The human colon contains a community of microbial species, mostly bacteria, which is often referred to as the gut microbiota. The community is considered essential to human well-being by conferring additional energy-harvesting capacity, niche exclusion of pathogens, and molecular signaling activities that are integrated into human physiological processes.
KEYWORDS: gut microbiota, plant glycans
ABSTRACT
The human colon contains a community of microbial species, mostly bacteria, which is often referred to as the gut microbiota. The community is considered essential to human well-being by conferring additional energy-harvesting capacity, niche exclusion of pathogens, and molecular signaling activities that are integrated into human physiological processes. Plant polysaccharides (glycans, dietary fiber) are an important source of carbon and energy that supports the maintenance and functioning of the gut microbiota. Therefore, the daily quantity and quality of plant glycans consumed by the human host have the potential to influence health. Members of the gut microbiota differ in ability to utilize different types of plant glycans. Dietary interventions with specific glycans could modulate the microbiota, counteracting ecological perturbations that disrupt the intricate relationships between microbiota and host (dysbiosis). This review considers prospects and research options for modulation of the gut microbiota by the formulation of diets that, when consumed habitually, would correct dysbiosis by building diverse consortia that boost functional resilience. Traditional “prebiotics” favor bifidobacteria and lactobacilli, whereas dietary mixtures of plant glycans that are varied in chemical complexity would promote high-diversity microbiotas. It is concluded that research should aim at improving knowledge of bacterial consortia that, through shared nourishment, degrade and ferment plant glycans. The consortia may vary in composition from person to person, but functional outputs will be consistent in a given context because of metabolic redundancy among bacteria. Thus, the individuality of gut microbiotas could be encompassed, functional resilience encouraged, and correction of dysbiosis achieved.
INTRODUCTION
WHY MODULATE THE GUT MICROBIOTA OF HUMANS?
A microbial community largely composed of bacterial species, often referred to as the gut microbiota, inhabits the human colon (1). Early in life, when milk (especially human breast milk) is the sole source of nutrients for the infant host, the gut microbiota has a relatively simple taxonomic composition (low alpha-diversity), consisting predominantly of bifidobacterial species (2). Assembly of a much more complex microbiota occurs once solid (weaning, complementary) foods are introduced into the diet from 4 to 6 months of age. Plant glycans in fruit and vegetables that are indigestible by human processes in the gut pass to the colon, where they become available as growth substrates for bacteria (3, 4). Plant glycans are included in the category of dietary components known as dietary fiber (Text Box 1) (5). Ingestion of increasing amounts of plant glycans during the first year of life is accompanied by increasing diversity of bacterial species until, by about 12 months of age, a gut microbiota with many similarities to that of adults has developed (6, 7). Members of the bacterial families Lachnospiraceae and Ruminococcaceae, as well as Bacteroidaceae, are then the predominant taxa within the microbiota in terms of relative abundances (8). However, a mature microbiota in which characteristic taxa are maintained temporally in relatively constant proportions may not be reached until children are 3 to 4 years of age or even older (9, 10). All of these developments are subject to influences associated with mode of delivery at birth, ethnicity, microbial dispersion, antibiotic administration, human genetics, and nourishment (11–17). It is nevertheless clear that the formation of the natural assemblage of bacterial species is influenced by two major factors, one exogenous, i.e., host diet components (such as plant glycans), and the other endogenous, i.e., host secretions (such as glycans associated with mucus) (3, 18). Practical manipulation of mucus production is likely to be difficult if not impossible to achieve, but the amount and kinds of plant glycans (dietary fiber) delivered to the colon can be controlled by dietary intake. Thus, dietary intervention with plant glycans provides a means of modulating the gut microbiota of adult humans.
TEXT BOX 1.
TEXT BOX 1 CODEX definition of dietary fiber (5)
Dietary fiber means carbohydrate polymersa with 10 or more monometric unitsb which are not hydrolyzed by the endogenous enzymes in the small intestine of humans and belong to the following categories:
-
1.
Edible carbohydrate polymers naturally occurring in the food as consumed.
-
2.
Carbohydrate polymers, which have been obtained from food raw material by physical, enzymatic, or chemical means and which have been shown to have a physiological effect of benefit to health, as demonstrated by generally accepted scientific evidence to competent authorities.
-
3.
Synthetic carbohydrate polymers, which have been shown to have a physiological effect of benefit to health, as demonstrated by generally accepted scientific evidence to competent authorities.
aWhen derived from a plant origin, dietary fiber may include fractions from lignan and/or other compounds associated with polysaccharides in the plant cell walls. These compounds also may be measured by a certain analytical method(s) for dietary fiber.
bDecision on whether to include carbohydrates of 3 to 9 monomeric units should be left up to national authorities.
Modulation (exerting a controlling influence) of the microbiota in a targeted manner might be useful in the treatment or prevention of human diseases and medical conditions. Although the rudiments of gut microbiota composition and its principal emergent properties were known long ago, DNA-based technology has provided detailed information about the taxonomic composition and functional capacity of the gut microbiota (19, 20). There is evidence from such studies that microbiota composition is altered in association with certain human diseases and conditions in comparison to the microbiota of well people (21–49). This evidence of “dysbiosis” (taxa imbalance; community perturbation leading to disturbance of microbe/host relationship) in sick humans has understandably become a major driving force for gut microbiota research. Although most evidence of dysbiosis is based on statistical correlation, and accepting that attempts at testing Koch’s postulates using human microbiota rodents may be flawed (49, 50), investigations into the modulation of the gut microbiota using plant glycans nevertheless remain a valid goal in gut microbiota research due to the need to alleviate the enormous burden placed on health services by noncommunicable diseases.
WHAT ARE PLANT GLYCANS?
Plant cells contain polymeric compounds that consist of a large number of monosaccharides with glycosidic linkages: plant glycans. Two groups of glycans (starches and hemicelluloses) that comprise dietary fiber have received particular attention in the case of the human gut microbiota (8). Starch is an energy storage polymer produced by many plants and consists of two types of molecules: linear and helical amylose and branched amylopectin. The proportions of amylose and amylopectin in starch are variable according to plant source. Amylose is more resistant to digestion by human amylase than is amylopectin. Moreover, starch may be inaccessible to digestion if trapped within seeds and grains. When cooked, starch undergoes gelatinization, in which the crystal structure is broken down and the molecules become accessible to amylase. However, cooling, drying, and freezing starch cause changes in starch structure (retrogradation), which transforms it into an indigestible form (Table 1) (51–53). The second major group of plant glycans, the hemicelluloses, provides the matrix in which cellulose fibrils are embedded within the plant cell wall (Table 1). They include mannans, xylans, arabinogalactans, glucans, and pectins that vary greatly in chemical composition and structure, even within specific glycan groups when they are derived from different plant sources. For example, xylans consist of a xylopyranose backbone onto which methylglucuronic acid groups, O-acetyl groups, or sugars such as arabinose can be substituted in greater or lesser complexity. Ferulic acid can be attached to these arabinose molecules (54). Arabinoxylans have backbones substituted in places with arabinose and are common in cereal grains (wheat, barley, maize, rice). They form part of the diet of humans worldwide. Hence, there has been considerable interest in the microbiota response to ingesting arabinoxylan oligosaccharides (AXOS) derived by hydrolysis of arabinoxylans (55–62). A consistent finding is that short-chain fatty acid (SCFA) (usually butyrate) concentrations rise during consumption of arabinoxylan/AXOS-supplemented diets, but the increases are small and individualistic (55, 63). Since therapeutic or prophylactic levels of butyrate have not been established, the significance of these augmentations to human health is unknown, but they show promise because they indicate that modulating the metabolic output of the microbiota is possible.
TABLE 1.
Examples of starch and nonstarch plant glycans
| Plant glycan | Descriptiona |
|---|---|
| Cellulose | A linear chain of several hundred to many thousands of β(1→4)-linked d-glucose units. |
| Hemicelluloses | Heteropolymers, such as arabinoxylans, mixed-link (β1-3, 1-4) glucans, and glucomannan. |
| Pectin | A complex polysaccharide rich in galacturonic acid and composed of heterogeneous branched components such as HG, RGI, and RGII. HG is a linear polymer of (1–4)-linked α-d-GalpA. RGI consists of a repeating disaccharide [-4)-α-d-GalpA-(1–2)-α-l-Rhap-(1-] with arabinan, galactan, and/or arabinogalactans attached to rhamnose residues. RGII has a backbone of HG to which are attached complex side chains (51). Pectins represent the major “soluble” fiber in many fruits and vegetables. |
| Starch | Two forms of glucose polymers—amylose (linear, helical) and amylopectin (branched). Some forms of starch are more resistant to hydrolysis by human pancreatic amylase: RS1, physically inaccessible, such as that trapped in partly milled grains and seeds; RS2, native granular structure (β-crystalline), such as that in raw potato and banana; RS3, retrograded due to cooking and cooling, which produces new crystalline matrices (potato, bread, cornflakes); RS4, starch that has been chemically modified to resist digestion through chemical cross-linking; RS5, amylose-lipid complexes, slow digestion, not true resistant starch (52, 53). |
HG, homogalacturonan; RG, rhamnogalacturonan; RS, resistant starch.
The chemical diversity that exists among plant glycans has the potential to differentially influence particular attributes of the microbiota, because only bacteria that possess the appropriate, specialized biochemical machinery will metabolize them. If all other growth-limiting nutrients are available, these species will replicate and perhaps have increased cell numbers and/or metabolic output. Earlier analyses of genomic data of cultured members of the gut microbiota, as well as metagenomic investigations of whole communities, have revealed biochemical pathways by which the bacteria can degrade a wide variety of plant glycans (64, 65). Genomic and biochemical investigations of cultured members of the gut microbiota have revealed molecular details of how the specialized bacteria degrade plant glycans for growth (66–69). In general, they comprise highly specific, colocalized, coregulated complexes of proteins at cell surfaces that recognize, sequester, and degrade glycans and transfer hydrolytic products resulting from their degradation into the bacterial cell (Table 2).
TABLE 2.
Examples of molecular modules associated with plant glycan degradation by human gut bacteriaa
| Metabolic module | Components | Reference(s) |
|---|---|---|
| Polysaccharide utilization loci (PULs; e.g., Bacteroides ovatus; Roseburia/Eubacterium rectale group [gpPULs]) | Genetic loci that encode proteins for the highly specific capture, degradation, and importation of specific glycans. Each kind of PUL targets a different glycan structure. The archetypal PUL is the starch utilization system (Sus) that contains eight colocalized and coregulated genes. gpPULs are not close homologs of Bacteroides PULs and vary between Firmicutes in terms of specific glycosyl hydrolase genes. | 66–70 |
| Cellulosome (e.g., Ruminococcus champanellensis) and amylosome (e.g., Ruminococcus bromii) | Complexes of molecules that bring hydrolytic enzymes and carbohydrate domains in substrates together at the cell surface, facilitated by dockerins and cohesins. The amylosome is simpler than a cellulosome but is associated with the exceptional ability of bacteria to degrade particulate-resistant starches. | 71–74 |
| Cell surface-associated complex (e.g., Monoglobus pectinilyticus) | A combination of enzymes that degrade pectin. Esterases remove methyl and acetyl groups from pectins, allowing pectic lyases access to HG and RG backbones. S-layer proteins may anchor enzymes to the cell surface. | 75 |
| Xylan degradation genes (e.g., Bacteroides ovatus, Bacteroides xylanisolvens) | Utilization of xylan by B. ovatus and B. xylanisolvens is associated with one large PUL and one small PUL (remnant) that are transcriptionally linked. The small PUL is linked to the utilization of xylans of simple composition, whereas the large PUL is associated with the degradation of structurally complex xylans. A key feature of xylan degradation appears to be endo-1,4-β-xylanases belonging to glycosyl hydrolase family 10. Of the Bacteroidetes, only B. ovatus and B. xylanisolvens have these PULs. | 76–79 |
gpPULs, Gram-positive polysaccharide utilization loci; HG, homogalacturonan; RG, rhamnogalacturonan.
Plant glycans might be said to belong to the category of food supplements known as “prebiotics,” i.e., “a substrate that is selectively utilized by host microorganisms conferring a health benefit” (80). However, “prebiotic” commonly refers to inulin, fructooligosaccharides (FOS), and galactooligosaccharides (GOS), which are used as food additives with the aim of producing an augmentation of the numbers of bifidobacteria and lactic acid bacteria in the gut. The use of glycans with greater structural complexity is more likely to target the bacterial species that are characteristically members of the normally predominant families of bacteria of the mature gut microbiota of adult humans. Therefore, the FOS (derived from hydrolysis of inulin or by chemical synthesis) and galactose-containing substances (GOS; synthesized from lactose), although widely used in the food industry, will not be considered further in this review. There are already many published articles contributing information on this topic (81–87).
IS THERE EVIDENCE FOR DIFFERENTIAL USE OF PLANT GLYCANS BY GUT BACTERIA?
A differential effect on the members of the microbiota is implicit in the concept of modulation using dietary fiber. Preferential utilization of particular glycans by different bacterial taxa is the specific basis of the modulatory effect. Culture-based studies confirm the evidence from genetic studies that specialization of substrate utilization by gut bacteria occurs, even to the level of species and strains (77, 88–93). Much of this work relates to commonly detected members of the gut microbiota that belong to the genus Bacteroides, which can serve as an exemplar. In general, Bacteroides species have relatively large genomes that encode numerous carbohydrate-active enzymes (CAZymes) conferring the ability of the bacteria to synthesize, recognize, or metabolize complex carbohydrates (94). The results of laboratory studies with cultured Bacteroides species show, indeed, that the species differ in ability to use particular glycans as growth substrates (for example, barley β-d-glucan) (Fig. 1A). Biochemically complex molecules, such as flax xylan, narrow the number of species that, in pure culture, can utilize the substrate (Fig. 1B). In some instances, clustering of species according to growth in medium containing pectic substrates occurs (Fig. 1C), indicating that several species can potentially occupy the same fundamental niche in the gut community while others have unique roles. Similar growth profiles may reflect interchangeable members of bacterial consortia involved in the degradation and fermentation of specific glycans. Additionally, individual species have preferences to degrade particular regions of complex glycans. For example, several Bacteroides species can grow using the homogalacturonan backbone of pectins, but relatively few can utilize the rhamnogalacturonan portion and only after the removal of arabinan, galactan, and arabinogalactan side chains by other bacteria. Utilization of these sugars for growth is also differential among Bacteroides ovatus, Bacteroides finegoldii, Bacteroides cellulosyliticus, and Bacteroides intestinalis (91). Moreover, bacteria prioritize glycans as to order of utilization for growth. Bacteroides ovatus, for example, utilizes less complex, smaller polysaccharides before using branched, higher-molecular-weight polymers (approximate order, β-glucan, pectin, xyloglucan, and arabinoxylan [100, ∼300, 1,028, and 232 kDa, respectively]) when presented with a mixture of these substrates (92, 95–97). Bacteroides thetaiotaomicron, when provided with a mixture of 12 glycans, prioritizes the use of some over others, as evidenced by measurement of the transcription of polysaccharide utilization loci (PULs) (98). Chemical linkages in glycans also influence the specificity of utilization. For example, the specific import of β2-6 fructan, relative to β2-1 fructan, into the periplasm of Bacteroides thetaiotaomicron is determined by cell surface-associated degradation and binding machinery (99). The structural complexity of the molecules and the corresponding need for coordinated actions of several enzymes in the degradation of substrates are important determinants of prioritization (100). The human host is likely to consume a mixture of glycans from different plant sources (fruits and vegetables) each day, so knowledge of how the bacterial taxa react to this in terms of the prioritization of use of specific glycans, and the extent to which each is used, will be critical to the success of efforts to modulate the gut microbiota. For example, testing taxa to utilize specific plant glycans relies on the culture of fastidious bacteria under laboratory conditions, a skill that needs to be encouraged despite an emphasis in recent decades on culture-independent (nucleic acid-based) studies of the gut microbiota. Metagenomic studies indicate that a large proportion of the taxa detected in gut microbiota studies do not yet have a cultivated representative, but a proportion of these genomes may originate in gut transients, not inhabitants (101). However, the common, predominant members of the microbiota have been cultivated and are extant in culture collections and hence available for use in screening complex glycans as growth substrates (102, 103). Pure culture studies are a starting point, but multiplex cocultures will be necessary to reveal the interactive networks by which complex substrates are degraded by consortia of bacterial species. Thus, trophic patterns need to be elucidated and used in the design of food interventions in human studies. Continuous cocultures in chemostats, and in multistage fermentors simulating the gut, will provide useful knowledge in this respect, because the bacteria can be maintained as stable populations (87, 104–108). Chemostats provide steady-state conditions (growth occurs at a constant rate and in a constant environment) under which all of the bacterial cells have a similar metabolic state under conditions of carbon and energy limitation. These conditions relate to bacterial growth in the colon, where competition for growth substrates is intense due to the large, complex microbial community. This is apparent by observation of the reduced breadth of realized niches (actual activity in the community) relative to that of fundamental niches (potential capacity measured in vitro) of gut bacteria (109). However, the colon is probably not exactly a continuous-culture system, because the digesta passes through the gut in boluses and the ileocecal valve controls passage of digesta into the large bowel. Hence, the colon environment may provide a mixture of batch and continuous culture conditions that are difficult to replicate in vitro.
FIG 1.
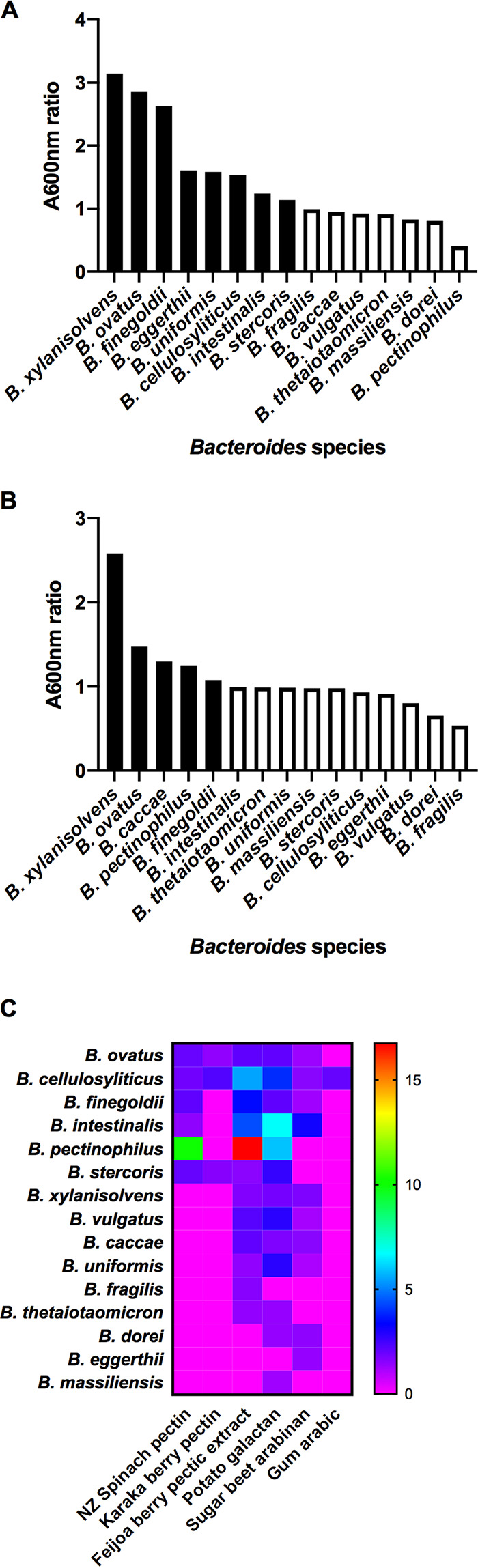
Differential utilization of plant glycans by Bacteroides species. (A) Barley β-glucan as a growth substrate; (B) New Zealand flax xylan as a growth substrate; (C) pectic polysaccharides as growth substrates. (A and B) Mean A600 ratios after 24 h of incubation of cultures of Bacteroides species in basal medium containing plant glycans (0.2%, wt/vol) relative to basal medium. Significantly different values (P < 0.05) from controls are shown as black bars. (C) Significantly (P < 0.05) augmented growth relative to the control (A600 ratios) in the form of a heat map. Nonsignificant differences in growth are plotted as zero. Data are taken from references 89, 91, and 92.
It will be important to recognize and understand the functioning of bacterial consortia associated with the degradation and fermentation of specific plant glycans. Dietary intervention with plant glycans will be most likely to modulate interactive bacterial species rather than individual target species that act alone. Microbial consortia can be thought of as clusters of taxa that cooperate metabolically to degrade and ferment complex substrates. These consortia are based on shared nourishment of the members (syntrophy). Polysaccharide backbones of complex substrates may be prepared for hydrolysis by removal of sugars linked to the polymer. Other species capable of hydrolyzing the substrate (all or in part) “leak” potential nutrients to taxa that, themselves, cannot degrade the intact substrate. It is assumed that there will be a quid pro quo in that the nonhydrolytic members of the consortium will help provide an environment that enhances life for the whole entity (19). For example, this could be by production of growth-limiting nutrients, such as vitamins (92, 110–112). Metabolic cross-feeding of hydrogen (interspecies hydrogen transfer) is likely to be important because maintenance of a low partial pressure of hydrogen thermodynamically favors reoxidation of NADH resulting from fermentative reactions. This allows even more fermentation, and therefore increased growth of saccharolytic bacteria, to occur (113). Acetogens, methanogens, and sulfate-reducing bacteria are important hydrogenotrophs, even though they are present at relatively low abundances in the human colon (114). Metabolic cross-feeding of fermentation acids is also notable in gut microbiotas (115, 116). For example, succinate produced by Bacteroides species is then transformed into propionate by Veillonella parvula (97, 117). It will therefore be important to identify robust clusters of taxa in metagenomic studies of the microbiota in assessing the modulatory effects of dietary modification.
CHOOSING THE RIGHT DIETARY FIBER TO MODULATE THE GUT MICROBIOTA
Modulation of the gut microbiota presupposes that suitable choices of glycans that have differential effects can be found and then administered in conjunction with knowledge of what is normally consumed. Food frequency questionnaires (FFQs) are widely used in nutritional studies to gain insight into the variety of food groups and associated nutrients that are consumed over a prescribed period, usually 7 to 30 days (118). FFQs collect information on the frequency of consumption of foods but usually not on the quantity consumed. When FFQs do include questions about quantity, quantity is typically based on standard portion sizes rather than direct weight or use of household utensils. A drawback is that FFQs rely on recall of what was eaten, and recall bias is likely to increase with longer periods of recall (119). The weighed food record (WFR) provides the most accurate information about dietary intake (120–122). It requires that all food and beverage portions be weighed at the time of consumption; plate waste is recorded, along with a description of the food, preparation methods, and brand names. The WFR can be used to validate the accuracy of an FFQ. Information from FFQs and/or WFRs enables habitual dietary patterns to be constructed, and individuals can be graded according to adherence/nonadherence to a pattern (123). Dietary patterns have been used to investigate associations between diet and disease and should be of increasing benefit in understanding dietary impacts on the maturation of the gut microbiota (124, 125). Taken a step further, measurement of adherence to dietary patterns in association with microbiota data might reveal deficits in specific plant glycan intake, which could be rectified by dietary correction of natural food intake or the use of diet supplements. This assumes that nutritional analysis is equal to the task; there are concerns about the estimation of specific kinds of dietary fiber. Definitions of food categories change, and the kinds of technical assays that are used in analysis affect the completeness and accuracy of food composition databases (126, 127). Once candidate dietary glycans are identified, procedures to achieve increased consumption of specific types of dietary fiber in attractive and palatable ways will need to be devised with consideration of daily caloric intake. Dose-response studies will be needed to determine and standardize efficacy. Large doses of complex glycans, relative to current intakes, may be required for efficacy, and this could repel consumers in Western countries, perhaps even globally, who need considerable persuasion to consume even 15 to 20 g of dietary fiber per day (128, 129). While laboratory culture studies use purified substrates extracted from plants, substances used in interventions may require cooking, which can alter chemical structures and hence might affect efficacy in the gut (130). Therefore, it will be wise to test processed dietary fiber, as well as “raw” ingredients, in screening studies.
WHICH ASPECT OF THE GUT MICROBIOTA SHOULD WE MODULATE?
Thought needs to be given to which aspect of the gut microbiota will be used as a measure of efficacy. The concept of “dysbiosis” is broadly based on a perceived imbalance in the taxa comprising the microbiota. Therefore, taxonomic analysis would be appropriate from this point of view, and compositional results of dysbiotic and treated microbiotas could be compared. Methods for accomplishing this work are readily available, but in order to obtain reliable species-level data, metagenomic analysis will be required rather than only sequencing PCR amplicons of variable nucleotide regions of 16S rRNA genes. Indeed, given the biochemical variation of strains within species, much more in-depth analysis of the composition of microbiotas will be necessary (131–134). Identification of enterotypes (robust clusters of taxa) and construction of microbial (correlation) networks from sequencing data may facilitate understanding of community trophic structures. Networks show individual microbes as nodes (hub species) and species cooccurrence or mutual exclusion as feature-feature pairs (edges); an edge may imply a biological or biochemical relationship between features. Microbes that benefit each other may be positively correlated, whereas microbes that compete for the same niche may be negatively correlated (16, 135–138). Such kinds of analysis may reveal consortia of bacterial species involved in the utilization of particular substrates. The prevalence of biochemical pathways in microbiotas can be deduced from metagenomic data, but these data, which show only metabolic capacity, will not necessarily change as a result of diet interventions, given the stability of genotypes associated with “function” among individuals with microbiotas of markedly different taxonomic compositions (25). This stability reflects the metabolic redundancy characteristic of gut microbiotas in which the same biochemical function is exerted by more than one bacterial species (139, 140). Transcriptome studies might be useful in monitoring the impact of dietary intervention, because clues to the dynamics of intercommensal relationships can be obtained using transcriptomic analysis; it potentially “eavesdrops” on the molecular conversations of members of a community (92, 141, 142). The use of stable-isotope-labeled substrates to probe the microbial utilization of specific substances has also gained attention in microbial ecology and has potential in investigating the flow of carbon from plant carbohydrates to bacterial consortia in the gut. Stable-isotope probing (SIP) can provide insights into both microbial phylogeny and metabolic activity by tracking stable-isotope-labeled atoms from substrates to incorporation in bacterial 16S rRNA (RNA-SIP) (143–146). Technically simpler and less expensive options of monitoring could be directed toward the emergent properties of the microbiota. The most obvious are the proportions of SCFAs (principally acetate, propionate, and butyrate) produced by the microbiota as a result of the collective activities of the community in degrading and fermenting exogenous and endogenous substrates (147). SCFAs are ligands of receptors in the gut mucosa and, as signaling molecules, affect immune and physiological processes (such as satiety) (148–150). Measurement of other biochemical molecules that have the potential to signal in otherwise human processes (for example the gut-brain axis) may be even more rewarding (151–153).
EVIDENCE THAT THE GUT MICROBIOTA OF HUMANS RESPONDS TO DIETARY DIFFERENCES
Compelling evidence of dietary modulation of the gut microbiota comes from studies of the compositions and emergent properties of the gut community of nonwesternized humans in Africa compared to those of westerners. For example, children living in a rural village in Burkina Faso, Africa, eating food high in dietary fiber relative to the “modern Western diet” of urban Italian children had plentiful Bacteroidetes (57.7% compared to 22.4% relative abundance) in feces, whereas Firmicutes were depleted (27.3% versus 63.7%) (154). Members of the genera Prevotella and Xylanibacter that had the genetic capacity to hydrolyze cellulose and xylan were abundant in the African children’s microbiota but were lacking in the microbiota of Italian children. As a consequence of fiber degradation and fermentation, there was a higher concentration of SCFAs in the feces of Burkina Faso children. These observations of the fecal microbiotas of rural African children reflected consumption of a polysaccharide-rich diet from which they were able to achieve maximal energy harvest through bacterial fermentation.
Similarly, the Hadza, African hunter-gatherers, have a gut microbiota that has greater taxonomic diversity of bacteria than that of Italians (155, 156). The microbiota includes many Bacteroidetes, Clostridiales, Bacteroidales, and Lachnospiraceae that do not resemble previously detected families and genera. Moreover, the Hadza microbiota differs between men and women, reflecting the different diets of the groups stemming from different community roles: women tend to consume more dietary fiber than men. The dietary fiber is sourced especially from tubers, which is associated with increases in the relative abundance of spirochetes (Treponema) in the microbiotas of women. These bacteria are rare in Western microbiotas. Hadza men consume more meat while on hunting expeditions. Their microbiota has increased abundances of Eubacterium and Blautia relative to that of women. No such gender differentiation of Italian microbiotas was apparent. Emergent properties of the microbiotas differed between the Hadza and Italians, reflecting the relative amounts and types of dietary fiber that were consumed. Italians had more butyrate in the feces than the Hadza, who had more propionate. Lower concentrations of butyrate were associated with lower content of Clostridium clusters IV and XIVa (butyrate producers), whereas more propionate was associated with greater relative abundance of Prevotella.
These studies, together with studies in which participants undertook major changes in diet under experimental conditions (“plant-based” diet, i.e., rich in grains, legumes, fruits, and vegetables, or “animal-based” diet, i.e., rich in meats, eggs, and cheese), provide clear evidence that the microbiota and its function can be modulated by habitual dietary intake (157). Under experimental conditions, the altered dietary intakes of the participants did not affect the variety of bacteria present (alpha-diversity), but changes in microbiota composition based on differences in phylogeny, relative abundances of “species,” and degree of similarity of microbiotas (beta-diversity) could be detected between the animal-based diet group and other groups just 1 day after the food reached the colon. The most common clusters associated with the animal-based diet were composed of bile-resistant bacteria, which correlated with the known increase of bile acids in the gut when there is a high fat intake. Feces from participants consuming the animal-based diet had much lower concentrations of SCFAs originating from the fermentation of carbohydrates (acetate and butyrate) but higher concentrations of SCFAs originating from the fermentation of amino acids (isobutyrate, isovalerate), relative to baseline and plant-based diet samples.
OTHER CONSIDERATIONS LEARNED FROM DIETARY INTERVENTION STUDIES
Important knowledge associated with modulation of the gut microbiota has emerged from more subtle dietary interventions.
A high-diversity microbiota may not respond to dietary intervention.
Microbiotas that are already relatively high in diversity do not respond to increased intake of dietary fiber even at a level of 40 g per day (128). Thus, screening the gut microbiotas of potential participants in intervention studies to identify individuals with low-diversity communities has been suggested. There would then be some hope of obtaining “improvement” (158–160). High-diversity microbiota compositions are considered preferable to low-diversity communities (139, 161–163). This is because communities with high taxonomic diversity have high functional diversity associated with metabolic redundancy among taxa. This gives resilience to communities, because the loss of some members of the microbiota will not necessarily mean the total loss of a particular function (140). Dysbiotic microbiotas may lack this functional resilience and hence do not perform the usual interactive relationship with the human host. These low-diversity microbiotas are clearly a target for dietary intervention, because plant glycans are the main nutritional driver of the gut microbiota (3).
Among unselected human subjects, only some will have microbiotas that respond to a particular dietary intervention.
For example, Walker and colleagues investigated the effect of dietary intervention in overweight men (159). A diet containing resistant starch (RS) was included in the study. Increases in the numbers of a small number of taxa (Ruminococcus bromii-like, Oscillibacter-like, and Eubacterium rectale-like bacteria) were detected by quantitative PCR when the RS diet was consumed, but there was considerable interindividual variation. In two human subjects, greater than 60% of RS passed through the gut undigested, probably related to the paucity of R. bromii-like bacteria in the feces. Baxter and colleagues reached similar conclusions in a more recent study (164). Correcting a society-wide deficiency (e.g., in butyrate concentration) will be difficult, because some humans will not respond and therefore benefit. However, in correcting a dysbiosis associated with disease, the aim is not to provide a “one-diet-fits-all” approach to improving general health but must target specific aspects of the microbiota that are associated with particular diseases or conditions. So, a realistic aim is to target dysbiosis-associated diseases but not to reach as far as personalized nutrition, unless a critical factor for microbiota regeneration is absent in particular individuals. This requires a different mindset from that of traditional prebiotics, because it requires a more pharmaceutical approach (restoring, correcting, or modifying specific organic functions) rather than general wellness. Bacteria detected in the gut microbiotas of pastoral and hunter-gatherer societies (Succinivibrionaceae, Paraprevotellaceae, Spirochaetaceae, Prevotellaceae), which are dependent on seasonal diet, but not in the microbiotas of westerners might be necessary “probiotic” adjuncts to dietary modification in order to supply “missing microbes” (165).
The quantity of dietary fiber above baseline level that is required to produce consistently measurable results.
The amount of dietary fiber that might need to be consumed daily, long term, in order to produce a sustainable outcome could be beyond the tolerance of modern-day humans in Western countries. Daily intakes of 80 to 150 g of dietary fiber per day would equate to that consumed by the Hadza (155). Changing quality rather than quantity of dietary fiber might be a more attractive proposition. Substantial knowledge of the differential utilization of plant glycans by bacterial consortia in the gut, including comparisons of what is ingested and what is excreted, is required to enhance the value of dietary intervention studies. Dose-response experiments are allied with this work because the minimum efficacious amount of glycan in relation to potential adverse effects should be known (for example, excessive flatus production). In the case of the gut microbiota, the dose response is conditional because extensive metabolic interactions within the microbiota are likely. Several of these aspects were investigated by Deehan and colleagues, who found that different forms of chemically modified starch with small structural differences produced differential effects in microbiota composition and SCFA production and that augmentation of these effects by increased intake was limited because a plateau was reached at 35 g per day (166).
Testing the long-term effects of dietary intervention.
Limitations of investigations of the gut microbiota include the relatively small numbers of human subjects that are recruited to studies and the analysis of a single fecal sample per subject. With dietary interventions, it seems crucial to conduct temporal studies during which dietary modification is performed in double cross-over trials that are months in duration. We do not know accurately the extent of seasonal variation on microbiota composition in Western societies or whether modulation is sustainable in the long term. Even if dietary intervention is maintained, changes in microbiota characteristics may soon disappear and revert to baseline. This aspect requires extensive research (167).
Incorporating the glycans as a “package” of substrates with differential effects.
Dietary fads should not be encouraged, and all dietary interventions need to be considered rigorously for potential to cause harm. Ways need to be devised to encourage the consumption of a balanced diet that contains mixtures of glycans that will support the development and maintenance of a highly diverse and functionally resilient gut microbiota. In large part, this will succeed only if ways to increase intake of particular dietary fibers in a palatable form can be found. For example, the “Mediterranean diet,” inspired by the eating habits of people in Greece, southern Italy, and Spain in the 1940s and 50s, is characterized by fruit, vegetables, fish, and whole grains and limited saturated fats (168–170). While most interest has focused on the reduced risk of heart disease associated with this diet, Italians who adhere closely to the diet have higher SCFA concentrations in feces than in those with lesser adherence. This indicates an impact of a diet containing complex carbohydrates on the emergent properties of the microbiota. Specifying the types of fruits and vegetables in a diet on the basis of the amount and types of plant glycans that will be delivered to the microbiota would be an advance.
POTENTIAL DYSBIOTIC TARGETS
Inflammatory bowel diseases (IBD) (Crohn’s disease [CD] and ulcerative colitis [UC]) are disorders that have genetic predispositions, environmental modifiers, and chronic immune-mediated tissue damage (171, 172). The gut microbiota has been invoked as a major environmental factor in IBD mainly because of evidence obtained from experimental animal models of gut inflammation. Colitis occurs only if genetically predisposed germfree animals are colonized by a gut microbiota (173). A single bacterial species has not been identified as the causative agent of IBD. Rather, human studies indicate that the diversity of species comprising the microbiota in CD in particular is less than in health. There is generally a depletion of members of clostridial cluster IV (Clostridium leptum cluster) that characteristically produce butyrate. Interest has also been placed on Faecalibacterium prausnitzii and Roseburia species (clostridial cluster XIVa, Clostridium coccoides cluster), because there is an inverse correlation between abundance of these species and disease activity (22, 24, 30, 174–176). Although a causative effect of dysbiosis has not been established in IBD, butyrate is an energy source for colonocytes and has anti-inflammatory effects. Thus, correction of the dysbiosis by modulation of the microbiota to produce more butyrate remains an attractive treatment option. IBD patients tend to have low fiber intakes (https://www.crohnscolitisfoundation.org/diet-and-nutrition/what-should-i-eat). This lower intake of dietary fiber may be the actual reason for reduced production of butyrate. Increasing dietary fiber intake may therefore be beneficial for some IBD patients but may not be appropriate for patients with bowel strictures where there is potential for blockage.
The best current nutrition-based treatment for IBD (mainly pediatric CD) is exclusive enteral nutrition (EEN), in which patients consume a liquid diet that provides all of their energy and nutrient needs (177–179). Ironically, EEN results in lower butyrate production by the gut microbiota because the diet does not contain dietary fiber (180). Perhaps the importance of butyrate in gut health has been overstated, but further research may show that smaller quantities can still be therapeutic. These observations point to the need to develop threshold criteria for microbial populations and metabolites. How big an increase/decrease in butyrate concentration, for example, is needed to produce a therapeutic or prophylactic outcome?
Metabolic syndrome encompasses a cluster of conditions (elevated blood pressure, elevated blood glucose concentration, excess body fat around the waist, abnormal blood cholesterol levels) that, when occurring together, increase the risk of heart disease, stroke, and diabetes. These noncommunicable diseases are of increasing concern throughout the world and are clearly associated with lower intakes of dietary fiber (129). Therefore, metabolic syndrome provides the best target of dietary modulation of the gut microbiota. Millions of people will be benefited, and costs for health services will be reduced. As mentioned above, SCFAs produced by the microbiota, as well as having caloric value, are important signaling molecules in the gut (148, 181–185). Additionally, propionate has anorectic properties by influencing gut hormones (peptide YY [PYY] and glucagon-like peptide 1 [GLP-1]) and thus reducing energy intake (186). Bile acids are also of interest in relation to metabolic syndrome. Bile acids are formed in the liver and released into the small bowel in bile. The bile acids are required at critical concentrations to assist in the emulsification of dietary lipids to facilitate hydrolysis by digestive enzymes and for absorption of fatty acids by the small bowel mucosa. Highly efficient recycling of bile acids occurs (the enterohepatic circulation), but about 5% of bile acids reach the colon, where members of the Bacteroidetes phylum in particular carry out chemical transformations that result in the secondary bile acids, deoxycholate and lithocholate (187, 188). The deconjugation of primary bile acids is the first step in the transformation process, whereby amino acids (glycine or taurine) are removed. This step is catalyzed by bile salt hydrolases produced by the bacteria (187). Experiments with poultry and mice, as well as observations in human gastric bypass patients, indicate that bile salt hydrolase activity in the bowel can modify lipid absorption, blood levels of bile acids, blood lipid chemistry, and weight loss (188–191). These effects probably involve signaling through bile salt receptors in the gut mucosa. Thus, the effects of dietary modulation of the gut microbiota could, in this case, be determined indirectly using metabolomic measurements of blood (plasma) constituents. In addition to standard diagnostic tests based on blood samples (for example, concentrations of glucose, HbA1c, cholesterols, and insulin), the availability of kits that provide standardized technical means for targeted analysis using liquid chromatography-mass spectrometry (LC-MS) makes this a feasible option (192–194). Combined microbial and human profiling could be used to thoroughly monitor efficacy in dietary intervention studies.
DIFFERENCES IN SCIENTIFIC AND COMMERCIAL IMPERATIVES
Medical/scientific goals in modulation of the microbiota will likely focus on correcting specific dysbioses associated with particular diseases and to be allied, wherever possible, with the aim of increasing dietary fiber intake, known to promote general good health (129, 195). This approach may be satisfied by the recommendation to consume particular classes of fruits and vegetables that are known to contain glycans that can be degraded and fermented by specific bacterial taxa. The commercial imperative may be rather to extract (purify) specific glycans, especially from material that is currently considered waste (for example, peelings or seeds) so that increased value is extracted from crops. The extracted and processed glycans can be incorporated into capsules or packaged powders for consumption, making delivery of the “dietary fiber” to the human gut easy but, of course, somewhat more expensive because of processing costs. The medical/scientific and commercial approaches are not mutually exclusive and could be combined, but consideration needs to be given to the efficacy of administration of a single substrate (which may have been chemically altered during extraction) to the gut ecosystem rather than a complexity of glycans such as might be found in natural fruits and vegetables (unprocessed except by cooking in some instances). The relative efficacies of these two approaches need to be tested, as well as comparisons of ease of uptake and cost to the consumer, and should always be defined in relation to improved health.
THE RESEARCH JOURNEY AHEAD
Evaluation of the literature indicates that continuing to conduct dietary intervention trials of a speculative nature (feed it and see what happens to the microbiota) are not helpful. The trials do not offer clear solutions that will minimize the ravages of noncommunicable diseases, largely because the ecology of health-associated microbiotas has not yet been adequately defined. We know well who are the members of microbiotas and what they are capable of doing in a gross sense, but we cannot adequately predict why perturbations occur in relation to specific diseases. Nor are we able to produce a roadmap of how a perturbed ecosystem can be rectified in the long term. Clearly, we have to understand how the gut microbiota functions in health, including knowledge of thresholds of abundances of taxa or bioactive substances produced by the microbiota, above or below which disease are associated. Otherwise, changes in relative abundances of particular taxa and/or butyrate, for example, lack meaningful context. Modulations of the microbiota need to be considered in terms of the formation and function of metabolic consortia. A spatial perspective of the habitat is likely to be important; bacterial cells are associated with plant particulate material in the digesta, and these bacteria doubtless have a role in the hydrolysis and fermentation of plant polysaccharides in the gut (196, 197). Spatial associations on food particles of hydrolytic bacteria (producers) with other bacterial species that benefit from “leakage” of hydrolytic products that are potential growth substrates can be envisioned in the development of consortia in the gut ecosystem.
The next phase of research is thus to define the composition of bacterial consortia associated with the degradation and fermentation of plant glycans and the changing abundances of specific consortia associated with alterations to diet. The aim is to fit members of bacterial taxa together into consortia, as in fitting pieces together in a jigsaw puzzle, and to measure the kinetics of the integrated metabolism within each consortium. Different consortia will be formed around different glycans, which specifically fuel each consortium, so there will be many consortia with different functions in the microbiota when a complex mixture of plant glycans is available. The further aim is to test the hypothesis that consortia that carry out the same function can differ in terms of constituent bacterial species (phylogenetically diverse bacteria can encode the same functions). This phenomenon would explain the individuality of human gut microbiota compositions but a general similarity in overall community functions. A structured, habitual, but interesting diet would promote the formation of varied consortia, thus maintaining a high-diversity community with functional resilience. Although a seemingly simple proposition, implementation of the diverse-glycan diet will not be easy, given that dietary modulation needs to begin in early adulthood to produce consequences that will be appreciable only in the fifth and sixth decades of life when considered in relation to metabolic syndrome. It is unlikely that the “ancestral” gut microbiota detected in hunter-gatherers can be achieved in Western countries—the Hadza, too, prefer meat and honey to dietary fiber-laden tubers (165). The plea is for fundamental ecological research based on an understanding of the gut microbiota, in particular how high-diversity communities can be promoted by linking the intake of complex plant glycans of diverse chemistry with modulation of the gut microbiota.
ACKNOWLEDGMENT
Gerald W. Tannock is Professor Emeritus of the University of Otago, Dunedin, New Zealand, and is hosted by the Department of Microbiology and Immunology.
REFERENCES
- 1.Franzosa EA, Hsu T, Sirota-Madi A, Shafquat A, Abu-Ali G, Morgan XC, Huttenhower C. 2015. Sequencing and beyond: integrating molecular 'omics' for microbial community profiling. Nat Rev Microbiol 13:360–372. doi: 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cummings JH, Stephen AM. 1980. The role of dietary fibre in the human colon. Can Med Assoc J 123:1109–1114. [PMC free article] [PubMed] [Google Scholar]
- 4.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J, Jun W. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Jones JM. 2014. CODEX-aligned dietary fiber definitions help to bridge the 'fiber gap'. Nutr J 13:34. doi: 10.1186/1475-2891-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leong C, Haszard JJ, Lawley B, Otal A, Taylor RW, Szymlek-Gay EA, Fleming EA, Daniels L, Fangupo LJ, Tannock GW, Heath AM. 2018. Mediation analysis as a means of identifying dietary components that differentially affect the fecal microbiota of infants weaned by modified baby-led and traditional approaches. Appl Environ Microbiol 84:e00914-18. doi: 10.1128/AEM.00914-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White BA, Lamed R, Bayer EA, Flint HJ. 2014. Biomass utilization by gut microbiomes. Annu Rev Microbiol 68:279–296. doi: 10.1146/annurev-micro-092412-155618. [DOI] [PubMed] [Google Scholar]
- 9.Derrien M, Alvarez AS, de Vos WM. 2019. The gut microbiota in the first decade of life. Trends Microbiol 27:997–1010. doi: 10.1016/j.tim.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Leong C, Haszard JJ, Heath AM, Tannock GW, Lawley B, Cameron SL, Szymlek-Gay EA, Gray AR, Taylor BJ, Galland BC, Lawrence JA, Otal A, Hughes A, Taylor RW. 2020. Using compositional principal component analysis to describe children's gut microbiota in relation to diet and body composition. Am J Clin Nutr 111:70–78. doi: 10.1093/ajcn/nqz270. [DOI] [PubMed] [Google Scholar]
- 11.Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Caporaso JG, Knights D, Clemente JC, Nakielny S, Gordon JI, Fierer N, Knight R. 2013. Cohabiting family members share microbiota with one another and with their dogs. Elife 2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tannock GW, Lawley B, Munro K, Gowri Pathmanathan S, Zhou SJ, Makrides M, Gibson RA, Sullivan T, Prosser CG, Lowry D, Hodgkinson AJ. 2013. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl Environ Microbiol 79:3040–3048. doi: 10.1128/AEM.03910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Björkstén B, Engstrand L, Andersson AF. 2014. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 63:559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 14.Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB. 2014. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol 68:217–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich JK, Davenport ER, Clark AG, Ley RE. 2017. The relationship between the human genome and microbiome comes into view. Annu Rev Genet 51:413–433. doi: 10.1146/annurev-genet-110711-155532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawley B, Otal A, Moloney-Geany K, Diana A, Houghton L, Heath A-LM, Taylor RW, Tannock GW. 2019. Fecal microbiotas of Indonesian and New Zealand children differ in complexity and bifidobacterial taxa during the first year of life. Appl Environ Microbiol 85:e01105-19. doi: 10.1128/AEM.01105-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Lawley B, Wong G, Otal A, Chen L, Ying TJ, Lin X, Pang WW, Yap F, Chong YS, Gluckman PD, Lee YS, Chong MF, Tannock GW, Karnani N. 2020. Ethnic diversity in infant gut microbiota is apparent before the introduction of complementary diets. Gut Microbes 11:1362–1373. doi: 10.1080/19490976.2020.1756150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberton AM, Corfield AP. 1999. Mucin degradation and its significance in inflammatory conditions of the gastrointestinal tract, p 222–261. In Tannock GW (ed), Medical importance of the normal microflora. Kluwer Academic Publishers, Dordrecht, the Netherlands. [Google Scholar]
- 19.Tannock GW. 2017. Understanding the gut microbiota. Wiley, Hoboken, NJ. [Google Scholar]
- 20.Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall LI, McDonald D, Melnik AV, Morton JT, Navas J, Quinn RA, Sanders JG, Swafford AD, Thompson LR, Tripathi A, Xu ZZ, Zaneveld JR, Zhu Q, Caporaso JG, Dorrestein PC. 2018. Best practices for analysing microbiomes. Nat Rev Microbiol 16:410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 21.Tvede M, Rask-Madsen J. 1989. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet 1:1156–1160. doi: 10.1016/s0140-6736(89)92749-9. [DOI] [PubMed] [Google Scholar]
- 22.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Fölsch UR, Timmis KN, Schreiber S. 2004. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 24.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flint HJ, Scott KP, Louis P, Duncan SH. 2012. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 27.Finegold SM, Downes J, Summanen PH. 2012. Microbiology of regressive autism. Anaerobe 18:260–262. doi: 10.1016/j.anaerobe.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 28.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker AW, Lawley TD. 2013. Therapeutic modulation of intestinal dysbiosis. Pharmacol Res 69:75–86. doi: 10.1016/j.phrs.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Cao Y, Shen J, Ran ZH. 2014. Association between Faecalibacterium prausnitzii reduction and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Gastroenterol Res Pract 2014:1–7. doi: 10.1155/2014/872725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Böhm J, Brunetti F, Habermann N, Hercog R, Koch M, Luciani A, Mende DR, Schneider MA, Schrotz-King P, Tournigand C, Tran Van Nhieu J, Yamada T, Zimmermann J, Benes V, Kloor M, Ulrich CM, von Knebel Doeberitz M, Sobhani I, Bork P. 2014. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol 10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shanahan F, Quigley EM. 2014. Manipulation of the microbiota for treatment of IBS and IBD: challenges and controversies. Gastroenterology 146:1554–1563. doi: 10.1053/j.gastro.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA, Jr, Ahmed T, Gordon JI. 2014. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tilg H, Moschen AR. 2014. Microbiota and diabetes: an evolving relationship. Gut 63:1513–1521. doi: 10.1136/gutjnl-2014-306928. [DOI] [PubMed] [Google Scholar]
- 35.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, Mohn WW, Turvey SE, Finlay BB, CHILD Study Investigators. 2015. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 36.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Pedersen HK, Arumugam M, Kristiansen K, Voigt AY, Vestergaard H, Hercog R, Costea PI, Kultima JR, Li J, Jørgensen T, Levenez F, Dore J, Nielsen HB, Brunak S, Raes J, Hansen T, Wang J, Ehrlich SD, Bork P, Pedersen O, MetaHIT Consortium. 2015. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujimura KE, Lynch SV. 2015. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe 17:592–602. doi: 10.1016/j.chom.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajilić-Stojanović M, Jonkers DM, Salonen A, Hanevik K, Raes J, Jalanka J, de Vos WM, Manichanh C, Golic N, Enck P, Philippou E, Iraqi FA, Clarke G, Spiller RC, Penders J. 2015. Intestinal microbiota and diet in IBS: causes, consequences, or epiphenomena? Am J Gastroenterol 110:278–287. doi: 10.1038/ajg.2014.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brüssow H. 2016. Biome engineering—2020. Microb Biotechnol 9:553–563. doi: 10.1111/1751-7915.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazidi M, Rezaie P, Kengne AP, Mobarhan MG, Ferns GA. 2016. Gut microbiome and metabolic syndrome. Diabetes Metab Syndr 10:S150–S157. doi: 10.1016/j.dsx.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 41.Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Jansson JK, Dorrestein PC, Knight R. 2016. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 535:94–103. doi: 10.1038/nature18850. [DOI] [PubMed] [Google Scholar]
- 42.Sze MA, Schloss PD. 2016. Looking for a signal in the noise: revisiting obesity and the microbiome. mBio 7:e01018-16. doi: 10.1128/mBio.01018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hooks KB, O’Malley MA. 2017. Dysbiosis and Its discontents. mBio 8:e01492-17. doi: 10.1128/mBio.01492-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker WA. 2017. The importance of appropriate initial bacterial colonization of the intestine in newborn, child, and adult health. Pediatr Res 82:387–395. doi: 10.1038/pr.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arrieta MC, Arévalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, Vaca M, Boutin RCT, Morien E, Jin M, Turvey SE, Walter J, Parfrey LW, Cooper PJ, Finlay B. 2018. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol 142:424–434.e10. doi: 10.1016/j.jaci.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duvallet C. 2018. Meta-analysis generates and prioritizes hypotheses for translational microbiome research. Microb Biotechnol 11:273–276. doi: 10.1111/1751-7915.13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neville BA, Forster SC, Lawley TD. 2018. Commensal Koch's postulates: establishing causation in human microbiota research. Curr Opin Microbiol 42:47–52. doi: 10.1016/j.mib.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Wu Y, Wan J, Choe U, Pham Q, Schoene NW, He Q, Li B, Yu L, Wang TTY. 2019. Interactions between food and gut microbiota: impact on human health. Annu Rev Food Sci Technol 10:389–408. doi: 10.1146/annurev-food-032818-121303. [DOI] [PubMed] [Google Scholar]
- 49.Brüssow H. 2020. Problems with the concept of gut microbiota dysbiosis. Microb Biotechnol 13:423–434. doi: 10.1111/1751-7915.13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter J, Armet AM, Finlay BB, Shanahan F. 2020. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell 180:221–232. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 51.Willats WGT, Knox JP, Mikkelsen JD. 2006. Pectin: new insights into an old polymer are starting to gel. Trends Food Sci Technol 17:97–104. doi: 10.1016/j.tifs.2005.10.008. [DOI] [Google Scholar]
- 52.Englyst HN, Kingman SM, Cummings JH. 1992. Classification and measurement of nutritionally important starch fractions. Eur J Clin Nutr 46 Suppl 2:S33–S50. [PubMed] [Google Scholar]
- 53.DeMartino P, Cockburn DW. 2020. Resistant starch: impact on the gut microbiome and health. Curr Opin Biotechnol 61:66–71. doi: 10.1016/j.copbio.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 54.Mendis M, Martens EC, Simsek S. 2018. How fine structural differences of xylooligosaccharides and arabinoxylooligosaccharides regulate differential growth of Bacteroides species. J Agric Food Chem 66:8398–8405. doi: 10.1021/acs.jafc.8b01263. [DOI] [PubMed] [Google Scholar]
- 55.Walton GE, Lu C, Trogh I, Arnaut F, Gibson GR. 2012. A randomised, double-blind, placebo controlled cross-over study to determine the gastrointestinal effects of consumption of arabinoxylan-oligosaccharides enriched bread in healthy volunteers. Nutr J 11:36. doi: 10.1186/1475-2891-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rakoff-Nahoum S, Coyne MJ, Comstock LE. 2014. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol 24:40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duncan SH, Russell WR, Quartieri A, Rossi M, Parkhill J, Walker AW, Flint HJ. 2016. Wheat bran promotes enrichment within the human colonic microbiota of butyrate-producing bacteria that release ferulic acid. Environ Microbiol 18:2214–2225. doi: 10.1111/1462-2920.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hemsworth GR, Thompson AJ, Stepper J, Sobala ŁF, Coyle T, Larsbrink J, Spadiut O, Goddard-Borger ED, Stubbs KA, Brumer H, Davies GJ. 2016. Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut. Open Biol 6:160142. doi: 10.1098/rsob.160142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanegas SM, Meydani M, Barnett JB, Goldin B, Kane A, Rasmussen H, Brown C, Vangay P, Knights D, Jonnalagadda S, Koecher K, Karl JP, Thomas M, Dolnikowski G, Li L, Saltzman E, Wu D, Meydani SN. 2017. Substituting whole grains for refined grains in a 6-wk randomized trial has a modest effect on gut microbiota and immune and inflammatory markers of healthy adults. Am J Clin Nutr 105:635–650. doi: 10.3945/ajcn.116.146928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aoe S, Nakamura F, Fujiwara S. 2018. Effect of wheat bran on fecal butyrate-producing bacteria and wheat bran combined with barley on Bacteroides abundance in Japanese healthy adults. Nutrients 10:1980. doi: 10.3390/nu10121980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moro Cantu-Jungles T, do Nascimento GE, Zhang X, Iacomini M, Cordeiro LMC, Hamaker BR. 2019. Soluble xyloglucan generates bigger bacterial community shifts than pectic polymers during in vitro fecal fermentation. Carbohydr Polym 206:389–395. doi: 10.1016/j.carbpol.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 62.Nguyen NK, Deehan EC, Zhang Z, Jin M, Baskota N, Perez-Muñoz ME, Cole J, Tuncil YE, Seethaler B, Wang T, Laville M, Delzenne NM, Bischoff SC, Hamaker BR, Martínez I, Knights D, Bakal JA, Prado CM, Walter J. 2020. Gut microbiota modulation with long-chain corn bran arabinoxylan in adults with overweight and obesity is linked to an individualized temporal increase in fecal propionate. Microbiome 8:118. doi: 10.1186/s40168-020-00887-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jefferson A, Adolphus K. 2019. The effects of intact cereal grain fibers, including wheat bran on the gut microbiota composition of healthy adults: a systematic review. Front Nutr 6:33. doi: 10.3389/fnut.2019.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 65.Zou Y, Xue W, Luo G, Deng Z, Qin P, Guo R, Sun H, Xia Y, Liang S, Dai Y, Wan D, Jiang R, Su L, Feng Q, Jie Z, Guo T, Xia Z, Liu C, Yu J, Lin Y, Tang S, Huo G, Xu X, Hou Y, Liu X, Wang J, Yang H, Kristiansen K, Li J, Jia H, Xiao L. 2019. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol 37:179–185. doi: 10.1038/s41587-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cho KH, Salyers AA. 2001. Biochemical analysis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J Bacteriol 183:7224–7230. doi: 10.1128/jb.183.24.7224-7230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cockburn DW, Koropatkin NM. 2016. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J Mol Biol 428:3230–3252. doi: 10.1016/j.jmb.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 68.Grondin JM, Tamura K, Déjean G, Abbott DW, Brumer H. 2017. Polysaccharide utilization loci: fueling microbial communities. J Bacteriol 199:e00860-16. doi: 10.1128/JB.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwalm ND, Groisman EA. 2017. Navigating the gut buffet: control of polysaccharide utilization in Bacteroides spp. Trends Microbiol 25:1005–1015. doi: 10.1016/j.tim.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 70.Sheridan PO, Martin JC, Lawley TD, Browne H, Harris HM, Bernalier-Donadille A, Duncan SH, O'Toole PW, Scott K, Flint HJ. 2016. Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes. Microb Genom 2:e000043. doi: 10.1099/mgen.0.000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bayer EA, Belaich JP, Shoham Y, Lamed R. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol 58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 72.Ben David Y, Dassa B, Borovok I, Lamed R, Koropatkin NM, Martens EC, White BA, Bernalier-Donadille A, Duncan SH, Flint HJ, Bayer EA, Moraïs S. 2015. Ruminococcal cellulosome systems from rumen to human. Environ Microbiol 17:3407–3426. doi: 10.1111/1462-2920.12868. [DOI] [PubMed] [Google Scholar]
- 73.Moraïs S, David YB, Bensoussan L, Duncan SH, Koropatkun NM, Martens EC, Flint HJ, Bayer EA. 2015. Enzymatic profiling of cellulosomal enzymes from the human gut bacterium, Ruminococcus champanellensis, reveals a fine-tuned system for cohesin-dockerin recognition. Environ Microbiol 18:542–546. doi: 10.1111/1462-2920.13047. [DOI] [PubMed] [Google Scholar]
- 74.Ze X, Ben David Y, Laverde-Gomez JA, Dassa B, Sheridan PO, Duncan SH, Louis P, Henrissat B, Juge N, Koropatkin NM, Bayer EA, Flint HJ. 2015. Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic Firmicutes bacterium Ruminococcus bromii. mBio 6:e01058-15. doi: 10.1128/mBio.01058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim CC, Healey GR, Kelly WJ, Patchett ML, Jordens Z, Tannock GW, Sims IM, Bell TJ, Hedderley D, Henrissat B, Rosendale DI. 2019. Genomic insights from Monoglobus pectinilyticus: a pectin-degrading specialist bacterium in the human colon. ISME J 13:1437–1456. doi: 10.1038/s41396-019-0363-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang M, Chekan JR, Dodd D, Hong PY, Radlinski L, Revindran V, Nair SK, Mackie RI, Cann I. 2014. Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes. Proc Natl Acad Sci U S A 111:E3708–3717. doi: 10.1073/pnas.1406156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Baslé A, Morland C, Day AM, Zheng H, Rogers TE, Thompson P, Hawkins AR, Yadav MP, Henrissat B, Martens EC, Dupree P, Gilbert HJ, Bolam DN. 2015. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun 6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Despres J, Forano E, Lepercq P, Comtet-Marre S, Jubelin G, Chambon C, Yeoman CJ, Miller MEB, Fields CJ, Martens E, Terrapon N, Henrissat B, White BA, Mosoni P. 2016. Xylan degradation by the human gut Bacteroides xylanisolvens XB1AT involves two distinct gene clusters that are linked at the transcriptional level. BMC Genomics 17:326. doi: 10.1186/s12864-016-2680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leth ML, Ejby M, Workman C, Ewald DA, Pedersen SS, Sternberg C, Bahl MI, Licht TR, Aachmann FL, Westereng B, Abou Hachem M. 2018. Differential bacterial capture and transport preferences facilitate co-growth on dietary xylan in the human gut. Nat Microbiol 3:570–580. doi: 10.1038/s41564-018-0132-8. [DOI] [PubMed] [Google Scholar]
- 80.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. 2017. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol 14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 81.Oozeer R, van Limpt K, Ludwig T, Ben Amor K, Martin R, Wind RD, Boehm G, Knol J. 2013. Intestinal microbiology in early life: specific prebiotics can have similar functionalities as human-milk oligosaccharides. Am J Clin Nutr 98:561S–571S. doi: 10.3945/ajcn.112.038893. [DOI] [PubMed] [Google Scholar]
- 82.Vulevic J, Juric A, Tzortzis G, Gibson GR. 2013. A mixture of trans-galactooligosaccharides reduces markers of metabolic syndrome and modulates the fecal microbiota and immune function of overweight adults. J Nutr 143:324–331. doi: 10.3945/jn.112.166132. [DOI] [PubMed] [Google Scholar]
- 83.Vulevic J, Juric A, Walton GE, Claus SP, Tzortzis G, Toward RE, Gibson GR. 2015. Influence of galacto-oligosaccharide mixture (B-GOS) on gut microbiota, immune parameters and metabonomics in elderly persons. Br J Nutr 114:586–595. doi: 10.1017/S0007114515001889. [DOI] [PubMed] [Google Scholar]
- 84.Liu F, Li P, Chen M, Luo Y, Prabhakar M, Zheng H, He Y, Qi Q, Long H, Zhang Y, Sheng H, Zhou H. 2017. Fructooligosaccharide (FOS) and galactooligosaccharide (GOS) increase Bifidobacterium but reduce butyrate producing bacteria with adverse glycemic metabolism in healthy young population. Sci Rep 7:11789. doi: 10.1038/s41598-017-10722-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rigo-Adrover MDM, van Limpt K, Knipping K, Garssen J, Knol J, Costabile A, Franch À, Castell M, Pérez-Cano FJ. 2018. Preventive effect of a synbiotic combination of galacto- and fructooligosaccharides mixture with Bifidobacterium breve M-16V in a model of multiple rotavirus infections. Front Immunol 9:1318. doi: 10.3389/fimmu.2018.01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tandon D, Haque MM, Gote M, Jain M, Bhaduri A, Dubey AK, Mande SS. 2019. A prospective randomized, double-blind, placebo-controlled, dose-response relationship study to investigate efficacy of fructo-oligosaccharides (FOS) on human gut microflora. Sci Rep 9:5473. doi: 10.1038/s41598-019-41837-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sims IM, Tannock GW. 2020. Galacto- and fructo-oligosaccharides utilized for growth by cocultures of bifidobacterial species characteristic of the infant gut. Appl Environ Microbiol 86:e00214-20. doi: 10.1128/AEM.00214-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lawley B, Sims IM, Tannock GW. 2013. Whole-transcriptome shotgun sequencing (RNA-seq) screen reveals upregulation of cellobiose and motility operons of Lactobacillus ruminis L5 during growth on tetrasaccharides derived from barley β-glucan. Appl Environ Microbiol 79:5661–5669. doi: 10.1128/AEM.01887-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Centanni M, Hutchison JC, Carnachan SM, Daines AM, Kelly WJ, Tannock GW, Sims IM. 2017. Differential growth of bowel commensal Bacteroides species on plant xylans of differing structural complexity. Carbohydr Polym 157:1374–1382. doi: 10.1016/j.carbpol.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 90.Bell TJ, Draper SL, Centanni M, Carnachan SM, Tannock GW, Sims IM. 2018. Characterization of polysaccharides from feijoa fruits (Acca sellowiana Berg.) and their utilization as growth substrates by gut commensal Bacteroides species. J Agric Food Chem 66:13277–13284. doi: 10.1021/acs.jafc.8b05080. [DOI] [PubMed] [Google Scholar]
- 91.Centanni M, Carnachan SM, Bell TJ, Daines AM, Hinkley SFR, Tannock GW, Sims IM. 2019. Utilization of complex pectic polysaccharides from New Zealand plants (Tetragonia tetragonioides and Corynocarpus laevigatus) by gut Bacteroides species. J Agric Food Chem 67:7755–7764. doi: 10.1021/acs.jafc.9b02429. [DOI] [PubMed] [Google Scholar]
- 92.Centanni M, Sims IM, Bell TJ, Biswas A, Tannock GW. 2020. Sharing a β-glucan meal: transcriptomic eavesdropping on a Bacteroides ovatus-Subdoligranulum variabile-Hungatella hathewayi consortium. Appl Environ Microbiol 86:e01651-20. doi: 10.1128/AEM.01651-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tuncil YE, Thakkar RD, Arioglu-Tuncil S, Hamaker BR, Lindemann SR. 2020. Subtle variations in dietary-fiber fine structure differentially influence the composition and metabolic function of gut microbiota. mSphere 5:e00180-20. doi: 10.1128/mSphere.00180-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 95.Hamaker BR, Tuncil YE. 2014. A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J Mol Biol 426:3838–3850. doi: 10.1016/j.jmb.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 96.Centanni M, Bell TJ, Sims IM, Tannock GW. 2020. Preferential use of plant glycans for growth by Bacteroides ovatus. Anaerobe 66:102276. doi: 10.1016/j.anaerobe.2020.102276. [DOI] [PubMed] [Google Scholar]
- 97.Liu Y, Heath AL, Galland B, Rehrer N, Drummond L, Wu XY, Bell TJ, Lawley B, Sims IM, Tannock GW. 2019. Substrate use prioritization by a coculture of five species of gut bacteria fed mixtures of arabinoxylan, xyloglucan, β-glucan, and pectin. Appl Environ Microbiol 86:e01905-19. doi: 10.1128/AEM.01905-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rogers TE, Pudlo NA, Koropatkin NM, Bell JS, Moya Balasch M, Jasker K, Martens EC. 2013. Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol Microbiol 88:876–890. doi: 10.1111/mmi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, Baslé A, Cartmell A, Terrapon N, Stott K, Lowe EC, McLean R, Shearer K, Schückel J, Venditto I, Ralet M-C, Henrissat B, Martens EC, Mosimann SC, Abbott DW, Gilbert HJ. 2018. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 3:210–219. doi: 10.1038/s41564-017-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Almeida A, Nayfach S, Boland M, Strozzi F, Beracochea M, Shi ZJ, Pollard KS, Sakharova E, Parks DH, Hugenholtz P, Segata N, Kyrpides NC, Finn RD. 2020. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol doi: 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, Bork P, Ehrlich SD, Wang G. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. 2011. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A 108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Macfarlane GT, Macfarlane S, Gibson GR. 1998. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol 35:180–187. doi: 10.1007/s002489900072. [DOI] [PubMed] [Google Scholar]
- 105.Duncan SH, Scott KP, Ramsay AG, Harmsen HJM, Welling GW, Stewart CS, Flint HJ. 2003. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl Environ Microbiol 69:1136–1142. doi: 10.1128/aem.69.2.1136-1142.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hoskisson PA, Hobbs G. 2005. Continuous culture—making a comeback? Microbiology (Reading) 151:3153–3159. doi: 10.1099/mic.0.27924-0. [DOI] [PubMed] [Google Scholar]
- 107.Kettle H, Louis P, Holtrop G, Duncan SH, Flint HJ. 2015. Modelling the emergent dynamics and major metabolites of the human colonic microbiota. Environ Microbiol 17:1615–1630. doi: 10.1111/1462-2920.12599. [DOI] [PubMed] [Google Scholar]
- 108.Centanni M, Ferguson SA, Sims IM, Biswas A, Tannock GW. 2019. Bifidobacterium bifidum ATCC 15696 and Bifidobacterium breve 24b metabolic interaction based on 2'-O-fucosyl-lactose studied in steady-state cultures in a Freter-style chemostat. Appl Environ Microbiol 85:e02783-18. doi: 10.1128/AEM.02783-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tannock GW, Wilson CM, Loach D, Cook GM, Eason J, O'Toole PW, Holtrop G, Lawley B. 2012. Resource partitioning in relation to cohabitation of Lactobacillus species in the mouse forestomach. ISME J 6:927–938. doi: 10.1038/ismej.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Degnan PH, Taga ME, Goodman AL. 2014. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab 20:769–778. doi: 10.1016/j.cmet.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Putnam EE, Goodman AL. 2020. B vitamin acquisition by gut commensal bacteria. PLoS Pathog 16:e1008208. doi: 10.1371/journal.ppat.1008208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Soto-Martin EC, Warnke I, Farquharson FM, Christodoulou M, Horgan G, Derrien M, Faurie JM, Flint HJ, Duncan SH, Louis P. 2020. Vitamin biosynthesis by human gut butyrate-producing bacteria and cross-feeding in synthetic microbial communities. mBio 11:e00886-20. doi: 10.1128/mBio.00886-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wolin MJ. 1974. Metabolic interactions among intestinal microorganisms. Am J Clin Nutr 27:1320–1328. doi: 10.1093/ajcn/27.11.1320. [DOI] [PubMed] [Google Scholar]
- 114.Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, Gordon JI. 2010. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem 285:22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Flint HJ, Duncan SH, Scott KP, Louis P. 2015. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc 74:13–22. doi: 10.1017/S0029665114001463. [DOI] [PubMed] [Google Scholar]
- 116.Wang SP, Rubio LA, Duncan SH, Donachie GE, Holtrop G, Lo G, Farquharson FM, Wagner J, Parkhill J, Louis P, Walker AW, Flint HJ. 2020. Pivotal roles for pH, lactate, and lactate-utilizing bacteria in the stability of a human colonic microbial ecosystem. mSystems 5:e00645-20. doi: 10.1128/mSystems.00645-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP, Flint HJ, Louis P. 2014. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J 8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kirkpatrick SI, Reedy J, Butler EN, Dodd KW, Subar AF, Thompson FE, McKinnon RA. 2014. Dietary assessment in food environment research: a systematic review. Am J Prev Med 46:94–102. doi: 10.1016/j.amepre.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Coates J, Colaiezzi B, Fiedler JL, Wirth J, Lividini K, Rogers B. 2012. A program needs-driven approach to selecting dietary assessment methods for decision-making in food fortification programs. Food Nutr Bull 33:S146–S156. doi: 10.1177/15648265120333S202. [DOI] [PubMed] [Google Scholar]
- 120.Carlsen MH, Lillegaard IT, Karlsen A, Blomhoff R, Drevon CA, Andersen LF. 2010. Evaluation of energy and dietary intake estimates from a food frequency questionnaire using independent energy expenditure measurement and weighed food records. Nutr J 9:37. doi: 10.1186/1475-2891-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Alemayehu AA, Abebe Y, Gibson RS. 2011. A 24-h recall does not provide a valid estimate of absolute nutrient intakes for rural women in southern Ethiopia. Nutrition 27:919–924. doi: 10.1016/j.nut.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 122.Nightingale H, Walsh KJ, Olupot-Olupot P, Engoru C, Ssenyondo T, Nteziyaremye J, Amorut D, Nakuya M, Arimi M, Frost G, Maitland K. 2016. Validation of triple pass 24-hour dietary recall in Ugandan children by simultaneous weighed food assessment. BMC Nutr 2:56. doi: 10.1186/s40795-016-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hu FB. 2002. Dietary pattern analysis: a new direction in nutritional epidemiology. Curr Opin Lipidol 13:3–9. doi: 10.1097/00041433-200202000-00002. [DOI] [PubMed] [Google Scholar]
- 124.Hodge A, Bassett J. 2016. What can we learn from dietary pattern analysis? Public Health Nutr 19:191–194. doi: 10.1017/S1368980015003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schulze MB, Martínez-González MA, Fung TT, Lichtenstein AH, Forouhi NG. 2018. Food based dietary patterns and chronic disease prevention. BMJ 361:k2396. doi: 10.1136/bmj.k2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Westenbrink S, Brunt K, van der Kamp JW. 2013. Dietary fibre: challenges in production and use of food composition data. Food Chem 140:562–567. doi: 10.1016/j.foodchem.2012.09.029. [DOI] [PubMed] [Google Scholar]
- 127.Rainakari A-I, Rita H, Putkonen T, Pastell H. 2016. New dietary fibre content results for cereals in the Nordic countries using AOAC 2011.25 method. J Food Comp Anal 51:1–8. doi: 10.1016/j.jfca.2016.06.001. [DOI] [Google Scholar]
- 128.Tap J, Furet JP, Bensaada M, Philippe C, Roth H, Rabot S, Lakhdari O, Lombard V, Henrissat B, Corthier G, Fontaine E, Doré J, Leclerc M. 2015. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol 17:4954–4964. doi: 10.1111/1462-2920.13006. [DOI] [PubMed] [Google Scholar]
- 129.Reynolds A, Mann J, Cummings J, Winter N, Mete E, Te Morenga L. 2019. Carbohydrate quality and human health: a series of systematic reviews and meta-analyses. Lancet 393:434–445. doi: 10.1016/S0140-6736(18)31809-9. [DOI] [PubMed] [Google Scholar]
- 130.Smith C, Van Haute MJ, Rose DJ. 2020. Processing has differential effects on microbiota-accessible carbohydrates in whole grains during in vitro fermentation. Appl Environ Microbiol 86:e01705-20. doi: 10.1128/AEM.01705-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li SS, Zhu A, Benes V, Costea PI, Hercog R, Hildebrand F, Huerta-Cepas J, Nieuwdorp M, Salojärvi J, Voigt AY, Zeller G, Sunagawa S, de Vos WM, Bork P. 2016. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science 352:586–589. doi: 10.1126/science.aad8852. [DOI] [PubMed] [Google Scholar]
- 132.Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. 2016. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res 26:1612–1625. doi: 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vatanen T, Plichta DR, Somani J, Münch PC, Arthur TD, Hall AB, Rudolf S, Oakeley EJ, Ke X, Young RA, Haiser HJ, Kolde R, Yassour M, Luopajärvi K, Siljander H, Virtanen SM, Ilonen J, Uibo R, Tillmann V, Mokurov S, Dorshakova N, Porter JA, McHardy AC, Lähdesmäki H, Vlamakis H, Huttenhower C, Knip M, Xavier RJ. 2019. Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat Microbiol 4:470–479. doi: 10.1038/s41564-018-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Van Rossum T, Ferretti P, Maistrenko OM, Bork P. 2020. Diversity within species: interpreting strains in microbiomes. Nat Rev Microbiol 18:491–506. doi: 10.1038/s41579-020-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, MetaHIT Consortium, et al. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bauer E, Thiele I. 2018. From network analysis to functional metabolic modeling of the human gut microbiota. mSystems 3:e00209-17. doi: 10.1128/mSystems.00209-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Röttjers L, Faust K. 2018. From hairballs to hypotheses: biological insights from microbial networks. FEMS Microbiol Rev 42:761–780. doi: 10.1093/femsre/fuy030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cheng M, Ning K. 2019. Stereotypes about enterotype: the old and new ideas. Genomics Proteomics Bioinformatics 17:4–12. doi: 10.1016/j.gpb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mori AS, Furukawa T, Sasaki T. 2013. Response diversity determines the resilience of ecosystems to environmental change. Biol Rev Camb Philos Soc 88:349–364. doi: 10.1111/brv.12004. [DOI] [PubMed] [Google Scholar]
- 140.Moya A, Ferrer M. 2016. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol 24:402–413. doi: 10.1016/j.tim.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 141.Moran MA, Satinsky B, Gifford SM, Luo H, Rivers A, Chan L-K, Meng J, Durham BP, Shen C, Varaljay VA, Smith CB, Yager PL, Hopkinson BM. 2013. Sizing up metatranscriptomics. ISME J 7:237–243. doi: 10.1038/ismej.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Plichta DR, Juncker AS, Bertalan M, Rettedal E, Gautier L, Varela E, Manichanh C, Fouqueray C, Levenez F, Nielsen T, Doré J, Machado AM, de Evgrafov MC, Hansen T, Jørgensen T, Bork P, Guarner F, Pedersen O, Metagenomics of the Human Intestinal Tract (MetaHIT) Consortium, Sommer MO, Ehrlich SD, Sicheritz-Pontén T, Brunak S, Nielsen HB. 2016. Transcriptional interactions suggest niche segregation among microorganisms in the human gut. Nat Microbiol 1:16152. doi: 10.1038/nmicrobiol.2016.152. [DOI] [PubMed] [Google Scholar]
- 143.Radajewski S, Ineson P, Parekh NR, Murrell JC. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- 144.Egert M, de Graaf AA, Maathuis A, de Waard P, Plugge CM, Smidt H, Deutz NEP, Dijkema C, de Vos WM, Venema K. 2007. Identification of glucose-fermenting bacteria present in an in vitro model of the human intestine by RNA-stable isotope probing. FEMS Microbiol Ecol 60:126–135. doi: 10.1111/j.1574-6941.2007.00281.x. [DOI] [PubMed] [Google Scholar]
- 145.Manefield M, Whiteley AS, Griffiths RI, Bailey MJ. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol 68:5367–5373. doi: 10.1128/aem.68.11.5367-5373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tannock GW, Lawley B, Munro K, Sims IM, Lee J, Butts CA, Roy N. 2014. RNA-stable-isotope probing shows utilization of carbon from inulin by specific bacterial populations in the rat large bowel. Appl Environ Microbiol 80:2240–2247. doi: 10.1128/AEM.03799-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cummings JH, Macfarlane GT. 1991. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol 70:443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 148.Thorburn AN, Macia L, Mackay CR. 2014. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity 40:833–842. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 149.Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian McKenzie C, Hijikata A, Wong C, Binge L, Thorburn AN, Chevalier N, Ang C, Marino E, Robert R, Offermanns S, Teixeira MM, Moore RJ, Flavell RA, Fagarasan S, Mackay CR. 2015. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun 6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 150.Kumar J, Rani K, Datt C. 2020. Molecular link between dietary fibre, gut microbiota and health. Mol Biol Rep 47:6229–6237. doi: 10.1007/s11033-020-05611-3. [DOI] [PubMed] [Google Scholar]
- 151.Caspani G, Swann J. 2019. Small talk: microbial metabolites involved in the signaling from microbiota to brain. Curr Opin Pharmacol 48:99–106. doi: 10.1016/j.coph.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 152.Strandwitz P, Kim KH, Terekhova D, Liu JK, Sharma A, Levering J, McDonald D, Dietrich D, Ramadhar TR, Lekbua A, Mroue N, Liston C, Stewart EJ, Dubin MJ, Zengler K, Knight R, Gilbert JA, Clardy J, Lewis K. 2019. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol 4:396–403. doi: 10.1038/s41564-018-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, Claes S, Van Oudenhove L, Zhernakova A, Vieira-Silva S, Raes J. 2019. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol 4:623–632. doi: 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- 154.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, Fiori J, Gotti R, De Bellis G, Luiselli D, Brigidi P, Mabulla A, Marlowe F, Henry AG, Crittenden AN. 2014. Gut microbiome of the Hadza hunter-gatherers. Nat Commun 5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, Dominguez-Bello MG, Sonnenburg JL. 2017. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357:802–806. doi: 10.1126/science.aan4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Martínez I, Kim J, Duffy PR, Schlegel VL, Walter J. 2010. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One 5:e15046. doi: 10.1371/journal.pone.0015046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. 2011. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, Shields-Cutler RR, Kim AD, Shmagel AK, Syed AN, Personalized Microbiome Class Students, Walter J, Menon R, Koecher K, Knights D. 2019. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe 25:789–802.e5. doi: 10.1016/j.chom.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 161.Konopka A. 2009. What is microbial community ecology? ISME J 3:1223–1230. doi: 10.1038/ismej.2009.88. [DOI] [PubMed] [Google Scholar]
- 162.Coyte KZ, Schluter J, Foster KR. 2015. The ecology of the microbiome: networks, competition, and stability. Science 350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 163.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcón Ó, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G, Dominguez-Bello MG. 2015. The microbiome of uncontacted Amerindians. Sci Adv 1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. 2019. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio 10:e02566-18. doi: 10.1128/mBio.02566-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sonnenburg ED, Sonnenburg JL. 2019. The ancestral and industrialized gut microbiota and implications for human health. Nat Rev Microbiol 17:383–390. doi: 10.1038/s41579-019-0191-8. [DOI] [PubMed] [Google Scholar]
- 166.Deehan EC, Yang C, Perez-Muñoz ME, Nguyen NK, Cheng CC, Triador L, Zhang Z, Bakal JA, Walter J. 2020. Precision microbiome modulation with discrete dietary fiber structures directs short-chain fatty acid production. Cell Host Microbe 27:389–404.e6. doi: 10.1016/j.chom.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 167.Leeming ER, Johnson AJ, Spector TD, Le Roy CI. 2019. Effect of diet on the gut microbiota: rethinking intervention duration. Nutrients 11:2862. doi: 10.3390/nu11122862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, Serrazanetti DI, Di Cagno R, Ferrocino I, Lazzi C, Turroni S, Cocolin L, Brigidi P, Neviani E, Gobbetti M, O'Toole PW, Ercolini D. 2016. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 65:1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 169.Ghosh TS, Rampelli S, Jeffery IB, Santoro A, Neto M, Capri M, Giampieri E, Jennings A, Candela M, Turroni S, Zoetendal EG, Hermes GDA, Elodie C, Meunier N, Brugere CM, Pujos-Guillot E, Berendsen AM, De Groot L, Feskins EJM, Kaluza J, Pietruszka B, Bielak MJ, Comte B, Maijo-Ferre M, Nicoletti C, De Vos WM, Fairweather-Tait S, Cassidy A, Brigidi P, Franceschi C, O'Toole PW. 2020. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut 69:1218–1228. doi: 10.1136/gutjnl-2019-319654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Meslier V, Laiola M, Roager HM, De Filippis F, Roume H, Quinquis B, Giacco R, Mennella I, Ferracane R, Pons N, Pasolli E, Rivellese A, Dragsted LO, Vitaglione P, Ehrlich SD, Ercolini D. 2020. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 69:1258–1268. doi: 10.1136/gutjnl-2019-320438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Macdonald TT, Monteleone G. 2005. Immunity, inflammation, and allergy in the gut. Science 307:1920–1925. doi: 10.1126/science.1106442. [DOI] [PubMed] [Google Scholar]
- 172.Xavier RJ, Podolsky DK. 2007. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 173.Sartor RB. 2005. Role of commensal enteric bacteria in the pathogenesis of immune-mediated intestinal inflammation: lessons from animal models and implications for translational research. J Pediatr Gastroenterol Nutr 40(Suppl 1):S30–S31. doi: 10.1097/00005176-200504001-00018. [DOI] [PubMed] [Google Scholar]
- 174.Sokol H, Lay C, Seksik P, Tannock GW. 2008. Analysis of bacterial bowel communities of IBD patients: what has it revealed? Inflamm Bowel Dis 14:858–867. doi: 10.1002/ibd.20392. [DOI] [PubMed] [Google Scholar]
- 175.Kostic AD, Xavier RJ, Gevers D. 2014. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Laserna-Mendieta EJ, Clooney AG, Carretero-Gomez JF, Moran C, Sheehan D, Nolan JA, Hill C, Gahan CGM, Joyce SA, Shanahan F, Claesson MJ. 2018. Determinants of reduced genetic capacity for butyrate synthesis by the gut microbiome in Crohn's disease and ulcerative colitis. J Crohns Colitis 12:204–216. doi: 10.1093/ecco-jcc/jjx137. [DOI] [PubMed] [Google Scholar]
- 177.Heuschkel RB, Menache CC, Megerian JT, Baird AE. 2000. Enteral nutrition and corticosteroids in the treatment of acute Crohn's disease in children. J Pediatr Gastroenterol Nutr 31:8–15. doi: 10.1097/00005176-200007000-00005. [DOI] [PubMed] [Google Scholar]
- 178.Lewis JD, Abreu MT. 2017. Diet as a trigger or therapy for inflammatory bowel diseases. Gastroenterology 152:398–414.e6. doi: 10.1053/j.gastro.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 179.MacLellan A, Connors J, Grant S, Cahill L, Langille M, Van Limbergen J. 2017. The impact of exclusive enteral nutrition (EEN) on the gut microbiome in Crohn's disease: a review. Nutrients 9:0447. doi: 10.3390/nu9050447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gerasimidis K, Bertz M, Hanske L, Junick J, Biskou O, Aguilera M, Garrick V, Russell RK, Blaut M, McGrogan P, Edwards CA. 2014. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn's disease during enteral nutrition. Inflamm Bowel Dis 20:861–871. doi: 10.1097/MIB.0000000000000023. [DOI] [PubMed] [Google Scholar]
- 181.Cani PD. 2014. Metabolism in 2013: the gut microbiota manages host metabolism. Nat Rev Endocrinol 10:74–76. doi: 10.1038/nrendo.2013.240. [DOI] [PubMed] [Google Scholar]
- 182.Louis P, Hold GL, Flint HJ. 2014. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 183.Chambers ES, Preston T, Frost G, Morrison DJ. 2018. Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr Nutr Rep 7:198–206. doi: 10.1007/s13668-018-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Võsa U, Mujagic Z, Masclee AAM, Jonkers DMAE, Oosting M, Joosten LAB, Netea MG, Franke L, Zhernakova A, Fu J, Wijmenga C, McCarthy MI. 2019. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet 51:600–605. doi: 10.1038/s41588-019-0350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Blaak EE, Canfora EE, Theis S, Frost G, Groen AK, Mithieux G, Nauta A, Scott K, Stahl B, van Harsselaar J, van Tol R, Vaughan EE, Verbeke K. 2020. Short chain fatty acids in human gut and metabolic health. Benef Microbes 11:411–455. doi: 10.3920/BM2020.0057. [DOI] [PubMed] [Google Scholar]
- 186.Chambers ES, Viardot A, Psichas A, Morrison DJ, Murphy KG, Zac-Varghese SE, MacDougall K, Preston T, Tedford C, Finlayson GS, Blundell JE, Bell JD, Thomas EL, Mt-Isa S, Ashby D, Gibson GR, Kolida S, Dhillo WS, Bloom SR, Morley W, Clegg S, Frost G. 2015. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64:1744–1754. doi: 10.1136/gutjnl-2014-307913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. 2008. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A 105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ma H, Patti ME. 2014. Bile acids, obesity, and the metabolic syndrome. Best Pract Res Clin Gastroenterol 28:573–583. doi: 10.1016/j.bpg.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Guban J, Korver DR, Allison GE, Tannock GW. 2006. Relationship of dietary antimicrobial drug administration with broiler performance, decreased population levels of Lactobacillus salivarius, and reduced bile salt deconjugation in the ileum of broiler chickens. Poult Sci 85:2186–2194. doi: 10.1093/ps/85.12.2186. [DOI] [PubMed] [Google Scholar]
- 190.Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan CG. 2014. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U S A 111:7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Penney NC, Kinross J, Newton RC, Purkayastha S. 2015. The role of bile acids in reducing the metabolic complications of obesity after bariatric surgery: a systematic review. Int J Obes (Lond) 39:1565–1574. doi: 10.1038/ijo.2015.115. [DOI] [PubMed] [Google Scholar]
- 192.Dudley JT, Butte AJ. 2009. Identification of discriminating biomarkers for human disease using integrative network biology. Pac Symp Biocomput 2009:27–38. [PMC free article] [PubMed] [Google Scholar]
- 193.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, Sauk JS, Wilson RG, Stevens BW, Scott JM, Pierce K, Deik AA, Bullock K, Imhann F, Porter JA, Zhernakova A, Fu J, Weersma RK, Wijmenga C, Clish CB, Vlamakis H, Huttenhower C, Xavier RJ. 2019. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tong TYN, Koulman A, Griffin JL, Wareham NJ, Forouhi NG, Imamura FA. 2020. Combination of metabolites predicts adherence to the Mediterranean diet pattern and its associations with insulin sensitivity and lipid homeostasis in the general population: The Fenland Study, United Kingdom. J Nutr 150:568–578. doi: 10.1093/jn/nxz263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cummings JH, Engineer A. 2018. Denis Burkitt and the origins of the dietary fibre hypothesis. Nutr Res Rev 31:1–15. doi: 10.1017/S0954422417000117. [DOI] [PubMed] [Google Scholar]
- 196.Walker AW, Duncan SH, Harmsen HJM, Holtrop G, Welling GW, Flint HJ. 2008. The species composition of the human intestinal microbiota differs between particle-associated and liquid phase communities. Environ Microbiol 10:3275–3283. doi: 10.1111/j.1462-2920.2008.01717.x. [DOI] [PubMed] [Google Scholar]
- 197.Patnode ML, Beller ZW, Han ND, Cheng J, Peters SL, Terrapon N, Henrissat B, Le Gall S, Saulnier L, Hayashi DK, Meynier A, Vinoy S, Giannone RJ, Hettich RL, Gordon JI. 2019. Interspecies competition impacts targeted manipulation of human gut bacteria by fiber-derived glycans. Cell 179:59–73.e13. doi: 10.1016/j.cell.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]


