This study showed the genetic diversity and population structure of S. aureus presenting the same spa type, t899, but belonging to different STs. Our findings revealed that these isolates vary deeply in their core and accessory genomes, contrary to what is regularly inferred from studies using spa typing only.
KEYWORDS: MRSA, t899, spa type, MLST, cgMLST, whole-genome sequencing
ABSTRACT
Methicillin-resistant Staphylococcus aureus (MRSA) presenting spa type t899 is commonly associated with sequence type 9 (ST9) but is also increasingly linked to ST398. This study provides genomic insight into the diversity of t899 isolates using core genome multilocus sequence typing (cgMLST), single nucleotide polymorphism (SNP)-based phylogeny, and the description of selected antimicrobial resistance and virulence markers. The SNP-based phylogenic tree showed that isolates sharing the same spa type (t899) but different STs highly diverged in their core and accessory genomes, revealing discriminant antimicrobial resistance (AMR) and virulence markers. Our results highlighted the idea that in a surveillance context where only spa typing is used, an additional multiplex PCR for the detection of the tet(M), sak, and seg genes would be valuable in helping distinguish ST9 from ST398 isolates on a routine basis.
IMPORTANCE This study showed the genetic diversity and population structure of S. aureus presenting the same spa type, t899, but belonging to different STs. Our findings revealed that these isolates vary deeply in their core and accessory genomes, contrary to what is regularly inferred from studies using spa typing only. Given that identical spa types can be associated with different STs and that spa typing only is not appropriate for S. aureus isolates that have undergone major recombination events which include the passage of the spa gene (such as in t899-positive MRSA), the combination of both MLST and spa typing methods is recommended. However, spa typing alone is still largely used in surveillance studies and basic characterization. Our data suggest that additional markers, such as tet(M), sak, and seg genes, could be implemented in an easy and inexpensive manner in order to identify S. aureus lineages with a higher accuracy.
INTRODUCTION
Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) was first described in 2005 (1, 2) and rapidly gained importance because of its capacity to colonize and infect humans, particularly pig farmers. Clonal complex 398 (CC398) has been referred to as the predominant LA-MRSA lineage in Europe (3, 4), but other lineages, including CC9 and CC5, have also been detected (3–7).
spa type t899 originally belongs to CC9 (sequence type 9 [ST9]), the predominant LA-MRSA genotype reported in Asia, and was identified in pigs and their associated human workers (8). More recently, spa type t899 was also reported in a CC9/CC398 hybrid strain in Europe (9, 10). This hybrid is a unique genotype with a CC398 chromosomal backbone and smaller CC9 region with a spa gene (9). CC9 differs from the European pig-associated CC398 with regard to clonal type, staphylococcal cassette chromosome mec element (SCCmec) content, and resistance profile (4).
CC398 (ST398) isolates belonging to spa type t899 have received specific attention because they cause sporadic illness in humans (9, 11–13). This lineage notably harbors the staphylococcal complement inhibitor (scn), staphylokinase (sak), and chemotaxis inhibitory protein (chp) genes associated with the ϕSa3 immune evasion cluster (IEC), allowing adaptation to the human host (10). spa types t011 and t034 (which display close tandem repeat successions) are still the main types associated with CC398, but t899 is increasingly described in the literature both in human and veterinary medicine (14–19).
The CC9/CC398 hybrid strain has been identified from livestock (9, 10) and related personnel (11, 20) in several European countries. Furthermore, two CC398 LA-MRSA spa type t899 isolates were recently reported as incidental findings during a clinical investigation from turkey and pheasant in the United Kingdom (10, 21). These two isolates were shown to belong to the CC9/C398 hybrid genotype and were quite similar to the clone that was reported from continental Europe (10, 13, 22). Interestingly, t899 has been increasingly associated with several single-locus variants (SLVs) of CC398 and CC9 (16–18).
Overall, the occurrence of the same spa types in distant lineages has been reported, resulting from either convergent evolution or genetic recombination (23, 24). As an example, CC239 MRSA is a hybrid strain of CC30 (founder, ST30) and CC8 (founder, ST8) (23). ST34 and ST42 backgrounds have also been suggested to be of hybrid origin (24). Recently, t304 isolates belonging to ST6, ST1649, ST8, and ST4290 were reported by Bartels et al. (25).
Since spa typing is still largely used as the unique typing method, for example in large surveillance studies and in low-income countries, our aim was to characterize t899 isolates using single nucleotide polymorphism (SNP)-based phylogeny on publicly available genomic data and associated metadata in order to identify markers that could be implemented in an easy and inexpensive manner in order to identify LA-MRSA lineages with a higher accuracy.
RESULTS
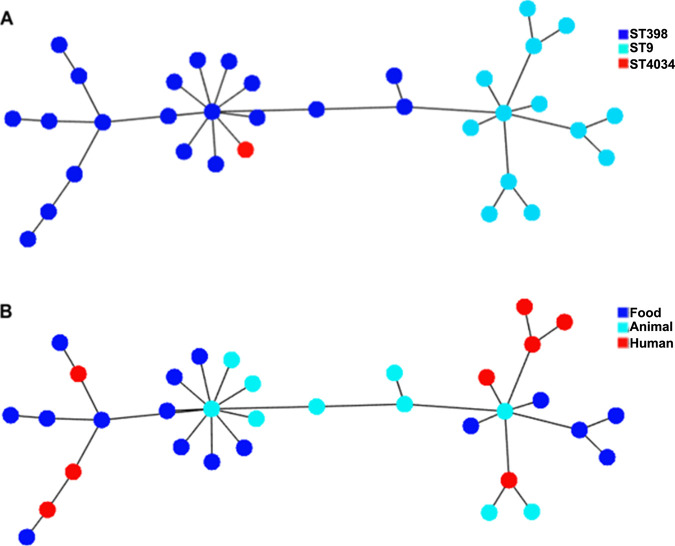
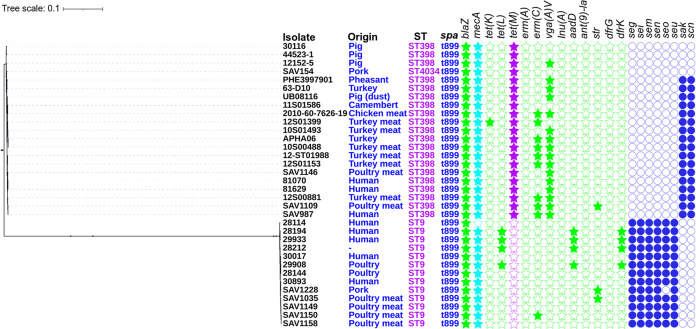
Thirty-four LA-MRSA genomes of t899 isolates were analyzed, of which 20 belonged to ST398, 13 to ST9, and 1 to ST4034 (a single-locus variant [SLV] of ST398, differing by one substitution [A294T] in the arcC gene). Metadata associated with the selected isolates were recorded (Table 1). All t899 isolates harbored the mecA gene on a SCCmec IVa(2B) element, except for two which presented either the SCCmec V element or an undefined cassette. Both the spanning tree and the SNP-based phylogenetic tree (Fig. 1B and 2) confirmed a strong clustering according to the ST, with the ST4034/t899 isolate differing from ST398/t899 by only 41 core alleles. Other characteristics, including matrix/sample origin, did not appear to cluster in this SNP analysis.
TABLE 1.
Genotypic features of 34 methicillin-resistant Staphylococcus aureus spa type t899 isolatesa
| Accession no. | Isolate ID | Yr of isolation | Geographic origin | Isolate source | MLST | SCCmec | Tetracycline resistance gene |
|---|---|---|---|---|---|---|---|
| SRR1290901 | 28114 | 2013 | Germany | Human | ST9 | IVa(2B) | ND |
| SRR1290895 | 28144 | 2013 | Germany | Poultry | ST9 | ND | ND |
| SRR1303238 | 28194 | 2010 | Netherlands | Human | ST9 | IVa(2B) | tet (L) |
| SRR1290877 | 28212 | — | Germany | Animal | ST9 | IVa(2B) | tet (L) |
| SRR1303551 | 29908 | — | Germany | Poultry | ST9 | IVa(2B) | tet (L) |
| SRR1303423 | 29933 | 2010 | Netherlands | Human | ST9 | IVa(2B) | tet (L) |
| SRR1303430 | 30017 | 2011 | Netherlands | Human | ST9 | IVa(2B) | ND |
| ERR594184 | 30893 | 2014 | Germany | Human | ST9 | IVa(2B) | ND |
| SRR7825591 | SAV1035 | 2017 | Poland | Poultry meat | ST9 | IVa(2B) | ND |
| SRR7825590 | SAV1149 | 2017 | Germany | Poultry meat | ST9 | IVa(2B) | ND |
| SRR7825587 | SAV1150 | 2017 | Germany | Poultry meat | ST9 | IVa(2B) | ND |
| SRR7825588 | SAV1158 | 2017 | Germany | Poultry meat | ST9 | IVa(2B) | ND |
| SRR7825595 | SAV1228 | 2017 | Czech Republic | Pork | ST9 | IVa(2B) | ND |
| ERR2442746 | APHA06 | 2016 | England | Turkey | ST398 | IVa(2B) | tet(M) |
| ERR2562460 | PHE3997901 | 2017 | Scotland | Pheasant | ST398 | IVa(2B) | tet(M) |
| SRR1218618 | 10S00488 | 2010 | Germany | Turkey meat | ST398* | IVa(2B) | tet(M) |
| SRR1290866 | 81070 | — | Germany | Human | ST398 | IVa(2B) | tet(M) |
| SRR1290867 | 11S01586 | 2011 | Germany | Camembert | ST398* | IVa(2B) | tet(M) |
| SRR1290868 | 12-ST01988 | 2012 | Germany | Turkey meat | ST398* | IVa(2B) | tet(M) |
| SRR1290875 | 12S01399 | 2012 | Germany | Turkey meat | ST398* | IVa(2B) | tet(M), tet(K) |
| SRR1300909 | 63-D10 | 2012 | France | Turkey | ST398* | IVa(2B) | tet(M) |
| SRR1303281 | 12S00881 | 2012 | Germany | Turkey meat | ST398* | IVa(2B) | tet(M) |
| SRR1303432 | 2010-60-7626-19 | 2010 | Denmark | Chicken meat | ST398* | IVa(2B) | tet(M) |
| SRR1303468 | 12S01153 | 2012 | Germany | Turkey meat | ST398* | IVa(2B) | tet(M) |
| SRR1303550 | 81629 | — | Denmark | Human | ST398 | IVa(2B) | tet(M) |
| SRR1303558 | 10S01493 | 2010 | Germany | Turkey meat | ST398* | IVa(2B) | tet(M) |
| SRR445027 | 12152-5 | 2008 | Italy | Pig | ST398* | IVa(2B) | tet(M) |
| SRR445029 | 30116 | 2008 | Italy | Pig | ST398* | V(5C2&5) | tet(M) |
| SRR445030 | 44523-1 | 2008 | Italy | Pig | ST398* | IVa(2B) | tet(M) |
| SRR445060 | UB08116 | 2008 | France | Pig (dust) | ST398 | IVa(2B) | tet(M) |
| SRR7825589 | SAV1146 | 2017 | Germany | Poultry meat | ST398 | IVa(2B) | tet(M) |
| SRR7825592 | SAV1109 | 2017 | Poland | Poultry meat | ST398 | IVa(2B) | tet(M) |
| SRR7825593 | SAV0154 | 2013 | Czech Republic | Pork | ST4034 | IVa(2B) | tet(M) |
| SRR7825594 | SAV0987 | 2017 | Czech Republic | Human | ST398 | IVa(2B) | tet(M) |
*, CC398/CC9 hybrid strain; tet, tetracycline resistance gene; —, not available; ND, not detected. Isolates with SAV numbers are unique to this study.
FIG 1.
Minimum spanning tree based on the concatenated core genome of 34 S. aureus t899 strains with color annotation based on (A) MLST and (B) matrix. Visualization was realized using PHYLOViZ.
FIG 2.
Maximum-likelihood phylogenetic tree based on SNP analyses of 34 S. aureus t899 strains obtained by CSI Phylogeny from the Center for Genomic Epidemiology web-based platform. Sequence type (ST), spa type (spa), isolate origin, the presence of antimicrobial resistance, and virulence markers are indicated. The filled stars and circles indicate confirmed antimicrobial resistance and virulence markers, respectively, as extracted from ResFinder and Virulence Finder. Visualization was realized using iTOL. Isolates with SAV numbers are unique to this study. Genes encoding resistance to beta-lactams (blaZ, mecA), tetracycline [tet(K), tet(M), tet(L)], macrolide-lincosamide-streptogramin B [erm(A), erm(C)], pleuromutilin-lincosamide-streptogramin A [vga(A)V], lincosamides [lnu(A)], aminoglycosides [aadD, ant(9)-la, str], and trimethoprim (dfrK, dfrG) and those encoding staphylococcal enterotoxins (seg, sei, sem, sen, seo, seu), staphylokinase (sak), and staphylococcal complement inhibitor (scn) are shown.
The phylogenetic tree (Fig. 2) also showed major divergences in the antimicrobial resistance and virulence patterns depending on the ST. All ST398 isolates carried the tet(M) gene, while none of the ST9 isolates carried this tetracycline resistance gene (Fig. 2). Concerning virulence markers, most ST398 spa type t899 isolates harbored the scn and sak genes, indicating the presence of the IEC cluster, while ST9 isolates were devoid of the IEC cluster but systematically harbored the seg, sei, sem, sen, and seu genes, encoding enterotoxin-like proteins (Fig. 2).
DISCUSSION
In this study, the SNP-based phylogeny analysis was consistent with the core genome multilocus sequence typing (cgMLST) analysis, with t899 isolates clustering apart based on STs. Although spa typing has a remarkable predictive power over clonal relationships, predicting genetic relatedness based on spa type does not appear appropriate for isolates that have undergone major recombination events, including spa gene passages (26–31). This important genomic recombination is not frequent in S. aureus, and the major representatives of such events are ST239, ST34, and ST42. The CC9/CC398 hybrid is another important example, giving rise to t899 isolates which largely diverge from their original CC9 genetic backgrounds and which mediate human diseases given their arsenal of virulence factors.
Here, we characterized t899 isolates from different STs using whole-genome-sequencing (WGS)-based approaches, together with epidemiological data, antimicrobial resistance genes, and virulence markers. This analysis revealed the differential occurrence of genes that can be used to further characterize t899 isolates. ST398-t899 isolates harbored the IEC cluster, which is crucial for disrupting the normal function of the human immune system (22, 32–34). Among the 34 t899 isolates tested, all ST398 representatives harbored the tet(M) gene, which is either transposon located or chromosomal, while ST9 representatives either were susceptible or carried the plasmid-located tet(L) gene. The tetracycline resistance gene tet(M) is a common feature of LA-MRSA ST398, while it is absent from MRSA ST9 (35–37). In contrast, ST9 isolates carried staphylococcal enterotoxin (SE) genes, which were not detected in ST398 isolates. This clear discrepancy between the two lineages would be useful to refine the LA-MRSA characterization when only spa typing is used and indicates the presence of t899 isolates.
Overall, investigations into S. aureus populations using WGS would be useful for future molecular epidemiology studies and for more closely examining the global evolution of S. aureus lineages. WGS also helps to assess the performances of classical typing methods by comparison. According to David et al. (38), two genotyping methods examining distinct genetic loci will not consistently provide identical results in classifying MRSA isolates, mostly because these methods assess genetic differences that can evolve independently. Classification systems often employed for epidemiological research have created competing nomenclatures that are useful for assessing the relatedness of isolates but are unfortunately not always directly comparable. This study emphasizes that spa typing is not sufficient to characterize t899-positive LA-MRSA. Accordingly, this study suggests the usefulness of an additional genomic marker to assign t899-positive MRSA isolates to the ST9 or ST398 clone, which may include tet(M), sak, and/or seg genes. Of course, this analysis should be refined when new t899 isolates belonging to other STs are sequenced and characterized.
MATERIALS AND METHODS
Bacterial collection.
Thirty-four t899 S. aureus isolates were found in the publicly available databases, and their corresponding characteristics (MLST, matrix [human, food, and animal origins], and geographical origin) were recorded. Raw reads were downloaded from NCBI, reads were quality checked with FastQC v.0.65, and low-quality reads were trimmed using Trimmomatic v.0.36.4 (39). Subsequently, contigs were generated using the SPAdes ve.3.5.0 algorithm (40), and those whose length exceeded 200 bp were retained in the assembly. In the literature, spa type t899 was also found to belong to 15 other SLVs and multilocus variants (MLVs) of ST9 and ST398 (see Table S1 in the supplemental material). These isolates could unfortunately not be included in our analysis because of the absence of associated WGS data.
cgMLST analyses.
Isolates were subjected to cgMLST analyses. Genome-wide gene-by-gene microbial typing was performed using Ridom SeqSphere+ S. aureus cgMLST analysis with default parameters (41). The cgMLST data contain 1,861 coding loci representing the core genome (41). Once an allelic profile was assigned to each genome, a minimum spanning tree was constructed from the concatenated core genome sequences and visualized using the online tool PHYLOViZ. cgMLST loci with no allele calls were ignored in the pairwise comparison during the tree construction. The minimum spanning tree constructed on the basis of cgMLST data illustrates clusters by ST, spa type, or matrix (Fig. 1).
SNP analysis and phylogenetic tree.
A phylogenetic tree was constructed based on single nucleotide polymorphism (SNP) analysis (9, 10, 22). SNPs were identified by mapping reads against the ST398 reference genome (strain S0385; GenBank accession no. AM990992). The maximum-likelihood phylogenetic tree was established in CSI Phylogeny using default settings (42). The phylogenetic tree visualization was realized using iTOL (Interactive Tree of Life) (43).
Detection of resistance genes and selected virulence markers using WGS data.
The online tools ResFinder v.3.2 (44) and Virulence Finder v. 2.0 (45) from the Center for Genomic Epidemiology web-based platform were used to detect genes encoding potential resistance to antimicrobials and virulence markers, respectively. For a hit to be reported by the two programs, it had to cover at least 60% of the length of the gene sequence in the database with sequence identities of 60% and 90%, respectively. WGS-assembled data were used to perform the analysis.
Data availability.
The sequence information for isolates SAV1035, SAV1149, SAV1150, SAV1158, and SAV1228 has been deposited in the SRA database under study accession number SRP161670. Individual accession numbers are listed in Table 1.
Supplementary Material
ACKNOWLEDGMENTS
We thank the service Transversal Activities in Applied Genomics from Sciensano for the paired-end sequencing reactions and for the development and maintenance of the in-house instance of the Galaxy workflow management system and Andrew D. Miller for his help in proofreading the manuscript. We thank Mirko Rossi for his valuable input in the course of designing this study and Beatriz Guerra and Antonio Rinaldi for cgMLST analysis.
This study was supported by the Ministry of Education, Youth and Sports, project no. CZ.1.05./2.1.00/19.0385. Funding sources did not affect the design of this study, data collection, data analysis, decisions on publication, or preparation of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Armand-Lefevre L, Ruimy R, Andremont A. 2005. Clonal comparison of Staphylococcus from healthy pig farmers, human controls, and pigs. Emerg Infect Dis 11:711–714. doi: 10.3201/eid1105.040866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis 11:1965–1966. doi: 10.3201/eid1112.050428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butaye P, Argudín MA, Smith TC. 2016. Livestock-associated MRSA and its current evolution. Curr Clin Micro Rpt 3:19–31. doi: 10.1007/s40588-016-0031-9. [DOI] [Google Scholar]
- 4.Ye X, Wang X, Fan Y, Peng Y, Li L, Li S, Huang J, Yao Z, Chen S. 2016. Genotypic and phenotypic markers of livestock-associated methicillin-resistant Staphylococcus aureus CC9 in humans. Appl Environ Microbiol 82:3892–3899. doi: 10.1128/AEM.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui S, Li J, Hu C, Jin S, Li F, Guo Y, Ran L, Ma Y. 2009. Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J Antimicrob Chemother 64:680–683. doi: 10.1093/jac/dkp275. [DOI] [PubMed] [Google Scholar]
- 6.Frana TS, Beahm AR, Hanson BM, Kinyon JM, Layman LL, Karriker LA, Ramirez A, Smith TC. 2013. Isolation and characterization of methicillin-resistant Staphylococcus aureus from pork farms and visiting veterinary students. PLoS One 8:e53738. doi: 10.1371/journal.pone.0053738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hau SJ, Allué-Guardia A, Rusconi B, Haan JS, Davies PR, Frana TS, Eppinger M, Nicholson TL. 2018. Single nucleotide polymorphism analysis indicates genetic distinction and reduced diversity of swine-associated methicillin resistant Staphylococcus aureus(MRSA) ST5 isolates compared to clinical MRSA ST5 isolates. Front Microbiol 9:2078. doi: 10.3389/fmicb.2018.02078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guardabassi L, O'Donoghue M, Moodley A, Ho J, Boost M. 2009. Novel lineage of methicillin-resistant Staphylococcus aureus, Hong Kong. Emerg Infect Dis 15:1998–2000. doi: 10.3201/eid1512.090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsen J, Stegger M, Andersen PS, Petersen A, Larsen AR, Westh H, Agersø Y, Fetsch A, Kraushaar B, Käsbohrer A, Feβler AT, Schwarz S, Cuny C, Witte W, Butaye P, Denis O, Haenni M, Madec JY, Jouy E, Laurent F, Battisti A, Franco A, Alba P, Mammina C, Pantosti A, Monaco M, Wagenaar JA, De Boer E, Van Duijkeren E, Heck M, Domínguez L, Torres C, Zarazaga M, Price LB, Skov RL. 2016. Evidence for human adaptation and foodborne transmission of livestock-associated methicillin-resistant Staphylococcus aureus. Clin Infect Dis 63:1349–1352. doi: 10.1093/cid/ciw532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma M, AbuOun M, Nunez-Garcia J, Rogers J, Welchman D, Teale C, Anjum MF, Kearns AM, Pichon B, Foster G, Robb A, McMillan M. 2018. MRSA spa type t899 from food animals in the UK. Vet Rec 182:697–698. doi: 10.1136/vr.k2576. [DOI] [PubMed] [Google Scholar]
- 11.Pan A, Battisti A, Zoncada A, Bernieri F, Boldini M, Franco A, Giorgi M, Iurescia M, Lorenzotti S, Martinotti M, Monaci M, Pantosti A. 2009. Community-acquired methicillin-resistant Staphylococcus aureus ST398 infection, Italy. Emerg Infect Dis 15:845–847. doi: 10.3201/eid1505.081417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monaco M, Pedroni P, Sanchini A, Bonomini A, Indelicato A, Pantosti A. 2013. Livestock-associated methicillin-resistant Staphylococcus aureus responsible for human colonization and infection in an area of Italy with high density of pig farming. BMC Infect Dis 13:258. doi: 10.1186/1471-2334-13-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen J, Petersen A, Larsen AR, Sieber RN, Stegger M, Koch A, Aarestrup FM, Price LB, Skov RL, Johansen HK, Westh H, Pedersen M, Jensen US, Jensen MLS, Chen M, Strøbæk S, Østergaard C, Lomborg S, Ellermann-Eriksen S, Ripadal P, Danish MRSA Study Group. 2017. Emergence of livestock-associated methicillin-resistant Staphylococcus aureus bloodstream infections in Denmark. Clin Infect Dis 65:1072–1076. doi: 10.1093/cid/cix504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Köck R, Apel J, Gerth C, Gundlach S, Lambrecht C, Josef TS. 2010. Prevalence of MRSA CC398 in pig holdings and human hospital patients in North Rhine-Westphalia, Germany. Methods 3:2010. [Google Scholar]
- 15.Köck R, Schaumburg F, Mellmann A, Köksal M, Jurke A, Becker K, Friedrich AW. 2013. Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) as causes of human infection and colonization in Germany. PLoS One 8:e55040. doi: 10.1371/journal.pone.0055040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz J, Boklund A, Toft N, Halasa T. 2018. Drivers for livestock-associated methicillin-resistant Staphylococcus aureus spread among Danish pig herds—a simulation study. Sci Rep 8:1–11. doi: 10.1038/s41598-018-34951-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tegegne HA, Koláčková I, Karpíšková R. 2017. Diversity of livestock associated methicillin-resistant Staphylococcus aureus. Asian Pac J Trop Med 10:929–931. doi: 10.1016/j.apjtm.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Pirolo M, Gioffrè A, Visaggio D, Gherardi M, Pavia G, Samele P, Ciambrone L, Di Natale R, Spatari G, Casalinuovo F, Visca P. 2019. Prevalence, molecular epidemiology, and antimicrobial resistance of methicillin-resistant Staphylococcus aureus from swine in southern Italy. BMC Microbiol 19:1–12. doi: 10.1186/s12866-019-1422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battisti A, Franco A, Merialdi G, Hasman H, Iurescia M, Lorenzetti R, Feltrin F, Zini M, Aarestrup FM. 2010. Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Vet Microbiol 142:361–366. doi: 10.1016/j.vetmic.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O'Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan H-L, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone K. 2017. Risk assessment on methicillin-resistant Staphylococcus aureus (MRSA), with a focus on livestock-associated MRSA, in the UK food chain. Food Standards Agency, London, United Kingdom. [Google Scholar]
- 22.Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, ŻMudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3:e00305-11. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto T, Takano T, Higuchi W, Iwao Y, Singur O, Reva I, Otsuka Y, Nakayashiki T, Mori H, Reva G, Kuznetsov V, Potapov V. 2012. Comparative genomics and drug resistance of a geographic variant of ST239 methicillin-resistant Staphylococcus aureus emerged in Russia. PLoS One 7:e29187. doi: 10.1371/journal.pone.0029187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas JC, Godfrey PA, Feldgarden M, Robinson DA. 2012. Draft genome sequences of Staphylococcus aureus sequence type 34 (ST34) and ST42 hybrids. J Bacteriol 194:2740–2741. doi: 10.1128/JB.00248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartels MD, Worning P, Andersen LP, Bes M, Enger H, Ås CG, Hansen TA, Holzknecht BJ, Larssen KW, Laurent F, Mäkitalo B, Pichon B, Svartström O, Westh H. 2020. Repeated introduction and spread of the MRSA clone t304/ST6 in northern Europe. Clin Microbiol Infect doi: 10.1016/j.cmi.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Asadollahi P, Farahani NN, Mirzaii M, Khoramrooz SS, van Belkum A, Asadollahi K, Dadashi M, Darban-Sarokhalil D. 2018. Distribution of the most prevalent spa types among clinical isolates of methicillin-resistant and -susceptible Staphylococcus aureus around the world: a review. Front Microbiol 9:163. doi: 10.3389/fmicb.2018.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosch T, Pluister GN, van Luit M, Landman F, van Santen-Verheuvel M, Schot C, Witteveen S, van der Zwaluw K, Heck MEOC, Schouls LM. 2015. Multiple-locus variable number tandem repeat analysis is superior to spa typing and sufficient to characterize MRSA for surveillance purposes. Future Microbiol 10:1155–1162. doi: 10.2217/fmb.15.35. [DOI] [PubMed] [Google Scholar]
- 28.Darban-Sarokhalil D, Khoramrooz SS, Marashifard M, Malek Hosseini SAA, Parhizgari N, Yazdanpanah M, Gharibpour F, Mirzaii M, Sharifi B, Haeili M. 2016. Molecular characterization of Staphylococcus aureus isolates from southwest of Iran using spa and SCCmec typing methods. Microb Pathog 98:88–92. doi: 10.1016/j.micpath.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol 42:792–799. doi: 10.1128/jcm.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Hara FP, Suaya JA, Ray GT, Baxter R, Brown ML, Mera RM, Close NM, Thomas E, Amrine-Madsen H. 2016. spa typing and multilocus sequence typing show comparable performance in a macroepidemiologic study of Staphylococcus aureus in the United States. Microb Drug Resist 22:88–96. doi: 10.1089/mdr.2014.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruppitsch W, Indra A, Stöger A, Mayer B, Stadlbauer S, Wewalka G, Allerberger F. 2006. Classifying spa types in complexes improves interpretation of typing results for methicillin-resistant Staphylococcus aureus. J Clin Microbiol 44:2442–2448. doi: 10.1128/JCM.00113-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuny C, Wieler L, Witte W. 2015. Livestock-associated MRSA: the impact on humans. Antibiotics (Basel) 4:521–543. doi: 10.3390/antibiotics4040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarthy AJ, Witney AA, Gould KA, Moodley A, Guardabassi L, Voss A, Denis O, Broens EM, Hinds J, Lindsay JA. 2011. The distribution of mobile genetic elements (MGEs) in MRSA CC398 is associated with both host and country. Genome Biol Evol 3:1164–1174. doi: 10.1093/gbe/evr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hau SJ, Sun J, Davies PR, Frana TS, Nicholson TL. 2015. Comparative prevalence of immune evasion complex genes associated with β-hemolysin converting bacteriophages in MRSA ST5 isolates from swine, swine facilities, humans with swine contact, and humans with no swine contact. PLoS One 10:e0142832. doi: 10.1371/journal.pone.0142832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J, Kearns AM, Pichon B, Young B, Wilson DJ, Llewelyn MJ, Paul J, Peto TEA, Crook DW, Walker AS, Golubchik T. 2014. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 52:1182–1191. doi: 10.1128/JCM.03117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B, Pop M. 2009. ARDB—antibiotic resistance genes database. Nucleic Acids Res 37:443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossney AS, Shore AC, Morgan PM, Fitzgibbon MM, O'Connell B, Coleman DC. 2007. The emergence and importation of diverse genotypes of methicillin-resistant Staphylococcus aureus (MRSA) harboring the panton-valentine leukocidin gene (pvl) reveal that pvl is a poor marker for community-acquired MRSA strains in Ireland. J Clin Microbiol 45:2554–2563. doi: 10.1128/JCM.00245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.David MZ, Taylor A, Lynfield R, Boxrud DJ, Short G, Zychowski D, Boyle-Vavra S, Daum RS. 2013. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for Panton-Valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol 51:814–819. doi: 10.1128/JCM.02429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. 2014. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Letunic I, Bork P. 2019. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence information for isolates SAV1035, SAV1149, SAV1150, SAV1158, and SAV1228 has been deposited in the SRA database under study accession number SRP161670. Individual accession numbers are listed in Table 1.