Listeria monocytogenes is a foodborne bacterial pathogen responsible for listeriosis. Whole-genome sequencing has been extensively used in public health and food industries to characterize circulating Listeria isolates, but genomic data on isolates occurring in natural environments and wild animals are still scarce.
KEYWORDS: Listeria monocytogenes, wild animal, tonsil, virulence, cgMLST, WGS
ABSTRACT
Listeria monocytogenes is a major human and animal foodborne pathogen. However, data from environmental reservoirs remain scarce. Here, we used whole-genome sequencing to characterize Listeria species isolates recovered over 1 year from wild animals in their natural habitats in Spain. Three different Listeria spp. (L. monocytogenes [n = 19], Listeria ivanovii subsp. londoniensis [n = 4], and Listeria innocua [n = 3]) were detected in 23 animal tonsils (9 deer, 14 wild boars) and 2 feeding troughs. No Listeria species was detected in feces. L. monocytogenes was detected in tonsils of 44.4% (8 out of 18) of deer and 40.7% (11 out of 27) of wild boars. L. monocytogenes isolates belonged to 3 different core genome multilocus sequence typing (cgMLST) types (CTs) of 3 distinct sublineages (SL1, SL387, and SL155) from lineages I and II. While cgMLST type L1-SL1-ST1-CT5279 (IVb; clonal complex 1 [CC1]) occurred only in one animal, types L1-SL387-ST388-CT5239 (IVb; CC388) and L2-SL155-ST155-CT1170 (IIa; CC155) were retrieved from multiple animals. In addition, L1-SL387-ST388-CT5239 (IVb; CC388) isolates were collected 1 year apart, revealing their long-term occurrence within the animal population and/or environmental reservoir. The presence of identical L. monocytogenes strains in deer and wild boars suggests contamination from a common food or environmental source, although interhost transmission cannot be excluded. Pathogenicity islands LIPI-1, LIPI-3, and LIPI-4 were present in 100%, 5%, and 79% of the L. monocytogenes isolates, respectively, and all L. monocytogenes lineage II isolates (n = 3) carried SSI-1 stress islands. This study highlights the need for monitoring L. monocytogenes environmental contamination and the importance of tonsils as a possible L. monocytogenes intrahost reservoir.
IMPORTANCE Listeria monocytogenes is a foodborne bacterial pathogen responsible for listeriosis. Whole-genome sequencing has been extensively used in public health and food industries to characterize circulating Listeria isolates, but genomic data on isolates occurring in natural environments and wild animals are still scarce. Here, we show that wild animals carry pathogenic Listeria and that the same genotypes can be found at different time points in different host species. This work highlights the need of Listeria species monitoring of environmental contamination and the importance of tonsils as a possible L. monocytogenes intrahost reservoir.
INTRODUCTION
Listeria monocytogenes is a foodborne pathogen that causes listeriosis. Although rare, human listeriosis has one of the highest case fatality rates (20 to 30%) and hospitalization rates (>97%) among foodborne pathogens (1, 2). In domestic ruminants, L. monocytogenes can cause rhombencephalitis, septicemia, and abortion, but animals tend to be asymptomatic carriers that shed the bacterium in their feces (3–6). In pigs, listeriosis manifests mainly as septicemia, with encephalitis reported less frequently and abortions rarely (7).
Previous studies have detected the presence of L. monocytogenes in a small percentage of wild boar (0 to 6.1%) (8–12) or deer (1 to 5.4%) (11, 12) feces samples. Likely as a consequence of animal fecal excretion, L. monocytogenes is frequently found in soil, decaying vegetables, and river and canal waters, leading to direct contamination of raw food materials, which are then conveyed to food processing industries (13). In tonsils, which are lymphoid tissue aggregates situated at the entrance of the digestive and respiratory tracts and known sites of multiplication of L. monocytogenes (14), the detection of L. monocytogenes has been reported to be high (8.2 to 35% in wild boars; 13% in deer) (9, 10, 15).
L. monocytogenes can also persist in food processing plants for months to decades (16–18). It has been shown that L. monocytogenes may travel along the bird migration route, leading to transmission over a large geographical distance by black-headed gull (19). Persistent carriage has also been shown for other bacteria, such as Staphylococcus aureus, where its long-term nasal carriage has been associated with high pathogen burden and more extensive spreading of staphylococci into the environment (20).
The L. monocytogenes population is heterogeneous and can be classified into lineages (21), PCR genoserogroups (22), sequence types (STs), and clonal complexes (CCs or clones), as defined by multilocus sequence typing (MLST) (23), and sublineages (SLs) and core genome MLST (cgMLST) types (CTs), as defined by cgMLST (24). This heterogeneity also reflects different pathogenic potential among L. monocytogenes isolates, with lineage I been more frequently isolated from clinical cases (25).
Whole-genome sequencing (WGS) permits an unprecedented subtyping resolution and strain characterization, being a powerful tool for national and international surveillance and outbreak investigations in clinical and food production settings (26, 27). However, genomic data on Listeria spp. occurring in natural environments and wild animals are still scarce (28). Here, we used WGS to subtype and characterize Listeria species isolates recovered during 1 year from wild boars and deer hunted in a region of the southeast of Spain and their environments.
RESULTS AND DISCUSSION
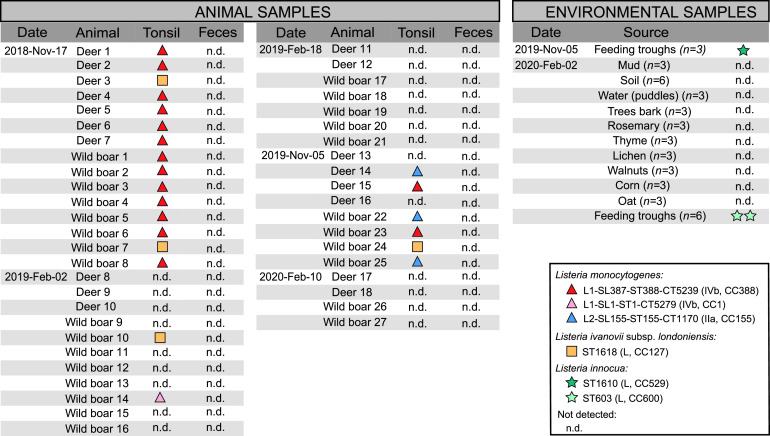
A total of 90 tonsil and feces samples were collected from 45 animals in the same 600-hectare hunting area in the southeast of Spain over a period of 15 months. Two different Listeria spp. were detected in 23 animals (Fig. 1). L. monocytogenes and L. ivanovii were isolated from 19 (42.2%) and 4 (8.8%) out of 45 wild animal tonsils, respectively (Fig. 2). Eight out of 18 (44.4%) deer and 11 out of 27 (40.7%) wild boars were found to be positive for L. monocytogenes in tonsils. One deer (5.5%) and 3 wild boars (11.1%) were positive for L. ivanovii in tonsils. No L. monocytogenes or L. ivanovii could be detected in the feces samples from the same wild animals (50% limit of detection [LOD50] in feces, 0.2 CFU/g; LOD50 in tonsils, 0.4 CFU/g). Accordingly, other studies detected L. monocytogenes in feces in a small percentage of wild boar (1.5%, 1.3%, 6.1%, 0%, and 0%) (8–12) or deer (5.4% and 1%) (11, 12) feces samples. These results suggest that wild animals are frequent carriers of L. monocytogenes and L. ivanovii in tonsils, but shedding in feces occurs rarely, confirming previous results that reported L. monocytogenes in 8.2% (6/73) and 1.4% (1/73) of wild boar tonsils and feces, respectively (9). Similar results have been reported in domestic pigs, where 45% of examined animals (n = 103) harbored L. monocytogenes in the tonsils and only 3% were fecal excretors (29). Experimental oral infection studies in sheep (14) showed that L. monocytogenes can be isolated in large numbers in the tonsils even after fecal excretion is stopped, concluding that tonsils are a long-lasting site of L. monocytogenes colonization. Altogether, these results suggest that animals can silently carry L. monocytogenes in tonsils even without L. monocytogenes fecal shedding.
FIG 1.
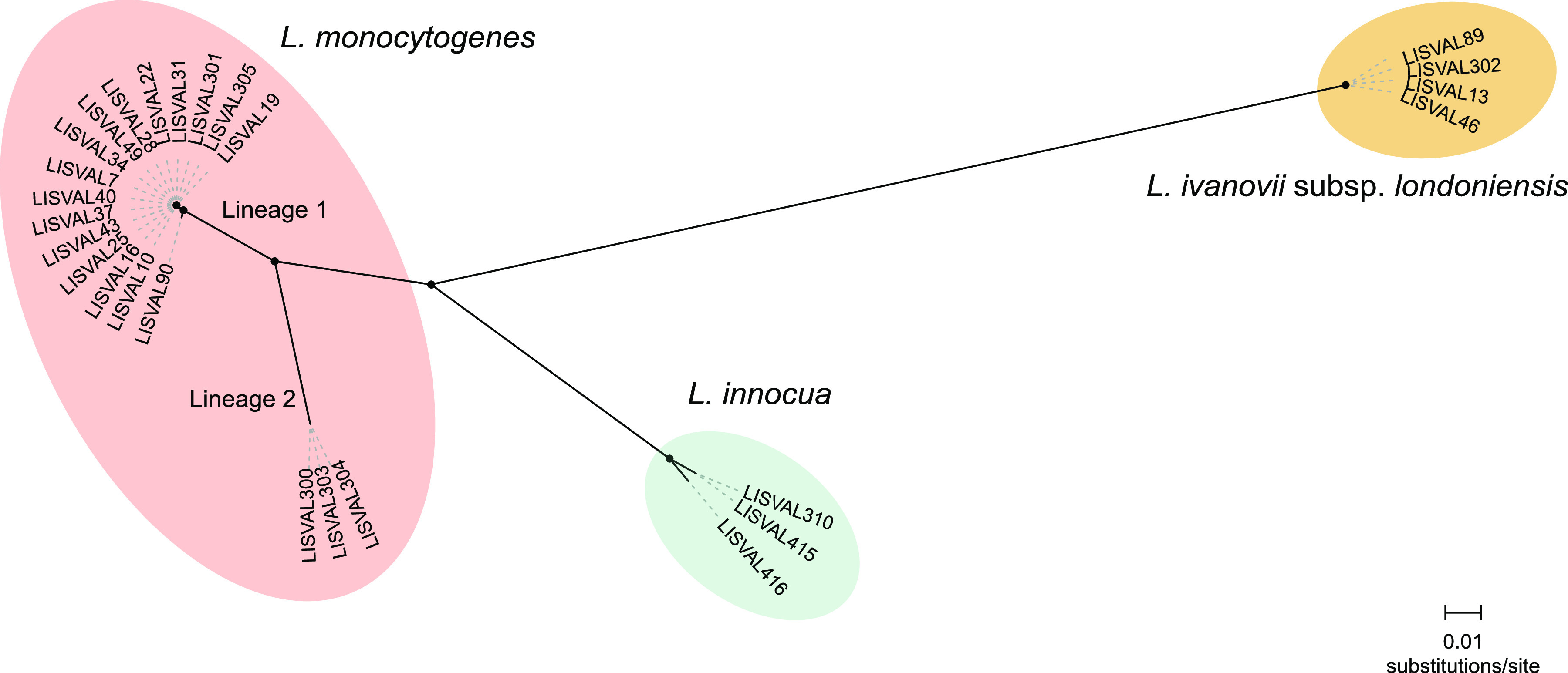
Unrooted maximum-likelihood phylogeny based on the whole-genome alignment (764,193 nucleotide positions) of the Listeria isolates collected in this study (n = 26). Circles at the nodes denote bootstrap support of >90 based on 1,000 replicates.
FIG 2.
Listeria species genotypes detected in this study.
In an attempt to track the ecological source of the Listeria species isolated from the wild animals, 42 environmental samples were further collected from the hunting area of the Mediterranean forest (Fig. 2). These samples included soil and decaying vegetation, since they are considered the primary habitat of Listeria spp. (30, 31), and samples from feeding troughs (including soil, oats, and corn) in the soil. Three L. innocua isolates were obtained from the food troughs in the forest, but no other Listeria spp. could be isolated from the environmental samples analyzed (Fig. 1 and 2).
Genome-based genotyping analysis showed that L. monocytogenes isolates retrieved from the wild animals belonged to phylogenetic lineages I (n = 16; 84.2%) and II (n = 3; 15.8%) and genoserogroups IVb (n = 16; 84.2%) and IIa (n = 3; 15.8%). Importantly, lineage I is often prevalent in clinical cases (25) and has been associated with long-term gut colonization in in vivo studies (32). On the basis of 7-locus MLST, isolates were assigned to ST1 (n = 1), ST388 (n = 15), and ST155 (n = 3), previously reported in Europe. ST388 (IVb; CC388) was reported in 2019 meat industry and retail products in the biggest listeriosis outbreak in Spain (33). ST155 (IIa; CC155) was reported in 2017 from fish samples and an outbreak of invasive listeriosis in Austria (34). ST1 (IVb; CC1) was shown to be hypervirulent and neurotropic (25) and has been reported to be predominant in rhombencephalitis cases in ruminants (35).
At the level of the core genome, isolates belonged to 3 different CTs from sublineages SL1 (n = 1), SL387 (n = 15), and SL155 (n = 3). Isolates within cgMLST type L1-SL387-ST388-CT5239 (n = 15; from wild boars and deer tonsils) were collected 1 year apart in different wild animals from the same geographic area. Isolates differed by only 0 to 2 cgMLST allelic differences (Fig. 3) and 0 to 3 whole-genome single-nucleotide polymorphisms (wgSNPs), confirming its close phylogenetic relationship and highlighting a suspected common contamination source. Isolates within cgMLST type L2-SL155-ST155-CT1170 (n = 3) differed by 0 to 4 cgMLST allelic differences (0 to 5 wgSNPs) and were found in two different wild animal species (deer and wild boars), suggesting contamination from a food or environmental source shared by both species, although interhost transmission cannot be excluded.
FIG 3.
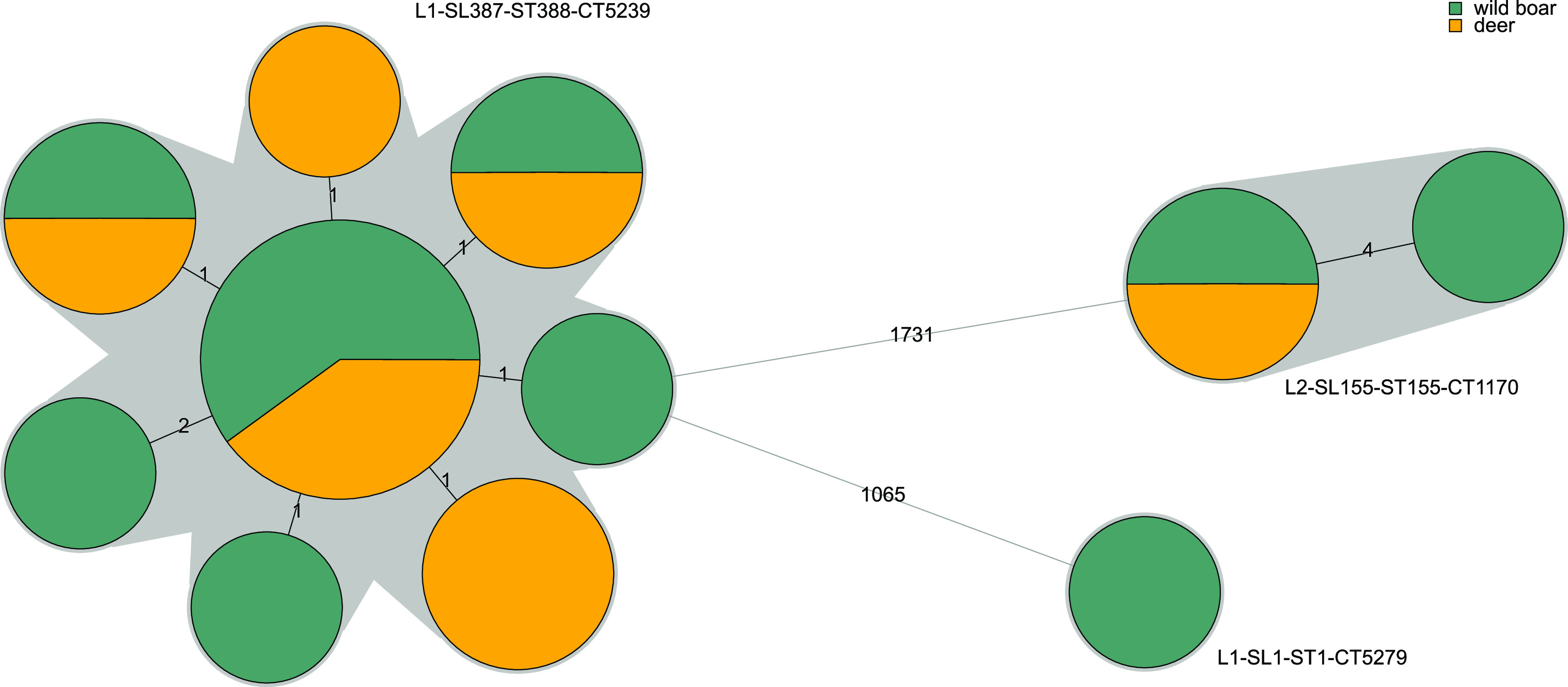
Minimum spanning tree based on the 1,748-locus cgMLST profiles of the L. monocytogenes isolates collected in this study (n = 19). Profiles are represented by colored circles, where size is proportional to the number of isolates and gray zones denote profiles belonging to the same cgMLST type. The number of allelic differences between profiles is indicated on the branches.
Previous studies have shown a low genetic diversity of L. monocytogenes isolated from wild animals (rodents and marine organisms) (36). The successful multiplication of L. monocytogenes in infected animals may favor the selection of invasive clones and their further transmission, as previously suggested (32, 36). Our data are consistent with the hypothesis that wild environments provide opportunities for the selection of invasive L. monocytogenes clones adapted for survival in wild animals and that tonsils constitute a reservoir of L. monocytogenes asymptomatic carriage. These findings are of public health concern, since these animals may favor the transmission of pathogenic Listeria through the food chain to domestic animals and humans.
The L. monocytogenes intracellular survival pathogenicity island LIPI-1 and the inlAB internalization locus were present in all L. monocytogenes isolates (Fig. 4). Consistent with L. monocytogenes phylogeny, isolate L1-SL1-ST1-CT5279 carried LIPI-3. LIPI-3 encodes listeriolysin S, a bacteriocin highly expressed in the intestine of orally infected mice that alters the host intestinal microbiota and promotes intestinal colonization by L. monocytogenes (37–39). Isolates within cgMLST type L1-SL387-ST388-CT5239 encoded LIPI-4, which has been shown to increase L. monocytogenes neural and placental tropism (25).
FIG 4.
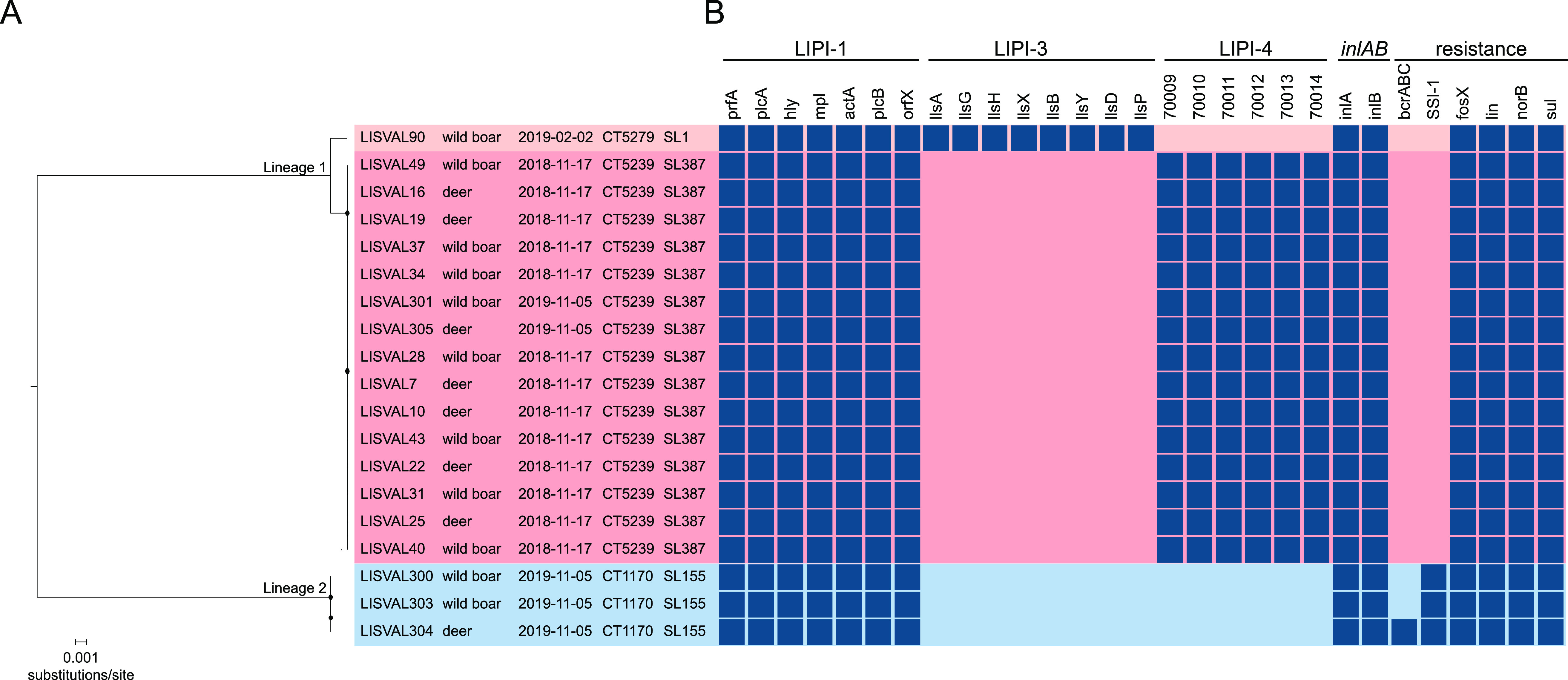
Virulence and resistance gene profiles of the Listeria species isolates sequenced in this study. (A) Midpoint-rooted maximum-likelihood phylogeny based on the whole-genome alignment (2,475,309 nucleotide positions). Circles at the nodes denote bootstrap support of >90 based on 1,000 replicates. Isolate’s name, sample origin, sampling date (yyyy-mm-dd), sublineage (SL), and cgMLST type (CT) are indicated in the tips. (B) Resistance and virulence gene patterns. Color-filled boxes represent the presence of different genetic traits. Empty boxes represent genes with truncations leading to premature stop codons.
Intrinsic antibiotic resistance genes (fosX, Iin, norB, and sul) were present in all L. monocytogenes isolates (Fig. 4). All lineage II L. monocytogenes isolates also carried SSI-1, and one isolate (LISVAL304; L2-SL155-ST155-CT1170) harbored the benzalkonium chloride resistance cassette (bcrABC) (Fig. 4). The SSI-1 stress survival islet has been shown to contribute to L. monocytogenes survival and growth in low pH and high salt concentrations, facilitating the adaptation and persistence in natural and food processing environments (40).
Our results represent one of the first contributions to understanding the genomic diversity and epidemiology of Listeria spp. in wild animals and their natural habitat in the southeast of Spain. By integrating epidemiological data and comparative genomics, we have uncovered the long occurrence of virulent L. monocytogenes strains in wild animals and demonstrate that tonsils are an important reservoir of pathogenic Listeria spp. Wild animals may contaminate crops or the environment where domestic animals graze, which then get contaminated and can pass L. monocytogenes to humans via food. These findings can contribute to guide public health authorities to better control the health hazards associated with L. monocytogenes.
MATERIALS AND METHODS
Animals and sampling.
In the time frame of November 2018 to February 2020, 27 wild boars (Sus scrofa) and 18 deer (Cervus elaphus hispanicus) were hunted for consumption in a 600-hectare hunting area of Mediterranean forest within regulation measures in the southeast of Spain. The postmortem management of hunt-harvested wild boars was conducted according to the European Regulation (EC) no. 853/2004 of the European Parliament and of the Council of 29 April 2004. The animals were hunted by stand hunting. The animals were observed, including for signs of central nervous system disorders, by the hunters before they were shot. Hunted animals were transported from the hunting area to an evisceration room, and samples were taken by veterinarians within 6 h after hunting. Before dissecting the head for tonsil sampling, the skin was disinfected with 70% ethanol. Tonsils were removed before thoracic and abdominal cavities were opened. Tonsils were removed and stored in a sterile bag. When opening the carcasses, no macroscopic abnormalities were found in the thoracic and abdominal cavities, assuming hunted animals were healthy. Fecal specimens from the rectum of all the animals were taken and stored in a sterile bag. To avoid contamination, new and independent sterile scalpels were used for each animal and sample (tonsils and rectum). All samples were collected using disposable gloves by aseptic conditions. The entire sample material was stored at 5 ± 3°C and transported to the laboratory within 24 h.
Additionally, three soil samples from distinct soil types (humus, clay, and sand) were collected from the area where the wild animals live and were hunted. The same types of soil samples (n = 3) were collected from areas enriched with oats and corn grains (spread in the soil to feed animals). Soil samples were taken from the surface to a depth of 5 cm downwards. Additionally, three clay samples from the soil next to the drinking troughs were obtained. Twenty-one vegetation samples were taken, including wild rosemary (n = 3), wild thyme (n = 3), pine tree bark (used by wild animals as scrapers) (n = 3), lichen (n = 3), moldy walnuts (n = 3), corn (from the bag used to feed wild animals) (n = 3), and oat grains (from the bag used to feed wild animals) (n = 3). Moreover, 40 ml of water samples (n = 3) from puddles was taken aseptically in polypropylene tubes. Finally, 9 samples from feeding troughs (including soil, oats, and corn) in the soil (3 different locations, 3 samples per location) were also collected. All samples were collected using disposable gloves. The entire sample material was stored at 5 ± 3°C and transported to the laboratory within 24 h.
Listeria species isolation and identification.
For the isolation of Listeria spp., 8 g of rectal fecal samples, 5 g of tonsils (first minced with a scalpel into approximately 1- to 2-mm pieces and subsequently mechanically disrupted by grinding), or 8 g of environmental samples was diluted 1/10 in half Fraser broth (Scharlab, Spain). Samples were homogenized and incubated for 24 h at 30°C for enrichment. One hundred microliters of the incubated suspension was transferred to 10 ml of Fraser broth (Scharlab, Spain) and incubated at 37°C for 24 h. After the second enrichment, 100 μl of the culture and two 10-fold dilutions in phosphate-buffered saline were transferred to RAPID′L. mono plates (Bio-Rad, USA) and incubated at 37°C for 24 h. Characteristic colonies presumed as Listeria spp. (colonies were blue or white, with or without a yellow halo, round, convex, 1 to 2 mm) were confirmed in selective Oxford agar plates for Listeria (Scharlab, Spain) (colonies were gray-green in color with a black sunken center and a black halo, 1 to 2 mm) and Columbia CNA agar with 5% sheep blood agar plates (colonies were opaque, flat, 1 to 2 mm). From each positive sample, one isolate colony was obtained and preserved in glycerol at −80°C for the following analysis.
Listeria isolates referred to the World Health Organization Collaborating Center for Listeria (Institut Pasteur, Paris) were identified with matrix-assisted laser desorption ionization–time of flight mass spectrometry using the MicroFlex LT system with last MBT library DB-7854 (Bruker Daltonics, Bremen, Germany), as previously described (41).
Genome sequencing and assembly.
Genomic DNA was extracted using the DNeasy blood and tissue extraction kit (Qiagen, Denmark) as described previously (24). DNA quality and concentration were measured by Qubit 3 (Thermo Fisher Scientific, USA). The DNA library was prepared with the Nextera XT DNA library preparation kit (Illumina, USA) according to the manufacturer’s instructions. Paired-end sequencing (2× 150 bp) was performed using NextSeq 550 (Illumina) according to standard Illumina sequencing protocols. Raw reads were trimmed with fqCleaner v.3.0 as previously described (26) and assembled with SPAdes v.3.11 (42) with automatic kmer selection. To confirm species identification, the pairwise genomic average nucleotide BLAST identity was determined by comparing each assembly against the genomes of Listeria species type strains using the enveomics package (43), as previously described (44).
Molecular typing and phylogenetic analysis.
Genoserogrouping (22), MLST (7 loci [23]), cgMLST profiles (1,748 loci [24]), and virulence and resistance profiles (124 loci [24]) were obtained from the assemblies using the BLASTN algorithm (45) implemented at the BIGSdb-Lm platform (24, 46), with minimum nucleotide identity and alignment length coverage of 70% and word size of 10, as previously described (24). MLST profiles were classified into sequence types (STs) and grouped into clonal complexes (CCs) as previously described (23). cgMLST profiles were grouped into cgMLST types (CTs) and sublineages (SLs), using the cutoffs of 7 and 150 allelic mismatches, respectively, as previously described (24). Minimum spanning trees were built from cgMLST profiles using BioNumerics 7.6 software (Applied Maths, Belgium). wgSNP-based alignments were built from trimmed reads with the Snippy v.4.1.0 pipeline (https://github.com/tseemann/snippy), using LISVAL90 (for genus and L. monocytogenes mappings), LISVAL301, or LISVAL300 (for L1-SL387-ST387-CT5239 and L2-SL155-ST155-CT1170 intracluster mappings, respectively) as reference genomes. Maximum likelihood phylogenies were inferred from the whole-genome alignments using IQ-tree v.2.0.6 (47) under the best-fit nucleotide substitution model (GTR+F+I+G) (48) determined by ModelFinder (49) and using an ultrafast bootstrapping of 1,000 replicates (50). Trees were visualized and annotated with iTol v.4.2 (51).
Data availability.
All sequences are publicly available at the European Nucleotide Archive (BioProject no. PRJEB41942) and BIGSdb-Lm (https://bigsdb.pasteur.fr/listeria). The accession numbers are detailed in Table S1 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hélène Bracq-Dieye, Nathalie Tessaud-Rita, Pierre Thouvenot, and Guillaume Vales (Institut Pasteur, Paris, France) for DNA extraction and the P2M platform (Institut Pasteur, Paris, France) for genome sequencing.
This work was supported by Generalitat Valenciana (project reference GV/2018/A/183) and by the Spanish Ministry of Science and Innovation (project reference PID2019-110764RA-I00) (J.J.Q.), Institut Pasteur, Inserm, and Santé Publique France. J. J. Quereda is supported by a “Ramón y Cajal” contract of the Spanish Ministry of Science, Innovation and Universities (RYC-2018-024985-I). C. Palacios-Gorba is supported by a predoctoral contract from the Universidad Cardenal Herrera-CEU. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Charlier C, Perrodeau E, Leclercq A, Cazenave B, Pilmis B, Henry B, Lopes A, Maury MM, Moura A, Goffinet F, Dieye HB, Thouvenot P, Ungeheuer MN, Tourdjman M, Goulet V, de Valk H, Lortholary O, Ravaud P, Lecuit M, MONALISA Study Group . 2017. Clinical features and prognostic factors of listeriosis: the MONALISA national prospective cohort study. Lancet Infect Dis 17:510–519. doi: 10.1016/S1473-3099(16)30521-7. [DOI] [PubMed] [Google Scholar]
- 2.EFSA, ECDC. 2019. The European Union One Health 2018 zoonoses report. EFSA J 17:e05926. doi: 10.2903/j.efsa.2019.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts AJ, Wiedmann M. 2003. Pathogen, host and environmental factors contributing to the pathogenesis of listeriosis. Cell Mol Life Sci 60:904–918. doi: 10.1007/s00018-003-2225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leclercq A. 2018. Listeria monocytogenes. In Manual of diagnostic tests and vaccines for terrestrial animals. World Organization for Animal Health, Paris, France. [Google Scholar]
- 5.Ho AJ, Ivanek R, Grohn YT, Nightingale KK, Wiedmann M. 2007. Listeria monocytogenes fecal shedding in dairy cattle shows high levels of day-to-day variation and includes outbreaks and sporadic cases of shedding of specific L. monocytogenes subtypes. Prev Vet Med 80:287–305. doi: 10.1016/j.prevetmed.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Esteban JI, Oporto B, Aduriz G, Juste RA, Hurtado A. 2009. Faecal shedding and strain diversity of Listeria monocytogenes in healthy ruminants and swine in Northern Spain. BMC Vet Res 5:2. doi: 10.1186/1746-6148-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein H, Stessl B, Brunthaler R, Loncaric I, Weissenbock H, Ruczizka U, Ladinig A, Schwarz L. 2018. Listeriosis in fattening pigs caused by poor quality silage–a case report. BMC Vet Res 14:362. doi: 10.1186/s12917-018-1687-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashidani H, Kanzaki N, Kaneko Y, Okatani AT, Taniguchi T, Kaneko K, Ogawa M. 2002. Occurrence of yersiniosis and listeriosis in wild boars in Japan. J Wildl Dis 38:202–205. doi: 10.7589/0090-3558-38.1.202. [DOI] [PubMed] [Google Scholar]
- 9.Wacheck S, Fredriksson-Ahomaa M, Konig M, Stolle A, Stephan R. 2010. Wild boars as an important reservoir for foodborne pathogens. Foodborne Pathog Dis 7:307–312. doi: 10.1089/fpd.2009.0367. [DOI] [PubMed] [Google Scholar]
- 10.Weindl L, Frank E, Ullrich U, Heurich M, Kleta S, Ellerbroek L, Gareis M. 2016. Listeria monocytogenes in different specimens from healthy red deer and wild boars. Foodborne Pathog Dis 13:391–397. doi: 10.1089/fpd.2015.2061. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida T, Sugimoto T, Sato M, Hirai K. 2000. Incidence of Listeria monocytogenes in wild animals in Japan. J Vet Med Sci 62:673–675. doi: 10.1292/jvms.62.673. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki Y, Goshima T, Mori T, Murakami M, Haruna M, Ito K, Yamada Y. 2013. Prevalence and antimicrobial susceptibility of foodborne bacteria in wild boars (Sus scrofa) and wild deer (Cervus nippon) in Japan. Foodborne Pathog Dis 10:985–991. doi: 10.1089/fpd.2013.1548. [DOI] [PubMed] [Google Scholar]
- 13.Walland J, Lauper J, Frey J, Imhof R, Stephan R, Seuberlich T, Oevermann A. 2015. Listeria monocytogenes infection in ruminants: is there a link to the environment, food and human health? A review. Schweiz Arch Tierheilkd 157:319–328. doi: 10.17236/sat00022. [DOI] [PubMed] [Google Scholar]
- 14.Zundel E, Bernard S. 2006. Listeria monocytogenes translocates throughout the digestive tract in asymptomatic sheep. J Med Microbiol 55:1717–1723. doi: 10.1099/jmm.0.46709-0. [DOI] [PubMed] [Google Scholar]
- 15.Stella S, Tirloni E, Castelli E, Colombo F, Bernardi C. 2018. Microbiological evaluation of carcasses of wild boar hunted in a hill area of northern Italy. J Food Prot 81:1519–1525. doi: 10.4315/0362-028X.JFP-18-077. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira V, Wiedmann M, Teixeira P, Stasiewicz MJ. 2014. Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J Food Prot 77:150–170. doi: 10.4315/0362-028X.JFP-13-150. [DOI] [PubMed] [Google Scholar]
- 17.Keto-Timonen R, Tolvanen R, Lunden J, Korkeala H. 2007. An 8-year surveillance of the diversity and persistence of Listeria monocytogenes in a chilled food processing plant analyzed by amplified fragment length polymorphism. J Food Prot 70:1866–1873. doi: 10.4315/0362-028x-70.8.1866. [DOI] [PubMed] [Google Scholar]
- 18.Carpentier B, Cerf O. 2011. Review–persistence of Listeria monocytogenes in food industry equipment and premises. Int J Food Microbiol 145:1–8. doi: 10.1016/j.ijfoodmicro.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Gan L, Cao X, Wang Y, Wang Y, Jiang H, Lan R, Xu J, Ye C. 2019. Carriage and potential long distance transmission of Listeria monocytogenes by migratory black-headed gulls in Dianchi Lake, Kunming. Emerg Microbes Infect 8:1195–1204. doi: 10.1080/22221751.2019.1647764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White A, Hemmerly T, Martin MP, Knight V. 1959. Studies on the origin of drug-resistant staphylococci in a mental hospital. Am J Med 27:26–39. doi: 10.1016/0002-9343(59)90058-0. [DOI] [PubMed] [Google Scholar]
- 21.Wiedmann M, Bruce JL, Keating C, Johnson AE, McDonough PL, Batt CA. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun 65:2707–2716. doi: 10.1128/IAI.65.7.2707-2716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog 4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, Tarr C, Bjorkman JT, Dallman T, Reimer A, Enouf V, Larsonneur E, Carleton H, Bracq-Dieye H, Katz LS, Jones L, Touchon M, Tourdjman M, Walker M, Stroika S, Cantinelli T, Chenal-Francisque V, Kucerova Z, Rocha EP, Nadon C, Grant K, Nielsen EM, Pot B, Gerner-Smidt P, Lecuit M, Brisse S. 2016. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol 2:16185. doi: 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maury MM, Tsai YH, Charlier C, Touchon M, Chenal-Francisque V, Leclercq A, Criscuolo A, Gaultier C, Roussel S, Brisabois A, Disson O, Rocha EP, Brisse S, Lecuit M. 2016. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet 48:308–313. doi: 10.1038/ng.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moura A, Tourdjman M, Leclercq A, Hamelin E, Laurent E, Fredriksen N, Van Cauteren D, Bracq-Dieye H, Thouvenot P, Vales G, Tessaud-Rita N, Maury MM, Alexandru A, Criscuolo A, Quevillon E, Donguy MP, Enouf V, de Valk H, Brisse S, Lecuit M. 2017. Real-time whole-genome sequencing for surveillance of Listeria monocytogenes. Emerg Infect Dis 23:1462–1470. doi: 10.3201/eid2309.170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Walle I, Bjorkman JT, Cormican M, Dallman T, Mossong J, Moura A, Pietzka A, Ruppitsch W, Takkinen J, European Listeria Wgs Typing Group . 2018. Retrospective validation of whole genome sequencing-enhanced surveillance of listeriosis in Europe, 2010 to 2015. Euro Surveill 23:1700798. doi: 10.2807/1560-7917.ES.2018.23.33.1700798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitman KJ, Bono JL, Clawson ML, Loy JD, Bosilevac JM, Arthur TM, Ondrak JD. 2020. Genomic-based identification of environmental and clinical Listeria monocytogenes strains associated with an abortion outbreak in beef heifers. BMC Vet Res 16:70. doi: 10.1186/s12917-020-2276-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buncic S. 1991. The incidence of Listeria monocytogenes in slaughtered animals, in meat, and in meat products in Yugoslavia. Int J Food Microbiol 12:173–180. doi: 10.1016/0168-1605(91)90067-Y. [DOI] [PubMed] [Google Scholar]
- 30.Vivant AL, Garmyn D, Piveteau P. 2013. Listeria monocytogenes, a down-to-earth pathogen. Front Cell Infect Microbiol 3:87. doi: 10.3389/fcimb.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linke K, Ruckerl I, Brugger K, Karpiskova R, Walland J, Muri-Klinger S, Tichy A, Wagner M, Stessl B. 2014. Reservoirs of Listeria species in three environmental ecosystems. Appl Environ Microbiol 80:5583–5592. doi: 10.1128/AEM.01018-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maury MM, Bracq-Dieye H, Huang L, Vales G, Lavina M, Thouvenot P, Disson O, Leclercq A, Brisse S, Lecuit M. 2019. Hypervirulent Listeria monocytogenes clones' adaption to mammalian gut accounts for their association with dairy products. Nat Commun 10:2488. doi: 10.1038/s41467-019-10380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.WHO. 16 September 2019. Listeriosis–Spain. https://www.who.int/csr/don/16-september-2019-listeriosis-spain/en/.
- 34.Cabal A, Pietzka A, Huhulescu S, Allerberger F, Ruppitsch W, Schmid D. 2019. Isolate-based surveillance of Listeria monocytogenes by whole genome sequencing in Austria. Front Microbiol 10:2282. doi: 10.3389/fmicb.2019.02282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dreyer M, Aguilar-Bultet L, Rupp S, Guldimann C, Stephan R, Schock A, Otter A, Schupbach G, Brisse S, Lecuit M, Frey J, Oevermann A. 2016. Listeria monocytogenes sequence type 1 is predominant in ruminant rhombencephalitis. Sci Rep 6:36419. doi: 10.1038/srep36419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaytseva E, Ermolaeva S, Somov GP. 2007. Low genetic diversity and epidemiological significance of Listeria monocytogenes isolated from wild animals in the far east of Russia. Infect Genet Evol 7:736–742. doi: 10.1016/j.meegid.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 37.Cotter PD, Draper LA, Lawton EM, Daly KM, Groeger DS, Casey PG, Ross RP, Hill C. 2008. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog 4:e1000144. doi: 10.1371/journal.ppat.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quereda JJ, Dussurget O, Nahori MA, Ghozlane A, Volant S, Dillies MA, Regnault B, Kennedy S, Mondot S, Villoing B, Cossart P, Pizarro-Cerda J. 2016. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc Natl Acad Sci U S A 113:5706–5711. doi: 10.1073/pnas.1523899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quereda JJ, Nahori MA, Meza-Torres J, Sachse M, Titos-Jimenez P, Gomez-Laguna J, Dussurget O, Cossart P, Pizarro-Cerda J. 2017. Listeriolysin S is a streptolysin S-like virulence factor that targets exclusively prokaryotic cells in vivo. mBio 8:e00259-17. doi: 10.1128/mBio.00259-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan S, Begley M, Hill C, Gahan CG. 2010. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J Appl Microbiol 109:984–995. doi: 10.1111/j.1365-2672.2010.04726.x. [DOI] [PubMed] [Google Scholar]
- 41.Thouvenot P, Vales G, Bracq-Dieye H, Tessaud-Rita N, Maury MM, Moura A, Lecuit M, Leclercq A. 2018. MALDI-TOF mass spectrometry-based identification of Listeria species in surveillance: a prospective study. J Microbiol Methods 144:29–32. doi: 10.1016/j.mimet.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 42.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez-R L, Konstantinidis K. 2014. Bypassing cultivation to identify bacterial species. Microbe 9:111–118. doi: 10.1128/microbe.9.111.1. [DOI] [Google Scholar]
- 44.Quereda JJ, Leclercq A, Moura A, Vales G, Gomez-Martin A, Garcia-Munoz A, Thouvenot P, Tessaud-Rita N, Bracq-Dieye H, Lecuit M. 2020. Listeria valentina sp. nov., isolated from a water trough and the faeces of healthy sheep. Int J Syst Evol Microbiol 70:5868–5879. doi: 10.1099/ijsem.0.004494. [DOI] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 46.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tavaré S. 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. American Mathematical Society, Providence, RI. [Google Scholar]
- 49.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences are publicly available at the European Nucleotide Archive (BioProject no. PRJEB41942) and BIGSdb-Lm (https://bigsdb.pasteur.fr/listeria). The accession numbers are detailed in Table S1 in the supplemental material.