Abstract
Objectives:
Vascular endothelial cells (ECs) play a critical role in maintaining vascular homeostasis. Aberrant EC metabolism leads to vascular dysfunction and metabolic diseases. Transcription factor EB (TFEB), a master regulator of lysosome biogenesis and autophagy, has protective effects on vascular inflammation and atherosclerosis. However, the role of endothelial TFEB in metabolism remains to be explored. In this study, we sought to investigate the role of endothelial TFEB in glucose metabolism and underlying molecular mechanisms.
Approach and Results:
To determine whether endothelial TFEB is critical for glucose metabolism in vivo, we utilized EC-selective TFEB knockout (EC-TFEB KO) and transgenic mice (EC-TFEB Tg) fed a high-fat diet (HFD). EC-TFEB KO mice exhibited significantly impaired glucose tolerance compared with control mice. Consistently, EC-TFEB Tg mice showed improved glucose tolerance. In primary human ECs, small interfering RNA-mediated TFEB knockdown blunts Akt signaling. Adenovirus-mediated overexpression of TFEB consistently activates Akt and significantly increases glucose uptake in ECs. Mechanistically, TFEB upregulates insulin receptor substrate 1 and 2 (IRS1 and IRS2). TFEB increases IRS2 transcription measured by reporter gene and chromatin immunoprecipitation assays. Furthermore, we found that TFEB increases IRS1 protein via downregulation of microRNAs (miR-335, miR-495 and miR-548o). In vivo, Akt signaling in the skeletal muscle and adipose tissue was significantly impaired in EC-TFEB KO mice and consistently improved in EC-TFEB Tg mice on HFD.
Conclusions:
Our data revealed a critical role of TFEB in endothelial metabolism and suggest that TFEB constitutes a potential molecular target for the treatment of vascular and metabolic diseases.
Keywords: TFEB, endothelial cells, IRS1, IRS2, microRNA, insulin resistance
Subject terms: Vascular Disease, Cell Signaling/Signal Transduction, Metabolism, Mechanisms Or
Graphical Abstract
INTRODUCTION
Endothelial cells (ECs) line the lumen surface of blood vessels and maintain vascular homeostasis. EC dysfunction is prevalent in cardiovascular disease (CVD) and leads to the pathogenesis of atherosclerosis, hypertension and retinopathy. Emerging studies suggest a causal role for ECs in systemic metabolic dysregulation1. EC metabolism is involved in vessel sprouting2 and systemic metabolism1. Modulation of EC metabolism can markedly affect systemic glucose and lipid homeostasis, underscoring the critical role of endothelium in the regulation of whole-body metabolism1.
Transcription factor EB (TFEB) is a transcription factor containing basic helix-loop-helix (HLH) and leucine zipper (LZ) domains3, which was characterized as a master regulator of autophagy and lysosomal biogenesis4, 5. TFEB improves metabolic syndrome via autophagy-dependent regulation of lipid and glucose metabolism in the adipose tissue and liver6-10.
TFEB is required for vascularization in the placenta during mouse embryo development11. Recently, others and we revealed that TFEB inhibits endothelial inflammation12, 13, positively regulates postischemic angiogenesis14 and controls vascular development by promoting EC proliferation15. However, whether endothelial TFEB can regulate systemic metabolism remains unknown.
In this study, utilizing EC-TFEB knockout (KO) mice and EC-TFEB transgenic (Tg) mice, we found that endothelial TFEB improves glucose tolerance under the high-fat diet (HFD) conditions. We demonstrated that TFEB activates Akt signaling and glucose uptake via upregulation of IRS1 and IRS2 in ECs.
MATERIALS AND METHODS
The data that support the findings of this study are available from the corresponding author on reasonable request. An extended version of this section is available as Supplemental methods in the SUPPLEMENTAL MATERIAL.
Animal experiments
All animal experiments were performed according to the guidelines of the University of Michigan Animal Care and Use Committee. The EC-selective TFEB transgenic (mTie2 promoter-driven) mice (EC-TFEB Tg) (C57BL/6J background) and EC-selective TFEB knockout (floxed Tfeb/Ve-Cadherin Cre +) mice (EC-TFEB KO) (C57BL/6N background) were described as previously 14. Littermate wild-type mice were used as control mice for the EC-TFEB Tg mice and littermate floxed Tfeb mice were used as control mice for the EC-TFEB KO mice. Mice were fed a normal laboratory diet (22.5% protein, 11.8% fat, and 52% carbohydrate by mass; Cat# 5LOD, LabDiet), or high-fat diet (60% fat, 20% carbohydrate, 20% protein; Cat# D12492, Research Diets, New Brunswick, NJ, USA). Sex difference in insulin resistance was reported in humans and rodents previously. Females are more insulin sensitive than males16. In the present study, insulin sensitivity was altered only in the male EC-TFEB transgenic mice but not in the female mice. Therefore, we used male mice to determine the role of endothelial TFEB in glucose metabolism in this study.
Small RNA-Seq and microarray analysis
RNA was isolated with miRNeasy Mini Kit (Cat# 217004, Qiagen). The small RNA library preparation and deep sequencing were performed by the Advanced Genomics Core at the University of Michigan. Briefly, RNA quality was assessed by TapeStation RNA ScreenTape (Cat # 5067-5576, Agilent), and RNA abundance was quantified by Qubit RNA BR Assay Kit (Cat #Q10210, Thermo Fisher). Indexed small RNA libraries were prepared with NEB Small RNA kit with Size Selection Using Pippin Prep per protocol (NEB #E7300S) and sequenced on a NovaSeq 6000 platform (Illumina). Pair-end 51 bp reads were generated according to the manufacturer’s recommendation. On average, ~13 million reads were obtained for each sample. The read quality check was performed with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Small RNA-Seq data were then analyzed using OASIS 2.017 (https://oasis.ims.bio/index.php) with the default settings. Differentially expressed miRNAs were identified by comparing two groups with absolute fold change ≥ 1.5 and adjusted p-value < 0.05. Microarray analysis of TFEB-regulated genes in human umbilical vein endothelial cells (HUVECs) (GEO, accession number GSE108384), and RNA seq analysis of glucose transporters in human coronary artery endothelial cells (HCAECs) (GEO, accession number, GSE124522) were published previously14, 18.
miRNAs targeting IRS1 prediction
miRNAs targeting IRS1 was predicted by online bioinformatics databases: miRDB19, TargetScan 20 and DIANA-microT web server v5.0 21. The miRNA candidates predicted by at least two databases were selected for further analysis. The miRNAs that are both regulated by TFEB with the basal read counts above 60 from small RNA-Seq and are predicted as IRS1 targeting miRNAs were merged in R (version 3.6.1) to identify the potential TFEB-regulated miRNAs targeting IRS1.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using the SimpleChIP Enzymatic Chromatin IP Kit with Magnetic Beads (Cat# 9003S, Cell Signaling Technology) in accordance with the manufacturer’s protocol. The primer sequences are shown in the Major Resources Table in the Supplemental Materials.
ChIP-Seq data analysis
ChIP-Seq data were obtained from the GEO database (accession number GSE88894). Reads from the dataset were mapped to the human genome (NCBI GRCh38) with bowtie2. The peak calling was done with MACS2 and the reads from input served as a control. Samples were normalized to adjust for sequencing depth. The peaks were visualized in Integrative Genomics Viewer (IGV).
Statistical Analysis
All statistical analyses were performed using GraphPad Prism version 7.0 (GraphPad Software, San Diego, CA). Data were analyzed for normality and equal variance. Student t-test was used to compare two groups. One-way ANOVA was used for comparisons among >2 groups. Two-way ANOVA was used for comparisons among >2 groups and >2 conditions. The Bonferroni post hoc test was applied for multiple comparisons. Data are presented as mean ± SE of mean (SEM). A P value < 0.05 was considered statistically significant.
Data Sharing Statement
MicroRNA data have been deposited in NCBI Gene Expression Omnibus and are accessible through GEO series accession Number GSE148026 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148026).
RESULTS
Endothelial TFEB attenuates systemic glucose intolerance in mice on HFD.
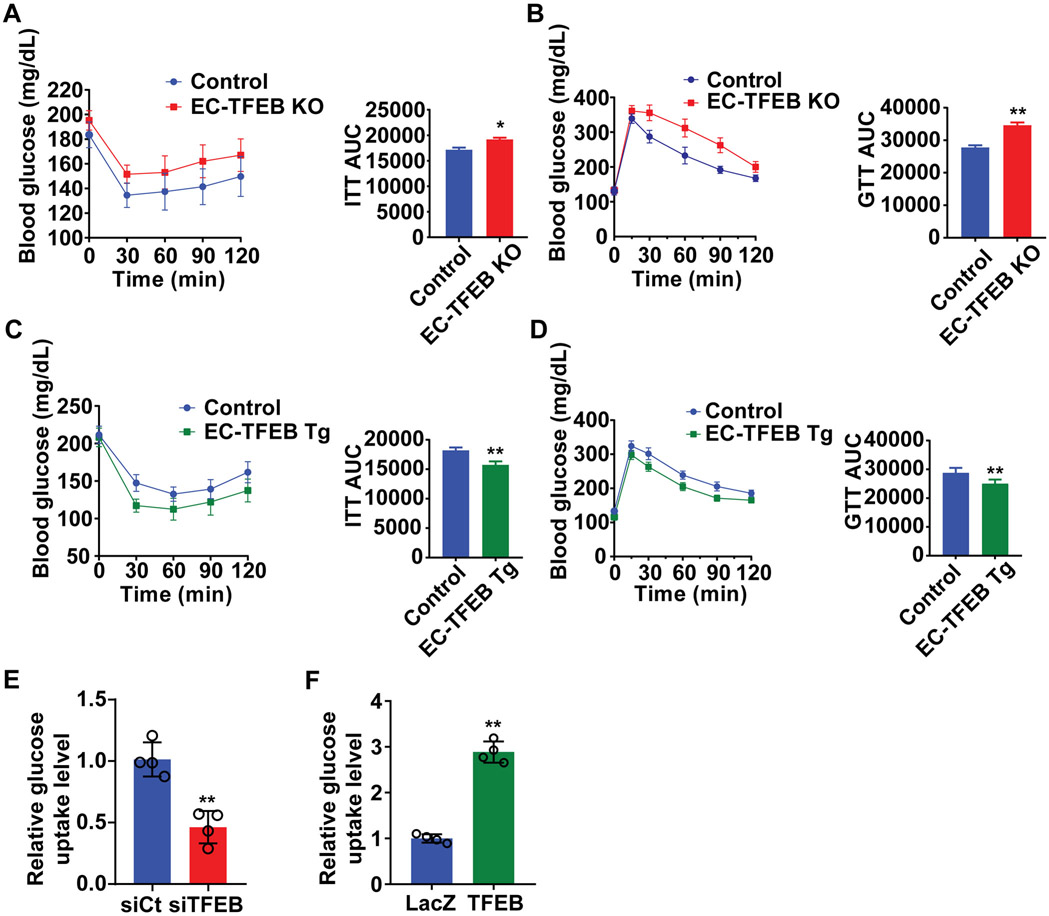
ECs regulate systemic glucose metabolism in vivo and EC dysfunction contributes to obesity and metabolic disorders1, 22. Using EC-TFEB KO mice and EC-TFEB Tg mice (Supplemental Figure I A-B), we found that there is no significant difference in glucose tolerance and insulin sensitivity in the male EC-TFEB KO mice and EC-TFEB Tg mice on normal laboratory diet when compared with their respective control mice (Supplemental Figure II A-D). The high-fat diet (HFD) mouse model has become one of the most important tools for understanding the interplay of high fat diets with obesity, hyperglycemia and insulin resistance23. EC-TFEB KO mice on HFD exhibited worsened glucose tolerance and insulin sensitivity, assessed by glucose tolerance test (GTT) and insulin tolerance test (ITT) (Figure 1A-B). Consistently, EC-TFEB Tg mice on HFD showed improved glucose tolerance and insulin sensitivity compared with the control group (Figure 1C-D). However, there was no difference in female mice on HFD between EC-TFEB KO mice and control mice (Supplemental Figure II E-F). Our in vivo data indicate that endothelial TFEB improves glucose dysregulation upon HFD challenge, especially in male mice. In human coronary artery endothelial cells (HCAECs), TFEB knockdown reduced glucose uptake to 51 ± 9% compared with the siRNA-control (siCt) group (Figure 1E). Moreover, adenovirus-mediated overexpression of TFEB increased glucose uptake up to 2.89 ± 0.12-fold (Figure 1F), indicating a critical role of TFEB in endothelial glucose metabolism.
Figure 1. Endothelial transcription factor EB (TFEB) improves systemic glucose tolerance in male mice on high fat diet (HFD).
Male endothelial TFEB genetically engineered transgenic mice on HFD were subjected to insulin tolerance test (ITT) or glucose tolerance test (GTT). A-B, EC-TFEB knockout (KO) and control mice were fed an HFD for 26 weeks. A, ITT (insulin, 1 U/kg) and its area under the curve (AUC) (n=7-9/each group). B, GTT (glucose, 1 g/kg) via intraperitoneal injection and its AUC (n=7-9/each group). C-D, EC-TFEB Tg mice and control mice were fed HFD for 20 weeks. C, ITT (insulin, 1 U/kg) and its AUC (n=11/each group). D, GTT (glucose, 1 g/kg) and its AUC in (n=11/each group). E, Human coronary artery endothelial cells (HCAECs) were transfected with siRNA-control (siCt, 25 nM) or siRNA-TFEB (siTFEB, 25 nM) for 48 hours, and then incubated with 1mM 2-deoxyglucose for 40 minutes. Glucose uptake in ECs was measured by Glucose Uptake-Glo assay (n=4/each group). F, HCAECs were infected with adenovirus encoding LacZ (Ad-LacZ; multiplicity of infection [MOI], 10) or TFEB (Ad-TFEB, 10 MOI). Forty-eight hours later, glucose uptake was measured as in E (n=4/each group). Data are presented as mean ± SEM. Left panels in A, B, C and D were analyzed by 2-way ANOVA followed by Bonferroni test. Right panels for AUC in A, B, C and D used unpaired Student’s t-test; Data in E and F used unpaired Student’s t-test. *P <0.05, **P <0.01.
TFEB increases glucose uptake via activation of Akt in ECs.
Insulin-activated Akt signaling in ECs is involved in the regulation of systemic glucose metabolism22,24. We tested whether TFEB regulates Akt signaling in ECs. In HCAECs, TFEB knockdown reduced the phosphorylation of Akt at both S473 and T308 to an average of 40% and 46%, respectively, when compared with siCt group (Figure 2A). Consistently, adenovirus-mediated overexpression of TFEB increased Akt phosphorylation at S473 and T308 up to 2.06 ± 0.30-fold and 1.73 ± 0.10-fold (Figure 2B). Palmitic acid (PA), a very common saturated fatty acid found in many diets, induces insulin resistance and type 2 diabetes in vivo25. In the presence of PA, TFEB still increased Akt phosphorylation at both S473 and T308 up to 1.60 ± 0.20-fold and 1.54 ± 0.10-fold, respectively (Figure 2B), suggesting a promoting effect of TFEB on Akt signaling in ECs. Akt activation promotes glucose uptake by inducing GLUT1 plasma membrane translocation 26, 27. We found that LY294002, an inhibitor of phosphoinositide 3-kinase (PI3K)/Akt pathway, significantly attenuated TFEB-induced glucose uptake (Figure 2C), suggesting Akt is essential for TFEB to increase glucose uptake. GLUT1 is the major glucose transporter in ECs28, 29. We analyzed RNA-seq data from HCAECs available in the Gene Expression Omnibus (GEO) database (accession number: GSE124522)18, and revealed that GLUT1 is the major glucose transporter in HCAECs (Supplemental Figure III A). Although TFEB did not change GLUT1 expression at the protein level (Supplemental Figure III B-C), TFEB-dependent glucose uptake was partially abolished by GLUT1 knockdown (Figure 2D-E). Thus, our data suggest that Akt-GLUT1 mediates the effect of TFEB on EC glucose uptake.
Figure 2. Transcription factor EB (TFEB) activates AKT serine/threonine kinase (Akt) signaling in endothelial cells (ECs).
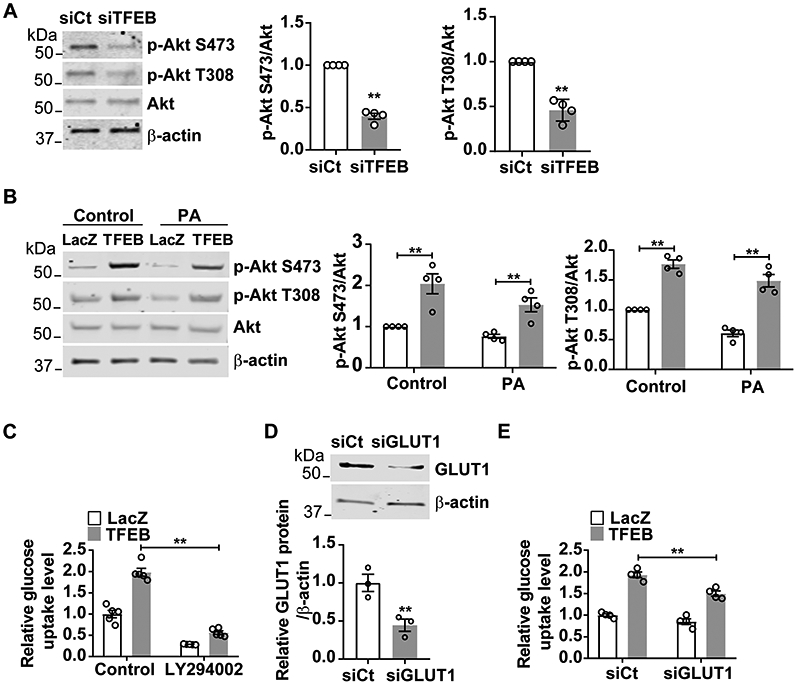
A, Human coronary artery endothelial cells (HCAECs) were transfected with siRNA-control (siCt; 25 nM) or siRNA-TFEB (siTFEB; 25 nM) for 48 hours. The phosphorylation of Akt was determined by Western blot from four independent experiments. B, HCAECs were infected with Ad-LacZ (10 MOI) or Ad-TFEB (10 MOI). After twenty-four hours, cells were treated with palmitic acid (PA; 200 μM). Twenty-four hours later, the phosphorylation of Akt was determined by Western blot from four independent experiments. C, HCAECs were infected with Ad-LacZ (10 MOI) or Ad-TFEB (10 MOI). After thirty-six hours, cells were treated with LY294002 (50 μM) for 12 hours. Glucose uptake was measured by Glucose Uptake-Glo Assay (n=5/each group). D-E, HCAECs were transfected with siCt (25 nM) or siRNA- glucose transporter 1 (siGLUT1; 25nM). Twenty-four hours later, cells were infected with Ad-LacZ or Ad-TFEB (10 MOI) for forty-eight hours. D, GLUT1 protein was determined by Western blot (n=3/each group). E, glucose uptake was measured by Glucose Uptake-Glo Assay (n=4/each group). Data are presented as mean ± SEM. Data in A, B and D used unpaired Student’s t-test. Data in C and E were analyzed by 2-way ANOVA followed by Bonferroni test. *P <0.05, **P <0.01.
TFEB activates Akt signaling via upregulation of IRS1 and IRS2.
To explore how TFEB activates Akt signaling, we re-analyzed the microarray data from HUVECs overexpressing TFEB14. Among the TFEB-regulated genes, insulin receptor substrate 2 (IRS2), which is the critical node of the insulin-Akt signaling30, was upregulated by TFEB in ECs (Supplemental Figure IV). Our qRT-PCR and Western blot data demonstrated that TFEB upregulated both mRNA and protein of IRS2 in ECs (Figure 3A-B). Furthermore, although the mRNA level of insulin receptor substrate 1 (IRS1), another important member of IRS family30 was not changed by TFEB overexpression (Figure 3A), IRS1 protein was significantly upregulated (Figure 3B). Consistently, there was a reduction of IRS2 at both mRNA and protein levels but only a reduction of IRS1 protein in TFEB-knockdown ECs (Figure 3C-3D).
Figure 3. Transcription factor EB (TFEB) activates the AKT serine/threonine kinase (Akt) signaling pathway by upregulating insulin receptor substrate 1 (IRS1) and insulin receptor substrate 2 (IRS2).
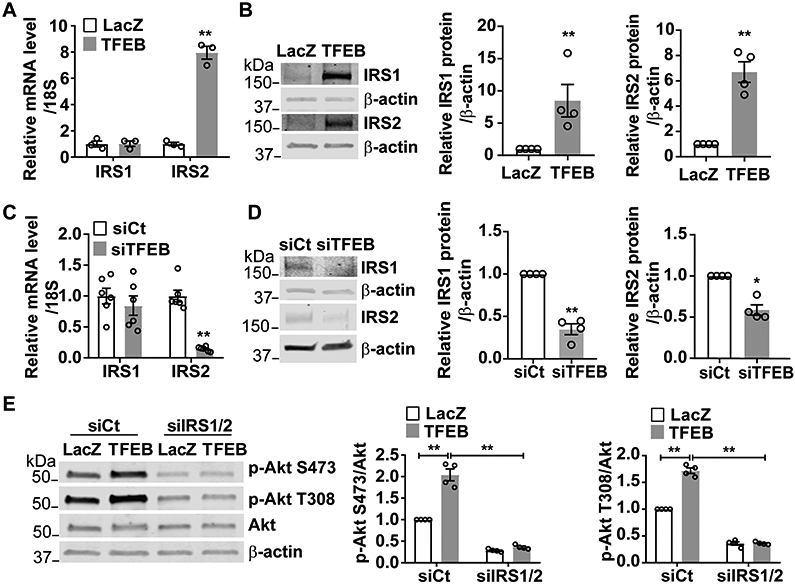
A-B, Human coronary artery endothelial cells (HCAECs) were infected with Ad-LacZ (10 MOI) or Ad-TFEB (10 MOI). Forty-eight hours later, mRNA and protein of IRS1 and IRS2 were determined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) (A, n=3/each group) and Western blot (B; four independent experiments), respectively. C-D, HCAECs were transfected with siCt (25 nM) or siTFEB (25 nM) for 48 hours. The relative mRNA level and protein abundance of IRS1 and IRS2 were determined by qRT-PCR (C; n=6/each group) and Western blot (D; four independent experiments), respectively. E, HCAECs were transfected with siCt (50 nM) or co-transfected with siRNA-IRS1 (siIRS1, 25 nM) and siRNA-IRS2 (siIRS2, 25 nM). Twenty-four hours later, the cells were infected with Ad-LacZ (10 MOI) or Ad-TFEB (10 MOI). Forty-eight hours later, the phosphorylation of Akt was determined by Western blot from four independent experiments. Data are presented as mean ± SEM; Data in A, B, C, D used unpaired Student’s t-test. Data in E were analyzed by 2-way ANOVA followed by Bonferroni test. *P <0.05, **P <0.01.
To determine whether IRS1 and IRS2 mediate the TFEB-dependent activation of Akt, we knocked down IRS1 and IRS2 in ECs. IRS1 knockdown partially attenuated and IRS2 knockdown had no significant effect on the TFEB-induced Akt activation (Supplemental Figure V A-D). However, concurrent knockdown of IRS1 and IRS2 abolished the TFEB-induced Akt activation, indicating an essential role of both IRS1 and IRS2 in the TFEB-Akt signaling axis (Figure 3E). It is well recognized that tyrosine phosphorylation enhances IRS1 activity31. IRS1 was immunoprecipitated and tyrosine phosphorylation was detected in bovine aortic endothelial cells (BAECs) overexpressing TFEB. We found that TFEB overexpression did not increase the tyrosine phosphorylation of IRS1 (Supplemental Figure VI). Therefore, in ECs, TFEB activates IRS signaling at least partially via upregulation of IRS1/IRS2 expression.
TFEB upregulates IRS2 at the transcriptional level.
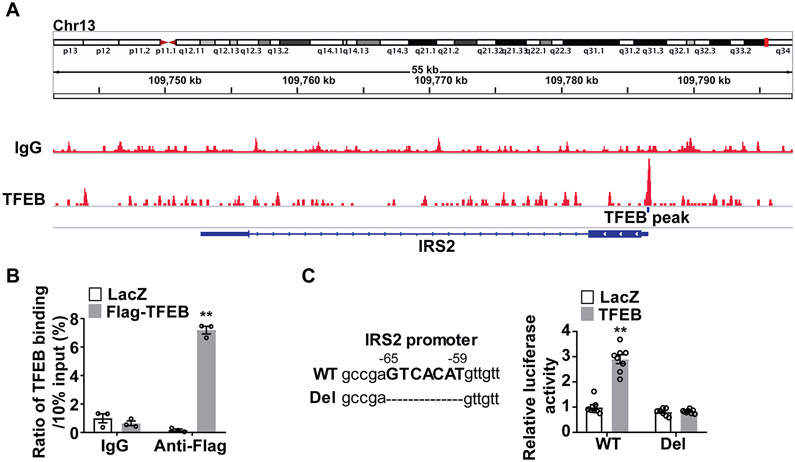
Next, we investigated the mechanisms mediating the TFEB-dependent upregulation of IRS2. We analyzed chromatin immunoprecipitation sequencing (ChIP-Seq) data obtained from HUVEC overexpressing TFEB in the Gene Expression Omnibus (GEO) database (accession number GSE88894)15 and revealed a putative TFEB binding site in IRS2 promoter region (−65bp~−59bp) (Figure 4A). The direct binding of TFEB to the promoter of human IRS2 was determined by Chromatin immunoprecipitation (ChIP) assays (Figure 4B). Using a luciferase reporter gene driven by a 500 bp IRS2 promoter region (−440bp~+59bp), we found that TFEB activates the IRS2 promoter activity. The deletion of the TFEB binding site (−65bp~−59bp) abolished the TFEB-induced luciferase activity (Figure 4C).
Figure 4. Transcription factor EB (TFEB) upregulates insulin receptor substrate 2 (IRS2) expression at the transcriptional level.
A, Chromatin immunoprecipitation sequencing (ChIP-seq) data were obtained from the Gene Expression Omnibus (GEO, accession number GSE88896) and were analyzed for TFEB binding peaks in the promoter of human IRS2 gene. B, Human coronary artery endothelial cells (HCAECs) were infected with Ad-LacZ (20 MOI) or Ad-Flag TFEB (20 MOI) for 48 hours. The binding of TFEB to the IRS2 promoter was measured by ChIP assays using the antibody against Flag (n=3/each group). C, Hela cells were used as a tool for the reporter gene assay. The cells were transfected with pGL4.10 Luciferase reporters driven by wild type (WT, 500 bp IRS2 promoter region: −440bp~+59bp) or deletion (del, −65 bp~−59 bp deletion corresponding to the TFEB binding site) form of IRS2 promoter for 6 hours, and then the cells were infected with Ad-LacZ (10 MOI) and Ad-TFEB (10 MOI). Forty-eight hours later, luciferase activity was measured and normalized by Renilla activity (n=8/each group). Data in B and C are presented as mean ± SEM; **P <0.01 using 2-way ANOVA followed by Bonferroni test.
TFEB increases IRS1 protein via miRNAs.
TFEB increases IRS1 protein but not its mRNA. ChIP-seq analysis suggests that there is no TFEB binding site in the human IRS1 promoter region (Supplemental Figure VII). To determine whether TFEB affects IRS1 protein stability, we treated the cells with cycloheximide (CHX) to inhibit de novo protein synthesis. Our data suggest that TFEB has no effect on the IRS1 protein stability in ECs (Supplemental Figure VIII A-B). MicroRNAs (miRNA) target mRNA and modulate its translation process32. We did small RNA sequencing and identified 176 small RNAs showing differential expression between TFEB overexpression and control in HCAECs, including 116 miRNAs, 31 Piwi-interacting RNA (piRNA), 6 predicted miRNAs and other small RNAs (Supplemental Figure IX A-B). Nine candidate miRNAs regulated by TFEB were further predicted to target IRS1 using miRDB prediction database19, TargetScan20 and DIANA-microT web server v5.021 (Figure 5A). Among the candidate miRNAs, only miR-335-3p, miR-495-3p and miR-548o-3p were consistently regulated by altered TFEB levels (overexpression or knockdown) in ECs, as determined by qRT-PCR (Figure 5B-C). To assess whether these miRNAs mediate the TFEB-dependent increase in IRS1 protein expression, we treated ECs with miRNA mimics. miR-335-3p, miR-495-3p and miR-548o-3p mimics significantly attenuated TFEB’s effect (Figure 5D-F). Noteworthy, miR-335-3p, miR-495-3p and miR-548o-3p mimics did not change IRS1 mRNA (Supplemental Figure IX C-E). We constructed a luciferase reporter containing the 3’ UTR fragment (1 kb) of IRS1 mRNA, which includes a binding site of miR-335-3p, miR-495-3p, miR-548o-3p, respectively. We found that miR-495-3p mimic can partially abolish the TFEB-dependent effects on IRS1 protein expression (Supplemental Figure X). Collectively, our data suggest that TFEB regulates IRS1 at a post-transcriptional level via miRNAs.
Figure 5. Transcription factor EB (TFEB) increases insulin receptor substrate 1 (IRS1) expression via microRNAs (miRNAs).
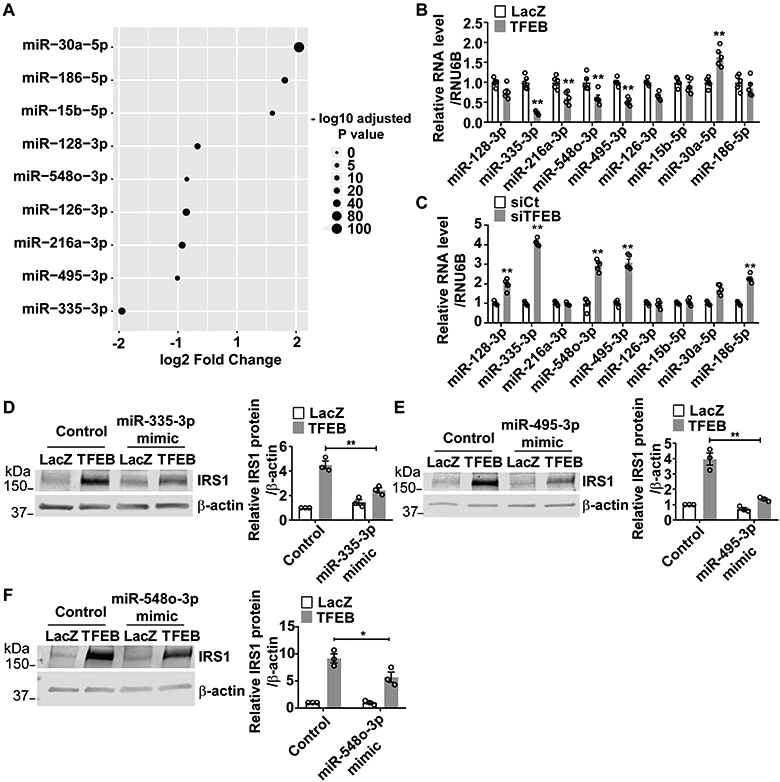
A, Human coronary artery endothelial cells (HCAECs) were infected with Ad-LacZ (10 MOI) or Ad-TFEB (10 MOI). Forty-eight hours later, miRNA abundance was determined by small RNA sequencing. The miRNAs targeting IRS1 were predicted by miRDB, TargetScan and DIANA-microT web server v5. The miRNAs that target IRS1, which are also regulated by TFEB, are shown in the dot plot (n=4/each group). B, HCAECs were infected with Ad-LacZ (10 MOI) or Ad-TFEB (10 MOI). Forty-eight hours later, the miRNAs targeting IRS1 were analyzed by quantitative reverse transcription-polymerase chain reaction (qRT-PCR; n=5/each group). C, HCAECs were transfected with siCt (25 nM) or siTFEB (25 nM) for 48 hours. The miRNAs targeting IRS1 were analyzed by qRT-PCR (n=5/each group). D-F, HCAECs were transfected with miR-335-3p, miR-495-3p or miR-548o-3p mimics (30 nM/each siRNA). Twenty-four hours later, the cells were infected with Ad-LacZ (10 MOI) or Ad-TFEB (10 MOI). After 48 hours, the IRS1 protein was determined by Western blot and quantitatively analyzed from three independent experiments. Data are presented as mean ± SEM. Data in B and C used unpaired Student’s t-test. Data in D-F were analyzed by 2-way ANOVA followed by Bonferroni test. *P <0.05, **P <0.01.
Endothelial TFEB increases insulin sensitivity in the skeletal muscle and adipose tissue.
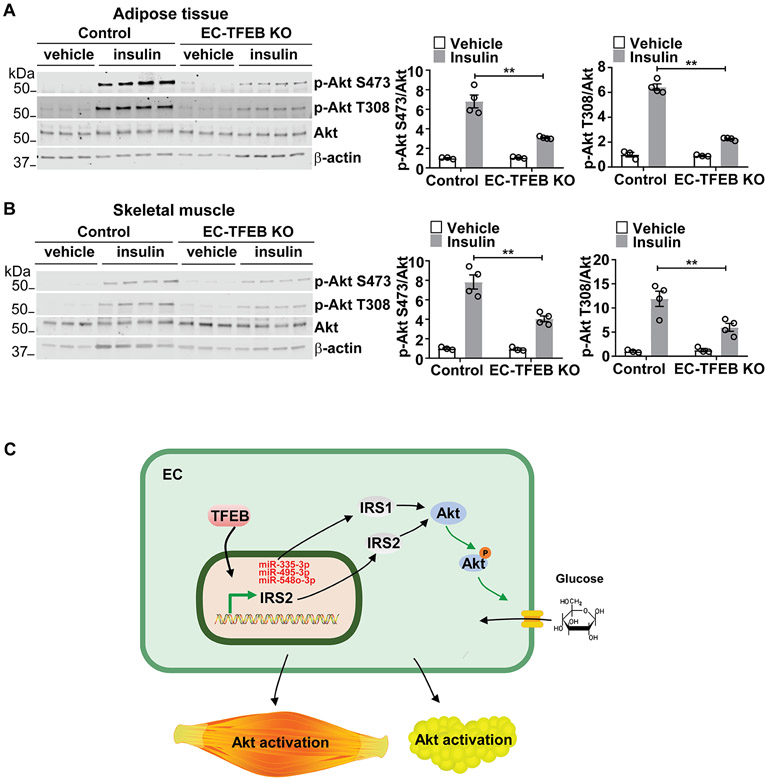
Insulin/Akt signaling in ECs affects insulin signaling in peripheral tissues, particularly in metabolic tissues, thereby regulating systemic insulin sensitivity and glucose tolerance in the whole body33, 34. We tested Akt signaling in the liver, adipose tissue and skeletal muscle. Akt phosphorylation was significantly impaired in the adipose tissue and skeletal muscle from the EC-TFEB KO mice compared with the control mice on HFD (Figure 6A-6B). Consistently, Akt signaling was improved in the adipose tissue and skeletal muscle from the EC-TFEB Tg mice on HFD (Supplemental Figure XI A-B). However, there was no significant difference in Akt phosphorylation in the liver from EC-TFEB KO and Tg mice on HFD (Supplemental Figure XI C-D). Endothelial IRS2 was reported to promote Insulin secretion35. However, there was no significant changes in serum insulin concentration in EC TFEB KO mice on HFD when compared with control mice (P =0.0847, Supplemental Figure XII).
Figure 6. Endothelial transcription factor EB (TFEB) KO impairs AKT serine/threonine kinase (Akt) signaling in the skeletal muscle and adipose tissue.
Male EC-TFEB KO mice were fed a high fat diet (HFD) for 26 weeks. After fasting overnight, the mice were administered insulin (5 U/kg, i.p.). Ten minutes later, tissues were harvested for the analysis of Akt signaling. A-B, the phosphorylation of Akt in the adipose tissue (A) and skeletal muscle (B) of EC-TFEB KO mice was determined by Western blot and quantitatively analyzed (n=3-4/each group). Data in A-B are presented as mean ± SEM; **P <0.01 using 2-way ANOVA followed by Bonferroni test. C, Schematic diagram: endothelial TFEB upregulates insulin receptor substrate 2 (IRS2) at the transcriptional level and increases insulin receptor substrate 1 (IRS1) protein via downregulation of miRNAs, leading to activation of Akt signaling and glucose uptake in ECs. Endothelial TFEB improve systemic glucose tolerance in vivo.
Insulin flux across ECs to muscle cells is essential for insulin action in vivo36-38. To explore whether TFEB influences insulin transport across ECs, we measured fluorescein isothiocyanate-labeled insulin (FITC-insulin) uptake and transport in ECs in vitro. We found that TFEB significantly increases not only insulin uptake in ECs (Supplemental Figure XIII A) but also insulin transport across ECs (Supplemental Figure XIII B). Our data indicate that endothelial TFEB may increase insulin sensitivity of skeletal muscles and adipose tissues via enhancing insulin transport across ECs.
We also determined the expression of IRS1, IRS2, miR-335-3p and miR-495-3p in skeletal muscle and epidydimal adipose tissue from EC-TFEB KO and EC-TFEB Tg mice on high-fat diet. In adipose tissue and skeletal muscle, there was no significant difference in the expression of miR-335-3p and miR-495-3p (Supplemental Figure XIV) and IRS1 and IRS2 (Supplemental Figures XV and XVI) in EC-TFEB KO and EC-TFEB Tg mice when compared with their respective control mice. miR-548o has no homolog in mice39 for which its expression could not be addressed in these mouse samples. Moreover, no significant differences in the expression of PGC1α, GLUT4 and the ratio of LC3 II /LC3 I were observed in the skeletal muscle from EC-TFEB KO mice, EC-TFEB Tg mice when compared with their respective control mice (Supplementary Figure XVI). Increased oxidative stress is associated with insulin resistance, particularly in skeletal muscle40, 41. We measured the ROS levels in mouse skeletal muscle by dihydroethidium (DHE) staining. EC-TFEB KO did not significantly alter the ROS levels in skeletal muscle (Supplemental Figure XVII). We also measured IRS1 phosphorylation at Ser307, which is responsive to ROS stimulation42 and promotes insulin sensitivity in mice43. In skeletal muscle, EC-TFEB KO did not affect the Ser307 phosphorylation of IRS1 (Supplemental Figure XVIII).
DISCUSSION
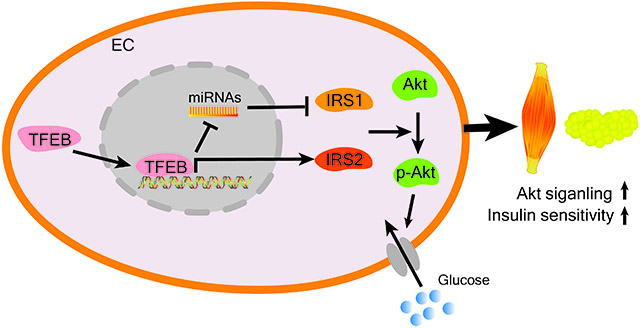
In the present study, we demonstrated that TFEB enhances Akt signaling in ECs and metabolically active tissues and improves glucose tolerance in vivo. TFEB activates Akt signaling via the upregulation of IRS1 and IRS2 through distinct mechanisms in ECs. (Figure 6C).
TFEB regulates genes involved in lipid catabolism through peroxisome proliferator-activated receptor alpha (PPAR-α) and peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) and reduces lipid accumulation in an autophagy-dependent manner in the liver6. Physical activity induces TFEB translocation into the myonuclei and further increases glucose uptake and mitochondrial biogenesis in skeletal muscles44. Moreover, TFEB activators ameliorate metabolic syndrome via autophagy in the liver and adipose tissue8, 9. Conversely, TFEB activity can be regulated by metabolic conditions. TFEB is induced to translocate into nuclei by starvation in the liver6 and TFEB activity is decreased in human islets from T2D donors12, suggesting a crosstalk between TFEB and metabolism in metabolically active tissues.
There are more than one trillion ECs in human body, directly contacting with blood and mediating the nutrient sensing of organs1. The endothelium is a metabolically active organ that plays a crucial role in both vascular homeostasis and systemic metabolism45. In endothelial cells, numerous genes, including CD36, PPARγ, IRS2, have been proven to regulate systemic glucose level1, 22, 33, 46, 47. At this moment, it is a challenge to dissect detailed mechanisms by which altered EC function affects glucose metabolism in peripheral tissues in vivo33, 47-49. We will explore the role and detailed mechanisms of TFEB in the crosstalk between ECs and metabolically active tissues in follow-up studies. TFEB is responsive to environmental nutrient changes. Under diabetic conditions, TFEB is phosphorylated and its activity was found to be decreased in ECs12. That may be the reason why the phenotype can only be observed in mice after high-fat diet feeding in our study.
Insulin/Akt signaling in ECs is of importance in the regulation of metabolism in peripheral tissues22. In the present study, we found that TFEB activates Akt phosphorylation and increases glucose uptake in ECs. On the other hand, Akt can phosphorylate TFEB at Ser467 and inhibit TFEB nuclear translocation in human cervical cancer cell line Hela50. Another study found that under glucose deprivation conditions, mTORC2-Akt-mediated inactivation of GSK3β leads to TFEB nuclear retention in human colorectal adenocarcinoma cell line HT2951. Therefore, there might be a cross talk between TFEB and Akt, through which cells adapt to varying environmental changes and maintain homeostasis. Loss of endothelial insulin receptor (IR) causes a functional delay of systemic insulin action34. Likewise, endothelial IRS1 and IRS2 double KO mice showed impaired Akt signaling in skeletal muscles33. In the present study, we found that endothelial TFEB activates Akt signaling via the upregulation of IRS1 and IRS2 and enhances insulin/Akt signaling in skeletal muscles and adipose tissues. IRS1 and IRS2 are the critical mediators for insulin signaling pathway26. Loss of IRS1 or IRS2 in different tissues will result in metabolic disorders in mice30. In ECs, IRS1 or IRS2 KO causes insulin resistance and accelerates atherosclerosis33, 52. IRS1 overexpression in ECs enhances wound healing in diabetes53. Collectively, endothelial IRS1 and IRS2 have critical roles in both metabolic disorders and vascular diseases.
In endothelial IRS2 KO mice, insulin/Akt/eNOS signaling is impaired in ECs, which reduces capillary recruitment, insulin delivery and insulin secretion33. In our study, IRS1/IRS2/Akt signaling was enhanced by TFEB in ECs. However, insulin concentration was not significantly reduced in EC-TFEB KO mice after glucose challenge. Insulin flux across ECs to muscle cells is a rate-limiting process essential for insulin action in vivo36-38. Endothelial Akt activation increases insulin transport from the vascular lumen to the tissue interstitium37, 38, 54. Our data show that TFEB activates Akt phosphorylation in ECs (Figure 2A-B). Meanwhile, TFEB significantly increases insulin uptake in ECs and its transport across ECs (Supplemental Figure XIII), revealing a mechanism that mediates the improved insulin sensitivity in peripheral tissues induced by endothelial TFEB. The contribution of interstitial insulin in skeletal muscle and adipose tissues warrant future studies.
In the liver, we did not find significant changes in Akt signaling (Supplemental Figure XI C-D). Hepatic endothelial cells are fenestrated, forming the multiple gaps in the capillaries. In comparison, endothelium in adipose tissue and skeletal muscle is continuous1. Therefore, insulin may freely shuttle from circulation to the interstitium of the liver, resulting in no difference in Akt signaling in the liver between genetically engineered EC-TFEB transgenic mice and control mice.
GLUT1 appears to be the predominant glucose transporter in ECs28, 29. Our data demonstrated that GLUT1 knockdown significantly attenuates TFEB-induced glucose uptake in HCAECs (Figure 2D-E), indicating that GLUT1 is essential for TFEB-dependent glucose uptake. A previous study demonstrated that TFEB in skeletal muscles upregulates GLUT1 expression44. However, in our study, we found that TFEB does not affect GLUT1 protein expression in ECs (Supplemental Figure III B-C). GLUT1 translocation to the plasma membrane could be a potential mechanism mediating the TFEB-dependent glucose uptake, which warrants further investigation.
IRS2 is regulated by multiple transcription factors, including transcription factor E3 (TFE3)55, sterol regulatory element-binding proteins (SREBPs)56, hypoxia-inducible factor-2α (HIF-2α)57, cAMP-responsive element binding protein(CREB)58 and forkhead box O 3a (FoxO3a)59. We found that TFEB regulates IRS2 expression at the transcriptional level. Both TFE3 and TFEB belong to MITF family60 and TFEB interacts with TFE3 in adipocytes61. Thus, TFEB may form heterodimers with TFE3 to regulate IRS2 expression in ECs.
Currently, it is already known that TFEB can regulate numerous protein-coding genes, like autophagy and lysosome genes and anti-oxidant genes4, 5, 10, 13, 62. However, whether TFEB regulates miRNAs remains largely unknown. In the present study, we performed small RNA sequencing in ECs and identified 122 miRNAs regulated by TFEB. It has been reported that IRS1 protein expression can be regulated by several miRNAs63, 64. In our study, we found that TFEB decreases miR-335-3p, miR-495-3p and miR-548o-3p, thereby mediating the TFEB-dependent upregulation of IRS1 protein. We further found that at least miR-495-3p mimic can antagonize the effects of TFEB on the expression of the IRS1 3’UTR-luciferase reporter (Supplemental Figure X). Using miRDB prediction, we found that miR-495-3p only has one binding site (251-257bp), miR-335-3p has 2 binding sites (498-504bp,1486-1492bp) and miR-548o-3p has 3 binding sites (470-476bp, 2310-2316bp, 2341-2347bp) within 4960 bp in the IRS1 3’UTR. We cannot exclude that miR-335-3p and miR-548o-3p may bind to multiple sites in the 3’UTR of IRS1 mRNA to repress IRS1 translation. Of note, from the small RNA seq analysis, miR-216-3p, miR-21-5p and let-7i-5p are the top-three highly expressed miRNAs in ECs. In addition, we found that miR-486-5p is highly upregulated by TFEB with high abundance in ECs. miR-486-5p from human endothelial colony forming cell-derived exomes attenuates ischemic kidney injury in mice65. Whether this miRNA mediates the effects of TFEB on ECs warrants future investigation.
Although TFEB upregulates IRS1, IRS2 and LC3-II/LC3-I14, and suppresses microRNAs (miR-335-3p, miR-495-3p and miR-548o-3p) in ECs, altered endothelial TFEB did not change the expression of these genes in skeletal muscle and adipose tissue. ECs are only a fraction of the cells within these tissues. The gene expression changes in ECs may not significantly affect the gene expression in the tissue. In addition, endothelial TFEB-dependent activation of Akt in adipose tissue and skeletal muscle may result from phosphorylation of IRS1/IRS2 in these tissues. Although no effect of TFEB on IRS1 Ser307 was observed, we cannot exclude a possible impact of TFEB on other IRS1 phosphorylation sites. The underlying mechanisms through which EC-TFEB regulates IRS1/IRS2 phosphorylation in skeletal muscle and adipose tissues will require extensive future studies.
Sex differences in insulin resistance were reported in humans and rodents previously. Females are more insulin sensitive than males16. In the current study, we observed improved insulin sensitivity in male EC-TFEB Tg mice and impaired insulin sensitivity in male EC-TFEB KO mice when compared with their respective control mice. However, there was no significant difference in insulin sensitivity in the genetically engineered EC-TFEB female mice. Higher insulin sensitivity, different energy partitioning and the features of glucose or lipid metabolism in the female may contribute to the sex differences16. Sex steroid hormone estrogen protects against insulin resistance through reducing food intake, attenuating inflammatory response and promoting lipolysis, which may also partially explain the sex differences66. The mechanism mediating the TFEB-dependent sex differences and possible contribution of estrogen to the insulin sensitivity in female mice warrant future investigation.
In this study, we used EC-TFEB Tg and KO mice to test the hypothesis. VE-Cadherin and mTie2 are not only expressed in ECs, but also expressed in blood monocytes and bone marrow-derived hematopoietic cells67, 68. Of note, our previous study suggests that there is no significant increase in TFEB expression in peritoneal or bone marrow-derived macrophages in EC-TFEB Tg mice13. Altogether, our studies give a new insight into the roles of endothelial TFEB in the systemic glucose regulation and the interaction between ECs and the peripheral metabolic tissues.
Supplementary Material
Highlights.
Endothelial TFEB improves systemic glucose tolerance in mice.
TFEB activates Akt signaling via upregulation of IRS1 and IRS2.
TFEB increases IRS2 at the transcriptional level and upregulates IRS1 protein through downregulation of miRNAs.
Acknowledgements
J. Sun and H. Lu performed experiments and analyzed results. J. Sun and Y. Fan wrote the manuscript. W. Liang, G. Zhao, L. Ren, D. Hu, Z. Chang, Y. Liu, M. T. Garcia-Barrio and J. Zhang provided the technical support and contributed to the discussion of the project. Y. E. Chen and Y. Fan designed research and discussed results. All authors edited and approved the manuscript.
Sources of Funding
This study is supported by National Institutes of Health Grants R01HL138094 and R01HL145176 to Y.F., R01HL137214, R01HL068878, and R01HL134569 to Y.E.C., and R01HL138139 to J.Z.
Nonstandard Abbreviations and Acronyms
- Akt
AKT serine/threonine kinase
- AUC
area under the curve
- ECs
endothelial cells
- EC-TFEB KO
endothelial cell-selective TFEB knockout
- EC-TFEB Tg
endothelial cell-selective TFEB transgenic
- GLUT1
glucose transporter 1
- GTT
glucose tolerance test
- HCAECs
human coronary artery endothelial cells
- HFD
high fat diet
- IRS1
insulin receptor substrate 1
- IRS2
insulin receptor substrate 2
- ITT
insulin tolerance test
- miRNA
microRNA
- MOI
multiplicity of infection
- TFEB
Transcription factor EB
Footnotes
Disclosures
None.
REFERENCES
- 1.Graupera M, Claret M. Endothelial cells: New players in obesity and related metabolic disorders. Trends Endocrinol Metab. 2018;29:781–794 [DOI] [PubMed] [Google Scholar]
- 2.Li X, Sun X, Carmeliet P. Hallmarks of endothelial cell metabolism in health and disease. Cell Metab. 2019;30:414–433 [DOI] [PubMed] [Google Scholar]
- 3.Fisher DE, Carr CS, Parent LA, Sharp PA. Tfeb has DNA-binding and oligomerization properties of a unique helix-loop-helix/leucine-zipper family. Genes Dev. 1991;5:2342–2352 [DOI] [PubMed] [Google Scholar]
- 4.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477 [DOI] [PubMed] [Google Scholar]
- 5.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. Tfeb links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, Wollenberg AC, Di Bernardo D, Chan L, Irazoqui JE, Ballabio A. Tfeb controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao X, Wang S, Zhao K, Li Y, Williams JA, Li T, Chavan H, Krishnamurthy P, He XC, Li L, Ballabio A, Ni HM, Ding WX. Impaired tfeb-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology. 2018;155:865–879 e812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim H, Lim YM, Kim KH, Jeon YE, Park K, Kim J, Hwang HY, Lee DJ, Pagire H, Kwon HJ, Ahn JH, Lee MS. A novel autophagy enhancer as a therapeutic agent against metabolic syndrome and diabetes. Nat Commun. 2018;9:1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C, Niederstrasser H, Douglas PM, et al. Small-molecule tfeb pathway agonists that ameliorate metabolic syndrome in mice and extend c. Elegans lifespan. Nat Commun. 2017;8:2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans TD, Zhang X, Jeong SJ, He A, Song E, Bhattacharya S, Holloway KB, Lodhi IJ, Razani B. Tfeb drives pgc-1alpha expression in adipocytes to protect against diet-induced metabolic dysfunction. Sci Signal. 2019;12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG. The bhlh-zip transcription factor tfeb is essential for placental vascularization. Development. 1998;125:4607–4616 [DOI] [PubMed] [Google Scholar]
- 12.Song W, Zhang CL, Gou L, He L, Gong YY, Qu D, Zhao L, Jin N, Chan TF, Wang L, Tian XY, Luo JY, Huang Y. Endothelial tfeb (transcription factor eb) restrains ikk (ikappab kinase)-p65 pathway to attenuate vascular inflammation in diabetic db/db mice. Arterioscler Thromb Vasc Biol. 2019;39:719–730 [DOI] [PubMed] [Google Scholar]
- 13.Lu H, Fan Y, Qiao C, Liang W, Hu W, Zhu T, Zhang J, Chen YE. Tfeb inhibits endothelial cell inflammation and reduces atherosclerosis. Sci Signal. 2017;10 [DOI] [PubMed] [Google Scholar]
- 14.Fan Y, Lu H, Liang W, Garcia-Barrio MT, Guo Y, Zhang J, Zhu T, Hao Y, Zhang J, Chen YE. Endothelial tfeb (transcription factor eb) positively regulates postischemic angiogenesis. Circ Res. 2018;122:945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doronzo G, Astanina E, Cora D, Chiabotto G, Comunanza V, Noghero A, Neri F, Puliafito A, Primo L, Spampanato C, Settembre C, Ballabio A, Camussi G, Oliviero S, Bussolino F. Tfeb controls vascular development by regulating the proliferation of endothelial cells. EMBO J. 2019;38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macotela Y, Boucher J, Tran TT, Kahn CR. Sex and depot differences in adipocyte insulin sensitivity and glucose metabolism. Diabetes. 2009;58:803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahman RU, Gautam A, Bethune J, Sattar A, Fiosins M, Magruder DS, Capece V, Shomroni O, Bonn S. Oasis 2: Improved online analysis of small rna-seq data. BMC Bioinformatics. 2018;19:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu H, Sun J, Liang W, Zhang J, Rom O, Garcia-Barrio MT, Li S, Villacorta L, Schopfer FJ, Freeman BA, Chen YE, Fan Y. Novel gene regulatory networks identified in response to nitro-conjugated linoleic acid in human endothelial cells. Physiol Genomics. 2019;51:224–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Wang X. Mirdb: An online database for prediction of functional microrna targets. Nucleic Acids Res. 2020;48:D127–D131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microrna target sites in mammalian mrnas. Elife. 2015;4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. Diana-microt web server v5.0: Service integration into mirna functional analysis workflows. Nucleic Acids Res. 2013;41:W169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubota T, Kubota N, Kadowaki T. The role of endothelial insulin signaling in the regulation of glucose metabolism. Rev Endocr Metab Disord. 2013;14:207–216 [DOI] [PubMed] [Google Scholar]
- 23.Wang CY, Liao JK. A mouse model of diet-induced obesity and insulin resistance. Methods Mol Biol. 2012;821:421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang XL, Zhang L, Youker K, Zhang MX, Wang J, LeMaire SA, Coselli JS, Shen YH. Free fatty acids inhibit insulin signaling-stimulated endothelial nitric oxide synthase activation through upregulating pten or inhibiting akt kinase. Diabetes. 2006;55:2301–2310 [DOI] [PubMed] [Google Scholar]
- 25.Palomer X, Pizarro-Delgado J, Barroso E, Vazquez-Carrera M. Palmitic and oleic acid: The yin and yang of fatty acids in type 2 diabetes mellitus. Trends Endocrinol Metab. 2018;29:178–190 [DOI] [PubMed] [Google Scholar]
- 26.Manning BD, Toker A. Akt/pkb signaling: Navigating the network. Cell. 2017;169:381–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beg M, Abdullah N, Thowfeik FS, Altorki NK, McGraw TE. Distinct akt phosphorylation states are required for insulin regulated glut4 and glut1-mediated glucose uptake. Elife. 2017;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann GE, Yudilevich DL, Sobrevia L. Regulation of amino acid and glucose transporters in endothelial and smooth muscle cells. Physiol Rev. 2003;83:183–252 [DOI] [PubMed] [Google Scholar]
- 29.Park JL, Heilig CW, Brosius FC 3rd. Glut1-deficient mice exhibit impaired endothelium-dependent vascular relaxation. Eur J Pharmacol. 2004;496:213–214 [DOI] [PubMed] [Google Scholar]
- 30.Kubota T, Kubota N, Kadowaki T. Imbalanced insulin actions in obesity and type 2 diabetes: Key mouse models of insulin signaling pathway. Cell Metab. 2017;25:797–810 [DOI] [PubMed] [Google Scholar]
- 31.Gual P, Le Marchand-Brustel Y, Tanti JF. Positive and negative regulation of insulin signaling through irs-1 phosphorylation. Biochimie. 2005;87:99–109 [DOI] [PubMed] [Google Scholar]
- 32.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mrna translation and stability by micrornas. Annu Rev Biochem. 2010;79:351–379 [DOI] [PubMed] [Google Scholar]
- 33.Kubota T, Kubota N, Kumagai H, et al. Impaired insulin signaling in endothelial cells reduces insulin-induced glucose uptake by skeletal muscle. Cell Metab. 2011;13:294–307 [DOI] [PubMed] [Google Scholar]
- 34.Konishi M, Sakaguchi M, Lockhart SM, Cai W, Li ME, Homan EP, Rask-Madsen C, Kahn CR. Endothelial insulin receptors differentially control insulin signaling kinetics in peripheral tissues and brain of mice. Proc Natl Acad Sci U S A. 2017;114:E8478–E8487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashimoto S, Kubota N, Sato H, Sasaki M, Takamoto I, Kubota T, Nakaya K, Noda M, Ueki K, Kadowaki T. Insulin receptor substrate-2 (irs2) in endothelial cells plays a crucial role in insulin secretion. Diabetes. 2015;64:876–886 [DOI] [PubMed] [Google Scholar]
- 36.Yang YJ, Hope ID, Ader M, Bergman RN. Insulin transport across capillaries is rate limiting for insulin action in dogs. J Clin Invest. 1989;84:1620–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King GL, Johnson SM. Receptor-mediated transport of insulin across endothelial cells. Science. 1985;227:1583–1586 [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Liu Z, Li G, Barrett EJ. The vascular endothelial cell mediates insulin transport into skeletal muscle. Am J Physiol Endocrinol Metab. 2006;291:E323–332 [DOI] [PubMed] [Google Scholar]
- 39.Liang T, Guo L, Liu C. Genome-wide analysis of mir-548 gene family reveals evolutionary and functional implications. J Biomed Biotechnol. 2012;2012:679563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henriksen EJ, Diamond-Stanic MK, Marchionne EM. Oxidative stress and the etiology of insulin resistance and type 2 diabetes. Free Radic Biol Med. 2011;51:993–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Meo S, Iossa S, Venditti P. Skeletal muscle insulin resistance: Role of mitochondria and other ros sources. J Endocrinol. 2017;233:R15–R42 [DOI] [PubMed] [Google Scholar]
- 42.Bloch-Damti A, Potashnik R, Gual P, Le Marchand-Brustel Y, Tanti JF, Rudich A, Bashan N. Differential effects of irs1 phosphorylated on ser307 or ser632 in the induction of insulin resistance by oxidative stress. Diabetologia. 2006;49:2463–2473 [DOI] [PubMed] [Google Scholar]
- 43.Copps KD, Hancer NJ, Opare-Ado L, Qiu W, Walsh C, White MF. Irs1 serine 307 promotes insulin sensitivity in mice. Cell Metab. 2010;11:84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mansueto G, Armani A, Viscomi C, et al. Transcription factor eb controls metabolic flexibility during exercise. Cell Metab. 2017;25:182–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theodorou K, Boon RA. Endothelial cell metabolism in atherosclerosis. Front Cell Dev Biol. 2018;6:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanda T, Brown JD, Orasanu G, Vogel S, Gonzalez FJ, Sartoretto J, Michel T, Plutzky J. Ppargamma in the endothelium regulates metabolic responses to high-fat diet in mice. J Clin Invest. 2009;119:110–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Son NH, Basu D, Samovski D, et al. Endothelial cell cd36 optimizes tissue fatty acid uptake. J Clin Invest. 2018;128:4329–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang X, Miao Y, Luo Y, et al. Suppression of endothelial ago1 promotes adipose tissue browning and improves metabolic dysfunction. Circulation. 2020;142:365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yokoyama M, Okada S, Nakagomi A, Moriya J, Shimizu I, Nojima A, Yoshida Y, Ichimiya H, Kamimura N, Kobayashi Y, Ohta S, Fruttiger M, Lozano G, Minamino T. Inhibition of endothelial p53 improves metabolic abnormalities related to dietary obesity. Cell Rep. 2014;7:1691–1703 [DOI] [PubMed] [Google Scholar]
- 50.Palmieri M, Pal R, Nelvagal HR, et al. Mtorc1-independent tfeb activation via akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun. 2017;8:14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li L, Friedrichsen HJ, Andrews S, Picaud S, Volpon L, Ngeow K, Berridge G, Fischer R, Borden KLB, Filippakopoulos P, Goding CR. A tfeb nuclear export signal integrates amino acid supply and glucose availability. Nat Commun. 2018;9:2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rask-Madsen C, Li Q, Freund B, et al. Loss of insulin signaling in vascular endothelial cells accelerates atherosclerosis in apolipoprotein e null mice. Cell Metab. 2010;11:379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katagiri S, Park K, Maeda Y, Rao TN, Khamaisi M, Li Q, Yokomizo H, Mima A, Lancerotto L, Wagers A, Orgill DP, King GL. Overexpressing irs1 in endothelial cells enhances angioblast differentiation and wound healing in diabetes and insulin resistance. Diabetes. 2016;65:2760–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H, Wang AX, Liu Z, Barrett EJ. Insulin signaling stimulates insulin transport by bovine aortic endothelial cells. Diabetes. 2008;57:540–547 [DOI] [PubMed] [Google Scholar]
- 55.Nakagawa Y, Shimano H, Yoshikawa T, et al. Tfe3 transcriptionally activates hepatic irs-2, participates in insulin signaling and ameliorates diabetes. Nat Med. 2006;12:107–113 [DOI] [PubMed] [Google Scholar]
- 56.Ide T, Shimano H, Yahagi N, Matsuzaka T, Nakakuki M, Yamamoto T, Nakagawa Y, Takahashi A, Suzuki H, Sone H, Toyoshima H, Fukamizu A, Yamada N. Srebps suppress irs-2-mediated insulin signalling in the liver. Nat Cell Biol. 2004;6:351–357 [DOI] [PubMed] [Google Scholar]
- 57.Wei K, Piecewicz SM, McGinnis LM, et al. A liver hif-2alpha-irs2 pathway sensitizes hepatic insulin signaling and is modulated by vegf inhibition. Nat Med. 2013;19:1331–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jhala US, Canettieri G, Screaton RA, Kulkarni RN, Krajewski S, Reed J, Walker J, Lin X, White M, Montminy M. Camp promotes pancreatic beta-cell survival via creb-mediated induction of irs2. Genes Dev. 2003;17:1575–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsunekawa S, Demozay D, Briaud I, McCuaig J, Accili D, Stein R, Rhodes CJ. Foxo feedback control of basal irs-2 expression in pancreatic beta-cells is distinct from that in hepatocytes. Diabetes. 2011;60:2883–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor tfeb links mtorc1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salma N, Song JS, Kawakami A, Devi SP, Khaled M, Cacicedo JM, Fisher DE. Tfe3 and tfeb transcriptionally regulate peroxisome proliferator-activated receptor gamma2 expression in adipocytes and mediate adiponectin and glucose levels in mice. Molecular and cellular biology. 2017;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lynch MR, Tran MT, Ralto KM, et al. Tfeb-driven lysosomal biogenesis is pivotal for pgc1alpha-dependent renal stress resistance. JCI Insight. 2019;5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Hu C, Cheng J, Chen B, Ke Q, Lv Z, Wu J, Zhou Y. Microrna-145 suppresses hepatocellular carcinoma by targeting irs1 and its downstream akt signaling. Biochem Biophys Res Commun. 2014;446:1255–1260 [DOI] [PubMed] [Google Scholar]
- 64.Zhang J, Du YY, Lin YF, Chen YT, Yang L, Wang HJ, Ma D. The cell growth suppressor, mir-126, targets irs-1. Biochem Biophys Res Commun. 2008;377:136–140 [DOI] [PubMed] [Google Scholar]
- 65.Vinas JL, Burger D, Zimpelmann J, Haneef R, Knoll W, Campbell P, Gutsol A, Carter A, Allan DS, Burns KD. Transfer of microrna-486–5p from human endothelial colony forming cell-derived exosomes reduces ischemic kidney injury. Kidney Int. 2016;90:1238–1250 [DOI] [PubMed] [Google Scholar]
- 66.Tramunt B, Smati S, Grandgeorge N, Lenfant F, Arnal JF, Montagner A, Gourdy P. Sex differences in metabolic regulation and diabetes susceptibility. Diabetologia. 2020;63:453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Payne S, De Val S, Neal A. Endothelial-specific cre mouse models. Arterioscler Thromb Vasc Biol. 2018;38:2550–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murdoch C, Tazzyman S, Webster S, Lewis CE. Expression of tie-2 by human monocytes and their responses to angiopoietin-2. J Immunol. 2007;178:7405–7411 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.