Although the budding yeast S. cerevisiae, which is used in the production of alcoholic beverages and bioethanol, is highly tolerant of ethanol, high concentrations of ethanol are also stressful to the yeast and cause various adverse effects, including protein denaturation. A pretreatment with mild stress improves the ethanol tolerance of yeast cells; however, it currently remains unclear whether it increases PQC activity and reduces the levels of denatured proteins.
KEYWORDS: Saccharomyces cerevisiae, acquired stress resistance, severe ethanol stress, proteostasis, protein quality control, insoluble proteins, bichaperone system, aggregase, Lsg1
ABSTRACT
Acute severe ethanol stress (10% [vol/vol]) damages proteins and causes the intracellular accumulation of insoluble proteins in Saccharomyces cerevisiae. On the other hand, a pretreatment with mild stress increases tolerance to subsequent severe stress, which is called acquired stress resistance. It currently remains unclear whether the accumulation of insoluble proteins under severe ethanol stress may be mitigated by increasing protein quality control (PQC) activity in cells pretreated with mild stress. In the present study, we examined the induction of resistance to severe ethanol stress in PQC and confirmed that a pretreatment with 6% (vol/vol) ethanol or mild thermal stress at 37°C significantly reduced insoluble protein levels and the aggregation of Lsg1, which is prone to denaturation and aggregation by stress, in yeast cells under 10% (vol/vol) ethanol stress. The induction of this stress resistance required the new synthesis of proteins; the expression of proteins comprising the bichaperone system (Hsp104, Ssa3, and Fes1), Sis1, and Hsp42 was upregulated during the pretreatment and maintained under subsequent severe ethanol stress. Since the pretreated cells of deficient mutants in the bichaperone system (fes1Δ hsp104Δ and ssa2Δ ssa3Δ ssa4Δ) failed to sufficiently reduce insoluble protein levels and Lsg1 aggregation, the enhanced activity of the bichaperone system appears to be important for the induction of adequate stress resistance. In contrast, the importance of proteasomes and aggregases (Btn2 and Hsp42) in the induction of stress resistance has not been confirmed. These results provide further insights into the PQC activity of yeast cells under severe ethanol stress, including the brewing process.
IMPORTANCE Although the budding yeast S. cerevisiae, which is used in the production of alcoholic beverages and bioethanol, is highly tolerant of ethanol, high concentrations of ethanol are also stressful to the yeast and cause various adverse effects, including protein denaturation. A pretreatment with mild stress improves the ethanol tolerance of yeast cells; however, it currently remains unclear whether it increases PQC activity and reduces the levels of denatured proteins. In the present study, we found that a pretreatment with mild ethanol upregulated the expression of proteins involved in PQC and mitigated the accumulation of insoluble proteins, even under severe ethanol stress. These results provide novel insights into ethanol tolerance and the adaptive capacity of yeast. They may also contribute to research on the physiology of yeast cells during the brewing process, in which the concentration of ethanol gradually increases.
INTRODUCTION
The budding yeast Saccharomyces cerevisiae efficiently produces ethanol through alcoholic fermentation, and ethanol concentrations ultimately increase to higher than 10% (vol/vol) during the typical brewing process of wine and Japanese sake. However, high concentrations of ethanol are toxic, even for yeast cells, and exert a number of adverse effects on yeast cells as severe ethanol stress. More than 10% (vol/vol) ethanol inhibits the transport systems of glucose and amino acids (1, 2), prevents the nuclear export of bulk poly(A)+ mRNA (3), and induces the strong repression of overall protein synthesis (4). Additionally, long-term exposure to severe ethanol stress leads to persistent actin depolarization and the aberrant localization of septins (5). We recently identified the intracellular accumulation of denatured proteins as another adverse effect of severe ethanol stress on yeast cells; severe ethanol stress (10% [vol/vol]) as well as thermal stress at 42°C significantly increased the levels of insoluble proteins and ubiquitinated proteins in living yeast cells (6).
The accumulation of denatured proteins often vitiates cellular homeostatic functions and has been implicated in the pathogenesis of neurodegenerative disorders, such as Alzheimer’s disease, Huntington’s disease, and Parkinson’s disease (7–9). The protein quality control (PQC) system in the proteostasis network plays a critical role in mitigating the toxicity of denatured proteins. The PQC system primarily suppresses or scavenges denatured proteins using molecular chaperones, the ubiquitin-proteasome system (UPS), and autophagy in eukaryotic cells (10–15). Additionally, excess denatured proteins are not scattered in cells; they assemble at intracellular deposition sites, such as CytoQ, IPODs (Insoluble PrOtein Deposits), and INQs/JUNQs (INtranuclear Quality/JUxta-Nuclear Quality control compartments) (16–19). The formation of these deposition sites is useful for the sequestration of toxic denatured proteins, the generation of denatured protein-free progeny via asymmetric inheritance, the efficient refolding/degradation of denatured proteins, and the prevention of Hsp70 overload (20–22).
In S. cerevisiae, Hsp104, Hsp70 (Ssa1 to -4), and Fes1 constitute the bichaperone system, which plays a role in the refolding and reactivation of aggregated proteins (10, 22, 23). Fes1 promotes the dissociation of denatured proteins from Hsp70 in the refolding process (24, 25). Hsp104 functions as a disaggregase for the dissolution of protein aggregates, together with other molecular chaperones (18, 26–29). Since Hsp104 binds to the surfaces of denatured protein aggregates and forms granules, Hsp104-GFP (green fluorescent protein) is used as a representative marker of the deposition sites of denatured proteins (19, 27, 30, 31). Btn2 and Hsp42 play important roles as aggregases (sequestrases) in the assembly of deposition sites together with Sis1 (22, 28, 30, 32–35). Since these factors play important roles in yeast proteostasis, various deficient strains have been reported to exhibit abnormal phenotypes due to defects in the PQC system (18, 22, 29). We also confirmed that hsp104Δ and btn2Δ mutants showed the delayed clearance of denatured proteins during the recovery process from severe ethanol stress (6).
Cells preexposed to mild stress conditions develop enhanced resistance to subsequent severe stress conditions. This phenomenon is referred to as “acquired stress resistance,” and a well-known example is increased tolerance to heat shock (36). A pretreatment with mild thermal stress has been shown to increase resistance to subsequent lethal temperatures in yeast cells through the upregulated expression of stress-responsive genes, including PQC-related genes (36–39). Regarding acquired resistance to severe ethanol stress, genome-wide gene expression was previously examined and the findings obtained revealed that alterations in H+-ATPase activity and the lipid composition of the plasma membrane caused by a mild ethanol pretreatment contributed to the acquisition of resistance to high concentrations of ethanol (40–44). However, limited information is currently available on adaptive responses to severe ethanol stress in yeast proteostasis, and it remains unclear whether the activity of the PQC system is enhanced by a pretreatment with mild ethanol stress.
Therefore, the present study investigated whether a pretreatment with mild stress affects proteostasis in yeast cells under subsequent severe ethanol stress. We confirmed that a pretreatment with 6% (vol/vol) ethanol or mild thermal stress at 37°C significantly reduced insoluble protein levels in yeast cells under subsequent 10% (vol/vol) ethanol stress, suggesting that the activity of the PQC system was enhanced by the pretreatment. The acquisition of enhanced PQC activity required the new synthesis of proteins, and the pretreated cells of the ssa2Δ ssa3Δ ssa4Δ and fes1Δ hsp104Δ mutants did not induce sufficient reductions in insoluble protein levels under subsequent severe ethanol stress. These results provide novel insights into the acquired resistance of yeast cells to severe ethanol stress.
RESULTS
The pretreatment with mild ethanol stress mitigated the accumulation of insoluble proteins.
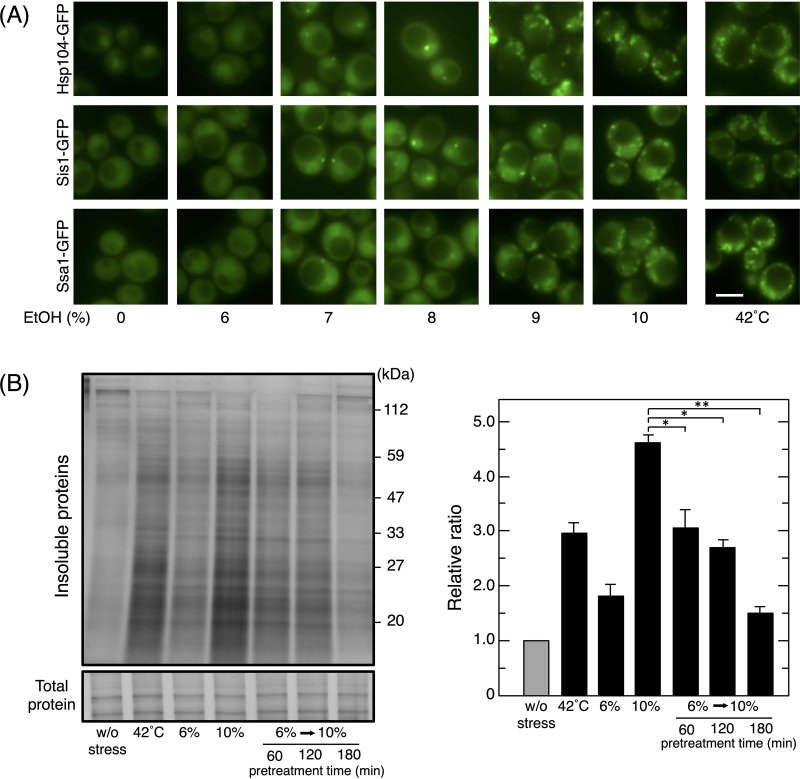
To establish the pretreatment conditions needed to enhance the activity of PQC, we initially examined the concentration-dependent effects of ethanol on the formation of the deposition sites of denatured proteins. Since Hsp104 binds to denatured protein aggregates and forms granules, Hsp104-GFP is a frequently used marker of deposition sites (27, 30, 31). The formation of Hsp104-GFP foci was induced by a treatment with thermal stress at 42°C and >7% (vol/vol) ethanol stress, but not 6% (vol/vol) ethanol (Fig. 1A). Another molecular chaperone, Ssa1 (Hsp70), and an Hsp40 cochaperone (Sis1) also formed foci under >7% (vol/vol) ethanol stress and thermal stress at 42°C.
FIG 1.
Reduction in denatured protein levels in cells pretreated with mild ethanol (EtOH) stress. (A) Yeast cells expressing GFP-tagged proteins were treated with various concentrations of ethanol for 180 min or thermal stress at 42°C for 60 min in SD medium. Representative images are shown. Bar, 5 μm. (B) Cells were pretreated with 6% (vol/vol) ethanol for 60 to 180 min and then exposed to 10% (vol/vol) ethanol for 180 min. Intracellular levels of insoluble proteins were assayed. Samples were separated using a 10% SDS-polyacrylamide gel and visualized by silver staining. Representative images are shown in the left panel and quantified data in the right panel. Relative ratios were calculated from three independent experiments. Each value is expressed as the mean ± SD of fold changes in the staining levels of insoluble proteins relative to those in nonstressed cells. *, P < 0.05; **, P < 0.01 by the Student t test.
Since 6% (vol/vol) ethanol did not induce the formation of the deposition sites of denatured proteins, it was used as a mild dose of ethanol stress, and we investigated whether a pretreatment with 6% (vol/vol) ethanol mitigated the accumulation of denatured proteins under severe ethanol stress (10% [vol/vol]). In the present study, insoluble protein levels were measured as an indicator of denatured protein accumulation. Consistent with previous findings (6), the treatment with 10% (vol/vol) ethanol for 180 min and thermal stress at 42°C for 60 min caused the significant accumulation of insoluble proteins, whereas the treatment with 6% (vol/vol) ethanol for 180 min induced only a slight increase in insoluble protein levels (Fig. 1B). Although the pretreatment with 6% (vol/vol) ethanol for 60 or 120 min partly mitigated the accumulation of insoluble proteins during the subsequent treatment with 10% (vol/vol) ethanol, the 180-min pretreatment with 6% (vol/vol) ethanol adequately suppressed the accumulation of insoluble proteins (Fig. 1B). These results suggested that yeast cells acquired enhanced PQC activity after the pretreatment with 6% ethanol for 180 min.
New protein synthesis was essential for the acquisition of enhanced PQC activity.
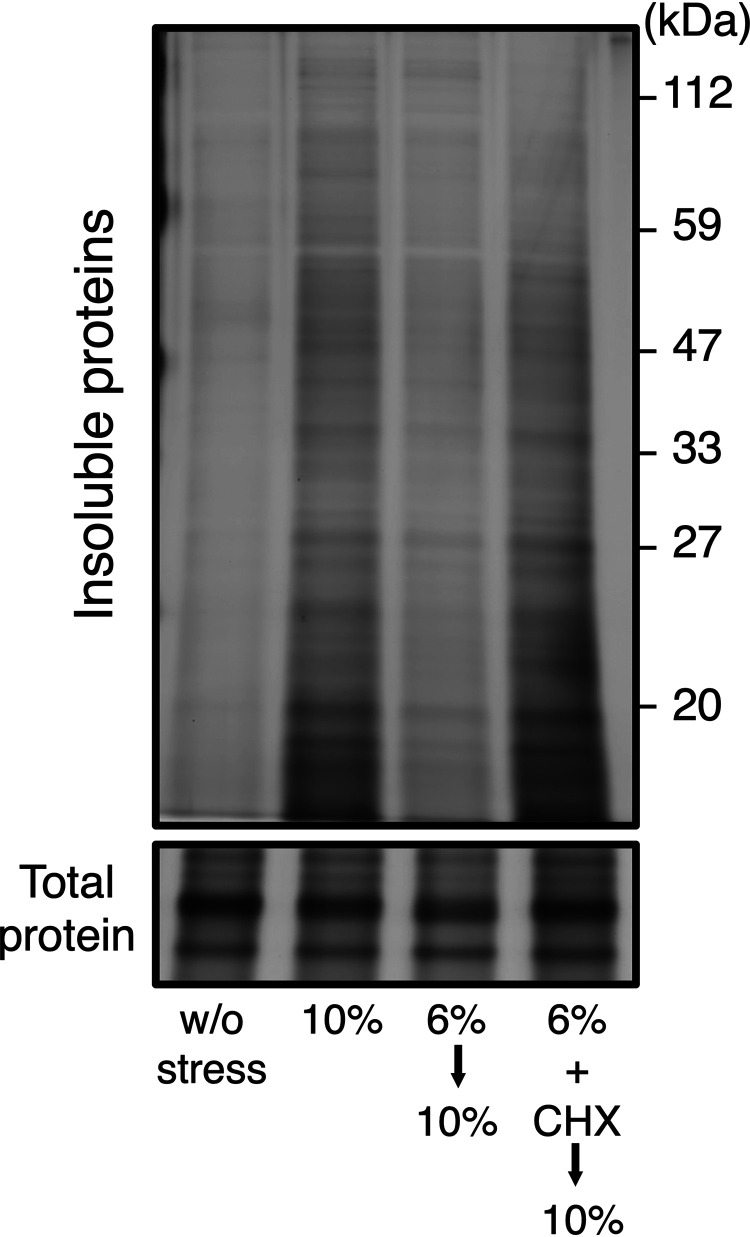
A previous study reported that cycloheximide (CHX), a representative inhibitor of translation elongation (45), blocked the acquisition of tolerance to severe ethanol stress (43). We also examined whether CHX blocked the acquisition of enhanced PQC activity caused by the pretreatment with 6% (vol/vol) ethanol. As shown in Fig. 2, the pretreatment with 6% (vol/vol) ethanol plus CHX did not mitigate the accumulation of insoluble proteins under subsequent 10% (vol/vol) ethanol stress, demonstrating that new protein synthesis during the pretreatment was required for enhanced PQC activity.
FIG 2.
Acquired resistance to severe ethanol stress required new protein synthesis. Yeast cells were pretreated with 6% (vol/vol) ethanol for 180 min with or without 200 μg/ml cycloheximide (CHX) and then exposed to 10% (vol/vol) ethanol stress for 180 min. Intracellular levels of insoluble proteins were assayed by the silver staining of SDS-polyacrylamide gels.
The pretreatment with mild thermal stress induced cross-protection against protein denaturation caused by severe ethanol stress.
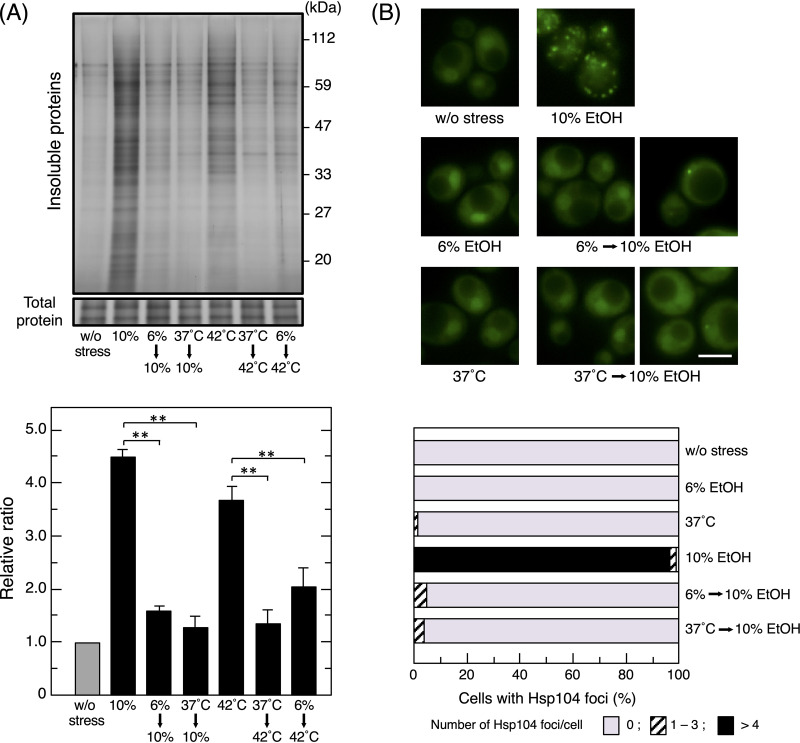
A pretreatment with a certain stress has been shown to induce cross-protection and enhance resistance to different types of stress (39, 46–48). We examined the effects of cross-protection induced by the pretreatment with mild thermal stress at 37°C on protein denaturation caused by severe ethanol stress. Insoluble protein levels induced by 10% (vol/vol) ethanol were significantly lower in cells pretreated with 37°C than in those without the pretreatment (Fig. 3A). In contrast, the pretreatment with 6% (vol/vol) ethanol also reduced the levels of insoluble proteins that accumulated under severe thermal stress at 42°C.
FIG 3.
Cross-protection between ethanol (EtOH) stress and thermal stress. (A) Cells were pretreated with or without mild stress (6% ethanol for 180 min or 37°C for 60 min) and then exposed to severe stress (10% ethanol for 180 min or thermal stress at 42°C for 60 min). Intracellular levels of insoluble proteins were assayed. **, P < 0.01 by the Student t test. (B) The formation of Hsp104-GFP foci was monitored under each condition. Representative images are shown in the upper panel. The quantification of cells containing foci is shown in the lower panel. One hundred cells under each condition were examined, and experiments were repeated 3 times (300 cells in total were examined). Bar, 5 μm.
We investigated the effects of the pretreatment on the formation of Hsp104-GFP foci. The pretreatment with 6% (vol/vol) ethanol or 37°C had a negligible effect on the formation of Hsp104-GFP foci (Fig. 3B). As shown in Fig. 1A, the majority of yeast cells directly challenged with 10% (vol/vol) ethanol stress formed multiple foci of Hsp104-GFP. In contrast, when challenged with 10% (vol/vol) ethanol stress after the pretreatment, only a small percentage of cells formed Hsp104-GFP foci, and when they did, only a few foci per cell were observed (Fig. 3B). These results clearly demonstrated that the pretreatment with 6% (vol/vol) ethanol or 37°C reduced protein insolubilization under subsequent severe ethanol stress.
The pretreatment with mild stress affected protein expression under severe stress.
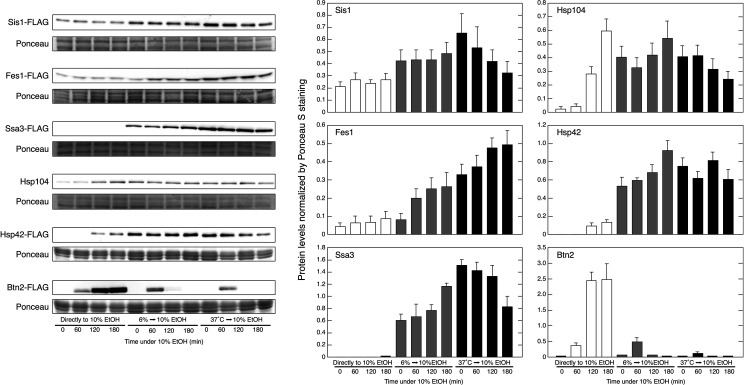
We examined whether pretreatments with mild stress affected the levels of proteins involved in PQC under severe ethanol stress (Fig. 4). Despite their transcriptional activation (see Fig. S1 in the supplemental material), neither Ssa3 (Hsp70) nor Sis1 protein levels were markedly increased by the direct exposure to 10% (vol/vol) ethanol for 180 min. However, these levels were markedly elevated during the pretreatment with 6% (vol/vol) ethanol or 37°C, and Ssa3 levels continued to increase during the subsequent treatment with 10% (vol/vol) ethanol in cells pretreated with 6% (vol/vol) ethanol.
FIG 4.
A pretreatment with mild stress affected protein expression under severe ethanol (EtOH) stress. Sis1-FLAG, Ssa3-FLAG, Fes1-FLAG, Hsp42-FLAG, Hsp104, and Btn2-FLAG levels under stress conditions were assayed by Western blotting. Yeast cells were pretreated with or without mild stress (6% ethanol stress for 180 min or thermal stress at 37°C for 60 min) and then exposed to 10% (vol/vol) ethanol stress for 0, 60, 120, or 180 min. Ponceaus S staining was performed to confirm equal loading and the transfer of all proteins. Protein levels were normalized by Ponceau S staining, and data are expressed as the mean ± SD (n = 3).
Fes1, Hsp104, and Hsp42 protein levels were gradually increased by the direct exposure to 10% (vol/vol) ethanol. The synthesis of these proteins was also promoted during the pretreatment with 6% (vol/vol) ethanol or 37°C, allowing the pretreated cells to confront subsequent 10% ethanol stress with significantly enhanced levels of Fes1, Hsp104, and Hsp42.
In contrast, Btn2 showed a different expression pattern from the proteins described above under severe ethanol stress. Consistent with previous findings (4), Btn2 protein levels markedly increased with time when cells were directly exposed to 10% (vol/vol) ethanol. On the other hand, Btn2 levels in cells pretreated with 6% (vol/vol) ethanol or 37°C were transiently elevated and then disappeared after 120 min of the subsequent treatment with 10% (vol/vol) ethanol, while the protein levels of Ssa3, Sis1, Fes1, Hsp104, and Hsp42 were strongly maintained during the subsequent treatment with 10% (vol/vol) ethanol. The turnover of Btn2 by the UPS is generally rapid but is suppressed by stress (32, 49). Therefore, the proteasomal turnover of Btn2 may have been restored, even under severe ethanol stress, in pretreated cells.
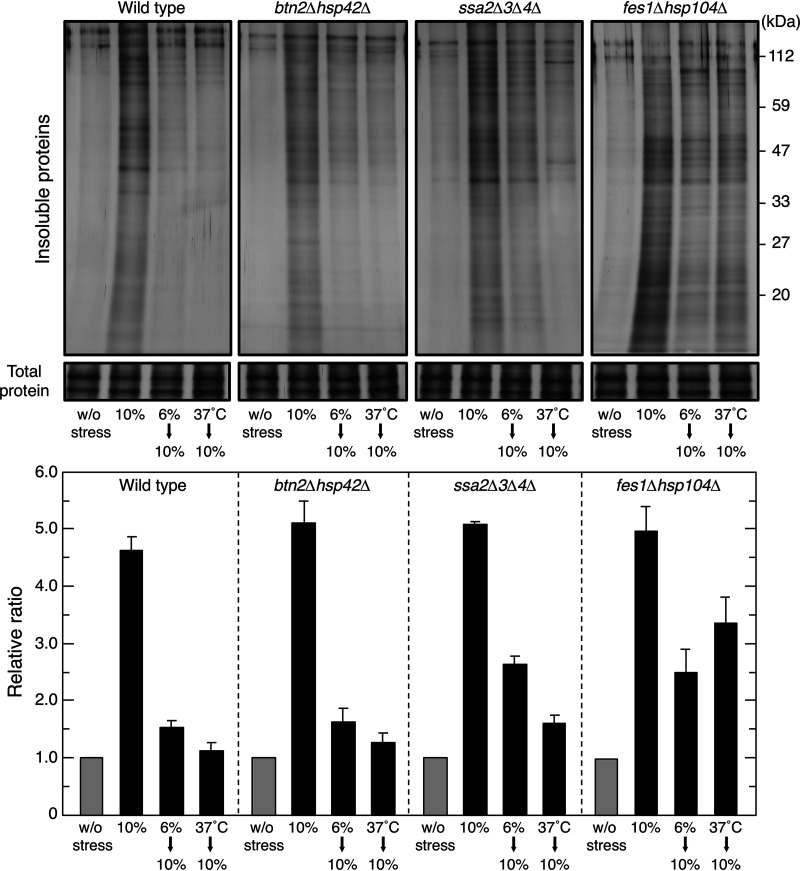
To verify the importance of the induced expression of PQC-related proteins, we used various mutants deficient in PQC-related genes to examine their acquired resistance to severe ethanol stress. Regarding the reductions observed in insoluble protein levels by the pretreatments, btn2Δ hsp42Δ cells behaved similarly to wild-type cells. On the other hand, the accumulation of insoluble proteins was not fully mitigated by the pretreatments in fes1Δ hsp104Δ cells and ssa2Δ ssa3Δ ssa4Δ cells (22, 50, 51) (Fig. 5). Since insoluble protein levels were similar among fes1Δ hsp104Δ, ssa2Δ ssa3Δ ssa4Δ, and wild-type cells upon a direct challenge with 10% (vol/vol) ethanol, the induced expression of Fes1, Hsp104, and Hsp70 during the mild stress pretreatment appeared to be important for reducing insoluble protein levels under subsequent 10% (vol/vol) ethanol stress.
FIG 5.
PQC activity in knockout mutants. Insoluble aggregated protein levels in various strains were assayed. Cells were treated with 10% (vol/vol) ethanol for 180 min after the pretreatment with or without mild stress (6% ethanol stress for 180 min or thermal stress at 37°C for 60 min). Representative images are shown in the upper panel, and quantified data are shown in the lower panel. Data are expressed as the mean ± SD (n = 3).
The pretreatment with mild stress prevented Lsg1 aggregation under severe ethanol stress.
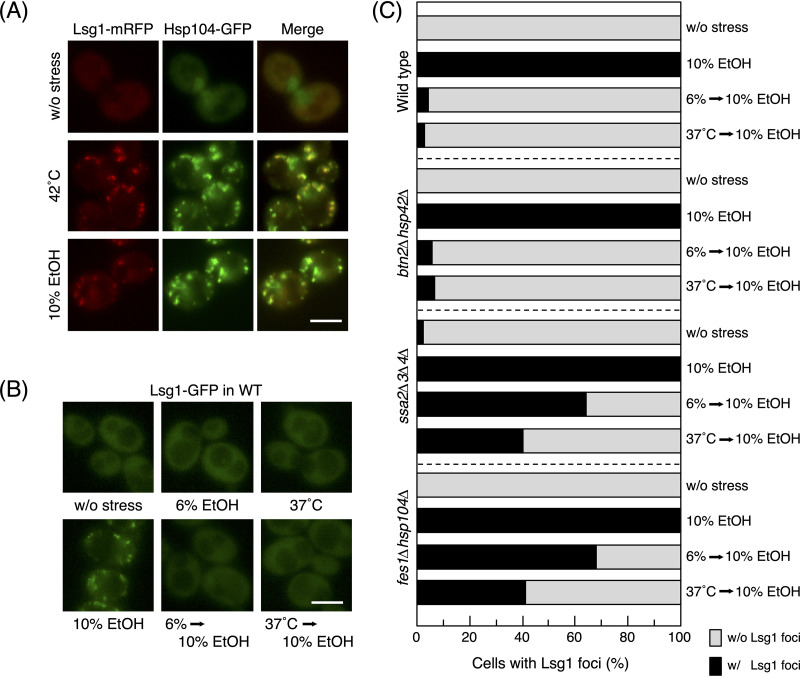
The denaturation of a specific protein was also examined to clarify whether it was consistent with the results of the silver staining of insoluble proteins. Lsg1 was previously identified as a cytosolic protein that is prone to denaturation and aggregation by thermal stress at 42°C (52). We verified that the treatment with 42°C induced the formation of multiple foci of Lsg1 and found that 10% (vol/vol) ethanol stress exerted similar effects (Fig. 6A). Furthermore, Lsg1-mRFP (monomeric red fluorescent protein) foci showed very similar localization with Hsp104-GFP foci after the treatments with 10% (vol/vol) ethanol and 42°C. These results suggested that severe ethanol stress also caused the aggregation of Lsg1, similar to thermal stress at 42°C.
FIG 6.
Aggregation of Lsg1 in response to severe ethanol (EtOH) stress. (A) Cells expressing Lsg1-mRFP and Hsp104-GFP were treated with thermal stress at 42°C for 60 min or 10% (vol/vol) ethanol stress for 180 min. Bar, 5 μm. (B and C) Cells expressing Lsg1-GFP were pretreated with or without mild stress (6% ethanol for 180 min or 37°C for 60 min) and then exposed to 10% (vol/vol) ethanol for 180 min. Representative images of wild-type cells under each condition are shown in panel B. The quantification of cells containing Lsg1-GFP foci in various mutants is shown in panel C. One hundred cells under each condition were examined, and experiments were repeated 3 times (300 cells in total were examined).
Although the mild stress treatment with 6% (vol/vol) ethanol or 37°C did not induce the aggregation of Lsg1, it significantly mitigated the formation of Lsg1 foci under subsequent 10% (vol/vol) ethanol stress in wild-type cells (Fig. 6B and C). This mitigating effect was also noted in btn2Δ hsp42Δ cells, but not in ssa2Δ ssa3Δ ssa4Δ or fes1Δ hsp104Δ cells (Fig. 6C). These results were consistent with those obtained from the silver staining of insoluble proteins (Fig. 5).
Cells pretreated with mild stress strongly inhibited the formation of insoluble proteins.
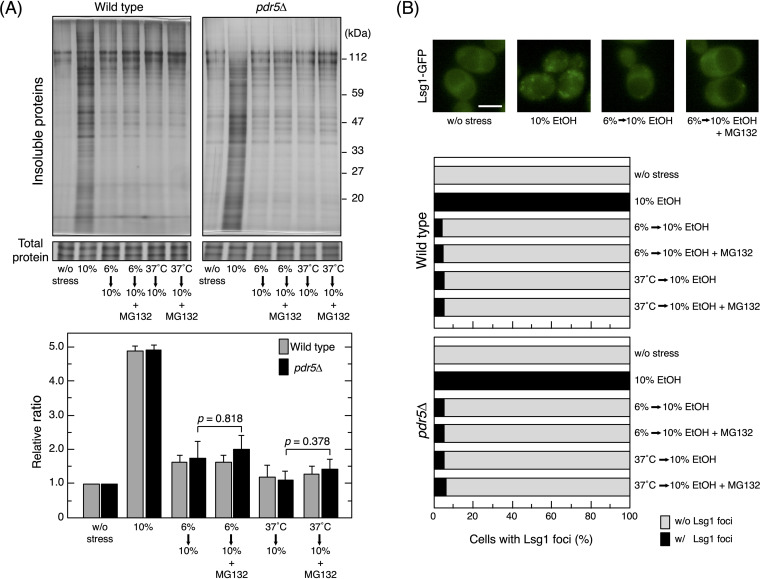
Insoluble protein levels may be reduced by the activation of insoluble protein removal and the suppression of insoluble protein generation. However, it is currently unclear whether the removal efficiency of insoluble proteins was enhanced in pretreated cells. To clarify the contribution of insoluble protein removal, proteasome activity was inhibited by MG132, an efficient cell-permeative proteasome inhibitor (12, 53), after the pretreatment with 6% (vol/vol) ethanol or 37°C. We confirmed that the treatment with 100 μM MG132 blocked proteasome activity under our experimental conditions based on the increases observed in Btn2 levels in cells at 37°C (see Fig. S2 in the supplemental material). In pretreated wild-type cells, the MG132 treatment did not markedly affect the levels of insoluble proteins or Lsg1 aggregation under subsequent severe ethanol stress (Fig. 7A and B). We confirmed that MG132 did not significantly affect the levels of insoluble proteins or Lsg1 aggregation using pdr5Δ cells, which are less likely to excrete MG132 and are commonly used in proteasome inhibition assays (54, 55). These results suggested that the contribution of proteasomes to reducing the accumulation of insoluble proteins in pretreated cells was not significant.
FIG 7.
Effects of MG132 on insoluble protein levels in pretreated cells. Pretreated cells (wild-type and pdr5Δ) with 6% (vol/vol) ethanol (EtOH) for 180 min or at 37°C for 60 min were exposed to 10% (vol/vol) ethanol for 180 min in the presence or absence of 100 μM MG132, a potent cell-permeative inhibitor of proteasomes. (A) The intracellular levels of insoluble proteins were analyzed. Representative images are shown in the upper panel and quantified data in the lower panel. Relative ratios were calculated from three independent experiments. Each value is expressed as the mean ± SD (n = 3). (B) The quantification of cells containing Lsg1-GFP foci under each condition is shown. One hundred cells under each condition were examined, and experiments were repeated 3 times (300 cells in total were examined).
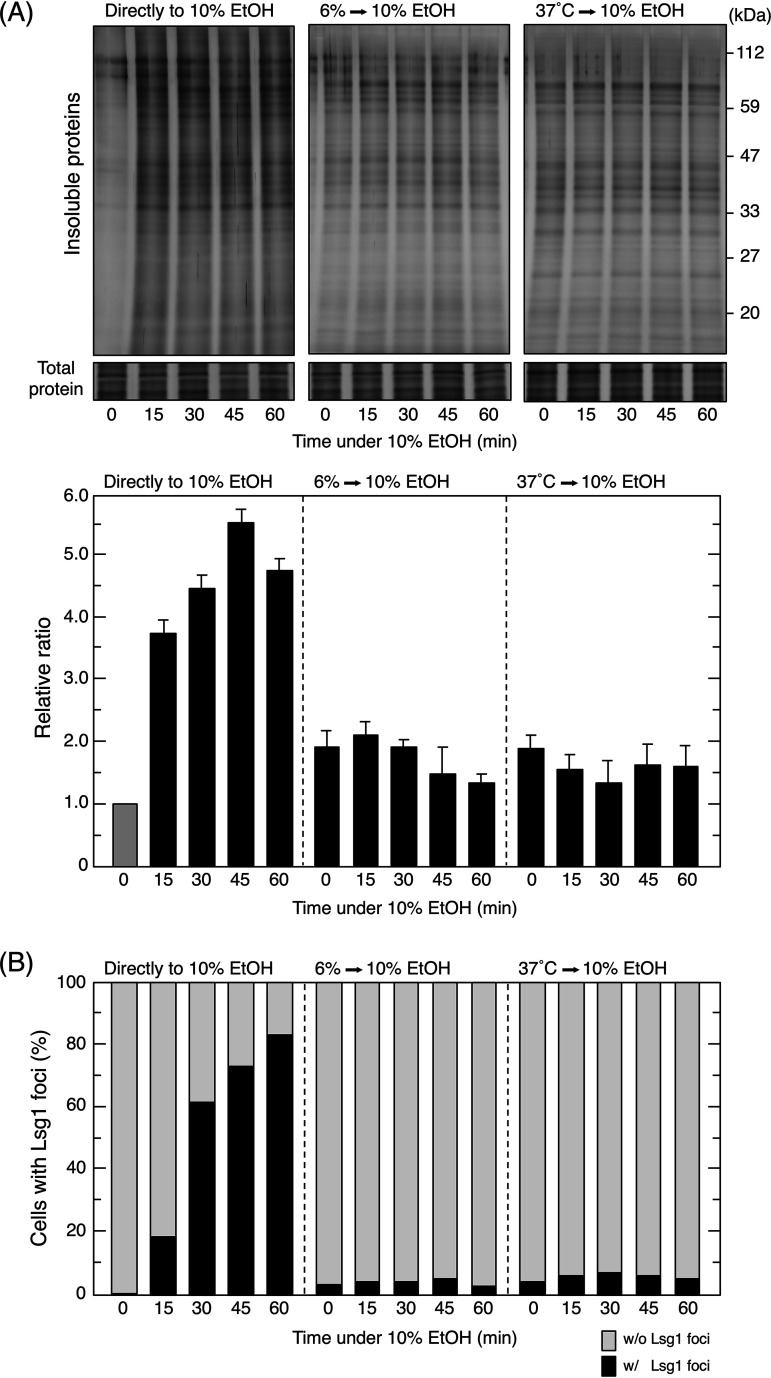
Furthermore, to clarify whether insoluble protein generation is suppressed in pretreated cells, we examined intracellular insoluble protein levels during the early stages of the treatment with 10% (vol/vol) ethanol. As shown in Fig. 8A, insoluble proteins gradually accumulated with time in cells directly exposed to 10% (vol/vol) ethanol stress. In contrast, insoluble protein levels did not increase in pretreated cells during the early stages. Although the initial levels of insoluble proteins were higher in pretreated cells than in nonpretreated cells (time zero min under 10% ethanol stress), insoluble protein levels in pretreated cells slightly decreased over time. Similar results were observed for the formation of Lsg1 foci (Fig. 8B). Therefore, the pretreatment with mild stress, either 6% (vol/vol) ethanol or thermal stress at 37°C, repressed the generation of insoluble proteins throughout the subsequent treatment with 10% (vol/vol) ethanol.
FIG 8.
The generation of insoluble proteins was suppressed in pretreated cells. The intracellular levels of insoluble proteins (A) and Lsg1-GFP foci (B) were analyzed every 15 min for the first 60 min after exposure to severe ethanol (EtOH) stress (10% [vol/vol]). (A) The intracellular levels of insoluble proteins were analyzed. Representative images are shown in the upper panel and quantified data in the lower panel. Relative ratios were calculated from three independent experiments. Each value is expressed as the mean ± SD (n = 3). (B) The quantification of cells containing Lsg1-GFP foci under each condition is shown. One hundred cells under each condition were examined, and experiments were repeated 3 times (300 cells in total were examined).
DISCUSSION
We here demonstrated that the accumulation of insoluble proteins and Lsg1 aggregation in yeast cells pretreated with 6% (vol/vol) ethanol or mild thermal stress at 37°C was mitigated under subsequent severe ethanol stress, clearly indicating that PQC activity was enhanced by the pretreatment. This enhancement in PQC activity by the pretreatment is regarded as a type of acquired resistance by yeast cells to severe ethanol stress.
The induction of enhanced PQC activity required the novel synthesis of proteins during the pretreatment, and pretreated cells maintained elevated expression levels of various proteins under subsequent 10% (vol/vol) ethanol stress. Since 10% (vol/vol) ethanol markedly inhibits translation activity, the synthesis of most proteins is strongly suppressed, except for several exceptions, such as Btn2 (4, 56). Despite the significant increases observed in the mRNA levels of SIS1 and SSA3, the respective protein levels were not markedly affected when cells were directly exposed to 10% (vol/vol) ethanol stress. On the other hand, the treatment with 6% (vol/vol) ethanol or 37°C did not strongly inhibit translation and increased the expression levels of various proteins (57–60). The levels of Fes1, Hsp104, and Ssa3, which constitute the bichaperone system (10, 22), were markedly upregulated in pretreated cells. Therefore, pretreated cells may tolerate severe ethanol stress due to an enhanced bichaperone system via newly synthesized proteins, whereas cells directly exposed to 10% (vol/vol) ethanol have to cope with protein denaturation using a weaker bichaperone system. Additionally, the insufficient acquired stress resistance of fes1Δ hsp104Δ and ssa2Δ ssa3Δ ssa4Δ cells indicated the importance of enhanced bichaperone systems for increased PQC activity in pretreated cells. The enhancement of the bichaperone system may have contributed to a reduction in insoluble protein accumulation in pretreated cells.
We cannot exclude the possibility that other mechanisms besides the bichaperone system were enhanced in pretreated cells to prevent the accumulation of insoluble proteins. Previous studies demonstrated that 6% (vol/vol) ethanol and thermal stress activated general stress pathways, resulting in increased levels of intracellular trehalose (39, 57, 61, 62). Since trehalose stabilizes protein structures and prevents protein denaturation and aggregation (63, 64), increased levels of intracellular trehalose due to the pretreatments may also contribute to the suppression of insoluble protein generation. Other molecular chaperones may also contribute to the mitigation of insoluble proteins in pretreated cells (24, 55).
In pretreated cells, insoluble protein levels did not increase immediately after the exposure to secondary stress, and the effects of proteasome inhibition by MG132 were very limited. These results suggest that the reduction observed in insoluble protein levels in pretreated cells was not due to the more efficient degradation of insoluble proteins. This interpretation appears to be supported by Rpn4, a transcription activator of proteasome-related genes, not markedly affecting the acquisition of increased resistance to severe ethanol (43). Severe protein denaturation, i.e., the generation of heavily damaged proteins that cannot be reactivated and refolded, may have been less likely to occur in pretreated cells due to the activation of the bichaperone system and other mechanisms. Since the generation of heavily damaged proteins was effectively suppressed, proteasomes appeared to be less important in pretreated cells. Additionally, a previous study demonstrated that in contrast to many exogenous thermolabile proteins, endogenous proteins are not destined to be degraded when severely aggregated by heat shock (65). Even under severe ethanol stress, denatured endogenous proteins may not be readily degraded.
Regarding aggregases, we were unable to confirm their distinct importance for enhanced PQC activity because btn2Δ hsp42Δ cells and wild-type cells were able to suppress the accumulation of insoluble proteins induced by the pretreatment. However, the contribution of aggregases to acquired stress resistance did not appear to be entirely absent. Hsp42 was maintained at high levels in pretreated cells during the exposure to 10% (vol/vol) ethanol. Hsp42 also functions as a molecular chaperone and interacts with partially damaged proteins to prevent protein aggregation (14, 22, 66); therefore, it may contribute to the prevention of severe protein denaturation, similar to the bichaperone system and trehalose. In contrast to Hsp42, Btn2 levels were not maintained under the subsequent 10% (vol/vol) ethanol stress in pretreated cells but were markedly elevated in cells directly exposed to 10% ethanol stress (Fig. 4). Since the turnover of Btn2 by the UPS is very effective (32, 49), our results suggest that the function of the UPS was maintained, even under 10% ethanol stress, in pretreated cells but not in nonpretreated cells.
Due to a lack of information, it currently remains unclear whether insoluble proteins accumulate in yeast cells during the brewing process of wine and Japanese sake. In typical wine and Japanese sake brewing processes, ethanol concentrations gradually increase and finally exceed 10% (vol/vol). Therefore, yeast cells are progressively exposed to mild to severe ethanol stress in the wine must or sake mash. Since the pretreatment with mild ethanol stress enhanced PQC activity in the present study, the accumulation of insoluble proteins may be suppressed due to the enhancement of PQC activity during the brewing process. A previous study reported that the expression of Btn2 was not induced during the wine-making process until the final stage (67). This may also reflect the long-term prevention of the accumulation of insoluble proteins during the wine-making process. We plan to collaborate with brewers in order to obtain a more detailed understanding of protein damage in yeast cells during the wine-making and sake brewing processes.
A mild ethanol pretreatment was previously shown to induce markedly stronger resistance to severe ethanol stress in the majority of wild yeast strains, including wine and sake yeasts, than in S288c-derived common laboratory strains, including BY4742, which was used in the present study (43). On the other hand, modern sake yeast strains commonly show defective stress responses due to mutations in MSN4 (msn4C15540A), PPT1 (Δppt1::Ty2), and RIM15 (rim155055insA) (68–70). Differences in the capacity to increase PQC activity between laboratory and brewer’s strains are an interesting subject for future research.
MATERIALS AND METHODS
Strains and medium.
The parental wild-type strain BY4742 (MATα his3Δ1 ura3Δ0 leu2Δ0 lys2Δ0) and its isogenic gene-deletion mutants, hsp42Δ::kanMX, hsp104Δ::kanMX, and pdr5Δ::kanMX were obtained from Open Biosystems Inc. (AL, USA). To disrupt the BTN2 gene, a DNA fragment (2.0 kb) containing btn2Δ::CgHIS3 was amplified using the primers btn2-F/R (Table 1) and pCgHIS3 as a template (71). The amplicon was introduced into hsp42Δ::kanMX to construct the btn2Δ hsp42Δ double-knockout mutant. To disrupt the FES1 gene, a DNA fragment (1.8 kb) encoding fes1Δ::CgURA3 was amplified using the primers fes1-F/R (Table 1) and pCgURA3 as a template (71). The amplicon was introduced into hsp104Δ::kanMX to construct the fes1Δ hsp104Δ double-knockout mutant. pCgHIS3 and pCgURA3 were provided by the National BioResource Project, Japan. JN516 (ssa2Δ ssa3Δ ssa4Δ) was provided by E. A. Craig (51).
TABLE 1.
List of primers used in knockout mutant and plasmid construction
| Name | Sequence |
|---|---|
| FES1-F1 | 5′-AAATATCTAGAGACAAGACAAAGCCACTCG-3′ |
| FES1-R1 | 5′-CGGACCTCGAGATAATACATACTTTACGGC-3′ |
| FES1-F2 | 5′-ATTATCTCGAGCTGACTACAAGGATGACGATGACAAGTGATTACGTCCGTAGAAATAAAATGATA-3′ |
| FES1-R2 | 5′-AAGATGGTACCAACTGTTTACTTGTACTGA-3′ |
| SIS1-F1 | 5′-GGTCCGAGCTCTGGTCCTGGTGGTCCTGGC-3′ |
| SIS1-R1 | 5′-GGATTCTCGAGAAAAATTTTCATCTATAGC-3′ |
| SIS1-F2 | 5′-TTTTTCTCGAGCTGACTACAAGGATGACGATGACAAGTAATAGTAATCCTAAGCAAATATAATTA-3′ |
| SIS1-R2 | 5′-TTTGAGGATCCCTACGTAATGACTGTTCCT-3′ |
| SSA1-F1 | 5′-TGCTGGTGGTGTCTAGACCAAGTTGATTCC-3′ |
| SSA1-R1 | 5′-CACCAATTGGCTCGAGCAACTTCTTCAACG-3′ |
| SSA3-F1 | 5′-GGATGCAGGAACTAGTGCAGGGATGAACGT-3′ |
| SSA3-R1 | 5′-AGAAGAATACTCGAGCAACCTCTTCCACTG-3′ |
| SSA3-F2 | 5′-AGTTGCTCGAGCCGACTACAAGGATGACGATGACAAGTGATTATTCTTCTATAGTGTTCT-3′ |
| SSA3-R2 | 5′-ACGTCATCTTTACCCGTGGTACCTAATTTC-3′ |
| LSG1-F1 | 5′-ACCGCGGTGGCGGCCGCTCTAGAGCGCTTGCTAAGGATTTGATTGTTCCA-3′ |
| LSG1-R1 | 5′-CTCCTTTGCTAGCCATAGCTCGAGAATTATTTTCAATGCTAAAAACTTTGCTTTTCGCATTTTTAC-3′ |
| btn2-F | 5′-AGTTCTTGGCGAAGTAAAGTGGCAAAACAAATGGAAGATCTATTGCATTAGTTGTAAAACGACGGCCAGT-3′ |
| btn2-R | 5′-TCCCTTGGGAGATCTGCTTAGGGACTCGTTGTATCTGTCAACTTCCTATCACAGGAAACAGCTATGACC-3′ |
| fes1-F | 5′-AGAGCACTCATCGTCAGTCAGAAAGCCATTACCTTTCAACGAAAGAGTGTTGTAAAACGACGGCCAGT-3′ |
| fes1-R | 5′-TGGTTTGGCGGTGTTATCACTTAATACAGGTGCTATGCAGTCGAGCCCCACAGGAAACAGCTATGACC-3′ |
Stress treatment.
Yeast cells were cultured in SD medium (2% glucose, 0.67% yeast nitrogen base without amino acids, 20 mg/liter uracil, 30 mg/liter l-lysine HCl, 100 mg/liter l-leucine, and 20 mg/liter l-histidine HCl) with reciprocal shaking (120 rpm) at 28°C. The seed culture was prepared by inoculating cells from agar plates in 3 ml SD medium. After cultivation for 12 h, cells from the seed culture were diluted in 250 ml SD medium (the initial optical density was adjusted to an optical density at 600 nm [OD600] of 0.05) in an Erlenmeyer flask (500 ml) and cultured further for more than eight generations to reset the cellular memory of acquired stress resistance (72). Exponentially growing cells were harvested at an OD600 of 0.5 to 0.6. Regarding the treatment with ethanol, harvested cells were transferred into fresh SD medium containing ethanol and incubated in a water bath at 28°C with shaking (120 rpm). In the thermal stress treatment, 25 ml of a cell culture in a 50-ml plastic centrifuge tube (catalog no. 227261; Greiner Bio-One International GmbH, Kremsmünster, Austria) was incubated in a water bath (37 or 42°C) with shaking (120 rpm). Proteasome activity was blocked by 100 μM MG132 (A11043; AdooQ Bioscience, CA, USA) (53).
Plasmids.
The primers used for plasmid construction are listed in Table 1. All FLAG tag-fused proteins were expressed using integrative plasmids, and the transcription of each gene was under the control of its own promoter.
(i) YIp-FES1-FLAG.
A 0.4-kbp fragment encoding a part of the open reading frame (ORF) of FES1 was amplified by PCR using the primers FES1-F1/R1 and cloned into the XbaI/XhoI sites of pJK67 (73) to construct YIp-FES1-GFP. A 0.3-kbp fragment encoding a FLAG tag sequence, stop codon, and the 3′-flanking region of FES1 was amplified using the primers FES1-F2/R2 and cloned into the XhoI/KpnI sites of YIp-FES1-GFP to construct YIp-FES1-FLAG. To integrate FES1-FLAG at the chromosomal FES1 locus, YIp-FES1-FLAG was linearized by HindIII and then introduced into yeast cells. This plasmid allowed the expression of FLAG-tagged Fes1 short form (Fes1 S) (74) to be visualized by Western blotting.
(ii) YIp-SIS1-GFP and YIp-SIS1-FLAG.
A 0.8-kbp fragment encoding a part of the ORF of SIS1 was amplified by PCR using the primers SIS1-F1/R1 and cloned into the SacI/XhoI sites of pJK67 to construct YIp-SIS1-GFP. A 0.3-kbp fragment encoding a FLAG tag sequence, stop codon, and the 3′-flanking region of SIS1 was amplified using the primers SIS1-F2/R2 and cloned into the XhoI/BamHI sites of YIp-SIS1-GFP to construct YIp-SIS1-FLAG. To integrate SIS1-GFP and SIS1-FLAG at the chromosomal SIS1 locus, YIp-SIS1-GFP and YIp-SIS1-FLAG were linearized by XbaI and then introduced into yeast cells.
(iii) YIp-SSA1-GFP.
A 0.7-kbp fragment encoding a part of the ORF of SSA1 was amplified by PCR using the primers SSA1-F1/R1 and cloned into the XbaI/XhoI sites of pJK67 to construct YIp-SSA1-GFP. To integrate SSA1-GFP at the chromosomal SSA1 locus, YIp-SSA1-GFP was linearized by SalI and then introduced into yeast cells.
(iv) YIp-SSA3-FLAG.
A 1.5-kbp fragment encoding a part of the ORF of SSA3 was amplified by PCR using the primers SSA3-F1/R1 and cloned into the SpeI/XhoI sites of pJK67 to construct YIp-SSA3-GFP. A 0.5-kbp fragment encoding a FLAG tag sequence, stop codon, and the 3′-flanking region of SSA3 was amplified using the primers SSA3-F2/R2 and cloned into the XhoI/KpnI sites of YIp-SSA3-GFP to construct YIp-SSA3-FLAG. To integrate SSA3-FLAG at the chromosomal SSA3 locus, YIp-SSA3-FLAG was linearized through its digestion with ClaI and then introduced into yeast cells.
(v) YIp-LSG1-GFP and YIp-LSG1-mRFP.
A 1.5-kbp fragment encoding a part of the ORF of LSG1 was amplified by PCR using the primers LSG1-F1/R1 and cloned into the XbaI/XhoI sites of pJK67 and YIp-DCP2-mRFP (56) to construct YIp-LSG1-GFP and YIp-LSG1-mRFP, respectively. To integrate LSG1-GFP or LSG1-mRFP at the chromosomal LSG1 locus, YIp-LSG1-GFP and YIp-LSG1-mRFP were linearized by HpaI and then introduced into yeast cells. To construct YIp-HIS3-LSG1-GFP, the LSG1-GFP region of YIp-LSG1-GFP was cloned in pRS303, which is an integrative plasmid with the HIS3 marker (75).
The construction of YIp-HSP42-FLAG, YIp-HSP104-GFP, and YIp-BTN2-FLAG was described in our previous studies (4, 31, 76). To construct YIp-LEU2-HSP104-GFP, the HSP104-GFP region of YIp-HSP104-GFP was cloned in pRS305, which is an integrative plasmid with the LEU2 marker (75).
qRT-PCR.
Quantitative reverse transcription-PCR (qRT-PCR) was conducted to assess the mRNA expression levels of the BTN2, FES1, HSP42, HSP104, SIS1, and SSA3 genes using previously described methods (4). Comparisons of mRNA expression levels were performed by normalizing the mRNA level of each gene to that of the reference gene, ACT1. The oligonucleotide sequences of the primers used in qRT-PCR are listed in Table 2.
TABLE 2.
List of primers used in qRT-PCR
| Name | Sequence |
|---|---|
| FES1-F | 5′-AGAACTACGTGCTGCTGCTT-3′ |
| FES1-R | 5′-ACGTCGAGTGGCTTTGTCTT-3′ |
| HSP42-F | 5′-GGACCAACCAACAGGCAAAC-3′ |
| HSP42-R | 5′-GTGGTCTCGACGATTCCTCC-3′ |
| HSP104-F | 5′-GGCCATCAAGCAACAAGCTC-3′ |
| HSP104-R | 5′-GCGGTCTTACCGATACCTGG-3′ |
| SIS1-F | 5′-GCATCAAGCTCTCCCACGTA-3′ |
| SIS1-R | 5′-CATGTGGGCCCTTTCTTCCA-3′ |
| SSA3-F | 5′-ATGTTGCGCCATTGTCCCTA-3′ |
| SSA3-R | 5′-ATTTGAGGCACACCTCTGGG-3′ |
| BTN2-F | 5′-TTTCCGAAGGTGGCATCAAC-3′ |
| BTN2-R | 5′-CTTTCGCTTTCTCCGCTTCTTC-3′ |
| ACT1-F | 5′-TTGGATTCCGGTGATGGTGTTACT-3′ |
| ACT1-R | 5′-TGAAGAAGATTGAGCAGCGGTTTG-3′ |
Protein analysis.
Insoluble aggregated protein levels were analyzed by the method of Koplin et al. (77) with a slight modification (6, 78). Briefly, after a treatment with Zymolyase-20T (2.5 mg/ml) (Nacalai Tesque, Kyoto, Japan) at 25°C for 20 min, cells were disrupted by vortexing with glass beads in lysis buffer (50 mM potassium phosphate buffer, 1.0 mM EDTA, and 5% glycerol, pH 7.0). Unbroken cells and debris were removed by centrifugation (200 × g for 20 min), and the total protein concentration of each sample was then measured and normalized. Insoluble aggregated proteins were collected by centrifugation (16,000 × g for 20 min), washed twice with lysis buffer containing 2% NP-40, and solubilized with urea buffer (50 mM Tris–HCl, 6.0 M urea, and 5% SDS, pH 7.5). Samples were subjected to 10% polyacrylamide gel electrophoresis and visualized by silver staining using Sil-Best Stain One (Nacalai Tesque).
Although the majority of proteins are resistant to glass bead-induced aggregation, some yeast proteins were aggregated during glass bead lysis (79). To investigate the potential artifacts of glass bead lysis, we performed mortar-pestle lysis to collect insoluble aggregated proteins using the method of Roth et al. (80). Since similar results were obtained by mortar-pestle lysis and glass bead lysis (see Fig. S3 in the supplemental material), the level of artifacts induced by glass bead lysis was considered to be negligible, and relative changes in insoluble protein levels were clearly observed with our method using glass bead lysis.
FLAG-tagged protein and Hsp104 levels were monitored by Western blotting using the procedure described previously (4, 6). An anti-FLAG M2 antibody (F1804; Sigma-Aldrich, MO, USA); anti-Hsp104 antibody (ADI-SPA-1040-D ENZ; Enzo Life Sciences, Inc., NY, USA); anti-mouse IgG, horseradish peroxidase (HRP)-linked antibody (7076S; Cell Signaling Technology, MA, USA); and anti-rabbit IgG, HRP-linked antibody (7074S; Cell Signaling Technology) were used for Western blotting. The bands on silver staining and Western blots were quantified using ImageJ software (http://imagej.nih.gov/ij/) and normalized by total proteins or Ponceau S staining, respectively. The significance of differences was evaluated by an unpaired two-tailed Student t test.
Fluorescence microscopic analysis.
A fluorescence microscope system (Leica AF6500; Leica Microsystems Vertrieb GmbH, Wetzlar, Germany) was used in the fluorescence microscopic analysis. The percentage of cells containing Hsp104-GFP or Lsg1-GFP foci was calculated by examining 100 cells under each condition and shown as the mean ± standard deviation (SD). Experiments were repeated 3 times (300 cells in total were examined under each condition).
Supplementary Material
ACKNOWLEDGMENTS
We thank E. A. Craig and the National BioResource Project, Japan, for providing yeast strains and plasmids.
The present study was supported by the Japan Society for the Promotion of Science to S.I. under grant numbers 17H03795, 19H02884, and 20H02900.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Leāo C, van Uden N. 1982. Effects of ethanol and other alkanols on the glucose transport system of Saccharomyces cerevisiae. Biotechnol Bioeng 24:2601–2604. doi: 10.1002/bit.260241124. [DOI] [PubMed] [Google Scholar]
- 2.Alexandre H, Plourde L, Charpentier C, Francois J. 1998. Lack of correlation between trehalose accumulation, cell viability and intracellular acidification as induced by various stresses in Saccharomyces cerevisiae. Microbiology 144:1103–1111. doi: 10.1099/00221287-144-4-1103. [DOI] [PubMed] [Google Scholar]
- 3.Takemura R, Inoue Y, Izawa S. 2004. Stress response in yeast mRNA export factor: reversible changes in Rat8p localization are caused by ethanol stress but not heat shock. J Cell Sci 117:4189–4197. doi: 10.1242/jcs.01296. [DOI] [PubMed] [Google Scholar]
- 4.Yamauchi Y, Izawa S. 2016. Prioritized expression of BTN2 of Saccharomyces cerevisiae under pronounced translation repression induced by severe ethanol stress. Front Microbiol 23:1319. doi: 10.3389/fmicb.2016.01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Homoto S, Izawa S. 2018. Persistent actin depolarization caused by ethanol induces the formation of multiple small cortical septin rings in yeast. J Cell Sci 131:jcs217091. doi: 10.1242/jcs.217091. [DOI] [PubMed] [Google Scholar]
- 6.Kato S, Yoshida M, Izawa S. 2019. Btn2 is involved in the clearance of denatured proteins caused by severe ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res 19:foz079. doi: 10.1093/femsyr/foz079. [DOI] [PubMed] [Google Scholar]
- 7.Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. 2011. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 8.Eisele YS, Monteiro C, Fearns C, Encalada SE, Wiseman RL, Powers ET, Kelly JW. 2015. Targeting protein aggregation for the treatment of degenerative diseases. Nat Rev Drug Discov 14:759–780. doi: 10.1038/nrd4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jouanne M, Rault S, Voisin-Chiret A. 2017. Tau protein aggregation in Alzheimer’s disease: an attractive target for the development of novel therapeutic agents. Eur J Med Chem 139:153–167. doi: 10.1016/j.ejmech.2017.07.070. [DOI] [PubMed] [Google Scholar]
- 10.Glover JR, Lindquist S. 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 11.Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature 475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 12.Amm I, Sommer T, Wolf DH. 2014. Protein quality control and elimination of protein waste: the role of the ubiquitin–proteasome system. Biochim Biophys Acta 1843:182–196. doi: 10.1016/j.bbamcr.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka K, Matsuda N. 2014. Proteostasis and neurodegeneration: the roles of proteasomal degradation and autophagy. Biochim Biophys Acta 1843:197–204. doi: 10.1016/j.bbamcr.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Ungelenk S, Moayed F, Ho CT, Grousl T, Scharf A, Mashaghi A, Tans S, Mayer MP, Mogk A, Bukau B. 2016. Small heat shock proteins sequester misfolding proteins in near-native conformation for cellular protection and efficient refolding. Nat Commun 7:13673. doi: 10.1038/ncomms13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dikic I. 2017. Proteasomal and autophagic degradation systems. Annu Rev Biochem 86:193–224. doi: 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- 16.Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald M, Lindquist S, Cyr DM. 2008. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci U S A 105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaganovich D, Kopito R, Frydman J. 2008. Misfolded proteins partition between two distinct quality control compartments. Nature 454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escusa-Toret S, Vonk WIM, Frydman J. 2013. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol 15:1231–1243. doi: 10.1038/ncb2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sontag EM, Samant RS, Frydman J. 2017. Mechanisms and functions of spatial protein quality control. Annu Rev Biochem 86:97–122. doi: 10.1146/annurev-biochem-060815-014616. [DOI] [PubMed] [Google Scholar]
- 20.Chen B, Retzlaff M, Roos T, Frydman J. 2011. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol 3:a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou C, Slaughter BD, Unruh JR, Guo F, Yu Z, Mickey K, Narkar A, Ross RT, McClain M, Li R. 2014. Organelle-based aggregation and retention of damaged proteins in asymmetrically diving cells. Cell 159:530–542. doi: 10.1016/j.cell.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho CT, Grousl T, Shatz O, Jawed A, Ruger-Herreros C, Semmelink M, Zahn R, Richter K, Bukau B, Mogk A. 2019. Cellular sequestrases maintain basal Hsp70 capacity ensuring balanced proteostasis. Nat Commun 10:4851. doi: 10.1038/s41467-019-12868-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle SM, Wickner S. 2009. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci 34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Mir SS, Fiedler D, Cashikar AG. 2009. Ssd1 is required for thermotolerance and Hsp104-mediated protein disaggregation in Saccharomyces cerevisiae. Mol Cell Biol 29:187–200. doi: 10.1128/MCB.02271-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gowda NKC, Kaimal JM, Kityk R, Daniel C, Liebau J, Öhman M, Mayer MP, Andréasson C. 2018. Nucleotide exchange factors Fes1 and HspBP1 mimic substrate to release misfolded proteins from Hsp70. Nat Struct Mol Biol 25:83–89. doi: 10.1038/s41594-017-0008-2. [DOI] [PubMed] [Google Scholar]
- 26.Parsell DA, Kowal AS, Singer MA, Lindquist S. 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 27.Zhou C, Slaughter BD, Unruh JR, Eldakak A, Rubinstein B, Li R. 2011. Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell 147:1186–1196. doi: 10.1016/j.cell.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill SM, Hanzén S, Nyström T. 2017. Restricted access: spatial sequestration of damaged proteins during stress and aging. EMBO Rep 18:377–391. doi: 10.15252/embr.201643458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaimal JM, Kandasamy G, Gasser F, Andréasson C. 2017. Coordinated Hsp110 and Hsp104 activities power protein disaggregation in Saccharomyces cerevisiae. Mol Cell Biol 37:e00027-17. doi: 10.1128/MCB.00027-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Specht S, Miller SBM, Mogk A, Bukau B. 2011. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol 195:617–629. doi: 10.1083/jcb.201106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itooka K, Takahashi K, Izawa S. 2016. Fluorescence microscopic analysis of antifungal effects of cold atmospheric pressure plasma in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 100:9295–9304. doi: 10.1007/s00253-016-7783-2. [DOI] [PubMed] [Google Scholar]
- 32.Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. 2012. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell 23:3041–3056. doi: 10.1091/mbc.E12-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller SB, Ho CT, Winkler J, Khokhrina M, Neuner A, Mohamed MY, Guilbride DL, Richter K, Lisby M, Schiebel E, Mogk A, Bukau B. 2015. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J 34:778–797. doi: 10.15252/embj.201489524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller SB, Mogk A, Bukau B. 2015. Spatially organized aggregation of misfolded proteins as cellular stress defense strategy. J Mol Biol 427:1564–1574. doi: 10.1016/j.jmb.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Nussbaum I, Weindling E, Jubran R, Cohen A, Bar-Nun S. 2014. Deteriorated stress response in stationary-phase yeast: Sir2 and Yap1 are essential for Hsf1 activation by heat shock and oxidative stress, respectively. PLoS One 9:e111505. doi: 10.1371/journal.pone.0111505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verghese J, Abrams J, Wang Y, Morano KA. 2012. Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev 76:115–158. doi: 10.1128/MMBR.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hahn JS, Neef DW, Thiele DJ. 2006. A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol Microbiol 60:240–251. doi: 10.1111/j.1365-2958.2006.05097.x. [DOI] [PubMed] [Google Scholar]
- 39.Berry DB, Gasch AP. 2008. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell 19:4580–4587. doi: 10.1091/mbc.e07-07-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aguilera F, Peinado RA, Millán C, Ortega JM, Mauricio JC. 2006. Relationship between ethanol tolerance, H+-ATPase activity and the lipid composition of the plasma membrane in different wine yeast strains. Int J Food Microbiol 110:34–42. doi: 10.1016/j.ijfoodmicro.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Dinh TN, Nagahisa K, Hirasawa T, Furusawa C, Shimizu H. 2008. Adaptation of Saccharomyces cerevisiae cells to high ethanol concentration and changes in fatty acid composition of membrane and cell size. PLoS One 3:e2623. doi: 10.1371/journal.pone.0002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dinh TN, Nagahisa K, Yoshikawa K, Hirasawa T, Furusawa C, Shimizu H. 2009. Analysis of adaptation to high ethanol concentration in Saccharomyces cerevisiae using DNA micro-array. Bioprocess Biosyst Eng 32:681–688. doi: 10.1007/s00449-008-0292-7. [DOI] [PubMed] [Google Scholar]
- 43.Lewis JA, Elkon IM, McGee MA, Higbee AJ, Gasch AP. 2010. Exploiting natural variation in Saccharomyces cerevisiae to identify genes for increased ethanol resistance. Genetics 186:1197–1205. doi: 10.1534/genetics.110.121871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis JA, Broman AT, Will J, Gasch AP. 2014. Genetic architecture of ethanol-responsive transcriptome variation in Saccharomyces cerevisiae strains. Genetics 198:369–382. doi: 10.1534/genetics.114.167429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obrig TG, Culp WJ, McKeehan WL, Hardesty B. 1971. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J Biol Chem 246:174–181. [PubMed] [Google Scholar]
- 46.Święciło A. 2016. Cross-stress resistance in Saccharomyces cerevisiae yeast—new insight into an old phenomenon. Cell Stress Chaperones 21:187–200. doi: 10.1007/s12192-016-0667-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDaniel EA, Stuecker TN, Veluvolu M, Gasch AP, Lewis JA. 2018. Independent mechanisms for acquired salt tolerance versus growth resumption induced by mild ethanol pretreatment in Saccharomyces cerevisiae. mSphere 3:e00574-18. doi: 10.1128/mSphere.00574-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stuecker TN, Scholes AN, Lewis JA. 2018. Linkage mapping of yeast cross protection connects gene expression variation to a higher-order organismal trait. PLoS Genet 14:e1007335. doi: 10.1371/journal.pgen.1007335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Storey AJ, Hardman HE, Byrum SD, Mackintosh SG, Edmondson RD, Wahls WP, Tackett AJ, Lewis JA. 2020. Accurate and sensitive quantitation of the dynamic heat shock proteome using tandem mass tags. J Proteome Res 19:1183–1195. doi: 10.1021/acs.jproteome.9b00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gowda NK, Kandasamy G, Froehlich MS, Dohmen RJ, Andréasson C. 2013. Hsp70 nucleotide exchange factor Fes1 is essential for ubiquitin-dependent degradation of misfolded cytosolic proteins. Proc Natl Acad Sci U S A 110:5975–5980. doi: 10.1073/pnas.1216778110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Becker J, Walter W, Yan W, Craig EA. 1996. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol 16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruan L, Zhou C, Jin E, Kucharavy A, Zhang Y, Wen Z, Florens L, Li R. 2017. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543:443–446. doi: 10.1038/nature21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braun HA, Umbreen S, Groll M, Kuckelkorn U, Mlynarczuk I, Wigand ME, Drung I, Kloetzel PM, Schmidt B. 2005. Tripeptide mimetics inhibit the 20 S proteasome by covalent bonding to the active threonines. J Biol Chem 280:28394–28401. doi: 10.1074/jbc.M502453200. [DOI] [PubMed] [Google Scholar]
- 54.Dobzinski N, Chuartzman SG, Kama R, Schuldiner M, Gerst JE. 2015. Starvation-dependent regulation of Golgi quality control links the TOR signaling and vacuolar protein sorting pathways. Cell Rep 12:1876–1886. doi: 10.1016/j.celrep.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 55.Kandasamy G, Andréasson C. 2018. Hsp70–Hsp110 chaperones deliver ubiquitin-dependent and -independent substrates to the 26S proteasome for proteolysis in yeast. J Cell Sci 131:jcs210948. doi: 10.1242/jcs.210948. [DOI] [PubMed] [Google Scholar]
- 56.Kato K, Yamamoto Y, Izawa S. 2011. Severe ethanol stress induces assembly of stress granules in Saccharomyces cerevisiae. Yeast 28:339–347. doi: 10.1002/yea.1842. [DOI] [PubMed] [Google Scholar]
- 57.van Voorst F, Houghton-Larsen J, Jønson L, Kielland-Brandt MC, Brandt A. 2006. Genome-wide identification of genes required for growth of Saccharomyces cerevisiae under ethanol stress. Yeast 23:351–359. doi: 10.1002/yea.1359. [DOI] [PubMed] [Google Scholar]
- 58.Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B. 2013. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol 23:2452–2462. doi: 10.1016/j.cub.2013.09.058. [DOI] [PubMed] [Google Scholar]
- 59.Yamamoto Y, Izawa S. 2013. Adaptive response in stress granule formation and bulk translational repression upon a combined stress of mild heat shock and mild ethanol stress in yeast. Genes Cells 18:974–984. doi: 10.1111/gtc.12090. [DOI] [PubMed] [Google Scholar]
- 60.Mühlhofer M, Berchtold E, Stratil CG, Csaba G, Kunold E, Bach NC, Sieber SA, Haslbeck M, Zimmer R, Buchner J. 2019. The heat shock response in yeast maintains protein homeostasis by chaperoning and replenishing proteins. Cell Rep 29:4593–4607.e8. doi: 10.1016/j.celrep.2019.11.109. [DOI] [PubMed] [Google Scholar]
- 61.Auesukaree C. 2017. Molecular mechanisms of the yeast adaptive response and tolerance to stresses encountered during ethanol fermentation. J Biosci Bioeng 124:133–142. doi: 10.1016/j.jbiosc.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 62.Dunayevich P, Baltanás R, Clemente JA, Couto A, Sapochnik D, Vasen G, Colman-Lerner A. 2018. Heat-stress triggers MAPK crosstalk to turn on the hyperosmotic response pathway. Sci Rep 8:15168. doi: 10.1038/s41598-018-33203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singer MA, Lindquist S. 1998. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol Cell 1:639–648. doi: 10.1016/s1097-2765(00)80064-7. [DOI] [PubMed] [Google Scholar]
- 64.Ma M, Liu ZL. 2010. Mechanisms of ethanol tolerance in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 87:829–845. doi: 10.1007/s00253-010-2594-3. [DOI] [PubMed] [Google Scholar]
- 65.Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, Airoldi EM, Pan T, Budnik BA, Drummond DA. 2015. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162:1286–1298. doi: 10.1016/j.cell.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee HY, Chao JC, Cheng KY, Leu JY. 2018. Misfolding-prone proteins are reversibly sequestered to an Hsp42-associated granule upon chronological aging. J Cell Sci 131:jcs220202. doi: 10.1242/jcs.220202. [DOI] [PubMed] [Google Scholar]
- 67.Kato S, Yamauchi Y, Izawa S. 2018. Protein synthesis of Btn2 under pronounced translation repression during the process of alcoholic fermentation and wine-making in yeast. Appl Microbiol Biotechnol 102:9669–9677. doi: 10.1007/s00253-018-9313-x. [DOI] [PubMed] [Google Scholar]
- 68.Noguchi C, Watanabe D, Zhou Y, Akao T, Shimoi H. 2012. Association of constitutive hyperphosphorylation of Hsf1p with a defective ethanol stress response in Saccharomyces cerevisiae sake yeast strains. Appl Environ Microbiol 78:385–392. doi: 10.1128/AEM.06341-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watanabe D, Wu H, Noguchi C, Zhou Y, Akao T, Shimoi H. 2011. Enhancement of the initial rate of ethanol fermentation due to dysfunction of yeast stress components Msn2p and/or Msn4p. Appl Environ Microbiol 77:934–941. doi: 10.1128/AEM.01869-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watanabe D, Araki Y, Zhou Y, Maeya N, Akao T, Shimoi H. 2012. A loss-of-function mutation in the PAS kinase Rim15p is related to defective quiescence entry and high fermentation rates of Saccharomyces cerevisiae sake yeast strains. Appl Environ Microbiol 78:4008–4016. doi: 10.1128/AEM.00165-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakumoto N, Matsuoka I, Mukai Y, Ogawa N, Kaneko Y, Harashima S. 2002. A series of double disruptants for protein phosphatase genes in Saccharomyces cerevisiae and their phenotypic analysis. Yeast 19:587–599. doi: 10.1002/yea.860. [DOI] [PubMed] [Google Scholar]
- 72.Guan Q, Haroon S, Bravo DG, Will JL, Gasch AP. 2012. Cellular memory of acquired stress resistance in Saccharomyces cerevisiae. Genetics 192:495–505. doi: 10.1534/genetics.112.143016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kahana JA, Schlenstedt G, Evanchuk DM, Geiser JR, Hoyt MA, Silver PA. 1998. The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol Biol Cell 9:1741–1756. doi: 10.1091/mbc.9.7.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gowda NKC, Kaimal JM, Masser KA, Kang W, Friedländer MR, Andréasson C. 2016. Cytosolic splice isoform of Hsp70 nucleotide exchange factor fes1 is required for the degradation of misfolded proteins in yeast. Mol Biol Cell 27:1210–1219. doi: 10.1091/mbc.E15-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Izawa S, Kita T, Ikeda K, Inoue Y. 2008. Heat shock and ethanol stress provoke distinctly different responses in 3’-processing and nuclear export of HSP mRNA in Saccharomyces cerevisiae. Biochem J 414:111–119. doi: 10.1042/BJ20071567. [DOI] [PubMed] [Google Scholar]
- 77.Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. 2010. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide associated complex on ribosomes. J Cell Biol 189:57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Itooka K, Takahashi K, Kimata Y, Izawa S. 2018. Cold atmospheric pressure plasma causes protein denaturation and endoplasmic reticulum stress in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 102:2279–2288. doi: 10.1007/s00253-018-8758-2. [DOI] [PubMed] [Google Scholar]
- 79.Papanayotou I, Sun B, Roth AF, Davis NG. 2010. Protein aggregation induced during glass bead lysis of yeast. Yeast 27:801–816. doi: 10.1002/yea.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roth AF, Feng Y, Chen L, Davis NG. 2002. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol 159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.