Abstract
Background
Personalized risk assessment provides opportunities for tailoring treatment, optimizing healthcare resources and improving outcome. The aim of this study was to develop a 90-day mortality-risk prediction model for identification of high- and low-risk patients undergoing surgery for colorectal cancer.
Methods
This was a nationwide cohort study using records from the Danish Colorectal Cancer Group database that included all patients undergoing surgery for colorectal cancer between 1 January 2004 and 31 December 2015. A least absolute shrinkage and selection operator logistic regression prediction model was developed using 121 pre- and intraoperative variables and internally validated in a hold-out test data set. The accuracy of the model was assessed in terms of discrimination and calibration.
Results
In total, 49 607 patients were registered in the database. After exclusion of 16 680 individuals, 32 927 patients were included in the analysis. Overall, 1754 (5.3 per cent) deaths were recorded. Targeting high-risk individuals, the model identified 5.5 per cent of all patients facing a risk of 90-day mortality exceeding 35 per cent, corresponding to a 6.7 times greater risk than the average population. Targeting low-risk individuals, the model identified 20.9 per cent of patients facing a risk less than 0.3 per cent, corresponding to a 17.7 times lower risk compared with the average population. The model exhibited discriminatory power with an area under the receiver operating characteristics curve of 85.3 per cent (95 per cent c.i. 83.6 to 87.0) and excellent calibration with a Brier score of 0.04 and 32 per cent average precision.
Conclusion
Pre- and intraoperative data, as captured in national health registries, can be used to predict 90-day mortality accurately after colorectal cancer surgery.
Early prediction of postoperative mortality is essential for tailoring postoperative treatment and care. In this nationwide study including 32 927 patients undergoing surgery for colorectal cancer, the discriminatory power of the model yielded an AUROC of 85.3% (95% CI, 83.6 to 87.0) and excellent calibration. Pre- and intraoperative phenotypic data can be used to accurately predict and identify patients at risk of early adverse surgical outcome.
Introduction
Improvements in 30-day mortality rates have made surgery for colorectal cancer (CRC) a feasible treatment option for the majority of patients despite an increase in baseline risk of patients1–4. However, recent studies have questioned the appropriateness of 30-day mortality as a quality metric of CRC surgery, as mortality rates nearly double at 90 days5–7. Accurate identification of distinct patient trajectories could facilitate early risk stratification, target clinical interventions and optimize care pathways to improve outcomes. Prediction of 90-day mortality could be a useful tool for tailoring postoperative treatment and surgical care.
Recent advances in data science have challenged the boundaries of conventional clinical decision making and personalized risk profiling8,9. Individualized risk assessment provides opportunities for tailoring treatment strategies, optimizing healthcare resources and improving outcomes10,11. Data sources often differ by design, purpose and coding, limiting utilization in clinical practice12,13. Harmonization of data structures and vocabularies enables standardized analytical tools to be developed, that have proven efficient in producing reliable, transparent and reproducible patient-level risk-prediction models14,15.
This study aimed to develop and standardize a multivariable patient-level model for prediction of 90-day mortality after CRC surgery, utilizing supervised machine learning on standardized nationwide CRC quality-assurance data.
Methods
Data sources
Data were acquired from the Danish Colorectal Cancer Group (DCCG.dk)16 database via a formal application from the Danish National Clinical Registry (www.rkkp.dk). This is a nationwide database containing information on all patients diagnosed with primary CRC in Denmark since 1 May 2001. A full description of the data source is provided in Table S1.
Target population
The target population was defined as all patients with a diagnosis of CRC undergoing major curative surgery in Denmark between 1 January 2004 and 31 December 2015. A complete list of the surgical procedures included in the study is available in Table S2. Patients undergoing palliative or intended compromised surgery were excluded, as were those diagnosed with CRC who died on the day of surgery. CRC was defined as adenocarcinoma of the colon or rectum, including histological subtypes.
Outcome
The outcome was all-cause mortality, with a time at risk of 90 days. All patients experienced the outcome of interest before 90 days of follow-up or contributed with at least 90 days of follow-up after surgery. No patients were lost to follow-up. The modelling index (t = 0) was set to postoperative day 1 to be able to include pre- and intraoperative variables in a surgical ‘gloves-off’ design targeting postoperative patient-level risk prediction.
Predictors
Pre- and intraoperative variables were obtained from the DCCG.dk including prior medical history within 10 years of CRC diagnosis. A description of the source variables is provided in Table S3. Before analysis, all source variables were mapped to the Observational Medical Outcomes Partnership (OMOP) common vocabulary, using the SNOMED-CT, LOINC and CPT4 classification systems. After mapping, data were stored in a relational database structured according to the OMOP Common Data Model (CDM)12. Initial mapping from Danish source concepts to the OMOP common vocabulary was conducted by health taxonomy experts, using the Observational Health Data Science and Informatics (OHDSI) Natural Language Processing tools and manual curation. A multidisciplinary team of medical practitioners and data scientists performed quality control on the procedure mappings and evaluated the validity of links between source concepts and their equivalents in the OMOP common vocabulary. Granularity loss in the initial mappings was corrected by linking the source concepts with more precise terms from the OMOP common vocabulary. Where no equivalent could be found between source and OMOP concepts, the source concepts were populated in the CDM as custom concepts. Using the OHDSI Patient-Level Prediction framework, predictors were selected among pre- or intraoperative variables17. A total of 121 OMOP concepts participated as potential predictors in the model-training process. All concepts were classified within the OMOP domains of gender, conditions, measurements, observations and procedures. All continuous numerical values were categorized, except for height and weight measurements. A full list of the OMOP predictors is provided in Table S4.
Missing data
Standard machine learning practice was followed, encoding the categorical variable classes using one-hot encoding18 (1-of-K encoding19, binary encoding). During one-hot encoding, binary predictors were constructed for each variable class, indicating the presence or absence of the class in the source data. Thus, missing values were indicated in the CDM by the recorded absence of all classes for a single categorical variable.
Missing values of two continuous numerical variables affected by missing data, height and weight were imputed by zero.
Statistical analysis
Data were randomly divided into two data sets, one for model training using 75 per cent of patients and one for model testing containing 25 per cent. A least absolute shrinkage and selection operator (LASSO) logistic regression model20 was then developed for the prediction of 90-day mortality. The variance was optimized by maximizing the out-of-sample likelihood by three-fold cross-validation in the training set. Model performance in terms of discrimination was assessed by area under the receiver operating characteristics curve (AUROC) and area under the precision recall curve (average precision). Model calibration was evaluated by inspection of a standard calibration plot. Calibration was performed by dividing patients into deciles based on the predicted risk. The average predicted risk was calculated and plotted against the observed risk. Finally, a linear model was fitted, and the intercept and slope calculated to give a summary of the model calibration. Model prediction accuracy was evaluated by the Brier score as a measure of distance between the actual outcome and the predicted probability assigned to the outcome for each observation. Brier scores range between 0.0 and 1.0 with low values being desirable. The model results were benchmarked versus a Gradient Boosting Machine (GBM)21 model using the same predictor variables.
Reporting of this study was in adherence to the TRIPOD guidelines22. No risk groups were prespecified, and no external or temporal validation was planned for this nationwide study. The Regional Data Protection Committee approved the study (REG-071-2018). Ethical approval was not required according to Danish law23.
ATLAS version 2.7.2 (OHDSI) was used for cohort definition and data analysis. ATLAS is an open source application developed as a part of OHDSI intended to provide a unified interface to patient level data and analytics. The analysis backend of ATLAS was R version 3.4.3 (R Foundation for Statistical Computing). R is a language and environment for statistical computing and graphics. R is available as Free Software under the terms of the Free Software Foundation’s GNU General Public License in source code form. The R Foundation is seated in Vienna, Austria and currently hosted by the Vienna University of Economics and Business. It is a registered association under Austrian law and active worldwide. The R Foundation can be contacted by e-mail to R-foundation at r-project.org or The R Foundation for Statistical Computing c/o Institute for Statistics and Mathematics Wirtschaftsuniversität Wien Welthandelsplatz 1 1020 Vienna, Austria. The R packages used in this study were: OhdsiRTools (version 1.7.0), Cyclops (version 2.0.2), and PatientLevelPrediction (version 3.0.5). OHDSI is a multi-stakeholder, interdisciplinary collaborative to bring out the value of health data through large-scale analytics. All solutions are open-source. OHDSI has established an international network of researchers and observational health databases with a central coordinating center housed at Columbia University. Packages were obtained from the OHDSI Methods Library (https://github.com/OHDSI). A full description of the standardized data science framework is available in Table S1.
Results
Participants
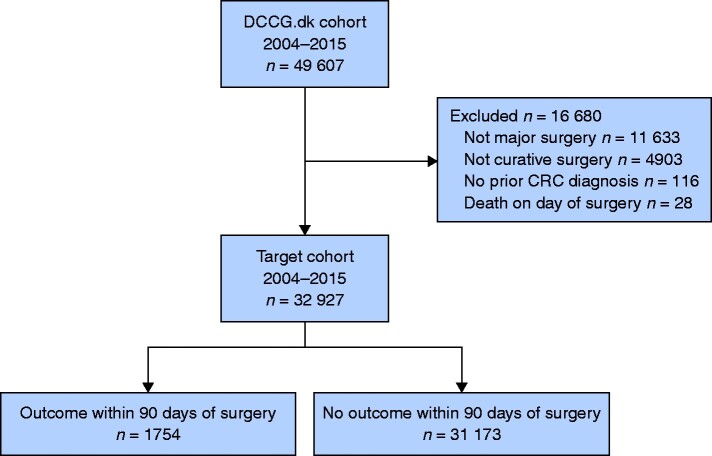
Data from 49 607 Danish patients were retrieved. After exclusions, 32 927 patients were included in the analysis (Fig. 1). Mean(s.d.) patient age was 70.3(10.5) years; 15 340 (46.6 per cent) were female; 20 756 (63 per cent) had no concurrent co-morbidities (Charlson Comorbidity Index 0); 30 112 (91.5 per cent) underwent elective surgery; and 17 659 (53.6 per cent) were operated by a minimally invasive technique. Death within 90 days of surgery occurred in 1754 (5.3 per cent) patients. Baseline characteristics by outcome status are summarized in Table 1.
Fig. 1.
Cohort definition
Description of the study cohort, including cohort exclusions with reasons. The DCCG.dk cohort included all Danish patients registered in the nationwide Danish Colorectal Cancer Group (DCCG.dk) database with a diagnosis of colorectal cancer (CRC) between 1 January 2004 and 31 December 2015
Table 1.
Baseline characteristics of the study cohort
| Characteristic | Study cohort (n = 32 927) | Missing | Alive 90 days after surgery (n = 31 173) | Deceased within 90 days after surgery (n = 1754) |
|---|---|---|---|---|
| Demographic | ||||
| Sex, female | 15 340 (46.6) | 0 (0) | 14 563 (46.7) | 777 (44.3) |
| Age (years)* | 70.3(10.5) | 0 | 69.9(10.4) | 79. (8.8) |
| Height (cm)* | 170.83(9.4) | 4741(14.4) | 170.89(9.4) | 169.26(10.0) |
| Weight (kg)* | 75.75(16.8) | 4596(14.0) | 75.92(16.8) | 71.61(17.7) |
| Smoking status ‡ | 5693 (17.3) | |||
| Non-smoker | 10 214 (37.5) | – | 9864 (37.7) | 350 (32.5) |
| Ex-smoker | 11 533 (42.4) | – | 11 078 (42.4) | 455 (42.3) |
| Current smoker | 5487 (20.2) | – | 5215 (19.9) | 272 (25.3) |
| Alcohol consumption | 5568 (16.9) | |||
| Consumer, 1–14 drinks/week | 16 176 (59.1) | – | 15 643 (59.5) | 533 (49.1) |
| Consumer, 15–21 drinks/week | 2037 (7.45) | – | 1972 (7.5) | 65 (6.0) |
| Consumer, >21 drinks/week | 1964 (7.2) | – | 1879 (7.2) | 85 (7.8) |
| Non-consumer | 7182 (26.3) | – | 6779 (25.8) | 403 (37.1) |
| Clinical characteristics | ||||
| ASA score | 460 (1.4) | |||
| I | 7418 (22.9) | – | 7316 (23.8) | 102 (5.9) |
| II | 18 149 (55.9) | – | 17 496 (56.9) | 653 (38.1) |
| III | 6516 (20.1) | – | 5683 (18.5) | 833 (48.5) |
| IV | 368 (1.1) | – | 245 (0.8) | 123 (7.2) |
| V | 16 (0.0) | – | 11 (0.0) | 5 (0.3) |
| Charlson Comorbidity Index | 0 (0) | |||
| 0 | 20 756 (63.0) | – | 20 079 (64.4) | 677 (38.6) |
| 1-2 | 9124 (27.7) | – | 8438 (27.1) | 686 (39.1) |
| ≥3 | 3047 (9.3) | – | 2656 (8.5) | 391 (22.3) |
| Co-morbidities§ | 0 (0) | |||
| Chronic obstructive lung disease | 2619 (8.0) | – | 2314 (7.4) | 305 (17.4) |
| Dementia | 296 (0.9) | – | 230 (0.7) | 66 (3.8) |
| Hemiplegia | 52 (0.2) | – | 44 (0.1) | 8 (0.5) |
| Cerebrovascular disease | 2576 (7.8) | – | 2276 (7.3) | 300 (17.1) |
| Peripheral vascular disease | 1465 (4.5) | – | 1297 (4.2) | 168 (9.6) |
| Heart failure | 1424 (4.3) | – | 1214 (3.9) | 210 (12.0) |
| Myocardial infarction | 1150 (3.5) | – | 1030 (3.3) | 120 (6.8) |
| Secondary malignant disease | 373 (1.1) | – | 322 (1.0) | 51 (2.9) |
| Widespread metastatic malignant disease | 589 (1.8) | – | 533 (1.7) | 56 (3.2) |
| Liver disease, mild | 224 (0.7) | – | 196 (0.6) | 28 (1.6) |
| Liver disease, moderate to severe | 63 (0.2) | – | 48 (0.2) | 15 (0.9) |
| Diabetes mellitus without complications | 2733 (8.3) | – | 2513 (8.1) | 220 (12.5) |
| Diabetes mellitus with complications | 1195 (3.6) | – | 1088 (3.5) | 107 (6.1) |
| Renal insufficiency | 702 (2.1) | – | 581 (1.9) | 121 (6.9) |
| Peptic ulcer | 1012 (3.1) | – | 902 (2.9) | 110 (6.3) |
| Connective tissue disorder | 824 (2.5) | – | 759 (2.4) | 65 (3.7) |
| Surgical characteristics | ||||
| Surgical urgency | 5 (0.0) | |||
| Emergency | 2810 (8.5) | – | 2321 (7.5) | 489 (27.9) |
| Elective | 30 112 (91.5) | – | 28 847 (92.6) | 1265 (72.1) |
| Surgical approach | 0 (0) | |||
| Laparotomy | 15 299 (46.5) | – | 14 070 (45.1) | 1229 (70.1) |
| Laparoscopy | 16 732 (50.8) | – | 16 227 (52.1) | 505 (28.8) |
| TaTME | 106 (0.3) | – | 103 (0.3) | 3 (0.2) |
| Robotic-assisted surgery | 821 (2.5) | – | 802 (2.6) | 19 (1.1) |
| Conversion of procedure ¶ | 2068 (6.3) | 218 (0.7) | 1966 (6.4) | 102 (5.8) |
| Intraoperative blood loss | 100 (50–300)† | 892 (2.7) | 100 (50–300)† | 250 (100–600)† |
| Intraoperative blood transfusion # | 5107 (15.6) | 110 (0.3) | 4424 (14.2) | 683 (39.1) |
| Intraoperative complications** | 1254 (6.2) | 12 828 (39.0) | 1134 (5.9) | 120 (13.9) |
| Tumour characteristics | ||||
| Cancer type †† | 0 (0) | |||
| Colon | 22 140 (67.2) | – | 20 769 (66.6) | 1371 (78.2) |
| Rectum | 10 787 (32.8) | – | 10 404 (33.4) | 383 (21.8) |
| Tumor perforation present | 1675 (5.1) | 56 (0.2) | 1447 (4.7) | 228 (13.0) |
| Treatment characteristics | ||||
| Preoperative oncological treatment | 0 (0) | |||
| Chemoradiationtherapy | 2545 (7.7) | – | 2497 (8.0) | 48 (2.7) |
| Chemotherapy | 2854 (8.7) | – | 2772 (8.9) | 82 (4.7) |
| Radiotherapy | 2460 (7.5) | – | 2408 (7.7) | 52 (3.0) |
| Preoperative surgical treatment | 13 002 (39.5) | |||
| SEMS | 1195 (6.0) | – | 1133 (5.9) | 62 (7.7) |
| Damage-control surgery‡‡ | 3 (0.0) | – | 0 (0) | 3 (0.4) |
| Preoperative MDT assessment§§ | 11 957 (61.4) | 13 449 (40.8) | 11 599 (62.1) | 358 (45.7) |
Values in parentheses are percentages, except where indicated otherwise. *Values are mean(s.d.); †values are median (i.q.r.). ‡Non-smoker, never smoked tobacco; ex-smoker, smoking cessation for ≥8 weeks before surgery; current smoker, currently smoking. §Diagnoses of co-morbidity registered up to 10 years before colorectal cancer diagnosis. ¶Conversion from minimally invasive surgery to open surgery. Transfusion of any blood product during surgery. **Any iatrogenic injury during surgery to undefined anatomical structure, urinary bladder, the duodenum, the gallbladder, the colon, the liver, the spleen, the pancreas, the sacral nerves, the small intestine, the ureter, the urethra, the vagina or the ventricle. ††Rectal cancer, tumours located within 15 cm of the anal verge; colon cancer, tumours located more than 15 cm from the anal verge. ‡‡. §§Multidisciplinary team (MDT) was formally registered since 1 January 2010 (quality control indicator since 1 January 2014). TaTME, transanal total mesorectal excision; SEMS, self-expanding metal stent. Damage Control Surgery: Two-stage surgical procedure due to acute presentation.
Model development
The data set included 121 candidate predictors corresponding to 14.5 events per predictor variable. Utilizing the standardized OHDSI Patient-Level Prediction framework for model development and validation, the data were split into 24 695 training patients and 8231 test patients. The training set was further divided into three equally sized data sets of 8233, 8231 and 8231 patients to perform three-fold cross-validation for hyperparameter selection. The training and test data sets included 1316 and 438 outcome events, respectively. In total, 65 predictors were selected in the final model. Among the selected model co-variables, increasing age, smoking status, the combined co-morbidity status (Charlson Comorbidity index) and individual co-morbid conditions at time of surgery, such as cerebrovascular disease, chronic obstructive lung disease, heart failure and peripheral vascular disease, were more frequent among patients who died within 90 days of surgery. In addition, clinical factors associated with emergency surgery, including open surgery, tumour perforation and perioperative blood transfusion, were more frequent among patients who died within 90 days of surgery. A full description of the model predictors is provided in Table S5.
Model performance
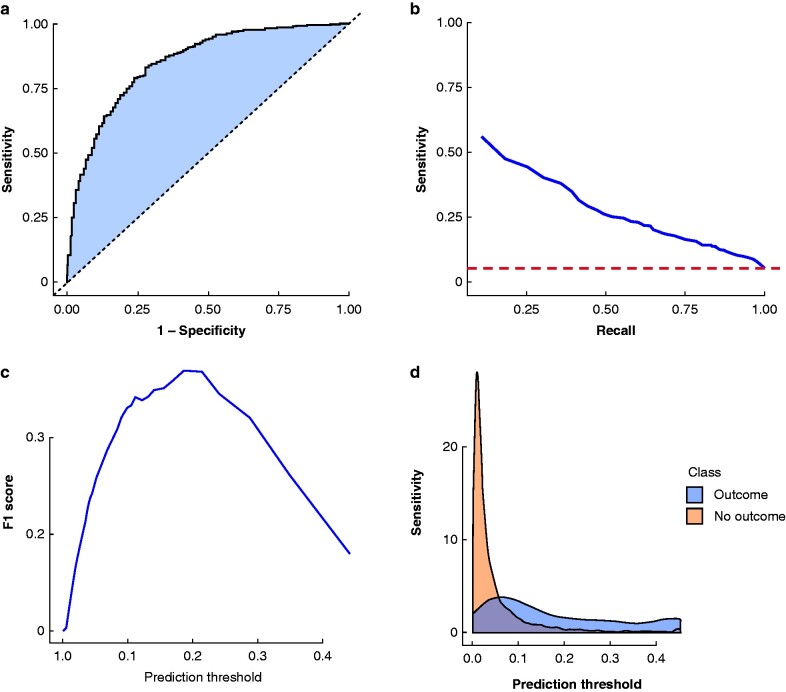
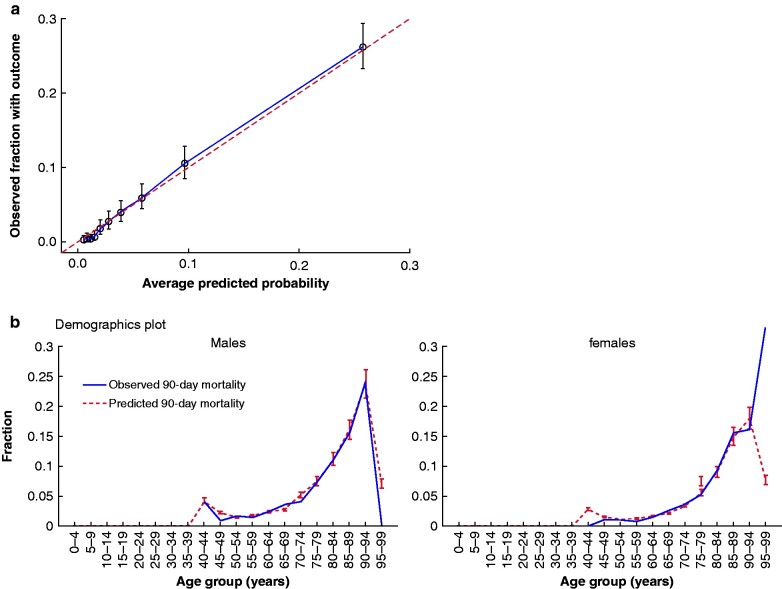
The model's test-set AUROC was 85.3 (95 per cent c.i. 83.6 to 87.0) for prediction of 90-day mortality following surgery for colorectal cancer. The model showed excellent calibration with a Brier score of 0.04 and 0.32 average precision. The model discrimination characteristics are presented in Fig. 2, with age, gender and overall model calibrations in Fig. 3. The intercept of the linear model fitted on the calibration results was -0.00 with a calibration gradient of 1.04. Clinical model performance characteristics for identifying high- and low-risk patients are detailed in Tables 2 and 3.
Fig. 2.
Model discrimination characteristics
a Receiver operating characteristic curve for the least absolute shrinkage and selection operator regression model test set (n = 8231). Area under the receiver operating characteristic (AUROC) curve = 85.28 (95% c.i. 83.55 to 85.01). The false-positive rate is 1 – specificity. The diagonal line indicates the neutral predictive value (AUROC = 50.0). b Precision-recall plot with an area under the precision-recall curve of 31.40. Recall (sensitivity) and precision (positive predictive value) are shown on the x- and y-axes, respectively. The horizontal line indicates the neutral predictive value (positive predictive value of 5.3, i.e., average population risk of death). c F1 score versus prediction threshold for 90-day mortality after colorectal cancer surgery. The F1 score represents a balanced measure of overall classifier performance, combining both precision and recall with equal weights. The classifier performance of the model was optimal at model threshold of 0.2. d Model prediction score distribution. The prediction score distribution represents the predicted risk distribution for those with and without the outcome. The more these curves overlap, the worse the model discrimination performance
Fig. 3.
Model calibration
a Calibration plot for the least absolute shrinkage and selection operator regression model test set (n = 8231). Bars indicate 95% confidence intervals. Calibration gradient = 1.04. Intercept = -0.00. b Demographic calibration plots for across gender and age groups of 5-year intervals
Table 2.
Model performance characteristics: identification of high-risk patients at various model thresholds
| Model risk threshold | Proportion of the patients flagged as high-risk by the model (%) | Sensitivity | Specificity | Risk of 90-day mortality (%)* | Relative risk compared with the population | |
|---|---|---|---|---|---|---|
| 0.5 | 0.6 | 7.1 | 99.7 | 60.8 | 11.4 | |
| 0.45 | 0.9 | 10.0 | 99.6 | 57.9 | 10.9 | |
| 0.4 | 1.3 | 13.5 | 99.3 | 53.6 | 10.1 | |
| 0.35 | 2.0 | 17.6 | 98.9 | 47.8 | 9.0 | |
| 0.3 | 2.8 | 23.5 | 98.4 | 44.8 | 8.4 | |
| 0.25 | 3.8 | 29.0 | 97.6 | 40.7 | 7.7 | |
| 0.2 | 5.5 | 36.7 | 96.3 | 35.7 | 6.7 | |
| 0.15 | 8.3 | 45.0 | 93.7 | 28.7 | 5.4 | |
| 0.1 | 14.0 | 60.3 | 88.6 | 23.0 | 4.3 | |
| 0.05 | 28.1 | 79.5 | 79.5 | 15.0 | 2.8 | |
| 0.025 | 48.5 | 92.2 | 53.9 | 10.1 | 1.9 | |
| 0.01 | 79.1 | 98.6 | 22.0 | 6.6 | 1.2 | |
| 0.005 | 95.8 | 99.8 | 4.4 | 5.5 | 1.0 | |
Model risk threshold: continuous model coefficient scale ranging from 0.0–1.0. * Positive predictive value. Classification of high-risk patients with an above-average risk of 90-day mortality according to specific model risk threshold settings. The average population risk of 90-day mortality after colorectal cancer surgery was 5.3%.
Table 3.
Model performance characteristics: identification of low-risk patients at various model thresholds
| Model risk threshold | Proportion of patients flagged as low-risk by the model (%) | Specificity | Sensitivity | Risk of 90-day mortality (per cent)* | Relative risk compared with the population |
|---|---|---|---|---|---|
| 0.5 | 99.4 | 99.7 | 7.1 | 5.0 | 0.9 |
| 0.45 | 99.1 | 99.6 | 10.0 | 4.8 | 0.9 |
| 0.4 | 98.7 | 99.3 | 13.5 | 4.7 | 0.9 |
| 0.35 | 98.0 | 98.9 | 17.6 | 4.5 | 0.9 |
| 0.3 | 97.2 | 98.4 | 23.5 | 4.2 | 0.8 |
| 0.25 | 96.2 | 97.6 | 29.0 | 3.9 | 0.7 |
| 0.2 | 94.5 | 96.3 | 36.7 | 3.6 | 0.7 |
| 0.15 | 91.6 | 93.7 | 45.0 | 3.2 | 0.6 |
| 0.1 | 86.0 | 88.6 | 60.3 | 2.5 | 0.5 |
| 0.05 | 71.9 | 79.5 | 79.5 | 1.5 | 0.3 |
| 0.025 | 51.5 | 53.9 | 92.2 | 0.8 | 0.2 |
| 0.01 | 20.9 | 22.0 | 98.6 | 0.3 | 0.1 |
| 0.005 | 4.2 | 4.4 | 99.8 | 0.3 | 0.1 |
Model risk threshold: continuous model coefficient scale ranging from 0.0–1.0. *Negative predictive value. Classification of low-risk patients with a below-average risk of 90-day mortality according to specific model risk threshold settings. The average population risk of 90-day mortality after colorectal cancer surgery was 5.3%.
The discriminatory performance of the GBM benchmark model was slightly lower with AUROC 84.63 (95 per cent c.i. 82.83 to 86.42). Calibration intercept was -0.00 and calibration gradient 1.04 with 0.04 Brier score and 0.32 average precision. The performance characteristics of the GBM model is presented in Figs S1 and S2.
Discussion
Based on nationwide CRC quality-assurance data, a supervised machine-learning model predicting 90-day mortality after CRC surgery was successfully developed and internally validated. The model was developed by adhering to the OMOP CDM and the OHDSI framework for data standardization and patient-level prediction14 resulting in high discriminatory power with AUROC of 85.28 per cent and excellent calibration with robust predictions across age and sex distributions. The model index was set on postoperative day 1, incorporating prior medical history and the intraoperative course of each patient into a postoperative ‘gloves-off’ prediction model. From a clinical perspective, many of the high-risk groups identified by the model have the potential to participate in personalized patient treatment. A characteristic example was the identification of 5.5 per cent of all patients undergoing CRC surgery who faced more than a 35 per cent risk of 90-day mortality, corresponding to a relative risk 6.7 times higher than the average population risk (5.3 per cent). The ability to personalize patients' risk and identify patients at high risk enables closer and prolonged observation in order to prevent complications, especially those that have the potential to result in death. Identification of such patients can also optimize healthcare-resource utilization, allowing more resources can be allocated for patients at high risk to ensure targeted rehabilitation and closer follow-up. Similarly, the patients identified as low risk can gain benefit by treatment personalization. For example, 20.9 per cent of patients identified by the model with low risk (0.3 per cent) of death within 90 days of CRC surgery, were facing 17.6 times decreased risk of death compared with the average risk of the CRC population. A patient with such a low-risk profile may benefit from tailored care, including early discharge or early referral to adjuvant chemotherapy when indicated.
The standardized machine learning approach maximized the predictive accuracy of deep phenotypic, nationwide quality-assurance data for early postoperative prediction of mortality after CRC surgery. This approach contrasts with previous models targeting postoperative short-term outcome predictions by limited or intermediate sets of prespecified predictors24–30. Limiting the number of predictor variables has been widely advocated to reduce model complexity and make models applicable in bedside clinical settings, but such approaches conflict with the current digital reality of clinical medicine, where patients generate abundant and potentially predictive data31. Although the performance of the model may be considered high in terms of discriminatory power and calibration, the clinical interpretation of single underlying predictors, for instance open versus minimally invasive surgery, should be made with caution as they do not represent causal relationships per se32. Modifying potentially reversible predictors or targeting postoperative interventions for high- or low-risk individuals should call for testing in randomized clinical trials33.
Previous models have primarily targeted prediction of mortality within 30 days after surgery; the most prominent being the Surgical Risk Calculator developed by the American College of Surgeons24. The Surgical Risk Calculator model achieved a high accuracy with a C-index of 0.91 and Brier score 0.029 in predicting 30-day mortality by incorporating 21 preoperative predictor variables into the prediction model. Data from the American College of Surgeons National Surgical Quality Improvement Program is, however, limited by a short follow-up of only 30 days. Currently, models targeting 90-day mortality predictions after colorectal cancer surgery are scarce and do not target clinical use34.
This study has several limitations. Although using the full spectrum of readily available quality-assurance data maximized the predictive power of the model, this also increased model complexity. Integration of the prediction model directly into an electronic health record could provide clinicians with support for accurate postoperative patient-risk stratification and limit resources spent on collecting data for real-time clinical applications.
The model was derived from a nationwide patient cohort, including data on all patients diagnosed with CRC in Denmark. As the data may be unique to the Danish CRC setting, external validation should be performed, to see if these findings can be generalized to other countries. This external validation would also determine if the discriminatory power of this model was robust for populations and data-capture processes in other healthcare systems, so that the extent of recalibration required to provide accurate predictions in these other contexts could be identified. The present study group aims to perform this validation within the European Health Data and Evidence Network (EHDEN) and the international OHDSI community by conducting network studies. A significant limitation of most previous models includes considerable risk of spectrum bias. Using nationwide data from a quality-assurance registry containing more than 95 per cent of all CRC patients35 with data collected contemporaneously by the operating surgeon at each institution using a standardized web-based application, this risk is considered low for the present study.
The model showed a tendency towards over-estimation in low- and high-probability female patients, reflecting the nature of the dataset that included only limited numbers of patients in both extremes of the age distribution. Of note, the model accurately predicted mortality outcomes in both sexes undergoing either acute or elective surgery for either colon or rectal cancer in individuals from 50 to 95 years of age. In general, the classification characteristics of the model showed a moderate average precision. A class imbalance may explain these findings in the dataset, which conferred the model to assign the majority non-outcome class higher weight. A feasible option for future studies would be to investigate the predictive power in external data sets and clinical trial settings to assess how transportable the model is to other data sets, populations and clinical practice. The model presented here was not intended to support clinical decision making during preoperative planning, but exclusively targeted postoperative risk prediction.
The model provides accurate identification of high- and low-risk patients early in the postoperative period that can provide clinicians with a powerful tool for personalized patient trajectories and the opportunity to improve outcomes after surgery for CRC.
Supplementary Material
Acknowledgements
The Regional Data Protection Committee approved the study (REG-071-2018). Ethical approval was not required according to Danish law. The study was not preregistered. R.P.V. affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned have been explained. The manuscript guarantors had full access to all the data in the study, take responsibility for the integrity of the data and the accuracy of the data analysis, and had final responsibility for the decision to submit for publication.
The lead author R.P.V. attests that all listed authors meet the authorship criteria and that no others meeting the criteria have been omitted.
Concept and design: R.P.V., R.D.B., E.R.H., A.O., F.B., L.C., C.G., J.A.Z., E.A., H.H.R., I.D., N.D., P.B.R., P.R., I.G.; analysis and interpretation of data: R.P.V., I.D., N.D., P.B.R., P.R., I.G.; drafting of the manuscript: R.P.V., R.D.B., I.D., N.D.; critical revision of the manuscript: R.D.B., E.R.H., A.O., F.B., L.C., C.G., J.A.Z., E.A., H.H.R., I.D., N.D., P.B.R., P.R., I.G.; statistical analyses: R.P.V., I.D., N.D., P.B.R., P.R., I.G.; study guarantors: R.P.V., I.D., I.G. P.R.R. and I.G. shared last authorship.
Disclosure. All authors have completed the ICMJE uniform disclosure form at www.icmje.org/coi_disclosure.pdf and declare: E.A. was a full-time employee of Odysseus Data Services Inc.; P.B.R. was a full-time employee of Janssen Research & Development LLC at the time the study was conducted and reports ownership of stocks, stock options and pension rights from Janssen Research & Development LLC. P.R. has received unconditional research grants from Boehringer-Ingelheim, GSK, Janssen Research & Development, Novartis, Pfizer, Yamanouchi, and Servier; I.G. has received unrestricted research grants from Pharmacosmos, Reponex Pharmaceuticals A/S, Perfusion Tech, Intuitive Surgical, and consultancy fees from Medtronic and Ethicon; there are no other relationships or activities that could appear to have influenced the submitted work.
Data sharing and data accessibility
This study was based on Danish national register data. These data do not belong to the authors but to the Danish Health Data Authority and the authors are not permitted to share them, except in aggregate (e.g., a publication).
Funding
This project received support from the EHDEN project. EHDEN received funding from the Innovative Medicines Initiative 2 Joint undertaking (JU) under grant agreement No 806968. The JU receives support from the European Union's Horizon 2020 research and innovation program and EFPIA. The funders played no role in study design, data collection, data analysis, data interpretation or reporting.
Supplementary material
Supplementary material is available at BJS Open online.
Contributor Information
R P Vogelsang, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark.
R D Bojesen, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark; Department of Surgery, Slagelse Hospital, Slagelse, Denmark.
E R Hoelmich, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark.
A Orhan, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark.
F Buzquurz, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark.
L Cai, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark.
C Grube, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark.
J A Zahid, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark.
E Allakhverdiiev, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark; Odysseus Data Services Inc., Cambridge, Massachusetts, USA.
H H Raskov, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark.
I Drakos, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark.
N Derian, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark.
P B Ryan, Department of Medical Informatics, Janssen Research & Development LLC, Raritan, New Jersey, USA; Columbia University, New York, New York, USA.
P R Rijnbeek, Department of Medical Informatics, Erasmus University Medical Centre, Rotterdam, The Netherlands.
I Gögenur, Center for Surgical Science, Department of Surgery, Zealand University Hospital, Koege, Denmark; Department of Clinical Medicine, University of Copenhagen, Copenhagen, Denmark.
References
- 1. Degett TH, Dalton SO, Christensen J, Søgaard J, Iversen LH, Gögenur I et al. Mortality after emergency treatment of colorectal cancer and associated risk factors – a nationwide cohort study. Int J Colorectal Dis 2019;34:85–95 [DOI] [PubMed] [Google Scholar]
- 2. Sjo OH, Larsen S, Lunde OC, Nesbakken A. Short term outcome after emergency and elective surgery for colon cancer. Colorectal Dis 2009;11:733–739 [DOI] [PubMed] [Google Scholar]
- 3. Iversen LH, Ingeholm P, Gögenur I, Laurberg S. Major reduction in 30-day mortality after elective colorectal cancer surgery: a nationwide population-based study in Denmark 2001–2011. Ann Surg Oncol 2014;21:2267–2273 [DOI] [PubMed] [Google Scholar]
- 4. Iversen LH, Green A, Ingeholm P, Østerlind K, Gögenur I. Improved survival of colorectal cancer in Denmark during 2001–2012 – the efforts of several national initiatives. Acta Oncol 2016;55:10–23 [DOI] [PubMed] [Google Scholar]
- 5. Adam MA, Turner MC, Sun Z, Kim J, Ezekian B, Migaly J et al. The appropriateness of 30-day mortality as a quality metric in colorectal cancer surgery. Am J Surg 2018;215:66–70 [DOI] [PubMed] [Google Scholar]
- 6. Visser BC, Keegan H, Martin M et al. Death after colectomy: it's later than we think. Arch Surg 2009;144:1021–1027 [DOI] [PubMed] [Google Scholar]
- 7. Damhuis RAM, Wijnhoven BPL, Plaisier PW, Kirkels WJ, Kranse R, van Lanschot JJ et al. Comparison of 30-day, 90-day and in-hospital postoperative mortality for eight different cancer types. Br J Surg 2012;99:1149–1154 [DOI] [PubMed] [Google Scholar]
- 8. Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med 2019;380:1347–1358 [DOI] [PubMed] [Google Scholar]
- 9. Nielsen AB, Thorsen-Meyer H-C, Belling K, Nielsen AP, Thomas CE, Chmura PJ et al. Survival prediction in intensive-care units based on aggregation of long-term disease history and acute physiology: a retrospective study of the Danish National Patient Registry and electronic patient records. Lancet Digit Heal 2019;1:e78–89 [DOI] [PubMed] [Google Scholar]
- 10. Parikh RB, Schwartz JS, Navathe AS. Beyond genes and molecules – a precision delivery initiative for precision medicine. N Engl J Med 2017;376:1609–1612 [DOI] [PubMed] [Google Scholar]
- 11. Mitka M. Data-based risk calculators becoming more sophisticated – and more popular. JAMA 2009;302:730–731 [DOI] [PubMed] [Google Scholar]
- 12. Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform 2015;216:574–578 [PMC free article] [PubMed] [Google Scholar]
- 13. Hripcsak G, Ryan PB, Duke JD, Shah NH, Park RW, Huser V et al. Characterizing treatment pathways at scale using the OHDSI network. Proc Natl Acad Sci U S A 2016;113:7329–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reps JM, Schuemie MJ, Suchard MA, Ryan PB, Rijnbeek PR. Design and implementation of a standardized framework to generate and evaluate patient-level prediction models using observational healthcare data. J Am Med Informatics Assoc 2018;25:969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Q, Reps JM, Kostka KF, Ryan PB, Zou Y, Voss EA et al. Development and validation of a prognostic model predicting symptomatic hemorrhagic transformation in acute ischemic stroke at scale in the OHDSI network. PLoS One 2020;15:e0226718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Danish Colorectal Cancer Group (DCCG.dk). https://dccg.dk/. (accessed 20 August 2020)
- 17. Reps JM, Rijnbeek PR, Ryan PB. Identifying the DEAD: development and validation of a patient-level model to predict death status in population-level claims data. Drug Saf 2019;42:1377–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gori M. Deep architectures. In: Katey Birtcher, Steve Merken, Peter Jardim (eds) Machine Learning: A Constraint-Based Approach (1st edn). Burlington, MA: Morgan Kaufmann Publishers, 2017, 236–238 [Google Scholar]
- 19. Bishop PM. In: Jordan M, Kleinberg J, Schölkopf B (eds), Pattern Recognition and Machine Learning (1st edn). New York, NY: Springer Science+Buisness Media, 2006, 359–418. [Google Scholar]
- 20. Suchard MA, Simpson SE, Zorych I, Ryan P, Madigan D. Massive parallelization of serial inference algorithms for a complex generalized linear model. ACM Trans Model Comput Simul 2013;23:1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD '16). New York, NY, USA: Association for Computing Machinery; 2016: 785–794. [Google Scholar]
- 22. Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 2015;350:g7594. [DOI] [PubMed] [Google Scholar]
- 23. National Comitee on Health Research Ethics . What to Notify? https://en.nvk.dk/how-to-notify/what-to-notify (accessed 6 October 2020)
- 24. Bilimoria KY, Liu Y, Paruch JL, Zhou L, Kmiecik TE, Ko CY et al. Development and evaluation of the universal ACS NSQIP surgical risk calculator: a decision aid and informed consent tool for patients and surgeons. J Am Coll Surg 2013;217:833–842.e1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stefani LC, Gutierrez CDS, Castro S. M D J, Zimmer RL, Diehl FP, Meyer LE et al. Derivation and validation of a preoperative risk model for postoperative mortality (SAMPE model): an approach to care stratification. PLoS One 2017;12:e0187122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gawande AA, Kwaan MR, Regenbogen SE, Lipsitz SA, Zinner MJ. An Apgar score for surgery. J Am Coll Surg 2007;204:201–208 [DOI] [PubMed] [Google Scholar]
- 27. Tekkis PP, Prytherch DR, Kocher HM, Senapati A, Poloniecki JD, Stamatakis JD et al. Development of a dedicated risk-adjustment scoring system for colorectal surgery (colorectal POSSUM). Br J Surg 2004;91:1174–1182 [DOI] [PubMed] [Google Scholar]
- 28. Copeland GP, Jones D, Walters M. POSSUM: a scoring system for surgical audit. Br J Surg 1991;78:355–360 [DOI] [PubMed] [Google Scholar]
- 29. de Vries S, Jeffe DB, Davidson NO, Deshpande AD, Schootman M. Postoperative 30-day mortality in patients undergoing surgery for colorectal cancer: development of a prognostic model using administrative claims data. Cancer Causes Control 2014;25:1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fazio VW, Tekkis PP, Remzi F, Lavery IC. Assessment of operative risk in colorectal cancer surgery: the Cleveland Clinic Foundation colorectal cancer model. Dis Colon Rectum 2004;47:2015–2024 [DOI] [PubMed] [Google Scholar]
- 31. Goldstein BA, Navar AM, Pencina MJ, Ioannidis JPA. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. JAMA 2017;24:198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doshi-Velez F, Perlis RH. Evaluating machine learning articles. JAMA 2019;322:1777–1779 [DOI] [PubMed] [Google Scholar]
- 33. Kappen TH, van Klei WA, van Wolfswinkel L, Kalkman CJ, Vergouwe Y, Moons KGM et al. Evaluating the impact of prediction models: lessons learned, challenges, and recommendations. Diagn Progn Res 2018;2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walker K, Finan PJ, Van Der Meulen JH. Model for risk adjustment of postoperative mortality in patients with colorectal cancer. Br J Surg 2015;102:260–280 [DOI] [PubMed] [Google Scholar]
- 35. Ingeholm P, Gögenur I, Iversen LH. Danish colorectal cancer group database. CLEP 2016;8:465–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study was based on Danish national register data. These data do not belong to the authors but to the Danish Health Data Authority and the authors are not permitted to share them, except in aggregate (e.g., a publication).