Abstract
Glioblastoma is a primary brain cancer with a near 100% recurrence rate. Upon recurrence, the tumour is resistant to all conventional therapies, and because of this, 5-year survival is dismal. One of the major drivers of this high recurrence rate is the ability of glioblastoma cells to adapt to complex changes within the tumour microenvironment. To elucidate this adaptation's molecular mechanisms, specifically during temozolomide chemotherapy, we used chromatin immunoprecipitation followed by sequencing and gene expression analysis. We identified a molecular circuit in which the expression of ciliary protein ADP-ribosylation factor-like protein 13B (ARL13B) is epigenetically regulated to promote adaptation to chemotherapy. Immuno-precipitation combined with liquid chromatography-mass spectrometry binding partner analysis revealed that that ARL13B interacts with the purine biosynthetic enzyme inosine-5′-monophosphate dehydrogenase 2 (IMPDH2). Further, radioisotope tracing revealed that this interaction functions as a negative regulator for purine salvaging. Inhibition of the ARL13B-IMPDH2 interaction enhances temozolomide-induced DNA damage by forcing glioblastoma cells to rely on the purine salvage pathway. Targeting the ARLI3B-IMPDH2 circuit can be achieved using the Food and Drug Administration-approved drug, mycophenolate mofetil, which can block IMPDH2 activity and enhance the therapeutic efficacy of temozolomide. Our results suggest and support clinical evaluation of MMF in combination with temozolomide treatment in glioma patients.
Keywords: glioblastoma, chemoresistance, purine biosynthesis, cellular plasticity
Glioblastoma has a near 100% recurrence rate, and upon recurrence the tumour is resistant to conventional therapies. Shireman et al. show that this adaptation is driven in part by an interaction between ciliary protein ARL13B and a purine biosynthetic enzyme. Targeting this interaction could boost therapeutic efficacy.
Introduction
Glioblastoma multiforme (GBM) is a universally lethal primary brain tumour that will take the lives of roughly 20 000 people this year in the USA alone.1 Despite an aggressive standard of care therapy, the median survival of a patient with GBM is just 20 months.2 This unfavourable prognosis is mainly due to the high recurrence rate, as recurrent GBMs are often unresponsive to all avenues of therapy. Although it has become a significant focus of recent research, the true mechanistic underpinnings of resistance to therapies are yet to be elucidated. The heterogeneous nature of GBM tumours combined with the existence of resistant subpopulations, such as cancer stem cells (CSCs), are considered to be significant drivers of the adaptation to therapy.3-5
GBM cells are known for their adaptive or ‘plastic’ nature, which allows for the acquisition of stem cell characteristics (CSC-like state) in response to appropriate microenvironmental cues such as hypoxia or therapeutic stress.6-9 Research has shown that such plasticity is regulated by changes in permissive epigenetic states, which allow cells to use rapid context-dependent regulation of genes that may be necessary for conferring fitness during therapy.10-12 Unfortunately, therapeutically actionable targets whose inhibition would limit CSC adaptation have proven challenging to uncover.
A canonical driver of context-dependent epigenetic plasticity is the polycomb repressor complex (PRC) and its catalytic subunit enhancer of zeste homologue 2 (EZH2). The PRC2/EZH2 complex not only required for different neurodevelopmental processes but also associated with disease pathogenesis, including GBM.13-20 EZH2 impacts global gene expression via deposition of silencing H3K27me3 marks throughout the genome and has been shown to alter STAT3 signalling as well as play a critical role in CSC maintenance.16,19 Tumour cells can also adapt to treatment via rapid alteration of core metabolic pathway activity.21,22 Druggable targets for both of these plasticity pathways are lacking and thus make therapeutic intervention largely impossible.
In this study, we show that resistance to temozolomide (TMZ), as part of the standard of care for newly diagnosed GBM, is associated with EZH2/PRC2-regulated ADP-ribosylation factor-like protein 13B (ARL13B) expression. Further, we demonstrated an interaction between ARL13B and inosine-5′-monophosphate dehydrogenase 2 (IMPDH2), a rate-limiting enzyme of purine biosynthesis, that impacts GBM’s adaptive response to TMZ by inhibiting purine salvaging. Disruption of IMPDH2 activity by using an FDA approved compound, mycophenolate mofetil (MMF), significantly increased the therapeutic efficacy of TMZ. MMF extends the survival of patient-derived xenograft (PDX) models of mice across all GBM subtypes. Our study, therefore, provides evidence of a rapidly clinically translatable opportunity to enhance the efficacy of alkylating agents in GBM.
Materials and methods
Animal studies
Athymic nude mice [NU(NCr)-Foxn1nu; Charles River Laboratory] were maintained according to all Institutional Animal Care and Use Committee guidelines. In compliance with all applicable federal and state statutes governing animal use in biomedical research, the mice were housed before and during the study in a temperature- and humidity-controlled room following a strict 12-h light/dark cycle. For all applicable animal experiments sex differences were kept consistent across groupings with males and females equally represented in 50/50 group splits.
Cell culture
PDX GBM specimens were obtained from Dr C. David James (Northwestern University). These PDX lines were maintained in vivo serially passaging in the flank of immunocompromised nu/nu mice according to previously published protocol.23 For in vitro experiments were performed in Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher Scientific) supplemented with low 1% foetal bovine serum (FBS; Atlanta Biologicals) and 1% Antibiotic Antimycotic Solution (Corning) according to established protocols. These PDX models have been extensively characterized at the molecular level and identified as EGFR amplified (VIII), TERT mutation (C228T), IDH wild-type, MGMT unmethylated and classical subtype for GBM6; for GBM43, this line has p53 mutant, p16 null, TERT mutation (C228T), IDH wild-type, MGMT unmethylated, and it is a proneural subtype; and GBM5 is a mesenchymal subtype with TERT mutation (C228T), IDH wild-type and is MGMT methylated.23,24
For the generation of shRNA knockdown lines, lentivirus particles were made using HEK293 cells (ATCC) transfected with second generation packaging/envelope plasmids (Dr Yasuhiro Ikeda, Mayo Clinic) and shRNA clones (GeneCopoeia). U251 cells were obtained from American Type Cell Culture and maintained for in vitro experiments in DMEM supplemented with 10% FBS and 1% Antibiotic Antimycotic Solution. CRISPR knockout of U251 cells was created by direct transfection with Cas9 nuclease and sgRNA targeting ARL13B (Dharmacon).
Mass spectroscopy
Samples were run on an SDS-PAGE gel, and a gel band was subject to in-gel digestion. The gel band was washed in 100 mM ammonium bicarbonate/acetonitrile (ACN) and reduced with 10 mM dithiothreitol at 50°C for 30 min. Cysteines were alkylated with 100 mM iodoacetamide in the dark for 30 min at room temperature. The gel band was washed in 100 mM ammonium bicarbonate/ACN before adding 600 ng trypsin for overnight incubation at 37°C. The supernatant containing peptides was saved into a new tube. The gel was washed at room temperature for 10 min with gentle shaking in 50% ACN/5% formic acid, and the supernatant was saved to peptide solution. Washing was repeated each by 80% ACN/5% formic acid, and 100% ACN, and all supernatant was saved into a peptide solution then subjected to SpeedVacTM drying. After lyophilization, peptides were reconstituted with 5% ACN/0.1% formic acid in water and injected onto a trap column (150 µm internal diameter × 3 cm in-house packed with ReproSil C18, 3 µm) coupled with an analytical column (75 µm internal diameter × 10.5 cm, PicoChip column packed with ReproSil C18, 3 µm) (New Objectives, Inc). Samples were separated using a linear gradient of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in ACN) over 120 min using a Dionex UltiMateTM 3000 Rapid Separation nanoLC (Thermo Fisher Scientific). Mass spectrometry data were obtained on an Orbitrap Elite Mass Spectrometer (Thermo Fisher Scientific). Data were analysed using Mascot (Matrix Science, Boston, MA) v.2.5.1 against the Swiss-Prot Human database (2019), and results were reported at 1% false discovery rate (FDR) in Scaffold v.4.8.4 (Proteome Software, Portland, OR).
Immunohistochemistry
Slides were placed in an oven at 60°C for 1 h; then paraffin was removed using xylene (3 × 5 min), 100% ethanol (2 × 3 min), and 95% ethanol for 3 min. Slides were then placed into a retrieval solution (pH 6) and incubated in a Biocare Medical Decloaking Chamber set to 110°C for 5 min. After cooling to room temperature, slides were washed three times with PBS and liquid blocked with a marker. Next, 200 µl of peroxidase (covering the tissue) was added and incubated for 10 min before being washed off with PBS. Protein block background (with background sniper from Biocore Medical) was added and incubated for 15 min at room temperature. Slides were rinsed in PBS for 1–2 min. Slides were then blocked for 60 min with goat serum, and the primary antibody was then added according to manufactures recommendations and incubated overnight. After overnight incubation, slides were washed with TBST/Tween solution three times for 1 min. The secondary antibody was added and incubated at room temperature for 35 min after which it was washed off three times with PBS. DAB chromogen (diluted 1:2 in DAB buffer) was added and incubated for 3 min. Haematoxylin counterstain was then applied for 1 min and removed. Slides were then dehydrated using graded alcohol and xylazine (95% ethanol 3 min, 100% ethanol 2 × 3 min, xylene 3 × 5 min), and a coverslip was applied using a xylene-based mounting medium.
Chromatin immunoprecipitation-sequencing and bioinformatics
Cells exposed to either dimethyl sulphoxide (DMSO) or TMZ for the desired amount of time were washed twice with sterile PBS and then exposed to 1% paraformaldehyde (Sigma) for crosslinking. After crosslinking for 15 min, the reaction was stopped with 2.5 M glycine (Sigma), and cells were rocked for an additional 5 min. Cells were then collected by scraping using minimal amounts of PBS to wash cells off the dish. The scrapings were collected, and pellets of cells were flash-frozen and sent to Zymogen Research to undergo chromatin immunoprecipitation (ChIP)-sequencing according to their established protocols. Sequencing data were returned to us and processed bioinformatically after FastQC determined sequencing to be satisfactory. Alignment to the reference genome Hg38 was done using Tophat2, and peak calling were done using MACS2 software with a P-value threshold set to 0.05. Peaks were initially visualized using IGV, and graphical visualizations were made by using a combination of both ChIPSeeker2 and pygenometracks.25
Metabolite isolation and liquid chromatography-mass spectrometry profiling
To determine the relative abundances of intracellular metabolites, extracts were prepared and analysed by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS). Briefly, for targeted steady-state samples, metabolites were extracted on dry ice with 4 ml 80% methanol (−80°C), as described previously.26 Insoluble material was pelleted by centrifugation at 3000g for 5 min, followed by two subsequent extractions of the insoluble pellet with 0.5 ml 80% methanol, with centrifugation at 20 000g for 5 min. The 5-ml metabolite extract from the pooled supernatants was dried down under nitrogen gas using an N-EVAP (Organomation Associates). Next, 50% acetonitrile was added to the dried metabolite pellets for reconstitution. The sample solutions were then centrifuged for 15 min at 20 000g, 4°C. The supernatant was collected for LC-MS analysis.
Hydrophilic metabolite profiling
Samples were analysed by high-performance liquid chromatography and high-resolution mass spectrometry and tandem mass spectrometry (HPLC-MS/MS). Specifically, the system consisted of a Thermo Q ExactiveTM in line with an electrospray source and an Ultimate 3000 (Thermo) series HPLC consisting of a binary pump, degasser, and auto-sampler outfitted with an Xbridge Amide column (Waters; dimensions of 4.6 mm × 100 mm and a 3.5 µm particle size). The mobile phase A contained 95% (vol/vol) water, 5% (vol/vol) ACN, 20 mM ammonium hydroxide, 20 mM ammonium acetate, pH 9.0; B was 100% ACN. The gradient was as follows: 0 min, 15% A; 2.5 min, 30% A; 7 min, 43% A; 16 min, 62% A; 16.1–18 min, 75% A; 18–25 min, 15% A with a flow rate of 400 μl/min. The capillary of the ESI source was set to 275°C, with sheath gas at 45 arbitrary units, auxiliary gas at five arbitrary units, and the spray voltage at 4.0 kV. In positive/negative polarity switching mode, an m/z scan range from 70 to 850 was chosen, and MS1 data were collected at a resolution of 70 000. The automatic gain control (AGC) target was set at 1 × 106, and the maximum injection time was 200 ms. The top five precursor ions were subsequently fragmented, in a data-dependent manner, using the higher energy collisional dissociation (HCD) cell set to 30% normalized collision energy in MS2 at a resolution power of 17 500. Data acquisition and analysis were carried out by Xcalibur 4.1 software and Tracefinder 4.1 software, respectively (both Thermo Fisher Scientific).
In vivo metabolite tracing
Using bioluminescence imaging, it was determined that mice had well-established tumours. Mice were anaesthetized using isoflurane and placed into a tail vein exposure rig (Braintree Scientific). Using butterfly 27-gauge needles, the vein was accessed, and the isotope of choice was administered to the animal. The isotope solution was dissolved into sterile saline, and the infusion was given as an initial bolus of ∼100 µl over 90 s, and a final infusion of ∼250 µl infused at 3 µl/min rate for 2 h using an infusion pump (Braintree Scientific). Tails of the mice were observed during the entire procedure to ensure needle placement within the vein was maintained. Once the infusion was complete, the mice were immediately sacrificed, desired tissue was then excised, and flash frozen until metabolite extraction could take place according to above protocol.
U-14C-glycine, 3H-hypoxanthine, 14C-guanosine and 3H-guanine incorporation into RNA and DNA
Cells (∼80% confluent) were incubated for 15 h, then treated and labelled as indicated in the respective figures. Cells were labelled with 2 μCi of either U-14C-glycine, 3H-hypoxanthine, and 3H-guanine. Cells were harvested, and RNA or DNA was isolated using Allprep DNA/RNA kits according to the manufacturer’s instructions and quantified using a spectrophotometer. Next, 70 μl of eluted DNA or 30 μl of eluted RNA were added to scintillation vials, radioactivity was measured by liquid scintillation counting and then normalized to the total DNA or RNA concentrations, respectively. All conditions were analysed with biological triplicates and representative of at least two independent experiments.
Statistics
All statistics were performed by accompanying analysis software where indicated or by GraphPad Prism software version 8.0. Survival statistics were compared between multiple groups using Bonferroni correction applied to log-rank tests. Generally, t-tests and ANOVAs (one- and two-way) were used to preform analyses and all P-values reported are adjusted for multiple comparisons where appropriate.
Data availability
Raw data were generated at Northwestern university. Derived data supporting the findings of this study are available from the corresponding author on request.
Results
EZH2 expression is increased in the cancer stem cell compartment during chemotherapy
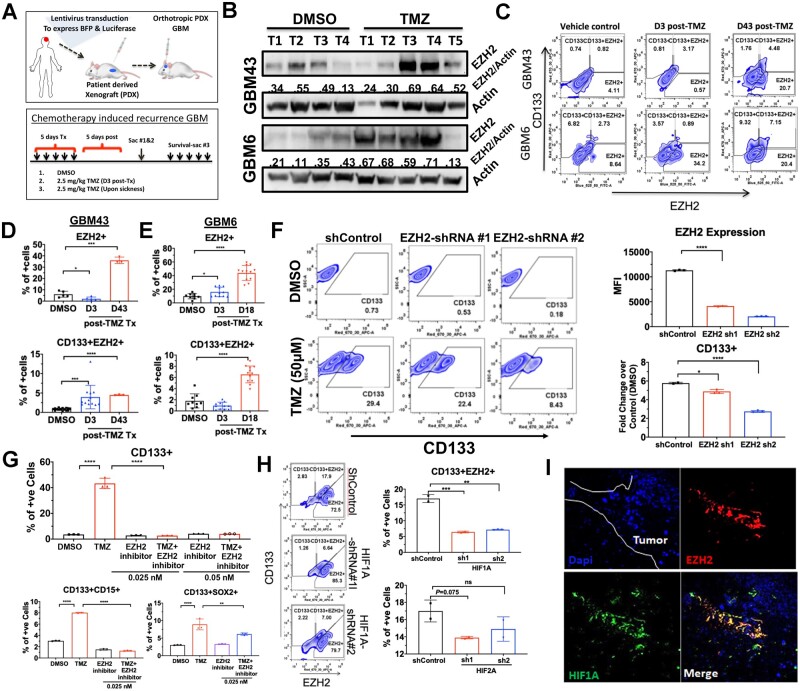
Several reports have demonstrated the significance of PRC2/EZH2 activity in gliomagenesis and the maintenance of the CSC niche.16,27 Furthermore, during normal neuronal development, it is well known that EZH2 aids in allowing state-dependent epigenetic transitions within cell types.13,28 In light of our research demonstrating that chemotherapeutic stress can promote plasticity induced CSC niche in GBM,8,29,30 we now chose to investigate the role of EZH2 in promoting chemotherapy-induced plasticity and chemoresistance in GBM. Fluorescence-activated cell sorting (FACS) analysis revealed that a clinically relevant dose31–33 of TMZ increased the EZH2 expressing cells, the CSC frequency as defined by CD133 expression, as well as CD133-positive CSCs also expressing EZH2 in a time-dependent manner in vitro (Supplementary Fig. 1A and B). Immunoblot analysis of tumour tissues isolated from two different subtypes of PDX tumours confirms elevated EZH2 expression during therapy (Fig. 1B) as did FACS analysis. (Fig. 1C–E; one-way ANOVA adjusted P-values, *P < 0.05, ***P < 0.001). Although the per cent of EZH2-positive (+) cells was increased ∼8-fold in GBM43 PDX and 5-fold in GBM6 post-therapy recurrent GBM (Fig. 1D), we saw the most considerable increase in the CD133+ GBM cells co-expressing EZH2 compartment both during (Day 3; Fig. 1D and E) and post TMZ therapy recurrent GBM (Days 43 and 18; Fig. 1D and E, respectively). A short hairpin (sh)RNA-mediated EZH2 knockdown PDX line shows that in the absence of EZH2, TMZ-mediated increase of the CD133+ CSCs was significantly inhibited (Fig. 1F; one-way ANOVA adjusted P-values shControl versus shRNA#1 P < 0.05, shControl versus shRNA#2 P < 0.001). We have previously demonstrated that post-TMZ CD133+ cells in our PDX model behave like CSC-like cells both in vitro and in vivo.8,21,29,30
Figure 1.
EZH2 expression is increased in the CSC compartment during TMZ-based chemotherapy. (A) Schematic showing establishment of a blue fluorescent protein (BFP) and luciferase expressing PDX line as well as the treatment regimen (left) for establishment of recurrent tumours. PDXs were established for 7 days and treated with TMZ (2.5 mg/kg). Animals were sacrificed and analysed by FACS during therapy (Day 3) and after recurrent (Days 17–18 for GBM6 and Days 43–44 for GBM43). (B) Western blot analysis on tumour tissue isolated from in vivo intracranial PDX tumour treated with DMSO or TMZ during therapy. Numbers are relative pixel intensity of EZH2 bands normalized to actin (n = 4–5). (C) Representative FACS plots for intracranial PDX tumour analysed by FACS with HLA staining for human cells, CSC marker CD133, and EZH2 during therapy (Day 3: D3) and in recurrence (Day 43: D43). (D) Bar graphs displaying per cent of cells staining for either EZH2 alone or EZH2+CD133+ double-positive cells in DMSO (vehicle control) and post TMZ (2.5 mg/kg) at Days 3 and 43 for GBM43 (n = 3). (E) Same as before but post TMZ recurrent at Day 17 for GBM6 (n = 3). (F) Representative FACS plots from GBM6 transduced with lenti-vectors carrying scramble control (shControl) and two different shRNAs for EZH2. Top bar graph represents the EZH2 knockdown efficiency analysed by FACS and represented by mean florescent intensity. Bottom bar graph representing per cent of cells residing in CD133+ gate quantified by fold change over DMSO condition when treated with EZH2 shRNAs. (G) Bar graph representing FACS data displaying per cent of cells showing CD133+, CD133 and CD15+, or CD133 and SOX2+ double-positive, GBM43 PDX cells treated with DMSO, TMZ (2.5 mg/kg), EZH2 inhibitor 3-DZnep (0.05 nM), or combination in vivo flank model as EZH2 inhibitor did not cross the blood–brain barrier. (H) Representative FACS analysis plots and bar graph displaying CD133 and EZH2 staining across shControl and HIF1A or HIF2A knockdown with two shRNAs in GBM43. Numbers represent per cent of cells residing within specific gate. Bar graphs represent FACS data showing per cent of cells expressing both CD133 and EZH2 across the shRNA treatment with either HIF1A or HIF2A knockdown using multiple shRNA. (I) Representative images from orthotopic PDX tumour GBM43 stained for HIF1A and EZH2. All error bars in graphs depict three technical FACS replicates for each animal, each analysis is validated in at least three different animals and represent mean ± standard deviation (SD). *P < 0.05 ***P < 0.001.
To validate this finding in vivo, we used 3-deazaneplanocin (DZNep), an adenosylhomocysteine inhibitor previously reported to inhibit EZH2.16 In the subcutaneous PDX model, DZNep significantly reduced post-therapy CD133+ cells, the double-positive CD15+CD133+ cells (Fig. 1G; two-way ANOVA adjusted P-values CD133+ DMSO versus TMZ P < 0.001, CD133+CD15+ DMSO versus TMZ P < 0.001; Supplementary Fig. 1C), as well as CD133+ cells expressing the CSC-specific transcription factor, SOX2 (Fig. 1G; two-way ANOVA adjusted P-values CD133+SOX2+ TMZ versus EZH2 inhibitor P < 0.001, TMZ versus TMZ+EZH2 inhibitor P < 0.001).
We have previously shown that TMZ treatment-associated increases in CSCs are HIF dependent.20,29 Now we show that the TMZ increases HIF1A in the CD133+ CSC compartment (Supplementary Fig. 1D; ***P < 0.001). HIF knockdown cells were developed to investigate this relationship (Supplementary Fig. 2A; **P < 0.01, ***P < 0.001). Knocking down HIF1A, but not HIF2A, resulted in a significant reduction in the TMZ-associated CSC expansion as well as stemness (Fig. 1H; shControl_HIF1 versus sh1 P < 0.001; shControl_HIF2 versus sh3 P = not significant; Supplementary Fig. 1D). Moreover, HIF1A expression was positively correlated with EZH2 expression in patient GBM samples (Supplementary Fig. 2B; GlioVis34 CGGC Dataset Pearsons correlation r = 0.42, Rembrandt Dataset r = 0.52). Analysis of the Anatomical Structure RNA-Seq database from the Ivy Glioblastoma Atlas Project revealed regional gene expression patterns of HIF1 and EZH2 in the patients’ GBM tissue were also similar, with expression lowest in the leading edge of the tumour and significantly higher in the perinecrotic zone and pseudopalisading cells when compared (Supplementary Fig. 2C; Tukey’s pairwise comparisons). Finally, immunofluorescent analysis of mouse brain with PDX revealed co-localization of HIF1A and EZH2 expression (Fig. 1I). From this, we conclude that EZH2 is essential for TMZ-induced expansion of brain tumour-initiating cells, and HIF1A may play a regulatory role.
ARL13B is a downstream target of EZH2 and its expression negatively correlates with GBM prognosis
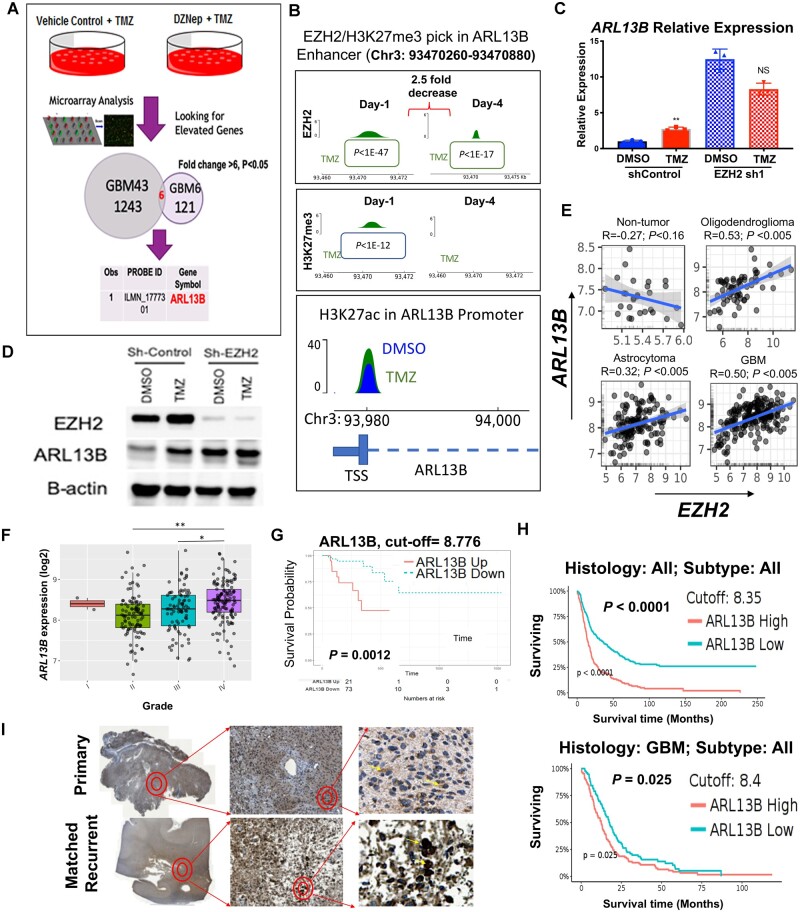
We postulate the PRC2/EZH2 complex alters the expression of genes necessary for TMZ-induced cellular plasticity by modulating the chromatin landscape, ultimately driving therapeutic adaptation. To identify potential targets of PRC2/EZH2, we used gene expression analysis with two subtypes of PDX treated with TMZ in the presence or absence of EZH2 inhibitor. The top five EZH2 targeted genes common between the two subtypes of PDX were further evaluated for their clinical significance by using the CCGA data and GlioVis visualization tools34 (Supplementary Fig. 3). The most highly enriched target was ARL13B, as its expression was altered over 6-fold in the presence of DZNep and TMZ as opposed to TMZ treatment alone (Fig. 2A;P < 0.05). ARL13B is an ADP-ribosylation factor family small GTPase primarily described as a ciliary protein; however, its role in gliomagenesis is understudied.
Figure 2.
Ciliary protein ARL13B is a downstream target of EZH2 and its expression correlates negatively with GBM patient prognosis. (A) Schematic diagram of experimental design where GBM43 and GBM6 were treated with either vehicle control or DZNep (0.05 nM) in combination with TMZ (50 µM) for 48 h. Gene expression analysis between the different subtypes of GBM identified ARL13B as one of the top genes for which expression was altered in the absence of EZH2 activity (fold change >6, P < 0.05). (B) Genome-wide ChIP-Seq analysis was performed on GBM43 cells at Days 1 and 4 after TMZ (50 µM) for enrichment of EZH2, H3K27me3 and H3K27ac marks. P-values from Macs2 software called over input control. (C) Quantitative PCR analysis demonstrating that ARL13B expression increases after 4 days of TMZ treatment as compared to vehicle control. Error bars represent mean ± SD of three technical qPCR replicates. Each experiment repeated at least twice. (D) Immunoblot analysis corroborating qPCR data from C at the protein level at 4 days post TMZ treatment. (E) TCGA data demonstrating significant positive correlation between EZH2 expression and ARL13B expression in oligodendroglioma, astrocytoma, and GBM but not in normal brain tissue. (F) TCGA data showing mRNA expression of ARL13B increasing significantly with tumour grade; (G) probability of survival decreasing with higher expression of ARL13B; and (H) time to median survival significantly decreasing with high expression of ARL13B in all brain tumour (top) and only in GBM (bottom). Data were gathered using optimal cut-off. (I) GBM patient tissue samples stained for ARL13B in matched pairs of primary and recurrent tumours demonstrating increased nuclear staining for ARL13B upon recurrence (n = 10). *P < 0.05, **P < 0.01.
Given EZH2’s global regulatory role, we carried out a genome-wide ChIP-seq analysis for EZH2 and H3K27me3 marks during therapy (Supplementary Fig. 4A). These data indicate a possible interaction with the Stat3 pathway, an established oncogenic driver of gliomagenesis (Supplementary Fig. 4B–D).35 As the EZH2-Stat3 axis has been previously reported to be an important contributor to gliomagenesis,19,36 this observation serves as validation for our screen. The screen also indicated EZH2 could directly bind to one of the established ARL13B enhancer regions (Ch3:93470260-93470889)37 and this interaction was significantly decreased 4 days post-TMZ therapy as compared to control (Fig. 2B; 2.5-fold decrease in expression Day 1: P < 1 × 10−47, Day 4: P < 1 × 10−17). The presence of H3K27me3, EZH2/PRC2’s canonical silencing indicator, correlated both spatially and temporally with the binding of EZH2 within the ARL13B enhancer. These binding release dynamics correlated with an increase in H3K27ac at the transcription start site of ARL13B 4 days post-TMZ therapy (Fig. 2B, bottom and Supplementary Fig. 4E; 2.1-fold enrichment TMZ/DMSO P < 2.1 × 10−22, q < 2.6 × 10−18), which resulted in increased ARL13B at both the transcript (Fig. 2C; one-way ANOVA adjusted P = 0.0069, n = 3 replicates) and protein levels during TMZ therapy (Fig. 2D and Supplementary Fig. 4F). These data indicated that EZH2 could directly bind to the ARL13B enhancer region, change the surrounding chromatin landscape, and negatively regulate its expression, as the shRNA knockdown of EZH2 also elevated the levels of ARL13B mRNA and protein [Fig. 2C, D and Supplementary Fig. 4B; one-way ANOVA adjusted P = 0.0121 (DMSO), adjusted P = 0.0288 (TMZ)]. This binding is decreased after 4 days post-TMZ therapy, allowing GBM cells to induce ARL13B expression in a controlled manner (Fig. 2B–D).
To investigate the clinical significance of ARL13B in gliomagenesis, we next examined the relationship between EZH2 and ARL13B in the context of GBM using GlioVis visualization tools and show no correlation in non-tumour tissues, but a positive correlation between ARL13B and EZH2 expression in GBM, astrocytoma, and oligodendroglioma (Fig. 2E; R = 0.5, P < 0.005; R = 0.32, P < 0.005; R = 0.53, P < 0.005). Moreover, ARL13B expression increased concurrently with tumour grade (Fig. 2F; TCGA Rembrandt, ANOVA test grade II versus IV adjusted P < 0.0001; grade III versus IV adjusted P < 0.0001), and its elevated expression is negatively correlated with overall survival (Fig. 2G; right, log-rank cut-off = 8.776 P = 0.0012). Importantly, high ARL13B expression was associated with accelerated relapse and recurrence (Fig. 2G and H; log-rank cut-off 8.35 P < 0.0001, cut-off 8.4 P < 0.025). Finally, we profiled 10 matched primary and recurrent GBM cases from the Northwestern Nervous System Tumor Bank and compared ARL13B expression via immunohistochemistry. Our board-certified neuropathologist analysis revealed that ARL13B is well expressed in primary and recurrent tumours, but a nuclear localization shift seemed to occur in the matched recurrent slides (Fig. 2I and Supplementary Fig. 5C).
ARL13B regulates cellular plasticity and contributes to tumour engraftment in vivo
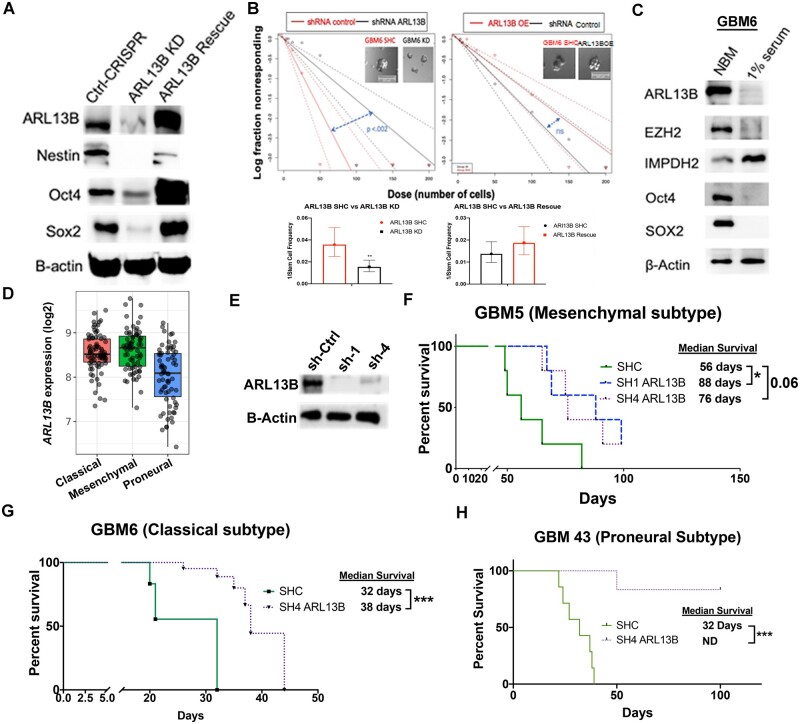
As data indicated that during TMZ therapy, EZH2 promotes plasticity and regulates ARL13B expression, we next investigated if ARL13B itself can be involved in promoting plasticity in GBM. Knocking down ARL13B significantly reduced the expression of several stemness transcription factors such as SOX2 and Oct4 (Fig. 3A). Such knockdown also reduced stemness in GBM cell measured by extreme limiting dilution assay (ELDA) (CSC frequency for control 1/28, ARL13B knockdown 1/65.1, P < 0.002; Fig. 3B, left and Supplementary Fig. 6B). The loss of stemness can be rescued by re-expressing ARL13B in the knockdown cells (CSC frequency for knockdown 1/72.8, rescue 1/53.6 P > 0.05; Fig. 3B, right and Supplementary Fig. 6C). To examine if the cell fate state influenced the expression of ARL13B, we cultured PDX lines in stem cell media (Neurobasal media with 10 ng/ml EGF and FGF) or differentiation condition media containing 1% foetal bovine serum.29,38 Immunoblot analysis indicated that when PDX lines (GBM5, GBM6, GBM43) cultured in Neurobasal media, the expression of ARL13B as well as stemness transcription factors Sox2 were significantly elevated in three different subtypes of GBM (Fig. 3C and Supplementary Fig. 6). However, we observed some subtype-specific differences. For example, Oct4 expression was elevated in the Neurobasal media condition for GBM6 and GBM43 but downregulated in GBM5.
Figure 3.
ARL13B regulates cellular plasticity and is required for tumour engraftment in PDX models. (A) Western blot analysis of canonical stem cell markers in U251 GBM cells with wild-type expression, CRISPR-mediated deletion, and lentiviral-mediated rescue of ARL13B. (B) Extreme limiting dilution analysis assay on cells with CRISPR knockdown (KD) of ARL13B or lentiviral rescue of ARL13B compared to non-targeted guide RNA. Error bars demonstrate 95% confidence interval (CI) of stem cell frequency P-value determined from 12-well replicates of plate counting in provided software. (C) Immunoblot analysis showing differential expression of ARL13B, EZH2 an IMPDH2 as well as canonical stem cell markers SOX2 and Oct4 in the neurobasal (10 ng/ml EGF, 10 ng/ml FGF) or differentiation condition media (1% FBS). (D) TCGA data demonstrating mRNA expression of ARL13B across GBM subtypes. (E) Immunoblot analysis showing shRNA mediated knockdown efficiency of ARL13B in GBM5 PDX line. (F–H) In vivo engraftment analysis after removal of ARL13B using shRNA from mesenchymal (F, GBM5), classical (G, GBM6) and proneural (H, GBM43) subtypes of PDX GBM (n = 6 mice per group, 50% male and female mice). *P < 0.05, **P < 0.01, ***P < 0.005.
In GBM patient samples, ARL13B mRNA expression was similar in the classical and mesenchymal subtype. However, the proneural subtype expression was significantly lower (Fig. 3D; GlioVis; one-way ANOVA adjusted P < 0.0001). To investigate the role of ARL13B in GBM growth in vivo, the shRNA approach was employed to knock down its expression in all three subtypes of PDX GBM (Fig. 3E). Knocking down of ARL13B resulted in a 57% improvement of median survival in the mesenchymal subtype [Fig. 3F; log-rank adjusted for multiple comparisons control (shC) versus sh1 P = 0.02, shC versus sh4 P = 0.06]; a 72% improvement of median survival in classical subtype GBM6 (Fig. 3G; log-rank P-adjusted = 0.0002); and completely blocked tumour formation in proneural GBM43 subtype (Fig. 3H; log-rank P-adjusted = 0.0004). Taken together, these data indicate that ARL13B is a downstream target of EZH2/PRC2 regulation that may contribute to disease progression in patients with GBM.
Knockdown of ARL13B sensitizes GBM cells to temozolomide
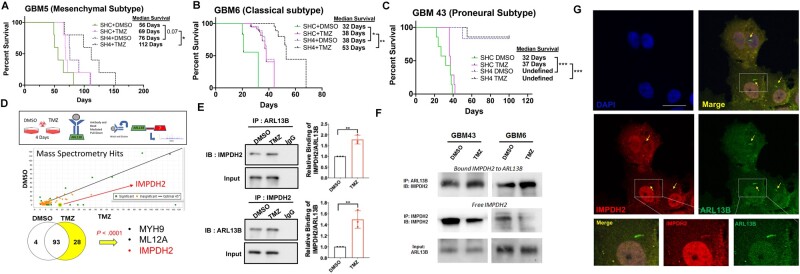
Our initial experiments identified ARL13B as a downstream target of EZH2 during TMZ therapy; therefore, we next sought to assay if the removal of ARL13B sensitized GBM to TMZ therapy in vivo. We established PDX lines (GBM5, GBM6, GBM43) with shRNA-mediated ARL13B knockdown and intracranially injected them into nude mice. Following tumour engraftment (7 days post-implantation), animals were treated with a 2.5 mg/kg dose of TMZ.39 Removal of ARL13B significantly sensitized mesenchymal PDX GBM5 with ∼25% increase in median survival (Fig. 4A; shC+DMSO versus sh4+DMSO adjusted log-rank P = 0.07; shC+TMZ versus sh4+TMZ adjusted log-rank P = 0.05; sh4+DMSO versus sh4+TMZ adjusted log-rank P = 0.102), as well as classical PDX GBM6, ∼21% increase in median survival, demonstrating a role for ARL13B in response to TMZ therapy (Fig. 4B; shC+DMSO versus sh4+DMSO adjusted log-rank P = 0.025; shC+TMZ versus sh4+TMZ adjusted log-rank P = 0.004; sh4+DMSO versus sh4+TMZ adjusted log-rank P = 0.0021). Knocking down nearly abolished the engraftment capacity of the proneural subtype GBM43 (Fig. 4C; shC+DMSO versus sh4+DMSO adjusted log-rank P = 0.0004, shC+TMZ versus sh4+TMZ adjusted log-rank P = 0.0003), preventing an assessment of the effect of TMZ. Taken together, we conclude that ARL13B contributes to gliomagenesis by modulating stemness and engraftment capacity.
Figure 4.
Loss of ARL13B increases in vivo sensitivity to TMZ and ARL13B interacts with IMPDH2 during TMZ therapy. (A–C) Kaplan-Meier curves for end point survival analysis evaluating the role of ARL13B in TMZ sensitivity. ShRNA-mediated knockdown of PDX lines (A) GBM5, (B) GBM6, and (C) GBM43. PDXs are engrafted intracranially and treated with TMZ (2.5 mg/kg) for five consecutive days beginning 7 days after tumour implantation (n = 6/group, 50% male and 50% female mice) and mice are evaluated for end point survival. (D) Schematic representing experimental flow of antibody pulldown mass spectroscopy analysis. P-values determined by Scaffold proteomics software. (E) Immunoprecipitation (IP) using anti-IMPDH2 or anti-ARL13B antibodies with an IgG control during DMSO or TMZ therapy to evaluate ARL13B IMPDH2 interaction in GBM6. Error bars represent densitometry quantification from biological triplicate immunoprecipitations. (F) Immunofluorescent co-staining with antibodies against IMPDH2 and ARL13B in the GBM6 line demonstrating the co-localization in the ciliary structure. (G) Representative immunoprecipitation experiment demonstrating the levels of free and bound IMPDH2 protein during TMZ therapy across GBM6 and 43. Samples were serially immunoprecipitated as described in the ‘Materials and methods’ section to assay free and bound forms of the protein.
IMPDH2 interacts with ARL13B during temozolomide therapy
ARL13B is known to localize around primary cilia, a sensory organelle found on eukaryotic cells known to function as a hub for cellular signalling, including sonic hedgehog signalling (SHH).40,41 SHH signalling is known to contribute to various human malignancies, including the childhood brain tumour medulloblastoma.42–44 In PDX GBM, the length of ARL13B-positive cilia, as well as the number of ciliated GBM cells, increased significantly after TMZ exposure (Supplementary Fig. 7A–D). However, total SHH expression, as well as the suppressor of SHH expression SUFU, remained unchanged (Supplementary Fig. 7E). The loss of ARL13B reduced the cilia formation in GBM (Supplementary Fig. 8A). However, the elimination of cilia by pharmacological inhibitor ciliobrevin45 did not yield a decrease in cancer cell proliferation in combination with TMZ (Supplementary Fig. 8B). These data indicated to us a possible alternative mechanism of ARL13B-mediated TMZ resistance in GBM independent of SHH.
To examine other potential mechanisms, we performed immunoprecipitation using the ARL13B antibody, followed by LC-MS (Supplementary Table 1). We analysed these genes based on their involvement in gliomagenesis using the TCGA and CCGA database available in GlioVis. This analysis identified IMPDH2, a key rate-limiting enzyme for purine biosynthesis,46,47 as a novel interacting partner of ARL13B (Fig. 4D;P < 0.0001 calculated by Scaffold Viewer software, n = 2 replicates). Time course analysis of post-therapy expression indicated subtype-specific increases is IMPDH2, EZH2, and ARL13B during therapy with pronounced increases at Day 8 in GBM43 and GBM6 (Supplementary Fig. 9A and B).
The IMPDH2-ARL13B interaction is significantly augmented during TMZ therapy in GBM43 (Fig. 4E;n = 3 biological replicates ARL13B immunoprecipitation: P = 0.0028, IMPDH2 immunoprecipitation: P = 0.0059). The interaction was further validated in different breast cancer cells (Supplementary Fig. 9C) as well as in PDX lines (Supplementary Fig. 9D). Immunofluorescent analysis in PDX lines revealed the co-localization of both proteins in the cytoplasm and around primary cilia (Fig. 4F). Finally, sequential immunoprecipitation was performed to examine the dynamics of this interaction and observed that within 24 h after TMZ exposure, the free IMPDH2 level was reduced as the ARL13B-IMPDH2 interaction is increased (Fig. 4G). Here, we identified a novel interaction between ARL13B and IMPDH2 and demonstrated that TMZ could play a role in influencing this interaction.
ARL13B and IMPDH2 interaction regulates purine synthesis through the consumption of hypoxanthine
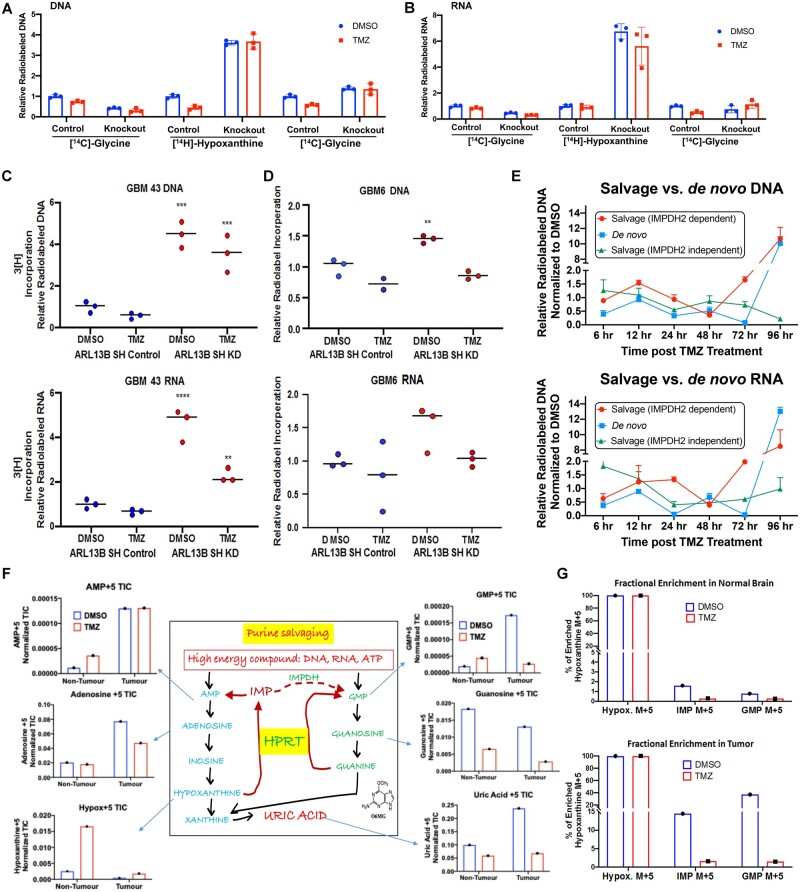
Purines can be built in the cell using one of two pathways: (i) the energy-demanding de novo pathway where the purine is generated within the cell from a ribose ring48; and (ii) by a more energy-efficient salvage pathway where purines can be recycled from the microenvironment.49 All cells can perform both types of synthesis; however, the brain typically utilizes the energy-efficient salvage pathway over the energetically taxing de novo pathway.50 IMPDH2 is the rate-limiting enzyme for IMP-XMP-GMP conversion, essential for both de novo and salvage pathways. As the IMPDH2-ARL13B interaction has never been reported in the literature, we hypothesized that this binding event might influence the ability of GBM cells to regulate purine biosynthesis. Our initial ChIP-seq data revealed significant changes among common purine biosynthetic enzymes in both relative expressions, as well as the transcription activation mark H3K27ac at the corresponding transcription start site after 4 days of TMZ therapy (Supplementary Fig. 10). To investigate the role of the IMPDH2-ARL13B interaction, we carried out radiolabel tracing in nucleic acids using 14C-glycine to measure the de novo purine pathway: 3H-hypoxanthine to provide a direct measurement of de novo GMP synthesis through IMPDH; and 14C-guanosine, which is directly converted to GMP independently of IMPDH activity via the purine salvage enzyme hypoxanthine guanine phosphoribosyltransferase (HGPRT). Measurement of radiolabelled isotope incorporation within DNA and RNA indicated that TMZ decreased the amount of salvage activity (3H-hypoxanthine incorporation) while having no significant effect on de novo synthesis (14C-glycine) in DNA (Fig. 5A; two-way ANOVA Tukey adjusted P = 0.003). However, in cells lacking ARL13B, there was a 4–6-fold increase in salvage activity (3H-hypoxanthine incorporation), which was unaffected by TMZ treatment. Knockdown of ARL13B also significantly reduced the amount of de novo synthesis (14C-glycine incorporation; Fig. 5A and B; DNA Salvage Tukey adjusted P < 0.001, RNA Salvage Tukey adjusted P < 0.0001, DNA de novo Tukey adjusted P = 0.002). Conversely, IMPDH2-independent salvage (14C-guanosine incorporation) was increased in response to ARL13B knockdown, although to a much lesser extent than IMPDH2-dependent salvage (3H-hypoxanthine) (Fig. 5A and B; DNA Tukey adjusted P < 0.0001, RNA Tukey adjusted P = 0.6). These results indicated two possibilities: (i) ARL13B may function as a negative regulator of salvage purine biosynthesis, and removing it increased purine salvaging; and (ii) that ARL13B impairs the ability of GBM cells to use de novo synthesis, instead of forcing salvage pathway utilization. These observations were next validated in PDX lines GBM43 and GBM6 using shRNA-based knockdown of ARL13B, and we observed similar results (Fig. 5C and D; one-way adjusted ANOVA GBM43 DNA P = 0.0003, P = 0.0008, GBM43 RNA P > 0.0001, P = 0.006, GBM6 DNA P = 0.004).
Figure 5.
ARL13B and IMPDH2 interaction functions as a negative regulator of purine salvaging and TMZ therapy alters purine biosynthesis pathways. (A) Radiolabelled tracing of the three major biosynthetic pathways measured by the corresponding radiolabelled metabolite incorporation at the DNA level with or without ARL13B, in the absence and presence of TMZ (50 µM for 48 h) in U251 cells where ARL13B is knocked out with CRISPR-Cas9. Graph depicts relative incorporation of specific radiolabel normalized to control DMSO condition. (B) Same experiments as described before but measuring incorporation in RNA. (C) Radiolabelled tracing of ARL13B knockdown GBM43 PDX treated with TMZ (50 µM) for 48 h. Graph depicts relative 3H incorporation in the DNA across cells with and without ARL13B during TMZ therapy in GBM43. (D) Radiolabelled tracing of ARL13B knockdown GBM6 PDX as described before. (E) Radiolabelled tracing of different purine biosynthesis pathways after 6, 12, 24, 48, 72 and 96 h post TMZ (50 µM) exposure in GBM43. Data presented as normalized to DMSO incorporation from matched time point. (F) Mice bearing GBM43 PDX tumours were infused with labelled hypoxanthine (13C5) for 2 h through systemic tail vein injection. Tissues were collected from intracranial tumour, contralateral normal brain, and subject to analysis by LC-MS. Bar graphs represent in vivo tracing of metabolites from mice bearing a GBM43 PDX tumour. All counts are represented as normalized total ion counts (TIC) and were isolated from tumour or non-tumour tissue ± TMZ. (G) Fractional enrichment for hypoxanthine, IMP, and GMP represented as a bar graphs measured from samples of normal brain tissue. Data measured in per cent enrichment of the mass (M) +5 due to detected m/z shift because of five C13’s in labelled hypoxanthine used. (H) Fractional enrichment in tumour tissue for the experiment described above. Enrichment was calculated from protein-normalized total ion counts and normalized to 13C hypoxanthine incorporation. All error bars in experiments represent technical replicates and display mean ± SD. **P < 0.01 ***P< 0.001 ****P < 0.0001.
We next examined how purine biosynthesis was altered in response to a physiological dose of TMZ over time. We observed a higher utilization of salvage synthesis (3H-hypoxanthine incorporation) carrying up to 48 h where de novo synthesis (14C-glycine incorporation) briefly outpaced it.31,32,51 After 48 h, both IMPDH2-dependent de novo synthesis and IMPDH2-dependent salvage synthesis rebounded and even drastically increased ∼10-fold by 96 h. However, IMPDH2-independent salvage synthesis (14C-guanosine incorporation) remained relatively low [Fig. 5E two-way ANOVA adjusted for multiple comparisons: 96-h salvage (IMPDH2 independent) versus salvage (IMPDH2 dependent) P < 0.0001, 96-h salvage (IMPDH2 independent) versus de novo P < 0.0001].
Temozolomide downregulates purine salvage in vivo
Our data strongly suggested that TMZ is altering the purine synthesis pathways used in glioma. To analyse in vivo effects, we used heavy isotope-labelled hypoxanthine (13C5) to quantify the purine biosynthesis flux through the IMPDH2-dependent salvage pathway. Mice bearing GBM43 PDX tumours were infused with labelled hypoxanthine for 2 h through systemic tail vein infusion. LC-MS examination was conducted post-infusion for isotope incorporation in tumour tissue, contralateral normal brain, and liver tissue (Supplementary Fig. 11A and B). Results demonstrated that the tumour region contained a higher level of labelled AMP, adenosine, and uric acid as compared to cells from the non-tumour area (Fig. 5F). Furthermore, mice harbouring subcutaneous GBM43 demonstrated increased metabolic fractional enrichment when compared back to normal brain and tumour brain within the same mouse, indicating that the microenvironment plays a role in modulating purine biosynthesis (Supplementary Fig. 11C).
Interestingly, the levels of labelled hypoxanthine, the first metabolite injected, were much lower within the tumour when compared to the non-tumour brain, possibly because of rapid utilization. GMP levels, which rely on IMPDH2-mediated conversion of IMP to XMP, are increased in the tumour compared to the normal brain (Fig. 5F). We next carried out a similar experiment in vitro using ARL13B knockdown cells and steady-state metabolomics, which also demonstrated an increase in salvage pathway metabolites detected once ARL13B was lost (Supplementary Fig. 12).
Remarkably, mice that received TMZ therapy showed decreased levels of GMP as compared to normal brain. Fractional enrichment analysis revealed that both IMP and GMP levels were higher in the tumour tissue as compared to the normal brain but that TMZ therapy decreased IMP and GMP levels within the tumour significantly compared to the normal brain (Fig. 5G). Analysis of purine transporter expression revealed similar equilibrative nucleotide transporter (ENT) family levels with or without ARL13B knockdown indicating that ARL13B does not play a role in altering the cellular nucleotide transporter expression level, but rather impacts de novo and salvage pathway regulation directly (Supplementary Fig. 13A).
ARL13B knockdown increases salvage pathway mediated recycling of damaged environmental purines
Based on our in vitro and in vivo data, we propose that GBM cells downregulate salvage pathway synthesis during TMZ therapy, relying instead on de novo purine synthesis. Because TMZ derives its therapeutic efficacy by alkylating purines, we hypothesized that the inhibitory effect of ARL13B on the salvage pathway might allow GBM cells to avoid recycling damaged purines from the tumour microenvironment during TMZ therapy. Moreover, because GBM cells must maintain their nucleotide pools in order to survive and proliferate, de novo purine synthesis is maintained.
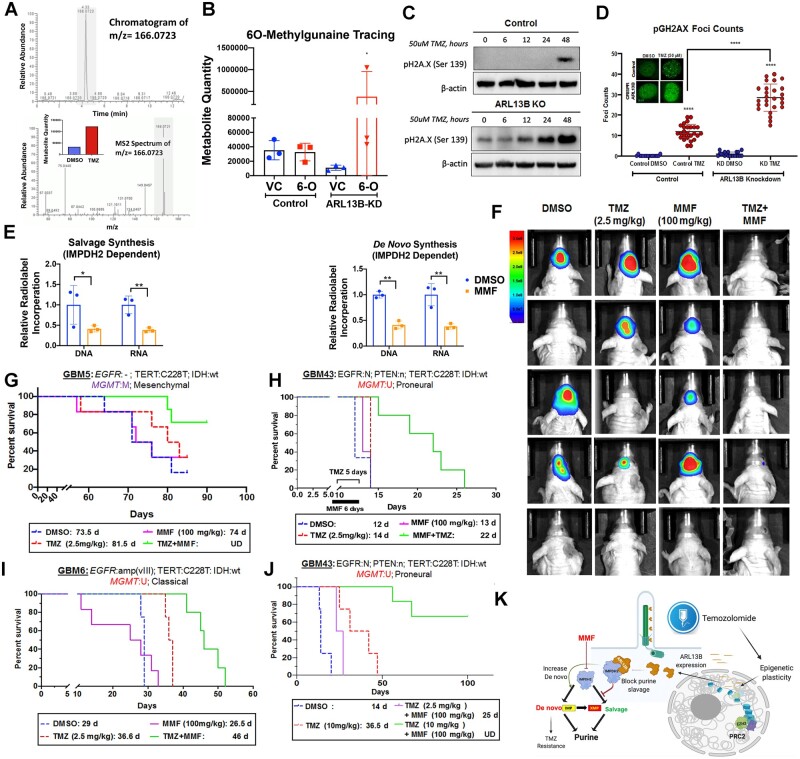
To address whether GBM cells increase de novo purine synthesis in order to avoid utilization of TMZ-damaged purines, but still maintain their nucleotide pools, we first used a modified LC-MS approach for the detection of O6-methylguanine (O6-MG), the key nucleotide damage caused by TMZ treatment, in the genomic DNA of GBM cells. As shown in Fig. 6A, the O6-MG mark increases within 24 h post-exposure to TMZ, validating a positive control (m/z = 166.0.723; Fig. 6A). We then analysed ARL13B wild-type and knockdown cells treated with commercially synthesized O6-MG compound, with results showing that the O6-MG mark is incorporated from the environment and recycled using salvage pathway synthesis into the DNA of ARL13B knockdown cells significantly more than the GBM cells with wild-type ARL13B (Fig. 6B;P = 0.025). This demonstrates that in the absence of ARL13B, a possible negative regulator of purine salvage, GBM cells salvage and incorporate damaged environmental purines into their DNA. This observation also supports the notion that ARL13B knockdown cells showed an increased propensity towards DNA damage in response to alkylating agent-based chemotherapy such as TMZ and bis-chloroethylnitrosourea (BCNU). (Fig. 6C, D and Supplementary Fig. 13B; one-way adjusted ANOVA P < 0.0001). In vitro data also suggested that blocking IMPDH2 activity with MMF and combining with TMZ was superior in producing DNA damage and cell killing in GBM43 and GBM6. However, overexpression of ARL13B could rescue the cells from sensitivity to the MMF+TMZ combination (Supplementary Fig. 14A–E).
Figure 6.
IMPDH2 inhibitor MMF increases PDX GBM sensitivity towards TMZ in vivo. (A) Chromatograms depicting peak enrichment for O6-methylguanine (O6-MG) marks in GBM (U251) cell DNA by mass spectroscopy. Graph inset demonstrates ion count of O6-MG in cells treated with either DMSO or TMZ for 48 h. (B) Graph depicting O6-MG detection in samples with and without ARL13B exposed to vehicle control (VC) or O6-MG (6-O). Counts are represented as ion count with technical triplicates. (C) Western blots analysis on cells with or without ARL13B treated with TMZ for 0, 6, 12, 24 and 48 h and stained with pH2A.X to evaluate the extend of DNA damage. (D) pH2A.X foci counting in cells with and without ARL13B and treated with either DMSO or TMZ (n = 35). (E) Radiolabelled tracing of IMPDH2-dependent salvage, de novo, and IMPDH2-independent salvage synthesis in GBM43 exposed to DMSO control or MMF (5 µM). DNA and RNA relative incorporations are shown from two biological replicates. (F) Bioluminescence (BLI) measurement of engrafted tumour burden from the mesenchymal GBM5 line followed weekly after receiving DMSO, 2.5 mg/kg TMZ for 5 days, 100 mg/kg MMF for 6 days, or TMZ and MMF combination (n = 6). To address sex as a biological variable we used male and female mice equally. (G) Kaplan-Meier curves showing survival of mice implanted with mesenchymal subtypes of PDX GBM5 treated with DMSO, 2.5 mg/kg TMZ for 5 days, 100 mg/kg MMF for 6 days, or TMZ and MMF combination. Animals were monitored for end point survival. (H) Same experiments as above but with proneural subtype GBM43, and (I) classical subtype GBM6. (J) Kaplan-Meier curve of mice with GBM43 PDX tumour treated with 10 mg/kg TMZ in combination with 100 mg/kg MMF along with above controls. (K) The overall mechanism of therapy induced cellular plasticity can promote expression of ciliary protein ARL13B. ARL13B can interact with IMPDH2 to block purine salvaging and contributes to TMZ-based chemotherapy adaptation. Bar graph and images show biological replicates. Image from Week 3 shown. All error bars represent technical experimental replicates unless noted otherwise and display mean ± SD. *P < 0.05 ** P < 0.01 ****P < 0.0001.
Blocking IMPDH2 activity with mycophenolate mofetil sensitizes GBM cells to temozolomide in vivo
We hypothesize that GBM cells need to maintain de novo synthesis in order to remain proliferative and cope with the downregulation of the salvage pathway in response to therapy. Our data support this and indicate a translational therapeutic opportunity if a way can be found to inhibit de novo purine synthesis during TMZ therapy, forcing GBM cells to salvage alkylated purines from the environment. Unfortunately, there is no pharmacological compound that can inhibit ARL13B to replicate our phenotype. However, IMPDH2 activity can be effectively targeted by blood–brain barrier-permeable compounds such as MMF or mizoribine, both of which are in the clinic for transplant setting.52 We first tested mizoribine and MMF in our mouse models and determined MMF to be better tolerated and superior to mizoribine (Supplementary Fig. 15). Because of concerns about the potential clinical side effects of using an immunosuppressive agent in combination with TMZ, we also carried out an extensive safety profile in mice using an MMF and TMZ dose combination greater than any achieved in our survival study (150 μm) (Supplementary Fig. 16C). These data demonstrated MMF+TMZ was no more immune suppressive than TMZ alone and that mice showed no lasting effects of toxicity from either agent (Supplementary Fig. 16). MMF reduced de novo purine biosynthesis, causing an increase in salvage synthesis. (Fig. 6E; multiple t-tests IMPDH2 dependent salvage: DNA P = 0.05, RNA P = 0.01, IMPDH2 dependent de novo: DNA P = 0.001, RNA P = 0.01, IMPDH2 independent salvage RNA P = 0.006). This reduction of de novo synthesis forces GBM cells to uptake alkylated nucleotides via purine salvage, recapitulating the ARL13B knockdown phenotype. To validate if combining MMF with TMZ can enhance the therapeutic efficacy of TMZ in vivo, a dose-finding experiment was done with varying MMF concentrations in combination with TMZ in which 100 mg/kg was found to be the superior dose (Supplementary Fig. 14B). Three different GBM subtypes of PDX (GBM5, GBM6, and GBM43) were tested following the intracranial injection of tumour cells in mice. After 7 days post-implantation animals were treated with TMZ (2.5 mg/kg), 24 h following MMF administration (100 mg/kg). In the classical GBM subtype model, this combination therapy improved median survival by 25%, 57%, and 10% in classical, proneural, and mesenchymal models, respectively (Fig. 6G–I; P = 0.004, 0.01, and 0.02, respectively). The mice engrafted with GBM5, a mesenchymal line, were followed weekly with bioluminescence imaging to track tumour response to treatment, with results showing that MMF+TMZ is superior in slowing tumour growth (Fig. 6J). To assess the efficacy of this schema, we conducted another survival experiment with GBM43 using the same MMF dosing protocol (100 mg/kg 24-h lead-in) but now with a higher dose of TMZ (10 mg/kg). This significantly enhanced the survival benefit over the TMZ alone group and may indicate that frontloading a higher TMZ dose may be more efficacious in this combination of therapy (Fig. 6J; TMZ alone TMZ+MMF 10 mg/kg log-rank adjusted P = 0.001).
Discussion
Plasticity-mediated adaptation to therapy has been hypothesized to play a significant role in GBM recurrence.12,18,53 However, delineation of direct mechanisms, and thus actionable targets, is lacking. In this study, we have uncovered a mechanism of metabolic adaptation to alkylating chemotherapies driven by epigenetic plasticity in GBM. We show that in a PDX model, EZH2 influences therapy-resistant CSC frequency during alkylating-based chemotherapy by regulating ciliary protein ARL13B. We also establish that during TMZ therapy, ARL13B interacts with IMPDH2 to modulate purine biosynthesis in order to reduce the effectiveness of chemotherapy. By blocking IMPDH2 activity using an FDA approved drug, we show that the effects of alkylating agents are significantly enhanced across all subtypes of GBM in multiple PDX models.
EZH2/PRC2 is a well-studied epigenetic regulator whose activity contributes to different malignancies, including GBM. Recent research demonstrates that CSCs rely on EZH2/PRC2 mediated genome-wide reposition of repressive histone marks in order to adapt and resist targeted kinase inhibitor therapy.10 This could also be true during TMZ therapy, and in this work, we have demonstrated that EZH2 activity is involved in the formation of CSCs. To identify targets resulting from the global action of EZH2 during therapy, we performed an unbiased screen and identified ARL13B as a downstream target. Our results indicated that during TMZ therapy, reducing EZH2 binding within an ARL13B enhancer region could cause activation of ARL13B gene expression rather than silencing, representing an epigenetic mechanism for control of ARL13B expression dynamics.
A key challenge in overcoming epigenetic plasticity in GBM is identifying an actionable target of the plastic responses critical for tumour adaptation. As we delved into how the EZH2-ARL13B axis contributes to GBM’s response to chemotherapy, we found a novel interaction between ARL13B and IMPDH2, a critical rate-limiting enzyme of the de novo purine biosynthesis pathway. Our data indicate that without therapeutic pressure, GBM tissue in the PDX model maintains a high level of purine recycling salvage pathway utilization compared to normal brain tissue. When exposed to TMZ, this heavy reliance on the salvage pathway was decreased via interaction between ARL13B and IMPDH2, which causes GBM cells to switch to de novo purine biosynthesis to meet their nucleotide demand. We hypothesized that the switch in the purine biosynthetic pathway could be due to GBM cells trying to bypass the salvage pathway to avoid salvage incorporation of purines alkylated during TMZ chemotherapy. Our data support this notion by demonstrating when ARL13B is removed from GBM cells and GBM cells are exposed to alkylated purines in their media, salvage-mediated uptake of the alkylated purine occurs, causing damage within DNA. Moreover, all subtypes of GBM are more sensitive to TMZ therapy when they lack ARL13B. Disrupting the ARL13B-IMPDH2 interaction by pharmacologically inhibiting IMPDH2 rather than viral knockout of ARL13B also leads to increased efficacy of TMZ in all PDX subtypes in vivo. Pharmacological inhibition of IMPDH2 is possible with several FDA approved compounds indicating translation of these results into the clinical setting quickly is possible.
Metabolic adaption within tumours is becoming a highly explored topic, and along with the current study, crucial new work has highlighted the importance of both purine biosynthesis broadly, as well as an IMPDH2 specifically, in the growth of GBM. Wang and colleagues54 demonstrate that the synthesis of purines utilizing the de novo pathway is especially important in maintaining CSC cells, and that targeting this can disrupt their growth potential.54 Our results corroborate this by demonstrating reduced stem cell forming capacity when ARL13B is lost as well as the loss of expression of key stem cell markers that can ultimately be rescued by restoration of ARL13B expression. Kofuji and colleagues55 also show the importance of IMPDH2 and GTP biosynthesis in general for the growth of glioma.55 They show that GBM relies on de novo GTP biosynthesis over salvage GTP biosynthesis, which dovetails with our findings that forcing salvage purine biosynthesis alone exploits a therapeutic weakness in GBM. Importantly, this is one of the first studies to examine these metabolic dynamics under chemotherapeutic pressure, which is a vital consideration when studying tumours that develop resistance to therapies.
A significant question raised by our research is how GBM cells are able to sense alkylated purines in their environment. We hypothesize that this could have some dependence on the nature of ARL13B as a GTPase; in fact, recent research has tied the IMP/GTP balance to the IMPDH2 function.56 Furthermore, cells under stress in starvation conditions can form rod and ring structures termed purinosomes,48 which are similar physically to cilia, possibly pointing to an avenue for a new biological process contained in cilia or the need for ARL13B to aid in forming the purinosome itself. Furthermore, recent research proposed dynamic functional crosstalk between DNA damage response and cilia-associated proteins.57,58 Indeed, these primary cilia structures have also been documented to occur in GBM patient tissue.59 Thus, it is conceivable that TMZ-induced DNA damage may influence ciliogenesis and allow GBM cells to mount an adaptive response to promote resistance. Although our data strongly suggest the ARL13B-IMPDH2 axis can promote therapeutic adaptation in GBM, further research will be required to elucidate the precise mechanism of such resistance.
In conclusion, we present a mechanism of epigenetic plasticity that can influence purine biosynthesis pathways and allow GBM to resist alkylating-based chemotherapy. Our work also identifies a druggable target that can be inhibited by an FDA approved compound in order to enhance the efficacy of standard of care therapy. Overall, a better understanding of these epigenetic driven metabolic adaptation processes will be essential for developing effective therapeutic strategies against GBM.
Funding
This work was supported by the National Institute of Neurological Disorders and Stroke grant 1R01NS096376, 1R01NS112856 the American Cancer Society grant RSG-16-034-01-DDC (to A.U.A.) grant, R01NS095642 (to C.D.J.), National Institute of Cancer grant R00CA194192, National Institute of Medicine grant R01GM135587 (to I.B.-S.) and R35CA197725 (to M.S.L), P50CA221747 SPORE for Translational Approaches to Brain Cancer. The work in this manuscript is dedicated to the memory of Ashad Uddin Ahmed.
Competing interests
J.M.S., C.H. and A.U.A. report a patent repurposing MMF for GBM therapy, which is pending.
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Glossary
- CSC
cancer stem cells
- FACS
fluorescence-activated cell sorting
- GBM
glioblastoma multiforme
- LC-MS
liquid chromatography mass spectrometry
- MMF
mycophenolate moefitil
- PDX
patient-derived xenograft
- TMZ
temozolomide
References
- 1. Fabian D, Guillermo Prieto Eibl M, Alnahhas I, et al. Treatment of glioblastoma (GBM) with the addition of tumor-treating fields (TTF): A review. Cancers (Basel). 2019;11:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Taillibert S, Kanner A, et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. JAMA. 2017;318:2306-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fidoamore A, Cristiano L, Antonosante A, et al. Glioblastoma stem cells microenvironment: The paracrine roles of the niche in drug and radioresistance. Stem Cells Int. 2016;2016:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharifzad F, Ghavami S, Verdi J, et al. Glioblastoma cancer stem cell biology: Potential theranostic targets. Drug Resist Updat. 2019;42:35-45. [DOI] [PubMed] [Google Scholar]
- 5. Bernstock JD, Mooney JH, Ilyas A, et al. Molecular and cellular intratumoral heterogeneity in primary glioblastoma: Clinical and translational implications. J Neurosurg. 2019;133:1-9. [DOI] [PubMed] [Google Scholar]
- 6. Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN.. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8:3274-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaffer CL, Brueckmann I, Scheel C, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci USA. 2011;108:7950-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Auffinger B, Tobias AL, Han Y, et al. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ. 2014;21:1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dahan P, Martinez Gala J, Delmas C, et al. Ionizing radiations sustain glioblastoma cell dedifferentiation to a stem-like phenotype through survivin: Possible involvement in radioresistance. Cell Death Dis. 2014;5:e1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liau BB, Sievers C, Donohue LK, et al. Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell. 2017;20:233-246.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Safa AR, Saadatzadeh MR, Cohen-Gadol AA, Pollok KE, Bijangi-Vishehsaraei K.. Glioblastoma stem cells (GSCs) epigenetic plasticity and interconversion between differentiated non-GSCs and GSCs. Genes Dis. 2015;2:152-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sheahan AV, Ellis L.. Epigenetic reprogramming: A key mechanism driving therapeutic resistance. Urol Oncol. 2018;36:375-379. [DOI] [PubMed] [Google Scholar]
- 13. Qi L, Jing‐Li C, Hu Y, et al. The dynamics of polycomb group proteins in early embryonic nervous system in mouse and human. Int J Dev Neurosci. 2013;31:487-495. [DOI] [PubMed] [Google Scholar]
- 14. Zhao L, Li J, Ma Y, et al. Ezh2 is involved in radial neuronal migration through regulating Reelin expression in cerebral cortex. Sci Rep. 2015;5:15484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deevy O, Bracken AP.. PRC2 functions in development and congenital disorders. Development. 2019;146:dev181354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suvà M-L, Riggi N, Janiszewska M, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211-9218. [DOI] [PubMed] [Google Scholar]
- 17. Orzan F, Pellegatta S, Poliani PL, et al. Enhancer of Zeste 2 (EZH2) is up-regulated in malignant gliomas and in glioma stem-like cells. Neuropathol Appl Neurobiol. 2011;37:381-394. [DOI] [PubMed] [Google Scholar]
- 18. Natsume A, Ito M, Katsushima K, et al. Chromatin regulator PRC2 is a key regulator of epigenetic plasticity in glioblastoma. Cancer Res. 2013;73:4559-4570. [DOI] [PubMed] [Google Scholar]
- 19. Kim E, Kim M, Woo D-H, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang P, Wan W, Xiong S, et al. HIF1alpha regulates glioma chemosensitivity through the transformation between differentiation and dedifferentiation in various oxygen levels. Sci Rep. 2017;7:7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caragher SP, Shireman JM, Huang M, et al. Activation of dopamine receptor 2 prompts transcriptomic and metabolic plasticity in glioblastoma. J Neurosci. 2019;39:1982-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oizel K, Chauvin C, Oliver L, et al. Efficient mitochondrial glutamine targeting prevails over glioblastoma metabolic plasticity. Clin Cancer Res. 2017;23:6292-6304. [DOI] [PubMed] [Google Scholar]
- 23. Sarkaria JN, Carlson BL, Schroeder MA, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12:2264-2271. [DOI] [PubMed] [Google Scholar]
- 24. Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramírez F, Bhardwaj V, Arrigoni L, et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 2018;9:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ben-Sahra I, Howell JJ, Asara JM, Manning BD.. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim S-H, Joshi K, Ezhilarasan R, et al. EZH2 protects glioma stem cells from radiation-induced cell death in a MELK/FOXM1-dependent manner. Stem Cell Reports. 2015;4:226-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neo WH, Yap K, Lee SH, et al. MicroRNA miR-124 controls the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression. J Biol Chem. 2014;289:20788-20801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee G, Auffinger B, Guo D, et al. Dedifferentiation of glioma cells to glioma stem-like cells by therapeutic stress-induced HIF signaling in the recurrent GBM model. Mol Cancer Ther. 2016;15:3064-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hasan T, Caragher SP, Shireman JM, et al. Interleukin-8/CXCR2 signaling regulates therapy-induced plasticity and enhances tumorigenicity in glioblastoma. Cell Death Dis. 2019;10:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beier D, Röhrl S, Pillai DR, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Research Support, Non-U.S. Cancer Res. 2008;68:5706-5715. [DOI] [PubMed] [Google Scholar]
- 32. Rosso L, Brock CS, Gallo JM, et al. A new model for prediction of drug distribution in tumor and normal tissues: Pharmacokinetics of temozolomide in glioma patients. Cancer Res. 2009;69:120-127. [DOI] [PubMed] [Google Scholar]
- 33. Ostermann S, Csajka C, Buclin T, et al. Plasma and cerebrospinal fluid population pharmacokinetics of temozolomide in malignant glioma patients. Clin Cancer Res. 2004;10:3728-3736. [DOI] [PubMed] [Google Scholar]
- 34. Bowman RL, Wang Q, Carro A, Verhaak RG, Squatrito M.. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017;19:139-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang N, Ahn SH, Kong DS, Lee HW, Nam DH.. The role of STAT3 in glioblastoma progression through dual influences on tumor cells and the immune microenvironment. Mol Cell Endocrinol. 2017;451:53-65. [DOI] [PubMed] [Google Scholar]
- 36. Zhang J, Chen L, Han L, et al. EZH2 is a negative prognostic factor and exhibits pro-oncogenic activity in glioblastoma. Cancer Lett. 2015;356:929-936. [DOI] [PubMed] [Google Scholar]
- 37. Gao T, He B, Liu S, Zhu H, Tan K, Qian J.. EnhancerAtlas: A resource for enhancer annotation and analysis in 105 human cell/tissue types. Bioinformatics. 2016;32:3543-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391-403. [DOI] [PubMed] [Google Scholar]
- 39. Tobias AL, Thaci B, Auffinger B, et al. The timing of neural stem cell-based virotherapy is critical for optimal therapeutic efficacy when applied with radiation and chemotherapy for the treatment of glioblastoma. Stem Cells Transl Med. 2013;2:655-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jeng K-S, Chang C-F, Lin S-S.. Sonic hedgehog signaling in organogenesis, tumors, and tumor microenvironments. Int J Mol Sci. 2020;21:758. doi:10.3390/ijms21030758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bay SN, Long AB, Caspary T.. Disruption of the ciliary GTPase Arl13b suppresses Sonic hedgehog overactivation and inhibits medulloblastoma formation. Proc Natl Acad Sci USA. 2018;115:1570-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mariani LE, Bijlsma MF, Ivanova AA, Suciu SK, Kahn RA, Caspary T.. Arl13b regulates Shh signaling from both inside and outside the cilium. MBoC. 2016;27:3780-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Larkins CE, Aviles GD, East MP, Kahn RA, Caspary T.. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. MBoC. 2011;22:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gajjar AJ, Robinson GW.. Medulloblastoma—translating discoveries from the bench to the bedside. Nat Rev Clin Oncol. 2014;11:714-722. [DOI] [PubMed] [Google Scholar]
- 45. Girardet L, Bernet A, Calvo E, et al. Hedgehog signaling pathway regulates gene expression profile of epididymal principal cells through the primary cilium. FASEB J. 2020;34:7593-7609. [DOI] [PubMed] [Google Scholar]
- 46. Camici M, Garcia-Gil M, Pesi R, Allegrini S, Tozzi MG.. Purine-metabolising enzymes and apoptosis in cancer. Cancers. 2019;11:1354. doi:10.3390/cancers11091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hedstrom L. IMP dehydrogenase: Structure, mechanism, and inhibition. Chem Rev. 2009;109:2903-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pedley AM, Benkovic SJ.. A new view into the regulation of purine metabolism: The purinosome. Trends Biochem Sci. 2017;42:141-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Micheli V, Camici M, G. Tozzi M, et al. Neurological disorders of purine and pyrimidine metabolism. Curr Top Med Chem. 2011;11:923-947. [DOI] [PubMed] [Google Scholar]
- 50. Seegmiller JE, Rosenbloom FM, Kelley WN.. Enzyme defect associated with a sex-linked human neurological disorder and excessive purine synthesis. Science. 1967;155:1682-1684. [DOI] [PubMed] [Google Scholar]
- 51. Brada M, Judson I, Beale P, et al. Phase I dose-escalation and pharmacokinetic study of temozolomide (SCH 52365) for refractory or relapsing malignancies. Clinical Trial Clinical Trial, Phase I Research Support, Non-U.S. Gov't. Br J Cancer. 1999;81:1022-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Allison AC, Eugui EM.. Purine metabolism and immunosuppressive effects of mycophenolate mofetil (MMF). Clin Transplant. 1996;10:77-84. [PubMed] [Google Scholar]
- 53. Das PK, Pillai S, Rakib MA, et al. Plasticity of cancer stem cell: Origin and role in disease progression and therapy resistance. Stem Cell Rev Rep. 2020;16:397-412. doi:10.1007/s12015-019-09942-y. [DOI] [PubMed] [Google Scholar]
- 54. Wang X, Yang K, Xie Q, et al. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat Neurosci. 2017;20:661-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kofuji S, Hirayama A, Eberhardt AO, et al. IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat Cell Biol. 2019;21:1003-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Keppeke GD, Chang CC, Peng M, et al. IMP/GTP balance modulates cytoophidium assembly and IMPDH activity. Cell Div. 2018;13:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Villumsen BH, Danielsen JR, Povlsen L, et al. A new cellular stress response that triggers centriolar satellite reorganization and ciliogenesis. EMBO J. 2013;32:3029-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Johnson CA, Collis SJ.. Ciliogenesis and the DNA damage response: A stressful relationship. Cilia. 2016;5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sarkisian MR, Siebzehnrubl D, Hoang-Minh L, et al. Detection of primary cilia in human glioblastoma. J Neurooncol. 2014;117:15-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data were generated at Northwestern university. Derived data supporting the findings of this study are available from the corresponding author on request.