Abstract
The brain signature concept aims to characterize brain regions most strongly associated with an outcome of interest. Brain signatures derive their power from data-driven searches that select features based solely on performance metrics of prediction or classification. This approach has important potential to delineate biologically relevant brain substrates for prediction or classification of future trajectories. Recent work has used exploratory voxel-wise or atlas-based searches, with some using machine learning techniques to define salient features. These have shown undoubted usefulness, but two issues remain. The preponderance of recent work has been aimed at categorical rather than continuous outcomes, and it is rare for non-atlas reliant voxel-based signatures to be reported that would be useful for modelling and hypothesis testing. We describe a cross-validated signature region model for structural brain components associated with baseline and longitudinal episodic memory across cognitively heterogeneous populations including normal, mild impairment and dementia. We used three non-overlapping cohorts of older participants: from the UC Davis Aging and Diversity cohort (n = 255; mean age 75.3 ± 7.1 years; 128 cognitively normal, 97 mild cognitive impairment, 30 demented and seven unclassified); from Alzheimer’s Disease Neuroimaging Initiative (ADNI) 1 (n = 379; mean age 75.1 ± 7.2; 82 cognitively normal, 176 mild cognitive impairment, 121 Alzheimer’s dementia); and from ADNI2/GO (n = 680; mean age 72.5 ± 7.1; 220 cognitively normal, 381 mild cognitive impairment and 79 Alzheimer’s dementia). We used voxel-wise regression analysis, correcting for multiple comparisons, to generate an array of regional masks corresponding to different association strength levels of cortical grey matter with baseline memory and brain atrophy with memory change. Cognitive measures were episodic memory using Spanish and English Neuropsychological Assessment Scales instruments for UC Davis and ADNI-Mem for ADNI 1 and ADNI2/GO. Performance metric was the adjusted R2 coefficient of determination of each model explaining outcomes in two cohorts other than where it was computed. We compared within-cohort performances of signature models against each other and against other recent signature models of episodic memory. Findings were: (i) two independently generated signature region of interest models performed similarly in a third separate cohort; (ii) a signature region of interest generated in one imaging cohort replicated its performance level when explaining cognitive outcomes in each of other, separate cohorts; and (iii) this approach better explained baseline and longitudinal memory than other recent theory-driven and data-driven models. This suggests our approach can generate signatures that may be easily and robustly applied for modelling and hypothesis testing in mixed cognition cohorts.
Keywords: brain signature, cross-validation, episodic memory, grey matter density, longitudinal atrophy
See Jolly and Hampshire (doi:10.1093/brain/awab140) for a scientific commentary on this article.
Fletcher et al. test the efficacy of an exploratory signature region of interest approach for delineating brain resources associated with episodic memory and memory change. They demonstrate the robustness of this approach and suggest potential applications in areas such as cognitive reserve and brain maintenance.
See Jolly and Hampshire (doi:10.1093/brain/awab140) for a scientific commentary on this article.
Introduction
This paper lays out a method for quantifying structural brain components associated with baseline and longitudinal episodic memory. We focus on this cognitive domain because episodic memory change is a key marker both of ageing and incipient Alzheimer’s disease.1 However, measuring direct associations between brain structure and episodic memory is not completely straightforward because memory relies on a variety of cognitive processes and brain circuits.2 Medial temporal structures (entorhinal, perirhinal and parahippocampal cortices and hippocampus) are known to be associated with cross-sectional episodic memory.3 Longitudinal measurements of memory and brain change may show stronger associations than do baseline measurements.4 For example, Murphy et al.5 found that 6-month atrophy rates in medial temporal structures were associated with episodic memory decline. Previous work from our group has demonstrated that hippocampal volume is associated with cross-sectional episodic memory6 and that memory change is associated with global grey matter atrophy and incrementally with temporal lobe grey matter atrophy.7
Analyses of brain resources associated with episodic memory baseline and change have frequently used theory-driven lists comprising prespecified structures, such as medial temporal, precuneus and global cortical grey volumes, yielding models of undoubted explanatory power.8-10 However, recent work has also examined signature region of interest approaches for computing brain features associated with various outcomes, free of prior suppositions. The signature approach encompasses exploratory, data-driven methods for selecting brain features that are strongly associated with outcomes, either from a high dimensional space such as brain voxel locations, or from a lower dimensional space such as a list of regions of interest.11 Feature selection techniques include machine learning,11 voxel aggregation into signature regions12,13 and systematic testing and selection from a list of regions of interest.14-16 Efforts differ by their goals of signatures for classification into discrete categories or predicting continuous outcomes. Machine learning techniques like support vector machines (SVM)17 that are designed to learn discriminative predictors of binary outcomes,18 have been applied e.g. to binary classification of diagnosis19 or cognitive decline.20,21 For brain signatures of continuous outcomes, relevant vector regression (RVR) has been applied to predict continuous variables such as cognitive test scores22-24 or brain age.25,26
Among recent signature approaches, few to our knowledge have used voxel-based exploratory methods to compute and report signature regions not associated with collections of atlas-based components. This approach has the potential to delineate ‘non-standard’ (i.e. not-conforming to prespecified atlas parcellations) regions most associated with outcomes of interest. Such regions may more accurately reflect relevant brain architecture. Furthermore, they are easily computed as masks in a brain template space; if shown to be robustly applicable in a range of cohorts, they could be widely useful for model building and hypothesis testing. One voxel-aggregation model is the Dickerson-Bakkour signature of very early Alzheimer’s disease,12,13 but when examined as a marker for episodic memory27 the signature was implemented by eight regions of interest from the Freesurfer atlas.28 A machine learning study used RVR to generate voxel-based weight maps predicting memory decline24 in a cognitively normal cohort of two groups separated by amyloid-β positivity; again, though, this work reported regional results as percentages of standard atlas regions covered by the weight maps. Another relevant study was not based on machine-learning or voxel aggregation, instead systematically searching the FreeSurfer atlas28 to find brain regions most associated with memory subtasks and to episodic memory itself.15 They reported only the effect sizes and P-values for associations of individual regions of interest with a latent episodic memory variable.
This paper aims to implement a signature approach to continuous episodic memory that is ‘performant’ (explaining a near optimal amount of outcome variance from the features available, and therefore useful in models of cognition); ‘informative’ (delineating brain regions that provide insights into brain relation with memory); and ‘economical’, containing relatively few components that are easily generated. We aim to fill a gap in the burgeoning literature of computational efforts to delineate brain-cognition relations. We outline a unified algorithmic voxel-aggregation approach for brain signature region of interest models of baseline and longitudinal episodic memory outcomes that is designed to be applicable in cohorts of older participants encompassing the range of cognitive diagnosis from normal to demented. We then test the hypothesis that (i) this approach will better explain baseline and longitudinal memory than theory-driven ‘standard’ models based on pre-selected regions and it will perform at least as well as other data-driven models based on sophisticated feature selection such as the recent works cited. Like machine-learning approaches, voxel aggregation entails ‘learning’ a signature meta-region of interest from a particular ‘training’ cohort, requiring validation in independent cohorts to demonstrate that it can robustly generalize. Therefore, we also hypothesize that (ii) a signature region of interest generated in one imaging cohort will replicate its performance level of R2 association when explaining cognitive outcomes in other, separate cohorts. Finally, two independently generated signature region of interest models will perform similarly (i.e. reproducibly) in a third separate cohort.
Materials and methods
Imaging cohorts
We used three imaging cohorts of older adults: (i) 255 participants from the UC Davis Aging and Diversity Cohort (ADC) with MRI acquired on a 1.5 T scanner; (ii) 379 participants drawn from Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu) ADNI 1 with 1.5 T imaging; and (iii) 680 participants from ADNI GO/ADNI2 database with 3 T MRI scans. To ensure validity of cross-validation, all cohorts were non-overlapping. Each cohort included individuals with normal cognition, mild cognitive impairment (MCI), and dementia. All subjects had cognitive evaluations for at least two study visits and at least two structural MRI scans with over 1-year interscan separation (see Table 1 for participant characteristics at baseline).
Table 1.
Demographic profiles of the three cohorts
| n | Age, years | Gender | Education, years | Race/ethnicity | Clinical diagnosis | |
|---|---|---|---|---|---|---|
| ADC | 255 | 75.3 (7.1) |
M = 104 F = 151 |
13.2 (4.5) |
Asian 8 Hispanic 70 African American 61 White 116 |
CN 128 MCI 97 Demented 30 Not classified 7 |
| ADNI 1 | 379 | 75.1 (7.2) |
M = 217 F = 162 |
15.5 (3.0) |
Hispanic/Latino 7 Not H/L 368 Not classified 4 |
CN 82 Late MCI 176 AD 121 |
| ADNI2/GO | 680 | 72.5 (7.1) |
M = 359 F = 321 |
16.4 (2.6) |
Hispanic/Latino 21 Not H/L 654 Not classified 5 |
CN 220 Early MCI 249 Late MCI 132 AD 79 |
Data are presented as means (standard deviation, SD). AD = Alzheimer’s disease; CN = cognitively normal.
One of the aims of the ADC cohort was to explore heterogeneity in cognitive trajectories of ageing associated with mixed pathologies and diverse ethno-racial identities. It consists of 1450 subjects with structural neuroimaging and cognitive assessments. It is ethno-racially heterogeneous, including large Hispanic and African American representations, recruited over a period of 20 years from the East Bay and Sacramento regions of Northern California.
ADNI was launched by the National Institute of Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies, and non-profit organizations in 2003 as a public-private partnership. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early Alzheimer’s disease. The principal investigator of ADNI is Michael Weiner, MD, VA Medical Center and University of California, San Francisco. For current information on ADNI, see www.adni-info.org.
Inclusion criteria for data from all our cohorts were availability of two or more structural MRI scans separated by at least 1 year, and two or more cognitive evaluations with dates close to those of the MRI scans.
Cognitive assessment
Cognitive measurements were Spanish and English Neuropsychological Assessment Scales (SENAS) verbal episodic memory29,30 for the ADC cohort and ADNI-Mem31,32 for both ADNI cohorts. These measures assess the cognitive construct of episodic memory for verbal information. Both use list learning tasks, although ADNI-Mem also includes other types of memory tasks such as the Alzheimer’s Disease Assessment Scale-Cognitive Subscale (ADAS-Cog) and Mini-Mental State Examination (MMSE). Both are sensitive to individual differences across the full range of episodic memory performance. We determined intercepts and slopes from memory data from assessments performed over multiple study visits using random effects models.
MRI data
We used baseline scans for grey matter density computations and baseline plus latest scans for atrophy computations. MRI acquisition parameters for ADC33 and ADNI34 have been reported previously.
MRI image processing
MRI whole head structural scans were processed by in-house pipelines developed in our laboratory,35 producing brain extraction, tissue segmentation including grey matter and white matter hyperintensity quantification, followed by B-spline registration36 with an age-appropriate template structural brain image37 for use in voxelwise analysis, as reported previously.35 All visualization of brain image data was performed using viewer tools developed in-house.
Grey matter density quantification
We operationalized cross-sectional brain resources for episodic memory via grey matter density computations. Grey matter density quantification was performed at the voxel level in each native space image as part of our pipeline, using the DiReCT diffeomorphism-based application38 applied to the segmented grey matter mantle. DiReCT and the popular Freesurfer package39 each compute grey matter thickness measures. DiReCT is ‘volume-based’ (i.e. assigning grey matter density to individual 3D voxel volume locations) whereas Freesurfer is ‘surface-based’ (i.e. calculating vertex-wise distances between inner and outer grey matter 2D surface meshes).40 Our decision to use DiReCT in our model depended first on our need for voxelwise measures, and second on a comparison between the methods showing greater statistical predictive power for age and gender for DiReCT measures.41 Tissue segmentation input to the DiReCT application were generated by our in-house pipeline as described above. Native space grey matter density maps were deformed to template space via B-spline parameters previously computed in our pipeline.
Longitudinal volume change quantification
Voxel-level estimates of tissue atrophy were computed based upon registration of a single pair of serial MRI scans—the baseline and latest scans for each subject having interscan separation of at least 1 year. We carried out image pair registration via an in-house tensor-based morphometry (TBM) application designed to enhance sensitivity to change by estimating structural edge probabilities to minimize spurious change indications derived from noise, as reported previously.7,42,43 Volume change, consisting of tissue atrophy and CSF expansion rates, was computed at each voxel by the log-transform of the determinant of the 3 × 3 Jacobian matrix of the TBM deformation field, which we refer to as log-Jacobians. The log transformation produces a distribution centred at 0 with negative values indicating atrophy and positive values indicating expansions; when the magnitudes are small, the log-Jacobian is roughly the percentage change at each voxel. We used the log-Jacobian as it is quite sensitive to volume change at the voxel level.42,43 This then enabled us to aggregate voxel clusters of significant atrophy associated to outcome without recourse to predefined regional masks, in accordance with our exploratory approach. We normalized each subject’s log-Jacobian map by the appropriate multiplicative factor to yield a representation of change over a 2-year interscan interval. For voxelwise analysis, all native-space log-Jacobian maps were deformed to template space.
Data analysis
Our data analysis consisted of voxelwise and region of interest-based analyses, all conducted in the template space.
The signature region of interest model computation
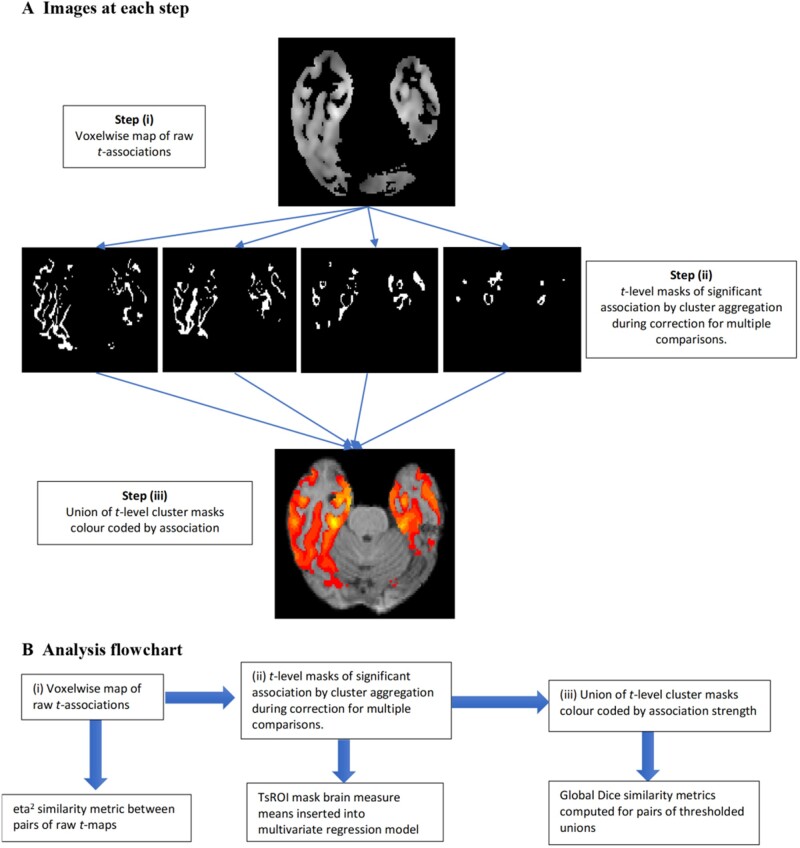
We developed signature regions of interest based upon t-values of voxelwise regressions for brain characteristics (grey matter density at baseline, log-Jacobians for atrophy rates) predicting episodic memory outcomes (baseline and longitudinal change). This consisted of three steps, illustrated in Fig. 1 and described in more detail below: (i) regression voxelwise t-maps; (ii) array of disjoint cluster maps of significant associations indexed by bins corresponding to discrete levels of t-association values; and (iii) assembled signature region of interest models of episodic memory.
Figure 1.
Steps leading to TsROIs. (A) Illustration of three steps in the signature region of interest approach. (i) Generation of raw t-map associations between voxel predictors and episodic memory; (ii) generation of significant t-level association cluster masks (TsROIs); (iii) assembly of TsROIs into a combined map for visual and global similarity metric comparisons. (B) Sequence of analyses associated with each step. Top row: Sequence of steps from A. Bottom row: Associated analyses.
Regression t-maps
Our first step was to generate a t-map derived from the association of cognitive outcome to brain parameters at each relevant voxel (grey matter mask for baseline, brain tissue mask for longitudinal). We performed multivariate regressions of episodic memory on single voxel measures (i.e. grey matter density for baseline and log-Jacobian atrophy rate for longitudinal) co-varying for participant age, education and gender. Future work to achieve full models of brain resources supporting memory will need to include other brain measures such as white matter integrity.
T-value signature association regions of interest
To correct for multiple comparisons over the large number of voxels and aggregate the voxels of brain associations into significant clusters, we performed non-parametric t-value cluster significance computations44 separately for t-values in distinct bins of width 1 over a range of values, from t = 3 to 8 for baseline and somewhat higher for longitudinal tests. The bin size width of 1 is a parameter of convenience. We tested bin widths of 0.5 and found little difference in outcomes (data not shown). For the range of t-level bins, the maximum was the highest t-value at which a significant cluster size could be found. The minimum was based on an assessment that for 100+ df (all cohorts had more than 100 participants) the critical t-value (P < 0.001) was ∼3. We used this minimum value for baseline models. In longitudinal models, t-values were shifted higher and we judged that the clusters at level 3 were too broad and did not add much power to the regression models. In that setting our lowest t-value was 4.
The t-value significant clusters are computed from tests of the null hypothesis of no relation between outcome and individual voxel brain measures. If the null hypothesis is true, then permuting the association between brain image and cognitive outcome should generate maximal associated cluster sizes of voxels similar to those of the actual regressions when aggregated over the image.44 To compute these significant clusters, we performed 10 000 iterations, randomly permuting cognitive outcomes under the assumption of a null association. We retained clusters from the original regressions whose size was in the top fifth percentile of the size distribution. In practice, most regions of interest were in the highest 0.1%. The resulting t-value significant clusters formed our array of signature regions of interest (TsROIs) indexed by strength of association to outcome.
Signature region of interest models of outcome
For our final regression model, the TsROI means of grey matter density (for baseline) or log-Jacobian (for longitudinal) were inserted as independent variables into a regression predicting cognition, controlling for age, gender and education. Thus, each TsROI was a separate predictor in the model of cognitive outcome (Equation 1). To avoid bias, the TsROIs generated in one cohort were applied to generate means in each of the other two cohorts.
Model performance evaluation
We tested our final regression models for consistency and optimality, using a cross-validation design. Model performance was measured by the adjusted R2 coefficient of determination that penalizes the fit score according to the number of independent variables. Each array of TsROIs generated in cohort A (e.g. ADC) was used to sample brain means in cohorts B (e.g. ADNI1) and C (e.g. ADNI2/GO). We will refer to the TsROIs generated in A as ‘eligible’ for cohorts B and C. Our final regression model thus had the form:
| (1) |
controlling for age, gender and education, where cognition and brain voxel means were sampled in a given cohort (e.g. B or C) using TsROIs generated in an eligible cohort (e.g. A). Cognition refers to episodic memory intercept at baseline or episodic memory slope for longitudinal models, and each TsROI samples grey matter density mean (baseline) or log-Jacobian mean (longitudinal). The TsROIs were separately generated for baseline grey matter and longitudinal log-Jacobian data. The effect size β-estimates are computed in each target cohort rather than being predetermined, in order to accommodate image data variability between cohorts.
Similarity and consistency
We tested the similarity and consistency of our models using three criteria. First, we used a numerical similarity metric, eta2 to evaluate the similarity or closeness of the raw t-maps generated in all three cohorts for baseline and longitudinal regressions. The value is 1 for maps that are identical. Use of this similarity metric is relevant because the t-maps were the starting point to generate the final model via computation of the TsROIs. The eta2 metric has been used previously to evaluate image association map similarity45,46 and is preferable to the cross-correlation as a voxel-based metric because the latter is insensitive to global differences in magnitude. Thus, unlike the cross-correlation, the eta2 metric is lower if there are systematic magnitude differences in the t-maps, which are important for indicating strengths of regression association. Second, we compared the aggregate of all TsROIs generated from separate cohorts using Dice scores.47 Next, for model performance we evaluated ‘replicability’, meaning that a single eligible TsROI model should perform comparably in different cohorts (i.e. replicate the results) according to the coefficient of determination (R2) measure of the cognitive variance it explains. And finally, we examined ‘reproducibility’, meaning that two eligible TsROI models should perform similarly by the adjusted R2 measure when applied to a third cohort.
Comparing TsROI signature with other model performances
We compared the TsROI models eligible in a given cohort with other models for predicting episodic memory in that cohort. For comprehensive tests, we used representative models from theory-driven as well as data-driven approaches. For baseline memory, the comparisons were a theory-driven model involving hippocampal atrophy,48 and three data-driven models: first, based on the cortical Alzheimer’s disease signature27; second, based on an approach selecting regions of interest from the Freesurfer atlas that had significant effects on baseline episodic memory when tested individually15; and third, a search for regions of interest whose cortical thickness or volume measures best yield an Alzheimer’s disease classification signature.14 Although surface-based Freesurfer thickness measures were used in two of the comparison models,15,27 they do not generate the voxelwise grey matter measures used in our model. We sought comparisons of regional effectiveness using the same grey matter measures for all models. This and earlier findings that DiReCT is more reliable14 and more strongly predictive41 with the same underlying regions of interest, conditioned our decision to use DiReCT when evaluating the comparison models. For longitudinal memory change our theory-driven comparison was temporal grey matter atrophy rates7; the data-driven comparison was an RVR approach to explaining longitudinal cognitive decline.24 All of these sources, including those that were explicitly voxel-based, listed standard atlas component regions of interest associated with their models. We implemented each model in our datasets using corresponding elements of the Mindboggle atlas (https://mindboggle.info/), which is a second generation version of the Desikan-Killiany-Tourville atlas of grey matter parcellation.49,50 Most selection choices were unambiguous. For the model of Epelbaum et al.,15 however, their aim was to delineate cortical regions associated with three component subscores of the Free and Cued Selective Reminding Test. Episodic memory itself was modelled using a latent class approach. To obtain an implementation for comparison with our signature models, we incorporated grey matter region of interest elements whose single association with episodic memory was reported by Epelbaum et al.15 to have false discovery rate (FDR)-corrected P-values < 0.05. For the RVR model predicting episodic memory decline,24 the voxel-based grey matter locations were not available, but a list was provided of component brain areas having the highest percentage overlap with the regions of that study and we used this list for our model implementation. All comparison models were implemented as the union of their component brain areas, yielding a single index as the average brain measure computed over this union. This follows the approach explicitly stated in some of the references.14,24,27
Data availability
Derived data supporting the findings of this study, and resulting signature region of interest masks, are available from the corresponding author upon reasonable request.
Results
Demographic profiles
Participant characteristics of our three cohorts are displayed in Table 1. With respect to sex distributions, ADC was different (P < 0.05) than both ADNI cohorts, but the two ADNI cohorts did not differ. For age, ADC and ADNI 1 participants were older than ADNI GO/ADNI 2 (P < 0.01) but ADC and ADNI 1 were not different. For years of education, ADC had less than ADNI 1 (P < 0.01) and ADNI2/GO (P < 0.01), while ADNI 1 was less than ADNI2/GO (P < 0.01). The ADC cohort exhibited a wide racial/ethnic diversity, with the largest group being European ancestry (45.4%) but sizeable representations of Hispanic (27.5%) and African American (23.9%) groups. ADNI cohorts were predominantly white (https://adni.ucsd.edu/perl/intra/psi/Rreports/adni/adnidsum.rnw).
All three datasets had sizable representations of clinical diagnoses from clinically normal to MCI to dementia. The ADC cohort had a higher percentage of cognitively normal than either ADNI cohort (P < 0.01 in both cases), but the ADNI cohorts did not differ in proportions of normal participants. ADC had a smaller fraction of MCI than either of the ADNI cohorts (P < 0.01 for both comparisons) but the ADNI cohorts did not differ in MCI. Finally, the proportion of ADC participants with dementia did not differ from ADNI2/GO Alzheimer’s disease, but both differed from that for ADNI 1 Alzheimer’s disease (P < 0.01). Thus, the ADC cohort had the greatest proportion of cognitively normal and ADNI 1 the greatest proportion of demented subjects. The ADNI cohort diagnoses are principally in the Alzheimer’s spectrum, whereas the ADC demented category encompassed vascular as well as Alzheimer’s disease dementias. ADNI specifies two levels of MCI (early and late MCI) and Alzheimer’s disease dementia, whereas ADC participants were classified simply as MCI or demented, without reference to Alzheimer’s disease. For the purposes of comparisons in these results, we combined early and late MCI into a single MCI category for the ADNI cohorts.
Similarity evaluations of the signature regions of interest
T-map eta2 similarity across cohorts
As previously described, our approach starts with generating raw t-maps of voxel associations with episodic memory composite scores in each cohort. This includes grey matter density measures associated to baseline memory scores (ADNI-Mem or ADC SENAS verbal) or tissue log-Jacobian atrophy rates associated with longitudinal memory change. Results for t-map similarity measures are presented in Table 2.
Table 2.
Pairwise similarity measures
| mem ∼ Grey matter density: t-map similarity (eta2) | |||||||
|---|---|---|---|---|---|---|---|
| ADC baseline | ADNI 1 baseline | ADNI2/GO | |||||
| ADC | 1 | 0.788 | 0.747 | ||||
| ADNI 1 | 1 | 0.816 | |||||
|
ADNI2/GO |
|
|
1 |
||||
|
Δmem ∼ log-Jacobian atrophy: t-map similarity (eta2) | |||||||
|
|
ADC |
ADNI 1 |
ADNI2/GO |
||||
| ADC | 1 | 0.932 | 0.872 | ||||
| ADNI 1 | 1 | 0.935 | |||||
|
ADNI2/GO |
|
|
1 |
||||
| mem ∼ Grey matter density: union cluster ROI similarity (Dice) | |||||||
|
| |||||||
| ADC | ADNI 1 | ADNI2/GO | Epelbaum et al. 15 | Busovaca et al. 27 | Schwarz et al. 14 | ||
|
| |||||||
| ADC | 1 | 0.360 | 0.444 | 0.252 | 0.232 | 0.508 | |
| ADNI 1 | 1 | 0.610 | 0.605 | 0.449 | 0.381 | ||
|
ADNI2/GO |
|
|
1 |
0.443 |
0.326 |
0.427 |
|
| Δmem ∼ log-Jacobian atrophy: union cluster ROI similarity (Dice) | |||||||
|
| |||||||
| ADC | ADNI 1 | ADNI2/GO | Caballero et al. 24 | ||||
|
| |||||||
| ADC | 1 | 0.765 | 0.709 | 0.151 | |||
| ADNI 1 | 1 | 0.830 | 0.204 | ||||
| ADNI2/GO | 1 | 0.193 | |||||
Pairwise eta2 similarity measures for grey matter t-map associations to baseline episodic memory; pairwise eta2 similarity measures for longitudinal tissue atrophy associations witd memory change; pairwise Dice score similarities for baseline combined (union) region of interest (ROI) models; and pairwise Dice similarities for longitudinal associations.
The eta2 similarity measures were higher for t-maps of longitudinal associations, but both sets of pairwise measures were high. For example, the baseline measurements were higher than the eta2 similarity ranges (0.57–0.65) reported by Bakkour et al.46 showing that four independently generated Alzheimer’s disease signature grey matter atrophy maps were ‘reasonably similar’. And the longitudinal similarity coefficients were close to or exceeded those reported for maps that were reported as ‘very similar’ (0.92) in the same reference. Pairwise Dice scores are reported in Table 2 for the union masks of all signature models against each other and the comparison model masks. All of the TsROI longitudinal models had high Dice similarities that were also much higher than similarities with the Caballero model mask. For baseline, the two ADNI TsROI models had a high Dice similarity but the ADC similarity with each of them was lower.
Similarity of combined TsROI arrays from each cohort
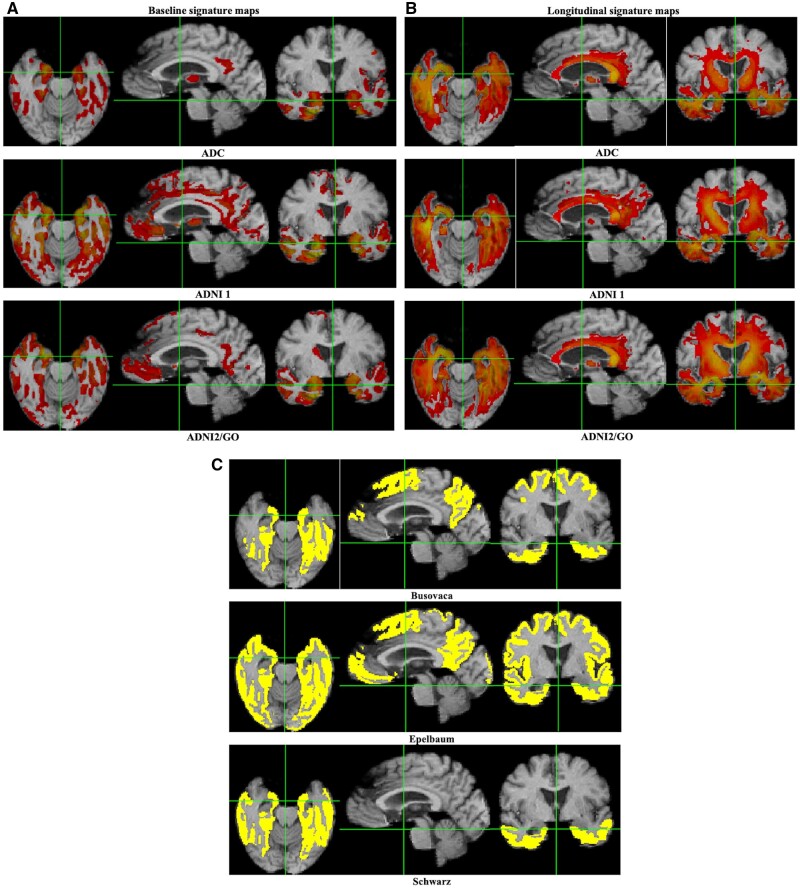
Combined cluster maps for baseline and longitudinal associations are presented in Fig. 2. Overall configurations show good visual similarity across cohorts, with greater similarity seen among the longitudinal maximal cluster maps than for baseline. Baseline associations of grey matter density with memory (Fig. 2A) exhibit highest strengths in medial temporal areas including entorhinal cortex and hippocampus (orange to yellow colours), with some involvement of lateral and posterior temporal regions and substantial portions of the caudate (the latter regions are not shown). Baseline t-values generated in all cohorts ranged from 3 to 8. Longitudinal associations (Fig. 2B) of cognitive change to atrophy rates were also heavily represented in temporal lobe regions including white matter, but in addition more dorsally in portions of the corpus callosum including the splenium; the thalamus; and extensive white matter regions parallel to the lateral ventricles. Interestingly, the hippocampus was not well represented in maps of longitudinal associations. Longitudinal t-values ranged from a minimum t-level of 4 in all cohorts to a maximum of 10 in ADC, 12 for ADNI 1 and 17 for ADNI GO2, which was a much larger dataset. Colours are not to the same scale across cohorts due to different ranges of t-thresholds in each cohort.
Figure 2.
Visualizing the signature regions of interest. Combined t-level cluster signature maps for associations of brain measures and episodic memory composite scores. (A) Baseline associations of grey matter density to memory intercept. (B) Longitudinal associations of brain atrophy rates with memory change. Colour maps are on the same scale of 3–8 (red to yellow) for baseline. Longitudinally, minimum values are 4 (red) and maximum values (yellow) vary by cohort. (C) Corresponding coordinate plane slices for comparison baseline model union regions of interest from Busovaca et al.,27 Epelbaum et al.15 and Schwarz et al.14
Model performance measures
We present results for performance evaluations of our signature regions of interest in models explaining episodic memory. We use the adjusted R2 measure of regression model fit to evaluate how much memory variance is explained by grey matter density measured in our signature regions of interest. As outlined above, we tested replicability (whether a model using signature regions of interest developed in cohort A will replicate its performance in cohorts B and C) and reproducibility (whether signature regions of interest generated in B and C reproduce the same results in cohort A).
Summary of TsROI model results across cohorts
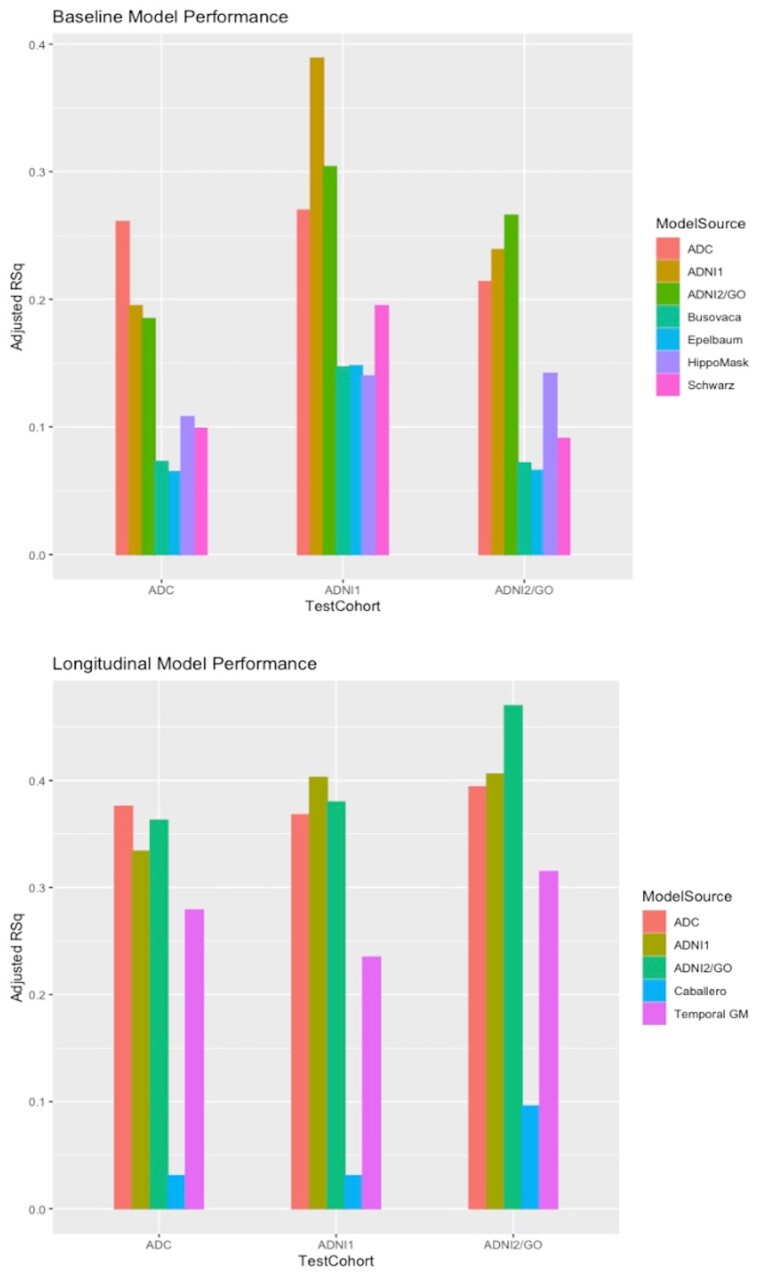
As shown in Table 3, replicability (comparing R2 fits in a row) and reproducibility (R2 fits in a column) are good to excellent for the cohort signature region of interest models. For context, we also evaluated the R2 fits of each model applied in the cohort where generated (italicized diagonal entries); these are uniformly higher than fits for other signature models, suggesting that the self-generated models may benefit from a favourable bias in their own cohort by adapting to idiosyncratic elements present there. However, the performance of the other models is generally close to that of the cohorts in which the models were derived, while exceeding by a larger degree the comparison standard brain models shown in the bottom rows. Of note is that the reproducibility and R2 fits are higher for the longitudinal (Table 3) than for the baseline models.
Table 3.
Summary of replicability and reproducibility
| Baseline TsROI models across cohorts: adjusted R2 fits to episodic memory baseline | ||||
|---|---|---|---|---|
| Derivation cohort | Num ROIs | Cross-validation: target cohorts |
||
| ADC | ADNI 1 | ADNI2/GO | ||
| ADC | 5 | 0.261 | 0.270 | 0.214 |
| ADNI 1 | 6 | 0.195 | 0.389 | 0.239 |
| ADNI2/GO | 6 | 0.185 | 0.304 | 0.266 |
|
| ||||
|
Longitudinal TsROIs across cohorts: adjusted R2 fits to episodic memory change
| ||||
|
| ||||
| Derivation cohort | Num ROIs |
Cross-validation: target cohorts |
||
|
|
|
ADC |
ADNI 1 |
ADNI2/GO |
| ADC | 7 | 0.376 | 0.368 | 0.394 |
| ADNI 1 | 9 | 0.334 | 0.403 | 0.406 |
| ADNI2/GO | 14 | 0.363 | 0.380 | 0.470 |
Table shows a summary of replicability (rows) and reproducibility (columns). Entries are R2 fits of each derivation cohort in appropriate regression models applied to target cohort. Diagonal entries in bold present performance of models in cohort where derived, for comparison and context. Top: Models of baseline memory by grey matter density. Bottom: Models of longitudinal memory change by atrophy rates. ROI = region of interest.
Comparison with other models
Adjusted R2 fits of all models are graphed in Fig. 3. As expected, each TsROI model performed better than the others in its own cohort. All three models outperformed the alternatives in every test cohort. Alternative comparison model evaluations using ‘multi-region of interest’ independent variables are shown in Supplementary Fig. 1. Visual representations of recent comparison model union signatures are provided in Fig. 2C. These display widely varying extents but a common inclusion of medial and ventral temporal regions.
Figure 3.
Model performances by adjusted R2. Bar graph comparisons of adjusted R2 model performance. Top: For baseline episodic memory. Bottom: For memory change. GM = grey matter; RSq = R2.
Discussion
Summary of method and performance tests
We have described and tested a method to generate signature region of interest models of brain measures for explaining interindividual variance in baseline and longitudinal change in episodic memory, among older adults across the spectrum from normal cognition to Alzheimer’s disease. Our method involved an algorithmic (step-by-step) approach for computing signature regions of interest in an arbitrary cohort. It was followed by rigorous demonstrations of three key performance properties. First, a single model performed at similar levels explaining baseline or longitudinal episodic memory in two cohorts that were separate from the one where it was generated. Second, two such independently created models performed similarly to one another in a third separate cohort. Finally, signature regions of interest performed better than other brain models including both data- and theory-driven models applied in a given cohort.
Relations of signature regions of interest to previous models
Our programme shares an exploratory data-driven approach with several previously described signature region of interest models.12-15,24,27,51 Some of these relied on location-wise searches13,24,51 while the others performed searches for best-performing average summary measures for regions in an atlas of regions of interest.
Striking differences are seen in the performance (adjusted R2 or amount of variance explained) by our multiple independent predictor TsROIs compared to that of single averages over a union of regions in other models (Fig. 3). To our knowledge, the multiple independent predictor approach has not been previously proposed. For completeness, we also computed the performances of other models when their regions of interest are used as independent predictors, with results close to those of our models though generally lower (Supplementary material). That being said, prior regression models using data-driven approaches are generally less efficient, with more independent variables (Supplementary material), suggesting our models may be more economical and informative.
In previous local approaches,13,27,51 locations were selected using an uncorrected probability threshold for significant association of local grey matter measures to outcome. In our approach, voxel measures showing strong association to memory outcome had to pass a further test of membership in contiguous clusters all exceeding specified t-value thresholds, computed by non-parametric correction for multiple comparisons on size of the clusters.44 Our aggregation step thus incorporated a rigorous test for significance based upon cluster size membership that has no counterpart in the previously described signature regions of interest.
Differences aside, it is striking that the union region of interest masks for comparison baseline models14,15 have moderate to strong Dice similarity measures with ours (Table 2), and that those models show ‘multi-region of interest’ performance generally closer to our baseline models than is the theory-driven hippocampal model (Supplementary material). The overlaps of all of these baseline data-driven signatures may suggest a core set of temporal lobe and possibly posterior cingulate/precuneus locations (Fig. 2A and C) underlying memory. Data-driven signatures implemented in different ways may therefore be converging on a consensus of brain regions that optimally model brain association with baseline episodic memory function. By contrast, there were lower Dice similarities of ours versus the competing longitudinal model even as our three longitudinal models were highly similar (Table 2). This suggests an area needing further exploration.
Issues raised by differences in cohorts
Of the cohorts we used to generate signature region of interest models, ADC was ethno-racially diverse, whereas the two ADNI cohorts were not. The ADNI cohorts also focused primarily on Alzheimer’s disease pathology, whereas the ADC also contained extensive occurrence of cerebrovascular disease (white matter hyperintensities). The three cohorts also exhibited varying age and education distributions. To account for these differences, we controlled for age, gender and education. Combining groups having disparate ethno-racial backgrounds has important advantages. It allows identification of brain-cognition relationships that are common across wide variations in ancestry, health, education and social economic status. However, prior work by our group also suggests that ethno-racial differences exist in brain-cognition relations,9 pointing to a fruitful avenue for future research elucidating similarities and differences.
Levels of pathophysiology associated with cognitive impairment may also affect the brain-behaviour signatures. Testing these hypotheses is beyond our current scope but constitutes a promising area of future research based upon our current models. We did not control for or stratify by diagnosis as we were specifically interested in quantifying the relationship of brain measures to cognitive outcome along the entire range of cognitive ability. To achieve broader variance of brain and cognitive measures for our proof-of-concept, we included a mix of diagnoses by design. Subcomponents of the identified regions may have differing effects on memory for individuals with no significant cognitive impairment, MCI or a dementia syndrome, suggesting avenues for future research.
Relations to concepts of cognitive reserve
Results of the current project have potential applications to the concepts of cognitive reserve, brain reserve and brain maintenance. Cognitive reserve is conceived as a factor that actively sustains cognition in the face of brain insults due to ageing or pathology.52 It is invoked to explain discrepancies between actual versus predicted cognition based on available brain variables.53 Brain reserve is conceived as a ‘passive’ factor consisting of brain resources that allow some individuals to maintain normal cognition for longer than others.52,53 For example, greater cortical grey matter thickness may be able to absorb more neuron loss before cognitive impairment becomes measurable. Finally, brain maintenance is the concept of minimal brain change over time supporting minimally declining trajectories of cognitive change.1
Our approach suggests the potential for defining more specific models of brain reserve and brain maintenance than are found in the standard brain models invoked in previous publications. This is because we have found some brain regions that are highly associated with aspects of cognitive performance, while others are not. Therefore, the brain may be able to tolerate more loss in some regions than in others before global or domain-specific cognitive impairment occurs. Our results also have the potential to sharpen cognitive reserve definitions by making clearer the specific brain regions associated with measures of cognition. For example, the reserve-as-residual approach8,54 operationalizes cognitive reserve as the residual of cognitive variance in a model involving brain reserve and demographic factors. However, the residual defining cognitive reserve might contain a mixture of actual cognitive reserve with effects of unaccounted brain reserve. To the extent that our signature model outperforms standard models explaining baseline episodic memory, it could implicitly reduce the overall residual by removing some of those brain effects, thus producing a better or ‘purer’ measure of cognitive reserve itself. A question for future research is how residual measures of cognitive reserve will change (and diminish) when currently unaccounted factors are identified and transferred to brain reserve using the approach outlined here.
Limitations
The signature region of interest approach embodies important strengths with its hypothesis-free voxelwise computation of volumetric brain regions most strongly related to cognitive outcomes. This supports our aim for an accounting of brain resources explaining more memory variance than other models (Fig. 3). Since the signature region of interest depends on native-to-template space deformations, however, it is necessarily subject to the limitations of such deformations. The deformation of some brain regions to template space is inherently noisier than others; hence our approach may have been biased towards prioritizing regions that are more accurately mapped (i.e. with less noise or standard deviation) into template space. Perhaps most salient are difficulties in hippocampal matching; these might be addressed in future modifications by using hybrid models incorporating native-space hippocampal volumes with voxelwise search in template space.
Another concern may be that competing signature models used measures different from ours to generate signature regions. We tested performance using the same measures for all; it is conceivable that using surface-based measures for the competing models would have produced different comparisons. This was beyond the scope of the current project but may make for informative future investigations.
For simplicity, we restricted our focus to brain grey matter measures at baseline and to longitudinal tissue atrophy. Although these explained a substantial amount of variance in episodic memory, eventual structural models more fully accounting for relevant brain attributes to memory should include additional measures, such as white matter structural integrity55,56 and structural connectivity.57 Incorporating a broader array of brain measurements could both increase explanatory power and help to mitigate the differences in fit performances.
Conclusion
In conclusion, we have implemented an algorithmic approach for generating robust signature region of interest models, identifying essential brain regions that support baseline and longitudinal episodic memory in cognitively heterogeneous cohorts. These models outperformed other models previously used, and our approach together with other recent data-driven signature models may signal a convergence towards the maximal amount of memory variance that these brain measures can account for, providing useful biomarkers having clinical and theoretical applications. Our approach needs to be extended by including a more complete array of appropriate brain measurements to more fully explain variation in cognitive performance. But given the unattainability of a complete mapping of cognition and brain, a comprehensive model will probably always need to also consider the judicious inclusion of non-brain factors. We have included age, gender and education here. The consistent accounting for large amounts of cognitive variance in the current models suggests that this approach merits further development.
Supplementary Material
Acknowledgements
We thank the UC Davis ADC and ADNI for making this study possible. We thank the Advanced Psychometrics Methods in Cognitive Aging Research Conference (https://alzheimer.ucdavis.edu/fhpsych/) collaborative groups for invaluable support and advice. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Funding
This work was supported by several grants from the National Institute on Aging. These were: R01 AG 031563, D.M., PI; P30 AG10129 and R01 AG047827, C.D., PI.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- ADC =
UC Davis Aging and Diversity Cohort
- ADNI =
Alzheimer’s Disease Neuroimaging Initiative
- MCI =
mild cognitive impairment
- TsROI =
t-value signature association region of interest
References
- 1. Nyberg L, Lövdén M, Riklund K, Lindenberger U, Bäckman L.. Memory aging and brain maintenance. Trends Cogn Sci. 2012;16:292-305. [DOI] [PubMed] [Google Scholar]
- 2. Head D, Kennedy KM, Rodrigue KM, Raz N.. Age differences in perseveration: cognitive and neuroanatomical mediators of performance on the Wisconsin Card Sorting Test. Neuropsychologia. 2009;47:1200-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ranganath C. A unified framework for the functional organization of the medial temporal lobes and the phenomenology of episodic memory. Hippocampus. 2010;20:1263-1290. [DOI] [PubMed] [Google Scholar]
- 4. Rodrigue KM, Raz N.. Shrinkage of the entorhinal cortex over five years predicts memory performance in healthy adults. J. Neurosci. 2004;24:956-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murphy EA, , Holland D, , Donohue M, et al. Six-month atrophy in MTL structures is associated with subsequent memory decline in elderly controls. Neuroimage. 2010;53:1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mungas D, Harvey D, Reed BR, et al. Longitudinal volumetric MRI change and rate of cognitive decline. Neurology. 2005;65:565-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fletcher E, Gavett B, Harvey D, et al. Brain volume change and cognitive trajectories in aging. Neuropsychology. 2018;32:436-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reed BR, Mungas D, Farias ST, et al. Measuring cognitive reserve based on the decomposition of episodic memory variance. Brain. 2010;133:2196–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gavett BE, Fletcher E, Harvey D, et al. Ethnoracial differences in brain structure change and cognitive change. Neuropsychology. 2018;32:529-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bettcher BM, Gross AL, Gavett BE, et al. Dynamic change of cognitive reserve: associations with changes in brain, cognition, and diagnosis. Neurobiol Aging. 2019;83:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cuingnet R, Gerardin E, Tessieras J, et al. Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage. 2011;56:766-781. [DOI] [PubMed] [Google Scholar]
- 12. Bakkour A, Morris JC, Dickerson BC.. The cortical signature of prodromal AD: regional thinning predicts mild AD dementia. Neurology. 2009;72:1048-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dickerson BC, Bakkour A, Salat DH, et al. The cortical signature of Alzheimer’s disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb. Cortex. 2009;19:497-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwarz CG, Gunter JL, Wiste HJ, et al. A large-scale comparison of cortical thickness and volume methods for measuring Alzheimer s disease severity. NeuroImage Clin. 2016;11:802-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Epelbaum S, Bouteloup V, Mangin JF, et al. Memento Study group, et al. Neural correlates of episodic memory in the Memento cohort. Alzheimer’s Dement. Transl Res Clin Interv. 2018;4:224-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rondina JM, Ferreira LK, de Souza Duran FL, et al. Selecting the most relevant brain regions to discriminate Alzheimer’s disease patients from healthy controls using multiple kernel learning: A comparison across functional and structural imaging modalities and atlases. NeuroImage Clin. 2018;17:628-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fan Y, Shen D, Davatzikos C. Classification of structural images via high-dimensional image warping, robust feature extraction, and SVM. In: Duncan JS, Gerig G, eds. Lecture notes in computer science. Springer, Berlin, Heidelberg;2005:1-8. [DOI] [PubMed] [Google Scholar]
- 18. Fan Y, Shen D, Gur RC, Gur RE, Davatzikos C.. COMPARE: classification of morphological patterns using adaptive regional elements. IEEE Trans Med Imaging. 2007;26:93-105. [DOI] [PubMed] [Google Scholar]
- 19. Klöppel S, Stonnington CM, Chu C, et al. Automatic classification of MR scans in Alzheimer’s disease. Brain. 2008;131:681-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Plant C, Teipel SJ, Oswald A, et al. Automated detection of brain atrophy patterns based on MRI for the prediction of Alzheimer’s disease. Neuroimage. 2010;50:162-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clark VH, Resnick SM, Doshi J, et al. Longitudinal imaging pattern analysis (SPARE-CD index) detects early structural and functional changes before cognitive decline in healthy older adults. Neurobiol Aging. 2012;33:2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stonnington CM, Chu C, Klöppel S, et al. Predicting clinical scores from magnetic resonance scans in Alzheimer’s disease. Neuroimage. 2010;51:1405-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y, Goh JO, Resnick SM, Davatzikos C.. Imaging-based biomarkers of cognitive performance in older adults constructed via high-dimensional pattern regression applied to MRI and PET. PLoS One. 2013;8:e85460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Araque Caballero MÁ, Klöppel S, Dichgans M, Ewers M; Alzheimer☳ Disease Neuroimaging Initiative. For the Alzheimer’s Disease Neuroimaging Initiative. Spatial patterns of longitudinal gray matter change as predictors of concurrent cognitive decline in amyloid positive healthy subjects. J Alzheimers Dis. 2016;55:343-358. [DOI] [PubMed] [Google Scholar]
- 25. Franke K, Ziegler G, Klöppel S, Gaser C; Alzheimer's Disease Neuroimaging Initiative. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage. 2010;50:883-892. [DOI] [PubMed] [Google Scholar]
- 26. Luders E, Cherbuin N, Gaser C.. Estimating brain age using high-resolution pattern recognition: younger brains in long-term meditation practitioners. Neuroimage. 2016;134:508-513. [DOI] [PubMed] [Google Scholar]
- 27. Busovaca E, Zimmerman ME, Meier IB, et al. Is the Alzheimer’s disease cortical thickness signature a biological marker for memory? Brain Imaging Behav. 2016;10:517-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Desikan RS, Ségonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968-980. [DOI] [PubMed] [Google Scholar]
- 29. Mungas D, Reed BR, Crane PK, Haan MN, González H.. Spanish and English Neuropsychological Assessment Scales (SENAS): further development and psychometric characteristics. Psychol Assess. 2004;16:347-359. [DOI] [PubMed] [Google Scholar]
- 30. Mungas D, Reed BR, Tomaszewski Farias S, DeCarli C.. Criterion-referenced validity of a neuropsychological test battery: equivalent performance in elderly Hispanics and non-Hispanic Whites. J Int Neuropsychol Soc. 2005;11:620-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crane PK, Carle A, Gibbons LE, et al. For the Alzheimer’s Disease Neuroimaging Initiative, et al. Development and assessment of a composite score for memory in the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Brain Imaging Behav. 2012;6:502-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mungas D, Crane PK, Gibbons LE, Manly JJ, Glymour MM, Jones RN.. Advanced psychometric analysis and the Alzheimer’s Disease Neuroimaging Initiative: reports from the 2011 Friday Harbor conference. Brain Imaging Behav. 2012;6:485-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fletcher E, Villeneuve S, Maillard P, et al. β-amyloid, hippocampal atrophy and their relation to longitudinal brain change in cognitively normal individuals. Neurobiol Aging. 2016;40:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jack CR JrBarnes J, Bernstein MA, et al. Magnetic resonance imaging in Alzheimer’s Disease Neuroimaging Initiative 2. Alzheimers Dement. 2015;11:740-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fletcher E, Carmichael O, Pasternak O, Maier-Hein KH, DeCarli C.. Early brain loss in circuits affected by alzheimer’s disease is predicted by fornix microstructure but may be independent of gray matter. Front Aging Neurosci. 2014;6:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rueckert D, Aljabar P, Heckemann RA, Hajnal JV, Hammers A.. Diffeomorphic registration using B-splines. Med Image Comput Comput Assist Interv. 2006;702-709. [DOI] [PubMed] [Google Scholar]
- 37. Kochunov P, Lancaster JL, Thompson P, et al. Regional spatial normalization: toward and optimal target. J Comput Assist Tomogr. 2001;25:805-816. [DOI] [PubMed] [Google Scholar]
- 38. Das SR, Avants BB, Grossman M, Gee JC.. Registration based cortical thickness measurement. Neuroimage. 2009;45:867-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fischl B. FreeSurfer. Neuroimage. 2012;62:774-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fischl B, Dale AM.. Measuring the thickness of the human cerebral cortex. Proc Natl Acad Sci USA. 2000;97:11050-11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tustison NJ, Cook P. A, Klein A, et al. Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. Neuroimage. 2014;99:166-179. [DOI] [PubMed] [Google Scholar]
- 42. Fletcher E, Knaack A, Singh B, et al. Combining boundary-based methods with tensor-based morphometry in the measurement of longitudinal brain change. IEEE Trans Med Imaging. 2013;32:223-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fletcher E. Using prior information to enhance sensitivity of longitudinal brain change computation. In: CH Chen, ed. Frontiers of medical imaging. World Scientific;2014:63-81. [Google Scholar]
- 44. Nichols T, Holmes AP.. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15:1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cohen AL, Fair DA, Dosenbach NUF, et al. Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage. 2008;41:45-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bakkour A, Morris JC, Wolk D. A, Dickerson BC.. The effects of aging and Alzheimer’s disease on cerebral cortical anatomy: specificity and differential relationships with cognition. Neuroimage. 2013;76:332-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26:297-302. [Google Scholar]
- 48. Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:280-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Klein A, Tourville J.. 101 labeled brain images and a consistent human cortical labeling protocol. Front Neurosci. 2012;6:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Klein A, Ghosh SS, Bao FS, et al. Mindboggling morphometry of human brains. PLoS Comput Biol. 2017;13:e1005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gross AL, Manly JJ, Pa J, et al. Cortical signatures of cognition and their relationship to Alzheimer’s disease. Brain Imaging Behav. 2012;6:584-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stern Y. What is cognitive reserve? Theory and research application of the reserve concept. J Int Neuropsychol Soc. 2002;8:448-460. [PubMed] [Google Scholar]
- 53. Stern Y, Arenaza-Urquijo EM, Bartrés-Faz D, et al. Whitepaper: defining and investigating cognitive reserve, brain reserve, and brain maintenance. Alzheimers Dement. 2018;16:1305-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zahodne LB, Manly JJ, Brickman AM, Siedlecki KL, Decarli C, Stern Y.. Quantifying cognitive reserve in older adults by decomposing episodic memory variance: replication and extension. J Int Neuropsychol Soc. 2013;19:854-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lockhart SN, Mayda ABV, Roach AE, et al. Episodic memory function is associated with multiple measures of white matter integrity in cognitive aging. Front Hum Neurosci. 2012;6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Seiler S, Fletcher E, Hassan-Ali K, et al. Cerebral tract integrity relates to white matter hyperintensities, cortex volume, and cognition. Neurobiol Aging. 2018;72:14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fjell AM, Sneve MH, Storsve AB, Grydeland H, Yendiki A, Walhovd KB.. Brain events underlying episodic memory changes in aging: a longitudinal investigation of structural and functional connectivity. Cereb Cortex. 2016;26:1272–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Derived data supporting the findings of this study, and resulting signature region of interest masks, are available from the corresponding author upon reasonable request.