstream regulators, including the quorum-sensing and TfoX-dependent regulator QstR and the TfoX-controlled cytidine regulator CytR, that contribute to competence control (13–15).
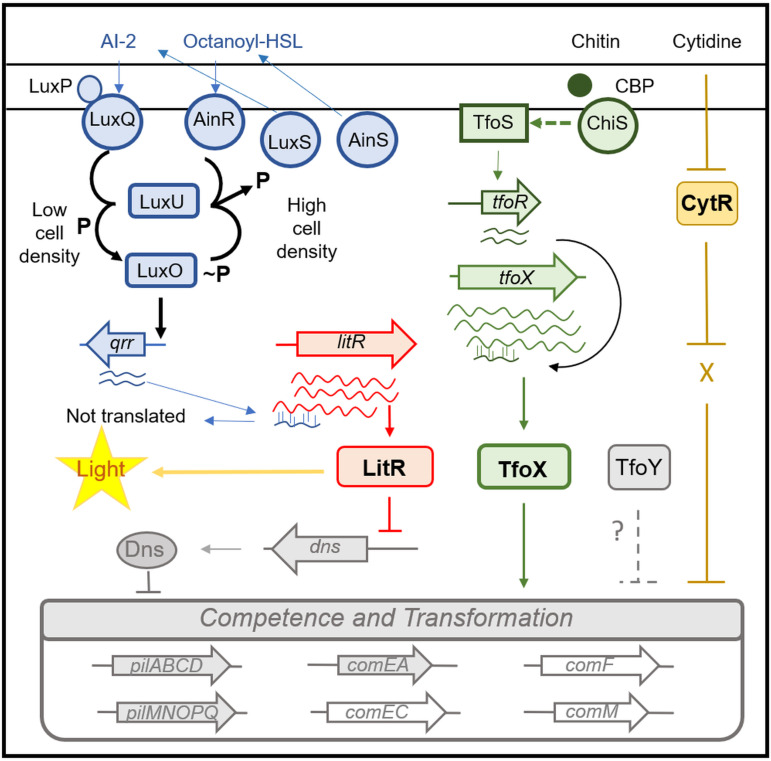
FIG 1.
Model for competence and transformation of V. fischeri. Major regulatory pathways and other factors predicted, based on studies of V. cholerae, to control competence and transformation of V. fischeri are shown. In the top left (blue), quorum sensing regulators are shown. At low cell density, two kinases, LuxQ and AinR, function as kinases, promoting phosphorylation, via LuxU, of the response regulator LuxO, which, in turn, activates transcription of the gene for small RNA Qrr1. Qrr1 activity results in decreased levels of LitR (orange), a positive regulator of competence that promotes light production; in V. cholerae, the LitR homolog HapR controls transcription of dns, which encodes a nuclease that interferes with DNA uptake. At high cell densities, autoinducers made by LuxS and AinS switch the activity of LuxQ and AinR to phosphatases, thereby decreasing Qrr1 levels and increasing LitR levels and light production (yellow). Another major regulator of competence is TfoX (middle, in green). In V. cholerae, TfoX is controlled at the level of translation by the sRNA TfoR, which, in turn, is controlled by chitin-sensing regulators ChiS and TfoS. In V. fischeri, competence is readily induced by overproduction of TfoX, even in the absence of chitin. Another important regulator appears to be the cytidine-responsive regulator CytR (right, in brown), which activates competence via an unknown mechanism. The role of TfoY, if any, in competence remains unknown. At the bottom (in gray and white), a few of the structural genes known to be important for competence in V. cholerae are shown; gray coloring indicates factors assessed in this study.
The V. cholerae competence genes are largely conserved in other Vibrio species, including Vibrio fischeri, an important model for mutualistic microbe-host associations, bioluminescence, quorum sensing, and biofilm formation (5, 7, 8, 16). In 2010, Pollack-Berti et al. reported that V. fischeri could be made competent by growth in the presence of chitin oligosaccharides and/or by overexpression of the competence regulatory factor TfoX (17). While the reported frequency of transformation is somewhat variable, ranging as much as 100-fold between experiments (2, 17), this ability to directly introduce “naked” DNA into V. fischeri has facilitated numerous genetic manipulations. For example, V. fischeri researchers have used TfoX overexpression to (i) map a deletion in a lab strain (18), (ii) backcross mutations of interest (for an example, see reference 19), and (iii) readily make strains containing multiple mutations (for an example, see reference 20). Combined with the recent development of tools to rapidly make unmarked deletions using splicing by overlap extension PCR (SOE PCR) and FLP (flippase)-dependent recombination technology, the approach of using TfoX-induced competence has advanced the development of V. fischeri as a model genetic organism by facilitating the rapid and targeted manipulation of this microbe (17, 18, 21, 22).
Beyond the finding that TfoX overexpression promotes competence, however, little is known about the regulatory pathway(s) that controls competence in V. fischeri. Given the conservation of competence genes, we hypothesized that the competence pathway of V. fischeri is similar to that of V. cholerae (Fig. 1). Indeed, we found that transformation of V. fischeri depends on the HapR homolog LitR (23), as well as on genes predicted to encode structural proteins required for competence. We also identified a role in transformation for the putative cytidine-responsive regulator CytR and determined that the use of specific conditions or plasmids impacts transformation frequency. Finally, we developed a new dual tfoX and litR overexpression plasmid that could induce transformation about 100- to 1,000-fold higher than the original tfoX plasmid in a strain of V. fischeri with low genetic tractability.
RESULTS
LitR is required for V. fischeri competence.
Our interest in V. fischeri competence was prompted when two independently generated litR mutants failed to incorporate a marked allele into their genomes following multiple transformation attempts. These results suggested that litR might be required for competence of V. fischeri, as is the case for the LitR homolog HapR in V. cholerae (2, 23). However, because the initial study on V. fischeri competence reported a wide range in transformation frequencies (17), we first sought to obtain consistent and reliable data by optimizing and standardizing the conditions used to obtain transformants. As described in Materials and Methods, we used fresh minimal medium made from the component reagents to avoid precipitation of the component parts and thus altered growth. We found that reducing the overnight growth of the tfoX-overproducing cells to approximately 14 h (rather than a more typical 16 h or longer) substantially diminished the subsequent lag phase following subculture on the next day, resulting in a more consistent outgrowth with cells generally reaching an optical density (OD) of about 0.5 in 5 or 6 h. Finally, we used a consistent amount (500 ng) of genomic DNA (gDNA) and a consistent recovery time of 90 min to permit better comparisons between experiments.
These optimized conditions resulted in a relatively consistent transformation frequency for the control strains but did not yield any transformants for the litR mutants (Fig. 2A). Complementation using litR expressed from a plasmid restored transformation frequencies to the level achieved by the control strain (Fig. 2B). Together, these data confirm a role for LitR in controlling transformation of V. fischeri.
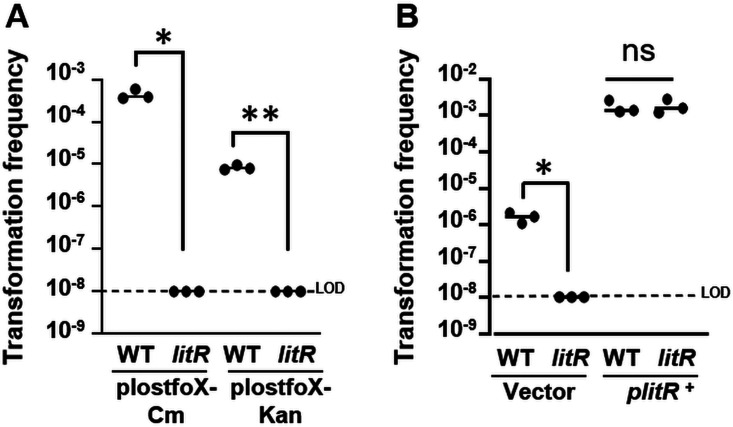
FIG 2.
LitR is required for transformation of V. fischeri. Shown is the transformation frequency (total Trimr CFU/viable CFU) of V. fischeri cells exposed to gDNA from KV8300 (fliQ::Trim). (A) Transformation frequencies of plostfoX-Cm- or plostfoX-Kan-containing ES114 (WT) and litR strains PMF8 (litR::Kan) and JB19 (litR::Erm) containing plostfoX-Cm and plostfoX-Kan, respectively. (B) Transformation frequencies of ES114 and JB19 containing plostfoX-Kan and either vector control (pVSV105) or litR expression plasmid plitR (pPMF5). Both litR mutants PMF8 and JB19 exhibited transformation frequencies below the limit of detection (LOD). *, P < 0.05; **, P < 0.005; ns, not significant.
These experiments also revealed an unexpected finding: V. fischeri exhibited over a 10-fold higher transformation frequency when it overproduced TfoX from plostfoX (Cmr) (termed plostfoX-Cm here for clarity) (17) relative to plostfoX-Kan (Kanr) (21) (Fig. 2A). The cause of this discrepancy is unknown, as the plasmids are identical except for the antibiotic resistance cassette, and resequencing of the respective tfoX cassettes revealed no differences. Thus, it is possible that selection for Cmr maintains the plasmid better than does selection for Kanr. In support of this possibility, we observed a similar difference in transformation frequencies when we evaluated newly generated plasmids (pJJC2 [Cmr] and pJJC3 [Kanr]) in which we substituted tfoX for an arabinose-inducible tfoX cassette in the same two backbones (see Fig. S1 in the supplemental material). As a result of the differential transformation frequencies that result from the use of plostfoX-Cm and plostfoX-Kan, we generally chose to use plostfoX-Cm for experiments in which a high transformation frequency was needed (for example, to evaluate the roles of structural factors expected to have a severe competence defect), while we used plostfoX-Kan to better assess subtler defects that might be overcome by the use of plostfoX-Cm.
Putative competence genes required for V. fischeri transformation.
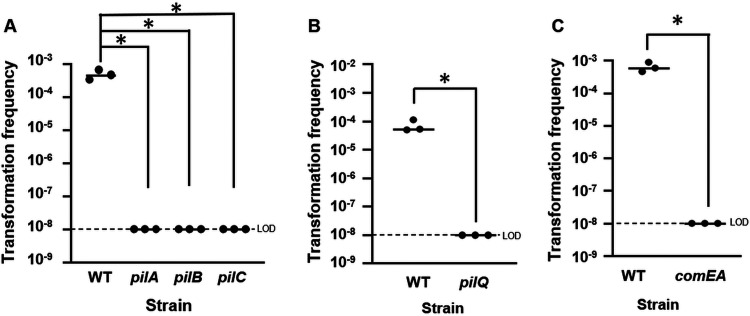
The finding that LitR is a critical factor in competence prompted us to determine the requirement of other genes predicted in the literature to function in competence in V. fischeri and/or known to be important for V. cholerae competence (Fig. 1) (for examples, see references 5, 7, and 8). Namely, we tested the requirement for the following putative pilus genes, homologous to V. cholerae type IV pili components: pilA, pilB, pilC, and pilQ (VF_2185 to VF_2187 and VF_2293, respectively), for which transposon (Tn) insertion mutants were available. We also generated a deletion mutant for putative competence gene comEA (VF_0801) and tested its role. Whereas tfoX-overexpressing wild-type cells were transformed at high levels (a transformation frequency of ∼10−3 to 10−4, or about 1 transformant per 1,000 to 10,000 CFU), tfoX-overexpressing mutants defective for the pil genes pilA, pilB, pilC, and pilQ failed to be transformed (Fig. 3A and B and Fig. S2A). Similarly, the comEA mutant produced no transformants (Fig. 3C and Fig. S2B). These data suggest that the structural proteins demonstrated as competence factors in V. cholerae (7) likely function in the same manner in V. fischeri.
FIG 3.
V. fischeri transformation requires pil and comEA genes. Shown are transformation frequencies (Trimr CFU/viable CFU) of plostfoX-containing wild-type and mutant V. fischeri cells exposed to gDNA from KV8300 (fliQ::Trim). (A) Transformation frequencies of plostfoX-Cm-containing ES114 (WT) and pil Tn mutant strains KV8875 (pilA), KV8876 (pilB), and KV8877 (pilC). (B) Transformation frequencies of plostfoX-Kan-containing ES114 and KV8879 (pilQ). (C) Transformation frequencies of plostfoX-Cm-containing ES114 and KV9219 (comEA). The pilA, pilB, pilC, pilQ, and comEA mutants each exhibited a transformation frequency below the LOD. *, P < 0.05.
Neither TfoR nor TfoY is required when TfoX is overexpressed.
We next tested roles for two putative competence factors, TfoR and TfoY. In V. cholerae, TfoX translation is activated by the sRNA TfoR, which binds the 5′ end of the tfoX mRNA (12). We thus hypothesized that TfoR activity could be necessary for optimal TfoX production in V. fischeri, even when the latter is overproduced from a multicopy plasmid. However, we observed no impact of tfoR deletion on transformation frequency (Fig. S3).
TfoY is a distant homolog of TfoX whose role in competence has been evaluated in two studies (17, 24). In the original study of TfoX-mediated competence in V. fischeri, Pollack-Berti et al. reported that chitobiose-induced competence depended on TfoY (17). A more recent report, however, found no role for TfoY in TfoX-dependent activities in V. fischeri or V. cholerae. Instead, the authors concluded that the two proteins function independently of one another and that TfoY functioned primarily in a type VI secretion pathway responsible for releasing extracellular DNA by killing competitor bacteria in the environment (25). In an attempt to address this discrepancy, we generated a deletion mutant of tfoY and determined the transformation frequency. In multiple independent experiments, we observed either no difference or a small but statistically insignificant increase in the transformation frequency of the tfoY mutant relative to the wild-type control (Fig. S4). Thus, under our TfoX overproduction conditions, TfoY appears to play no role in the competence pathway.
In V. cholerae, TfoY also controls motility (25, 26). Thus, we assessed the impact of tfoY deletion on motility of V. fischeri; we found that it caused a severe decrease in migration of this organism through soft agar (Fig. S4). We conclude that V. fischeri TfoY appears to function similarly in the two Vibrio species with respect to motility.
Role for CytR.
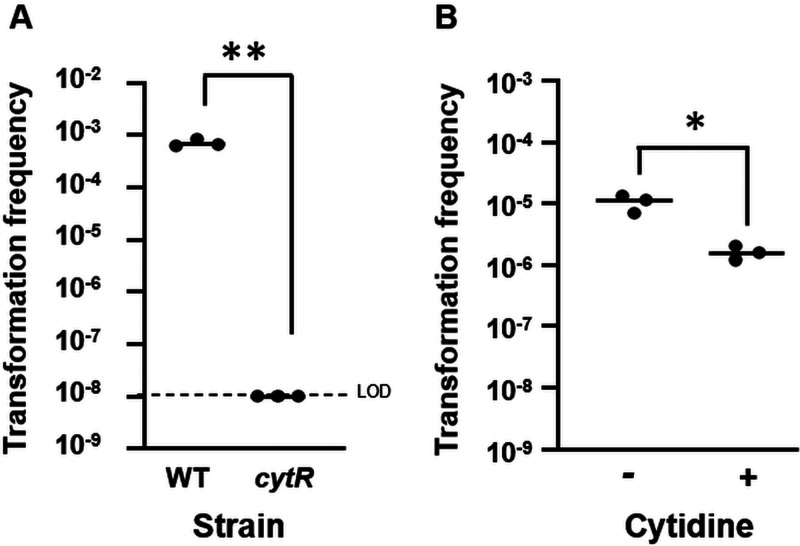
We next assessed the role of CytR, a putative cytidine-responsive regulator that promotes V. cholerae competence through an as-yet-unknown mechanism (Fig. 1) (14, 15). To test the role of CytR in V. fischeri, we generated a cytR deletion mutant. This strain failed to produce transformants (Fig. 4A and Fig. S5). Because CytR is inhibited by cytidine (14), these data indicate that V. fischeri competence may be negatively controlled by cytidine. To test this possibility, cytidine was added to the cells together with DNA at the start of the 30-min incubation period. The presence of cytidine decreased the transformation frequency about 10-fold (Fig. 4B). Thus, cytidine is inhibitory, indicating that V. fischeri and V. cholerae share this aspect of competence control.
FIG 4.
V. fischeri transformation is activated by CytR and inhibited by cytidine. (A) Transformation frequencies (total Trimr CFU/viable CFU) of plostfoX-Cm-containing wild-type strain ES114 (WT) and ΔcytR mutant (KV8840) cells exposed to gDNA from KV8300 (fliQ::Trim). (B) Transformation frequencies of plostfoX-Kan-containing ES114 (WT) exposed to 100 mM cytidine or left untreated during incubation with KV8300 gDNA. The cytR mutant exhibited a transformation frequency below the LOD. *, P < 0.05; **, P < 0.005.
Transformation is controlled by regulators of litR.
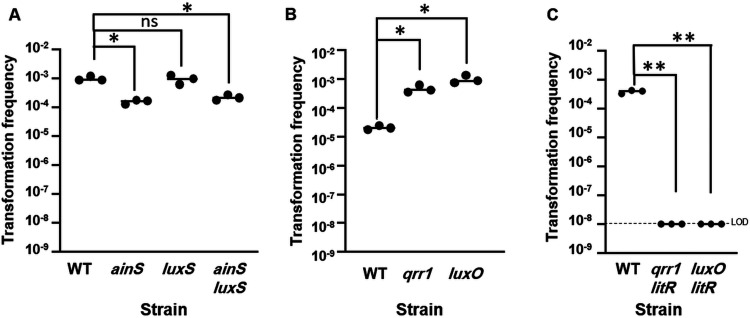
LitR production and thus activity is controlled by quorum-sensing regulators (Fig. 1) (27–30). Thus, we hypothesized that disruption of regulators that positively and negatively control LitR would exert opposite effects on transformation. Indeed, a double mutant lacking the autoinducer synthases AinS and LuxS, which positively control litR expression, exhibited decreased transformation (Fig. 5A). A similar result was seen for an ainS single mutant, while a luxS single mutant was indistinguishable from the wild-type strain (Fig. 5A). These data parallel those seen for the control of V. fischeri bioluminescence, in which AinS plays the more important role (27, 31, 32).
FIG 5.
Quorum-sensing regulators control V. fischeri transformation. (A) Transformation frequencies (Trimr CFU/viable CFU) of plostfoX-Cm-containing wild-type (WT) and ainS (NL60), luxS (CL39), and ainS luxS (KV9420) mutant strains. (B) Transformation frequencies of plostfoX-Kan-containing wild-type (WT) and qrr1 (TIM305) and luxO (KV5467) mutant strains. (C) Transformation frequencies of plostfoX-Cm-containing wild-type and qrr1 litR (KV8790) and luxO litR (KV8791) mutant strains. All strains were exposed to gDNA from KV8300 (fliQ::Trim). The qrr1 litR and luxO litR mutants each exhibited a transformation frequency below the LOD. *, P < 0.05; **, P < 0.005.
LitR is negatively regulated by sRNA Qrr1, which in turn is activated by LuxO, and thus mutation of either qrr1 or luxO results in increased LitR levels (29) (Fig. 1). Consistent with their roles in controlling LitR levels, the loss of Qrr1 or LuxO caused as much as a 20-fold increase in competence (Fig. 5B). This increase did not overcome the necessity for litR, as qrr1 litR and luxO litR double mutants failed to transform (Fig. 5C). Together, these data demonstrate that LitR and its regulators control competence, similar to HapR of V. cholerae (2), and reveal strains (Δqrr1 and ΔluxO) with increased competence that can be used as tools when a high transformation frequency is critical.
Requirement for LitR can be partially suppressed by nuclease mutation.
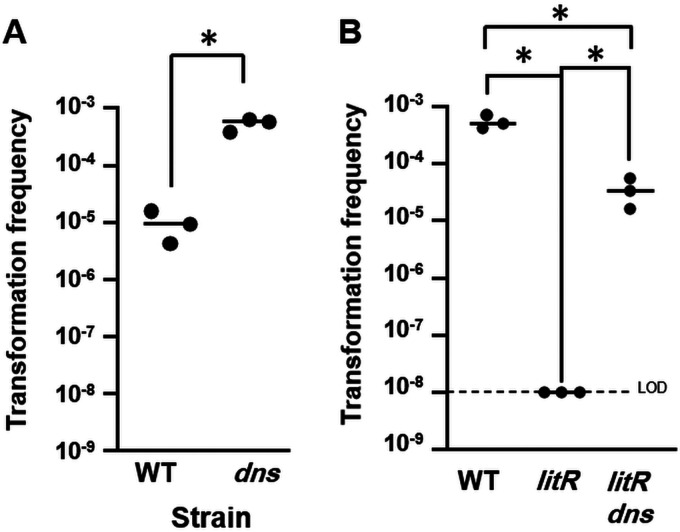
In V. cholerae, a key role for HapR is to inhibit transcription of the gene encoding Dns, a nuclease that degrades extracellular DNA, preventing its uptake (6). Deletion of dns is sufficient to largely restore competence to a hapR mutant. To test whether the same is true for V. fischeri, we identified and deleted the dns homolog, VF_0437, and found that the loss of Dns significantly increased the transformation frequency (∼50-fold) (Fig. 6A). We then combined the dns mutation with a litR mutation and found that the resulting double mutant exhibited over a 3-log increase in transformation frequency relative to that of the litR mutant (Fig. 6B). The ability of the dns mutation to suppress the litR transformation defect suggests that the requirement for LitR can be largely attributed to the activity of Dns, which presumably mediates degradation of extracellular DNA.
FIG 6.
Dns inhibits V. fischeri transformation. Shown are transformation frequencies (Trimr CFU/viable CFU) of strains exposed to gDNA from KV8300 (fliQ::Trim). (A) plostfoX-Kan-containing wild-type (WT; ES114) and dns mutant (KV8807) strains. (B) plostfoX-Cm-containing wild-type (WT; ES114) and litR (PMF8) and litR dns (KV9352) mutant strains. The litR mutant, but not the litR dns mutant, exhibited a transformation frequency below the LOD. *, P < 0.05.
tfoX-induced competence peaks at mid-exponential growth.
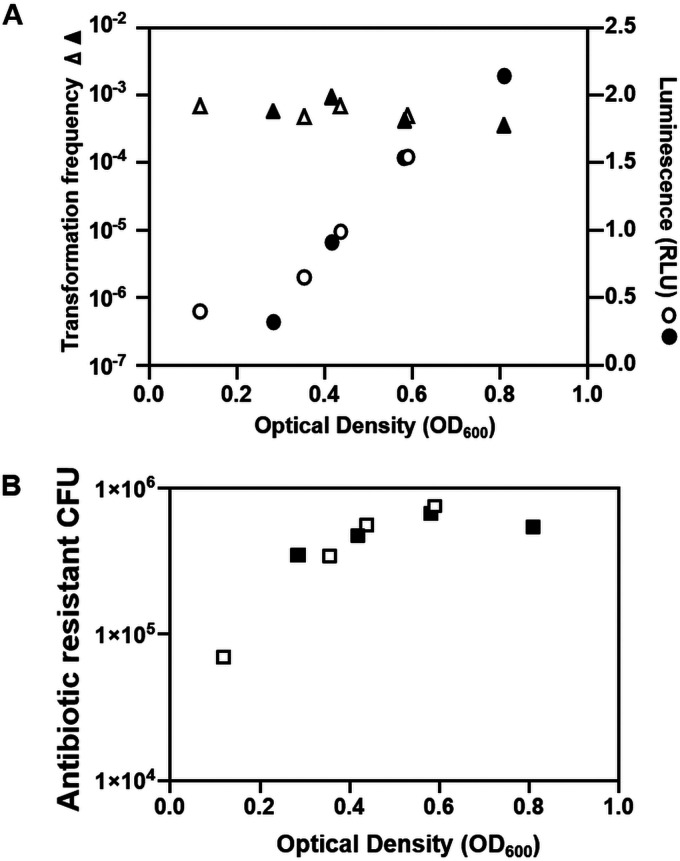
Because quorum-sensing regulation changes over time, we sought to determine if a specific point in the cell cycle was optimal for transformation by assessing transformation frequencies at different times during liquid growth. Furthermore, we reasoned that another quorum-sensing-controlled phenotype, luminescence, could provide an easy marker for LitR activation (Fig. 1): increased LitR levels should correspond to increased luminescence and increased competence, and thus, luminescence measurements might have predictive value. Surprisingly, however, under our standard TfoX overproduction conditions, the transformation frequencies were consistently at the same approximate level regardless of the cell density (Fig. 7A and Fig. S6). Furthermore, competence could not be predicted by light production: while relatively little light was produced under these minimal medium conditions (which are not the typical conditions used for assessing light production by V. fischeri), it did increase modestly over time. However, even at low cell densities when there was little light production, the cells achieved high competence. These data indicate that levels of LitR are only one factor governing competence in TfoX-overexpressing cells and/or that additional levels of control over luminescence (e.g., LuxR and ArcA [33, 34]) impact the timing of luminescence relative to that of competence. We note, however, that with increased OD and thus increased cell number, increased numbers of transformants were obtained (i.e., the overall numbers of transformants, not normalized for total cell number) (Fig. 7B). Thus, the OD of culture used for transformations may ultimately affect the ability to obtain a desired mutant.
FIG 7.
Transformation frequencies during V. fischeri growth. (A) Transformation frequencies (Trimr CFU/viable CFU) and relative light units (RLU) of plostfoX-Cm-containing ES114 assessed at the indicated optical densities during growth of V. fischeri in minimal medium. (B) The same transformation experiment as shown in panel A, plotted instead as numbers of transformants at each optical density (not normalized to total cell number). Cells were exposed to gDNA from KV8300 (fliQ::Trim). This experiment was performed by sampling from two flasks, indicated by the open and closed symbols.
Co-overexpression of TfoX and LitR promotes KB2B1 transformation.
Finally, we sought to apply the knowledge gleaned in this study to further increase transformation frequencies of V. fischeri. We hypothesized that dual overexpression of the two major regulators, TfoX and LitR, might result in higher levels of transformation. To test this possibility, we generated pJJC4, which contains both genes in the Cmr vector backbone. When we compared the transformation frequencies of pJJC4- versus plostfoX-Cm-containing cells of wild-type V. fischeri strain ES114, however, we observed no difference (Fig. S7). Because ES114 is already highly transformable with overexpression of tfoX alone, we turned our attention to KB2B1, another isolate of V. fischeri (35) that, in our hands, is only poorly transformable. We compared the transformation frequencies of KB2B1 derivatives that carried either plostfoX-Cm or pJJC4. We found that there was over a 100-fold increase upon dual overexpression of LitR and TfoX (Fig. 8). Thus, competence and/or transformation of otherwise less transformable V. fischeri isolates can be further induced by co-overexpression of these two positive regulators.
FIG 8.
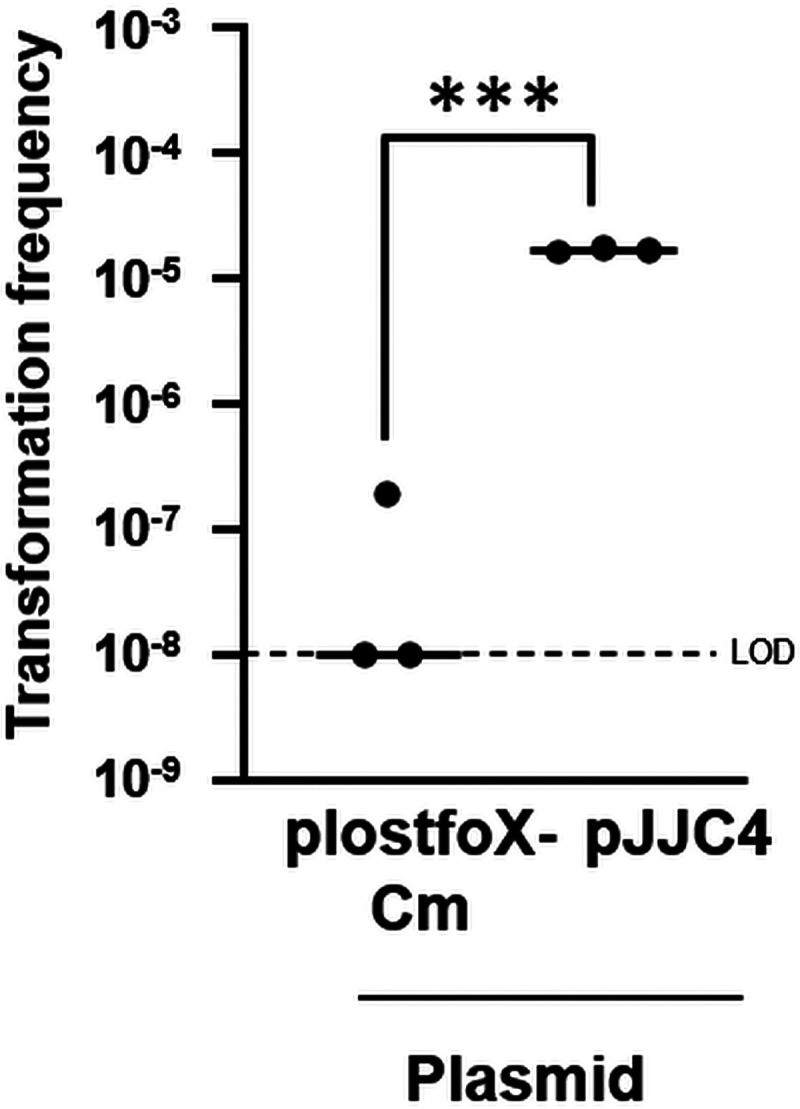
Co-overexpression of tfoX and litR promotes transformation of KB2B1. Shown are transformation frequencies (Trimr CFU/viable CFU) of plostfoX-Cm- and pJJC4-containing KB2B1 strains exposed to gDNA from KV8300 (fliQ::Trim). Two of the three replicate samples of KB2B1 resulted in transformation frequencies below the LOD. ***, P < 0.0005.
DISCUSSION
Chitin- and TfoX-inducible competence of V. fischeri was first described in 2010 (17), yet to date, little has been reported about the mechanisms underlying this phenomenon in this bacterium. Bioinformatic analysis by Antonova and Hammer (5) indicated that V. fischeri possesses homologs for 22 of 23 genes known or predicted to be important for V. cholerae transformation. While our work did not analyze all of those homologs, our results support the model that V. fischeri uses similar factors and at least some underlying mechanisms (Fig. 1), providing a foundation for future work characterizing transformation in this microbe.
Our finding that LitR functions as a key regulator of competence in V. fischeri may not be considered surprising given the importance of its homolog in controlling competence in V. cholerae. However, recent work studying competence in other Vibrio species demonstrated that the LitR/HapR homolog OpaR is not required for competence of V. parahaemolyticus and that the V. campbellii homolog LuxR is not critical for transformation of at least one strain of that species (36). That work underscores the need to test predictions based on bioinformatic analyses, as we have begun to do in this study. It is of note that one V. cholerae homolog known to be missing in V. fischeri is the gene for QstR, an intermediate positive regulator found in V. cholerae (13, 37). The lack of a qstR homolog in the genome signifies that V. fischeri has made some adjustments to its competence pathway that will be of interest to uncover.
This new role for LitR in V. fischeri competence expands our understanding of this regulator, whose function has been primarily associated with luminescence control and, to a lesser extent, motility (23, 29, 32). Due to the substantial role of LitR in controlling luminescence, we had hypothesized that we could use luminescence, which is readily measured in real time, to predict when cells might be competent. Unfortunately, our work indicated no relationship between light production and competence under the conditions used, namely, TfoX overproduction and growth in a minimal medium (Fig. 7). It was perhaps a naive view that competence and light production would correspond, given the known complex control over light production that would prevent such a straightforward assessment of LitR induction and thus competence. Alternatively, or in addition, the use of tfoX overexpression could disconnect competence from light production. Regardless, researchers in the field who wish to use this method to generate mutants may benefit from the knowledge that while TfoX-induced transformation frequency in V. fischeri is not OD dependent, the numbers of transformants increase with increasing OD (Fig. 7); thus, performing the transformation at higher ODs may be superior for obtaining a rare transformant due to, for example, a low concentration of DNA containing the marker/mutation of interest.
We had also hoped to resolve the controversy in the field over the requirement for TfoY. We found that the loss of TfoY failed to significantly impact transformation frequencies, consistent with the findings obtained by others (17, 24). In the original report of competence in V. fischeri, TfoY was shown to be necessary for full competence (17, 24). The discrepancy could lie in the use by Pollack-Berti et al. of chitohexose, which was omitted in this study. In V. cholerae, TfoY appears to function to induce a type VI secretion system (T6SS), which, in turn, promotes killing of other bacteria and thus release of free DNA in the environment (24). To date, no T6SS-mediated killing capability has been observed for ES114 (38), leaving unanswered the question of if/how TfoY contributes to competence in this microbe. Of note, TfoY does appear to play a critical, positive role in motility in V. cholerae, and it has been suggested to play a similar role in V. fischeri by overexpression studies (25, 26), which we confirmed in this study. This gene was not discovered in a wide-scale transposon mutagenesis search for genes involved in motility and chemotaxis (39), possibly because its relatively small size (588 bp) may limit insertions at that location. Alternatively, differences in how the motility experiment was conducted in this study relative to that of Brennan et al. (39) could account for this discrepancy; in V. cholerae, decreasing the nutrient content of the growth medium alters the motility phenotype of the tfoY mutant (26).
A major underlying goal of this work was to increase transformation frequencies to facilitate genetic manipulations of the classical wild-type strain ES114, with the expectation that lessons learned might also be leveraged to promote studies of less genetically tractable V. fischeri strains. Indeed, our work revealed mutant strains, namely, qrr1, luxO, and dns mutants, with increased transformation frequencies that can be used as tools for low-efficiency transformations, such as when the DNA derives from an SOE PCR approach, which can yield low levels of the correct product. Given that a marked mutation of interest can be subsequently moved from, for example, a qrr1 mutant into a wild-type (or other) strain background by transformation, these strains are valuable tools. In some instances, however, it may be necessary or desirable to have a high transformation frequency of a specific strain. Thus, it was of interest that the two published versions of the tfoX expression plasmid resulted in different transformation efficiencies, with plasmid carrying the Cmr marker yielding more transformants than those obtained with the Kanr plasmid. Because we observed a similar phenomenon using derivatives with a newly introduced, arabinose-inducible tfoX gene, we conclude that the difference depends on the resistance marker and/or the stronger selective pressure of the Cm antibiotic. The disparity is substantial, and those seeking optimal conditions for transformation will thus need to consider the vector “backbone” as a factor—and whether these plasmids behave the same way when carried by a distinct isolate as they do in ES114. Finally, we developed pJJC4, which overexpresses tfoX and litR in the context of the better Cmr plasmid, to further enhance transformation frequency. While this plasmid did not increase transformation of ES114, it permitted a higher level of transformation of KB2B1, a V. fischeri isolate that is less genetically tractable. KB2B1 is of interest due to its “dominance” in colonizing its squid host (35). Consequently, these findings may be expected to advance the investigation of this strain and potentially other V. fischeri strains that are less amenable to genetic manipulation.
In summary, this work forms the foundation for the construction of a competence pathway by testing hypotheses based on bioinformatics and identifying roles for litR and other putative competence factors. Furthermore, it delivers insight into conditions, strains, and plasmids that promote transformation of V. fischeri. We anticipate that these findings will accelerate the construction of mutants in various strain backgrounds and promote the study not only of competence but also of other phenomena of interest.
MATERIALS AND METHODS
Strains and media.
V. fischeri strains, plasmids, and primers used in this study are listed in Tables 1 to 3, respectively. Escherichia coli strains TAM1 or TAM1 λ pir (Active Motif, Carlsbad, CA), π3813 (40), GT115 (InvivoGen, San Diego, CA), and DH5α were used for plasmid maintenance, cloning, and conjugation purposes. For routine growth, maintenance, or selection for transformants, V. fischeri was grown in the complex medium LBS (41, 42), which contains 1% tryptone, 0.5% yeast extract, 2% sodium chloride, and 50 mM Tris (pH 7.5). For transformations, V. fischeri was grown in Tris minimal medium (TMM), which contains 300 mM sodium chloride, 50 mM magnesium sulfate, 0.33 mM potassium phosphate dibasic, 10 μM ferrous ammonium sulfate, 0.1% ammonium chloride, 10 mM N-acetylglucosamine, 10 mM potassium chloride, 10 mM calcium chloride, and 100 mM Tris (pH 7.5). This medium was made fresh from concentrated and autoclaved stocks, with the exception that Tris was added from a concentrated, filter-sterilized stock. E. coli was grown in the complex medium LB (43), which contains 1% tryptone, 0.5% yeast extract, and 1% sodium chloride. Agar (1.5%) was used for solid media. As appropriate, antibiotics were added as follows (final concentrations in parentheses): for V. fischeri, chloramphenicol (Cm; 1 μg ml−1 for single-copy selection or 5 μg ml−1 for plasmid maintenance), erythromycin (5 μg ml−1), kanamycin (Kan; 100 μg ml−1), and trimethoprim (Trim; 10 μg ml−1); for E. coli, ampicillin (100 μg ml−1), Cm (12.5 μg ml−1), and Kan (50 μg ml−1).
TABLE 1.
Strains constructed and/or used in this study
| Strain | Genotype | Derivationa | Reference |
|---|---|---|---|
| CL39 | luxS::Kan | NA | 27 |
| ES114 | Wild type | NA | 50 |
| JB19 | litR::Erm | NA | 34 |
| KV5467 | ΔluxO | Conjugation of ES114 with pVAR36-containing E. coli | This study |
| KV8300 | ΔfliQ::FRT-Trim | NA | 22 |
| KV8790 | Δqrr1 litR::Erm | TT TIM305 with gJB19 | This study |
| KV8791 | ΔluxO litR::Erm | TT KV5467 with gJB19 | This study |
| KV8807 | Δdns::FRT-Erm | TT ES114 with SOE PCR with primers 2504 and 2505 (ES114), 2089 and 2090 (pKV494), and 2506 and 2507 (ES114) | This study |
| KV8840 | ΔcytR::FRT-Erm | TT ES114 with SOE PCR with primers 2512 and 2513 (ES114), 2089 and 2090 (pKV494), and 2514 and 2515 (ES114), amplified with primers 2512 and 2515 | This study |
| KV8875 | pilA::mTn5 | mTn5 mutagenesis of ES114, backcrossed into ES114 | This study |
| KV8876 | pilB::mTn5 | mTn5 mutagenesis of ES114, backcrossed into ES114 | This study |
| KV8877 | pilC::mTn5 | mTn5 mutagenesis of ES114, backcrossed into ES114 | This study |
| KV8879 | pilQ::mTn5 | mTn5 mutagenesis of ES114, backcrossed into ES114 | This study |
| KV9219 | ΔcomEA::FRT-Erm | TT ES114 with SOE PCR with primers sets 2628 and 2629 (ES114), 2089 and 2090 (pKV494), and 2630 and 2631 (ES114) | This study |
| KV9352 | Δdns::FRT-Erm litR::Kan | TT KV8807 with gPMF8 | This study |
| KV9420 | ΔainS luxS::Kan | TT NL60 with gCL39 | This study |
| NL60 | ΔainS | NA | 51 |
| PMF8 | litR::Kan | NA | 23 |
| TIM305 | Δqrr1 | NA | 29 |
TfoX-induced transformation (TT) was performed with tfoX overexpression versions of the indicated strains using either SOE PCR DNA or genomic DNA (gDNA) from the indicated strain. NA, not applicable; mTn5, mini-Tn5.
TABLE 2.
Plasmids constructed and/or used in this study
| Plasmid | Description | Resistance marker | Reference |
|---|---|---|---|
| pEVS104 | Conjugal plasmid | Kan | 44 |
| pJJC4 | plostfoX-Cm + litR | Cm | This study |
| pKV363 | Suicide vector | Cm | 47 |
| pKV494 | Carries FRT-Ermr cassette | Ap, Erm | 22 |
| pLostfoX-Cma | pEVS79 + tfoX | Cm | 17 |
| pLostfoX-Kan | plostfoX with Kanr in place of Cmr | Kan | 21 |
| pVAR36 | pKV363 + sequences flanking luxO | Cm | 52 |
| pVSV105 | Stable Vibrio plasmid | Cm | 48 |
| pPMF5 | pVSV105 containing litR | Cm | This study |
TABLE 3.
Primers used in this study
| Primer no. | Purpose | Sequencea |
|---|---|---|
| 908 | Check Tn5 insertions | GCACTGAGAAGCCCTTAGAGCC |
| 1319 | Delete luxO | taggcggccgcacttagtatgGGAAGCAGTATCTTCTACCAT |
| 1320 | Delete luxO | catactaagtgcggccgcctaTGGAATGAAAGATAAGGGGAC |
| 1344 | Delete luxO | CGTAAAGTTGTTGCACCTAAG |
| 1345 | Delete luxO | GCAGGTAAGATGGATCATAGG |
| 2089 | Amplify antibiotic resistance cassettes by PCR | CCATACTTAGTGCGGCCGCCTA |
| 2090 | Amplify antibiotic resistance cassettes by PCR | CCATGGCCTTCTAGGCCTATCC |
| 2504 | Delete dns | ACCCCACTTCAACAAGCATC |
| 2505 | Delete dns | taggcggccgcactaagtatggGAGCCAATAGCGATAATGCT |
| 2506 | Delete dns | ggataggcctagaaggccatggTGTCGCAAATAGACATAATTAGAG |
| 2507 | Delete dns | TCAGGAACAGTATTACGTCCAC |
| 2512 | Delete cytR | GTGCAGGGCGTGCAAGTAAA |
| 2513 | Delete cytR | taggcggccgcactaagtatggTCCTGCCGATAAAGCAACGT |
| 2514 | Delete cytR | ggataggcctagaaggccatggGTTAGAGAGAGTACGGCGTCC |
| 2515 | Delete cytR | CACTGCATAATGTAAGGGATCTC |
| 2531 | Coexpress tfoX and litR | ggtaccGATTAAGGAAGAGCTGTTAAC |
| 2532 | Coexpress tfoX and litR | ggtaccGCTGCGGAAGTATTTGAAGG |
| 2626 | Check putative pilQ::Tn5 insertion | ggtaccCACCTAAAGGGAGTGTGATTAAAGT |
| 2627 | Check putative pilQ::Tn5 insertion | gagctcGGCACCATATTCCTCACAGATAT |
| 2628 | Delete comEA | GGAAGCCCTAGAACTTGCTC |
| 2629 | Delete comEA | taggcggccgcactaagtatggACTGAGTAAAGTGAGTATGCG |
| 2630 | Delete comEA | ggataggcctagaaggccatggATTGGGGATAAACTGGTTGAG |
| 2631 | Delete comEA | AGTTAAATTTGCGATGGCTC |
| 2859 | Check putative pilA::Tn5 insertion | TCACCCGATTCATCTTGGAG |
| 2860 | Check putative pilA::Tn5 insertion | GATAGAGGCGAGGTTGGTGTGC |
| 2861 | Check putative pilB::Tn5 insertion | TATCCCAGGCTGTCAAGGCC |
| 2862 | Check putative pilB::Tn5 insertion | CTGAACTGTTAATGCCACGCCA |
| 2863 | Check putative pilC::Tn5 insertion | GAAGATGGCACCACCAGCTTAC |
| 2864 | Check putative pilC::Tn5 insertion | GGGGCAAGATCTGGAAAGCA |
| litR-ndeI F | Amplify litR and ligate into pVSV105 | ccccatatgGATTAAGGAAGAGCTGTTAACGG |
| litR-ndeI R | Amplify litR and ligate into pVSV105 | ccccatatgGCTGCGGAAGTATTTGAAGG |
Lowercase letters indicate primer “tails.”
Strain construction.
Mutant V. fischeri strains were generated by introducing a tfoX overexpression plasmid, such as plostfoX-Cm (17) or plostfoX-Kan (21), from E. coli by triparental conjugation with pEVS104-containing (44) E. coli and the appropriate V. fischeri recipient as described previously (45). The resulting strains were used in transformations with DNA derived from SOE PCR (46) amplifications with primers as indicated in Tables 1 and 3. High-fidelity PCR enzyme EMD Millipore KOD DNA polymerase was used for PCR amplifications to generate fragments containing a central antibiotic resistance cassette and ∼500-bp flanking sequences homologous to regions up- and downstream of the target gene as described previously (22). As a result of using the often-inefficient SOE PCR to obtain a desired PCR product, the amount of the DNA added during these strain construction transformations was variable and generally much less than described in “V. fischeri transformation” below. In addition, no assessments of total CFU were performed during strain constructions. Putative mutants that arose on selective medium were purified by streaking, first on the same selective medium and then nonselectively to permit loss of the tfoX-containing plasmid. The presence of the desired mutation was confirmed by PCR with outside primers. In some cases, the mutation was moved from an original recipient strain to a different background by isolating genomic DNA and using it to transform a new strain of interest. The unmarked ΔluxO strain KV5467 was generated as previously described (40, 47) using pVAR36.
Plasmid construction and analysis.
Previously unpublished plasmids pJJC4 and pPMF5 (Table 2) were used in this study and were constructed as follows. For plasmid pJJC4, a PCR product containing litR with flanking KpnI sequences (obtained with primers 2531 and 2532) was first cloned into pJET1.2 using the Thermo Scientific CloneJET PCR cloning kit, prior to subcloning into KpnI-digested plostfox-Cm (retaining tfoX). pPMF5 was generated by amplifying litR using PCR with primers litR-NdeI F and litR-NdeI R and cloning NdeI-digested DNA into pVSV105 (48). To evaluate potential differences in the tfoX gene between plostfoX-Cm and plostfoX-Kan, the plasmids were purified and the tfoX gene was sequenced using M13 forward and reverse primers supplied by ACGT, Inc. (Wheeling, IL). No differences in sequence between the two plasmids were observed.
V. fischeri transformation.
V. fischeri strains carrying a tfoX overexpression plasmid were grown in TMM, made as described in “Strains and media” above, containing 100 μg ml−1 of Kan or 5 μg ml−1 of Cm. Cultures of ES114 derivatives were grown overnight in 5 ml of medium in 18- by 150-mm tubes at 28°C with shaking for 13 to 16 h, then subcultured by diluting the culture to a starting culture density (optical density at 600 nm [OD600]) of 0.05 in 20 ml of the same medium in 125-ml baffled flasks, and grown with shaking at 24°C; for KB2B1 derivatives, cultures were subcultured to a starting OD600 of 0.1. For most experiments, cultures were grown until the OD600 was approximately equal to 0.5. Then, in triplicate, a 0.5-ml aliquot was transferred into a 2-ml Eppendorf tube. The cells were then exposed to DNA, or left unexposed for the negative control, and then incubated for 30 min at room temperature. For assessing transformation frequencies for the experiments, 500 ng of genomic DNA derived from strain KV8300 (ΔfliQ::FRT-Trim) (22) was used. Genomic DNA was prepared using the Quick-DNA Miniprep Plus kit (Zymo Research, obtained from Genesee Scientific) and normalized to a concentration of 100 ng per μl. Following the incubation period, a 0.5-ml aliquot of LBS broth was added to each culture and the 1-ml mixtures were transferred to 18- by 150-mm test tubes. The test tubes were incubated with shaking at 28°C for 90 min to permit recovery and expression of the antibiotic resistance gene. At that time, the cells were (i) diluted (or left undiluted) and spread onto selective medium (LBS plus trimethoprim) to assess the number of transformants that had recombined the DNA of interest and (ii) diluted and spotted in 10-μl aliquots on a nonselective medium (LBS) to determine the total CFU. In a few cases, cells that had completed the 90-min recovery were held on ice for a short period prior to dilution and plating; we observed no substantial difference in numbers of transformants or total CFU before or after ice exposure. Transformation frequency was calculated as the number of antibiotic-resistant CFU (multiplied by the dilution factor) divided by the number of total CFU (multiplied by the dilution factor). A similar calculation was performed to determine the limit of detection (LOD), using an approximate average total CFU of 1 × 109. Occasionally, selection for Trimr yielded spontaneously resistant colonies; these colonies appeared more translucent than true transformants and retained an ability to swim in soft agar, unlike true transformants, which became nonmotile due to acquiring the mutation in flagellar gene fliQ (ΔfliQ::FRT-Trimr). The phenotype of each mutant strain was evaluated in the manner described above at least twice in duplicate or triplicate, with similar outcomes.
Transformation and light production.
For measurements of transformation frequency over time, and corresponding luminescence levels, we used the following procedure. V. fischeri strain ES114 carrying plostfoX-Cm was grown in TMM containing Cm overnight with shaking at 28°C for no longer than 14 h. The cells were then subcultured into each of two 125-ml baffled flasks to an OD of 0.05 in 20 ml of fresh medium (TMM containing Cm) and incubated with shaking at 24°C. The OD600 of the samples were measured at 1-h intervals until an OD of 0.2 was reached. Then, samples were taken every 30 min to assess growth, luminescence, and transformation, until an OD of about 0.4 was reached, and then every 15 min for the remainder of the experiment, which we performed until the cells reached an OD600 of around 1, which took about 6 or 7 h. In this medium, the cells reach a final OD600 of between 1.2 and 1.5. Samples were taken from alternating flasks throughout the experiment to reduce the decrease in culture volume from any one flask. At these time points, 1 ml was pipetted into a sterile scintillation vial, aerated in a consistent manner by pipetting before using a Turner Designs TD-20/20 luminometer to measure relative light units (RLU). A portion (0.5 ml) was then transferred to an Eppendorf tube containing DNA for transformation, as outlined above, while the remaining 0.5 ml was transferred to a cuvette to measure the OD600. The remainder of the transformation experiment was carried out as described above in “V. fischeri transformation.”
pil mutants.
The pil mutants were derived from a transposon (mini-Tn5 (mTn5) mutant library collection. To reduce the concern that an unlinked mutation could be responsible for any observed phenotype, the pil mutants (pilA::mTn5, pilB::mTn5, pilC::mTn5, and pilQ::mTn5) were backcrossed into tfoX-containing ES114. In addition, the position of the Tn insertion was verified both by sequencing (49) and by applying PCR to amplify a segment of DNA using a Tn-specific primer and a gene-specific primer.
Statistics.
Statistics were performed on GraphPad using Student’s t test with Welch’s correction.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eric Stabb for sending mini-Tn5 mutants, Val Ray for generating the unmarked luxO deletion mutant, and David Christensen and members of the Visick lab for help with culturing strains and for valuable scientific discussions and reviewing the manuscript.
We gratefully acknowledge funding from NIGMS with grants R01 GM114288 and R35 GM130355 awarded to K.L.V., the Wheaton College Aldeen Grant to J.T., and a Cal Poly Research, Scholarly, and Creative Activities Grant to P.M.F.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lipp EK, Huq A, Colwell RR. 2002. Effects of global climate on infectious disease: the cholera model. Clin Microbiol Rev 15:757–770. doi: 10.1128/cmr.15.4.757-770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 3.Dalia AB, McDonough E, Camilli A. 2014. Multiplex genome editing by natural transformation. Proc Natl Acad Sci U S A 111:8937–8942. doi: 10.1073/pnas.1406478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayes CA, Dalia TN, Dalia AB. 2017. Systematic genetic dissection of PTS in Vibrio cholerae uncovers a novel glucose transporter and a limited role for PTS during infection of a mammalian host. Mol Microbiol 104:568–579. doi: 10.1111/mmi.13646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonova ES, Hammer BK. 2015. Genetics of natural competence in Vibrio cholerae and other vibrios. Microbiol Spectr 3:VE-0010-2014. doi: 10.1128/microbiolspec.VE-0010-2014. [DOI] [PubMed] [Google Scholar]
- 6.Blokesch M, Schoolnik GK. 2008. The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae. J Bacteriol 190:7232–7240. doi: 10.1128/JB.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seitz P, Blokesch M. 2013. DNA-uptake machinery of naturally competent Vibrio cholerae. Proc Natl Acad Sci U S A 110:17987–17992. doi: 10.1073/pnas.1315647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y, Bernardy EE, Hammer BK, Miyashiro T. 2013. Competence and natural transformation in vibrios. Mol Microbiol 89:583–595. doi: 10.1111/mmi.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antonova ES, Hammer BK. 2011. Quorum-sensing autoinducer molecules produced by members of a multispecies biofilm promote horizontal gene transfer to Vibrio cholerae. FEMS Microbiol Lett 322:68–76. doi: 10.1111/j.1574-6968.2011.02328.x. [DOI] [PubMed] [Google Scholar]
- 10.Ball AS, Chaparian RR, van Kessel JC. 2017. Quorum sensing gene regulation by LuxR/HapR master regulators in vibrios. J Bacteriol 199:e00105-17. doi: 10.1128/JB.00105-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suckow G, Seitz P, Blokesch M. 2011. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J Bacteriol 193:4914–4924. doi: 10.1128/JB.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto S, Izumiya H, Mitobe J, Morita M, Arakawa E, Ohnishi M, Watanabe H. 2011. Identification of a chitin-induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae. J Bacteriol 193:1953–1965. doi: 10.1128/JB.01340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo Scrudato M, Blokesch M. 2013. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res 41:3644–3658. doi: 10.1093/nar/gkt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonova ES, Bernardy EE, Hammer BK. 2012. Natural competence in Vibrio cholerae is controlled by a nucleoside scavenging response that requires CytR-dependent anti-activation. Mol Microbiol 86:1215–1231. doi: 10.1111/mmi.12054. [DOI] [PubMed] [Google Scholar]
- 15.Watve SS, Thomas J, Hammer BK. 2015. CytR is a global positive regulator of competence, Type VI secretion, and chitinases in Vibrio cholerae. PLoS One 10:e0138834. doi: 10.1371/journal.pone.0138834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stabb EV, Visick KL. 2013. Vibrio fischeri: a bioluminescent light-organ symbiont of the bobtail squid Euprymna scolopes, p 497–532. In Rosenberg E, DeLong EF, Stackebrandt E, Lory S, Thompson F (ed), The prokaryotes. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 17.Pollack-Berti A, Wollenberg MS, Ruby EG. 2010. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ Microbiol 12:2302–2311. doi: 10.1111/j.1462-2920.2010.02250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks JF, II, Gyllborg MC, Kocher AA, Markey LE, Mandel MJ. 2015. TfoX-based genetic mapping identifies Vibrio fischeri strain-level differences and reveals a common lineage of laboratory strains. J Bacteriol 197:1065–1074. doi: 10.1128/JB.02347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson CM, Marsden AE, Tischler AH, Koo J, Visick KL. 2018. Vibrio fischeri biofilm formation prevented by a trio of regulators. Appl Environ Microbiol 84:e01257-18. doi: 10.1128/AEM.01257-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tischler AH, Lie L, Thompson CM, Visick KL. 2018. Discovery of calcium as a biofilm-promoting signal for Vibrio fischeri reveals new phenotypes and underlying regulatory complexity. J Bacteriol 200:e00016-18. doi: 10.1128/JB.00016-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks JF, Gyllborg MC, Cronin DC, Quillin SJ, Mallama CA, Foxall R, Whistler C, Goodman AL, Mandel MJ. 2014. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci U S A 111:17284–17289. doi: 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Visick KL, Hodge-Hanson KM, Tischler AH, Bennett AK, Mastrodomenico V. 2018. Tools for rapid genetic engineering of Vibrio fischeri. Appl Environ Microbiol 84:e00850-18. doi: 10.1128/AEM.00850-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fidopiastis PM, Miyamoto CM, Jobling MG, Meighen EA, Ruby EG. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol Microbiol 45:131–143. doi: 10.1046/j.1365-2958.2002.02996.x. [DOI] [PubMed] [Google Scholar]
- 24.Metzger LC, Stutzmann S, Scrignari T, Van der Henst C, Matthey N, Blokesch M. 2016. Independent regulation of type VI secretion in Vibrio cholerae by TfoX and TfoY. Cell Rep 15:951–958. doi: 10.1016/j.celrep.2016.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metzger LC, Matthey N, Stoudmann C, Collas EJ, Blokesch M. 2019. Ecological implications of gene regulation by TfoX and TfoY among diverse Vibrio species. Environ Microbiol 21:2231–2247. doi: 10.1111/1462-2920.14562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pursley BR, Maiden MM, Hsieh ML, Fernandez NL, Severin GB, Waters CM. 2018. Cyclic di-GMP regulates TfoY in Vibrio cholerae to control motility by both transcriptional and posttranscriptional mechanisms. J Bacteriol 200:e00578-17. doi: 10.1128/JB.00578-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lupp C, Ruby EG. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J Bacteriol 186:3873–3881. doi: 10.1128/JB.186.12.3873-3881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyamoto CM, Dunlap PV, Ruby EG, Meighen EA. 2003. LuxO controls luxR expression in Vibrio harveyi: evidence for a common regulatory mechanism in Vibrio. Mol Microbiol 48:537–548. doi: 10.1046/j.1365-2958.2003.03453.x. [DOI] [PubMed] [Google Scholar]
- 29.Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. 2010. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol 77:1556–1567. doi: 10.1111/j.1365-2958.2010.07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyamoto CM, Lin YH, Meighen EA. 2000. Control of bioluminescence in Vibrio fischeri by the LuxO signal response regulator. Mol Microbiol 36:594–607. doi: 10.1046/j.1365-2958.2000.01875.x. [DOI] [PubMed] [Google Scholar]
- 31.Lupp C, Urbanowski M, Greenberg EP, Ruby EG. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol Microbiol 50:319–331. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- 32.Lupp C, Ruby EG. 2005. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J Bacteriol 187:3620–3629. doi: 10.1128/JB.187.11.3620-3629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol 182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bose JL, Kim U, Bartkowski W, Gunsalus RP, Overley AM, Lyell NL, Visick KL, Stabb EV. 2007. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol Microbiol 65:538–553. doi: 10.1111/j.1365-2958.2007.05809.x. [DOI] [PubMed] [Google Scholar]
- 35.Bongrand C, Koch EJ, Moriano-Gutierrez S, Cordero OX, McFall-Ngai M, Polz MF, Ruby EG. 2016. A genomic comparison of 13 symbiotic Vibrio fischeri isolates from the perspective of their host source and colonization behavior. ISME J 10:2907–2917. doi: 10.1038/ismej.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson CA, Podicheti R, Rusch DB, Dalia AB, van Kessel JC. 2019. Diversity in natural transformation frequencies and regulation across Vibrio species. mBio 10:e02788-19. doi: 10.1128/mBio.02788-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaskolska M, Stutzmann S, Stoudmann C, Blokesch M. 2018. QstR-dependent regulation of natural competence and type VI secretion in Vibrio cholerae. Nucleic Acids Res 46:10619–10634. doi: 10.1093/nar/gky717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Speare L, Cecere AG, Guckes KR, Smith S, Wollenberg MS, Mandel MJ, Miyashiro T, Septer AN. 2018. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci U S A 115:E8528–E8537. doi: 10.1073/pnas.1808302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennan CA, Mandel MJ, Gyllborg MC, Thomasgard KA, Ruby EG. 2013. Genetic determinants of swimming motility in the squid light-organ symbiont Vibrio fischeri. Microbiologyopen 2:576–594. doi: 10.1002/mbo3.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Roux F, Binesse J, Saulnier D, Mazel D. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol 73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graf J, Dunlap PV, Ruby EG. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol 176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stabb EV, Reich KA, Ruby EG. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD(+)-glycohydrolases. J Bacteriol 183:309–317. doi: 10.1128/JB.183.1.309-317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis RW, Botstein D, Roth JR. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 44.Stabb EV, Ruby EG. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol 358:413–426. doi: 10.1016/s0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- 45.DeLoney CR, Bartley TM, Visick KL. 2002. Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. J Bacteriol 184:5121–5129. doi: 10.1128/jb.184.18.5121-5129.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horton RM. 1997. In vitro recombination and mutagenesis of DNA. SOEing together tailor-made genes. Methods Mol Biol 67:141–149. doi: 10.1385/0-89603-483-6:141. [DOI] [PubMed] [Google Scholar]
- 47.Shibata S, Visick KL. 2012. Sensor kinase RscS induces the production of antigenically distinct outer membrane vesicles that depend on the symbiosis polysaccharide locus in Vibrio fischeri. J Bacteriol 194:185–194. doi: 10.1128/JB.05926-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol 72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoudenmire JL, Black M, Fidopiastis PM, Stabb EV. 2019. Mutagenesis of Vibrio fischeri and other marine bacteria using hyperactive Mini-Tn5 derivatives. Methods Mol Biol 2016:87–104. . doi: 10.1007/978-1-4939-9570-7_9. [DOI] [PubMed] [Google Scholar]
- 50.Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lyell NL, Dunn AK, Bose JL, Stabb EV. 2010. Bright mutants of Vibrio fischeri ES114 reveal conditions and regulators that control bioluminescence and expression of the lux operon. J Bacteriol 192:5103–5114. doi: 10.1128/JB.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ray VA, Visick KL. 2012. LuxU connects quorum sensing to biofilm formation in Vibrio fischeri. Mol Microbiol 86:954–970. doi: 10.1111/mmi.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.