The initial bacterial colonization of human infants is crucial for lifelong health. Understanding the factors driving this colonization will therefore be of great importance.
KEYWORDS: infant gut microbiota
ABSTRACT
The nutritional drivers for mother-child sharing of bacteria and the corresponding longitudinal trajectory of the infant gut microbiota development are not yet completely settled. We therefore aimed to characterize the mother-child sharing and the inferred nutritional utilization potential for the gut microbiota from a large unselected cohort. We analyzed in depth gut microbiota in 100 mother-child pairs enrolled antenatally from the general population-based Preventing Atopic Dermatitis and Allergies in Children (PreventADALL) cohort. Fecal samples collected at gestational week 18 for mothers and at birth (meconium), 3, 6, and 12 months for infants were analyzed by reduced metagenome sequencing to determine metagenome size and taxonomic composition. The nutrient utilization potential was determined based on the Virtual Metabolic Human (VMH, www.vmh.life) database. The estimated median metagenome size was ∼150 million base pairs (bp) for mothers and ∼20 million bp at birth for the children. Longitudinal analyses revealed mother-child sharing (P < 0.05, chi-square test) from birth up to 6 months for 3 prevalent Bacteroides species (prevalence, >25% for all age groups). In a multivariate analysis of variance (ANOVA), the mother-child-shared Bacteroides were associated with vaginal delivery (1.7% explained variance, P = 0.0001). Both vaginal delivery and mother-child sharing were associated with host-derived mucins as nutrient sources. The age-related increase in metagenome size corresponded to an increased diversity in nutrient utilization, with dietary polysaccharides as the main age-related factor. Our results support host-derived mucins as potential selection means for mother-child sharing of initial colonizers, while the age-related increase in diversity was associated with dietary polysaccharides.
IMPORTANCE The initial bacterial colonization of human infants is crucial for lifelong health. Understanding the factors driving this colonization will therefore be of great importance. Here, we used a novel high-taxonomic-resolution approach to deduce the nutrient utilization potential of the infant gut microbiota in a large longitudinal mother-child cohort. We found mucins as potential selection means for the initial colonization of mother-child-shared bacteria, while the transition to a more adult-like microbiota was associated with dietary polysaccharide utilization potential. This knowledge will be important for a future understanding of the importance of diet in shaping the gut microbiota composition and development during infancy.
INTRODUCTION
Despite recent major discoveries concerning the role of the human gut microbiota in health and disease, we still lack detailed knowledge about the nutritional factors driving the initial colonization of the infant (1). There are indications of mother to child transmission of the gut microbiota during delivery (2–7). Nutritional sources that are involved in the microbiota selection in the gut, however, have not yet been analyzed comprehensively, limiting our mechanistic understanding of the colonization process during human infancy. Most of the knowledge, including the potential of the microbiota to induce host-derived nutrients (8) and the balance between host-derived and dietary nutrients, (9) has been derived from murine systems. The main knowledge gap for the human infant gut microbiota is to what extent host-derived nutrients select bacteria shared between mothers and their children and how dietary nutrients drive the diversification of the microbiota during weaning (10).
There are still technical hurdles to deducing the nutrient utilization potential of the gut microbiota. Current approaches in microbiota research are limited by the lack of sequencing depth of low-abundance microbiota for shotgun analyses (11), while 16S rRNA gene sequencing is limited by taxonomic resolution (12). The limitations of the current methods highlight the need for high-resolution approaches that also cover the low-abundance microbiota. Recently, reduced metagenome sequencing (RMS) was developed, which combines sequencing depth with taxonomic resolution (13, 14) and secures the needs for matching with functional databases such as the Virtual Metabolic Human (VMH, www.vmh.life) database, which is the first comprehensive database linking nutrient utilization potential to species (15).
The Preventing Atopic Dermatitis and Allergies in children (PreventADALL) study is a longitudinal general population-based mother-child birth cohort study (16) aimed at testing strategies to prevent the development of allergic disease, as well as to identify early life factors associated with other noncommunicable diseases.
To derive a model for nutrients as a driver for gut colonization, the aim of the current study was to characterize in depth both the mother-child sharing and the nutrient utilization potential of the microbiota from the longitudinal PreventADALL cohort using a combination of RMS and VMH analyses.
RESULTS
A total of 266,800,000 high-quality RMS sequencing reads were obtained from the 500 analyzed samples, with a mean sequencing depth of 533,600 per sample (see Fig. S1 in the supplemental material).
Metagenome size and diversity.
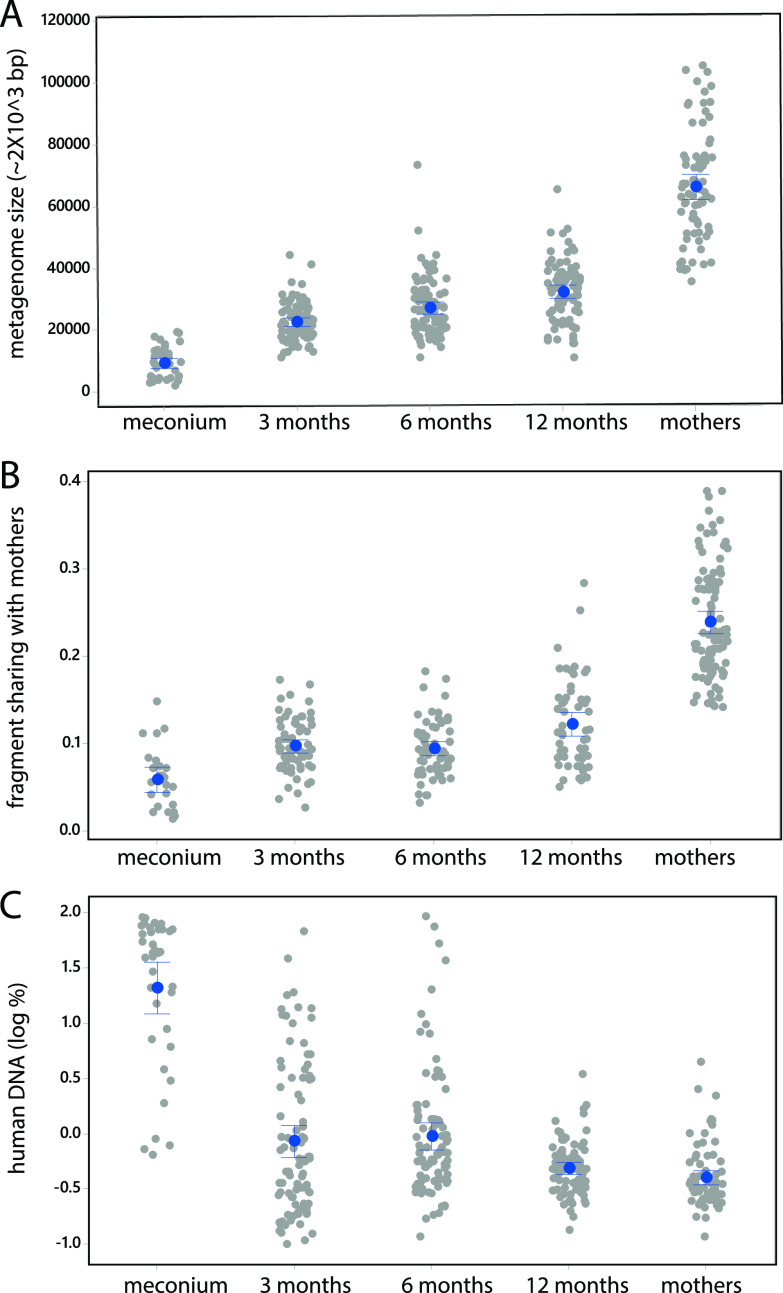
Based on the sequencing, we identified 1.6 million unique RMS fragments by clustering at 97% using VSEARCH. From the RMS fragment distribution, an average metagenome size of ∼150 million bp was estimated for mothers and about ∼20 million for newborns (meconium), with a gradual increase from 3 to 12 months of infant age of from ∼50 to ∼70 million bp (Fig. 1A).
FIG 1.
Diversity analyses based on RMS data. (A) Estimated metagenome size based on the number of unique fragments (assumption of one RMS fragment per 2,000 bp). (B) Fraction of fragments shared with mothers for each infant age group. (C) Portion of fragments assigned to human DNA for each infant age group. Each gray dot represents one sample, while the blue dots represent the mean values, and the error bars represent the 95% confidence interval.
The median fragment sharing across mothers was 22%. The mothers did not show significantly higher fragment sharing with their own child compared to the other children at any age (P > 0.05, Kruskal-Wallis test). The overall level of fragment sharing between mothers and infants, however, increased with the infant’s age from 5% for meconium, 9% at 3 and 6 months, to 11% sharing at 12 months (Fig. 1B). The amount of human DNA in stool samples declined from >10% in meconium to <1% in samples from mothers and infants at 3, 6, and 12 months of age (Fig. 1C).
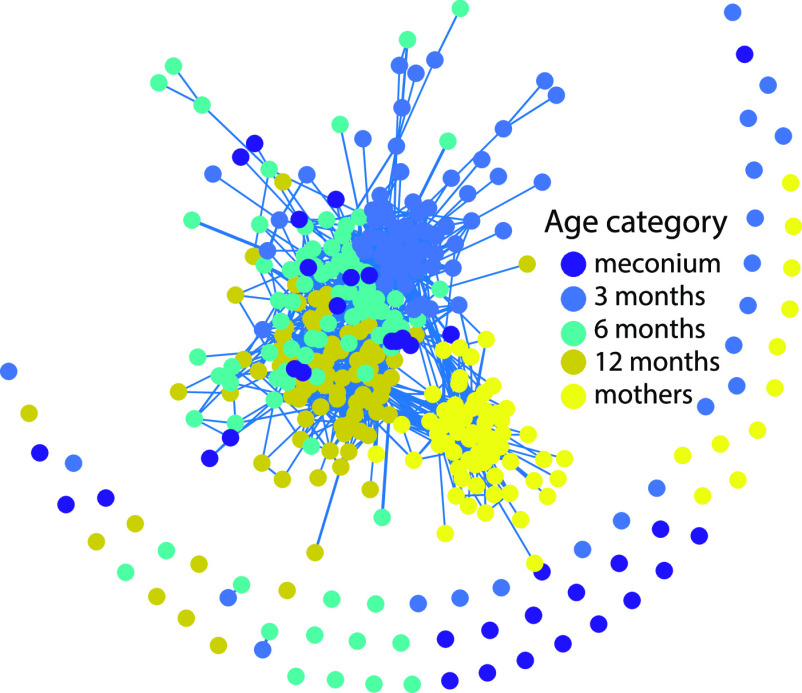
Using correlation analyses for the RMS fragment distribution, we identified a clear clustering for mothers, while clustering patterns at 3 to 12 months were less distinct. Meconium samples did not show distinct clustering patterns (Fig. 2).
FIG 2.
Correlation network across samples based on RMS fragment distribution. Each dot represents one sample, while the lines indicate samples with positive FDR-corrected (P < 0.05) Spearman correlations. Samples represent newborn (meconium) and 3/6/12 months of infant age, while mothers’ samples were from midpregnancy.
Taxonomic composition and nutrient utilization potential.
Kraken2 with the standard RefSeq database classified 72% of the RMS fragments across all the samples, while the customized HumGut database classified 82% of the RMS fragments belonging to a total of 391 species. We observed a relative increase of Bifidobacterium between newborns (meconium) and 3 months of infant age, with a decline from 6 to 12 months of age (Fig. 3). As Bifidobacterium declined, the relative abundance of Lachnospiraceae increased.
FIG 3.
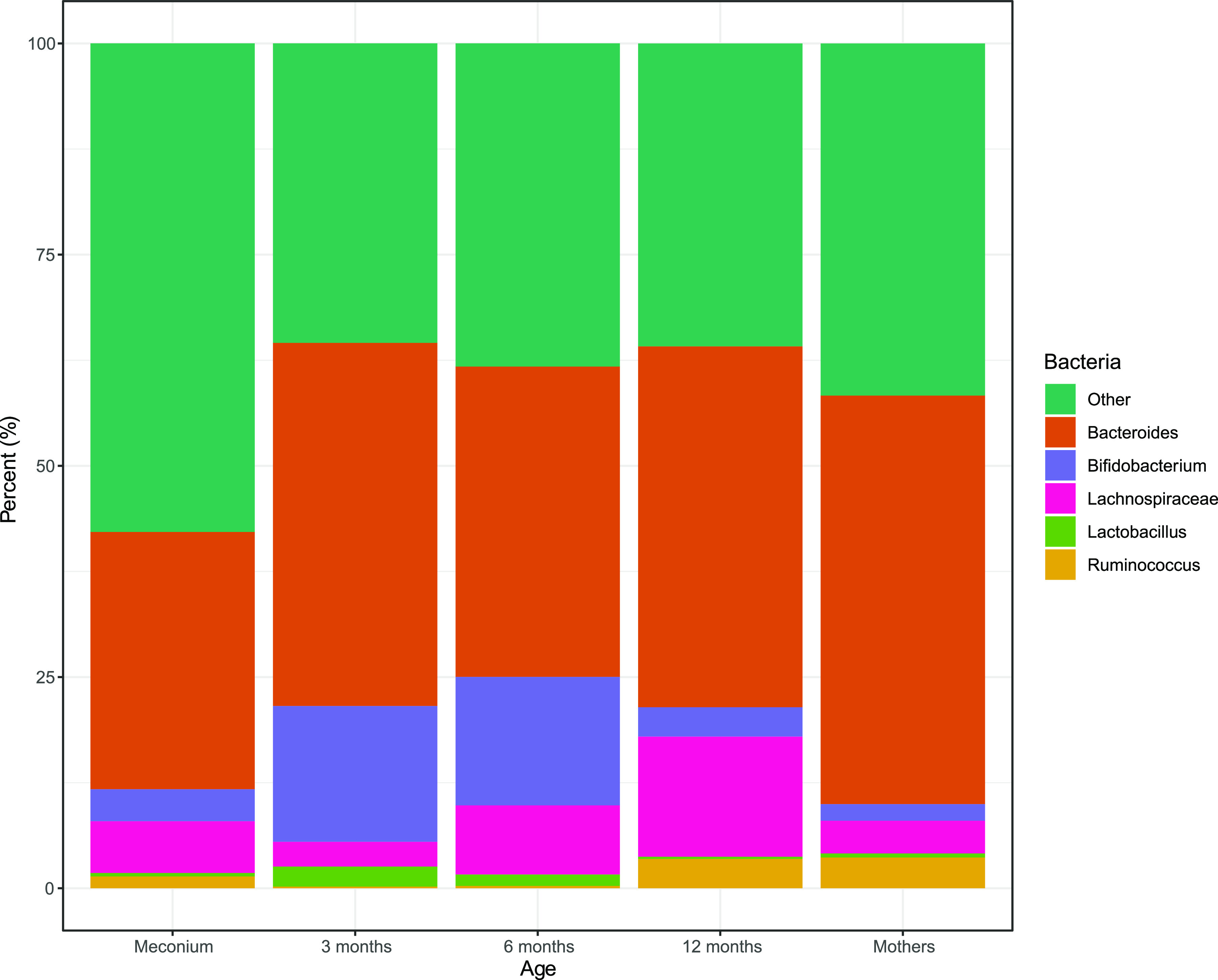
Relative gut microbiota composition for the different age groups. The relative abundances are presented with color codes for the different age groups, meconium (newborns), 3 months, 6 months, and 12 months of infant age, and mothers.
In addition, the relative abundance of the Bacteroides genus was observed to be high in all age groups.
The overall highest nutrient utilization potential was identified for both simple and complex sugars, while amino acids showed the lowest. There was an overall trend with an increase in the utilization potential of complex sugars and a decrease in mucus-derived sugars from 3 months of age (Fig. 4).
FIG 4.
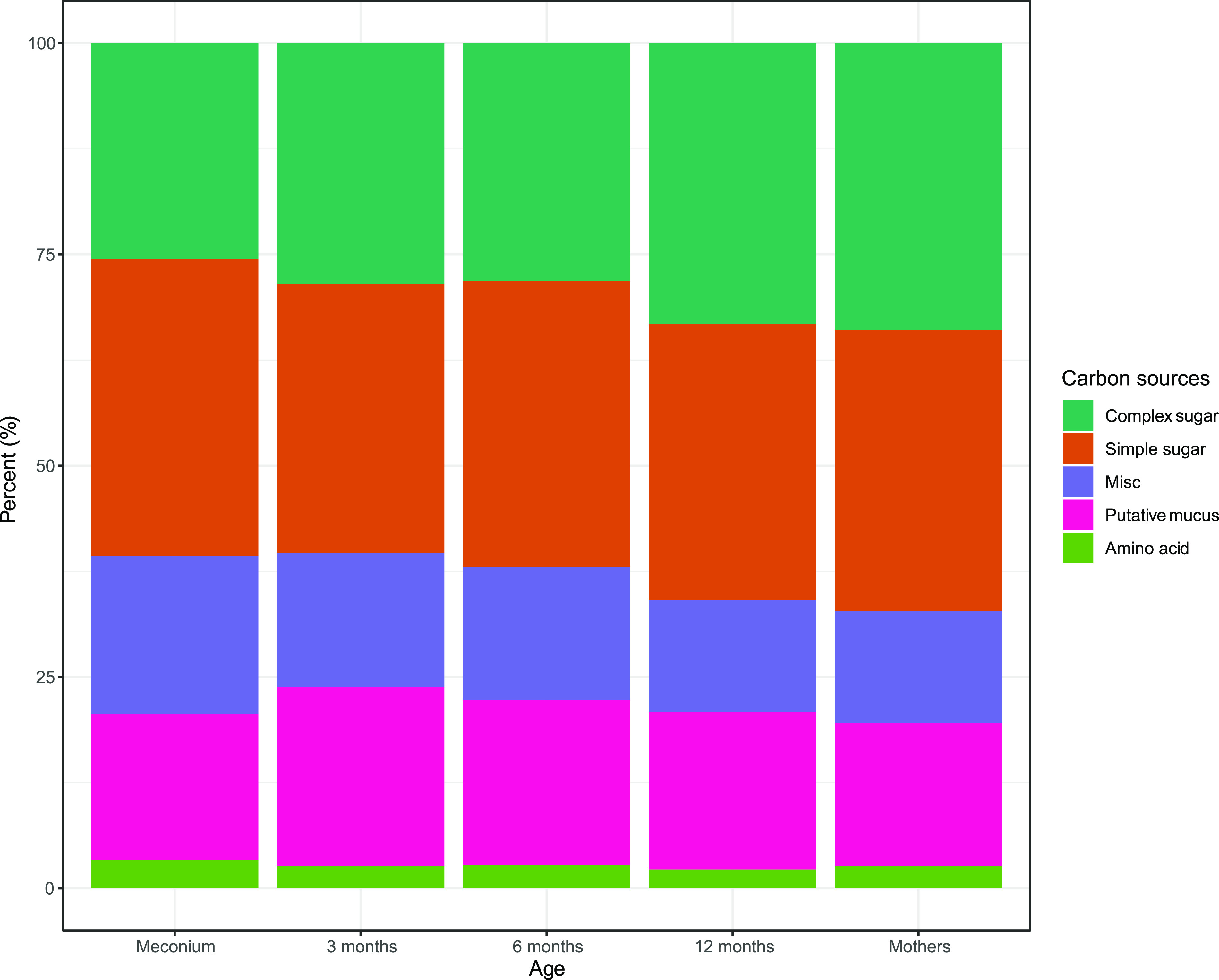
Deduced carbon sources for the different age groups. The bar chart represents the relative abundances of potential carbon sources derived from the VMH database, based on the bacterial species. The carbon sources are divided into the five groups simple and complex sugars, putative mucus, amino acids, and miscellaneous (Misc). They are represented with color codes for the different age groups.
Age-related associations.
Grouping based on offspring age and samples from mothers explained 6.9% of the variance in the bacterial species composition (analysis of variance-simultaneous component analysis [ASCA-ANOVA], P < 0.0001). These analyses revealed 3 main factors, with the first factor being related to increasing age (Fig. 5A). At the species level, Escherichia coli and Bifidobacterium longum showed the strongest associations with newborns (respective loadings of −0.061 and −0.063), while Coprococcus comes and Coprococcus catus showed the strongest association with increasing infant age and samples from mothers (loadings of 0.14 for both). At the genus level, the genera Bifidobacterium, Enterobacter, and Citrobacter were associated with young infant age, while Ruminococcus and Alistipes were associated with mother samples (Fig. 5A). The second factor was related to bacteria abundant in 12-month-old infants, with Lachnospiraceae showing the strongest association (Fig. 5B). The third component indicates that Bifidobacterium was associated with samples from 3- and 6-month-old infants (Fig. 5C).
FIG 5.
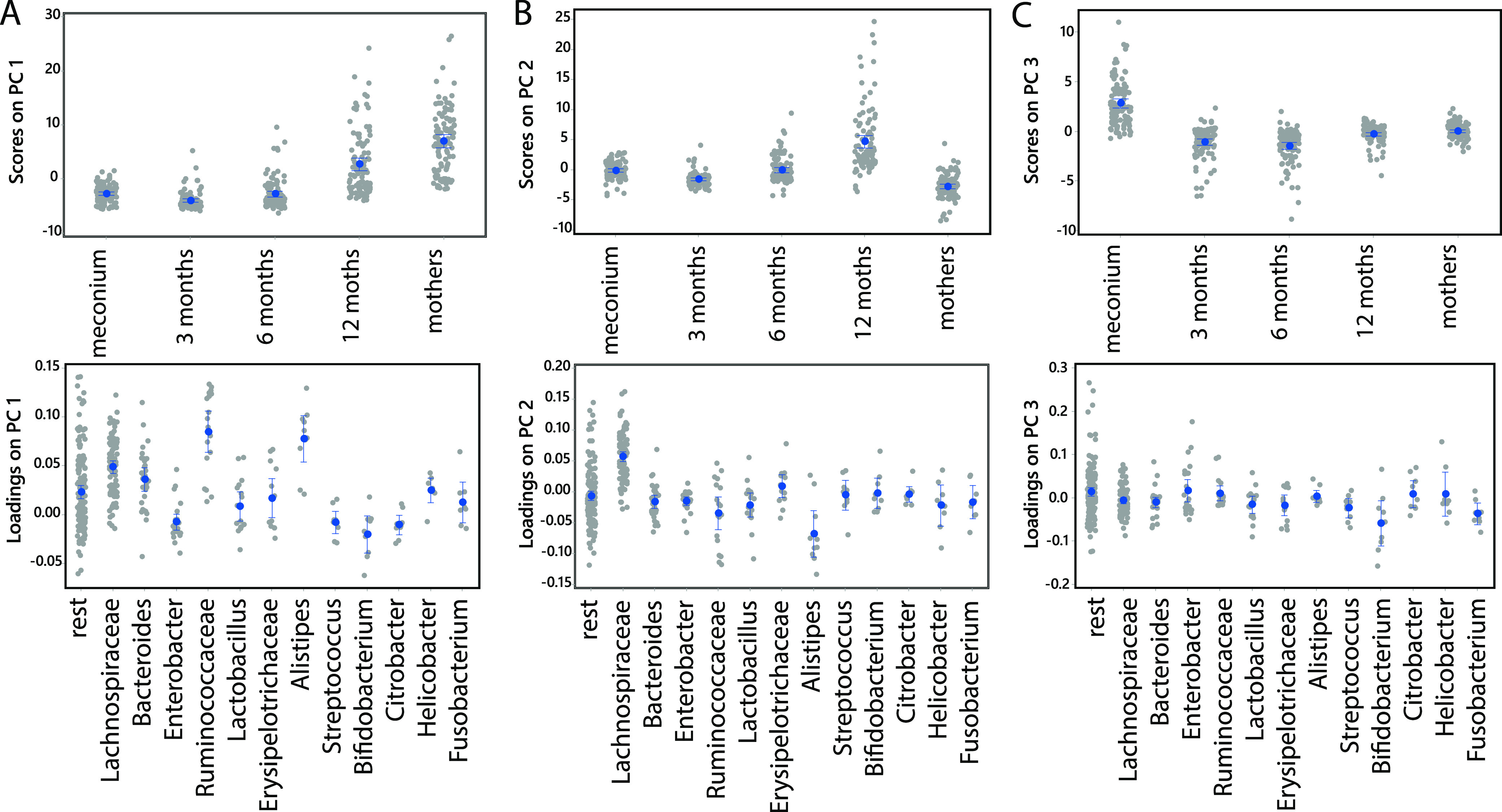
Association between age groups and microbiota. The association between age groups and microbiota was determined by ASCA-ANOVA analyses for the first three principal components. (A to C) The first principal component (A) explained 64.8% of the variance, the second component (B) explained 23.8% of the variance, and the third component (C) explained 8.4% of the variance. The top panels represent the scores of the samples, while the bottom panels represent the loading (importance) of the different taxonomic groups.
There were significant age-related compositional differences in nutrient utilization potential, with age groups explaining 8.6% of the variance (ASCA-ANOVA, P < 0.0001). We identified two main age-related factors associated with carbon source utilization potential. The first factor was mainly manifested at 3 months of age with loadings associated with sugars and oligosaccharides (Fig. 6A). The second factor was mainly connected with age, where dietary polysaccharides were associated with increased age while miscellaneous carbon sources, simple sugars, and putative mucus were related to low age (Fig. 6B).
FIG 6.
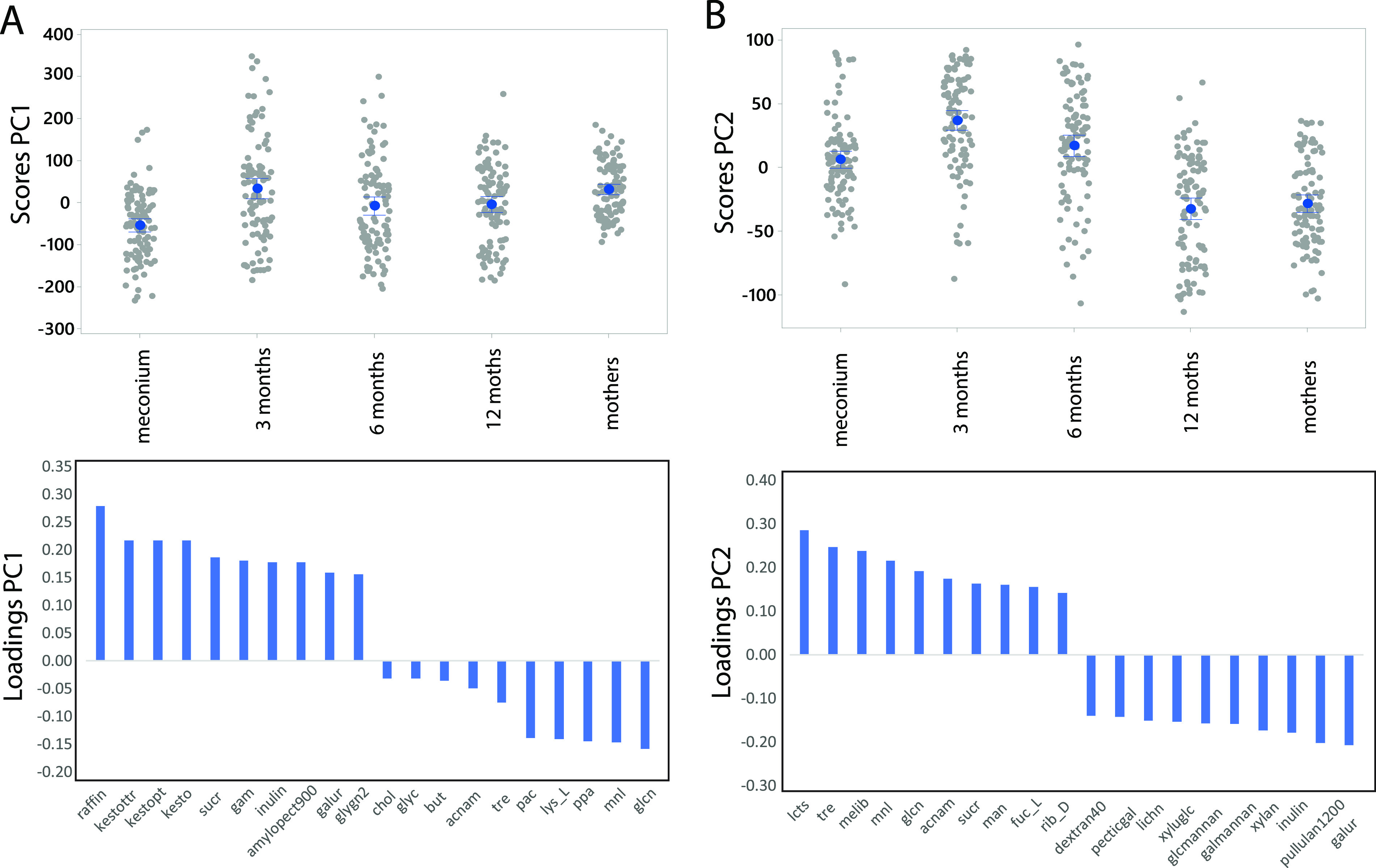
Association between age groups and carbon sources. The association between age groups and microbiota was determined by ASCA-ANOVA. Results from the two first principal components are shown in panels A and B, respectively. For both components, the top panels represent the score of the samples while the lower panels represent the loading (importance) of the different taxonomic groups for the 10 variables with the highest and the lowest loadings. Abbreviations are defined in Table S2.
Mother-child associations.
Using ASCA-ANOVA, we identified that samples coming from the same mother-child pairs (mother, 18th week of pregnancy; meconium, 3, 6, and 12 months for the child) explained 21.1% of the variance in bacterial composition collectively for all age groups (P = 0.005). Of the 49 bacterial species showing a significant false-discovery rate (FDR)-corrected mother-child association by a Kruskal-Wallis test, 33.3% belonged to the genus Bacteroides. Five species (four Bacteroides and one Parabacteroides) were significantly (P < 0.05, chi-square test) shared between mother and infant, lasting up to 6 months of age, while the association was not seen at 12 months (Fig. 7).
FIG 7.
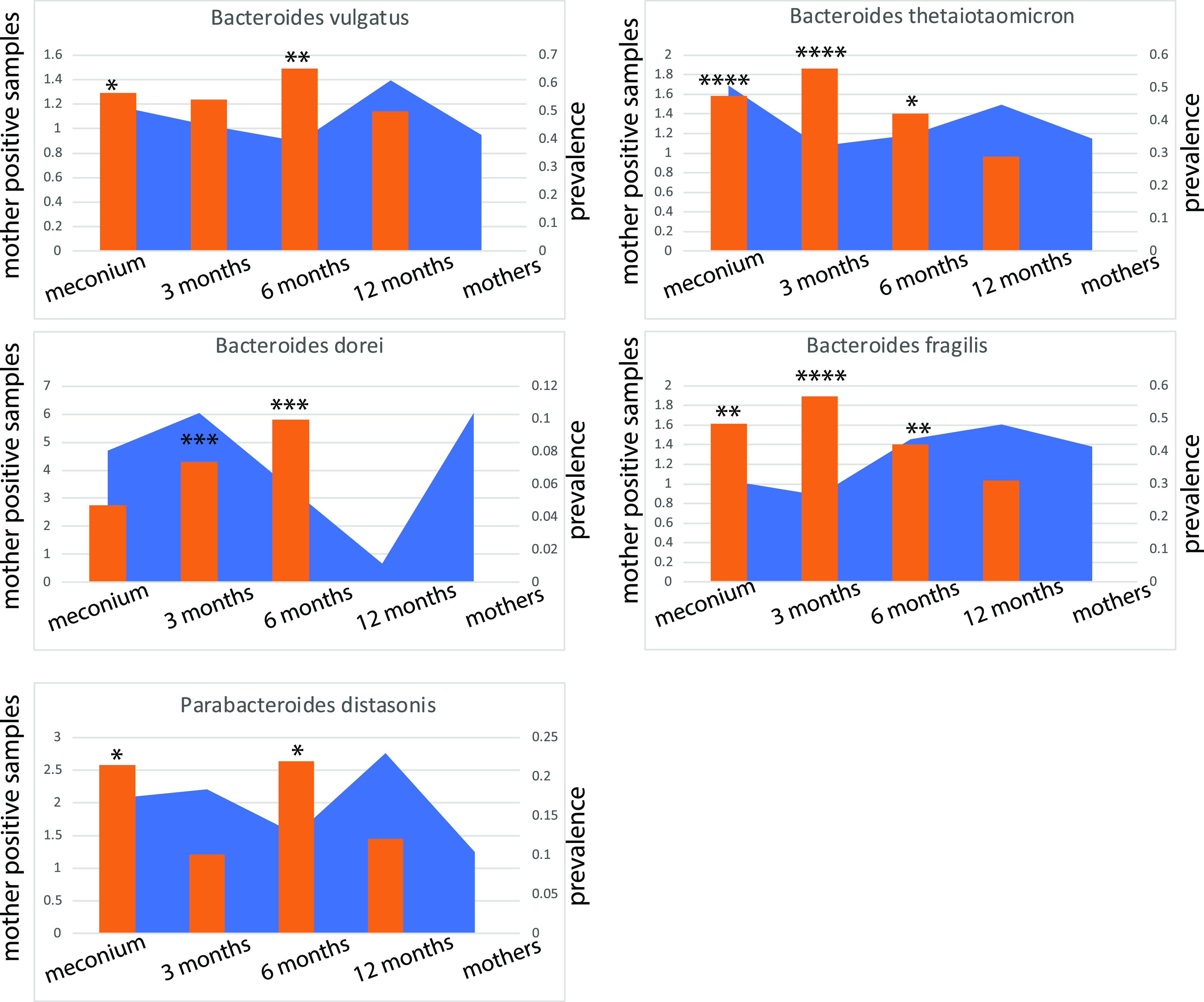
Mother-child-associated bacteria. Orange bars represent the prevalence in infants whose mothers have positive samples divided by the total prevalence across all mother-child pairs for each age group. The blue graph represents the overall prevalence for each age group. Asterisks represent significance levels for chi-square tests; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ***, P < 0.0001.
Delivery mode explained 1.7% of the overall microbiota variance (P = 0.0001, ASCA-ANOVA). Bacteroides fragilis showed the strongest association with vaginal delivery with a loading of 0.69, followed by Bacteroides thetaiotaomicron and Bacteroides uniformis with respective loadings of 0.30 and 0.38. For bacteria associated with caesarian section, Enterococcus faecalis and Bifidobacterium breve showed the strongest associations, with negative loadings of −0.24 for both.
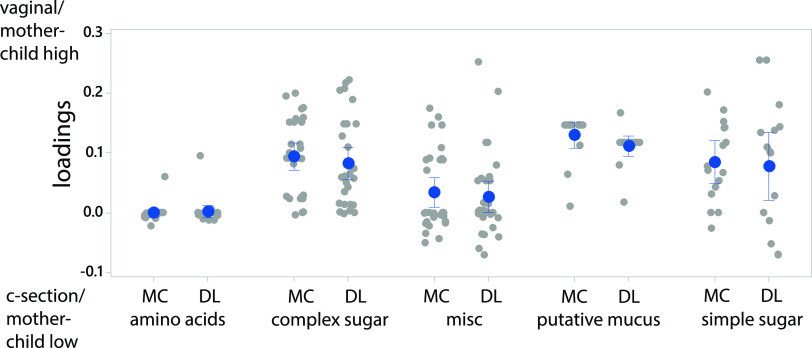
Mother-child pairs explained 30.6% of the variance while delivery mode explained 3.1% (ASCA-ANOVA, P < 0.001 for both) for the carbon source utilization potential. There was a relatively strong positive correlation (Spearman rho = 0.89, P < 0.0005) between the loadings for both mother-child association and vaginal delivery mode, with mucins being the most important for mother-child-associated bacteria in vaginally delivered infants (Fig. 8).
FIG 8.
Correlation between nutrients important for mother-child association and delivery mode (x axis). The importance of the nutrients was determined by ASCA-ANOVA analyses. The figure illustrates loadings on the y axis and mother-child associations (MC) and delivery mode (DL) for the different nutrients.
Dietary associations.
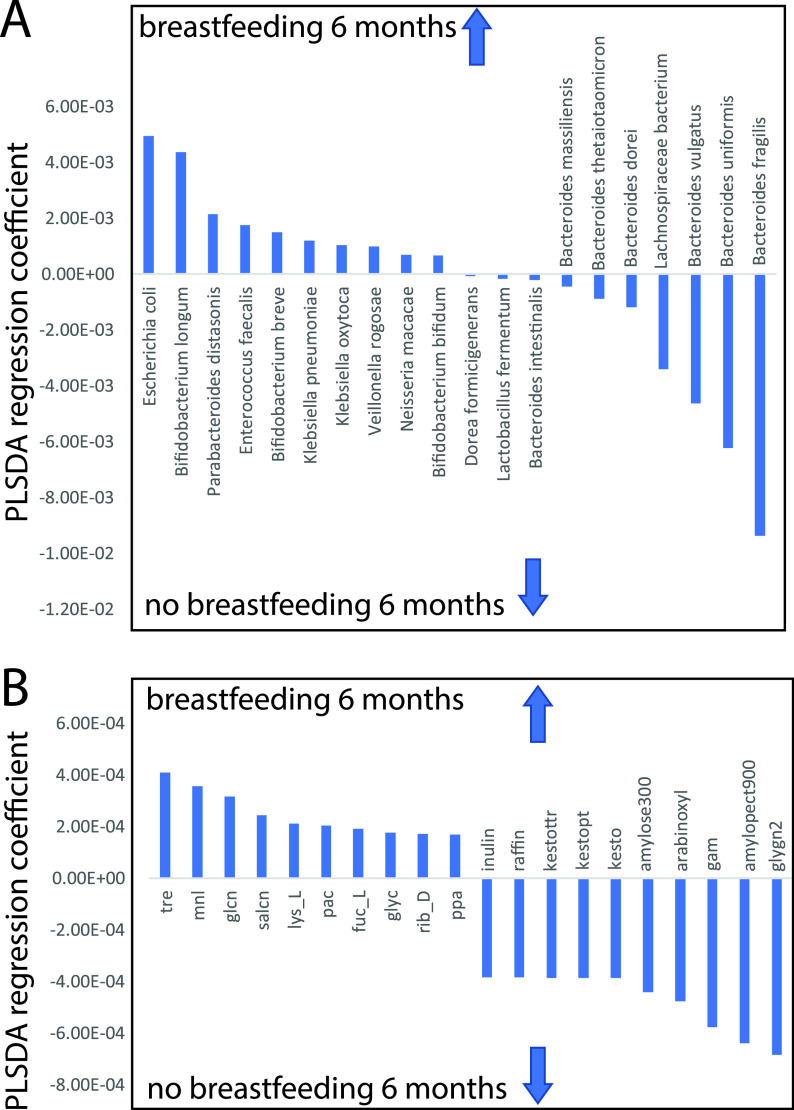
We investigated the relation of dietary intake to both microbiota species composition and nutrition utilization potential based on questionnaire information about breastfeeding and introduction of solid food and dairy products. Overall, we only found significant association for Citrobacter freundii between the species composition and breastfeeding at 6 months for the whole data set (P < 0.05, FDR corrected Spearman). We therefore analyzed the 6-month samples separately, together with breastfeeding information at 6 months. Partial least-squares discriminant analyses (PLSDA) revealed an accuracy of classification of 0.69 (cross-validated) in predicting species composition at 6 months by breastfeeding, while the accuracy of classification was 0.66 for the nutrient utilization potential. The regression vector revealed that E. coli and Bifidobacterium longum showed the strongest association with breastfeeding, while B. uniformis and B. fragilis showed an association with lack of breastfeeding at 6 months (Fig. 9A). For the carbon source utilization potential, breastfeeding showed the strongest association with trehalose, while a lack of breastfeeding showed the strongest association with glycogen (Fig. 9B).
FIG 9.
Regression coefficients connected to partial least-squares (PLS) discriminant analysis for breastfeeding at 6 months. (A and B) Coeffects for (A) species composition and (B) carbon sources. The arrows represent the direction of the associations. For the carbon sources, abbreviations are provided in Table S2.
DISCUSSION
In murine models, it has been shown that Bacteroides can induce mucin production from gut epithelial cells as their own nutrient source (8). Here, we also show that mucins as nutrients can be important for transmission of mother-child-associated bacteria. Taken together, this indicates positive host selection as a possible mechanism for securing mother to child transmission. The main mother-child-associated bacteria, B. vulgatus, B. thetaiotaomicron, and B. fragilis, all showed mucin degradation potential (15). Therefore, direct transmission combined with positive selection is a likely mechanism for securing transmission of important bacteria from mother to child.
Dietary compounds such as pectin, inulin, and pullulan represent the main components associated with the age of the child. This indicates diet as a main driver for the establishment of the adult-like gut microbiota. Interestingly, pullulans are commonly used in edible films, but very little is known about the effect on the gut microbiota (17). Pectin, on the other hand, is a well-known modulator of the gut microbiota composition (18), while inulin stimulates Faecalibacterium prausnitzii, which is one of the most abundant bacteria in the human adult gut (19).
For the age-related colonization patterns, we observed an overrepresentation of Lachnospiraceae and underrepresentation of Alistipes at 12 months, while Alistipes and Ruminococcaceae were strongly associated with mothers at midpregnancy, suggesting that the colonization of the infant gut with these bacteria is delayed compared to Lachnospiraceae. Alistipes was the genus that showed the largest difference between 12-month-old children and mothers at midpregnancy. In children, Alistipes has been associated with abdominal pain (20), while in adults Alistipes is considered beneficial by increasing the mucosal barrier (21).
We found the same mother-child-associated Bacteroides species and their association with vaginal delivery as previously found in the TEDDY study (3) and the BBC study (4). These findings suggest that an early transmission between mother and offspring takes place and that colonization occurs before passing of the first stool, potentially after rupture of the amniotic membrane (5).
The mother-child association until 6 months of age for different species of Bacteroides may have an impact on the infant’s immunological development later in life. We previously showed that colonization by B. fragilis without E. coli colonization is associated with IgE sensitization (22). In a study of Finnish and Estonian children, a link between Bacteroides ovatus and type I diabetes was identified (23), while a recent study suggests that B. vulgatus protects against Vibrio cholerae infections (24). Several other studies also reported on important immunomodulatory properties of Bacteroides (25). Furthermore, the high levels of Bacteroides, in particular, B. vulgatus, correspond to a recent meta-study of all publicly available shotgun libraries of the human gut microbiota (26).
The associations of both microbial composition and function with the questionnaire data were weak. Overall, we only found a significant association for C. freundii with breastfeeding at 6 months. However, when using only the data from 6 months in predicting breastfeeding, we found both E. coli and B. longum to be positively associated, while B. uniformis and B. fragilis were negatively associated. These results are consistent with previous findings of Bifidobacteria suppressing Bacteroides (27), with mother’s milk selecting Bifidobacteria (28).
For the nutrient utilization potential, trehalose did show the strongest association with breastfeeding at 6 months. This association is difficult to explain based on the current knowledge. A potential explanation could be that trehalose represents an important component of opportunistic fungal pathogens (29), so trehalose degradation could represent a potential protection against fungal pathogen infection by mother’s milk-selected bacteria. This explanation, however, needs further experimental verification.
Taken together, our results support a mother-to-child transmission model where host-derived carbon sources secure the initial transmission while dietary fiber selects adult-like gut bacteria. Furthermore, the fact that dietary polysaccharides are associated with an adult-like gut microbiota supports the hypothesis that they protect the mucosa by driving the microbiota to utilize dietary carbon sources rather than mucosal ones (9).
MATERIALS AND METHODS
Sample material.
The sample material is from the general population-based PreventADALL, which is a mother-child birth cohort recruited at 18 weeks of pregnancy (16). The study enrolled 2,697 nonselected pregnant women and their 2,396 infants in Oslo, Østfold (both Norway), and Sweden. We selected the first recruited 100 mother-child pairs with fecal samples for at least four of the following five time points: maternal sample at approximately 18 weeks of pregnancy, offspring meconium samples from birth, and fecal samples from the infant at 3, 6, and 12 months of age. The included children were born at 39.6 ± 1.6 weeks of gestation with a birth weight of 3,577 ± 529 g, with 22 infants delivered via caesarean section and 78 delivered vaginally. Immediately after sampling, the stool samples were collected in sterile extraction plants and diluted 1:10 in DNA stabilization buffer to avoid degradation of DNA (PSP spin stool DNA sampling tube; Nordic Biolabs, Taby, Sweden), followed by storage at −80°C. Informed written consent was obtained from all of the pregnant mothers upon inclusion and again from both parents upon inclusion of the newborn child. The PreventADALL study was approved by the Regional Ethical Committee (REK) for Medical and Health Research Ethics in South-Eastern Norway (2014/518) as well as in Sweden (2014/2242-31/4) by the Regional Ethics Committee of Stockholm. The study is registered as NCT02449850 at www.clinicaltrials.gov.
Lysis and DNA extraction.
We used a combination of chemical and mechanical lysis, with 300 μl sample material being mixed with acid-washed glass beads of three different sizes (0.2 g [<106 μm], 0.2 g [425 to 600 μm], and 2 × 2.5 to 3.5 mm) of beads (Sigma-Aldrich, Germany). The samples were processed twice using a FastPrep 96 instrument (MP Biomedicals, USA) at 1,800 rpm for 40 s, with subsequent centrifugation at 13,000 rpm for 5 min. DNA extraction was carried out using the MagMidi kit on the KingFisher Flex robot (Thermo Fisher Scientific, USA) following the manufacturer’s recommendations (LCG Genomics, UK).
Generation and sequencing of RMS libraries.
Approximately 10 ng genomic DNA was cut for 1 h at 37°C using a combination of 8 U EcoRI and 4 U MseI prior to adapter ligation for 3 h at 37°C using T4 DNA ligase (New England Biolabs, USA). Adapters were made of equal amounts of forward strands (EcoRI, 5′-CTCGTAGACTGCGTACC-3′; MseI, 5′-GACGATGAGTCCTGAG-3′) and reverse strands (EcoRI, 5′-AATTGGTACGCAGTCTAC-3′; MseI, 5′-TACTCAGGACTCAT-3′).
Adapter ligated genomic DNA was amplified with specific EcoRI forward (5′-GACTGCGTACCAATTC-3′) and MseI reverse (5′-GATGAGTCCTGAGTAA-3′) primer pairs using 1× Hotfire Pol Blend master mix ready to load (Solis Biodyne, Estonia). The samples were amplified using 25 cycles of denaturation at 95°C for 30 s, hybridization at 55°C for 1 min, and elongation at 72°C for 1 min, before a final step at 72°C for 7 min. The number of cycles was increased to 30 for meconium (newborn) samples. All PCR amplifications were conducted on a 2720 thermal cycler (Applied Biosystems, USA). The PCR products were purified using an AMPure XP system (Beckman Coulter, USA) according to the manufacturer’s recommendations.
Indexing was done using 1× Firepol master mix ready to load (Solis Biodyne, Estonia), with Illumina TruSeq indexes. We used the following PCR thermocycles for indexing: 95°C for 5 min, followed by 10 cycles of denaturation at 95°C for 30 s, hybridization at 56°C for 1 min, and elongation at 72°C for 1 min, before a final step of 72°C for 7 min. The samples were mixed in equimolar concentrations after amplification, with a final AMPure purification prior to sequencing. The sequencing was performed using the HiSeq 3000 sequencer at the Norwegian Sequencing Center (University of Oslo, Oslo, Norway).
Data processing.
We used Cutadapt to demultiplex raw data files into sample-wise files (30). Each read pair was taxonomically classified using Kraken2 with default settings. We used both the standard Kraken2 database and a customized genome database representing the human gut pan-taxa (26). The number of RMS fragments varies a lot between species, and read counts from Kraken2 must be normalized for this effect in order to reflect species abundances. For all genomes in our database, we counted the number of RMS fragments in silico. From this, we retrieved the per-species average RMS fragment count and divided that by the Kraken2 read count for a species to obtain normalized read counts reflecting the abundance of the species. The normalized data for all samples were presented in a species table.
In addition to the species table, we also generated a table showing RMS fragment cluster read counts across samples, similar to an OTU table from 16S amplicons, using VSEARCH software (31). The data processing is similar to a standard 16S-based pipeline, with the exception of some RMS fragments being too long to produce overlapping reads, in which case reads were joined, i.e., pasted together, with a section of N’s in between. After merging/joining, all reads were clustered at 97% identity, producing the read count table.
Functional assignments.
The functional assignments for nutrient (carbon source) utilization potential were determined by assuming similar functions for bacterial species identified by RMS in the current study, as for those in the Virtual Metabolic Human (VMH, www.vmh.life) database (15). This was done by matching the RMS assigned species names with those for the VMH database. Each carbon source quantification was achieved by adding together the relative abundance for each bacterial species containing the given function. Functions were treated statistically in the same manner as for taxonomic composition.
Statistical analyses.
Analysis of variance-simultaneous component analysis (ASCA-ANOVA), a multivariate statistical variant of ANOVA (32), was used to investigate the relation both between age and mother-child pair and the microbiota. Partial least-squares discriminant analyses (PLSDA) were used for investigating the predictability of microbiota composition and carbon sources based on categorical metadata such as the dietary information (Table S1). A nonparametric Kruskal-Wallis test was conducted to test statistically significant differences in the relative amounts of bacterial species connected to mother-child pairs, while Spearman correlation was used to determine the age-related associations of the microbiota. We used a chi-square test to test the association between dichotomized variables. All P values were false-discovery corrected by Benjamini-Hochberg multiple test corrections, with a significance threshold of 0.05.
Data availability. The centroid sequences and the corresponding RMS OTU table have been deposited in the OSF repository (osf.io) under the project name RMS_infant:Gut. The raw sequencing data are available from the corresponding author (knut.rudi@nmbu.no) upon request.
Supplementary Material
ACKNOWLEDGMENTS
We sincerely thank all the study participants and all the individuals involved in facilitating and running the PreventADALL study. Special thanks are given to the study group (Oslo-universitetssykehus.no/avdelinger/barne-og-ungdomsklinikken/preventadall).
This work was financially supported by the Norwegian Asthma and Allergy Foundation and the Research Council of Norway through project no. 301364, “UnveilMe: Unveiling the Role of Microbial Metabolites in Human Infant Development.”
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wang S, Ryan CA, Boyaval P, Dempsey EM, Ross RP, Stanton C. 2020. Maternal vertical transmission affecting early-life microbiota development. Trends Microbiol 28:28–45. doi: 10.1016/j.tim.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Yassour M, Jason E, Hogstrom LJ, Arthur TD, Tripathi S, Siljander H, Selvenius J, Oikarinen S, Hyoty H, Virtanen SM, Ilonen J, Ferretti P, Pasolli E, Tett A, Asnicar F, Segata N, Vlamakis H, Lander ES, Huttenhower C, Knip M, Xavier RJ. 2018. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe 24:146–154.e4. doi: 10.1016/j.chom.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni H, Metcalf GA, Muzny D, Gibbs RA, Vatanen T, Huttenhower C, Xavier RJ, Rewers M, Hagopian W, Toppari J, Ziegler A-G, She J-X, Akolkar B, Lernmark A, Hyoty H, Vehik K, Krischer JP, Petrosino JF. 2018. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, Kumar N, Stares MD, Rodger A, Brocklehurst P, Field N, Lawley TD. 2019. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574:117–121. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rehbinder EM, Lodrup Carlsen KC, Staff AC, Angell IL, Landro L, Hilde K, Gaustad P, Rudi K. 2018. Is amniotic fluid of women with uncomplicated term pregnancies free of bacteria? Am J Obstet Gynecol 219:289.e1–289.e12. doi: 10.1016/j.ajog.2018.05.028. [DOI] [PubMed] [Google Scholar]
- 6.Korpela K, Costea P, Coelho LP, Kandels-Lewis S, Willemsen G, Boomsma DI, Segata N, Bork P. 2018. Selective maternal seeding and environment shape the human gut microbiome. Genome Res 28:561–568. doi: 10.1101/gr.233940.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, Armanini F, Truong DT, Manara S, Zolfo M, Beghini F, Bertorelli R, De Sanctis V, Bariletti I, Canto R, Clementi R, Cologna M, Crifò T, Cusumano G, Gottardi S, Innamorati C, Masè C, Postai D, Savoi D, Duranti S, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Milani C, Mangifesta M, Anzalone R, Viappiani A, Yassour M, Vlamakis H, Xavier R, Collado CM, Koren O, Tateo S, Soffiati M, Pedrotti A, Ventura M, Huttenhower C, Bork P, Segata N. 2018. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24:133–145. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bry L, Falk PG, Midtvedt T, Gordon JI. 1996. A model of host-microbial interactions in an open mammalian ecosystem. Science 273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 9.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Núñez G, Martens EC. 2016. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167:1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avershina E, Lundgard K, Sekelja M, Dotterud C, Storro O, Oien T, Johnsen R, Rudi K. 2016. Transition from infant- to adult-like gut microbiota. Environ Microbiol 18:2226–2236. doi: 10.1111/1462-2920.13248. [DOI] [PubMed] [Google Scholar]
- 11.Malmstrom RR, Eloe-Fadrosh EA. 2019. Advancing genome-resolved metagenomics beyond the shotgun. mSystems 4:e00118-19. doi: 10.1128/mSystems.00118-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avershina E, Angell IL, Simpson M, Storro O, Oien T, Johnsen R, Rudi K. 2018. Low maternal microbiota sharing across gut, breast milk and vagina, as revealed by 16S rRNA gene and reduced metagenomic sequencing. Genes (Basel) 9:231. doi: 10.3390/genes9050231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravi A, Avershina E, Angell IL, Ludvigsen J, Manohar P, Padmanaban S, Nachimuthu R, Snipen L, Rudi K. 2018. Comparison of reduced metagenome and 16S rRNA gene sequencing for determination of genetic diversity and mother-child overlap of the gut associated microbiota. J Microbiol Methods 149:44–52. doi: 10.1016/j.mimet.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 15.Noronha A, Modamio J, Jarosz Y, Guerard E, Sompairac N, Preciat G, Danielsdottir AD, Krecke M, Merten D, Haraldsdottir HS, Heinken A, Heirendt L, Magnusdottir S, Ravcheev DA, Sahoo S, Gawron P, Friscioni L, Garcia B, Prendergast M, Puente A, Rodrigues M, Roy A, Rouquaya M, Wiltgen L, Zagare A, John E, Krueger M, Kuperstein I, Zinovyev A, Schneider R, Fleming RMT, Thiele I. 2019. The Virtual Metabolic Human database: integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res 47:D614–D624. doi: 10.1093/nar/gky992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lødrup Carlsen KC, Rehbinder EM, Skjerven HO, Carlsen MH, Fatnes TA, Fugelli P, Granum B, Haugen G, Hedlin G, Jonassen CM, Landro L, Lunde J, Marsland BJ, Nordlund B, Rudi K, Sjoborg K, Soderhall C, Staff AC, Vettukattil R, Carlsen KH, The Study Group . 2018. Preventing Atopic Dermatitis and ALLergies in Children: the PreventADALL study. Allergy 73:2063–2070. doi: 10.1111/all.13468. [DOI] [PubMed] [Google Scholar]
- 17.Chlebowska-Smigiel A, Gniewosz M, Kieliszek M, Bzducha-Wrobel A. 2017. The effect of pullulan on the growth and acidifying activity of selected stool microflora of human. Curr Pharm Biotechnol 18:121–126. doi: 10.2174/1389201017666161229154324. [DOI] [PubMed] [Google Scholar]
- 18.Larsen N, Bussolo de Souza C, Krych L, Barbosa Cahú T, Wiese M, Kot W, Hansen KM, Blennow A, Venema K, Jespersen L. 2019. Potential of pectins to beneficially modulate the gut microbiota depends on their structural properties. Front Microbiol 10:223–223. doi: 10.3389/fmicb.2019.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. 2008. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 101:541–550. doi: 10.1017/S0007114508019880. [DOI] [PubMed] [Google Scholar]
- 20.Saulnier DM, Riehle K, Mistretta T-A, Diaz M-A, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J. 2011. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gavin PG, Mullaney JA, Loo D, Cao K-AL, Gottlieb PA, Hill MM, Zipris D, Hamilton-Williams EE. 2018. Intestinal metaproteomics reveals host-microbiota interactions in subjects at risk for type 1 diabetes. Diabetes Care 41:2178–2186. doi: 10.2337/dc18-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudi K, Storro O, Oien T, Johnsen R. 2012. Modelling bacterial transmission in human allergen-specific IgE sensitization. Lett Appl Microbiol 54:447–454. doi: 10.1111/j.1472-765X.2012.03229.x. [DOI] [PubMed] [Google Scholar]
- 23.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hamalainen AM, Peet A, Tillmann V, Uibo R, Mokurov S, Dorshakova N, Ilonen J, Virtanen SM, Szabo SJ, Porter JA, Lahdesmaki H, Huttenhower C, Gevers D, Cullen TW, Knip M, Xavier RJ, DIABIMMUNE Study Group . 2016. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.You JS, Yong JH, Kim GH, Moon S, Nam KT, Ryu JH, Yoon MY, Yoon SS. 2019. Commensal-derived metabolites govern Vibrio cholerae pathogenesis in host intestine. Microbiome 7:132. doi: 10.1186/s40168-019-0746-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wexler AG, Goodman AL. 2017. An insider’s perspective: Bacteroides as a window into the microbiome. Nat Microbiol 2:17026–17026. doi: 10.1038/nmicrobiol.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiseni P, Rudi K, Wilson RC, Hegge FT, Snipen L. 2020. HumGut: a comprehensive human gut prokaryotic genomes collection filtered by metagenome data. bioRxiv doi: 10.1101/2020.03.25.007666:2020.03.25.007666. [DOI] [PMC free article] [PubMed]
- 27.Shiba T, Aiba Y, Ishikawa H, Ushiyama A, Takagi A, Mine T, Koga Y. 2003. The suppressive effect of Bifidobacteria on Bacteroides vulgatus, a putative pathogenic microbe in inflammatory bowel disease. Microbiol Immunol 47:371–378. doi: 10.1111/j.1348-0421.2003.tb03368.x. [DOI] [PubMed] [Google Scholar]
- 28.LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. 2010. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol 76:7373–7381. doi: 10.1128/AEM.00675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perfect JR, Tenor JL, Miao Y, Brennan RG. 2017. Trehalose pathway as an antifungal target. Virulence 8:143–149. doi: 10.1080/21505594.2016.1195529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 31.Rognes T, Flouri T, Nichols B, Quince C, Mahe F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smilde AK, Jansen JJ, Hoefsloot HC, Lamers RJ, van der Greef J, Timmerman ME. 2005. ANOVA-simultaneous component analysis (ASCA): a new tool for analyzing designed metabolomics data. Bioinformatics 21:3043–3048. doi: 10.1093/bioinformatics/bti476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.