Abstract
In the last two decades, various therapies have been introduced for lung carcinoma patients, including tyrosine-kinase inhibitors for different mutations. While some of them are specific to specific tumor types, others, like NTRK1–3 fusions, are found in various solid tumors. The occurrence of an NTRK1,2 or 3 fusion acts as a biomarker for efficient treatment with NTRK inhibitors, irrespectively of the tumor type. However, the occurrence of the NTRK1–3 fusions in lung carcinomas is extremely rare. We performed a retrospective analysis to evaluate the applicability of immunohistochemistry with the pan-TRK antibody in the detection of NTRK fusions in lung carcinomas. The study cohort included 176 adenocarcinomas (AC), 161 squamous cell carcinomas (SCC), 31 large-cell neuroendocrine carcinomas (LCNEC), and 19 small cell lung carcinomas (SCLC). Immunohistochemistry (IHC) was performed using the pan-TRK antibody (clone EPR17341, Ventana) on tissue microarrays, while confirmation for all positive cases was done using RNA-based Archer FusionPlex MUG Lung Panel. On IHC staining, 12/387 samples (3.1%) demonstrated a positive reaction. Ten SCC cases (10/161, 6.2%), and two LCNEC cases (2/31, 6.5%) were positive. Positive cases demonstrated heterogeneous staining of tumor cells, mostly membranous with some cytoplasmic and in one case nuclear pattern. RNA-based sequencing did not demonstrate any NTRK1–3 fusion in our patients’ collective. Our study demonstrates that pan-TRK expression in lung carcinoma is very low across different histologic types. NTRK1–3 fusions using an RNA-based sequencing approached could not be detected. This stresses the importance of confirmation of immunohistochemistry results by molecular methods.
Subject terms: Cancer, Molecular biology, Biomarkers, Molecular medicine
Introduction
Lung cancer is one of the most common malignancies worldwide and the leading cause of cancer-related death1. However, according to the recent literature, as a result of anti-tobacco campaigns, screening and new therapy options, the mortality in the USA has decreased both in men and women, by 48% and 23%, respectively2. In the last couple of years, the real “game-changer” in the therapy of lung carcinoma is immunotherapy. Nevertheless, since the first tyrosine-kinase inhibitors against activating EGFR mutations were introduced a little bit more than a decade ago, there has been an improvement in understanding molecular mechanisms responsible for lung cancer development including the detection of targetable mutations. Although these targetable mutations are present in a small proportion of all lung carcinomas, the number of druggable mutations increases every year. According to the latest international recommendations from 20183, preferred way of predictive testing for advanced non-squamous non-small cell lung carcinomas (NSCLC) is multigene testing including EGFR, ALK, ROS1, BRAF, MET, HER2, KRAS, and RET. Very recently, Food and Drug Administration (FDA) and European Medicine Agency (EMA) have approved treatment for tumors harboring neurotrophic tyrosine receptor kinase (NTRK) gene fusions, as a first-line or subsequent-line of therapy4.
Three NTRK genes (NTRK 1, NTRK 2, and NTRK 3) encode the three transmembrane neurotrophin receptors TrkA (NTRK1), TrkB (NTRK2), and TrkC (NTRK3)5. TRK receptors play a significant role in the development and functioning of the central and peripheral nervous system6–8. However, chromosomal rearrangements of these genes with different partners may cause activation and/or overexpression of TRK receptors resulting in tumor development9, 10. NTRK fusions are characteristically found in several rare tumors, like congenital infantile fibrosarcoma, congenital “cellular” mesoblastic nephroma, secretory breast carcinoma, or mammary analogue secretory carcinoma of the salivary glands11–18. The most common fusion found in about 90% of these cases is ETV6-NTRK311–18. Unfrequently, NTRK fusions are described in other rare tumors, like soft tissue neoplasms, but also in common solid tumors, like NSCLC, colorectal carcinoma, gastrointestinal stromal tumors, papillary thyroid carcinoma, glioma, malignant melanoma, and pancreatic adenocarcinoma4, 9, 16, 19–27. Overall incidence of NTRK fusions in all solid tumors is very low, accounting for less than 1%. According to the published data, NTRK fusions in NSCLC are found in 0.1–1% of cases8, 28, 29. However, although rare, targeted therapy induces a response in the vast majority of patients harboring these fusions, and their identification is crucial for further treatment4, 20, 30, 31.
The most reliable method to identify NTRK fusions is RNA-based massive parallel sequencing (MPS). However, it is not available in every institution, it is time-consuming and expensive. Immunohistochemistry, using a pan-TRK antibody, is an affordable and easily available technique in most pathology laboratories. Therefore, this method has been suggested as an optimal screening tool for a TRK fusion protein expression, which if positive should be confirmed with MPS32, 33. Nevertheless, the staining pattern is not uniform and there is no standardized approach for scoring and interpretation of IHC expression23, 33–35.
To evaluate the patterns of staining and the applicability of immunohistochemistry with the pan-TRK antibody in the detection of NTRK fusions, we performed a retrospective analysis on a lung carcinoma cohort including different tumor subtypes and tested all positive samples with MPS.
Material and methods
Study cohort
From the archives of the Diagnostic and Research Institute of Pathology, 387 lung carcinoma cases diagnosed between 1993 and 2012 were selected for this retrospective analysis. All cases were re-evaluated according to WHO 2015 criteria36 to confirm a diagnosis and to select adequate tissue areas for the tissue microarray (TMA) construction. Furthermore, all cases were re-staged according to the UICC/AJCC staging from 201737. At the time of the study all patients have passed away, therefore we were not able to obtain informed consents. This study conforms to the principles outlined in the Declaration of Helsinki and was approved by the Ethics Committee of the Medical University of Graz (24-135 ex11/12).
The cohort included 176 adenocarcinomas (AC), 161 squamous cell carcinomas (SCC), 31 large-cell neuroendocrine carcinomas (LCNEC), and 19 small cell lung carcinomas (SCLC). All patients underwent surgery and the resection material was used for further analysis. Clinicopathological data are summarized in Table 1. When looking at individual cancer subtypes, in AC median age was 64 years (range 41–84). The majority of the patients in this groups were males (110/176, 62.5%), and according to UICC/AJCC 80/176 (45.5%) were in stage I, 63/176 (35.8%) in stage II, 29/176 (16.5%) in stage III and 3/176 (1.7%) in stage IV. The remaining single case could not be staged due to a lack of data. The median age in SCC was 65 years (range 41–89). The vast majority of patients with SCC were male (141/161, 87.6%). 52/161 (32.3%) were stage I carcinomas, 82/161 (50.9%) were stage II, 23/161 (14.3%) stage III, and 1 (0.6%) stage IV. For three patients data were not available for further staging. In the LCNEC patients’ group, the median age was 64 (range 37–89), 19/31 (61.3%) patients were male. According to UICC/AJCC classification, 7/31 cases (22.6%) were stage I, 13 (41.9%) stage II, 7/31 (22.6%) stage III. For 4 cases we were not able to determine the stage due to a lack of data. The median age in the SCLC group was 65 years (range 52–86), with male predominance (15/19, 78.9%). The majority of patients (12/19, 63.2%) were in stage II, 5/19 (26.3%) were stage III and 2 (10.5%) were stage IV.
Table 1.
Study cohort with the results of immunohistochemistry with a pan-TRK antibody.
| n | % | |
|---|---|---|
| Histology | ||
| Adenocarcinoma | 176 | 45.5 |
| Squamous cell carcinoma | 161 | 41.6 |
| Large-cell neuroendocrine carcinoma | 31 | 8.0 |
| Small cell lung carcinoma | 19 | 4.9 |
| Age at diagnosis | ||
| Median | 64 | |
| Range | 37–89 | |
| Gender | ||
| Male | 285 | 73.6 |
| Female | 102 | 26.4 |
| Stage at diagnosis (UICC 2017) | ||
| I | 139 | 35.9 |
| II | 170 | 43.9 |
| III | 64 | 16.5 |
| IV | 6 | 1.6 |
| Undefined (lack of data) | 8 | 2.1 |
| NTRK positive | 12 | 3.1 |
| Adenocarcinoma | 0/176 | 0 |
| Squamous cell carcinoma | 10/161 | 6.2a |
| Large-cell neuroendocrine carcinoma | 2/31 | 6.5a |
| Small cell lung carcinoma | 0/19 | 0 |
a% of the number of each histologic subtype, not of the whole study cohort.
Immunohistochemical analysis
For TMA construction, four 0.6 mm cores were used from each tumor sample, which was formalin-fixed and paraffin-embedded (FFPE), using TMA Grand Master (3DHistech, Budapest, Hungary). For immunohistochemical (IHC) analysis, 4 µm-thick TMA sections were used. Pan-TRK immunohistochemical staining (rabbit monoclonal antibody, clone EPR17341, RTU, Roche, Ventana) was performed on the Benchmark Ultra using iVIEW DAB Detection Kit (both from Ventana Medical Systems, Tucson, AZ). As the positive controls, normal appendix and brain samples were used. Furthermore, one tumor with NGS-proven NTRK fusion was used as additional control of the staining, in this tumor, 80% of tumor cells showed cytoplasmic positivity. The evaluation of staining included a percentage of positive tumor cells, intensity of staining (weak, moderate, strong), and localization of staining (cytoplasmic, membranous, nuclear). Any staining stronger than a background in ≥ 1% of tumor cells, regardless of localization, was regarded as positive23. The evaluation was performed by three authors (IB, LB, SS) and was expressed as a mean value of all cores available for analysis per patient. Any discrepancies were resolved by joint discussion. Whole sections of positive cases were also stained with pan-TRK antibody to investigate the presence of intratumoral heterogeneity.
Molecular analysis
All cases with positive IHC reactions were sent for further analysis using RNA-based Archer FusionPlex MUG Lung Panel (ArcherDX, Boulder, CO). RNA was isolated from the 5–8, 10 μm thick, FFPE sections cut from a representative block using macrodissection and the Maxwell RSC RNA FFPE kit. RNA quantification was performed using ribogreen fluorescence, and 250 ng total RNA was used. NGS libraries were sequenced on Ion S5 (Ion Torrent, Thermo Fischer, Waltham, MA) using the Ion PI Hi-Q Sequencing 200 kit (Thermo Fischer, Waltham, MA). ArcherDX Analysis software Version 5.1.3. (ArcherDX, Boulder, CO) was used for data analysis.
Ethics approval
The study was approved by the Ethics Committee of the Medical University of Graz (24-135 ex11/12), which granted the waiver for the informed consent for this specific study, since, unfortunately, at the time of this study all patients whose samples were used have already passed away.
Results
Immunohistochemical analysis
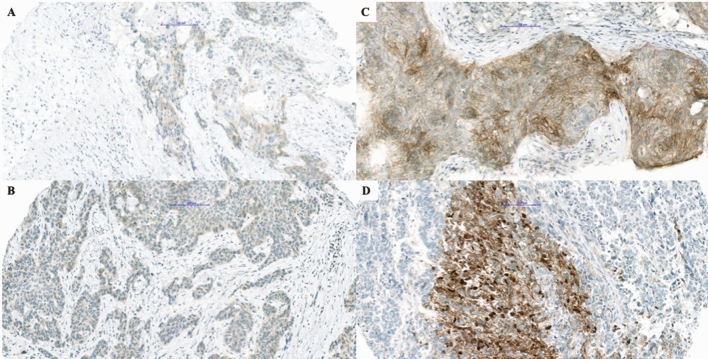
On IHC staining, 12/387 samples (3.1%) demonstrated positive reaction, including ten SCC cases (10/161, 6.2%), and two cases in the LCNEC group (2/31, 6.5%). 8/10 SCC (80%) showed weak cytoplasmic and 2/10 (20%) strong membranous staining pattern, with no more than 10% of positive tumor cells in all but one SCC which demonstrated strong membranous staining in 70% of tumor cells (Fig. 1). Strong pan-TRK expression was found in one (50%) LCNEC with cytoplasmic staining in 60% of tumor cells. The second positive LCNEC case presented strong cytoplasmic and focal nuclear positivity in 25% of tumor cells (Fig. 1). None of the analyzed AC and SCLC showed a positive IHC reaction.
Figure 1.
Presentation of positive immunohistochemical reactions. Different patterns of immunohistochemical staining with pan-Trk antibody in squamous cell carcinoma (A–C) and large cell neuroendocrine carcinoma (D). In (A,B) there is a weak cytoplasmic reaction, while one can appreciate a strong membranous reaction in (C), and nuclear and membranous reaction in (D) (bar = 100 µm).
Furthermore, the stained whole sections of all positive tumor samples demonstrated clear heterogeneity in the staining pattern.
Molecular analysis
In seven cases no NTRK1–3 fusions could be proven. Five cases could not be evaluated due to insufficient RNA quality, despite repeated analysis using different tumor tissue blocks. Interestingly, the age of the blocks did not play a role in this rather high failure rate.
Discussion
Results of our study analyzing pan-TRK expression in lung carcinoma demonstrate very low positivity across different histologic types, without any confirmed NTRK1–3 fusions using an RNA-based sequencing method. This stresses the importance of confirmation of immunohistochemistry results by molecular methods.
Our results are in concordance with recent studies that also clearly demonstrated a very low occurrence of these mutations in lung carcinoma. In one of the first studies looking at the oncogenic and drug-sensitive NTRK rearrangements in the lung AC, Vaishnavi et al. have found NTRK1 fusions in 3/91 patients with lung AC (Table 2)29. Such a high incidence (3.3%) was very promising, however, later studies were not able to confirm this finding. Of note, this study had a selection bias, since only tumors without any other already known oncogenic driver mutations were included. In 2018, Gatalica et al. presented results of 11,502 solid tumors´ samples submitted for molecular profiling, using RNA-based ArcherDx FusionPlex Assay for fusion detection. Among other tumor types, 4,073 NSCLC were included, and in 4 cases NTRK1–3 fusions (0.10%) were found (see Table 2)38. A multicentric study in 2018 by Farago et al. found 11 NSCLC (0.23%) harboring NTRK1 and NTRK3 fusions (listed in Table 2) in 4872 screened cases, using also RNA-based MPS10. The majority of positive cases were AC (9), with one SCC and one LCNEC. Very recently, the largest cohort of RNA-based NTRK1–3 fusion analysis was performed on 38,095 solid tumor samples, including 3,993 lung AC13. Interestingly, they found the same incidence of 0.23% of NTRK fusion-positive AC (9/3,993). In summary, all these studies together included 27 lung carcinomas harboring NTRK1–3 fusions, 25 being AC10, 13, 29, 38. Among these, the NTRK1 gene was the most common fusion partner (17/25, 68%), followed by NTRK3 (6/25, 24%), and only rarely NTRK2. One published SCC and one LCNEC harbored an NTRK3 gene fusion10.
Table 2.
Distribution of fusion partners according to the histologic type in published studies.
| Study | Histologic type | Number of analyzed lung carcinoma | Positive cases (histologic type) | Percentage of positive cases | Detected fusions |
|---|---|---|---|---|---|
| Vaishnavi et al., 2013 | AC | 91 | 3 (AC) | 3.3% |
CD74-NTRK1 MPRIP-NTRK1 NTRK1a |
| Gatalica et al., 2018 | NSCLC | 4073 | 4 (AC) | 0.10% |
TPM3-NTRK1 SQSTM1-NTRK2 ETV6-NTRK3 ETV6-NTRK3 |
| Farago et al. 2018 | NSCLC | 4872 | 9 (AC) | 0.23% |
IRF2BP2-NTRK1 IRF2BP2-NTRK1 MPRIP-NTRK1 SQSTM1-NTRK1 SQSTM1-NTRK1 TPM3-NTRK1 TPR-NTRK1 ETV6-NTRK3 ETV6-NTRK3 |
| 1 (SCC) | ETV6-NTRK3 | ||||
| 1 (NE) | SQSTM1-NTRK3 | ||||
| Solomon et al., 2020 | AC | 3993 | 9 (AC) | 0.23% |
EPS15-NTRK1 EPS15-NTRK1 F11-NTRK1 IRF2BP2-NTRK1 TFG-NTRK1 TPM3-NTRK1 STRN-NTRK2 RBPMS-NTRK3 SQSTM1-NTRK3 |
AC adenocarcinoma, NSCLC non-small cell lung carcinoma, SCC squamous cell carcinoma, NE neuroendocrine carcinoma.
aThis fusion was detected using a break-apart FISH probe detecting different NTRK1 fusions.
Current recommendations suggest that RNA-based MPS technologies are the golden standard to detect NTRK gene fusions in all solid tumors33, 39. However, RNA-based MPS methods are not available in all pathology laboratories, are very expensive and time-consuming. Therefore, immunohistochemistry is used as a screening method to search for pan-TRK protein expression that may be caused by NTRK-fusions. In contrast to MPS immunohistochemistry is widely available, does not require as much tumor tissue as molecular methods, is fast and cheap. Currently, there is no consensus about the best anti-NTRK antibody to be used. There are monoclonal antibodies detecting specific proteins, for example, rabbit TrkA (clone ab76291, Abcam), rabbit TrkB (clone J9.777.7 Thermo Fisher or clone EPR 17805-146 from Abcam), or the ones detecting overexpression of all NTRK1–3 proteins (rabbit pan-TRK antibody, clone EPR17341 from Roche/Ventana or Abcam) and A7H6R (Cell Signaling). As far as we know, there is only one report comparing two different clones, EPR17341 (both from Ventana and Abcam) and A7H6R (Cell Signaling), demonstrating comparable performance in different laboratories40. Both previously mentioned studies (from Gatalica et al. and Solomon et al.) used clone EPR17341 from Abcam. The first study by Gatalica showed an overall sensitivity of 75%, with 95.9% specificity38. Solomon et al. demonstrated lower specificity (81.1%) and nicely showed that sensitivity is not the same for NTRK1, NTRK2, and NTRK3 gene fusions, being 96.2%, 100%, and 79.4% respectively13. These results are in contrast to other published studies where the sensitivity of 95.2% and 97%, and very high specificity, 100%, and 98%, respectively were found23, 34. An additional study using also EPR17341 from Abcam demonstrated high sensitivity, but lower specificity35. All mentioned studies have demonstrated that IHC is much better at the detection of NTRK1 and NTRK2 gene fusions, but lacks sensitivity for the detection of NTRK3 gene fusions. What is the reason for NTRK1–3 expression, other than fusion in NTRK1–3 genes, is not completely clear. An explanation is probably in genetic and/or epigenetic changes, like activating mutations which are found in some lung neuroendocrine carcinomas41. When we combine these facts with the previously mentioned incidence of NTRK1–3 gene fusions in lung carcinoma, it is more than possible that a certain proportion of samples harboring NTRK fusions (especially NTRK3) are being missed using IHC as a screening method. This is a crucial point to have in mind when deciding which method of testing to use.
For this study, we have used pan-TRK ready-to-use assay (clone EPR17341 Roche/Ventana) and analyzed protein expression not only in lung AC but also in SCC, LCNEC, and SCLC. Overall, we have found 12 positive samples out of 387 cases (3.1%). Interestingly, 6.5% of analyzed LCNEC were positive, as well as 6.2% of SCC. Using an RNA-based MPS approach, no NTRK1–3 fusions were detected.
In a very recent study, Leal et al. used a cocktail of pan-TRK (clone A7H6R, Cell Signaling), ALK, and ROS-1 antibodies on TMA and found a positive reaction in 0.4% of NSCLC (2/522), while all SCLC (105) were negative for this antibody cocktail. After RNA sequencing, two positive NSCLC demonstrated ALK fusions. In this study, no NSCLC or SCLC with NTRK1–3 gene fusions has been detected as well42. Using the same antibody as in our study, with some protocol modifications, Elfving et al. evaluated the expression of pan-TRK in 617 NSCLC43. They have found a weak positive reaction in 17 cases (2.8%) and in an additional 1.8% of cases (11/617) moderate to strong staining was observed. The majority of IHC positive cases, like in our study, were SCC. They have also found the staining pattern to be heterogeneous, and rarely more than 80% of tumor cells showed positive staining. This study is similar to ours concerning the case selection, since both studies used surgical material to construct TMAs, and the majority of patients included in the study were in lower clinical stages, in comparison to other published data. Analogous to our study, none of the cases demonstrated NTRK1–3 gene fusion using RNA-based sequencing.
Our study has several limitations. First, it is a single-center, retrospective study, using older FFPE tissue. This can theoretically influence IHC results, or like in some of our cases make RNA-based molecular analysis impossible, although the average age of our blocks did not differ between successfully tested and failed samples. However, the influence of preanalytical variables can here not be excluded. Altogether, the number of cases was not very high, especially for some histologic groups (LCNEC and SCLC), which might explain the rather high incidence of IHC positive cases in SCC, LCNEC, and SCLC groups. Moreover, the IHC analysis was performed using a TMA-based approach, which, although 4 cores from each tumor were used, cannot compensate for the staining heterogeneity of tumors. This, on the other hand, represents the real-life situation where we are dealing with small biopsies in a large number of patients. Lastly, all tumors included in this study were not analyzed using RNA-sequencing and the rate of the IHC false-negative cases could not be evaluated.
Our study has confirmed that protein expression does not imply the presence of NTRK1–3 gene fusions and has, therefore, to be verified, ideally by RNA-based MPS. Furthermore, NTRK1–3 fusions occur infrequently in lung carcinomas. However, whether the protein expression is also important for the therapeutic effect, even without fusion, and the real number of cases harboring these rare fusions that we miss using immunohistochemistry as a screening should be clarified in further studies.
Author contributions
L.B., I.B., and B.L.A. designed the study; L.B., I.B., H.P., and S.S. analyzed the cases. J.L. provided clinical data and administrative support. All authors contributed to the draft manuscript writing and approved the final version.
Funding
No funding was obtained for this study.
Data availability
Available upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lung Source: Globocan 2020 Number of New Cases in 2020, Both Sexes, All Ages. https://gco.iarc.fr/today. Accessed 28 December 2020 (2020).
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Thorac. Oncol. 2018;13:323–358. doi: 10.1016/j.jtho.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N. Engl. J. Med. 2018;378:731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amatu A, Sartore-Bianchi A, Bencardino K, et al. Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann. Oncol. 2019;30:VIII5–VIII15. doi: 10.1093/annonc/mdz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reichardt LF. Neurotrophin-regulated signaling pathways. Philos. Trans. R. Soc. B Biol. Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang EJ, Reichardt LF. Trk receptors: Roles in neuronal signal transduction. Annu. Rev. Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 8.Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015;5:25–34. doi: 10.1158/2159-8290.CD-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khotskaya YB, Holla VR, Farago AF, et al. Targeting TRK family proteins in cancer. Pharmacol. Ther. 2017;173:58–66. doi: 10.1016/j.pharmthera.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Farago AF, Taylor MS, Doebele RC, et al. Clinicopathologic features of non-small-cell lung cancer harboring an NTRK gene fusion. JCO Precis. Oncol. 2018 doi: 10.1200/PO.18.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knezevich SR, McFadden DE, Tao W, et al. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat. Genet. 1998;18:184–187. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- 12.Knezevich SR, Garnett MJ, Pysher TJ, et al. ETV6-NTRK3 gene fusions and trisomy 11 establish a histogenetic link between mesoblastic nephroma and congenital fibrosarcoma. Cancer Res. 1998;58:5046–5048. [PubMed] [Google Scholar]
- 13.Solomon JP, Linkov I, Rosado A, et al. NTRK fusion detection across multiple assays and 33,997 cases: Diagnostic implications and pitfalls. Mod. Pathol. 2020;33:38–46. doi: 10.1038/s41379-019-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourgeois JM, Knezevich SR, Mathers JA, Sorensen PHB. Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am. J. Surg. Pathol. 2000;24:937–946. doi: 10.1097/00000478-200007000-00005. [DOI] [PubMed] [Google Scholar]
- 15.Davis JL, Lockwood CM, Albert CM, et al. Infantile NTRK-associated mesenchymal tumors. Pediatr. Dev. Pathol. 2018;21:68–78. doi: 10.1177/1093526617712639. [DOI] [PubMed] [Google Scholar]
- 16.Church AJ, Calicchio ML, Nardi V, et al. Recurrent EML4-NTRK3 fusions in infantile fibrosarcoma and congenital mesoblastic nephroma suggest a revised testing strategy. Mod. Pathol. 2018;31:463–473. doi: 10.1038/modpathol.2017.127. [DOI] [PubMed] [Google Scholar]
- 17.Tognon C, Knezevich SR, Huntsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 18.Skálová A, Vanecek T, Sima R, et al. Mammary analogue secretory carcinoma of salivary glands, containing the etv6-ntrk3 fusion gene: A hitherto undescribed salivary gland tumor entity. Am. J. Surg. Pathol. 2010;34:599–608. doi: 10.1097/PAS.0b013e3181d9efcc. [DOI] [PubMed] [Google Scholar]
- 19.Lezcano C, Shoushtari AN, Ariyan C, et al. Primary and metastatic melanoma with NTRK fusions. Am. J. Surg. Pathol. 2018;42:1052–1058. doi: 10.1097/PAS.0000000000001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amatu A, Sartore-Bianchi A, Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open. 2016;1:1. doi: 10.1136/esmoopen-2015-000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doebele RC, Davis LE, Vaishnavi A, et al. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov. 2015;5:1049–1057. doi: 10.1158/2159-8290.CD-15-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshihara K, Wang Q, Torres-Garcia W, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hechtman JF, Benayed R, Hyman DM, et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am. J. Surg. Pathol. 2017;41:1547–1551. doi: 10.1097/PAS.0000000000000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SJ, Li GG, Kim ST, et al. NTRK1 rearrangement in colorectal cancer patients: Evidence for actionable target using patient-derived tumor cell line. Oncotarget. 2015;6:39028–39035. doi: 10.18632/oncotarget.5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang S, Cotzia P, Hyman DM, et al. NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am. J. Surg. Pathol. 2018;42:791–798. doi: 10.1097/PAS.0000000000001055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landman Y, Ilouze M, Wein S, et al. Rapid response to larotrectinib (LOXO-101) in an adult chemotherapy-naive patients with advanced triple-negative secretory breast cancer expressing ETV6-NTRK3 fusion. Clin. Breast Cancer. 2018;18:e267–e270. doi: 10.1016/j.clbc.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 27.Brčić I, Godschachner TM, Bergovec M, et al. Broadening the spectrum of NTRK rearranged mesenchymal tumors and usefulness of pan-TRK immunohistochemistry for identification of NTRK fusions. Mod. Pathol. 2020 doi: 10.1038/s41379-020-00657-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farago AF, Le LP, Zheng Z, et al. Durable clinical response to entrectinib in NTRK1-rearranged non-small cell lung cancer. J. Thorac. Oncol. 2015;10:1670–1674. doi: 10.1097/01.JTO.0000473485.38553.f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat. Med. 2013;19:1469–1472. doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drilon A, Ou SHI, Cho BC, et al. Repotrectinib (Tpx-0005) is a next-generation ros1/trk/alk inhibitor that potently inhibits ros1/trk/alk solvent-front mutations. Cancer Discov. 2018;8:1227. doi: 10.1158/2159-8290.CD-18-0484. [DOI] [PubMed] [Google Scholar]
- 31.Hong DS, DuBois SG, Kummar S, et al. Larotrectinib in patients with TRK fusion-positive solid tumours: A pooled analysis of three phase 1/2 clinical trials. Lancet Oncol. 2020;21:531–540. doi: 10.1016/S1470-2045(19)30856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Demetri GD, Antonescu CR, Bjerkehagen B, et al. Diagnosis and management of tropomyosin receptor kinase (TRK) fusion sarcomas: Expert recommendations from the World Sarcoma Network. Ann. Oncol. 2020;31:1506–1517. doi: 10.1016/j.annonc.2020.08.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchiò C, Scaltriti M, Ladanyi M, et al. ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann. Oncol. 2019;30:1417–1427. doi: 10.1093/annonc/mdz204. [DOI] [PubMed] [Google Scholar]
- 34.Rudzinski ER, Lockwood CM, Stohr BA, et al. Pan-Trk immunohistochemistry identifies NTRK rearrangements in pediatric mesenchymal tumors. Am. J. Surg. Pathol. 2018;42:927–935. doi: 10.1097/PAS.0000000000001062. [DOI] [PubMed] [Google Scholar]
- 35.Hung YP, Fletcher CDM, Hornick JL. Evaluation of pan-TRK immunohistochemistry in infantile fibrosarcoma, lipofibromatosis-like neural tumour and histological mimics. Histopathology. 2018;73:634–644. doi: 10.1111/his.13666. [DOI] [PubMed] [Google Scholar]
- 36.Travis, W.D., Brambilla, E., Burke, A., Marx, A., Nicholson, G. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. (International Agency for Research on Cancer, 2015). [DOI] [PubMed]
- 37.Brierley, J.D., Gospodarowicz, M.K., Wittekind, C. TNM classification of malignant tumours-8th edition. Union Int. Cancer Control (2017).
- 38.Gatalica Z, Xiu J, Swensen J, Vranic S. Molecular characterization of cancers with NTRK gene fusions. Mod. Pathol. 2019;32:147–153. doi: 10.1038/s41379-018-0118-3. [DOI] [PubMed] [Google Scholar]
- 39.Penault-Llorca F, Rudzinski ER, Sepulveda AR. Testing algorithm for identification of patients with TRK fusion cancer. J. Clin. Pathol. 2019 doi: 10.1136/jclinpath-2018-205679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Winne K, Sorber L, Lambin S, et al. Results of a first panTRK IHC ringtrial. Ann. Oncol. 2019;30:11. [Google Scholar]
- 41.Marchetti A, Felicioni L, Pelosi G, et al. Frequent mutations in the neurotrophic tyrosine receptor kinase gene family in large cell neuroendocrine carcinoma of the lung. Hum. Mutat. 2008;29:609–616. doi: 10.1002/humu.20707. [DOI] [PubMed] [Google Scholar]
- 42.Leal JL, Peters G, Szaumkessel M, et al. NTRK and ALK rearrangements in malignant pleural mesothelioma, pulmonary neuroendocrine tumours and non-small cell lung cancer. Lung Cancer. 2020;146:154–159. doi: 10.1016/j.lungcan.2020.05.019. [DOI] [PubMed] [Google Scholar]
- 43.Elfving H, Broström E, Moens LNJ, et al. Evaluation of NTRK immunohistochemistry as a screening method for NTRK gene fusion detection in non-small cell lung cancer. Lung Cancer. 2020 doi: 10.1016/j.lungcan.2020.11.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Available upon reasonable request.