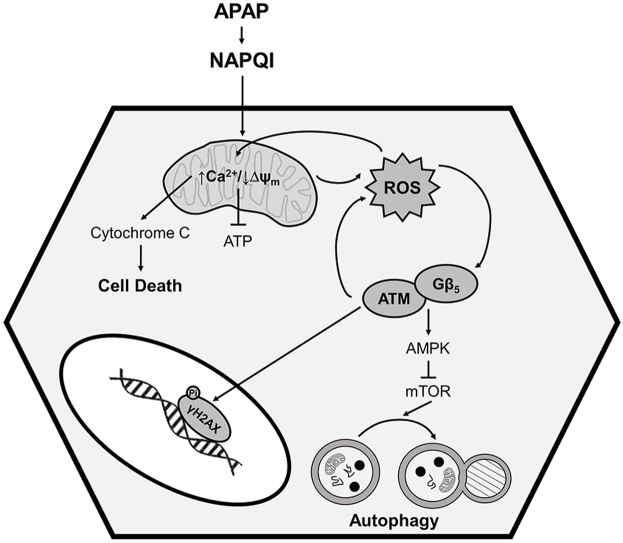
Abstract
Excessive ingestion of the common analgesic acetaminophen (APAP) leads to severe hepatotoxicity. Here we identify G protein β5 (Gβ5), elevated in livers from APAP overdose patients, as a critical regulator of cell death pathways and autophagic signaling in APAP-exposed liver. Liver-specific knockdown of Gβ5 in mice protected the liver from APAP-dependent fibrosis, cell loss, oxidative stress, and inflammation following either acute or chronic APAP administration. Conversely, overexpression of Gβ5 in liver was sufficient to drive hepatocyte dysfunction and loss. In hepatocytes, Gβ5 depletion ameliorated mitochondrial dysfunction, allowed for maintenance of ATP generation and mitigated APAP-induced cell death. Further, Gβ5 knockdown also reversed impacts of APAP on kinase cascades (e.g. ATM/AMPK) signaling to mammalian target of rapamycin (mTOR), a master regulator of autophagy and, as a result, interrupted autophagic flux. Though canonically relegated to nuclear DNA repair pathways, ATM also functions in the cytoplasm to control cell death and autophagy. Indeed, we now show that Gβ5 forms a direct, stable complex with the FAT domain of ATM, important for autophosphorylation-dependent kinase activation. These data provide a viable explanation for these novel, G protein-independent actions of Gβ5 in liver. Thus, Gβ5 sits at a critical nexus in multiple pathological sequelae driving APAP-dependent liver damage.
Keywords: Acetaminophen, Drug-induced liver injury, G protein β5, ATM, Autophagy, Oxidative stress
Graphical abstract
Highlights
-
•
Gβ5 is robustly up-regulated in livers from human patients with a history of chronic APAP use.
-
•
Gβ5is both necessary and sufficient for APAP-dependent oxidative stress and cell loss in liver.
-
•
Gβ5 knockdown allows for maintenance of hepatic function, decreases fibrosis and inflammation, and mitigates lethality following APAP exposure.
-
•
Gβ5 forms a direct complex with ATM to drive ROS generation and trigger cell death.
Abbreviations
- Acetaminophen
APAP
- acute liver failure
ALF
- N-acetyl-p-benzoquinone imine,
NAPQI
- glutathione
GSH
- G protein β5
Gβ5
- ataxia telangiectasia mutated
ATM
- AMP-dependent protein kinase
AMPK
- mammalian target of rapamycin
mTOR
- N-acetyl-l-cysteine
NAC
- Regulator of G protein Signaling
RGS
- GTPase activating protein
GAP
- N-acetyl-p-benzoquinone imine
NAPQI
- reactive oxygen species
ROS
- small hairpin RNA
shRNA
- knockdown
KD
- overexpression
OE
- Terminal deoxynucleotidyl transferase dUTP Nick-End Labelling
TUNEL;
- hematoxylin and eosin
H&E
- alanine aminotransferase
ALT
- aspartate aminotransferase
AST
- total bilirubin
TBIL;
- mitochondrial membrane potential
Δψm
- hydrogen peroxide
H2O2
- adenosine triphosphate
ATP
- FRAP/ATM/TRRAP
FAT
- drug-induced liver injury
DILI
- proliferating cell nuclear antigen
PCNA
- α smooth muscle actin
αSMA
- c-Jun N-terminal kinase
JNK
- transforming growth factor β1
TGF-β1
- non-alcoholic fatty liver disease
NAFLD
- ns
not significant
- mitochondrial permeability transition pore
mPTP
- wild type
WT
- transmission electron microscopy
TEM
1. Introduction
Acetaminophen (acetyl-para-aminophenol, APAP) is an active component of numerous prescription and over-the-counter medications used in the treatment of mild pain and fever. Though generally considered safe and effective, APAP overdose, whether intentional or accidental, is the leading cause of acute liver failure (ALF) in the U.S. and Europe [1]. Limiting APAP dosing to no more than 4000 mg per diem is typically sufficient to prevent severe liver injury. However, factors such as age, genetics, malnutrition, alcohol consumption, and underlying liver disease can modify the maximally tolerated APAP dose [2,3].
Though research on APAP-induced hepatic toxicity has spanned decades, the underlying mechanism(s) remain poorly understood. The vast majority of ingested APAP is glucuronidated or sulfated and promptly excreted. A small percentage, however, is metabolized by hepatic cytochrome P450 enzymes into the highly reactive intermediate N-acetyl-p-benzoquinone imine (NAPQI), which is typically detoxified by glutathione (GSH) conjugation. In the initial stages of APAP liver injury, NAPQI depletes GSH stores and adds sulfhydryl adducts to cellular proteins [4]. The resulting oxidative stress, mitochondrial uncoupling, adenosine triphosphate (ATP) depletion and activation of c-Jun N-terminal (JNK) ultimately leads to hepatocyte necrosis [5,6]. Following acute injury, autophagic removal of damaged proteins and organelles is recruited to maintain cell viability and permit liver regeneration [7]. Indeed, pharmacological induction of autophagy ameliorates APAP-induced hepatotoxicity [7,8]. Currently, GSH replacement with N-acetyl-l-cysteine (NAC) remains the only effective treatment for APAP overdose. However, this therapy is only effective within 8–12 h following APAP ingestion and provides little relief for patients presenting later or for whom high levels of APAP were consumed over an extended period [9]. Thus, investigations into later-stage, downstream pathological mechanisms driving APAP-mediated hepatic damage are essential to development of more efficacious therapies.
The atypical G protein β 5 (Gβ5) forms co-stabilizing complexes with members of the R7 sub-family of regulator of G protein signaling (RGS) proteins (RGS6, RGS7, RGS9, RGS11), which function as GTPase activating proteins (GAPs) forGα subunits [10]. Though best characterized as G protein regulators, Gβ5-R7 complexes also perform G protein-independent functions. Indeed, in heart Gβ5 promotes reactive oxygen species (ROS)-dependent cardiotoxicity following exposure to multiple cancer chemotherapeutics [11]. Expression of Gβ5 is highest in excitable cell types, though a potential liver-intrinsic function for Gβ5 has been proposed due to the observed hepatic hypertrophy and altered lipid deposition observed in Gβ5+/− and Gβ5−/− mice [12]. Similarly, a role for Gβ5-interacting protein RGS6 has been demonstrated in alcoholic hepatosteatosis [13].
Here, we identify hepatic Gβ5 as a critical driver of APAP-induced hepatotoxicity. We detected robust Gβ5 protein upregulation in murine tissue and cells, human hepatocytes and liver tissue samples from patients exposed to toxic levels of APAP. Knockdown of Gβ5 impacted several key cellular processes driving APAP-dependent liver damage including mitochondrial function, oxidative stress, autophagy and cell death. Thus, Gβ5 emerges as a viable druggable target in APAP-mediated liver injury.
2. Materials & methods
2.1. Antibodies and reagents
Tables with full details regarding antibodies (Table S2), reagents (Table S1), assay kits (Table S3), and cell lines (Table S4) can be found in the Supplementary Tables 1–4. This includes catalog information for kits used for detection of alanine aminotransferase (ALT), aspartate aminotransferase (AST), triglycerides, tissue collagen, hydroxyproline, calcium (Ca2+), albumin, mitochondrial isolation, mitochondrial and total ATP, transforming growth factor β1 (TGF-β1), and cell death (cytoplasmic histone-associated DNA fragments). Samples were collected and analyses performed according to the manufacturer's instructions. Also available are catalog information and dilutions for all antibodies used in immunohistochemistry and western blotting.
2.2. Animals
Mouse experiments were performed at the Geethanjali College of Pharmacy, Hyderabad, India (Ref No: 1648/PO/Re/S/12/CPCSEA/50) in associtaion with the Department of Pharmaceutical Sciences, Babasaheb Bhimrao Ambedkar University, Lucknow. Animals were procured from Biological E. Limited (Registration #38/99/CPCSEA) and were handled following the Guide for the Care and Use of Laboratory Animals. Male Swiss albino mice (22–30 g, 8–10 weeks of age) were maintained on a balanced laboratory diet as per NIN, Hyderabad, India and given tap water ad libitum. Animal housing facilities were kept at 20 ± 2 °C, 65–70% humidity, and day/night cycle (12 h/12 h). Animals were group housed 2–5 mice/cage.
2.3. Gβ5 gene silencing via small hairpin RNA (shRNA) delivery in vivo
shRNA against Gβ5was purchased from Santacruz Biotechnology (Paso Robles, CA, USA). A scrambled shRNA served as the control. A commercially available transfection reagent (In vivofectamine 3.0, Thermo Fisher Scientific, Waltham, MA, USA) was used to deliver the shRNA via intravenous (tail vein) injection. Briefly, 7–8 μg of selected shRNA was diluted in 60 μL of 5% glucose and mixed with the transfection reagent. The mixture was incubated for 15 min at room temperature to allow the complexes to form. After optimizing a published protocol [14], mice received 3 injections with a 3-day interval between injections. The in vivo delivery efficiency of shRNA was assessed by Western blot. Gβ5 shRNA1 had the highest Gβ5 silencing efficiency and was selected for further experimentation. Following shRNA administration, body weight (1X/week) and food intake (1–2X/week) were monitored. After administration of Gβ5 shRNA, body weight was slightly reduced, but food intake was not significantly changed compared with that of the scramble shRNA injection group (data not shown). The drop in body weight was modest when compared to changes observed in Gβ5 knockout mice [12].
2.4. APAP treatment regimens
Two APAP treatment regimens were employed to generate data in this study. First, we utilized a range of toxic APAP doses (200–400 mg/kg, i.p.) to assess the impact of Gβ5 knockdown on acute APAP-induced liver injury [15]. A dose of 350 mg/kg, i.p resulted in an optimal time window (96 h) for endpoint monitoring and intervention and was selected for further experiments (Fig. S1). 10 Control and 10 Gβ5 KD mice divided into two independent cohorts were given a single APAP bolus (350 mg/kg, i.p.) 10 days following administration of shRNA. Unless otherwise noted, animals were sacrificed 48 h after dosing and samples isolated for downstream histological and biochemical analyses. In a separate cohort, mice (n = 10/group) were administered vehicle (control) or APAP (350 mg/kg, i.p.) with or without concurrent NAC treatment (100 mg/kg, i.p.) at 1 or 6 h post-APAP administration. These time points were chosen as they sit before (1 h) and after (6 h) a known cut off time for NAC efficacy in APAP-induced toxicity [16]. Animals were sacrificed after 24 h for sample collection. Finally, control and Gβ5 KD (n = 10/group) mice were given a single dose of APAP (350 mg/kg, i.p.) with simultaneous leupeptin (Leu; 40 mg/kg, i.p.), Torin1 (Tor; 2 mg/kg, i.p.), or vehicle administration. Samples were collected after 6 h for endpoint analyses [7]. A second treatment regimen utilized subtoxic APAP doses comparable to a typical therapeutic schedule [17]. APAP (2, 4 or 6 mg/kg, i.p., biweekly) was administered to control (n = 20) or Gβ5 KD (n = 20) mice divided into two independent cohorts over a period of 12 weeks. Samples were taken at 6 weeks or 12 weeks post-initiation of drug treatment where indicated. Unless otherwise indicated, data presented were derived from animals given a dose of 4 mg/kg, i.p., biweekly for 6 weeks. Animals were sacrificed via cervical dislocation; blood was collected; and tissues dissected and subdivided for histological and biochemical analyses.
2.5. Histology & immunohistochemistry
Formalin-fixed, paraffin-embedded mouse and human liver tissue sections were stained with Hematoxylin and Eosin (H&E) to determine macroscopic changes in tissue architecture. The lysochromediazo dye, oil red o (Sigma, St. Louis, MO, USA), was used for staining of neutral triglycerides and lipids in liver tissue sections. The Masson trichrome staining kit (Sigma) and Sirius red stain were used to detect collagen deposition indicative of liver fibrosis according to the manufacturers’ protocol. Detection of cytotoxicity in liver tissue was achieved using a Terminal deoxynucleotidyltransferasedUTP Nick-End Labelling (TUNEL) kit from Biovision (Milpitas, CA, USA). Immunohistochemical staining of both mouse and human liver tissue sections was performed as per a standard protocol. Briefly, sections were dewaxed in xylene (2 times, 15 min each), treated with graded series of alcohol solutions for 10 min, immersed in 3% hydrogen peroxide in methanol to block endogenous peroxide activity and washed with distilled water (2 times, 5 min each). For antigen retrieval, slides were dipped in citrate buffer for 15 min at 100 °C. Then they were washed with 1X PBS buffer for 5 min and blocking was done with 5% BSA in PBS for 1 h at room temperature. Next, slides were incubated overnight at 4 °C with primary antibodies in 3% BSA in PBS (1:500) within a moist chamber. Following washing (3 × 10 min) in PBS at room temperature, sections were incubated for 1 h at room temperature with peroxidase-conjugated secondary antibodies (1:1000). A positive reaction was detected by exposure to stable diaminobenzidene (Abcam, Cambridge, UK) for 3 min. The sections were counterstained in Harris hematoxylin, treated with graded series of alcohol for 10 min each grade (30%, 50%, 70%, 90% and 100%) and dipped in xylene overnight. Slides were then mounted with DPX (Sigma) and observed under the microscope. For proliferating cell nuclear antigen (PCNA), F4/80 and TUNEL staining, 7–10 sections were stained from each animal with 5 pictures randomly selected from each slide, scored for the positive stained (brown color) nuclei and averaged. For Gβ5 staining in mouse and human liver sections, the histoscore was determined from the average stain intensity using Image J (NIH) for 7–10 slides/animal, 5 images per slide.
2.6. Isolation and culture of murine hepatocytes
Primary adult hepatocytes were isolated from 2-month-old mice according to a standard collagenase perfusion protocol. The liver was perfused first with EGTA solution (5.4 mM KCl, 0.44 mM KH2PO4, 140 mM NaCl, 0.34 mM Na2HPO4, 0.5 mM EGTA, and 25 mM Tricine, pH 7.2) and then with DMEM (Gibco) containing 0.075% type I collagenase (Sigma), followed by an additional digestion step (0.009% collagenase at 37 °C with agitation for 15 min) and centrifugation as described previously [7,18]. The isolated hepatocytes were then cultured in hepato-ZYME-SFM media (Thermo Fisher Scientific) on collagen-coated plates and maintained at 37 °C in a humidified cell culture incubator (5% CO2). Cells were not disturbed for at least 16–24 h before drug treatment. Post isolation, cells were treated with APAP (5 mM) in the presence or absence of NAC (5 mM), RU360 (50 μM), cyclosporine A (0.2 μM), ATMi KU-55933 (5 μM) or GW78838 (5 μM) where indicated. Cells were transduced with lentiviral vectors encoding Gβ5 shRNA or control shRNA (Santa Cruz Biotech) according to the manufacturer's instructions.
2.7. Isolation and culture of primary human hepatocytes
Human livers were removed at autopsy from deceased individuals within 1–4 h after cessation of cardiac function. Samples were acquired from individuals with no reported medical complications (hepatic or cardiac pathology, diabetes, or kidney disorders). Medical history was collected from the relatives of the deceased through a questionnaire. Relatives were briefed about the goal and design of the study and written consent was obtained. The condition of organs was further confirmed by studying gross architectural changes by H&E during autopsy. Primary human hepatocytes were isolated from human liver tissue essentially as previously described [19,20]. Briefly, liver tissue samples were collected on ice in MEME solution with 0.5% fatty acid free BSA by the clinician of the Forensic Medicine Department, Sagore Dutta Medical College & Hospital, Kolkata, West Bengal. Immediately, under aseptic conditions, tissues were first diced, washed in cold HBSS and then minced thoroughly in MEME. EGTA (0.5 mM) was added in the cell slurry and it was placed in a shaking water bath for 10–15 min at 37 °C. After centrifugation, the cell slurry was washed twice in MEME. Pre-warmed digestion buffer (HBSS, 0.05% collagenase IV, 0.5% fatty acid free BSA, 10 mM CaCl2) was added and the slurry placed in a shaking water bath again for 30 min at 37 °C. BSA was included in the digestion process to minimize cell damage & prevent hemolysis of red blood cells. The solution was gently vortexed by repeated pipetting and passed through a metal strainer to remove lumps. The resultant supernatant was filtered again through a 100 μm cell strainer and then placed on ice. Then cell suspensions were centrifuged (3000 rpm for 5 min, 4 °C) and the supernatant was discarded. The hepatocyte pellet was gently re-suspended in minimal amount of MEME and RBC lysis buffer added to completely remove the RBC from the cell suspension. After 3 min, cells were centrifuged again (3000 rpm for 5 min, 4 °C), washed with MEME twice and finally resuspended in William's E medium. Cells were counted for viability and diluted to 1 million cells per ml in medium containing supplements (1% non-essential amino acids, 1% GlutaMAX™, 2% human serum, 100 nM dexamethasone, 100 nM insulin and 0.375% fatty acid free BSA). Isolated hepatocytes were plated on type 1 collagen coated plates, at a density of 250,000/cm2. After adherence (overnight undisturbed), cells were transfected with Gβ5 or scrambled shRNA using a Neon electroporator. Cells were then treated with drugs (as for mouse hepatocytes) or exposed to media containing 10% serum collected from patients with reported APAP liver toxicity (or control serum).
2.8. Gβ5 cloning and construct generation
The full-length GNB5 coding sequence (isoform A) was amplified by PCR from human blood cDNA using Phusion Hot Start II High-Fidelity PCR Master Mix (Thermo Fisher) with compatible cloning sites (XhoI/HindIII). RNA was isolated from human blood using Trizol (Invitrogen, Carlsbad, CA, USA) and cDNA was prepared by reverse transcription of RNA using a cDNA synthesis kit (Thermo Fisher) following the manufacturers’ protocol. The resultant PCR product was loaded onto a 1% agarose gel, the gel was visualized under UV light gel doc (UVP chemStudio, Analytik Jena, Jena, Germany), and a band was observed, subsequently cut, and eluted using a gel extraction kit (Qiagen, Hilden, Germany). Following amplification, a 1059bp band was resolved using agarose gel electrophoresis, extracted (Qiagen Gel Extraction kit), and a second PCR performed (Platinum Taq High Fidelity) to generate overhangs suitable for ligation into the pMD20-T vector (Takara Bio, Kyoto, Japan). Subcloning into the pEGFP-N1 vector was performed by double digestion of the vector and plasmid (pMD20T + Insert) with XhoI/HindIII (New England Biolabs, NEB, Ipswich, MA, USA). The vector and insert were ligated using T4 DNA ligase (NEB) in an overnight reaction at 4 °C temperature. The ligation product was then transformed again into DH5α cells, plated on LB agar contained (50 mg/ml Kanamycin), and incubated at 37 °C overnight. Clones were picked and grown in LB medium with antibiotic (kanamycin. Plasmids were isolated and restriction digestion was performed for identification of positive clones. Primers used to generate full length and Gβ5 deletion constructs are listed in Supplemental Table 5.
The full-length mouse Gβ5 sequence was isolated from mouse brain and cloned into the PMD20 vector as above. The lentiviral vector for mGβ5 was generated via subcloning into the pLenti CMV Puro DEST cloning vector (Addgene, Watertown, MA, USA) and packaged using the pMD2.G VSV-G envelope expressing plasmid (Addgene) and psPAX2 (Addgene). Lentiviral particles were generated in HEK293 cells as per a standard protocol. 70 μL of lentivirus containing 2 × 108 particles of either mGβ5-Lenti or a control empty vector virus with the addition of invivofectamine was injected into the tail vein of mice. Two weeks after lentiviral injection, the mice were subjected to APAP treatment (4 mg/kg, i.p. biweekly). After 4 weeks of treatment mice, were euthanized by cervical dislocation and blood/multiple tissues were collected for downstream analysis.
2.9. Cell transfection
Prior to transfection, cells were plated at low density (approximately 1 × 105 cells/60 mm dish) and allowed to grow to 60%–70% confluence (24–36 h after seeding). Cells were transfected using lipofectamine 3000 (Thermo Fisher Scientific) or via electroporation (Neon Electroporator, Thermo Fisher Scientific) following the manufacturer's protocols.
2.10. Generation of Gβ5 KO HepaRG cells using CRISPR/Cas9
Guide RNA (gRNA) targeting human GNB5 gene exon2 were designed using tools available from Integrated DNA technologies (IDT, Newark, NJ, USA). High on target and low off target gRNAs were chosen without a PAM sequence, cloned into the PX459 CRISPR system plasmid (Addgene) using standard methods [21] and confirmed via sequencing. Briefly, 10 μg of Px459 plasmid was digested with 5 μl (10 units) of BbsI restriction enzyme (NEB) for 5 h at 37 °C, run on 1% agarose gel and the digested product was then eluted with a gel extraction kit (Qiagen). The product was then dissolved in 30 μl of nuclease free water. Oligos for this experiment were resuspended in nuclease free water to a final concentration of 100 μM. The reaction mixture was 5 μl of each oligo, 5 μl 10X T4 DNA ligation buffer, 2.5 μl T4 PNK and nuclease free water to make the volume up to 50 μl. Annealing of oligos was performed at 37 °C for 30 min to add the 5'phosphate and reactions then incubated at 95 °C for 5min followed by a ramp down to 25 °C at 5 °C per minute. Annealed oligos were diluted at 1:50 in nuclease free water and ligated with PX459 crispr plasmid by taking 100 μg of digested plasmid, 2 μl of annealed oligos, 1 μl 10X T4 ligation buffer and 1 μl of T4 DNA ligase enzyme (NEB) in total volume of 10 μl and incubated in 16 °C in a master cycler and then overnight at 4 °C. 11 μl of the ligation mixture was transformed into 90 μl of DH5α competent cells and plated in ampicillin containing LB agar plates. Positive clones were selected and validated by PCR with U6 forward primer and each oligos antisense strand. The resulting construct was transfected into HepaRG cells using lipofectamine 3000 (ThermoFisher). Cells were re-plated 48 h post-transfection and subjected to puromycin selection. After 14 days puromycin selected colonies were plated at 1 cell/well. Nine colonies were picked and each colony was pelleted down separately for subsequent genomic DNA isolation by phenol/chloroform/isoamyl alcohol extraction for sequencing and protein detection by western blotting. We successfully knocked out Gβ5 in two colonies (colony 8 & 9) and used colony 8 for the subsequent experiments. The T7 endonuclease 1 (T7E1) mismatch detection assay was used for validation.
2.11. Cellular fractionation
Subcellular fractionation from mouse liver tissue was performed following published protocols [7,22] with slight modifications. Control and Gβ5 liver KD male mice were given APAP (350 mg/kg, i.p.) for 6 h. Mice were sacrificed, livers were minced in 0.25 M sucrose and homogenized. The homogenate was then centrifuged at 2000 g for 5 min. Both the supernatant and pellet were kept for further processing. The pooled supernatants of 3 mice were then centrifuged at 17,000 g for 15 min. The resulting pellet was re-suspended in 1.9 ml 0.25 M sucrose with 2.8 ml of 85% Nycodenz. The layers from bottom to top were as follows: mitochondrial-lysosomal suspension and differential concentrations of Nycodenz. After centrifugation at 27,000 g for 3 h, the interphase fractions of autophagosome and lysosomes were collected by a syringe. Each fraction was diluted again in 0.25 M sucrose, centrifuged at 24,000×g for 10 min and final pellets were collected and lysed with RIPA buffer for Western blot analysis.
2.12. HepG2/HUVEC Co-culture experiment
To establish conditions for phenotypically stable co-cultures [23], HUVECs and HepG2 were first cultured separately in HiEndoXLTM Endothelial Cell Expansion Medium (Himedia, Mumbai, India) and MEME (Sigma), respectively, for 3 days. Subsequently, HUVECs and HepG2 were seeded in MEME in 6-well and 12-well plates at an initial density of 21,000 cells/cm2 for mono-cultures and 21,000 cells/cm2 at a ratio of 1:1 in co-cultures. For co-cultures, HUVECs were seeded 4–5 h before the addition of HepG2 to allow them to adhere to tissue culture plates. Cells were then cultured for 1–2 days prior to drug treatment. Mono- and co-cultures were incubated for 24 h in culture medium containing 5 mM APAP with or without GW788388 pre-treatment (1 h) where indicated.
2.13. Immunoprecipitation
HepaRG cells (3 × 106) were treated with APAP (5 mM) for 24 h, lysed and protein concentration measured using standard protocol. 600 μg of protein was equilibrated with IP lysis buffer (50 mM Tris, 5 mM EDTA, 250 mM NaCl and 0.1% Triton X-100) and bait antibodies (ATM or control mouse IgG) for 12 h on a rotor at 4 °C. 30 μl of Protein G sepharose beads (Abcam) were pre-cleared, equilibrated and then added to lysate after 12 h. After a 2-h incubation, protein extracts were immunoprecipitated with respective antibodies followed by washing 3 X with IP buffer. Eluted immuno-complexes were subjected to immunoblotting with prey antibody (Gβ5, Millipore). Sample preparation for immunoblotting was done under non-reducing conditions.
2.14. MitoSox staining
Post APAP treatment, mouse primary hepatocytes were washed thoroughly, loaded with 5 μM of MitoSox solution (1–2 ml to cover the whole dish), incubated for 15 min at 37 °C, washed 3X with PBS in the dark, mounted using vectashield with DAPI and visualized by fluorescence microscopy. The number of MitoSox + cells (red stained) were counted on each coverslip.
2.15. Measurement of ROS generation
ROS generation was estimated in tissues and primary cells using the cell-permeable oxidation-sensitive probe, CM-H2DCFDA (DCFDA, Sigma). Cells were harvested by centrifugation, washed three times with ice-cold PBS, re-suspended in PBS and incubated with 5 μM CM-H2DCFDA for 20 min at 37 °C. After incubation cells were again washed and lysed in PBS with 1% Tween 20. Tissue samples were homogenized in ice-cold PBS followed by the addition of 5 μM CM-H2DCFDA dissolved in PBS and for 20 min at 37 °C.The ROS level in cell lysates or tissue homogenates was determined at the ratio of dichlorofluorescein excitation at 480 nm to emission at 530 nm. We should note here that the CM-H2DCFDA assay is utilized here as a general oxidative stress indicator and not as a detector of a specific oxidant due to known limitations of the probe [24].
2.16. Acidic vesicle detection
Using acridine orange we detected & quantified the formation of acidic vesicles during autophagy through spectrofluorometry. HepaRG cells were transfected with Gβ5 or scrambled shRNA and then treated with APAP (5 mM) for 24 h. Post-trypsinization, cells were washed twice with PBS and then stained with acridine orange (1 μg/μl) for 30 min in the dark. Cells were washed again with PBS twice and lysed. The resultant supernatant was processed through the spectrofluorometer with excitation at 460 nm and emission at 530 nm.
2.17. Immunoblotting
Tissues were promptly dissected and flash frozen using liquid nitrogen. Tissue homogenates and cell pellets were prepared in 1X RIPA buffer containing protease (p8340) and phosphatase (#3) inhibitor cocktails (Sigma) and protein content quantified by BCA assay. 20 μg of protein per sample was subjected to SDS-PAGE and transferonto nitrocellulose membranes. Membranes were washed 1X in TBST and blocked for 1 h with 5% BSA in TBST. After one wash with 1X TBST for 5 min, the membranes were incubated overnight in primary antibodies dissolved in 3% BSA in TBST at 4 °C. The next day, membranes were washed 3Xin TBST at room temperature and probed with respective horseradish peroxidase-labelled secondary antibodies dissolved in 3% BSA in TBST (1:5000). Membranes were washed three times using 1X TBST for 5 min duration at room temperature. Immunoblots were developed (UVP chemStudioAnalytik Jena) using the chemiluminescence method and densitometric quantification of immunoblot bands was performed using Image J software (U.S. NIH). For quantification of immunoblots, protein levels were first normalized to β-Actin loading controls and then expressed relative to control samples. More specifically, an average of scores for controls samples was generated and used as a common dividing factor to determine relative levels of proteins of interest.
2.18. Electron microscopy
Murine tissues were fixed using 2% glutaraldehyde at room temperature and processed for the transmission electron microscopy (TEM) study as per a standard method. Images were obtained at the High-Resolution Electron Microscopy Facility (JEOL, Peabody, MA) at SGPGI, Lucknow, India.
2.19. Immunostaining and fluorescence microscopy
HepaRG were seeded on a glass coverslip in a 6 well-plate. Using the Neon electroporator, cells were co-transfected with the LC3-GFP plasmid together with plasmids encoding scramble or Gβ5 shRNA. 16 h after transfection, cells were challenged with APAP (5 mM) for 6 h. Post treatment, cells were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 2 h at room temperature, mounted with Vectashield and DAPI (Invitrogen) and kept at −20 °C for microscopy. Pictures were taken using a fluorescence microscope (Optika, Italy) with a 40× objective. >15 cells/coverslip were counted randomly to quantify the autophagic puncta per cell. The LC3-GFP plasmid was the kind gift from Dr. Santosh Chauhan, Institute of Life Science, Bhubaneswar, India.
2.20. Collagen formation assay
The collagen specific dye Sirius red was utilized to quantitate collagen from lysates of cells treated with vehicle or APAP (5 mM, 24 h) essentially as previously described [25]. Briefly, a solution of 5 μg/ml Sirius red was prepared by dissolving Sirius red in saturated picric acid. After 1 h, cells were washed with PBS twice and lysed in 0.1 M sodium hydroxide at room temperature. The supernatant was collected and colorimetric measurement of done at 530 nm.
2.21. Bioinformatics study
A bioinformatics approach was used to understand the interaction between protein kinase ataxia-telangiectasia mutated (ATM) of Human (Homo sapiens) and Guanine nucleotide-binding protein subunit beta-5 (GNB5) of house mouse (Mus musculus). For the bioinformatic analysis we used the human GNB5 isoform-1 protein sequence from UniProt database [26] to do a NCBI protein blast [27] search in the RCSB protein structure database [28]. We found 99.43% identity and 89% query coverage (from amino acid 43 to 395) with 2PBI_B protein of Mus musculus. Protein structures were downloaded from protein data bank https://www.rcsb.org/with PDB ID: 5 NP1, structural resolution of 5.7 Å for ATM and PDB ID: 2PBI, resolution 1.95 Å for GNB5. Human ATM shows two states as closed and open dimers in a dynamic equilibrium [29]. Because the open dimer lacks the intermolecular interactions that block the peptide-binding site in the closed dimer it is considered the more active conformation and was selected for this study [29]. GNB5 protein with PDB ID: 2PBI has 4 chains: A, B, C and D. A is identical to C and B is identical to D. The D chain was selected for the interaction study. The energy minimization was done for the ATM and GNB5 D chain to remove structural constraints with GROMACS version 2019.3 under periodic boundary conditions in a 2.0-nm cubic box. In the first step, genion command was used to neutralize the charged proteins. Then energy minimization was performed using GROMOS96 43a1 force field with the steepest descent algorithm at 50,000 steps. The ‘editconf’ command was used to generate pdb file from gro file post energy minimization. The protein files were further modified by pymol 2.2.0 to remove SOL from energy minimized structures. The top 10 interaction models were considered for further Molecular Dynamics (MD) Simulations. Based on initial biochemical screening, the interaction of ATM with the WD40 domain of GNB5 was selected for further processing. We employed the GROMACS software to illustrate the setup, energy minimization, conductance, and analysis of protein intermolecular interactions and step-by-step MD simulations between these proteins. Constant temperature MD simulation was performed to study the thermodynamics and structure dynamics characteristics of ATM-GNB5 complex. Other software packages were also used for the study: 1) protein visualization programs, i.e., PyMOL or VMD, 2) Python for general data analysis, 3) Python libraries specifically designed to analyze MD trajectories, i.e., MDAnalysis and MDTraj, and 4) a molecule packing optimization software, PACKMOL. The protein topology was first generated using pdb2gmx tool at standard pH 7.0 amino acid protonation state and CHARMM27 all-atom force field was used for the simulation. The input structure was solvated with the extended single - point charge (SPC) water model in a cubic box with 2.0 nm space around the solute. The net charge of the system was neutralized by genion. MD simulations for the complex of ATM and GNB5 were achieved by using GROMACS 2019.3. The protein topology was generated by using pdb2gmx tool at standard pH 7.0 amino acid protonation state. CHARMM27 all-atom force field was used for the simulation. The input structure was solvated with the extended single - point charge (SPC) water model and the net charge of the system was neutralized by genion. The MD simulation was done for 10 ns to identify the stable interaction between the ATM and GNB5. The protein files were further modified by pymol 2.4 and Discovery studio visualizer to predict the interacting amino acids.
2.22. Human samples
Post-mortem human tissue samples (control and liver injury; tissue and serum) were acquired after obtaining the ethical clearance from the Centre of Biomedical Research Ethics Committee (Ref: IEC/CBMR/Corr/2018/14/3). All the experiments have been performed in collaboration with Department of Surgery and Department of Forensic Medicine, Sagore Dutta Medical College & Hospital, Kolkata, West Bengal. Control samples were approximate age matched and confirmed free of liver pathology. Summarized and individual demographic, health history and liver function test data for patients can be found in Supplementary Tables 6 and 7, respectively. Tissue samples were categorized as “APAP-associated Injury” for individuals with a history of chronic APAP use.
2.23. Data acquisition and statistical analyses
Murine physiology experimental data was generated from two independent animal cohorts. Cell culture experiments were performed with a minimal experimental N of 3. Data were analyzed by student's t-test, one-, or two-way ANOVA with the post hoc adjustments as appropriate. Dunnett's and Sidak's corrections for multiple comparisons were used for one- and two-way ANOVA, respectively. Statistical analyses were performed using GraphPad Prism Software (La Jolla, CA, USA). For Kaplan–Meier plots of mouse survival, statistical significance was analyzed by the log-rank (Mantel–Cox) test. Results were considered significantly different at P < 0.05. Values are expressed as means ± S.E.M.
3. Results
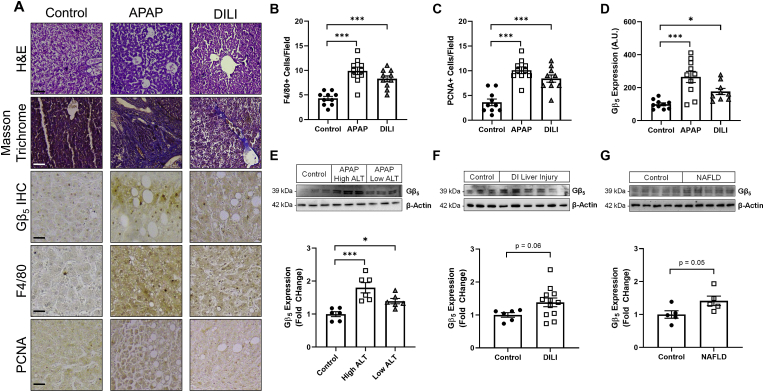
Gβ5 is up-regulated in human APAP-induced liver injury – We collected liver tissue and serum samples from human subjects with a history of drug-induced liver injury (DILI) and/or APAP-induced liver injury (Supplementary Table S7). All patients exhibit elevated ALT, AST, and total bilirubin (TBIL) (Supplementary Table S6). Histological analysis revealed detectable liver fibrosis and inflammation (Fig. 1A and B) as well as ongoing regeneration (Fig. 1A and C) in APAP and DILI samples. We noted robust Gβ5 up-regulation in APAP-induced liver injury samples via both immunohistochemistry (Fig. 1D) and Western blot (Fig. 1E) particularly in following severe damage (high ALT). A trend for increased Gβ5 protein was also found in DILI (Fig. 1F) and non-alcoholic fatty liver disease (NAFLD) (Fig. 1G). Notably, a doublet of Gβ5 immunoreactive bands (~39 kDa and ~44 kDa) was detectable in liver indicating the potential existence of multiple splice forms as occurs in the vertebrate retina [30].
Fig. 1.
Gβ5 is up-regulated in human patients with a history of APAP-induced liver injury. (A) Human liver autopsy samples were subjected to histological analysis of gross architecture (H&E), fibrosis (Masson Trichrome), inflammation (F4/80), proliferation (PCNA) and Gβ5 expression [scale bar = 100 μm]. Quantification (n = 10) of (B) F4/80+ cells, (C) PCNA + cells and (D) Gβ5 expression from histological analyses. (E) Liver tissue samples from APAP-induced liver injury patients were stratified based on injury severity and probed for expression of Gβ5 (n = 6). Gβ5 protein expression in (F) DILI (n = 6–10) and (G) NAFLD (n = 5). β-Actin serves as a loading control for all immunoblots. ns = not significant. *P < 0.05, ***P < 0.001 via one-way ANOVA with Dunnett's post-hoc test. Data are presented as mean ± SEM.
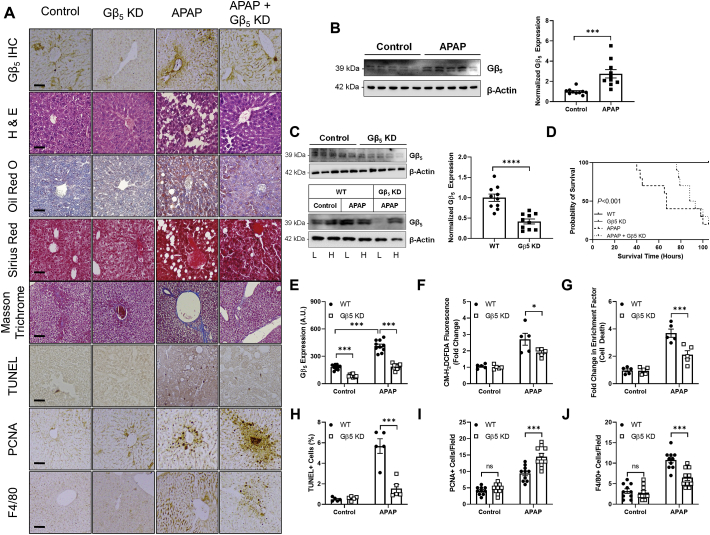
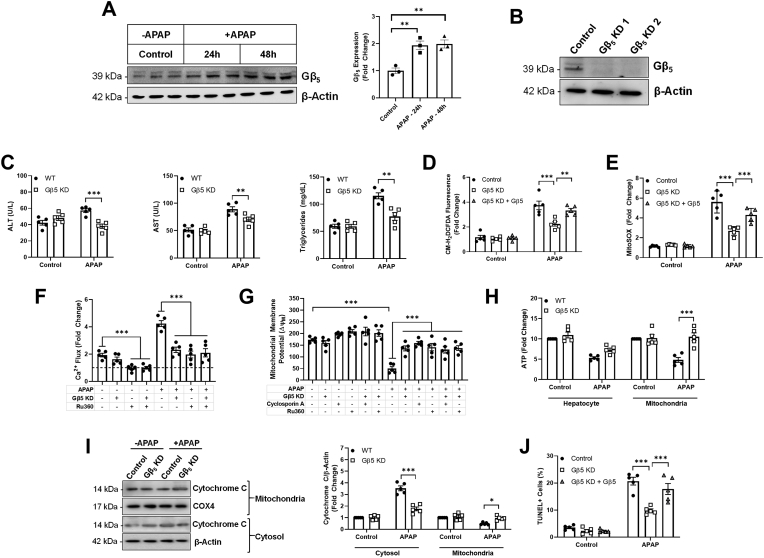
Gβ5 is up-regulated following acute APAP exposure and contributes to APAP-dependent pathological sequalae in liver – In order to demonstrate a functional role for Gβ5 in APAP-induced liver damage in vivo, we next moved to the murine model system exploiting the conservation of liver microarchitecture across mammalian species. To identify a lethal APAP dose with an optimized intervention window, animals were given a single APAP bolus (200–400 mg/kg) and survival monitored over time (Fig. S1). A 350 mg/kg dose resulted in near complete lethality within 96 h, and this dose was utilized for acute APAP toxicity experiments. Induction of Gβ5 protein was detectable within 48 h following APAP administration (Fig. 2B). Because global Gβ5 knockout results in neurodevelopmental abnormalities [31], we utilized a shRNA-based strategy to knockdown of Gβ5 expression specifically in liver (Gβ5 KD). We could reproducibly achieve >50% knockdown selectively in liver (Fig. 2C). Notably, APAP-dependent Gβ5 up-regulation was confined to liver and not observed in cardiac tissue (Fig. 2C). In APAP-exposed liver, our shRNA prevented Gβ5 up-regulation (Fig. 2E). Gβ5 knockdown (Gβ5 KD) was sufficient to prolong animal survival (Fig. 2D) and ameliorate hepatic lipid accumulation and fibrosis (Fig. 2A), oxidative stress (Fig. 2F), cell death (Fig. 2A, G, 2H) and F4/80+ Kupffer cell recruitment (Fig. 2A and J). Loss of Gβ5 also drove enhanced liver regeneration in APAP-exposed mice as indicated by dramatic increases in levels of PCNA (Fig. 2I).
Fig. 2.
Gβ5 knockdown protects against acute APAP hepatotoxicity (350 mg/kg, i.p., 48 h).(A) Histological characterization of Gβ5 expression, gross liver architecture (H&E), hepatic lipid deposition (Oil Red O), hepatic fibrosis (Sirius Red & Masson Trichrome), cell death (TUNEL), cellular proliferation (PCNA) and inflammation (F4/80)[scale bar = 100 μm]. (B) Immunoblotting for Gβ5 (n = 10). (C) Immunoblotting for Gβ5 was performed to determine the efficacy (n = 10) and specificity of Gβ5 knockdown in liver (L = liver; H = heart). WT (scramble shRNA) and Gβ5 KD (Gβ5 shRNA) mice were given APAP (350 mg/kg, i.p.) for 48 h.(D) Kaplan-Meier survival curve (n = 10). (E) Quantification of Gβ5immunohistochemistry (n = 10). (F) CM-H2DCFDA fluorescence (total ROS; n = 5). (G) Cell death (fold increase in histone-associated DNA fragments; n = 5). Quantification (n = 10) of (H) TUNEL + cells, (I) PCNA+ (proliferating) cells, and (J) F4/80+ (inflammatory) cells. β-Actin serves as a loading control for all immunoblots. *P < 0.05, ***P < 0.001, ****P < 0.0001 via student's t-test or two-way ANOVA with Sidak's post-hoc test. Data are presented as mean ± SEM.
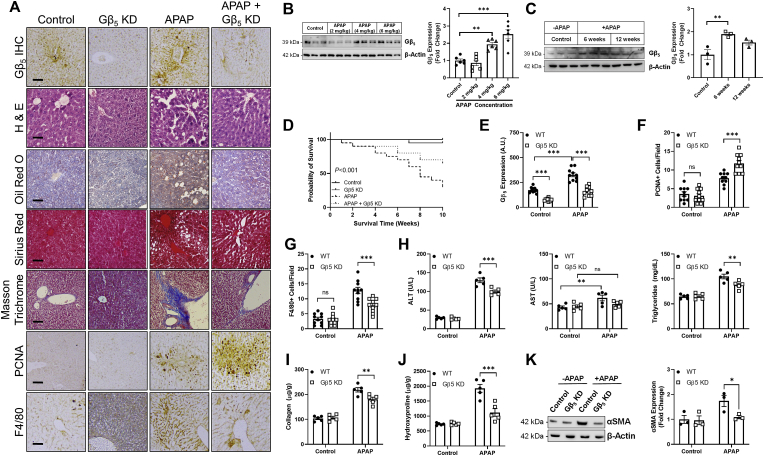
Gβ5 KD improves pathological endpoints following chronic APAP treatment – Chronic APAP treatment (2X per week) at lower doses (2–6 mg/kg) also increased Gβ5 protein in liver (Fig. 3B). Peak Gβ5 expression was observed 6 weeks following APAP exposure (4 mg/kg) (Fig. 3C). At this dose and time point, Gβ5 KD in liver (Fig. 3E) improved mouse survival (Fig. 3D) and partially reversed the pathological impact of APAP on liver function tests (Fig. 3H). Gβ5 KD mitigated APAP-dependent inflammation (Fig. 3A and G) and enhanced liver regeneration (Fig. 3A and F). Fibrotic remodeling (Fig. 3A) including increases in α smooth muscle actin (αSMA) (Fig. 3K), total collagen deposition (Fig. 3I) and accumulation of hydroxyproline (Fig. 3J) were also partially reversed in livers lacking APAP-driven Gβ5 up-regulation.
Fig. 3.
Gβ5 depletion ameliorates APAP-induced liver damage following chronic exposure. APAP (2, 4 or 6 mg/kg, i.p., biweekly) was administered to WT (scramble shRNA) or Gβ5 KD (Gβ5shRNA) mice over a period of 12 weeks. (A) Histological characterization of Gβ5 expression, gross liver architecture (H&E), hepatic lipid deposition (Oil Red O), hepatic fibrosis (Sirius Red & Masson Trichrome), cellular proliferation (PCNA) and inflammation (F4/80) [scale bar = 100 μm] 6 weeks post-initiation of APAP dosing (4 mg/kg). (B) Dose response relationship for APAP-dependent hepatic Gβ5up-regulation (6 weeks, n = 6). (C) Time course of Gβ5 up-regulation in liver (4 mg/kg, n = 3).(D) Kaplan-Meier survival curve (4 mg/kg, n = 20). For the remaining experiments, a dose of 4 mg/kg, i.p. was utilized and analyses performed at 6 weeks. (E) Quantification of Gβ5 immunohistochemistry (n = 10). Quantification of (F) PCNA and (G) F4/80 positive cells (n = 10). (H) Serum ALT, AST and triglycerides (n = 5). Hepatic (I) collagen (n = 10), (J) hydroxyproline (n = 5), and (K) αSMA expression (n = 3). β-Actin serves as a loading control for all immunoblots. ns = not significant. *P < 0.05, **P < 0.01, ***P < 0.001 via one-way or two-way ANOVA with Dunnett'sor Sidak's post-hoc test, respectively. Data are presented as mean ± SEM.
Gβ5 drives mitochondrial dysfunction, oxidative stress and cell death in APAP-treated murine hepatocytes – Consistent with results obtained in vivo, APAP triggered rapid and robust up-regulation of Gβ5 in cultured murine hepatocytes (Fig. 4A). Gβ5 KD (Fig. 4B) ameliorated elevations in ALT, AST, and triglycerides following APAP treatment (Fig. 4C). APAP decreased proliferation (Fig. S2A) and increased F4/80 levels (Fig. S2B), effects reversed via Gβ5 KD. A high ROS burden is known to activate the mitochondrial cell death pathway [32], regulated by Gβ5 in heart [11]. Thus, we investigated the impact of Gβ5 KD on APAP-induced oxidative stress. Indeed, hepatocytes lacking APAP-induced Gβ5 up-regulation displayed decreased total (Fig. 4D) and mitochondrial (Fig. 4E) ROS that could be restored by restoring Gβ5 protein expression. Irrespective of APAP exposure, Gβ5 KD was sufficient to decrease mitochondrial Ca2+ flux to the same level as Ru360, a blocker of the mitochondrial Ca2+ uniporter (Fig. 4F). Similarly, decreasing Gβ5 expression allowed for maintenance of the mitochondrial membrane potential (Δψm) (Fig. 4G) and ATP output (Fig. 4H). Further, APAP triggered release of cytochrome C from the mitochondria into the cytosol, a process reversed via Gβ5 KD (Fig. 4I). Finally, consistent with maintenance of mitochondrial integrity, Gβ5 KD also partially prevented APAP-induced toxicity in isolated hepatocytes (Fig. 4J, S2C, S2D).
Fig. 4.
Gβ5 promotes mitochondrial dysfunction and cell death in isolated murine hepatocytes. Primary hepatocytes were isolated from WT (scramble shRNA) or Gβ5 KD (Gβ5 shRNA) mice and exposed to APAP in culture (5 mM). For a subset of experiments, Gβ5 expression was restored via transient transfection. (A) Gβ5protein expression (n = 3). (B) shRNA validation. Unless otherwise noted, hepatocytes were exposed to APAP for 24 h. (C) ALT, AST, and triglycerides (n = 5). (D) CM-H2DCFDA fluorescence (total ROS, n = 5). (E) Mitochondrial superoxide (n = 5). (F) Mitochondrial Ca2+ content (n = 5) ± Ru360 pre-treatment (50 μM, 1 h) to block Ca2+ uptake. (G) Mitochondrial membrane potential (ΔψM; n = 5) ± Ru360 or mPTP blocker cyclosporin A (0.2 μM) pre-treatment (1 h). (H) Hepatocyte and mitochondrial ATP levels (12 h APAP, n = 5). (I) Cytosolic and mitochondrial cytochrome C levels (n = 5). (J) TUNEL + cells. β-Actin serves as a loading control for all immunoblots. ns = not significant. *P < 0.05, **P < 0.01, ***P < 0.001 via one-way or two-way ANOVA with Dunnett's or Sidak's post-hoc test, respectively. Data are presented as mean ± SEM.
APAP-dependent Gβ5 up-regulation depends on ROS – Given that Gβ5/R7 complexes have been identified as critical mediators of ROS generation in multiple tissues [11,33,34], we hypothesized thatROS might, in turn, influence Gβ5 expression. Indeed, hydrogen peroxide (H2O2) treatment led to Gβ5 up-regulation in HepaRG cells, a line of mature, metabolically functional human hepatocytes that express Gβ5 (Fig. S3A), comparable to results obtained with APAP (Fig. S3B). Conversely, scavenging of either hydrogen peroxide or superoxide precluded APAP-driven Gβ5 induction (Fig. S3C). In the absence of APAP, superoxide scavenging increased Gβ5 levels indicating that dynamic perturbations in ROS homeostasis may stimulate Gβ5-dependent intracellular signaling.
Gβ5 influences autophagic flux in APAP-exposed liver cells and intact tissue – While bolstering ROS buffering capacity with the glutathione donor NAC remains the only clinically approved treatment for APAP overdose, in our hands the beneficial impact of NAC was temporally restricted appearing if NAC was administered 1 h after APAP but largely absent at 6 h comparable to prior reports [16]. Even this small delay in NAC administration was sufficient to significantly impair the efficacy of this intervention in amelioration of APAP-induced free radical production (Fig. S4A), lethality (Fig. S4B), and compromised liver function (Fig. S4C, S4D). Further, in HepaRG cells, Gβ5 KD was more effective than NAC in mitigation of APAP-induced ROS accumulation (Fig. S5B) and cell death (Fig. S5C). Thus, we hypothesized that APAP-mediated pathological sequelae modulated by Gβ5 might involve mechanisms independent of ROS centric pathways targeted by NAC.
Effective APAP detoxification requires both antioxidant-mediated NAPQI neutralization as well as clearance of damaged proteins and organelles via autophagy. Gβ5 up-regulation in liver samples from APAP-induced liver injury patients was associated with increased phosphorylation of AMP-activated protein kinase (AMPK), depletion of autophagic vesicle receptor p62 and accumulation of autophagy marker LC3-II (Fig. S6A). Further, knockdown of Gβ5 expression in primary human hepatocytes was sufficient to prevent APAP-induced phosphorylation of AMPK and JNK; down-regulation of mammalian target of rapamycin (mTOR) effectors phospho-S6 and 4EBP1; and alterations in p62 and LC3-II (Fig. S6B). These data led us to hypothesize that Gβ5 might promote APAP-dependent liver damage by modulating autophagy.
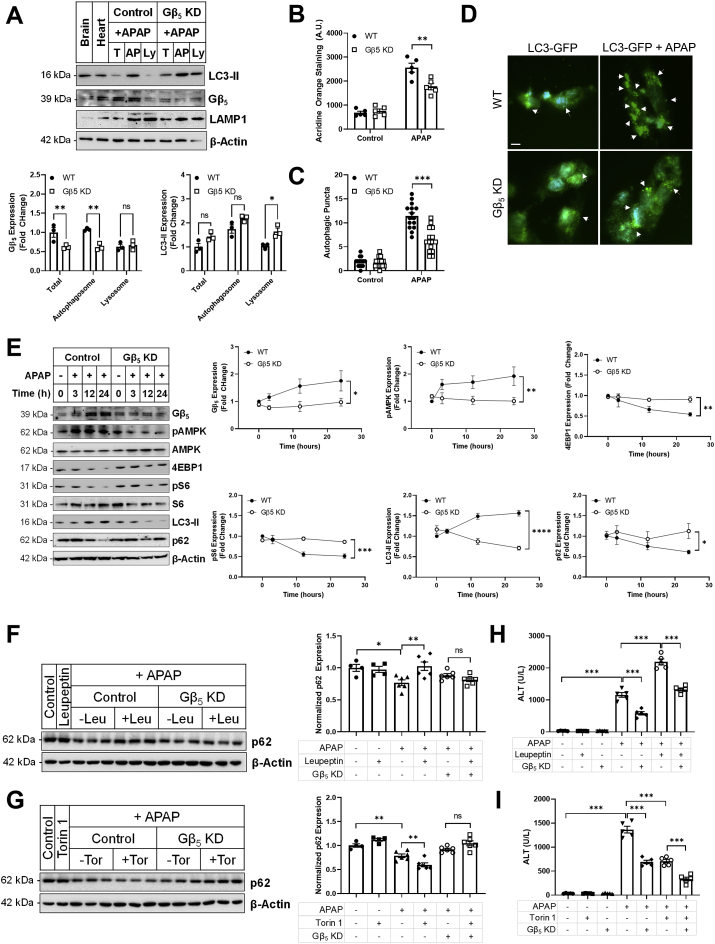
In liver, subcellular fractionation revealed significant concentration of Gβ5 protein in the autophagosome compartment (Fig. 5A) and Gβ5 KD resulted in accumulation of the structural autophagosome membrane protein LC3-II in the lysosomal fraction (Fig. 5A). APAP increased staining of acidic vacuoles in human HepaRG cells, an effect that was partially reversed via Gβ5 KD (Fig. 5B). As acridine orange fluorescence is not selective for autophagosomes, we next looked directly at cytoplasmic puncta formed by processing and recruitment of LC3-GFP to the autophagosome membrane. Here, Gβ5 depletion decreased APAP-mediated autophagosome formation (Fig. 5C and D). Changes in autophagosome formation were also evident in the livers of Gβ5 KD mice by TEM (Fig. S7). In murine hepatocytes, a lack of Gβ5 up-regulation translated into maintenance of autophagosomal marker p62 and decreased LC3-II levels (Fig. 5E). Gβ5 KD prevented APAP-induced AMPK phosphorylation as well as down-regulation of mTOR effectors 4EBP1 and pS6 (Fig. 5E). Together, these data indicate that manipulation of Gβ5 levels alters autophagic flux.
Fig. 5.
Gβ5 influences autophagic flux post-APAP exposure. (A) Gβ5, LC3-II (autophagosome marker), and Lamp-1 (lysosome marker) protein expression in the total lysate (T), autophagosome (AP) and lysosomal (Ly) sub-cellular compartments. Lysates were isolated from WT (control, scramble shRNA) and Gβ5 KD (Gβ5 shRNA) mice (350 mg/kg APAP, 6 h). (B) Lysates from primary murine hepatocytes (n = 3) treated with APAP (5 mM, 0–24 h) were subjected to immunoblotting for Gβ5, autophagosome markers (p62, LC3-II), kinase activation (AMPK) and mTOR signaling (4EBP1, S6). HepaRG cells were treated with APAP (5 mM, 18 h) and (C) acridine orange staining for acidic vacuoles (n = 5) and (D) LC3-GFP puncta formation (n = 15) [scale bar = 100 μm]. WT (control) and Gβ5 KD mice were treated with APAP (350 mg/kg, i.p.) ± leupeptin (Leu; 40 mg/kg, i.p.), Torin1 (Tor; 2 mg/kg, i.p.) or vehicle for 6 h p62 expression was evaluated in (F) leupeptin and (G) Torin1 treated mice (n = 4–6). Serum ALT analyzed in (H) leupeptin and (I) Torin1 treated mice (n = 5). β-Actin serves as a loading control for all immunoblots. ns = not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 via one-way or two-way ANOVA with Dunnett's or Sidak's post-hoc test, respectively. Data are presented as mean ± SEM.
Inhibition of autophagy via blockade of lysosomal proteases with leupeptin exacerbates APAP-induced liver injury while activation of autophagy via inhibition of mTOR with Torin1 is protective [7]. In vivo, leupeptin and Torin1 have opposing consequences on p62 in liver following APAP exposure. However, Gβ5 KD rendered tissue insensitive to pharmacological manipulations by either leupeptin (Fig. 5F) or Torin1 (Fig. 5G). Though this molecular marker of autophagic flux was not impacted, concomitant leupeptin or Torin1 treatment did increase (Fig. 5H) or decrease (Fig. 5I) ALT levels, respectively, indicating that, even in the absence of Gβ5 up-regulation there remains systemic autophagic tone and/or parallel Gβ5-independent signaling cascades sensitive to leupeptin and Torin1.
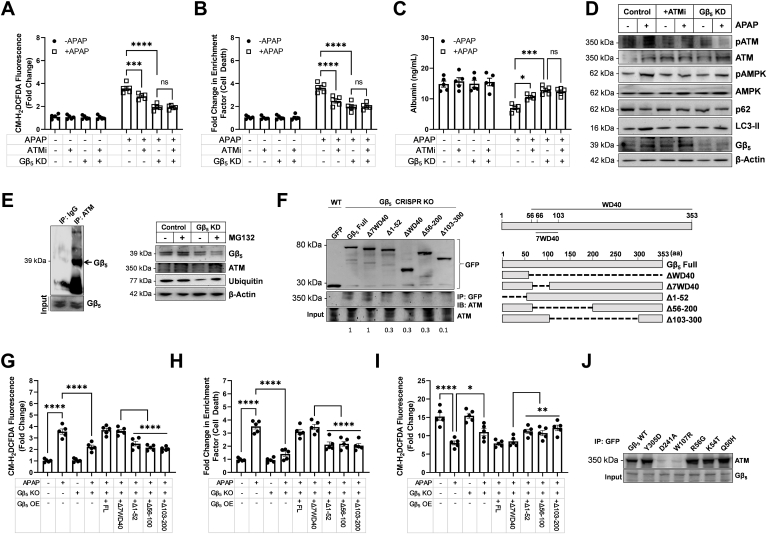
Gβ5 directly interacts with ATM in liver and Gβ5 KD phenocopies ataxia telangiectasia mutated (ATM) kinase inhibition in APAP-exposed hepatocytes – We next sought to provide mechanistic insight into how Gβ5, canonically believed to participation in GPCR regulation, simultaneously controls two G protein-independent processes. Previous reports demonstrated that Gβ5-interacting protein RGS6 controls ATM-dependent DNA damage signaling in cancer cells [35]. We noted that Gβ5 expression was highly correlated with levels of the multifunctional kinase ATM (Fig. S8A), extensively studied for its role in nuclear DNA damage repair and mitochondrial-dependent cell death and more recently implicated in autophagy, mitophagy and pexophagy [[36], [37], [38], [39]]. Indeed, expression of ATM and its substrate γH2AX were increased in murine liver following acute (Fig. S8B) or chronic (Fig. S8C) APAP exposure. Similarly, we noted that APAP-induced ATM activation could be mitigated in primary human hepatocytes via knockdown of Gβ5 (Fig. S6B). Thus, we hypothesized that ATM might represent a critical node linking Gβ5 to autophagic and cell death signaling cascades.
To test this hypothesis, we first turned to a pharmacological approach. Inhibition of ATM's kinase activity resulted in modest reductions in ROS generation (Fig. 6A), cell death (Fig. 6B) and albumin production (Fig. 6C) in APAP-treated HepaRG cells, actions that were non-additive with Gβ5 KD. Similarly, ATMi phenocopied the impact of Gβ5 KD on APAP-dependent AMPK phosphorylation, p62 depletion and LC3-II accumulation (Fig. 6D). Together, these data indicate that ATM and Gβ5 may function in the same pathway to support APAP-dependent oxidative stress, cell death, and autophagy. To further strengthen our argument, we now show that ATM and Gβ5 form a co-immunoprecipitatable complex in hepatocytes (Fig. 6E). Mathematical modeling of ATM and Gβ5 crystal structures revealed the potential for a highly stable direct complex supported by key residues in the FRAP/ATM/TRRAP (FAT) domain of ATM and spread throughout Gβ5 (Fig. S9, Table S8, S9). Because Gβ5 functions to stabilize other known interacting proteins [10], we hypothesized that it performed a similar role for ATM. However, Gβ5 KD failed to impact ATM protein levels in the presence or absence of proteasome inhibition with MG132 (Fig. 6E). We were intrigued to note, however, that Gβ5 depletion was sufficient to trigger a corresponding decrease in total ubiquitin indicating that Gβ5 may play a role in protein catabolism in liver (Fig. 6E). To screen for critical domains of Gβ5 required for ATM binding we first established Gβ5 knockout HepaRG cells utilizing CRISPR/Cas9 technology (Fig. S10). Next, we attempted to pulldown ATM with various Gβ5 deletion constructs. Removal of either the Gβ5N-terminus or portions of the WD40 domain significantly attenuated ATM-Gβ5 binding (Fig. 6F). Importantly, though introduction of full length Gβ5 or mutants retaining full ATM interaction were able to completely restore APAP-dependent ROS generation (Fig. 6G), cell death (Fig. 6H) or albumin depletion (Fig. 6I) in cells lacking endogenous Gβ5, this effect was attenuated for mutants with decreased ATM binding. Finally, our simulation identified 6 Gβ5 residues predicted to support the Gβ5-ATM complex. Mutation of 2 residues in Gβ5 (D241A and W107R) almost completely abolished ATM- Gβ5 co-immunoprecipitation (Fig. 6J).
Fig. 6.
Gβ5-ATM complexes drive APAP-dependent oxidative stress, autophagy and cell death in human hepatocytes. (A–D) HepaRG cells were transfected with scramble (control) or Gβ5 shRNA and treated with APAP (5 mM, 24 h) ± 1 h pre-treatment ATM inhibitor KU-55933 (ATMi, 5 μM). (A) CM-H2DCFDA fluorescence (total ROS, n = 5). (B) Cell death (fold increase in histone-associated DNA fragments; n = 5). (C) Albumin released into culture medium (n = 5). (D) Immunoblotting for pATM/ATM, pAMPK/AMPK, Gβ5, and autophagy markers LC3-II and p62.(E) ATM was immunoprecipitated from hepatocyte lysates and the resultant pull-downs probed for Gβ5 (right). Immunoblotting for Gβ5, ATM, and total ubiquitin in HepaRG cell lysates ± proteasome inhibitor MG132 (1 μM) (left).(F)GFP-tagged Gβ5 constructs were transfected into HepaRG cells in which Gβ5 expression was eliminated via CRIPSR/Cas9-dependent excision (Gβ5 KO). GFP was immunoprecipitated and resultant complexes probed for ATM. (G) ROS (n = 5), (H) cell death (n = 5) and (I) albumin production (n = 5) from Gβ5 KO HepaRG cells transfected with various Gβ5 deletion constructs. (J) ATM immunoprecipitation with Gβ5 point mutants transfected into Gβ5 KO HepaRG cells. β-Actin serves as a loading control for all immunoblots. ns = not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 via one-way or two-way ANOVA with Dunnett's or Sidak's post-hoc test, respectively. Data are presented as mean ± SEM.
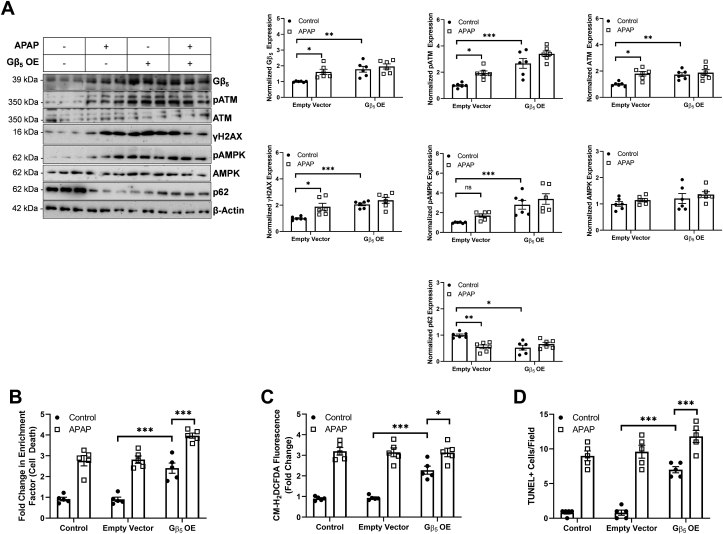
Gβ5 overexpression (OE) in liver is sufficient to drive ATM activation as well as cell death and autophagic signaling – Given that APAP exposure triggered robust up-regulation of Gβ5 in liver, we hypothesized that Gβ5 overexpression might be sufficient to drive APAP-dependent hepatotoxicity. Thus, we expressed Gβ5 in liver via viral transduction and noted that Gβ5 overexpression alone resulted in ATM, γH2AX and AMPK phosphorylation as well as p62 downregulation comparable to levels observed in APAP-exposed livers (Fig. 7A). Similarly, cell death (Fig. 7B and D) and ROS generation (Fig. 7C) were detectable in the liver of Gβ5 OE animals. APAP treatment did result in further elevations of both cytotoxicity and oxidative stress possibly due to actions of endogenous Gβ5.
Fig. 7.
Gβ5 overexpression (OE) in liver phenocopies the impact of APAP. Exogenous Gβ5 expression was introduced into the livers of mice followed by APAP exposure (4 mg/kg i.p., biweekly, 4 weeks). (A) Immunoblotting for Gβ5, ATM and its effectors, and markers of autophagy in liver (n = 6) with quantification. (B) Cell death (fold increase in cytoplasmic histone fragments, n = 5), (C) ROS generation (CM-H2DCFDA fluorescence, n = 5) and (D) TUNEL + cells (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001 via two-way ANOVA with Sidak's post-hoc test. Data are presented as mean ± SEM.
Paracrine factors promote Gβ5-dependent, APAP-induced hepatocyte dysfunction – In addition to direct APAP treatment, the exposure of primary human hepatocytes to serum from patients presenting with APAP associated liver damage resulted in rapid and robust up-regulation of Gβ5, activation of DNA damage signaling (ATM, γH2AX) and alterations in signaling impacting autophagy including the AMPK, mTOR and JNK cascades (Fig. S11). Importantly, introduction of Gβ5 shRNA largely reversed these changes (Fig. S11). These data suggest that APAP triggers release of hepatotoxic factors into the systemic circulation.
To further investigate paracrine mechanism(s) of Gβ5-mediated, APAP-driven liver damage we next exploited an observation from the literature regarding the apparent protective impact of organotypic co-culture systems on APAP cytotoxicity [23]. In mono-cultures of either the human hepatocyte cell line HepG2 or human umbilical vein endothelial cells (HUVEC), APAP triggered robust Gβ5 up-regulation that was absent in co-cultures cells (Fig. S12A). The lack of APAP-induced oxidative stress (Fig. S12B) and cell death (Fig. S12C) could be restored in co-cultured cells by Gβ5 overexpression. This phenomenon afforded us the opportunity to investigate additional factors that might influence Gβ5 expression in hepatocytes. Indeed, APAP-dependent release of the anti-inflammatory cytokine transforming growth factor β1 (TGF-β1) was unique to the co-culture system (Fig. S12D). Inhibition of TGF-β1 signaling with the receptor blocker GW788388 prevented Gβ5 up-regulation in APAP-treated HepG2 cells without impacting expression in HUVEC cells indicating that, in co-cultured cells, TGF-β1 likely restricts APAP-dependent mechanism(s) that drive Gβ5 recruitment (Fig. S12E).
We also note that Gβ5 expression is not restricted to hepatocytes in liver but also detectable in stellate cells (Fig. S13A), which play a key role in APAP-induced hepatic fibrosis and represent a key source of TGF-β1 in liver [40,41]. Knockdown of Gβ5in the hepatic stellate cell line LX2 prevented APAP-dependent molecular changes (Fig. S13A), oxidative stress (Fig. S13B), cell death (Fig. S13C) and collagen depletion (Fig. S13D) indicating that Gβ5 might function via hepatocyte extrinsic mechanisms to promote APAP-induced liver damage.
4. Discussion
Hepatotoxicity limits the clinical utility of APAP, which, nevertheless remains amongst the most ubiquitous analgesic and antipyretic drugs worldwide. Here we provide novel insight into heretofore undelimited mechanisms driving APAP-induced liver injury demonstrating that Gβ5 is both necessary and sufficient to drive APAP-dependent hepatotoxicity. In primary murine and human hepatocytes, human hepatocyte cell lines, and in vivo, APAP exposure triggers rapid and robust up-regulation of Gβ5. Further, in liver samples isolated from APAP overdose patients, Gβ5 protein was elevated particularly in patients with severe injury. Consistent with a critical role for Gβ5 as a driver of APAP-dependent hepatotoxicity, liver specific knockdown of Gβ5 in mice ameliorated APAP-induced hepatic fibrosis, lipid accumulation, and inflammation and improved the balance between hepatocyte proliferation and cell death. Similarly, Gβ5 overexpression in liver was sufficient to trigger cytotoxicity and induce oxidative stress. The concordance of datasets obtain in human and murine systems underscores the utility of the mouse as model of APAP-induced liver injury and a means to understand the role of Gβ5 therein.
The cellular response to APAP exposure balances machinations aimed at mitigating oxidative stress and removing damaged proteins and organelles with initiation of cell death signaling cascades in the face of insurmountable injury. In early stages, depletion of antioxidant stores and formation of toxic protein adducts by the reactive APAP metabolite NAPQI results in oxidative stress, mitochondrial dysfunction, and ATP depletion. Mitochondrial proteins such as the adenosine triphosphate (ATP) synthase α-subunit and complex I/II of mitochondrial electron transport chain are direct targets of NAPQI and damage to these proteins results directly in production of superoxide that can then be dismutated by manganese superoxide dismutase to H2O2 [42]. Subsequent depletion of glutathione stores can then lead to accumulation of additional oxidative and nitrative species [42]. In murine hepatocytes, Gβ5 KD was sufficient to decrease mitochondrial ROS, maintain basal mitochondrial Ca2+ flux, prevent loss of mitochondrial membrane potential, restore mitochondrial ATP production, and prevent cell death. Scavenging of hydrogen peroxide or superoxide prevented Gβ5 up-regulation in APAP exposed hepatocytes indicating that ROS function as an initial trigger to initiate a feed-forward loop by which Gβ5 accumulation facilitates further oxidative stress. Thus, our data are consistent with a model whereby NAPQI generation results in the formation of mitochondrial protein adducts that damage the mitochondria, impair organelle function, and increase oxidative stress. Gβ5 up-regulation is triggered by ROS accumulation and acts in a feed forward manner to exacerbate oxidative stress, promote mitochondrial dysfunction, and activate additional pro-death intracellular signaling cascades. What results is a self-perpetuating cycle of cellular dysfunction that can be ameliorated by preventing Gβ5 up-regulation.
In liver, Gβ5 is enriched in the autophagosome fraction leading us to hypothesize that it could play a role in cellular autophagy, a critical process whereby damaged proteins and organelles are removed and known to be essential in preventing APAP-induced liver damage [7,8]. mTOR complex 1 (mTORC1) integrates signaling through multiple cascades (e.g. JNK, AMPK) to initiate autophagy in response to environmental stressors and is essential for maintaining hepatic integrity [43]. Gβ5KD impaired formation of autophagic puncta and alterations in autophagy markers p62 and LC3-II in APAP exposed human and murine hepatocytes and was associated with complete loss of APAP-dependent AMPK/JNK phosphorylation and mTOR-mediated 4EBP1 and phospho-S6 depletion. p62 levels were also rendered insensitive to modulation by autophagy inhibitor leupeptin or autophagy activator Torin1 in Gβ5 KD livers, though these drugs functioned additively with Gβ5 KD to exacerbate or improve liver function, respectively. It is important to note here that the protective impacts of Gβ5KD coupled to decreased APAP-induced autophagy seem counterintuitive considering evidence demonstrating mitigation of APAP-dependent liver damage following induction of autophagy [7,8]. Time course data from APAP-treated hepatocytes shed some light on this paradox emphasizing that the impact of Gβ5 on autophagic markers depends on the length of APAP exposure. Several signaling cascades responsible for sensing different modalities of cellular stress converge on autophagic markers. Gβ5 appears to act upstream of AMPK, also targeted by ATM [38], allowing for bidirectional modification of autophagy through parallel processes. Thus, we propose a model wherein Gβ5 up-regulation represents a watershed event pushing the cell toward a catastrophic fate in face of insurmountable cellular stress. By preventing Gβ5 induction, the cell is given time to recruit survival mechanisms and restore cellular functionality without initiating cell death.
In our analysis of liver samples from APAP exposed patients, we noted a molecular signature associated with high Gβ5 expression and characterized by ATM up-regulation, activation of the DNA damage marker γH2AX, AMPK phosphorylation and down-regulation of autophagy marker p62. Notably, ATM and Gβ5 levels were highly correlated in DILI samples, a phenomenon consistent across species and cell types. In addition to its canonical function as an initiator of the DNA damage response, ATM also localizes to the mitochondria where it controls respiration [44] and mitophagy [36] and the cytoplasm where it regulates autophagy via an AMPK- and mTORC1-dependent mechanism [38]. Gβ5 interactor RGS6 was previously shown to control doxorubicin-induced ATM activation in cancer cells via a ROS-dependent mechanism [35] leading us to postulate that the ability of Gβ5to simultaneously control mitochondrial function and autophagy might derive from regulation of ATM, activated directly via oxidation [45]. Indeed, ATM inhibition phenocopies the impact of Gβ5KD on APAP-induced markers of autophagy. Further, no additive benefit can be achieved by combining the two interventions providing evidence that these proteins may function in the same pathway to modulate APAP-dependent liver damage. We should note here that the impact of ATM inhibition on APAP-induced ROS accumulation and cell death in hepatocytes was not as robust as that obtained following Gβ5 knockdown. Though perhaps the consequence of insufficient inhibitor concentration, Gβ5likely possess ATM-independent cytotoxic actions in liver.
Strengthening a functional link between ATM and Gβ5 is our evidence that Gβ5 and ATM form a complex in hepatocytes. In silico modeling identified 3 N-terminal residues (Q50, K54, and R56) and 3 residues in Gβ5's WD40 domain (W107, D241, Y305) supporting the Gβ5-ATM complex. Indeed, deletion of these domains compromised ATM binding. However, mutation of W107 or D241 were sufficient to completely abolish ATM- Gβ5 co-immunoprecipitation. These data are consistent with the proposed function of WD40 domains in G protein β subunits, which act as interfaces for protein-protein scaffolding [46]. Gβ5-interacting residues on ATM were localized to the FAT domain, critical for ATM activation via autophosphoylation as well as substrate interactions in humans. More specifically, the FAT domain blocks substrate access to the kinase domain preventing ATM activity in the absence of DNA damage [47]. We propose 3 potential mechanism(s) whereby Gβ5-ATM complex formation might influence ATM activity: 1) by anchoring ATM to key sub-cellular compartments; 2) by facilitating substrate recruitment or 3) by directly activating ATM through interactions with the FAT domain. Importantly, Gβ5 might represent a new means to activate ATM downstream of ROS and independent of DNA damage. Future work will seek to further delineate the molecular determinants and functional importance of the Gβ5/ATM interaction.
In addition to hepatocyte intrinsic mechanism(s), our data demonstrate that cytokines present serum from APAP overdose patients are sufficient to up-regulate Gβ5 protein and trigger Gβ5-dependent cellular dysfunction. Co-culture experiments emphasized that factors (e.g. TGF-β1) released from hepatocyte-adjacent cell types may also function to maintain hepatocyte functionality, at least in part, by preventing Gβ5 up-regulation. Further, Gβ5 KD in hepatic stellate cells, drivers of hepatic fibrosis, also improved APAP-dependent oxidative stress and cell survival and decreased fibrotic markers. Together these data indicated that autocrine, paracrine, and even endocrine processes likely also contribute to APAP-induced, Gβ5-mediated liver injury.
Though APAP is a particularly potent inducer of Gβ5 expression in liver, Gβ5 up-regulation was detectable across DILI samples. This observation compounded with the unique ability of Gβ5 to simultaneously modulate several key cellular signaling cascades indicates that Gβ5 may play a more universal role in promoting liver damage upon accumulation in hepatocytes and surrounding cells. Disrupting Gβ5-mediated impacts on autophagy and cell death might represent a viable means, therefore, to maintain liver function following exposure to APAP or other hepatotoxic factors. As the current therapy for APAP overdose, NAC, is only effective in a narrow window 8–12 h following APAP exposure [9], characterization of novel participants in the pathogenesis of APAP-dependent liver damage represents an important step toward the formulation of new, more efficacious interventions.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgements
Our sincere gratitude to Ramalingaswami Fellowship and Dr. Meenakshi Munshi, Department of Biotechnology (DBT) for the necessary help. We also thank Dr. Santosh Chauhan, Institute of Life Science, Bhubaneswar, India for the kind gift of LC3-GFP plasmid and Dr. Sougata Roy Chowdhury, INSPIRE Faculty, University of Calcutta for the intellectual help.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101965.
Financial Support
This work was supported by Centre of Biomedical Research (CBMR) internal research funds and Department of Biotechnology, India (BT/RLF/Re-entry/15/2013) and BT/PR21156/MED/30/1753/2016 to BM. AP thanks CSIR, India for the predoctoral fellowship (09/028/0956/2015-EMR-1).
Author contributions
Conception and design: A. Pramanick, S. Chakraborti, A. Stewart, B. Maity; Acquisition of data: A. Pramanick, S. Chakraborti, T. Mahata, M. Basak, A.S. Sengar, P. Kumar, S. Saha, S.K. Verma, P.K. Singh, B. Bhattacharya, S. Biswas, B. Maity; Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): A. Pramanick, S. Chakraborti, T. Mahata, M. Basak, S. Saha, S.K. Verma, P.K. Singh, S. Biswas, P.B. Pal, S. Sarkar, D. Nath, V. Agrawal, S. Chatterjee, A. Stewart, B. Maity; Writing, review, and/or revision of the manuscript: A. Pramanick, S. Chakraborti, A. Stewart, B. Maity; Study supervision: D. Nath, A. Stewart, B. Maity.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bernal W., Wendon J. Acute liver failure. N. Engl. J. Med. 2013;369(26):2525–2534. doi: 10.1056/NEJMra1208937. [DOI] [PubMed] [Google Scholar]
- 2.Mayoral W., Lewis J.H., Zimmerman H. Drug-induced liver disease. Curr. Opin. Gastroenterol. 1999;15(3):208–216. doi: 10.1097/00001574-199905000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Khandelwal N., James L.P., Sanders C., Larson A.M., Lee W.M., Acute Liver Failure Study G. Unrecognized acetaminophen toxicity as a cause of indeterminate acute liver failure. Hepatology. 2011;53(2):567–576. doi: 10.1002/hep.24060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu Y., Benet L.Z., Burlingame A.L. Identification of the hepatic protein targets of reactive metabolites of acetaminophen in vivo in mice using two-dimensional gel electrophoresis and mass spectrometry. J. Biol. Chem. 1998;273(28):17940–17953. doi: 10.1074/jbc.273.28.17940. [DOI] [PubMed] [Google Scholar]
- 5.Donnelly P.J., Walker R.M., Racz W.J. Inhibition of mitochondrial respiration in vivo is an early event in acetaminophen-induced hepatotoxicity. Arch. Toxicol. 1994;68(2):110–118. doi: 10.1007/s002040050043. [DOI] [PubMed] [Google Scholar]
- 6.Masubuchi Y., Suda C., Horie T. Involvement of mitochondrial permeability transition in acetaminophen-induced liver injury in mice. J. Hepatol. 2005;42(1):110–116. doi: 10.1016/j.jhep.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Ni H.M., McGill M.R., Chao X., Du K., Williams J.A., Xie Y. Removal of acetaminophen protein adducts by autophagy protects against acetaminophen-induced liver injury in mice. J. Hepatol. 2016;65(2):354–362. doi: 10.1016/j.jhep.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni H.M., Bockus A., Boggess N., Jaeschke H., Ding W.X. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology. 2012;55(1):222–232. doi: 10.1002/hep.24690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smilkstein M.J., Knapp G.L., Kulig K.W., Rumack B.H. Efficacy of oral N-acetylcysteine in the treatment of acetaminophen overdose. Analysis of the national multicenter study (1976 to 1985) N. Engl. J. Med. 1988;319(24):1557–1562. doi: 10.1056/NEJM198812153192401. [DOI] [PubMed] [Google Scholar]
- 10.Chen C.K., Eversole-Cire P., Zhang H., Mancino V., Chen Y.J., He W. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc. Natl. Acad. Sci. U. S. A. 2003;100(11):6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakraborti S., Pramanick A., Saha S., Roy S.S., Chaudhuri A.R., Das M. Atypical G protein beta5 promotes cardiac oxidative stress, apoptosis, and fibrotic remodeling in response to multiple cancer chemotherapeutics. Canc. Res. 2018;78(2):528–541. doi: 10.1158/0008-5472.CAN-17-1280. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q., Levay K., Chanturiya T., Dvoriantchikova G., Anderson K.L., Bianco S.D. Targeted deletion of one or two copies of the G protein beta subunit Gbeta5 gene has distinct effects on body weight and behavior in mice. Faseb. J. 2011;25(11):3949–3957. doi: 10.1096/fj.11-190157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart A., Maity B., Anderegg S.P., Allamargot C., Yang J., Fisher R.A. Regulator of G protein signaling 6 is a critical mediator of both reward-related behavioral and pathological responses to alcohol. Proc. Natl. Acad. Sci. U. S. A. 2015;112(7):E786–E795. doi: 10.1073/pnas.1418795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J.Y., Park K.J., Hwang J.Y., Kim G.H., Lee D., Lee Y.J. Activating transcription factor 3 is a target molecule linking hepatic steatosis to impaired glucose homeostasis. J. Hepatol. 2017;67(2):349–359. doi: 10.1016/j.jhep.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 15.Mossanen J.C., Tacke F. Acetaminophen-induced acute liver injury in mice. Lab. Anim. 2015;49(1 Suppl):30–36. doi: 10.1177/0023677215570992. [DOI] [PubMed] [Google Scholar]
- 16.James L.P., McCullough S.S., Lamps L.W., Hinson J.A. Effect of N-acetylcysteine on acetaminophen toxicity in mice: relationship to reactive nitrogen and cytokine formation. Toxicol. Sci. 2003;75(2):458–467. doi: 10.1093/toxsci/kfg181. [DOI] [PubMed] [Google Scholar]
- 17.Hinson J.A., Roberts D.W., James L.P. Mechanisms of acetaminophen-induced liver necrosis. Handb. Exp. Pharmacol. 2010;(196):369–405. doi: 10.1007/978-3-642-00663-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ni H.M., Du K., You M., Ding W.X. Critical role of FoxO3a in alcohol-induced autophagy and hepatotoxicity. Am. J. Pathol. 2013;183(6):1815–1825. doi: 10.1016/j.ajpath.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green C.J., Charlton C.A., Wang L.M., Silva M., Morten K.J., Hodson L. The isolation of primary hepatocytes from human tissue: optimising the use of small non-encapsulated liver resection surplus. Cell Tissue Bank. 2017;18(4):597–604. doi: 10.1007/s10561-017-9641-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu I.C., Lipsky M.M., Cole K.E., Su C.H., Trump B.F. Isolation and culture of hepatocytes from human liver of immediate autopsy. Vitro Cell Dev. Biol. 1985;21(3 Pt 1):154–160. doi: 10.1007/BF02621352. [DOI] [PubMed] [Google Scholar]
- 21.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larson A.M. Acetaminophen hepatotoxicity. Clin. Liver Dis. 2007;11(3):525–548. doi: 10.1016/j.cld.2007.06.006. vi. [DOI] [PubMed] [Google Scholar]
- 23.Nelson L.J., Navarro M., Treskes P., Samuel K., Tura-Ceide O., Morley S.D. Acetaminophen cytotoxicity is ameliorated in a human liver organotypic co-culture model. Sci. Rep. 2015;5:17455. doi: 10.1038/srep17455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalyanaraman B., Darley-Usmar V., Davies K.J., Dennery P.A., Forman H.J., Grisham M.B. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic. Biol. Med. 2012;52(1):1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majumder S., Piguet A.C., Dufour J.F., Chatterjee S. Study of the cellular mechanism of Sunitinib mediated inactivation of activated hepatic stellate cells and its implications in angiogenesis. Eur. J. Pharmacol. 2013;705(1–3):86–95. doi: 10.1016/j.ejphar.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 26.UniProt C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Rose Y., Duarte J.M., Lowe R., Segura J., Bi C., Bhikadiya C. RCSB protein data bank: architectural advances towards integrated searching and efficient access to macromolecular structure data from the PDB archive. J. Mol. Biol. 2020:166704. doi: 10.1016/j.jmb.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baretic D., Pollard H.K., Fisher D.I., Johnson C.M., Santhanam B., Truman C.M. Structures of closed and open conformations of dimeric human ATM. Sci. Adv. 2017;3(5) doi: 10.1126/sciadv.1700933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson A.J., Aragay A.M., Slepak V.Z., Simon M.I. A novel form of the G protein beta subunit Gbeta5 is specifically expressed in the vertebrate retina. J. Biol. Chem. 1996;271(45):28154–28160. doi: 10.1074/jbc.271.45.28154. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J.H., Pandey M., Seigneur E.M., Panicker L.M., Koo L., Schwartz O.M. Knockout of G protein beta5 impairs brain development and causes multiple neurologic abnormalities in mice. J. Neurochem. 2011;119(3):544–554. doi: 10.1111/j.1471-4159.2011.07457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Redza-Dutordoir M., Averill-Bates D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta. 2016;1863(12):2977–2992. doi: 10.1016/j.bbamcr.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 33.Yang J., Maity B., Huang J., Gao Z., Stewart A., Weiss R.M. G-protein inactivator RGS6 mediates myocardial cell apoptosis and cardiomyopathy caused by doxorubicin. Canc. Res. 2013;73(6):1662–1667. doi: 10.1158/0008-5472.CAN-12-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maity B., Yang J., Huang J., Askeland R.W., Bera S., Fisher R.A. Regulator of G protein signaling 6 (RGS6) induces apoptosis via a mitochondrial-dependent pathway not involving its GTPase-activating protein activity. J. Biol. Chem. 2011;286(2):1409–1419. doi: 10.1074/jbc.M110.186700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J., Yang J., Maity B., Mayuzumi D., Fisher R.A. Regulator of G protein signaling 6 mediates doxorubicin-induced ATM and p53 activation by a reactive oxygen species-dependent mechanism. Canc. Res. 2011;71(20):6310–6319. doi: 10.1158/0008-5472.CAN-10-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valentin-Vega Y.A., Kastan M.B. A new role for ATM: regulating mitochondrial function and mitophagy. Autophagy. 2012;8(5):840–841. doi: 10.4161/auto.19693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tripathi D.N., Chowdhury R., Trudel L.J., Tee A.R., Slack R.S., Walker C.L. Reactive nitrogen species regulate autophagy through ATM-AMPK-TSC2-mediated suppression of mTORC1. Proc. Natl. Acad. Sci. U. S. A. 2013;110(32):E2950–E2957. doi: 10.1073/pnas.1307736110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexander A., Cai S.L., Kim J., Nanez A., Sahin M., MacLean K.H. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. U. S. A. 2010;107(9):4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J., Tripathi D.N., Jing J., Alexander A., Kim J., Powell R.T. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 2015;17(10):1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen K., Chang W., Gao X., Wang H., Niu W., Song L. Depletion of activated hepatic stellate cell correlates with severe liver damage and abnormal liver regeneration in acetaminophen-induced liver injury. Acta Biochim. Biophys. Sin. 2011;43(4):307–315. doi: 10.1093/abbs/gmr005. [DOI] [PubMed] [Google Scholar]
- 41.Fu R., Wu J., Ding J., Sheng J., Hong L., Sun Q. Targeting transforming growth factor betaRII expression inhibits the activation of hepatic stellate cells and reduces collagen synthesis. Exp. Biol. Med. 2011;236(3):291–297. doi: 10.1258/ebm.2010.010231. [DOI] [PubMed] [Google Scholar]
- 42.Yan M., Huo Y., Yin S., Hu H. Mechanisms of acetaminophen-induced liver injury and its implications for therapeutic interventions. Redox Biol. 2018;17:274–283. doi: 10.1016/j.redox.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho C.S., Kowalsky A.H., Namkoong S., Park S.R., Wu S., Kim B. Concurrent activation of growth factor and nutrient arms of mTORC1 induces oxidative liver injury. Cell Discov. 2019;5:60. doi: 10.1038/s41421-019-0131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valentin-Vega Y.A., Maclean K.H., Tait-Mulder J., Milasta S., Steeves M., Dorsey F.C. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119(6):1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo Z., Kozlov S., Lavin M.F., Person M.D., Paull T.T. ATM activation by oxidative stress. Science. 2010;330(6003):517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 46.Schapira M., Tyers M., Torrent M., Arrowsmith C.H. WD40 repeat domain proteins: a novel target class? Nat. Rev. Drug Discov. 2017;16(11):773–786. doi: 10.1038/nrd.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marechal A., Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013;5(9) doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.