With increasing environmental concerns about the exhausting use of fossil fuels, development of a sustainable biofuel production platform has been attracting significant public attention. Ethanologenic Z. mobilis species are endowed with an efficient ethanol fermentation capacity that surpasses, in several respects, that of baker’s yeast (Saccharomyces cerevisiae), the most-used microorganism for ethanol production.
KEYWORDS: Alphaproteobacteria, Zymomonas mobilis, cell division, cell growth, cellular heterogeneity, chromosome organization, ftsZ
ABSTRACT
The alphaproteobacterium Zymomonas mobilis exhibits extreme ethanologenic physiology, making this species a promising biofuel producer. Numerous studies have investigated its biology relevant to industrial applications and mostly at the population level. However, the organization of single cells in this industrially important polyploid species has been largely uncharacterized. In the present study, we characterized basic cellular behavior of Z. mobilis strain Zm6 under anaerobic conditions at the single-cell level. We observed that growing Z. mobilis cells often divided at a nonmidcell position, which contributed to variant cell size at birth. However, the cell size variance was regulated by a modulation of cell cycle span, mediated by a correlation of bacterial tubulin homologue FtsZ ring accumulation with cell growth. The Z. mobilis culture also exhibited heterogeneous cellular DNA content among individual cells, which might have been caused by asynchronous replication of chromosome that was not coordinated with cell growth. Furthermore, slightly angled divisions might have resulted in temporary curvatures of attached Z. mobilis cells. Overall, the present study uncovers a novel bacterial cell organization in Z. mobilis.
IMPORTANCE With increasing environmental concerns about the use of fossil fuels, development of a sustainable biofuel production platform has been attracting significant public attention. Ethanologenic Z. mobilis species are endowed with an efficient ethanol fermentation capacity that surpasses, in several respects, that of baker’s yeast (Saccharomyces cerevisiae), the most-used microorganism for ethanol production. For development of a Z. mobilis culture-based biorefinery, an investigation of its uncharacterized cell biology is important, because bacterial cellular organization and metabolism are closely associated with each other in a single cell compartment. In addition, the current work demonstrates that the polyploid bacterium Z. mobilis exhibits a distinctive mode of bacterial cell organization, likely reflecting its unique metabolism that does not prioritize incorporation of nutrients for cell growth. Thus, another significant result of this work is to advance our general understanding in the diversity of bacterial cell architecture.
INTRODUCTION
Over the last 2 decades, there has been significant progress in uncovering the complexity of bacterial cellular organization (1). These developments owe a great deal to recent advances in visualization techniques and genetic tools, enabling us to scrutinize bacterial cells. However, mostly due to the availability of tools, strains, and established protocols, our knowledge of bacterial cellular organization is still limited to major model organisms, such as Escherichia coli, Bacillus subtilis, Caulobacter crescentus, and others (1, 2).
It has been shown that several bacteria do not necessarily follow the canonical modes of growth and cell division displayed in model bacterial species (2–4). For example, species of the animal symbiont bacterium Thiosymbion divide longitudinally, exhibiting a striking contrast to the canonical vertical division (5). Polarized cell division in the intracellular pathogen Chlamydia trachomatis is another example of an unusual mode of propagation (6). Thus, although knowledge obtained from the bacterial models is immensely useful and covers common key mechanisms, it is unclear to what extent the studied mechanisms can be applied to other bacteria with specific metabolisms, physiologies, or living strategies.
The facultative anaerobic bacterium Zymomonas mobilis is a natural ethanologen. Previous studies have extensively characterized its physiology with relation to industrial applications such as biofuel production (7). Z. mobilis is renowned for its relatively simple and very fast catabolism involving the Entner-Doudoroff (ED) pathway coupled to the high activity of pyruvate carboxylase and alcohol dehydrogenase, which allows Z. mobilis to convert glucose to ethanol nearly at theoretical yields under anaerobic conditions (7, 8). Thus, only small amounts of substrate carbon are incorporated as biomass in Z. mobilis, making this species an attractive biocatalyst in refinery systems. Recent advances in metabolic engineering of Z. mobilis have broadened its substrate spectrum and increased its stress tolerance (9).
Besides being the most efficient ethanol producer among bacteria, Z. mobilis is also known for its aerobic respiration, which yields very little or no energy (8, 10). Z. mobilis carries a complete electron transport chain (ETC) which is constitutively expressed under anaerobic conditions (8, 10, 11). Despite its high rate of oxygen respiration, surpassing that of E. coli, and the possession of FoF1-type ATPase (12), there is no evidence of oxidative phosphorylation for energy generation in aerobically growing Z. mobilis cells (8). Z. mobilis appears to produce energy solely through substrate phosphorylation by the ED pathway. Interestingly, aerobically growing Z. mobilis produces acetaldehyde in the culture as respiration withdraws the electron donor NADH from ethanol synthesis reactions (8, 13). Accumulated acetaldehyde in the culture is toxic to the cells, and thus, aeration perturbs the cell growth (14). The biological significance of aerobic respiration in Z. mobilis remains elusive (15–17).
Despite its industrially appealing physiology, the cellular organization of Z. mobilis has never been addressed. In contrast to other industrial workhorses, such as E. coli, B. subtilis, and Saccharomyces cerevisiae, in which cell biology has been thoroughly scrutinized, only limited investigations have been done in Z. mobilis cells. This lack of understanding of Z. mobilis cell biology may be a bottleneck for manipulation of its metabolism to be fully exploited, considering that cell growth and division are a consequence of the glycolysis that also produces ethanol as the major end product. Thus, better understanding in the regulation of cell geometry is needed to guide rational metabolic engineering for Z. mobilis-based biorefineries.
Z. mobilis belongs to the class Alphaproteobacteria, a bacterial group exhibiting a wide range of cell shapes and living strategies (18). Recent reports suggest that Z. mobilis cells are polyploid (19, 20). Although polyploidy is prevalent across the kingdom of life, it has been explored very little in bacteriology, as major model species are monoploids (21). Thus, investigating Z. mobilis polyploidy and relevant cell biological features would deepen our understanding of the diverse repertoire of bacterial cell organization.
In the present study, we discovered and characterized the heterogeneous nature of Z. mobilis cells.
RESULTS AND DISCUSSION
Growth and division of single cells of Z. mobilis strain Zm6.
Growth kinetics of Z. mobilis cells under various conditions are well characterized at the population level (22). In contrast, single-cell growth of Z. mobilis has not been examined before, and therefore, important features of Z. mobilis biology may have been overlooked.
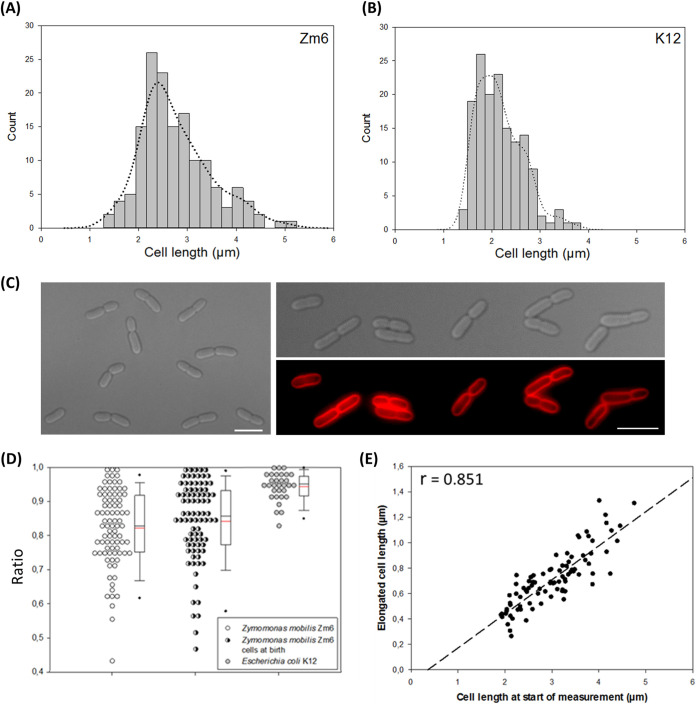
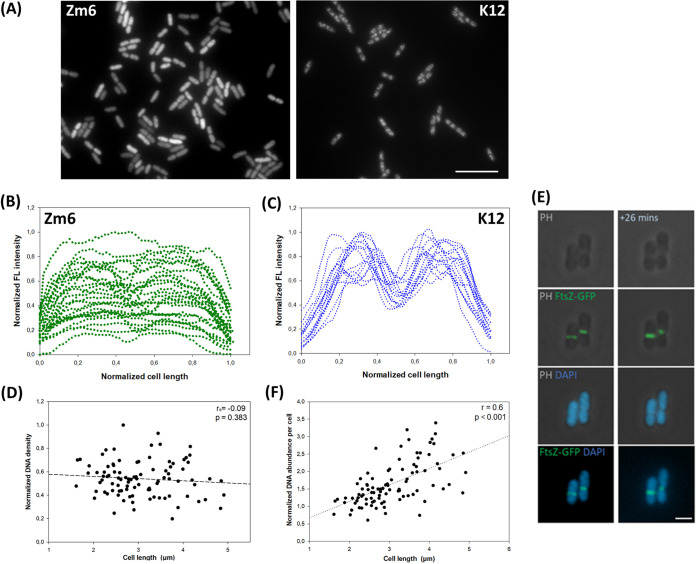
We first determined cell size and shape of growing Z. mobilis strain Zm6 in complex medium under anaerobic conditions and compared them with those of E. coli strain K-12 MG1655 (referred to here as K-12) growing aerobically in LB medium. Microscopy imaging showed that volumes of growing Zm6 cells were generally larger than those of growing E. coli K-12 cells (Fig. 1A and B), the mean cellular lengths and widths being 2.81 ± 0.71 μm and 1.28 ± 0.08 μm, respectively, for Zm6 and 2.16 ± 0.47 μm and 0.93 ± 0.06 μm, respectively, for K-12. The Zm6 cells also exhibited a wider distribution of cell lengths than K-12 cells (Fig. 1A and B). The Zm6 cells at later growth phases (optical density at 600 nm [OD600] = 1.5) in the same culture had longer cells (3.22 ± 0.75 μm; n = 315), possibly caused by the toxicity of accumulated ethanol in the culture (2.9 g/liter). The doubling times for Zm6 and K-12 cells in the actively dividing phase were 105 min and 28 min, respectively.
FIG 1.
Cell size, morphology, and single-cell growth profile of Z. mobilis strain Zm6. (A and B) Histogram of cell length in growing Z. mobilis strain Zm6 (OD600 = 0.44; n = 150) and growing E. coli strain K-12 MG1655 (OD600 = 0.7; n = 150). The Zm6 cell population exhibited a wider range of cell lengths than K-12 cells. Estimated kernel density was plotted by a bandwidth of 0.25 for both histograms. (C) Phase-contrast images of growing Zm6 cells (left and right top) and fluorescence image of Zm6 stained with the membrane dye FM4-64 (right bottom). Zm6 was grown in the complex medium (OD600 = 1) under anaerobic conditions, stained with FM4-64 (20 μg/ml) for 15 min, and subsequently washed with PBS before being mounted on an agarose pad for imaging. Note that attached daughter cells often showed different cell lengths. Red, Fm4-64. Bar, 5 μm. (D) Dot density plot of cell length ratio between attached paired cells (length of short cell to length of long cell). Data for Zm6 pairs at a random cell cycle stage (n = 85) (left plot), Zm6 pairs at birth (n = 87) (middle plot), and K-12 pairs at random cell cycle stage (n = 31) (right plot) are shown. Box-and-whisker plots are marked with means (red) and medians (blue). (E) Plot of elongated-cell length during 40 min of growth by a function of cell length at the start of measurement. r, Pearson's correlation coefficient. P < 0.001. n = 88.
Zm6 cells formed rod shapes with elliptical ends (Fig. 1C). We also frequently observed sibling Z. mobilis cells attached as pairs (Fig. 1C), as previously reported (23). Chained cells were occasionally observed as well. Conversely, E. coli cells did not show such an attachment at the septum in most growing cells, implying that septal hydrolysis in Z. mobilis cells was not as active as in E. coli cells. Interestingly, the attached paired Z. mobilis cells often showed different cell lengths within a pair (Fig. 1C). We found this variant cell size between sibling cells intriguing and further examined the cause and consequence of this variance.
We considered the possibility that apical growth, the growth mode employed by several Alphaproteobacteria species (18), might be a potential cause of the different lengths of sibling cells. To test if Zm6 cells grow apically, we stained the growing cells with fluorescent d-amino acids to visualize sites of active peptidoglycan synthesis (24). The staining demonstrated that Z. mobilis cells incorporated new cell wall materials at the division site and lateral walls (see Fig. S1 in the supplemental material). Thus, Zm6 cells grow at the septum and the lateral cell wall, excluding an apical growth as the cause.
We then performed time-lapse imaging to monitor the growth of single cells for approximately two generations. Growing Zm6 cells were mounted on an agarose pad, sealed, and incubated for about 30 min before imaging. This was done to minimize microaerobic stress during the transfer, to ensure that Zm6 cells would continue to grow anaerobically during image acquisition. The imaging series revealed that the constrictions took place often around, but not exactly at, midcell positions. Interestingly, division site selection was not strictly regulated to the midcell position in Zm6 cells, exhibiting a sharp contrast to accurate midcell positioning found in many bacterial species.
Furthermore, when the cell length ratio of attached pairs at birth (short to long) was compared with the ratio of paired cells at random cell cycle stages, the mean ratio was slightly higher in the pairs at birth (Fig. 1D). This indicates that another mechanism might have contributed to the different cell lengths within pairs during growth. We then measured elongation of cells for 40 min in randomly selected Zm6 cells and plotted it as a function of cell length at the start of measurement. The plot showed a positive simple linear correlation (Fig. 1E), demonstrating that the cell growth capacity per fixed cell volume was almost constant between individual cells.
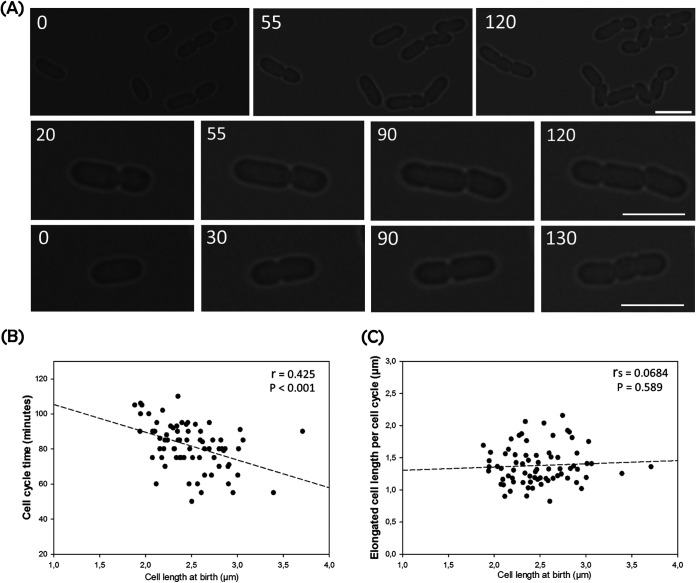
The combination of two mechanisms, nonmidcell division and size-dependent growth, could result in production of long, filamentous cells. However, this was not the case, and we therefore looked for an explanation. By following the growth of paired sibling cells, we observed that longer cells divided earlier than the shorter ones (Fig. 2A). Among paired cells with a length difference of more than 25% (i.e., a short-long daughter cell length ratio of <0.8), 74% of pairs (n = 27) showed that the longer cells in the pairs started constriction 20 min earlier than the shorter cells. To determine whether this was sibling specific, we plotted cell cycle time versus cell size at birth for individual cells, showing a negative regression (Fig. 2B). Hence, longer Zm6 cells tended to divide earlier than shorter ones, regardless of pairing. This was an intriguing paradox, as we had observed that the cells actively generated variant cell size over divisions but, at the same time, deployed a strategy to limit size variance by modulating their cell cycle time.
FIG 2.
Time-lapse imaging of growing Zm6 cells. Growing Zm6 cells in liquid cultures (OD600, 0.5 to 0.75) were directly mounted on complex-medium–agarose pads and incubated for about 30 min before time-lapse imaging. Cell growth and division were analyzed using ImageJ (NIH). (A) Time-lapse imaging of growing Zm6 cells. Images are displayed over time, from left to right. Representative examples of early division by long sibling cells are shown in the middle and bottom panels. Numbers indicate minutes after imaging started. Bars, 3.9 μm. (B) The cell cycle time of individual cells is plotted against cell size at birth. Negative correlation is shown by Pearson's r, with a P value of <0.001; n = 73. (C) Plot of elongated cell length over one generation by a function of birth size. No correlation was found by Spearman's rank correlation test. P = 0.589; n = 73.
These observations reflected questions raised about bacterial size regulation, which has been a subject of intense debate (25–27). Several reports using single-cell quantitative approaches have demonstrated that several bacterial species grow to a fixed cell volume per cell cycle during steady-state growth, regardless of cell size at birth (28–32). The addition of a constant cell volume, described by the adder model (28–32), results in less cell size variation over cell cycles. However, the rule was not applied in E. coli under slow-growth conditions (31, 33) or in Mycobacterium species that grow asymmetrically by apical growth (34).
We then attempted to examine whether Z. mobilis cells applied the constant increment regulation. We measured ΔCL, the elongated cell length over a cell cycle in individual cells, and plotted it against cell length at birth (Fig. 2C). It should be noted that several small cells generated during imaging did not complete division due to the time limit of the method, and these were not included in the analysis.
The analysis displayed dispersed plotting, indicating that ΔCL did not appear to be strictly constant at the individual cell level under our experimental conditions. The relative standard deviation of ΔCL (21.2%) was significantly higher than those of other bacteria previously characterized (28, 35). The regression of plotting, however, did not show a slope over cell length, and no correlation was found (Fig. 2C). At the population level, the cell growth may have been governed by the constant ΔCL principle, despite the fact that this principle was not strictly executed by individual cells. Further analysis using a more defined imaging method, combined with mathematical models, is required to identify the specific governing law of cell size homeostasis in Z. mobilis.
Taken together, the data presented here provide evidence that variant Z. mobilis cell size is generated by nonmidcell, asymmetric cell division and further enhanced by size-dependent growth of single cells. The generated long cells execute division roughly timed according to added cell volume or other unknown cues. Consequently, cells that are short at birth have longer cell cycle spans than cells that are longer at birth. These cellular organizations appear to maintain variant cell size in Z. mobilis cell populations while preventing excessively large cells.
Subcellular localization of FtsZ in Z. mobilis cells.
To gain further insights into Z. mobilis cell division, we fused Z. mobilis FtsZ (ZZ6_0455) to green fluorescent protein (GFP) to visualize its subcellular localization. The bacterial tubulin homologue FtsZ plays a central role in bacterial cytokinesis by recruiting proteins required for division (1). We constructed a pBBR1 plasmid derivative carrying ftsZ-gfp with the native promoter that allowed expression in strain Z6KF2, into which it was introduced (Fig. 3). Our analysis of the effect of the fusion (Fig. S2) led us to conclude that the possibility of artifacts with the FtsZ-GFP fusion is limited, and we used strain Z6KF2 for further investigations.
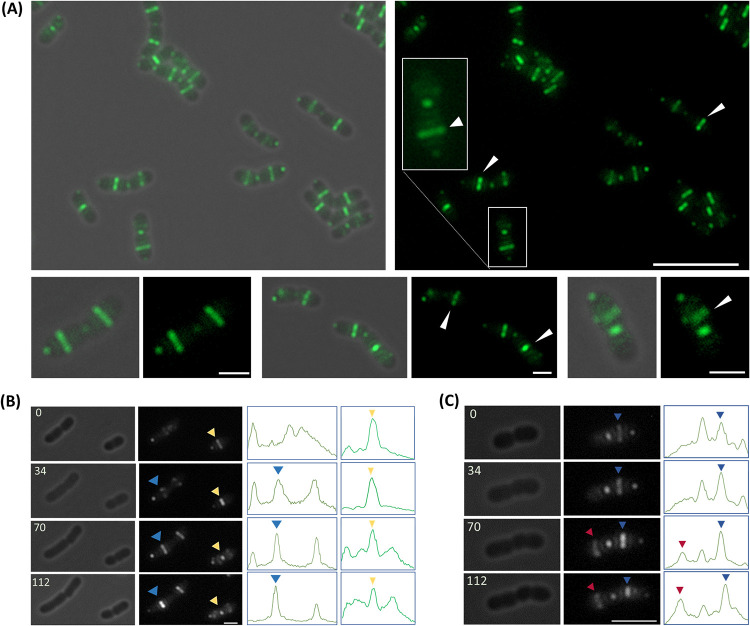
FIG 3.
Subcellular localization of FtsZ-GFP in Z. mobilis strain Z6KF2. (A) FtsZ-GFP localizes as a Z-ring in the majority of Z6KF2 cells. (Left) Overlays of phase-contrast and fluorescence images (green, FtsZ-GFP). (Right) Fluorescence images. Bottom panels show enlarged images of single cells. White arrowheads indicate cells that had earlier Z-ring formation or more abundant FtsZ in the Z-ring in one of the paired cells. Bars, 10 μm (top) and 2 μm (bottom). (B and C) Time-lapse imaging of growing Z6KF2 cells. Phase-contrast images (left) are compared with fluorescence images (middle) (white, FtsZ-GFP). Line profiles (right) show the relative FL intensities of GFP along the long axis of each cell. Colored arrowheads indicate FtsZ-GFP fluorescence signals and corresponding peaks in line profiles. Numbers in phase-contrast images indicate minutes after commencement of imaging. Earlier Z-ring formation (indicated by a blue arrowhead at 34 min in panel C) was linked to earlier division (indicated by a blue arrowhead at 70 min in panel C) in paired cells. Bars, 2 μm (B) and 4 μm (C).
Fluorescence imaging showed that FtsZ-GFP localizes as a Z-ring in Z6KF2 cells (Fig. 3). Z-rings position at division sites in Z. mobilis cells, as Z-rings and division sites along the long axis of cells were found very close to each other (Fig. 1D; Fig. S3). FtsZ-GFP foci were also found at the free cell poles of many cells (Fig. 3), a phenomenon previously observed in other Alphaproteobacteria (36–38).
Interestingly, some attached pairs exhibited different FtsZ-GFP patterns within a pair; typically, the fluorescence (FL) signal intensity of the Z-ring was generally higher in the larger sibling cell than the smaller cell (Fig. 3A and B). We also observed some pairs in which a Z-ring had already formed in the larger daughter cell, while only signal clusters were present in the smaller cell (Fig. 3A and C).
A recent report suggested that cellular FtsZ concentration is not constant among E. coli cells during the cell cycle under slow-growth conditions (39). The observed abundance of FtsZ as a ring variant likely reflected different cell cycle stages of Z. mobilis cells. We found that 51.2% (n = 82) of the paired cells showed more than a 1.5-fold difference in FtsZ ring FL intensity peaks within a pair. Thus, temporal regulation of FtsZ expression often varied within paired cells, despite the fact that that pairs were born at the same time. Therefore, not only its spatial regulation but also temporal regulation of FtsZ appear to be heterogeneous events among Zm6 cells.
Early Z-ring formation was possibly linked to early constriction in Zm6 cells, but polyploid bacteria might have checkpoints for chromosomal organizations that may delay the division. The Zm6 cells also divide at noncentral positions, which might have a destabilizing effect on the Z-ring. We then performed time-lapse imaging to visualize FtsZ-GFP localization over the cell cycle. Imaging was performed at intervals of 15 to 20 min to minimize the effects of phototoxicity and bleaching. The imaging of attached paired cells showed that formation of the Z-ring 20 min earlier in one of the paired cells resulted in constriction 20 min earlier in that cell (Fig. 3C), for all observed pairs (n = 15; 100%). Thus, our data indicate that there were possibly no major checkpoints to delay constriction after formation of the Z-ring.
Furthermore, we found a positive correlation between FL peak intensity of FtsZ ring and cell size (Fig. S4). The plot shows that FtsZ accumulation at the Z-ring generally increases with cell size, suggesting that large cells accumulate FtsZ at the Z-ring faster than smaller cells, taking into account the faster growth of large cells (Fig. 1E). Thus, it appears that size- or growth-correlated FtsZ ring accumulation at the division site is likely a key fundamental for temporal control of Z. mobilis division.
Interestingly, a recent study suggested that balanced biosynthesis and a threshold of accumulation of FtsZ are the key mechanisms for accomplishing the adder rule in E. coli cells growing at steady state (30). Although it is not clear if Z. mobilis uses the constant increment rule for size regulation, the principle underlying the E. coli adder rule may be applied in the size correction mechanism of Z. mobilis cells.
Spatial FtsZ regulation in Z. mobilis cells.
Next, to understand spatial regulation of Z. mobilis FtsZ, we conducted an in silico survey for Z. mobilis homologs of known cell cycle and FtsZ regulators in other bacteria. We found that MinCDE complex and SlmA, prototypical negative regulators of FtsZ, are not encoded in the Z. mobilis genome (40, 41). Instead, the Z. mobilis Zm6 chromosome encodes a C. crescentus mipZ homologue (ZZ6_0908). MipZ is a P-loop ATPase that negatively regulates FtsZ positioning in C. crescentus by acting as an antagonist of Z-ring formation (42). Notably, a recent report suggests that MipZ also regulates cell septation by localizing at midcell in Rhodobacter sphaeroides, a member of the Alphaproteobacteria that carries two different chromosomes (43). Thus, Z. mobilis MipZ might possess an adapted function in polyploidy, in addition to its function in the spatial regulation of FtsZ.
To gain additional insights into FtsZ spatial regulation, we set out to monitor the division process of stress-induced filamentous Z. mobilis cells. A previous report demonstrated that Min proteins accurately regulate positioning of multiple Z-rings in stress-induced filamentous E. coli cells, which ensures cell size control of daughter cells (44). We exploited this type of stress response to visualize Z. mobilis FtsZ positioning regulation at the cellular level.
We first attempted a cephalexin treatment (40 μg/ml) to induce filamentation. However, we found that Z. mobilis has a high intrinsic tolerance to cephalexin, as it does to most commonly used antibiotics (45). We sought an alternative approach and learned that the salt NaCl at a mild concentration induces filamentation and a bulge of a single pole in Z. mobilis cells (46). Utilizing this response, we induced ∼30-μm-long filamentous Z6KF2 cells and monitored the subcellular localization of FtsZ-GFP.
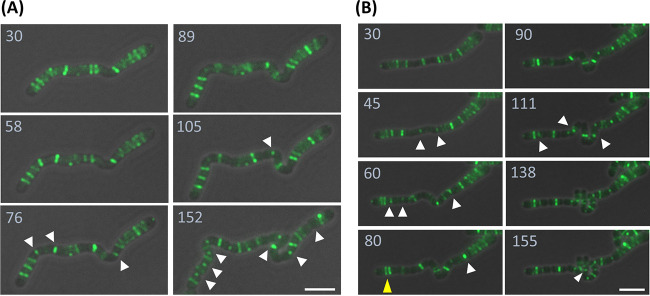
During the growth under saline stress (0.2 M NaCl), FtsZ-GFP formed foci or ambiguous structures in the filamented Z6KF2 cells (Fig. S5). After the cells were transferred to the identical medium without salt, the FtsZ-GFP assemblies were reorganized to several ladder-like structures, as the cells started to regrow and divide (Fig. 4A and B). During the recovery phase, we observed randomized FtsZ-GFP positioning, for example, in proximate double FtsZ-GFP rings (Fig. 4B, yellow arrowhead at 80 min). Also, the timing of cell division was not coordinated among cells, as shown by simultaneous ladder-like divisions found in a part of cells (Fig. 4A, 152 min). The Z. mobilis FtsZ spatiotemporal regulation appeared not to be under tight control during recovery, in contrast to one-by-one divisions organized by equally spaced FtsZ rings in the filamentous E. coli cells (44).
FIG 4.
Subcellular localization of FtsZ-GFP in filamentous Z6KF2 cells during recovery from stress. Growing Zm6 cells were subcultured in the complex medium supplemented with 0.2 M NaCl and grown for 5 h. The cells were then transferred to an agarose pad containing growth medium without additional salt. After 30 min of an incubation that allowed recovery and regrowth of the transferred cells, imaging commenced. Time-lapse imaging of two long filamentous cells is displayed. White arrowheads indicate the positions where cell division was completing or just completed. Note that positioning of FtsZ-GFP was not tightly controlled in the filamentous Zm6 cells. (A) A ladder-like FtsZ-GFP structure resulted in simultaneous divisions at 152 min. (B) Proximate double FtsZ-GFP rings, indicated by a yellow arrowhead, were observed. All images are overlays of phase-contrast and fluorescence images (green, FtsZ-GFP). Numbers indicate minutes after cells were mounted on the complex-medium–agarose pad. Bars, 5 μm.
Importantly, the filamentous Z. mobilis cells managed to divide and regrow, in spite of the random positioning of division sites and timing, indicating that tightly coordinated division was not necessary for cell growth. These observations implied that Z. mobilis might have evolved to produce different-sized daughter cells during stress response, for better survival after recovery from stress.
DNA content in Z. mobilis cells.
Previously, a transposon mutagenesis study using Z. mobilis strain Zm4 showed that the transposon insertion mutants exhibited heterozygous genotypes, suggesting that Z. mobilis is a polyploid organism (20). The copy number of the Zm4 chromosome (2.05 Mbp) was estimated to be >50, using quantitative PCR (qPCR) (19). Extractions of genomic DNA resulted in a similar ratio of extracted-DNA amount to extracted-cell numbers between strains Zm6 and Zm4 (1.24 ± 0.03 μg/108 cells for Zm6; 1.38 ± 0.02 μg/108 cells for Zm4). Based on similar genomic content between two Z. mobilis strains, it is likely that Zm6 also possesses a similar polyploidy feature. We therefore asked how multiple chromosomes organize in relation to growth and cell cycle in Zm6 cells.
We visualized nucleoids by staining live Zm6 cells with the DNA dye Syto 9. The staining showed dispersed nucleoid localization throughout the cells, and no obvious condensation of nucleoids was found (Fig. 5A). Interestingly, the average FL intensities within a cell compartment significantly varied between individual cells, implying that cellular DNA crowdedness or density was heterogeneous in the culture (Fig. 5A and B). We observed no anucleate Zm6 cells.
FIG 5.
Analysis of stained nucleoids in Z. mobilis cells. (A) Fluorescence images of DNA staining in growing cells of Z. mobilis strain Zm6 (left) and E. coli strain K-12 (right). Zm6 cells were stained with Syto 9 at 0.55 μM for 10 min. K-12 cells were stained with 5 μg/ml DAPI for 15 min. Both fluorescence signals are white in the images. Note that FL signal intensities varied between Z. mobilis cells. Bar, 10 μm. (B) FL signal profiling of 30 Zm6 cells (n = 30) over the long axis of each cell. FL signal and cell length were normalized. Profiles show that the intensity varied between Zm6 cells. (C) FL signal profiling of K-12 cells (n = 15) over the long axis of each cell. FL signal and cell length were normalized. K-12 cells exhibited similar FL profiles. (D) Plot of normalized average of FL intensity within Zm6 cells, as a function of cell length. There is no correlation between the two factors (Spearman's rank correlation test; P = 0.589, n = 96). (E) Newly dividing Z6KF2 cells labeled with DAPI. Panels (top to bottom) show phase-contrast images, overlays of phase-contrast and fluorescence images (green), overlays of phase-contrast and fluorescence images (blue), and overlays of two fluorescence images (green and blue). Right images were taken 26 min after left images. Green, FtsZ-GFP; blue, DAPI. Bar, 2 μm. (F) Plot of normalized DNA abundance per cell compartment by the cell length of Zm6 cells. A positive correlation (Pearson's r = 0.6) was found. n = 96.
For comparison, we stained E. coli strain K-12 by using the DNA dye DAPI (4′,6-diamino-2-phenylindole) and observed localization patterns of E. coli nucleoids similar to those previously reported (47, 48). Nucleoid occlusion and condensation were observed in all E. coli cells (Fig. 5A). The FL intensity of stained DNA was approximately the same between different K-12 cells (Fig. 5A and C).
We initially suspected that the variant signal intensities in Zm6 cells might be an artifact from the staining procedure. We therefore attempted to exclude this possibility by employing different staining methods, and similar results were obtained (Fig. S6). As Z. mobilis is polyploid, we concluded that the growing Zm6 culture exhibited heterogeneous cellular DNA crowdedness/density among individual cells.
Interestingly, 1- to 3-fold FL intensity variance was found between similar-sized Zm6 cells (Fig. 5D). Non-midcell division was a potential suspect for causing uneven distribution of macromolecules to daughter Z. mobilis cells. Nevertheless, most of the attached sibling cells, including newly dividing pairs, showed very similar FL intensity within a pair (Fig. 5E; Fig. S7). This demonstrated that division was not the main cause of the large variance in intensity.
Here, another interpretation of the equal signal intensities within pairs is that chromosome numbers per single-cell compartment varied between sibling cells, roughly proportional to cell size difference (Fig. 1D). Thus, interestingly, the multiple copies of chromosomes in parental cells appeared to be unequally distributed between the daughter cells, according to their birth sizes, to ensure that chromosome numbers per cell volume, i.e., DNA crowdedness, became roughly equal between daughter cells. This type of chromosome distribution resembles the observation made in the minD mutant of the polyploid cyanobacterium Synechococcus elongatus (49), and thus, there may be a conserved mechanism for spatial organization of multiple chromosomes among polyploid bacteria. However, the S. elongatus minD mutant produces anucleate cells, whereas Z. mobilis Zm6 strain does not.
If it is not due to nonmidcell divisions, what would cause the variant DNA crowdedness between Zm6 cells? DNA crowdedness within a cell was found not to correlate with its cell length in the Zm6 culture (Fig. 5D). However, DNA abundance per cell compartment (average FL intensity integrated over the cell length) showed a modest positive correlation with cell size (Fig. 5F). This correlation resembles cell size-to-genome copy number coordination in polyploid bacterial species. For example, stepwise, asynchronous replication was observed in S. elongatus to ensure a positive correlation between chromosome numbers and cell volume (49, 50). A similar correlation was also found in the symbiont bacterium Sinorhizobium meliloti during its differentiation to the bacteroid form (51, 52) and in the gigantic bacterium Epulopiscium species (53). Thus, asynchronous chromosome replication coordinated with cell volume expansion may be a shared trait among bacteria with multiple copies of the same chromosome.
In this context, we speculate that asynchronous chromosome replication might be loosely coordinated with cell growth in Z. mobilis and that miscoordination may have caused the heterogeneous DNA crowdedness over generations. However, this should be experimentally verified by live imaging of chromosome numbers in growing Z. mobilis cells.
Overall, our data suggestively argue that the polyploid bacterium Z. mobilis possesses a unique mode of chromosome organization. The heterogeneity of cellular DNA crowdedness observed in Z. mobilis cells appears to be unique even among polyploid bacteria, because cell size is well correlated with DNA content in polyploid bacterial species such as S. elongatus and Epulopiscium species (53, 54), and these bacteria likely exhibit homogeneous DNA density among individual cells.
Cell curvature of Z. mobilis cells.
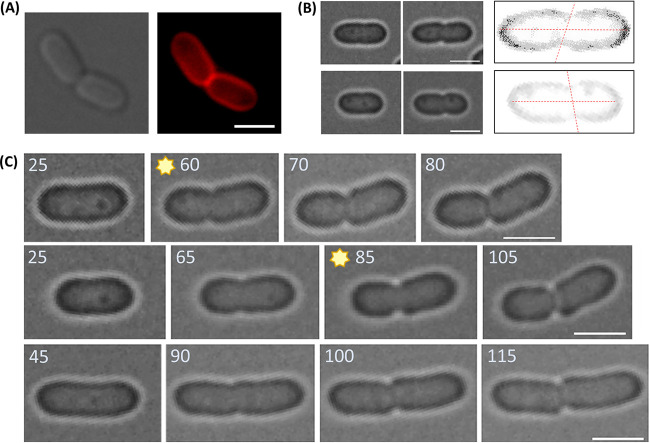
Growing Zm6 cells often existed as attached pairs (Fig. 1C), constituting 36% of the growing population (OD600, 0.8; n = 430). It was eye-catching that, in some attached pairs, extended long cell axes from the daughter cells crossed each other, instead of being in line with each other (Fig. 1C and 6A). In other words, a rim shape of two attached cells is curved or bent, despite the semisymmetric shape of each single cell.
FIG 6.
Curvature of attached Z. mobilis sibling cells. (A) FM4-64 labeling of sibling Zm6 cells that exhibit attachment-based curvature. The images are phase-contrast (left) and fluorescence (right) images. Red, FM4-64. (B) Angled division in Zm6 cells. Phase-contrast images of Zm6 cells before (left) and after (middle) constrictions took place. In the right image, the rim of a cell was made more pronounced by adjusting the contrast of the image. Two red dotted lines, showing the constriction axis and the long axis of the cell, show that the division is not perpendicular to the long axis of the cell. (C) Time-lapse imaging of single Zm6 cells showing oblique division. Division of three cells over time is displayed separately (left to right). Numbers indicate minutes after commencement of imaging. Images highlighted with stars show the curved cells before completion of division. Note that cells in the upper two panels showed the attachment curvature, while the cell in the bottom panel did not exhibit the curvature despite the inclined division. Bars, 2 μm.
These jointly curved cells were fractional in growing cultures, constituting 10.4% of the total population (pairs with an angle between the long axes of >20°; n = 430). The Zm6 cells did not separate right after division, likely due to temporary insufficient activity of peptidoglycan hydrolase, but how was attachment-based curvature generated?
Uneven cell wall hydrolysis activity could be one cause of the curvature, but it might require an extremely delicate control. Another possibility is that the curvature was mechanically introduced during the division process. With this in mind, we observed that constrictions in dividing Zm6 cells were often found slightly inclined at the short axes of cells, i.e., constriction occurred at a slightly oblique angle (Fig. 6B). This is in accordance with our observation of slanted Z-rings in some growing Z6KF2 cells (Fig. S8). For clarification of the angled division, we examined Z. mobilis cells using electron microscopy. The electron micrographs suggested that constriction was slightly inclined in some Z. mobilis cells (Fig. S9).
We then followed the fate of cells that divided in a slightly oblique manner. We noticed that the curvature/bend of a single cell was occasionally generated upon slanted constriction, but before completion of septum closure. (Fig. 6C, panels highlighted with stars). When the division was completed, the two attached cells exhibited the mentioned curvature (Fig. 6A and C). When the cells divided without an obvious angle from the short axis of the cell, bending of cells was not observed (Fig. 2A, bottom panels). These observations led us to speculate that an angled division might be a cause of the observed curvature of attached cells.
However, it should be noted that the timing of separation of attached cells appeared to be random among Z. mobilis cells. A collision of two separated elongating sibling cells, regardless of angle of the division that had just happened, could result in an appearance similar to the attached-curve phenotype observed during time-lapse imaging. This was the source of our initial skepticism regarding the speculation. However, the growth of colliding cells eventually resulted in sliding of cells, which was distinguishable from the attached curved cells. The curvature was also observed in chemically fixed cells as well, in which colliding cells were excluded in imaging. More importantly, the curvature of single cells was generated before the completion of division (Fig. 6C), suggesting that the division process by itself could generate the curve by altering the vector of turgor in cells.
Also, it should be noted that an angled constriction did not always introduce attached curvature to cells (e.g., bottom panels of Fig. 6C). Our observations suggest that a combination of nonmidcell division and angled constriction led to the curvature, but neither mechanism alone did. Further examination of the mechanical properties of Z. mobilis cells in relation to positioning and angle of divisions is required to understand the mechanism of curve generation.
While an oblique spindle orientation has been shown to be an important mechanism for generating complexity in eukaryotic cells (55–57), the orientation of the division septum in rod-shaped bacteria has not been addressed. Our literature survey found one observed bacterial diagonal-like division, which is associated with branching formation in a multicellular cyanobacterium (58). Considering its fast catabolism with inefficient growth, some Z. mobilis cells may maintain an attached-curved state for a long time in their natural habitats. Further investigations of the biological significance of Z. mobilis temporal cell curvature might afford novel insights into bacterial cell biology.
Cellular heterogeneity in Z. mobilis cell population: why and how?
Z. mobilis is well known for its extremely efficient homoethanol fermentation of simple sugars, such as glucose. Glucose is passively transported into Z. mobilis cells by the glucose facilitator proteins without an energy requirement (8, 59). This unusual transportation of glucose allows Z. mobilis cells to take up glucose approximately 8 times faster than actively growing E. coli cells do (60). This high flux of the ED pathway, known as the catabolic highway, coupled with the active pyruvate decarboxylase and the truncated tricarboxylic acid (TCA) cycle, allocates only less than 5% of carbon substrate for building biomass in Z. mobilis cells (22, 59, 61). In such a bacterium, with rapid catabolism and low efficiency of anabolism and energy production, expansive growth and frequent, accurate division might not be a master plan for surviving in nature.
The present work shows that Z. mobilis spatiotemporal cellular organization is distinctively different from those found in the model bacterial species, where active and reproducible propagations are the main drivers. This study provides evidence that Z. mobilis cells favor heterogeneity in their division sites, cell sizes and DNA content among individual cells, under laboratory nutrient-rich anaerobic growth conditions.
The observed heterogeneity might resemble a bacterial bet-hedging strategy of preparation for unpredictable environmental stress (62, 63). In fact, Z. mobilis cells are tolerant to a broad range of pHs, high ethanol content (22), and high doses of different antibiotics (45). It is tempting to speculate that the robust resilience to certain stresses by Z. mobilis, likely facilitated by its cellular heterogeneity, is conceptually compatible with its main biological activity, the rapid production of ethanol. Both strategies do not prioritize growth but rather invest in available nutrients in the defense and preparation for fluctuating environments and competitors. Further investigations on the enigmatic ecology of Z. mobilis would shed light on the implication of its cellular heterogeneity (64).
How Z. mobilis achieves its heterogenetic phenotype at the molecular and cellular levels deserves further elucidation, especially with regard to FtsZ behavior. Intriguingly, FtsZ spatial regulation contributes to variant Z. mobilis cell size at birth (Fig. 1D), while temporal FtsZ regulation works to reduce cell size variance (Fig. 2B and Fig. S3). Notably, more than 50% of total soluble protein content in Z. mobilis cell is enzymes responsible for fermentation (8, 65). Considering the simplicity and quantity of metabolism in Z. mobilis, particular ED-intermediate metabolites or related enzymes might directly bind to cell-structural proteins to dictate cell organization in Z. mobilis cells, as direct connections of particular metabolites to morphogenic proteins were previously observed in other bacteria (66–68).
Despite available genetic tools being relatively limited, there are several exciting questions regarding Z. mobilis cell biology. How Z. mobilis unique metabolism is related to its cellular organization and the biological significance of its polyploidy are currently the most intriguing to us.
MATERIALS AND METHODS
Strains, constructs, growth conditions, and analytical measurements.
The strains and primers used in this study are listed in Table 1. The medium for growing Z. mobilis was a complex medium containing glucose (20 g/liter), yeast extract (5 g/liter), NH4SO4 (1 g/liter), KH2PO4 (1 g/liter), and MgSO4 (0.5 g/liter). The Z. mobilis cultures (12 ml) were grown in capped test tubes shaken at 200 rpm under anaerobic conditions at 30°C throughout this study. The E. coli culture was grown in LB medium in baffled flasks shaken at 200 rpm at 37°C.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or sequence (5′-3′) | Source or reference |
|---|---|---|
| Strains | ||
| Zymomonas mobilis | ||
| Zm6 | ATCC 29191 | Lab stock |
| Zm4 | ATCC 31821 | Lab stock |
| Z6KF1 | Zm6 carrying pZKF1 | This study |
| Z6KF2 | Zm6 carrying pZKF2 | This study |
| Escherichia coli K-12 MG1655 | ATCC 700926 | Lab stock |
| Plasmids | ||
| pBBR1MCS-2 | Kmr | 69 |
| pZKF1 | pBBR1-MCS2 harboring ftsZ gene with its native promoter | This study |
| pZKF2 | pBBR1-MCS2 harboring ftsZ-gfp fusion gene with ftsZ promoter | This study |
| Primers | ||
| ZftsZF | ATTTTATCTAGATGCGTCGGAACCCTTTGATC | |
| ZftsZR_ws | ATTTTAAAGCTTGGAATTACCCTGACGATTCA | |
| ZftsZR | ATTTTAAAGCTTGGGTGCCGATAAATTCGGAC | |
| GFPF | ATTTTAAAGCTTGTACCAAGTAAAGGAGAAG | |
| GFPR | ATTTTACTCGAGTTATTTGTATAGTTCATCCATGC | |
The plasmid pZKF1 was constructed as follows. Zm6 ftsZ (ZZ6_0455) was amplified with Phusion polymerase (Thermo Scientific Fisher) using the primers ZftsZF and ZftsZR and genomic DNA of Zm6 as a template. Genomic DNA was extracted combining a lysozyme method and the DNeasy blood and tissue kit (Qiagen). The product was purified by the PCR purification kit (Qiagen) and cut with XbaI and HindIII (New England Biolabs). The plasmid pBBR1MCS-2 was digested with XbaI and HindIII. The cut plasmid and PCR product were then purified and ligated with T4 DNA ligase (New England Biolabs). For constructing pZKF2, Zm6 ftsZ was amplified using the primers ZftsZF and ZftsZR_ws. The product was purified and digested with XbaI and HindIII. The gfpmut3 was amplified from our lab plasmid pSB-M1g with the primers GFPF and GFPR by Phusion polymerase. The product was purified and digested with XbaI and XhoI. The plasmid pBBR1MCS-2 was cut with XhoI and HindIII. All the products were purified and ligated with T4 DNA ligase, yielding pZKF2 carrying ftsZ-gfp with the native ftsZ upstream promoter. Sanger sequencing (Eurofins) confirmed that all the constructions were correctly made. Transformation into Z. mobilis was done by electroporation (2.5 kV) using disposable electroporation cuvettes with a 0.2-cm gap (VWR) and a MicroPulser instrument (Bio-Rad). The transformants were selected with kanamycin (300 μg/ml).
To observe FtsZ-GFP localizations in Z6KF2 cells, the Z6KF2 culture was grown in the medium with kanamycin (300 μg/ml) overnight. The fully grown cultures were then subcultured in the medium without kanamycin (starting OD600, 0.05) and grown to an OD600 of 0.4 to 0.8. The cells were then mounted on the complex-medium–agarose pad (1% [wt/vol]) for imaging. According to our pilot experiments, the plasmid appeared to be stable without a selection marker for at least 30 generations.
For the salt-induced-filamentation study, growing Z6KF2 cultures in the regular complex medium with kanamycin (300 μg/ml) were subcultured to the complex medium supplemented with additional 0.2 M NaCl but without kanamycin. The cultures were grown for 5 h, the cells were transferred to the agarose pad of the identical medium without NaCl, and imaging was performed.
Determination of ethanol concentrations in the cultures was performed using a Waters 2695e Alliance high-performance liquid chromatograph (HPLC) (Waters) with a Hi-plex column (300 by 7.7 mm; Agilent). The running condition was 0.05 M sulfuric acid as a mobile phase at a flow rate of 0.8 ml/min. The ethanol concentrations of samples were determined from a linear external standard curve.
The concentration and purity of genomic DNA were measured using a NanoDrop One spectrophotometer (Thermo Scientific).
Light microscopy.
A Zeiss Axio Imager Z2 microscope was used with an AxioCam MR R3 (Zeiss) camera for capturing fluorescence and phase-contrast imaging. A Nikon Ti Eclipse epifluorescence microscope equipped with a Hamamatsu C11440-22C camera was used for time-lapse imaging. A Nikon 60× oil (numerical aperture [NA], 1.4) Plan Apo microscope objectives was used for imaging the cells.
For capturing phase-contrast images of Zm6 cells, a growing culture of Zm6 was directly mounted on the agarose pad (1% [wt/vol]) containing the identical medium and sealed with a cover glass for imaging. This procedure was basically used for time-lapse imaging as well. The cells were incubated in the sealed state at room temperature for about 30 min before time-lapse imaging.
For membrane staining, FM4-64 (20 μg/ml) was added to the growing culture, and the cells were incubated for 15 min at room temperature. The stained cells were washed with PBS once before imaging. Fluorescent images were analyzed with ZEN pro software (Zeiss). Statistical significance was calculated using SigmaPlot (Systat Software).
For DNA staining, an actively growing Zm6 culture was incubated with Syto 9 green fluorescent nucleic acid stain (Thermo Fisher Scientific) at a concentration of 0.5 or 11 μM for 10 min or with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) (Thermo Fisher Scientific) at a concentration of 50 μg/ml for 20 min at room temperature. The stained cells were washed in PBS before mounting on a PBS-agarose pad for imaging. We also imaged stained cells mounted on the agarose pad made of the complex medium used for liquid culture and obtained results similar to those obtained with the PBS-agarose pad. For fixation of cells, Z. mobilis cells were fixed with 10% formalin (vol/vol) for half an hour. The fixed cells were washed with PBS 3 times before imaging. For visualizing E. coli DNA, the growing cells in LB supplemented with 0.1% glucose were stained with DAPI at a concentration of 5 μg/ml for 15 min at room temperature. The stained cells were transferred to a PBS-agarose pad for imaging.
To visualize growing sites in Z. mobilis, HADA (Tocris) was used at 500 μM. Zm6 cultures were incubated with HADA at 30°C for 20 min. The stained cells were washed with PBS twice and mounted on a PBS-agarose pad for imaging.
Image analysis.
Cell length was used as a proxy for cell volume in Z. mobilis throughout the study. For Fig. 1A and B, cell length and width were measured using ZEN pro software. For Fig. 1E, elongated cell length was measured using Image J (NIH) and ZEN pro. Several cells started to constrict and slightly curved during the 40 min of measurement during the time-lapse imaging. In this case, the long axes of two cell compartments were measured separately, and the combined length was measured for calculating elongated cell length in Fig. 1E.
For Fig. 2, the time-lapse imaging was analyzed using ImageJ. Cell cycle time was defined as time between completions of division, and the completion of division was judged by the visible line of the septum between two cells (Fig. 2B and C). Our time-lapse imaging was time limited due to the use of the agarose pad method, and the several small cells (cell length < 1.5 μm) that were generated during imaging did not complete division during the time-lapse imaging; these were not included in the analysis (Fig. 2B and C).
For Fig. 3 and 5, fluorescence (FL) signal intensity profile was measured using the profile function of Zen 2.3 pro. Estimation of background fluorescence signals was done by measuring FL intensity of cell-free backgrounds close to measured cells. The background FL intensity was subtracted from the measured FL intensity of FtsZ-GFP and nucleoids for the plot and FL profiles (Fig. 3 and 5; Fig. S4). For Fig. 5D and F, the data set from Zm6 cells stained with 0.5 μM Syto 9 was used to obtain the plot.
For measurement of FL intensities within single cells, the measured cells were selected on the basis of the phase-contrast images, in order to avoid possible biasing (Fig. 5B to D and F; Fig. S7). FL intensities at all measured points over the long cell axis were averaged to calculate the average FL intensity (Fig. 5D). The data set was normalized, with the highest intensity being 1. The average FL intensity was interpreted as crowdedness of DNA, due to dispersed nucleoid localization in Zm6 cells. To calculate an approximate abundance of DNA per single-cell compartment in Zm6 cells, the normalized FL intensity was multiplied by the cell length (Fig. 5F). Visualization of nucleoids in the strain Z6KF2 was performed with DAPI staining at 30 μg/ml for 15 min and washing with PBS before mounting on the complex-medium–agarose pad for imaging (Fig. 5E).
Electron microscopy.
Three microliters of mid-exponential-phase Z. mobilis Zm4 or Zm6 cells was applied to glow-discharged holey carbon grids (Quantifoil R3.5/1; Cu; 200 mesh) and plunge-frozen in liquid ethane using a Vitrobot Mark IV vitrification device (Thermo Fisher Scientific). Image acquisition was performed using a Titan Krios transmission electron microscope (Thermo Fisher Scientific) operated at liquid nitrogen temperature at 300 keV and equipped with a Falcon 3 direct electron detector (Thermo Fisher Scientific). Cells were imaged at a magnification of ×11,000 in low-dose mode with a 1- to 2-s exposure.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Elina Balodite and Uldis Kalnenieks for their advice on handling Z. mobilis culture and for donation of the plasmid pBBR1MCS-2. We thank Rhami Lale for donation of the plasmid pSB-M1g.
The study was supported by Norges Forskningsråd (grant number 258657).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Surovtsev IV, Jacobs-Wagner C. 2018. Subcellular organization: a critical feature of bacterial cell replication. Cell 172:1271–1293. doi: 10.1016/j.cell.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eswara PJ, Ramamurthi KS. 2017. Bacterial cell division: nonmodels poised to take the spotlight. Annu Rev Microbiol 71:393–411. doi: 10.1146/annurev-micro-102215-095657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang C, Caccamo PD, Brun YV. 2015. Mechanisms of bacterial morphogenesis: evolutionary cell biology approaches provide new insights. Bioessays 37:413–425. doi: 10.1002/bies.201400098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.den Blaauwen T. 2018. Is longitudinal division in rod-shaped bacteria a matter of swapping axis? Front Microbiol 9:822. doi: 10.3389/fmicb.2018.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pende N, Wang J, Weber PM, Verheul J, Kuru E, Rittmann SKMR, Leisch N, VanNieuwenhze MS, Brun YV, den Blaauwen T, Bulgheresi S. 2018. Host-polarized cell growth in animal symbionts. Curr Biol 28:1039–1051.E5. doi: 10.1016/j.cub.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdelrahman Y, Ouellette SP, Belland RJ, Cox JV. 2016. Polarized cell division of Chlamydia trachomatis. PLoS Pathog 12:e1005822. doi: 10.1371/journal.ppat.1005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, He Q, Yang Y, Wang J, Haning K, Hu Y, Wu B, He M, Zhang Y, Bao J, Contreras LM, Yang S. 2018. Advances and prospects in metabolic engineering of Zymomonas mobilis. Metab Eng 50:57–73. doi: 10.1016/j.ymben.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Kalnenieks U. 2006. Physiology of Zymomonas mobilis: some unanswered questions. Adv Microb Physiol 51:73–117. doi: 10.1016/S0065-2911(06)51002-1. [DOI] [PubMed] [Google Scholar]
- 9.Yang S, Fei Q, Zhang Y, Contreras LM, Utturkar SM, Brown SD, Himmel ME, Zhang M. 2016. Zymomonas mobilis as a model system for production of biofuels and biochemicals. Microb Biotechnol 9:699–717. doi: 10.1111/1751-7915.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalnenieks U, Balodite E, Rutkis R. 2019. Metabolic engineering of bacterial respiration: high vs. low P/O and the case of Zymomonas mobilis. Front Bioeng Biotechnol 7:327. doi: 10.3389/fbioe.2019.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balodite E, Strazdina I, Galinina N, McLean S, Rutkis R, Poole RK, Kalnenieks U. 2014. Structure of the Zymomonas mobilis respiratory chain: oxygen affinity of electron transport and the role of cytochrome c peroxidase. Microbiology (Reading) 160:2045–2052. doi: 10.1099/mic.0.081612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyes L, Scopes RK. 1991. Membrane-associated ATPase from Zymomonas mobilis; purification and characterization. Biochim Biophys Acta 1068:174–178. doi: 10.1016/0005-2736(91)90207-O. [DOI] [PubMed] [Google Scholar]
- 13.Kalnenieks U, Balodite E, Strähler S, Strazdina I, Rex J, Pentjuss A, Fuchino K, Bruheim P, Rutkis R, Pappas KM, Poole RK, Sawodny O, Bettenbrock K. 2019. Improvement of acetaldehyde production in Zymomonas mobilis by engineering of its aerobic metabolism. Front Microbiol 10:2533. doi: 10.3389/fmicb.2019.02533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa H, Nobayashi H, Tanaka H. 1990. Mechanism of fermentation performance of Zymomonas mobilis under oxygen supply in batch culture. J Ferment Bioeng 70:34–40. doi: 10.1016/0922-338X(90)90027-T. [DOI] [Google Scholar]
- 15.Jones-Burrage SE, Kremer TA, McKinlay JB. 2019. Cell aggregation and aerobic respiration are important for Zymomonas mobilis ZM4 survival in an aerobic minimal medium. Appl Environ Microbiol 85:e00193-19. doi: 10.1128/AEM.00193-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi T, Kato T, Watakabe S, Song W, Aikawa S, Furukawa K. 2015. The respiratory chain provides salt stress tolerance by maintaining a low NADH/NAD+ ratio in Zymomonas mobilis. Microbiology (Reading) 161:2384–2394. doi: 10.1099/mic.0.000195. [DOI] [PubMed] [Google Scholar]
- 17.Rutkis R, Strazdina I, Balodite E, Lasa Z, Galinina N, Kalnenieks U. 2016. The low energy-coupling respiration in Zymomonas mobilis accelerates flux in the Entner-Doudoroff pathway. PLoS One 11:e0153866. doi: 10.1371/journal.pone.0153866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randich AM, Brun YV. 2015. Molecular mechanisms for the evolution of bacterial morphologies and growth modes. Front Microbiol 6:580–580. doi: 10.3389/fmicb.2015.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brenac L, Baidoo EEK, Keasling JD, Budin I. 2019. Distinct functional roles for hopanoid composition in the chemical tolerance of Zymomonas mobilis. Mol Microbiol 112:1564–1575. doi: 10.1111/mmi.14380. [DOI] [PubMed] [Google Scholar]
- 20.Skerker JM, Leon D, Price MN, Mar JS, Tarjan DR, Wetmore KM, Deutschbauer AM, Baumohl JK, Bauer S, Ibáñez AB, Mitchell VD, Wu CH, Hu P, Hazen T, Arkin AP. 2013. Dissecting a complex chemical stress: chemogenomic profiling of plant hydrolysates. Mol Syst Biol 9:674. doi: 10.1038/msb.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soppa J. 2014. Polyploidy in Archaea and Bacteria: about desiccation resistance, giant cell size, long-term survival, enforcement by a eukaryotic host and additional aspects. J Mol Microbiol Biotechnol 24:409–419. doi: 10.1159/000368855. [DOI] [PubMed] [Google Scholar]
- 22.Swings J, De Ley J. 1977. The biology of Zymomonas. Bacteriol Rev 41:1–46. doi: 10.1128/BR.41.1.1-46.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Vuuren HJJ, Priest FG. 2003. Gram-negative brewery bacteria, p 219–245. In Priest FG, Campbell I (ed), Brewing microbiology. Springer, Boston, MA. [Google Scholar]
- 24.Kuru E, Tekkam S, Hall E, Brun YV, Van Nieuwenhze MS. 2015. Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat Protoc 10:33–52. doi: 10.1038/nprot.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jun S, Taheri-Araghi S. 2015. Cell-size maintenance: universal strategy revealed. Trends Microbiol 23:4–6. doi: 10.1016/j.tim.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Robert L. 2015. Size sensors in bacteria, cell cycle control, and size control. Front Microbiol 6:515. doi: 10.3389/fmicb.2015.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaechter M, MaalØe O, Kjeldgaard NO. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J Gen Microbiol 19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 28.Campos M, Surovtsev IV, Kato S, Paintdakhi A, Beltran B, Ebmeier SE, Jacobs-Wagner C. 2014. A constant size extension drives bacterial cell size homeostasis. Cell 159:1433–1446. doi: 10.1016/j.cell.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amir A. 2014. Cell size regulation in bacteria. Phys Rev Lett 112:208102. doi: 10.1103/PhysRevLett.112.208102. [DOI] [Google Scholar]
- 30.Si F, Le Treut G, Sauls JT, Vadia S, Levin PA, Jun S. 2019. Mechanistic origin of cell-size control and homeostasis in bacteria. Curr Biol 29:1760–1770.E7. doi: 10.1016/j.cub.2019.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nieto C, Arias-Castro J, Sánchez C, Vargas-García C, Pedraza JM. 2020. Unification of cell division control strategies through continuous rate models. Phys Rev E 101:e022401. doi: 10.1103/PhysRevE.101.022401. [DOI] [PubMed] [Google Scholar]
- 32.Deforet M, van Ditmarsch D, Xavier JB. 2015. Cell-size homeostasis and the incremental rule in a bacterial pathogen. Biophys J 109:521–528. doi: 10.1016/j.bpj.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallden M, Fange D, Lundius EG, Baltekin Ö, Elf J. 2016. The synchronization of replication and division cycles in individual E. coli cells. Cell 166:729–739. doi: 10.1016/j.cell.2016.06.052. [DOI] [PubMed] [Google Scholar]
- 34.Logsdon MM, Ho P-Y, Papavinasasundaram K, Richardson K, Cokol M, Sassetti CM, Amir A, Aldridge BB. 2017. A parallel adder coordinates mycobacterial cell-cycle progression and cell-size homeostasis in the context of asymmetric growth and organization. Curr Biol 27:3367–3374.E7. doi: 10.1016/j.cub.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taheri-Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, Paulsson J, Vergassola M, Jun S. 2015. Cell-size control and homeostasis in bacteria. Curr Biol 25:385–391. doi: 10.1016/j.cub.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiu S-W, Roberts MAJ, Leake MC, Armitage JP. 2013. Positioning of chemosensory proteins and FtsZ through the Rhodobacter sphaeroides cell cycle. Mol Microbiol 90:322–337. doi: 10.1111/mmi.12366. [DOI] [PubMed] [Google Scholar]
- 37.Zupan JR, Cameron TA, Anderson-Furgeson J, Zambryski PC. 2013. Dynamic FtsA and FtsZ localization and outer membrane alterations during polar growth and cell division in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A 110:9060–9065. doi: 10.1073/pnas.1307241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goley ED, Yeh Y-C, Hong S-H, Fero MJ, Abeliuk E, McAdams HH, Shapiro L. 2011. Assembly of the Caulobacter cell division machine. Mol Microbiol 80:1680–1698. doi: 10.1111/j.1365-2958.2011.07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Männik J, Walker BE, Männik J. 2018. Cell cycle-dependent regulation of FtsZ in Escherichia coli in slow growth conditions. Mol Microbiol 110:1030–1044. doi: 10.1111/mmi.14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortiz C, Natale P, Cueto L, Vicente M. 2016. The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol Rev 40:57–67. doi: 10.1093/femsre/fuv040. [DOI] [PubMed] [Google Scholar]
- 41.Desiniotis A, Kouvelis VN, Davenport K, Bruce D, Detter C, Tapia R, Han C, Goodwin LA, Woyke T, Kyrpides NC, Typas MA, Pappas KM. 2012. Complete genome sequence of the ethanol-producing Zymomonas mobilis subsp. mobilis centrotype ATCC 29191. J Bacteriol 194:5966–5967. doi: 10.1128/JB.01398-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thanbichler M, Shapiro L. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 43.Dubarry N, Willis CR, Ball G, Lesterlin C, Armitage JP. 2019. In vivo imaging of the segregation of the 2 chromosomes and the cell division proteins of Rhodobacter sphaeroides reveals an unexpected role for MipZ. mBio 10:e02515-18. doi: 10.1128/mBio.02515-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wehrens M, Ershov D, Rozendaal R, Walker N, Schultz D, Kishony R, Levin PA, Tans SJ. 2018. Size laws and division ring dynamics in filamentous Escherichia coli cells. Curr Biol 28:972–979.E5. doi: 10.1016/j.cub.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 45.Walia SK, Carey VC, All BP, Ingram LO. 1984. Self-transmissible plasmid in Zymomonas mobilis carrying antibiotic resistance. Appl Environ Microbiol 47:198–200. doi: 10.1128/AEM.47.1.198-200.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vriesekoop F, Rasmusson M, Pamment NB. 2002. Respective effects of sodium and chloride ions on filament formation and growth and ethanol production in Zymomonas mobilis fermentations. Lett Appl Microbiol 35:27–31. doi: 10.1046/j.1472-765x.2002.01137.x. [DOI] [PubMed] [Google Scholar]
- 47.Bakshi S, Choi H, Rangarajan N, Barns KJ, Bratton BP, Weisshaar JC. 2014. Nonperturbative imaging of nucleoid morphology in live bacterial cells during an antimicrobial peptide attack. Appl Environ Microbiol 80:4977–4986. doi: 10.1128/AEM.00989-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gray WT, Govers SK, Xiang Y, Parry BR, Campos M, Kim S, Jacobs-Wagner C. 2019. Nucleoid size scaling and intracellular organization of translation across bacteria. Cell 177:1632–1648.E20. doi: 10.1016/j.cell.2019.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain IH, Vijayan V, O'Shea EK. 2012. Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc Natl Acad Sci U S A 109:13638–13643. doi: 10.1073/pnas.1211144109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohbayashi R, Nakamachi A, Hatakeyama TS, Watanabe S, Kanesaki Y, Chibazakura T, Yoshikawa H, Miyagishima S-Y. 2019. Coordination of polyploid chromosome replication with cell size and growth in a cyanobacterium. mBio 10:e00510-19. doi: 10.1128/mBio.00510-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maróti G, Kondorosi É. 2014. Nitrogen-fixing Rhizobium-legume symbiosis: are polyploidy and host peptide-governed symbiont differentiation general principles of endosymbiosis? Front Microbiol 5:326. doi: 10.3389/fmicb.2014.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mergaert P, Uchiumi T, Alunni B, Evanno G, Cheron A, Catrice O, Mausset A-E, Barloy-Hubler F, Galibert F, Kondorosi A, Kondorosi E. 2006. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium–legume symbiosis. Proc Natl Acad Sci U S A 103:5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mendell JE, Clements KD, Choat JH, Angert ER. 2008. Extreme polyploidy in a large bacterium. Proc Natl Acad Sci U S A 105:6730–6734. doi: 10.1073/pnas.0707522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen AH, Afonso B, Silver PA, Savage DF. 2012. Spatial and temporal organization of chromosome duplication and segregation in the cyanobacterium Synechococcus elongatus PCC 7942. PLoS One 7:e47837. doi: 10.1371/journal.pone.0047837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peyre E, Morin X. 2012. An oblique view on the role of spindle orientation in vertebrate neurogenesis. Dev Growth Differ 54:287–305. doi: 10.1111/j.1440-169X.2012.01350.x. [DOI] [PubMed] [Google Scholar]
- 56.Chalmers AD, Strauss B, Papalopulu N. 2003. Oriented cell divisions asymmetrically segregate aPKC and generate cell fate diversity in the early Xenopus embryo. Development 130:2657–2668. doi: 10.1242/dev.00490. [DOI] [PubMed] [Google Scholar]
- 57.Lancaster MA, Knoblich JA. 2012. Spindle orientation in mammalian cerebral cortical development. Curr Opin Neurobiol 22:737–746. doi: 10.1016/j.conb.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nürnberg DJ, Mariscal V, Parker J, Mastroianni G, Flores E, Mullineaux CW. 2014. Branching and intercellular communication in the section V cyanobacterium Mastigocladus laminosus, a complex multicellular prokaryote. Mol Microbiol 91:935–949. doi: 10.1111/mmi.12506. [DOI] [PubMed] [Google Scholar]
- 59.Sprenger GA. 1996. Carbohydrate metabolism in Zymomonas mobilis: a catabolic highway with some scenic routes. FEMS Microbiology Lett 145:301–307. doi: 10.1111/j.1574-6968.1996.tb08593.x. [DOI] [Google Scholar]
- 60.Fuhrer T, Fischer E, Sauer U. 2005. Experimental identification and quantification of glucose metabolism in seven bacterial species. J Bacteriol 187:1581–1590. doi: 10.1128/JB.187.5.1581-1590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doelle HW, Kirk L, Crittenden R, Toh H, Doelle MB. 1993. Zymomonas mobilis—science and industrial application. Crit Rev Biotechnol 13:57–98. doi: 10.3109/07388559309069198. [DOI] [PubMed] [Google Scholar]
- 62.Yang DC, Blair KM, Salama NR. 2016. Staying in shape: the impact of cell shape on bacterial survival in diverse environments. Microbiol Mol Biol Rev 80:187–203. doi: 10.1128/MMBR.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thattai M, van Oudenaarden A. 2004. Stochastic gene expression in fluctuating environments. Genetics 167:523–530. doi: 10.1534/genetics.167.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weir PM. 2016. The ecology of Zymomonas: a review. Folia Microbiol (Praha) 61:385–392. doi: 10.1007/s12223-016-0447-x. [DOI] [PubMed] [Google Scholar]
- 65.Algar EM, Scopes RK. 1985. Studies on cell-free metabolism: ethanol production by extracts of Zymomonas mobilis. J Biotechnol 2:275–287. doi: 10.1016/0168-1656(85)90030-6. [DOI] [Google Scholar]
- 66.Weart RB, Lee AH, Chien A-C, Haeusser DP, Hill NS, Levin PA. 2007. A metabolic sensor governing cell size in bacteria. Cell 130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monahan LG, Hajduk IV, Blaber SP, Charles IG, Harry EJ. 2014. Coordinating bacterial cell division with nutrient availability: a role for glycolysis. mBio 5:e00935-14. doi: 10.1128/mBio.00935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sperber AM, Herman JK. 2017. Metabolism shapes the cell. J Bacteriol 199:e00039-17. doi: 10.1128/JB.00039-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kovach ME, Elzer PH, Steven Hill D, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.