Rieske oxygenases are involved in the degradation of various aromatic compounds. These dioxygenases usually carry out hydroxylation of hydrophobic aromatic compounds and supply substrates with hydroxyl groups for extradiol/intradiol dioxygenases to cleave rings, and have been extensively studied. Salicylate 5-hydroxylase NagGH is a novel Rieske monooxygenase with high similarity to Rieske dioxygenases, and also shares reductase and ferredoxin similarity with a Rieske dioxygenase naphthalene 1,2-dioxygenase (NagAcAd) in Ralstonia sp. strain U2.
KEYWORDS: polycyclic aromatic hydrocarbon degradation, salicylate 5-monooxygenase, crystal structure, product regio-selectivity, Rieske nonheme iron oxygenase
ABSTRACT
Rieske nonheme iron oxygenases (ROs) catalyze the oxidation of a wide variety of substrates and play important roles in aromatic compound degradation and polycyclic aromatic hydrocarbon degradation. Those Rieske dioxygenases that usually act on hydrophobic substrates have been extensively studied and structurally characterized. Here, we report the crystal structure of a novel Rieske monooxygenase, NagGH, the oxygenase component of a salicylate 5-monooxygenase from Ralstonia sp. strain U2 that catalyzes the hydroxylation of a hydrophilic substrate salicylate (2-hydroxybenzoate), forming gentisate (2, 5-dihydroxybenzoate). The large subunit NagG and small subunit NagH share the same fold as that for their counterparts of Rieske dioxygenases and assemble the same α3β3 hexamer, despite that they share low (or no identity for NagH) sequence identities with these dioxygenase counterparts. A potential substrate-binding pocket was observed in the vicinity of the nonheme iron site. It featured a positively charged residue Arg323 that was surrounded by hydrophobic residues. The shift of nonheme iron atom caused by residue Leu228 disrupted the usual substrate pocket observed in other ROs. Residue Asn218 at the usual substrate pocket observed in other ROs was likewise involved in substrate binding and oxidation, yet residues Gln316 and Ser367, away from the usual substrate pocket of other ROs, were shown to play a more important role in substrate oxidation than Asn218. The unique binding pocket and unusual substrate-protein hydrophilic interaction provide new insights into Rieske monooxygenases.
IMPORTANCE Rieske oxygenases are involved in the degradation of various aromatic compounds. These dioxygenases usually carry out hydroxylation of hydrophobic aromatic compounds and supply substrates with hydroxyl groups for extradiol/intradiol dioxygenases to cleave rings, and have been extensively studied. Salicylate 5-hydroxylase NagGH is a novel Rieske monooxygenase with high similarity to Rieske dioxygenases, and also shares reductase and ferredoxin similarity with a Rieske dioxygenase naphthalene 1,2-dioxygenase (NagAcAd) in Ralstonia sp. strain U2. The structure of NagGH, the oxygenase component of salicylate 5-monooxygenase, gives a representative of those monooxygenases and will help us understand the mechanism of their substrate binding and product regio-selectivity.
INTRODUCTION
In the bioremediation of persistent and toxic aromatic compounds that pollute the environment, Rieske nonheme iron oxygenases (ROs) have stimulated sustained interest. They catalyze a wide variety of oxidative transformations in a range of catabolic and biosynthetic pathways, such as dioxygenation, monooxygenation, oxidative cyclization, and desaturation. Therefore, the substrate pockets of ROs are amenable to a wide variety of substrates. The well-studied Rieske dioxygenases, including naphthalene dioxygenase (NDO), biphenyl dioxygenase (BPDO), nitrobenzene dioxygenase (NBDO), cumene dioxygenase, carbazole-1,9α-dioxygenase, toluene dioxygenase, and benzene dioxygenase, usually initiate the degradation of hydrophobic aromatic compounds, along with polycyclic aromatic hydrocarbons (PAHs), via the hydroxylation of the substrates, where the hydroxyl groups are recognized by the enzymes involved in the lower degradation pathways. Also, some Rieske dioxygenases are involved in the initial degradation of aromatic compounds with carboxyl groups, such as benzoate dioxygenase, phthalate dioxygenase, anthranilate dioxygenase, terephthalate 1,2-dioxygenase, and halobenzoate dioxygenases. However, biochemical and structural studies on Rieske monooxygenases are very limited.
Previous research indicated that the gap between Rieske monooxygenases and dioxygenases seems bridgeable. Interestingly, several Rieske oxygenases were identified as monooxygenases but shared considerable sequence identities with dioxygenases, including salicylate 5-monooxygenase from Ralstonia sp. strain U2 (1), salicylate 1-monooxygenase from Sphingomonas sp. strain CHY-1 (2), 2-oxoquinoline monooxygenase from Pseudomonas putida strain 86 (3), and 4-androstene-3,17-dione monooxygenases from Mycobacterium tuberculosis (4). On the other hand, some dioxygenases also act as monooxygenases. The toluene-2,3-dioxygenase from Pseudomonas putida UV4 catalyzes both the dihydroxylation of aromatic rings as well as the monohydroxylation of cycloalkane rings (5). An NDO (NDO9816-4) from Pseudomonas sp. strain NCIB 9816-4 can catalyze both dioxygenation using naphthalene, many related 2- and 3-ring aromatic and hydroaromatic compounds as the substrates, and monooxygenation using toluene, ethylbenzene, acetophenone, and 2-hydroxyacetophenone as the substrates (6). When using 2-nitrotoluene as a substrate, NDO9816-4 catalyzes a monooxygenation reaction, producing a nitrobenzyl alcohol, while the NBDO (NBDOJS765) from Comamonas sp. strain JS765 catalyzes a dioxygenation reaction, producing a catechol (7). Through structural comparison, the difference between Asn258 of NBDOJS765 and Val260 of NDO9816-4 at the same position was identified. Mutant NBDOJS765-N258V conducts a 2-nitrotoluene monooxygenation reaction, forming mostly nitrobenzyl alcohol as does NDO9816-4 (8, 9). These results clearly imply that the orientation of the substrate in a preferred manner can determine the regio-specificity of product formation (10).
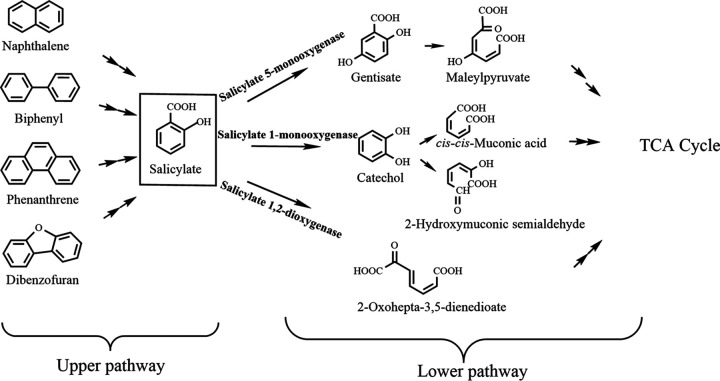
During bacterial degradation of various PAHs through naphthalene catabolic pathway, the hydroxylation of salicylate is an important step and links the so-called upper and lower degradation pathways (Fig. 1). Salicylate 1-monooxygenase, converting salicylate to catechol, has been well characterized, but only few salicylate 5-monooxygenases that convert salicylate to gentisate have been reported previously (1, 11–13). Notably, Rieske monooxygenases are found to be involved in this process, which is exemplified by salicylate 5-monooxygenase in the naphthalene degradation pathway of Ralstonia sp. strain U2 (1). Three components constitute this hydroxylase system, a monooxygenase component NagGH, a ferredoxin reductase NagAa, and a ferredoxin NagAb. The monooxygenase component, NagGH, is made up of two types of subunits, large subunit NagG and small subunit NagH. NagG is proposed as the region where the hydroxylation occurs. In the naphthalene degradation pathway of Ralstonia sp. strain U2 (via gentisate) (14), salicylate 5-monooxygenase shares electron transport proteins NagAa and NagAb with NDO (1, 15). Moreover, NagG shares a 30% sequence identity with the large subunit of NDO9816-4. NagGH also shows identities with a halobenzoate dioxygenase from Pseudomonas aeruginosa strain 142 (16), an anthranilate dioxygenase from Burkholderia cepacia strain DBO1 (17), and a terephthalate 1,2-dioxygenase from Comamonas sp. strain E6 (18) of 42%, 39%, and 39% for the large subunits, respectively, and 27%, 33%, and 24% for the small subunits, respectively. A previous work showed NagGH with an interesting substrate range, but the mechanism remains elusive (15). Where those dioxygenases recognize substrates mainly via hydrophobic interactions between enzymes and substrates, substrate recognition for NagGH appears to involve hydrophilic groups.
FIG 1.
Catabolic pathways for the degradation of some PAHs involving salicylate as an intermediate. The hydroxylation of salicylate links the so-called upper and lower degradation pathways of naphthalene, biphenyl, phenanthrene, and dibenzofuran.
To illustrate how the substrates were recognized by Rieske monooxygenases, we herein solved the crystal structure of NagGH and identified the residues potentially involved in the formation of the substrate-binding pocket. Mutagenesis assays suggested that residues Arg323 and Ser367 in the substrate-binding pocket are essential for protein folding and catalysis, respectively. We also propose the molecular and structural basis for substrate recognition and specificity determination. Our work provides clues to the different substrate-binding modes between NagGH and Rieske dioxygenases.
RESULTS
NagGH assembles a typical architecture of ROs.
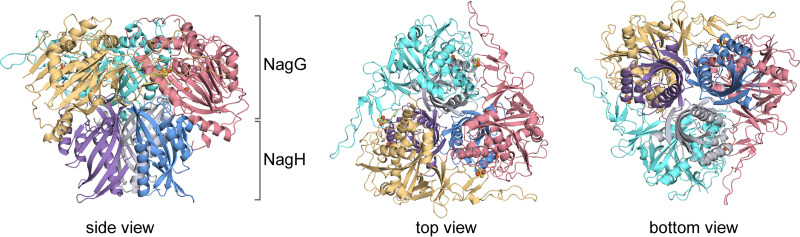
The monooxygenase component (NagGH) of salicylate 5-monooxygenase from the naphthalene degradation pathway of Ralstonia sp. strain U2 consists of three large subunits, NagG, and three small ones, NagH. The crystal structure of NagGH was determined at a resolution of 2.6 Å with an Rwork of 0.191 and Rfree of 0.235. Crystallographic statistics are summarized in Table S1 in the supplemental material. The crystal structure indicates that three copies of NagG and NagH assemble an α3β3 hexamer, which is the same as the previously identified oligomerization of NagGH (15). The hexamer assembles as a mushroom-shaped quaternary structure with 3-fold symmetry (Fig. 2), where NagG and NagH locate at the upper and bottom, respectively. This oligomeric organization is usually observed in the structures of previously reported oxygenase components of ROs, such as NDO9816-4 (19, 20), NBDOJS765 (8), and BPDOLB400 (BPDO from Burkholderia xenovorans strain LB400) (21).
FIG 2.
Overall structure of NagGH. The crystal structure of NagGH is an α3β3 hexamer. Three α subunits (NagG) and three β subunits (NagH) are at the top and bottom, respectively, and each subunit is shown in a different color. They assemble into a mushroom-shaped organization (side view).
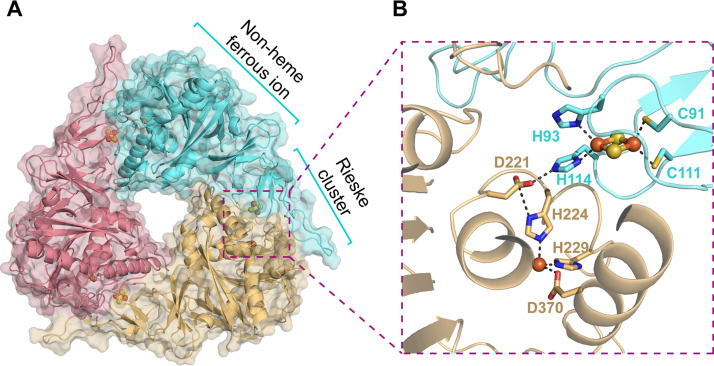
NagG shares 31% and 30% sequence identities to the large subunits of NDO9816-4 and BPDOLB400, respectively (Fig. S1), and likewise folds into two domains (Fig. S2B), including an N-terminal Rieske iron-sulfur cluster domain that binds a Rieske-type [2Fe-2S] iron-sulfur cluster and a C-terminal catalytic domain that binds a nonheme ferrous ion (Fig. 3A). In the Rieske domain, four residues coordinate the iron-sulfur cluster. Residues Cys91 and Cys111 bind to one iron atom of the iron-sulfur cluster and residues His93 and His114 bind to the other (Fig. 3B). In the C-terminal domain, residues His224, His229, and Asp370 assemble a nonheme iron-binding triad that binds a ferrous ion (Fig. 3B). In the α3β3 hexamer, a nonheme iron center in one NagG molecule is adjacent to the [2Fe-2S] cluster of a neighbor subunit, with a distance of 12 Å between the nonheme iron center and one iron atom of the [2Fe-2S] cluster. Residue Asp221 that bridges two histidines, His114 and His224, is proposed to shuttle the electron from the [2Fe-2S] cluster to the nonheme iron (Fig. 3B). Residues involved in the coordination of Rieske cluster (Cys91, Cys111, His93, and His114) and mononuclear iron (His224, His229, and Asp370), along with the aspartic acid (Asp221) that bridges the Rieske cluster of one subunit and the nonheme iron of another of two NagG, are completely conserved in all of the structures of ROs determined to date.
FIG 3.
The catalytic center formed by the Rieske-type [2Fe-2S] cluster and the nonheme iron center. (A) The trimer organization of NagG. Each NagG consists of a Rieske-type [2Fe-2S] cluster and a nonheme iron center. (B) The Rieske-type [2Fe-2S] cluster in one NagG molecule (cyan) and the nonheme iron center in an adjacent molecule (yellow) together form a full catalytic center. The nonheme iron and sulfur atoms are shown as orange and yellow spheres, respectively. The residues that coordinate [2Fe-2S] and nonheme iron are shown as sticks.
NagH folds into a cystatin-like protein fold where a beta-sheet grooves a helix (22), as do the small subunits of NDO9816-4, NBDOJS765, and BPDOLB400. Though NagH does not share any sequence identity to the small subunit of NDO9816-4, their overall structures are well superposed to each other, with a Cα root-mean-square deviation (RMSD) of 2.3 Å, except there is an additional N-terminal alpha-helix in the latter (Fig. S2C). Notably, once NagH is well superposed to the small subunit of NDO9816-4, the overall structure of NagGH is also well superposed to that of NDO9816-4, and vice versa (Fig. S2C). This observation suggests the high structural similarity between NagGH and other known ROs, and confirms that the hexamer acts as a functional unit, similar to other ROs.
Potential substrate-binding pocket features a positively charged residue Arg323 surrounded by hydrophobic residues.
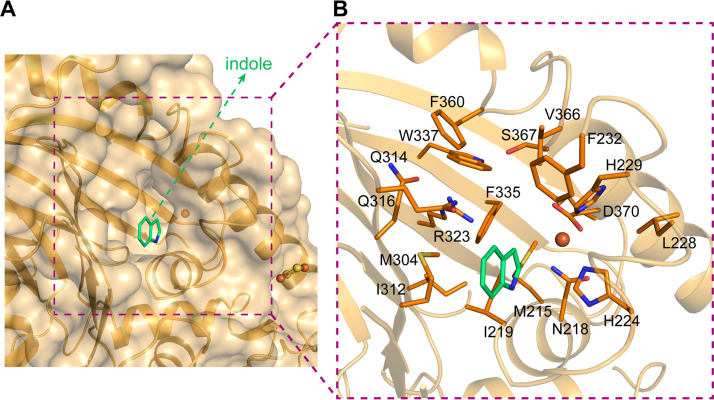
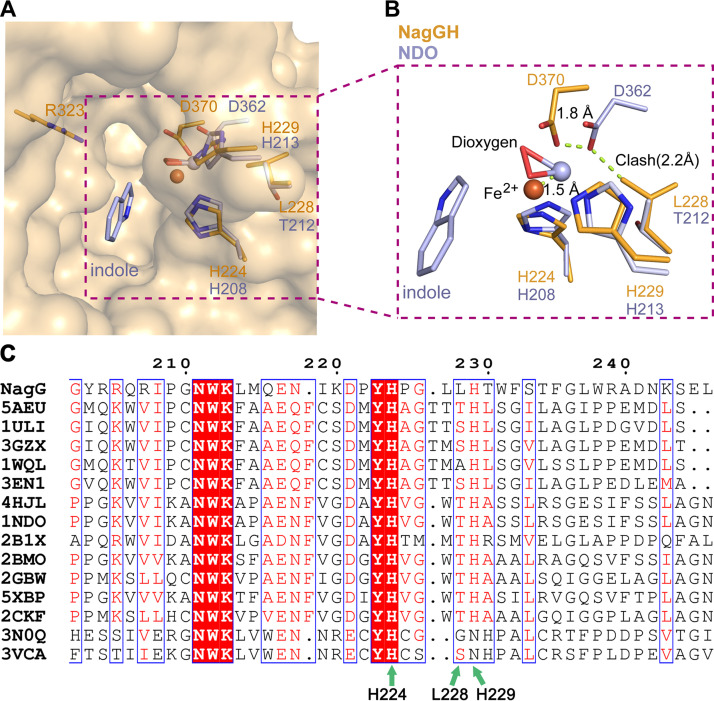
A large pocket in the vicinity of the mononuclear iron center was observed (Fig. 4A) and proposed to accommodate the substrate salicylate. Residues Met215 and Ile219 constitute the bottom of this pocket. The catalytic triad (residues His224, His229, and Asp370) and residue Asn218 surround this pocket from one side, with residues Met304, Ile312, Arg323, Phe335, Gln316, Trp337, Phe360, Val366, and Phe232 from the other side (Fig. 4B). Interestingly, this pocket includes a positively charged residue, Arg323, and two neutrally charged hydrophilic residues, Gln316 and Ser367. The side chain of Arg323 is surrounded by the side chains of some hydrophobic residues Phe335 (4.0 Å), Trp337 (3.2 Å), Met304 (4.3 Å), Ile312 (4.5 Å), and also bonds to Gln314 via a hydrogen bond with a distance of 2.4 Å. Thus, its side chain is well fixed and points to the iron center, with a distance of 6.6 Å. Side chains of Ser367 and Gln316 extend to the space where Arg323 locates. This kind of space arrangement makes these three hydrophilic residues assemble a hydrophilic pattern that locates in the center of a hydrophobic region. This pocket covers the space that should accommodate substrates in other ROs and is much larger than the binding space needed for substrate and dioxygen. In contrast, the substrate indole in NDO9816-4 (PDB ID 1O7N) binds at a hydrophobic site that corresponds to the space surrounded by residues Met215, Ile219, Met304, Ile312, and Phe335 of NagG. Therefore, these three residues surrounded by hydrophobic residues are proposed to be involved in the catalytic reaction and substrate binding.
FIG 4.
The potential substrate-binding pocket around the nonheme iron center of NagG. (A and B) The substrate indole in NDO9816-4 is superposed to NagGH and shown in green to indicate the usual substrate-binding site of Rieske dioxygenases. The nonheme iron atom and the residues in NagGH that constitute the substrate pocket are shown as sphere and stick representations, respectively.
Rearranged nonheme iron center disrupts the substrate pocket observed in other ROs.
In NagGH, residues His224, His229, and Asp370 coordinate the iron atom via three atoms of their side chains with distances of 2.29 Å, 2.11 Å, and 2.34 Å, respectively (Fig. 5A and B, and Table 1). The other oxygen atom (O2) of the carboxyl group in Asp370 is also probably involved in coordinating the iron center with a significantly long distance, 3.07 Å (Fig. 5B, Table 1). These distances make the iron coordination mode similar to BPDOLB400, yet different from NDO9816-4 and NBDOJS765, where the iron is coordinated by four atoms from the triad with distances of 2.1 to 2.4 Å (Table 1).
FIG 5.
A shift of the nonheme iron atom of NagGH, in comparison with NDO9816-4. (A and B) The superposition of the nonheme iron-binding motifs as well as some substrate-binding residues. NagGH and NDO9816-4 are shown in orange and gray, respectively. The shifts of residue Asp370 and the iron atom are indicated by dash lines. The close distance between Asp362 in NDO9816-4 and Leu228 in NagG is indicated as “clash.” (C) Sequence alignment of NagG and its homologs. NagG, AAD12607.1; 5AEU, WP_011494299.1; 1ULI, WP_011599002.1; 3GZX, Q46372.1; 1WQL, WP_039594666; 3EN1, WP_012052601.1; 4HJL, WP_011117400.1; 1NDO, WP_011117400.1; 2B1X, WP_087561958.1; 2BMO, AAL76202.1; 2GBW, WP_037508582.1; 5XBP, AGH09226.1; 2CKF, WP_037485720.1; 3N0Q, WP_011536837.1; 3VCA, WP_010967390.1. The sequence number of NagG is labeled on the top of the protein sequences. Green arrows indicate three residues potentially important for iron binding.
TABLE 1.
The distances between the nonheme iron center and its coordinating residues
A coordinating geometry with restricted space at the nonheme iron site highly relates to the regio-selectivity of product formation in ROs. Related to this, the geometry near the nonheme iron is critical for the substrate range and the product regio-selectivity. The superposition of NagGH, NDO9816-4, NBDOJS765, and BPDOLB400 revealed that the 2-His-1-carboxylate facial triad of NagGH does not match well with those of other ROs, though NagG shares high sequential and structural similarities with other ROs. The superposition of NagGH and NDO9816-4 was then performed via the two histidine residues in the triad and indicated a 1.5 Å shift for the nonheme iron and a 1.8 Å shift for the carboxyl group of Asp370 (Fig. 5B). This shift was caused by the difference of a residue that preceded the second histidine in the triad: Thr212 in NDO9816-4, Thr210 in NBDOJS765, Ser229 in BPDO from Rhodococcus sp. strain RHA1 (23), Ala239 in cumene dioxygenase from Pseudomonas fluorescens IP01 (24), and Leu228 in NagGH (Fig. 5C). Notably, this residue is highly conserved as threonine in other structure-known proteins other than NagG (Fig. 5C). The corresponding residue Thr212 in the NDO9816-4 (1O7N) is in the vicinity of the aspartate residues of the triad, with a distance of 3.66 Å (the distance is 3.71 Å between Thr210 and Asp360 in NBDOJS765 [2BMQ]). This distance could not tolerate the substitution of threonine to a residue with bulky sidechain, otherwise triggering a structural rearrangement. Residue Leu228 has a bulkier sidechain than threonine, thus causing the shift of the sidechain of Asp370 as well as the nonheme ferrous iron. Besides, the nearest distance between the side chain of Ile219 and ion is 5.3 Å, apparently shorter compared with other ROs, such as 5.8 Å for NDO9816-4 (20) and 5.7 Å for 2-oxoquinoline 8-monooxygenase (3). Thus, this short distance further hampered salicylate’s accommodation and pushed it toward Arg323.
As a result, the nonheme iron center moved toward the position where substrates are located in other ROs. The shift of the nonheme iron center narrowed the potential pocket that accommodated substrates and dioxygen. We thus propose that the salicylate and dioxygen were unable to be accommodated by this pocket (Fig. 5B). In other words, NagGH could not bind to salicylate and dioxygen in the same manner as do other ROs.
Residues Arg323 and Ser367 are potentially essential for enzymatic activity.
To investigate the enzymatic mechanism, the roles of those hydrophilic residues in the substrate-binding pocket were assessed via mutagenesis assays. In the apparent steady-state kinetic assay, salicylate 5-monooxygenase oxidized the substrate salicylate with a Km of 22.4 μM and a kcat of 130.2 × 10−3 · s−1 (Fig. S3). After the mutation of Arg323 to alanine, NagGH folded improperly and could not be purified in a soluble form. This observation suggested the essential role of Arg323 in protein folding and substrate binding. Mutant S367A retained only about 10% of the enzymatic ability of wild type and caused the Km and Vmax values to be unable to be determined, suggesting that this residue is involved in substrate binding and/or substrate oxidation. In contrast, mutants N218A and Q316A catalyzed the oxidation of salicylate with a Km of 39.8 μM and 23.6 μM, respectively, and a kcat of 40.4 × 10−3 · s−1 and 52.9 × 10−3 · s−1, respectively, which are comparable to those of wild-type protein. These kinetic parameters suggested that residues N218 and Q316 are also involved in, but not essential for, the catalytic function.
The carboxyl group of salicylate is required for binding to NagGH.
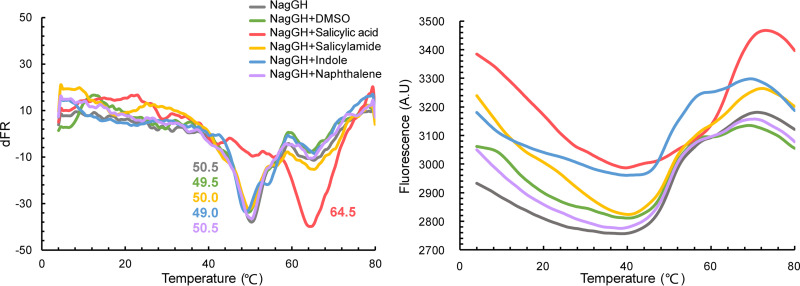
To access the interaction between NagGH and substrate, the isothermal titration calorimetry assay was performed, yet was not successful. The differential scanning fluorimetric (DSF) assay was then conducted and it was found that the melting temperature of NagGH was approximately 50°C. The presence of 1 mM salicylate resulted in a Tm increase of about 14°C (Tm = 64.5°C), indicating that the salicylate binding enhances the overall stability of NagGH (Fig. 6). To compare the substrate difference of NagGH and NDO, naphthalene was then assayed since it, not indole, is the exact substrate of NDO. Naphthalene was not found to shift the melting temperature of NagGH, suggesting that NagGH did not bind it. The result revealed that a hydrophobic molecule could not be a substrate for NagGH. Salicylamide, a salicylate homolog molecule, was unable to bind with NagGH either, indicating that the carboxyl group of salicylate is required for NagGH recognition.
FIG 6.
Differential scanning fluorimetric (DSF) assay of NagGH in the presence of different chemicals. The Tm values from the derivative of the fluorescent signals (left) and the fluorescence of NagGH with different chemicals (right) are shown. The Tm values are labeled in the left panel.
DISCUSSION
Mononuclear nonheme iron oxygenases, widely involved in the degradation of diverse aromatic pollutants, have been recognized to catalyze a wide range of reactions (25). These enzymes include extradiol dioxygenases, intradiol dioxygenases, ROs, catechol dioxygenases, lipoxygenases, α-ketoglutarate-dependent oxygenases, and carotenoid cleavage dioxygenases. ROs usually carry out dihydroxylation or monohydroxilation reactions in the biodegradation of aromatic compounds, followed by ring cleavage by extradiol/intradiol dioxygenases. The substrate-binding mode and the dioxygen activation mechanism for the dioxygenation function of some ROs have been extensively studied and well elucidated (20). Their substrates generally contain at most one hydrophilic group, such as a hydroxyl or carboxyl group, or none. Lacking hydrophilic groups for recognition, they usually bind in a hydrophobic pocket. In contrast, the substrates of extradiol/intradiol dioxygenases contain at least two hydrophilic groups that are necessary for substrate binding near the nonheme iron.
Salicylate 5-monooxygenase NagGH in this study is a novel and rare RO which catalyzes the mono-hydroxylation as a monooxygenase does but acts on a substrate salicylate that contains two hydrophilic groups for recognition, as is found for extradiol/intradiol dioxygenases (22). In addition, ROs have been reported with the activities of monooxygenases whose substrates usually contain one hydrophilic group, different from the substrates of salicylate 5-monooxygenases as well as extradiol/intradiol dioxygenases. Different substrates of ROs, including NDO9816-4 and BPDOLB400, were observed to bind in similar pockets, and their substrate-binding orientations were proposed to determine the substrate selectivity and regio-specificity of product formation. Though salicylate 5-monooxygenase has been discovered for two decades, its structural and enzymatic mechanisms still remain elusive. Its unique properties have led us to investigate how salicylate 5-hydroxylase recognizes the substrate and how the regio-specificity of product is determined. The structure revealed in this study provides an opportunity to address these questions.
The structure of the large subunit NagG is similar to those of large subunits of previously reported NDO9816-4 and BPDOLB400, since it shares 31% and 30% sequence identities with NDO9816-4 and BPDOLB400, respectively. As expected, a Rieske type iron-sulfur cluster of one NagG molecule and a nonheme mononuclear iron of another adjacent subunit in the NagG trimer assemble a typical RO catalytic motif. Interestingly, the structure of the small subunit NagH is also similar to other small subunits, although NagH and those small subunits do not share obvious sequence similarity. As a result, the architecture of NagGH is the same as those of NDO9816-4 and BPDOLB400. This fact suggests that structure folding of the small subunit is more essential for enzymatic function than its protein sequence. Under this scenario, NagH probably plays an assistance role for the enzymatic activity of NagG. It is proposed that NagH can maintain the overall structure and oligomeric organization of NagGH, making sure the Rieske cluster of one protomer is close to the nonheme iron center of another protomer, thus maintaining the entire catalytic center. It can then recruit reductase NagAa and ferridoxin NagAb to reduce the oxidized NagGH, thus keeping the enzyme always in an active form.
The most significant structural difference between NagGH and previously reported ROs comes from the substrate-binding pocket, in line with their different substrates. Other ROs usually target hydrophobic substrates, such as biphenyl (6), indole (26), and naphthalene (27), and have hydrophobic substrate-binding pockets to accommodate these kinds of hydrophobic substrates. Meanwhile, NDO also acts on some substrates with some hydrophilic moieties, such as naphthoic acid (28) and 2-nitronaphthalene (29). In contrast, NagGH exhibits a substrate-binding pocket formed by both hydrophobic and hydrophilic residues and features a positively charged residue Arg323. The arrangement of this pocket may relate to the presence of a negatively charged carboxyl group and a hydrophobic phenyl group of salicylate.
Another difference between NagGH and most other known ROs is that the rearrangement of the nonheme iron triad caused by the substitution of conserved residues T212 in NDO9816-4 to Leu228 in NagGH. This rearrangement resulted in the shift of the iron center toward the substrate pocket accommodating naphthalene or indole in NDO9816-4, producing a narrowed pocket that could not accommodate salicylate and dioxygen simultaneously. Moreover, if NagGH binds to salicylate directly at the nonheme iron center, the 5′-carbon atom of salicylate will be too far away from the iron, making it difficult to donate an electron to dioxygen, and could not be attacked by the dioxygen bound to the iron. Thus, we prefer the hypothesis that NagGH binds to salicylate at a different site from other ROs, in agreement with the proposition in a recent study (30). The mutation of residue Ser367, which is located away from the corresponding substrate-binding pockets of NDO9816-4 and BPDOLB400, decreased the enzymatic activities and substrate-binding ability much more than the mutation of residue Asn218, suggesting that NagGH could not bind to salicylate via the similar pockets in NDO9816-4 and BPDOLB400. NagGH probably recognizes the substrate via hydrogen bonding, not via hydrophobic interaction as observed in NDO9816-4 and BPDOLB400. Considering the role of Ser367 and Arg323 observed in the mutagenesis assay, we proposed that salicylate binds to the featured pocket mainly via hydrogen bond interaction with residues Arg323 and Ser367. Regrettably, the essential role of Arg323 in protein folding makes it impossible to evaluate its role in substrate binding via mutagenesis assays. We therefore assessed the role of the carboxyl group of salicylate in substrate binding and found it is essential for binding to NagGH, which provided a clue that only the positively charged residue Arg323 at the potential pocket was involved in carboxyl group recognition. We compared the protein sequences of those ROs acting on substrates with one carboxyl group, such as anthranilate dioxygenase from Burkholderia cepacia DBO1 (17), terephthalate 1,2-dioxygenase from Comamonas sp. strain E6 (18), halobenzoate dioxygenase from P. aeruginosa 142 (16), ortho-halobenzoate 1,2-dioxygenase from Achromobacter xylosoxidans strain NH44784-1996 (6), and cinnamic acid dioxygenase from Comamonadaceae bacterium (7), and it is evident that those hydrophobic residues involved in pocket formation in NagGH are conserved as hydrophobic ones across its homologs (Fig. S4). Interestingly, Arg323 is conserved whereas Ser367 is not. The former was then proposed to directly recognize the carboxyl group in those ROs acting on a substrate with a carboxyl group, whereas the latter was proposed to only contribute to the substrate recognition in NagGH.
A model of how NagGH binds to salicylate is proposed here (Fig. 7). In this model, residue Arg323 interacts with the negatively charged carboxyl group of salicylate via hydrogen bonds, as well as electrostatic interaction. Two hydrogen bonds between the guanidine group and carboxyl group are proposed, resulting in the two groups located at the same plane. Ser367 forms a hydrogen bond with the hydroxyl group of salicylate to distinguish salicylate from those compounds without a hydroxyl group. Gln316 forms a hydrogen bond with one oxygen atom of the carboxyl group, assisting the anchoring of salicylate. Those hydrophobic residues are proposed to facilitate the entrance and binding of salicylate through its phenyl group.
FIG 7.
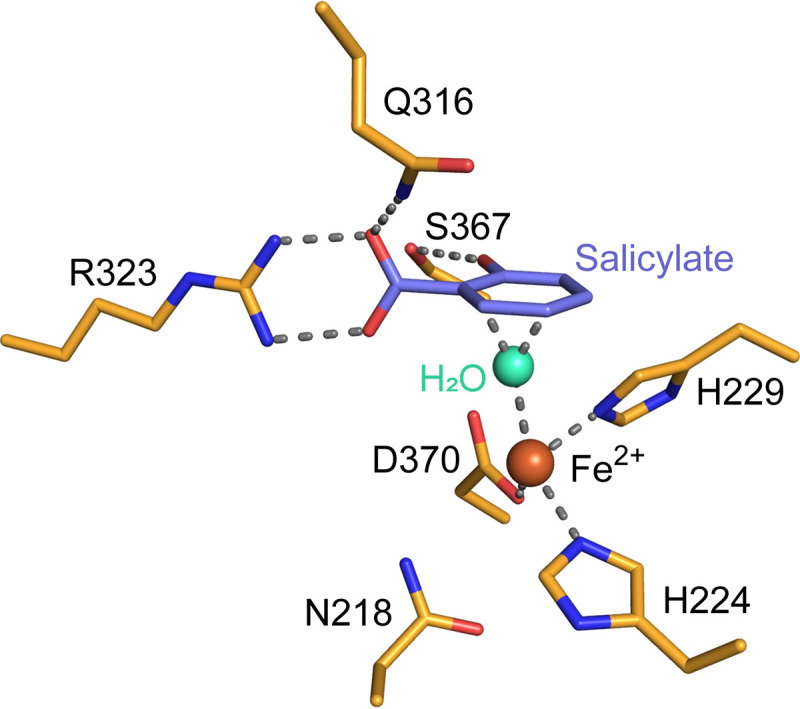
The proposed substrate-binding model of NagGH. The salicylate molecule is manually modeled and shown in blue. The nonheme iron atom and a water molecular in the active center are shown as orange and cyan spheres, respectively. The hydrogen bonds involved in binding salicylate and coordinating the iron atom are shown as dashed lines.
How the dioxygen molecule binds to the enzymes and is activated is a key for illustrating the enzymatic mechanism. A recent study described the dioxygen activation that followed the substrate binding (30). The authors proposed that an Fe (III)-superoxo species attacked the C5 atom of salicylate, with a similar mechanism described for the cis-dihydrodiol-forming Rieske dioxygenases (31, 32). In this case, a substrate radical-forming reaction was involved in the catalytic mechanism. In a previous study, of all the salicylate analogs tested (3-hydroxybenzoate, 4-hydroxybenzoate, 2,4-dihydroxybenzoate, 2,6-dihydroxybenzoate, 3,4-dihydroxybenzoate, catechol, 2-nitrobenzoate, 2-nitrophenol, 3-nitrophenol, and 4-nitrophenol), only three of them could be oxidized by NagGH (a salicylate structurally similar compound, 2-nitrophenol, and two substituted salicylates, 2,4-dihydroxybenzoate and 2,6-dihydroxybenzoate) (15). NagGH showed a 3-fold increase in Km for 2-nitrophenol over that for salicylate, which implied that in terms of substrate affinity, a carboxyl group was preferred over the nitro group, which may be related to the hydrogen bond interaction between Arg323 and substrates, whereas NagGH showed a slight increase in kcat for 2-nitrophenol as compared to salicylate. A nitro group can help stabilize the substrate radical intermediate, which may account for this phenomenon. In addition, the aforementioned substrate specificity of NagGH demonstrated its requirement for the presence of both a hydroxyl and its ortho-carboxy/carboxymethyl/nitro group, as indicated in our enzyme-substrate interaction model. NagGH showed a slightly lower Km value for 2,4-dihydroxybenzoate and a significant higher apparent Km for 2,6-dihydroxybenzoate than for salicylate, which indicated that an additional hydroxyl group was allowed but its location greatly affected the Km.
In summary, we reported the crystal structure of Rieske monooxygenase NagGH, revealed a rearrangement of the nonheme iron center, and identified the residues potentially essential for substrate binding. Together with the previous studies, it can be concluded that Rieske monooxygenases and dioxygenases bind to substrates in different manners, though they may recruit the same mechanism to oxygenate substrates. This study also deepened our understanding of a less-well-studied Rieske monooxygenase and may provide guidance in modification of the substrate specificity for these enzymes.
MATERIALS AND METHODS
Protein purification.
Genes nagAa, nagAb, and nagGH were amplified from Ralstonia sp. strain U2 and subcloned into pET28a(+) (Novagen, Madison, WI) to obtain pET28a-nagAa, pET28a-nagAb, and pET28a-nagGH, respectively. These plasmids encoding N-terminal His-tagged NagAa, NagAb, and NagGH were transformed into E. coli strain BL21(DE3) (Novagen, Madison, WI). The cells were cultured in LB medium supplied with additional 0.1 mM ferrous citrate for ∼2 to 3 h at 37°C. After 0.4 mM IPTG (isopropyl-β-D-thiogalactopyranoside) was added to the medium, the cells were cultured for another ∼4 to 5 h and then harvested. The pellet was resuspended in buffer containing 20 mM Tris-HCl (pH 8.0), 300 mM NaCl, 10 mM imidazole, and 5 mM β-mercaptoethanol and lysed by sonication. After centrifugation, the supernatant was loaded onto a Ni-NTA affinity column and the protein was eluted by buffer containing 250 mM imidazole. The protein was further purified using a Superdex200 gel filtration chromatography column (GE Healthcare Life Sciences, Pittsburgh, PA) and then ultrafiltered to the required concentration for subsequent catalytic analysis or crystallization (8 mg/ml).
Crystallization, data collection, and structure determination.
The crystal was obtained from the buffer containing 2% tascimate, 0.1 M imidazole (pH 7.0), and 10% (wt/vol) PEG3350 using the handing drop vapor diffusion method. The diffraction data were collected on the beamline BL17 at KEK photon factory. The data were integrated with XDS package (33) and scaled with Aimless (34). The phases were calculated by the Phenix via MR-SAD method using an NagG model that was built by Modeller (35) as the searching model. The model was automatically built by Phenix (36) and manually done in Coot (37) and refined by Phenix (36). The structural figures were prepared using PyMol (https://pymol.org).
Steady-state kinetics.
The NagAa, NagAb, and NagGH proteins used for enzyme assays were purified as described previously (15). The kinetic parameters of salicylate 5-monooxygenase and its mutants were measured with some modifications as follows. Enzyme activity was measured by monitoring NADH oxidation at 340 nm and the molar extinction coefficient for NADH was taken as 6.220 cm−1 · M−1. All assays were routinely measured in 50 mM 2-N-morpholinoethanesulfonic acid buffer (MES) (pH 6.4) at an ambient temperature. Each reaction mixture contained 0.25 μM NagGH, 3.75 μM NagAb, 0.25 μM NagAa, 0.5 μM FAD, 0.3 mM NADH, 0.1 mM ferrous ammonium sulfate, and 0.5 mM salicylate in 50 mM MES buffer (pH 6.4). Sets of salicylate 5-monooxygenase assays were carried out using at least eight substrate concentrations between ∼10 to 500 μM. Kinetic data were evaluated by nonlinear regression analysis with the Michaelis–Menten equation (υ = Vmax × [S]/[Km+[S]]), using the Windows-based prism software. The catalytic constant kcat was calculated using the equation Vmax = kcat × [E], where [E] = total enzyme concentration.
Differential scanning fluorimetry.
Differential scanning fluorimetry (DSF) experiments were carried out using 0.1 mg/ml protein in 20 mM Tris pH 8.0, 150 mM NaCl with Sypro Orange (5× final concentration, Invitrogen catalog number S6650). The substrates were dissolved in dimethyl sulfoxide (DMSO) with the concentration of 100 mM, and diluted to the final concentration of 1 mM in the reactions. The mixture was monitored in hard-shell 96-well skirted PCR plates (Bio-Rad, catalog number HSP-9601) using a 40 μl volume and a Bio-Rad CFX96 RT-PCR system (excitation wavelength, 450 to 490 nm; emission wavelength, 560 to 590 nm).
Data availability.
The protein structure and associated diffraction data have been deposited in the Protein Data Bank under the accession number PDB 7C8Z.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by the National Key R&D Program of China (2016YFA0601102 and 2019YFA0905500), the Science and Technology Commission of Shanghai Municipality (17JC1403300), and the National Natural Science Foundation of China (31700661).
We declare no conflicts of interest with the contents of this article.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Zhou NY, Al-Dulayymi J, Baird MS, Williams PA. 2002. Salicylate 5-hydroxylase from Ralstonia sp. strain U2: a monooxygenase with close relationships to and shared electron transport proteins with naphthalene dioxygenase. J Bacteriol 184:1547–1555. doi: 10.1128/jb.184.6.1547-1555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jouanneau Y, Micoud J, Meyer C. 2007. Purification and characterization of a three-component salicylate 1-hydroxylase from Sphingomonas sp. strain CHY-1. Appl Environ Microbiol 73:7515–7521. doi: 10.1128/AEM.01519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martins BM, Svetlitchnaia T, Dobbek H. 2005. 2-Oxoquinoline 8-monooxygenase oxygenase component: active site modulation by Rieske-[2Fe-2S] center oxidation/reduction. Structure 13:817–824. doi: 10.1016/j.str.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Capyk JK, D'Angelo I, Strynadka NC, Eltis LD. 2009. Characterization of 3-ketosteroid 9α-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis. J Biol Chem 284:9937–9946. doi: 10.1074/jbc.M900719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derek R B, Narain D S, Paul J S, Jagdeep C, David J G, Howard D. 1991. Bacterial oxidation of benzocyloalkenes to yield monol, diol and triol metabolites. Tetrahedron Lett 32:3887–3890. doi: 10.1016/S0040-4039(00)79405-6. [DOI] [Google Scholar]
- 6.Resnick SM, Lee K, Gibson DT. 1996. Diverse reactions catalyzed by naphthalene dioxygenase from Pseudomonas sp strain NCIB 9816. J Industrial Microbiology 17:438–457. doi: 10.1007/BF01574775. [DOI] [Google Scholar]
- 7.Lessner DJ, Johnson GR, Parales RE, Spain JC, Gibson DT. 2002. Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl Environ Microbiol 68:634–641. doi: 10.1128/aem.68.2.634-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friemann R, Ivkovic-Jensen MM, Lessner DJ, Yu CL, Gibson DT, Parales RE, Eklund H, Ramaswamy S. 2005. Structural insight into the dioxygenation of nitroarene compounds: the crystal structure of nitrobenzene dioxygenase. J Mol Biol 348:1139–1151. doi: 10.1016/j.jmb.2005.03.052. [DOI] [PubMed] [Google Scholar]
- 9.Ju KS, Parales RE. 2006. Control of substrate specificity by active-site residues in nitrobenzene dioxygenase. Appl Environ Microbiol 72:1817–1824. doi: 10.1128/AEM.72.3.1817-1824.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferraro DJ, Gakhar L, Ramaswamy S. 2005. Rieske business: structure-function of Rieske non-heme oxygenases. Biochem Biophys Res Commun 338:175–190. doi: 10.1016/j.bbrc.2005.08.222. [DOI] [PubMed] [Google Scholar]
- 11.Cho O, Choi KY, Zylstra GJ, Kim YS, Kim SK, Lee JH, Sohn HY, Kwon GS, Kim YM, Kim E. 2005. Catabolic role of a three-component salicylate oxygenase from Sphingomonas yanoikuyae B1 in polycyclic aromatic hydrocarbon degradation. Biochem Biophys Res Commun 327:656–662. doi: 10.1016/j.bbrc.2004.12.060. [DOI] [PubMed] [Google Scholar]
- 12.Costa DMA, Gómez SV, de Araújo SS, Pereira MS, Alves RB, Favaro DC, Hengge AC, Nagem RAP, Brandão TAS. 2019. Catalytic mechanism for the conversion of salicylate into catechol by the flavin-dependent monooxygenase salicylate hydroxylase. Int J Biol Macromol 129:588–600. doi: 10.1016/j.ijbiomac.2019.01.135. [DOI] [PubMed] [Google Scholar]
- 13.Izmalkova TY, Sazonova OI, Nagornih MO, Sokolov SL, Kosheleva IA, Boronin AM. 2013. The organization of naphthalene degradation genes in Pseudomonas putida strain AK5. Res Microbiol 164:244–253. doi: 10.1016/j.resmic.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Zhou NY, Fuenmayor SL, Williams PA. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J Bacteriol 183:700–708. doi: 10.1128/JB.183.2.700-708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang T, Zhou NY. 2014. Purification and characterization of salicylate 5-hydroxylase, a three-component monooxygenase from Ralstonia sp. strain U2. Appl Microbiol Biotechnol 98:671–679. doi: 10.1007/s00253-013-4914-x. [DOI] [PubMed] [Google Scholar]
- 16.Tsoi TV, Plotnikova EG, Cole JR, Guerin WF, Bagdasarian M, Tiedje JM. 1999. Cloning, expression, and nucleotide sequence of the Pseudomonas aeruginosa 142 ohb genes coding for oxygenolytic ortho dehalogenation of halobenzoates. Appl Environ Microbiol 65:2151–2162. doi: 10.1128/AEM.65.5.2151-2162.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang HK, Mohseni P, Zylstra GJ. 2003. Characterization and regulation of the genes for a novel anthranilate 1,2-dioxygenase from Burkholderia cepacia DBO1. J Bacteriol 185:5871–5881. doi: 10.1128/jb.185.19.5871-5881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasoh M, Masai E, Ishibashi S, Hara H, Kamimura N, Miyauchi K, Fukuda M. 2006. Characterization of the terephthalate degradation genes of Comamonas sp. strain E6. Appl Environ Microbiol 72:1825–1832. doi: 10.1128/AEM.72.3.1825-1832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kauppi B, Lee K, Carredano E, Parales RE, Gibson DT, Eklund H, Ramaswamy S. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 20.Karlsson A, Parales JV, Parales RE, Gibson DT, Eklund H, Ramaswamy S. 2003. Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron. Science 299:1039–1042. doi: 10.1126/science.1078020. [DOI] [PubMed] [Google Scholar]
- 21.Dhindwal S, Gomez-Gil L, Neau DB, Pham TT, Sylvestre M, Eltis LD, Bolin JT, Kumar P. 2016. Structural basis of the enhanced pollutant-degrading capabilities of an engineered biphenyl dioxygenase. J Bacteriol 198:1499–1512. doi: 10.1128/JB.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murzin AG, Brenner SE, Hubbard T, Chothia C. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 23.Furusawa Y, Nagarajan V, Tanokura M, Masai E, Fukuda M, Senda T. 2004. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J Mol Biol 342:1041–1052. doi: 10.1016/j.jmb.2004.07.062. [DOI] [PubMed] [Google Scholar]
- 24.Dong X, Fushinobu S, Fukuda E, Terada T, Nakamura S, Shimizu K, Nojiri H, Omori T, Shoun H, Wakagi T. 2005. Crystal structure of the terminal oxygenase component of cumene dioxygenase from Pseudomonas fluorescens IP01. J Bacteriol 187:2483–2490. doi: 10.1128/JB.187.7.2483-2490.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Li J, Liu A. 2017. Oxygen activation by mononuclear nonheme iron dioxygenases involved in the degradation of aromatics. J Biol Inorg Chem 22:395–405. doi: 10.1007/s00775-017-1436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allen CCR, Boyd DR, Dalton H, Sharma ND, Brannigan I, Kerley NA, Sheldrake GN, Taylor SC. 1995. ChemInform abstract: enantioselective bacterial biotransformation routes to cis-diol metabolites of monosubstituted benzenes, naphthalene and benzocycloalkenes of either absolute configuration. ChemInform 26. doi: 10.1002/chin.199524037. [DOI] [Google Scholar]
- 27.Jeffrey AM, Yeh HJ, Jerina DM, Patel TR, Davey JF, Gibson DT. 1975. Initial reactions in the oxidation of naphthalene by Pseudomonas putida. Biochemistry 14:575–584. doi: 10.1021/bi00674a018. [DOI] [PubMed] [Google Scholar]
- 28.Knackmuss HJ, Beckmann W, Otting W. 1976. Microbiological synthesis of (+)-cis-1,2-dihydroxy-1,2-dihydronaphthalene-2-carboxylic acid. Angew Chem Int Ed Engl 15:549. doi: 10.1002/anie.197605491. [DOI] [PubMed] [Google Scholar]
- 29.Mahajan MC, Phale PS, Vaidyanathan CS. 1994. Evidence for the involvement of multiple pathways in the biodegradation of 1- and 2-methylnaphthalene by Pseudomonas putida CSV86. Arch Microbiol 161:425–433. doi: 10.1007/BF00288954. [DOI] [PubMed] [Google Scholar]
- 30.Rogers MS, Lipscomb JD. 2019. Salicylate 5-hydroxylase: intermediates in aromatic hydroxylation by a Rieske monooxygenase. Biochemistry 58:5305–5319. doi: 10.1021/acs.biochem.9b00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivard BS, Rogers MS, Marell DJ, Neibergall MB, Chakrabarty S, Cramer CJ, Lipscomb JD. 2015. Rate-determining attack on substrate precedes Rieske cluster oxidation during cis-dihydroxylation by benzoate dioxygenase. Biochemistry 54:4652–4664. doi: 10.1021/acs.biochem.5b00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutherlin KD, Rivard BS, Böttger LH, Liu LV, Rogers MS, Srnec M, Park K, Yoda Y, Kitao S, Kobayashi Y, Saito M, Seto M, Hu M, Zhao J, Lipscomb JD, Solomon EI. 2018. NRVS studies of the peroxide shunt intermediate in a Rieske dioxygenase and its relation to the native FeII O2 reaction. J Am Chem Soc 140:5544–5559. doi: 10.1021/jacs.8b01822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webb B, Sali A. 2016. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The protein structure and associated diffraction data have been deposited in the Protein Data Bank under the accession number PDB 7C8Z.