Abstract
Background
Q.Clear is a Bayesian penalized likelihood (BPL) reconstruction algorithm that presents improvements in signal-to-noise ratio (SNR) in clinical positron emission tomography (PET) scans. Brain studies in research require a reconstruction that provides a good spatial resolution and accentuates contrast features however, filtered back-projection (FBP) reconstruction is not available on GE SIGNA PET-Magnetic Resonance (PET-MR) and studies have been reconstructed with an ordered subset expectation maximization (OSEM) algorithm. This study aims to propose a strategy to approximate brain PET quantitative outcomes obtained from images reconstructed with Q.Clear versus traditional FBP and OSEM.
Methods
Contrast recovery and background variability were investigated with the National Electrical Manufacturers Association (NEMA) Image Quality (IQ) phantom. Resolution, axial uniformity and SNR were investigated using the Hoffman phantom. Both phantoms were scanned on a Siemens Biograph 6 TruePoint PET-Computed Tomography (CT) and a General Electric SIGNA PET-MR, for FBP, OSEM and Q.Clear. Differences between the metrics obtained with Q.Clear with different β values and FBP obtained on the PET-CT were determined.
Results
For in plane and axial resolution, Q.Clear with low β values presented the best results, whereas for SNR Q.Clear with higher β gave the best results. The uniformity results are greatly impacted by the β value, where β < 600 can yield worse uniformity results compared with the FBP reconstruction.
Conclusion
This study shows that Q.Clear improves contrast recovery and provides better resolution and SNR, in comparison to OSEM, on the PET-MR. When using low β values, Q.Clear can provide similar results to the ones obtained with traditional FBP reconstruction, suggesting it can be used for quantitative brain PET kinetic modelling studies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40658-021-00386-3.
Keywords: PET-MR, Reconstruction, Bayesian, Brain imaging
Background
Positron emission tomography (PET) is an imaging technique that allows for non-invasive quantitative measurement of biological processes in vivo. Image reconstruction methods can broadly be divided into analytical and iterative algorithms. Whereas analytical reconstruction algorithms (e.g. filtered back-projection, FBP) assume continuous data and introduce a discrete character to it a posteriori, iterative reconstruction algorithms (e.g. ordered subset expectation maximization, OSEM) assume discretely sampled data. Although iterative reconstruction algorithms are routinely used in the clinical setting, where image quality and lesion contrast are of great importance, analytical reconstruction algorithms are still used in research for accurate PET data quantification via kinetic modelling [1].
The block sequential regularized expectation maximization (BSREM) algorithm is a Bayesian penalized likelihood (BPL) method that uses prior knowledge as a relative difference penalty term in the cost function, weighted by a penalization parameter β [2]. Unlike expectation-maximization (EM) algorithms that typically become noisy as the number of iterations is increased, the penalty term suppresses noise allowing the BSREM algorithm to iterate to convergence, in principle increasing the accuracy of the quantitative image measurements [2, 3]. Although BPL algorithms are not new, their use in clinical and research settings has been limited due to the computational cost involved and lack of availability in clinical systems [2]. Recently, General Electric (GE) Healthcare has released the BSREM penalized likelihood reconstruction algorithm under the product name of Q.Clear. However, due to its recent release, its impact in clinical use and research applications is still being evaluated [2]. The FBP reconstruction is not available for clinical use on the GE SIGNA PET-MR scanner; hence, OSEM reconstructions have been used for processing brain studies. In smaller regions, such as the ones that can be found in the brain, the convergence rate of OSEM process must be stopped early in order to not compromise image quality due to excessive noise [4, 5]. Although OSEM is being used for processing of both whole-body and brain scans, studies such as the ones conducted by Reilhac et al. [6] and Walker et al. [7] have reported a positive bias in regions with low activity and a negative bias in regions of high activity in low-count scans which had been reconstructed with this algorithm. Jian et al. [8] however found a negative bias in both high-count and low-count regions, in scans which had been acquired and reconstructed under a similar paradigm as described above [6–8]. This is of particular importance with radiotracers which are mass dependent due to the potential of pharmacological effects. The restricted injected dose limits may therefore result in noisy imaging data with low count statistics. Despite multiple advances in iterative methods of quantification (e.g. OSEM and BPL), FBP is still used as method of choice for accurate brain PET kinetic modelling studies due to its linear response. The impact of using non-FBP methods for reconstruction of quantitative brain studies is poorly understood and with latest PET-MR technology rapidly gaining momentum in the field of brain clinical research, studies are needed to assess and minimise the gap between traditional PET-CT kinetic modelling studies with data reconstructed using FBP versus PET-MRI OSEM and Q.Clear approaches.
Furthermore, brain PET imaging plays a critical role in clinical diagnosis of dementia and other neurological disorders. Despite that, to date, studies looking to assess Q.Clear performance in clinical PET have been primarily focused on whole-body analysis and fluorinated radiotracers [9–13]; therefore, there is a need to assess the performance of this framework in the context of neuroimaging and with different PET isotopes. This study aimed to evaluate the performance of the Q.Clear, against that of the widely used OSEM and the FBP algorithms in brain phantom images acquired on a clinical PET-CT and on a clinical PET-MR system using 18F- and 11C-labelled radiotracers. We hypothesise that despite differences in scanner design and performance as well as reconstruction frameworks, brain PET quantitative outcomes can be approximated by assessing the performance of different reconstruction algorithms and identifying those that result in least impact on successful quantitative PET-MR brain studies.
Materials and methods
The PET-CT and PET-MR data reported here was collected at a single site. The primary source for radiation measurements performed in the department is with a 137Cs source that is used for the daily quality control procedures on the dose calibrators. The nominal activity of this source was previously adjusted as part of a cross calibration exercise, to the secondary standard ionisation chamber at the National Physical Laboratory, in the UK. The remaining measurement equipment including the PET-CT and PET-MR scanners are then calibrated using measurements made from the dose calibrator and a cylindrical phantom filled with 18F or 11C tracer. Additionally, and for the purposes of this single-centre study, a large phantom volume-of-interest (VOI) for 18F and 11C was used, prior to starting the reconstruction comparison [14].
PET-CT and PET-MR phantom data acquisition and reconstruction
The National Electrical Manufacturers Association (NEMA) Image Quality (IQ) phantom was prepared by adding [18F]BCPP-EF (49.5 ± 5.4 MBq, mean ± SD, n = 2) solution to the phantom, ensuring that the hot spheres contained a concentration four times that of the background (22.4 kBq/mL versus 5.6 kBq/mL) [15]. The two larger spheres were filled with non-radioactive water, henceforth referred to as cold spheres. This phantom was scanned for 40 min once in the department single-centre benchmark PET-CT scanner (Siemens 6 Biograph TruePoint, Siemens Healthcare, Germany; detector size 4.0 × 4.0 × 20 mm3 (transverse, axial, depth directions) and NEMA NU 2–2007 full-width half maximum at 1 cm from centre of 4.1 mm transverse and 4.7 mm axial [16]) and once in the department single-centre benchmark PET-MR scanner (GE SIGNA, GE Healthcare, USA; detector size 4.0 × 5.3 × 25 mm3 and NEMA NU 2–2007 full-width half maximum at 1 cm from centre of 4.05 mm transverse and 6.08 mm axial [17]). In both scanners the data was acquired in listmode and a matrix of 128 × 128 was used for reconstruction.
The Hoffman phantom was prepared by mixing 29.6 MBq of [18F]BCPP-EF, or 34.4 MBq of [11C]SA4503, or 36.4 MBq of [11C]UCB-J in water and then filling the phantom, ensuring the removal of large air bubbles. The 18F phantom was scanned for 40 min in the PET-CT scanner, reconstructed with a matrix of 256 × 256 and for 40 min in the PET-MR scanner, reconstructed with a matrix of 384 × 384, in order to keep the voxel size as similar as possible across all PET datasets. The matrix size on z-direction for Hoffman scans acquired in the PET-MR is 89, for Hoffman scans acquired in the PET-CT is 109, for NEMA IQ acquired in the PET-MR is 89 and for NEMA IQ acquired in the PET-CT is 111. The voxel size for the Hoffman scans acquired in the PET-MR is 1 × 1 × 2.78 mm3, for the Hoffman scans acquired in the PET-CT is 1.02 × 1.02 × 2.03 mm3, for the NEMA IQ acquired in PET-MR is 4.69 × 4.69 × 2.78 mm3 and for the NEMA IQ acquired in the PET-CT is 5.35 × 5.35 × 5 mm3. Due to the short half-life of 11C, the Hoffman phantom was filled with [11C]SA4503 solution and scanned in the PET-MR and subsequently filled with [11C]UCB-J solution and scanned in the PET-CT. The duration of the acquisition and acquisition parameters were the same as for the 18F phantom and the data was acquired in listmode for both the 11C and 18F phantoms.
Each NEMA and Hoffman phantom scans acquired on the PET-CT scanner was reconstructed 6 times and each NEMA and Hoffman phantoms acquired on the PET-MR scanner was reconstructed 13 times, as can be observed in Table 1. The FBP reconstructions were only performed on the PET-CT scanner and the time of flight (TOF with time resolution of < 386 ps) Q.Clear reconstructions were only performed on the PET-MR. The three-dimensional (3D) OSEM reconstructions were performed on the PET-CT and TOF-OSEM reconstructions were performed on the PET-MR. OSEM with 4 iterations and 16 subsets was selected based on previously reported data comparing TOF and non-TOF measurements in different PET systems [5, 18–20]. Furthermore, the Q.Clear algorithm has been devised to improve image quality, without increasing noise, by using a penalty function. This penalty function behaves as a noise suppression term. To estimate correspondence of Q.Clear β value (up to 1000) and the size of the FBP and OSEM filter kernel for two different isotopes and brain phantoms in a variety of outcome measures (e.g. resolution, noise and uniformity), a wide range of filter from 5 to 15 mm was used in this study. Attenuation correction on the PET-CT was performed with a low-dose attenuation correction CT scan performed prior to the PET acquisition (NEMA phantom: 30 mAs, 130 kV, 5 mm slice, 1.5 pitch and 1.5 s rotation time; Hoffman phantom: 30 mAs, 130 kV, 3 mm slice, 0.55 pitch and 0.8 s rotation time). Attenuation correction on the PET-MR was performed with a GE CT-based template of the respective phantoms. All images acquired in the PET-CT and in the PET-MR have been reconstructed with random and scatter correction. These protocols were designed based on centre benchmark during this single-centre project and based on previous literature as detailed above.
Table 1.
Summary of methods used for reconstructing the NEMA and Hoffman phantom datasets
| Reconstruction method | Nomenclature | 18F NEMA PET-CT | 18F NEMA PET-MR | 18F Hoffman PET-CT | 18F Hoffman PET-MR | 11C Hoffman PET-CT | 11C Hoffman PET-MR |
|---|---|---|---|---|---|---|---|
| FBP with 5 mm filter (PET-CT) | FBP_5mm | x | x | x | |||
| FBP with 10 mm filter (PET-CT) | FBP_10mm | x | x | x | |||
| FBP with 15 mm filter (PET-CT) | FBP_15mm | x | x | x | |||
| 3D OSEM 4 iterations, 8 subsets, 5 mm filter (PET-CT) | OSEM_4i8s5mm | x | |||||
| 3D OSEM 4 iterations, 8 subsets, 10 mm filter (PET-CT) | OSEM_4i8s10mm | x | |||||
| 3D OSEM 4 iterations, 8 subsets, 15 mm filter (PET-CT) | OSEM_4i8s15mm | x | |||||
| 3D OSEM 4 iterations, 16 subsets, 5 mm filter (PET-CT) | OSEM_4i16s5mm | x | x | ||||
| 3D OSEM 4 iterations, 16 subsets, 10 mm filter (PET-CT) | OSEM_4i16s10mm | x | x | ||||
| 3D OSEM 4 iterations, 16 subsets, 15 mm filter (PET-CT) | OSEM_4i16s15mm | x | x | ||||
| ToF 3D OSEM 4 iterations, 8 subsets, 5 mm filter (PET-MR) | OSEM_4i8s5mm | x | |||||
| ToF 3D OSEM 4iterations, 8 subsets, 10 mm filter (PET-MR) | OSEM_4i8s10mm | x | |||||
| ToF 3D OSEM 4 iterations, 8 subsets, 15 mm filter (PET-MR) | OSEM_4i8s15mm | x | |||||
| ToF 3D OSEM 4 iterations, 16 subsets, 5 mm filter (PET-MR) | OSEM_4i16s5mm | x | x | ||||
| ToF 3D OSEM 4 iterations, 16 subsets, 10 mm filter (PET-MR) | OSEM_4i16s10mm | x | x | ||||
| ToF 3D OSEM 4 iterations, 16 subsets, 15 mm filter (PET-MR) | OSEM_4i16s15mm | x | x | ||||
| ToF 3D Q.Clear with β100 (PET-MR) | QClear100 | x | x | x | |||
| ToF 3D Q.Clear with β200 (PET-MR) | QClear200 | x | x | x | |||
| ToF 3D Q.Clear with β300 (PET-MR) | QClear300 | x | x | x | |||
| ToF 3D Q.Clear with β400 (PET-MR) | QClear400 | x | x | x | |||
| ToF 3D Q.Clear with β500 (PET-MR) | QClear500 | x | x | x | |||
| ToF 3D Q.Clear with β600 (PET-MR) | QClear600 | x | x | x | |||
| ToF 3D Q.Clear with β700 (PET-MR) | QClear700 | x | x | x | |||
| ToF 3D Q.Clear with β800 (PET-MR) | QClear800 | x | x | x | |||
| ToF 3D Q.Clear with β900 (PET-MR) | QClear900 | x | x | x | |||
| ToF 3D Q.Clear with β1000 (PET-MR) | QClear1000 | x | x | x |
Data analysis
The NEMA phantom scans were analysed using a customised Interactive Data Language (IDL ®) program according to NEMA standards [21, 22]. Circular regions of interest (ROIs), equal in diameter to each sphere, and 60 adjacent background ROIs were drawn. Contrast and background variability were calculated using the NEMA NU 2-2012 equations [21].
The percentage contrast for each hot sphere was calculated according to Eq. 1:
| 1 |
where CH is the average of the counts found in the ROI for a hot sphere, CB is the average of the background counts in the background ROI for the same sphere, aH and aB the activity concentration in the hot sphere and in the background, respectively [21].
The Percentage contrast for each cold sphere was calculated according to Eq. 2:
| 2 |
where CC represents the average of counts in the ROI for a cold sphere and CB represents the average of the 60 background ROI counts for the same sphere size [21].
For the background variability, the standard deviation of the background ROI counts for each sphere size was calculated according to Eq. 3,
| 3 |
where k equals the 60 background ROI counts and the background variability was calculated according to Eq. 4:
| 4 |
The Hoffman phantom data were analysed using the VivoQuant® software version 3.5 patch 2 (inviCRO LLC, USA) [23]. The resolution (expressed as full-width-half-maximum, FWHM) was determined by correlation of the acquired images with a digital version of the Hoffman phantom convolved with different Gaussian filters. This allowed for comparing estimated in-plane and axial resolutions (VivoQuant Hoffman Phantom Analysis Workflow Working Instruction Document, unpublished). The axial uniformity metric was determined by drawing a VOI in the right putamen (size of 2400 mm3) and calculating the percentage standard deviation according to Eq. 5 [24]:
| 5 |
Where CP is the average counts in the VOI and σp the standard deviation.
The signal-to-noise ratio (SNR) was determined by drawing a VOI in the right putamen and a VOI in the background “white matter” region of the Hoffman phantom (devoid of radioactivity) and it was calculated according to Eq. 6:
| 6 |
Where CP is the average counts in the VOI for the putamen, CW is the average counts in the VOI placed in a uniform area in the background and σW the standard deviation in the background [25].
Differences in contrast, background variability, resolution, uniformity and SNR were calculated relative to the FBP reconstruction with 5 mm FWHM Gaussian filter, the standard FBP reconstruction for the department. Bland-Altman plots were used to investigate the quantitative differences between the FBP with 5 mm FWHM Gaussian filter (obtained in the PET-CT) and the TOF-OSEM with 4 iterations, 8 subsets and 5 mm filter (obtained in the PET-MR) versus Q.Clear with different β values.
GraphPad Prism version 8.1.0 for Windows (GraphPad Software, USA) was used for statistical analysis and graphical representation [26].
Results
NEMA and Hoffman phantom results with 18F-solution
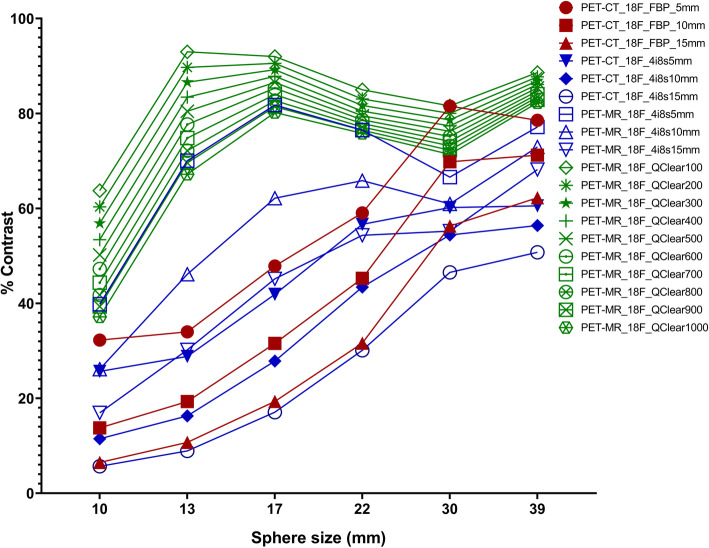
The Q.Clear reconstructions (varying β values) from the PET-MR provided consistently higher percentage contrast compared to OSEM reconstructions on the PET-CT and the PET-MR, as well as the FBP on the PET-CT. For all reconstruction methods, the percentage contrast was highest for large diameter spheres of the NEMA phantom and reduced with sphere size (Fig. 1). The largest variability in the percentage contrast across all reconstruction methods was measured for the 13 mm sphere (mean 55.7%, standard deviation 29.4%, median 69.6% and coefficient of variation 52.8%) compared to the smallest variability for the 30 mm sphere (mean 69.0%, standard deviation 10.5%, median 72.3% and coefficient of variation 15.2%). The lowest quantitative differences were found for Q.Clear with β1000 when comparing with FBP with a 5 mm kernel and TOF-OSEM with 4 iterations, 8 subsets and 5 mm kernel (13.5 and 0.36, respectively) (Supplementary Files 1 and 2).
Fig. 1.
NEMA phantom measured percentage contrast recovery for all reconstruction methods when using 18F-solution. Note highest percentage contrast of Q.Clear methods compared with OSEM and FBP methods
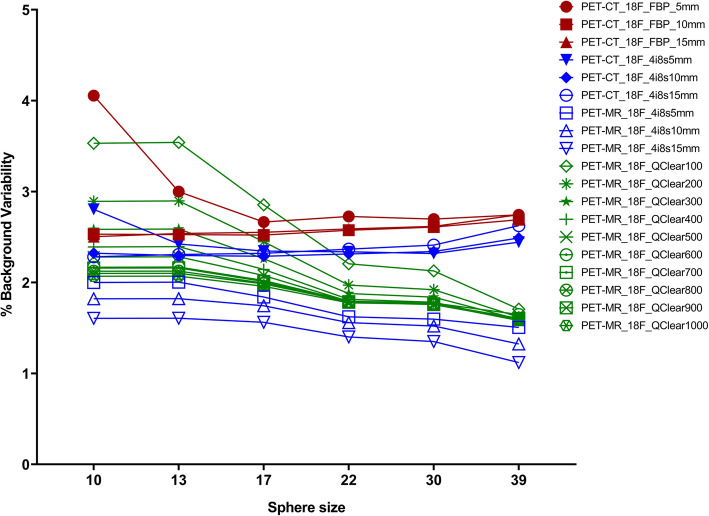
Analysis of the NEMA phantom background showed the OSEM on the PET-MR resulted in the smallest background variability of all methods (Fig. 2). The largest background variability was measured for FBP with the smallest filter kernel, followed by the Q.Clear method with the lowest β value of 100. For each sphere size, the measured mean background variability dropped from 2.43 % (10 mm sphere) to 1.89% (39 mm sphere). The same trend was observed for the standard deviation (0.58 to 0.53 %) and median (2.28 to 1.61%), while the coefficients of variation were relatively stable at 23.8%, 18.6%, 15.1%, 19.0%, 20.1% and 28.4% for the 10, 13, 17, 22, 30, and 39 mm sphere, respectively. The lowest quantitative difference was found for Q.Clear with β100 (0.32) when comparing with FBP with a 5-mm kernel and for Q.Clear with β1000 (0.11) when TOF-OSEM with 4 iterations, 8 subsets and 5 mm kernel (Supplementary Files 3 and 4).
Fig. 2.
NEMA phantom measured background variability for all reconstruction methods when using 18F-solution. Note OSEM reconstructions performed on the PET-MR scanner resulted in the lowest background variability of all methods
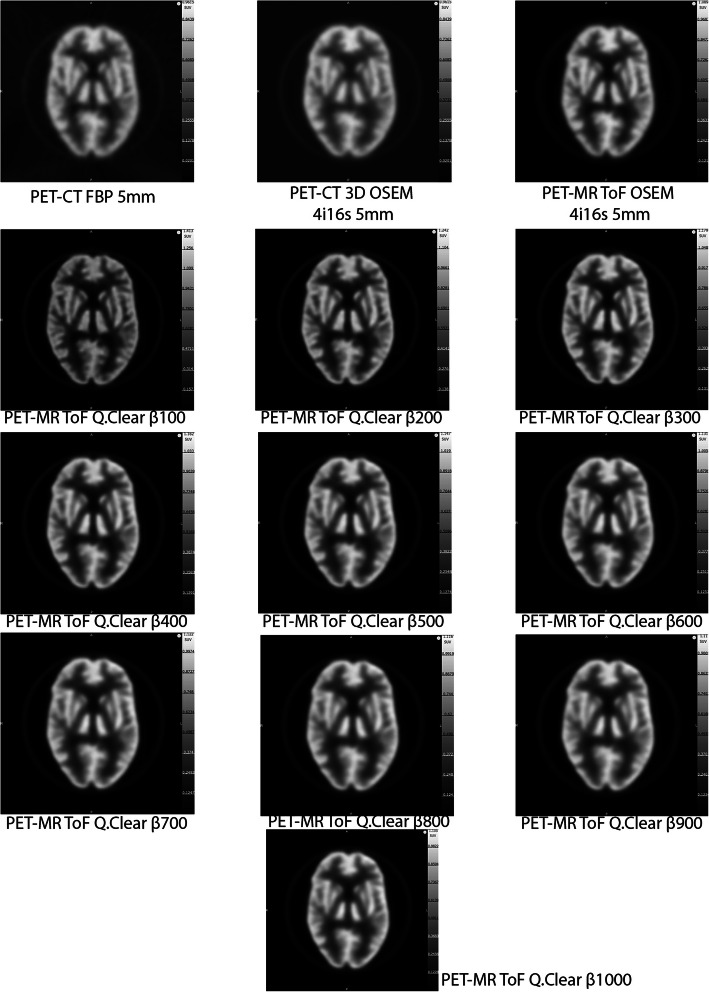
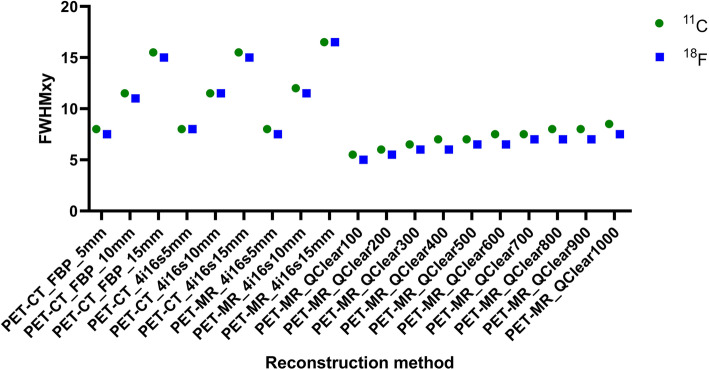
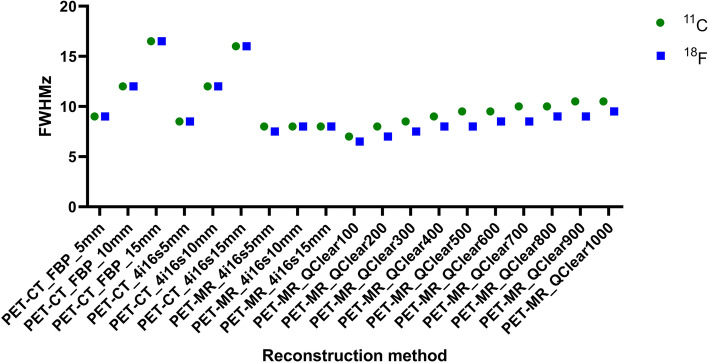
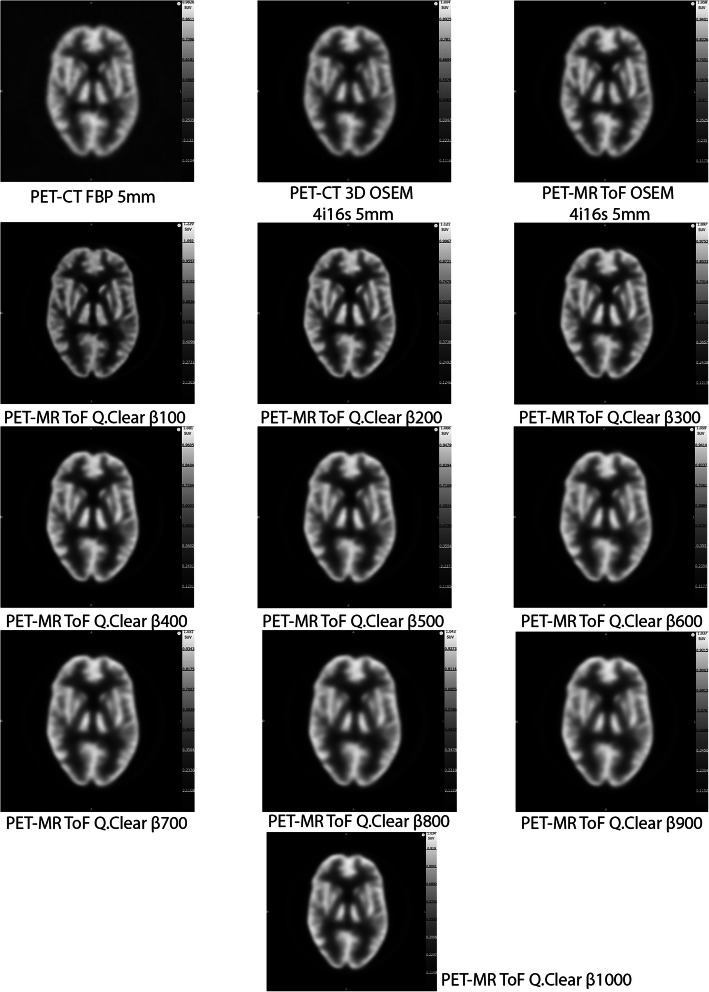
Images of the Hoffman phantom filled with the 18F solution and reconstructed with different methods are presented in Fig. 3. The highest FWHM (x,y) (worst transaxial spatial resolution) of 16.5 mm observed for the Hoffman phantom, was with 18F in the PET-MR, for OSEM 4 iterations, 16 subsets and a 15 mm filter (Fig. 4). The lowest FWHM of 5 mm was for Q.Clear with β of 100. A FWHM of 7.5 mm was measured for FBP with 5 mm filter. Relative to the FBP with 5 mm filter reconstruction, the largest difference (− 9.0 mm) was for PET-MR OSEM 4 iterations, 16 subsets and 15 mm filter; while the smallest difference (0.0 mm) was for PET-MR OSEM 4 iterations, 16 subsets and 5 mm filter together with Q.Clear β value of 1000.
Fig. 3.
Hoffman phantom filled with 18F-BCPP in the PET-CT and PET-MR. FBP and 3D OSEM 4 iterations, 16 subsets, 5 mm filter obtained in the PET-CT are displayed. TOF OSEM 4 iterations, 16 subsets, 5 mm filter and TOF Q.Clear β100 to 1000 obtained in the PET-MR are also displayed. Note the visual differences in image quality for the Q.Clear reconstructions as β increases
Fig. 4.
Hoffman phantom measured FWHM (x,y) for all reconstruction methods when using a 11C and a 18F-solution. Note the best resolution was obtained with the Q.Clear with β100, for both radionuclides
The highest FWHM (z) (worst z-axis spatial resolution) of 16.5 mm was observed for FBP with a 15-mm filter (Fig. 5). The lowest FWHM (z) of 6.5 mm was for Q.Clear reconstruction with β of 100. Relative to FBP with a 5-mm filter, the largest difference was for FBP with a 15-mm filter (− 7.5 mm), while the smallest was for the Q.Clear with β800 or 900 (0.0 mm).
Fig. 5.
Hoffman phantom measured FWHM (z) for all reconstruction methods when using a 11C and a 18F-solution. Note the best resolution was obtained with the Q.Clear with β100, for both radionuclides
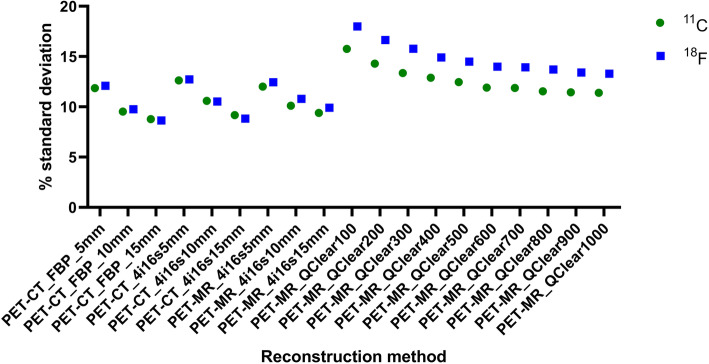
The Q.Clear with β of 100 yielded the poorest uniformity of 18.0%, while the best uniformity was measured for FBP with a 15-mm filter (8.6%) (Fig. 6). Relative to FBP with 5 mm filter, the largest difference (− 5.9) was for PET-MR Q.Clear with β of 100, while the smallest difference (− 0.3) was for PET-MR OSEM 4 iterations, 16 subsets, 5 mm filter.
Fig. 6.
Hoffman phantom measured uniformity for all reconstruction methods when using a 11C and a 18F-solution. Note the best uniformity was obtained with FBP with a 15-mm filter
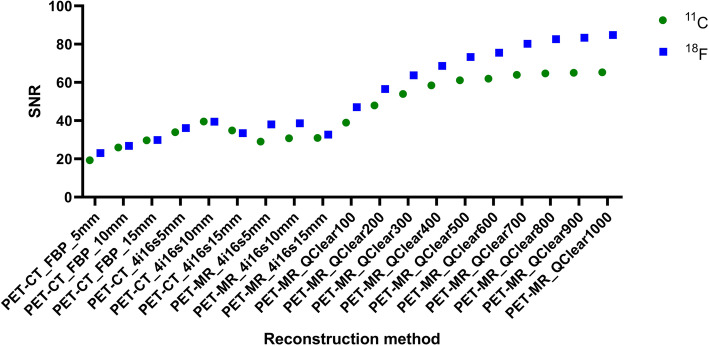
For SNR, the largest value (84.8) was for Q.Clear with β of 1000 (Fig. 7). The poorest SNR was for FBP with 5 mm filter (23.0). Relative to FBP with 5 mm filter, the largest difference (− 61.8) was for Q.Clear with β of 1000, while the smallest difference (− 3.8) was for FBP with 10 mm filter.
Fig. 7.
Hoffman phantom measured signal-to-noise for all reconstruction methods when using a 11C and a 18F-solution. Note the best signal-to-noise was obtained with Q.Clear with β1000
Hoffman phantom results with 11C-solution
Images of the Hoffman phantom filled with the 11C solutions and reconstructed with different methods are presented in Fig. 8. The highest FWHM (x,y) was 16.5 mm for PET-MR OSEM reconstruction with 4 iterations, 16 subsets and 15 mm filter (Fig. 4). The lowest FWHM of 5.5 mm was for Q.Clear with β of 100. A FWHM of 8 mm was measured for FBP with 5 mm filter. Relative to this, the largest difference (− 8.5 mm) was for PET-MR OSEM 4 iterations, 16 subsets and 15 mm filter, while the smallest difference (0.0 mm) was for PET-CT OSEM 4 iterations, 16 subsets and 5 mm filter together with PET-MR OSEM 4 iterations, 16 subsets and 5 mm filter and Q.Clear with β800 and 900.
Fig. 8.
Hoffman phantom filled with 11C-SA4503 and 11C-UCB-J in the PET-CT and PET-MR. FBP and 3D OSEM 4 iterations, 16 subsets, 5 mm filter obtained in the PET-CT are displayed. TOF OSEM 4iterations 16 subsets, 5 mm filter, and TOF Q.Clear β100 to 1000 obtained in the PET-MR are also displayed. Note the visual differences in image quality for the Q.Clear reconstructions as β increases
The highest FWHM (z) was 16.5 mm for FBP and 15 mm filter (Fig. 5). The lowest FWHM of 7.0 mm was for Q.Clear with β of 100. A FWHM of 9 mm was measured for FBP with 5 mm filter. Relative to this, the largest difference was measured for FBP with a 15-mm filter (− 7.5 mm), while the smallest was for Q.Clear with β400 (0.0 mm).
The Q.Clear reconstruction with β of 100 yielded the poorest uniformity of 15.8%, while the highest uniformity was for FBP with a filter of 15 mm (8.8%) (Fig. 6). Relative to the FBP with 5 mm filter, the largest difference (− 3.9) was for Q.Clear with β of 100; while the smallest difference (− 0.03) was for Q.Clear with β of 700.
For SNR, the highest value (65.3) was for Q.Clear with β of 1000 (Fig. 7). The poorest SNR was for FBP with 5 mm filter (19.3). Relative to this, the largest difference (− 45.9) was for Q.Clear with β of 1000, while the smallest difference (− 6.6) was for FBP with 10 mm filter.
Discussion
This study investigated the performance of the Q.Clear reconstruction algorithm using a PET-MR against OSEM (PET-MR and PET-CT) and FBP (PET-CT) algorithms on general use and brain phantom data. Different isotopes were used to characterise noise, uniformity, SNR and quantitative bias outcomes, and the Hoffman brain phantom was also selected to simulate radioisotope distribution in the grey and white matter of the brain.
Carbon-11 and Fluorine-18 tracers are used in clinical and research PET not only because of their short half-life but also due to the short-range of the positrons in tissue [27]. Our study demonstrates that the results obtained for the spatial resolution, signal-to-noise and axial uniformity metrics, present very similar patterns when using the Hoffman phantom filled with 18F or 11C. This data is in accordance with Conti et al.’s [27] findings using the NEMA phantom filled with pure β+ emitters and scanned up until 200 million net true counts were obtained. In their study, the 18F and 11C images presented very similar radial profiles.
Our NEMA phantom data demonstrate that as the Q.Clear β value increases, the contrast recovery and background variability decrease. Using the same phantom filled with 18F-FDG and a GE Discovery 690 PET/CT scanner, Teoh et al. also found that when Q.Clear β values increased, the contrast recovery and background variability decreased [28]. Furthermore, our data shows that the contrast recovery results obtained are lower for the FBP and OSEM reconstructions (performed on the PET-CT and on the PET-MR) than for the Q.Clear reconstructions. This is also in line with Teoh et al.’s findings, as the group reported the lowest contrast recovery results when using the OSEM reconstruction versus Q.Clear reconstructions. As expected, our data shows that as the sphere diameter increases from 10 to 17 mm (hot spheres) and from 30 to 39 mm (cold spheres), the contrast recovery also increases, in line with previous work [28].
The background variability results are higher for Q.Clear than for OSEM when reconstructing data on the PET-MR. This is in contrast with Teoh et al.’s findings in the PET-CT scanner as in the study mentioned above the group reported OSEM background variability results higher or equal to the background variability results obtained with Q.Clear with β > 200 [28]. This may be partly due to the differences in the width of filter used (2 mm and 6.4 mm in Teoh et al.’s study vs 5 mm, 10 mm and 15 mm used in our study) and to the use of Point Spread function modelling in Teoh et al.’s study [28]. The FBP and OSEM background results on the PET-CT are very similar. Interestingly, unlike the OSEM background variability results obtained in the PET-CT which present a slight upwards trend, the OSEM PET-MR results present a downwards trend, as the sphere diameter increases. This downwards trend is consistent with the findings from Caribé et al., who scanned an 18F-filled phantom in the GE Signa PET-MR and reconstructed the acquired dataset with TOF-OSEM with 4 iterations and 28 subsets. The team obtained a background variability of 6.1% for the sphere with 10 mm decreasing with the increase in sphere diameter to 2.7% for the 37 mm sphere [17]. Reynés-Llompart et al. scanned a 18F-filled NEMA phantom on a GE Discovery IQ PET-CT scanner. They found that as β values increased, the background variability and the contrast recovery coefficients decreased [29].
The FWHM(x,y) and FWHM(z) results show that the Q.Clear reconstructions with different β values on the PET-MR are more closely related to the FBP reconstruction, with a 5-mm kernel, rather than the FBP reconstructions with the 10 mm and 15 mm kernel in the PET-CT. The FWHM(x,y) results obtained for the Q.Clear reconstructions in the PET-MR are lower although still related to the results obtained for the FBP reconstruction with 5 mm filter in the PET-CT. The FWHM(z) results obtained for the Q.Clear reconstructions with β < 400 are considerably lower than the ones obtained for the FBP and OSEM reconstructions performed in the PET-CT. These metrics indicate an improvement in the in plane and axial resolution with this algorithm. This is consistent with the data obtained by Rogasch et al., who scanned a NEMA phantom during 30 min in a GE Discover MI PET-CT system and reconstructed the data with TOF-OSEM 4 iterations, 16 subsets and 2 mm filter, TOF-OSEM 2 iterations, 17 subsets and 2 mm filter, TOF-OSEM 2 iterarions, 8 subsets and 6.4 mm filter, Q.Clear β150, Q.Clear β300 and Q.Clear β450 [20]. The group reconstructed the spatial resolution from the radial activity profiles of the 37 mm sphere and found that all the Q.Clear reconstructions resulted in better spatial resolution results than TOF-OSEM [20].
Uniformity is strongly dependent on the β value and for β < 600, it can be worse than the uniformity obtained with the FBP reconstruction. Additionally, as the β value increases, so does the signal-to-noise and the difference to the FBP reconstructions. This data matches the visual image quality and is consistent with reports from clinical scans and other studies [30–34]. The uniformity and SNR results are explained by the fact that the β value acts as a noise suppression term and penalizes the differences in image intensity between bordering pixels [34].
Overall, Q.Clear with lower β levels improves FWHM(x,y) and FWHM(z), whereas Q.Clear with higher β levels improves uniformity and SNR. The findings in our study which was conducted in a GE Signa PET-MR scanner are consistent with those obtained by Reynés-Llompart et al. on a GE Discovery IQ PET-CT scanner. The team conducted a clinical evaluation of torso and brain acquisition and found that, after subjective quality assessment, β values between 300 and 400 are recommended for reconstructing torso acquisitions and β values between 100 and 200 are recommended for brain acquisitions [29].
Conclusion
Q.Clear improves contrast recovery on the PET-MR in comparison to OSEM. Moreover, Q.Clear also provides better in plane, axial resolution and signal-to-noise; however, its effect on image uniformity requires further investigations. For brain PET studies, in which spatial resolution is paramount, the Q.Clear reconstruction with β value of 100 will provide the best results based on our novel data with the Hoffman phantom, albeit with lower SNR compared with β value of 1000 and equivalent values to FBP.
Supplementary Information
Additional file 1: Supplementary file 1. Fig. 1 Bland-Altman plots assessing agreement between FBP with 5mm and Q.Clear with different β values, for the contrast recovery data obtained from the NEMA phantom. FBP 5mm and Q.Clear β100 (A); FBP 5mm and Q.Clear β200 (B); FBP 5mm and Q.Clear β300 (C); FBP 5mm and Q.Clear β400 (D); FBP 5mm and Q.Clear β500 (E). Fig. 2 Bland-Altman plots assessing agreement between FBP with 5mm and Q.Clear with different β values, for the contrast recovery data obtained from the NEMA phantom. FBP 5mm and Q.Clear β600 (F); FBP 5mm and Q.Clear β700 (G); FBP 5mm and Q.Clear β800 (H); FBP 5mm and Q.Clear β900 (I); FBP 5mm and Q.Clear β1000 (J).
Additional file 2: Supplementary file 2. Fig. 1 Bland-Altman plots assessing agreement between TOF-OSEM 4iteration, 8subsets with 5mm (4i8s5mm) and Q.Clear with different β values, for the contrast recovery data obtained from the NEMA phantom. TOF-OSEM 4i8s5mm and Q.Clear β100 (A); TOF-OSEM 4i8s5mm and Q.Clear β200 (B); TOF-OSEM 4i8s5mm and Q.Clear β300 (C); TOF-OSEM 4i8s5mm and Q.Clear β400 (D); TOF-OSEM 4i8s5mm and Q.Clear β500 (E). Fig. 2 Bland-Altman plots assessing agreement between TOF-OSEM 4iteration, 8subsets with 5mm (4i8s5mm) and Q.Clear with different β values, for the contrast recovery data obtained from the NEMA phantom. TOF-OSEM 4i8s5mm and Q.Clear β600 (F); TOF-OSEM 4i8s5mm and Q.Clear β700 (G); TOF-OSEM 4i8s5mm and Q.Clear β800 (H); TOF-OSEM 4i8s5mm and Q.Clear β900 (I); TOF-OSEM 4i8s5mm and Q.Clear β1000 (J).
Additional file 3: Supplementary file 3. Fig. 1 Bland-Altman plots assessing agreement between FBP with 5mm and Q.Clear with different β values, for the background variability data obtained from the NEMA phantom. FBP 5mm and Q.Clear β100 (A); FBP 5mm and Q.Clear β200 (B); FBP 5mm and Q.Clear β300 (C); FBP 5mm and Q.Clear β400 (D); FBP 5mm and Q.Clear β500 (E). Fig. 2 Bland-Altman plots assessing agreement between FBP with 5mm and Q.Clear with different β values, for the background variability data obtained from the NEMA phantom. FBP 5mm and Q.Clear β600 (F); FBP 5mm and Q.Clear β700 (G); FBP 5mm and Q.Clear β800 (H); FBP 5mm and Q.Clear β900 (I); FBP 5mm and Q.Clear β1000 (J).
Additional file 4: Supplementary file 4. Fig. 1 Bland-Altman plots assessing agreement between TOF-OSEM 4iteration, 8subsets with 5mm (4i8s5mm) and Q.Clear with different β values, for the background variability data obtained from the NEMA phantom. TOF-OSEM 4i8s5mm and Q.Clear β100 (A); TOF-OSEM 4i8s5mm and Q.Clear β200 (B); TOF-OSEM 4i8s5mm and Q.Clear β300 (C); TOF-OSEM 4i8s5mm and Q.Clear β400 (D); TOF-OSEM 4i8s5mm and Q.Clear β500 (E). Fig. 2 Bland-Altman plots assessing agreement between TOF-OSEM 4iteration, 8subsets with 5mm (4i8s5mm)and Q.Clear with different β values, for the background variability data obtained from the NEMA phantom. TOF-OSEM 4i8s5mm and Q.Clear β600 (F); TOF-OSEM 4i8s5mm and Q.Clear β700 (G); TOF-OSEM 4i8s5mm and Q.Clear β800 (H); TOF-OSEM 4i8s5mm and Q.Clear β900 (I); TOF-OSEM 4i8s5mm and Q.Clear β1000 (J).
Acknowledgements
AAST is funded by the British Heart Foundation (BHF/FS/19/34/34354) and is a recipient of a Wellcome Trust award (221295/Z/20/Z).
The authors would like to thank Nicholas Keat (Invicro UK) for the excellent support with the NEMA phantom analysis and Gabrielle Azzopardi (Invicro UK) for the valuable contribution with phantom preparation.
Code availability
All analyses were done using custom made code at inviCRO, a Konica Minolta Company and is subject to proprietary restrictions on sharing. However, details on fundamentals of this software are described in the manuscript.
Abbreviations
- PET
Positron emission tomography
- FBP
Filtered back-projection
- OSEM
Ordered subset expectation maximization
- BSREM
Block sequential regularized expectation maximization
- BPL
Bayesian penalized likelihood
- EM
Expectation-maximization
- GE
General Electric
- CT
Computed tomography
- MR
Magnetic resonance
- NEMA
National Electrical Manufacturers Association
- IQ
Image quality
- kBq
Kilobecquerel
- mL
Millilitre
- TOF
Time of flight
- 3D
Three-dimensional
- IDL
Interactive Data Language
- ROI
Regions of interest
- FWHM
Full-width half maximum
- VOI
Volume of interest
- SNR
Signal-to-noise ratio
- cc
Cubic centimetre
Authors’ contributions
DR, WH and AAST are responsible for study conception and design. DR was also responsible for data collection and analysis. All authors contributed equally to data interpretation and manuscript drafting. All authors read and approved the final manuscript.
Author’s information
DR is a Senior Research PET Technologist at Invicro, London. DR holds a BSc in Nuclear Medicine and a MSc in Neuroimaging for Research. DR is currently serving has a portfolio Assessor for The Register of Clinical Technologists and has acted as a reviewer for the European Society of Molecular Imaging (ESMI) and World Molecular Imaging Congress (WMIC) congresses.
Dr. WH is head of imaging physics at the Invicro imaging centre in London. Dr. WH’s doctorate is in experimental atomic physics, but WH has worked in medical imaging for over 25 years, in the NHS, academia, and industry, and for the last 20 years has specialised in positron emission tomography. Recent research interests include motion correction, dosimetry, dose optimisation and PET-MR.
Dr. AAST is a Senior Research Fellow in Positron Emission Tomography (PET) imaging at the University of Edinburgh. Dr. AAST serves as reviewer for various journals, funding bodies and international congresses, including the World Molecular Imaging Congress (WMIC). Dr. AAST is the co-chair of the STANDARD group of the European Society of Molecular Imaging (ESMI), founder of the “PET is Wonderful” group and member of the Molecular Imaging Committee of the Scottish Imaging Network: A Platform for Scientific Excellence (SINAPSE).
Funding
Not applicable.
Availability of data and materials
The datasets generated and analysed during the current study are not publicly available due to proprietary restrictions but are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tarantola G, Zito F, Gerundini P. PET instrumentation and reconstruction algorithms in whole-body applications. J Nucl Med. 2003;44(5):756–769. [PubMed] [Google Scholar]
- 2.te Riet J, Rijnsdorp S, Roef MJ, Arends AJ. Evaluation of a Bayesian penalized likelihood reconstruction algorithm for low-count clinical 18F-FDG PET/CT. EJNMMI Phys. 2019;6(1):32. doi: 10.1186/s40658-019-0262-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chilcott AK, Bradley KM, McGowan DR. Effect of a Bayesian penalized likelihood PET reconstruction compared with ordered subset expectation maximization on clinical image quality over a wide range of patient weights. Am J Roentgenol. 2018;210(1):153–157. doi: 10.2214/AJR.17.18060. [DOI] [PubMed] [Google Scholar]
- 4.Deidda D, Efthimiou N, Manber R, et al. 2016 IEEE Nuclear Science Symposium, Medical Imaging Conference and Room-Temperature Semiconductor Detector Workshop (NSS/MIC/RTSD) Strasbourg: IEEE; 2016. Comparative evaluation of image reconstruction methods for the siemens PET-MR scanner using the stir library; pp. 1–6. [Google Scholar]
- 5.Morey AM, Kadrmas DJ. Effect of varying number of OSEM subsets on PET lesion detectability. J Nucl Med Technol. 2013;41(4):268–273. doi: 10.2967/jnmt.113.131904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reilhac A, Tomeï S, Buvat I, Michel C, Keheren F, Costes N. Simulation-based evaluation of OSEM iterative reconstruction methods in dynamic brain PET studies. NeuroImage. 2008;39(1):359–368. doi: 10.1016/j.neuroimage.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 7.Walker MD, Asselin M-C, Julyan PJ, Feldmann M, Talbot PS, Jones T, Matthews JC. Bias in iterative reconstruction of low-statistics PET data: benefits of a resolution model. Phys Med Biol. 2011;56(4):931–949. doi: 10.1088/0031-9155/56/4/004. [DOI] [PubMed] [Google Scholar]
- 8.Jian Y, Planeta B, Carson RE. Evaluation of Bias and Variance in Low-count OSEM List Mode Reconstruction. Phys Med Biol. 2015;60(1):15–29. doi: 10.1088/0031-9155/60/1/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyrzykowski M, Siminiak N, Kaźmierczak M, et al. Impact of the Q.Clear reconstruction algorithm on the interpretation of PET/CT images in patients with lymphoma. EJNMMI Res. 2020;10:99. doi: 10.1186/s13550-020-00690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Texte E, Gouel P, Thureau S, et al. Impact of the Bayesian penalized likelihood algorithm (Q.Clear®) in comparison with the OSEM reconstruction on low contrast PET hypoxic images. EJNMMI Phys. 2020;7:28. doi: 10.1186/s40658-020-00300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teoh EJ, McGowan DR, Schuster DM, et al. Bayesian penalised likelihood reconstruction (Q.Clear) of 18F-fluciclovine PET for imaging of recurrent prostate cancer: semi-quantitative and clinical evaluation. Br J Radiol. 2018;91(1085):20170727. doi: 10.1259/bjr.20170727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witkowska-Patena E, Budzyńska A, Giżewska A, Dziuk M, Walęcka-Mazur A. Ordered subset expectation maximisation vs Bayesian penalised likelihood reconstruction algorithm in 18F-PSMA-1007 PET/CT. Ann Nucl Med. 2020;34(3):192–199. doi: 10.1007/s12149-019-01433-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshii T, Miwa K, Yamaguchi M, et al. Optimization of a Bayesian penalized likelihood algorithm (Q.Clear) for 18F-NaF bone PET/CT images acquired over shorter durations using a custom-designed phantom. EJNMMI Phys. 2020;7:56. doi: 10.1186/s40658-020-00325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huizing DMV, Koopman D, van Dalen JA, Gotthardt M, Boellaard R, Sera T, et al. Multicentre quantitative 68Ga PET/CT performance harmonisation. EJNMMI Phys. 2019;6:19. doi: 10.1186/s40658-019-0253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medican Imaging Phantsom. Data Spectrum Corporation. https://www.spect.com/. Accessed 16 Apr 2020.
- 16.Jakoby BW, Bercier Y, Watson CC, Bendriem B, Townsend DW. Performance characteristics of a new LSO PET/CT scanner with extended axial field-of-view and PSF reconstruction. IEEE Trans Nucl Sci. 2009;56(3):633–639. doi: 10.1109/TNS.2009.2015764. [DOI] [Google Scholar]
- 17.Caribé PRRV, Koole M, D’Asseler Y, Deller TW, Van Laere K, Vandenberghe S. NEMA NU 2–2007 performance characteristics of GE Signa integrated PET/MR for different PET isotopes. EJNMMI Physics. 2019;6(1):11. doi: 10.1186/s40658-019-0247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conti M, Bendriem B, Casey M, Chen M, Kehren F, Michel C, Panin V. First experimental results of time-of-flight reconstruction on an LSO PET scanner. Phys Med Biol. 2005;50(19):4507–4526. doi: 10.1088/0031-9155/50/19/006. [DOI] [PubMed] [Google Scholar]
- 19.Kadrmas DJ, Casey ME, Conti M, Jakoby BW, Lois C, Townsend DW. Impact of time-of-flight on PET tumor detection. J Nucl Med. 2009;50(8):1315–1323. doi: 10.2967/jnumed.109.063016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogasch JM, Suleiman S, Hofheinz F, et al. Reconstructed spatial resolution and contrast recovery with Bayesian penalized likelihood reconstruction (Q.Clear) for FDG-PET compared to time-of-flight (TOF) with point spread function (PSF) EJNMMI Physics. 2020;7(1):2. doi: 10.1186/s40658-020-0270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Electrical Manufacturers Association (NEMA), Standards Publication NU 2-2012, Performance Measurements of Positron Emission Tomographs (NEMA, Rosslyn, VA, 2012). https://www.nema.org/standards/view/Performance-Measurements-of-Positron-Emission-Tomographs. Accessed 28 June 2020.
- 22.IDL Software - Interactive Data Visualization Solution. https://www.harrisgeospatial.com/Software-Technology/IDL. Accessed 16 Apr 2020.
- 23.Software. Invicro. https://invicro.com/capabilities/software/. Accessed 16 Apr 2020.
- 24.Liu H, Wang K, Tian J. Postreconstruction filtering of 3D PET images by using weighted higher-order singular value decomposition. Biomed Eng Online. 2016;15(1). 10.1186/s12938-016-0221-y. [DOI] [PMC free article] [PubMed]
- 25.Ziegler S, Jakoby BW, Braun H, Paulus DH, Quick HH. NEMA image quality phantom measurements and attenuation correction in integrated PET/MR hybrid imaging. EJNMMI Physics. 2015;2(1):18. doi: 10.1186/s40658-015-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prism - GraphPad. https://www.graphpad.com/scientific-software/prism/. Accessed 28 June 2020
- 27.Conti M, Eriksson L. Physics of pure and non-pure positron emitters for PET: a review and a discussion. EJNMMI Phys. 2016;3(1):8. doi: 10.1186/s40658-016-0144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teoh EJ, McGowan DR, Macpherson RE, Bradley KM, Gleeson FV. Phantom and clinical evaluation of the Bayesian penalized likelihood reconstruction algorithm Q.Clear on an LYSO PET/CT system. J Nucl Med. 2015;56(9):1447–1452. doi: 10.2967/jnumed.115.159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynés-Llompart G, Gámez-Cenzano C, Vercher-Conejero JL, Sabaté-Llobera A, Calvo-Malvar N, Martí-Climent JM. Phantom, clinical, and texture indices evaluation and optimization of a penalized-likelihood image reconstruction method (Q.Clear) on a BGO PET/CT scanner. Med Phys. John Wiley & Sons, Ltd. 2018;45:3214–3222. doi: 10.1002/mp.12986. [DOI] [PubMed] [Google Scholar]
- 30.Parvizi N, Franklin JM, McGowan DR, Teoh EJ, Bradley KM, Gleeson FV. Does a novel penalized likelihood reconstruction of 18F-FDG PET-CT improve signal-to-background in colorectal liver metastases? Eur J Radiol. 2015;84(10):1873–1878. doi: 10.1016/j.ejrad.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 31.Sah B-R, Stolzmann P, Delso G, et al. Clinical evaluation of a block sequential regularized expectation maximization reconstruction algorithm in 18F-FDG PET/CT studies. Nucl Med Commun. 2017;38(1):57–66. doi: 10.1097/MNM.0000000000000604. [DOI] [PubMed] [Google Scholar]
- 32.Passalaqua S, Kappadath S, Branch D, et al. Qualitative and quantitative evaluation of regularized PET image reconstruction. J Nucl Med. 2014;55(supplement 1):579. [Google Scholar]
- 33.Ahn S, Ross SG, Asma E, Miao J, Jin X, Cheng L, Wollenweber SD, Manjeshwar RM. Quantitative comparison of OSEM and penalized likelihood image reconstruction using relative difference penalties for clinical PET. Phys Med Biol. 2015;60(15):5733–5751. doi: 10.1088/0031-9155/60/15/5733. [DOI] [PubMed] [Google Scholar]
- 34.O’Doherty J, DR MG, Abreu C, Barrington S. Effect of Bayesian-penalized likelihood reconstruction on [13N]-NH3 rest perfusion quantification. J Nucl Cardiol. 2017;24(1):282–290. doi: 10.1007/s12350-016-0554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary file 1. Fig. 1 Bland-Altman plots assessing agreement between FBP with 5mm and Q.Clear with different β values, for the contrast recovery data obtained from the NEMA phantom. FBP 5mm and Q.Clear β100 (A); FBP 5mm and Q.Clear β200 (B); FBP 5mm and Q.Clear β300 (C); FBP 5mm and Q.Clear β400 (D); FBP 5mm and Q.Clear β500 (E). Fig. 2 Bland-Altman plots assessing agreement between FBP with 5mm and Q.Clear with different β values, for the contrast recovery data obtained from the NEMA phantom. FBP 5mm and Q.Clear β600 (F); FBP 5mm and Q.Clear β700 (G); FBP 5mm and Q.Clear β800 (H); FBP 5mm and Q.Clear β900 (I); FBP 5mm and Q.Clear β1000 (J).
Additional file 2: Supplementary file 2. Fig. 1 Bland-Altman plots assessing agreement between TOF-OSEM 4iteration, 8subsets with 5mm (4i8s5mm) and Q.Clear with different β values, for the contrast recovery data obtained from the NEMA phantom. TOF-OSEM 4i8s5mm and Q.Clear β100 (A); TOF-OSEM 4i8s5mm and Q.Clear β200 (B); TOF-OSEM 4i8s5mm and Q.Clear β300 (C); TOF-OSEM 4i8s5mm and Q.Clear β400 (D); TOF-OSEM 4i8s5mm and Q.Clear β500 (E). Fig. 2 Bland-Altman plots assessing agreement between TOF-OSEM 4iteration, 8subsets with 5mm (4i8s5mm) and Q.Clear with different β values, for the contrast recovery data obtained from the NEMA phantom. TOF-OSEM 4i8s5mm and Q.Clear β600 (F); TOF-OSEM 4i8s5mm and Q.Clear β700 (G); TOF-OSEM 4i8s5mm and Q.Clear β800 (H); TOF-OSEM 4i8s5mm and Q.Clear β900 (I); TOF-OSEM 4i8s5mm and Q.Clear β1000 (J).
Additional file 3: Supplementary file 3. Fig. 1 Bland-Altman plots assessing agreement between FBP with 5mm and Q.Clear with different β values, for the background variability data obtained from the NEMA phantom. FBP 5mm and Q.Clear β100 (A); FBP 5mm and Q.Clear β200 (B); FBP 5mm and Q.Clear β300 (C); FBP 5mm and Q.Clear β400 (D); FBP 5mm and Q.Clear β500 (E). Fig. 2 Bland-Altman plots assessing agreement between FBP with 5mm and Q.Clear with different β values, for the background variability data obtained from the NEMA phantom. FBP 5mm and Q.Clear β600 (F); FBP 5mm and Q.Clear β700 (G); FBP 5mm and Q.Clear β800 (H); FBP 5mm and Q.Clear β900 (I); FBP 5mm and Q.Clear β1000 (J).
Additional file 4: Supplementary file 4. Fig. 1 Bland-Altman plots assessing agreement between TOF-OSEM 4iteration, 8subsets with 5mm (4i8s5mm) and Q.Clear with different β values, for the background variability data obtained from the NEMA phantom. TOF-OSEM 4i8s5mm and Q.Clear β100 (A); TOF-OSEM 4i8s5mm and Q.Clear β200 (B); TOF-OSEM 4i8s5mm and Q.Clear β300 (C); TOF-OSEM 4i8s5mm and Q.Clear β400 (D); TOF-OSEM 4i8s5mm and Q.Clear β500 (E). Fig. 2 Bland-Altman plots assessing agreement between TOF-OSEM 4iteration, 8subsets with 5mm (4i8s5mm)and Q.Clear with different β values, for the background variability data obtained from the NEMA phantom. TOF-OSEM 4i8s5mm and Q.Clear β600 (F); TOF-OSEM 4i8s5mm and Q.Clear β700 (G); TOF-OSEM 4i8s5mm and Q.Clear β800 (H); TOF-OSEM 4i8s5mm and Q.Clear β900 (I); TOF-OSEM 4i8s5mm and Q.Clear β1000 (J).
Data Availability Statement
The datasets generated and analysed during the current study are not publicly available due to proprietary restrictions but are available from the corresponding author on reasonable request.