Abstract
Campylobacter spp. have been a predominant cause of bacterial foodborne gastroenteritis worldwide, causing substantial costs to public healthcare systems. This study aimed to assess the invasion and pro-inflammatory cytokine production capacity of Campylobacter coli strains isolated in Brazil. A total of 50 C. coli isolated from different sources in Brazil were analyzed for their capacity of invasion in Caco-2 and U-937 cell lines. The production of pro-inflammatory cytokines was quantitatively measured in response to C. coli. All the strains studied showed invasion percentage ≥ 40% in polarized Caco-2 cells. In U-937 cells assay, 35 of 50 C. coli strains studied showed invasion percentage ≥ 50%. A significant increase in IL-8 production by infected U-937 cells was observed for 17.5% of the C. coli isolates. The high percentages of invasion in Caco-2 and U-937 cells observed for all studied strains, plus the increased production of IL-8 by U-937 cells against some strains, highlighted the pathogenic potential of the C. coli studied and bring extremely relevant data since it has never been reported for strains isolated in Brazil and there are a few data for C. coli in the literature.
Keywords: Foodborne pathogens, Caco-2 and U-937 cell lines, Cytokines, Brazil
Introduction
Campylobacter has been a predominant cause of bacterial diarrheal disease in humans. According to Centers for Disease Control and Prevention (CDC), this bacterium causes an estimated 1.5 million illnesses each year in the USA being Campylobacter jejuni and Campylobacter coli the two main species reported in these cases [1–4]. However, in Brazil, the campylobacteriosis has been underreported and underdiagnosed, and there is a lack of sufficient data to access the incidence of this pathogen in the country [5–10].
Campylobacter species can be isolated from humans and animals, such as birds, mammals, reptiles, and, also from mollusks and protozoans. However, 50–80% of the reservoirs of this pathogen have been attributed to birds [11–13]. The transmission of this bacterial species occurs mainly via consumption and handling of improperly prepared poultry meat and consumption of unpasteurized milk [1, 14]. Campylobacteriosis is usually a self-limiting disease with symptoms such as fever, headache, abdominal cramps, and bloody diarrhea. However, in some cases, infections can lead to hospitalizations, post-infection sequelae, and death [15, 16].
Despite the high incidence of campylobacteriosis around the world, the knowledge of the mechanisms related to gut colonization and pathogenesis of C. coli species is more limited compared with that available for other foodborne pathogens [14, 17, 18]. Although the gut colonization is intrinsically related to the processes of adhesion and invasion to intestinal epithelium cells, this process is not completely clear for C. coli and the majority of the published studies were carried out with C. jejuni species [17, 19–21].
In general, human campylobacteriosis is a multifactorial process involving the interaction of bacteria to host epithelial cells and the response of the host immune system. Thus, in vitro models of human and animal cell culture and in vivo animal models have helped to clarify the mechanisms related to the infection process of emerging species such as C. coli [14].
In addition, studies have shown that Campylobacter is able to induce acute inflammatory enteritis [22–24] due to pro-inflammatory cytokine induction and subsequent recruitment of defense cells [25, 26]. In vitro experiments in human-derived cell lines [23, 26, 27] have shown that C. jejuni induced interleukin-8 (IL-8) which have also been found in stools of patients with campylobacteriosis [28, 29]. Moreover, secretion of IL-6 and tumor necrosis factor alpha (TNF-α) induced by C. jejuni and C. coli was also reported [23, 24, 30, 31].
Therefore, studies that can help to characterize and elucidate the pathogenesis of C. coli strains are of utmost importance since there is a paucity of data on this globally important pathogen and due to the fact that the majority of studies were performed with C. jejuni species.
The aims of this study were to assess the capacity of invasion to human colorectal adenocarcinoma cells (Caco-2) and human macrophages (U-937) of C. coli strains isolated in Brazil. Furthermore, the pro-inflammatory cytokine production was verified.
Material and methods
Bacterial strains
The 50 C. coli strains studied were selected from the collection of the Campylobacter Reference Laboratory of the Oswald Cruz Institute of Rio de Janeiro (FIOCRUZ) as described by Gomes et al [10]. These strains were isolated from human feces (12 strains), animals (15 strains), the environment (15 strains), and chicken meat (8 strains) from some cities in the Rio de Janeiro, São Paulo, and Minas Gerais States in the southeast region of Brazil between 1995 and 2011 (Table 1). Moreover, the Salmonella typhimurium ATCC 14028 and Campylobacter jejuni ATCC 33291 strains were used as controls in cell experiments.
Table 1.
Source, state, year, and IL-8 production induced of the 50 Campylobacter coli strains studied
| Strains | Source | State | Year | IL-8 Concentration in U-937 (pg/mL) | IL-8 Concentration in Caco-2 (pg/mL) |
|---|---|---|---|---|---|
| CCAMP 771 | Sewage | RJ | 1995 | 2719.48 | - |
| CCAMP 840 | Sewage | RJ | 1995 | - | - |
| CCAMP 821 | Monkey | RJ | 1995 | - | - |
| CCAMP 820 | Monkey | RJ | 1995 | - | 0.00 |
| CCAMP 787 | Sewage | RJ | 1996 | - | - |
| CCAMP 819 | Sewage | RJ | 1996 | 3004.62 | - |
| CCAMP 765 | Sewage | RJ | 1996 | 2954.05 | - |
| CCAMP 767 | Sewage | RJ | 1996 | - | - |
| CCAMP 761 | Sewage | RJ | 1996 | - | 4.57 |
| CCAMP 764 | Sewage | RJ | 1996 | - | 0.00 |
| CCAMP 791 | Monkey | RJ | 1997 | - | - |
| CCAMP 818 | Sewage | RJ | 1997 | 24.48 | 9.41 |
| CCAMP 775 | Sewage | RJ | 1997 | - | - |
| CCAMP 495 | Human | RJ | 1998 | - | - |
| CCAMP 494 | Human | RJ | 1998 | - | - |
| CCAMP 490 | Human | RJ | 1998 | - | - |
| CCAMP 841 | Monkey | RJ | 1998 | 2490.68 | 0.00 |
| CCAMP 502 | Human | RJ | 1999 | - | 0.07 |
| CCAMP 498 | Human | RJ | 1999 | - | 0.00 |
| CCAMP 975 | Monkey | RJ | 1999 | 83.66 | - |
| CCAMP 726 | Monkey | RJ | 2000 | - | 0.00 |
| CCAMP 503 | Human | RJ | 2000 | - | - |
| CCAMP 834 | Water | RJ | 2000 | - | 0.23 |
| CCAMP 595 | Human | RJ | 2001 | - | - |
| CCAMP 667 | Monkey | RJ | 2002 | 2427.20 | 1.75 |
| Cc 01 | Human | SP | 2002 | 4.51 | - |
| Cc 10 | Human | SP | 2003 | 3504.63 | 0.00 |
| Cc 04 | Human | SP | 2003 | 21.60 | 0.23 |
| CCAMP 182 | Monkey | RJ | 2003 | - | - |
| CCAMP 165 | Monkey | RJ | 2003 | 2185.62 | 0.00 |
| Cc03 | Human | SP | 2003 | 1800.63 | - |
| Cc05 | Human | SP | 2003 | - | - |
| CCAMP 446 | Monkey | RJ | 2004 | 4023.50 | - |
| CCAMP 463 | Potable Water | MG | 2004 | 17.57 | 4.51 |
| CCAMP 469 | Potable Water | MG | 2004 | 11.76 | - |
| CCAMP 464 | Potable Water | MG | 2004 | - | - |
| CCAMP 394 | Monkey | RJ | 2004 | - | - |
| CCAMP 769 | Sewage | MG | 2004 | 27.29 | 0.00 |
| CCAMP 392 | Monkey | RJ | 2007 | - | 0.31 |
| CCAMP 1010 | Monkey | RJ | 2007 | 2863.22 | 0.12 |
| CCAMP 1000 | Monkey | RJ | 2007 | 23.50 | - |
| CCAMP 1117 | Monkey | RJ | 2009 | - | 0.90 |
| CCAMP 1062 | Chicken wing | RJ | 2010 | 22.60 | 0.11 |
| CCAMP 1067 | Chicken liver | RJ | 2010 | - | - |
| CCAMP 1063 | Chicken gizzard | RJ | 2010 | - | - |
| CCAMP 1064 | Chicken wing | RJ | 2010 | 35.64 | 0.11 |
| CCAMP 1066 | Chicken liver | RJ | 2010 | 25.06 | - |
| CCAMP 1071 | Chicken wing | RJ | 2011 | 22.36 | 0.00 |
| CCAMP 1075 | Chicken liver | RJ | 2011 | 22.44 | - |
| CCAMP 1073 | Chicken wing | RJ | 2011 | 2279.46 | - |
MG Minas Gerais, RJ Rio de Janeiro, RS Rio Grande do Sul, SP São Paulo
The genus and species confirmation were performed by PCR to detect the specific regions of the 16S rRNA, ceuE, and mapA genes as described by Gomes et al [10].
Cell cultures
Caco-2 cells were grown in Dulbecco’s modified Eagle medium (DMEM) (Sigma-Aldrich, Arklow, Ireland) with 10% fetal bovine serum (Life Technologies, CA, USA) and 1% antibiotic/antimycotic (Life Technologies) in 75-cm2 tissue culture flasks at 37 °C in a 5% CO2 atmosphere until cell layers were confluent. The cells were seeded into 12-well tissue culture plates at a concentration of 1x105 cells per well, and plates were incubated in a 5% CO2 atmosphere at 37 °C for 12 days to provide the polarization of the cells [32, 33].
The U-937 were grown in Roswell Park Memorial Institute medium (RPMI) (Sigma-Aldrich) supplemented with 10% bovine fetal serum (Life Technologies) in a 5% CO2 atmosphere at 37 °C until confluent. The cells were seeded into 24-well tissue culture plates at a concentration of 1 × 105 cells per well, and 10 nM of phorbol myristate acetate (PMA; Sigma) was added in the medium in order to differentiate monocytes in macrophages. The plates were incubated in a 5% CO2 atmosphere at 37 °C for 24 h. Thereafter, the medium was changed, and the plates were incubated for additional 24 h without PMA in a 5% CO2 atmosphere at 37 °C [33].
Caco-2 invasion assay
The C. coli strains were grown at 42 °C overnight and the inoculum was obtained as described by Gomes et al [10]. Bacterial growth was adjusted to an OD600 = 0.1 which correspond to approximately 8 log10 CFU/mL [20], and an aliquot was plated to verify the number of viable cells. Subsequently, the bacterial growth was centrifuged at 8000×g for 5 min and resuspended in 1-mL DMEM cell culture medium without antibiotic and/or fetal bovine serum. The OD600 = 0.2 was used to S. typhimurium ATCC 14028 control strain [34].
Each bacterial strain was tested in triplicate, and wells without bacteria were used as blank controls. The multiplicity of infection (MOI) of 100:1 was used to infect the Caco-2 cells with C. coli, and the bacterium-cell interactions occurred for 90 min in a 5% CO2 atmosphere at 37 °C, as previously described by [35–40], with modifications. After incubation, the wells were washed with phosphate buffered saline (PBS 1X) and treated with 1 mL of the DMEM containing 30 μg/mL of gentamicin. The plates were incubated additional 90 min; the supernatant was seeded to ensure antibiotic activity and subsequently washed once with PBS 1X. After that, 1 mL of the DMEM with no antibiotics and no fetal bovine serum was added to each well that was incubated for 3 h in a 5% CO2 atmosphere at 37 °C. The cells were washed with PBS 1X followed by a cell lysis with a 1% Triton X-100 solution for 5 min.
The CFU/mL were determined by serial dilutions in 0.8% saline solution and plating on Muller Hinton supplemented with 5% sheep’s blood. The percentages of invasion in Caco-2 cells were determined by dividing the Log CFU/mL value after the cell lysis by log 107 (initial inoculum), and the results were multiplied by 100.
U-937 invasion assay
The inoculum was obtained according to what is described in item 2.3. After adjusting the optical density, an aliquot was plated to verify the number of viable cells, and 1 mL of stationary phase cells was centrifuged at 8000×g for 5 min and subsequently washed with PBS 1X for three times. The C. coli strains were opsonized with 20% of mouse serum (Sigma-Aldrich) at 37 °C for 15 min. After that, cells were centrifuged at 8000×g for 5 min and subsequently resuspended in 1 mL of RPMI cell culture medium without antibiotic and/or fetal bovine serum.
Each bacterial strain was tested in triplicate, and wells without bacteria were used as blank controls. The MOI of 100:1 was used to infect the U-937 cells with C. coli, and the bacterium-cell interactions occurred for 30 min in a 5% CO2 atmosphere at 37 °C, as previously described by [40, 41], with modifications. After incubation, the wells were washed with PBS 1X and treated with 1 mL of the RPMI containing 30 μg/mL of gentamicin. The plates were incubated additional 90 min; the supernatant was seeded to ensure antibiotic activity and subsequently washed with PBS 1X. After that, 1 mL of the RPMI with no antibiotics and no fetal bovine serum was added to each well that was incubated for 3 h in a 5% CO2 atmosphere at 37 °C. The cells were washed with PBS 1X followed by a cell lysis with a 1% Triton X-100 solution for 5 min.
The CFU/mL were determined by serial dilutions in 0.8% saline solution and plating on Muller Hinton supplemented with 5% sheep’s blood. The percentages of invasion in U-937 cells were determined according to what is described in item 2.3.
Quantification of pro-inflammatory cytokine production in Caco-2 and U-937 cells
The production of pro-inflammatory cytokines IL-8, IL-1β, IL-6, IL-10, TNF, and IL-12p70 were quantitatively measured by the CBA Human Inflammatory Cytokines Kit (BD Biosciences, CA, USA) in response to C. coli. A total of 45 C. coli strains isolates from different year and sources were selected for this assay of which 24 C. coli strains showed different invasion percentages in U-937 cells and 21 C. coli strains showed different invasion percentages in Caco-2 cells.
The plates were prepared as described in item 2.2, and the inoculum was obtained according to what is described in item 2.3. To start the assays, each bacterial suspension was added to each well of plate with a MOI ratio of 100:1. Then, the plates were incubated in a 5% CO2 atmosphere at 37 °C for 90 min for the bacterium-cell interactions [40, 41]. After the incubation, the supernatant of each well was collected, transferred to a properly labeled sterile tube, and stored in a freezer −80 °C until the quantification of pro-inflammatory cytokine assay.
In the assay day, the supernatant was processed using CBA Human Inflammatory Cytokines Kit according to the manufacturer’s instructions and subsequently submitted to the BD LSRFortessa-1 flow cytometer.
Results
Caco-2 invasion assay
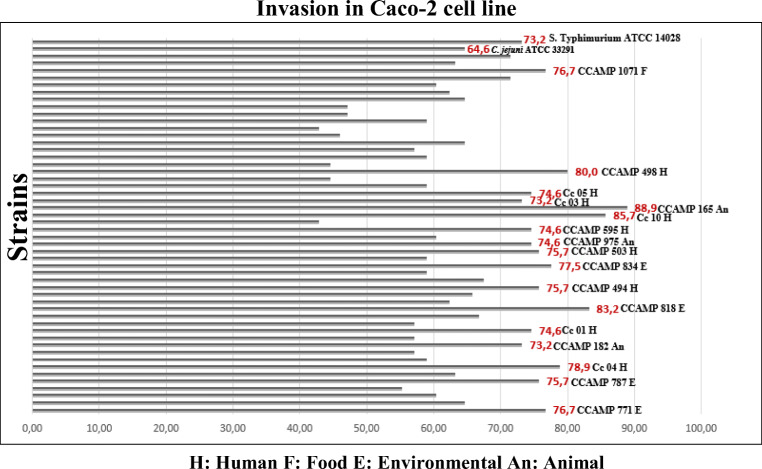
All the 50 C. coli strains studied (Table 1) showed invasion percentages higher than 40% in Caco-2 polarized cells (Fig. 1). Seventeen C. coli strains including nine isolated from humans (Cc 10 (85.7%), CCAMP 498 (80%), Cc 04 (78.9%), CCAMP 494 (75.7%), CCAMP 503 (75.7%), Cc 01 (74.6%), Cc 05 (74.6%), CCAMP 595 (74.6%), and Cc 03 (73.2%)), four isolated from the environmental (CCAMP 818 (83.2%), CCAMP 834 (77.5%), CCAMP 771 (76.7%), and CCAMP 787 (75.7%)), three isolated from animals (CCAMP 165 (88.9%), CCAMP 975 (74.6%), and CCAMP 182 (73.2%)), and one isolated from food (CCAMP 1071 (76.7%)) showed invasion percentages greater than or equal to the percentage showed by S. typhimurium ATCC 14028 (73.2%). The CCAMP 165 isolated from animal showed the highest invasion percentage (88.9%) when compared with the other C. coli strains studied (Fig. 1). Furthermore, 50% of the studied strains showed percentages of invasion greater than or equal to the percentage presented by C. jejuni ATCC 33291 (64.6%).
Fig. 1.
Invasion percentages of 50 Campylobacter coli strains studied and the controls (Campylobacter jejuni ATCC 33291 and Salmonella Typhimurium ATCC 14028) in Caco-2 epithelial cells
U-937 invasion assay
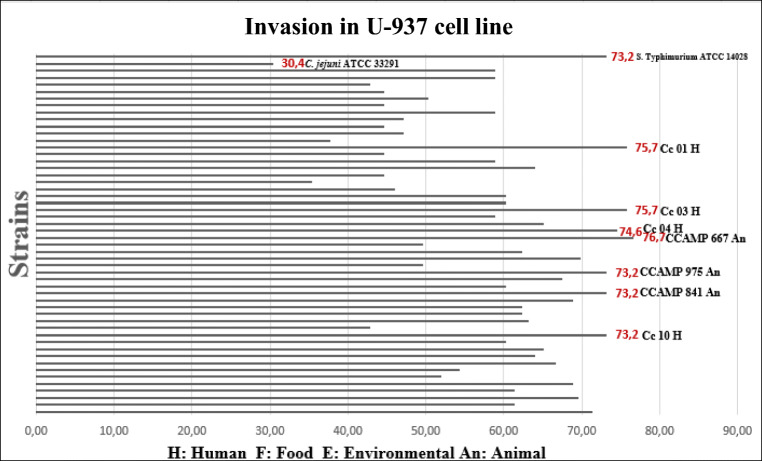
Thirty-six of 50 C. coli strains studied (Table 1) plus the S. typhimurium ATCC 14028 control strain showed the invasion percentages greater than or equal to 50% (Fig. 2). The CCAMP 667 strain isolated from animal showed the highest invasion percentage (76.7%) when compared with the others 49 C. coli strains studied. Seven C. coli strains including four isolated from human (Cc 03 (75.7%), Cc 01 (75.7%), Cc 04 (74.6%), and Cc 10 (73.2%)) and three isolates from animal (CCAMP 667 (76.7%), CCAMP 975 (73.2%), and CCAMP 841 (73.2%)) showed percentages of invasion greater than or equal to the percentage showed by S. typhimurium ATCC 14028 (73.2%). All the strains studied showed percentages of invasion greater than C. jejuni ATCC 33291 (30.4 %) (Fig. 2).
Fig. 2.
Invasion percentages of 50 Campylobacter coli strains studied and the controls (Campylobacter jejuni ATCC 33291 and Salmonella typhimurium ATCC 14028) in U-937 macrophages
Quantification of pro-inflammatory cytokine production in CaCo-2 and U-937 cells
The quantitative analysis of pro-inflammatory cytokine production in U-937 cells in response to the presence of some C. coli strains after 90 min of bacterium-cell interactions showed high concentrations of IL-8 production (Table 1). The CCAMP 446 strain isolated from animal induced the highest IL-8 production in U-937 cells (4023.00 pg/mL). The CCAMP 819 strain isolated from the environmental and the strain Cc 10 isolated from human induced the IL-8 production in concentrations more than 3000.00 pg/mL. In contrast, quantitative analysis of pro-inflammatory cytokine production in Caco-2 cells after 90 min of bacterium-cell interactions did not present significant values (Table 1).
The IL-12p70, TNF, IL-10, IL-6, and IL-1β cytokines analyzed against U-937, and Caco-2 cells did not show expressive values in response to the presence of some C.coli strains after 90 min of bacterium-cell interactions (data not shown).
Discussion
Campylobacter spp. have been a predominant cause of bacterial foodborne gastroenteritis worldwide, causing substantial costs to public healthcare systems [3, 4, 42]. Despite this, the exact mechanisms of C. coli infections are still not completely understood [14, 17, 18]. The gut colonization is intrinsically related to the processes of adhesion and invasion in intestinal epithelium, and the clinical manifestations of campylobacteriosis in humans have been directly linked to the virulence potential of different isolates together with the immunological predisposition of the host [43–48].
In the present study, we access the invasion and the inflammatory effect of C. coli strains, isolated from different sources in Brazil, in phagocytic and in intestinal human cells. Several models have been used to study the ability of Campylobacter to colonize and invade the gastrointestinal tract [30, 43]. Among them, Caco-2 epithelial cell model is the most commonly used being useful to mimic the behavior of Campylobacter in human gut [49, 50].
The invasion percentages in Caco-2 U-937 cells were greater than or equal to the percentage showed by S. typhimurium ATCC (73.2%), which has its invasion and pathogenicity mechanism extensively studied [51–54]. Specifically, the results herein obtained for C. coli strains isolated from humans suggested that these strains have a higher invasion capacity compared with non-clinical strains. Furthermore, 50% of the strains studied showed invasion percentages higher than or equal to the C. jejuni ATCC 33291 (64.6%) which has been often used in studies involving Campylobacter [55, 56].
To the generation of appropriate defense responses from the host, leukocyte recruitment to inflammatory sites is essential [57, 58]. In the invasion assay in U-937 human macrophage, 35 of 50 C. coli strains studied showed an invasion percentage higher than or equal to 50% what demonstrated a high survivability of the strains analyzed against human host defense cells (Fig. 2).
Of note, seven C. coli strains being four isolated from humans and three isolated from animals showed invasion percentages higher than or equal to the S. typhimurium ATCC 14028 (73.2%); in other words, strains isolated from clinical sources showed higher invasion percentages in human macrophages compared with strains isolated from food and the environment (Fig. 2). All the strains studied showed invasion percentages higher than the one presented by C. jejuni ATCC 33291 (30.4 %).
Although the molecular mechanisms of Campylobacter are poorly understood when compared with other foodborne pathogens, studies have shown that there are correlations between the severity of clinical symptoms and the invasiveness and survival capability of Campylobacter spp. [59–61]. Moreover, a great variability in invasion and survival capacity among Campylobacter isolates has been previously described [62–64].
It was shown that the invasion and survival capacity in both Caco-2 epithelial cells and U-937 phagocytic cells was strain-dependent [65–67]. Interestingly, the high invasion percentages observed in the present study was different from published data by Pan and colleagues and Zheng and colleagues [61, 65] that observed a remarkably low rate of adherence and invasion which reinforced the pathogenic potential of the C. coli herein studied and brings extremely relevant data that has never been reported for strains isolated in Brazil and contributed to the worldwide information once there is a few data for C. coli in the literature [27, 62–65, 67]. In previous studies of our research group, high growth and survival rates were also observed for the majority of those strains under different stress conditions such as tolerance to temperature variations, survivability in 7.5% of NaCl, survivability under acid, and oxidative stresses [10].
The high levels of pro-inflammatory cytokines in the early stage of Campylobacter infection suggest the development of an inflammatory cascade that is responsible for diarrhea during intestinal infection [68]. Adhesion and invasion to intestinal cells by C. jejuni induce secretion of IL-8, IL-1, IL-6, interferon-gamma (IFN-γ), TNF-α, and IL-4 among other cytokines that promote the recruitment of macrophages and neutrophils [25, 26]. In addition, studies have demonstrated that Campylobacter and Lactobacillus acidophilus interaction with Caco-2 cells induced the production of the pro-inflammatory cytokine interleukin IL-8, but this response was strain-dependent which could justify the absence of expressive values observed in this study [69–71].
The high concentration of IL-8 production observed in U-937 cells exposed for 90 min to the presence of C. coli strains selected from this study (Cc 03, Cc10, CCAMP 165, CCAMP 446, CCAMP 667, CCAMP 765, CCAMP 771, CCAMP 819, CCAMP 841, CCAMP 1010, and CCAMP 1073) demonstrated the immunogenic potential of some strains studied (Table 1). In addition, the results obtained in the pro-inflammatory cytokines analyses suggest that some studied C. coli, despite having high invasion percentages in both Caco-2 and U-937 cells, did not induce the production of the analyzed cytokines, which is an interesting fact as these strains have a previously demonstrated pathogenic potential, but they do not cause an immune response in the host.
It is important to mention that previous studies showed that C. jejuni can invade and evade cells without causing necrosis, apoptosis, cell lysis, and inflammation in the intestinal wall [72]. Thus, further studies to assess gene expression should be carried out in order to evidence more clearly the pathogenic potential of the studied strains.
In conclusion, the high percentages of invasion in Caco-2 and U-937 cells observed for all studied strains, plus the increased production of IL-8 by U-937 cells against some strains highlighted the pathogenic potential of the C. coli studied, a data that has not been reported before.
Funding
This work was financially supported by the São Paulo Research Foundation (FAPESP) (Process 2016/24716-3) and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001". During the course of this work, Gomes, C. N. was supported by FAPESP (Proc. 2015/23408-0), and Falcão, J. P. received a productive fellowship (Proc. CNPq 304399/2018-3) from the National Council for Scientific and Technological Development (CNPq).
Declaration
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Silva J, Leite D, Fernandes M, Mena C, Gibbs PA, Teixeira P. Campylobacter spp. as a foodborne pathogen: a review. Front Microbiol. 2011;2:1–12. doi: 10.3389/fmicb.2011.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaakoush ON, Castaño-Rodríguez N, Mitchell HM, Man SM. Global epidemiology of Campylobacter infection. Clin Microbiol Rev. 2015;28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control) The European Union One Health 2018 Zoonoses Report. EFSA J. 2019;17(12):5926. doi: 10.2903/j.efsa.2019.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC- Centers for Disease Control and Prevention (2019) Preliminary incidence and trends of infections with pathogens transmitted commonly through food — Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2015–2018. [DOI] [PMC free article] [PubMed]
- 5.Aquino MH, Pacheco AP, Ferreira MC, Tibana A. Frequency of isolation and identification of thermophilic Campylobacters from animals in Brazil. Vet J. 2002;164:159–161. doi: 10.1053/tvjl.2001.0698. [DOI] [PubMed] [Google Scholar]
- 6.Aquino MH, Filgueiras AL, Matos R, Santos KR, Ferreira T, Ferreira MC, Teixeira LM, Tibana A. Diversity of Campylobacter jejuni and Campylobacter coli genotypes from human and animal sources from Rio de Janeiro, Brazil. Res Vet Sci. 2010;88:214–217. doi: 10.1016/j.rvsc.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Fica A, Seelmann D, Porte L, Eugenin D, Gallardo R. A case of myopericarditis associated to Campylobacter jejuni infection in the southern hemisphere. Braz J Infect Dis. 2012;16:294–296. [PubMed] [Google Scholar]
- 8.Gomes CN, Souza RA, Passaglia J, Duque SS, Medeiros MIC, Falcão JP. Genotyping of Campylobacter coli strains isolated in Brazil suggests possible contamination amongst environmental, human, animal and food sources. J Med Microbiol. 2016;65:80–90. doi: 10.1099/jmm.0.000201. [DOI] [PubMed] [Google Scholar]
- 9.Frazão MR, Medeiros MIC, Duque SS, Falcão J. Pathogenic potential and genotypic diversity of Campylobacter jejuni: a neglected food-borne pathogen in Brazil. J Med Microbiol. 2017;66:350–359. doi: 10.1099/jmm.0.000424. [DOI] [PubMed] [Google Scholar]
- 10.Gomes CN, Passaglia J, Vilela FP, Pereira da Silva FMHS, Duque SS, Falcão JP. High survival rates of Campylobacter coli under different stress conditions suggest that more rigorous food control measures might be needed in Brazil. Food Microbiol. 2018;73:327–333. doi: 10.1016/j.fm.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald C, Nachamkin I. Campylobacter and Arcobacter. In: Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, editors. Manual of Clinical Microbiology. Washington: ASM PRESS; 2007. pp. 933–942. [Google Scholar]
- 12.Havelaar AH, Mangen MJJ, De Koeijer AA, Bogaardt MJ, Evers EG, Jacobs-Reitsma WF, Van Pelt W, Wagenaar JA, De Wit GA, Vander Der Zee H, Nauta MJ. Effectiveness and efficiency of controlling Campylobacter on broiler chicken meat. Risk Analysis. 2007;27(4):831–844. doi: 10.1111/j.1539-6924.2007.00926.x. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald C. Campylobacter. Clin Lab Med. 2015;35(2):289–298. doi: 10.1016/j.cll.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Man SM. The clinical importance of emerging Campylobacter species. Nat Rev Gastroenterol Hepatol. 2011;8:669e685. doi: 10.1038/nrgastro.2011.191. [DOI] [PubMed] [Google Scholar]
- 15.Moore JE, Corcoran D, Dooley JSG, Fanning S, Lucey B, Matsuda M, McDowell DA, Mégraud F, Millar BC, O'Mahony R, O'Riordan L, O'Rourke M, Rao JR, Rooney PJ, Sails A, Whyte P. Campylobacter. Vet Res. 2005;36:351–382. doi: 10.1051/vetres:2005012. [DOI] [PubMed] [Google Scholar]
- 16.Hodges LM, Carrillo CD, Upham JP, Borza A, Eisebraun M, Kenwell R, Mutschall SK, Haldane D, Scheihauf E, Taboada EN. A strain comparison of Campylobacter isolated from retail poultry and human clinical cases in Atlantic Canada. PLoS ONE. 2019;14(5):e0215928. doi: 10.1371/journal.pone.0215928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5:665e679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 18.Gao B, Vorwerk H, Huber C, Lara-Tejero M, Mohr J, Goodman AL, Eisenreich W, Galan JE, Hofreuter D. Metabolic and fitness determinants for in vitro growth and intestinal colonization of the bacterial pathogen Campylobacter jejuni. PLoS Biol. 2017;15:1e37. doi: 10.1371/journal.pbio.2001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid AN, Palyada K, Naikare H, Stintzi A. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl Environ Microbiol. 2008;74:1583e1597. doi: 10.1128/AEM.01507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birk T, Wik MT, Lametsch R, Knochel S. Acid stress response and protein induction in Campylobacter jejuni isolates with different acid tolerance. BMC Microbiol. 2012;12:13. doi: 10.1186/1471-2180-12-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar-Phillips GS, Hanning I, Slavik M. Influence of acid-adaptation of Campylobacter jejuni on adhesion and invasion of INT 407 cells. Foodb Pathog Dis. 2013;10:1037e1043. doi: 10.1089/fpd.2013.1544. [DOI] [PubMed] [Google Scholar]
- 22.Baqar S, Rice B, Lee L, Bourgeois AL, El Din AN, Tribble DR, Heresi GP, Mourad AS, Murphy JR. Campylobacter jejuni enteritis. Clin Infect Dis. 2001;33(6):901–905. doi: 10.1086/322594. [DOI] [PubMed] [Google Scholar]
- 23.Jones MA, Totemeyer S, Maskell DJ, Bryant CE, Barrow PA. Induction of pro-inflammatory responses in the human monocytic cell line THP-1 by Campylobacter jejuni. Infect Immun. 2003;71(5):2626–2633. doi: 10.1128/IAI.71.5.2626-2633.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu L, Bray MD, Osorio M, Kopecko DJ. Campylobacter jejuni induces maturation and cytokine production in human dendritic cells. Infect Immun. 2006;74(5):2697–2705. doi: 10.1128/IAI.74.5.2697-2705.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Salloom F. Campylobacter-stimulated INT407 cells produce dissociated cytokine profiles. J Infect. 2003;47(3):217–224. doi: 10.1016/s0163-4453(03)00076-8. [DOI] [PubMed] [Google Scholar]
- 26.Mac-Callum AJ, Harris D, Haddock G, Everest PH. Campylobacter jejuni-infected human epithelial cell lines vary in their ability to secrete interleukin-8 compared to in vitro-infected primary human intestinal tissue. Microbiol. 2006;152(12):3661–3665. doi: 10.1099/mic.0.29234-0. [DOI] [PubMed] [Google Scholar]
- 27.Zheng J, Meng J, Zhao S, Singh R, Song W. Campylobacter-induced interleukin-8 secretion in polarized human intestinal epithelial cells requires Campylobacter-secreted cytolethal distending toxin- and Toll-like receptor-mediated activation of NF-kappaB. Infect Immun. 2008;76(10):4498–4508. doi: 10.1128/IAI.01317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Islam D, Ruamsap N, Aksomboon A, Khantapura P, Srijan A, Mason CJ. Immune responses to Campylobacter (C. jejuni or C. coli) infections: a two-year study of US forces deployed to Thailand. APMIS. 2014;122(11):1102–1113. doi: 10.1111/apm.12266. [DOI] [PubMed] [Google Scholar]
- 29.Hamza E, Kittl S, Kuhnert P. Temporal induction of pro-inflammatory and regulatory cytokines in human peripheral blood mononuclear cells by Campylobacter jejuni and Campylobacter coli. PLoS ONE. 2017;12(2):e0171350. doi: 10.1371/journal.pone.0171350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friis LM, Keelan M, Taylor DE. Campylobacter jejuni drives MyD88-independent interleukin-6 secretion via toll-like receptor 2. Infect Immun. 2009;77(4):1553–1560. doi: 10.1128/IAI.00707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klančnik A, Pogačar MS, Raspor P, Abram M, Mozina SS, Vučković D (2016) Virulence genes and cytokine profile in systemic murine Campylobacter coli infection. Virulence:2150–5594 [DOI] [PMC free article] [PubMed]
- 32.Everest PH, Goossens H, Butzler JP, Lloyd D, Knutton S, Ketley JM, Williams PH. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J Med Microbiol. 1992;37:319–332. doi: 10.1099/00222615-37-5-319. [DOI] [PubMed] [Google Scholar]
- 33.Campioni F, Gomes CN, Bergamini AMM, Rodrigues DP, Tiba-Casas P, Falcão JP (2020) Comparison of cell invasion, macrophage survival and inflammatory cytokines profiles between Salmonella enterica serovars Enteritidis and Dublin from Brazil. J Appl Microbiol:1–24 [DOI] [PubMed]
- 34.Shah J, Desai PT, Chen D, Stevens JR, Weimer BC. Preadaptation to cold stress in Salmonella enterica Serovar Typhimurium increases survival during subsequent acid stress exposure. Appl Environ Microbiol. 2013;79(23):7281–7289. doi: 10.1128/AEM.02621-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagle BR, Upadhyay A, Arsi K, Shrestha S, Venkitanarayanan K, Donoghue AM, Donoghue DJ (2017) Application of β-Resorcylic acid as potential antimicrobial feed additive to reduce Campylobacter colonization in broiler chickens. Front Microbiol 8 [DOI] [PMC free article] [PubMed]
- 36.Upadhyay A, Arsi K, Wagle BR, Upadhyaya I, Shrestha S, Donoghue AM, Donoghue DJ. Trans-cinnamaldehyde, carvacrol, and eugenol reduce Campylobacter jejuni colonization factors and expression of virulence genes in vitro. Front Microbiol. 2017;8:713. doi: 10.3389/fmicb.2017.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagle BR, Donoghue AM, Shrestha S, Upadhyaya I, Arsi K, Gupta A, Liyanage R, Rath NC, Donoghue DJ, Upadhyay A. Carvacrol attenuates Campylobacter jejuni colonization factors and proteome critical for persistence in the chicken gut. Poult Sci. 2020;99:4566–4577. doi: 10.1016/j.psj.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fierer J, Eckmann L, Fang F, Pfeifer C, Finlay BB, Guiney D. Expression of the Salmonella virulence plasmid gene spvB in cultured macrophages and non- phagocytic cell. Infect Immun. 1993;61:5231–5236. doi: 10.1128/iai.61.12.5231-5236.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeifer CG, Marcus SL, Steele-Mortimer O, Knodler A, Finlay BB. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect Immun. 1999;67:5690–5698. doi: 10.1128/iai.67.11.5690-5698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreira CG, Weinshenker D, Sperandio V. QseC mediates Salmonella enterica serovar Typhimurium virulence in vitro and in vivo. Infect Immun. 2010;78:914–926. doi: 10.1128/IAI.01038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Detweiler CS, Monack DM, Brodsky IE, Mathew H, Falkow S. VirK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol Microbiol. 2003;48:385–400. doi: 10.1046/j.1365-2958.2003.03455.x. [DOI] [PubMed] [Google Scholar]
- 42.Buchanan CJ, Webb AL, Mutschall SK, Kruczkiewicz P, Barker DOR, Hetman BM, Gannon VPJ, Abbott DM, Thomas JE, Inglis GD, Taboada EN. A genome-wide association study to identify diagnostic markers for human pathogenic Campylobacter jejuni strains. Front Microbiol. 2017;8:1224. doi: 10.3389/fmicb.2017.01224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. Experimental Campylobacter jejuni infection in humans. J Infect Dis. 1988;157(3):472–479. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
- 44.Babakhani FK, Joens LA. Primary swine intestinal-cells as a model for studying Campylobacter jejuni invasiveness. Infect Immun. 1993;61:2723–2726. doi: 10.1128/iai.61.6.2723-2726.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allos BM. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis. 2001;32(8):1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 46.Janssen R, Krogfelt KA, Cawthraw SA, Van Pelt W, Wagenaar JA, Owen RJ. Host-pathogen interactions in Campylobacter infections: the host perspective. Clin Microbiol Rev. 2008;21:505–518. doi: 10.1128/CMR.00055-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dasti JI, Tareen AM, Lugert R, Zautner AE, Gross U. Campylobacter jejuni: A brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int J Med Microbiol. 2010;300:205–211. doi: 10.1016/j.ijmm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 48.Poly F, Guerry P. Pathogenesis of Campylobacter. Curr Opin Gastroenterol. 2008;24:27–31. doi: 10.1097/MOG.0b013e3282f1dcb1. [DOI] [PubMed] [Google Scholar]
- 49.Hänel I, Muller J, Muller W, Schulze E. Correlation between invasion of Caco-2 eukaryotic cells and colonization ability in the chick gut in Campylobacter jejuni. Vet Microbiol. 2004;101:75–82. doi: 10.1016/j.vetmic.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Gilbert CD, Slavik MF. Evaluation of attachment and penetration abilities of Campylobacter jejuni isolates obtained from humans and chicken carcasses during processing and at retail. J Food Safety. 2005;25:209–223. [Google Scholar]
- 51.Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella Typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci U. S. A. 1986;83(14):5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gulig PA, Curtiss R. Plasmid-associated virulence of Salmonella Typhimurium. Infect Immun 1987. 1987;55(12):2891–2901. doi: 10.1128/iai.55.12.2891-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harrington KA, Hormaeche CE. Expression of the innate resistance gene Ity in mouse Kupffer cells infected with Salmonella typhimurium in vitro. Microb Pathog. 1986;1:269–274. doi: 10.1016/0882-4010(86)90051-3. [DOI] [PubMed] [Google Scholar]
- 54.García-Quintanilla M, Casadesús J. Virulence plasmid interchange between strains ATCC 14028, LT2, and SL1344 of Salmonella enterica serovar Typhimurium. Plasmid. 2011;65(2):169–175. doi: 10.1016/j.plasmid.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Allen KJ, Griffiths MW. Use of luminescent Campylobacter jejuni ATCC 33291 to assess eggshell colonization and penetration in fresh and retail eggs. J Food Prot. 2011;64(12):2058–2062. doi: 10.4315/0362-028x-64.12.2058. [DOI] [PubMed] [Google Scholar]
- 56.The AHT. Lee SM, Dykes GA. Association of some Campylobacter jejuni with Pseudomonas aeruginosa biofilms increases attachment under conditions mimicking those in the environment. Plos One. 2019;14(4):e0215275. doi: 10.1371/journal.pone.0215275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebnet K, Vestweber D. Molecular mechanisms that control leukocyte extravasation: the selectins and the chemokines. Histochem Cell Biol. 1999;112:1–23. doi: 10.1007/s004180050387. [DOI] [PubMed] [Google Scholar]
- 58.Coupade C, Solito E, Levine JD. Dexamethasone enhances interaction of endogenous annexin 1 with L-selectin and triggers shedding of L-selectin in the monocytic cell line U-937. Br J Pharmacol 2003. 2003;140:133–145. doi: 10.1038/sj.bjp.0705413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fauchere JL, Rosenau A, Veron M, Moyen EN, Richard S, Pfister A. Association with HeLa cells of Campylobacter jejuni and Campylobacter coli isolated from human feces. Infect Immun. 1986;54:283–287. doi: 10.1128/iai.54.2.283-287.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Russell RG, O’donnoghue M, Blake DC, Jr, Zulty J, Detolla LJ. Early colonic damage and invasion of Campylobacter jejuni in experimentally challenged infant Macaca mulatta. J Infect Dis. 1993;168(1):210–215. doi: 10.1093/infdis/168.1.210. [DOI] [PubMed] [Google Scholar]
- 61.Pan H, Ge Y, Xu H, Zhang J, Kuang D, Yang X, Xudong S, Huang Z, Shi X, Xu X, Meng J. Molecular characterization, antimicrobial resistance and Caco-2 cell invasion potential of Campylobacter jejuni/coli from young children with diarrhea. Pediatr Infect Dis J. 2016;35(3):330–334. doi: 10.1097/INF.0000000000001016. [DOI] [PubMed] [Google Scholar]
- 62.Newell DG, Pearson A. The invasion of epithelial cell lines and the intestinal epithelium of infant mice by Campylobacter jejuni/coli. J Diarrhoeal Dis Res. 1984;2(1):19–26. [PubMed] [Google Scholar]
- 63.Malagón I, García S, Heredia N. Adherence, invasion, toxigenic, and chemotactic properties of Mexican Campylobacter strains. J Food Protect. 2010;73:2093–2098. doi: 10.4315/0362-028x-73.11.2093. [DOI] [PubMed] [Google Scholar]
- 64.Murphy H, Cogan T, Hughes R, Humphrey T. Porcine intestinal epithelial responses to Campylobacter infection. Comp Immunol Microbiol Infect Dis. 2001;34:489–495. doi: 10.1016/j.cimid.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 65.Zheng J, Meng J, Zhao S, Singh R, Song W. Adherence to and invasion of human intestinal epithelial cells by Campylobacter jejuni and Campylobacter coli isolates from retail meat products. J Food Protect. 2006;69:768–774. doi: 10.4315/0362-028x-69.4.768. [DOI] [PubMed] [Google Scholar]
- 66.Fearnley C, Manning G, Bagnall M, Javed MA, Wassenaar TM, Newell DG. Identification of hyperinvasive Campylobacter jejuni strains isolated from poultry and human clinical sources. J Med Microbiol. 2008;57:570–580. doi: 10.1099/jmm.0.47803-0. [DOI] [PubMed] [Google Scholar]
- 67.Habib I, Louwen R, Uyttendaele M, Houf K, Vandenberg O, Nieuwenhuis EE, Miller WG, Van Belkum A, De Zutter L. Correlation between genotypic diversity, lipooligosaccharide gene locus class variation, and Caco-2 cell invasion potential of Campylobacter jejuni isolates from chicken meat and humans: contribution to virulotyping. Appl Environ Microbiol. 2009;75:4277–4288. doi: 10.1128/AEM.02269-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sestak K, Merritti CK, Borda J, Schwamberger SR, Cogswell F, Didier S, Didier PJ, Plauche G, Bohm RP, Aye PP, Alexa P, Ward R, Andrew A, Saylor E, Didier ES, Lackner A. Infectious agent and immune response characteristics of chronic enterocolitis in captive. Infect Immun. 2003;71:4079–4086. doi: 10.1128/IAI.71.7.4079-4086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hickey TE, McVeigh AL, Scott DA, Michielutti RE, Bixby A, Carroll SA, Bourgeois AL, Guerry P. Campylobacter jejuni cytolethal distending toxin mediates release of interleukin-8 from intestinal epithelial cells. Infect Immun. 2000;68:6535–6541. doi: 10.1128/iai.68.12.6535-6541.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang Y, Lü X, Man C, Han L, Shan Y, Qu X, Liu Y, Yang S, Xue Y, Zhang Y. Lactobacillus acidophilus induces cytokine and chemokine production via NF-κB and p38 mitogen-Activated Protein Kinase Signaling Pathways in Intestinal Epithelial Cells. Clin Vaccine Immunol. 2012;19(4):603–608. doi: 10.1128/CVI.05617-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zeuthen LH, Fink LN, Metzdorff SB, Kristensen MB, Licht TR, Nellemann C, Frøkiær H (2010). Lactobacillus acidophilus induces a slow but more sustained chemokine and cytokine response in naïve foetal enterocytes compared to commensal Escherichia coli.BMC Immunol 11(1): 1-10, 2. [DOI] [PMC free article] [PubMed]
- 72.Deun KV, Pasmans F, Ducatelle R, Flahou B, Vissenberg K, Martel A, Broeck WV, Immerseel FV, Haesebrouck F. Colonization strategy of Campylobacter jejuni results in persistent infection of the chicken gut. Vet Microbiol. 2008;130:285–297. doi: 10.1016/j.vetmic.2007.11.027. [DOI] [PubMed] [Google Scholar]