Abstract
Purpose
Cystic fibrosis (CF), caused by pathogenic variants in the CF transmembrane conductance regulator (CFTR), affects multiple organs including the exocrine pancreas, which is a causal contributor to cystic fibrosis–related diabetes (CFRD). Untreated CFRD causes increased CF-related mortality whereas early detection can improve outcomes.
Methods
Using genetic and easily accessible clinical measures available at birth, we constructed a CFRD prediction model using the Canadian CF Gene Modifier Study (CGS; n = 1,958) and validated it in the French CF Gene Modifier Study (FGMS; n = 1,003). We investigated genetic variants shown to associate with CF disease severity across multiple organs in genome-wide association studies.
Results
The strongest predictors included sex, CFTR severity score, and several genetic variants including one annotated to PRSS1, which encodes cationic trypsinogen. The final model defined in the CGS shows excellent agreement when validated on the FGMS, and the risk classifier shows slightly better performance at predicting CFRD risk later in life in both studies.
Conclusion
We demonstrated clinical utility by comparing CFRD prevalence rates between the top 10% of individuals with the highest risk and the bottom 10% with the lowest risk. A web-based application was developed to provide practitioners with patient-specific CFRD risk to guide CFRD monitoring and treatment.
INTRODUCTION
Genome-wide association studies (GWAS) have been successful at identifying genetic contributors to disease,1 however, clinical utility of GWAS findings has been slow to follow. One explanation is that the genetic architecture of complex phenotypes is multifaceted and individual GWAS findings have small effect sizes that limit their potential alone as predictors of disease.2 Although GWAS has provided us with important mechanistic insight into disease, further defining genetic markers for risk prediction could have significant impact on personalized medicine. Here, we investigate genomic-based risk prediction for cystic fibrosis–related diabetes (CFRD).
Cystic fibrosis (CF) is a life-limiting genetic disease caused by loss-of-function pathogenic variants in the cystic fibrosis transmembrane conductance regulator (CFTR) and affects multiple organs including the exocrine pancreas. Pancreatic damage and the resulting exocrine pancreatic insufficiency (PI) contribute to CFRD,3 which is seen in 19% of adolescents and 40~50% of CF individuals by age 40.4 CFRD is associated with increased morbidity due to worsening lung and nutritional status, which often precedes CFRD diagnosis, and increased mortality if CFRD remains untreated.4 Early identification could improve clinical outcomes and reduce mortality.5 Current guidelines recommend annual CFRD screening with 2-hour oral glucose tolerance testing (OGTT) after 10 years of age; however, there is poor adherence with screening rates reported below 50%.6 Identifying individuals at greatest risk of developing CFRD as early as possible could improve adherence.
CFRD occurs predominantly in individuals with severe CFTR pathogenic variants that result in PI.7 Thus, currently the best predictor of CFRD risk is whether an individual has CFTR pathogenic variants associated with PI (85% of the CF population8); however, we expect variation in risk even within individuals that are PI. In addition to the CFTR contribution, GWAS has identified genetic modifiers of CFRD at SLC26A9 and several established type 2 diabetes susceptibility loci.9,10 Consistent with PI CFTR variants’ elevating risk for CFRD, recent studies have suggested a major cause of CFRD to be prenatal and early postnatal damage to the exocrine pancreas.3 The degree of pancreatic damage and reduction in acinar tissue are reflected by circulating immunoreactive trypsinogen (IRT), which is partially encoded by serine protease 1 (PRSS1). Newborn-screened (NBS) IRT and its longitudinal measures in the first 2 years of life have been shown to associate with CFRD risk in two independent samples.3 However, routine longitudinal measurement of IRT is not standard of care for young CF individuals and is unavailable for older CF individuals who were diagnosed later in life but are at greatest CFRD risk today. Therefore, this study aims to identify biomarkers that can predict CFRD onset using genetic and easily accessible clinical measures early in life. With the Canadian CF Gene Modifier study (CGS), we developed a prediction model to identify individuals at highest risk of CFRD at different ages and validated our prediction in an independent CF cohort from France.
MATERIALS AND METHODS
Demographics, genotyping, and phenotyping
Two independent population-based cohorts were included in this study: the CGS (n = 1,958) and the French CF Gene Modifier Study (FGMS, n = 1,003). CGS was used to develop the predictive model while FGMS was used to validate the predictions. Ninety-seven percent of the CGS participants included in this study were diagnosed by characteristic clinical manifestations of CF and subsequently genotyped on genome-wide Illumina microarrays.11 We included 1,958 individuals from the CGS who have CFTR variants associated with PI or have a CFTR genotype carried by individuals diagnosed with CFRD in the CGS. Specifically, CFRD was seen in CGS participants who had a PI pathogenic variant and one of the following “mild” CFTR alleles: 2789+5G>A, A455E, G85E, and IVS8(5T). Thus, we included ten individuals without a CFRD diagnosis but with these same CFTR genotypes.
Recorded clinical measures available early in life included sex, body mass index (BMI), and meconium ileus (MI), an intestinal obstruction at birth found in ~15% of CF individuals. Although BMI was shown to associate with type 2 diabetes in the general population,12 we did not find time-varying BMI to be a strong predictor of future CFRD risk and we removed it from the analyses.
Dramatic improvements in median survival over the last few decades13 have been met with increased rates of CFRD diagnosis that previously did not have time to manifest or went undetected. The first consensus guidelines for CFRD screening were not established until 1990.14 Therefore, CF individuals born before 1970 were not subject to uniform CFRD screening during adolescence. Not surprisingly, we discovered significant cohort effects within the CGS and FGMS data sets in which different generations of CF individuals have different CFRD prevalence rates. To account for these differences, we defined cohort based on the decade in which an individual was born and adjusted for cohort effects when constructing the prediction model. For instance, individuals born in the 1970s or the 1980s were grouped into separate cohorts. Moreover, we excluded French and Canadian participants born before 1970 for all subsequent analyses.
In CF, the standard of care is to employ annual OGTT testing to conclude the presence of CFRD, but there is poor adherence to this time-consuming test that requires an overnight fast.15 In the CGS, CFRD status was determined using a combination of chart review and the Canadian CF patient registry.9 Patients diagnosed with CFRD had a physician’s diagnosis, were not reported to have type 1 or type 2 diabetes (T1DM; T2DM), and satisfied one of the following:
Daily treatment with insulin or oral diabetes medication
2-hour glucose level exceeding 11.1 mmol/L (200 mg/dL) during OGTT
HbA1c of at least 7%
Individuals without CFRD were censored at the last clinic visit or year of organ transplant. Individuals with post-transplant diabetes, gestational diabetes, and steroid-induced diabetes were removed from analysis.
In the FGMS, CF individuals were recruited from 48 French CF centers. Inclusion and diagnostic criteria used in the FGMS were the same as defined in the CGS. Genotyping design was reported previously.11
The two cohorts did not differ by sex or MI prevalence (Table 1). However, CF individuals in the CGS were slightly older than the FGMS participants. Given that CFTR pathogenic variants are indicators of exocrine pancreatic disease severity,16 we constructed a CFTR severity score based on the combination of CFTR pathogenic variants from both alleles, with details provided in Appendix A.
Table 1.
Characteristics of cystic fibrosis (CF) individuals across the discovery (Canadian GMS; CGS) and the validation (French GMS; FGMS) data set.
| Variable | Canadian GMS (n = 1,958) | French GMS (n = 1,003) |
|---|---|---|
| CFRD (cases) | 619 (31.6%) | 374 (37.3%) |
| Sex (females) | 926 (47.3%) | 480 (47.9%) |
| Meconium ileus | 334 (17.1%) | 141 (14.1%) |
| Newborn screened | 58 (3.0%) | 415 (42.5%)a |
| CFTRvariant score | ||
| 5 | 51 (2.6%) | 14 (1.4%) |
| 4 | 389 (19.9%) | 201 (20.0%) |
| 3 | 1185 (60.5%) | 667 (66.5%) |
| 2 | 170 (8.7%) | 68 (6.8%) |
| 1 | 163 (8.3%) | 53 (5.3%) |
| Age cohort (year of birth) | ||
| 1970s | 336 (17.2%) | 128 (12.8%) |
| 1980s | 634 (32.4%) | 317 (31.6%) |
| 1990s | 737 (37.6%) | 392 (39.1%) |
| After 2000 | 251 (12.8%) | 166 (16.6%) |
Individuals enrolled in the FGMS are less likely to carry a mild CFTR pathogenic variant compared with participants in the CGS.
CFRD cystic fibrosis–related diabetes, GMS Gene Modifier Study.
aTwenty-seven French GMS individuals were missing information for newborn screening. A higher proportion of French individuals were newborn screened since nationwide newborn screening was implemented in France in 200237, earlier than that in all Canadian provinces and territories.
For the predictive model we evaluated a set of 3,984 single-nucleotide polymorphisms (SNPs) that were annotated to genes previously identified as CF modifiers. These included genes that code for proteins residing at the apical plasma membrane alongside CFTR;17,18 variants identified as genetic modifiers of CFRD9 or SNPs associated with other common CF comorbidities including MI11 and lung function decline.19
To address the potential for population stratification in the CGS training data, we used KING20 to perform principal component analysis (PCA). SNPs with minor allele frequency greater than 0.05 and with low pairwise linkage disequilibrium (r2 < 0.2) were included. The Tracy–Widom test determined that ten principal components (PCs) were statistically significant (p < 0.01) in the CGS and were incorporated as predictors in feature selection and model fitting (Appendix B). The lack of differences in model performance with and without adjustment for the PCs (Appendix C) suggests limited confounding due to population structure in the CGS. Moreover, both studies are ethnically homogeneous (>94% Europeans) with non-Europeans defined as >3 SD from the center of the 1000 Genomes European cluster (Appendix D).
The variables included in model training consisted of the 3,984 preselected SNPs, MI, sex, CFTR severity score, and the first ten PCs.
Developing risk scores for CFRD
With the goal of predicting CFRD, all 1,958 individuals in the CGS were included to construct a prediction model that was then validated on the independent FGMS cohort (n = 1,003). To compare model performance across the two independent studies, we performed internal cross-validation within the CGS to reduce overfitting. Since using a single pair of training and validation sets can produce overly optimistic results, we randomly partitioned 1,958 participants into a training (n = 1,300) and a validation set (n = 658) and repeated this partition 500 times. Model fitting was based solely on the training sets while the validation sets were used to assess model performance. We also calculated 95% confidence intervals (CI) for predictive accuracy at specified ages.
CFRD risk was modeled in a three-stage approach: (1) hierarchical clustering to remove highly correlated SNPs; (2) stability selection21 and component-wise gradient boosting22 to rank variable importance by their selected frequencies, with a 50% cutoff used to select predictors most strongly associated with CFRD risk; and (3) Cox proportional hazards (Cox PH) model was used to re-estimate overpenalized effect sizes23 (Appendix J).
We compared our three-stage approach to a univariate, pruning, and thresholding polygenic risk score (PRS) analysis24,25 with different p value cutoffs (0.05, 0.01, 0.005, 0.001, 0.0005, 0.0001; Appendix F). The PRS analysis included CFTR severity score, ten PCs, and the clinical variables sex, MI, and cohort to ensure a fair comparison.
Evaluating CFRD risk scores
Time-dependent area under the curve (AUC(t)) was evaluated to compare model performance and its change over time. Given the paucity of CFRD events at early ages, we investigated our model’s capability to accurately predict CFRD risk between 15 and 35 years, with emphasis on early detection. The AUC(t) curves were plotted for both CGS and FGMS cohorts, to compare performance between the studies.
We calculated age-dependent positive predictive values (PPV) and negative predictive values (NPV) using different CFRD risk score thresholds (Appendix H). This provides a comprehensive display of model performance with flexibility in modifying risk thresholds for CFRD screening. To assess model performance using a more clinically relevant measure, we compared CFRD prevalence rates among individuals with the highest and the lowest 10% risk. Since individuals at the tails of the risk distribution are most affected by clinical decisions,26 clinicians could emphasize the need for more frequent OGTT testing for the high-risk individuals.
RESULTS
We calculated the CFRD-free probabilities and their 95% CIs at different ages for Canadians (CGS) with different CFTR severity scores (Appendix E). CFRD-free probabilities for individuals with the least severe CFTR score (Supplementary Fig. 4, red curve) are higher than the other groups across all ages. In contrast, CFRD-free probabilities for individuals with other CFTR scores either overlap extensively (scores 2 to 4) or cannot be reliably estimated due to the smaller sample size (score 5). To avoid excess uncertainty in the fitted model, we dichotomized CFTR scores into a high (scores 2 to 5) and a low (score 1) group for all subsequent analyses rather than using an ordinal scale; this choice had little impact on the final model performance (Appendix G).
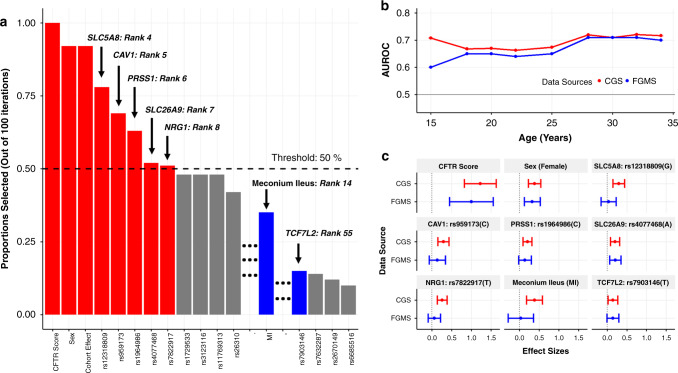
We ranked variable importance by stability selection using all individuals in the CGS (Fig. 1a). Eight variables exceed the 50% threshold (red, Fig. 1a). The CFTR severity score is by far the strongest predictor (hazard ratio [HR] 95% CI: [2.01, 4.54]), selected in 100% of the stability selection subsets. Sex and cohort effect are the second and third most important variables for predicting CFRD risk, both chosen in 92% of the subsets. SNPs annotated to genes that contribute to exocrine pancreatic disease severity are also ranked highly as predictors including rs4077468 annotated to the previously identified MI and CFRD modifier SLC26A9 (HR 95% CI: [1.07,1.34]) and rs1964986 annotated to PRSS1 (HR 95% CI: [1.09,1.38]). PRSS1 encodes cationic trypsinogen and had not been reported to associate with CFRD, although it has been previously associated with MI in CF.11
Fig. 1. Feature selection and model performance for the cystic fibrosis–related diabetes (CFRD) prediction model.
(a) Stability selection and component-wise gradient boosting with 100 iterations. Black dashed line: predefined threshold at 50% of iterations. Red: predictors exceeding stability selection threshold. Blue: meconium ileus (MI) and rs7903146 (TCF7L2), previously shown to be associated with immunoreactive trypsinogen (IRT) at birth and type 2 diabetes, respectively, ranked highly among the predictors. Over 96% of the 2,488 predictors were chosen in <10% of the 100 iterations; they are not shown. (b) Model performance in the Canadian CF Gene Modifier Study (CGS) and French CF Gene Modifier Study (FGMS) calculated by area under the receiver operating characteristic curve (AUROC) as a function of age in years. Model was trained and internally cross-validated in the CGS and externally validated in the FGMS cohort. The 95% confidence intervals of the average AUC(t) are shown in the CGS through bars. (c) Forest plots depicting univariate log hazard ratios estimated from the CGS and FGMS studies. The vertical dotted line represents a log hazard ratio equal to 0.
In addition to the predictors that exceed the predefined threshold (Fig. 1a, red), we further included known CFRD risk factors or confounders to construct the final prediction model. These include the ten PCs to adjust for population structure; rs7903146 (TCF7L2; Fig. 1a, blue), an established type 2 diabetes gene27 that was ranked highly among the predictors even if it did not exceed the 50% threshold; and another highly ranked predictor, MI (Fig. 1a, rank 14, blue). MI is also correlated with exocrine pancreatic disease severity3,11 and was previously shown to be a marker of the known but not widely measured CFRD risk factor, NBS IRT.11 Although MI is associated with exocrine pancreatic disease severity, it remains associated with increased CFRD risk after adjusting for CFTR severity score in our model. Both MI and rs7903146 surpassed the majority of the SNPs not shown in the figure as greater than 96% of the SNPs evaluated were selected in less than 10% of the iterations.
Table 2 lists the HRs and the corresponding 95% CIs fitted in a multivariate Cox PH model after adjusting for cohort effects and the 10 PCs in the CGS. The risk allele or risk group is noted in parentheses. As expected, CF individuals carrying more severe pathogenic variants (higher CFTR scores) have much higher risk of CFRD. Females and individuals born with MI also exhibit higher CFRD risk. For the SLC26A9 SNP rs4077468, the A allele is associated with increased CFRD risk while CF individuals carrying the T allele at rs7903146 also show greater susceptibility to CFRD. The results indicate both genetic and clinical characteristics contribute to CFRD risk, with genotype information beyond CFTR improving the model’s explained variation in CFRD risk from 12% to 18% in the CGS.
Table 2.
Effect sizes (hazard ratios) and the 95% confidence intervals fitted using a multivariate Cox proportional hazard (PH) model in the CGS.
| Gene annotation | Predictor | Hazard ratio | 95% CI |
|---|---|---|---|
| CFTR | CFTR variant score | 3.02 | (2.01, 4.54) |
| – | Sex (female) | 1.48 | (1.26, 1.74) |
| SLC5A8 | rs12318809 (G) | 1.35 | (1.16, 1.57) |
| CAV1 | rs959173(C) | 1.27 | (1.10, 1.47) |
| PRSS1 | rs1964986(C) | 1.23 | (1.09, 1.38) |
| SLC26A9 | rs4077468 (A) | 1.20 | (1.07, 1.34) |
| NRG1 | rs7822917 (T) | 1.31 | (1.16, 1.48) |
| – | Meconium ileus (MI) | 1.29 | (1.05, 1.59) |
| TCF7L2 | rs7903146 (T) | 1.18 | (1.05, 1.34) |
CGS Canadian Cystic Fibrosis Gene Modifier Study, CI confidence interval.
Risk allele/risk group noted in parentheses after the listed predictor.
Fig. 1b shows the time-dependent accuracy measure, AUC(t), for CGS and FGMS. The age-dependent model defined in the CGS shows excellent agreement when validated in the FGMS, demonstrating that our approach has selected stable predictors generalizable to other populations. The risk classifier also shows slightly better performance at predicting CFRD risk later in life (e.g., AUC = 0.71, age = 28 in FGMS) in both study cohorts. Of note, our model outperforms univariate PRS regardless of the chosen p value cutoff (Appendix F).
To further investigate model performance between CGS and FGMS, we plotted univariate log HR and the 95% CI for each selected predictor (Fig. 1c). Increase in CFRD risk for females and those with at least one copy of the type 2 diabetes risk allele (rs7903146[T]) show good agreement in both studies. Those with at least one copy of the PRSS1 (rs1964986(C)) and those with at least one copy of the SLC26A9 risk variant (rs4077468[A]) also show similar increases in CFRD risk in both independent data sets. However, several predictors including MI, the variants rs12318809 (SLC5A8), rs7822917 (NRG1), and rs959173 (CAV1) have much weaker effects in the FGMS. The effect size of the CFTR score is comparable in the FGMS and CGS, albeit with a wider CI for the FGMS since relatively fewer individuals carry mild CFTR pathogenic variants in the FGMS. Wider CIs can also be observed for other predictors due to a smaller sample size in FGMS. Consequently, the ability of our model to stratify CFRD risk based on the CFTR score may be underutilized in the FGMS and leads to underestimated performance at younger ages. Winner’s curse, in which the associations of selected predictors in the training data set are more likely to be overestimated, might also be a contributing factor.28
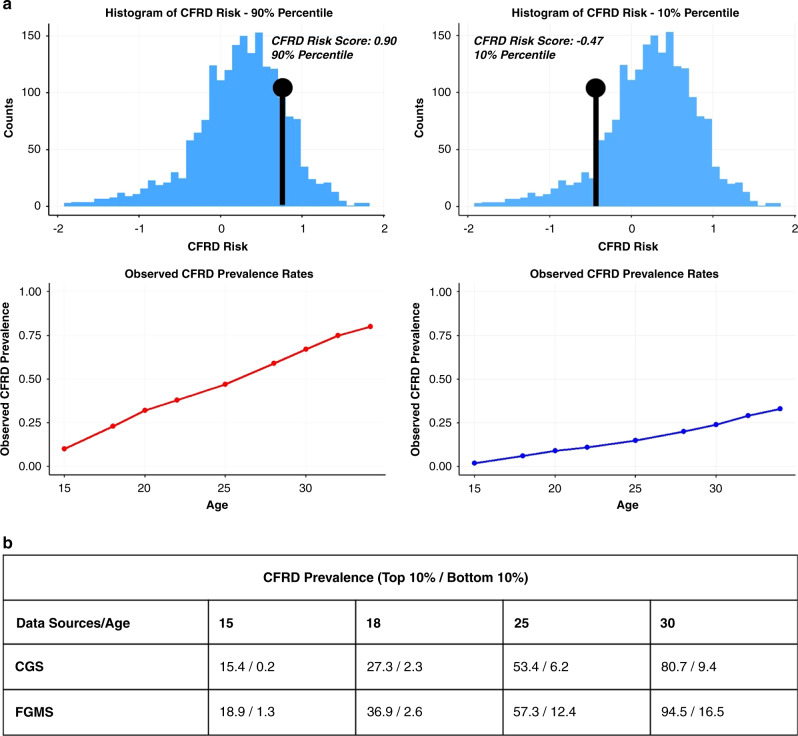
Since AUC(t) only measures a model’s ability to rank individuals based on their estimated risk, we further evaluated a more clinically relevant metric by comparing CFRD prevalence rates between individuals with the highest and lowest 10% risk. Figure 2b shows the CFRD prevalence rates at specified ages for both independent cohorts. Individuals with the highest/lowest CFRD risk in the FGMS were identified using the model trained on the CGS, while internal validation was used for assessing CFRD prevalence in the CGS. At age 18, 37% of the highest-risk individuals would have developed CFRD in FGMS, compared with less than 3% among the lowest-risk individuals. At age 25, 53% of the highest-risk individuals would have developed CFRD in CGS, compared to 6% of the lowest-risk individuals. In both data sets, the highest-risk individuals have much higher CFRD prevalence rates than the lowest-risk individuals. Age-dependent PPVs and NPVs (Appendix H) further demonstrate successful differentiation between high-risk and low-risk individuals across a wider range of risk scores. Using a 70% cutoff (Supplementary Fig. 7, dark blue, PPV), we expect >80% of individuals with the highest estimated risk (top 30%) to be diagnosed with CFRD by their early 30s. Similarly, the model also demonstrates considerable differentiation for the NPVs between individuals with varying CFRD risk (Supplementary Fig. 7).
Fig. 2. Cystic fibrosis–related diabetes (CFRD) prediction model stratifies high-risk and low-risk individuals.
(a) Web-based application for clinical use. The percentile of a CF individual’s estimated CFRD risk and the observed CFRD prevalence rates across ages are returned to facilitate downstream clinical decision making. The figure showcases a high-risk individual with CFRD score in the 90th percentile, and another low-risk individual with CFRD score in the 10th percentile. (b) CFRD prevalence (top 10%/bottom 10%) at different ages for both independent data sets. Prevalence for individuals with the highest and lowest 10% CFRD risk scores are listed. CGS Canadian CF Gene Modifier Study, FGMS French CF Gene Modifier Study.
To facilitate clinical use of the model, we have developed an application (https://predictcfrd.research.sickkids.ca/) that allows users to enter their genetic and clinical measurements and returns the estimated age-dependent CFRD risk (Appendix I). Fig. 2a demonstrates the information returned for CF individuals with different estimated risk. For a CF individual with a risk score of 0.90, which falls in the 90th percentile of the risk distribution, observed CFRD prevalence rates (Fig. 2a, left) demonstrate that ~10% of individuals in this percentile will be diagnosed with CFRD by the age of 15 and nearly 50% by the age of 25. Conversely, we expect <15% of individuals that fall in the 10th percentile of risk (Fig. 2a, right) to be diagnosed with CFRD by their mid-20s.
DISCUSSION
We developed a model to estimate an individual’s CFRD risk using genetic and clinical measures available at birth. The final model can differentiate individuals with varying CFRD risk with reasonable accuracy across different ages. The selected variables that are among the strongest predictors of CFRD risk—CFTR severity score, MI, and the genetic variants annotated to PRSS1 and SLC26A9—suggest that measures of exocrine pancreatic disease severity are major predictors of CFRD. These results are supported by findings from earlier studies that showed increased risk in those born with MI,9 and that SNPs annotated to SLC26A9 are associated with CFRD9 through their impact on exocrine pancreatic damage.3,11 The SLC26A9 variant (rs4077468) and MI were shown to associate with CFRD in a previous study using partially overlapping individuals from the CGS.9 However, the results were confirmed in our study using 555 (28%) new participants from the CGS and an independent French population cohort (FGMS) not included in the initial study.9 Investigating other factors independent of those associated with exocrine pancreatic damage, we found that females exhibit higher CFRD risk, consistent with previous findings;7,29 and the type 2 diabetes gene, TCF7L2, also ranks highly among the predictors.
Our application (https://predictcfrd.research.sickkids.ca/) can assist clinicians in determining an individual’s CFRD risk across the age spectrum from measures obtained one time as early as birth. The Cystic Fibrosis Foundation recommends universal annual screening for CFRD. Findings here should not impact the recommended annual screening, even for those predicted to have the lowest risk, as less frequent monitoring would likely have a negative impact, regardless of risk category. Poor adherence to annual screening has, however, hindered its efficacy. Providing a percentile of an individual’s risk estimate and the CFRD prevalence rates across ages would highlight individuals at greater risk earlier in their disease course and could motivate improved adherence to regular OGTT measurements, or perhaps greater frequency, for the high-risk subgroup at the discretion of their care provider.
We compared CFRD prevalence between individuals with the highest and lowest 10% risk since those at the tails of the risk distribution are most affected by clinical decision making.26 The model is capable of identifying individuals most susceptible to CFRD at different ages while maintaining a reliable estimation for those at low risk. In addition to age-distributed CFRD prevalence rates for each CF individual, age-dependent PPVs and NPVs using different thresholds for the CFRD high-risk category (Appendix H) serve to showcase the efficacy of the model and provide additional information to facilitate clinical decision making. Moreover, the results also demonstrate the benefit of genotyping modifiers in addition to the CFTR common causal variants in newborn screening programs, as incorporating modifier genotype information in addition to CFTR and clinical measurements (e.g., sex, MI, cohort) significantly increased the explained variation in CFRD risk (12% to 18%) in the CGS.
Despite taking extra precautions to avoid overfitting in our training data, winner’s curse might still contribute to overestimated effect sizes and lead to predictors being less robust in the validation cohort.30 The comparable predictive performance between the CGS and FGMS, however, provides some reassurance that our model is capturing a robust component of the genetic predisposition to CFRD. Moreover, by leveraging both Canadian and French cohorts, we provide further assurance that our model can be generalized outside of the population on which it was trained.31
In both the CGS and FGMS, the CFRD diagnosis data came from individual physicians. As most diagnoses are supported with OGTT, we do not expect significant impact from adopting a 7% cutoff for HbA1c compared with the general guideline of 6.5%.4 However, it is plausible that the use of a higher HbA1c cutoff in this study resulted in underdiagnosis in our analyzed cohorts. Moreover, although CFRD presents differently than T1DM, and T1DM and other forms such as maturity onset diabetes of the young (MODY) are rare in CF, it is possible that a small number of individuals may have been misrepresented as having CFRD.
We note a few limitations of this study, especially for the model’s use in clinical settings. The tool is designed to serve as an additional piece of information to enhance clinical care for CFRD and requires discretion by the clinical care provider to dichotomize CF individuals into high and low-risk groups based on the reported age-distributed prevalence. The CF gene modifiers are not routinely genotyped on CFTR diagnostic panels, and this change is needed to enable clinical use. The proposed model is constructed from measures obtained one time, as early as birth, and does not update risk predictions based on a patient’s current age or other longitudinal factors. Although a conditional risk model would be of interest, given the limited sample size and the corresponding stability of the model, we chose to focus on leveraging genetic and clinical measurements available at birth to emphasize early detection.
Although the model shows clinically relevant performance in stratifying CFRD risk among individuals in the Canadian and French studies, its clinical utility for future CF individuals relies upon the assumption that the CFRD diagnosis guidelines and prevalence remain static. Highly effective CFTR modulators could potentially affect the natural history of CFRD and reduce its prevalence in the modulator-treated population,32 although the impact of current therapies on pancreatic morbidity in CF remains unknown.33 Trikafta™ has been approved for 90% of CF individuals, yet variability in its effectiveness has been reported.34 Moreover, it remains unavailable in many countries including Canada. Clinical utility in patients on highly effective CFTR modulators will need to be reinvestigated in future work.
Conclusion
CFRD is associated with poor prognosis in individuals with CF while early diagnosis and aggressive treatment contribute to improvements in survival.4 Thus, annual CFRD screening from 10 years of age is recommended.35 Despite these recommendations, compliance with testing is low.36 We have developed a model that estimates an individual’s CFRD risk at different ages over the course of their disease. The risk estimates can be used by clinical care providers to improve adherence to recommended annual screening or to trigger increased testing frequency. The hope is that improved adherence or more frequent testing will lead to earlier diagnosis and contribute to further gains in median survival that the CF population have been realizing over the last few decades.
Supplementary information
Acknowledgements
The authors thank the patients and families who participated in the CGS and the FGMS in the contributing CF centers across Canada and France. We also express our gratitude to the clinical research assistants, collaborators, and principal investigators involved in both the CGS and FGMS. The study is indebted to the group of FGMS investigators that make external validation of the tool possible. Funding was provided by Cystic Fibrosis Foundation STRUG17PO; Canadian Institutes of Health Research (MOP 258916, MOP 117978, MOP 388348, MOP167282), Cystic Fibrosis Canada (2626), and the CFIT Program funded by the SickKids Foundation and CF Canada; Natural Sciences and Engineering Research Council of Canada (RGPIN-2015- 03742, 250053-2013); This work was funded by the Government of Canada through Genome Canada (OGI-148) andsupported by a grant from the Government of Ontario; and Institut National de la Santé et de la Recherche Medicale, Assistance Publique Hopitaux de Paris, Université Pierre et Marie Curie Paris, Agence Nationale de la Recherche (R09186DS), DGS, Association Vaincre La Mucoviscidose, Chancellerie des Universite´s (Legs Poix), Association Agir Informer Contre la Mucoviscidose, GIS-Institut des Maladies Rares. The funders of the study play no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Y.L. is a trainee and funding recipient of the CANSSI Ontario STAGE (Strategic Training for Advanced Genetic Epidemiology) program at the University of Toronto.
Author contributions
Conceptualization: L.J.S., J.M.R Data curation: K.K., J.G., N.P., J.A., F.L., D.A., P.B., S.B., Y.B., L.B., C.B, J.B., C.B., M.C., R.A., G.C., A.D., C.D., L.F., K.G., N.H., A.H., D.H, S.I., A.I., M.J., E.K., L.K., L.L., W.L., V.L., E.M., D.M., V.M., M.M., N.M., M.P., J.P., A.P., B.Q., J.R., C.S., M.J.S, N.V., D.V., T.V., P.W., R.W. E.B., H.C. Formal analysis: Y.L., L.J.S. Funding acquisition: L.J.S. Investigation: Y.L., L.J.S. Methodology: Y.L., L.J.S. Project administration: L.J.S. Resources and software: Y.L. Supervision: L.J.S. Visualization: Y.L. Writing: Y.L., L.J.S. Writing—review & editing: all authors.
Data availability
Genotype data are available by application to the CF Canada National Data Registry for access to confidential clinical data for the purpose of CF research.
Code availability
Code is available from the authors upon request.
Ethics Declaration
The study was reviewed and approved by the Research Ethics Boards (REBs) at each participating study site including the Research Ethics Board of the Hospital for Sick Children, and the French ethical committee (CPP number 2004/15) with information collection approved by CNIL (number 04.404). The detailed list of REBs can be found in Appendix K in the Supplementary Materials. Informed consent for study participation was obtained from each participant and documented using REB-approved consent forms, which are stored at the respective study sites.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this article was revised: the risk alleles of rs1964986 (PRSS1) and rs959173 (CAV1) were flipped, which should be the C allele for both variants instead of the A and T alleles listed in the paper. In addition, rs1964986(A) should be changed to rs1964986(C).
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/13/2021
A Correction to this paper has been published: 10.1038/s41436-021-01281-z
Supplementary information
The online version of this article (10.1038/s41436-020-01073-x) contains supplementary material, which is available to authorized users.
References
- 1.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soave D, et al. Evidence for a causal relationship between early exocrine pancreatic disease and cystic fibrosis-related diabetes: a Mendelian randomization study. Diabetes. 2014;63:2114–2119. doi: 10.2337/db13-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran A, Dunitz J, Nathan B, Saeed A, Holme B, Thomas W. Cystic fibrosis-related diabetes: current trends in prevalence, incidence, and mortality. Diabetes Care. 2009;32:1626–1631. doi: 10.2337/dc09-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franck Thompson, E., Watson, D., Benoit, C. M., Landvik, S. & McNamara, J. The association of pediatric cystic fibrosis-related diabetes screening on clinical outcomes by center: a CF patient registry study. J Cyst. Fibros. 19, 316–320 (2020). [DOI] [PubMed]
- 6.Boudreau V, et al. Variation of glucose tolerance in adult patients with cystic fibrosis: What is the potential contribution of insulin sensitivity? J. Cyst. Fibros. 2016;15:839–845. doi: 10.1016/j.jcf.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Lewis C, et al. Diabetes-related mortality in adults with cystic fibrosis. Role of genotype and sex. Am. J. Respir. Crit. Care Med. 2015;191:194–200. doi: 10.1164/rccm.201403-0576OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson-Corley KN, Meyerholz DK, Engelhardt JF. Pancreatic pathophysiology in cystic fibrosis. J. Pathol. 2016;238:311–320. doi: 10.1002/path.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackman SM, et al. Genetic modifiers of cystic fibrosis-related diabetes. Diabetes. 2013;62:3627–3635. doi: 10.2337/db13-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aksit, M. A. et al. Genetic modifiers of cystic fibrosis–related diabetes have extensive overlap with type 2 diabetes and related traits. J. Clin. Endocrinol. Metab.105, 1401–1415 (2020). [DOI] [PMC free article] [PubMed]
- 11.Gong J, et al. Genetic association and transcriptome integration identify contributing genes and tissues at cystic fibrosis modifier loci. PLoS Genet. 2019;15:e1008007. doi: 10.1371/journal.pgen.1008007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganz ML, Wintfeld N, Li Q, Alas V, Langer J, Hammer M. The association of body mass index with the risk of type 2 diabetes: a case-control study nested in an electronic health records system in the United States. Diabetol. Metab. Syndr. 2014;6:50. doi: 10.1186/1758-5996-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephenson AL, Stanojevic S, Sykes J, Burgel PR. The changing epidemiology and demography of cystic fibrosis. Presse Med. 2017;46:e87–e95. doi: 10.1016/j.lpm.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Moran A, et al. Diagnosis, screening and management of cystic fibrosis related diabetes mellitus: a consensus conference report. Diabetes Res. Clin. Pract. 1999;45:61–73. doi: 10.1016/S0168-8227(99)00058-3. [DOI] [PubMed] [Google Scholar]
- 15.McLean M, Lambert C, Gevers E, Cowlard J, Chaudry R, Nwokoro C. 12 years too late? Rethinking CFRD screening. J. Cyst. Fibros. 2015;14:S104. doi: 10.1016/S1569-1993(15)30356-8. [DOI] [Google Scholar]
- 16.Ooi CY, et al. type of CFTR mutation determines risk of pancreatitis in patients with cystic fibrosis. Gastroenterology. 2011;140:153–161. doi: 10.1053/j.gastro.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 17.Sun L, et al. Multiple apical plasma membrane constituents are associated with susceptibility to meconium ileus in individuals with cystic fibrosis. Nat. Genet. 2012;44:562–569. doi: 10.1038/ng.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soave D, et al. A joint location-scale test improves power to detect associated SNPs, gene sets, and pathways. Am. J. Hum. Genet. 2015;97:125–138. doi: 10.1016/j.ajhg.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corvol H, Blackman SM, Boëlle PY, Cutting GR, Knowles MR. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat. Commun. 2015;6:8382. doi: 10.1038/ncomms9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meinshausen N, Bühlmann P. Stability selection. J. R. Stat. Soc. Series B Stat. Methodol. 2010;72:417–473. doi: 10.1111/j.1467-9868.2010.00740.x. [DOI] [Google Scholar]
- 22.He K, Li Y, et al. Component-wise gradient boosting and false discovery control in survival analysis with high-dimensional covariates. Bioinformatics. 2016;32:50–57. doi: 10.1093/bioinformatics/btw249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meinshausen N. Relaxed lasso. Comput. Stat. Data Anal. 2007;52:374–393. doi: 10.1016/j.csda.2006.12.019. [DOI] [Google Scholar]
- 24.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soave D, Strug LJ. Testing calibration of Cox survival models at extremes of event risk. Front. Genet. 2018;9:177. doi: 10.3389/fgene.2018.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng S, Zhu Y, Lu B, Xu F, Li X, Lai M. TCF7L2 gene polymorphisms and type 2 diabetes risk: a comprehensive and updated meta-analysis involving 121,174 subjects. Mutagenesis. 2013;28:25–37. doi: 10.1093/mutage/ges048. [DOI] [PubMed] [Google Scholar]
- 28.Sun L, et al. BR-squared: a practical solution to the winner’s curse in genome-wide scans. Hum. Genet. 2011;129:545–552. doi: 10.1007/s00439-011-0948-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adler AI, Shine BS, Chamnan P, Haworth CS, Bilton D. Genetic determinants and epidemiology of cystic fibrosis-related diabetes: results from a British cohort of children and adults. Diabetes Care. 2008;31:1789–1794. doi: 10.2337/dc08-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao R, Boehnke M. Quantifying and correcting for the winner’s curse in genetic association studies. Genet. Epidemiol. 2009;33:453–462. doi: 10.1002/gepi.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi SW, Mak TS, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 2020;15:2759–2772. doi: 10.1038/s41596-020-0353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volkova N, Bilton D, et al. Disease progression in patients with cystic fibrosis treated with ivacaftor: data from national US and UK registries. J. Cyst. Fibros. 2020;19:68–79. doi: 10.1016/j.jcf.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Thomassen JC, Mueller MI, Alejandre Alcazar MA, Rietschel E, van Koningsbruggen-Rietschel S. Effect of Lumacaftor/Ivacaftor on glucose metabolism and insulin secretion in Phe508del homozygous cystic fibrosis patients. J. Cyst. Fibros. 2018;17:271–275. doi: 10.1016/j.jcf.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 34.Shteinberg M, Taylor-Cousar JL. Impact of CFTR modulator use on outcomes in people with severe cystic fibrosis lung disease. Eur. Respir. Rev. 2020;29:190112. doi: 10.1183/16000617.0112-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moran A, et al. Clinical care guidelines for cystic fibrosis-related diabetes: a position statement of the American Diabetes Association and a clinical practice guideline of the Cystic Fibrosis Foundation, endorsed by the Pediatric Endocrine Society. Diabetes Care. 2010;33:2697–2708. doi: 10.2337/dc10-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdulhamid I, Guglani L, Bouren J, Moltz KC. Improving screening for diabetes in cystic fibrosis. Int. J. Health Care Qual. Assur. 2015;28:441–451. doi: 10.1108/IJHCQA-05-2014-0059. [DOI] [PubMed] [Google Scholar]
- 37.Sarles J, et al. Neonatal screening for cystic fibrosis: comparing the performances of IRT/DNA and IRT/PAP. J. Cyst. Fibros. 2014;13:384–390. doi: 10.1016/j.jcf.2014.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotype data are available by application to the CF Canada National Data Registry for access to confidential clinical data for the purpose of CF research.
Code is available from the authors upon request.