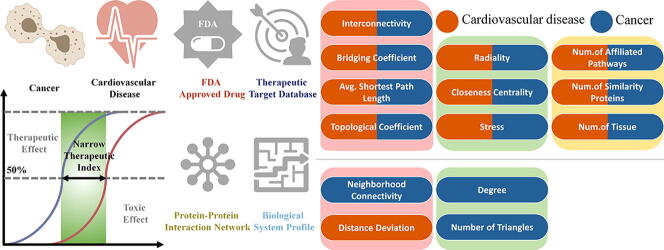
Graphical abstract
Keywords: Narrow therapeutic index, Cancer, Cardiovascular disease, Artificial intelligence, Drug mechanism
Abstract
An appropriate therapeutic index is crucial for drug discovery and development since narrow therapeutic index (NTI) drugs with slight dosage variation may induce severe adverse drug reactions or potential treatment failure. To date, the shared characteristics underlying the targets of NTI drugs have been explored by several studies, which have been applied to identify potential drug targets. However, the association between the drug therapeutic index and the related disease has not been dissected, which is important for revealing the NTI drug mechanism and optimizing drug design. Therefore, in this study, two classes of disease (cancers and cardiovascular disorders) with the largest number of NTI drugs were selected, and the target property of the corresponding NTI drugs was analyzed. By calculating the biological system profiles and human protein–protein interaction (PPI) network properties of drug targets and adopting an AI-based algorithm, differentiated features between two diseases were discovered to reveal the distinct underlying mechanisms of NTI drugs in different diseases. Consequently, ten shared features and four unique features were identified for both diseases to distinguish NTI from NNTI drug targets. These computational discoveries, as well as the newly found features, suggest that in the clinical study of avoiding narrow therapeutic index in those diseases, the ability of target to be a hub and the efficiency of target signaling in the human PPI network should be considered, and it could thus provide novel guidance in the drug discovery and clinical research process and help to estimate the drug safety of cancer and cardiovascular disease.
1. Introduction
A narrow therapeutic index (NTI) of a drug implies that a tiny variation in the dosage of the drug might lead to treatment failure or severe adverse drug reactions [1], [2], [3], [4], [5]. It also hampers drug development since researchers have to conduct additional studies [6] to modify the compound structure, and some failures in drug research [7] are caused by the NTI of drug candidates. In the course of clinical research, some NTI drugs pose great risks in clinical use due to the lack of clear dose adjustment recommendations [4], [8]. It is essential to start mitigation methods to avoid unfavorable traits or to potentially alter resources to alternative candidates by gaining an early consideration of the likely TI value of a certain drug [9], [10]. Moreover, this is critical for avoiding clinical trials because TI with low indication specificity may be considered morally unacceptable [2]. Therefore, the molecular mechanisms of NTI drugs play a prominent role in pharmaceutical discovery and clinical research and help to estimate drug safety and efficacy [11].
However, it is complicated to determine and interpret the TI of a drug because this depends not only on the stage of development that affects the available data but also on the properties of the indications for which the drug is being developed [2], [12]. A widely used concept of TI is the quantitative relationship between pharmacology and safety toxicology, but the definition of a therapeutic or toxic effect is highly dependent on different therapeutic and toxic effect types [3]. For example, imatinib can allow more toxicity with a smaller TI value when used in cancer in pursuit of higher pharmacological exposure, but there must be a larger and more reasonable TI value when used for pulmonary hypertension [2], [13]. This adds complexity to the understanding of the molecular mechanisms of NTI drugs. In fact, of the 161 NTI drugs currently FDA approved, almost half of them belong to cancer and cardiovascular disease. Cancer is a group of diseases characterized by uncontrolled cell growth. The cardiovascular disease usually involves narrowed or blocked blood vessels, which can contribute to heart attack, angina, or stroke and is characterized by acute onset, critical condition, and rapid progression [14]. These observations suggest that there may be disease-specific pathology, resulting in different types of the disease each with its characteristics [15], and the molecular mechanisms of NTI drugs in different diseases may also exhibit large variations. Therefore, when designing drugs and conducting clinical research on these two types of diseases, it is necessary to consider the different molecular mechanisms of NTI drugs between them.
To enhance the understanding of TI, a variety of studies and some approaches have been developed to enhance the ability to reveal the mechanism underlying NTI drugs, such as the exposure-centric TI approach [2], preclinical pharmacology model [16], [17], [18], assessment of off-target safety margins [19]. Recently, an article was published in Frontiers in Pharmacology [20] using a target-based approach, combining the profiles of human protein–protein interaction (PPI) network, and biological systems to find features or feature groups that can be used to indicate the drug’s narrow TI. It identified 8 features that could collectively indicate that NTI drug targets are tremendously connected and centralized and are related to target druggability in all diseases. Agnieszka Potęga, et al. [21] have shown that this target-based approach to study the mechanisms underlying NTI drugs is important to indicate a well-balanced profile between efficacy and safety. However, no studies have revealed the underlying mechanism behind the complex definition and interpretation of TI in different diseases, and what significantly limits NTI drug design and clinical studies for both specific diseases, and this needs to be solved urgently.
Therefore, in this study, the underlying mechanisms of NTI drugs aimed at cancer and cardiovascular disease were analyzed based on not only the human PPI network features but also the biological system profiles. To discover this underlying mechanism, the NTI and NNTI drug targets were divided into three groups: (i) NTI drug targets of cancer, (ii) NTI drug targets of cardiovascular disease, and (iii) NNTI drug targets for all indications. Next, through the comparative analysis of the target groups (i) and (iii) and the target groups (ii) and (iii), several essential features that could distinguish the two groups were identified, and further studies revealed similarities and differences in the characteristics of cancer and cardiovascular disease. Overall, these findings combined with the newly recognized features can indicate the underlying mechanisms of NTI drugs targeting cancer and cardiovascular disease, respectively, which offer certain guidance in assessing the risks and benefits of drug candidates, as well as drug discovery and clinical research in cancer and cardiovascular disease.
2. Materials and methods
2.1. NTI drugs collection and associated targets and indications identification
The NTI approved drugs and their related drug targets and indications were obtained through the following steps. First, 1,921 FDA approved drugs with their related indications were systematically collected and identified from the orange book of the US FDA [72]. Then, all the corresponding diseases were standardized by the ICD-11 codes (the latest version of the International Classification of Diseases) [73]. Next, the corresponding targets of the approved drugs were authorized by the therapeutic target database (TTD) [74], and 506 corresponding targets of the approved drugs were confirmed. Third, a systematic literature review of all these drugs was performed to confirm their TI value by searching the PubMed database using such keyword combinations as “Drug Name/Synonym” + “Therapeutic ranges” / ‘‘Therapeutic index” / ‘‘Therapeutic ration” / ‘‘Therapeutic window”. Consequently, 36 NTI drugs targeting cancer and 18 NTI drugs targeting cardiovascular disease are discovered, which account for approximately half of all NTI drugs. Moreover, 29 NNTI drugs targeting all indications are also distinguished. The FDA-approved NTI drugs for cancer and cardiovascular disease together with their standardized indication, ICD-11 codes, and targets are provided in Table 1, and the NNTI drugs for all indication together with their standardized indication, ICD-11 codes, and targets are provided in Table 2.
Table 1.
FDA approved NTI drugs of cancer and cardiovascular disease together with their standardized indication, ICD-11 code, and target. ADRA1: Adrenergic receptor alpha 1; ADRA2: Adrenergic receptor alpha 2; ADRB1: Adrenergic receptor beta-1; ADRB2: Adrenergic receptor beta-2; ADRB3: Adrenergic receptor beta-3; ATIII: Antithrombin-III; BCL-2: Apoptosis regulator BCL-2; F2: coagulation factor II; F10: Activated coagulation factor X; DHFR: Dihydrofolate reductase; TOP1: DNA topoisomerase I; TOP2: DNA topoisomerase II; EGFR: Epidermal growth factor receptor; ESR: Estrogen receptor; hDNA: Human deoxyribonucleic acid; IMPDH1: Inosine-5′-monophosphate dehydrogenase 1; IFNA2: Interferon-alpha 2; NET: Norepinephrine transporter; PDGFRB: Platelet-derived growth factor receptor; RET: Proto-oncogene c-Ret; RRM2: Ribonucleoside-diphosphate reductase M2; mTOR: Serine/threonine-protein kinase mTOR; SPT ATPase: Sodium/potassium-transporting ATPase; TMP1: Thymidylate synthase; TUB: Tubulin; c-Kit: Tyrosine-protein kinase Kit; KDR: Vascular endothelial growth factor receptor 2; VKORC1: Vitamin K epoxide reductase complex 1; SCN5A: Voltage-gated sodium channel alpha Nav1.5; SCN5A: Voltage-gated sodium channel alpha Nav1.5; SCN11A: Voltage-gated sodium channel alpha Nav1.9.
| FDA Approved Drug (Reference for NTI) | Time of Approval | FDA Approved Indication | ICD-11 Code | Disease Class | Target Name |
|---|---|---|---|---|---|
| Argatroban [22] | 2000 | Intracardiac thrombosis | BC46 | Cardiovascular | F2 |
| Axitinib [23] | 2012 | Rectum cancer | 2B92 | Cancer | KDR |
| Busulfan [24] | 1954 | Chronic myeloid leukemia | 2B33 | Cancer | hDNA |
| Capecitabine [25] | 1998 | Breast cancer | 2C60 | Cancer | TMP1 |
| Carboplatin [26] | 1989 | Ovary cancer | 2C73 | Cancer | hDNA |
| Cisplatin [27] | 1978 | Ovary cancer | 2C73 | Cancer | hDNA |
| Clonidine [28] | 1974 | Hypertension | BA00 | Cardiovascular | ADRA2 |
| Cyclophosphamide [29] | 1959 | Acute myeloid leukemia | 2A60 | Cancer | hDNA |
| Dalteparin Sodium [22] | 1994 | Deep vein thrombosis | BD71 | Cardiovascular | ATIII |
| Digitoxin [30] | 1982 | Heart failure | BD10 | Cardiovascular | SPT ATPase |
| Digoxin [31] | 1954 | Heart failure | BD10 | Cardiovascular | SPT ATPase |
| Disopyramide Phosphate [32] | 1977 | Ventricular tachyarrhythmia | BC71 | Cardiovascular | SCN5A |
| Docetaxel [33] | 1996 | Breast cancer | 2C60 | Cancer | TUB |
| Doxorubicin HCl [34] | 1974 | Breast cancer | 2C60 | Cancer | TOP2 |
| Epinephrine [35] | 1951 | Coronary artery disease | BA80 | Cardiovascular | ADRB1 |
| Epirubicin HCl [36] | 1999 | Axillary node cancer | 2D60 | Cancer | TOP2 |
| Etoposide [37] | 1983 | Testis cancer | 2C80 | Cancer | TOP2 |
| Etoposide Phosphate [26] | 1996 | Ovary cancer | 2C73 | Cancer | TOP2 |
| Everolimus [38] | 2009 | Renal cell carcinoma | 2C90 | Cancer | mTOR |
| Flecainide Acetate [39] | 1985 | Arrhythmic | BC64 | Cardiovascular | SCN5A |
| Fluorouracil [40] | 1962 | Colorectal cancer | 2B91 | Cancer | TMP1 |
| Fondaparinux Sodium [41] | 2001 | Deep vein thrombosis | BD71 | Cardiovascular | F10 |
| Gefitinib [42] | 2003 | Lung cancer | 2C25 | Cancer | EGFR |
| Gemcitabine HCl [43] | 1996 | Pancreatic cancer | 2C10 | Cancer | RRM2 |
| Guanethidine Monosulfate [22] | 1960 | Hypertensive crisis | BA03 | Cardiovascular | NET |
| Interferon Alfa-2B [44] | 1986 | Melanoma | 2C30 | Cancer | IFNA2 |
| Irinotecan HCl [34] | 1996 | Colorectal cancer | 2B91 | Cancer | TOP1 |
| Lidocaine [45] | 1948 | Ventricular tachyarrhythmia | BC71 | Cardiovascular | SCN11A |
| Mercaptopurine [46] | 1953 | Acute lymphocytic leukemia | 2A82 | Cancer | IMPDH1 |
| Methotrexate Sodium [26] | 1953 | Breast cancer | 2C60 | Cancer | DHFR |
| Mitomycin [47] | 1981 | Stomach cancer | 2B72 | Cancer | hDNA |
| Mitotane [48] | 1970 | Adrenal gland cancer | 2D11 | Cancer | ESR |
| Oxaliplatin [49] | 2002 | Colorectal cancer | 2B91 | Cancer | hDNA |
| Paclitaxel [50] | 1992 | Kaposi sarcoma | 2B57 | Cancer | TUB; BCL-2 |
| Pazopanib HCl [23] | 2009 | Renal cell carcinoma | 2C90 | Cancer | c-Kit; KDR; PDGFRB |
| Pemetrexed [51] | 2004 | Pleura cancer | 2C26 | Cancer | DHFR; TMP1 |
| Pemetrexed Disodium [51] | 2004 | Pleura cancer | 2C26 | Cancer | DHFR; TMP1 |
| Phenprocoumon [52] | 1957 | Intracardiac thrombosis | BC46 | Cardiovascular | VKORC1 |
| Prazosin HCl [22] | 1976 | Hypertension | BA00 | Cardiovascular | ADRA1 |
| Procainamide HCl [53] | 1950 | Ventricular tachyarrhythmia | BC71 | Cardiovascular | BTX-activated cardiac channel |
| Propafenone HCl [22] | 1989 | Atrial fibrillation | BC81 | Cardiovascular | ADRB1; ADRB2; ADRB3 |
| Quinidine [22] | 1950 | Ventricular tachyarrhythmia | BC71 | Cardiovascular | SCN5A |
| Regorafenib [23] | 2012 | Gastrointestinal stromal cancer | 2B5B | Cancer | c-Kit; KDR; RET |
| Sorafenib Tosylate [23] | 2005 | Adrenal gland cancer | 2D11 | Cancer | EGFR; c-Kit; KDR; PDGFRB |
| Sotalol HCl [22] | 1992 | Ventricular tachyarrhythmia | BC71 | Cardiovascular | ADRB1 |
| Sunitinib Malate [23] | 2006 | Gastrointestinal stromal cancer | 2B5B | Cancer | KDR |
| Teniposide [26] | 1992 | Acute lymphocytic leukemia | 2A82 | Cancer | TOP2 |
| Thioguanine [46] | 1966 | Acute myeloid leukemia | 2A60 | Cancer | hDNA |
| Topotecan HCl [26] | 1996 | Ovary cancer | 2C73 | Cancer | TOP1 |
| Vandetanib [23] | 2011 | Thyroid gland cancer | 2D10 | Cancer | EGFR; KDR; RET |
| Vinblastine Sulfate [39] | 1965 | Hodgkin lymphoma | 2B30 | Cancer | TUB |
| Vincristine Sulfate [54] | 1963 | Acute lymphocytic leukemia | 2A82 | Cancer | TUB |
| Vinorelbine Tartrate [55] | 1994 | Lung cancer | 2C25 | Cancer | TUB |
| Warfarin Sodium [31] | 1954 | Pulmonary thromboembolism | BB00 | Cardiovascular | VKORC1 |
Table 2.
FDA approved NNTI drugs of all indications together with their standardized indication, ICD-11 code, and target. ABAT: GABA transaminase; ACE: Angiotensin-converting enzyme; CACNA1G: Voltage-gated calcium channel alpha Cav3.1; CACNA2D1: Voltage-gated calcium channel alpha-2/delta-1; CACNA2D2: Voltage-gated calcium channel alpha-2/delta-2; CACNA2D3: Voltage-gated calcium channel alpha-2/delta-3; CYSLTR1: Leukotriene CysLT1 receptor; D2R: Dopamine D2 receptor; DPP4: Dipeptidyl peptidase 4; DPYSL2: Dihydropyrimidinase related protein 2; F10: Activated coagulation factor X; GABRA1: GABA(A) receptor alpha-1; GABRG3: GABA(A) receptor gamma-3; GAR: Gamma-aminobutyric acid receptor; GRIA: Glutamate receptor AMPA; GRIK1: Glutamate receptor ionotropic kainate 1; hDNA: Human deoxyribonucleic acid; HIV RT: Human immunodeficiency virus Reverse transcriptase; KCNQ2: Voltage-gated potassium channel Kv7.2; KCNQ3: Voltage-gated potassium channel Kv7.3; NET: Norepinephrine transporter; NMDAR: N-methyl-D-aspartate receptor; PPP3CA: Calcineurin; SCN11A: Voltage-gated sodium channel alpha Nav1.9.; SCN1A: Sodium channel protein type 1 subunit alpha; SV2A: Synaptic vesicle glycoprotein 2A; TACR1: Substance-P receptor; TOP2: DNA topoisomerase II.
| FDA Approved Drug (Reference for NTI) | Time of Approval | FDA Approved Indication | ICD-11 Code | Disease Class | Target Name |
|---|---|---|---|---|---|
| Apixaban [56] | 2012 | Deep vein thrombosis | BD71 | Cardiovascular | F10 |
| Aripiprazole [57] | 2002 | Schizophrenia | 6A20 | Mental disorder | D2R |
| Atomoxetine HCl [58] | 2002 | ADHD | 6A05 | Mental disorder | NET |
| Clobazam [59] | 2011 | Epilepsy or seizures | 8A60 | Nervous system | GABRA1; GABRG3 |
| Clonazepam [22] | 1975 | Epilepsy or seizures | 8A60 | Nervous system | GABRA1 |
| Enalapril Maleate [60] | 1985 | Hypertension | BA00 | Cardiovascular | ACE |
| Ethosuximide [22] | 1960 | Epilepsy or seizures | 8A60 | Nervous system | CACNA1G |
| Ezogabine [59] | 2011 | Epilepsy or seizures | 8A60 | Nervous system | KCNQ2; KCNQ3 |
| Felbamate [22] | 1993 | Epilepsy or seizures | 8A60 | Nervous system | NMDAR |
| Gabapentin [61] | 1993 | Epilepsy or seizures | 8A60 | Nervous system | CACNA2D2; CACNA2D3 |
| Gabapentin Enacarbil [61] | 1993 | Epilepsy or seizures | 8A60 | Nervous system | CACNA2D2; CACNA2D3 |
| Lacosamide [62] | 2008 | Epilepsy or seizures | 8A60 | Nervous system | DPYSL2 |
| Lamivudine [63] | 1995 | HIV infection | 1C62 | Infection | HIV RT |
| Lamotrigine [22] | 1994 | Bipolar disorders | 6A60 | Mental disorder | SCN11A |
| Levetiracetam [22] | 1999 | Epilepsy or seizures | 8A60 | Nervous system | SV2A |
| Linagliptin [64] | 2011 | Type 2 diabetes mellitus | 5A11 | Metabolic disease | DPP4 |
| Mechlorethamine HCl [65] | 1949 | Mature T-cell lymphoma | 2A90 | Cancer | hDNA |
| Mitoxantrone HCl [66] | 1987 | Multiple sclerosis | 8A40 | Nervous system | TOP2 |
| Montelukast Sodium [67] | 1998 | Asthma | CA23 | Respiratory system | CYSLTR1 |
| Oxcarbazepine [22] | 2000 | Epilepsy or seizures | 8A60 | Nervous system | SCN11A |
| Perampanel [56] | 2012 | Epilepsy or seizures | 8A60 | Nervous system | GRIA |
| Pimecrolimus [68] | 2001 | Atopic eczema | EA80 | Skin disease | PPP3CA |
| Pregabalin [69] | 2004 | Epilepsy or seizures | 8A60 | Nervous system | CACNA2D1 |
| Rivaroxaban [59] | 2011 | Deep vein thrombosis | BD71 | Cardiovascular | F10 |
| Rolapitant HCl [70] | 2015 | Nausea or vomiting | DD90 | Digestive system | TACR1 |
| Rufinamide [62] | 2008 | Epilepsy or seizures | 8A60 | Nervous system | N.A. |
| Topiramate [22] | 1996 | Epilepsy or seizures | 8A60 | Nervous system | GABRA1 |
| Vigabatrin [71] | 2009 | Types of seizures | 8A68 | Nervous system | ABAT |
| Zonisamide [22] | 2000 | Epilepsy or seizures | 8A60 | Nervous system | SCN1A |
2.2. Assessing the profile of human PPI network properties and biological systems for corresponding therapeutic targets
The human PPI network properties studied in this research consisted of 15,554 proteins and 642,304 interactions between these proteins, and these were created via the information furnished by the STRING database [75]. Only those protein interactions with confidence above 0.95 were selected for further analysis to guarantee the dependability of the analytical data [76], [77]. Thus, in this study, a subnetwork consisting of 8,509 proteins, and 40,468 interactions between these proteins was developed for subsequent study. Additionally, the PPI network characteristics of corresponding therapeutic targets were obtained by the PROFEAT [78] and the tool Network Analyzer of Cytoscape [79], [80]. In summary, 32 PPI network properties were calculated for further analysis, as shown in Table 3 (six features that are all zeroes were deleted, which are: ‘closeness centrality sum’, ‘bridging centrality’, ‘eigenvector centrality’, ‘page rank centrality’, ‘number of selfloops’, and ‘current flow closeness’). Then, the additional four features of each corresponding target in the biological system profile were estimated.
Table 3.
The calculated mean and median values of 30 properties in the human PPI network and biological system profiles.
| Feature | Cancer |
Cardiovascular Disease |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean Value of Targets of the NTI | Median Value of Targets of the NTI | Mean Value of Targets of the NNTI | Median Value of Targets of the NNTI | Mean Value of Targets of the NTI | Median Value of Targets of the NTI | Mean Value of Targets of the NNTI | Median Value of Targets of the NNTI | ||
| Connectivity/adjacency-based | Bridging coefficient | 1.02E−01 | 4.10E−02 | 6.79E−01 | 6.10E−02 | 7.64E−01 | 1.40E−01 | 3.70E+00 | 3.23E−01 |
| Clustering coefficient | 1.32E−01 | 9.90E−02 | 1.84E−01 | 7.60E−02 | 2.85E−01 | 3.17E−01 | 2.72E−01 | 1.00E−01 | |
| Degree | 2.12E+01 | 1.50E+01 | 2.07E+01 | 1.30E+01 | 5.66E+00 | 5.50E+00 | 4.92E+00 | 3.00E+00 | |
| Degree centrality | 2.74E−03 | 2.00E−03 | 2.64E−03 | 2.00E−03 | 7.50E−04 | 1.00E−03 | 5.49E−04 | 0.00E+00 | |
| Interconnectivity | 2.64E−01 | 2.55E−01 | 4.13E−01 | 2.69E−01 | 4.83E−01 | 4.38E−01 | 5.28E−01 | 5.00E−01 | |
| Neighborhood connectivity | 3.49E+01 | 2.77E+01 | 2.59E+01 | 2.04E+01 | 1.75E+01 | 1.47E+01 | 1.81E+01 | 1.13E+01 | |
| Number of triangles | 3.00E+01 | 1.70E+01 | 4.60E+01 | 1.50E+01 | 2.53E+00 | 2.00E+00 | 2.50E+00 | 1.00E+00 | |
| Scaled degree | 2.83E−02 | 2.00E−02 | 2.75E−02 | 1.70E−02 | 7.50E−03 | 7.50E−03 | 6.56E−03 | 4.00E−03 | |
| Topological coefficient | 9.63E−02 | 1.01E−01 | 1.70E−01 | 1.13E−01 | 2.57E−01 | 2.84E−01 | 2.76E−01 | 2.06E−01 | |
| Z-score | 3.41E−02 | 1.50E−02 | 3.23E−02 | 9.00E−03 | −1.33E−02 | −1.35E−02 | −1.58E−02 | −2.20E−02 | |
| Shortest path length-based | Average shortest path length | 3.90E+00 | 4.01E+00 | 4.41E+00 | 4.06E+00 | 4.80E+00 | 4.79E+00 | 4.95E+00 | 5.04E+00 |
| Betweenness centrality | 2.14E−03 | 1.00E−03 | 2.04E−03 | 0.00E+00 | 5.00E−04 | 0.00E+00 | 4.02E−04 | 0.00E+00 | |
| Average closeness centrality | 2.58E−01 | 2.50E−01 | 2.36E−01 | 2.46E−01 | 2.10E−01 | 2.09E−01 | 2.06E−01 | 1.99E−01 | |
| Current flow betweenness | 4.33E−03 | 3.00E−03 | 3.86E−03 | 2.00E−03 | 1.25E−03 | 1.00E−03 | 1.13E−03 | 1.00E−03 | |
| Deviation | 9.89E+03 | 1.07E+04 | 1.39E+04 | 1.11E+04 | 1.70E+04 | 1.69E+04 | 1.82E+04 | 1.89E+04 | |
| Distance deviation | 7.30E+03 | 6.47E+03 | 7.04E+03 | 6.61E+03 | 3.06E+03 | 3.89E+03 | 4.35E+03 | 4.16E+03 | |
| Distance sum | 3.11E+04 | 3.19E+04 | 3.51E+04 | 3.23E+04 | 3.82E+04 | 3.81E+04 | 3.94E+04 | 4.01E+04 | |
| Eccentric | 1.08E+00 | 1.34E+00 | 1.10E+00 | 1.34E+00 | 8.23E−01 | 6.60E−01 | 6.32E−01 | 6.60E−01 | |
| Eccentricity | 1.03E+01 | 1.00E+01 | 1.06E+01 | 1.00E+01 | 1.13E+01 | 1.20E+01 | 1.14E+01 | 1.10E+01 | |
| Eccentricity dentrality | 9.77E−02 | 1.00E−01 | 9.53E−02 | 1.00E−01 | 8.91E−02 | 8.30E−02 | 8.82E−02 | 9.10E−02 | |
| Harmonic closeness centrality | 2.21E+03 | 2.12E+03 | 2.00E+03 | 2.10E+03 | 1.76E+03 | 1.75E+03 | 1.72E+03 | 1.64E+03 | |
| Load centrality | 2.11E−03 | 1.00E−03 | 2.11E−03 | 0.00E+00 | 5.00E−04 | 0.00E+00 | 4.02E−04 | 0.00E+00 | |
| Normalized betweenness | 4.64E−03 | 2.00E−03 | 4.46E−03 | 1.00E−03 | 1.28E−03 | 1.00E−03 | 9.71E−04 | 0.00E+00 | |
| Residual closeness centrality | 6.77E+02 | 6.16E+02 | 5.49E+02 | 5.98E+02 | 3.78E+02 | 3.66E+02 | 3.58E+02 | 3.00E+02 | |
| Radiality | 8.18E−01 | 8.12E−01 | 7.87E−01 | 8.09E−01 | 7.63E−01 | 7.63E−01 | 7.53E−01 | 7.48E−01 | |
| Stress | 2.59E+06 | 9.00E+05 | 2.33E+06 | 4.81E+05 | 4.80E+05 | 3.25E+05 | 4.73E+05 | 2.08E+05 | |
| Biological system-based | Number of affiliated pathways | 3.70E+00 | 3.00E+00 | 2.65E+00 | 3.00E+00 | 2.00E+00 | 2.00E+00 | 1.65E+00 | 2.00E+00 |
| Number of similarity proteins | 1.35E+01 | 1.20E+01 | 9.55E+00 | 9.00E+00 | 2.33E+01 | 1.60E+01 | 1.05E+01 | 9.00E+00 | |
| Number of target expression | 1.48E−01 | 7.14E−04 | 2.79E−01 | 2.65E−01 | 4.23E−01 | 1.91E−01 | 2.81E−01 | 2.08E−01 | |
| Number of tissue | 3.69E+00 | 4.00E+00 | 3.06E+00 | 3.00E+00 | 3.78E+00 | 4.00E+00 | 2.63E+00 | 2.00E+00 | |
The first feature is the number of target-affiliated pathways that were collected from the KEGG database [81]. This feature was confirmed by two aspects. On the one hand, the pathway of the corresponding drug targets should be necessary for life not only for patients but also in healthy individuals. On the other hand, the therapeutic target should be upstream and have the ability to regulate the biological function of the pathway. The second feature is the number of each therapeutic drug target distributed in human tissues, which was offered in the TissueDistributionDBs [82] and UniProt [83] databases. The determination of this feature depends on a higher level of total protein (>5%) distributed in a particular tissue or a higher target concentration in that tissue than the average protein concentration. To explore the off-target collateral effect, the third feature was adopted, which is the number of human similarity proteins. This was determined by counting the number of similar proteins that are outside the target protein family for the studied drug target [84], [85]. This was calculated using BLAST similarity screening with the cutoff value of e-value < 0.005 [86], [87] for the human proteome method furnished in the UniProt database [83]. The differential expression of the target is the fourth feature, which is capable of reflecting the expression differences of the corresponding target between diseased and healthy populations for specific diseases [74], [88], [89]. The expression data were gathered from TTD [90] and calculated by using the HG-U133 Plus 2.0 platform which was determined by the Gene Expression Omnibus database [91].
Collectively, these 36 features are valuable and meaningful in revealing human protein–protein interaction data for a given target, including their connectivity, organization, robustness, and stability in the human PPI network [92], [93], [94] and the on-target and off-target pharmacology of the studied targets [85], [95]. These two aspects are key to enhancing potency for characterizing the underlying mechanisms of NTI drugs [2], [96]. In previous publications, including our previous analysis [20], a series of analyses have been performed by these 36 features. And these 36 features (30 features are described in Table 3, excluding the 6 features that the calculated values are zero) are still adopted in this study to further explore the different features of NTI drug targets between two representative disease classes (cancers and cardiovascular diseases). Their calculation formulas and biological descriptions are separately reflected in Supplementary Table S1.
2.3. NTI drug characteristic identification in two diseases by an artificial intelligence-based algorithm
Artificial intelligence (AI) has seen significant advancement in recent decades for aiding drug treatment [97], [98], [99], [100], [101], predicting drug-target or drug-drug interactions [5], [102], [103], and optimizing treatment protocols [104], [105], [106], including machine learning algorithms [107], [108], [109], deep learning methods [110], [111], [112], and cognitive-computing [113]. In this study, to better understand the underlying mechanisms of NTI drugs, one of the most widely used artificial intelligence algorithms, Boruta, which was based on a random forest classifier [18], [114], was adopted. This method compares the correlation between real features and random probes to determine the extension of the correlation [115]. The Boruta algorithm was built by an AI-based method (machine learning), which is particularly suitable for low-dimensional data sets in other available strategies because of its strong stability in variable selection [116], [117]. Then, the different characteristics between NTI and NNTI drug targets of cancer and cardiovascular disease were determined by the R package Boruta, respectively [118]. Notably, assessing the profile of human PPI network properties and the biological system for each target was conducted using the Boruta algorithm in the R environment and setting the parameters as follows: holdHistory and mcAdj = TRUE, getImp = getImpRfZ, maxRuns = 100, doTrace = 2, p-value < 0.05. Eventually, the features that could elucidate the essential factors indicating narrow TI of drugs in cancer and cardiovascular disease were respectively selected.
3. Results and discussion
3.1. Merging the human PPI network and biological system properties for artificial intelligence-based algorithm
The drug risk-to-benefit ratio (RBR) is mainly determined by the drug target profile of the network properties and biological system [84], [119], [120], [121]. Network characteristics are inherent to drug targets in human PPI networks, and biological system properties can mirror the pharmacology of on-target and off-target. In this paper, the most comprehensive sets of characteristics belong to the human PPI network properties and biological system profiles were chosen to further explore the different features of NTI drug targets between two representative diseases (cancer and cardiovascular disease). Their calculation formulas and biological descriptions are separately reflected in Supplementary Table S1. The average and median values of 30 features for cancer NTI drug targets, cardiovascular disease NTI drug targets, and NNTI drug targets were also calculated (removing six characteristics equal to 0), as shown in Table 3. These 30 features were classified into three categories according to the attributes inherent in each feature, that is, the connectivity/adjacency-based properties, the shortest path length-based properties, and the human biological system properties, as also shown in Table 3.
The mean and median values between the two groups of targets (NTI and NNTI drug targets) for each disease in Table 3 show a significant difference between the two groups of targets in many features. However, none of them can be used separately as an indicator to distinguish between NTI drug targets and NNTI drug targets. Only through collective combination can NTI drug targets be more effectively distinguished from NNTI drug targets [20]. Therefore, in the next part of the study, we integrated the feature selection method based on artificial intelligence to select some important indexes from these features that can be combined to determine the drug targets of NTI and the drug targets of NNTI. However, this approach seems to introduce a very strong bias when 36 features are directly used for feature selection because of the significant dependence between 19 of these features [20]. Therefore, after a thorough investigation of 36 features, the 19 features were eventually merged into five features due to their innate interdependence. Considering the remaining 17 relatively independent features, a total of 22 features for each target were applied for further feature selection. The method of feature integration referred to previous research by our group [20], as well as the biological description and equation of those 36 properties in human PPI networks and biological system profiles, provided in Supplementary Table S1.
3.2. Revealing the essential properties of NTI drug targets in cancer by an artificial intelligence-based algorithm
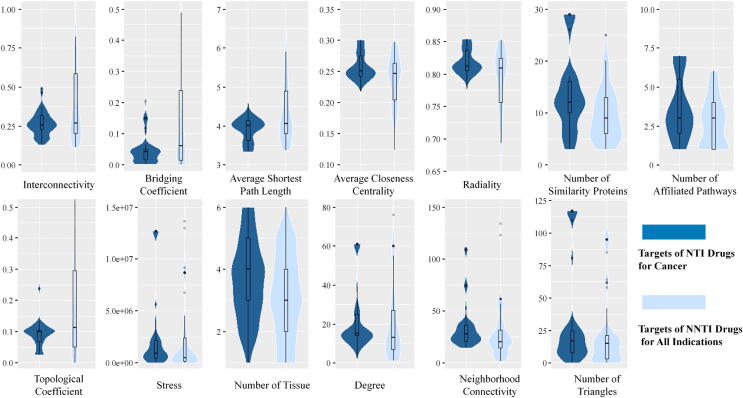
The Boruta algorithm was built by an artificial intelligence method, which is particularly suitable for low-dimensional data sets compared to other available strategies because of its strong stability in variable selection [20], [116]. Specifically, by setting the key parameters (described in detail in the Materials and Methods section), the R package Boruta was used to selecting the key difference features from 22 target profiles. The feature selection result is shown in Fig. 1, which means that 13 features were selected to collectively reflect the underlying mechanism of cancer NTI drugs, including ‘interconnectivity’, ‘bridging coefficient’, ‘average shortest path length’, ‘average closeness centrality’, ‘radiality’, ‘topological coefficient’, ‘number of affiliated pathways’, ‘number of similarity proteins’, ‘stress’, ‘number of tissues’, ‘degree’, ‘neighborhood connectivity’, and ‘number of triangles’. The violin districts colored dark blue and light blue refer to the NTI drug targets in cancer and NNTI drug targets in all indications, respectively. Among these 13 selected features, some important features displayed an upward trend from NTI to NNTI drug targets (such as ‘interconnectivity’), while others showed a downward trend (such as ‘average closeness centrality’). In particular, the ‘average closeness centrality’ is defined as the reciprocal of the average shortest path length of the studied target. It measures how fast information spreads from a studied drug target to other reachable proteins in the PPI network [122], and the ‘interconnectivity’ is a connection metric that indicates the quality or status of the studied targets connected [123]. It was reported that a higher value of ‘average closeness centrality’ and a higher level (lower value) of ‘interconnectivity’ of the target demonstrated a greater lethality risk [20], [124], which meant that a protein with tremendous centrality and connectivity carries a greater lethality risk. The results from our study proved that the capabilities of the applied Boruta algorithms in determining essential features of cancer NTI drug targets were due to the trends of the values of features in NTI and NNTI in Fig. 1, in agreement with these previous studies. Moreover, what we found also suggested that some features could be indirectly relevant to the drug risk-to-benefit ratio [124], [125], and NTI drug targets of cancer in the biological network were not only inclined to be hub proteins [126] but also to have high centrality and connectivity.
Fig. 1.
Violin plot of the 13 features identified in cancer. For each feature, dark blue represents the targets of NTI drugs for cancer, and light blue represents the targets of NNTI drugs for all indications. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Discovering the basic characteristics of NTI drug targets in cardiovascular disease by artificial intelligence-based algorithm
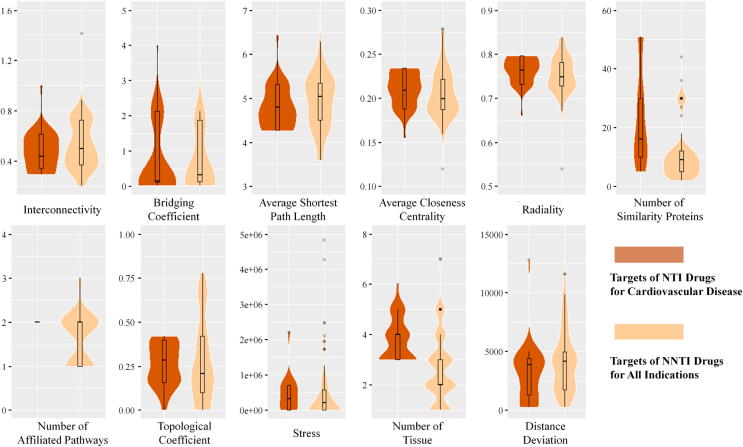
To identify the features of NTI drugs treating cardiovascular disease, the Boruta algorithm was adopted. The results are shown in Fig. 2. Eleven features were selected to collectively reflect the underlying mechanism of cardiovascular disease NTI drugs, including ‘interconnectivity’, ‘bridging coefficient’, ‘average shortest path length’, ‘average closeness centrality’, ‘radiality’, ‘number of affiliated pathways’, ‘number of similarity proteins’, ‘topological coefficient’, ‘stress’, ‘number of tissues’, and ‘distance deviation’. In Fig. 2, the violin districts colored dark orange and light orange refers to the NTI drug targets in cardiovascular disease and NNTI drug targets in all indications, respectively. Similar to drug targets in cancer, some important features displayed an upward trend from NTI to NNTI drug targets (such as ‘average shortest path length’). In contrast, some displayed a downward trend (such as ‘radiality’). The ‘average shortest path length’ describes the average length of shortest paths between the studied drug target and all other proteins in the studied PPI network [127], and the ‘radiality’ is the reachability level of the studied nodes through diverse shortest paths throughout the network [128]. Moreover, the trend of these features in NTI and NNTI drug targets meant that NTI drug targets of cardiovascular disease were likely to have more links with other proteins. [129].
Fig. 2.
Violin plot of the 11 features identified in cardiovascular disease. For each feature, dark orange represents the targets of NTI drugs for cardiovascular disease, and light orange represents the targets of NNTI drugs for all indications. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Exploring the shared/differential characteristics of NTI drug targets between cancers and cardiovascular diseases
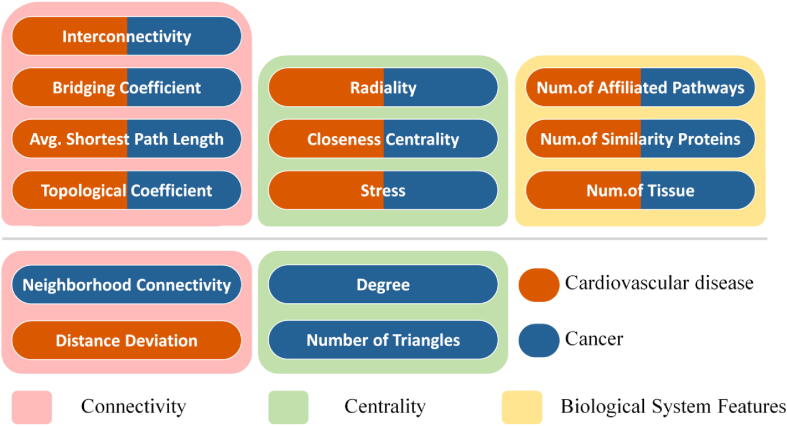
The shared/differential features of NTI drug targets between cancers and cardiovascular diseases identified in the study are provided in Fig. 3. The boxes of pink background are the feature class of Connectivity, the boxes of light green background are the feature class of Centrality, and the box of yellow background provides the feature class of Biological System Profile. Besides, the dark blue bars indicated the characteristics of NTI drug targets for cancers, and the orange bars denoted the characteristics of NTI drug targets for cardiovascular diseases. Those 10 features in the first layer are shared by both cancers and cardiovascular diseases. Seven of these 10 features are the same as those identified by the previous report [20], which include ‘average shortest path length’, ‘bridging coefficient’, ‘closeness centrality’, ‘interconnectivity’, ‘number of affiliated pathways’, ‘number of similarity proteins’ and ‘radiality’. These features indicated that the NTI drug targets of cancers and cardiovascular diseases were greatly connected and centralized in human PPI networks, and shared a biological system of the large number of similar proteins and target-affiliated pathways [20]. Moreover, these results validated the capability of Boruta [114] in determining the features of NTI drug targets in different disease classes.
Fig. 3.
Classification of the key features of cancer and cardiovascular disease determined in this study into three feature groups.
The second layer is unique characteristics identified for cancers (dark blue) and cardiovascular diseases (organge). For cancers, the identified features included ‘neighborhood connectivity’, ‘degree’, and ‘number of triangles’. For cardiovascular diseases, the discovered feature was ‘distance deviation’. As reported, drug targets tend to have a lower ‘clustering coefficient’ in cancer [15]. The ‘clustering coefficient’ denoted neighborhood connectivity [130], and the ‘clustering coefficient’ decreased with the increase in the number of interacting proteins [131]. Therefore, a lower ‘clustering coefficient’ indicates higher ‘neighborhood connectivity’. The ‘degree’ means the total number of edges connected to the studied node, and the ‘number of triangles’ that referred to the percentage of the triangle between a node and its neighbors. The higher the ‘degree’ and ‘number of triangles’, the higher the centrality of the drug target, and the more likely it is to lead to adverse drug reactions [132]. Thus, the computational discoveries of our result that cancer has a higher value of ‘degree’, ‘number of triangles’, and ‘neighborhood connectivity’ are consistent with literature reports. These phenomena indicate that the molecular mechanisms underlying of NTI drug targets in cancer require greater attention for the higher level of ‘degree’, ‘number of triangles’, and ‘neighborhood connectivity’.
3.5. Clinical implication of the identified features underlying NTI drug targets
Based on the above analyses, a total of ten features from three feature groups were identified as common features for both disease classes (cancers & cardiovascular diseases) in distinguishing NTI from NNTI drug targets, and there were another 4 features from two feature groups that were singled out by one of those 2 disease classes. Those shared feature groups identified in this study were consistent with our previous publication [20], which reaffirmed the importance of these shared features in differentiating NTI drug targets from NNTI ones. Since the vast majority of all NTI drugs were from those two disease classes (cancers & cardiovascular diseases), it was not surprising to have such similarity in the shared feature groups. Such shared features could also provide a new direction for optimizing the drug efficacy-safety balance [20]. Particularly, the importance of these shared features in the prediction of drug-induced hepatotoxicity has already been reported by the previous publication [133].
More importantly, those features that were unique in different disease classes were concentrated in two feature classes of Connectivity and Centrality. Particularly, the features unique to cancer included ‘degree’, ‘neighborhood connectivity’, and ‘number of triangles’. The ‘degree’ denoted the number of proteins in the human PPI network that interacted with the studied drug target [134]. The ‘neighborhood connectivity’ indicated the average number of interacting proteins of all the studied drug target’s neighbors [135]. The ‘number of triangles’ showed the number of triangles that included the studied target as a vertex [136], and this triangular relationship includes the studied drug target and its interacting proteins, as well as the interactions among the interacting proteins. In fact, these three features could collectively represent whether the studied drug target acted as a hub in the human PPI network, the higher their values the stronger the core position of the studied targets [137]. These findings represent an emphasis by cancers in differentiating NTI drug targets with respect to the target's ability to be a hub in the PPI network. In other words, the narrow therapeutic index of an anticancer drug may originate from its interaction with hub protein [138], [139]. To improve the situation of the narrow therapeutic index of anticancer drugs, it is necessary to impose more requirements on target selection. The hub proteins in the human PPI network should be avoided when designing anticancer drugs.
Different from cancers, the unique feature singled out in cardiovascular diseases is ‘distance deviation’, which belongs to the connectivity feature group. The ‘distance deviation’ indicated the absolute difference between the sum of all shortest paths starting from the studied target to all other proteins and the mean shortest path length of all the proteins in the human PPI network [135]. This implied an emphasis on the efficiency of inter-target signaling in NTI drug targets for cardiovascular diseases [135], [140], which may indicate the needs for a more in-depth study of target signaling pathways when designing drugs for cardiovascular diseases. All in all, this study identified the key target features indicating the NTI drugs for cancers and cardiovascular diseases, which has great clinical implications in the drug designs for both disease classes.
4. Conclusion
This work is the first practice to reveal the underlying mechanism behind the complex definition and interpretation of NTI between different disease classes. Ten shared and four unique features were identified for both disease classes (cancers & cardiovascular diseases) to distinguish NTI drug targets from NNTI ones. This work suggested that in the clinical study of avoiding narrow therapeutic index in those diseases, the ability of target to be a hub and the efficiency of target signaling in the human PPI network should be especially considered.
Author contributions
Feng ZHU, Su ZENG conceived the idea and supervised the work. Jiayi YIN, Xiaoxu LI performed the research. Jiayi YIN, Xiaoxu LI and Fengcheng LI prepared and analyzed the data. Jiayi YIN, Xiaoxu LI, Yinjing LU, wrote the manuscript. All authors have read and approved this manuscript.
Funding
Funded by National Natural Science Foundation of China (81872798 & U1909208); Natural Science Foundation of Zhejiang Province (LR21H300001); National Key R&D Program of China (2018YFC0910500); Leading Talent of the ‘Ten Thousand Plan’ - National High-Level Talents Special Support Plan of China; Fundamental Research Fund for Central Universities (2018QNA7023); Key R&D Program of Zhejiang Province (2020C03010). This work was supported by Alibaba-Zhejiang University Joint Research Center of Future Digital Healthcare; Alibaba Cloud; Information Technology Center of Zhejiang University.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.04.035.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Yu L.X., Jiang W., Zhang X., Lionberger R., Makhlouf F. Novel bioequivalence approach for narrow therapeutic index drugs. Clin Pharmacol Ther. 2015;97(3):286–291. doi: 10.1002/cpt.28. [DOI] [PubMed] [Google Scholar]
- 2.Muller P.Y., Milton M.N. The determination and interpretation of the therapeutic index in drug development. Nat Rev Drug Discov. 2012;11(10):751–761. doi: 10.1038/nrd3801. [DOI] [PubMed] [Google Scholar]
- 3.Tamargo J., Le Heuzey J.Y., Mabo P. Narrow therapeutic index drugs: a clinical pharmacological consideration to flecainide. Eur J Clin Pharmacol. 2015;71(5):549–567. doi: 10.1007/s00228-015-1832-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krens S.D., Lassche G., Jansman F.G.A., Desar I.M.E. Dose recommendations for anticancer drugs in patients with renal or hepatic impairment. Lancet Oncol. 2019;20(4):e200–e207. doi: 10.1016/S1470-2045(19)30145-7. [DOI] [PubMed] [Google Scholar]
- 5.Chen L., Chu C., Zhang Y.-H., Zheng M., Zhu L. Identification of drug-drug interactions using chemical interactions. Curr Bioinform. 2017;12(6):526–534. [Google Scholar]
- 6.Han H., Jain A.D., Truica M.I., Izquierdo-Ferrer J., Anker J.F. Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell. 2019;36(5):483–497. doi: 10.1016/j.ccell.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sang S.B., Guo X., Wang J.Z., Li H.M., Ma X.Y. Real-time and label-free detection of VKORC1 genes based on a magnetoelastic biosensor for warfarin therapy. J Mater Chem B. 2020;8(29):6271–6276. doi: 10.1039/d0tb00354a. [DOI] [PubMed] [Google Scholar]
- 8.Subbiah V., Yang D., Velcheti V., Drilon A., Meric-Bernstam F. State-of-the-art strategies for targeting RET-dependent cancers. J Clin Oncol. 2020;38(11):1209–1221. doi: 10.1200/JCO.19.02551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaykov A.N., Mayer J.P., DiMarchi R.D. Pursuit of a perfect insulin. Nat Rev Drug Discov. 2016;15(6):425–439. doi: 10.1038/nrd.2015.36. [DOI] [PubMed] [Google Scholar]
- 10.Moreno L., Pearson A.D.J., Paoletti X., Jimenez I., Geoerger B. Early phase clinical trials of anticancer agents in children and adolescents - an ITCC perspective. Nat Rev Clin Oncol. 2017;14(8):497–507. doi: 10.1038/nrclinonc.2017.59. [DOI] [PubMed] [Google Scholar]
- 11.Dixit V.A. A simple model to solve a drug toxicity problem. Toxicol Res. 2019;8(2):157–171. doi: 10.1039/c8tx00261d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jakhar R., Dangi M., Khichi A., Chhillar A.K. Relevance of molecular docking studies in drug designing. Curr Bioinform. 2020;15(4):270–278. [Google Scholar]
- 13.ten Freyhaus H., Dumitrescu D., Berghausen E., Vantler M., Caglayan E. Imatinib mesylate for the treatment of pulmonary arterial hypertension. Expert Opin Investig Drugs. 2012;21(1):119–134. doi: 10.1517/13543784.2012.632408. [DOI] [PubMed] [Google Scholar]
- 14.Cai Y., Zhang X., Shen J., Jiang B., Hu D. Heparin-binding protein: a novel biomarker linking four different cardiovascular diseases. Cardiol Res Pract. 2020;2020:9575373. doi: 10.1155/2020/9575373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J., Zhu K., Zheng W., Xu H. A comparative study of disease genes and drug targets in the human protein interactome. BMC Bioinf. 2015;16(Suppl 5):S1. doi: 10.1186/1471-2105-16-S5-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinrichs M.J.M., Ryan P.M., Zheng B., Afif-Rider S., Yu X.Q. Fractionated dosing improves preclinical therapeutic index of pyrrolobenzodiazepine-containing antibody drug conjugates. Clin Cancer Res. 2017;23(19):5858–5868. doi: 10.1158/1078-0432.CCR-17-0219. [DOI] [PubMed] [Google Scholar]
- 17.Bottino D.C., Patel M., Kadakia E., Zhou J., Patel C. Dose pptimization for anticancer drug combinations: maximizing therapeutic index via clinical exposure-toxicity/preclinical exposure-efficacy modeling. Clin Cancer Res. 2019;25(22):6633–6643. doi: 10.1158/1078-0432.CCR-18-3882. [DOI] [PubMed] [Google Scholar]
- 18.Zhao X., Chen L., Guo Z.-H., Liu T. Predicting drug side effects with compact integration of heterogeneous networks. Curr Bioinform. 2019;14(8):709–720. [Google Scholar]
- 19.Zhang L., Fang Y., Kopecek J., Yang J. A new construct of antibody-drug conjugates for treatment of B-cell non-Hodgkin's lymphomas. Eur J Pharm Sci. 2017;103:36–46. doi: 10.1016/j.ejps.2017.02.034. [DOI] [PubMed] [Google Scholar]
- 20.Li X.X., Yin J., Tang J., Li Y., Yang Q. Determining the balance between drug efficacy and safety by the network and biological system profile of its therapeutic target. Front Pharmacol. 2018;9:1245. doi: 10.3389/fphar.2018.01245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potega A., Zelaszczyk D., Mazerska Z. Electrochemical and in silico approaches for liver metabolic oxidation of antitumor-active triazoloacridinone C-1305. J Pharm Anal. 2020;10(4):376–384. doi: 10.1016/j.jpha.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drugs F. Pharmacometric approach to define narrow therapeutic index (NTI) drugs & evaluate bioequivalence (BE) criteria for NTI drugs. Drugs@FDA. 2018;1(1):1–25. [Google Scholar]
- 23.Bretagne M., Boudou-Rouquette P., Huillard O., Thomas-Schoemann A., Chahwakilian A. Tyrosine kinase inhibiting the VEGF pathway and elderly people: Tolerance, pre-treatment assessment and side effects management. Bull Cancer. 2016;103(3):259–272. doi: 10.1016/j.bulcan.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Lin Y.S., Kerr S.J., Randolph T., Shireman L.M., Senn T. Prediction of intravenous busulfan clearance by endogenous plasma biomarkers using global pharmacometabolomics. Metabolomics. 2016;12(10):161. doi: 10.1007/s11306-016-1106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciftci R., Tas F., Karabulut S., Ciftci S. Combination of capecitabine and phenytoin may cause phenytoin intoxication: a case report. Am J Ther. 2015;22(1):e17–e19. doi: 10.1097/MJT.0b013e318293b10a. [DOI] [PubMed] [Google Scholar]
- 26.Rousseau A., Marquet P., Debord J., Sabot C., Lachatre G. Adaptive control methods for the dose individualisation of anticancer agents. Clin Pharmacokinet. 2000;38(4):315–353. doi: 10.2165/00003088-200038040-00003. [DOI] [PubMed] [Google Scholar]
- 27.Onoda J.M., Nelson K.K., Taylor J.D., Honn K.V. Cisplatin and nifedipine: synergistic antitumor effects against an inherently cisplatin-resistant tumor. Cancer Lett. 1988;40(1):39–47. doi: 10.1016/0304-3835(88)90260-1. [DOI] [PubMed] [Google Scholar]
- 28.Romano M.J., Dinh A. A 1000-fold overdose of clonidine caused by a compounding error in a 5-year-old child with attention-deficit/hyperactivity disorder. Pediatrics. 2001;108(2):471–472. doi: 10.1542/peds.108.2.471. [DOI] [PubMed] [Google Scholar]
- 29.Ekhart C., Gebretensae A., Rosing H., Rodenhuis S., Beijnen J.H. Simultaneous quantification of cyclophosphamide and its active metabolite 4-hydroxycyclophosphamide in human plasma by high-performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;854(1–2):345–349. doi: 10.1016/j.jchromb.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 30.Gozalpour E., Greupink R., Bilos A., Verweij V., van den Heuvel J.J. Convallatoxin: a new P-glycoprotein substrate. Eur J Pharmacol. 2014;744:18–27. doi: 10.1016/j.ejphar.2014.09.031. [DOI] [PubMed] [Google Scholar]
- 31.Henderson J.D., Esham R.H. Generic substitution: issues for problematic drugs. South Med J. 2001;94(1):16–21. [PubMed] [Google Scholar]
- 32.Valdes R., Jr., Jortani S.A., Gheorghiade M. Standards of laboratory practice: cardiac drug monitoring. National academy of clinical biochemistry. Clin Chem. 1998;44(5):1096–1109. [PubMed] [Google Scholar]
- 33.Eckmann K., Michaud L.B., Rivera E., Madden T.L., Esparza-Guerra L. Pilot study to assess toxicity and pharmacokinetics of docetaxel in patients with metastatic breast cancer and impaired liver function secondary to hepatic metastases. J Oncol Pharm Pract. 2014;20(2):120–129. doi: 10.1177/1078155213480536. [DOI] [PubMed] [Google Scholar]
- 34.Han Y.L., Yu H.L., Li D., Meng X.L., Zhou Z.Y. Inhibitory effects of limonin on six human cytochrome P450 enzymes and P-glycoprotein in vitro. Toxicol In Vitro. 2011;25(8):1828–1833. doi: 10.1016/j.tiv.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 35.Simons F.E. Epinephrine (adrenaline) in the first-aid, out-of-hospital treatment of anaphylaxis. Novartis Found Symp. 2004;257:228–243. [PubMed] [Google Scholar]
- 36.Cersosimo R.J., Hong W.K. Epirubicin: a review of the pharmacology, clinical activity, and adverse effects of an adriamycin analogue. J Clin Oncol. 1986;4(3):425–439. doi: 10.1200/JCO.1986.4.3.425. [DOI] [PubMed] [Google Scholar]
- 37.Najar I.A., Johri R.K. Pharmaceutical and pharmacological approaches for bioavailability enhancement of etoposide. J Biosci. 2014;39(1):139–144. doi: 10.1007/s12038-013-9399-3. [DOI] [PubMed] [Google Scholar]
- 38.van Gelder T., Fischer L., Shihab F., Shipkova M. Optimizing everolimus exposure when combined with calcineurin inhibitors in solid organ transplantation. Transplant Rev. 2017;31(3):151–157. doi: 10.1016/j.trre.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Padhi D., Harris R. Clinical pharmacokinetic and pharmacodynamic profile of cinacalcet hydrochloride. Clin Pharmacokinet. 2009;48(5):303–311. doi: 10.2165/00003088-200948050-00002. [DOI] [PubMed] [Google Scholar]
- 40.Saif M.W., Diasio R.B. Benefit of uridine triacetate (vistogard) in rescuing severe 5-fluorouracil toxicity in patients with dihydropyrimidine dehydrogenase (DPYD) deficiency. Cancer Chemother Pharmacol. 2016;78(1):151–156. doi: 10.1007/s00280-016-3063-1. [DOI] [PubMed] [Google Scholar]
- 41.Fareed J., Hoppensteadt D.A., Bick R.L. Management of thrombotic and cardiovascular disorders in the new millenium. Clin Appl Thromb Hemost. 2003;9(2):101–108. doi: 10.1177/107602960300900202. [DOI] [PubMed] [Google Scholar]
- 42.Thirukkumaran C., Morris D.G. Oncolytic viral therapy using reovirus. Methods Mol Biol. 2015;1317:187–223. doi: 10.1007/978-1-4939-2727-2_12. [DOI] [PubMed] [Google Scholar]
- 43.Mercier C., Raynal C., Dahan L., Ortiz A., Evrard A. Toxic death case in a patient undergoing gemcitabine-based chemotherapy in relation with cytidine deaminase downregulation. Pharmacogenet Genomics. 2007;17(10):841–844. doi: 10.1097/FPC.0b013e32825ea6e3. [DOI] [PubMed] [Google Scholar]
- 44.Pogue S.L., Taura T., Bi M., Yun Y., Sho A. Targeting attenuated interferon-alpha to myeloma cells with a CD38 antibody induces potent tumor regression with reduced off-target activity. PLoS ONE. 2016;11(9) doi: 10.1371/journal.pone.0162472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker B.M., Cusack B.J., Vestal R.E. Pharmacokinetic optimisation of drug therapy in elderly patients. Drugs Aging. 1995;7(1):10–18. doi: 10.2165/00002512-199507010-00002. [DOI] [PubMed] [Google Scholar]
- 46.Sahasranaman S., Howard D., Roy S. Clinical pharmacology and pharmacogenetics of thiopurines. Eur J Clin Pharmacol. 2008;64(8):753–767. doi: 10.1007/s00228-008-0478-6. [DOI] [PubMed] [Google Scholar]
- 47.Shah I.A., Lindup W.E., McCulloch P.G. Variability of mitomycin C adsorption by activated charcoal. J Pharm Pharmacol. 1998;50(3):251–256. doi: 10.1111/j.2042-7158.1998.tb06857.x. [DOI] [PubMed] [Google Scholar]
- 48.Theile D., Haefeli W.E., Weiss J. Effects of adrenolytic mitotane on drug elimination pathways assessed in vitro. Endocrine. 2015;49(3):842–853. doi: 10.1007/s12020-014-0517-2. [DOI] [PubMed] [Google Scholar]
- 49.Hwang S.J., Park J.W., Lee S.D., Kim G.J., Sin C.H. Capecitabine and oxaliplatin (XELOX) for the treatment of patients with metastatic gastric cancer and severe liver dysfunction. Korean J Intern Med. 2006;21(4):252–255. doi: 10.3904/kjim.2006.21.4.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minhas S., Setia N., Pandita S., Saxena R., Verma I.C. Prevalence of CYP2C8 polymorphisms in a North Indian population. Genet Mol Res. 2013;12(3):2260–2266. doi: 10.4238/2013.July.8.7. [DOI] [PubMed] [Google Scholar]
- 51.Posada M.M., Bacon J.A., Schneck K.B., Tirona R.G., Kim R.B. Prediction of renal transporter mediated drug-drug interactions for pemetrexed using physiologically based pharmacokinetic modeling. Drug Metab Dispos. 2015;43(3):325–334. doi: 10.1124/dmd.114.059618. [DOI] [PubMed] [Google Scholar]
- 52.Umer Usman M.H., Raza S., Raza S., Ezekowitz M. Advancement in antithrombotics for stroke prevention in atrial fibrillation. J Interv Card Electrophysiol. 2008;22(2):129–137. doi: 10.1007/s10840-008-9210-9. [DOI] [PubMed] [Google Scholar]
- 53.Kuhn M.A. Herbal remedies: drug-herb interactions. Crit Care Nurse. 2002;22(2):22–28. [PubMed] [Google Scholar]
- 54.Said R., Tsimberidou A.M. Pharmacokinetic evaluation of vincristine for the treatment of lymphoid malignancies. Expert Opin Drug Metab Toxicol. 2014;10(3):483–494. doi: 10.1517/17425255.2014.885016. [DOI] [PubMed] [Google Scholar]
- 55.Ibrahim N.K., Valero V., Rahman Z., Theriault R.L., Walters R.S. Phase I-II vinorelbine (navelbine) by continuous infusion in patients with metastatic breast cancer: cumulative toxicities limit dose escalation. Cancer Invest. 2001;19(5):459–466. doi: 10.1081/cnv-100103844. [DOI] [PubMed] [Google Scholar]
- 56.Mullard A. 2012 FDA drug approvals. Nat Rev Drug Discov. 2013;12(2):87–90. doi: 10.1038/nrd3946. [DOI] [PubMed] [Google Scholar]
- 57.Carstairs S.D., Williams S.R. Overdose of aripiprazole, a new type of antipsychotic. J Emerg Med. 2005;28(3):311–313. doi: 10.1016/j.jemermed.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 58.Spencer T., Biederman J., Wilens T. Nonstimulant treatment of adult attention-deficit/hyperactivity disorder. Psychiatr Clin North Am. 2004;27(2):373–383. doi: 10.1016/j.psc.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 59.Mullard A. 2011 FDA drug approvals. Nat Rev Drug Discov. 2012;11(2):91–94. doi: 10.1038/nrd3657. [DOI] [PubMed] [Google Scholar]
- 60.Verbeeck R.K., Kanfer I., Lobenberg R., Abrahamsson B., Cristofoletti R. Biowaiver monographs for immediate-release solid oral dosage forms: enalapril. J Pharm Sci. 2017;106(8):1933–1943. doi: 10.1016/j.xphs.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 61.Walia K.S., Khan E.A., Ko D.H., Raza S.S., Khan Y.N. Side effects of antiepileptics–a review. Pain Pract. 2004;4(3):194–203. doi: 10.1111/j.1533-2500.2004.04304.x. [DOI] [PubMed] [Google Scholar]
- 62.Hughes B. 2008 FDA drug approvals. Nat Rev Drug Discov. 2009;8(2):93–96. doi: 10.1038/nrd2813. [DOI] [PubMed] [Google Scholar]
- 63.Strauch S., Jantratid E., Dressman J.B., Junginger H.E., Kopp S. Biowaiver monographs for immediate release solid oral dosage forms: lamivudine. J Pharm Sci. 2011;100(6):2054–2063. doi: 10.1002/jps.22449. [DOI] [PubMed] [Google Scholar]
- 64.Graefe-Mody U., Retlich S., Friedrich C. Clinical pharmacokinetics and pharmacodynamics of linagliptin. Clin Pharmacokinet. 2012;51(7):411–427. doi: 10.2165/11630900-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 65.Eklund J.W., Trifilio S., Mulcahy M.F. Chemotherapy dosing in the setting of liver dysfunction. Oncology (Williston Park) 2005;19(8):1057–1063. [PubMed] [Google Scholar]
- 66.Saletan S. Mitoxantrone: an active, new antitumor agent with an improved therapeutic index. Cancer Treat Rev. 1987;14(3–4):297–303. doi: 10.1016/0305-7372(87)90021-1. [DOI] [PubMed] [Google Scholar]
- 67.Knorr B., Franchi L.M., Bisgaard H., Vermeulen J.H., LeSouef P. Montelukast, a leukotriene receptor antagonist, for the treatment of persistent asthma in children aged 2 to 5 years. Pediatrics. 2001;108(3):E48. doi: 10.1542/peds.108.3.e48. [DOI] [PubMed] [Google Scholar]
- 68.Eichenfield L.F., Beck L. Elidel (pimecrolimus) cream 1%: a nonsteroidal topical agent for the treatment of atopic dermatitis. J Allergy Clin Immunol. 2003;111(5):1153–1168. doi: 10.1067/mai.2003.1492. [DOI] [PubMed] [Google Scholar]
- 69.Hot drugs 2004. Nat Rev Drug Discov. 2004;Suppl:S3-40. [PubMed]
- 70.Wang X., Zhang Z.Y., Arora S., Hughes L., Wang J. Effects of rolapitant administered intravenously or orally on the pharmacokinetics of digoxin (P-glycoprotein substrate) and sulfasalazine (breast cancer resistance protein substrate) in healthy volunteers. J Clin Pharmacol. 2018;58(2):202–211. doi: 10.1002/jcph.1005. [DOI] [PubMed] [Google Scholar]
- 71.Hughes B. 2009 FDA drug approvals. Nat Rev Drug Discov. 2010;9(2):89–92. doi: 10.1038/nrd3101. [DOI] [PubMed] [Google Scholar]
- 72.Schwartz L.M., Woloshin S., Zheng E., Tse T., Zarin D.A. ClinicalTrials.gov and Drugs@FDA: a comparison of results reporting for new drug approval trials. Ann Intern Med. 2016;165(6):421–430. doi: 10.7326/M15-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.The L. ICD-11. Lancet. 2019;393(10188):2275. doi: 10.1016/S0140-6736(19)31205-X. [DOI] [PubMed] [Google Scholar]
- 74.Wang Y., Zhang S., Li F., Zhou Y., Zhang Y. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48(D1):D1031–D1041. doi: 10.1093/nar/gkz981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghosh S., Kumar G.V., Basu A., Banerjee A. Graph theoretic network analysis reveals protein pathways underlying cell death following neurotropic viral infection. Sci Rep. 2015;5:14438. doi: 10.1038/srep14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu K., Wang Z.Q., Wang S.J., Liu P., Qin Y.H. Hyaluronic acid-tagged silica nanoparticles in colon cancer therapy: therapeutic efficacy evaluation. Int J Nanomedicine. 2015;10:6445–6454. doi: 10.2147/IJN.S89476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang P., Tao L., Zeng X., Qin C., Chen S. A protein network descriptor server and its use in studying protein, disease, metabolic and drug targeted networks. Brief Bioinform. 2017;18(6):1057–1070. doi: 10.1093/bib/bbw071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas S., Bonchev D. A survey of current software for network analysis in molecular biology. Hum Genomics. 2010;4(5):353–360. doi: 10.1186/1479-7364-4-5-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kogenaru S., del Val C., Hotz-Wagenblatt A., Glatting K.H. TissueDistributionDBs: a repository of organism-specific tissue-distribution profiles. Theor Chem Acc. 2010;125(3–6):651–658. [Google Scholar]
- 83.UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zheng C.J., Han L.Y., Yap C.W., Ji Z.L., Cao Z.W. Therapeutic targets: progress of their exploration and investigation of their characteristics. Pharmacol Rev. 2006;58(2):259–279. doi: 10.1124/pr.58.2.4. [DOI] [PubMed] [Google Scholar]
- 85.Zhu F., Han L., Zheng C., Xie B., Tammi M.T. What are next generation innovative therapeutic targets? Clues from genetic, structural, physicochemical, and systems profiles of successful targets. J Pharmacol Exp Ther. 2009;330(1):304–315. doi: 10.1124/jpet.108.149955. [DOI] [PubMed] [Google Scholar]
- 86.Song L., Xu W., Li C., Li H., Wu L. Development of expressed sequence tags from the bay scallop. Argopecten irradians irradians Mar Biotechnol (NY) 2006;8(2):161–169. doi: 10.1007/s10126-005-0126-4. [DOI] [PubMed] [Google Scholar]
- 87.Singh S., Malik B.K., Sharma D.K. Choke point analysis of metabolic pathways in E.histolytica: a computational approach for drug target identification. Bioinformation. 2007;2(2):68–72. doi: 10.6026/97320630002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yin J., Sun W., Li F., Hong J., Li X. VARIDT 1.0: variability of drug transporter database. Nucleic Acids Res. 2020;48(D1):D1042–D1050. doi: 10.1093/nar/gkz779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yin J., Li F., Zhou Y., Mou M., Lu Y. INTEDE: interactome of drug-metabolizing enzymes. Nucleic Acids Res. 2021;49(D1):D1233–D1243. doi: 10.1093/nar/gkaa755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Y.H., Yu C.Y., Li X.X., Zhang P., Tang J. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018;46(D1):D1121–D1127. doi: 10.1093/nar/gkx1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(D1):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Higueruelo A.P., Jubb H., Blundell T.L. Protein-protein interactions as druggable targets: recent technological advances. Curr Opin Pharmacol. 2013;13(5):791–796. doi: 10.1016/j.coph.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 93.Rao HB, Zhu F, Yang GB, Li ZR, Chen YZ. Update of PROFEAT: a web server for computing structural and physicochemical features of proteins and peptides from amino acid sequence. Nucleic Acids Res. 2011;39(Web Server issue):W385-90. [DOI] [PMC free article] [PubMed]
- 94.Xu J., Wang P., Yang H., Zhou J., Li Y. Comparison of FDA approved kinase targets to clinical trial ones: insights from their system profiles and drug-target interaction networks. Biomed Res Int. 2016;2016:2509385. doi: 10.1155/2016/2509385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Y.H., Li X.X., Hong J.J., Wang Y.X., Fu J.B. Clinical trials, progression-speed differentiating features and swiftness rule of the innovative targets of first-in-class drugs. Brief Bioinform. 2020;21(2):649–662. doi: 10.1093/bib/bby130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang L.H., He Q.S., Liu K., Cheng J., Zhong M.D. ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018;46(D1):D911–D917. doi: 10.1093/nar/gkx899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Issa N.T., Stathias V., Schurer S., Dakshanamurthy S. Machine and deep learning approaches for cancer drug repurposing. Semin Cancer Biol. 2021;68:132–142. doi: 10.1016/j.semcancer.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu J., Lian X., Liu F., Yan X., Cheng C. Identification of novel key targets and candidate drugs in oral squamous cell carcinoma. Curr Bioinform. 2020;15(4):328–337. [Google Scholar]
- 99.Yang Q., Li B., Tang J., Cui X., Wang Y. Consistent gene signature of schizophrenia identified by a novel feature selection strategy from comprehensive sets of transcriptomic data. Brief Bioinform. 2020;21(3):1058–1068. doi: 10.1093/bib/bbz049. [DOI] [PubMed] [Google Scholar]
- 100.Tang J., Fu J., Wang Y., Li B., Li Y. ANPELA: analysis and performance assessment of the label-free quantification workflow for metaproteomic studies. Brief Bioinform. 2020;21(2):621–636. doi: 10.1093/bib/bby127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang Q., Wang Y., Zhang Y., Li F., Xia W. NOREVA: enhanced normalization and evaluation of time-course and multi-class metabolomic data. Nucleic Acids Res. 2020;48(W1):W436–W448. doi: 10.1093/nar/gkaa258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Romm E.L., Tsigelny I.F. Artificial intelligence in drug treatment. Annu Rev Pharmacol Toxicol. 2020;60:353–369. doi: 10.1146/annurev-pharmtox-010919-023746. [DOI] [PubMed] [Google Scholar]
- 103.Tang J., Mou M., Wang Y., Luo Y., Zhu F. MetaFS: Performance assessment of biomarker discovery in metaproteomics. Brief Bioinform. 2020 doi: 10.1093/bib/bbaa105. [DOI] [PubMed] [Google Scholar]
- 104.Marya N.B., Powers P.D., Fujii-Lau L., Abu Dayyeh B.K., Gleeson F.C. Application of artificial intelligence using a novel EUS-based convolutional neural network model to identify and distinguish benign and malignant hepatic masses. Gastrointest Endosc. 2020 doi: 10.1016/j.gie.2020.08.024. [DOI] [PubMed] [Google Scholar]
- 105.Li Z., Wang Y., Tai S., Wang J., Huang Y. APP medical diagnostic check-up consultation system based on speech recognition. Curr Bioinform. 2020;15(5):408–414. [Google Scholar]
- 106.Tang J., Fu J., Wang Y., Luo Y., Yang Q. Simultaneous improvement in the precision, accuracy, and robustness of label-free proteome quantification by optimizing data manipulation chains. Mol Cell Proteomics. 2019;18(8):1683–1699. doi: 10.1074/mcp.RA118.001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kwon E., Cho M., Kim H., Son H.S. A study on host tropism determinants of influenza virus using machine learning. Curr Bioinform. 2020;15(2):121–134. [Google Scholar]
- 108.Xue W., Yang F., Wang P., Zheng G., Chen Y. What contributes to serotonin-norepinephrine reuptake inhibitors' dual-targeting mechanism? The key role of transmembrane domain 6 in human serotonin and norepinephrine transporters revealed by molecular dynamics simulation. ACS Chem Neurosci. 2018;9(5):1128–1140. doi: 10.1021/acschemneuro.7b00490. [DOI] [PubMed] [Google Scholar]
- 109.Li F., Zhou Y., Zhang X., Tang J., Yang Q. SSizer: determining the sample sufficiency for comparative biological study. J Mol Biol. 2020;432(11):3411–3421. doi: 10.1016/j.jmb.2020.01.027. [DOI] [PubMed] [Google Scholar]
- 110.van der Burgh H.K., Schmidt R., Westeneng H.J., de Reus M.A., van den Berg L.H. Deep learning predictions of survival based on MRI in amyotrophic lateral sclerosis. Neuroimage Clin. 2017;13:361–369. doi: 10.1016/j.nicl.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seo S., Oh M., Park Y., Kim S. DeepFam: deep learning based alignment-free method for protein family modeling and prediction. Bioinformatics. 2018;34(13):i254–i262. doi: 10.1093/bioinformatics/bty275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang J., Wang H., Wang X., Chang H. Predicting drug-target interactions via FM-DNN learning. Curr Bioinform. 2020;15(1):68–76. [Google Scholar]
- 113.Krittanawong C., Zhang H., Wang Z., Aydar M., Kitai T. Artificial intelligence in precision cardiovascular medicine. J Am Coll Cardiol. 2017;69(21):2657–2664. doi: 10.1016/j.jacc.2017.03.571. [DOI] [PubMed] [Google Scholar]
- 114.Kursa M.B. Robustness of random forest-based gene selection methods. BMC Bioinf. 2014;15:8. doi: 10.1186/1471-2105-15-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pan Y., Wang Z., Zhan W., Deng L. Computational identification of binding energy hot spots in protein-RNA complexes using an ensemble approach. Bioinformatics. 2018;34(9):1473–1480. doi: 10.1093/bioinformatics/btx822. [DOI] [PubMed] [Google Scholar]
- 116.Degenhardt F., Seifert S., Szymczak S. Evaluation of variable selection methods for random forests and omics data sets. Brief Bioinform. 2019;20(2):492–503. doi: 10.1093/bib/bbx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tang J., Wang Y., Luo Y., Fu J., Zhang Y. Computational advances of tumor marker selection and sample classification in cancer proteomics. Comput Struct Biotechnol J. 2020;18:2012–2025. doi: 10.1016/j.csbj.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shang L., Liu C., Tomiura Y., Hayashi K. Machine-learning-based olfactometer: prediction of odor perception from physicochemical features of odorant molecules. Anal Chem. 2017;89(22):11999–12005. doi: 10.1021/acs.analchem.7b02389. [DOI] [PubMed] [Google Scholar]
- 119.Ragusa M., Avola G., Angelica R., Barbagallo D., Guglielmino M.R. Expression profile and specific network features of the apoptotic machinery explain relapse of acute myeloid leukemia after chemotherapy. BMC Cancer. 2010;10:377. doi: 10.1186/1471-2407-10-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guo R., Zhang X., Su J., Xu H., Zhang Y. Identifying potential quality markers of Xin-Su-Ning capsules acting on arrhythmia by integrating UHPLC-LTQ-orbitrap, ADME prediction and network target analysis. Phytomedicine. 2018;44:117–128. doi: 10.1016/j.phymed.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 121.Fathima A.J., Murugaboopathi G., Selvam P. Pharmacophore mapping of ligand based virtual screening, molecular docking and molecular dynamic simulation studies for finding potent NS2B/NS3 protease inhibitors as potential anti-dengue drug compounds. Curr Bioinform. 2018;13(6):606–616. [Google Scholar]
- 122.Ma B., Wang H.Z., Dsouza M., Lou J., He Y. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. Isme J. 2016;10(8):1891–1901. doi: 10.1038/ismej.2015.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Emig D., Ivliev A., Pustovalova O., Lancashire L., Bureeva S. Drug target prediction and repositioning using an integrated network-based approach. PLoS ONE. 2013;8(4) doi: 10.1371/journal.pone.0060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen L., Wang Q., Zhang L., Tai J., Wang H. A novel paradigm for potential drug-targets discovery: quantifying relationships of enzymes and cascade interactions of neighboring biological processes to identify drug-targets. Mol Biosyst. 2011;7(4):1033–1041. doi: 10.1039/c0mb00249f. [DOI] [PubMed] [Google Scholar]
- 125.Muhammad J., Khan A., Ali A., Fang L., Yanjing W. Network pharmacology: exploring the resources and methodologies. Curr Top Med Chem. 2018;18(12):949–964. doi: 10.2174/1568026618666180330141351. [DOI] [PubMed] [Google Scholar]
- 126.Wu C.C., Kannan K., Lin S., Yen L.S., Milosavljevic A. Identification of cancer fusion drivers using network fusion centrality. Bioinformatics. 2013;29(9):1174–1181. doi: 10.1093/bioinformatics/btt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang C., Gong B.S., Bushel P.R., Thierry-Mieg J., Thierry-Mieg D. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol. 2014;32(9):926–932. doi: 10.1038/nbt.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Scardoni G., Petterlini M., Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25(21):2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang L.C., Li X., Tai J.X., Li W., Chen L.N. Predicting candidate genes based on combined network topological features: a case study in coronary artery disease. PLoS ONE. 2012;7(6) doi: 10.1371/journal.pone.0039542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Modos D., Bulusu K.C., Fazekas D., Kubisch J., Brooks J. Neighbours of cancer-related proteins have key influence on pathogenesis and could increase the drug target space for anticancer therapies. NPJ Syst Biol Appl. 2017;3:2. doi: 10.1038/s41540-017-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kar G., Gursoy A., Keskin O. Human cancer protein-protein interaction network: a structural perspective. PLoS Comput Biol. 2009;5(12) doi: 10.1371/journal.pcbi.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ma X.J., Ma Y.H. The local triangle structure centrality method to rank nodes in networks. Complex. 2019;9057194 [Google Scholar]
- 133.Xu L., Liang G., Liao C., Chen G.D., Chang C.C. k-Skip-n-Gram-RF: a random forest based method for alzheimer's disease protein identification. Front Genet. 2019;10:33. doi: 10.3389/fgene.2019.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lawyer G. Understanding the influence of all nodes in a network. Sci Rep. 2015;5:8665. doi: 10.1038/srep08665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang P., Tao L., Zeng X., Qin C., Chen S.Y. PROFEAT update: a protein features web server with added facility to compute network descriptors for studying omics-derived networks. J Mol Biol. 2017;429(3):416–425. doi: 10.1016/j.jmb.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 136.Rubinov M., Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52(3):1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 137.Wheeler H., Woods R.L., Page J., Levi J. A phase II study of mitoxantrone in advanced squamous cell cancer of the head and neck. Invest New Drugs. 1990;8(1):109–111. doi: 10.1007/BF00216935. [DOI] [PubMed] [Google Scholar]
- 138.Jeong H., Mason S.P., Barabasi A.L., Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 139.Barabasi A.L., Oltvai Z.N. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 140.Embar V., Handen A., Ganapathiraju M.K. Is the average shortest path length of gene set a reflection of their biological relatedness? J Bioinform Comput Biol. 2016;14(6):1660002. doi: 10.1142/S0219720016600027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.