Abstract.
Purpose: Integrative analysis combining diagnostic imaging and genomic information can uncover biological insights into lesions that are visible on radiologic images. We investigate techniques for interrogating a deep neural network trained to predict quantitative image (radiomic) features and histology from gene expression in non-small cell lung cancer (NSCLC).
Approach: Using 262 training and 89 testing cases from two public datasets, deep feedforward neural networks were trained to predict the values of 101 computed tomography (CT) radiomic features and histology. A model interrogation method called gene masking was used to derive the learned associations between subsets of genes and a radiomic feature or histology class [adenocarcinoma (ADC), squamous cell, and other].
Results: Overall, neural networks outperformed other classifiers. In testing, neural networks classified histology with area under the receiver operating characteristic curves (AUCs) of 0.86 (ADC), 0.91 (squamous cell), and 0.71 (other). Classification performance of radiomics features ranged from 0.42 to 0.89 AUC. Gene masking analysis revealed new and previously reported associations. For example, hypoxia genes predicted histology (). Previously published gene signatures for classifying histology were also predictive in our model (). Gene sets related to the immune or cardiac systems and cell development processes were predictive () of several different radiomic features. AKT signaling, tumor necrosis factor, and Rho gene sets were each predictive of tumor textures.
Conclusions: This work demonstrates neural networks’ ability to map gene expressions to radiomic features and histology types in NSCLC and to interpret the models to identify predictive genes associated with each feature or type.
Keywords: radiogenomics, non-small cell lung cancer, gene expression, model interpretability, deep learning
1. Introduction
Precision medicine has driven the high-throughput profiling of both molecular and medical imaging data to identify detailed tumor subtypes that better predict survival and treatment outcomes. Radiogenomic studies attempt to integrate two complementary data types to explain tumor imaging patterns using molecular information and vice versa. For example, radiogenomic studies have shown that image features [e.g., appearance of a tumor on computed tomography (CT) or magnetic resonance imaging] predict molecular patterns (e.g., gene expression, gene mutation, or molecular subtypes).1–6 Radiogenomic studies support the derivation of tumors’ biological states from noninvasive imaging and the correlation of molecular information and imaging phenotypes to better understand cancer heterogeneity. However, radiogenomic studies are often limited by the high dimensionality of the data, the simplifying model assumptions (e.g., linearity), and the lack of validation datasets.7,8
Deep learning techniques have been widely used on molecular and imaging datasets given their ability to handle high-dimensional inputs without feature engineering and to represent nonlinear and hierarchical relationships between model inputs and outputs. Several studies have used deep learning models such as convolutional neural networks, generative adversarial networks, and autoencoders to uncover radiogenomic associations.5,6,9,10 However, while these works report accurate predictions of imaging phenotypes from genomic data, they do not attempt to provide a biological interpretation of what the model has learned. While high classification accuracy is important, the ability to interrogate the model is critical to validating the learned radiogenomic associations.
In our previous work, we addressed the model understanding challenge by presenting methods such as gene masking to interpret trained neural networks.11 We showed that the models were capable of learning radiogenomic associations that were consistent with prior work while also generating new associations for further consideration. A limitation of this prior work11 is that the analysis was performed using a single dataset from glioblastoma patients. We did not demonstrate the generalizability of our approach in other domains. Therefore, the purpose of this study is to investigate the ability of neural networks to model gene expression in a different cancer domain with multiple different histologies or stages, a variety of computationally derived image features (e.g., shape and texture), and an external validation or testing dataset.
Here, we present deep feedforward neural networks to model transcriptomes using two similarly derived radiogenomic datasets recently published in non-small cell lung cancer (NSCLC).12 As one of the few publicly released radiogenomic datasets available, the paper reported radiogenomic associations and provided a basis for comparison. First, we evaluate the ability of neural networks to independently predict two clinical traits (histology and stage) and 101 radiomic features using a transcriptome consisting of 21,766 gene expressions in a training dataset of 262 patients. Next, we demonstrate the generalizability of our neural network models in an independent validation dataset of 89 patients. Finally, we systematically probe the trained neural networks to define specific patterns of gene expression related to a clinical trait or radiomic feature. We compare the models’ learned relationships with previously reported associations.
2. Materials and Methods
2.1. Data
Clinical, imaging, and transcriptomic data were from 351 cases used in a prior study.12 The data consisted of two groups of patients, all of whom were diagnosed with primary tumors, had contrast-enhanced diagnostic CT, and underwent surgical resection. In one group, data were collected from 262 patients treated at the H. Lee Moffitt Cancer Center, Tampa, Florida, from 2006 to 2009 (Dataset1). The remaining 89 patients were treated at the Maastricht University Medical Centre, Maastricht, Netherlands (Dataset2). In this study, Dataset1 and Dataset2 were treated as training and testing datasets, respectively. Patient characteristics are compared between the two datasets in Table 1. The clinical stage referred to pathologic TNM staging and was represented using four categories: I, II, III, or other. Pathologic histology was captured using three categories: adenocarcinoma (ADC), squamous carcinoma (SCC), or “other.” CT scans were interpolated to have a voxel size of , and radiomic features were generated in prior work by the original study authors13 from regions identified using a semiautomated ensemble segmentation algorithm.14 Radiomic features were extracted from three-dimensional tumor volumes in contrast-enhanced presurgical CT scans to determine histogram statistics; morphology; textures, such as gray-level co-occurrence matrix (GLCM), gray-level run-length matrix (called RLGL), and gray-level size-zone matrix (GLSZM); Laplacian of Gaussian (LoG) transformations; and wavelet decompositions. Transcriptomes consisting of 21,766 gene expression levels were measured for all patients using the same Affymetrix microarray chip platform.
Table 1.
Patient characteristics, estimated or replicated from the source.12
| Training () | Testing () | ||
|---|---|---|---|
| Source | Country | USA | Netherlands |
| Gender | Male | 100 (38%) | 59 (66%) |
| Female | 124 (47%) | 28 (32%) | |
| N/A | 38 (15%) | 2 (2%) | |
| Histology | ADC | 129 (49%) | 42 (48%) |
| Squamous | 61 (23%) | 33 (38%) | |
| Other | 34 (13%) | 12 (14%) | |
| N/A | 38 (15%) | 0 (0%) | |
| Stage | I | 123 (47%) | 39 (44%) |
| II | 35 (13%) | 26 (29%) | |
| III | 46 (18%) | 12 (14%) | |
| Other | 20 (8%) | 10 (11%) | |
| N/A | 38 (14%) | 2 (2%) | |
| Smoking status | Current | 66 (25%) | N/A |
| Former | 141 (54%) | N/A | |
| None | 17 (7%) | N/A | |
| N/A | 38 (14%) | N/A | |
| Tumor site | Primary | 224 (86%) | 87 (98%) |
| N/A | 38 (14%) | 2 (2%) | |
| Status | Alive | 134 (51%) | 41 (46%) |
| Deceased | 90 (35%) | 46 (52%) | |
| N/A | 38 (14%) | 2 (2%) | |
| Follow-up | Median months | 32 | 31 |
In this study, each radiomic feature was a continuous variable that was transformed into binary classes using -means clustering in the training dataset. This approach was used to separate patients into two groups based on a single radiomic feature. If a radiomic feature was highly imbalanced (e.g., of the patients belong to one group), that feature was removed from the analysis. For example, the radiomic feature surface area to volume ratio was grouped into two clusters: cluster A had a mean ratio of 0.26, and cluster B had a mean ratio of 0.52. Each cluster represented one class. Clustering was performed via KMeans from sklearn. After filtering, 101 radiomic features out of the 636 features generated in the original study remained.12 These included seven types of radiomic features: 10 shape, 22 GLCM, 7 GLSZM, 10 RLGL, 12 stats, 9 LoG, and 31 wavelet features. The full names and class frequencies of the 101 radiomic features are provided in a GitHub repository (see Code Availability). LoG and wavelet features were selected from LoG_sigma_0_5_mm_2D and the HHH wavelet decomposition. Radiomic features with definitions that were not explicitly provided by the source12,13 were not considered during model interpretation. The same cluster boundaries discovered during training were used to binarize the testing data.
2.2. Radiogenomic Modeling
Dense feed-forward neural networks were used to map transcriptomes (which we defined as model inputs) to radiomic features (model outputs). Gene expression was standardized by mean subtraction and standard deviation division for each gene. Layers within the radiogenomic models were constructed with three or four hidden layers, where the first hidden layer had either 2000, 4000, or 6000 nodes. The number of hidden nodes was halved with each subsequent hidden layer. Neural networks were trained using cross-entropy loss, nonlinear activation functions, dropout, batch normalization, and early stopping by monitoring loss.
Performance was reported using area under the receiver operating characteristic curve (AUC). Hyperparameters were selected using a grid search based on the model that achieved the highest mean AUC in 10-fold cross-validation (CV) during training. During CV, the mean and standard deviation used for gene standardization were calculated using the training folds for each split; in testing, metrics were based on the entire training dataset. Training performance was averaged across CV folds. Accuracy was calculated based on a decision threshold of 0.5 class probability.
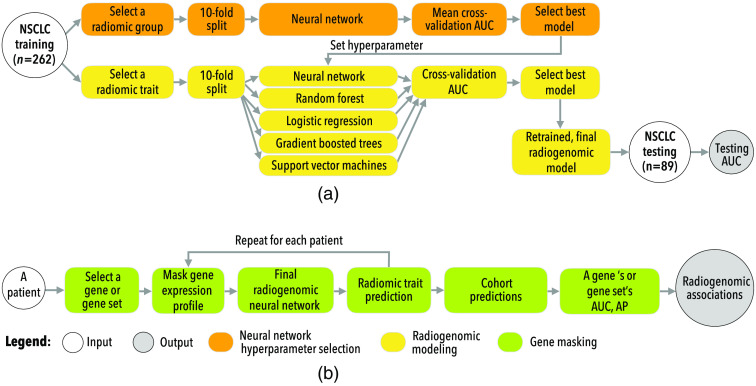
Figure 1(a) shows the overall procedure used to train the radiogenomic neural networks. To reduce the hyperparameter search for each radiomic feature, a grid search was performed using a neural network that predicted all features corresponding to one of the seven radiomic feature groups (previously defined as shape, GLCM, GLSZM, RLGL, stats, LoG, and wavelet). For example, one network would be trained to predict all 22 GLCM features as a multilabel classification task using the patient’s transcriptome as the input. Once a neural network was trained for each radiomic group, the hyperparameters used for the best performing network were then used to train a neural network that predicted a single radiomic feature within that group. Other classifiers, including logistic regression, support vector machines, random forest, and gradient boosted trees, were also implemented as a comparison. Each comparison model was trained to predict a single radiomic feature and evaluated against the neural network. The best performing model for each radiomic feature was then retrained on the entire training dataset to obtain the final model. Final models were evaluated on the testing dataset. Radiomic features with at least 0.70 test AUC were kept for further analysis.
Fig. 1.
An overview of this study’s approaches to (a) training and (b) interpretation radiogenomic neural networks.
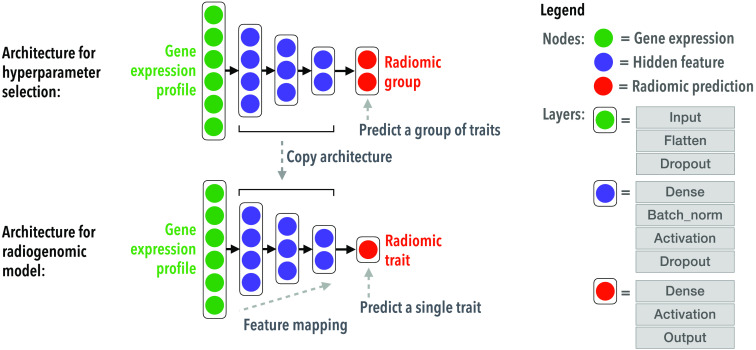
More details on the architecture of radiogenomic neural networks are shown in Fig. 2. All models and their hyperparameters are listed in Table 2.
Fig. 2.
The architecture and hyperparameter tuning of a radiogenomic neural network.
Table 2.
NSCLC radiogenomic models and hyperparameters. Grid search was used for selecting hyperparameters.
| Model type | # Models | Hyperparameter | Values |
|---|---|---|---|
| Logistic regression | 100 | Penalty type | Elastic net |
| L1 ratio | [0–1] | ||
| Solver | SAGA | ||
| Support vector machines | 400 | Kernel | Linear, poly, rbf, sigmoid |
| C penalty | |||
| Random forest | 120 | Trees | |
| Split criterion | Gini, entropy | ||
| Max. features | , | ||
| Max. depth | None | ||
| Gradient boosted trees | 150 | Trees | |
| Max. depth | [1–3] | ||
| Learning rate | |||
| Neural network | 48 | Hidden nodesa | [6000–250] |
| Hidden layers | 3, 4 | ||
| Optimizer | Nadam, Adadelta | ||
| Activation | Sigmoid, relu | ||
| Dropout | 0.4, 0.6 | ||
| Loss | Binary cross-entropy | ||
| Epochs (patience) | 500 (200) | ||
| Batch | 10 |
G: number of genes in the transcriptome
First hidden layers were either 2000, 4000, or 6000 nodes, where the number of hidden nodes for a layer was halved with each subsequent layer.
2.3. Modeling Tumor Histology and Stage
In addition to predicting radiomic features, we trained additional networks to predict histology and stage from the transcriptome. These networks have the same architecture as what was used for radiogenomic modeling. Histology and stage were each modeled as multilabel classification tasks (Table 1). The neural networks used categorical entropy loss and softmax activation in the prediction layer. Training scores were microaveraged across all classes and folds in CV. Test scores and model interpretation methods were based on one class versus all other classes unless otherwise noted. The methods for hyperparameter optimization and model selection were the same as previously described.
2.4. Using Interpretability Methods to Identify Gene Expression Patterns
Figure 1(b) shows the gene masking steps used to extract predictive gene expression patterns from trained neural networks. Gene masking was previously defined for radiogenomic neural networks11 and focused on a component of the model’s input using one predefined gene set at a time, a similar process to sensitivity analysis.15 For example, masking of genes related to hypoxia involved taking each patient’s gene expression profile, keeping only the hypoxia genes’ expressions, and setting all other gene expressions to zero (i.e., the input is masked). The masked input was then pushed through the model, and the model’s prediction of a radiomic feature was recorded. This process was repeated for the entire cohort, and classification performance was calculated via AUC and average precision (AP). Gene masking measured the model’s ability to predict radiomic features based on gene expression of a particular gene set, where the higher the performance is, the stronger the association in the cohort is. Masking resulted in “radiogenomic associations” learned by the model. The strength and generalizability of the learned radiogenomic associations were measured for each gene set by their performance (i.e., AUC) in the testing cohort. Predefined gene sets were used for gene masking. These included the “Hallmark” and “Gene Ontology” (GO) biological processes from Molecular Signature Database (MSigDB) v7.0.16 For simplicity, gene sets with a maximum of 500 genes were studied.
Relationships between gene expression and histology were also studied using gene masking. In prior work, a 42-gene signature was used to distinguish ADC from SCC.17 In another similar effort, a 75-probe set signature was found for ADC, SCC, and large cell carcinoma (LCC).18 The gene sets reported in these works were also examined in this work as a point of comparison.
3. Results
3.1. Predicting Histology or Stage from the Transcriptome
3.1.1. Deep neural networks outperform comparison approaches
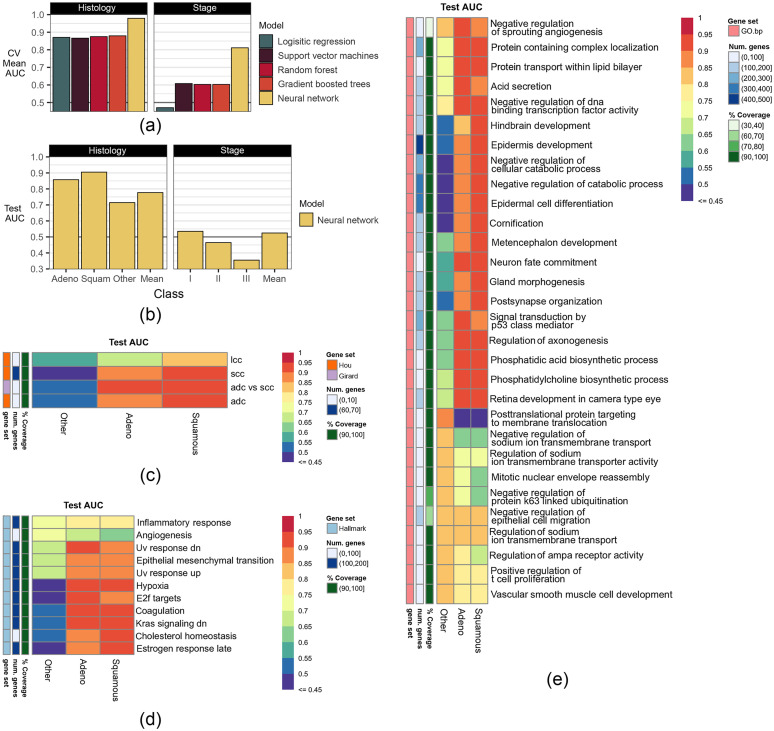
In predicting histology, deep neural networks outperformed other models by more than 0.10 AUC in 10-fold CV of the training dataset [Fig. 3(a)]. Performance of the histology model remained consistent in the testing dataset, achieving test scores of 0.86 AUC, 0.91 AUC, and 0.71 AUC in predicting ADC, SCC, or other histology, respectively. The neural network achieved an overall microaveraged test AUC of 0.77. In predicting stage, all models fared poorly, with the neural network still achieving the best performance. Given the poor performance in the predicting stage, only the neural network used to predict histology was analyzed using gene masking.
Fig. 3.
The ability of models to predict NSCLC histology and stage in (a) training and (b) testing. In training, models were evaluated using 10-fold CV, and models were compared using the mean AUC scores in CV. The top performing model was then retrained on the full training dataset and evaluated on the testing dataset. The testing performance scores are shown for each histology type and stage. (c)–(e) Gene masking of the histology neural network using gene sets from (c) published gene signatures for histology,17,18 (d) Hallmark (top five out of 50),19 and (e) Gene Ontology biological processes (GO.bp, top ten out of 7350).16 In (c)–(e), each column is a type of histology and each row is a gene set used to mask the trained model to inspect how well the model predicted a certain histology type. The color in a cell shows the model’s performance using a gene set to predict a histology type, where red denotes higher AUCs and purple denotes lower AUCs in the testing dataset. (b) The ability of the histology model to predict each histology type and the AUC score is based on using all genes in the gene expression profile. (c)–(e) The ability of a specific gene set in predicting a histology type. For more details on the gene sets used, see Sec. 2. Notation: lcc = gene signature for large cell carcinoma, scc = gene signature for squamous cell carcinoma, adc = gene signature for adenocarciomas, adc versus scc = gene signature for differentiating ADC from SCC.
3.1.2. Gene masking identifies gene sets that predict histology
Gene masking of the histology neural network showed agreement with previously published gene signatures for predicting NSCLC, ADC, and SCC.17,18 As shown in [Fig. 3(c)], the aforementioned gene signatures were also found to be predictive in our histology neural network in the testing dataset. In particular, the gene signature from Ref. 17 resulted in 0.93 test AUC in both ADC and SCC.
Hallmark gene sets were also predictive of histology [Fig. 3(d)]. Gene expression related to hypoxia, coagulation, and KRAS signaling predicted both ADC and SCC ( test AUC). Similar to the overall performance observed on the testing data, summarized in Fig. 3(b), the histology neural network was driven by accurate predictions in ADC and SCC, where the test AUC was about 0.20 higher than what was obtained when predicting the other histology class. Subsequently, this behavior was reflected in gene masking, where the most predictive gene sets for estimating other histology in testing were inflammatory response (0.73 AUC) and angiogenesis (0.72 AUC). Notably, angiogenesis was more predictive of other histologies (0.73 AUC) than ADC (0.66 AUC) or SCC (0.63 AUC) classes in testing.
The most predictive GO biological processes [Fig. 3(e)] were also associated with angiogenesis, epithelial mesenchymal transition, and hypoxia from the Hallmark gene sets. Of the gene sets considered in gene masking, negative regulation of DNA-binding transcription factor activity from GO had the best overall testing performance with the highest micro-averaged AUC of 0.79; the individual test AUCs were 0.93 in ADC, 0.91 in SCC, and 0.75 in other. The aforementioned gene set consisted of 170 genes, where 156 (91.2%) were in the gene expression profile. A summary of gene expression patterns is given in Table 3. Notably, KRAS is a major gene studied in NSCLC, and the KRAS Hallmark was found to be predictive of ACC and SCC.
Table 3.
Summary of predictive gene expression patterns in NSCLC histology.
| Histology | Transcriptomic pattern | Test | |||
|---|---|---|---|---|---|
| Themea | Gene set | Sourceb | AUC | AP | |
| ADC | Synthesis | Phosphatidylcholine biosynthetic process | G | 0.94 | 0.93 |
| Phosphatidic acid biosynthetic process | G | 0.94 | 0.86 | ||
| Transcription | Neg. regulation of dna-binding transcription factor activity | G | 0.93 | 0.92 | |
| Vasculature | Neg. regulation of sprouting angiogenesis | G | 0.93 | 0.92 | |
| Coagulation | H | 0.91 | 0.86 | ||
| Cell development | Retina development in camera-type eye | G | 0.93 | 0.87 | |
| Cell death | Hypoxia | H | 0.91 | 0.88 | |
| KRAS | KRAS signaling down | H | 0.91 | 0.84 | |
| UV | UV response down | H | 0.92 | 0.90 | |
| E2F | E2F targets | H | 0.92 | 0.87 | |
| Squamous cell | Cell development | Metencephalon development | G | 0.95 | 0.90 |
| Carcinoma | Differentiation | Epidermal cell differentiation | G | 0.94 | 0.94 |
| Neuron fate commitment | G | 0.92 | 0.87 | ||
| Catabolism | Neg. regulation of cellular catabolic process | G | 0.94 | 0.88 | |
| Cell death | Cornification | G | 0.94 | 0.93 | |
| Hypoxia | H | 0.91 | 0.90 | ||
| KRAS | KRAS signaling down | H | 0.93 | 0.93 | |
| Hormone | Estrogen response late | H | 0.92 | 0.91 | |
| Cholesterol | Cholesterol homeostasis | H | 0.92 | 0.83 | |
| Other | Transport | Posttranslational protein targeting to membrane, translocation | G | 0.86 | 0.35 |
| Regulation of sodium ion transmembrane transporter activity | G | 0.84 | 0.54 | ||
| AMPA receptor | Regulation of AMPA receptor activity | G | 0.85 | 0.44 | |
| Cell cycle | Mitotic nuclear envelope reassembly | G | 0.84 | 0.50 | |
| Ubiquitination | Neg. regulation of protein K63-linked ubiquitination | G | 0.84 | 0.38 | |
| Immune system | Inflammatory response | H | 0.73 | 0.37 | |
| Vasculature | Angiogenesis | H | 0.72 | 0.35 | |
Gene sets were categorized by comparing the top five gene sets in GO and Hallmark with at least 0.70 test AUC.
from MSigDB v7.0, where = Hallmark, = Gene Ontology, neg.= negative.
3.2. Predicting Radiomic Features from the Transcriptome
3.2.1. Overall performance
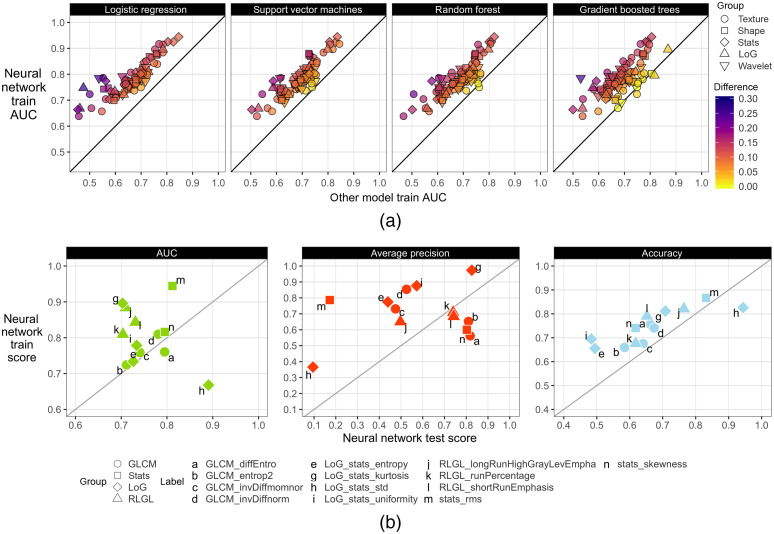
Neural networks were overall better at classifying radiomic features than all other models within the training dataset [Fig. 4(a)]. The only exceptions were in gradient boosted trees that had better performance in four radiomic features (differences below 0.026 AUC) and random forest in one radiomic feature (0.012 AUC difference). In testing, neural networks had 0.42 to 0.89 AUC, 0.45 to 0.94 accuracy, and 0.09 to 0.98 AP across all radiomic feature classifications. A subset of 13 radiomic features had at least 0.70 test AUC and was subsequently selected for interpretation. Figure 4(b) shows the neural network’s generalizability to classify the aforementioned 13 radiomic features in the testing dataset.
Fig. 4.
Radiogenomic modeling performance (a) between neural networks and other models in the training dataset. Neural network performance (b) in the training and testing datasets for the 13 radiomic features selected for further analysis. Train scores represent the averaged scores of the validation folds during 10-fold CV in the training dataset. The test scores are the model’s performance in the testing dataset after models were retrained on the full training dataset.
3.2.2. Gene masking identifies gene sets that predict radiomic features
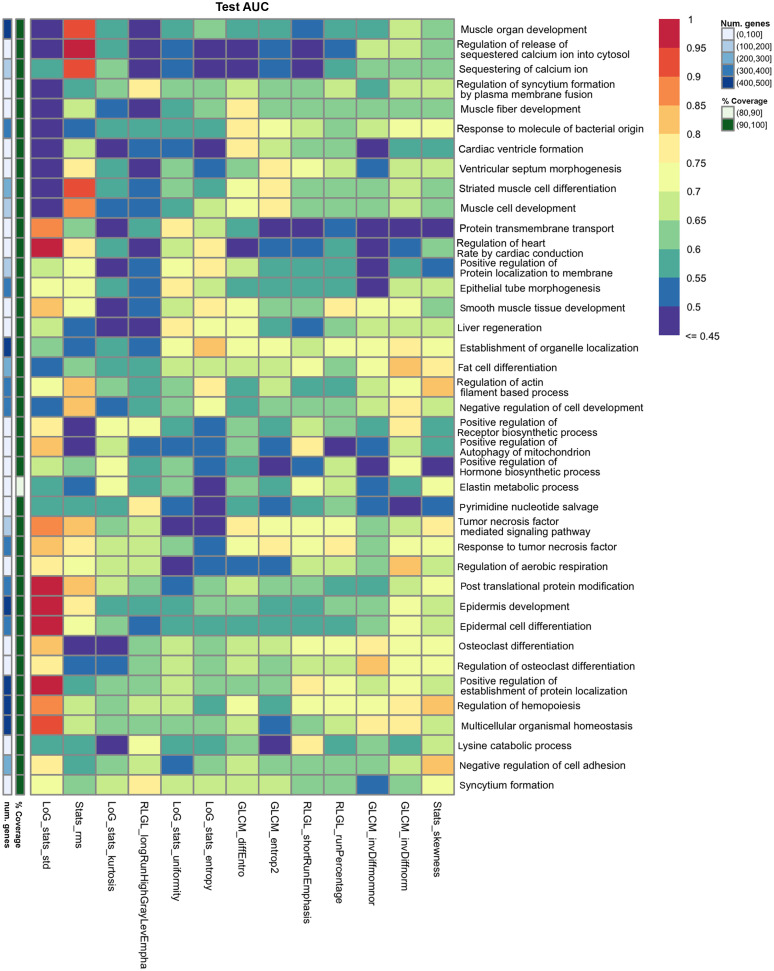
Figure 5 shows the top GO gene sets associated with predicting each radiomic feature. Overall, the results of gene masking suggest that the prediction of each radiomic feature was associated with a unique gene expression profile driven by different biological processes. None of the radiomic features had similar scores across all gene sets. Some gene sets were better for predicting one radiomic feature but not another. For example, the top two gene sets for predicting an imaging texture, RLGL_longRunHighGrayLevEmpha, were regulation of syncytium formation by plasma membrane fusion and pyrimidine nucleotide salvage, which had test AUCs above 0.75, but for all other 12 radiomic features these two gene sets were below 0.70 AUC and 0.65 AUC, respectively. Conversely, there were gene sets that could predict multiple radiomic features at once. For example, response to tumor necrosis factor (TNF) was predictive of two other imaging textures, GLCM_entrop2 (0.78 AUC, 0.81 AP) and RLGL_runPercentage (0.76 AUC, 0.76 AP), in testing. Hallmark gene sets were also applied to the radiomic models in gene masking analysis but were not as predictive as the GO gene sets.
Fig. 5.
Gene masking of the radiogenomics models with biological processes from GO. The top three gene sets ranked by test AUC for each radiomic feature are shown.
Radiogenomic associations were summarized for radiomic features related to histogram statistics of the tumor mask, the transformation of the mask (LoG features), and textures of tumors in Table 4. For example, the three gene sets that were most predictive of LoG_stats_std were related to post-translational protein modification, epidermis development, and DNA repair. Processes involving the immune system and cardiac system were the top predictors for several radiomic features. Many gene sets were related to cell development, varying from muscle, liver, epidermis, fat cell, and renal gene sets. AKT signaling, a targeted pathway in NSCLC therapy,20 was moderately predictive of RLGL_longRunHighGrayLevEmpha with 0.76 AUC but had an AP of 0.54 in testing. TNF was associated with RLGL_runPercentage and GLCM_entrop2. Rho signaling, associated with tumor suppressor activity and another targeted pathway in NSCLC,21,22 was associated with GLCM_diffEntro (0.75 AUC, 0.78 AP) and GLCM_invDiffmomnor (0.79 AUC, 0.55 AP). While GLCM_invDiffmomnor and invDiffnorm were correlated and clustered together, the two radiomic features had differing gene sets.
Table 4.
Summary of predictive gene expression patterns in NSCLC radiomic features.
| Radiomic feature | Transcriptomic pattern | test | ||
|---|---|---|---|---|
| Themea | Gene set (from GO) | AUC | AP | |
| Stats_skewness | Cytoskeleton | Reg. of actin filament-based process | 0.82 | 0.82 |
| Adhesion | Neg. Reg. off cell adhesion | 0.81 | 0.81 | |
| Neg. Reg. of cell-cell adhesion | 0.80 | 0.77 | ||
| Immune system | Reg. of hemopoiesis | 0.81 | 0.78 | |
| Reg. of leukocyte differentiation | 0.80 | 0.77 | ||
| Stats_rms | Transport | Reg. of release of sequestered calcium ion into cytosol | 0.95 | 0.65 |
| Sequestering of calcium ion | 0.93 | 0.30 | ||
| Development | Muscle organ development | 0.93 | 0.46 | |
| striated muscle cell differentiation | 0.93 | 0.32 | ||
| Actin filament-based movement | 0.92 | 0.26 | ||
| LoG_stats_std | Post-translational | Post-translational protein modification | 1.00 | 1.00 |
| Development | Epidermis development; epidermal cell differentiation | 1.00 | 1.00 | |
| DNA repair | 26-cm DNA double-strand break processing involved in repair via single-strand annealing | 0.99 | 0.79 | |
| Cell cycle | neg. reg. of mitotic cell cycle | 0.99 | 0.79 | |
| LoG_stats_uniformity | Cell development | Liver regeneration | 0.78 | 0.64 |
| Epithelial tube morphogenesis | 0.77 | 0.49 | ||
| Transport | Protein transmembrane transport | 0.77 | 0.64 | |
| Intracellular protein transmembrane transport | 0.76 | 0.61 | ||
| Catabolism | Organic acid catabolic process | 0.75 | 0.54 | |
| LoG_stats_entropy | Localization | Establishment of organelle localization | 0.82 | 0.47 |
| Pos. reg. of protein localization to membrane | 0.79 | 0.60 | ||
| Heart rate | Reg. of heart rate by cardiac conduction | 0.77 | 0.40 | |
| Cell mobility | Reg. of actin filament-based process | 0.77 | 0.46 | |
| External stimulus | Cellular response to mechanical stimulus | 0.77 | 0.44 | |
| LoG_stats_kurtosis | Connective tissue | Elastin metabolic process | 0.73 | 0.84 |
| Collagen metabolic process | 0.72 | 0.85 | ||
| Synthesis | Pos. reg. of receptor biosynthetic process | 0.73 | 0.81 | |
| Pos. reg. of hormone biosynthetic process | 0.73 | 0.84 | ||
| Immune system | Response-regulating cell surface receptor signaling pathway | 0.72 | 0.78 | |
| GLCM_diffEntro | Muscle | Muscle fiber development | 0.76 | 0.82 |
| Muscle cell differentiation | 0.75 | 0.79 | ||
| Cardiac ventricle formation | 0.76 | 0.77 | ||
| Bacteria | Response to molecule of bacterial origin | 0.76 | 0.73 | |
| Rho | Rho protein signal transduction | 0.75 | 0.78 | |
| GLCM_invDiffnorm | Cell development | Fat cell differentiation | 0.83 | 0.63 |
| Neg. reg. of cell development | 0.80 | 0.60 | ||
| Cell respiration | Reg. of aerobic respiration | 0.81 | 0.64 | |
| Immune system | Neg. reg. of lymphocyte activation | 0.80 | 0.63 | |
| Nervous system | Neuromuscular process controlling balance | 0.79 | 0.70 | |
| GLCM_invDiffmomnor | Immune system | Reg. of osteoclast differentiation | 0.81 | 0.58 |
| Osteoclast differentiation | 0.80 | 0.58 | ||
| Homeostasis | Multicellular organismal homeostasis(G) | 0.80 | 0.61 | |
| Tissue homeostasis | 0.79 | 0.57 | ||
| Rho | Reg. of Rho protein signal transduction | 0.79 | 0.55 | |
| GLCM_entrop2 | TNF | Response to TNF | 0.78 | 0.81 |
| Muscle | Muscle cell development | 0.77 | 0.80 | |
| Ventricular septum morphogenesis | 0.77 | 0.77 | ||
| Striated muscle cell differentiation | 0.76 | 0.78 | ||
| Drug response | Response to xenobiotic stimulus | 0.76 | 0.84 | |
| RLGL_shortRunEmphasis | Localization | Pos. regulation of establishment of protein localization | 0.79 | 0.77 |
| Catabolism | Lysine catabolic process | 0.79 | 0.80 | |
| Cell death | Pos. reg. of autophagy of mitochondrion | 0.77 | 0.78 | |
| Hormone | Pos. reg. of insulin secretion | 0.77 | 0.77 | |
| Cell mobility | Neuron projection guidance | 0.75 | 0.71 | |
| RLGL_longRunHighGrayLevEmpha | Syncytium | Syncytium formation | 0.78 | 0.54 |
| Reg. of syncytium formation by plasma membrane fusion | 0.77 | 0.47 | ||
| Synthesis | Pyrimidine nucleotide salvage | 0.76 | 0.50 | |
| AKT | Protein kinase B signaling | 0.76 | 0.54 | |
| Renal system | Renal sodium excretion | 0.76 | 0.44 | |
| RLGL_runPercentage | Muscle | Smooth muscle tissue development | 0.76 | 0.75 |
| TNF | Response to TNF | 0.76 | 0.76 | |
| TNF-mediated signaling pathway | 0.75 | 0.76 | ||
| Immune system | Myeloid leukocyte differentiation | 0.75 | 0.76 | |
| Renal system | Metanephros development | 0.74 | 0.80 | |
Shown are the top five GO gene sets ranked by AUC and with at least 0.50 AP; reg.= regulation; pos. = positive; neg. = negative.
To compare the results of our neural network with prior reported associations, we performed an analysis using radiogenomic modules. Radiogenomic modules, defined as a set of correlated radiomic features and gene expressions, were previously defined as part of the original study.12 Not all radiomic features were reported in the original study’s radiogenomic modules. In this paper, the radiomic models were masked with the same Reactome gene sets as Grossmann et al.12 using MSigDB v4.0. Table 5 summarizes the overlapping radiogenomic associations found in this study compared with the aforementioned work. The highest agreement was between LoG_stats_entropy and module 13, where three of the pathways in the module were also among the top ten most predictive gene sets in our radiogenomic model. Other comparisons did not have overlapping associations. For example, the authors reported a radiogenomic association between GLCM_diffEntro and the five pathways in module 2, while we found the most predictive pathway in module 2 was ranked 197 out of the 664 Reactome pathways used in gene masking. Thus, our model suggested that many other pathways were more predictive of the radiomic feature than the five pathways in module 2.
Table 5.
A comparison of the learned radiogenomic associations extracted from our neural networks and the modules previously identified in the same dataset.12 Each module consisted of a set of Reactome pathways and a set of image features. Shown are the modules that included the radiomic features used in this study. If any module’s set of pathways was ranked among the top 100 in gene masking, the top three pathways were listed.
| Radiomic trait | Reactome pathway | This study | Grossmann et al.12 | ||
|---|---|---|---|---|---|
| Test AUC | Ranka | Module | # Pathways | ||
| GLCM_diffEntro | Cross presentation of soluble exogenous antigens endosomes | 0.58 | 197 | 2 | 5 |
| Phase II conjugation | 0.69 | 9 | 12 | 35 | |
| Regulation of mitotic cell cycle | 0.66 | 21 | — | — | |
| ABCA transporters in lipid homeostasis | 0.65 | 28 | — | — | |
| LoG_stats_std | Regulation of ornithine decarboxylase | 0.94 | 15 | 2 | 5 |
| Cross presentation of soluble exogenous antigens endosomes | 0.82 | 89 | — | — | |
| Antigen processing cross presentation | 0.80 | 106 | — | — | |
| Cholesterol biosynthesis | 0.75 | 150 | 6 | 7 | |
| Signaling by TGF beta receptor complex | 0.83 | 81 | 8 | 17 | |
| Elongation arrest and recovery | 0.82 | 92 | — | — | |
| mRNA splicing | 0.75 | 142 | — | — | |
| LoG_stats_entropy | Elongation arrest and recovery | 0.73 | 1 | 7 | 8 |
| Mitochondrial protein import | 0.63 | 74 | — | — | |
| RNA pol III chain elongation | 0.58 | 180 | — | — | |
| Elongation arrest and recovery | 0.73 | 1 | 13 | 26 | |
| RNA pol II pre transcription events | 0.69 | 7 | — | — | |
| Formation of RNA pol II elongation complex | 0.69 | 9 | — | — | |
| Stats_skewness | Antigen processing cross presentation | 0.62 | 139 | 2 | 5 |
All Reactome pathways were ranked by test AUC.
4. Discussion
We demonstrated the ability of deep neural networks to learn associations between radiomic features or clinical traits and gene expression using two NSCLC cohorts. A relatively large training dataset of 262 patients was available. An independent test dataset of 89 patients allowed us to validate the generalizability of our neural network models. We showed that neural networks outperformed other machine learning models. While the overall test AUC was mixed across all 101 radiomic features (test AUC of 0.42–0.89), the thirteen radiomic features selected for gene masking and histology had an average test AUC above 0.70. We interpreted the models using gene masking and identified specific sets of gene expressions that were indicative of a trait or feature. Together, these results suggest that potential biological associations exist to explain the differences among histology classes and CT imaging characteristics of NSCLC patients.
A number of radiomic and radiogenomic studies have been performed with NSCLC patients.10,12,13,23–26 A recent study used the same dataset of 89 patients to train models to predict immune cell gene signatures of NSCLC tumors using CT radiomic features.27 Our model attempted to learn associations between high dimensional gene expression profiles and radiomic features. While deep neural networks were used to map CT image patches to tumor gene expression in Li et al.,10 the study did not report specific radiogenomic associations. Most related to our study was Grossmann et al.,12 which provided the source datasets. They used Gene Set Enrichment Analysis and the Iterative Signature Algorithm, a correlation and biclustering method, to define radiogenomic modules by grouping radiomic features with Reactome gene sets (i.e., pathways). The top ten most predictive Reactome pathways for a radiomic feature in our models overlapped with the radiogenomic modules defined by Grossmann et al.,12 but overall agreement was low. Differences in radiogenomic associations between our work and Grossmann et al.12 may relate to differences in methodology. Our study assessed the entire gene expression profile, whereas their study assessed correlations between radiomic features and subsets of genes. Moreover, the number of associations to be found was not predefined in our study, while Grossmann et al.12 specifically assessed 20 radiogenomic modules. Other studies12,24 have shown that their selected radiogenomic associations can differentiate between patients with longer versus shorter survival. We leave survival analysis using the radiogenomic associations found in our neural network models as future work.
We further explored the ability of neural networks to map transcriptomes to other relevant patient image features by training models to predict stage and histology. While neural networks were better at predicting stage and histology in the training dataset compared with other classifiers (based on scores from 10-fold CV), stage was poorly estimated in the testing dataset. However, the histology neural network has 0.77 test AUC when averaged across each histology type.
Several prior works for predicting NSCLC histology using radiomics have been based on differentiating ADC from SCC. For example, using five radiomic features, a study that trained a Naive Bayes model to distinguish ADC from SCC achieved a 0.72 AUC in a test set of 152 patients.28 In another similar study, a radiomic signature was able to distinguish ADC from SCC using a logistic regression model and 129 patients; the authors reported a 0.893 AUC in a test subset of 48 patients.29 More recent work reported logistic models that achieved 0.694, 0.780, 0.800, and 0.923 AUC for clinical, standard CT features, radiomics, or all three, respectively, to predict ADC versus SCC.30 The AUC scores were observed in a cohort of 93 patients but are likely overly optimistic given that there was no validation or test set to evaluate their models. In contrast, our neural network model used gene expression profiles to predict histology. In a test set of 89 patients, our model achieved a 0.86 test AUC when estimating ADC versus all other (SCC and other) histology types and a 0.91 test AUC when estimating SCC versus all other types (ADC and other). One notable study analyzed the association between CT-derived radiomic features and digital pathology-derived pathomic features to differentiate between two NSCLC subtypes.31
Additionally, there is a difference in our models’ performance when predicting histology compared with stage. The difference is likely because staging is based on factors such as tumor size, location, and spread to lymph nodes or metastasis in other sites, which is information that may not be readily seen in the gene expression of the collected tumor tissue. On the other hand, histology classification is based on the molecular and physical characterization of the collected sample, which is likely more related to the transcriptomic profiling of the tumor. Subsequently, we extracted the learned associations between gene expression and histology types with gene masking and compared with two previous studies that report gene expression signatures to predict NSCLC histology.17,18 The studies’ gene signatures were predictive in our models as well, indicating that our neural networks found similar associations as reported in prior work. In our model, other gene sets such as hypoxia (200 genes) and angiogenesis (36 genes) Hallmark gene sets and the neuronal (156 genes) GO gene set, are associated with histology and may have the potential to help automate or standardize the assignment of histology in the future.
This study has several notable limitations. The retrospective datasets used in this analysis were from two different sources with varying imaging protocols, which can add variance to radiomic feature values. A majority of patients were early-stage cancers, and thus late-stage patients were left out of the radiogenomic analysis. While sample size is an inherent concern of radiogenomic analysis, datasets with paired imaging and genomic data are difficult to obtain and limited in sample size. Interpreting the biological significance of radiomic features is challenging, and researchers are currently attempting to understand their correlation with tumor biology and other clinical traits. Radiomic features were binarized using clustering, and radiomic features with a minority class below 10% were removed. The tumor tissue samples used for transcriptome profiling are limited in that only one sample was acquired per patient, which may not fully capture tumor heterogeneity. These factors make it challenging to validate the relationships in radiogenomic models. The reported findings should be interpreted as possible associations and require further clinical or animal studies to validate.
In future work, the issues may be addressed by the standardization of imaging protocols to allow for consistent comparison of image features and maps such as genomic atlases to better characterize whole tumors. A larger sample size could result in a more complete representation of the general population and allow for modeling of radiomic features as continuous outputs. The ranked gene sets are interpreted based on GO descriptions. A more quantitative approach to compare the ranked gene sets between radiomic features, such as semantic similarity of GO terms,32 and a sensitivity analysis of resultant radiogenomic associations are needed. In addition to the transcriptome, there are likely other contributing factors to tumor image features, such as other molecular data (e.g., gene mutations and methylation) and patient covariates (e.g., smoking status). These factors could either be incorporated into the modeling process or used to stratify analyses. Additionally, the impact of knowing such radiogenomic associations at the time of tumor biopsy in relation to survival or treatment projection would be highly beneficial.
5. Conclusion
In this study, we present deep neural networks for mapping gene expressions to radiomic features or clinical traits in patients with non-small cell lung cancer. Our models are evaluated using public datasets. Neural networks are capable of modeling high-dimensional gene expression to predict tumor image features and lung cancer histology. We further interpret the models through gene masking and report the learned relationships between gene expression and a radiomic feature or histology type. We find that the network is capable of replicating previously reported associations while identifying new associations. The reported associations could be further studied to improve the automated classification of histology, predict specific gene expression profiles of patients presenting with an observable imaging phenotype, and develop a knowledge base of associations between imaging phenotypes to gene expression profiles that would be useful in informing individualized treatment planning.
Acknowledgments
This work is supported in part by the National Institutes of Health [F31CA221061 and T32EB016640 to N.F.S., U01CA196408 and R01CA210360 to D.R.A., W.H.], the National Science Foundation [#1722516 to W.H.], and the Department of Radiological Sciences through the Integrated Diagnostics (IDx) Shared Resource. Computational credit for use of Amazon Web Services was awarded through a partnership with the UCLA Department of Computational Medicine.
Biographies
Nova F. Smedley was a doctoral student in the Medical & Imaging Informatics group within the Department of Bioengineering at UCLA. Under the supervision of Dr. Hsu, her research focused on using machine learning approaches for improving the radiogenomic and prognostic analyses of cancer patients. She was a recipient of a National Institutes of Health fellowship, the Ruth L. Kirschstein Predoctoral Individual National Research Service Award.
Denise R. Aberle is professor and vice-chair of radiological sciences in the David Geffen School of Medicine and professor of bioengineering in the Samueli School of Engineering. She is board-certified in internal medicine and diagnostic radiology. She was the principal investigator of the American College of Radiology Imaging Network component of the National Lung Screening Trial ACRIN-NLST. Her research centers on lung cancer screening, early diagnosis, and prevention and screening implementation.
William Hsu is associate professor of radiological sciences and bioinformatics at the David Geffen School of Medicine at UCLA and associate professor of bioengineering in the Samueli School of Engineering. His research interests include multimodal data integration, machine learning, and imaging informatics. He directs the Integrated Diagnostics Shared Resource, which collects spatially registered radiology–pathology images along with clinical and molecular data to improve cancer screening and diagnosis.
Disclosures
The authors declare no potential conflicts of interest.
Contributor Information
Nova F. Smedley, Email: novasmedley@ucla.edu.
Denise R. Aberle, Email: DAberle@mednet.ucla.edu.
William Hsu, Email: whsu@mednet.ucla.edu.
Code Availability
Source code for the analysis described in this paper will be made available at https://github.com/novasmedley/deepRadiogenomics.
References
- 1.Segal E., et al. , “Decoding global gene expression programs in liver cancer by noninvasive imaging,” Nat. Biotechnol. 25, 675–680 (2007). 10.1038/nbt1306 [DOI] [PubMed] [Google Scholar]
- 2.Diehn M., et al. , “Identification of noninvasive imaging surrogates for brain tumor gene-expression modules,” Proc. Natl. Acad. Sci. U. S. A. 105, 5213–5218 (2008). 10.1073/pnas.0801279105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zinn P. O., et al. , “Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme,” PLoS One 6, e25451 (2011). 10.1371/journal.pone.0025451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colen R. R., et al. , “Imaging genomic mapping of an invasive MRI phenotype predicts patient outcome and metabolic dysfunction: a TCGA glioma phenotype research group project,” BMC Med. Genom. 7, 30 (2014). 10.1186/1755-8794-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang K., et al. , “Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging,” Clin. Cancer Res. 24, 1073–1081 (2018). 10.1158/1078-0432.CCR-17-2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ha R., et al. , “Predicting breast cancer molecular subtype with MRI dataset utilizing convolutional neural network algorithm,” J. Digital Imaging 32, 276–282 (2019). 10.1007/s10278-019-00179-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai H. X., et al. , “Imaging genomics in cancer research: limitations and promises,” Br. J. Radiol. 89, 20151030 (2016). 10.1259/bjr.20151030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avanzo M., et al. , “Machine and deep learning methods for radiomics,” Med. Phys. 47(5), e185–e202 (2020). 10.1002/mp.13678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korfiatis P., et al. , “Residual deep convolutional neural network predicts MGMT methylation status,” J. Digital Imaging 30, 622–628 (2017). 10.1007/s10278-017-0009-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S., et al. , “A novel radiogenomics framework for genomic and image feature correlation using deep learning,” in IEEE Int. Conf. Bioinf. Biomed., IEEE, pp. 1–8 (2018). 10.1109/BIBM.2018.8621432 [DOI] [Google Scholar]
- 11.Smedley N. F., El-Saden S., Hsu W., “Discovering and interpreting transcriptomic drivers of imaging traits using neural networks,” Bioinformatics 36(11), 3537–3548 (2020). 10.1093/bioinformatics/btaa126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossmann P., et al. , “Defining the biological basis of radiomic phenotypes in lung cancer,” Elife 6, e23421 (2017). 10.7554/eLife.23421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aerts H. J., et al. , “Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach,” Nat. Commun. 5, 4006 (2014). 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velazquez E. R., et al. , “A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists’ delineations and with the surgical specimen,” Radiother. Oncol. 105(2), 167–173 (2012). 10.1016/j.radonc.2012.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeiler M. D., Fergus R., “Visualizing and understanding convolutional networks,” in Eur. Conf. Comput. Vision, Springer, pp. 818–833 (2014). [Google Scholar]
- 16.Liberzon A., et al. , “Molecular signatures database (MSigDB) 3.0,” Bioinformatics 27, 1739–1740 (2011). 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girard L., et al. , “An expression signature as an aid to the histologic classification of non-small cell lung cancer,” Clin. Cancer Res. 22(19), 4880–4889 (2016). 10.1158/1078-0432.CCR-15-2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou J., et al. , “Gene expression-based classification of non-small cell lung carcinomas and survival prediction,” PLoS One 5, e10312 (2010). 10.1371/journal.pone.0010312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberzon A., et al. , “The molecular signatures database hallmark gene set collection,” Cell Syst. 1, 417–425 (2015). 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heavey S., O’Byrne K. J., Gately K., “Strategies for co-targeting the PI3K/AKT/mTOR pathway in NSCLC,” Cancer Treat. Rev. 40, 445–456 (2014). 10.1016/j.ctrv.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 21.Zhang B., “Rho GDP dissociation inhibitors as potential targets for anticancer treatment,” Drug Resist. Updates 9, 134–141 (2006). 10.1016/j.drup.2006.06.001 [DOI] [PubMed] [Google Scholar]
- 22.O’Hayre M., Degese M. S., Gutkind J. S., “Novel insights into G protein and G protein-coupled receptor signaling in cancer,” Curr. Opin. Cell Biol. 27, 126–135 (2014). 10.1016/j.ceb.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto S., et al. , “ALK molecular phenotype in non-small cell lung cancer: CT radiogenomic characterization,” Radiology 272, 568–576 (2014). 10.1148/radiol.14140789 [DOI] [PubMed] [Google Scholar]
- 24.Aerts H. J., et al. , “Defining a radiomic response phenotype: a pilot study using targeted therapy in NSCLC,” Sci. Rep. 6, 33860 (2016). 10.1038/srep33860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rizzo S., et al. , “CT radiogenomic characterization of EGFR, K-RAS, and ALK mutations in non-small cell lung cancer,” Eur. Radiol. 26, 32–42 (2016). 10.1007/s00330-015-3814-0 [DOI] [PubMed] [Google Scholar]
- 26.Gevaert O., et al. , “Predictive radiogenomics modeling of EGFR mutation status in lung cancer,” Sci. Rep. 7, 41674 (2017). 10.1038/srep41674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon H. J., et al. , “Deciphering the tumor microenvironment through radiomics in non-small cell lung cancer: correlation with immune profiles,” PLoS One 15, e0231227 (2020). 10.1371/journal.pone.0231227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu W., et al. , “Exploratory study to identify radiomics classifiers for lung cancer histology,” Front. Oncol. 6, 71 (2016). 10.3389/fonc.2016.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu X., et al. , “Radiomic signature as a diagnostic factor for histologic subtype classification of non-small cell lung cancer,” Eur. Radiol. 28, 2772–2778 (2018). 10.1007/s00330-017-5221-1 [DOI] [PubMed] [Google Scholar]
- 30.Digumarthy S. R., et al. , “Can CT radiomic analysis in NSCLC predict histology and EGFR mutation status?” Medicine (Baltimore) 98, e13963 (2019). 10.1097/MD.0000000000013963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alvarez-Jimenez C., et al. , “Identifying cross-scale associations between radiomic and pathomic signatures of non-small cell lung cancer subtypes: preliminary results,” Cancers 12(12) (2020). 10.3390/cancers12123663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G., et al. , “GOSemSim: an R package for measuring semantic similarity among GO terms and gene products,” Bioinformatics 26, 976–978 (2010). 10.1093/bioinformatics/btq064 [DOI] [PubMed] [Google Scholar]