Abstract
Saline environments are extreme habitats with a high diversity of microorganisms source of a myriad of biomolecules. These microorganisms are assigned as extremophiles recognized to be producers of new natural compounds, which can be synthesized by helping to survive under harshness and extreme conditions. In Brazil, in the saline and semi-arid region of Areia Branca (Caatinga biome), halotolerant bacteria (able to growth at high NaCl concentrations) were isolated from rhizosphere of native plants Blutaparon portulacoides and Spergularia sp. and their biopolymer production was studied. A total of 25 bacterial isolates were identified at genus level based on 16S rRNA gene sequence analysis. Isolates were mainly Gram-positive bacteria from Bacillaceae, Staphylococcaceae, Microbacteriaceae, and Bacillales XII incertae sedis families, affiliates to Bacillus, Staphylococcus, Curtobacterium, and Exiguobacterium genera, respectively. One of the Gram-negative isolates was identified as member of the Pseudomonadaceae family, genus Pseudomonas. All the identified strains were halotolerant bacteria with optimum growth at 0.6–2.0 M salt concentrations. Assays for biopolymer production showed that the halotolerant strains are a rich source of compounds as polyhydroxyalkanoates (PHA), biodegradable biopolymer, such as poly(3-hydroxybutyrate) (PHB) produced from low-cost substrates, and exopolysaccharides (EPS), such as hyaluronic acid (HA), metabolite of great interest to the cosmetic and pharmaceutical industry. Also, eight bacterial EPS extracts showed immunostimulatory activity, promising results that can be used in biomedical applications. Overall, our findings demonstrate that these biomolecules can be produced in culture medium with 0.6–2.0 M NaCl concentrations, relevant feature to avoid costly production processes. This is the first report of biopolymer-producing bacteria from a saline region of Caatinga biome that showed important biological activities.
Supplementary Information
The online version of this article (10.1007/s42770-021-00426-1) contains supplementary material, which is available to authorized users.
Keywords: Biopolymers, Halotolerant microorganisms, Polyhydroxyalkanoates (PHA), Exopolysaccharides (EPS), Immunostimulatory activity, Hyaluronic acid (HA), Caatinga biome
Introduction
Saline environments are rich source of natural compounds produced by halophilic microorganisms [1]; the research in these extreme habitats becomes important for discovering new molecules of biopolymers classes with innovative biotechnological applications. In Brazil, there are saline and semi-arid regions in the Caatinga biome that represent an exceptional understudied ecosystem. Caatinga is a unique biome located in northeastern Brazil (3–17° S to 35–45° W), with an area of approximately 900,000 km2 [2], and it is characterized by a semi-arid climate with vegetation adapted to high temperatures, low precipitation, and uneven rainfall distribution throughout the year [2, 3]. There are some studies on the biotechnological potential of microorganisms from this region [4].
Halotolerant microorganisms do not need salt for their growth, although tolerate high concentrations of NaCl (≥ 1 M) [5]. These microorganisms can be found in the three domains of life: Archaea, Bacteria, and Eukarya, and have been isolated from saline environments at different geographic areas such as lakes and salt marshes, deserts, and seas [6]. Halotolerant bacteria were isolated in the salt and semi-arid area of Areia Branca at Mossoró district located in Rio Grande do Norte state. These microorganisms are producers of several compounds for pharmaceutical, cosmetic, and food industry [1, 7, 8]. Specially, the biopolymers synthetized by halophilic bacteria are interesting macromolecules that have been used in biomedical and agro-industrial applications due to their biocompatibility and biodegradable properties [7, 9]. Also, macromolecules such as EPS, polymers of high molecular weight synthesized and accumulate at the surface cell, exhibited activity as antioxidants, immunostimulants, antiviral, and antitumoral [10, 11]. Hence, the present study aims to evaluate the biopolymer production by halotolerant bacteria isolated from rhizosphere of two native plants of the Caatinga biome. It is assumed that the natural polymers of extreme microorganisms may have novel or different chemical compositions from those already marketed and used by industry [8].
Material and methods
Isolates and bacterial identification
The bacterial isolates were kindly given by the Microorganisms Collection of the Brazilian Agricultural Research Corporation (Embrapa), Laboratory of Environmental Microbiology, at Jaguariúna, SP. These microorganisms were previously isolated from rhizosphere of two native plants Blutaparon portulacoides and Spergularia sp. (Supplementary material. Fig. S1) at a saline site of the Caatinga biome, Mossoró district, Rio Grande do Norte state, near Areia Branca saltworks, coordinates 4 57′ 22″ S–37 08′ 13″ W. Isolated bacteria grew under aerobic conditions in tryptone soya broth (TSB) medium and tryptone soya agar (TSA) medium at 30 °C.
Sequence analysis of the 16S rRNA gene and phylogenetic reconstruction
The bacterial isolates were identified by 16S rRNA gene sequencing. Genomic DNA was extracted using the Wizard® Genomic DNA kit (Promega) from 5 mL of an exponentially growing microbial culture that was centrifuged at 8000×g for 10 min. The 16S rRNA gene was amplified using the universal primers 27F (5′GAGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACT-3′) [12]. All PCRs (in a 50-μL total volume reaction) contained ≤ 0.1 μg of DNA template, 0.2 μM of each primer, 5X Master Mix® (Promega). The PCR program was an initial denaturation step at 95 °C for 2 min, followed by 40 cycles at 95 °C for 1 min, 60 °C for 30 s, 72 °C for 1 min, and a final elongation step of 10 min at 72 °C. Samples were purified using GFX MicroSpin Column. Sequencing was performed with four primers 1492R, 27F, 536f (5′-CAGCMGCCGCGGTAATWC-3′) and 782r (5′-ACCAGGGTATCTAATCCTGT-3′) for each sample. Sequencing analysis was achieved using ABI 3730 DNA Analyser. Raw sequence data were imported into ChromasPro 2.1.2 and manual curation was performed. Sequences were compared against sequences of type strains using EzBioCloud Database [13] (https://www.ezbiocloud.net/). A multiple alignment of obtained sequences with 16S rRNA sequences of type strains was carried out using the MUSCLE tool [14]. We constructed phylogenetic trees by the neighbor-joining method [15] using MEGA 7 [16] and maximum likelihood (ML) [17] based on the Tamura-Nei model [18] and performed using MEGA 7. Tree topologies were examined using bootstrap analysis with 1000 replications.
Bacterial growth in different NaCl concentrations
To evaluate the salinity tolerance, all the isolates were grown in TSB medium supplemented with four different concentrations of NaCl (M): 0.1, 0.6, 1.0, and 2.0, on plates of 96 wells. Cultures were incubated at 30 °C for 48 h. Bacterial growth was monitored by optical density (OD) at 600 nm on the Synergy H1 Microplate Reader (Biotek Instruments, Inc., USA). The experiments were performed in triplicate.
Polyhydroxyalkanoate production
To select PHA-producing strains, Caatinga isolates were grown in mineral salt medium (MSM) (Table S1) [19], supplemented with glucose 10 g/L and (NH4)2SO4 1 g/L as carbon and nitrogen sources, respectively, at pH 7.0. Solid media were obtained by adding agar (20 g/L). The plates were incubated at 30 °C for 48 h. PHA-producing strains were identified by dark blue color after Sudan Black B staining [20].
The PHA-producing strains selected after Sudan Black B staining on solid MSM were cultivated in shake flask experiments to confirm their PHA production capability. Bacterial strains were streaked on tryptone soya agar (TSA) for 24 h at 30 °C, with subsequent transferring of bacterial colonies to 250 mL Erlenmeyer flasks containing 100 mL tryptone soya broth (TSB) for 24 h, at 30 °C and 180 rpm. Aliquots of 1 mL from these cultivations were used to inoculate 250 mL Erlenmeyer flasks containing 100 mL MSM added to 10 g/L glucose and 1 g/L (NH4)2SO4, which were incubated for 72 h, at 30 °C and 180 rpm. Strains that showed ≥ 40% of PHA (% cell dry weight, %CDW) were selected and grown in MSM containing different carbon sources, such as glucose, glycerol, and xylose at 10 g/L, and different concentrations of NaCl, 0 M, 0.6 M, 1 M, and 2 M. The strains were incubated for 72 h, at 30 °C and 180 rpm.
Polyhydroxyalkanoate determination
PHA amount and composition were determined by propanolysis as described by Riis & Mai [21] and analyzed by GC method as described by Mendonça et al. [22]. Briefly, the polyesters were analyzed in an Agilent 7890A GC System (Agilent Technology, Santa Clara, California, USA) equipped with a HP5 capillary column after sample split (1:25). Helium (0.6 mL/min) was used as carrier gas. Injector and FID temperature were 250 °C and 300 °C, respectively. The oven was programmed to keep temperature at 100 °C for 3 min increasing temperature at a rate of 6 °C/min up to 240 °C, which was then maintained for 6 min. Benzoic acid was used as the internal standard. External standards were 3-hydroxybutyrate homopolymer [P(3HB)] (Sigma-Aldrich, Saint Louis, Missouri, USA), P3HB-co-3HV (Sigma-Aldrich, Saint Louis Missouri, USA) and PHAMCL produced by Pseudomonas putida ATCC 29347 from different fatty acids or by Pseudomonas sp. LFM046 from glucose.
Exopolysaccharide production
The qualitative determination of exopolysaccharide (EPS) production was performed according to Paulo et al. [23]. Caatinga isolates were inoculated onto 6-mm diameter paper discs disposed in an exopolysaccharide production medium (EPS-M) (Table S1) [24], at pH 7.5. The solid media were obtained by adding 15 g/L agar. The plates were incubated at 30 °C for 48 h. The production was characterized by the size of the halo produced and its slime appearance. The production of EPS was confirmed by mixing a portion of the mucoid substance in 2 mL of absolute ethanol, where the formation of a precipitate indicated the presence of EPS [23]. The bacterial isolates tested in solid EPS-M and selected as EPS-producing strains were cultivated in shake flask experiments to produce EPS. Bacterial strains were streaked on TSA for 24 h at 30 °C, with subsequent transferring of bacterial colonies to 50 mL Erlenmeyer flasks containing 10 mL EPS-M for 4 days, at 30 °C and 180 rpm.
Exopolysaccharide extraction
EPS extraction was performed according to the methodology described by Liang et al. [25]. Bacterial cultures were autoclaved at 121 °C for 20 min to reduce viscosity and culture medium was harvested by 20 min centrifugation at 10,000g and 4 °C. The supernatants were filtered with a 1.2-μm membrane and vacuum. The filtrates were mixed with two volumes of methanol (MeOH) and stirred vigorously to be kept overnight at 4 °C. After extraction, the MeOH extracts were centrifuged at 10,000g for 15 min to recover the precipitates. Subsequently, the precipitates were washed three times with sterile distilled water and lyophilized to obtain the extract called crude EPS. The crude EPS was dissolved in distilled water and stirred for 3 min at 80 °C. Then, the crude EPS was mixed in four volumes of ethanol (EtOH) and stirred vigorously to be kept overnight at 4 °C. After extraction, the extracts in EtOH were centrifuged at 10,000g for 15 min to recover the precipitates. The precipitates were dissolved in distilled water and deproteinized in 1/5 volumes of Sevag Reagent (CHCl3-BuOH, v/v = 5/1) seven times. Deproteinized solution was dialyzed against distilled water, concentrated on a rotary evaporator Concentrator plus (Eppendorf, Hamburg, Germany) at reduced pressure (45 °C) to approximately 5 mL of an aqueous suspension and lyophilized to obtain the deproteinized EPS. The deproteinized EPS were stored at 4 °C to be evaluated in immunostimulatory activity assays.
Exopolysaccharide immunostimulatory activity
The macrophage activation to proinflammatory phenotype was evaluated by Griess assay using murine macrophage cell line (RAW 264.7). The Griess reaction indirectly measures the NO levels, a proinflammatory metabolite. Griess reagent is a 1:1 mixture of an acetic acid solution (60%) (v/v) containing N-(1-Naftil)-ethylenediamine 0.1% (w/v) and sulfonamide 1% (w/v) in acetic acid (30%) [26]. The RAW264.7 cells were exposed to bacterial EPS at concentrations 1, 10, and 100 μg/mL or 100 ng/mL LPS (positive control) and sterile saline as negative control (C-) for 48 h. After incubation, medium samples (50 μL) were mixed with Griess reagent (50 μL) and incubated in dark at room temperature during 15 min. The absorbance was measured at 570 nm using a spectrophotometer. Nitrite production was calculated by interpolation of linear regression based on a nitrite standard curve. The differences between C- and other groups was evaluated by analysis of variance (ANOVA) followed by Tukey’s post-test. a and b values < 0.05 were considered statistically significant. The Laboratory of Marine Bioprospection and Biotechnology area was certified by National Technical Commission on Biosafety (CTNBio) on Brazil (license number 6.249/2018) to work with biosafety level 2 genetic modified organisms. The project to work with immunostimulating effects of RAW 264.7 cells exposed to molecules from bacteria was authorized by CTNBio under the license number 6.021/2018.
Hyaluronic acid production
Isolates were grown in 10 mL of liquid medium (HA-M) (Table S1), at pH 7.0. The cultures were incubated 7 days, at 30 °C and 180 rpm, using as positive control Streptococcus equi CBMAI 0265 (Farrow and Callins subsp. zooepidemicus ATCC 39920) HA-producing strain. For the quantification of HA, a protocol described by YU et al. [27] was used with modifications. In a 1.5-mL Eppendorf tube containing 550 μL of acetic acid 3% were added 400 μL of bacteria cultures followed by addition of 50 μL Alcian Blue 8GX solution, shaken in a vortex and wormer for 30 s in a microwave. Samples were cold down at room temperature for 2 h and centrifuged at 8000g for 2 min. Absorbance at 540 nm was read in a Synergy H1 microplate reader (Biotek Instruments, Inc., Winooski, USA). HA determination was getting through calibration curve of HA reference control [27].
Results
A group of 25 halotolerant bacteria isolated from the rhizosphere of two plants, Spergularia sp. and B. portulacoides, of a saline region from the Caatinga biome (Fig. S1); 24 isolates (96%) were Gram-positive, while 1 (4%) was Gram-negative. Colonies of 18 isolates were opaque with irregular borders, and the other 7 strains had colonies with yellow, orange, and cream pigments. All the isolates grew aerobically within 24 h at 30 °C on TSB medium.
Phylogenetic analysis of halotolerant bacteria based on 16S rRNA gene sequence
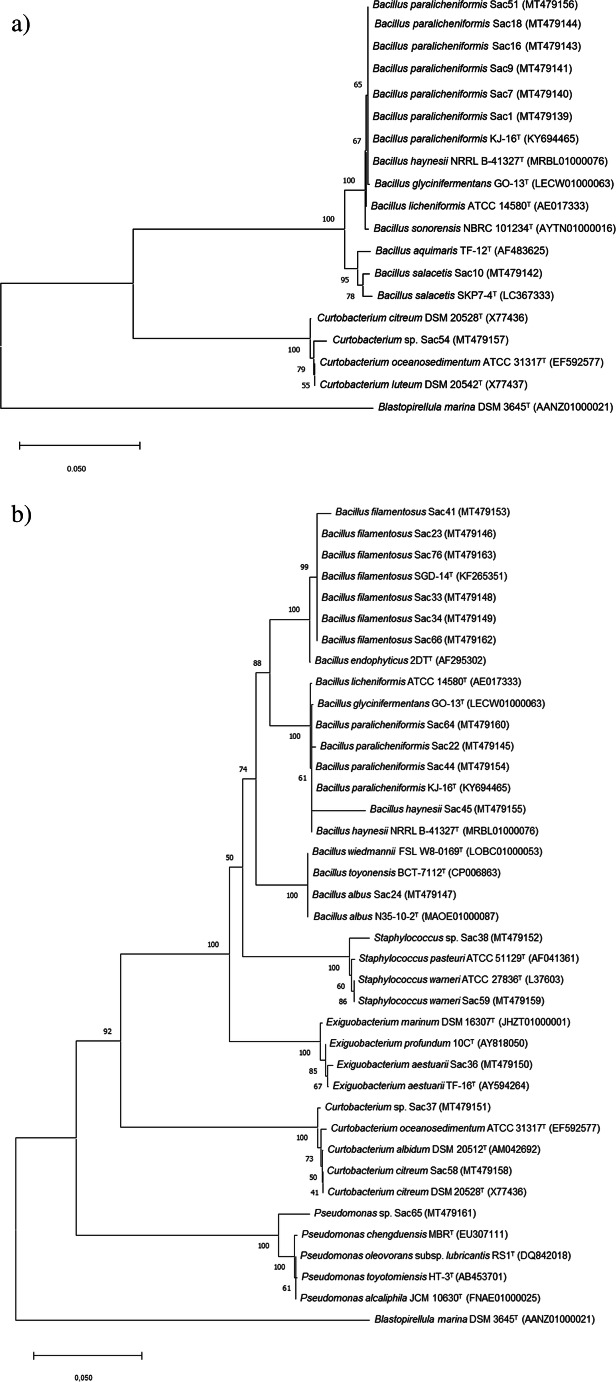
Sequence analysis of the 16S rRNA (1111–1400 bp) gene showed that strains belonged to the Firmicutes, Actinobacteria, and Proteobacteria phyla. The most abundant phylum was Firmicutes, represented by the Bacillaceae, Exiguobacteriaceae, and Staphylococcaceae families. Bacillus was the most abundant genus (18 strains), followed by Staphylococcus (2 strains) and Exiguobacterium (1 strain) (Table S2; Fig. 1). Actinobacteria and Proteobacteria phylum were represented by the Curtobacterium (3 strains) and Pseudomonas (1 strain) genera, respectively.
Fig. 1.
Phylogenetic tree based on 16S rRNA gene sequences of halotolerant bacterial isolates from the rhizosphere of Spergularia sp. (a), and B. portulacoides (b). Sequence accession numbers are given in brackets. The tree was constructed using neighbor-joining method. Blastopirellula marina was used as an outgroup
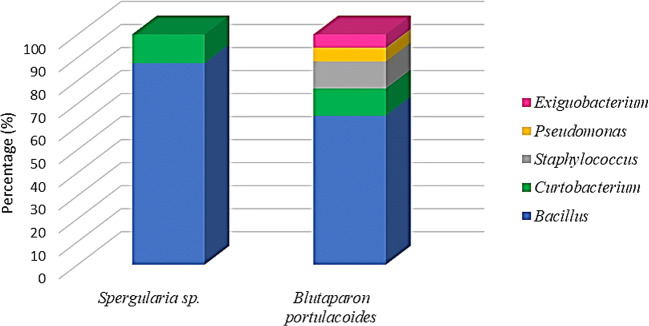
Bacillus and Curtobacterium were common genera isolated in both rhizospheric samples. Exiguobacterium, Staphylococcus, and Pseudomonas were genera only isolated from the rhizospheric sample of B. portulacoides plant (Table S2; Fig. 2).
Fig. 2.
Distribution of the relative abundance of the bacterial genera isolated from the rhizosphere of Spergularia sp. and B. portulacoides
Salinity tolerance
Salt tolerance of the Caatinga isolates has been evaluated and summarized in Fig. 3 (Table S3). From the rhizosphere of Spergularia sp., most of the strains could grow up to 2.0 M NaCl, and the growth was better at 1.0 M NaCl. Most of the strains from the rhizosphere of B. portulacoides could grow up to 2.0 M NaCl and showed better growth at that concentration. All bacterial strains were halotolerant.
Fig. 3.
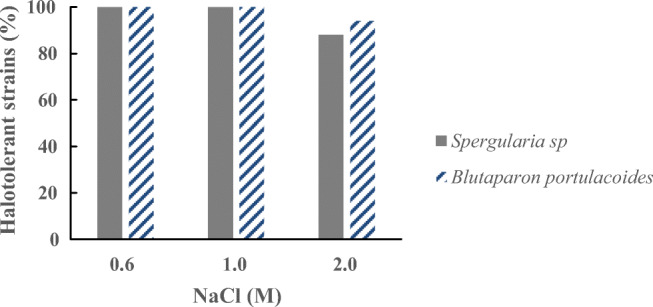
NaCl tolerance profile of halotolerant bacterial isolates from the rhizosphere of Spergularia sp. and B. portulacoides
Screening assays of microbial biopolymer production
Polyhydroxyalkanoate-producing isolates
Among our 25 isolates, 15 grew on mineral salt solid media containing glucose. However, only 4 isolates, Sac23, Sac33, Sac34, and Sac66, all belonging to the genus Bacillus and obtained from the rhizosphere of B. portulacoides, accumulated PHA as confirmed by Sudan black B staining reaction in solid MSM (Fig. S2).
The 4 PHA-producing isolates were cultivated in MSM containing glucose as sole carbon source to evaluate the amount and type of monomer composition on their PHA. After propanolysis reaction and consecutive polymer determination, a PHA constituted of 3-hydroxybutyrate homopolymer [P(3HB)] from glucose as sole carbon source was detected. PHA content produced by the isolates tested ranged from approximately 27 to 51% of cell dry weight (CDW) and among them, strains Sac33 and Sac34 (Bacillus filamentosus) showed ≥ 40% of PHA (%CDW) (Table 1). Based on these results, Sac33 and Sac34 were selected for further PHA production experiments.
Table 1.
PHA [P(3HB)] production by four halotolerant bacterial strains from the rhizosphere of B. portulacoides of a saline region of the Caatinga biome. Average and standard deviation of duplicate cultivation utilizing glucose as sole carbon source
| Bacterial isolate | Identified species | P(3HB) (%CDW) |
|---|---|---|
| Sac23 | B. filamentosus | 30.1 ± 5.1 |
| Sac33 | B. filamentosus | 40.5 ± 3.0 |
| Sac34 | B. filamentosus | 51.1 ± 2.9 |
| Sac66 | B. filamentosus | 26.9 ± 3.9 |
Both strains, Sac33 and Sac34, identified as B. filamentosus, were cultivated in MSM containing glucose, glycerol, and xylose as sole carbon source and supplemented with four different concentrations of NaCl (M). Polymer determination showed a PHA constituted of P(3HB) homopolymer from the three carbon sources evaluated (Table S4, Fig. 4).
Fig. 4.
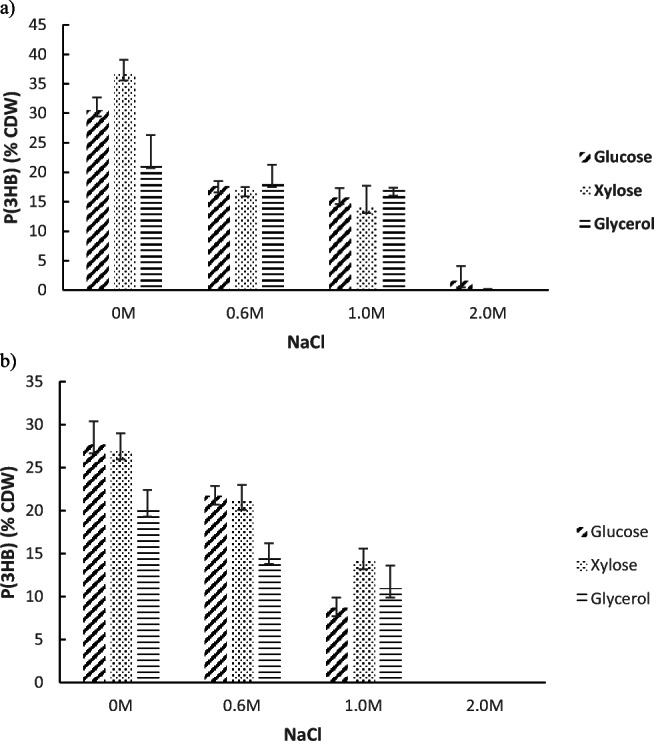
Accumulation profile of PHA [P(3HB)] by isolates Sac33 and Sac34 in MSM with NaCl and supplement with glucose, xylose and glycerol. B. filamentosus Sac33 (a) and B. filamentosus Sac34 (b). Average and standard deviation of triplicate cultivation after 72 h
Strains Sac33 and Sac34 were found to accumulate PHA in media containing 0.6 and 1.0 M of NaCl with any of the three carbon sources evaluated, obtaining concentrations of P(3HB) between 0.2 and 0.8 g/L, in both strains (Table S4). Nonetheless, both strains could not accumulate PHA in media supplemented with NaCl 2.0 M. The higher P(3HB) accumulation observed in both strains was 0.9 g/L (30.5% PHA) and 0.8 g/L (27.7% PHA) for Sac33 and Sac34, respectively.
Exopolysaccharide-producing isolates
A total of 12 halotolerant bacterial isolates were able to produce EPS. The presence of a translucent or creamy material involving a mucoid colony was indicative of EPS production potential (Fig. S3). The EPS-producing isolates were Sac65 (Pseudomonas sp.), Sac54 (Curtobacterium sp.), and ten isolates of the genus Bacillus: Sac1, Sac7, Sac9, Sac16, Sac18, Sac22, Sac51, Sac64 (B. paralicheniformis), Sac10 (B. salacetis), and Sac45 (B. haynesii). Most of the EPS-producing strains (67%) were isolated from the rhizosphere of Spergularia sp.
Immunostimulatory activity
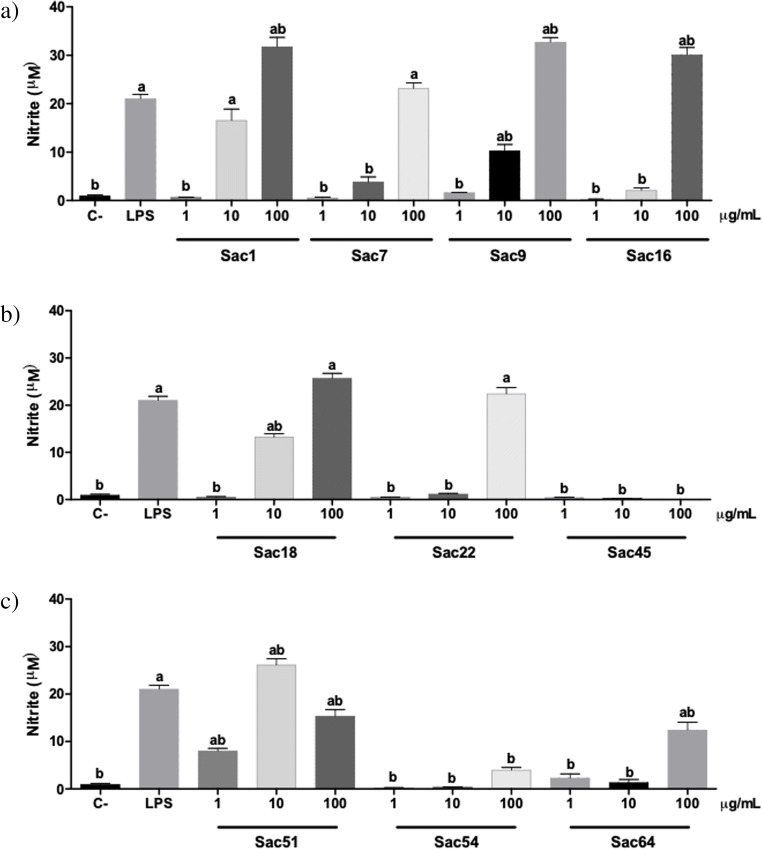
Ten of the twelve isolates with positive EPS production were selected and cultivated in EPS-M medium to produced exopolysaccharides. The EPS produced were extracted and used for evaluation of immunostimulatory activity by the Griess test that showed the pharmacological responses produced by murine macrophages (RAW 264.7) treated with the bacterial EPS (Fig. 5).
Fig. 5.
Evaluation of immunostimulatory activity by the Griess test. The figure shows the nitrite levels in the supernatant of murine macrophages (RAW264.7). Cells were exposed to sterile saline (negative control), 100 ng/mL lipopolysaccharide from Escherichia coli (LPS) (positive control), and the Caatinga bacterial exopolysaccharides (Sac) after 48 hours of incubation. The obtained absorbance values were transformed into nitrite concentration by interpolation and then evaluated by ANOVA followed by Tukey's post-test comparing all the samples. a = p <0,05 when compared with C-; and b = p <0,05 when compared with LPS. a) Assays carried out with strains belonged to B. paralicheniformis (Sac1, Sac7, Sac9 and Sac16) , b) B. paralicheniformis (Sac18 and Sac22) and B. haynesii (Sac45), c) B. paralicheniformis (Sac51 and Sac64) and C. oceanosedimentum (Sac54)
Bacterial EPS with activity detected on murine macrophages (RAW264.7) were produced, mostly, by strains belonging to the genus Bacillus that were isolated from the rhizosphere of Spergularia sp. (Sac1, Sac7, Sac9, Sac16, Sac18, and Sac51). Eight EPS increased nitrite (NO2−) concentration in the murine macrophage supernatant. These cells showed higher nitrite concentration in the Griess assay when were treated with 100 μg/mL of the bacterial EPS (Fig. 5). Four bacterial EPS of the strains identified as Bacillus paralicheniformis Sac1, Sac9, Sac16 (Fig. 5a), and Sac51 (Fig. 5c) increased NO2− concentration more than the positive control (LPS). Also, it should be noted that bacterial EPS in lower concentrations thereof, 1 and 10 μg/mL, obtained from strains Sac1, Sac9 (Fig. 5a), Sac18 (Fig. 5b), and Sac51 (Fig. 5c) increased the nitrite concentration in murine macrophages. The bacterial EPS from Sac45 (Fig. 5b) and Sac54 (Fig. 5c) strains did not stimulate NO release by macrophages.
Hyaluronic acid production
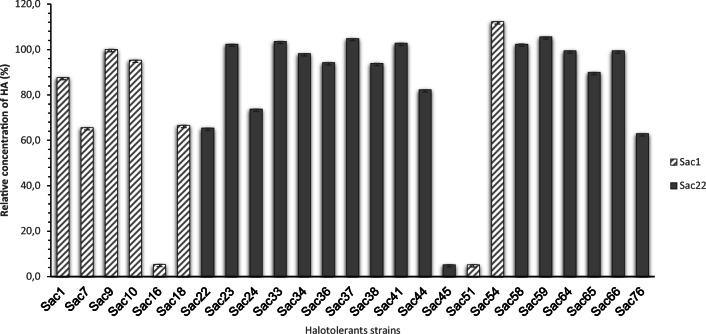
All the strains were able to produce hyaluronic acid (Table S5 and S6, Fig. 6).
Fig. 6.
Relative concentration of hyaluronic acid (%) obtained from halotolerant strains. Positive control (dashed line-100%), Streptococcus equi subsp. zooepidemicus ATCC 39920. Average and standard deviation of triplicate cultivation after 7 days
As shown in Fig. 6, the strain Sac54 identified as Curtobacterium sp. showed the highest relative concentration of HA compared to the control (line). It is also observed that the strains identified as B. filamentosus (Sac23, Sac33, and Sac41), Curtobacterium sp. (Sac37 and Sac58), and S. warneri (Sac59) showed higher HA production than the control. Most of the strains (68%) produced similar or higher relative concentrations than the positive control.
Discussion
To survive in saline environment, plants develop a number of physiological and biochemical mechanisms including synthesis of osmolytes, ion compartmentalization, activation of antioxidant enzyme, hormone modulation, ion exclusion, and blocking the entry of sodium ion (Na+) into the cell [28]. In addition to mechanisms used by the plant itself to tolerate salinity, the microbial population occurring in the soil also plays a key role to induce/enhance salinity tolerance in plants [28]. The rhizosphere is considered one of the most diverse microbial habitats with respect to species richness and community size, and harbors a variety of microorganisms that have the ability to promote plant growth by increasing the availability and uptake of carbon, nitrogen, and minerals from the soil [29]. The study of microorganisms from the rhizosphere plants of the Caatinga is extremely important since they are a means by which low-fertility soil and extreme environment conditions can become a powerful source of biotechnologically valuable metabolites [30]. In the present study, identification of halotolerant bacteria from the rhizosphere of Spergularia sp. and B. portulacoides, plants of a saline region of the Caatinga biome, was carried out. We also described the screening of biopolymers produced by these microorganisms.
Bacterial strains belonging to the genus Bacillus were the most abundant in the rhizosphere of Spergularia sp. and B. portulacoides. According to previous reports, Bacillus strains provide tolerance to host plants during abiotic stresses [31]. Several Bacillus-like halophilic bacteria have been isolated from extreme environments like marine sediments, salt mines, and dessert soils [32]. These groups of bacteria have high biotechnological importance due to the wide range of metabolites they synthesize [33, 34]. We focus in this study on the biopolymer synthesis by them.
Members of the genus Curtobacterium (Curtobacterium sp. and C. citreum) were identified from both rhizosphere samples. Some of these Actinobacteria strains have been isolated from desert ecosystems [35], from rocks of extreme arid regions [36], and from salt flats [37], which corresponds with the adverse conditions to the sites selected in Areia Branca for this study. Also, these genera are ubiquitous and mostly found associated with plants and soils [38], being described as halotolerant nitrogen-fixing endophytic bacteria [39] and halotolerant plant growth-promoting rhizobacteria (PGPR) [31], reports that show the potential of these bacteria as biofertilizers.
The genus Exiguobacterium was represented for only one isolated (E. aestuarii) from the rhizosphere of B. portulacoides. Members of this genus are interesting sources for microbial diversity studies in extreme environments due they are widespread in varied habitats, including cold environments such as Siberian permafrost and Antarctic ice [40], hot/hyperalkaline springs [41], freshwater, and marine waters [42]. The distinctive feature of these bacteria is their ability to grow under extreme environmental conditions, with temperatures ranging from − 12 to 55 °C under nutrient-limiting situations. The ability to grow over a wide range of temperatures makes this genus a suitable candidate for developing agriculturally, industrially, and environmentally relevant products such as extremozymes [43].
Member of the genus Staphylococcus (Staphylococcus sp. and S. warneri) were identified from the rhizosphere of B. portulacoides and they are halotolerant bacteria. Staphylococci are extensively spread in various niches [44], and have been isolated from hypersaline environments such as salt lakes [45], salt mines [46], and from salt rich fields [47]. Several strains of this genus proved themselves beneficial for plants by playing their role in growth promotion, protection from phytopathogens, and provision of different elemental nutrients such as nitrogen and phosphorus [48].
One Pseudomonas sp. strain was isolated from the rhizosphere of B. portulacoides. These bacteria have been isolated from different types of environments and many species are very frequent or dominant under extreme conditions [49]. They have also been reported to stimulate plant growth by the production of indole acetic acid (IAA), biofilm formation, or phosphate solubilization [31, 48]. Also, these genera are described as a PHA producer from diverse carbon sources [50].
All the strains grew on TSB media containing 0.1–1.0 M NaCl, and showed salt tolerance at 2.0 M NaCl, except the strain identified as E. aestuarii. All isolates are halotolerant bacteria. NaCl is not required for the growth of these bacteria but improves it at ≥ 0.1 M NaCl.
From the biopolymer screening carried out, it was found that four strains isolated from the rhizosphere of B. portulacoides and identified as B. filamentosus produced PHA constituted of P(3HB) homopolymer (short chain length PHA) from glucose as sole carbon source. It is a well-known fact that many of the Bacillus sp. have been reported to produce PHA utilizing a diverse group of carbon sources [9, 34]. Currently, the species B. filamentosus have not been reported as PHA producer; however in the genome of B. filamentosus hbe03 strain [51], an amino acid sequence coding for poly(R)-hydroxyalkanoic acid synthase subunit PhaC (class III) is annotated (Fig.S4). Comparing the sequence of the subunit PhaC of the B. filamentosus hbe603 strain with other sequences, it was observed that it is similar to those described in B. megaterium strains (Fig. S5), microorganism of this genus widely used and studied for the production of PHA [9, 52]. For future studies, it would be interesting to sequence the genome of these strains to elucidate the genes and metabolic pathways responsible for the synthesis of PHA in the Caatinga strains. The present study is the first report about B. filamentosus PHA producer strains.
During adverse environmental conditions, PHA support the growth, acting as a source of carbon, energy, and reducing power to the microbes and this notion can be practically feasible at industrial level to reduce the production cost of biopolymers commercially with environmental sustainability [53]. Among the 4 bacterial isolates, the strains B. filamentosus Sac33 and B. filamentosus Sac34 showed the highest cell content as PHA. They were selected to evaluate the effect of two carbon sources related with industrial residues and salt stress in the PHA production. The best cultured condition for strains B. filamentosus Sac33 and Sac34 to produce P(3HB) was in MSM containing glucose 10 g/L as sole carbon source and without NaCl (Table S4). Our results show that, despite growing and accumulating PHA, accumulation decreased with the increase of NaCl concentrations. Nevertheless, use of halotolerant bacteria on PHA production has been proposed worldwide, considering that high saline concentrations could prevent contaminants or even open bioreactors could be applied, depending of salt concentrations thus reducing costs arising from medium and bioreactor sterilization and also from maintaining process contaminant free as proposed in some industries based on the strain Halomonas bluephagenesis [54]. This strategy, coupled to relatively high pH values, has been successfully applied to grow halophilic bacteria in unsterile and continuous processes in seawater like medium for at least 1 month without any microbial contamination. Sac33 and Sac34 cultured in MSM at 0.6–1.0 M NaCl, despite not being the best condition for accumulation of these biopolymers, revealed the promising potential of B. filamentosus for PHA production in high concentrations of salt. On the other hand, both evaluated carbon sources, xylose and glycerol, are low value substrates and their utilization for biopolymer production is an integrated solution for bio-based industries [34, 55]. The use of Bacillus species for the production of PHA is due on three advantages that have over other bacterial genera, firts the absence of the lipopolysaccharide layer (LPS) which makes more easier the polymer extraction; second their ability to grow on cheap raw materials; and finally, they are generally recognized as safe (GRAS) microorganisms by the Food and Drug Administration (FDA) [56]. This assay was the first screening performed with halotolerant isolates from Caatinga biome. The PHA accumulated can still be improved in further experiments using different biotechnological approaches.
Strain Pseudomonas sp. Sac65 was negative in the screening carried out by Sudan black B staining reaction (Fig. S2), result that was not expected since different species of the genus are described as producing PHA [50, 57]. However, this strain would have the potential to accumulate PHA from unrelated carbon sources [58]. Further studies should be done with this strain to prove it.
In recent years, increased demand for natural polymers for food, pharmaceuticals, cosmetics, and other industrial applications has encouraged the interest for isolation and identification of new polysaccharides produced by microorganisms, which being isolated from extreme environments may have innovative applications or different characteristics to the traditional ones [1]. It is interesting the use of EPS in medical applications due to their specific biological activities, such as antioxidant activity, immunostimulating effects, antitumor effects, and antiviral activity [10]. Halotolerant strains belonging to the genus Bacillus (Sac1, Sac7, Sac9, Sac10, Sac16, Sac18, Sac22, Sac45, Sac51, and Sac64), Curtobacterium (Sac54), and Pseudomonas (Sac65) identified in this study showed EPS production. The genus Pseudomonas is a rich source of exopolysaccharides. The production of EPS by species of these genus has been reported in the literature [59], being the most studied those produced by P. aeruginosa species that include alginate, Psl, and Pel [60]. Curtobacterium species also are described as a producer of exopolysaccharides, metabolite that play an important role in plant adaptation [38]. The production of EPS by the genus Bacillus is also described in several studies [8], as well as the different applications of the polymer in the industries. It is interesting the use of these polymers as antiviral agents, antioxidants, immunostimulatory activity, and antibiofilm in different works [33, 61, 62]. With the objective of evaluate the immunostimulatory activity of the EPS produced by the Caatinga strains, we selected ten isolates for further experiments in murine macrophages. Macrophages are target cells for some immunomodulatory and antitumor agents that can be activated by different biologically active substances such as microbial polysaccharides. These products have been shown to have potent immunostimulating activity on cellular and humoral responses to antigens, invigorating natural killer cells, T cells, B cells, macrophages, and immune system responses [63]. The activation of macrophages to proinflammatory phenotype by the bacterial EPS obtained in the study was measured based on their ability to produce high levels of NO. The Griess test is an analytical test that indirectly measures the production of nitric oxide (NO), by detecting the presence of nitrite in solution. NO is an important molecule involved in the immune response in addition to other systems [64]. The results showed that eight among ten bacterial EPS evaluated activated murine macrophages (Fig. 5). Strains identified as Bacillus paralicheniformis (Sac1, Sac9, and Sac16 and Sac51), isolated from the rhizosphere of Spergularia sp., increased NO2− concentration, more than the positive control (LPS). Cheng et al. [33] described a strain of B. paralicheniformis (SR14) that synthesized an exopolysaccharide with antioxidant activity, and Abinaya et al. [61] described a B. licheniformis strain Dahb1 as producer of EPS with antimicrobial activity, and also exhibited effective larvicidal properties against malaria and Zika virus mosquito vectors. Our results are a first approximation to the immunostimulatory potential of the evaluated EPS, though; other confirmatory tests of the observed activity have to be performed, as well as the chemical characterization of bacterial exopolysaccharides.
Hyaluronic acid is a high-value biopolymer used in the biomedical, pharmaceutical, cosmetic, and food industries. Current methods of HA production, including extraction from animal sources and streptococcal cultivations, are associated with high costs and health risks [65]. Consequently, the search for other alternative microorganisms for the production of HA is of interest and desirable to the industry. Knowing the biotechnological potential of new strains that produce HA, it was evaluated the production of hyaluronic acid by halotolerant isolates from the Caatinga, performing cultures in liquid medium and quantification by the Alcian Blue staining reaction. Our results suggest that most of the halotolerant strains isolated from the rhizosphere of Spergularia sp. and B. portulacoides can produce HA. The isolate identified as Curtobacterium sp. (Sac54) produced the highest relative concentration of HA (Fig. 6) compared to the positive control, Streptococcus equi that is used industrially for the production of this biopolymer. To the best of our knowledge, there are no reports in the literature describing the genus Curtobacterium as a producer of HA. Also, strains identified as Bacillus (Sac23, Sac33, and Sac41) produced HA in higher concentration than the control. These Bacillus strains are attractive microorganisms for further experiments, previously mentioned, due they are GRAS and different studies have been development with these genera to enhance the production of HA [65]. Nevertheless, the results have yet to be evaluated by analytical methods because the dye can also measure another type of mucopolysaccharide [27]. Even so, the present study is the first report about screening possible producers of HA by halotolerant isolates from the Caatinga.
Conclusion
The results obtained in this study demonstrate the biotechnological potential and versatility of the strains as producers of biopolymers such as PHA and EPS that have attractive biological activities. Bacillus strains from the rhizosphere of B. portulacoides produced PHA from glucose, xylose, and glycerol as sole carbon source at high NaCl concentrations. Bacillus paralicheniformis strains from the rhizosphere of Spergularia sp. produce EPS that showed potential immunostimulatory activity in murine macrophages and also, could produce HA. This work will be the basis for the development of future research that aim, on one hand, to know and exploit the ability of these strains to use industrial waste as substrate for the production of PHAs, as well as the isolation, characterization, and purification of EPS with biological activity.
Supplementary information
(PDF 579 kb).
Authors’ contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Maria Paula Parada-Pinilla, Maria Alejandra Ferreira, Juan Camilo Roncallo, and Alexia Nathália Brígido Assef. The draft of the manuscript was written by Maria Paula Parada-Pinilla and Gabriel Padilla, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) with scholarships to MPPP, JCR, and MAF. Fellowship from National Council for Scientific and Technological Development CNPq to LFS (309086/2018-3). Fapesp Grant (Fundação de Apoio à Pesquisa do Estado de São Paulo) to GP (2009/52665-4; 2010/51458-9).
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
The authors declare that they were in agreement and remain in agreement in this publication.
Consent for publication
The authors declare authorization of all participants for publication.
Code availability
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Corral P, Amoozegar MA, Ventosa A (2020) Halophiles and their biomolecules: recent advances and future applications in biomedicine. Mar Drugs 18. 10.3390/md18010033 [DOI] [PMC free article] [PubMed]
- 2.Kavamura VN, Taketani RG, Ferreira C, de Melo IS, Mendes R. The role of species turnover in structuring bacterial communities in a local scale in the cactus rhizosphere. Plant Soil. 2018;425:101–112. doi: 10.1007/s11104-018-3570-4. [DOI] [Google Scholar]
- 3.Ricardo SDF, Coe HHG, Dias RR, de Sousa LOF, Gomes E. Reference collection of plant phytoliths from the Caatinga biome, Northeast Brazil. Flora Morphol Distrib Funct Ecol Plants. 2018;249:1–8. doi: 10.1016/j.flora.2018.09.003. [DOI] [Google Scholar]
- 4.Taketani RG, Lançoni MD, Kavamura VN, Durrer A, Andreote FD, Melo IS. Dry season constrains bacterial phylogenetic diversity in a semi-arid rhizosphere system. Microb Ecol. 2017;73:153–161. doi: 10.1007/s00248-016-0835-4. [DOI] [PubMed] [Google Scholar]
- 5.Hamedi J, Mohammadipanah F, Ventosa A. Systematic and biotechnological aspects of halophilic and halotolerant actinomycetes. Extremophiles. 2013;17:1–13. doi: 10.1007/s00792-012-0493-5. [DOI] [PubMed] [Google Scholar]
- 6.Gunde-Cimerman N, Plemenitaš A, Oren A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol Rev. 2018;42:353–375. doi: 10.1093/femsre/fuy009. [DOI] [PubMed] [Google Scholar]
- 7.Corinaldesi C, Barone G, Marcellini F et al (2017) Marine microbial-derived molecules and their potential use in cosmeceutical and cosmetic products. 1–21. 10.3390/md15040118 [DOI] [PMC free article] [PubMed]
- 8.Liu C, Baffoe DK, Zhan Y, Zhang M, Li Y, Zhang G. Halophile, an essential platform for bioproduction. J Microbiol Methods. 2019;166:1–8. doi: 10.1016/j.mimet.2019.105704. [DOI] [PubMed] [Google Scholar]
- 9.Schmid MT, Song H, Raschbauer M, Emerstorfer F, Omann M, Stelzer F, Neureiter M. Utilization of desugarized sugar beet molasses for the production of poly(3-hydroxybutyrate) by halophilic Bacillus megaterium uyuni S29. Process Biochem. 2019;86:9–15. doi: 10.1016/j.procbio.2019.08.001. [DOI] [Google Scholar]
- 10.Boujida N, Palau M, Charfi S, el Moussaoui N, Manresa A, Miñana-Galbis D, Skali Senhaji N, Abrini J. Isolation and characterization of halophilic bacteria producing exopolymers with emulsifying and antioxidant activities. Biocatal Agric Biotechnol. 2018;16:631–637. doi: 10.1016/j.bcab.2018.10.015. [DOI] [Google Scholar]
- 11.Radchenkova N, Boyadzhieva I, Atanasova N, Poli A, Finore I, di Donato P, Nicolaus B, Panchev I, Kuncheva M, Kambourova M. Extracellular polymer substance synthesized by a halophilic bacterium Chromohalobacter canadensis 28. Appl Microbiol Biotechnol. 2018;102:4937–4949. doi: 10.1007/s00253-018-8901-0. [DOI] [PubMed] [Google Scholar]
- 12.Powell JT, Chatziefthimiou AD, Banack SA, Cox PA, Metcalf JS. Desert crust microorganisms, their environment, and human health. J Arid Environ. 2015;112:127–133. doi: 10.1016/j.jaridenv.2013.11.004. [DOI] [Google Scholar]
- 13.Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 18.Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10. 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed]
- 19.Ramsay BA, Lomaliza K, Chavarie C, et al. Production of poly-(beta-hydroxybutyric-co-beta-hydroxyvaleric) acids. Appl Environ Microbiol. 1990;56:2093–2098. doi: 10.1016/S0922-338X(97)83009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlegel HG, Lafferty R, Krauss I. The isolation of mutants not accumulating poly-beta-hydroxybutyric acid. Arch Mikrobiol. 1970;71:283–294. doi: 10.1007/BF00410161. [DOI] [PubMed] [Google Scholar]
- 21.Riis V, Mai W. Gas chromatographic determination of poly-β-hydroxybutyric acid in microbial biomass after hydrochloric acid propanolysis. J Chromatogr A. 1988;445:285–289. doi: 10.1016/S0021-9673(01)84535-0. [DOI] [Google Scholar]
- 22.Mendonça TT, Tavares RR, Cespedes LG, Sánchez-Rodriguez RJ, Schripsema J, Taciro MK, Gomez JGC, Silva LF. Combining molecular and bioprocess techniques to produce poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) with controlled monomer composition by Burkholderia sacchari. Int J Biol Macromol. 2017;98:654–663. doi: 10.1016/j.ijbiomac.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Paulo EM, Vasconcelos MP, Oliveira IS, Affe HMJ, Nascimento R, Melo IS, Roque MRA, Assis SA. An alternative method for screening lactic acid bacteria for the production of exopolysaccharides with rapid confirmation. Food Sci Technol. 2012;32:710–714. doi: 10.1590/S0101-20612012005000094. [DOI] [Google Scholar]
- 24.Kavamura VN, Santos SN, da Silva JL, et al. Screening of Brazilian cacti rhizobacteria for plant growth promotion under drought. Microbiol Res. 2013;168:183–191. doi: 10.1016/j.micres.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Liang TW, Wang SL. Recent advances in exopolysaccharides from Paenibacillus spp.: production, isolation, structure, and bioactivities. Mar Drugs. 2015;13:1847–1863. doi: 10.3390/md13041847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green LC, Tannenbaum SR, Goldman P. Nitrate synthesis in the germfree and conventional rat. Science. 1981;212:56–58. doi: 10.1126/science.6451927. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, Tyo K, Alper H, Klein-Marcuschamer D, Stephanopoulos G. A high-throughput screen for hyaluronic acid accumulation in recombinant Escherichia coli transformed by libraries of engineered sigma factors. Biotechnol Bioeng. 2008;101:788–796. doi: 10.1002/bit.21947. [DOI] [PubMed] [Google Scholar]
- 28.Zahir ZA, Nadeem SM, Khan MY, Binyamin R. Role of halotolerant microbes in plant growth promotion under salt stress conditions. In: Kumar M, Etesami H, Kumar V, editors. Saline soil-based agriculture by halotolerant microorganisms. Singapore: Springer; 2019. pp. 209–253. [Google Scholar]
- 29.Mukhtar S, Mehnaz S, Mirza MS, Malik KA. Isolation and characterization of bacteria associated with the rhizosphere of halophytes (Salsola stocksii and Atriplex amnicola) for production of hydrolytic enzymes. Braz J Microbiol. 2019;50:85–97. doi: 10.1007/s42770-019-00044-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva-Lacerda GR, Santana RCF, Vicalvi-Costa MCV, Solidônio EG, Sena KXFR, Lima GMS, Araújo JM. Antimicrobial potential of actinobacteria isolated from the rhizosphere of the Caatinga biome plant Caesalpinia pyramidalis Tul. Genet Mol Res. 2016;15:1–12. doi: 10.4238/gmr.15017488. [DOI] [PubMed] [Google Scholar]
- 31.Abbas R, Rasul S, Aslam K, Baber M, Shahid M, Mubeen F, Naqqash T. Halotolerant PGPR: a hope for cultivation of saline soils. J King Saud Univ - Sci. 2019;31:1195–1201. doi: 10.1016/j.jksus.2019.02.019. [DOI] [Google Scholar]
- 32.López G, Diaz-Cárdenas C, Shapiro N, Woyke T, Kyrpides NC, David Alzate J, González LN, Restrepo S, Baena S. Draft genome sequence of Pseudomonas extremaustralis strain USBA-GBX 515 isolated from superparamo soil samples in Colombian Andes. Stand Genomic Sci. 2017;12:1–12. doi: 10.1186/s40793-017-0292-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Y, Xiao X, Li X, Song D, Lu Z, Wang F, Wang Y. Characterization, antioxidant property and cytoprotection of exopolysaccharide-capped elemental selenium particles synthesized by Bacillus paralicheniformis SR14. Carbohydr Polym. 2017;178:18–26. doi: 10.1016/j.carbpol.2017.08.124. [DOI] [PubMed] [Google Scholar]
- 34.Mohandas SP, Balan L, Jayanath G, Anoop BS, Philip R, Cubelio SS, Bright Singh IS. Biosynthesis and characterization of polyhydroxyalkanoate from marine Bacillus cereus MCCB 281 utilizing glycerol as carbon source. Int J Biol Macromol. 2018;119:380–392. doi: 10.1016/j.ijbiomac.2018.07.044. [DOI] [PubMed] [Google Scholar]
- 35.Bhatnagar A, Bhatnagar M. Microbial diversity in desert ecosystems. Curr Sci. 2005;89:91–100. doi: 10.1016/S1369-5274(02)00324-7. [DOI] [Google Scholar]
- 36.Kuhlman KR, Allenbach LB, Ball CL, Fusco WG, la Duc MT, Kuhlman GM, Anderson RC, Stuecker T, Erickson IK, Benardini J, Crawford RL. Enumeration, isolation, and characterization of ultraviolet (UV-C) resistant bacteria from rock varnish in the Whipple Mountains, California. Icarus. 2005;174:585–595. doi: 10.1016/j.icarus.2004.11.022. [DOI] [Google Scholar]
- 37.Cardinale M, Ratering S, Suarez C, Zapata Montoya AM, Geissler-Plaum R, Schnell S. Paradox of plant growth promotion potential of rhizobacteria and their actual promotion effect on growth of barley (Hordeum vulgare L.) under salt stress. Microbiol Res. 2015;181:22–32. doi: 10.1016/j.micres.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Bourles A, Guentas L, Chalkiadakis E, Majorel C, Juillot F, Cavaloc Y, Burtet-Sarramegna V, Medevielle V, Jourand P, Amir H. New caledonian ultramafic conditions structure the features of Curtobacterium citreum strains that play a role in plant adaptation. Can J Microbiol. 2019;65:880–894. doi: 10.1139/cjm-2019-0283. [DOI] [PubMed] [Google Scholar]
- 39.Alishahi F, Alikhani HA, Khoshkholgh-Sima NA, Etesami H. Mining the roots of various species of the halophyte Suaeda for halotolerant nitrogen-fixing endophytic bacteria with the potential for promoting plant growth. Int Microbiol. 2020;23:415–427. doi: 10.1007/s10123-019-00115-y. [DOI] [PubMed] [Google Scholar]
- 40.Chauhan H, Bagyaraj DJ, Selvakumar G, Sundaram SP. Novel plant growth promoting rhizobacteria—prospects and potential. Appl Soil Ecol. 2015;95:38–53. doi: 10.1016/j.apsoil.2015.05.011. [DOI] [Google Scholar]
- 41.Cabria GLB, Argayosa VB, Lazaro JEH, et al. Draft genome sequence of haloalkaliphilic Exiguobacterium sp. strain AB2 from Manleluag Ophiolitic Spring, Philippines. Genome Announc. 2014;2:2–3. doi: 10.1128/genomeA.00840-14.Copyright. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Mailem D, Eliyas M, Khanafer M, Radwan S. Culture-dependent and culture-independent analysis of hydrocarbonoclastic microorganisms indigenous to hypersaline environments in Kuwait. Microb Ecol. 2014;67:1–9. doi: 10.1007/s00248-014-0386-5. [DOI] [PubMed] [Google Scholar]
- 43.Kasana RC, Pandey CB. Exiguobacterium: an overview of a versatile genus with potential in industry and agriculture. Crit Rev Biotechnol. 2018;38:141–156. doi: 10.1080/07388551.2017.1312273. [DOI] [PubMed] [Google Scholar]
- 44.Giammarinaro P, Leroy S, Chacornac JP, Delmas J, Talon R. Development of a new oligonucleotide array to identify staphylococcal strains at species level. J Clin Microbiol. 2005;43:3673–3680. doi: 10.1128/JCM.43.8.3673-3680.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Máthé I, Borsodi AK, Tóth EM, Felföldi T, Jurecska L, Krett G, Kelemen Z, Elekes E, Barkács K, Márialigeti K. Vertical physico-chemical gradients with distinct microbial communities in the hypersaline and heliothermal Lake Ursu (Sovata, Romania) Extremophiles. 2014;18:501–514. doi: 10.1007/s00792-014-0633-1. [DOI] [PubMed] [Google Scholar]
- 46.Roohi A, Ahmed I, Khalid N, et al. Isolation and phylogenetic identification of halotolerant/halophilic bacteria from the salt mines of Karak, Pakistan. Int J Agric Biol. 2014;16:564–570. [Google Scholar]
- 47.Akram MS, Shahid M, Tariq M, Azeem M, Javed MT, Saleem S, Riaz S. Deciphering Staphylococcus sciuri SAT-17 mediated anti-oxidative defense mechanisms and growth modulations in salt stressed maize (Zea mays l.) Front Microbiol. 2016;7:1–14. doi: 10.3389/fmicb.2016.00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ali B. Functional and genetic diversity of bacteria associated with the surfaces of agronomic plants. Plants. 2019;8:91. doi: 10.3390/plants8040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vásquez-Ponce F, Higuera-Llantén S, Pavlov MS, Marshall SH, Olivares-Pacheco J. Phylogenetic MLSA and phenotypic analysis identification of three probable novel Pseudomonas species isolated on King George Island, South Shetland, Antarctica. Braz J Microbiol. 2018;49:695–702. doi: 10.1016/j.bjm.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venkateswar Reddy M, Mawatari Y, Onodera R, Nakamura Y, Yajima Y, Chang YC. Polyhydroxyalkanoates (PHA) production from synthetic waste using Pseudomonas pseudoflava: PHA synthase enzyme activity analysis from P. pseudoflava and P. palleronii. Bioresour Technol. 2017;234:99–105. doi: 10.1016/j.biortech.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 51.Jia N, Du J, Ding MZ, et al. Genome sequence of Bacillus endophyticus and analysis of its companion mechanism in the Ketogulonigenium vulgare-Bacillus strain consortium. PLoS One. 2015;10:1–17. doi: 10.1371/journal.pone.0135104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akdoğan M, Çelik E. Purification and characterization of polyhydroxyalkanoate (PHA) from a Bacillus megaterium strain using various dehydration techniques. J Chem Technol Biotechnol. 2018;93:2292–2298. doi: 10.1002/jctb.5572. [DOI] [Google Scholar]
- 53.Kumar M, Sundaram S, Gnansounou E, Larroche C, Thakur IS. Carbon dioxide capture, storage and production of biofuel and biomaterials by bacteria: a review. Bioresour Technol. 2018;247:1059–1068. doi: 10.1016/j.biortech.2017.09.050. [DOI] [PubMed] [Google Scholar]
- 54.Chen X, Yin J, Ye J, Zhang H, Che X, Ma Y, Li M, Wu LP, Chen GQ. Engineering Halomonas bluephagenesis TD01 for non-sterile production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) Bioresour Technol. 2017;244:534–541. doi: 10.1016/j.biortech.2017.07.149. [DOI] [PubMed] [Google Scholar]
- 55.Coutinho de Paula F, Kakazu S, Bilia Chimello de Paula C, et al. Burkholderia glumae MA13: a newly isolated bacterial strain suitable for polyhydroxyalkanoate production from crude glycerol. Biocatal Agric Biotechnol. 2019;20:101268. doi: 10.1016/j.bcab.2019.101268. [DOI] [Google Scholar]
- 56.Kumar P, Singh M, Mehariya S, Patel SKS, Lee JK, Kalia VC. Ecobiotechnological approach for exploiting the abilities of Bacillus to produce co-polymer of polyhydroxyalkanoate. Indian J Microbiol. 2014;54:151–157. doi: 10.1007/s12088-014-0457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urbina L, Wongsirichot P, Corcuera MÁ, Gabilondo N, Eceiza A, Winterburn J, Retegi A. Application of cider by-products for medium chain length polyhydroxyalkanoate production by Pseudomonas putida KT2440. Eur Polym J. 2018;108:1–9. doi: 10.1016/j.eurpolymj.2018.08.020. [DOI] [Google Scholar]
- 58.Anderson AJ, Haywood GW, Williams DR, Dawes EA (1990) The production of polyhydroxyalkanoates from unrelated carbon sources. In: Novel biodegradable microbial polymers. Springer, Netherlands, pp 119–129. 10.1007/978-94-009-2129-0_11
- 59.Vidhyalakshmi R, Valli Nachiyar C, Narendra Kumar G, Sunkar S, Badsha I. Production, characterization and emulsifying property of exopolysaccharide produced by marine isolate of Pseudomonas fluorescens. Biocatal Agric Biotechnol. 2018;16:320–325. doi: 10.1016/j.bcab.2018.08.023. [DOI] [Google Scholar]
- 60.Passos da Silva D, Matwichuk ML, Townsend DO, Reichhardt C, Lamba D, Wozniak DJ, Parsek MR. The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix. Nat Commun. 2019;10:1–11. doi: 10.1038/s41467-019-10201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abinaya M, Vaseeharan B, Divya M, Vijayakumar S, Govindarajan M, Alharbi NS, Khaled JM, al-anbr MN, Benelli G. Structural characterization of Bacillus licheniformis Dahb1 exopolysaccharide—antimicrobial potential and larvicidal activity on malaria and Zika virus mosquito vectors. Environ Sci Pollut Res. 2018;25:18604–18619. doi: 10.1007/s11356-018-2002-6. [DOI] [PubMed] [Google Scholar]
- 62.Hu X, Pang X, Wang PG, Chen M. Isolation and characterization of an antioxidant exopolysaccharide produced by Bacillus sp. S-1 from Sichuan Pickles. Carbohydr Polym. 2019;204:9–16. doi: 10.1016/j.carbpol.2018.09.069. [DOI] [PubMed] [Google Scholar]
- 63.Wang G, Zhu L, Yu B, Chen K, Liu B, Liu J, Qin G, Liu C, Liu H, Chen K. Exopolysaccharide from Trichoderma pseudokoningii induces macrophage activation. Carbohydr Polym. 2016;149:112–120. doi: 10.1016/j.carbpol.2016.04.093. [DOI] [PubMed] [Google Scholar]
- 64.Tripathi P. Nitric oxide and the immune response. Indian J Biochem Biophys. 2007;44:310–319. [PubMed] [Google Scholar]
- 65.Westbrook AW, Ren X, Moo-Young M, Chou CP. Engineering of cell membrane to enhance heterologous production of hyaluronic acid in Bacillus subtilis. Biotechnol Bioeng. 2018;115:216–231. doi: 10.1002/bit.26459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 579 kb).
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files).