Abstract
The core gut microbiome of adult honeybee comprises a set of recurring bacterial phylotypes, accompanied by lineage-specific, variable, and less abundant environmental bacterial phylotypes. Several mutual interactions and functional services to the host, including the support provided for growth, hormonal signaling, and behavior, are attributed to the core and lineage-specific taxa. By contrast, the diversity and distribution of the minor environmental phylotypes and fungal members in the gut remain overlooked. In the present study, we hypothesized that the microbial components of forager honeybees (i.e., core bacteria, minor environmental phylotypes, and fungal members) are compartmentalized along the gut portions. The diversity and distribution of such three microbial components were investigated in the context of the physico-chemical conditions of different gut compartments. We observed that changes in the distribution and abundance of microbial components in the gut are consistently compartment-specific for all the three microbial components, indicating that the ecological and physiological interactions among the host and microbiome vary with changing physico-chemical and metabolic conditions of the gut.
Subject terms: Microbiome, Microbial ecology
Introduction
Recent studies on the role of bacterial symbionts of the honeybee gut have shed light on their importance for host metabolism, biomass gain, pathogen resistance, and behavioral health1–4. The gut bacterial microbiome of adult Apis mellifera has extensively been investigated, with a particular emphasis on the taxonomy3,5,6, evolution2, genomics, and physiology of its bacterial members1,4,7–9, as well as on its modification under different environmental conditions, diet, and abiotic and biotic stressors10–15. The core gut microbiota of the adult honeybee is composed of five consistent, ubiquitous, and abundant bacterial taxa16: the beta proteobacterium Snodgrassella alvi (hereafter indicated as Snodgrassella), the gamma proteobacterium Gilliamella apicola (hereafter indicated as Gilliamella), Lactobacillus Firm-4, Lactobacillus Firm-5, and the actinobacterium Bifidobacterium3,5,17–19. In addition, the proteobacteria Frischella perrara and Bartonella apis (hereafter indicated as Frischella and Bartonella, respectively), together with Bombella, Commensalibacter, Apibacter, and Lactobacillus Firm-3, are present, albeit in a lower proportion (~1%–7% of the total bacterial community) in comparison with the former five5,17,20–22. These aerobic and facultative anaerobic phylotypes—rarely found outside the bee gut and hive environment23,24 and constituting the dominant members (95%–99% of the total bacterial community) of the honeybee gut microbiota—form spatially explicit patterns along the different gut compartments3,5,25.
Under natural conditions, bacteria form complex communities, and their evolution, functionality, and ecology are dictated by environmental factors and microbial interactions with host and among microbes18. Despite a simple structure and limited number of bacterial species, honeybee gut microbiota exhibit high genomic diversity18,26, which may act as a reservoir of metabolic functions that modulate host physiology and nutrition. This metabolic vicariance is encoded by microbial functional redundancy (i.e., each metabolic function can be performed by multiple coexisting and taxonomically distinct organisms) with respect to a multitude of functions, which could induce gut stability and allow bees to adapt to changing environmental conditions (i.e., disturbance)27,28. Although such genomic diversity and its localization along the gut are well known for the core microbiome18, they are not well characterized for the other microbiome components. For instance, bacteria of environmental origin (e.g., floral nectar and pollen29) found in small proportions (1%–5%) of the total bacterial community and having less prevalence (1%–20% of bacterial individuals) are often found in the honeybee gut5,17,30,31; however, their compartmentalization along the gut is unknown. Similarly, the compartmentalization of non-pathogenic fungal members occurring in the adult honeybee gut have been rarely investigated. Although several studies have demonstrated by cultivation-dependent and cultivation-independent approaches the presence of yeasts (subphylum Saccharomycotina) or other fungal phylotypes (members of Wickerhamomyces, Pleosporales, and Agaricales, among others)32–38, only one study analyzed their distribution along the gut38.
In the present study, we hypothesized that the honeybee gut compartments (i.e., the crop, midgut, ileum, and rectum) play an important role in mediating the distribution of non-core microbial members (i.e., other potential environmental bacteria and fungi), as they do for core microbial members, in the context of their spatially explicit patterns and different physico-chemical conditions (e.g., O2, pH, and redox) of gut. We characterized the microbiota (bacteria and fungi) diversity in the four gut compartments (the crop, midgut, ileum, and rectum) along with changes in gut metabolites, oxygen concentration, pH, and redox potential using a combination of molecular, biochemical, and microsensor approaches. Various analytical methods, including some that were previously used39, guided a focused investigation of honeybees collected from their environment (i.e., hives located in flowered prairies) to explore the natural microbial variability of the gut microbiome. This approach will allow us to confirm the presence and compartmentalization of the core bacteria that are consistently associated with the forager honeybee gut and reveal the way in which this spatially explicit pattern is conserved for bacteria and fungi acquired from the environment and not belonging to the core.
Results and discussion
Compartmentalization of the bacterial community present in the honeybee gut
Recent studies have investigated the diversity of the honeybee gut5,40 or body41 by focusing on the compartmentalization of the main bacterial phylotypes along the intestine38,42–44 and overlooking the remaining microbiome components (i.e., fungal and other-possibly environmental bacterial members). In our honeybees, magnified images of the gut indicated the presence of bacterial cells of different morphologies (primarily rod- and coccus-shaped bacteria), colonizing the gut wall and ingested pollen grain surface (Fig. 1a–d and Supplementary Fig. S1). Bacterial quantification (based on a normalized number of 16S rRNA gene copies) showed a significant compartmentalization of their load along the gut (Fig. 2a, b and Supplementary Table S1), with the highest densities observed in the distal compartments (ileum = 1.43 × 105 and rectum = 4.23 × 105 bacterial cells) than in the proximal compartments (crop = 1.83 × 104 and midgut = 1.87 × 104), thereby confirming the data reported in previous studies21,42. Furthermore, the gut compartments harbored significantly different bacterial communities (F3,16 = 6.6, p = 0.001; Fig. 2c and Supplementary Table S2) that were characterized by unique bacterial taxa assemblages (Fig. 2d and Supplementary Data 1) with similar alpha diversity (richness and Shannon diversity; F3,16 = 3.2, p = 0.06, and F3,16 = 0.55, p = 0.65, respectively; Supplementary Fig. S2a, b), resulting in a decrease in similarity with the increase in the gut distance (n = 190, p < 0.0001, R2 = 0.14; Supplementary Fig. S3a). The bacterial phylotypes belonging to the corbiculate core (i.e., Snodgrassella, Gilliamella, Lactobacillus Firm-4, Lactobacillus Firm-5, and Bifidobacterium) dominated the gut bacterial community (Fig. 2b, d), as confirmed via amplification of the total-active bacterial community from cDNA (Fig. 2e). The members of the corbiculate core were followed by “Apis-specific” Frischella and Bartonella phylotypes, along with the other corbiculate-associated Bombella, Commensalibacter, and Apibacter (Fig. 2d, e). Collectively, these taxonomic groups—represented by 32 operational taxonomic units (OTUs; Supplementary Fig. S4, Supplementary Data 1, and Supplementary Table S3)—accounted for 92%–98% of the total bacterial community (94%, 93%, 92%, and 98% in the crop, midgut, ileum, and rectum, respectively; Table 1). Consistent with the previous observations42–44, a compartment-specific distribution of three of the most abundant core phylotypes was observed (F3,16 = 10.1, p = 0.001); for instance, Snodgrassella, Gilliamella, and Frischella preferentially colonized the midgut and ileum, whereas Lactobacillus Firm-4 and Firm-5 mainly colonized the crop and rectum (Fig. 2b, d). Although the abundance, dominance, and ratios of core phylotypes were different8,17,23, they were consistently detected along the gut (Fig. 2b, d and Supplementary Table S1). Consequently, the differential richness of the bacterial component was the main determinant of the total beta-diversity pattern of the core gut microbiome of honeybees (Fig. 3a).
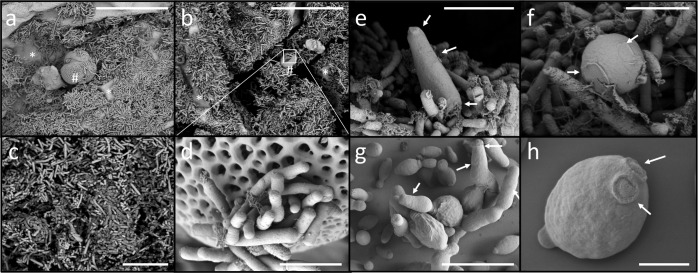
Fig. 1. Visualization of microorganisms inhabiting honeybee guts.
a–d Scanning electron microscope (SEM) micrographs showing bacterial cells present in a honeybee gut (i.e., rod-shape bacteria). Grains of pollen covered by bacteria are shown (see also Supplementary Fig. S1). e, f Fungal cells (i.e., yeast) associated with the gut wall of a honeybee. g, h Culture of yeast cells isolated from the honeybee gut. Cells belong to Hanseniaspora uvarum (strain L18) and Starmerella bombicola (strain L28; see also Supplementary Fig. S9 and Supplementary Table S7). Note the different scales on the SEM photographs. Honeybees used for SEM analysis were A. mellifera jemenitica from Saudi Arabia (Supplementary Table S6). Scale bars correspond to (a, b) 30 µm, (c) 3 µm, (d, f) 2 µm, (e, g) 10 µm, and (h) 1 µm.
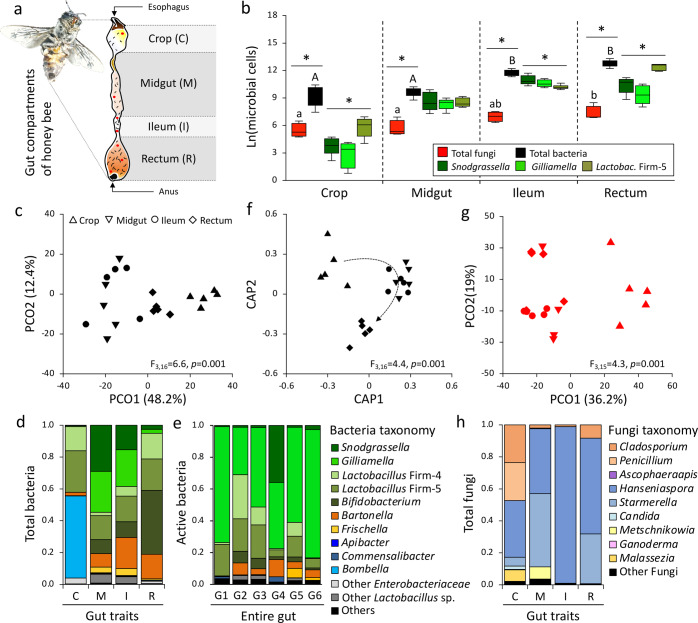
Fig. 2. Bacterial and fungal microbiota of the gut compartments in honeybees.
a Schematic representation of honeybee gut compartments (the crop, midgut, ileum, and rectum). Red circles and black rods indicate the relative distribution of total fungi and bacteria, respectively. b The abundance of the bacterial (black bars) and fungal (red bars) members in the honeybee gut compartments are indicated as ln-transformed number of bacterial cells (number of 16S rRNA gene copies measured are normalized for the number of 16S rRNA genes in a bacterial community, n = 4.7) and fungal cells (number of measured internal transcribed spacer [ITS] copies are normalized for the number of ITSs in a fungal community, n = 75.5). Significant differences among the bacterial and fungal cell abundance along gut tracts are indicated with capital and lower-case letters, respectively (Tukey’s multiple comparison test, p < 0.05), whereas the comparison among bacteria and fungi in each trait is indicated by an asterisk (*; t-test, p < 0.05). Abundance (ln-transformed) of Snodgrassella, Lactobacillus Firm-5, and Gilliamella in the different gut compartments are represented by shades of green and are expressed as the number of bacterial cells (number of 16S rRNA gene copies measured are normalized for the number of 16S rRNA gene in these phylotypes, n = 4). Significant differences among the above-mentioned bacterial species for each gut tract are indicated by an asterisk (*; ANOVA, p < 0.05). All the results are expressed per organ. Principal coordinates analysis (PCoA) showing the (c) bacterial and (g) fungal communities per each gut compartment, respectively. f Beta-diversity analysis of the other potential environmental bacteria portion. Samples were distributed following a “horseshoe shape” ordination (indicated by the arrow) in the space of the canonical analysis of principal coordinates (CAP). d, e Taxonomic affiliation of total (DNA) and active (cDNA) bacterial communities inhabiting the honeybee gut, respectively, along with (h) one for fungal communities (DNA). Abundance of bacterial taxa obtained from DNA are expressed as the number of reads normalized for the mean number of bacterial 16S rRNA genes and fungal ITSs available for each genus/class, respectively (for details, see “Material and methods”). b–d, f–h are referred to the gut compartments’ pools (each pool, n = 10) originating from Italian A. mellifera ligustica forager bees, whereas e is referred to the entire gut of Saudi Arabian A. mellifera jemenitica (n = 6). Compartmentalization of bacterial and fungal communities associated with Saudi Arabian forager bees is reported in Supplementary Fig. S5.
Table 1.
Occurrence of bacterial taxa along the gut compartments of A. mellifera ligustica forager bees from Italy.
| Bacterial categories | Bacterial taxa | N. OTUs in gut compartment (n = 5) | N. OTUs | Abund. (%) | |||
|---|---|---|---|---|---|---|---|
| Crop | Midgut | Ileum | Rectum | ||||
| Corbiculate core | Bifidobacterium | 1 | 2 | 2 | 3 | 3 | 15 |
| Gilliamella | 4 | 4 | 4 | 4 | 4 | 13.2 | |
| Firm-4 | 3 | 3 | 3 | 3 | 3 | 10.6 | |
| Firm-5 | 9 | 8 | 9 | 9 | 9 | 21.1 | |
| Snodgrassella | 3 | 3 | 3 | 2 | 4 | 12.7 | |
| Apis-specific | Bartonella | 2 | 2 | 2 | 3 | 3 | 10.4 |
| Frischella | 1 | 1 | 2 | 1 | 2 | 2.2 | |
| Other corbiculate associates | Commensalibacter | 2 | 2 | 2 | 2 | 2 | 0.2 |
| Apibacter | 1 | 1 | 1 | 1 | 1 | 0.2 | |
| Bombella | 1 | 1 | 1 | 1 | 1 | 8.8 | |
| Other potential environmental | Other Enterobacteriaceae | 9 | 16 | 15 | 13 | 16 | 3.4 |
| Other Lactobacillaceae | 20 | 11 | 9 | 10 | 21 | 1.9 | |
| Other Bacteria | 80 | 72 | 68 | 51 | 127 | 0.8 | |
Operational taxonomic units (OTUs) for each phylotype belong to the corbiculate core, Apis-specific, other corbiculate-associated, and other-possibly environmental categories along the four compartments. The total number of OTUs per bacterial taxa and their normalized-relative abundance (abund. %) in the total dataset are reported.
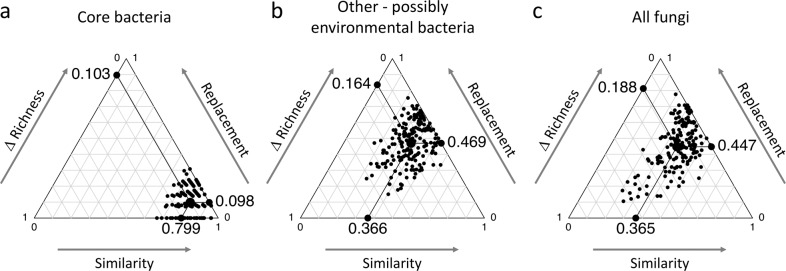
Fig. 3. Components of the microbial beta-diversity pattern.
Ternary plots are used to quantify the component of the assembly along the gut compartment for (a) core bacteria, (b) other-possibly environmental bacteria, and (c) fungi; note the dataset presented in Fig. 2c, f, g, respectively, were used to perform this analysis. Each point in the triangle plot was determined by a triplet of values from the similarity, replacement, and richness difference components. In each ternary, large central dots from which the lines start represent the centroid of the points; the lines represent the mean values of the three components (i.e., similarity, replacement, and richness difference).
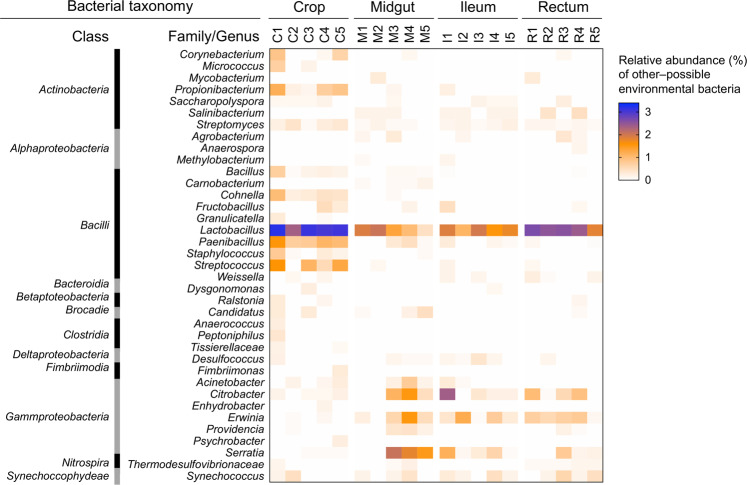
These dominant bacterial phylotypes co-existed alongside a high number (164 OTUs) of less-frequent bacterial OTUs (classified as other-possibly environmental bacteria, according to Kwong et al.17; see Supplementary Data 1). They were unevenly distributed along the gut compartments and were chiefly affiliated with Gammaproteobacteria (Erwinia, Serratia, Citrobacter, and Acinetobacter), Bacilli (Streptococcus, Paenibacillus, and Lactobacillus not belonging to Firm-4 or Firm-5), and Actinobacteria (Propionibacterium and Streptomyces; Table 1 and Fig. 4). These 164 other-possibly environmental bacterial OTUs were not amplified in the extraction blank and non‐template amplification controls (Supplementary Table S4), excluding their origin from laboratory work contaminations45. Although the acquisition of environmental bacteria may have been resulted from stochastic processes (e.g., nectar and pollen bacterial diversity46,47), their assembly was significantly affected by gut compartmentalization in terms of composition (i.e., niche partitioning; F3,16 = 4.4, p = 0.001; Figs. 2f and 4; n = 190, p < 0.0001, R2 = 0.14; Supplementary Fig. S3b) and richness (F3,16 = 4.7, p = 0.02; Supplementary Fig. S2c). Samples were distributed in a horseshoe shape ordination (arrow in Fig. 2f) in the space defined by the canonical analysis of principal coordinates (CAP), which indicated the presence of an ecological gradient. Moreover, this partitioning was confirmed by a higher contribution of OTUs turnover (i.e., species replacement48) to the beta-diversity pattern along the gut (Fig. 3b); specifically, environmental bacterial species tend to replace each other along the gut (Fig. 4), which is likely owing to the changing physico-chemical conditions of the gut (see Fig. 5 and its discussion) and biological correlations among members of the microbiome. For instance, the environmental bacterial species richness in the crop—the first compartment receiving environmental food source—was higher than that of the other gut portions (Supplementary Fig. S2c). In this compartment, environmental bacterial OTUs were unequally distributed across taxonomic groups (Supplementary Fig. S2d) and were dominated by Lactobacillus, Paenibacillus, Bacillus, Streptococcus, and Propionibacterium, the proportion of which was noted to be limited in other compartments (Fig. 4).
Fig. 4. Distribution of other-possibly environmental bacteria along the honeybee gut compartments.
Heat map represent the distribution of other-possibly environmental bacteria classes and families/genera detected from the analysis of the total bacterial communities (i.e., DNA; Fig. 2f) along the four gut compartments (the crop, midgut, ileum, and rectum); values are expressed as the relative abundance (%) of the normalized reads.
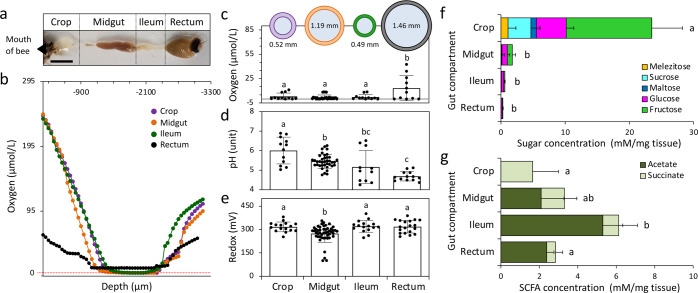
Fig. 5. Physico-chemical and metabolite characterization of the honeybee gut compartments.
a Honeybee gut compartments (the crop, midgut, ileum, and rectum) are depicted (bar = 0.5 cm). b–e Physico-chemical parameters measured by microsensors are reported for each gut compartment. b Representative radial profile of the oxygen concentration (μmol/l) along the gut compartments (i.e., the crop, midgut, ileum, and rectum) of honeybees. Depth refers to the distance covered by the electrode tip starting from the agarose surface (details in “Materials and methods”). c Schematic representation of the diameters of the gut compartments (circles at the top of the panel); diameters were expressed in mm, and their mean ± standard deviation (n = 12) are indicated by the internal and external circles, respectively. Details on the organ diameters are reported in Supplementary Table S12. The oxygen concentration measured in the central regions of the gut compartments (n = 11) are expressed as μmol/l. Microelectrode profiles of the (d) pH and (e) redox potential along the honeybee gut compartments (n = 12 and n = 16, respectively); values are expressed as unit and mV, respectively. Concentration (mM/mg of tissue) of (f) sugars and (g) short-chain fatty acids (SCFAs) in the gut compartments (n = 3); values are expressed as mean ± standard deviation. Letters show the results of Tukey’s multiple comparison test. Italian A. mellifera ligustica forager bees were considered for physico-chemical and metabolite analyses.
Our data regarding Italian A. mellifera ligustica (Fig. 2c, d, f) were confirmed by an analysis of A. mellifera jemenitica individuals collected in Saudi Arabia (Supplementary Fig. S5a, b and Supplementary Data 1). In this subspecies, we observed a significant compartmentalization of core and other-possibly environmental bacteria along the gut compartments (F3,16 = 2.69, p = 0.001 and F3,16 = 2.1, p = 0.009, respectively). Although the core bacteria followed the recognized trend of distribution and abundance (i.e., dominance of Snodgrassella and Gilliamella in the midgut and ileum and Lactobacillus Firm-4 and Firm-5 in the crop and rectum), other-possibly environmental bacteria showed a relative abundance ranging from 14%–17% in the initial portion of the gut to 4%–8% in the distal portion with a dominance of Enterobacteriaceae (Serratia, Klebsiella, and Shigella), Morganellaceae (Morganella, Providencia, and Proteus), Orbaceae, and Rhizobiaceae (Supplementary Data 1). Other-possibly environmental bacteria were also detected in Saudi Arabian nurse honeybees (Supplementary Table S5), following the same compartmentalization pattern observed for foragers (F3,15 = 3.54, p = 0.001; Supplementary Fig. S5e). Notably, the environmental bacteria detected in Saudi Arabian honeybees exhibited a markedly different taxonomic composition respect to the one in Italian honeybees, emphasizing the environmental origin of these members of the microbiome.
Because the culture-independent methods used in the present study cannot discriminate between non-viable dead bacterial cells (i.e., DNA from lysed bacterial cells) and live/active bacterial cells, we analyzed the cDNA synthetized from total RNA of active members of the bacterial community. Amplification of the 16S rRNA gene from cDNA confirmed that other-possibly environmental bacteria were present (relative abundance of 2%–6%) and metabolically active within the gut microbiome (Fig. 2e, Supplementary Fig. S6 and Supplementary Data 1). The abundance of these bacteria along the gut ranged from 2% to 8% (DNA), of which 2%–6% (cDNA) was metabolically active. The observed difference may occur owing to a part of such bacteria being filtered/selected by the gut environment as well as biotic interactions within the honeybee gut (i.e., anoxia and subacid pH), whereas the remaining bacteria are unable to survive and were thus detected as environmental DNA (∼2%). Their possible functional role was inferred from taxonomy by using the Tax4Fun2 platform (Supplementary Data 2). These bacteria were involved in (1) secondary metabolite biosynthesis, such as antibiotic synthesis, which can defend the insect host against parasites49,50 and/or regulate the microbiome balance via competitive interactions among members of the gut microbiome51,52; (2) xenobiotic biodegradation, which may aid the honeybee with detoxification of external substances from the diet53; and (3) glycan biosynthesis and degradation, which reveals their association with the gut and interaction with the host54,55. Furthermore, we observed that other-possibly environmental bacteria contribute to the metabolism of carbohydrates, amino acids, and lipids as well as the biosynthesis of cofactors and vitamins (Supplementary Data 2), thereby suggesting their ancillary contribution beyond that exerted by the core bacteria in degrading host-ingested substrates and distributing essential nutrients3,56.
An association between gut and environmental bacteria is evident in insects57, including honeybees5,17,29,43, but the magnitude and contribution to host function, performance, and/or health remains unelucidated. In the present study, adult forager honeybees were collected from the front of the hive following exposure to the natural environment (i.e., flowered prairie), thereby enabling the recruitment of main core bacteria from the hive and contact among mates and environmental bacteria during harvest3,29,44. Such recruitment processes are affected by the surrounding environment and pollination landscape (e.g., microbiome variation in nectar, pollen, and hive materials46,58), resulting in strong variation among the frequencies and biodiversity of bacteria associated with honeybees5,21,29,43. Although we anticipated that the type of environmental bacteria (non-gut bacteria, such as Actinomycetales, Alphaproteobacteria, Enterobacteriaceae, Pseudomonadales, Firmicutes, and Xanthomonadaceae43,58) would vary depending on environmental conditions (such as location, season, food source, and climate), their recruitment and distribution was determined by the interaction between the physico-chemical conditions and biological network of the honeybee gut. For instance, Kešnerová et al.5 found that minor community members (i.e., other-possibly environmental bacteria) detected in the short-lived forager and nurse honeybees (∼4 weeks) were present in lower concentrations in long-lived winter bees (~6 months) and can be transient colonizers that are mainly acquired from the surrounding environment during foraging. Winter bees feed strictly on food stored in the hive (i.e., aged pollen) and retain their feces all winter, affecting the physico-chemical conditions and the availability of nutrients in the gut, and consequently impacting the ecology of their gut microbiota5; despite it was quantified that winter bees had the highest bacterial loads, they were characterized by a lowest bacterial community alpha diversity, with dominance of Bartonella and Commensalibacter, reduction of opportunistic colonizers (Apibacter, Bombella, or L. kunkeii) and environmental bacteria (such as, Serratia and Klebsiella) possibly by the specific dietary conditions of these bees and the activation of host resistance mechanisms in place to favor their and overall colony survival5. Nevertheless, several studies have shown that non-core environmental bacteria can provide benefits: bacterial strains isolated from honeybee gut belonging to Lactobacillus kunkeei and Bacillus sp. were found to protect the host from pathogens such as Paenibacillus larvae and Melissococcus plutonius12,59, whereas Staphylococcaceae and Enterobacteriaceae members were able to metabolize plant derivates and sugars that are toxic to the host4. In natural environmental systems, members of the rare prokaryotic biosphere may represent a substantial source of genomic and functional features60. Similar to the importance of low-abundant bacteria in re-shaping ecosystems, they can enhance host immunity or prevent pathogen colonization in plant and animal associations60,61. Although generalist broad metabolic processes, such as respiration, metabolic potential, and cell yield are performed by dominant phylotypes, more specialized functions, such as the degradation of specific compounds, can be performed by low-abundant, niche-specialist phylotypes62. Currently, technical limitations preclude the ability to investigate the functioning of the in vivo ecosystem functioning of the rare biosphere in the gut interactome. For instance, absolute germ-free adult honeybee individuals do not exist and thus cannot be studied. However, published studies have reported a significant reduction in the total amount of bacterial cells in honeybee guts (i.e., < 105 bacterial cells per gut consisting of an erratic mix of bacterial species1,39) but never complete elimination, rendering this approach unsuitable for defining whether rare/environmental microbial components contribute to the maintenance of microbiota homeostasis in the honeybee gut.
Future investigations into other-possibly environmental bacteria, often masked by dominant bacterial phylotypes, will further elucidate microbial community recruitment, assembly, and functioning in the honeybee gut.
Compartmentalization of the mycobiota along the honeybee gut
The honeybee gut hosts microbial cells that are larger than bacterial cells, such as those of yeasts and other fungi (Fig. 1e, f and Supplementary Fig. S7). The number of fungi was consistently lower than that of bacteria; however, their abundance followed the same trend, with a progressive increment observed from the crop to the rectum (4.9 × 102, 5.8 × 102, 1.7 × 103, and 2.9 × 103 fungal cells in the crop, midgut, ileum, and rectum, respectively; data normalized according to Lofgren et al.63; Fig. 2b and Supplementary Table S1). Cultivation-dependent approaches were applied to three different honeybee species (i.e., A. mellifera ligustica, A. mellifera jemenitica, and A. florea) from beehives in five locations in two different countries (Italy and Saudi Arabia; Supplementary Table S6), which confirmed the presence and viability of the fungal cells (Table 2). The mean number of fungal colony-forming units (CFUs) isolated from the anterior (the crop and midgut) and posterior (the ileum and rectum) parts ranged from 2.7 × 102 to 1 × 104—consistent with quantitative polymerase chain reaction (qPCR) findings (note the single compartments reported in Fig. 2b). Among these isolates (90 in total) the most abundant genera retrieved were Starmerella (31%), Hanseniaspora (12%), Aspergillus, and Naganishia (both 11%); the genera Aureobasidium, Moniliella, Candida, and Penicillium were less abundant (Supplementary Table S7). Starmerella yeasts isolated from Italian honeybees (A. mellifera ligustica) clustered with the strains S. bombicola, whereas in the Saudi Arabian honeybees (A. mellifera jemenitica and A. florea), these isolates mainly clustered with Starmerella meliponinorum (Supplementary Fig. S8). All isolates of Hanseniaspora strains were identified as close relatives of H. uvarum (Supplementary Fig. S8). Using the size, shape, and morphology of Starmerella bombicola and Hanseniaspora uvarum isolates as a reference (scanning electron microscopy (SEM) micrographs, Fig. 1g, h and Supplementary Fig. S9, similar round- and club-shaped yeast cells were identified in the honeybee gut (Fig. 1e, f and Supplementary Fig. S7a–c). Moreover, other yeast-like and fungal-like morphologies were detected (Supplementary Fig. S7d–f).
Table 2.
Abundance of culturable fungi associated with two gut portions: anterior (i.e., the crop and midgut) and posterior (i.e., the ileum and rectum).
| Honeybee species | Location | Gut region | PDA (CFU per anterior organs) | YM (CFU per posterior organs) | ||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| A. mellifera ligustica | Milan | Anterior | 1.01 × 104 | 5.34 × 103 | 3.37 × 103 | 1.78 × 103 |
| Posterior | 9.50 × 103 | 5.25 × 103 | 3.17 × 103 | 1.75 × 103 | ||
| A. mellifera jemenitica | Jeddah | Anterior | 8.00 × 102 | 6.93 × 102 | 8.00 × 102 | 4.00 × 102 |
| Posterior | 1.47 × 103 | 1.22 × 103 | 1.07 × 103 | 8.33 × 102 | ||
| Makkah | Anterior | 2.67 × 102 | 1.15 × 102 | 3.00 × 102 | 1.15 × 102 | |
| Posterior | 1.40 × 103 | 5.77 × 102 | 3.27 × 103 | 2.94 × 103 | ||
| Madinah | Anterior | 4.76 × 103 | 4.05 × 103 | 7.64 × 103 | 7.88 × 103 | |
| Posterior | 8.67 × 102 | 1.15 × 103 | 2.36 × 103 | 1.91 × 103 | ||
| Apis florea | Thuwal | Anterior | 1.20 × 103 | 9.38 × 102 | 7.50 × 102 | 8.39 × 102 |
| Posterior | 2.95 × 103 | 2.88 × 103 | 3.53 × 103 | 5.77 × 103 | ||
| Total mean | Anterior | 3.43 × 103 | 4.13 × 103 | 2.57 × 103 | 3.08 × 103 | |
| Posterior | 3.24 × 103 | 3.59 × 103 | 2.68 × 103 | 1.03 × 103 | ||
Values are expressed as mean ± standard deviation (SD) of colony-forming unit (CFU) per gram of gut portion. Mean of all anterior and posterior compartments is also reported. The origin of individuals is specified in the table.
PDA potato dextrose agar, YM yeast medium.
An analysis of the fungal community yielded 118 OTUs (Supplementary Data 1). The crop hosted a fungal community that was significantly different from the following compartments of the gut (Fig. 2g and Supplementary Table S2), with a higher Shannon diversity but similar richness (Supplementary Fig. S2e, f). Similarity among communities significantly decreased from the crop to the rectum (n = 163, p < 0.0001, R2 = 0.34; Supplementary Fig. S3c), suggesting a niche partitioning and compartment specificity. Moreover, the beta-diversity pattern of the fungal community, similar to that of other-possibly environmental bacteria along the gut, was mainly explained by variations in species composition48 (species turnover; Fig. 3c), suggesting the presence of a well-established gradient along the gut. The genera Hanseniaspora (35%), Penicillium (24%), Cladosporium (9%), Malassezia (7%), Starmerella (5%), and Candida (2%) composed the crop mycobiota (Fig. 2h; data normalized according to Lofgren et al.63). Among these, Cladosporium, Penicillium, Candida, and Malassezia significantly reduced in abundance along the intestinal trait, thereby providing space for the proliferation of Saccharomycetales yeasts (Hanseniaspora and Starmerella), which subsequently become dominant (86%, 98%, and 91% relative abundance in the midgut, ileum, and rectum, respectively). The other detected fungi (Cladosporium, Penicillium, Malassezia, and Candida, among others; Fig. 2h, Supplementary Fig. S8, Supplementary Data 1, and Supplementary Table S7) were ubiquitous species found in varied habitats (including soil) as well as common plant pathogens and endophytes64 and animal associates65. Therefore, we consider fungi as microbiome members of environmental origin that were acquired by honeybees during food intake, particularly during foraging34,38. This was further confirmed by the fact that A. mellifera jemenitica forager honeybees from Saudi Arabia (Madinah) were colonized by complex communities dominated by Zygosaccharomyces—yeasts belonging to the Saccharomycetaceae family, which are often detected in hive material and bee guts34,38,66—along with members of Alternaria, Aspergillus, Eremothecium, and Fusarium (Supplementary Data 1). Notably, mycobiota members exhibited differential distribution across the gut compartments (F3,16 = 2.11, p = 0.01; Supplementary Fig. S5), confirming the trend that was previously observed in Italian honeybees (Fig. 2g). The presence of fungi, as gut-associated microorganisms, was also detected in nurse honeybee gut (Saudi Arabia) with a spatial-differentiation across the gut compartments (F3,16 = 3.2, p = 0.002). The mycobiota of nurse and forager honeybees from Saudi Arabia was dominated by the same main members belonging to Zygosaccharomyces (Supplementary Table S5 and Supplementary Data 1); it was not present in the Italian honeybees, presumably owing to the different surrounding environment (e.g., vegetation) of the sampling sites (Italy and Saudi Arabia). However, despite the elevated diversity of the mycobiota along the entire gut34,38, the dominant members of the mycobiota in the honeybees analyzed were consistently fermentative yeasts (Hanseniaspora and Starmerella in Italy, and Zygosaccharomyces in Saudi Arabia); based on their taxonomy and metabolic information available in literature, they can have a role in food digestion (i.e., sugar fermentation and nutrient recycling) and favor trophic and spatial interplay among gut microorganisms67–69 (Supplementary Data 2).
Fungal members have previously been detected in the honeybee gut, with their origin being mainly ascribed to food intake32–34,37,38,70. Yeasts are common gut inhabitants of insects, such as Drosophila fruit flies, wasps, and mosquitoes12,71–74, which may be owing to their ability to thrive in sugar-rich environments75. In previous metagenomics surveys of honeybee-associated microbes, sequences related to Saccharomycetales were retrieved among fungal components33–35,76,77. Indeed, species of the Starmerella and Hanseniaspora genera, along with those of the Saccharomyces, Candida, and Zygosaccharomyces genera, have been isolated from flowers, fruits, and flower-visiting insects72,78, including bees34,38,79. Therefore, we hypothesized that such yeasts are acquired by honeybees via their diet, from the surrounding environment, or mates, which is similar to what was observed for low-abundant bacteria17. Moreover, the dominance of Saccharomycetales yeasts in our foragers—i.e., Hanseniaspora and Starmerella in Italian bees and Zygosaccharomyces in Saudi Arabian bees (Supplementary Data 1)—was justified by the favorable niche that the honeybee gut provides for such sugar-fermentative microorganisms34,66,68. The frequent association between fungi (i.e., yeast-like) and the insect gut suggests that they are prominent in host homeostasis, physiology68, and development80, which possibly provides essential nutritional support, including enzymes, essential amino acids, vitamins, and sterols68. Conversely, yeasts benefit from the protected environment of the gut and a dispersion service provided by the insect host74,81. For example, S. cerevisiae spores from tetrads increase the outbreeding of the yeast82. Furthermore, yeasts such as Starmerella bacillaris are unable to grow in vitamin-free media, which is suggestive of their dependence on other gut inhabitants83.
In the gut, the fungal cells were consistently surrounded by bacteria (e.g., Starmerella and Hanseniaspora in Fig. 1e, f); this physical contact can establish interactions among gut-associated microorganisms and favor a trophic and spatial interplay among them (intra group) as well as with the core and non-core bacterial components. For example, in an artificial microbial community, the nitrogen overflow of S. cerevisiae reportedly enabled the survival of two Lactobacillus species84. This likely occurs in the honeybee gut microenvironment, where yeasts and Lactobacillus strains are both present in the same compartments. Moreover, considering the cell size (e.g., in Fig. 1e–h), fungal cells may model the spatial interactions and metabolic flow within the gut microbial community by designing the architecture of the gut and influencing the geometry of interactions among bacterial components—bacteria might exploit the surfaces made available by fungal cells to establish ecological interactions among them67,85.
Physico-chemical conditions along the honeybee gut compartments
Complex physico-chemical conditions characterize the insect gut ecosystem86. Compared with a previous study that focused on conventional honeybees with a reconstituted gut microbiota under laboratory conditions (sterile diet accompanied by hindgut homogenate of nurses)39, in the present study, we ascertained these conditions along the intestinal tract of forager honeybees (Fig. 5a and Supplementary Table S9), which were collected in their natural environment. Suboxic zones with positive redox potentials and subacid pH were confirmed along the entire honeybee digestive tract (Fig. 5 and Supplementary Fig. S10). The partial oxygen pressure (pO2) measured in the center of the gut lumen reached values that were close to zero (< 5 μmol/l) in the crop, midgut, and ileum, whereas they flattened to a significantly higher concentration in the rectum (range, 0–37.5 μmol/l; 0%–13% air saturation, and 0%–2.8% oxygen; F3,61 = 7.769, p = 0.0002; Fig. 5b, c; Supplementary Table S8). The micro-profiles revealed a significant decrement of subacid pH values along the digestive tract from the proximal to distal gut compartments (the crop: 6.1 ± 0.2, midgut: 5.6 ± 0.1, ileum: 5.3 ± 0.9, rectum: 4.8 ± 0.2; F3,70 = 15.4, p < 0.0001; Fig. 5d and Supplementary Fig. S11). This progressive reduction was significantly correlated with the increment of the microbial load detected by qPCR (Fig. 2b; Pearson’s product-moment correlation; t = −2.36, df = 18, p = 0.03; ρ = −0.49; Supplementary Fig. S11). Finally, although the gut was anoxic/strongly hypoxic, positive values of the redox potential were consistently detected along the gut (range, 215–370 mV; crop: 292 ± 7, midgut: 266 ± 4, ileum: 296 ± 9, rectum: 293 ± 8), with the midgut showing the lowest significant values (Fig. 5e and Supplementary Fig. S10). Based on these results, we confirmed that the four physical portions of the gut differed in size and shape (representation in Fig. 2a) as well as in their physico-chemical milieu (PERMANOVA: F3,61 = 10.756, p = 0.001; pairwise comparison in Supplementary Table S9a). Among these variables, the pH significantly explained core and other-possibly environmental bacterial, but not fungal, variations (Supplementary Table S10). Among the core bacteria, pH values controlled the abundance of Bartonella (OTU_3) and Bifidobacterium (OTU_2; Supplementary Fig. S12) with a higher relative abundance under the lower pH conditions that were created by the fermentative metabolisms of the same bacteria56,87,88. Conversely, the other-possibly environmental bacteria Paenibacillus (OTU_20), Propionibacterium (OTU_28), Cohnella (OTU_29), and SAR406 (OTU_21) were positively correlated with higher pH values within the crop (Supplementary Fig. S13), which is consistent with their pH optima near neutral conditions89–92.
The co-presence of anoxic conditions with positive redox potentials was observed in the intestinal tract of other arthropods and in honeybees39,86,93, and was caused by the production of oxidized products of the microbial metabolic activities during fermentation8. The creation of an anoxic/strongly hypoxic condition was mainly ascribed to the oxygen scavenger activity of gut wall-associated microorganisms, such as the obligate aerobe S. alvi94 (mainly detected in the ileum39) and acetic bacteria (Commensalibacter and Bombella; mainly found in the crop and midgut). Notably, Snodgrassella, a major oxygen scavenger, could be located in other gut compartments42,95; in our honeybees, it was consistently detected in the midgut, ileum, and rectum (Fig. 2b). Contrary to the anoxic condition detected by Zheng et al.39, a variable percentage of oxygen was observed in the rectum of the forager honeybees (Supplementary Table S8); this could be ascribed to the intrinsic dynamics of this gut compartment, in which different levels of pollen content, dietary habits, waste accumulation, fermentation processes, and microbial load can occur5,14,39, particularly for forager honeybees, which under field conditions supposedly consume less pollen and more honey and nectar compared with nurses. Such oxygen depletion creates a favorable environment for facultative anaerobes and microaerophilic fermentative bacteria (i.e., Gilliamella, Lactobacillus spp., and Bifidobacteium1,4,39) and anaerobic microorganisms inhabiting digestive tracts (e.g., coliforms, staphylococci, Bacillus spp.)96. The establishment and activity of these bacteria can determine a significant change in the pH39, as was observed in our study. Indeed, in vitro metabolic characterization showed the way in which strains isolated from the honeybee gut significantly reduced the pH of the cultural media owing to the release of acetic and lactic acids during fermentative processes26,39,94,97, thereby confirming the correlation between the bacterial load and pH reduction. Despite the consistent positive redox observed in the honeybee gut, overall higher values were measured in field-foraging honeybees compared to laboratory honeybees possessing a reconstituted microbiome39 (Supplementary Table S8).
Metabolic products along the honeybee gut compartments
Genomic and transcriptomic analyses, along with in vitro experiments, have shown that the predominant metabolic activity in the honeybee gut was the microbial fermentation of sugars and complex carbohydrates (e.g., pectin) into organic acids (among others4,8,9,39). In the present study, we evaluated the concentration of dietary metabolites (sugars) and their fermentation products (short-chain fatty acids [SCFAs]) along the gut compartments. Quantification of the metabolite milieu (SCFAs and sugars) along the intestinal tract confirmed the presence of a spatially explicit distribution pattern that contributed to gut compartmentalization25 (PERMANOVA: F3,8 = 15.123, p = 0.001; Supplementary Table S11; pairwise comparison in Supplementary Table S9b). In particular, a significant decrease was observed in sugar concentration along the gut (F3,8 = 14.35, p = 0.0014), with the main reduction occurring in fructose (from 13.32 ± 4.8 to 0.1 ± 0.06 mM/mg tissue in crop and rectum, respectively) and glucose (from 4.7 ± 1.2 to 0.2 ± 0.08 mM/mg tissue) concentrations as well as in the total consumption of sucrose, melezitose, and maltose from the crop to the ileum (Fig. 5f and Supplementary Table S11); maltotriose remained within the detection limit in all compartments. Notably, among the sugars, sucrose resulted in a significant variable that controlled the microbiome assembly (core bacteria, other-possibly environmental bacteria, and fungi) across the gut compartments (Supplementary Table S10). It negatively influenced the relative abundance of Bifidobacterium (OTU_2), Frischella (OTU_9), and Snodgrassella (OTU_4) members of the core phylotypes (Supplementary Fig. S12), which was consistent with their preferential localization in distal parts of the honeybee gut (the ileum and rectum; Fig. 2d), where the sucrose concentration was low (Fig. 5f); conversely, Lactobacillus Firm-5 (OTU_120) showed an opposite trend (Supplementary Fig. S12), which was expected based on its metabolic capability to utilize fructose derived from sucrose98. Furthermore, we detected a total of eight other-possibly bacterial OTUs associated with the presence of sucrose (Supplementary Fig. S13). These OTUs belonged to the Enterobacteriaceae family, and other Lactobacillus spp., showed both positive (OTU_10 and OTU_17) and negative (OTU_20, OTU_29, OTU_108, OTU_118, OTU_174, and OTU_258) relationships with this variable. Along with these results, sucrose positively influenced the presence of fungal members, such as Penicillium (OTU_41 and OTU_575), Ganoderma (OTU_168), and unclassified fungi (OTU_3 and OTU_29), which had higher relative abundance in the crop, whereas the fermentative yeast Hanseniaspora sp. (OTU_739) showed an increased abundance with the decrease of sucrose (Supplementary Fig. S14).
The two monosaccharides glucose and fructose and the disaccharide sucrose are the most common sugars of nectar and honey, which together with pollen, constitute the major components of the forager honeybee diet99,100. These simple sugars are, in part, passively absorbed via the host midgut wall101. Accordingly, these sugars were mainly detected in the crop, which was the first intestinal compartment following the honeybee mouth, and drastically reduced in the midgut. Unlike laboratory conditions, in a natural ecosystem, forager honeybees load their crop with a certain amount of honey before leaving the hive to sustain their flight energy cost102. The amount of honey loaded is adjusted according to several factors, such as distance and food source type that they intend to collect and carry to the hive102, which explain the high variability observed in terms of sugar concentration detected in the guts of these honeybees. In the absence or limited availability of nectar, honeybees can forage on honeydew (i.e., hemipteran waste product from plant phloem). In addition to nectar sugars, honeydew contains disaccharides (e.g., maltose) and trisaccharides (e.g., melezitose)103, which were also detected with high variability in forager honeybee guts in the present study (Fig. 5f). The type of sugar affects the relative abundance of dominant bacterial members in the microbiota of honeybee guts23,40, consequently leading to an alteration of the gut metabolic and physico-chemical conditions1,4,14,39, which was confirmed by comparing field-foragers bees with laboratory-reared bees (Supplementary Table S8).
Following consumption, the portion of sugars that is not directly assimilated by the host is further metabolized by members of the gut microbiome4,7,15,23, as clearly shown by Zheng et al., who used honeybees with a reduced bacterial load (< 105 bacterial cells per gut)39. As expected, the decrease in sugar concentration observed along the gut compartments (Fig. 5f) was paralleled by a gradual increase in organic acids and SCFAs (F3,8 = 7.44, p = 0.011; Fig. 5g, Supplementary Tables S11, and Supplementary Fig. S11). For instance, when acetic acid reached its highest concentration in the ileum, the high variability of succinic acid did not determine consistent trends; on the contrary, all the other organic acids and SCFAs that were measured (lactic, propionic, malic, formic, and butyric acids) were within the detection limit (< 0.01 mg/ml). SCFA accumulation in the honeybee ileum, followed by the rectum and midgut, can be ascribed to the higher bacterial load hosted by these compartments compared with the crop (Fig. 2b and Supplementary Fig. S11). In addition, we observed that among the SCFAs that were analyzed, acetic acid significantly affected the fungal distribution along the intestinal tract compartments (Supplementary Table S10). Members of the Penicillium genus (OTU_41, OTU_575, and OTU_458) and one unclassified fungus (OTU_20) were negatively correlated with the acetic acid concentration detected (Supplementary Fig. S14), which was possibly owing to the fact that organic acid, such as acetic acid, possessed antifungal activities104–106; it should be noted that we did not find any SCFAs that influenced the core or other-possibly environmental bacterial distributions (Supplementary Table S10).
Recent metagenomic, metatranscriptomic, metabolomic, and in vitro analyses have revealed that the breakdown and fermentation of host dietary macromolecules into alcohols, SCFAs, and other organic acids—useful to sustain host growth and development—are the predominant metabolic functions of the core gut microbiome of honeybees1,4,107. For instance, the production of SCFAs was mainly ascribed to the metabolic activity of the Lactobacillus Firm-5 clade (monocolonization, gnotobiotic, and reconstructed gut microbiota experiments1,39) and the digestion of pollen-derived compounds was mediated by Gilliamella and Bifidobacterium (e.g., pectin and hemicellulose)4,7–9,39. Notably, the presence of organic acids (e.g., succinate) in the crop—where these bacteria were limited (Fig. 2b)—can be ascribed to the fact that the honey load carried by foragers before leaving the hive102 naturally contains these molecules108.
Genes for sugar transport and carbohydrate-degrading enzymes for breaking down nectar sugars (and other potentially toxic carbohydrate) are specifically enriched in the Gilliamella, Bifidobacterium, and Lactobacillus Firm-4 and Firm-5 bacterial clades4,109. An in vitro study showed that bacterial isolates belonging to the Gilliamella, Lactobacillus Firm-5, and Bifidobacterium clades were associated with transcribing the genes encoding for acetate kinase and L-lactate dehydrogenase, thereby being responsible for acetic and lactic acid production, respectively, particularly in the ileum and rectum compartments4. Despite their capacity, genomic diversity and metabolic variations within each clade were observed, suggesting that each core microbiome member differentially contributed to honeybee metabolism4,18,26. Conversely, these fermentative metabolisms of main nectar sugars (glucose, fructose, and sucrose) were not consistently detected in isolates classified as non-core gut members (other-possibly environmental bacteria)4. In addition, more distantly related bacteria not belonging to the honeybee core (Staphylococcaceae, other Lactobacillales, and Enterobacteriaceae) could not utilize pollen-derived compounds, such as pectin, but could contribute to organic acid production by metabolizing secondary sugars that are common in the honeybee diet (mannose, rhamnose, and galactose)4.
Reduction of the gut bacterial load in laboratory-reared honeybees unequivocally revealed that bees with a depleted-microbiome (i.e., < 105 bacteria per gut consisting of an erratic mix of bacterial species) exhibited less accumulation of fermentative products (SCFAs) in the ileum and rectum gut portions with a significant reduction in body weight39. Interestingly, using the gut of honeybees monocolonized by individual bacterial strains of the corbiculate core, it was possible to demonstrate that most of the metabolic output of the bee gut microbiota can be ascribed to the metabolic activities of single microbial community members1. For instance, succinic acid was exclusively produced in honeybees colonized by Lactobacillus Firm-5 strains1, which possibly explain the presence of this metabolite in the midgut of honeybees studied in the present work where Firm-5 were also present.
Considering the increasing evidence regarding the effect of diet and environment on the microbial composition and metabolic output of the honeybee gut1,5,14,40, it is reasonable to deduce that the differences observed in sugars and SCFA concentrations between our study and that of Zheng et al.39 were likely owing to the different diet and environmental conditions to which the bees were subjected.
Conclusion
In the present study, we demonstrated that compartmentalization of the gut microbiota in honeybee occurs for main bacterial phylotypes as well as for non-core, less-prevalent environmental bacterial and fungal members. We comprehensively determined their distribution, diversity, and assembly for all four gut compartments. Compartmentalization of these microbial groups indicates that similar to the main bacterial phylotypes, they have an explicit pattern of distribution along the different conditions of the gut and are not simply acquired via diet and passively transited along the gut. Our results can provide the basis for further investigations regarding the ways in which fungi and environmental bacteria interact with the core microbiome to shed light on additional mechanisms that contribute to gut homeostasis. Elucidating the dynamics of the gut microbiome is crucial for understanding, explaining, and predicting the degree of host resilience while experiencing changes in environmental conditions and challenges by stressors and pathogens.
Methods
Honeybee sampling
Adult forager Apis mellifera ligustica (Hymenoptera: Apidae) bees (aged > 21 days) were collected from the front of experimental apiaries located at the Department of Agricultural, Forest and Food Sciences (DISAFA, Grugliasco, Torino, Italy) following exposure to the natural surrounding environment between September 2016 and October 2018 (Supplementary Table S6). Additionally, between May 2019 and January 2020, adult forager bees (A. mellifera jemenitica and A. florea) were obtained from sites across Saudi Arabia (Medina, Thuwal, Jeddah and Makkah; Supplementary Table S6). All collected specimens were immediately transported to the laboratory and processed. Animals used in our experiments were maintained and treated in compliance with the guidelines specified by the Italian Ministry of Forestry and Agriculture. In addition, all necessary permits were obtained when the research was conducted, and all experiments and procedures were approved by the University of Torino. The research at King Abdullah University of Science and Technology (KAUST) was performed in compliance with Saudi Arabian and International guidelines.
Honeybee gut dissections
Forager honeybees were chilled in a sterile Petri dish at 4 °C for 10 min, following which they were individually transferred to 10 ml sterile plastic tubes. They were surface sterilized by washing the individuals with 1% sodium hypochlorite and then 97% ethanol. Specimens were then rinsed three times with sterile water before dissection and extraction of the gut. For DNA-based and metabolite analysis, the whole intestine (attached to the thorax and the head of the insect; Fig. 5a) of a subset of samples (n = 50, A. mellifera ligustica from Italy and n = 50, A. mellifera jemenitica from Saudi Arabia, Supplementary Table S6) was extracted from the insect body, placed in a Petri dish, and frozen at –20 °C to avoid the release of internal gut content during dissection. Separation of the frozen guts was performed—starting from the crop, proceeding with the midgut, and concluding with separation of the ileum and rectum. Gut dissection was performed in Ringer’s solution under a stereomicroscope in sterile conditions using sterile forceps and needles, and the scalpel was sterilized between every cut. Specimens were discarded when any portion of the gut was released into the intestinal liquid during dissection of the fresh tissues or separation of the frozen tissues. The four gut compartments (i.e., the crop, midgut, ileum, and rectum) obtained were pooled (n = 10) in five separate tubes to obtain sufficient microbial DNA for molecular analysis. The pooled gut compartments were weighed using an analytical balance (Supplementary Table S12) and frozen at –80 °C for DNA and metabolite preservation.
Fungal counts and isolation from honeybee adult gut
For the fungal isolation procedures, samples collected from Italy (n = 9, A. mellifera ligustica) and Saudi Arabia (n = 15, A. mellifera jemenitica and A. florea) were used (Supplementary Table S6). In this case, following surface sterilization, the honeybee gut was divided into two parts: the anterior part, including the crop and midgut, and the hindgut region, including the ileum and rectum. The obtained portions were transferred to a tube containing 0.9% NaCl (400 and 900 µl for single and pool of gut portions, respectively) and were immediately homogenized. A serial dilution was prepared and plated on yeast mold (YM) agar (Conda, Milano, Italy) and potato dextrose (PD) agar (Conda, Milano, Italy) supplemented with 100 µg/ml chloramphenicol to inhibit bacterial growth. Plates were incubated at 30 °C for 48 h. The colonies were counted, and the total CFU/mg of gut portions were determined. A total of 90 colonies were randomly selected and purified in new plates. The fungal strains were further inoculated in liquid media (YM and PD); the pure cultures thus obtained were used to extract DNA by boiling lysis to prepare the glycerinates with 20% glycerol, which was stored at –80 °C. Internal transcribed spacer 1 (ITS1) fragment was amplified using primers ITS1f and 5.8s71. Amplicons were sequenced and then aligned against the NCBI database using the BLAST tool. Sequences were deposited into the European Nucleotide Archive under accession numbers LR746502-LR746517 and LR798000-LR798072.
Electron microscopy
All gut specimens used for SEM were dissected from fresh A. mellifera jemenitica obtained from Makkah and Medina regions of Saudi Arabia following surface sterilization. The whole guts dissected were fixed in a solution of 2.5% (v/v) glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) and stored at 4 °C. Additional steps for the sample preparation were reported in Supplementary Method S1. The honeybee gut was visualized using transmission electron microscopy (Supplementary Method S2). The yeasts (Hanseniaspora uvarum L18 and Starmerella bombicola L28; Supplementary Table S7) that were isolated from the guts of Italian A. mellifera ligustica forager honeybees were grown in 5 ml PD broth media for 24 h at 30 °C on a rotary shaker at 100 rpm. Yeast cells were collected by centrifugation at 800 rpm for 10 min and fixed as described in Supplementary Method S3. All coated samples (guts and yeast strains) were visualized using the SEM Quanta 600 FEI at the KAUST Imaging and Characterization Core Lab.
Extraction of nucleic acids
Overall, five pools of ten dissected gut compartments (a total of 50 crop, midgut, ileum, and rectum sections) were obtained from Italian and Saudi forager bees (A. mellifera ligustica and A. mellifera jemenitica, respectively; Supplementary Table S6); the compartments were homogenized in sterile 0.5 ml 1× TE (10 mM Tris-HCl pH 8, 1 mM EDTA pH 8) using sterile pestles for 1 min. After three freeze/thaw cycles (–80 °C for 10 min and 70 °C for 10 min), lysozyme (Sigma-Aldrich, St. Louis, MO, USA) was added to a final concentration of 0.5 mg/ml per sample, followed by incubation at 37 °C for 30 min. The obtained homogenizations were used for DNA extraction according to the protocol of sodium dodecyl sulfate–proteinase K-cethyltrimethyl ammonium bromide treatment by Vacchini et al.110. DNA was eluted in 1× TE [100 μl (midgut and rectum) or 50 μl (crop and ileum)] and stored at –20 °C. Further, sterile water was used as the control for the DNA extraction procedures to assess the presence of reagent contamination. The extracted DNA was used as the control in all the further molecular analyses. The concentration of DNA samples was measured using the Nanodrop ND-1000 spectrophotometer. The total RNA was extracted from single fresh gut sample of six forager bees (A. mellifera jemenitica) collected from Madinah (Saudi Arabia; Supplementary Table S6) and reverse transcribed to cDNA (Supplementary Method S4) to detect the transcriptionally active microbial cells that are associated with the gut microbiota.
Abundance of bacterial and fungal communities by qPCR
qPCRs were performed using the CFX Connect™ Real-Time PCR Detection System (Bio-Rad). All bacteria obtained from A. mellifera ligustica (Italy) were quantified by evaluating the 16S rRNA gene copies of bacteria with 357F and 907R primers111 using the following thermal protocol: 98 °C for 3 min, followed by 40 cycles of denaturation at 98 °C for 15 s, annealing at 58 °C for 30 s, and extension at 72 °C for 1 min. Finally, a melting curve analysis was performed from 65 to 95 °C (fluorescence was measured every 0.5 °C). To evaluate whether the selected bees had a healthy status comparable to the ones shown in previous studies42 and to exclude dysbiotic animals from the analyses, we evaluated the organ-specific distributions of three of the main bacterial core phylotypes—Snodgrassella, Lactobacillus Firm-5, and Gilliamella. Their presence across the gut compartment was quantified using the primer pairs Beta-1009-qtF/Beta-1115-qtR, Firm-5-81-qtF/Firm-5-183-qtR, and Gamma1-459-qtF/Gamma1-648-qtR, respectively42. The following thermal protocol was used for all reactions: 98 °C for 3 min; 40 cycles of 98 °C for 15 s, annealing at 55 °C for 15 s, and extension at 72 °C for 15 s. Finally, a melting curve analysis was performed from 65 to 95 °C (fluorescence was measured every 0.5 °C). For fungi, total copies of the fungal ITS1 were determined using ITS1f and 5.8s primers112. The PCR conditions consisted of an initial denaturation at 98 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min. Finally, a melting curve analysis was performed from 65 to 95 °C (fluorescence was measured every 0.5 °C). 16S rRNA gene and fungal ITS1 fragments cloned in pGEM®-T Easy Vector (Promega, Milan, Italy) and pCR™II-TOPO (Invitrogen, Milan, Italy) were used as standards111. The copy numbers of the obtained 16S rRNA gene were normalized via division to determine the mean 16S rRNA gene copy number (GCN) corresponding to the taxonomic variability present in the honeybee gut bacterial community (bacterial OTU table; Supplementary Data 1 based on rrnDB database113, n = 4.7), whereas for Snodgrassella, Lactobacillus Firm-5, and Gilliamella, the published genomes were used to obtain this information (16S rRNA gene copy, n = 48). Similarly, for fungi, the values of obtained ITS1 were divided by the mean number of ITS copies present in our community (fungal OTU table; Supplementary Data 1; based on Lofgren et al.63, n = 75.5). DNA extracted from sterile water was used as an additional control in the qPCRs. No amplification was detected in negative controls for all primers pairs (results in Supplementary Table S4).
We tested the difference in the normalized number of the total bacteria and fungi and considered it as an explanatory categorical variable in the “gut compartment” (the crop, midgut, ileum, and rectum) and the “taxon” (total bacteria and fungi), which were both fixed and orthogonal. Additionally, we evaluated the differences among the three bacterial species (Snodgrassella, Lactobacillus Firm-5, and Gilliamella) within each gut compartment. In this case, we considered only the factor “gut compartment” as an explanatory variable. A pairwise test was performed using the R package multicomp114.
Metabarcoding analysis of the bacterial 16S rRNA and fungal ITS of forager honeybee microbial community
Illumina libraries were prepared using the Illumina® Nextera XT Sample Prep Kit and amplifying the V3 and V4 variable regions of the 16S rRNA gene (341F and 785R primers115) from both DNA and cDNA as well as the internal transcribed spacer 2 (ITS2) region (ITS3F and ITS4R primers85) from DNA, following the protocol described in Marasco et al.115. All primers used contained an adapter for the sequencing platform and an 8-nucleotide barcode. A blank control of PCR and DNA/RNA extraction reagents was performed and used to incorporate the sequencing adapters. Amplification reactions were purified and normalized using the SequalPrep™ Normalization Plate Kit (Invitrogen). All tagged samples were pooled together and concentrated in a CentriVap DNA Concentrator (Labconco). The obtained 16S rRNA and fungal ITS2 libraries were sequenced using the MiSeq system with 2 × 300 base-pair read length from the Biological Core Lab at KAUST, Saudi Arabia. For both 16S rRNA and ITS fragment datasets, sequence analyses were performed using a combination of the UPARSE v8116 and QIIME version 1.9117. Raw forward and reverse reads for each sample were assembled into paired-end reads using the fastq-join method within QIIME (minimum overlapping of 30 nucleotides and maximum 1 mismatch within the region). The paired reads were then quality filtered (no ambiguous base calls with quality values of < 20 Phred Q score). The primer sequences were removed, and the individual sample files were merged into a single fasta file. OTUs were determined at 97% similarity threshold (OTU97) in UPARSE. OTUs with less than two observations (singletons) were eliminated; chimeras were removed using both de novo and reference “Gold” database detection. Representative sequences of each OTU97 were aligned with the database in QIIME using uclust118 and blast commands to search against the SILVA version 138119 and UNITE120 databases for bacteria and fungi, respectively. OTU97 tables were created (i.e., sample ID, OTU97 count matrix, and relative taxonomic affiliation of each OTU97) for bacteria (total and active values from DNA and cDNA, respectively) and fungi. Taxonomic affiliations among the core phylotypes were confirmed creating a phylogenetic tree using the sequence available in the public database (Supplementary Table S3). OTUs present in blank controls were removed from the dataset (Supplementary Table S4). Details on the raw read processing are provided in Supplementary Table S13. Raw sequences were deposited in the Sequence Read Archive of NCBI under BioProjects PRJNA422176, PRJNA632549, and PRJNA422177.
Abundance of reads obtained from DNA sequencing were normalized. The relative abundance of each bacterial OTU along the samples was divided for the mean 16S rRNA GCN of the corresponding taxonomic level available for that OTU, which was obtained from the rrnDB database113 and published genomes8 (Supplementary Data 1). For fungi, the ITS relative abundance scores were normalized using the copy number available in literature for the Ascomycota, Basidiomycota, and Zygomycota phyla63 (n = 58, n = 113, and n = 116, respectively); for unclassified fungal phyla, the reads where normalized for the mean number of ITS detected in all analyzed fungi63 (n = 99; Supplementary Data 1). Normalized OTU tables were used for further analysis.
To test the microbial (bacteria and fungi) compositional differences along the gut compartments of the bees, we performed a permutational multivariate analysis of the variance (PERMANOVA), considering the “gut compartment” fixed and orthogonal as categorical explanatory variables. Homogeneity of the dispersions among the categorical variables was a priori tested using PERMDISP. To visualize the beta-diversity of the normalized compositional Bray–Curtis matrices of bacteria and fungi, we used principal coordinates analysis (PCoA) and CAP. The statistical tests, PCoA, and CAP were performed using PRIMER v. 6.1 and PERMANOVA+ for PRIMER routines121. Correlations between microbial (core bacteria or other potential environmental bacteria and fungi), beta-diversity, and physico-chemical gut conditions were determined by performing a Mantel test using Spearman’s rank correlation with the R package vegan122. To explore the discriminant OTUs that were affected by changes in the physico-chemical conditions along the gut compartments, the manyglm() function of the R package mvabund123 was employed using the negative binomial error distribution. We selected the best model. Each discriminant OTU was detected using a significant univariate general linear model with a negative binomial error distribution for which the p values were adjusted for multiple testing using a step-down resampling procedure. We selected the response variable (i.e., OTU) with p value of <0.005 to detect the most significant OTU change among the variables responsible for the gut physico-chemical gradient (i.e., our explanatory variable). Further, we computed the beta-diversity components using the function beta.div.comp of the package adespatial124. The rate of decay of the similarity of the community (Bray–Curtis) along the gut compartments was evaluated for both bacterial and fungal communities; a linear regression statistical analysis was subsequently performed using a GraphPad Prism (GraphPad Software, La Jolla California USA, www.graphpad.com). Alpha diversity indices were calculated using routine DIVERSE in PRIMER121, and an ANOVA (Tukey’s multiple comparison test) was performed to test the differences in alpha diversity indices for the “gut compartment” using the GraphPad software. Finally, to examine the functional role of other-possibly environmental bacteria and fungi during their association with the honeybee gut, taxonomy was utilized to interfere with the bacterial metabolic functions using Tax4Fun2125 platform and the fungal functional guild (i.e., trophic mode) using the annotation tool FUNGuild v1.09126.
Microsensor measurements
The intact whole honeybee gut was dissected from adult forager bees as previously described and gently placed on the agarose surface of customized plastic chambers filled with 2% agarose and embedded in 0.5% agarose Ringer’s solution127. Microsensors (Unisense, Aarhus, Denmark) were used to measure the oxygen concentration, pH, and redox potential within the different gut compartments of A. mellifera ligustica (Italy). Overall, 11, 12, and 16 digestive tracts were dissected for the analysis of pO2, pH, and redox potential, respectively. Oxygen measurements were performed using oxygen microsensors with 50-µm-diameter tips (OX-50), following a calibration at pO2 of 0 and 21 kPa, as previously described127. pH microelectrodes (PH-50) with extremely sharp customized 50-µm-diameter tips were calibrated using standard solutions of pH 4.0, 7.0, and 9.0128. The redox microelectrodes (RD-50) with a 50-µm-diameter tip o were calibrated using saturated quinhydrone solutions in pH standard solutions of pH 4.0 and 7.0129. In both cases, the electrode potentials were measured against a reference electrode (REF-RM) and an open-ended Ag-AgCl electrode with a gel-stabilized electrolyte connected to a high-impedance millivolt-meter. During the entire duration of microsensor measurements, the Veho VMS-004D USB microscope was placed in front of the embedded organ. The points in which the tip of the microsensor enter and exit the gut tissue were registered, and their difference was used to estimate the diameter (mm) of each gut compartment39,127. The diameter was reported as the mean ± standard deviation (Supplementary Table S12). Tukey’s multiple comparison test was used to evaluate changes among gut the compartments.
Gut metabolites analysis
We measured the sugar and SCFA concentrations within the different gut compartments of A. mellifera ligustica (Italy) using high-performance liquid chromatography (HPLC), following the method reported by Zheng et al.39. The weights of the pooled gut compartments were measured using an analytical balance. Thereafter, six compartments were homogenized in 100 μl water, centrifuged at 15,000 × g for 5 min, and filtered with 0.22-μm microcentrifuge tube filters (Corning) for 10 min at 10,000 × g. The filtrate was acidified with H2SO4 to reach a final concentration of 10 mM. Sugars were determined by performing HPLC on 10 μl of sample using two Aminex HPX-87P columns in a series (300 mm i.d. × 7.8 mm) from Bio-Rad (Segrate, Italy) maintained at 75 °C. The HPLC consisted of an Alliance 2695 pump (Waters, Milford, MA) with a model 410 differential refractometer (Waters). Chromatographic analyses were performed using MilliQ water (Millipore, Darmstadt, Germany) as an eluent at a flow-rate of 0.60 ml/min. Sugar quantification was performed by the external standard method using aqueous solutions of melezitose, sucrose, maltose, glucose, and fructose. HPLC of organic acids was conducted by injecting 10 μl of each sample into the same HPLC apparatus equipped with a Waters 2487 UV detector. An Aminex HPX-87H column (300 mm i.d. × 7.8 mm, Bio-Rad) maintained at 50 °C was used. An eluting solvent (0.60 ml/min) contained 5 mM sulfuric acid. Quantitation of organic acids in extracts was performed at 210 nm using solutions of succinic, acetic, lactic, malic, formic, propionic, and butyric acids in 5 mM sulfuric acid as external standards. All sugars and organic acids were of analytical grade (Sigma-Aldrich, St. Louis, MO, USA). Analyses were performed in triplicate, and mean values (mM) per mg of tissue were reported. Data were processed using the Millennium 32 software (Waters). The concentration of ethanol was determined using an enzymatic assay kit (Roche, R-Biopharm Italia srl, Melegnano, Italy). PERMANOVA was used to evaluate the compartmentalization of gut metabolites as continuous response variables, whereas the gut portions were used as categorical explanatory variables that are fixed and orthogonal (four levels: crop, midgut, ileum, and rectum). Moreover, difference in each metabolite concentration along the gut was tested using ANOVA and Tukey’s multiple comparison test.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Sadaf Umer and Taskeen Begum for their support in organizing the laboratory work. We also thank Davide Cuttini, beekeeper University of Turin; Melanie Augustina, beekeeper in KAUST; Zohair Fatani, beekeeper in Jeddah; Mr. Walid Hindi Al-Hazmi, Chairman of Madinah Beekeepers association; Mr. Nawaf Hindi Al-Hazmi, member of Medinah Beekeeper association; and Mansour Hussain Al-Sharif, beekeeper in Makkah, for honeybee keeping and collection. We are grateful to Kohloud Seferji and Mr. Ziad Dawood for contact and transportation. D.D. acknowledges the financial support of King Abdullah University and Technology (KAUST) through the RSRC-CCF funding 2019-2020 for “The microbiome and stress adaptation of mangrove honeybee pollinators” and the Circular Carbon Economy initiative (grant number, REI/1/4483-01-01). E.C. acknowledges personal support from “Piano di Sostegno della Ricerca 2018: Linea 2” for the projects “Microbial symbionts of insects (MicroSYM)”.
Author contributions
E.C., M.F., R.M., and D.D. designed the study; M.C., R.M., E.G., I.D.N., and D.R. performed the experiments; E.C., M.F., and R.M. analyzed the data; E.C., S.B., and D.D. supported the research; E.C., M.F., R.M., and D.D. wrote the paper with contributions from the other authors; M.C., E.C., M.F., and R.M. contributed to the manuscript equally.
Data availability
All data required to evaluate the conclusions in the paper are present in the paper and/or the Additional Information; molecular data are submitted in the European Nucleotide Archive (ENA) under accession numbers LR746502-LR746517 and LR798000-LR798072 and in the Sequence Read Archive (SRA) of NCBI under BioProjects PRJNA422176, PRJNA632549, and PRJNA422177. All data are available upon request to the authors.
Code availability
The code used for data elaboration, graph creation, and statistical analysis related to this paper are available upon request to the authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Matteo Callegari, Elena Crotti, Marco Fusi, Ramona Marasco.
Contributor Information
Elena Crotti, Email: elena.crotti@unimi.it.
Daniele Daffonchio, Email: daniele.daffonchio@kaust.edu.sa.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-021-00212-9.
References
- 1.Kešnerová L, et al. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 2017;15:e2003467. doi: 10.1371/journal.pbio.2003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwong WK, Zheng H, Moran NA. Convergent evolution of a modified, acetate-driven TCA cycle in bacteria. Nat. Microbiol. 2017;2:17067. doi: 10.1038/nmicrobiol.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwong WK, Moran NA. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016;14:374–384. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee FJ, Miller KI, McKinlay JB, Newton ILG. Differential carbohydrate utilization and organic acid production by honey bee symbionts. FEMS Microbiol. Ecol. 2018;94:1–10. doi: 10.1093/femsec/fiy113. [DOI] [PubMed] [Google Scholar]
- 5.Kešnerová L, et al. Gut microbiota structure differs between honeybees in winter and summer. ISME J. 2020;14:801–814. doi: 10.1038/s41396-019-0568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinson VG, et al. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 2011;20:619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 7.Zheng H, et al. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc. Natl Acad. Sci. USA. 2019;116:25909–25916. doi: 10.1073/pnas.1916224116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwong WK, Engel P, Koch H, Moran NA. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc. Natl Acad. Sci. USA. 2014;111:11509–11514. doi: 10.1073/pnas.1405838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl Acad. Sci. USA. 2012;109:11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saelao P, Borba RS, Ricigliano V, Spivak M, Simone-Finstrom M. Honeybee microbiome is stabilized in the presence of propolis. Biol. Lett. 2020;16:2–6. doi: 10.1098/rsbl.2020.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motta EVS, Raymann K, Moran NA. Glyphosate perturbs the gut microbiota of honey bees. Proc. Natl Acad. Sci. USA. 2018;115:10305–10310. doi: 10.1073/pnas.1803880115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamdi C, et al. Gut microbiome dysbiosis and honeybee health. J. Appl. Entomol. 2011;135:524–533. doi: 10.1111/j.1439-0418.2010.01609.x. [DOI] [Google Scholar]
- 13.Muñoz-Colmenero, M. et al. Differences in honey bee bacterial diversity and composition in agricultural and pristine environments—a field study. Apidologie10.1007/s13592-020-00779-w (2020).
- 14.Ricigliano VA, Anderson KE. Probing the honey bee diet-microbiota-host axis using pollen restriction and organic acid feeding. Insects. 2020;11:291. doi: 10.3390/insects11050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seeburger VC, et al. The trisaccharide melezitose impacts honey bees and their intestinal microbiota. PLoS ONE. 2020;15:e0230871. doi: 10.1371/journal.pone.0230871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subotic S, et al. Honey bee microbiome associated with different hive and sample types over a honey production season. PLoS ONE. 2019;14:1–15. doi: 10.1371/journal.pone.0223834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwong WK, et al. Dynamic microbiome evolution in social bees. Sci. Adv. 2017;3:e1600513. doi: 10.1126/sciadv.1600513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellegaard, K. M. & Engel, P. Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 10, 446 (2019). [DOI] [PMC free article] [PubMed]
- 19.Zheng H, Steele MI, Leonard SP, Motta EVS, Moran NA. Honey bees as models for gut microbiota research. Lab. Anim. (NY) 2018;47:317–325. doi: 10.1038/s41684-018-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, E. A., Anderson, K. E., Corby-Harris, V., McFrederick, Q. S. & Newton, G. Reclassification of seven honey bee symbiont strains as Bombella apis. bioRxiv10.1101/2020.05.06.081802 (2020). [DOI] [PubMed]
- 21.Bonilla-Rosso G, et al. Acetobacteraceae in the honey bee gut comprise two distant clades with diverging metabolism and ecological niches. bioRxiv. 2019;41:1–50. [Google Scholar]
- 22.Siozios S, Moran J, Chege M, Hurst GDD, Paredes JC. Complete reference genome assembly for Commensalibacter sp. strain AMU001, an acetic acid bacterium Isolated from the gut of honey bees. Microbiol. Resour. Announc. 2019;8:1–2. doi: 10.1128/MRA.01459-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor MA, et al. The effect of carbohydrate sources: Sucrose, invert sugar and components of mānuka honey, on core bacteria in the digestive tract of adult honey bees (Apis mellifera) PLoS ONE. 2019;14:1–19. doi: 10.1371/journal.pone.0225845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch H, Abrol DP, Li J, Schmid-Hempel P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 2013;22:2028–2044. doi: 10.1111/mec.12209. [DOI] [PubMed] [Google Scholar]
- 25.Chomicki, G., Werner, G. D. A., West, S. A. & Kiers, E. T. Compartmentalization drives the evolution of symbiotic cooperation. Philos. Trans. R. Soc. B Biol. Sci. 375, 20190602 (2020). [DOI] [PMC free article] [PubMed]
- 26.Ellegaard KM, et al. Extensive intra-phylotype diversity in Lactobacilli and Bifidobacteria from the honey bee gut. BMC Genomics. 2015;16:284. doi: 10.1186/s12864-015-1476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louca S, et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018;2:936–943. doi: 10.1038/s41559-018-0519-1. [DOI] [PubMed] [Google Scholar]
- 28.Moya A, Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016;24:402–413. doi: 10.1016/j.tim.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Anderson KE, et al. Microbial ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera) PLoS ONE. 2013;8:e83125. doi: 10.1371/journal.pone.0083125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran NA. Genomics of the honey bee microbiome. Curr. Opin. Insect Sci. 2015;10:22–28. doi: 10.1016/j.cois.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horton MA, Oliver R, Newton IL. No apparent correlation between honey bee forager gut microbiota and honey production. PeerJ. 2015;3:e1329. doi: 10.7717/peerj.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tauber JP, Nguyen V, Lopez D, Evans JD. Effects of a resident yeast from the honeybee gut on immunity, microbiota, and Nosema disease. Insects. 2019;10:296. doi: 10.3390/insects10090296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ptaszyńska AA, Paleolog J, Borsuk G. Nosema ceranae infection promotes proliferation of yeasts in honey bee intestines. PLoS ONE. 2016;11:e0164477. doi: 10.1371/journal.pone.0164477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yun J-H, Jung M-J, Kim PS, Bae J-W. Social status shapes the bacterial and fungal gut communities of the honey bee. Sci. Rep. 2018;8:2019. doi: 10.1038/s41598-018-19860-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox-Foster DL, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 36.Lee FJ, Rusch DB, Stewart FJ, Mattila HR, Newton ILG. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ. Microbiol. 2015;17:796–815. doi: 10.1111/1462-2920.12526. [DOI] [PubMed] [Google Scholar]
- 37.Gilliam M, Prest DB. Fungi isolated from the intestinal contents of foraging worker honey bees, Apis mellifera. J. Invertebr. Pathol. 1972;20:101–103. doi: 10.1016/0022-2011(72)90087-0. [DOI] [Google Scholar]
- 38.Ludvigsen J, Andersen Å, Hjeljord L, Rudi K. The honeybee gut mycobiota cluster by season versus the microbiota which cluster by gut segment. Vet. Sci. 2021;8:4. doi: 10.3390/vetsci8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl Acad. Sci. USA. 2017;114:4775–4780. doi: 10.1073/pnas.1701819114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, et al. The different dietary sugars modulate the composition of the gut microbiota in honeybee during overwintering. BMC Microbiol. 2020;20:1–14. doi: 10.1186/s12866-019-1672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Regan T, et al. Characterisation of the British honey bee metagenome. Nat. Commun. 2018;9:4995. doi: 10.1038/s41467-018-07426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinson VG, Moy J, Moran NA. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 2012;78:2830–2840. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corby-Harris, V., Maes, P. & Anderson, K. E. The bacterial communities associated with honey bee (apis mellifera) foragers. PLoS ONE9, e95056 (2014). [DOI] [PMC free article] [PubMed]
- 44.Powell JE, Martinson VG, Urban-Mead K, Moran NA. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 2014;80:7378–7387. doi: 10.1128/AEM.01861-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weyrich LS, et al. Laboratory contamination over time during low-biomass sample analysis. Mol. Ecol. Resour. 2019;19:982–996. doi: 10.1111/1755-0998.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Arx M, Moore A, Davidowitz G, Arnold AE. Diversity and distribution of microbial communities in floral nectar of two night-blooming plants of the Sonoran Desert. PLoS ONE. 2019;14:5–7. doi: 10.1371/journal.pone.0225309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris MM, Frixione NJ, Burkert AC, Dinsdale EA, Vannette RL. Microbial abundance, composition, and function in nectar are shaped by flower visitor identity. FEMS Microbiol. Ecol. 2020;96:1–14. doi: 10.1093/femsec/fiaa003. [DOI] [PubMed] [Google Scholar]
- 48.Legendre P. Interpreting the replacement and richness difference components of beta diversity. Glob. Ecol. Biogeogr. 2014;23:1324–1334. doi: 10.1111/geb.12207. [DOI] [Google Scholar]
- 49.Daisley BA, et al. Novel probiotic approach to counter Paenibacillus larvae infection in honey bees. ISME J. 2020;14:476–491. doi: 10.1038/s41396-019-0541-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chevrette MG, et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-08438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Gutierrez E, Mayer MJ, Cotter PD, Narbad A. Gut microbiota as a source of novel antimicrobials. Gut Microbes. 2019;10:1–21. doi: 10.1080/19490976.2018.1455790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryu J-H, et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 53.Kikuchi Y, et al. Symbiont-mediated insecticide resistance. Proc. Natl Acad. Sci. USA. 2012;109:8618–8622. doi: 10.1073/pnas.1200231109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Comstock LE, Kasper DL. Bacterial glycans: key mediators of diverse host immune responses. Cell. 2006;126:847–850. doi: 10.1016/j.cell.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 55.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonilla-Rosso G, Engel P. Functional roles and metabolic niches in the honey bee gut microbiota. Curr. Opin. Microbiol. 2018;43:69–76. doi: 10.1016/j.mib.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 57.Santos-Garcia D, Mestre-Rincon N, Zchori-Fein E, Morin S. Inside out: microbiota dynamics during host-plant adaptation of whiteflies. ISME J. 2020;14:847–856. doi: 10.1038/s41396-019-0576-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson KE, et al. Hive-stored pollen of honey bees: many lines of evidence are consistent with pollen preservation, not nutrient conversion. Mol. Ecol. 2014;23:5904–5917. doi: 10.1111/mec.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Al-Ghamdi A, Ali Khan K, Javed Ansari M, Almasaudi SB, Al-Kahtani S. Effect of gut bacterial isolates from Apis mellifera jemenitica on Paenibacillus larvae infected bee larvae. Saudi J. Biol. Sci. 2018;25:383–387. doi: 10.1016/j.sjbs.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jousset A, et al. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 2017;11:853–862. doi: 10.1038/ismej.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benjamino J, Lincoln S, Srivastava R, Graf J. Low-abundant bacteria drive compositional changes in the gut microbiota after dietary alteration. Microbiome. 2018;6:86. doi: 10.1186/s40168-018-0469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rivett DW, Bell T. Abundance determines the functional role of bacterial phylotypes in complex communities. Nat. Microbiol. 2018;3:767–772. doi: 10.1038/s41564-018-0180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lofgren LA, et al. Genome‐based estimates of fungal rDNA copy number variation across phylogenetic scales and ecological lifestyles. Mol. Ecol. 2019;28:721–730. doi: 10.1111/mec.14995. [DOI] [PubMed] [Google Scholar]
- 64.Duarte APM, et al. Prevalence of the genus Cladosporium on the integument of leaf-cutting ants characterized by 454 pyrosequencing. Antonie Van. Leeuwenhoek. 2016;109:1235–1243. doi: 10.1007/s10482-016-0724-3. [DOI] [PubMed] [Google Scholar]
- 65.Elhady A, et al. Microbiomes associated with infective stages of root-knot and lesion nematodes in soil. PLoS ONE. 2017;12:e0177145. doi: 10.1371/journal.pone.0177145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Čadež N, Fülöp L, Dlauchy D, Péter G. Zygosaccharomyces favi sp. nov., an obligate osmophilic yeast species from bee bread and honey. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 2015;107:645–654. doi: 10.1007/s10482-014-0359-1. [DOI] [PubMed] [Google Scholar]
- 67.Warmink JA, Nazir R, Corten B, van Elsas JD. Hitchhikers on the fungal highway: the helper effect for bacterial migration via fungal hyphae. Soil Biol. Biochem. 2011;43:760–765. doi: 10.1016/j.soilbio.2010.12.009. [DOI] [Google Scholar]
- 68.Stefanini I. Yeast-insect associations: it takes guts. Yeast. 2018;35:315–330. doi: 10.1002/yea.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Engel P, Moran NA. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol. Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 70.Moubasher A. Yeasts and filamentous fungi inhabiting guts of three insect species in Assiut. Egypt Mycosphere. 2017;8:1297–1316. doi: 10.5943/mycosphere/8/9/4. [DOI] [Google Scholar]
- 71.Ricci I, et al. Different mosquito species host Wickerhamomyces anomalus (Pichia anomala): perspectives on vector-borne diseases symbiotic control. Antonie Van. Leeuwenhoek. 2011;99:43–50. doi: 10.1007/s10482-010-9532-3. [DOI] [PubMed] [Google Scholar]
- 72.Chandler JA, Eisen JA, Kopp A. Yeast communities of diverse Drosophila species: comparison of two symbiont groups in the same hosts. Appl. Environ. Microbiol. 2012;78:7327–7336. doi: 10.1128/AEM.01741-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niu L-H, et al. New insights into the fungal community from the raw genomic sequence data of fig wasp Ceratosolen solmsi. BMC Microbiol. 2015;15:27. doi: 10.1186/s12866-015-0370-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stefanini I, et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl Acad. Sci. USA. 2012;109:13398–13403. doi: 10.1073/pnas.1208362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lievens, B. et al. Microbiology of sugar-rich environments: diversity, ecology and system constraints. Environ. Microbiol. 10.1111/1462-2920.12570 (2014). [DOI] [PubMed]
- 76.Cornman RS, et al. Pathogen webs in collapsing honey bee colonies. PLoS ONE. 2012;7:e43562. doi: 10.1371/journal.pone.0043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kakumanu ML, Reeves AM, Anderson TD, Rodrigues RR, Williams MA. Honey bee gut microbiome is altered by in-hive pesticide exposures. Front. Microbiol. 2016;7:1–11. doi: 10.3389/fmicb.2016.01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vadkertiová R, Molnárová J, Vránová D, Sláviková E. Yeasts and yeast-like organisms associated with fruits and blossoms of different fruit trees. Can. J. Microbiol. 2012;58:1344–1352. doi: 10.1139/cjm-2012-0468. [DOI] [PubMed] [Google Scholar]
- 79.Daniel HM, et al. Starmerella neotropicalis f. a., sp. nov., a yeast species found in bees and pollen. Int. J. Syst. Evol. Microbiol. 2013;63:3896–3903. doi: 10.1099/ijs.0.055897-0. [DOI] [PubMed] [Google Scholar]
- 80.Cheng D-J, Hou RF. Determination and distribution of a female-specific protein in the brown planthopper, Nilaparvata lugens Stal (Homoptera: Delphacidae) Tissue Cell. 2005;37:37–45. doi: 10.1016/j.tice.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 81.Vega, F. E. & Dowd, P. F. The role of yeasts as insect endosymbionts. In Insect-Fungal Associations (eds. Vega, E. F. and Blackwell, M.): Ecology and Evolution 211–243 (Oxford University Press, 2005).
- 82.Reuter M, Bell G, Greig D. Increased outbreeding in yeast in response to dispersal by an insect vector. Curr. Biol. 2007;17:R81–R83. doi: 10.1016/j.cub.2006.11.059. [DOI] [PubMed] [Google Scholar]
- 83.Lemos Junior WJF, et al. Draft genome sequence of the yeast Starmerella bacillaris (syn., Candida zemplinina) FRI751 isolated from fermenting must of dried raboso grapes. Genome Announc. 2017;5:5–6. doi: 10.1128/genomeA.00224-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ponomarova O, et al. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 2017;5:345–357.e6. doi: 10.1016/j.cels.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Booth JM, et al. The role of fungi in heterogeneous sediment microbial networks. Sci. Rep. 2019;9:7537. doi: 10.1038/s41598-019-43980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chouaia B, et al. Developmental stages and gut microenvironments influence gut microbiota dynamics in the invasive beetle Popillia japonica Newman (Coleoptera: Scarabaeidae) Environ. Microbiol. 2019;21:4343–4359. doi: 10.1111/1462-2920.14797. [DOI] [PubMed] [Google Scholar]
- 87.Kešnerová L, Moritz R, Engel P. Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 2016;66:414–421. doi: 10.1099/ijsem.0.000736. [DOI] [PubMed] [Google Scholar]
- 88.Bottacini F, et al. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS ONE. 2012;7:e44229. doi: 10.1371/journal.pone.0044229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lazzeri AM, et al. Potential of novel food-borne Lactobacillus isolates against the honeybee pathogen Paenibacillus larvae. Biocontrol Sci. Technol. 2020;30:897–908. doi: 10.1080/09583157.2020.1769556. [DOI] [Google Scholar]
- 90.Koussémon M, Combet-Blanc Y, Ollivier B. Glucose fermentation by Propionibacterium microaerophilum: effect of pH on metabolism and bioenergetic. Curr. Microbiol. 2003;46:141–145. doi: 10.1007/s00284-002-3839-x. [DOI] [PubMed] [Google Scholar]
- 91.García-Fraile P, Velázquez E, Mateos PF, Martínez-Molina E, Rivas R. Cohnella phaseoli sp. nov., isolated from root nodules of Phaseolus coccineus in Spain, and emended description of the genus Cohnella. Int. J. Syst. Evol. Microbiol. 2008;58:1855–1859. doi: 10.1099/ijs.0.65468-0. [DOI] [PubMed] [Google Scholar]
- 92.Lee Y, Jeon CO. Cohnella algarum sp. Nov., isolated from a freshwater green alga Paulinella chromatophora. Int. J. Syst. Evol. Microbiol. 2017;67:4767–4772. doi: 10.1099/ijsem.0.002377. [DOI] [PubMed] [Google Scholar]
- 93.Šustr V, Stingl U, Brune A. Microprofiles of oxygen, redox potential, and pH, and microbial fermentation products in the highly alkaline gut of the saprophagous larva of Penthetria holosericea (Diptera: Bibionidae) J. Insect Physiol. 2014;67:64–69. doi: 10.1016/j.jinsphys.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 94.Kwong WK, Moran NA. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a memb. Int. J. Syst. Evol. Microbiol. 2013;63:2008–2018. doi: 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- 95.Maes PW, Rodrigues PAP, Oliver R, Mott BM, Anderson KE. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera) Mol. Ecol. 2016;25:5439–5450. doi: 10.1111/mec.13862. [DOI] [PubMed] [Google Scholar]
- 96.Rada V, Máchová M, Huk J, Marounek M, Dušková D. Microflora in the honeybee digestive tract: counts, characteristics and sensitivity. Apidologie. 1997;28:357–365. doi: 10.1051/apido:19970603. [DOI] [Google Scholar]
- 97.Engel P, Kwong WK, Moran NA. Frischella perrara gen. nov., sp. nov., a gammaproteobacterium isolated from the gut of the honeybee, Apis mellifera. Int. J. Syst. Evol. Microbiol. 2013;63:3646–3651. doi: 10.1099/ijs.0.049569-0. [DOI] [PubMed] [Google Scholar]
- 98.Olofsson TC, Alsterfjord M, Nilson B, Butler È, Vásquez A. Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and lactobacillus kullabergensis sp. nov., isol. Int. J. Syst. Evol. Microbiol. 2014;64:3109–3119. doi: 10.1099/ijs.0.059600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nepi M. New perspectives in nectar evolution and ecology: simple alimentary reward or a complex multiorganism interaction? Acta Agrobot. 2017;70:1–12. doi: 10.5586/aa.1704. [DOI] [Google Scholar]
- 100.Siddiqui AJ, Musharraf SG, Choudhary MI, Rahman A. Application of analytical methods in authentication and adulteration of honey. Food Chem. 2017;217:687–698. doi: 10.1016/j.foodchem.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 101.Crailsheim K. Intestinal transport of sugars in the honeybee (Apis mellifera L.) J. Insect Physiol. 1988;34:839–845. doi: 10.1016/0022-1910(88)90117-5. [DOI] [Google Scholar]
- 102.Harano K, Mitsuhata-Asai A, Konishi T, Suzuki T, Sasaki M. Honeybee foragers adjust crop contents before leaving the hive: effects of distance to food source, food type, and informational state. Behav. Ecol. Sociobiol. 2013;67:1169–1178. doi: 10.1007/s00265-013-1542-5. [DOI] [Google Scholar]
- 103.Shaaban B, Seeburger V, Schroeder A, Lohaus G. Sugar, amino acid and inorganic ion profiling of the honeydew from different hemipteran species feeding on Abies alba and Picea abies. PLoS ONE. 2020;15:e0228171. doi: 10.1371/journal.pone.0228171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cabo ML, Braber AF, Koenraad PMFJ. Apparent antifungal activity of several lactic acid bacteria against Penicillium discolor is due to acetic acid in the medium. J. Food Prot. 2002;65:1309–1316. doi: 10.4315/0362-028X-65.8.1309. [DOI] [PubMed] [Google Scholar]
- 105.Schillinger U, Villarreal JV. Inhibition of Penicillium nordicum in MRS medium by lactic acid bacteria isolated from foods. Food Control. 2010;21:107–111. doi: 10.1016/j.foodcont.2008.11.010. [DOI] [Google Scholar]
- 106.Guimarães A, Venancio A, Abrunhosa L. Antifungal effect of organic acids from lactic acid bacteria on Penicillium nordicum. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2018;35:1803–1818. doi: 10.1080/19440049.2018.1500718. [DOI] [PubMed] [Google Scholar]
- 107.Li F, et al. Studies on antibacterial activity and diversity of cultivable actinobacteria isolated from mangrove soil in futian and maoweihai of China. Evid. Based Complement. Altern. Med. 2019;2019:1–11. doi: 10.1155/2019/3476567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Consonni R, Bernareggi F, Cagliani LR. NMR-based metabolomic approach to differentiate organic and conventional Italian honey. Food Control. 2019;98:133–140. doi: 10.1016/j.foodcont.2018.11.007. [DOI] [Google Scholar]
- 109.Zheng H, et al. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. MBio. 2016;7:1–9. doi: 10.1128/mBio.01326-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vacchini V, et al. Bacterial diversity shift determined by different diets in the gut of the spotted wing fly Drosophila suzukii is primarily reflected on acetic acid bacteria. Environ. Microbiol. Rep. 2017;9:91–103. doi: 10.1111/1758-2229.12505. [DOI] [PubMed] [Google Scholar]
- 111.Favia G, et al. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc. Natl Acad. Sci. USA. 2007;104:9047–9051. doi: 10.1073/pnas.0610451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fierer N, Jackson JA, Vilgalys R, Jackson RRB. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 2005;71:4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt T. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015;43:D593–D598. doi: 10.1093/nar/gku1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biometrical J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 115.Marasco R, et al. Rhizosheath microbial community assembly of sympatric desert speargrasses is independent of the plant host. Microbiome. 2018;6:215. doi: 10.1186/s40168-018-0597-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 117.Caporaso JG, et al. QIIME allows analysis of high- throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 119.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Koljalg U, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2014;22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 121.Anderson, M. M. J. J., Gorley, R. N. R. N. & Clarke, K. R. R. PERMANOVA + for PRIMER: Guide to Software and Statistical Methods; PRIMER-E (PRIMER-E Ltd., 2008).
- 122.Oksanen J. Vegan: an introduction to ordination. Management. 2015;1:1–10. [Google Scholar]
- 123.Wang Y, Naumann U, Wright ST, Warton D. mvabund—an R package for model-based analysis of multivariate abundance data. Methods Ecol. Evol. 2012;3:471–474. doi: 10.1111/j.2041-210X.2012.00190.x. [DOI] [Google Scholar]
- 124.Dray, S. et al. adespatial: multivariate multiscale spatial analysis. 10.1890/11-1183.1 (2020).
- 125.Wemheuer F, et al. Tax4Fun2: prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome. 2020;15:11. doi: 10.1186/s40793-020-00358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nguyen NH, et al. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016;20:241–248. doi: 10.1016/j.funeco.2015.06.006. [DOI] [Google Scholar]
- 127.Brune A, Emerson D, Breznak JA. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 1995;61:2681–2687. doi: 10.1128/AEM.61.7.2681-2687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brune A, Kühl M. pH profiles of the extremely alkaline hindguts of soil-feeding termites (Isoptera: Termitidae) determined with microelectrodes. J. Insect Physiol. 1996;42:1121–1127. doi: 10.1016/S0022-1910(96)00036-4. [DOI] [Google Scholar]
- 129.Ebert A, Brune A. Hydrogen concentration profiles at the oxic-anoxic interface: a microsensor study of the hindgut of the wood-feeding lower termite Reticulitermes flavipes (Kollar) Appl. Environ. Microbiol. 1997;63:4039–4046. doi: 10.1128/AEM.63.10.4039-4046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data required to evaluate the conclusions in the paper are present in the paper and/or the Additional Information; molecular data are submitted in the European Nucleotide Archive (ENA) under accession numbers LR746502-LR746517 and LR798000-LR798072 and in the Sequence Read Archive (SRA) of NCBI under BioProjects PRJNA422176, PRJNA632549, and PRJNA422177. All data are available upon request to the authors.
The code used for data elaboration, graph creation, and statistical analysis related to this paper are available upon request to the authors.