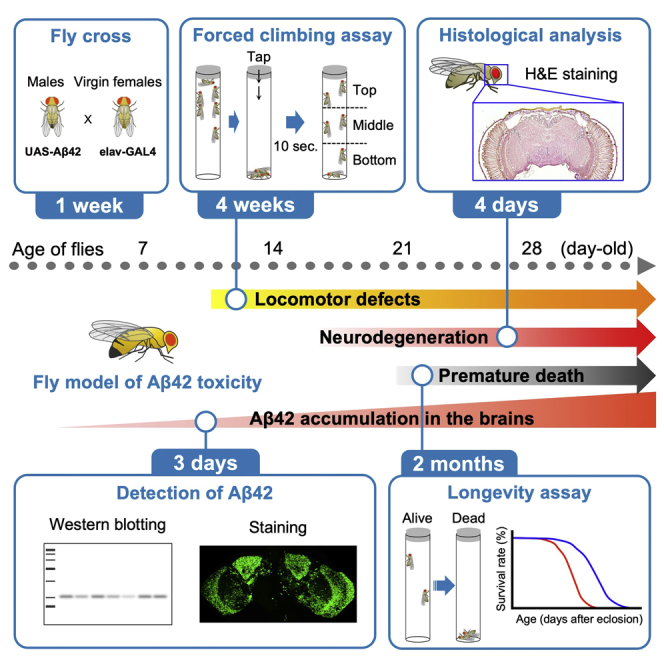
Summary
For decades, the fruit fly Drosophila melanogaster has been an efficient genetic model to investigate many aspects of human neurodegenerative diseases. Through genetic and pharmacologic approaches, these studies have revealed the molecular mechanisms underlying disease pathogenesis and provided therapeutic implications. Here, we describe a protocol for assessing Alzheimer's disease-related amyloid-β toxicity in a transgenic fly model through biochemical, histological, and behavioral analyses. We also discuss the advantages and limitations of our protocols.
For complete details on the use and execution of this protocol, please refer to Wang et al. (2021).
Subject areas: Behavior, Microscopy, Model Organisms, Neuroscience
Graphical abstract
Highlights
-
•
Drosophila is an efficient genetic model in neurodegenerative disease research
-
•
This protocol describes phenotypic analysis of a fly model of Alzheimer's disease
-
•
This protocol can be applied to genetic and pharmacologic screens using flies
For decades, the fruit fly Drosophila melanogaster has been an efficient genetic model to investigate many aspects of human neurodegenerative diseases. Through genetic and pharmacologic approaches, these studies have revealed the molecular mechanisms underlying disease pathogenesis and provided therapeutic implications. Here, we describe a protocol for assessing Alzheimer's disease-related amyloid-β toxicity in a transgenic fly model through biochemical, histological, and behavioral analyses. We also discuss the advantages and limitations of our protocols.
Before you begin
Preparation of fly food
Timing: 1 day
-
1.Fly food recipe is provided in “Materials and equipment”.
-
a.Add agar and sucrose to 8 L ddH2O in a pot and mix well.
-
b.Boil until the agar dissolves completely.
-
c.Add potassium tartrate, calcium chloride, and glucose to the pot and stir.
-
d.Add 2 L ddH2O, and then add corn meal and yeast while mixing.
-
e.Cook for 10 min until the mixture thickens.
-
f.Stop heating and cool down.
-
g.Cool to 65°C–70°C, measuring the temperature using a thermometer (we use a stainless-steel thermometer).
-
a.
-
2.
Add preservative (67 mL) to the fly food and mix well.
-
3.
Add acid mixture (117 mL) and mix well.
-
4.
Pour the fly food into plastic vials and bottles (5 mL/25 mm diameter vial, 40 mL/bottle).
-
5.
Cover the vials and bottles with cheese cloth and let them dry for 16–20 h.
-
6.
Cap the vials and bottles with plugs.
Optional: If you plan on performing drug feeding assay, add the required chemicals to the fly food after step 3. If a chemical is not water-soluble, use ethanol (final concentration < 1%) or DMSO (final concentration < 0.1%) as a vehicle.
Note: Cooling down the food is necessary to avoid volatilizing the acid mixture added in step 3. If you are adding the antibiotics or drugs (for drug feeding assay), you should cool down the fly food to prevent thermal degradation of those compounds.
Note: Fly foods can be stored at room temperature (20°C–25°C) for up to 1 week.
Preparation for forced climbing assay
Timing: < 30 min
-
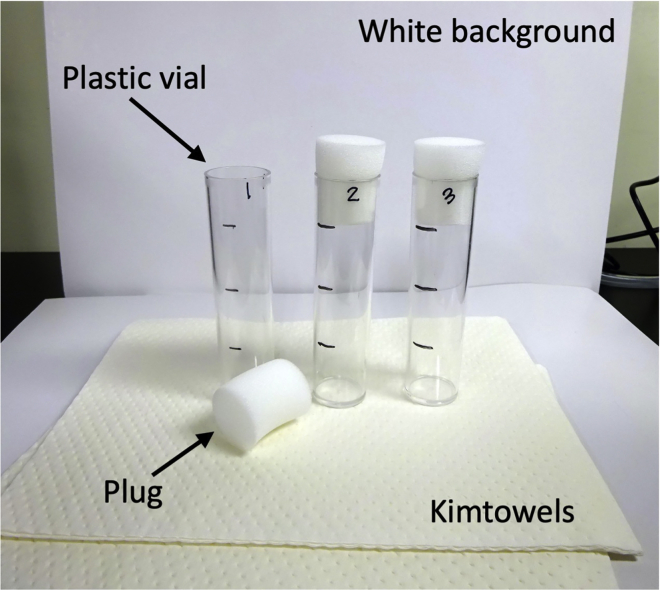
7.
Prepare the white board or white background, plastic vials, plugs, digital camera, and timer.
-
8.
Mark every inch from the bottom on the plastic vials and label the vials with the number for each genotype (Figure 1).
Figure 1.
Equipment for climbing assay
Preparation for histological analysis
To prepare 80% and 95% ethanol
Timing: < 10 min
To prepare 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) solution and 0.1% sodium bicarbonate solution
Timing: < 10 min, just before the assay
-
9.
Prepare 80% and 95% ethanol.
-
10.
Prepare 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) solution and 0.1% sodium bicarbonate solution.
(Refer to materials and equipment)
Preparation for detection of amyloid-β (Aβ42) by western blotting
To prepare the radioimmunoprecipitation (RIPA) buffer, Tris-Tricine running buffer, transfer buffer, PBS, Tris-buffered saline with 0.1% Tween 20 (TBST), and TBST+5% skim milk
Timing: 1 h
To prepare RIPA/SDS, 70% formic acid, and 2X Sample buffer
Timing: < 30 min, just before the assay
-
11.
Prepare the radioimmunoprecipitation (RIPA) buffer, Tris-Tricine running buffer, transfer buffer, PBS, Tris-buffered saline with 0.1% Tween 20 (TBST), and TBST+5% skim milk.
(Refer to materials and equipment)
-
12.
Prepare RIPA/SDS, 70% formic acid, and 2X Sample buffer.
(Refer to materials and equipment)
Preparation for 1-fluoro-2,5-bis(3-carboxy-4-hydroxystyryl)benzene (FSB) staining
To prepare PBS with 0.5% Triton X-100 (PBST), 50% glycerol/PBS, and 50% ethanol
Timing: < 30 min
To prepare 0.002% FSB solution
Timing: < 30 min, just before the assay
-
13.
Prepare PBS with 0.5% Triton X-100 (PBST), 50% glycerol/PBS, and 50% ethanol.
(Refer to materials and equipment)
-
14.Prepare 0.002% FSB solution.
-
a.Add 0.5 μL of 1% FSB/DMSO solution to 250 μL of 50% ethanol.
-
b.Protect from light.
-
a.
-
15.Prepare saturated lithium carbonate.
-
a.Add 10 mg lithium carbonate to 1 mL ddH2O and vortex well.
-
b.Spin down and stand for 5 min.
-
c.Use the supernatant as saturated lithium carbonate.
-
a.
Note: 0.002% FSB solution and the saturated lithium carbonate solution should be prepared fresh just before the assay.
Preparation for Aβ42 immunostaining
To prepare PBST and 50% glycerol/PBS
Timing: < 30 min
To prepare 10% formic acid
Timing: < 10 min, just before the primary antibody reaction
To prepare blocking solution
Timing: < 10 min, just before the primary and secondary antibody reactions
-
16.
Prepare PBST and 50% glycerol/PBS.
(Refer to materials and equipment)
-
17.
Prepare 10% formic acid.
(Refer to materials and equipment)
-
18.
Prepare blocking solution.
(Refer to materials and equipment)
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-β-amyloid, 1–16 antibody, clone 6E10 (mouse ascites) | BioLegend (Signet, Covance) | Cat# SIG-39300-1000; RRID: AB_662809 |
| Amyloid β (N) (82E1) Anti-Human Mouse IgG MoAb antibody | Immuno-Biological Laboratories | Cat# 10323; RRID: AB_10707424 |
| Sheep Anti-Mouse IgG, Whole Ab ECL Antibody, HRP Conjugated | GE Healthcare | Cat# NA931; RRID: AB_772210 |
| Goat Anti-Mouse IgG H&L (Alexa Fluor 488) preabsorbed Antibody | Abcam | Cat# ab150117; RRID: AB_2688012 |
| Chemicals, peptides, and recombinant proteins | ||
| Agar (for fly food) | Ina Food Industry Co., Ltd. | Cat# S-6 |
| Sucrose (for fly food) | Nissin Sugar Co., Ltd. | N/A |
| Potassium tartrate | Sigma-Aldrich | Cat# 24-5650-5 |
| Calcium chloride | Sigma-Aldrich | Cat# 05-0580-5 |
| Glucose | Nacalai Tesque | Cat# 16805-64 |
| Corn meal (for fly food) | Oriental Yeast Co., Ltd. | Cat# 02801500 |
| Yeast (for fly food) | Mitsubishi Tanabe Pharma | Product name: Ebios |
| Methyl 4-hydroxybenzoate | FUJIFILM Wako Pure Chemical Corporation | Cat# 132-02635 |
| Ethanol | FUJIFILM Wako Pure Chemical Corporation | Cat# 057-00451 |
| Phosphoric acid | Nacalai Tesque | Cat# 27618-55 |
| Propionic acid | FUJIFILM Wako Pure Chemical Corporation | Cat# 163-04726 |
| 16% Paraformaldehyde | Electron Microscopy Sciences | Cat# 15710 |
| Sodium bicarbonate | Sigma-Aldrich | Cat# 28-1850 |
| FSB (1-fluoro-2,5-bis(3-carboxy-4-hydroxystyryl)benzene, 1% DMSO solution) | Dojindo | Cat# F308 |
| Glycerol | FUJIFILM Wako Pure Chemical Corporation | Cat# 075-00616 |
| Lithium carbonate | Nacalai Tesque | Cat# 20619-42 |
| Formic acid | Kanto Chemical Co., Inc. | Cat# 16064-00 |
| Sodium chloride (NaCl) | Kanto Chemical Co., Inc. | Cat# 37144-86 |
| Potassium chloride (KCl) | Sigma-Aldrich | Cat# 24-4290 |
| Disodium hydrogen phosphate (Na2HPO4) | FUJIFILM Wako Pure Chemical Corporation | Cat# 193-02845 |
| Potassium dihydrogen phosphate (KH2PO4) | FUJIFILM Wako Pure Chemical Corporation | Cat# 169-04245 |
| Tris (hydroxymethyl) aminomethane (Tris) | Nacalai Tesque | Cat# 35434-21 |
| Hydrochloric acid (HCl) | FUJIFILM Wako Pure Chemical Corporation | Cat# 080-01066 |
| Sodium deoxycholate | FUJIFILM Wako Pure Chemical Corporation | Cat# 194-08311 |
| Polyoxyethylene (10) octylphenyl ether (Triton X-100) | FUJIFILM Wako Pure Chemical Corporation | Cat# 168-11805 |
| Tricine | Santa Cruz Biotechnology | Cat# SC-216103A |
| Sodium dodecyl sulfate | Nacalai Tesque | Cat# 31606-75 |
| Glycine | Nacalai Tesque | Cat# 17109-35 |
| Methanol | Kanto Chemical Co., Inc. | Cat# 25183-70 |
| Tween-20 | Kanto Chemical Co., Inc. | Cat# 40350-02 |
| Skim milk | Morinaga Milk | N/A |
| cOmplete Protease Inhibitor Cocktail | Merck | Cat# 11697498001 |
| 2-Mercaptoethanol | Nacalai Tesque | Cat# 21417-52 |
| Paraffine wax | Sakura Finetek Japan | Cat# 7810 |
| Multi mount 220 | Matsunami Glass Ind., Ltd | Cat# FM22001 |
| SlowFade™ Gold Antifade Mount | Thermo Fisher Scientific | Cat# S36936 |
| Normal goat serum | Jackson ImmunoResearch Laboratories | Cat# 005-000-121 |
| Xylene | Nacalai Tesque | Cat# 36612-93 |
| Critical commercial assays | ||
| ECL Prime Western Blotting Detection Reagents | GE Healthcare | Cat# RPN2236 |
| Novex™ Tricine SDS Sample Buffer (2X) | Thermo Fisher Scientific | Cat# LC1676 |
| Novex™ 10 to 20%, Tricine, 1.0 mm, Mini Protein Gel, 10-well | Thermo Fisher Scientific | Cat# EC6625BOX |
| Hematoxylin Solution, Mayer’s | Sigma-Aldrich | Cat# MHS16 |
| Eosin Y solution | Sigma-Aldrich | Cat# HT110132 |
| Experimental models: organisms/strains | ||
| D. melanogaster: UAS-Aβ42 | (Iijima et al., 2004; Iijima and Chiang, 2008) | N/A |
| D. melanogaster: elav-GAL4 | Bloomington Drosophila Stock Center | BDSC: 458; FlyBase: FBst0000458 |
| D. melanogaster: w1118 | Vienna Drosophila Resource Center | VDRC: 60000 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Prism 9 | GraphPad | https://www.GraphPad.com |
| Other | ||
| Auto tissue processor | Sakura Finetek Japan | Model: ETP-150C |
| Tissue Embedding Console System | Sakura Finetek Japan | Model: Tissue-Tek TEC |
| Rotary microtome | Yamato Kohki Industrial Co., Ltd. | Model: RX-860 |
| Microtome blade | Feather | Cat# S35 |
| Forceps | DUMONT | Cat# 0108-5-PO |
| Vacuum centrifuge concentrator | Eppendorf | Model: Vacufuge |
Materials and equipment
Equipment
We use the following equipment for preparation of paraffin sections.
Auto Tissue Processor (ETP-150C, Sakura Finetek).
Tissue Embedding Console System (Tissue-Tek TEC, Sakura Finetek).
Rotary microtome (RX-860, YAMATO KHOKI).
Microtome blade (S35, Feather).
Recipes
Fly food
| Reagent | Final concentration | Amount |
|---|---|---|
| Agar | 6.4 g/L | 70 g |
| Sucrose | 26.5 g/L | 292 g |
| Potassium tartrate | 7.4 g/L | 81 g |
| Calcium chloride | 0.6 g/L | 6.7 g |
| Glucose | 53 g/L | 583 g |
| Corn meal | 64.3 g/L | 707 g |
| Yeast | 27 g/L | 297 g |
| ddH2O | - | 10 L |
This recipe makes about 11 L of fly food.
Preservative for fly food
| Reagent | Amount |
|---|---|
| Methyl 4-hydroxybenzoate | 15 g |
| Ethanol | 67 mL |
Freshly prepare before use.
Acid mixture for fly food
| Reagent | Amount |
|---|---|
| Phosphoric acid | 5 mL |
| Propionic acid | 50 mL |
| ddH2O | 62 mL |
Freshly prepare before use.
1X PBS (pH 7.4)
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 137 mM | 8 g |
| KCl | 2.7 mM | 0.2 g |
| Na2HPO4 | 10 mM | 1.44 g |
| KH2PO4 | 1.8 mM | 0.24 g |
| ddH2O | n/a | Add up to 1 L |
| Total | n/a | 1 L |
Dissolve and store at room temperature (20°C–25°C) for a month.
4% PFA/PBS
| Reagent | Final concentration | Amount |
|---|---|---|
| 16% Paraformaldehyde | 4% | 100 μL |
| PBS | n/a | 300 μL |
| Total | n/a | 400 μL |
Freshly prepare before use.
CRITICAL: Paraformaldehyde is a hazardous chemical. Handle under fume hood and dispose of paraformaldehyde waste following proper procedure.
0.1% sodium bicarbonate solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium bicarbonate | 0.1% | 0.5 g |
| ddH2O | n/a | 500 mL |
| Total | n/a | 500 mL |
Freshly prepare before use.
1M Tris-HCl buffer (pH 8.0)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 1 M | 12.1 g |
| HCl | Titrate to pH 8.0 | n/a |
| ddH2O | n/a | Add up to 100 mL |
| Total | n/a | 100 mL |
Dissolve and titrate to pH 8.0 with HCl at room temperature (20°C–25°C) for a month.
RIPA buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 1M Tris-HCl buffer (pH 8.0) | 50 mM | 10 mL |
| Sodium deoxycholate | 0.5% | 1 g |
| Triton X-100 | 1% | 2 mL |
| NaCl | 150 mM | 1.75 g |
| ddH2O | n/a | Add up to 200 mL |
| Total | n/a | 200 mL |
Dissolve and store at 4°C for a month.
Tris-Tricine running buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 0.1 M | 12.1 g |
| Tricine | 0.1 M | 17.9 g |
| Sodium dodecyl sulfate | 0.1% | 1 g |
| ddH2O | n/a | Add up to 1 L |
| Total | n/a | 1 L |
Dissolve and store at room temperature (20°C–25°C) for a month.
CRITICAL: Sodium dodecyl sulfate is a hazardous chemical. Handle with care.
Transfer buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 96 mM | 7.2 g |
| Glycine | 12 mM | 1.46 g |
| Methanol | 20% | 200 mL |
| ddH2O | n/a | Add up to 1 L |
| Total | n/a | 1 L |
Dissolve and store at 4°C for 6 months. 20% methanol waste must be disposed following proper procedure.
1X TBST
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl, pH 7.4 | 96 mM | 10 mL |
| 5 M NaCl | 12 mM | 20 mL |
| Tween-20 | 0.1% | 1 mL |
| ddH2O | n/a | Add up to 1 L |
| Total | n/a | 1 L |
Dissolve and store at room temperature (20°C–25°C) for a week.
5% skim milk/TBST
| Reagent | Final concentration | Amount |
|---|---|---|
| Skim milk | 5% | 5 g |
| TBST | n/a | 100 mL |
| Total | n/a | 100 mL |
Freshly prepare before use.
RIPA/SDS
| Reagent | Final concentration | Amount |
|---|---|---|
| RIPA buffer | n/a | 9.1 mL |
| cOmplete Protease Inhibitor Cocktail (25X solution) | 1X | 0.4 mL |
| 20% Sodium dodecyl sulfate | 1% | 0.5 mL |
| Total | n/a | 10 mL |
Freshly prepare before use.
70% Formic acid
| Reagent | Final concentration | Amount |
|---|---|---|
| Formic acid (98%–99%) | 70% | 0.7 mL |
| ddH2O | n/a | 0.3 mL |
| Total | n/a | 1 mL |
Freshly prepare before use.
CRITICAL: Formic acid is a hazardous chemical. Handle under a fume hood.
2X Sample buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Novex™ Tricine SDS Sample Buffer (2X) | n/a | 910 μL |
| cOmplete Protease Inhibitor Cocktail (25X solution) | 1X | 40 μL |
| 2-mercaptoethanol | 5% | 50 μL |
| Total | n/a | 1 mL |
Freshly prepare before use.
0.5% PBST
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | n/a | 1 L |
| Triton X-100 | 0.5% | 5 mL |
| Total | n/a | 1 L |
Dissolve and store at room temperature (20°C–25°C) for a month.
50% Glycerol/PBS
| Reagent | Final concentration | Amount |
|---|---|---|
| Glycerol | 50% | 5 mL |
| PBS | n/a | Add up to 10 mL |
| Total | n/a | 10 mL |
Dissolve and store at room temperature (20°C–25°C) for 6 months.
10% Formic acid
| Reagent | Final concentration | Amount |
|---|---|---|
| Formic acid (98%–99%) | 10% | 0.1 mL |
| ddH2O | n/a | 0.9 mL |
| Total | n/a | 1 mL |
Freshly prepare before use.
CRITICAL: Formic acid is a hazardous chemical. Handle under a fume hood.
Blocking solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Normal goat serum | 10% | 50 μL |
| 20% Triton X-100 | 1% | 25 μL |
| PBS | n/a | 425 μL |
| Total | n/a | 500 μL |
Freshly prepare before use.
Step-by-step method details
Collection of virgin female flies
Timing: > 2 weeks
To obtain flies expressing Aβ42 pan-neuronally (experimental group: elavc155-GAL4/Y; UAS-Aβ42/+) or control flies (elavc155-GAL4/Y), we use the GAL4-UAS transgene expression system with the pan-neuronal elavc155-GAL4 driver (Bloomington stock #458). Because the elavc155-GAL4 transgene is located on the X chromosome, we collect virgin females from elavc155-GAL4 transgenic flies and cross them with male UAS-Aβ42 transgenic flies or parental w1118 flies to maximize the chance of obtaining the genetic targets.
-
1.
Estimate required number of experimental (elav-GAL4/Y; UAS-Aβ42/+) and control (elav-GAL4/Y) flies for the experiments (please refer to sections below for details) and write down the cross scheme to obtain a sufficient number of flies with the genetic targets.
-
2.
Prepare bottles of elavc155-GAL4 flies for collection of virgin females and bottles of UAS-Aβ42/CyO or w1118 flies for male collection. The number of bottles required depends strictly on the culture conditions in your lab. The culture condition in our lab is all under 24°C–25°C, 40%–50% humidity with 12 h light/dark cycle, if no further specification.
-
3.
(Optional) If necessary, transfer parental (F0) flies to new bottles for further virgin female collection.
-
4.
Adult flies will eclose around day 10–11. Collect virgin females of elavc155-GAL4 and males of UAS-Aβ42/CyO or w1118 until you obtain a sufficient number of flies for the cross.
-
5.
(Optional) Before starting the cross, place 15–20 virgin females or males in plastic vials (25 mm diameter) containing food and maintain the vials in a horizontal position for 4–5 days to make sure that no larva is hatched in the virgin female vials. Troubleshooting
CRITICAL: When collecting virgin females, please make sure not to select non-virgin females or male flies. If you are not confident, please add step 5 above to minimize a risk.
Set up the cross to obtain Aβ42 flies
Timing: 4–5 days
This section describes the cross used to obtain flies expressing Aβ42 pan-neuronally (experimental group: elavc155-GAL4/Y; UAS-Aβ42/+) or control flies (elavc155-GAL4/Y).
-
6.
Place ⁓30 virgin females of elavc155-GAL4 and ⁓15 males of UAS-Aβ42/CyO (experimental group) or w1118 (control group) into the bottles.
-
7.
Culture the bottles.
-
8.
After 4–5 days, depending on the culture conditions in your lab, discard or transfer the parental (F0) flies to new bottles to increase the number of bottles.
Collection of Aβ flies
Timing: < 10 days since the eclosion starts
From the crosses described above, we usually collect and use males for experiments because they express higher levels of Aβ42 peptides than females due to the dosage compensation of X chromosome. Consequently, adult males exhibit more robust phenotypes.
-
9.
Ten days after setting up the cross, start to collect F1 males from the experimental group (elavc155-GAL4/Y; UAS-Aβ42/+) or control group (elavc155-GAL4/Y). Anesthetize flies with CO2 gas and carefully sort them against the CyO balancer.
-
10.
Place 25 male flies in plastic vials (25 mm diameter) containing fly food and maintain the vials in a horizontal position. These flies are 0 day old.
-
11.
Change the vials every 3–4 days to obtain age-matched flies from the experimental and control groups for each experiment.
CRITICAL: When collecting flies from the bottle, please make sure to finish F1 fly collection within 10 days to avoid contamination by the subsequent F2 generation. Also, when separating flies under CO2 gas, minimize time and be careful not to physically damage the flies. These factors significantly affect observed phenotypes in flies.
Longevity assay
Timing: 2 months (age of flies: starting from eclosion and all through life)
To precisely control the age of flies, collect male flies for the experimental and control groups every day. To obtain consistent results, it is desirable to use 200 flies for each group.
-
12.Collect male flies from the experimental group (elavc155-GAL4/Y; UAS-Aβ42/+) or control group (elavc155-GAL4/Y), and place 25 flies into a plastic vial (25 mm diameter) containing fly food.
-
a.Collect more than 200 flies (25 flies/vial × 8 vials).
-
b.Maintain the vials in a horizontal position and age the flies.
-
a.
-
13.
Tap the flies onto the bottom of the food vials and transfer them to fresh food vials every 2 or 3 days. Count the number of dead flies at that time. You do not have to anesthetize flies in this step.
-
14.
Continue to score ages and the numbers of dead flies until the end of the experiment.
-
15.
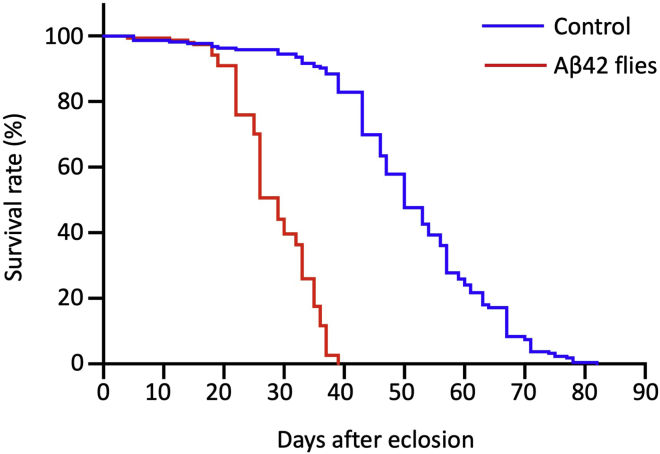
Analyze the survival of Aβ42 flies using the Kaplan–Meier method and compare with the survival of control flies (Figure 2).
Note: In general, longevity assays require at least 100 flies, but a larger number (100–200 flies) is preferable because it is more likely to yield consistent results.
CRITICAL: The lifespan of Drosophila is affected by temperature, diet, and many other factors. These environmental factors should be carefully controlled to obtain consistent results. In addition, the lifespan of Drosophila is strongly affected by its genetic background. To avoid any off-target effects, it is important to use a parental line for a transgenic or mutant line as a control to adjust genetic background. If you are not able to obtain an appropriate parental line, it is critical to outcross a transgenic or mutant line of interests to a control line used in your laboratory for 5 generations before starting the experiments. This is true for all of the phenotypic analyses described below.
Figure 2.
Survival curves of control and Aβ42 flies
Median lifespans (Ages, d, days after eclosion; Control; 50 d, Aβ42 flies; 29 d), p < 0.001 by Kaplan–Meier survival analyses with log-rank test (Control; n = 216, Aβ42 flies; n = 154). Experiments were repeated more than three times, and a representative result is shown (Iijima et al., 2004). The genotypes of the flies are as follows: (Control): elav-GAL4/Y, (Aβ42): elav-GAL4/Y; UAS-Aβ42/+.
Forced climbing assay
Timing: 3–4 weeks (age of flies: 1 week after eclosion)
Aβ42 flies exhibit age-dependent defects in climbing. We usually carry out a forced climbing assay once or twice a week, from 1 week old to 4 weeks old flies.
-
16.Day 1: Collect male flies from the experimental group (elavc155-GAL4/Y; UAS-Aβ42/+) or control group (elavc155-GAL4/Y) and age them.
-
a.Collect 125–150 flies and place 25 flies into a plastic vial (25 mm diameter) containing fly food. Prepare 5–6 vials.
-
b.Tap the flies to the bottom of the food vials and transfer them to fresh fly food vials every 2 or 3 days.
-
c.Perform a climbing assay once or twice per week, usually starting at 1 week old.
-
a.
-
17.
Day 7: Tap the flies onto the bottom of the food vials and transfer them to new empty plastic vials (25 mm diameter) marked with genotype and scale representing height from the bottom (Figure 1). If you plan to reuse the vials, gently wash them with warm water after each assay and dry them well to keep the vials clean. Please be careful not to scratch or damage the inside wall of the vials, which may affect the climbing performance of flies.
-
18.
To acclimate the flies to the environment, lay down the plastic vials containing the flies and leave them as they are for 5 min.
-
19.Perform the 1st trial.
-
a.Tap the vials against a foam pad or layered Kimtowels (3–4 vials at once) and knock the flies to the bottom.
-
b.Stand the vials in front of a white board. Healthy flies will immediately climb up along the wall of plastic vials (i.e., they will exhibit negative geotaxis).
-
c.After 10 sec, take a picture of the flies distributed in the vials (Figure 3). We set up the recording time as 10 sec because most control flies can climb to the top of the wall within this period of time. The recording time can be changed depending on your experimental conditions.
-
d.Place the vials in a horizontal position and leave them for 3–5 min.
-
a.
-
20.Perform the 2nd–5th trials.
-
a.Repeat steps 19-a–d.
-
a.
-
21.
After the assay, transfer the flies back into the original plastic vials containing fly food and maintain them.
-
22.
Transfer the flies to new plastic vials containing fresh fly food every 2 or 3 days until the next assay.
-
23.
Day 14: Repeat steps 17–22 once or twice per week. The endpoint of the climbing assay is when the Aβ42 flies stop climbing up the wall or when more than 40% of flies (10 in 25 flies) die in the food vial.
-
24.Analysis
-
a.Count the flies distributed in the top, middle, and bottom parts of the vials using the captured photo (Figure 3).
-
b.Calculate the percentages of flies distributed in each part of the vials and calculate the average of five trials for each vial.
-
c.Calculate the average percentages and standard errors of five or six vials for each genotype or treatment.
-
d.Compare percentages of flies that stayed in the bottom part of the vial among genotypes or treatments and perform statistical analyses using Student’s t-test (Figure 4).
-
a.
Note: The climbing assay requires 125–150 flies (n=5–6) to obtain consistent results. The endpoint may vary depending on the health and survival rate of the flies under the experimental conditions. From our experience, survival rates in each vial should be greater than 60% to obtain consistent results.
Figure 3.
Climbing assay
Vials containing about 25 flies were gently tapped to knock all flies to the bottom. After 10 sec, flies in the top, middle, and bottom thirds of the vial were imaged and counted.
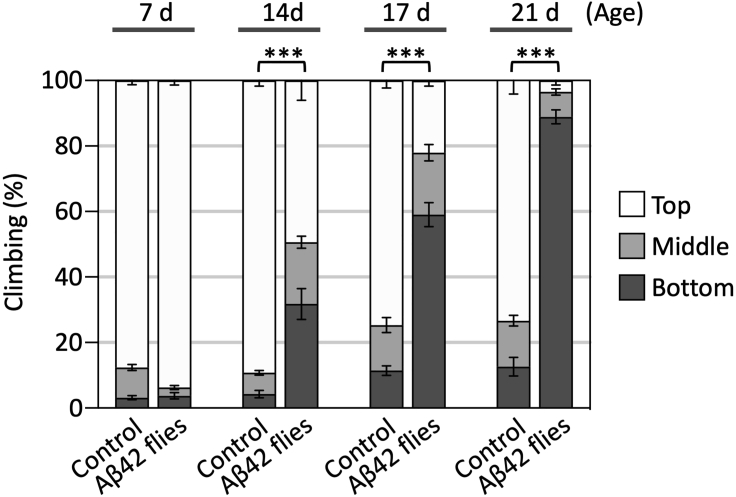
Figure 4.
Climbing assay data
Aβ42 flies exhibit age-dependent climbing deficits. Data show average percentages of flies that climbed to the top (white) or middle (light gray) of the vials or stayed at the bottom (dark gray). Ages (d, days after eclosion) are indicated on the top of the graph. Percentages of flies that stayed at the bottom were subjected to statistical analyses. Means ± SEM, n = 8; ∗∗∗p < 0.001 by Student’s t-test. The genotypes of the flies were as follows: (Control): elav-GAL4/Y, (Aβ42): elav-GAL4/Y; UAS-Aβ42/+.
Histological analysis
Timing: > 4 days (Age of flies: 3–4 weeks after eclosion)
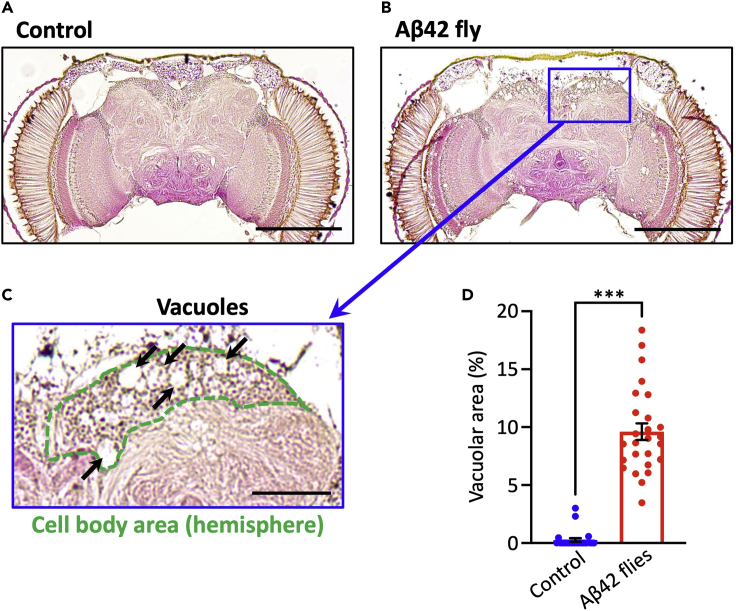
Under our experimental conditions, Aβ42 flies exhibit age-dependent neurodegeneration in the brain at the age of 4 weeks. Hematoxylin and Eosin (H&E) staining of paraffin section enable visualization of neurodegeneration in fly brains. Neuron loss and neuropil degeneration appear as vacuolation (Kretzschmar et al., 1997) in the cell body area (purple in H&E staining) or neuropil area (pink in H&E staining). We usually focus on the Kenyon cell body area, as neuron loss is evident when Aβ42 is expressed under the elavc155-GAL4 driver, and these neurons are important for learning and memory in Drosophila (Dubnau et al., 2001).
-
25.
Collect male flies from the experimental group (elavc155-GAL4/Y; UAS-Aβ42/+) or control group (elavc155-GAL4/Y) and age them until the age of 4 weeks.
-
26.Day 1: Dissect the flies.
-
a.Anesthetize the fly using CO2 gas.
-
b.Rinse the fly in 100% ethanol to wash out the wax, and place the fly in an embryo dish filled with PBS.
-
c.Under the stereomicroscope with light sources, keep the fly face-up using “No. 5” forceps (inox 08, DUMONT) and remove the proboscis.
-
d.Remove the air sac from the site of the proboscis to easily immerse the brain in fixative.
-
e.Remove the head from the abdomen and place it in the fixative.
-
a.
-
27.
Fix fly heads with 400 μL of 4% PFA/PBS in 1.5 mL microtube at 4°C for 16–20 h.
-
28.Day 2: Processing.
-
a.The next day, remove the fly heads from the 1.5 mL microtube using a disposable dropper and place them into the mesh bag (six fly heads/bag).
-
b.Remove the fixative and place the bag in the tissue processing cassette (Figure 5A).
-
c.Set the cassette in the tissue processor and begin processing (see Table 1 for the processing program). This takes 5–6 h.
-
a.
-
29.Embedding.
-
a.Turn on the cryo-plate to quickly cool the paraffin blocks.
-
b.Remove the processing cassettes from the tissue processor and keep them in a paraffin bath (paraffin embedding center) at 60°C.
-
c.Place the fly heads and melting paraffin into the stainless base mold on the heat block.
-
d.Align the fly heads, keeping the anterior side up, and transfer the mold containing the fly heads to the cold plate to hold the head in position.
-
e.Place the tissue cassette on top of the mold filled with paraffin.
-
f.Place the mold on the cooled cryo-plate until the paraffin completely solidifies (this takes 20–30 min).
-
g.After cooling, remove paraffin blocks from the mold and keep them at room temperature (20°C–25°C), (Figure 5B).
-
a.
-
30.Day 3: Sectioning.
-
a.Turn on the water bath filled with distilled water (38°C–40°C).
-
b.Trim the extra paraffin (Figure 5C) and soak the block in distilled water to avoid static electricity during sectioning.
-
c.Place the blade (S35, Feather) at an angle of 5°.
-
d.Cut sections of fly brain with a thickness of 6 μm serially from the posterior side to the anterior side of the fly head. Serial sections should be prepared as ribbons of sections (Figure 5D).
-
e.Cut the ribbons of sections (10–12 sections) and float them on warmed distilled water (38°C–40°C) in the water bath.
-
f.When the sections completely flatten out on the water, pick up the ribbon of sections using a slide glass (Figure 5E).
-
g.Stand the slide for more than 12 h at room temperature (20°C–25°C) to dry out the ribbon of sections (Figure 5F).
-
a.
-
31.Day 4: H&E staining.
-
a.Place the slides in a slide rack.
-
b.Soak the slides in the reagents for the appropriate time in the order listed in Table 2.
-
c.Remove the slide from xylene and place it in mounting media (Multi mount 220) under a coverslip.
-
d.Cover the fly heads sections.
-
a.
-
32.Analysis and Quantification (H&E-stained sections are analyzed using a microscope equipped with a camera (Axio Lab.A1, AxioCam 105 color; Zeiss)).
-
a.Observe all slides with serial sections of fly heads under the microscope.
-
b.Select the brain section with the most severe vacuolation in the same area of each brain, and capture an image of the section using the microscope camera (usually, this is done with a 10X objective).
-
c.Using the ImageJ software, analyze the total cell body area and all vacuole areas in the selected brain region.
-
d.Calculate the percentage of the vacuole area: 100∗ (vacuole area/total cell body).
-
e.Subject the percentage of vacuole area in Aβ42 flies and control flies to statistical analyses using Student’s t-test (Figure 6).
-
a.
Note: Analysis of neurodegeneration in Aβ42 fly brains requires at least 10 flies (n=10 × 2 hemispheres) to obtain consistent results. The severity of neurodegeneration may vary depending on the health and survival rate of flies under experimental conditions.
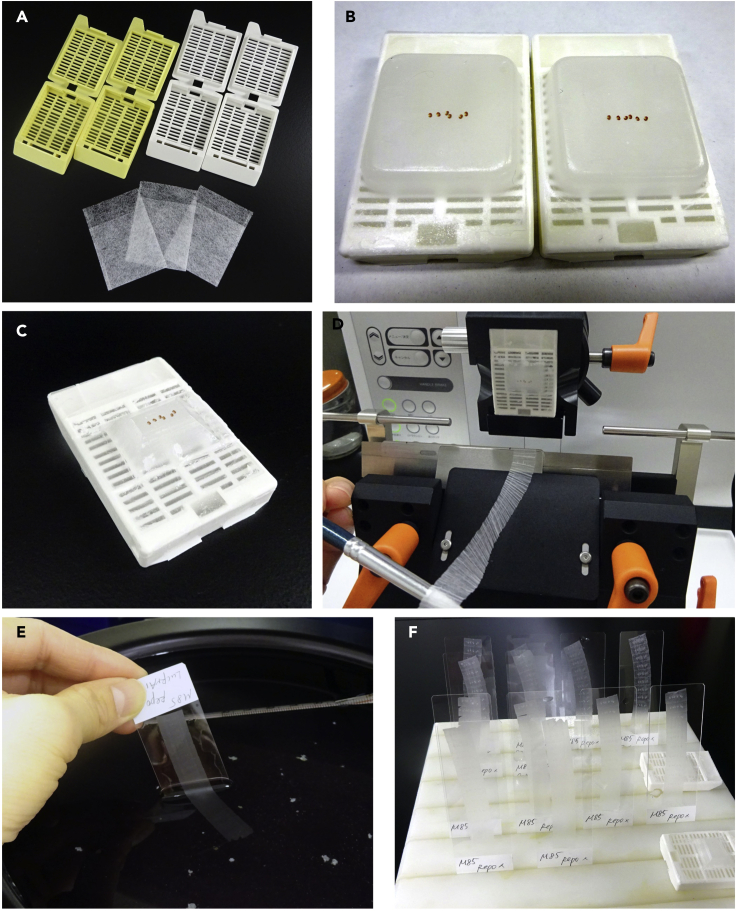
Figure 5.
Paraffin sectioning of fly heads
(A) Tissue processing cassettes and mesh bags.
(B) Paraffin-embedded fly heads.
(C) Trimming extra paraffin in the block.
(D) Making the ribbon of paraffin sections.
(E) Picking up the sections.
(F)Serial sections of fly heads on slide glasses (drying step).
Table 1.
Fly heads processing program for paraffin blocks
| Solution | Time | Temp. |
|---|---|---|
| 70% Ethanol | 20 min | 20°C–25°C |
| 80% Ethanol | 20 min | 20°C–25°C |
| 95% Ethanol | 20 min | 20°C–25°C |
| 100% Ethanol | 20 min | 20°C–25°C |
| 100% Ethanol | 20 min | 20°C–25°C |
| 100% Ethanol | 20 min | 20°C–25°C |
| Xylene (or Histo-clear) | 20 min | 20°C–25°C |
| Xylene (or Histo-clear) | 40 min | 20°C–25°C |
| Paraffin wax | 20 min | 60°C |
| Paraffin wax | 30 min | 60°C |
| Paraffin wax | 30 min | 60°C |
Table 2.
Hematoxylin and Eosin staining
| Step | Solution | Time | Remarks |
|---|---|---|---|
| Deparaffinization | Xylene (or Histo-clear) | 6 min | |
| Xylene (or Histo-clear) | 6 min | ||
| 100% Ethanol | 3 min | ||
| 100% Ethanol | 3 min | ||
| 95% Ethanol | 3 min | ||
| 80% Ethanol | 3 min | ||
| Distilled water | 6 min | ||
| Hematoxylin staining | Hematoxylin solution | 3–5 min | |
| Washing | Tap water | 1 min | |
| Tap water | 1 min | ||
| Tap water | 2 min | ||
| Coloring | 0.1% Sodium bicarbonate solution | 1 min | Do not agitate |
| Washing | Tap water | 1 min | |
| Distilled water | 2 min | ||
| Eosin staining | Eosin Y solution | 30–45 sec | |
| Washing | 95% Ethanol | 2 min | |
| 95% Ethanol | 2 min | ||
| 95% Ethanol | 2 min | ||
| Dehydration | 100% Ethanol | 3 min | Use solvents dehydrated by molecular sieve |
| 100% Ethanol | 3 min | ||
| Xylene (or Histo-clear) | 10 min | ||
| Xylene (or Histo-clear) | >10 min |
Figure 6.
Neurodegeneration in Aβ42 fly brains
Aβ flies exhibit age-dependent neurodegeneration in the cell body areas of the brains
(A–C) Representative images show paraffin-embedded, H&E-stained brain sections of control (A) and Aβ42 flies (B and C), respectively. Scale bars represent 200 μm (A and B) and 50 μm (C). Vacuolar areas are indicated by arrows, and the cell body area (hemisphere) is surrounded by a green dotted line.
(D) Percentages of vacuolar areas were analyzed. Means ± SEM, n = 24–26 hemispheres, ∗∗∗p < 0.001 by Student’s t-test. The genotypes of the flies are as follows: (Control) elav-GAL4/Y, (Aβ42): elav-GAL4/Y; UAS-Aβ42/+. Flies were 3 weeks old.
Detection of Aβ42 by western blot
Timing: > 3 days (Age of flies: 1–4 weeks after eclosion)
Aβ42 peptides expressed in fly heads aggregate and accumulate upon aging; however, it is not possible to distinguish between soluble Aβ42 and β-sheet–rich insoluble Aβ42 by conventional western blotting. Using this fractionation method for fly head lysates, soluble and insoluble Aβ42 can be quantified separately. Detergent-soluble and -insoluble Aβ42 can be sequentially extracted from fly heads by RIPA/SDS buffer followed by formic acid treatment.
-
33.
Collect male flies from the experimental group (elavc155-GAL4/Y; UAS-Aβ42/+) or control group (elavc155-GAL4/Y) and age them.
-
34.
At the age of interest, anesthetize flies with CO2 gas and place them in a 1.5 mL tube followed by flash-freezing in liquid nitrogen. The flash-frozen flies are stored at −80°C until use.
-
35.RIPA/SDS extraction of Aβ42 from fly heads.
-
a.Place the 1.5 mL tubes containing 20–25 frozen flies in a Dewar vessel containing liquid nitrogen.
-
b.Pick up a tube using forceps and quickly place it in a 50 mL conical centrifuge tube.
-
c.Shake the tubes to physically divide fly heads and abdomens.
-
d.Remove the contents of the 1.5 mL tube onto weighing paper (or a weighing dish) on ice and collect 20 fly heads in a new 1.5 mL tube.
-
e.Add 200 μL RIPA/SDS buffer to the tube and homogenize using a micro-pestle.
-
f.Incubate on ice for 1 h (vortex the tube every 20 min).
-
g.Spin the tubes at 16,000 × g for 20 min at 4˚C.
-
h.Transfer the supernatant (RIPA/SDS soluble fraction) to a new tube.
-
a.
Pause point: RIPA/SDS soluble fractions can be stored at −80°C for 1 week.
-
36.Formic acid extraction of Aβ42 from fly heads.
-
a.Add 400 μL RIPA/SDS buffer to the pellet and vortex (washing the pellet).
-
b.Spin the tubes at 16,000 × g for 20 min at 4˚C.
-
c.Carefully discard the supernatant and add 100 μL of 70% formic acid to the pellet.
-
d.Incubate on ice for 1 h, vortexing frequently.
-
e.Spin the tubes at 16,000 × g for 20 min at 4˚C.
-
f.Transfer the supernatant to new tubes.
-
g.Evaporate the formic acid using a vacuum centrifuge concentrator (Vacufuge, Eppendorf).
-
h.Resuspend the pellet in 40 μL DMSO (RIPA/SDS insoluble/formic acid-soluble fraction).
-
a.
Pause point: RIPA/SDS-insoluble/formic acid-soluble fractions can be stored at −80°C for 1 week.
-
37.Sample preparation for western blot.
-
a.Add 10 μL of 2X sample buffer to 10 μL RIPA/SDS soluble fraction per lane (1 head/lane).
-
b.Add 18 μL of 2X sample buffer to 2 μL RIPA/SDS insoluble fraction per lane (1 head/lane).
-
c.Heat the samples at 95˚C for 2 min to reduce and denature proteins and peptides.
-
d.Spin the tubes at 16,000 × g for 5 min at room temperature (20°C–25°C) and collect the supernatant.
-
a.
-
38.SDS-PAGE.
-
a.Run the gel using Novex Tris-Tricine 10%–20% gradient gel according to the manufacturer’s instructions. (https://tools.thermofisher.com/content/sfs/manuals/surelock_man.pdf).
-
a.
-
39.
Transfer the proteins onto nitrocellulose membrane using an XCell II™ Blot Module (Thermo Fisher Scientific). This takes 2 h at 100 mA constant current.
-
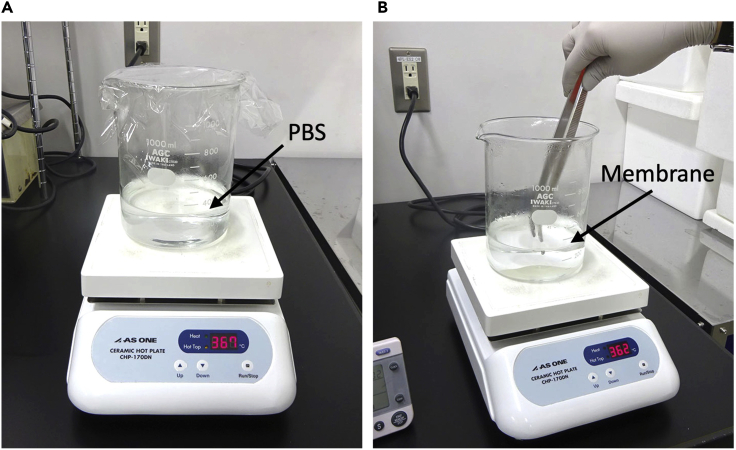
40.After the transfer, boil the membrane in PBS for 3 min. Troubleshooting
-
a.Boil 300 mL PBS in a 1 L beaker using a ceramic hot plate.
-
b.Put the nitrocellulose membrane in boiling PBS with the protein side down.
-
c.Boil the membrane for 3 min while holding it down with forceps (Figure 7).
-
a.
-
41.
Block the membrane with 5% skim milk in TBST for 1 h at room temperature (20°C–25°C).
-
42.
After washing the membrane with TBST for 5 min, incubate the membrane with anti-Aβ antibody (6E10, 1:4,000) at 4˚C for 16–20 h.
-
43.
After washing the membrane with TBST (3 × 5 min), incubate it for 2 h at room temperature (20°C–25°C) with anti-mouse IgG antibody conjugated to HRP (1:4,000).
-
44.
After washing the membrane with TBST (3 × 5 min), incubate it with ECL reagent.
-
45.
Detect the Aβ using a chemiluminescence image analyzer.
CRITICAL: Boiling the nitrocellulose membrane in PBS for 3 min is a critical step that increases the reactivity of Aβ42 to the anti-Aβ antibody (6E10).
Figure 7.
Boiling nitrocellulose membrane in PBS
(A) Boiling 300 mL PBS in 1 L beaker using ceramic hot plate.
(B) Boiling nitrocellulose membrane in PBS.
Visualization of Aβ42 in fly brains by whole mount immunostaining
Timing: > 3 days (Age of flies: 2–4 weeks after eclosion)
Aβ42 peptides expressed in fly brains can be visualized by whole mount immunostaining.
-
46.
Collect male flies from the experimental group (elavc155-GAL4/Y; UAS-Aβ42/+) or control group (elavc155-GAL4/Y) and age them.
-
47.Day 1: At the age of interest, dissect the brains (Methods video S1).
-
a.Anesthetize the fly using CO2 gas.
-
b.Rinse the fly in 100% ethanol to wash out the wax, and place the fly in an embryo dish filled with PBS.
-
c.Dissect the fly brain under a stereomicroscope with light sources.
-
d.Fix fly brains in 400 μL of 4% PFA/PBS in a 1.5 mL microtube for 1 h at room temperature (20°C–25°C).
-
a.
-
48.Primary antibody reaction.
-
a.After fixation, remove the fixative and rinse three times with PBST.
-
b.Wash with PBST (2 × 10 min) to permeabilize.
-
c.After removing PBST, add 200 μL of 10% formic acid and incubate with gentle shaking for 1 h at room temperature (20°C–25°C). Troubleshooting
-
d.Wash with PBST (2 × 10 min).
-
e.Remove the PBST and add 100 μL blocking solution (10% NGS/1% Triton X-100/PBS).
-
f.Incubate and gently shake for at least 10 min at room temperature (20°C–25°C).
-
g.Add 0.5 μL anti-Aβ antibody (82E1, 1:200) to the tube containing blocking solution and rotate at 4°C at least 16 h (this step can be 2–3 days).
-
a.
-
49.Day 2: Secondary antibody reaction.
-
a.Remove primary antibody reaction mixture and rinse three times with PBST.
-
b.Wash with PBST (3 × 15 min with gentle shaking).
-
c.Remove the PBST and add 100 μL blocking solution.
-
d.Incubate and gently shake for at least 10 min at room temperature (20°C–25°C).
-
e.Add 0.2 μL Alexa Fluor 488-conjugated anti-mouse IgG antibody (1:500) to the tube containing blocking solution and rotate in the dark at 4°C at least 16 h (this step can be 2–3 days).
-
a.
-
50.Day 3: Mount.
-
a.Remove secondary antibody reaction mixture and rinse three times with PBST.
-
b.Wash with PBST (3 × 15 min with gentle shaking).
-
c.Rinse with PBS.
-
d.Rinse with 50% glycerol/PBS.
-
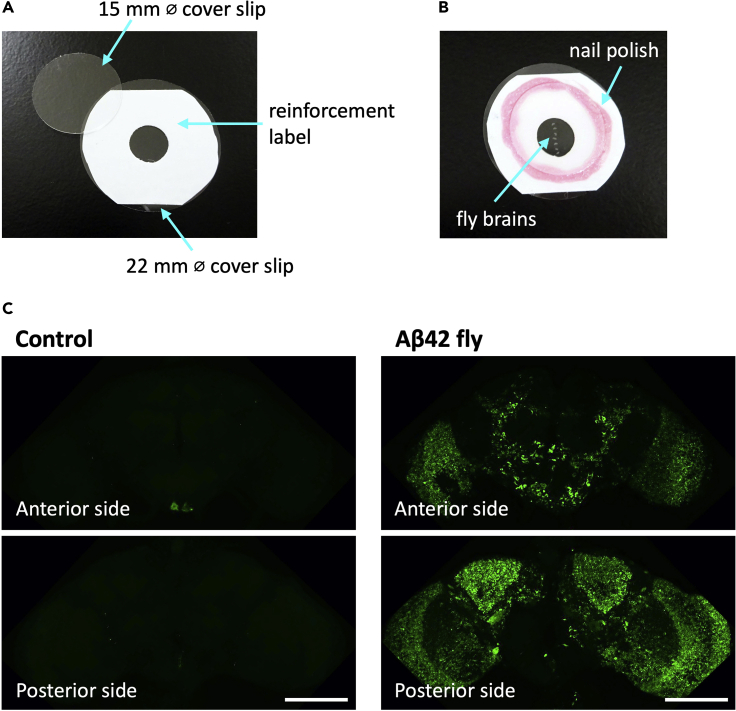
e.Prepare a 24 mm diameter round glass coverslip with a reinforcement label for a spacer (Figure 8A).
-
f.Transfer the fly brains to the center of the reinforcement label on the cover slip and align them, keeping the anterior side up.
-
g.Remove excess solution and fill with antifade mounting media (SlowFade Gold).
-
h.Cover the brains with a 15 mm diameter round glass cover slip (Figure 8B).
-
i.Seal the edge of the coverslip with nail polish.
-
a.
-
51.
Fly brains located between coverslips can be observed by confocal microscopy from both the anterior and posterior sides by flipping the slide.
-
52.
Acquire images by confocal microscopy (Figure 8C).
CRITICAL: Alexa Fluor 488 conjugated to anti-mouse IgG antibody should be protected from light. Therefore, steps 49–50 should be performed in the dark.
Optional: Fly brains can be fixed by incubating for 16–20 h at 4°C instead of for 1 h at room temperature (20°C–25°C). Either condition is suitable for immunostaining of Aβ.
Figure 8.
Whole mount immunostaining of Aβ42 in fly brains
(A) Glass coverslips used in a whole mount staining of fly brains.
(B) Fly brains were mounted between coverslips.
(C) Fly brains were stained with anti-Aβ antibody (green). Images were acquired by confocal microscopy using a 20× objective. Scale bars represent 100 μm. Genotypes of the flies are as follows: (Control): elav-GAL4/Y, (Aβ42): elav-GAL4/Y; UAS-Aβ42/+. Flies were 2 weeks old.
Visualization of Aβ42 in fly brains by FSB staining
Timing: 2 days (Age of flies: 2–4 weeks after eclosion)
Aβ42 peptides expressed in the fly brain aggregate and accumulate with aging. FSB fluorescent dye detects β-sheet structures and visualizes the aggregated form of Aβ42 peptides in the fly brain.
-
53.
Day 1: For collection, dissection, and fixation of fly brain, please refer to steps 46 and 47 above.
-
54.Day 2: FSB staining.
-
a.Aspirate away the fixative and wash with 500 μL PBST (2 × 5 min).
-
b.Remove the PBST and stain with 100–200 μL of 0.002% FSB in 50% ethanol solution for 30 min at room temperature (20°C–25°C) in the dark.
-
c.Remove the FSB solution and rinse twice with saturated lithium carbonate solution.
-
d.Remove the solution and rinse twice with 50% ethanol.
-
a.
-
55.
Mount: Please refer to the step 50c–i.
-
56.
Acquire images by confocal microscopy using the filter set for DAPI (Figure 9).
CRITICAL: The FSB reagent should be protected from light. Therefore, steps 54 and 55 should be done in the dark.
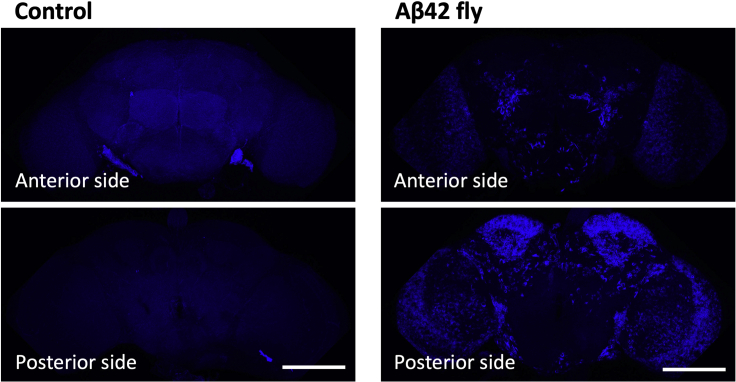
Figure 9.
FSB staining of Aβ42 fly brains
Fly brains were stained with FSB reagent (blue). Images were acquired using a 20× objective. Scale bars represent 100 μm. Genotypes of the flies are as follows: (Control): elav-GAL4/Y, (Aβ42 fly): elav-GAL4/Y; UAS-Aβ42/+. Flies were 3 weeks old.
Expected outcomes
A transgenic Drosophila expressing human amyloid-β42 (Aβ42) in neurons under the control of the pan-neuronal elavC155-GAL4 driver exhibits age-dependent locomotor deficits, neurodegeneration (vacuolation) in the brain, and premature death (Iijima et al., 2004; Iijima and Chiang, 2008). The locomotor deficits can be quantitatively assessed by the climbing assay. Climbing deficits in Aβ42 flies are evident 2–3 weeks after eclosion (Figure 4). Neurodegeneration in Aβ42 fly brains becomes prominent 4 weeks after eclosion (Figure 6). The maximum lifespan of Drosophila is around 80 days under normal growth conditions (25°C, 12 h light/dark cycle), whereas Aβ42 flies die prematurely (Figure 2, Median lifespan: control, 50 days; Aβ42 flies, 29 days). Aβ42 peptide expressed in fly brain aggregates and accumulates with aging; this can be quantitatively assessed by western blot, immunostaining, and FSB staining (Iijima et al., 2004, Figures 8C and 9).
Quantification and statistical analysis
All data analyses in each assay are described in the step-by-step method details. All experiments should be repeated at least twice, and the results from climbing assay or neurodegeneration should be expressed as means ± SEM. We use PRISM9 for the unpaired Student’s t-test and Kaplan–Meier survival analyses with log-rank tests.
Limitations
Well-conserved molecular pathways and sophisticated genetic tools make Drosophila a cost-effective and time efficient model organism for investigating molecular mechanisms underlying human diseases. In addition, recent progress in large-scale analysis of multi-omics data from patients revealed a number of candidate genes/pathways as therapeutic targets, and Drosophila models are becoming increasingly important components of validation platforms. Recently, we integrated transformative gene network modeling of Alzheimer’s disease (AD) patients with experimental validation in the Drosophila model; this analysis revealed critical gene networks responsible for AD pathogenesis, a potential therapeutic target, and a lead compound (Wang et al., 2021).
In this manuscript, we described the advantages of using our fly model in AD research. However, any animal models of human diseases have limitations. In the case of Drosophila, their superficial anatomy is quite different from human. However, many of the organs are remarkably conserved between fly and human in their origins, purposes, functions and regulatory mechanisms. For example, fly has a well-organized brain composed of several types of excitatory and inhibitory neurons and different types of glial cells similar to astrocytes, oligodendrocytes and schwann cells. Moreover, these fly glial cells cover some of important functions mediated by microglia and blood brain barrier in human brains. Thus, in order to best utilize fly models of human diseases, we believe that understanding the similarities and differences between fly and human at molecular, cellular, tissues, organ and system levels becomes important than ever before. We also expect that such cross-species analyses will yield logical therapeutic strategies to deal with various stressors, which are often associated with chronic human disease conditions. Researchers should perform a preliminary study of Drosophila biology and carefully consider whether their hypotheses and molecular targets can be validated or identified in Drosophila models.
Although fly is a simpler model organism than mouse, all phenotypes in Drosophila are affected by temperature, light, circadian rhythm, diet, and many other factors. To obtain consistent results, these environmental factors must be carefully controlled. In addition, not surprisingly, many phenotypes in Drosophila are sensitive to the genetic background. Therefore, it is crucial to set up appropriate genetic background controls to avoid any unwanted off-target effects.
Troubleshooting
Problem 1
No phenotype in Aβ42 flies (steps 15, 19–23, 32).
Potential solution
The UAS-Aβ42 or elavc155-GAL4 transgenic line may have a problem. Please check the expression levels of Aβ42 in fly heads by western blotting or immunostaining. If you do not detect Aβ42 expression, please check if each transgenic line carries expected transgene by PCR of genomic DNA.
Problem 2
The phenotypes are split in Aβ42 flies (steps 19–23, 32).
Potential solution
If you find that phenotypes are mixed or split in Aβ42 flies (for example, one half of Aβ42 flies show neurodegeneration, while the other half does not), check the expression levels of Aβ42 in both groups. If you find a difference in Aβ42 levels in two groups, there are two possible explanations for this. First, your cross may have been contaminated with non-virgin females from the elavc155-GAL4 transgenic line. In subsequent attempts, make sure to select only virgin females as described in step 5. Second, there may be a problem in the UAS-Aβ42 or elavc155-GAL4 transgenic lines (see potential solution for problem 1).
Problem 3
Unexpected premature death of control flies in the assay (steps 13 and 14).
Potential solution
Check the quality of fly food. The quality of fly food is an important factor to keep flies healthy and to produces consistent results. If fly food is not hardened completely, not dried enough during the preparation step or is stored at low temperature before use, it may produce water droplets on fly food and/or the inside wall of the vials, which may accidentally trap flies and cause premature death. Moreover, contamination by viruses, bacteria, or mold can also cause premature death. Therefore, it is critically important to keep your fly room and laboratory space clean.
Problem 4
Low or no Aβ42 signal in western blotting (step 45).
Potential solution
To detect Aβ42 using anti-Aβ antibody (6E10 or 82E1), it is critical to boil the membrane in PBS (step 40). Please DO NOT skip this step before the primary antibody reaction.
Problem 5
Low or no Aβ42 signal in whole mount immunostaining (step 52).
Potential solution
Treatment of fly brain with 10% formic acid before the primary antibody reaction (step 48c) significantly increases the Aβ42 signal (specifically, it improves the signal-to-noise ratio). It is important to use freshly prepared 10% formic acid. If the Aβ42 signal is still low, extend the incubation time up to 2 h.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Michiko Sekiya (mmsk@ncgg.go.jp) or Koichi M. Iijima (iijimakm@ncgg.go.jp).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate or analyze datasets or code.
Acknowledgments
We thank the Bloomington Drosophila Stock Center and the Vienna Drosophila Resource Center for fly stocks. This work was supported by the Japan Society for the Promotion of Science KAKENHI (JP18K07517 to M.S., JP20H03571 and JP20K20672 to K.M.I.), Research Funding for Longevity Science from the National Center for Geriatrics and Gerontology, Japan (grant numbers 19-49 to M.S., 19-7 to K.M.I., and 21-13 to M.S. and K.M.I.), and Takeda Science Foundation (to M.S. and to K.M.I.).
Author contributions
Conceptualization, methodology, and investigation, M.S. and K.M.I.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100501.
Contributor Information
Michiko Sekiya, Email: mmsk@ncgg.go.jp.
Koichi M. Iijima, Email: iijimakm@ncgg.go.jp.
References
- Iijima Koichi, Chiang Abeta42 mutants with different aggregation profiles induce distinct pathologies in Drosophila. PLoS One. 2008 doi: 10.1371/journal.pone.0001703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima K., Liu H.P., Chiang A.S., Hearn S.A., Konsolaki M., Zhong Y. Dissecting the pathological effects of human Abeta40 and Abeta42 in Drosophila: a potential model for Alzheimer’s disease. Proc. Natl. Acad. Sci. U S A. 2004;101:6623–6628. doi: 10.1073/pnas.0400895101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar D., Hasan G., Sharma S., Heisenberg M., Benzer S. The Swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J. Neurosci. 1997;17:7425–7432. doi: 10.1523/JNEUROSCI.17-19-07425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau J., Grady L., Kitamoto T., Tully T. Disruption of neurotransmission in Drosophila mushroom body blocks retrieval but not acquisition of memory. Nature. 2001;411:476–480. doi: 10.1038/35078077. [DOI] [PubMed] [Google Scholar]
- Schneider Caroline, A, Rasband, Eliceiri NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Li A., Sekiya M., D Beckmann N.D., Quan X., Schrode N., Fernando M.B., Yu A., Zhu L., Cao J. Transformative network modeling of multi-omics data reveals detailed circuits, key regulators, and potential therapeutics for Alzheimer's disease. Neuron. 2021;109:252–272. doi: 10.1016/j.neuron.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate or analyze datasets or code.