Abstract
Trueperella pyogenes (T. pyogenes) is a common opportunistic pathogen of many livestock and play an important regulation role during multibacterial infection and interaction with the host by its primary virulence factor pyolysin (PLO). The purpose of this study was to investigate the regulation role of PLO which serve as a combinational pathogen with lipopolysaccharide (LPS) during endometritis. In this study, the expression of bioactive recombinant PLO (rPLO) in a prokaryotic expression system and its purification are described. Moreover, we observed that rPLO inhibited the innate immune response triggered by LPS and that methyl-β-cyclodextrin (MBCD) abrogated this inhibitory effect in goat endometrium stromal cells (gESCs). Additionally, we show from pharmacological and genetic studies that rPLO-induced autophagy represses gene expression by inhibiting NLRP3 inflammasome activation. Importantly, this study reported that ATF6 serves as a primary regulator of the cellular inflammatory reaction to rPLO. Overall, these observations suggest that T. pyogenes PLO could create an immunosuppressive environment for other pathogens invasion by regulating cellular signaling pathways.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42770-021-00422-5.
Keywords: Pyolysin, LPS, Costimulation, Autophagy, ATF6
Introduction
Pyolysin, which is a primary virulence factor of T. pyogenes, is a secreted pore-forming toxin (PFT) that belongs to the cholesterol-dependent cytolysin (CDC) family [1]. T. pyogenes is an opportunistic pathogen, gram-positive, normally commensal pathogenic bacterium that can invade various mammals and nonmammals including livestock, wildlife animals, and even humans [2, 3]. The diseases suppurative arthritis, subcutaneous abscess, endocarditis, encephalitis, mastitis, metritis, and endometritis are related to T. pyogenes, which can also have serious economic consequences in agriculture [4–6]. The economic losses which caused by uterine disease is not a small number, it cost €1.411 billion in EU and $650 million in the USA every year [7]. Moreover, according to a investigate, the percentage of cows with endometritis was up to 52% (14/27) in a dairy farm in Beijing, China [6]. Previous research has investigated the genes plo, fimA, fimC, fimE, fimG, nanH, nanP, and cbpA in 71 T. pyogenes strains isolated from different animals and lesions; however, only plo was detected in all strains (71/71 = 100%); in contrast, 4 strains expressed fimG (4/71 = 5.6%) [8]. Therefore, various putative virulence genes exist, among which plo is the most common and the primary virulence factor of T. pyogenes [9–11].
Efforts have been made to characterize the role of PLO during T. pyogenes infection as part of an investigation into the molecular mechanisms of host protection against pathogenic invasion [12, 13]. After parturition, the uterus is the main target of T. pyogenes invasion; however, endometrial stromal cells are markedly more sensitive to PLO-mediated cytolysis than epithelial cells, since disruption of the endometrium epithelium is exploited by opportunistic pathogens we have mentioned [14]. Cholesterol-dependent cytolysin has a high affinity for cholesterol in the plasma of mammalian cells, and the same is true of PLO, as inhibition of FDFT1, a primary enzyme of the mevalonate pathway for the synthesis of cholesterol, significantly reduced the impact of PLO on cell viability [15]. Numerous studies determine whether CDC family members initiate and instruct innate immunity or adaptive immunity by pattern recognition receptors [16, 17], and different type of pathogen-associated molecular patterns (PAMPs) is investigated with LPS serving as a costimulatory ligands [18], but PLO has not been widely investigated in this field.
Autophagy is an essential intracellular lysosomal degradative pathway [19] that plays an important role in cellular homeostasis, cell differentiation, development, and diseases [20–22]. Importantly, autophagy serves as a primary antibacterial mechanism as it limits bacterial growth in the cytosol [23–26] in a process termed “xenophagy” [27]. Recent studies have demonstrated that sublytic concentrations of PLO can induce autophagy in eukaryotic cells, as determined by microtubule-associated protein 1 light chain 3 (LC3) cleavage, which produces LC3-II [28]. In addition, induction of autophagy by cannabinoid receptor 2 (CB2R) or the polyphenolic compound resveratrol could inhibit NLRP3 inflammasome-derived IL-1β secretion through the suppression of mitochondrial damage [29, 30]. The NLRP3 inflammasome is a pattern-recognition receptor (PRR) that functions to target pathogenic microbes and other endogenous or exogenous pathogens [31]. In addition to the NLRP3 protein, the adapter protein apoptosis-associated speck-like protein CARD domain (ASC) and caspase-1 are components of the NLRP3 inflammasome [32], and secretion of various pro-inflammatory cytokines, such as IL-1β, is mediated under stimulation [33]. The endoplasmic reticulum stress (ERs), also known as the unfolded protein response (UPR), is a highly conserved cellular process that plays increasingly recognized roles in cell growth, proliferation, survival, and prevention of pathogen invasion [34–36]. Accumulating evidence suggests that ERs/UPR can transcriptionally activate or regulate the expression of genes encoding autophagy components [37].
Previous studies have found that under the single treatment of sublytic concentration of PLO, there is no detectable accumulation of IL-1β, IL-6, or IL-8, which are pro-inflammatory cytokines, in the supernatant of bovine endometrial stromal and epithelial cells, though PLO is the mainly virulence factor of T. pyogenes and the stimulating factor of cellular autophagy [14]. However, the specific roles and molecular mechanism of PLO in combination with Escherichia coli LPS remain unknown. Therefore, the main objectives of the present study were to investigate the influence of PLO on gESCs under costimulation with LPS. Our results suggest that the expression of related cytokines and genes induced by LPS was blunted by PLO through suppression of the NLRP3 inflammasome. Importantly, autophagy and ATF6 are key regulators of this process.
Material and methods
Cell lines and bacterial strain
gESCs were immortalized by transfection with human telomerase reverse transcriptase (hTERT) and well stored in our laboratory [38, 39]. HEK 293T were obtained from The Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). gESCs and HEK 293T were cultured in Dulbecco’s modified Eagle medium: nutrient mixture F-12 (DMEM/F12; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco) and high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% FBS, respectively, in a humidified atmosphere with 5% CO2 at 37 °C. Cells were cultured for 24 h before treatment unless otherwise mentioned. T. pyogenes strain used in this study was isolated from a goat abscess by syringe sampling, and the goat’s life was not affected by the sampling. All animal procedures were approved under the control of the Guidelines for Animal Experiments by the Committee for the Ethics on Animal Care and Experiments of Northwest A&F University and performed under the control of the “Guidelines on Ethical Treatment of Experimental Animals” (2006) No. 398 set by the Ministry of Science and Technology, China. The identification of T. pyogenes strain was based on characteristics of the colony, sequence alignment of 16S rRNA and hemolysis that described previously [40]. T. pyogenes was cultured on Brain Heart Infusion (BHI; Difco) agar plates, which supplemented with 5% FBS, at 37 °C and 5% CO2 in a humidified incubator, or in BHI broth containing 5% FBS at 37 °C.
Antibodies and reagents
The following primary antibodies were used: rabbit anti-MAP1LC3B (Sigma, L7543, diluted 1:1000), rabbit anti-ATF6 (Abcam ab83504, 1:1000), rabbit anti-NLRP3 (Proteintech, 19771-1-AP, 1:1000), and mouse anti-β-actin (Tianjin Sanjian Biotech, 1:2000). The secondary antibodies were HRP-conjugated goat anti-rabbit (Zhongshan Golden Bridge Biotechnology, 1:5000) and HRP-conjugated goat anti-mouse (Zhongshan Golden Bridge Biotechnology, 1:5000). The reagent: methyl-β-cyclodextrin (Sigma-Aldrich, C4555) and chloroquine diphosphate salt (Sigma-Aldrich, C6628) were also used.
Hemolysis determination
In order to obtain sheep red blood cells (SRBCs), sheep blood was centrifuged at 1000 rpm for 10 min and removing supernatant, and then the red blood cells were resuspended with normal saline which process was repeat twice.
A hemolytic assay was performed nearly as previously described [13, 14]. Briefly, SRBCs were prewashed three times with PBS and diluted to 2% (v/v). For the first assay, MBCD (methyl-β-cyclodextrin) at different concentrations was incubated with SRBCs for 12 h, and then rPLO was administered for 1 h. To investigate cholesterol function in the process of hemolysis mediated by purified rPLO, rPLO was diluted to different concentrations and incubated with or without cholesterol at a concentration of 8 μg/μl for 1 h. Then, SRBCs were challenged with the mixture for 1 h at room temperature, the mixture was centrifuged at 1000 rpm for 10 min, and 150-μl samples of the supernatants were transferred into a 96-well polystyrene microplate. One arbitrary unit (AU) indicates rPLO at a final concentration of 2 ng/μl in this assay. The optical density at a wavelength of 405 nm was measured using a microplate reader. The experiments were performed in triplicate and repeated three times independently.
Small interference RNA transfection
A small interfering RNA (siRNA) targeting ATG5 (si-ATG5: GCUUCGAGAUGUGUGGUUUTT) was designed as previously described [41]. In this study, gESCs were cultured in a 6-well cell culture plate, and when the gESCs reached 70% confluence, siRNA oligo at a final concentration of 50 nM was used to transfect the cells with TurboFect transfection reagent (R0531, Thermo Fisher Scientific) supplemented with Opti-MEM (31985088, Gibco), and cells were cultured in the normal cell culture medium mentioned above for 48 h. Then, the cells were subjected to the corresponding processing. The negative control siRNA (si-NC: UUCUCCGAACGUGUCACGUTT) used in this experiment was provided by GenePharma (Shanghai, China).
Interference cell line construction
To knockdown expression of the ATF6 (GenBank accession number: XM_018046547.1) gene, three pairs of oligonucleotides against atf6 and a pair oligonucleotide that expressed a scrambled sequence (Table 1) were designed with a website (http://rnaidesigner.thermofisher.com/rnaiexpress) and synthesized by TSINGKE Biological Technology (Beijing, China). Sequences of the designed oligonucleotides are shown in Table 1. As described in a previously published article by our group [42], lentiviral vectors containing three short hairpin RNA (shRNA) sequences targeting the gene and a scrambled shRNA lentiviral vector were constructed. Next, the vectors above and lentivirus packaging vectors that encoded Gag-Pol, Rev, Tat, and a G protein were co-transfected into HEK 293T cells using TurboFect transfection reagent according to the manufacturer’s procedure. The culture medium was replaced after 12 h, and another 48 h was needed to package lentivirus; thereafter, the supernatant was collected and used to infect gESCs. Finally, the cells were harvested and used to determine ATF6 protein levels by Western blotting.
Table 1.
Primers for shRNA-ATF6
| shRNA | Sequences (loop in bold letters) (5′-3′) |
|---|---|
| shRNA-1 | GATCCGCAGTTGGATGCAGCAAATGATTCAAGAGATCATTTGCTGCATCCAACTGCTTTTTTG |
| AATTCAAAAAAGCAGTTGGATGCAGCAAATGATCTCTTGAATCATTTGCTGCATCCAACTGCG | |
| shRNA-2 | GATCCGCAGCACCCAAGACTCAAACATTCAAGAGATGTTTGAGTCTTGGGTGCTGCTTTTTTG |
| AATTCAAAAAAGCAGCACCCAAGACTCAAACATCTCTTGAATGTTTGAGTCTTGGGTGCTGCG | |
| shRNA-3 | GATCCGCTTAGAGGCAAGGTTAAAGGTTCAAGAGACCTTTAACCTTGCCTCTAAGCTTTTTTG |
| AATTCAAAAAAGCTTAGAGGCAAGGTTAAAGGTCTCTTGAACCTTTAACCTTGCCTCTAAGCG | |
| shRNA-NC | GATCCTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTTG |
| AATTCAAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAAG |
Western blotting
Whole-cell protein extraction from gESCs using a KGP2100 Kits (KeyGEN Biotech, China) was performed according to the manufacturer’s protocol. The protein concentrations of the cell lysates were determined using a bicinchoninic acid assay (KGPBCA; KeyGEN Biotech). The total cellular protein as degraded with 5× sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer by boiling in water for 10 min. Samples containing an equal quantity of total cellular protein were resolved by 12% SDS-PAGE, and the separated proteins were transferred onto PVDF membranes. The membranes were blocked with 5% nonfat milk in Tris-buffered saline containing 0.5% Tween 20 (TBST) for 1–2 h and incubated with the indicated primary antibodies diluted in TBST overnight at 4 °C. Then, horseradish peroxidase (HRP)-conjugated secondary antibodies were administered and incubated for 1 h at room temperature. Finally, the protein bands were visualized with a gel imaging system (Tanon Biotech, Shanghai, China) and quantified with Quantity One software (Bio-Rad Laboratories, Hercules, CA, USA).
Real-time quantitative PCR
Total RNA was extracted from cultured cells using RNAiso Plus (TaKaRa, China), and the same amount of total RNA was used to synthesize cDNA by reverse transcription using Evo M-MLV RT Kit with gDNA Clean for qPCR (Accurate Biotechnology Co., Ltd., Hunan, China). Then, qPCR was performed on a Bio-Rad CFX96 (Bio-Rad Laboratories, Inc.) system using SYBR® Premix Ex Taq II (TaKaRa, China) with a 20 μl reaction system. The melting peaks were determined by melting curve analysis to ensure product specificity, and the amplification efficiency of all specific primers was determined by standard curve, which suggested efficiencies of no less than 90%. The expression of each gene determined with three technical replicates and biological duplicates was quantified by 2−ΔΔCt method and is reported relative to expression of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primer sequences are listed in Table 2.
Table 2.
Primers for real-time quantitative PCR (RT-qPCR)
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| IL-1β | GGTGTTCTGCATGAGCTTCG | TTGTCCCTGATACCCAAGGC |
| TNF-α | CTCCTCATCCCCTTCTGGTTT | GGCCTCACTTCCCTACATCC |
| ICAM-1 | CGACTAGACCAGCGGGATTG | CTGGCCGTAGAGCACATTCA |
| SAA3 | GACATTCCTCAGGGAAGCTGG | TGAAGAGCCTCTCTGATCACTT |
| COX-2 | GAGTGTAGGATTCGACCAGTAT | CCTTGAAGTGGGTAAGTATGTAG |
| IL-6 | CCTCTTCACAAGCGCCTTCA | TGCTTGGGGTGGTGTCATTC |
| NLRP3 | CCGTCCTAAGCACCAACCAT | TTGTGGTGCAAGAGTCCCTC |
| ASC | CATGATGAGCAAGGGCGCTA | ATCAACGACTGTGACCCGTG |
| Caspase-1 | GGAGAGAAAGGACCGCACTC | AGTCCCACAGATCCCATCCA |
| GAPDH | GATGGTGAAGGTCGGAGTGAAC | GTCATTGATGGCAACGATGT |
Statistical analysis
The results are presented as the mean ± standard error of the mean (SEM), and each experiment was performed independently at least three times. GraphPad Prism 6.0 software (GraphPad Software Inc., San Diego, CA, USA) was used in this study for data analysis by two-way ANOVA or one-way ANOVA. The P < 0.05 value was considered to be significant.
Results
Cholesterol and methyl-β-cyclodextrin repressed the hemolytic activity of rPLO
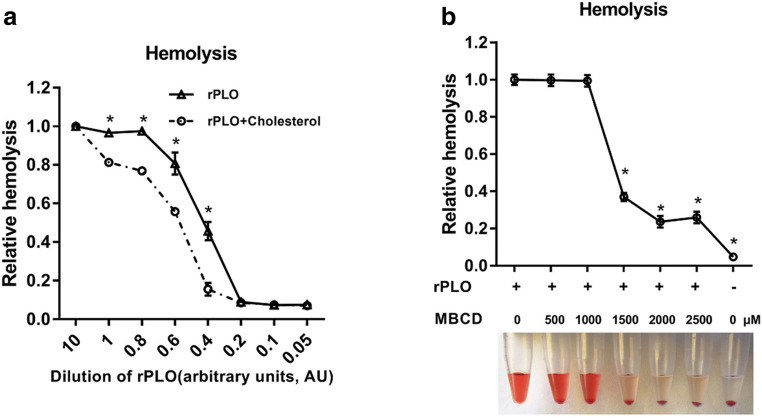
To authenticate the hemolytic activity of the purified pyolysin (PLO) (Fig. supplement), a hemolytic assay was performed. rPLO at different dilutions was incubated with or without cholesterol for 1 h, after which suspended sheep red blood cells were challenged with abovementioned mixtures for another 1 h to assess hemolysis (Fig. 1a). MBCD, which can bind cellular cholesterol, is a specific inhibitor of PLO-mediated lysis of immune cells [14]. To verify that the recombinant protein acted as a cholesterol-dependent cytolysin with another method, a sheep red blood cell suspension was pretreated with MBCD and then challenged with PLO. Hemolysis was measured with a microplate reader, which showed that hemolysis in the group treated with MBCD was significantly decreased compared to that in the group without MBCD treatment (Fig. 1b). Together with the results of sequencing analysis (not shown), this finding shows that the recombinant protein that we expressed and purified was T. pyogenes pyolysin.
Fig. 1.
Functional verification of purified rPLO. a rPLO at different dilution was incubated with or without 8 μg/μl cholesterol for 1 h, and a 2% sheep red blood cell suspension was challenged with the abovementioned mixture for 1 h. One AU indicates a final concentration of 2 ng/μl rPLO in this assay. b MBCD at different dilution was incubated with a 2% sheep red blood cell suspension for 12 h, after which rPLO was added and incubated with the mixture for 1 h. The optical density at a wavelength of 405 nm was measured using a microplate reader. Error bars represent SEM of three independent experiments, *P < 0.05
rPLO induced inflammatory mediators to different extents
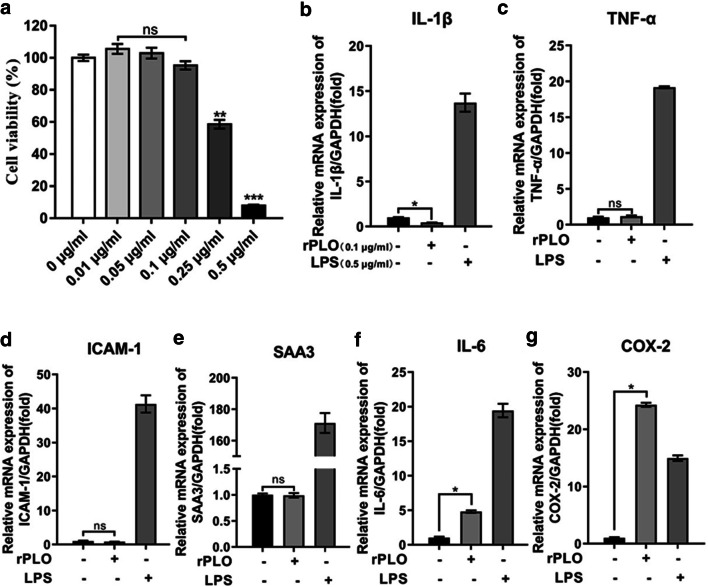
We next aimed to explore the responses of gESCs upon challenge with rPLO at sublytic concentrations. Therefore, gESCs were incubated with rPLO at different concentrations of 3 h, and cell proliferation was examined by the CCK-8 colorimetric assay to determine the experimental dose of rPLO (Fig. 2a) that does not induce significant cell death but carries out its biological function. Previous results have shown that PLO cannot induce upregulated IL-1β expression [14]; in this study, IL-1β expression was slightly downregulated by rPLO (Fig. 2b). Stimulation of gESCs with or without rPLO triggered similar changes in the expression of TNF-α, SAA3, and ICAM-1 (Fig. 2c–e). In contrast, IL-6 expression was slightly upregulated relative to that in the control group (Fig. 2f), and COX-2 expression was significantly elevated and even higher than that induced by LPS, which served as a positive control in this experiment (Fig. 2g). Together, these results indicated recombinantly expressed virulence factor from the opportunistic pathogen T. pyogenes, rPLO, could not induce a general inflammatory response. There are two possible explanations for this phenomenon: T. pyogenes does not activate the host inflammatory response to promote continued infection, or T. pyogenes may create an environment with low sensitivity for pathogen invasion.
Fig. 2.
Screening applicable working concentration of rPLO and relative inflammatory response genes expression under rPLO stimulation. a gESCs were challenged with control medium or medium containing rPLO at the indicated concentration in a gradient for 3 h. Cell viability was evaluated by CCK-8 assay. b–g. gESCs were challenged with control medium or medium containing 0.1 μg/ml rPLO or 0.5 μg/ml LPS for 3 h. mRNA levels of targets normalized to the levels of GAPDH were determined by RT-qPCR. Error bars represent the SEM of three independent experiments; ns indicates “no significance”; *P < 0.05, **P < 0.01, ***P < 0.001
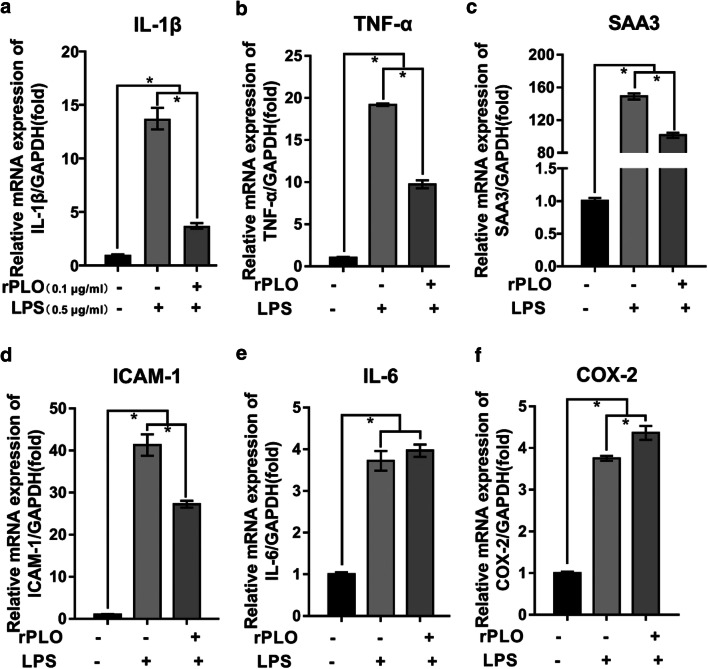
rPLO partially inhibited inflammatory mediator expression in the presence of LPS
As mentioned above, E. coli and T. pyogenes are two primary pathogenic microbes in postpartum endometritis. Therefore, we aimed to investigate the response of gESCs to these main virulence factors together. Intriguingly, rPLO at sublytic concentrations abrogated the upregulation of IL-1β, TNF-α, SAA3, and ICAM-1 mediated by LPS in gESCs (Fig. 3a–d). However, rPLO did not entirely repress induction of these genes by LPS, as we observed that their expression was significantly higher in the costimulation group than in the control group. However, the expression of IL-6 was similar in the costimulated group and the group stimulated with LPS alone (Fig. 3e). In contrast, the expression of COX-2 in the costimulated group was slightly elevated compared with that in the LPS-stimulated group (Fig. 3f).
Fig. 3.
The expression of relative inflammatory response genes under the costimulation of rPLO and LPS. a–f gESCs were challenged with control medium and medium containing 0.5 μg/ml LPS, or 0.1 μg/ml rPLO and 0.5 μg/ml LPS costimulation for 3 h. mRNA levels of targets normalized to the levels of GAPDH were determined by RT-qPCR. Error bars represent the SEM of three independent experiments, *P < 0.05
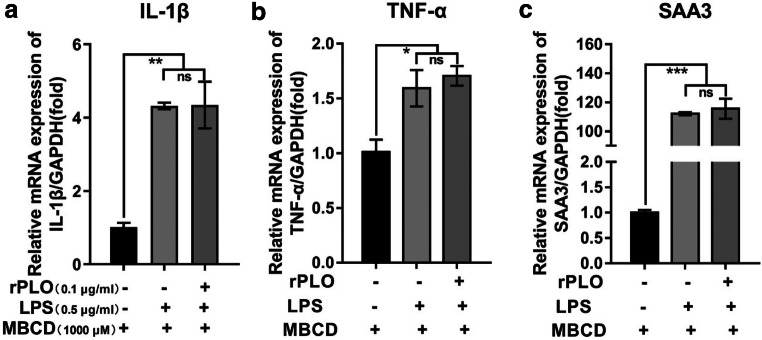
MBCD has been proven to be a specific inhibitor of PLO. Therefore, MBCD was used in this experiment to confirm the results described above. As previously described [15], MBCD was incubated with gESCs before LPS and rPLO stimulation. MBCD pretreatment rescued the downregulation of IL-1β, TNF-α, and SAA3 induced by rPLO (Fig. 4a–c). As shown for the first time, stimulation with rPLO and LPS together demonstrated that rPLO repressed innate immune response activation by LPS, and MBCD, a specific inhibitor of rPLO that bind the membrane, could abrogate this phenomenon, suggesting rPLO as an immunosuppressive factor to ensure multiple pathogen colonization within the host. Therefore, we speculate that the signaling pathway that regulates IL-1β was directly or indirectly by rPLO.
Fig. 4.
MBCD abrogated inhibitory effect of partly inflammatory response genes expression that rPLO induced. a–c gESCs were preincubated with MBCD for 12 h, after which cells were challenged with control medium and medium containing 0.5 μg/ml LPS, or 0.1 μg/ml rPLO and 0.5 μg/ml LPS costimulation for 3 h. mRNA levels of targets normalized to the levels of GAPDH were determined by RT-qPCR. Error bars represent the SEM of three independent experiments, *P < 0.05
rPLO repressed the NLRP3 inflammasome by autophagy
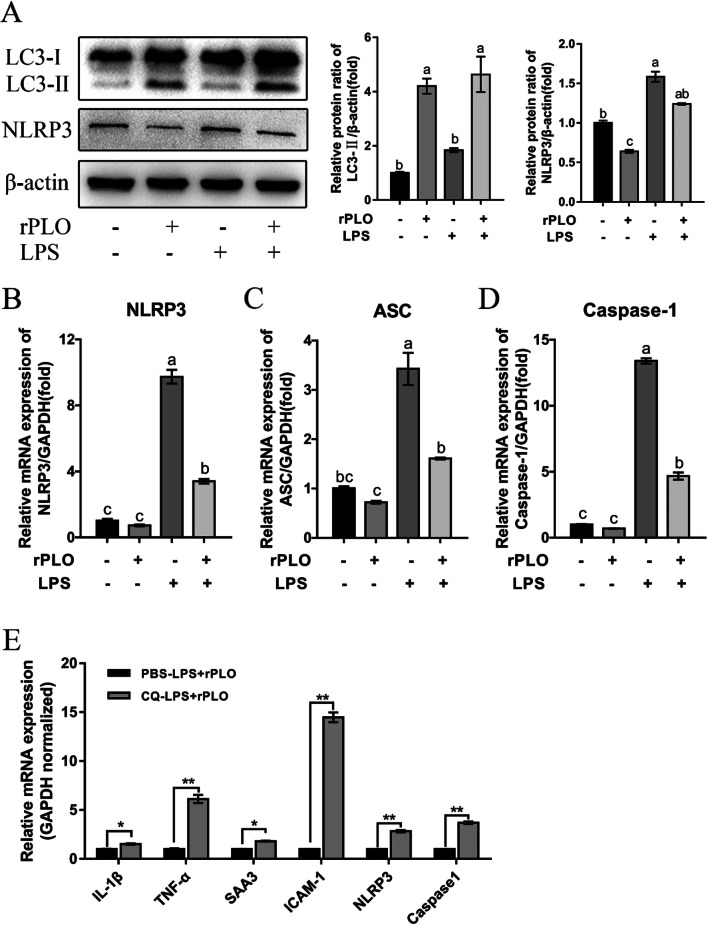
We then asked which pathway rPLO induces to regulate the effects of LPS on gESCs. As shown by the results of this study, we have demonstrated that the expression of IL-1β was inhibited (Fig. 3a); meanwhile, the NLRP3 inflammasome can be inhibited by autophagy, which then suppresses NLRP3 inflammasome-mediated IL-1β secretion in macrophages [29, 30]. Thus, we naturally speculated that rPLO suppresses IL-1β through inhibition of the NLRP3 inflammasome. In fact, NLRP3 was downregulated by rPLO (Fig. 5a–b), and the expression of ASC and Caspase1, components of multiprotein inflammasome complexes, was also downregulated at the mRNA level (Fig. 5b).
Fig. 5.
Autophagy and NLRP3 participated in regulating of rPLO induced inflammatory response genes expression. gESCs were challenged with control medium and medium containing 0.1 μg/ml rPLO, 0.5 μg/ml LPS, or costimulation for 3 h. a The cell samples were analyzed by Western blotting with anti-LC3, anti-NLRP3 and anti-β-actin (loading control) antibodies. The protein levels of target proteins relative to the protein level of β-actin were determined by densitometry. b–d mRNA levels of targets normalized to the levels of GAPDH were determined by RT-qPCR. P < 0.05. e gESCs were preincubated with CQ at 20 μM for 3 h, after which using rPLO and LPS costimulation for another 3 h, and relative mRNA levels were determined by RT-qPCR. Error bars represent the SEM of three independent experiments, *P < 0.05 and **P < 0.01
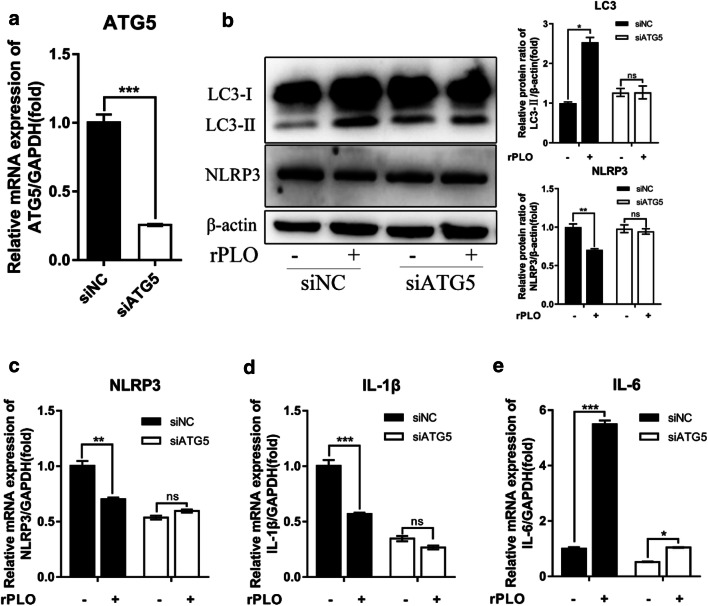
To further ascertain the key role of autophagy in this process, we chose chloroquine (CQ) as an inhibitor of autophagy. We pretreated gESCs with CQ at an appropriate dose and then costimulated with gESCs with rPLO and LPS as described above. These results were in line with our speculation, as the expression of IL-1β, TNF-α, SAA3, and ICAM-1 under CQ pretreatment was significantly upregulated (Fig. 5e). Meanwhile, to genetically prove the critical role of autophagy in this process, the siRNA technique was employed, and the expression of ATG5, which is involved in elongation and closure of the isolation membrane to form autophagosomes [43], was effectively suppressed (Fig. 6a). The upregulation of LC3-II and repression of NLRP3 occurred in an ATG5-dependent manner (Fig. 6b–d). Although IL-6 was still upregulated by rPLO in the siATG5-treated group, this effect was partially abrogated (Fig. 6e).
Fig. 6.
ATG5 participated in regulating of rPLO induced inflammatory response genes expression. gESCs were transfected with the small interfering RNAs (siRNAs) siNC and siATG5 and then challenged with control medium or medium containing 0.1 μg/ml rPLO for 3 h. a The efficiency of siATG5-mediated interference at the mRNA level was determined by RT-qPCR and normalized to the GAPDH level. b The cell samples were analyzed by Western blotting with anti-LC3, anti-NLRP3, and anti-β-actin (loading control) antibodies. The protein levels of target proteins relative to the protein level of β-actin were determined by densitometry. c–e. Targeted gene expression normalized to the GAPDH level were determined by RT-qPCR. Error bars represent the SEM of three independent experiments, *P < 0.05, **P < 0.01, and ***P < 0.001
Together, these results demonstrate the key role of the rPLO-triggered autophagy process in cell responses to rPLO repression of the NLRP3 inflammasome.
ATF6 is vital to the progression of rPLO-mediated inhibition of inflammatory mediators
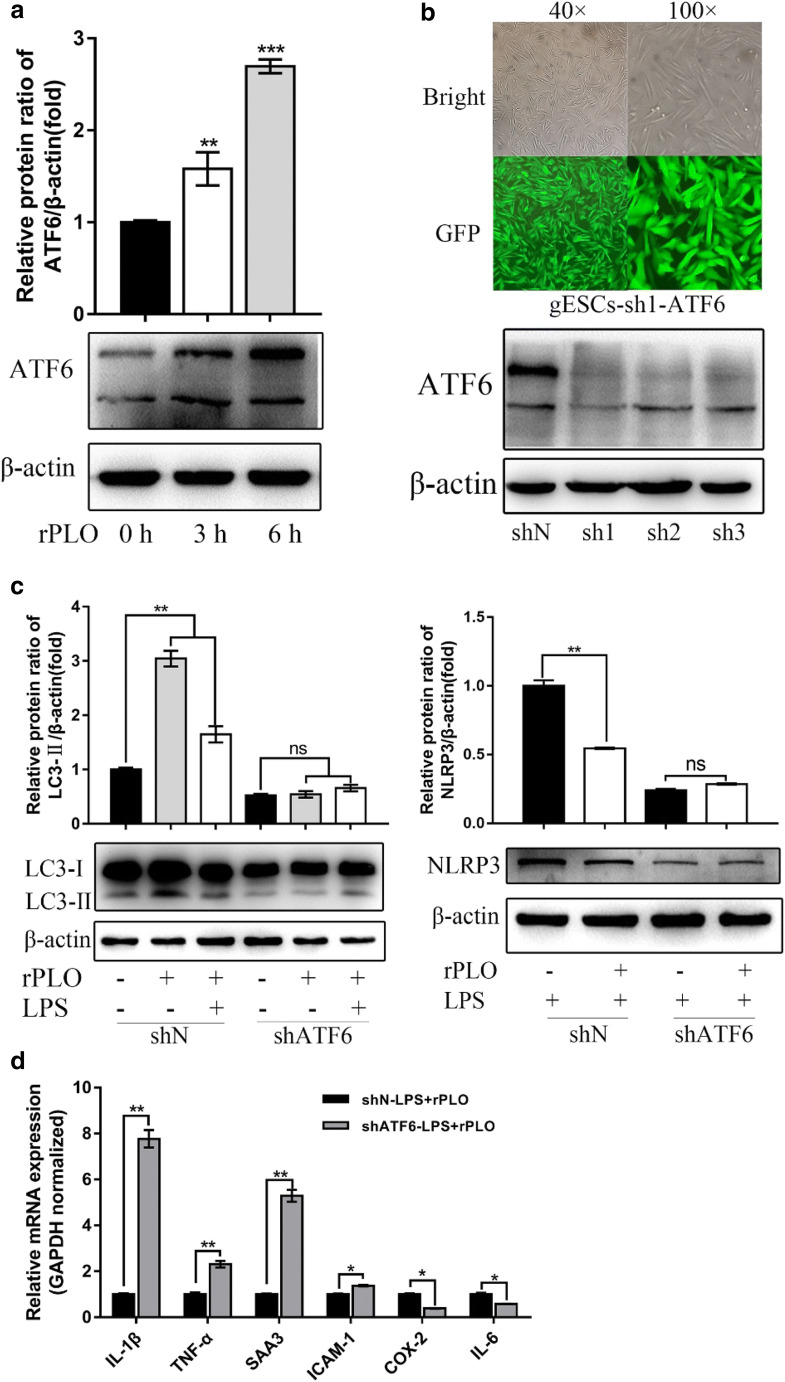
The ATF6 arm of the UPR is a vital ER stress transduction pathway that has previously been connected with pathogenic attack [44]. Therefore, it was naturally hypothesized in one study that ATF6 plays a role in protection against pore-forming toxins [44] and that ATF6 regulates the cellular response during rPLO attack. To test this hypothesis, we first investigated whether ATF6 was activated by rPLO. As shown, significant upregulation of ATF6 was observed in the group-administered rPLO administration (Fig. 7a). Cell lines expressing scramble control (shN) and short hairpin ATF6 (shATF6) with a GFP protein flag were constructed (Fig. 7b), and the efficiency of knockdown in the cell lines was determined by Western blotting (Fig. 7b). In ATF6 KD cells, LC3-II was not upregulated upon rPLO or rPLO and LPS stimulation compared to that in unstimulated cells (Fig. 7b). In contrast, LC3-II was significantly elevated in shN-expressing cells after the same treatment protocol (Fig. 7b). To further characterize the role of ATF6 in this process, we infected shN- and shATF6-expressing cells with LPS or LPS and rPLO together. In parallel with the change in LC3-II, rPLO-induced NLRP3 downregulation was dependent on ATF6 (Fig. 7c), and the result in shN cells was consistent with the data in Fig. 5a. Expression of the genes whose repression is shown in Fig. 3a–d displayed a similar tendency to be increased in shATF6-expressing cells (Fig. 7d), except IL-6 and COX-2, which were previously examined in this study and whose expression could not be inhibited by rPLO (Fig. 7d). Thus, these data suggest that ATF6 is essential for rPLO-mediated regulation of the related cell response by inhibition of autophagy and NLRP3 inflammasome activation.
Fig. 7.
ATF6 participated in regulating of rPLO induced inflammatory response genes expression. a gESCs were challenged with control medium and medium containing 0.1 μg/ml rPLO for 0 h, 3 h, and 6 h, and then the cells were harvested for Western blotting analysis with anti-ATF6 and anti-β-actin (loading control) antibodies. ATF6 levels relative to β-actin levels were determined by densitometry. b Lentivirus vector encoding ATF6 shRNA (pCD-513B-U6-shATF6) was constructed and transfected into HEK 293T cells. Then, the packaged shATF6 lentivirus was used to transfect gESCs after replacement of the complete culture medium with medium containing supplemental polybrene at 8mg/ml. Twelve hours later, complete culture medium was used to culture the cells for 48 h. As shown above, the cells were observed by inverted fluorescence microscopy; then, the cells were harvested for Western blotting to determine the efficiency of the lentivirus vector. C. gESCs-shN (native vector) and gESCs-shATF6 were treated with rPLO and LPS for 3 h, and then the cells were harvested for RT-qPCR. mRNA levels of target genes were normalized to those of GAPDH. Error bars represent the SEM of three independent experiments, *P < 0.05, **P < 0.01, and ***P < 0.001
Discussion
The virulence of most bacterial relies on secreted effectors that arose from the long evolutionary fight between host and pathogen, which modulate eukaryotic cell signal transduction. These effectors employ sophisticated strategies to manipulate host cellular signaling networks and thereby elude host defenses [45]. Mounting evidence has shown that PLO is a primary virulence effector during T. pyogenes infection. In addition, expression of the plo gene was widespread (100%) in T. pyogenes isolated from different domestic animals [8], which generates much interest in exploring this pathogenic mechanism of PLO. Because T. pyogenes is a common pathogenic bacterium of endometritis [6] and stromal cells are highly sensitive to PLO [14], endometrial stromal cells were naturally used in this study.
Here, we constructed a recombinant plasmid containing the full-length gene encoding mature PLO and used it to express and purify the rPLO protein. As indicated, purified rPLO induced SRBCs hemolysis in a concentration-dependent manner, and cholesterol preincubation with rPLO treatment or MBCD (methyl-β-cyclodextrin) preincubation with SRBCs inhibited its hemolytic activity. Although purified rPLO did not upregulate numerous cytokines, which is in line with studies demonstrating stromal and epithelial cell supernatants did not accumulate detectable amounts of cytokines during rPLO treatment [14], interestingly, the expression of cytokines induced by LPS was inhibited by rPLO. The large majority of research on this topic has found that multiple confounding postpartum conditions make it difficult to disentangle the mechanisms linking fertility with endometritis, and a model of endometritis using E. coli and T. pyogenes was developed [46]. However, as of yet, whether the two virulence factors of pathogen mentioned above engage in synergy or antagonism remains unknown. LPS is recognized by the pattern recognition receptor TLR4, after which a cellular signaling pathway is activated to induce IL-1β and other cytokine secretion [47]. Therefore, costimulation with pathogenic LPS was used in our study, and we suggest that rPLO inhibits IL-1β, TNF-α, SAA3, and ICAM-1 expression induced by LPS. We noted that delivery of OspF, an effector protein of the Shigella type III secretion system (TTSS) into host cells, modulated host signaling pathways and reduced cytokine secretion by dephosphorylating cellular mitogen-activated protein kinases (MAPKs) [48]. Drawing parallel the model of the immune preventive mechanism of OspF, it is plausible to assume that rPLO represses some signaling pathways to exert its virulence. A recent study suggested that autophagy is activated and that the NLRP3 inflammasome is inhibited as a protective mechanism in experimental autoimmune encephalomyelitis [30], which is in line with our findings that rPLO activates autophagy and represses the NLRP3 inflammasome at the mRNA and protein levels. Here, we used the autophagy inhibitor CQ, which blocks the fusion of autophagosomes and lysosomes to inhibit the autophagic process [49], to demonstrate that autophagy is employed by the effect of rPLO in inhibiting NLRP3 inflammasome activation and cytokine expression. This observation was not unexpected given that SdhA, a critical effector for Legionella pneumophila intracellular growth that functions to prevent host cell death, suppresses host innate immune detection by inhibiting AIM2 inflammasome activation and IL-1β secretion [50].
As highlighted in previous reports, a series of signaling pathways [28], such as the phosphorylation of ERK and p38 and autophagy, are activated by PLO. However, it is not yet clear whether the ERs-UPR axis, which affects the cytotoxic activity of LPS [51–53], is activated during rPLO stimulation of cells. In this study, we first demonstrate that ATF6, one pathway in the ERs-UPR axis, participates in regulating the function of PLO. Importantly, not only was ATF6 activated during rPLO stimulation but also rPLO-induced autophagy was affected in the ATF6 knockdown (KD) cell line. In our experiment, ATF6 was used as a regulator to control rPLO-induced autophagy; meanwhile, the NLRP3 repressive function of rPLO was also inhibited in the ATF6-KD cell line, and relative cytokine and gene expression profiles were also evidence of the effects of ATF6. Indeed, the link between the UPR and autophagy has been partly investigated [54], and the UPR can regulate autophagy through multiple pathways. Considering the change in LC3-II expression between the shN- and shATF6-expressing groups, ATF6 could influence one or all of processes in autophagy, including autophagosome formation, fusion of the autophagosome with the endosome or lysosome, and lysis of the autophagosome inner membrane. This observation is not unexpected given that the ERs-UPR is a conservative cellular process; however, ATF6 has emerged as a regulator of cell recognition of rPLO or signal transduction only in the pathogen invasion process.
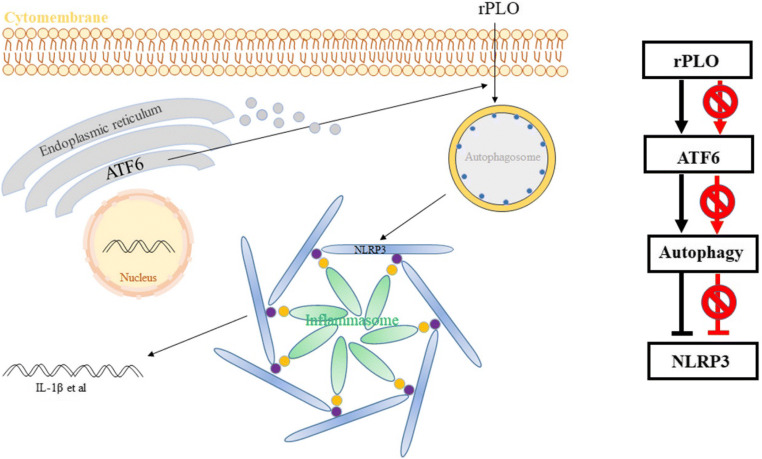
In conclusion, our results have characterized autophagy and the regulatory effect of ATF6 on the mechanism of PLO virulence in infected cells (Fig. 8). While PLO could employ cellular autophagy, we first demonstrated that autophagy and ATF6 jointly controlled cytokine and gene expression under costimulation with rPLO and LPS. Thus, these findings broaden our understanding of T. pyogenes pathogenesis and provide new insights into the mechanisms of rPLO-mediated infection of endometrial stromal cells. However, the mechanism by which PLO triggers autophagy and the mechanisms by which other signaling pathways participate in cellular reactions to PLO remain poorly understood. Fortunately, our team is engaged in much work.
Fig. 8.
Schematic model delineating the pathway of the responsiveness of the cells to rPLO regulated by ATF6 and autophagy
Supplementary information
Construction of the recombinant vector and purification of the fusion protein. A. The gene fragment encoding mature T. pyogenes PLO was cloned by PCR. B. The PLO gene fragment was connected to the prokaryotic expression vector pET-32a (+). Lane 1 shows PLO gene amplification from recombinant vector pET-32a-PLO. Lane 2 shows the positive control with the T. pyogenes genome as the PCR template. Lane 3 shows the negative control. C. Purified protein was authenticated by Coomassie brilliant blue staining (DOCX 370 kb)
Acknowledgments
We sincerely thank Professor Wei Liu and Associate Professor Keqiong Tang for providing help when we designed the study.
Funding
This study was supported by National Natural Science Foundation of China, Grant Number: 31772817.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Maozhen Qi and Jianguo Liu contributed equally to this work.
References
- 1.Billington SJ, Jost BH, Cuevas WA, Bright KR, Songer JG. The Arcanobacterium (Actinomyces) pyogenes hemolysin, pyolysin, is a novel member of the thiol-activated cytolysin family. J Bacteriol. 1997;179(19):6100–6106. doi: 10.1128/jb.179.19.6100-6106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jost BH, Billington SJ. Arcanobacterium pyogenes: molecular pathogenesis of an animal opportunist. Anton Leeuw Int J G. 2005;88(2):87–102. doi: 10.1007/s10482-005-2316-5. [DOI] [PubMed] [Google Scholar]
- 3.Ribeiro MG, Risseti RM, Bolanos CAD, Caffaro KA, de Morais ACB, Lara GHB, Zamprogna TO, Paes AC, Listoni FJP, Franco MMJ. Trueperella pyogenes multispecies infections in domestic animals: a retrospective study of 144 cases (2002 to 2012) Vet Q. 2015;35(2):82–87. doi: 10.1080/01652176.2015.1022667. [DOI] [PubMed] [Google Scholar]
- 4.Brodzki P, Bochniarz M, Brodzki A, Wrona Z, Wawron W. Trueperella pyogenes and Escherichia coli as an etiological factor of endometritis in cows and the susceptibility of these bacteria to selected antibiotics. Pol J Vet Sci. 2014;17(4):657–664. doi: 10.2478/pjvs-2014-0096. [DOI] [PubMed] [Google Scholar]
- 5.Rogovskyy AS, Lawhon S, Kuczmanski K, Gillis DC, Wu J, Hurley H, Rogovska YV, Konganti K, Yang CY, Duncan K. Phenotypic and genotypic characteristics of Trueperella pyogenes isolated from ruminants. J Vet Diagn Investig. 2018;30(3):348–353. doi: 10.1177/1040638718762479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang ML, Liu MC, Xu J, An LG, Wang JF, Zhu YH. Uterine microbiota of dairy cows with clinical and subclinical endometritis. Front Microbiol. 2018;9:2691. doi: 10.3389/fmicb.2018.02691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheldon IM, Cronin J, Goetze L, Donofrio G, Schuberth HJ. Defining postpartum uterine disease and the mechanisms of infection and immunity in the female reproductive tract in cattle. Biol Reprod. 2009;81(6):1025–1032. doi: 10.1095/biolreprod.109.077370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Risseti RM, Zastempowska E, Twaruzek M, Lassa H, Pantoja JCF, de Vargas APC, Guerra ST, Bolanos CAD, de Paula CL, Alves AC, Colhado BS, Portilho FVR, Tasca C, Lara GHB, Ribeiro MG. Virulence markers associated with Trueperella pyogenes infections in livestock and companion animals. Lett Appl Microbiol. 2017;65(2):125–132. doi: 10.1111/lam.12757. [DOI] [PubMed] [Google Scholar]
- 9.Billington SJ, Songer JG, Jost BH. Molecular characterization of the pore-forming toxin, pyolysin, a major virulence determinant of Arcanobacterium pyogenes. Vet Microbiol. 2001;82(3):261–274. doi: 10.1016/S0378-1135(01)00373-X. [DOI] [PubMed] [Google Scholar]
- 10.Dong WL, Liu L, Odah KA, Atiah LA, Gao YH, Kong LC, Ma HX. Antimicrobial resistance and presence of virulence factor genes in Trueperella pyogenes isolated from pig lungs with pneumonia. Trop Anim Health Prod. 2019;51(7):2099–2103. doi: 10.1007/s11250-019-01916-z. [DOI] [PubMed] [Google Scholar]
- 11.Jost BH, Songer JG, Billington SJ. An Arcanobacterium (Actinomyces) pyogenes mutant deficient in production of the pore-forming cytolysin pyolysin has reduced virulence. Infect Immun. 1999;67(4):1723–1728. doi: 10.1016/S0928-8244(98)00155-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin S, Healey GD, Sheldon IM. Isoprenoids increase bovine endometrial stromal cell tolerance to the cholesterol-dependent cytolysin from Trueperella pyogenes. Biol Reprod. 2018;99(4):749–760. doi: 10.1093/biolre/ioy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang WL, Wang HL, Wang B, Zhang Y, Hu YH, Ma B, Wang JW. Replacing the 238th aspartic acid with an arginine impaired the oligomerization activity and inflammation-inducing property of pyolysin. Virulence. 2018;9(1):1112–1125. doi: 10.1080/21505594.2018.1491256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amos MR, Healey GD, Goldstone RJ, Mahan SM, Duvel A, Schuberth HJ, Sandra O, Zieger P, Dieuzy-Labaye I, Smith DGE, Sheldon IM. Differential endometrial cell sensitivity to a cholesterol-dependent cytolysin links Trueperella pyogenes to uterine disease in cattle. Biol Reprod. 2014;90(3):54. doi: 10.1095/biolreprod.113.115972. [DOI] [PubMed] [Google Scholar]
- 15.Griffin S, Preta G, Sheldon IM. Inhibiting mevalonate pathway enzymes increases stromal cell resilience to a cholesterol-dependent cytolysin. Sci Rep. 2017;7:17050. doi: 10.1038/s41598-017-17138-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNeela EA, Burke A, Neill DR, Baxter C, Fernandes VE, Ferreira D, Smeaton S, El-Rachkidy R, McLoughlin RM, Mori A, Moran B, Fitzgerald KA, Tschopp J, Petrilli V, Andrew PW, Kadioglu A, Lavelle EC. Pneumolysin activates the NLRP3 inflammasome and promotes proinflammatory cytokines independently of TLR4. PLoS Path. 2010;6(11):e1001191. doi: 10.1371/journal.ppat.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malley R, Henneke P, Morse SC, Cieslewicz MJ, Lipsitch M, Thompson CM, Kurt-Jones E, Paton JC, Wessels MR, Golenbock DT. Recognition of pneumolysin by toll-like receptor 4 confers resistance to pneumococcal infection. Proc Natl Acad Sci U S A. 2003;100(4):1966–1971. doi: 10.1073/pnas.0435928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun P, Sun N, Yin W, Sun Y, Fan K, Guo J, Khan A, He Y, Li H. Matrine inhibits IL-1β secretion in primary porcine alveolar macrophages through the MyD88/NF-κB pathway and NLRP3 inflammasome. Vet Res. 2019;50(1):53. doi: 10.1186/s13567-019-0671-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 20.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang DL, Kang R, Vanden Berghe T, Vandenabeele P, Kroemer G. The molecular machinery of regulated cell death. Cell Res. 2019;29(5):347–364. doi: 10.1038/s41422-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagy defends cells against invading group a streptococcus. Science. 2004;306(5698):1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 24.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281(16):11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 25.Py BF, Lipinski MM, Yuan JY. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy. 2007;3(2):117–125. doi: 10.4161/auto.3618. [DOI] [PubMed] [Google Scholar]
- 26.Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10(11):1215–U1103. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 27.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120(2):159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Preta G, Lotti V, Cronin JG, Sheldon IM. Protective role of the dynamin inhibitor Dynasore against the cholesterol-dependent cytolysin of Trueperella pyogenes. FASEB J. 2015;29(4):1516–1528. doi: 10.1096/fj.14-265207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang YP, Ka SM, Hsu WH, Chen A, Chao LK, Lin CC, Hsieh CC, Chen MC, Chiu HW, Ho CL, Chiu YC, Liu ML, Hua KF. Resveratrol inhibits NLRP3 inflammasome activation by preserving mitochondrial integrity and augmenting autophagy. J Cell Physiol. 2015;230(7):1567–1579. doi: 10.1002/jcp.24903. [DOI] [PubMed] [Google Scholar]
- 30.Shao BZ, Wei W, Ke P, Xu ZQ, Zhou JX, Liu C. Activating cannabinoid receptor 2 alleviates pathogenesis of experimental autoimmune encephalomyelitis via activation of autophagy and inhibiting NLRP3 inflammasome. CNS Neurosci Ther. 2014;20(12):1021–1028. doi: 10.1111/cns.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao BZ, Xu ZQ, Han BZ, Su DF, Liu C. NLRP3 inflammasome and its inhibitors: a review. Front Pharmacol. 2015;6:262. doi: 10.3389/fphar.2015.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140(6):821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 33.Williams A, Flavell RA, Eisenbarth SC. The role of NOD-like receptors in shaping adaptive immunity. Curr Opin Immunol. 2010;22(1):34–40. doi: 10.1016/j.coi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bettigole SE, Glimcher LH. Endoplasmic reticulum stress in immunity. Annu Rev Immunol. 2015;33:107–138. doi: 10.1146/annurev-immunol-032414-112116. [DOI] [PubMed] [Google Scholar]
- 35.Oakes SA, Papa FR. The role of endoplasmic reticulum stress in human pathology. Annu Rev Pathol-Mech. 2015;10:173–194. doi: 10.1146/annurev-pathol-012513-104649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang KZ, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454(7203):455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senft D, Ronai ZA. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci. 2015;40(3):141–148. doi: 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang YY, Wang AH, Wu QX, Sheng HX, Jin YP. Establishment and characteristics of immortal goat endometrial epithelial cells and stromal cells with hTERT. J Anim Vet Adv. 2010;9(21):2738–2747. doi: 10.3923/javaa.2010.2738.2747. [DOI] [Google Scholar]
- 39.Yang DQ, Jiang TT, Liu JG, Hong J, Lin PF, Chen HT, Zhou D, Tang KQ, Wang AH, Jin YP. Interferon-tau regulates prostaglandin release in goat endometrial stromal cells via JAB1-unfolded protein response pathway. Theriogenology. 2018;113:237–246. doi: 10.1016/j.theriogenology.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 40.Sheldon IM, Noakes DE, Rycroft AN, Pfeiffer DU, Dobson H. Influence of uterine bacterial contamination after parturition on ovarian dominant follicle selection and follicle growth and function in cattle. Reproduction. 2002;123(6):837–845. doi: 10.1530/rep.0.1230837. [DOI] [PubMed] [Google Scholar]
- 41.Yang B, Xue QH, Guo JN, Wang XP, Zhang YM, Guo KK, Li W, Chen SY, Xue TX, Qi XF, Wang JY. Autophagy induction by the pathogen receptor NECTIN4 and sustained autophagy contribute to peste des petits ruminants virus infectivity. Autophagy. 2020;16(5):842–861. doi: 10.1080/15548627.2019.1643184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen FL, Lin PF, Li X, Sun J, Zhang Z, Du EQ, Wang AH, Jin YP. Construction and expression of lentiviral vectors encoding recombinant mouse CREBZF in NIH 3T3 cells. Plasmid. 2014;76:24–31. doi: 10.1016/j.plasmid.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Leidal AM, Levine B, Debnath J. Autophagy and the cell biology of age-related disease. Nat Cell Biol. 2018;20(12):1338–1348. doi: 10.1038/s41556-018-0235-8. [DOI] [PubMed] [Google Scholar]
- 44.Bischof LJ, Kao CY, Los FCO, Gonzalez MR, Shen ZX, Briggs SP, van der Goot FG, Aroian RV. Activation of the unfolded protein response is required for defenses against bacterial pore-forming toxin in vivo. PLoS Path. 2008;4(10):e1000176. doi: 10.1371/journal.ppat.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui JX, Shao F. Biochemistry and cell signaling taught by bacterial effectors. Trends Biochem Sci. 2011;36(10):532–540. doi: 10.1016/j.tibs.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Piersanti RL, Zimpel R, Molinari PCC, Dickson MJ, Ma ZX, Jeong KC, Santos JEP, Sheldon IM, Bromfield JJ. A model of clinical endometritis in Holstein heifers using pathogenic Escherichia coli and Trueperella pyogenes. J Dairy Sci. 2019;102(3):2686–2697. doi: 10.3168/jds.2018-15595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taniguchi K, Karin M. NF-kappa B, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 48.Li HT, Xu H, Zhou Y, Zhang J, Long CZ, Li SQ, Chen S, Zhou JM, Shao F. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315(5814):1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- 49.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ge JN, Gong YN, Xu Y, Shao F. Preventing bacterial DNA release and absent in melanoma 2 inflammasome activation by a Legionella effector functioning in membrane trafficking. Proc Natl Acad Sci U S A. 2012;109(16):6193–6198. doi: 10.1073/pnas.1117490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang Q, Chen S, Ren WK, Liu G, Yao K, Wu GY, Yin YL. Escherichia coli aggravates endoplasmic reticulum stress and triggers CHOP-dependent apoptosis in weaned pigs. Amino Acids. 2017;49(12):2073–2082. doi: 10.1007/s00726-017-2492-4. [DOI] [PubMed] [Google Scholar]
- 52.Quan K, Li SC, Wang DD, Shi Y, Yang ZX, Song JP, Tian YL, Liu YJ, Fan ZY, Zhu W. Berberine attenuates macrophages infiltration in intracranial aneurysms potentially through FAK/Grp78/UPR Axis. Front Pharmacol. 2018;9:565. doi: 10.3389/fphar.2018.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang HS, Chen Y, Fan L, Xi QL, Wu GH, Li XX, Yuan TL, He SQ, Yu Y, Shao ML, Liu Y, Bai CG, Ling ZQ, Li M, Liu Y, Fang J. The endoplasmic reticulum stress sensor IRE1 alpha in intestinal epithelial cells is essential for protecting against colitis. J Biol Chem. 2015;290(24):15327–15336. doi: 10.1074/jbc.M114.633560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Appenzeller-Herzog C, Hall MN. Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol. 2012;22(5):274–282. doi: 10.1016/j.tcb.2012.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Construction of the recombinant vector and purification of the fusion protein. A. The gene fragment encoding mature T. pyogenes PLO was cloned by PCR. B. The PLO gene fragment was connected to the prokaryotic expression vector pET-32a (+). Lane 1 shows PLO gene amplification from recombinant vector pET-32a-PLO. Lane 2 shows the positive control with the T. pyogenes genome as the PCR template. Lane 3 shows the negative control. C. Purified protein was authenticated by Coomassie brilliant blue staining (DOCX 370 kb)