Abstract
Congenital aortic valve stenosis (CAVS) affects up to 10% of the world population without medical therapies to treat the disease. New molecular targets are continually being sought that can halt CAVS progression. Collagen deregulation is a hallmark of CAVS yet remains mostly undefined. Here, histological studies were paired with high resolution accurate mass (HRAM) collagen-targeting proteomics to investigate collagen fiber production with collagen regulation associated with human AV development and pediatric end-stage CAVS (pCAVS). Histological studies identified collagen fiber realignment and unique regions of high-density collagen in pCAVS. Proteomic analysis reported specific collagen peptides are modified by hydroxylated prolines (HYP), a post-translational modification critical to stabilizing the collagen triple helix. Quantitative data analysis reported significant regulation of collagen HYP sites across patient categories. Non-collagen type ECM proteins identified (26 of the 44 total proteins) have direct interactions in collagen synthesis, regulation, or modification. Network analysis identified BAMBI (BMP and Activin Membrane Bound Inhibitor) as a potential upstream regulator of the collagen interactome. This is the first study to detail the collagen types and HYP modifications associated with human AV development and pCAVS. We anticipate that this study will inform new therapeutic avenues that inhibit valvular degradation in pCAVS and engineered options for valve replacement.
Subject terms: Congenital heart defects, Valvular disease, Peptides, Proteins, Proteomics, Protein-protein interaction networks, Mass spectrometry, Proteomic analysis, Cardiovascular biology, Congenital heart defects, Valvular disease
Introduction
Congenital aortic valve stenosis (CAVS) accounts for 10% of all congenital heart defect cases, affecting up to 6 of every 1000 newborns1. There are two distinct subsets of this disease: (1) pediatric end-stage, characterized by a rapid engorgement of the structure with extracellular matrix (ECM), and (2) the more frequently studied adult end-stage, characterized by fibrocalcific lesions2. These defects result in a narrowing of the aortic opening, obstructing aortic outflow and leading to left ventricular hypertrophy and heart failure. Despite the clinical significance, there are no available therapeutics. The patient is managed by “watchful waiting” until deteriorating cardiac hemodynamics indicates a need for surgical intervention, either through valve replacement or heart transplantation3. Bioengineered aortic valve replacements are especially limited for pediatric patients due to their inability to grow with the patient, lifelong requirements of anti-clotting agents, and serial surgeries to replace valves during somatic growth4. There is an ongoing need for identifying therapeutic targets, particularly for the pediatric population.
Collagens form the underlying scaffolding of valvular structure that influences valvular function and consequently cardiac function5–12. Collagen is developmentally regulated and is required for appropriate heart function during somatic growth12–14. The normal human aortic valve has an ECM trilayer structure defined by a high density collagenous fibrosa facing blood outflow, an interior spongiosa composed of primarily glycosaminoglycans, and the elastin-rich ventricularis7,12. In the mature human valve, a parallel alignment of collagen with valvular endothelium is critical for normal function7,13, highlighting collagen organization within the trilayer compartments as fundamental to valvular function. Pediatric CAVS produces excessive collagen deposition that mixes with other ECM proteins, creating a disorganized ECM structure, altering valvular function and leading to cardiac failure5,15,16. Although collagen deregulation is a hallmark of adult and pediatric valvular stenosis5,17,18 and a potential prognostic and therapeutic target19,20, very little is known about the complexities of collagen fiber regulation in the human valve.
In the current study, we test the hypothesis that pCAVS disease mechanisms also include dysregulation of collagen fiber arrangements with an altered post-translational regulation. A total of 20 human AV tissue specimens with clinical data on AV function were evaluated. Histopathological stains paired with microscopy methods were used to define collagen fiber changes and collagen variation within the valvular structure. A collagen-targeting21–24 high-resolution accurate mass (HRAM) proteomics approach was used to measure collagen type composition and PTM regulation within the collagen structure. Protein–protein interaction analyses were done to determine possible upstream regulators relevant to collagen deregulation observed in pCAVS. Upstream regulators were validated via immunohistochemistry and RT-PCR. Results of this study indicate that pCAVS shows fundamental differences in collagen fiber organization, collagen type regulation, and regulation of proline hydroxylation of collagens. These studies are the first to report specific collagen regulation due to pCAVS, and establish a foundation for new translational and post-translational collagen studies in fibrotic cardiovascular disease.
Results
Clinical and morphological characteristics of study cohort
Collagen from human aortic valve samples was studied across three patient categories: normal, pediatric end-stage congenital aortic valve stenosis (pCAVS), and aortic valve insufficiency (AVI) (Table 1). Age-matched pediatric samples ranged from Neonates (2 weeks) to Adolescent (18 years), with an average patient age of 6.75 ± 6.06 years. Gender was predominantly male (15, 57.7%). The race was primarily Caucasian (16, 61.5%). For each patient, serial tissue sections were evaluated for histopathological distribution of collagen by multiple staining methods and collagen fiber measurements. Translational and post-translational regulation of collagen was investigated by collagen targeted proteomics25 using HRAM proteomics on regions approximately 100 µm in thickness adjacent to histological sections. In summary, this cohort represents a suitable pathological variation towards probing ECM distribution in human aortic valve development and pediatric end-stage disease.
Table 1.
Descriptive statistics of patient characteristics for all pediatric aortic valve (AV) tissue samples featured in the study.
| SKU | Age (years) | Pediatric age group | Gender | Race/ethnicity | BSA (m2) | Leaflet morphology | Aortic valve function |
|---|---|---|---|---|---|---|---|
| Normal | |||||||
| DB017 | 0.5 | Infant | M | – | 0.38 | Trileaflet | Normal |
| DB051 | 0.66 | Infant | M | C | 0.34 | Trileaflet | Normal |
| NDRI | 2 | Young child | M | L | 0.63 | Trileaflet | Not recorded |
| DB85 | 9 | Child | F | C | 0.95 | Trileaflet | Normal |
| DB27 | 17 | Adolescent | M | C | 2.2 | Trileaflet | Normal |
| Normal Averages | 7.17 ± 7.5 | 75%M 25%F | 75%C 25%L | 1.03 | |||
| pCAVS | |||||||
| DB106 | 0.04 | Neonate | M | C | 0.21 | Bicuspid | Severe stenosis with AVI |
| DB43 | 0.15 | Infant | M | C | 0.24 | Trileaflet | Moderate stenosis with AVI* |
| DB40 | 0.75 | Infant | M | C | 0.37 | Bicuspid | Moderate stenosis |
| DB105 | 5 | Young child | M | C | 0.72 | Bicuspid | Moderate stenosis with AVI |
| DB119 | 5 | Young child | F | – | – | Bicuspid | Moderate stenosis with AVI |
| DB113 | 5 | Young child | – | – | – | – | Trivial stenosis; severe AVI |
| DB18 | 6 | Child | F | C | 0.7 | Bicuspid | Moderate stenosis with AVI |
| DB3 | 11 | Child | F | C | 1.5 | Bicuspid | Moderate stenosis with AVI |
| DB117 | 12 | Adolescent | M | – | Bicuspid | Moderate stenosis with AVI | |
| DB41 | 14 | Adolescent | M | C | 1.6 | Bicuspid | Moderate stenosis with AVI |
| DB16 | 17 | Adolescent | M | C | 2.1 | Bicuspid | Moderate stenosis with AVI |
| pCAVS Averages | 6.90 ± 5.8 | 70%M 30%F | 100%C | 0.93 | |||
| AVI | |||||||
| DB114 | 0.17 | Infant | F | – | – | Trileaflet | Trivial AVI |
| DB007 | 0.58 | Infant | – | – | – | Trileaflet | Mild nAVI |
| DB108 | 10 | Child | F | C | 1.3 | Trileaflet | Thickened with AVI |
| DB83 | 10 | Child | F | AA | 1.4 | Trileaflet | Trivial AVI |
| DB032 | 16 | Adolescent | – | – | – | Trileaflet | Mild nAVI |
| AVI Averages | 7.35 ± 6.8 | – | – | 1.35 | |||
Normal AVs are from cardiac transplant patients where normal AV function is define by 2D echocardiogram. 2D Echocardiogram mean gradients define pre-operative AV function where a normal gradient is defined as < 5 mmHg, mild stenosis as 5–25 mmHg, moderate stenosis as 25–50 mmHg, and severe stenosis as > 50 mmHg. BSA: body surface area; C: Caucasian; L: Latino; AA: African American; MD: Myxomatous Degeneration. DB43 was included in pCAVS due to stenosis, but main diagnosis was Hypertrophic Cardiomyopathy. Pediatric Age Groups are defined by National Institute of Child and Human Development: Neonate (0–30d), Infant (1 mo–2yrs), young Child (2–6 yrs), Child (6–12 yrs), Adolescent (12–18 yrs). Experiment information can be seen in Supplemental Table S5.
Pediatric CAVS valves trend to be collagen dominant at a younger age of end-stage
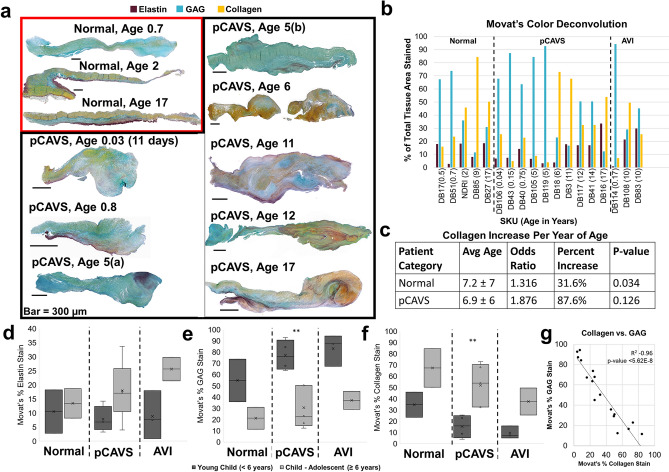
Color deconvolution of Movat’s Pentachrome on the valve cohort was performed to determine the dominant ECM type in each patient category (Fig. 1A, B, Supplementary Fig. S1). Movat’s Pentachrome stain showed an ECM trilayer in the normal valves with thickened, disorganized ECM due to pCAVS, consistent with previous literature reports on CAVS5,26. We stratified the pediatric tissues by neonatal to early childhood (0-including 5 years of age; rapid growth and development) and middle childhood to early adolescence (age 6–18 years of age). While differences in elastin can be seen from patient to patient, elastin content was not statistically significantly different between patient categories (Fig. 1B, D). Pediatric patients in all categories showed decreased glycosaminoglycan (GAG) staining over time relative to patient age (Fig. 1E). GAG decrease was significant within the pCAVS category compared between two patient age groups: young children (77.2 ± 14%; pediatric age < 6 years, n = 6) vs children to adolescents (30.6 ± 19%; pediatric age > 6 years, n = 5) (p-value 0.008) (Fig. 1E, F). Collagen content increased concomitant with decreasing GAG content over all patient groups (Spearman’s correlation − 0.96; p-value < 5.62E–8) (Fig. 1G). Interestingly, while collagen content progressively increased with age for each patient category, the likelihood of a pCAVS valve having a collagen dominant phenotype occurred at a younger age of end-stage. Histopathology evaluation revealed that the rate of collagen deposition increased by 56% in pCAVS compared to normal AV development. When accounting for age by multinomial logistical regression, analysis showed that the probability of a normal valve being in the collagen dominant ECM category increased by 32% (p-value 0.034, Fig. 1C) for every year of age. In contrast, pCAVS valves showed a trend to having a collagen dominant phenotype which increased by 88% (p-value 0.126, Fig. 1C) for every year of age. Collagen significantly increased in between pediatric patient age groups (34.7 ± 15.7% age < 6 years; 67.4 ± 24.1% age ≥ 6 years, p-value 0.0079) (Fig. 1F). These results show that the ECM disorganization of pCAVS involves a progressive collagen dominant phenotype concomitant with GAG decreases.
Figure 1.
Histology analysis identifies ECM mixing and collagen dominance in pCAVS valves at a young end-stage. (A) Examples Movat’s Pentachrome staining between normal and age matched pCAVS samples. Blue stain corresponds to glycosaminoglycan (GAG) regions, dark purple to elastin, and yellow to collagen. Normal valves show traditional trilayer structure, while pCAVS valves show lack of trilayer and areas of ECM mixing. (B) Quantification of Movat’s Staining via color deconvolution analysis. Blue = GAG, purple = Elastin, yellow = collagen. Shown as a percent of total Movat’s stain area. (C) Multinomial logistical regression analysis of Movat’s Pentachrome Color Deconvolution data shows pCAVS valves are collagen dominant at a younger age than normal patients. Variables: ECM dominant category (GAG, elastin, or collagen); Patient Category (normal or pCAVS); Age (0–18 years). (D–F) Quantification of Elastin, GAG, and Collagen (respectively) Movat’s Pentachrome color deconvolution results (Fig. 1B) comparing two pediatric age groups: young child (Newborn to < 6 years of age) and Child-Adolescent (6–18 years of age). **p-value 0.008, MWU test. pCAVS GAG decrease is concomitant with collagen increase across pediatric age categories (G) Scatterplot of all data points across Normal, pCAVS, and AVI samples for collagen vs. GAG stain percent of total area of Movat’s stain. Bar: 500 μm. Spearman’s correlation (R2) − 0.96 ( p-value < 5.62E–8). Normal n = 5; pCAVS n = 10; AVI n = 3.
Pediatric CAVS demonstrate high deposition of immature collagen compared to individual age-matched controls
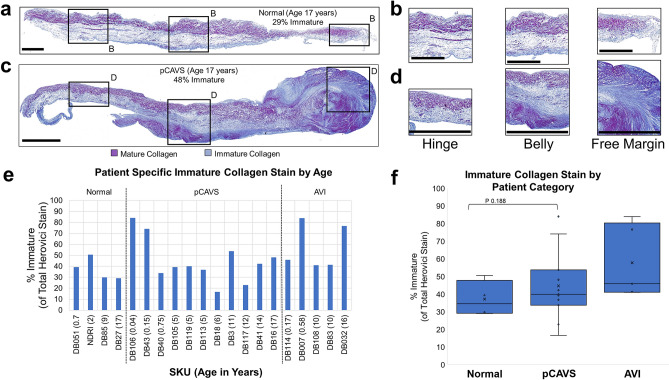
Herovici stain was used to assess valvular content of pre-collagen or immature collagen (blue) and mature collagen (purple) (Fig. 2, Supplementary Fig. S2; all valves studied)27. Immature collagen deposition varied by location on the valve structure and by pathology, suggestive of a link to mechanics of valve function. In trilayer regions close to the hinge region, normal valves and pCAVS showed immature collagen deposition in the spongiosa (Fig. 2A–D). Within the belly of the leaflet, immature collagen deposition aligned with regions of ventricularis delamination. However, pCAVS showed sites of pathological mixtures of immature and mature collagen. For example, in SKU DB16, immature collagen appears mixed with mature collagen along the ventricularis closer to the hinge. The free edge of this valve contains significant mixtures of both immature and mature collagen, suggestive of a dynamic production of collagen in this region. By Herovici stain, all valves showed new production of collagen (Fig. 2F). Averaged across ages at end-stage, there was no significant difference between normal and pCAVS levels of immature collagen. However, comparison of same age-matched patients showed increases in immature collagen (ex. 19% increase in SKU DB16 vs DB27) (Fig. 2E). Overall, the data show that both normal and pCAVS continually produce immature collagen with pCAVS showing pathological mixes of both immature and mature collagen.
Figure 2.
Pediatric CAVS demonstrate high deposition of immature collagen compared to age-matched controls. (A,B) Herovici staining of SKU DB27, a 17-year-old normal sample with regions of Hinge, Belly, and Free Margin 10 × zoom shown (B). Blue = immature. Purple-mature. Normal immature collagen production is localized to the ventricularis, while pCAVS immature production becomes increasing dysregulated towards the free margin. (C,D) Herovici staining of SKU DB16, an age-matched pCAVS sample with regions of Hinge, Belly, and Free Margin 10 × zoom shown in (D). (E) Patient specific breakdown of Herovici color deconvolution analysis. pCAVS patients show increased immature collagen production compared to age matched controls. (F) Boxplot showing quantification of Herovici color deconvolution, showing larger heterogeneity amongst pCAVS patients. Scale Bar: 300 μm. Normal n = 4; pCAVS n = 11; AVI n = 5.
Pediatric end-stage CAVS valves have greater numbers of collagen fibers with localized regions of collagen density
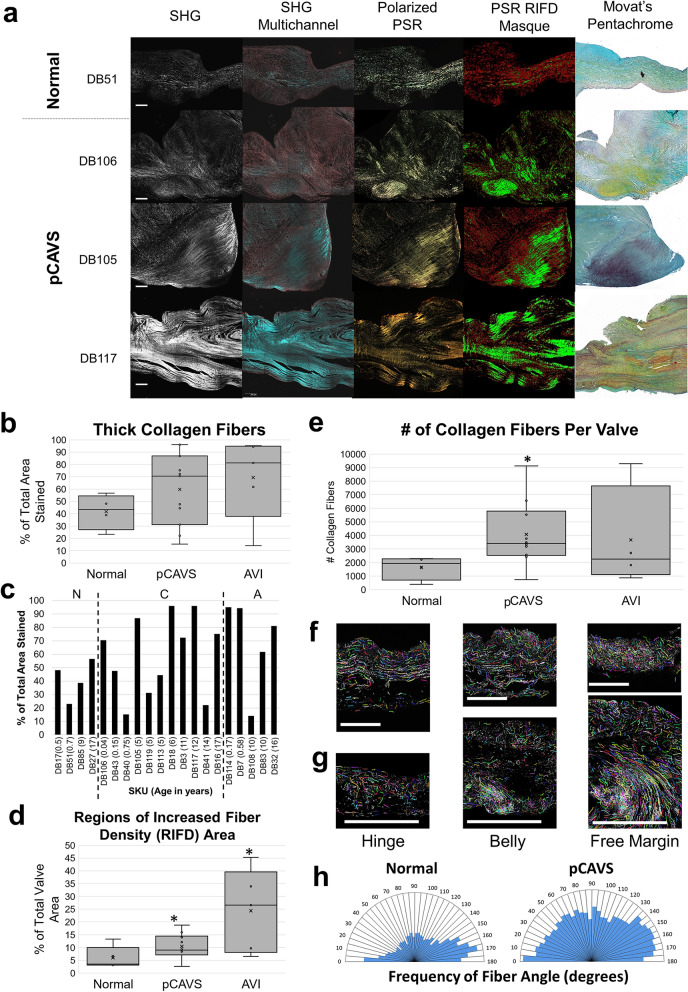
Collagen fiber measurements were done by Picrosirus Red (PSR) staining to characterize collagen fiber regulation (thickness, density, and alignment) (Fig. 3; Supplementary Fig. S3). Interestingly, all pCAVS valves showed localized areas of high-density collagen, confirmed by second harmonic generation (SHG) microscopy (Fig. 3A, Supplementary Fig. S3). Collagen fiber thickness was not significantly different between normal valves compared to pCAVS or AVI. However, larger interquartile ranges were observed in pCAVS (37–81% thick fibers) compared to normal valves (34–50% thick fibers), suggesting that pCAVS have increased heterogeneity of fiber thickness (Fig. 3B, C). Compared to normal valves, both pCAVS and AVI valves demonstrated a significant increase in regionalized collagen density normalized by area (p-value 0.024 and 0.019) (Fig. 3A, D). Localized areas of high-density collagen fibers did not appear to be specific to valvular anatomy and occurred at different locations within the leaflet. The amount of collagen fibers in pCAVS valves was significantly higher when compared to normal valves (2.5-fold more, p-value 0.014; normalized to cross-sectional area; Figure 3E). Picrosirius Red staining, in combination with CT-FIRE software, was used to determine the collagen fiber angle alignment relative to endothelium. Collagen fiber angles in normal valve structures were primarily between 150–180° relative to endothelium (SKU DB27, Fig. 3F, H), following previous observations in normal valve growth7,13,17. In comparison, the pCAVS valves showed a larger distribution of fiber angles (over 0–180° relative to the endothelium) along the length of the valve, suggestive of collagen fiber misalignment compared to normal controls (Fig. 3G, H). To summarize, the data shows that pCAVS valves are characterized by localization of high-density collagens and higher number of collagen fibers compared to normal valves.
Figure 3.
Pediatric end-stage CAVS valves have a greater number of collagen fibers with localized regions of collagen density. (A) Collagen fiber visualization of select pCAVS and normal valves. From left to right: Second Harmonic Generation (SHG); SHG multichannel; Polarized picrosirius red (PSR); PSR Regions on Increased Fiber Density (RIFD) Masque; Movat’s Pentachrome. Movat’s Pentachrome: blue = GAGs, yellow = collagen, and purple = elastin. Scale Bar = 200 μm. (B) Boxplot quantification of picrosirius red color deconvolution analysis, showing percent thick stain over total PSR stain. (C) Patient-specific data of PSR color deconvolution analysis, showing percent thick stain over total PSR stain. pCAVS and AVI patients show larger heterogeneity across age groups compared to normal. (D) Boxplot quantification of collagen RIFD areas normalized to total valve area. *p-value 0.024, MWU test. pCAVS and AVI patients have significantly more regions of high collagen fiber density compared to normal patients. (E) Boxplot showing quantification of total number of collagen fibers as measured by CT-FIRE analysis. *p-value 0.014, MWU test. pCAVS valves have significantly more collagen fibers than normal valves. (F,G) Hinge, Belly, and Free Margin (respectively) 10 × zooms of PSR-polarized light signal overlayed with CT-FIRE mask of collagen fibers for normal (F; SKU DB27) and pCAVS (G; SKU DB16) (Supplmentary Figure S3). Normal valves show stratified collagen fibers throughout the leaflet, while pCAVS valves show densely deposited fibers increasing in destratification from hinge to free margin. (H) Polar histogram showing frequency distribution of collagen fiber angles from 0–180°, across all normal valve samples (left) and pCAVS (right). pCAVS valves have a large distribution of fiber angels indicating destratification, while normal valve collagen fiber angles stratify to 0° and 180° relative to the endothelium. Normal n = 4; pCAVS n = 11; AVI n = 5. CT-FIRE V2.0 Beta was used (https://eliceirilab.org/software/ctfire/).
Proteomic analysis indicates collagen structure is post-translationally regulated in pCAVS
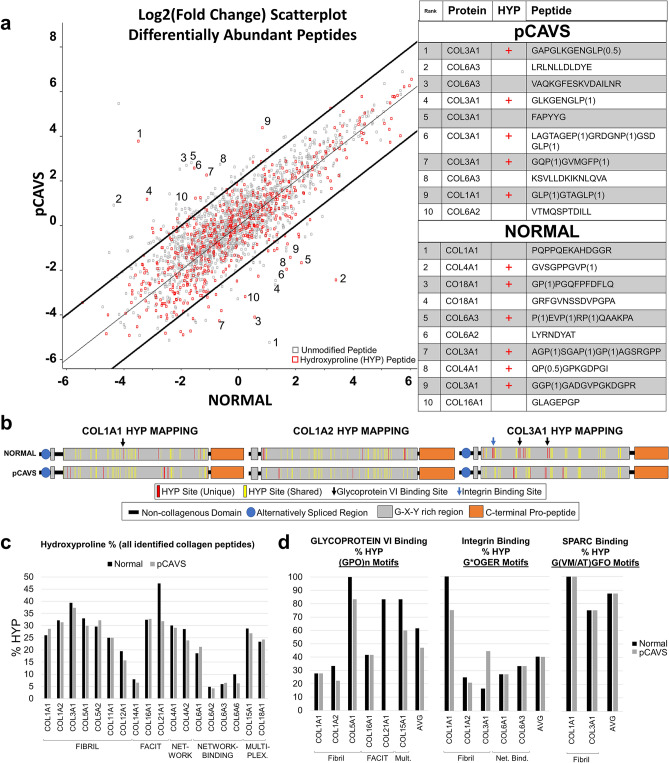
A collagen- and ECM protein-targeted proteomic analysis approach was next applied to the valve tissue cohort in order to understand how collagen fiber regulation corresponds with collagen protein structure regulation. The proteomics analysis workflow is summarized in Supplementary Figure S4 and described in Materials and Methods. A focus of the analysis was testing for hydroxylated proline (HYP) residues, a collagen post-translational modification essential to triple helical stability and collagen organization28,29. From a total of 2985 ECM peptides identified, 126 peptides were differentially expressed between normal and pCAVS by a minimum of twofold change (Fig. 4, Supplementary Tables S1, S2). The majority of altered peptides were from collagen type proteins (87/126; 69%) and these included site-specific modifications of hydroxylated proline (Fig. 4A). Hydroxyproline site mapping from age-matched normal and pCAVS valves suggested a strong potential for HYP site variation within the triple helical regions on primary fibril type collagens COL1A1, COL1A2 and COL3A1 (Fig. 4B). Site occupancy analytics showed 41 HYP sites in COL1A1 were shared between normal and pCAVS valves; the normal valve showed 5 unique sites of HYP modification while the pCAVS valve showed 8 sites of HYP modification that were not detected in normal valves (Fig. 4B). Consistent with data across the abundantly changed peptides, normal valve derived COL1A2 and COL3A1 contained more HYP sites than the age-matched pCAVS valve. In COL1A2, 41 sites were shared, with 9 unique HYP sites in normal valve and only 3 unique HYP sites in the age matched CAVS sample. However, for COL3A1, the normal AV (SKU DB27) showed an additional 16 unique HYP sites, with only 7 unique HYP sites identified in the pCAVS valve (SKU DB16; Fig. 4B). Among patient groups, the overall percent change in HYP content showed more subtle variation (Fig. 4C), yet it is likely that regulation at key HYP sites has greater effects on collagen structure organization28,30. specific HYP binding site motifs were evaluated for pCAVS related changes. These included motifs for glycoprotein VI (collagen-induced platelet adhesion and activation), integrin (cell-surface receptor for collagen)31 and SPARC (cell invasion) (Fig. 4D)30. Qualitatively, fibril type collagens COL1A2 and COL5A1 showed reduced HYP in glycoprotein VI binding sites. Similarly, COL1A1 demonstrated potential reduction in HYP content for integrin binding sites (Fig. 4D). COL21A1, which was recently found to be transcriptomically increased in cardiomyopathy remodeling32, showed large decreases in pCAVS HYP content; in pCAVS, HYPs in known binding motifs of glycoprotein VI were undetected in COL21A1 (Fig. 4D). The analysis of collagen structure variation by peptide analytics suggests that collagen HYP site regulation may contribute to human aortic valve status.
Figure 4.
Pediatric CAVS valves have reduced HYP, including in collagen-ECM binding sites. (A) A log2(fold change) scatterplot of pCAVS versus Normal peptides. Red squares are hydroxyproline containing peptides, gray squares are unmodified peptides. Bold lines indicate X + 2 and X − 2. Top 10 collagen peptides are annotated with corresponding sequence and protein shown (A, right). P(X) where X is between 0–1, indicated HYP probability at that site. Significantly regulated collagen subtypes differ between pCAVS and normal, with non-fibril types (multiplexin, FACIT) differentially regulated in normal. (B) Representative hydroxyproline site mapping for collagen 1A1, collagen 1A2, and collagen 3A1, shown for Normal and pCAVS patients (DB27 and DB16, respectively). Normal sample contained more HYP sites than age matched controls. Unique HYP sites in Normal (top, red) are found in glycoprotein vi and integrin binding motifs (black and blue arrows, respectively). HYP is required in these motifs for binding. (C) Quantification of hydroxyproline state of proteins in all peptides identified in normal (black) or pCAVS (gray). (D) Hydroxyproline percent in peptides containing glycoprotein vi, integrin, or SPARC binding motifs. Loss of HYP may indicate lack of collagen-ECM binding. FACIT: Fibril-associated collagen with interrupted triple helices; Net.: Network Collagen; Net. Bind: Network Binding Collagen; Mult.: Multiplexin (Multiple triple-helix domains with interruptions). Normal n = 4, pCAVS n = 11. MaxQuant and Perseus v.1.6.3.3. was used for this analysis (https://maxquant.net/maxquant/).
Valve status is characterized by a differential ECM proteome with primary activities of collagen interaction
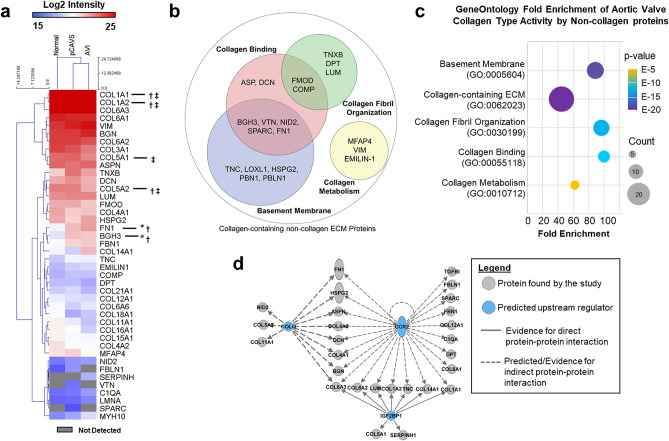
Protein level analysis was performed on the proteomic data (Fig. 5, Supplementary Table S3) to further evaluate how non-collagen ECM proteins contribute to collagen regulation. Proteomic analysis identified a total of 44 ECM proteins, including 18 collagen type proteins, many of which have not been previously associated with the human aortic valve (Fig. 5A). When measuring total protein abundance, most collagens showed very little change between normal and pCAVS, suggesting that regulation at the level of post-translational modifications may play a larger role in AV collagen organization. However, there were multiple ECM proteins associated with collagen regulation detected as differentially regulated. The abundance of the proteins fibronectin (FN1) and Transforming Growth Factor Beta Induced protein (BGH3; cell-collagen adhesions33) were significantly increased in pCAVS compared to normal AV (2.5-fold, MWUp-value 0.04; 3.4-fold, MWU p-value 0.02, respectively). In AVI, COL1A1 and FN1 increased compared to normal AV tissue (1.6-fold, MWU p-value 0.02; 4.6-fold, MWU p-value 0.03). Additionally, the proteomics identified SPARC and VTN proteins only in the pCAVS proteomes. Gene ontology analyses identified that 26 of the 29 non-collagen proteins were associated with collagen-containing extracellular matrix (Fig. 5B, C, Supplementary Table S4). Furthermore, very specific groups of proteins were involved in collagen binding, collagen fibril organization and collagen metabolism (Fig. 5B, C).
Figure 5.
Extracellular matrix proteomics implicates collagen binding and collagen organization pathways. (A) Aortic valve extracellular matrix proteins identified by the collagen-targeting proteomics. A total of 18 collagen proteins were identified with COL1A1, COL1A2 and COL6A3 being the most abundant collagens. Protein level quantification, which includes all peptides both unmodified and modified, revealed significant differences based on valve diagnoses in COL1A1, COL1A2, COL5A1, BGH3, FN1, and COL5A2. *Mann–Whitney U adj. p-value < 0.05 normal AV compared to pCAVS, †Mann–Whitney U adj. p-value < 0.05 normal AV compared to AVI, ‡Mann–Whitney U adj. p-value < 0.05 AVI compared to pCAVS. (B) Gene Ontology Functional classification of non-collagen type proteins identified in the proteomics dataset. (C) Gene Ontology Fold Enrichment analysis of collagen type function by non-collagenous proteins identified by proteomics, Fisher’s exactp-value ≤ 1.84E–5, false discover rate ≤ 6.66E–3. Proteins associated with collagen binding had the highest degree of fold enrichment, followed by collagen fibril organization. (D) Upstream analysis of the ECM proteome by Ingenuity Pathways Analysis to identify potential regulators that explain directional changes in aortic valve ECM proteins. Primary regulators affecting the proteome were C–C Motif Chemokine Receptor 2 (CCR2; Fisher’s Exactp-value 1.13E–39), Collagen Like Tail Subunit Of Asymmetric Acetylcholinesterase (COLQ; Fisher’s Exactp-value 9.67E-19), (Insulin Like Growth Factor 2 MRNA Binding Protein 1 (IGF2BP1, Fisher’s Exactp-value 1.05E–18). Normal n = 4, pCAVS n = 11.
To understand potential regulators of the dataset responsible for directional expression changes in pCAVS compared to normal, protein interaction networks were interrogated (Fig. 5D). While some regulators, such as TGFβ1, are known to be associated with ECM remodeling and deposition34, three novel upstream regulators of the collagen proteome were identified that could account for change in expression patterns. This included C–C Motif Chemokine Receptor 2 (CCR2; Fisher’s Exact p-value 1.13E–39), Collagen Like Tail Subunit Of Asymmetric Acetylcholinesterase (COLQ; Fisher’s Exact p-value 9.67E-19), and (Insulin Like Growth Factor 2 MRNA Binding Protein 1 (IGF2BP1, Fisher’s Exactp-value 1.05E–18). Both CCR2 and COLQ1 were implicated as inhibited in pCAVS based on directional changes35,36. On the whole, proteomics identified new collagen types in the human valve and demonstrated that changes in pCAVS structure may identify target regulators of the collagen proteome.
BAMBI as a potential novel ECM regulator of pediatric congenital aortic valve stenosis
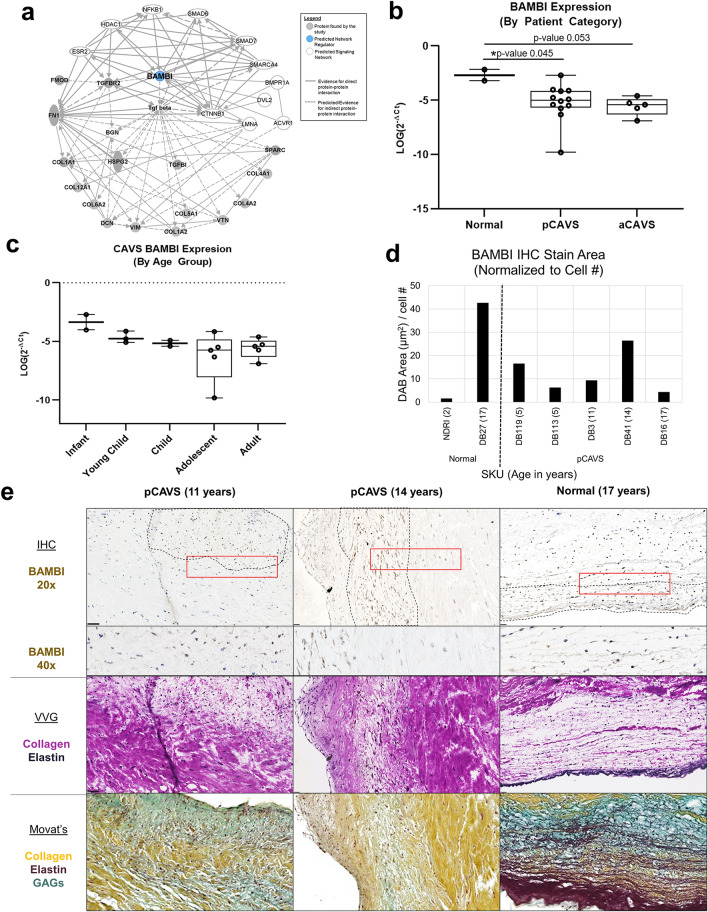
To identify master regulators that could be driving previously identified upstream regulators of the AV collagen proteome, causal network regulation analysis was performed. This analysis expands the scope of the upstream regulator analysis from Fig. 5 by integrating literature-observed cause-effect relationships (Fig. 6, Supplementary Figure S5). One such master regulator putatively identified was BMP and Activin Membrane-Bound Inhibitor homolog (BAMBI; p-value of overlap 1.16E–18) (Supplementary Fig. S5), previously identified as both developmentally regulated37 and a negative regulator of TGFβ138,39. Further studies have found that BAMBI may be biomechanically sensitive, and functions to protect murine heart from pressure overload through restraint of TGFβ signaling40. In this study, BAMBI was identified as having a high p-value of overlap, which unlike z-score-sorted regulators, is identified independently of the dataset’s regulation weight or direction41. This preliminary identification of BAMBI was confirmed to have both direct and indirect interaction with proteins implicated in valve disease and development, such as ACVR1, DVL2, and SMARCA4 (Fig. 6A; Supplementary Table S6) Confirmational transcription-level analysis was performed on a cohort of fresh-frozen pediatric and adult aortic valve samples (Supplementary Table S7, Fig. 6b, c, Supplemental Figure S5). Adult CAVS samples (aCAVS) used in this study showed no calcification by clinical annotation, with the except of SKU SB176. BAMBI expression was found to be significantly differentially regulated between patient categories of normal pediatric, pCAVS, and aCAVS (ANOVA p-value 0.013), with significant reduction in pCAVS (p-value 0.045) and trending reduction in aCAVS (p-value 0.053) compared to normal pediatric patients (Fig. 6b). Amongst CAVS patients, there is a suggestive trending decrease of BAMBI expression as a function of pediatric age categories (ANOVA p-value 0.096) (Fig. 6c).
Figure 6.
Proteomics based protein–protein interaction networks identify BAMBI as a potential master regulator of the collagen interactome in pCAVS. (A) Protein–Protein Interaction network showing relationship BAMBI (Supplemental Figure S5) relative to the ECM proteome and key valvular development proteins (Supplemental Table S6). (B) Quantification of qRT-PCR expression data on BAMBI (normalized to GAPDH), between patient categories. One-way ANOVA shows significant differential expression between patient categories (*p-value 0.013), with significant reduction in pCAVS (MWU *p-value 0.045) and trending reduction in aCAVS (MWU p-value 0.053) compared to normal pediatric patients. (Normal n = 2; pCAVS n = 12; aCAVS n = 5) C. Quantification of qRT-PCR expression data on BAMBI (normalized to GAPDH) of CAVS patients, compared between pediatric age categories (Infant n = 2; Young Child n = 3; Child n = 2; Adolescent n = 5; Adult n = 6). A suggestive trending decrease of BAMBI expression as a function of pediatric age categories is seen (ANOVA p-value 0.096) (Table 1; Supplemental Table S7). (D) DAB stain area quantification of BAMBI IHC of select valves in the cohort. (E) Representative immunohistochemistry staining of BAMBI in a selection of pCAVS and normal valves 20× (top) and 40 × zoom are shown. BAMBI positive cells appear highly localized, shown in dotted lines. Verhoeff van Geison (VVG) staining on the same valve section post-IHC study is shown (middle). Movat’s Pentachrome staining of a serial valve section (bottom). Positive BAMBI staining in pCAVS corresponds heavily to areas low in collagen staining (VVG) and high in GAG staining (Movat’s), while normal valve shows positive staining in collagen and elastin rich regions.
IHC staining of BAMBI was performed to visualize translational expression relative to ECM staining in age-matched samples (young child age 2–5; adolescent age 12–18) (Fig. 6D,E; Supplementary Figs. S6, S7). BAMBI was not abundantly detected in young normal valves (SKU NDRI, age 2) nor, in contrast, older adolescent pCAVS valves (SKU DB16, age 17) (Supplementary Fig. S6, S7). In the adolescent normal valve sample, BAMBI was observed in ventricularis endothelium and in regions of elastin delamination, as visualized by Verhoeff van Gieson (VVG) staining of the same tissue section (Fig. 6E, right). BAMBI was also detected in spatially localized regions of mixed collagen-GAG, seen in Movat’s Pentachrome staining (Fig. 6e). In pCAVS, only certain cells were BAMBI-positive and tended to be within the fibrosa-spongiosa mixed interstitium of GAG associated regions low in primary collagen (Fig. 6E, left and center columns). These data suggest that future studies on BAMBI may show its contribution to regional collagen deposition within the valve structure.
Discussion
Collagen forms the fundamental framework of the aortic valve starting from embryonic developmental stages onward; disease-driven changes deform the framework and ultimately lead to heart dysfunction10,14,42–44. In pCAVS, rapid and excessive production of ECM components increases leaflet thickness by up to eightfold, leading to pediatric heart failure5,16,45,46. Genomic studies, primarily characterized from adult end-stage phenotypes, show complex genetics with dissimilar patterns of inheritance, genetically heterogenous phenotypes, and multiple chromosomal regions that impact the ECM network directly or indirectly45,47–49. Gender may also play a role—while bicuspid aortic valve is three times as likely to be found in males, females have more collagen deposition and less calcification50,51. Surgical replacement of valves is an option in adults, but in children, valve replacements do not grow with the patient, leading to heart dysfunction and failure4,52. Directly or indirectly targeting collagen production in pediatric end-stage CAVS may be a viable therapy inhibiting the disease, but very little is known about translational and post-translational regulation collagen and ECM components that form the working structure of the maturing aortic valve. The goal of this study was to identify the dynamic collagen regulation that occurs in pCAVS, including collagen sub-type regulation, collagen fiber alignment, and regulation of hydroxylated proline residues associated with triple helical stability.
The ECM trilayer structure was lost in pCAVS and the rate of collagen deposition trended to increasing by up to 56% per year compared to normal AV development (Fig. 1). This coincides with previous reports that pCAVS involves overproduction of collagen to result in thickened valves5,10,16. Supporting this, immature collagen fibers were detected by staining in all ages of pCAVS localized to regions of mixed collagen and elastin, particularly valves that showed hypertrophic commissures. Significantly, the pCAVS valves showed that collagen fiber alignment with the endothelium is lost and resulted in a random fiber angle relative to endothelium that localized with stains for immature collagen fibers. An additional finding was that unique, pathologically confined regions of increased collagen fiber density were observed in all pCAVS structures (Fig. 3). This contrasted with measurements on normal valves in the study, where collagen fiber angles were parallel to the endothelium with the fibrosa staining for mature collagen fibers. Similar findings in normal human aortic growth have been reported, with emergence of collagen fiber alignment parallel to endothelium within maturing fibrosa7. Previous work in normal human aortic and pulmonary valve has shown that fibrosa collagen is circumferentially oriented, and that uniformly distributed, densely packed collagen fibers emerge with age13. Age-related increases in collagen density in pCAVS were attributed to an imbalance of normal collagen synthesis and remodeling required to maintain appropriate collagen stretch dependent on local hemodynamics13. We also observed that for valves with aortic valve insufficiency, higher but more variable levels of thick collagen fiber production were observed compared to either normal or pCAVS valves, supporting that valvular function is a balance of collagen production and collagen degradation10–12 with dependence on hemodynamics. Additional but unknown effects on collagen production and alignment may result from the cell type and density composition within the tissue microenvironment, in addition to the differential effects of hemodynamics throughout the pCAVS structure. Alterations in collagen fiber alignment are an important component of the pCAVS end-stage and are a useful target in mechanistic studies on the disease.
Collagen PTMs are numerous and include non-enzymatic glycation, lysine and proline crosslinking, N- and O-linked glycosylation, and tyrosine sulfation28,53,54. Here, we focused on hydroxylation of proline (HYP) to understand collagen regulation in pCAVS. HYP is a primary collagen PTM that regulates triple helical stability and consequently has a large influence on collagen organization in tissue28. Regulation of HYP sites within the collagen structure alters fiber conformation and integrity and works to promote or protect the collagen structure from degradation28,54–56. HYP is regulated by co-factors of iron, oxygen, and ascorbic acid, with dependency on nutritional status and oxidative stress 1,28,56–58. Cell-specific and disease-specific regulation of prolyl hydroxylases P4HA1, P4HA2, P4HA3 drive HYP modification on collagen58,59. Cell recognition of very specific sites of hydroxylated prolines along the collagen fiber results in signaling changes through integrins and the tyrosine kinase receptors discoidin domain receptors (DDRs)60–63. These cell-fiber interactions influence epithelial mesenchymal transition, cell proliferation survival and metastatic expansion61,62,64–66. The current study identified up to 15% reduction of HYP content in pCAVS, dependent on collagen subtype. However, HYP changes appeared to be site specific. It is possible that changes in HYP sites may contribute to the rapid collagen degradation and remodeling of pCAVS28. Further, the sites of HYP loss in the pCAVS group aligned with regions of glycoprotein VI and integrin binding, which require the presence of HYP within the sequence. Integrins play a crucial role in cell-ECM interactions, with recent data suggesting a key role in dynamic connective tissue remodeling events during wound healing67. Glycoprotein VI is a platelet specific glycoprotein that has been shown to have affinity for collagen in the dimeric form68. It has been reported that adult patients with AVS have decreased platelet function and aggregation, however the role of HYP content has not been explored as a potential mechanism69. This study identifies that HYP regulation on primary fibrillar collagens may contribute to intrinsic and extrinsic mechanisms of pCAVS and merits focused studies.
Histological studies identified localized regions of dense collagen formation in common with pCAVS. Protein level studies gave insight into the collagen composition of the pCAVS deregulated structures. Collagen alpha-2(V) (COL5A2), was found to have significant increases pCAVS compared to non-stenotic AVI valves. Importantly, COL5A2 has been reported within heterotypic Collagen Type 1 fibrils and plays an essential role in regulating fibrillogenesis and the size of collagen fibrils70. The histopathological finding of high-density collagen fiber regions in pCAVS may be reflective of the increased fibrillogenesis. However, to implicate COL5A2 directly, spatially oriented proteomics would be required to understand localization and regulation of the highly density collagen sites compared to collagen type composition. With localized molecular biomarker identification, these regions of dense collagen fibers provide a potential mechanism of early-stage AV fibrosis molecular marker detection in vivo71. Amongst non-collagen type proteins, the enrichment of proteins associated with collagen binding, collagen fibril organization, and basement membrane was consistent with our histopathological findings of collagen fiber misalignment and HYP regulation in binding sites. Interestingly, collagen chaperone SPARC was found to be enriched in pCAVS compared to normal AV and AVI patients, independent of age (Fig. 5a). While never explored in healthy or disease AV, the overexpression of SPARC has been reported with under-expression of COL4A1 and other network collagens in myocardium. Increases in SPARC has been linked to endothelial dysfunction, possibly via its binding to VCAM-1 and altered integrin interactions downstream72. Its abundance in pCAVS may suggest endothelial dysfunction. Research of healthy and diseased endothelial cell signaling in AV is ongoing, however direct correlation between VEC or VIC cell activation and corresponding collagen regulation remains to be explored16,73,74.
The study identified BAMBI as a novel candidate that may be involved in valvular biology. BAMBI is a negative regulator of TGFβ1 signaling38,39 and positively modulates Wnt/β-Catenin pathways by increasing interactions with frizzled 5 and disheveled segment polarity protein 2 (DVL2)37. While never studied directly in AV disease or development, recent studies suggest that BAMBI is regulated in the myocardium to restrain hypertrophy and fibrosis40. In the current study, network analysis highlighted BAMBI as a predicted node amongst proteins known to be involved in valvular development and disease. Interestingly, the initial protein-level staining studies demonstrated that the human aortic valve has cell-specific and localized expression of BAMBI, particularly within the glycosaminoglycan-rich spongiosa. Furthermore, by transcriptional analysis of total valvular content, BAMBI expression appeared to decrease in both pediatric and adult CAVS. It is conceivable that decreases in BAMBI may play a role in the excess ECM that is a hallmark of pCAVS. However, the role of BAMBI in regulating valvular biology remains undefined.
The sample size of the cohort was limited. Pediatric tissues inherently are difficult to obtain with a past national average of only 40% of families consenting for tissue use and donation75. Actual figures in specific regions may be lower76. This limitation similarly effects pediatric human aortic valve interstitial cell studies, as these require fresh samples for immediate cell isolation77,78 as opposed to the resected valves that are available through tissue banks. Nevertheless, this is the largest collagen study to date on valve tissue with pediatric end-stage aortic valve stenosis. Future investigations to extend studies from this small clinically-relevant cohort will be critical to understanding how developmental timing of valvular interstitial cell signaling influences spatial distribution of collagen fiber production.
Conclusions
This study is the first to identify a tissue associated collagen signature unique to pediatric end-stage aortic valve stenosis, which includes highly localized regions of dense and realigned collagen fibers within the valve structure. Additionally, this is the first quantitative and structural report of collagen hydroxyproline, or HYP, in valve development and disease. These data establish the groundwork for future studies examining the spatial regulation of collagen HYP within valvular structures and in relation to signaling factors driving collagen organization and deposition in pCAVS. We anticipate that this data will inform functional testing of cell-based mechanisms regulating collagen HYP, the generation of new mouse models of valve disease, and improvements in the development of collagen biomaterials used in in vitro valve disease models and bioengineered valve replacements.
Methods
Materials
Xylenes, 200 proof ethanol was from Fisher Scientific (Pittsburgh, PA, USA). Acetonitrile, Ammonium Bicarbonate, Calcium Chloride, Formic Acid, and Trizma Base were from Sigma-Aldrich (St. Louis, MO, USA). PNGase F Prime was purchased from N-Zyme Scientifics (Doylestown, PA). Collagenase type III (COLase3) (C. histolyticum) was purchased from Worthington Biochemical (Lakewood, NJ, USA).
Tissue procurement
Aortic valve tissue samples were procured through the Vanderbilt Core Laboratory for Translational and Clinical Research and the National Disease Research Interchange. The aortic valve tissues were collected under the Vanderbilt Pediatric Congenital Heart Disease Biorepository, written informed consent was obtained, and the project was approved by the Vanderbilt Institutional Review Board (IRB) and the IRB at the Medical University of South Carolina. De-identified tissues were obtained during reparative or transplant surgeries and characterized by pre-operative function into three patient categories: normal tricuspid (normal), pediatric CAVS bicuspid (pCAVS), and aortic valve insufficiency tricuspid (AVI). Valvular function is from de-identified complete 2D, color and spectral Doppler, or M-mode echocardiogram reports acquired prior to valve resection, replacement or heart transplant. The pCAVS patients were identified as being stenotic, predominantly bicuspid, and having mild aortic valve insufficiency. The AVI group only has aortic valve insufficiency, but are not stenotic and are all tricuspid. Adult CAVS samples (aCAVS) used in the qRT-PCR study showed no calcification by clinical annotation, with the except of SKU SB176. A complete list of studies performed on each tissue are listed in Supplementary Tables S5 and S7. Pediatric ages used to stratify the population are based in National Institute of Child Health and Human Development (NICHD) pediatric terminology defining pediatric age groups in terms of clinical treatment, considering rapid somatic growth stages79.
Histological staining
The 5-µm thick formalin-fixed, paraffin-embedded tissue sections were stained with Picrosirius Red (PSR; Polysciences, Inc., Warrington, PA, USA), Movat’s Pentachrome and Herovici (American Mastertech Scientific of StatLab, McKinney, TX, USA) following manufacturer’s protocols. Optical light whole tissue scans were done using a high-resolution slide scanner (Nanozoomer, Hamamatsu, Japan). Polarized light images of PSR were taking with a Spinning Disk CARV II microscopy (BD Biosciences, San Jose, CA, USA), and manually stitched together using Photoshop (Adobe, San Jose, CA, USA). Movat’s Pentachrome, PSR, and Herovici Color Deconvolution analysis was done using ImageJ80 adjusting for RGB hue threshold color (Movat: Elastin 0–19, 170–255; GAG 80–169; Collagen 20–79) (PSR: Thin 46–190; Thick 0.45, 191–255) (Herovici: Immature 0–167; Mature 168–255). Percent of each color threshold is represented as a percent of total area stained (RGB 0–255). RIFD (Region of Increased Collagen Fiber Density) analysis was done using ImageJ80. Briefly, polarized light images were converted to 8-bit and thresholded to include all signal. Images were processed with a mean filter of 1 μm and a look-up table of Red/Green was applied. Regions of increased collagen fiber density (RIFD) were quantified by thresholding to include the top 50% most dense signal (i.e., “green” component). Collagen fiber analysis (fiber length, width, angle) of PSR birefringent images were converted to 8-bit grayscale and analyzed using CT-FIRE81.
Second Harmonic Generation (SHG) microscopy was performed on the same tissue section used for PSR studies. SHG images were collected with a Spectra-Physics InSight X3 dual laser system and excited at 860 nm. SHG images and were collected at emission 432 nm with a 45 nm bandpass. SHG multichannel show overlay images collected at emission wavelengths: 432 nm (± 22.5 nm) (cyan; collagen SHG), 518 nm (± 22.5 nm) (green; collagen autofluorescence), 610 nm (± 35 nm) (red; PSR, elastin, and heme structures).
For immunohistochemistry, tissues were deparaffinized, antigen retrieved at pH 6 in sodium citate, then stained with HRP/DAB (ABC) Detection IHC Kit (Abcam, Cambridge, UK) according to manufacturer’s instructions with the following modifications: all incubation times were increased to 15 min. Primary antibody BAMBI (polyclonal) was diluted 1:100 (Millipore Sigma, St. Louis, MO, USA). IHC stained tissues were counterstained with Mayer’s hematoxylin (Electron Microscopy Sciences, Hatfield, PA, USA).
Proteomic tissue preparation
Proteomic studies used tissue sections adjacent to histological studies (~ 150 μm distance). Tissues were 5-µm thick and mounted on standard microscope slides (Tissue Tack, Polysciences, Inc., Warrington, PA, USA). For proteomic experiments, slides were heated at 60 °C for 1 h then dewaxed as follows: xylenes (3 min, two times), 200 proof ethanol (1 min, two times) 95% ethanol (1 min), 70% ethanol (1 min), HPLC water (3 min, two times). Valve surface area was used to determine equivalent amount of tissue was scraped with a razor off the slide into centrifuge tubes. 10 mM Tris pH 9 was added to each tube to cover the tissue. Tissues were ultrasonicated at 50% energy (Fisherbrand 120 sonic dismembrator;Fisher Scientific, Pittsburgh, PA, USA) for 2 min each and incubated at 60 °C for two hours to antigen retrieve formalin-fixed crosslinks82. Samples were buffer exchanged into water and deglycosylated with 2 μg of PNGaseF PRIME (N-zyme Scientifics, Doylestown, PA) for two hours at 38 °C at 450 rpm using a thermomixer (Eppendorf, Hamburg, Germany). Supernatant was removed, and samples were buffer exchanged into 10 mmol/L ammonium bicarbonate, 3 mmol/L CaCl2, pH 7.25 and treated with 4 μg of Collagenase Type III (COLase3) overnight, shaking at 38 °C. An additional 4 μg of COLase3 was then added to each sample and incubated for 5 h at 38 °C with mixing at 450 rpm. COLase3 activity of 3 Units/mL was confirmed prior to digestion by a colorimetric activity kit (ab196999, Abcam, Cambridge, MA). Samples were centrifuged at 16,000 g for 5 min at 4 °C and supernatant collected for proteomic analysis. Samples were purified by C18 STAGE tip83 (Pierce Biotechnology, Waltham, MA) followed by a C18 ZipTip (Millipore Scientific, Burlington, MA) according to manufacturer’s protocol. Solutions were modified for STAGE tip using 90% acetonitrile, 5% formic acid (initialize and elute); 5% acetonitrile, 5% formic acid (equilibrate and wash). STAGE-tip eluate was dried down via speed vac and resuspended in 0.1% TFA in water. An additional C18 ZipTip according to manufacturer’s protocol (Millipore Scientific) and sample resuspended in mobile phase A (2% acetonitrile, 0.2% formic acid).
Proteomics
Peptides were analyzed by data dependent acquisition on an Orbitrap Elite mass spectrometer equipped with a LC Packing U3000 nano-LC system (Thermo Scientific). Peptides were loaded onto a trap column and separated on a 75 μm × 30 cm classic pulled tip column (C18-Reprosil-AQ Pur RP 1.9 µm particles, Dr. Maisch, GmbH) at 60 °C. The gradient was from 5 to 40% solvent B over 180 min, where solvent A was 0.2% v/v formic acid in water and solvent B was 98% acetonitrile with 0.2% v/v formic acid. An FTMS survey scan was acquired over a mass range of 400–1700 m/z at a resolution of 60,000 with an automatic gain control (AGC) target value of 106, followed by CID MS/MS of the top 10 most intense ions in the ion trap. Dynamic exclusion was enabled with a repeat count of 1, repeat duration of 30 s, and exclusion duration of 180 s. Data were searched using MaxQuant version 1.6.3.384 against the human database (42,106 entries downloaded May 5, 2017) and a subset 1783 entries with keywords used (collagen, elastin, aggrecan, gelatin, osteonectin, perlecan, plasminogen, and fibronectin). Parameters included unspecified proteolytic enzyme, precursor mass tolerance of ± 20 ppm, and fragment mass tolerance ± 0.8 Da. Methionine oxidation, asparagine and glutamine deamidation were included as variable modifications. Proteins were identified with FDR ≤ 0.05 and at least two peptides. Hydroxylated proline modification site localization probabilities were obtained by re-searching MS/MS using a restricted database of identified proteins and allowing for dynamic modification proline oxidation. Perseus85 was used for protein and peptide level analyses.
Protein–protein interaction network analysis
Protein identifications were uploaded to Ingenuity Pathways Analysis (IPA; Qiagen Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) to identify putative regulators of the protein dataset. Networks were calculated using predicted relationships between regulators by p-value ≤ 0.01; a p-value < 1.0 × 10–12 was used for inclusion in the network. Upstream Regulator Networks were calculated using predicted relationships between regulators (CCR2, COLQ, IGF2BP1); a p-value < 1.0 × 10–18 was used for inclusion of the upstream regulator network. Causal Network analysis was done to identify Master Regulators of the data (BAMBI) and a p-value < 1.0 × 10–12 of overlap was used for inclusion in the network.
Proteomic data analysis and statistics
Perseus85 was used for protein and peptide level analyses. Hierarchical clustering was performed in Perseus on ANOVA significant peptide intensities via a permutation-based FDR of 0.05 and 250 randomizations. Clustering parameters: Euclidean distance, average linkage, and k-means pre-processing. Peptide level quantification was determined using peak intensities from peptides scoring ≥ 75 after median normalization and considered as significant with a minimum two-fold change. Relative changes in protein abundance based on normalized protein intensities were evaluated by ANOVA and Mann Whitney U test, correcting for multiple comparisons (IBM SPSS Statistics, version 25). A Type I error probability of ≤ 0.1 was used to evaluate the significance of the result. MWU-test was used in histological studies to calculate raw p-values. For this small clinical cohort, p-values of ≤ 0.1 are reported as trending to significance and p-values of ≤ 0.05 were used determine significant results.
GO enrichment was performed through Geneontology.org86,87. Genes were clustered to cellular component, biological process, or molecular function using Panther v1488 filtered by Fisher’s Exact (FE) p-values < 0.001 and false discovery rates (FDR) of < 0.005. Exported peak intensities are visualized as heatmaps after natural log transformation with MultiExperiment Viewer (http://www.tm4.org)89.
Real time quantitative reverse transcription PCR
Aortic valve (AV) specimens were collected at the time of surgery or from an explanted heart. Samples were immediately flash-frozen and stored at − 80 °C till used for RNA extraction. Total RNA from AV tissues was isolated using RNeasy Kit (Qiagen, Germantown, MD), according to the manufacturer’s instructions. The BAMBI gene expression level was validated by quantitative RT–PCR (qRT-PCR) using the Taqman method, as previously described90. Briefly, cDNA was generated using 100 ng of total RNA and High Capacity cDNA Synthesize Kit (Catalog # 4,368,814 Thermo Fisher Scientific). Predesigned BAMBI (catalog # Hs03044164) and GAPDH (catalog # Hs02758991) primers were obtained from Thermo Fisher Scientific and the qRT-PCR were run in triplicates according to the manufacturer’s instructions. All Ct values were reported below 40, indicating acceptable PCR efficiency91.
Supplementary Information
Acknowledgements
The authors appreciate IRB-approved access to tissues from the Vanderbilt Heart Registry and Biorepository. Collagen SHG imaging was conducted at the Multiphoton Intravital and Tissue Imaging Core (University of Nebraska Medical Center) which receives support through the Nebraska Center for Nanomedicine (NCN) Center for Biomedical Research Excellence (Institutional Development Award, National Institute of General Medical Sciences of the National Institutes of Health, P30GM127200), state funds from the Nebraska Research Initiative, and institutionally by the UNMC Office of the Vice Chancellor for Research. Work by CLC is supported by a T32 from NHLBI (HL007260) and an F31 from NHLBI (HL156524). The Orbitrap Elite used in this research was funded by S10 D010731 (NIH/OD) to LEB. Microscopy data supported by the South Carolina COBRE in Oxidants, Redox Balance and Stress Signaling (GM 103542). Additional support was provided by the South Carolina Centers of Economic Excellence SmartState Program to RRD and ASM. Additional support to RRD, ASM and PMA by U01CA242096 (NCI). PMA appreciates support by 16GRNT31380005 (American Heart Association), R21CA240148 (NCI), P20GM103542 (NIH/NIGMS). The authors thank Professor Gary Hardiman for informatics discussion.
Abbreviations
- AVS
Aortic valve stenosis
- pCAVS
Pediatric end-stage congenital aortic valve stenosis
- AVI
Aortic valve insufficiency
- ECM
Extracellular matrix
- PTM
Post-translational modification
- HYP
Hydroxyproline
- HRAM
High resolution accurate mass
- GAG
Glycosaminoglycan
Author contributions
YRS and DB specifically collected tissue specimens for use in these experiments. CLC and PMA conceived the experiments and drafted the manuscript. CLC performed the experiments. HJS did the second harmonic generation microscopy and analyzed results. CLC and PMA analyzed the data. JB, KNC, and SCW contributed to proteomic sample preparation and experimental design and evaluation of MaxQuant data. YRS contributed to qRT-PCR experiments. LEB, MAH, ASM, RRD, and PMA contributed experimental oversight. All authors reviewed the manuscript.
Data availability
Any datasets generated during the study (not already in supplemental materials) are available on reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE92 partner repository with the dataset identifier PXD021990. During the peer review process, this data can be accessed via reviewer_pxd021990@ebi.ac.uk (Password: YA2RlOK6).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-89164-w.
References
- 1.Hoffman JIE, Kaplan S, Liberthson RR. Prevalence of congenital heart disease. Am. Heart J. 2004;147:425–439. doi: 10.1016/j.ahj.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Kirklin JW, Barratt-Boyes BG. Congenital aortic stenosis. In Kirklin JW, Barratt-Boyes BG, eds. Cardiac Surgery. 2nd ed. New York: Churchill Livingstone, 1195–1237 (1993)
- 3.d'Arcy JL, Prendergast BD, Chambers JB, Ray SG, Bridgewater B. Valvular heart disease: the next cardiac epidemic. BMJ. 2010;97:91–93. doi: 10.1136/hrt.2010.205096. [DOI] [PubMed] [Google Scholar]
- 4.Sharabiani MTA, et al. Aortic valve replacement and the ross operation in children and young adults. J. Am. Coll. Cardiol. 2016;67:2858–2870. doi: 10.1016/j.jacc.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 5.Hinton RB, et al. Extracellular matrix remodeling and organization in developing and diseased aortic valves. Circ. Res. 2006;98:1431–1438. doi: 10.1161/01.RES.0000224114.65109.4e. [DOI] [PubMed] [Google Scholar]
- 6.Levay AK, et al. Scleraxis is required for cell lineage differentiation and extracellular matrix remodeling during murine heart valve formation in vivo. Circ. Res. 2008;103:948–956. doi: 10.1161/CIRCRESAHA.108.177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aikawa E, et al. Human semilunar cardiac valve remodeling by activated cells from fetus to adult: implications for postnatal adaptation, pathology, and tissue engineering. Circulation. 2006;113:1344–1352. doi: 10.1161/CIRCULATIONAHA.105.591768. [DOI] [PubMed] [Google Scholar]
- 8.Tsamis A, Krawiec JT, Vorp DA. Elastin and collagen fibre microstructure of the human aorta in ageing and disease: a review. J. R. Soc. Interface. 2013;10:20121004–20121004. doi: 10.1098/rsif.2012.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanson KP, et al. Spatial and temporal analysis of extracellular matrix proteins in the developing murine heart: a blueprint for regeneration. Tissue Eng. Part A. 2013;19:1132–1143. doi: 10.1089/ten.tea.2012.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinton RB, Yutzey KE. Heart valve structure and function in development and disease. Ann. Rev. Phys. 2011;73:29–46. doi: 10.1146/annurev-physiol-012110-142145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joyce EM, Liao J, Schoen FJ, Mayer JE, Sacks MS. Functional collagen fiber architecture of the pulmonary heart valve cusp. Ann. Thorac. Surg. 2009;87:1240–1249. doi: 10.1016/j.athoracsur.2008.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoen FJ. Evolving concepts of cardiac valve dynamics: the continuum of development, functional structure, pathobiology, and tissue engineering. Circulation. 2008;118:1864–1880. doi: 10.1161/CIRCULATIONAHA.108.805911. [DOI] [PubMed] [Google Scholar]
- 13.Oomen PJA, et al. Age-dependent changes of stress and strain in the human heart valve and their relation with collagen remodeling. Acta Biomater. 2016;29:161–169. doi: 10.1016/j.actbio.2015.10.044. [DOI] [PubMed] [Google Scholar]
- 14.Peacock JD, Lu Y, Koch M, Kadler KE, Lincoln J. Temporal and spatial expression of collagens during murine atrioventricular heart valve development and maintenance. Dev. Dyn. 2008;237:3051–3058. doi: 10.1002/dvdy.21719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockhart M, Wirrig E, Phelps A, Wessels A. Extracellular matrix and heart development. Birth Defects Res. A. 2011;91:535–550. doi: 10.1002/bdra.20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirrig EE, Hinton RB, Yutzey KE. Differential expression of cartilage and bone-related proteins in pediatric and adult diseased aortic valves. J. Mol. Cell. Cardiol. 2011;50:561–569. doi: 10.1016/j.yjmcc.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutson HN, et al. Calcific aortic valve disease is associated with layer-specific alterations in collagen architecture. PLoS ONE. 2016;11:e0163858. doi: 10.1371/journal.pone.0163858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eriksen HA, et al. Type I and type III collagen synthesis and composition in the valve matrix in aortic valve stenosis. Atherosclerosis. 2006;189:91–98. doi: 10.1016/j.atherosclerosis.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 19.Podlesnikar T, Delgado V, Bax JJ. Cardiovascular magnetic resonance imaging to assess myocardial fibrosis in valvular heart disease. Int. J. Cardiovasc. Imag. 2018;34:97–112. doi: 10.1007/s10554-017-1195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinkert MA, Hortensius RA, Ogle BM, Eliceiri KW. Cardiac Extracellular Matrix. Springer; 2018. pp. 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angel PM, et al. Mapping extracellular matrix proteins in formalin-fixed, paraffin-embedded tissues by MALDI imaging mass spectrometry. J. Proteome Res. 2018;17:635–646. doi: 10.1021/acs.jproteome.7b00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angel PM, et al. Extracellular matrix imaging of breast tissue pathologies by MALDI imaging mass spectrometry. Proteomics Clin. Appl. 2019;13:1700152. doi: 10.1002/prca.201700152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angel PM, et al. Extracellular matrix alterations in low grade lung adenocarcinoma compared to normal lung tissue by imaging mass spectrometry. J. Mass Spectrom. 2019;55:e4450. doi: 10.1002/jms.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angel PM, et al. Zonal regulation of collagen types and post-translational modifications in prostatic benign and cancer tissues by imaging mass spectrometry. Prostate. 2020;80:1071–1086. doi: 10.1002/pros.24031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angel PM, et al. Mapping extracellular matrix proteins in formalin-fixed, paraffin-embedded tissues by MALDI imaging mass spectrometry. J. Proteome Res. 2018 doi: 10.1021/acs.jproteome.7b00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wirrig EE, Hinton RB, Yutzey KE. Differential expression of cartilage and bone-realted proteins in pediatric and adult diseased aortic valves. J. Mol. Cell. Cardiol. 2012;50:561–569. doi: 10.1016/j.yjmcc.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herovici WG. A polychrome stain for differentiating precollagen from collagen. Stain Technol. 1963;38:204–206. [PubMed] [Google Scholar]
- 28.Shoulders MD, Raines RT. Collagen structure and stability. Annu. Rev. Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fidler AL, Boudko SP, Rokas A, Hudson BG. The triple helix of collagens–an ancient protein structure that enabled animal multicellularity and tissue evolution. J. Cell Sci. 2018;131:jcs203950. doi: 10.1242/jcs.203950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farndale RW. Collagen-binding proteins: insights from the collagen toolkits. Essays Biochem. 2019;63:337–348. doi: 10.1042/EBC20180070. [DOI] [PubMed] [Google Scholar]
- 31.Sipila KH, et al. Proline hydroxylation in collagen supports integrin binding by two distinct mechanisms. J. Biol. Chem. 2018;293:7645–7658. doi: 10.1074/jbc.RA118.002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carolina G-C, et al. New altered non-fibrillar collagens in human dilated cardiomyopathy: role in the remodeling process. PLoS ONE. 2016;11:e0168130. doi: 10.1371/journal.pone.0168130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skonier J, et al. βig-h3: a transforming growth factor-β-responsive gene encoding a secreted protein that inhibits cell attachment in vitro and suppresses the growth of CHO cells in nude mice. DNA Cell Biol. 1994;13:571–584. doi: 10.1089/dna.1994.13.571. [DOI] [PubMed] [Google Scholar]
- 34.Roberts AB, McCune BK, Sporn MB. TGF-beta: regulation of extracllular matrix. Kidney Int. 1992;41:557–559. doi: 10.1038/ki.1992.81. [DOI] [PubMed] [Google Scholar]
- 35.Kuroda N, Masuya M, Tawara I, Tsuboi J, Yoneda M, Nishikawa K, Kageyama Y, Hachiya K, Ohishi K, Miwi H, Yamada R, Hamada Y, Tanaka K, Kato T, Takei Y, Katayama N. Infiltrating CCR2+ monocytes and their progenies, fibrocytes, contribute to colon fibrosis by inhibiting collagen degradation through the production of TIMP-1. Nat. Sci. Rep. 2019;9:1–17. doi: 10.1038/s41598-019-45012-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling M, Luster A. Novel approach to inhibiting chemokine function. EMBO Mol. Med. 2011;3:510–512. doi: 10.1002/emmm.201100161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Z, et al. The pseudoreceptor BMP and activin membrane-bound inhibitor positively modulates Wnt/β-catenin signaling. J. Biol. Chem. 2008;283:33053–33058. doi: 10.1074/jbc.M804039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onichtchouk D, et al. Silencing of TGF-β signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- 39.Yan X, et al. Human BAMBI cooperates with Smad7 to inhibit transforming growth factor-β signaling. J. Biol. Chem. 2009;284:30097–30104. doi: 10.1074/jbc.M109.049304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villar AV, et al. BAMBI (BMP and activin membrane-bound inhibitor) protects the murine heart from pressure-overload biomechanical stress by restraining TGF-β signaling. Biochim. Biophys. Acta Mol. Basis Dis. 1832;323–335:2013. doi: 10.1016/j.bbadis.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Krä Mer A, Green J, Pollard J, Tugendreich S. Systems biology causal analysis approaches in ingenuity pathway analysis. BMC Genom. 2014 doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wirrig EE, Yutzey KE. Conserved transcriptional regulatory mechanisms in aortic valve development and disease. Arterioscler. Thromb. Vasc. Biol. 2014;34:737–741. doi: 10.1161/ATVBAHA.113.302071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nordquist E, LaHaye S, Nagel C, Lincoln J. Postnatal and adult aortic heart valves have distinctive transcriptional profiles associated with valve tissue growth and maintenance respectively. Front. Cardiovasc. Med. 2018;5:30. doi: 10.3389/fcvm.2018.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lincoln J, Garg V. Etiology of Valvular Heart Disease. Circ. J. 2014;78:1801–1807. doi: 10.1253/circj.cj-14-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinton, R. B. in Calcific Aortic Valve Disease (ed Elena Aikawa) Chapter 7. 173–203. 10.5772/55506 (InTech, Europe, Rijeka, Croatia, 2013).
- 46.Lewin MB, Otto CM. The bicuspid aortic valve: adverse outcomes from infancy to old age. Circulation. 2005;111:832–834. doi: 10.1161/01.CIR.0000157137.59691.0B. [DOI] [PubMed] [Google Scholar]
- 47.Hinton RB, et al. Hypoplastic left heart syndrome links to chromosomes 10q and 6q and is genetically related to bicuspid aortic valve. J. Am. Coll. Cardiol. 2009;53:1065. doi: 10.1016/j.jacc.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin LJ, et al. Evidence in favor of linkage to human chromosomal regions 18q, 5q and 13q for bicuspid aortic valve and associated cardiovascular malformations. Human Gen. 2007;121:275–284. doi: 10.1007/s00439-006-0316-9. [DOI] [PubMed] [Google Scholar]
- 49.Hitz MP, Brand T, Andelfinger G. Genetic regulation of heart valve development: clinical implications. Aswan Heart Centre Sci. Pract. Ser. 2011 [Google Scholar]
- 50.Simard L, et al. Sex-related discordance between aortic valve calcification and hemodynamic severity of aortic stenosis. Circ. Res. 2017;120:681–691. doi: 10.1161/CIRCRESAHA.116.309306. [DOI] [PubMed] [Google Scholar]
- 51.Voisine M, et al. Age, sex, and valve phenotype differences in fibro-calcific remodeling of calcified aortic valve. J. Am. Heart. Assoc. 2020 doi: 10.1161/JAHA.119.015610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henaine R, Roubertie F, Vergnat M, Ninet J. Valve replacement in children: a challenge for a whole life. Arch. Cardiovasc. Dis. 2012;105:517–528. doi: 10.1016/j.acvd.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. TRENDS Gen. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Bella J. Collagen structure: new tricks from a very old dog. Biochem. J. 2016;473:1001–1025. doi: 10.1042/BJ20151169. [DOI] [PubMed] [Google Scholar]
- 55.Krane SM. The importance of proline residues in the structure, stability and susceptibility to proteolytic degradation of collagens. Amino Acids. 2008;35:703–710. doi: 10.1007/s00726-008-0073-2. [DOI] [PubMed] [Google Scholar]
- 56.Wedemeyer WJ, Welker E, Scheraga HA. Proline cis-trans isomerization and protein folding. Biochemistry. 2002;41:14637–14644. doi: 10.1021/bi020574b. [DOI] [PubMed] [Google Scholar]
- 57.Tang L, et al. Global metabolic profiling identifies a pivotal role of proline and hydroxyproline metabolism in supporting hypoxic response in hepatocellular carcinoma. Clin. Cancer Res. 2018;24:474–485. doi: 10.1158/1078-0432.CCR-17-1707. [DOI] [PubMed] [Google Scholar]
- 58.Gorres KL, Raines RT. Prolyl 4-hydroxylase. Crit. Rev. Biochem. Mol. Biol. 2010;45:106–124. doi: 10.3109/10409231003627991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Annunen P, Autio-Harmainen H, Kivirikko KI. The novel type II prolyl 4-hydroxylase is the main enzyme form in chondrocytes and capillary endothelial cells, whereas the type I enzyme predominates in most cells. J. Biol. Chem. 1998;273:5989–5992. doi: 10.1074/jbc.273.11.5989. [DOI] [PubMed] [Google Scholar]
- 60.Hamaia S, Farndale RW. Integrin recognition motifs in the human collagens. Adv. Exp. Med. Biol. 2014;819:127–142. doi: 10.1007/978-94-017-9153-3_9. [DOI] [PubMed] [Google Scholar]
- 61.Ilamaran M, et al. A self-assembly and higher order structure forming triple helical protein as a novel biomaterial for cell proliferation. Biomater. Sci. 2019;7:2191–2199. doi: 10.1039/c9bm00186g. [DOI] [PubMed] [Google Scholar]
- 62.Davidenko N, et al. Selecting the correct cellular model for assessing of the biological response of collagen-based biomaterials. Acta Biomater. 2018;65:88–101. doi: 10.1016/j.actbio.2017.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leitinger B. Molecular analysis of collagen binding by the human discoidin domain receptors, DDR1 and DDR2 identification of collagen binding sites in DDR2. J. Biol. Chem. 2003;278:16761–16769. doi: 10.1074/jbc.M301370200. [DOI] [PubMed] [Google Scholar]
- 64.Sipilä KH, et al. Proline hydroxylation in collagen supports integrin binding by two distinct mechanisms. J. Biol. Chem. 2018;293:7645–7658. doi: 10.1074/jbc.RA118.002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamaia S, Farndale RW. I Domain Integrins 127–142. Berlin: Springer; 2014. [Google Scholar]
- 66.Wasinski B, et al. Discoidin domain receptors, DDR1b and DDR2, promote tumour growth within collagen but ddr1b suppresses experimental lung metastasis in HT1080 xenografts. Sci. Rep. 2020;10:1–21. doi: 10.1038/s41598-020-59028-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeltz C, Gullberg D. The integrin-collagen connection: a glue for tissue repair? J. Cell Sci. 2016;129:653–664. doi: 10.1242/jcs.180992. [DOI] [PubMed] [Google Scholar]
- 68.Miura Y, Takahashi T, Jung SM, Moroi M. Analysis of the interaction of platelet collagen receptor glycoprotein VI (GPVI) with collagen: a dimeric form of GPVI, but not the monomeric form, shows affinity to fibrous collagen. J. Biol. Chem. 2002;277:46197–46204. doi: 10.1074/jbc.M204029200. [DOI] [PubMed] [Google Scholar]
- 69.O'Brien JR, Etherington MD, Brant J, Watkins J. Decreased platelet function in aortic valve stenosis: high shear platelet activation then inactivation. Heart. 1995;74:641–644. doi: 10.1136/hrt.74.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leeming DJ, Karsdal MA. Type V Collagen. Biochemistry of Collagen, Laminins and Elastin. Academic Press; 2016. [Google Scholar]
- 71.Montesi, S. B., Désogère, P., Fuchs, B. C. & Caravan, P. in Journal of Clinical Investigation vol. 129 24–33 (American Society for Clinical Investigation, 2019). [DOI] [PMC free article] [PubMed]
- 72.Riley H, Bradshaw A. The influence of the extracellualr matrix in inflammation: findings from the SPARC null mouse. Anat. Rec. 2019;303:1624–1629. doi: 10.1002/ar.24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Armstrong EJ, et al. Heart valve development: endothelial cell signaling and differentiation. Circ. Res. 2004;95:459–470. doi: 10.1161/01.RES.0000141146.95728.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rabkin-Aikawa, E., Farber, M., Aikawa, M. & Schoen, F. J. in Journal of Heart Valve Disease Vol. 13 841–847 (2004). [PubMed]
- 75.Razek T, Olthoff K, Reilly PM. Issues in potential organ donor management. Surg. Clin. North Am. 2000;80:1021–1032. doi: 10.1016/s0039-6109(05)70111-0. [DOI] [PubMed] [Google Scholar]
- 76.Siebelink, M. J., Albers, M. J. I. J., Roodbol, P. F. & Van De Wiel, H. B. M. in Transplant International Vol. 25 265–271 (John Wiley & Sons, Ltd, 2012). [DOI] [PubMed]
- 77.Deng XS, et al. Adult aortic valve interstitial cells have greater responses to toll-like receptor 4 stimulation. Ann. Thorac. Surg. 2015;99:62–71. doi: 10.1016/j.athoracsur.2014.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duan B, Hockaday LA, Das S, Xu C, Butcher JT. Comparison of mesenchymal stem cell source differentiation toward human pediatric aortic valve interstitial cells within 3D engineered matrices. Tissue Eng. Part C Methods. 2015;21:795–807. doi: 10.1089/ten.tec.2014.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williams K, et al. Standard 6: age groups for pediatric trials. Pediatrics. 2012;129:S153–S160. doi: 10.1542/peds.2012-0055I. [DOI] [PubMed] [Google Scholar]
- 80.Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- 81.Bredfeldt JS, et al. Computational segmentation of collagen fibers from second-harmonic generation images of breast cancer. J. Biomed. Opt. 2014;19:016007. doi: 10.1117/1.JBO.19.1.016007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sprung RW, et al. Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis. Mol. Cell. Proteom. 2009;8:1988–1998. doi: 10.1074/mcp.M800518-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 84.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 85.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods. 2016;13:731. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 86.Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gene Ontology C. The gene ontology resource: 20 years and still GOing strong. Nucl. Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucl. Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 90.Su YR, Linton MF, Fazio S. Rapid quantification of murine ABC mRNAs by real time reverse transcriptase-polymerase chain reaction. J. Lipid Res. 2002;43:2180–2187. doi: 10.1194/jlr.d200020-jlr200. [DOI] [PubMed] [Google Scholar]
- 91.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 92.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucl. Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any datasets generated during the study (not already in supplemental materials) are available on reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE92 partner repository with the dataset identifier PXD021990. During the peer review process, this data can be accessed via reviewer_pxd021990@ebi.ac.uk (Password: YA2RlOK6).