Genomic analysis of families with unexplained portal hypertension reveals GIMAP5 as a critical regulator of liver sinusoidal endothelial cells and hepatic vascular homeostasis. This study identifies a novel genetic cause of portal hypertension and provides new insight into its pathogenesis.
Abstract
Portal hypertension is a major contributor to decompensation and death from liver disease, a global health problem. Here, we demonstrate homozygous damaging mutations in GIMAP5, a small organellar GTPase, in four families with unexplained portal hypertension. We show that GIMAP5 is expressed in hepatic endothelial cells and that its loss in both humans and mice results in capillarization of liver sinusoidal endothelial cells (LSECs); this effect is also seen when GIMAP5 is selectively deleted in endothelial cells. Single-cell RNA-sequencing analysis in a GIMAP5-deficient mouse model reveals replacement of LSECs with capillarized endothelial cells, a reduction of macrovascular hepatic endothelial cells, and places GIMAP5 upstream of GATA4, a transcription factor required for LSEC specification. Thus, GIMAP5 is a critical regulator of liver endothelial cell homeostasis and, when absent, produces portal hypertension. These findings provide new insight into the pathogenesis of portal hypertension, a major contributor to morbidity and mortality from liver disease.
Introduction
Despite major advances in the diagnosis and treatment of viral causes of hepatitis (Vilarinho and Lifton, 2016), the incidence of chronic liver disease continues to rise worldwide, affecting up to 1.5 billion people globally (GBD 2017 Disease and Injury Incidence and Prevalence Collaborators, 2018) and leading to ∼2 million deaths annually (Moon et al., 2020). Because the demand for liver transplantation far exceeds the supply of available donor organs, understanding the pathogenesis of advanced liver disease and its complications will be required to develop new therapies to reduce adverse disease outcomes. Portal hypertension—increased hepatic resistance to blood flow entering the liver—is a major contributor to the morbidity and mortality of liver disease owing to development of ascites, esophageal varices, hemorrhage, and hepatic encephalopathy (Vilarinho and Lifton, 2012). While portal hypertension is commonly regarded as a simple consequence of liver damage, phenotypic changes in hepatic endothelial cells can contribute to portal hypertension. Under physiological conditions, liver sinusoidal endothelial cells (LSECs), representing the major liver endothelial cell subpopulation, contain fenestrae (nondiaphragmed pores), lack a basement membrane, and do not express CD34 (DeLeve, 2015; Poisson et al., 2017). Preceding liver fibrosis, LSECs undergo a capillarization process that results in loss of fenestration and the development of an organized basement membrane (DeLeve, 2015; Schaffner and Poper, 1963), which contribute to portal hypertension. These changes are marked by increased expression of CD34. The pathophysiology and possible genetic basis of these changes is unknown. To gain new insight into the molecular pathogenesis of portal hypertension, we have sought to identify genetic causes of portal hypertension that occur in the absence of primary disease of the liver parenchyma.
Results and discussion
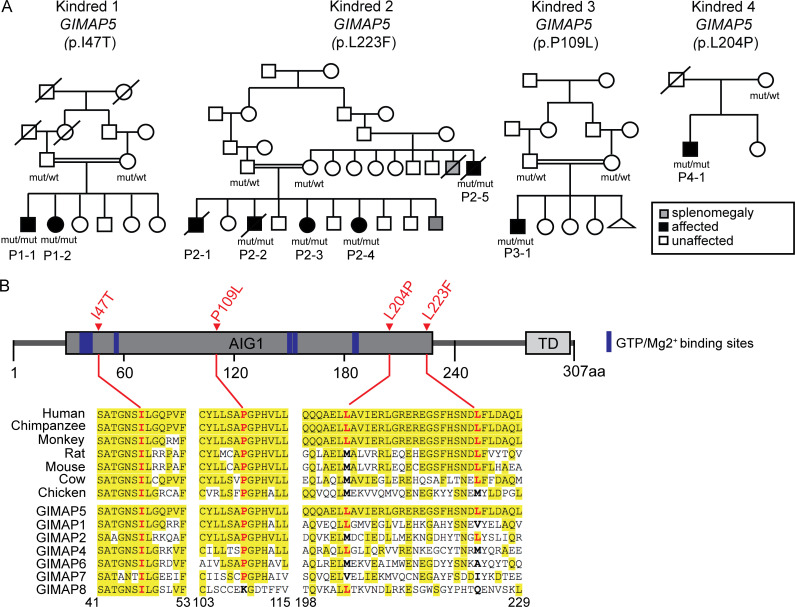
We report nine individuals from four unrelated families with unexplained portal hypertension (Fig. 1). All affected subjects have splenomegaly; thrombocytopenia, elevated transaminases, and esophageal varices were nearly universal (Table 1). Esophageal varices, a consequence of increased portal pressure, were confirmed in seven subjects by endoscopy. None had cirrhosis, splanchnic venous thrombosis, or other extrahepatic causes of portal hypertension. Notably, three affected individuals in kindred 2 are deceased; two of them died from complications of portal hypertension. Seven affected individuals from these families were subjected to whole-exome sequencing to high depth of coverage across all coding bases and flanking intronic segments, and variants were called. Variants of interest were genotyped by sequencing of PCR amplicons in all other available kindred members. Because affected members of three of the four families are consanguineous and none of the parents of affected subjects are themselves affected (Fig. 1), we sought potential causative variants under a recessive model of transmission.
Figure 1.
Rare recessive LOF mutations in GIMAP5 identified in four kindreds. (A) Pedigrees depict four unrelated families. Affected and unaffected subjects are shown as black-filled and white-filled symbols, respectively. Mutations (p.I47T in kindred 1, p.L223P in kindred 2, p.P109L in kindred 3, and p.L204P in kindred 4) are homozygous in the available affected subjects and heterozygous in the parents. Consanguineous unions are denoted by a double line. (B) Schematic representation of human GIMAP5 protein with AIG1 domain depicted in dark gray, where all four missense mutations are located. Conservation of Ile47, Pro109, Leu204, and Leu223 positions across orthologues and GIMAP family members are shown. Amino acid positions identical to the human reference are highlighted in yellow. AIG1, GTP-binding AIG1 homology domain; TD, transmembrane domain; mut, mutant; wt, wild-type.
Table 1. Summary of demographics, clinical presentation, genotype, and liver features of patients with GIMAP5 deficiency.
| Liver transaminases | Portal hypertension and hepatocellular carcinoma | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Age at presentation (yr) | Current age (yr) | Clinical presentation | Genotype, GIMAP5 | AST (U/L) | ALT (U/L) | Splenomegaly | Thrombocytopenia | Esophageal varices | Ascites | Hepatocellular carcinoma |
| P1-1 | 13 | 25 | Ecchymosis | p.I47T | 65 | 53 | ✓ | ✓ | ✓ | No | No |
| P1-2 | 11 | 23 | Hemoptysis | p.I47T | 86 | 70 | ✓ | ✓ | ✓ | No | No |
| P2-1 | 2 | Died at 17 | Recurrent infections | n/a | 49 | 54 | ✓ | ✓ | ✓ | No | No |
| P2-2 | 15 | Died at 22 | Fatigue, epistaxis | p.L223F | 85 | 247 | ✓ | ✓ | ✓ | ✓ | No |
| P2-3 | 17 | 33 | Abdominal and back pain | p.L223F | n/a | n/a | ✓ | ✓ | n/a | No | No |
| P2-4 | n/a | 31 | Recognized during screening of family members | p.L223F | n/a | n/a | ✓ | No | n/a | No | No |
| P2-5 | 35 | Died at 44 | Fatigue | p.L223F | 40 | 37 | ✓ | ✓ | ✓ | No | ✓a |
| P3-1 | 2 | 7 | Elevated transaminases and GGT, and hepatosplenomegaly | p.P109L | 113 | 76 | ✓ | ✓ | ✓ | ✓ | No |
| P4-1b | 14 | 21 | Petechia | p.L204P | 40 | 20 | ✓ | ✓ | ✓ | No | No |
ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, γ-glutamyl transferase; n/a, not available.
One 2-cm hepatic lesion with arterial enhancement and portal venous washout highly suspicious for hepatocellular carcinoma.
Patient was first described in Patterson et al. (2018).
All available affected members of these four kindreds shared different rare homozygous missense variants in the gene GIMAP5 predicted to be damaging by Polyphen-2 (Adzhubei et al., 2010) and had Combined Annotation Dependent Depletion (Kircher et al., 2014; Rentzsch et al., 2019) scores >20 (Fig. 1 and Table S1). GIMAP5 encodes GTPase of the immunity-associated protein (GIMAP) family member 5, a small GTPase predominantly expressed in lymphocytes that regulates lymphocyte survival (Barnes et al., 2010). In kindred 1, two affected siblings, the offspring of a first cousin union, were homozygous for a variant encoding a p.I47T substitution (Fig. 1). The I47T variant has an allele frequency of zero in gnomAD (Table S1). In kindred 2, there were four affected offspring of a second cousin union and an additional affected subject who is a sibling of one of the unaffected parents of this sibship. Four affected subjects were homozygous for the p.L223F variant, which also has an allele frequency of 0 in gnomAD. The genotype of P2-1 could not be confirmed, since he was already deceased. In kindred 3, a child of a first cousin union with noncirrhotic portal hypertension was homozygous for a p.P109L substitution. This variant allele is extremely rare, seen once among more than 251,400 alleles sequenced from diverse populations (Table S1; allele frequency in gnomAD of 3.982 × 10−6). Subject P4-1 is a 21-yr-old male with newly recognized noncirrhotic portal hypertension. He is the sole affected offspring of unrelated parents and has been previously reported with hemolytic anemia, thrombocytopenia, lymphopenia, splenomegaly, and GIMAP5 deficiency (Patterson et al., 2018). He is homozygous for a p.L204P substitution that has an allele frequency of 2.1 × 10−3 (Fig. 1, Table 1, and Table S1). Notably, all four mutations (p.I47T, p.P109L, p.L204P, and p.L223F) lead to loss of GIMAP5 protein expression, as shown in Patterson et al. (2018) and Park et al. (2021 Preprint).
The cosegregation of rare damaging homozygous genotypes that produce nonconservative substitutions at highly conserved amino acids with noncirrhotic portal hypertension is very unlikely to have occurred by chance. The probands of these kindreds each had only one to nine rare damaging homozygous genotypes, and no other gene with a homozygous damaging genotype was shared among all kindreds. These variants were rare in all populations examined. Parametric analysis of linkage in the three consanguineous kindreds specifying damaging GIMAP5 variants as rare, with very high penetrance of portal hypertension via recessive genotypes and very rare phenocopies, was performed. Logarithm of the odds scores in favor of linkage at a recombination fraction of 0 in kindreds 1–3 were, respectively, 1.8, >3.3 (the likelihood of the second allele in subject P2-6 being inherited by chance is indeterminate, and the genotype of deceased P2-1 could not be confirmed), and 1.2, providing a combined logarithm of the odds score >6.3 (odds 2 million to 1 in favor of linkage) and therefore very strong statistical support for the role of these rare recessive GIMAP5 genotypes in noncirrhotic portal hypertension.
Collectively, the identification of four families with independent rare damaging recessive genotypes in GIMAP5 that precisely cosegregate within families affected by noncirrhotic portal hypertension provides strong evidence that these genotypes cause portal hypertension in these families.
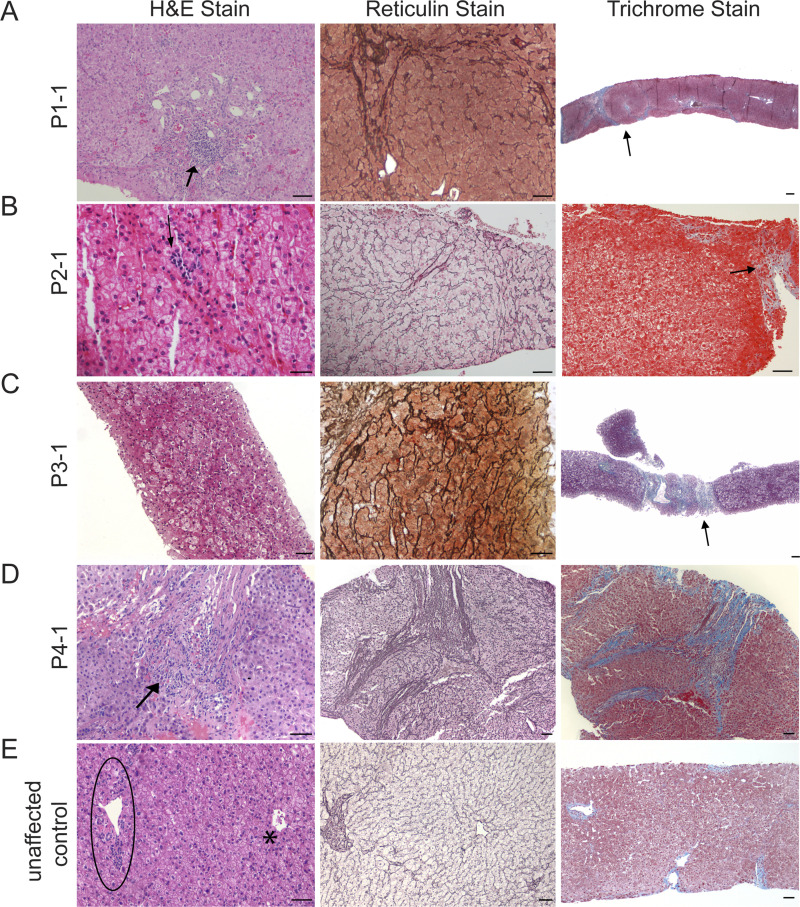
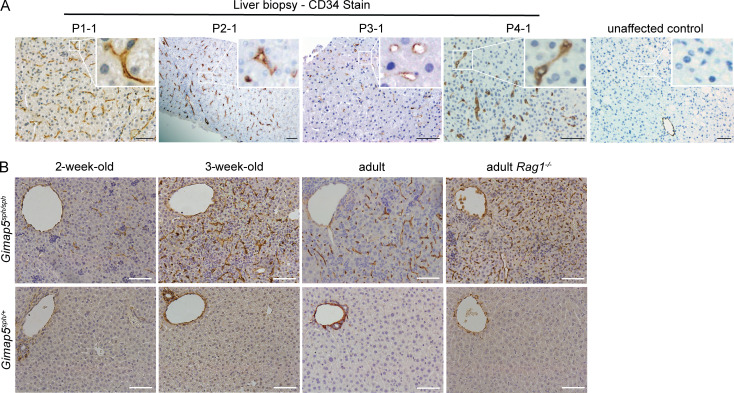
Liver biopsies were obtained from both affected siblings of kindred 1, P2-1 of kindred 2, and affected individuals in kindreds 3 and 4. Liver parenchyma from subjects P1-1 and P2-1 showed features consistent with nodular regenerative hyperplasia (Fig. S1). Importantly, we found marked CD34 immunostaining in the LSECs consistent with capillarization (Fig. 2 A). Like her sibling’s biopsy, the liver parenchyma of P1-2 lacked significant fibrosis or septa formation, and venules were absent in the portal areas, features consistent with idiopathic noncirrhotic portal hypertension (data not shown). Liver biopsy from subject P3-1 was small and showed no cirrhosis with a largely preserved architecture. This sample also showed increased CD34 staining in LSECs (Fig. 2 A), particularly near the portal tracts, with a heterogeneous staining pattern in the hepatic lobules. By contrast, in unaffected healthy individuals, CD34 is solely expressed in macrovascular hepatic vessels and absent on LSECs (Fig. 2 A). Subject P4-1’s liver biopsy showed features of nodular regenerative hyperplasia, no cirrhosis, and aberrant CD34 sinusoidal stain (Fig. 2 A and Fig. S1). This patient underwent direct portal venography, which showed increased portal pressure to 14 mm Hg (normal portal venous pressure, 5–10 mm Hg).
Figure S1.
Representative images of histological findings seen in liver biopsies from patients (P1-1, P2-1, P3-1, and P4-1) with biallelic mutations in GIMAP5. (A) P1-1’s liver biopsy shows lobular parenchyma devoid of any steatosis, hepatocytic ballooning, or acidophil necrosis. Minimal and focal lymphocytic infiltrates (arrow) are seen along with dilated channels at the periphery of the portal tracts, with some of them extending into the sinusoids (left panel; H&E stain). Middle panel reveals nodular architecture due to areas of regeneration consisting of thickened hepatic cell plates next to areas of atrophy without any intervening fibrosis (reticulin stain). Right panel shows few thin septa (in blue) in the left aspect of the picture (arrow) but lack of cirrhosis (trichrome stain). (B) P2-1’s liver biopsy shows largely unremarkable hepatocytes with occasional foci of lobular inflammation (arrow; left panel; H&E stain). Middle panel shows liver nodularity with zones of widened two-cell-thick plates consistent with hepatic regeneration bounded by narrow compressed liver cell plates (reticulin stain). Right panel shows periportal fibrosis (arrow; trichrome stain). (C) P3-1’s liver biopsy shows no significant abnormalities in the hepatocytes (left panel; H&E stain). Middle panel shows areas of regeneration consisting of thickened hepatic cell plates (reticulin stain). Right panel shows an area of hepatocyte loss with parenchymal collapse and fibrosis (arrow) but no evidence of cirrhosis (trichrome stain). (D) P4-1’s liver biopsy shows a portal tract with minimal and focal lymphocytic infiltrates (arrow). The lobular parenchyma is devoid of any steatosis, hepatocytic ballooning, or acidophil necrosis (left panel; H&E stain). Middle panel highlights an area of pericentral hepatocyte loss with parenchymal collapse and thin fibrous septa. The liver shows vague nodularity with areas of hepatic regeneration but no cirrhosis (reticulin stain). Right panel reveals few thin septa (blue) but lacks features of cirrhosis (trichrome stain). (E) Normal-appearing portal tract (oval) and a central venule (asterisk). The portal tract depicts a hepatic arteriole, bile duct, and portal venule. The portal venule is the largest structure with the widest lumen and a thin wall (H&E stain). Middle panel shows hepatic cords in a single thick cord plate. The central vein is seen near the center, and a portal tract is at the periphery (reticulin stain). Right panel shows normal architecture of the liver and no fibrosis (trichrome stain). Scale bars = 50 µm.
Figure 2.
Liver histology and CD34 expression in humans and mice with GIMAP5 deficiency. (A) Photomicrographs of abnormal CD34 immunostaining in liver sinusoids of four unrelated patients (P1-1, P2-1, P3-1, and P4-1) as compared with an unaffected control. (B) Photomicrographs of abnormal CD34 sinusoidal expression is shown in the liver sections from C57BL/6 Gimap5sph/sph mice at 2, 3, and >7 wk old and in adult C57BL/6 Gimap5sph/sphRag1−/− mice as compared with Gimap5sph/+ control mice. Histological data from 2-wk-old, 3-wk-old, adult Gimap5sph/sph, and adult Gimap5sph/sphRag1−/− mice was verified at least four, two, seven, and three independent times, respectively. Littermates were used as controls. Scale bar = 50 µm.
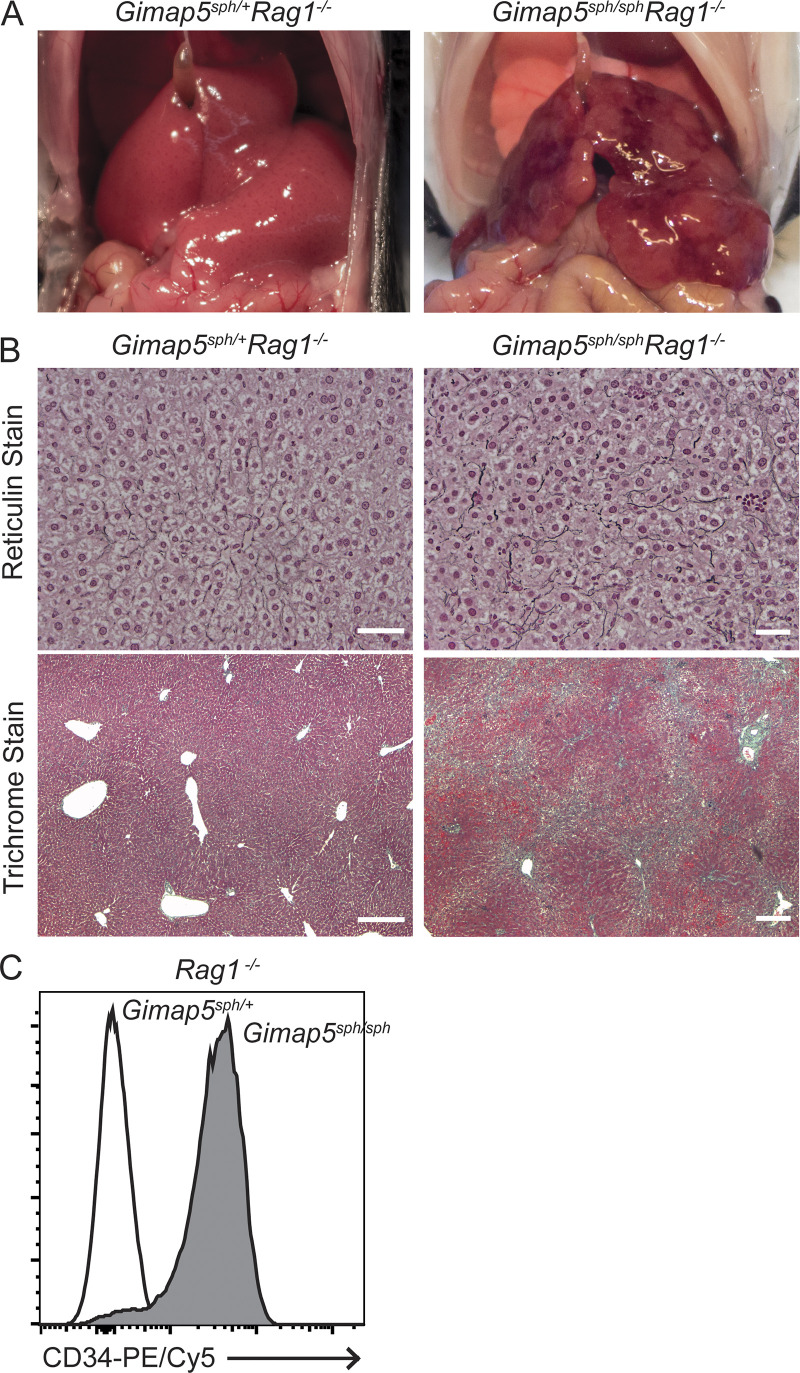
The liver abnormalities seen in these patients are reproduced in two independent GIMAP5-deficient mouse models. Both strains showed splenomegaly, nodular regenerative hyperplasia of the liver, and decreased life expectancy (Barnes et al., 2010; Schulteis et al., 2008). This distinctive finding provides strong confirmatory evidence that the recessive GIMAP5 genotypes found in humans result in loss of normal GIMAP5 function. Since liver disease and portal hypertension can be found in other immunodeficiencies and autoimmune syndromes (Azzu et al., 2019; Malamut et al., 2008), we tested whether the liver phenotype in GIMAP5 loss-of-function (LOF) mutant mice (Gimap5sph/sph) is a consequence of underlying defects in adaptive immune cells. We crossed the Gimap5sph/sph mice with Rag1−/− mice, which lack B and T cells and do not have evidence of portal hypertension, to produce mice homozygous for both Gimap5 LOF mutation and Rag1 deficiency. These mice showed the same liver abnormalities seen in Gimap5sph/sph mice (Fig. S2), indicating that the liver disease is not caused by aberrant B or T cell function. This result is consistent with prior reports showing that transplantation of Gimap5-deficient bone marrow cells into lethally irradiated WT recipient mice did not induce liver pathology (Barnes et al., 2010; Schulteis et al., 2008). In accord with our findings, Schulteis et al., (2008) also stated that liver pathology persisted in Gimap5−/− Rag2−/− double-knockout mice, but no data were shown in their study and therefore it was hard to judge how this phenotype was evaluated. Further examination of the livers of Gimap5sph/sph mutant mice showed aberrant expression of CD34 in sinusoidal endothelium, as seen in GIMAP5 mutant patients. CD34 overexpression could be detected as early as 2 wk of age in these mice and in mice with both Gimap5 and Rag1 deficiency (Fig. 2 B and Fig. S2). We therefore hypothesized that Gimap5 deficiency in sinusoidal endothelium could be important for liver disease and portal hypertension.
Figure S2.
Liver pathology of Gimap5sph/+Rag1−/− and Gimap5sph/sphRag1−/− mice. (A) Gross liver morphology of adult Gimap5sph/+Rag1−/− (smooth) and Gimap5sph/sphRag1−/− mice (nodular). (B) Reticulin and trichrome stains of liver sections from Gimap5sph/+Rag1−/− (left panels) and Gimap5sph/sphRag1−/− (right panels) show two-cell-thick plate and increased collagen (blue) deposition solely in mutant mice. (C) Flow cytometric analysis shows increased CD34 expression in liver endothelial cells (DAPI−CD45−CD31+) isolated from Gimap5sph/sphRag1−/− mice as compared with Gimap5sph/+Rag1−/− mice. Experimental data were verified in at least two independent experiments, and littermates were used as controls. Scale bars = 50 µm.
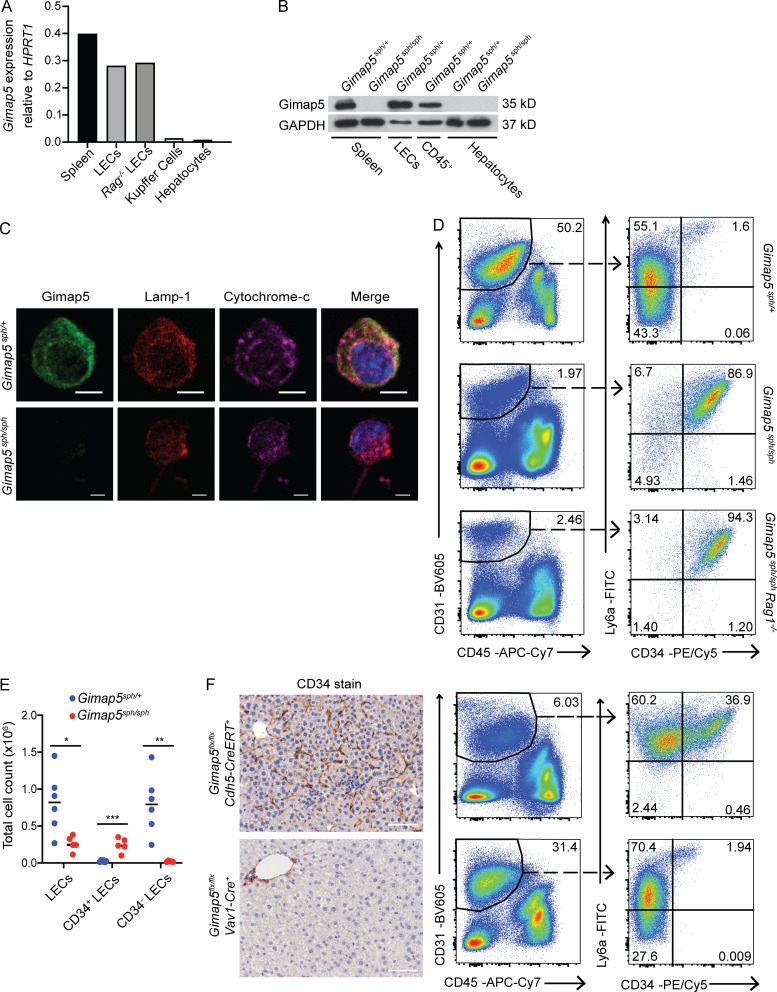
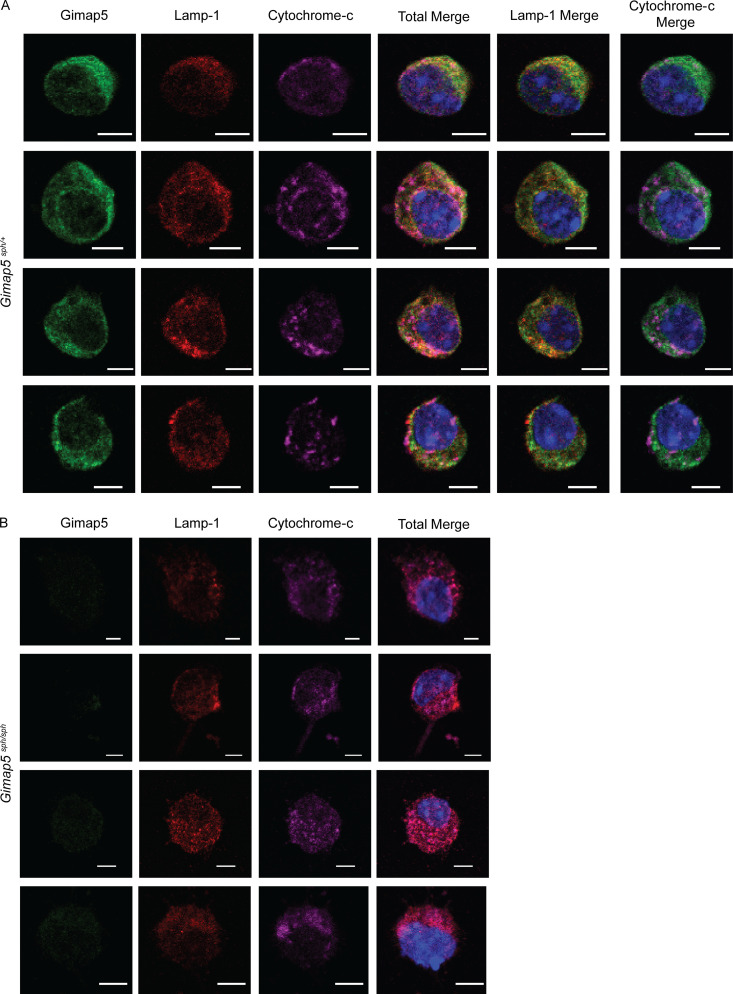
Because GIMAP5 is reported to be primarily expressed in lymphocytes (Nitta et al., 2006; Wong et al., 2010), we examined whether Gimap5 is expressed in the liver and, if so, in which cell types. We found that CD45−CD31+ liver endothelial cells, but not hepatocytes, express Gimap5 mRNA and protein (Fig. 3, A and B). Analysis of publicly available single-cell RNA sequencing (scRNA-seq) from mouse and human healthy livers confirmed Gimap5 gene expression in hepatic endothelial cells (Aizarani et al., 2019; Tabula Muris Consortium, 2018). Subsequently, we examined the subcellular localization of Gimap5 in CD45−CD31+ liver endothelial cells using confocal microscopy. We found that Gimap5 protein colocalizes with lysosomal marker Lamp1, supporting its localization within the lysosomes and associated compartments, although some signal could also be detected in the cytosol (Fig. 3 C and Fig. S3). This finding is consistent with studies using a monoclonal antibody against Gimap5 in Jurkat T cells (Wong et al., 2010), in CD4+ T cells (Patterson et al., 2018), and overexpression systems using lower plasmid amounts (Serrano et al., 2017). A bioinformatics tool (Blum et al., 2021) did not identify any signal peptides, but it predicted a type 4 transmembrane domain that would support Gimap5 localization to the lysosomal membrane.
Figure 3.
Genetic deficiency of GIMAP5 causes liver endothelial cell abnormalities. (A) Gimap5 mRNA expression in sorted liver endothelial cells (LECs; DAPI−CD45−CD31+) from C57BL/6 WT and Rag1−/− mice, and splenocytes, sorted Kupffer cells (DAPI−CD45+CD115+F4/80+), and hepatocytes from C57BL/6 mice. (B) Immunoblot for GIMAP5 in Gimap5sph/+ and Gimap5sph/sph splenocytes and hepatocytes and in sorted LECs and liver CD45+ cells from Gimap5sph/+ mice. GAPDH is shown as a loading control. (C) Confocal microscopy of LECs isolated from Gimap5sph/+ and Gimap5sph/sph and stained for GIMAP5 (green), Lamp-1 (red), and cytochrome-c (magenta) and counterstained with DAPI (blue). Scale bars = 5 µm. (D) LECs isolated from Gimap5sph/+, Gimap5sph/sph, and Gimap5sph/sphRag1−/− livers (left panels) and respective Ly6a and CD34 surface expression (right panels). (E) Absolute number of LECs that express CD34 or not in Gimap5sph/+ (n = 6) and Gimap5sph/sph (n = 5) livers. (F) Histological and flow cytometric analysis of sinusoidal CD34 expression in tamoxifen-treated adult Gimap5flx/flxxCdh5(PAC)-CreERT2 and Gimap5flx/flxxVav1-Cre mice (upper and bottom panels, respectively). Experimental data were verified in at least two independent experiments, and littermates were used as controls. Scale bars = 50 µm. Numbers depict the percentage of total cells. Student’s two-tailed t test was used. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. Adult mice, >7 wk old. APC, allophycocyanin; Cy, cyanine.
Figure S3.
Subcellular localization of Gimap5 in sorted liver endothelial cells (LECs; DAPI−CD45−CD31+). (A and B) Confocal microscopy of isolated LECs from Gimap5sph/+ (A) and Gimap5sph/sph (B) and stained for GIMAP5 (green), lysosomal marker Lamp-1 (red), and mitochondrial marker cytochrome-c (magenta) and counterstained with DAPI (blue). Experimental data were verified in at least two independent experiments. Scale bars = 5 µm. Images shown in Fig. 3 C are also included in the panels above.
Liver endothelial cells isolated from Gimap5sph/sph mutant mice in both WT and Rag1 deficient backgrounds showed markedly increased CD34 expression by flow cytometry as compared with hepatic endothelial cells from control mice (Fig. 3 D). Moreover, we also detected an approximately threefold reduction in the absolute number of CD45−CD31+ endothelial cells in Gimap5sph/sph livers as compared with heterozygous mice; and most of these cells express CD34 in contrast to low or absent CD34 staining in control mice (Fig. 3 E). These observations strongly suggest that intrinsic Gimap5 expression in liver endothelial cells is critical for preserving normal LSEC specification and identity, which in turn are critical for normal liver function.
To further test this conclusion, we crossed Gimap5flx/flx mice (Patterson et al., 2019) with Cdh5(PAC)-CreERT2 mice (Wang et al., 2010) or with Vav1-Cre mice. These approaches eliminate Gimap5 expression exclusively in endothelial cells or hematopoietic cells, respectively. We found that tamoxifen-induced Gimap5 deletion in endothelial cells led to aberrant liver sinusoidal CD34 expression, whereas constitutive Gimap5 deletion in Vav1+ hematopoietic cells caused no detectable liver abnormalities or abnormal CD34 expression in LSECs (Fig. 3 F). These findings revealed that the liver phenotype seen in Gimap5sph/sph mice results primarily from the lack of GIMAP5 in liver endothelial cells and is not the result of dysregulated GIMAP5-deficient immune cells.
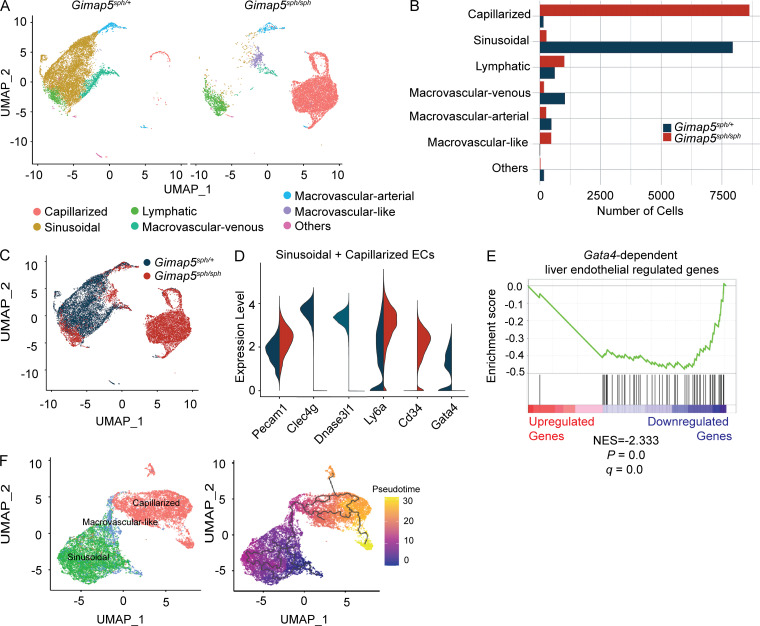
Given the important role of Gimap5 in liver endothelial cells, we performed scRNA-seq of CD45−CD31+ liver endothelial cells from mice that were either sufficient or deficient for Gimap5. Clustering of the results identified clearly distinct populations, with the vast majority representing either LSECs or capillarized endothelial cells (CECs; Fig. 4, A and B). Remarkably, the Gimap5sph/sph liver showed a near absence of LSECs and the emergence of a CEC population, a reduction in macrovascular venous and macrovascular arterial endothelial cells, and an expansion of lymphatic endothelial cells (Fig. 4, A and B). CECs present in GIMAP5-deficient liver were notable for the absence of GATA4 expression, a transcription factor required for LSEC specification that suppresses expression of genes required for capillarization of endothelial cells (Géraud et al., 2017; Winkler et al., 2021). Correspondingly, CECs also showed absence of classical LSEC markers (e.g., Clec4g, Dnase3l1), overexpression of markers associated with capillarized endothelium (e.g., Cd34, Pecam1; Fig. 4, C and D), and up-regulation of genes involved in extracellular matrix organization, such as Col4a2 and Sparc. Gene set enrichment analysis (GSEA) showed that GIMAP5-deficient CD45−CD31+ cells had markedly lower expression of genes that are Gata4 dependent in LSECs (Fig. 4 E). Collectively, these findings place GIMAP5 upstream of the LSEC-specifying GATA4 transcription factor. These data are consistent with previous observations that GATA4 expression was significantly decreased in human livers with advanced liver fibrosis and cirrhosis (Delgado et al., 2014), a setting in which endothelial cell capillarization is commonly observed. Notably, in addition to the expansion of CECs, Gimap5 deficiency also revealed the emergence of a macrovascular-like cell type that is not seen under homeostatic conditions (Fig. 4, A and B). To better understand the relationship between these cells, LSECs and CECs, we performed pseudotime trajectory analysis. This analysis suggests that LSECs transition into CECs via these macrovascular-like cells (Fig. 4 F).
Figure 4.
scRNA-seq analysis of liver endothelial cells from Gimap5sph/+ and Gimap5sph/sph mice. (A) Clustering of endothelial cells from Gimap5sph/+ and Gimap5sph/sph livers using the Louvain method with a resolution parameter 0.4. Uniform Manifold Approximation and Projection (UMAP). Cell identities were inferred from marker genes. (B) Absolute number of cells in each endothelial subpopulation. (C) Overlay of endothelial cell cluster maps annotated by genotype: Gimap5sph/+ and Gimap5sph/sph. (D) Pecam1, Clec4g, Dnase3l1, Ly6a, Cd34, and Gata4 expression in combined subpopulations of LSECs and CECs isolated from Gimap5sph/+ (blue) and Gimap5sph/sph (red) mice. ECs, endothelial cells. (E) Enrichment plot from GSEA of preranked list of genes differentially expressed in GIMAP5-deficient and sufficient liver endothelial cells as compared with a background list of mouse Gata4-dependent liver endothelial cell regulated genes. NES, normalized enrichment score. (F) Cluster of LSECs, CECs, and macrovascular-like cells with trajectory analysis performed in Monocle.
In several primary immunodeficiency syndromes, it has been proposed that accompanying liver disease results from hyperinflammation caused by a dysregulated immune system (Ben-Yakov et al., 2018; Coulter et al., 2017; Malamut et al., 2008; Ward et al., 2008). Here, we show that liver abnormalities in GIMAP5 deficiency are independent of immune cell function and result from liver endothelial cell dysfunction. Our findings are consistent with the possibility that nodular regenerative hyperplasia of the liver is primarily caused by a sinusoidal injury (Rothweiler et al., 2014). This study is a fascinating example of how specific gene expression in different cell types (circulating lymphocytes versus liver endothelial cells) may account for different organ involvement in certain monogenic multisystemic immunodeficiency and/or autoimmune syndromes. Importantly, modulation of the immune system by means of bone marrow transplantation and other methods will not correct the liver disease detected in these cases.
This study identifies GIMAP5 as a critical regulator of liver endothelial cell homeostasis, and, when absent, it produces portal hypertension. Liver endothelial cells, and in particular LSECs, are essential in regulating the hepatic vascular tone and maintenance of a regulated low portal pressure (Poisson et al., 2017). Under pathological conditions, such as liver fibrosis, LSECs de-differentiate into CECs (DeLeve, 2015; Iwakiri, 2012). It is presently unknown whether or how GIMAP5 activity is normally regulated and whether this regulation plays a continuous role in modulating function of LSECs. For example, it may change the degree of fenestration or synthesis of basement membrane, both characteristics of CECs. It is also possible that Gimap5 functions to promote the stability of pro-survival signals, such as Mcl-1 (Chen et al., 2011), besides controlling GATA4 expression and LSEC homeostasis. Since GIMAP5 is expressed in liver endothelial cells and not exclusively in LSECs, and because Gimap5 deficiency results in a reduction in macrovascular endothelial cells in addition to a nearly complete replacement of LSECs by CECs, further studies are required to determine the individual contributions of GIMAP5-expressing endothelial cell subsets to the liver abnormalities seen in both Gimap5-deficient mice and humans. The ability to induce capillarization by somatic loss of GIMAP5 raises the interesting possibility that GIMAP5-mediated signal transduction events are continuously required for the maintenance of LSEC identity. It will be of interest to determine whether signals resulting from normal or diseased liver alter GIMAP5 activity to promote or inhibit local perfusion via regulation of LSEC identity. This understanding might provide a means to prevent or mitigate development of portal hypertension in patients with advanced liver disease.
This study provides new cellular and molecular insights into the pathogenesis of portal hypertension, an important step toward uncovering new targeted therapeutic agents. Our previous work has shown that recurrent homozygous mutation in DGUOK, a deoxyguanosine kinase required for mitochondrial DNA replication, causes noncirrhotic portal hypertension (Vilarinho et al., 2016). In contrast to GIMAP5, DGUOK is ubiquitously expressed in parenchymal and nonparenchymal liver cells. Future studies will determine whether LOF mutations in DGUOK and GIMAP5 lead to portal hypertension via convergent or distinct pathways. Thus, genetic investigation provides new insights into severe liver disease that may lead to novel treatment approaches in the future.
Materials and methods
Human subjects
This study was performed in accordance with protocols approved by Yale University and National Institute of Allergy and Infectious Diseases institutional review boards. Study participants (or their legal guardians) provided written informed consent.
Exome sequencing analysis
Genomic DNA was obtained from peripheral blood leukocytes using standard methods. Exome sequence data were aligned to the reference human genome (build 19) using the Burrows-Wheeler Aligner, and variants were called using the Genome Analysis Toolkit. Variants with minor allele frequency <1% in the gnomAD databases (125,748 exome sequences of various ethnicities; last accessed in January 2020) were selected and annotated using ANNOVAR.
Sanger sequencing of genomic DNA
Sanger sequencing of the identified GIMAP5 variants was performed by PCR amplification of genomic DNA of all available affected individuals and their parents using the following forward and reverse primers: kindred 1, chr:7:150439367, 5′-AAGATAACTTGTCTGCAACACCA-3′ (forward) and 5′-GTAGCAGTCCCCGATGTTCT-3′ (reverse); kindred 2, chr7:150439894, 5′-GGGGAGGACGTTCATAGCTT-3′ (forward) and 5′-TGCTCCAGGGTCCAGAGAT-3′ (reverse); kindred 3, chr7:150439553, 5′-GATAACTTGTCTGCAACACCA-3′ (forward) and 5′-TGTCCTGAGCAGTGAAACGC-3′ (reverse); kindred 4, chr7:150439838, 5′-ACATGGAACGGGAGGAAAGTC-3′ (forward) and 5′-CACGAGGATCTGAGTCTGAATTG-3′ (reverse). Nomenclature of the GIMAP5 variant is based on National Center for Biotechnology Information reference sequence NM_018384.
Orthologues and other human GIMAPs
Full-length orthologues of GIMAP5 protein sequences from several species and related human GIMAP family members (GIMAPs 1, 2, 4, 6, 7, and 8) were obtained from GenBank. Protein sequences were aligned using the Clustal Omega algorithm.
Mice
All mice were maintained under specific pathogen–free conditions at Yale Animal Resources Center and used according to a protocol approved by the Yale University Institutional Animal Care and Use Committee. Gimap5sph/sph mice (Barnes et al., 2010) are available through the Mutant Mouse Resource & Research Centers (stock no. 030019-UCD). Rag1−/− mice were purchased from The Jackson Laboratory (stock no. 002216). Gimap5flx/flx mice (Patterson et al., 2019) were crossed with B6.Cg-Commd10Tg(Vav1-icre)A2Kio/J (purchased from The Jackson Laboratory; stock no. 008610) or with Cdh5(PAC)-CreERT2 strain (Wang et al., 2010). This strain was obtained with the permission of Dr. Ralf Adams (Max Planck Institute for Molecular Biomedicine, Muenster, Germany). Cdh5-ERT2 cre transgene expression was induced by administration of tamoxifen (Sigma-Aldrich; T5648). Tamoxifen was dissolved at a concentration of 10 mg/ml in corn oil (Sigma-Aldrich; C8267). Mice of both sexes were injected i.p. with 75 µg tamoxifen per gram of body weight daily for 5 consecutive days starting at 4 wk of age, followed by a 9-d pause, and treated for another round of five consecutive injections at 75 µg/g. Animals were analyzed within 1–4 wk after the final injection.
Liver endothelial and Kupffer cell isolation
Mice were euthanized in accordance with institutional animal care and use committee guidelines. The liver was flushed with PBS (1×, filtered) via the portal vein, followed by collagenase type 2 (Worthington Biochemical Corporation; LS004177) at a concentration of 1.5 mg/ml. The whole liver was excised, briefly rinsed in 1× PBS, then placed into 10 ml of prewarmed collagenase (1.5 mg/ml, 37°C). The liver was minced into small pieces, then digested at 37°C for 30 min. The minced liver was dissociated into single-cell suspension by gentle pipetting, then passed over a 40-µm filter. Hepatocytes were pelleted and removed by a series of low-speed centrifugations (60 g). Nonparenchymal cells remaining in the supernatant were pelleted by high-speed spin (350 g) and washed in RPMI media. Nonparenchymal cells were run through a Percoll gradient to enrich for endothelial cells. A density gradient was created by layering 5 ml of 50% Percoll, followed by 6.6 ml of 25% Percoll, then 3.3 ml of cell suspension in RPMI. The 100% Percoll stock was prepared by adding one part 10× PBS to nine parts Percoll (Sigma-Aldrich; P4937) and adjusting pH to 7.4. Cells were spun through the gradient at 900 g for 20 min with no brake to separate the endothelial cell fraction. The endothelial/Kupffer cell fraction was collected from the interface between the 50% and 25% Percoll layers, then washed in RPMI media and counted.
Murine histology
Liver tissue was collected and immediately fixed in 10% buffered formalin solution, followed by paraffin embedding. H&E, reticulin, trichrome, and CD34 staining were performed on 5-µm sections from the paraffin-embedded tissue blocks and imaged on an Olympus BX51 light microscope.
Flow cytometry and cell sorting
Endothelial cells were isolated as described above, and flow cytometry was performed on live cells. All antibody dilution and wash steps were performed using FACS buffer (2% FBS, 1 mM EDTA, 0.1% NaN3). Staining was performed on 96-well round-bottomed plates with ∼1 million cells/well using purified rat anti-mouse CD16/CD32 Mouse BD Fc Block (BD Pharmingen; 553141), followed by incubation in fluorochrome-conjugated primary antibodies for 20 min on ice. All the following antibodies were purchased from BioLegend and used at 1:100 dilution: allophycocyanin/cyanine 7 anti-mouse CD45 (103116), BV605 anti-mouse CD31 (102427), PE/Cy5 anti-mouse CD34 (119311), FITC anti-mouse Ly-6A (Sca-1; 108105), allophycocyanin anti-mouse F4/80 (123116), and Alexa Fluor 488 anti-mouse CD115 (CSF-1R; 135512). DAPI dye was used to determine viability. Liver endothelial cells were DAPI−CD45−CD31+, and these cells were stained for Sca-1 and Cd34. Of note, Sca-1 has been shown to be expressed in LSECs (Luna et al., 2004) and in bone marrow arterial and sinusoidal endothelial cells (Xu et al., 2018). Flow cytometry experiments were performed on the LSR II flow cytometer. Cell sorting was performed on a BD FACSAria II platform. Analysis was performed using FlowJo software.
Real-time PCR/TaqMan assay
RNA was isolated from endothelial cells, Kupffer cells, splenocytes, and hepatocytes using RNeasy Micro Plus or RNeasy Mini Plus kits. cDNA synthesis was performed using the SuperScript III cDNA Synthesis Kit (Invitrogen) and oligo(dT) 12–18 primers (Invitrogen) following a standard protocol. Real-time PCR was performed using FastStart Universal Probe Master Mix (Roche; 4913957001) and TaqMan assays for mouse Gimap5 (Thermo Fisher Scientific; Mm00658393_m1) and GAPDH (Thermo Fisher Scientific; Mm99999915_g1) on the ABI 7500 Real Time PCR System.
Immunoblotting
Protein lysates were prepared from cell pellets following direct isolation or cell sorting using Pierce radioimmunoprecipitation assay lysis buffer (Thermo Fisher Scientific; 89900) and standard methods. Lysates were run on 10% Mini PROTEAN TGX precast gels (Bio-Rad Laboratories) and transferred onto 0.2-µm nitrocellulose membranes (Bio-Rad Laboratories; 162-0146). Membranes were blocked in 5% milk and immunoblotted with primary antibody to GIMAP5 (MAC421; Wong et al., 2010; courtesy of Dr. Butcher) overnight at 4°C or with GAPDH (Invitrogen; AM4300) for 1 h at room temperature. Secondary antibodies used were ECL anti-rat IgG HRP-linked whole antibody (GE Healthcare; NA935) and ECL peroxidase-labeled anti-mouse antibody (GE Healthcare; NA931), respectively. Immunostaining was detected using Amersham ECL and ECL Prime Western Blotting detection reagents (GE Healthcare).
Confocal microscopy
Endothelial cells were isolated and sorted as described above. Cells were then spun onto microscope slides coated in 30% FBS at 800 revolutions per minute for 3 min using a cytocentrifuge (Thermo Scientific Cytospin 4). Cells were immediately fixed in 4% paraformaldehyde for 15 min. Background fluorescence was quenched with 100 mM ammonium chloride for 10 min and then permeabilized with 0.1% Triton X-100 for 15 min. PBS with 10% FBS was then used as a blocking solution for 1 h. Cells were then incubated overnight using anti-Gimap5 (MAC421; Wong et al., 2010; courtesy of Dr. Butcher) and anti-Lamp-1 (Abcam; ab208943). Secondary antibodies donkey anti-rat A488 (Jackson ImmunoResearch; 712-545-150) and donkey anti-rabbit A594 (Jackson ImmunoResearch; 711-585-152) were incubated for 1 h at room temperature. Cells were then stained for cytochrome-c using a directly conjugated Alexa Fluor 647 antibody (BD Biosciences; 558709) for 2 h at room temperature. DAPI was used to counterstain the nucleus, and slides were imaged on a Leica TCS SP8 confocal microscope.
scRNA-seq and analysis
Liver endothelial (DAPI−CD45−CD31+) cells isolated from one Gimap5sph/sph adult mouse and one Gimap5sph/+ adult mouse were sorted as outlined above and processed as follows: scRNA-seq library was generated using Chromium Single Cell 3′ Reagent Kits version 3 (10x Genomics) following the manufacturer’s protocol. Libraries were sequenced on an Illumina NovaSeq 6000 instrument using an S4 flow cell. Cell Ranger version 1.3 software (10x Genomics) was used to process Chromium Single Cell 3′ RNA-seq output data into a gene-cell data matrix. Seurat version 3.1.4 (Butler et al., 2018) was used for quality control, and cells with <200 expressed genes or >10% mitochondrial gene percentages were excluded to enrich high-quality cells. Cells were also discarded if Pecam1 was not expressed to elevate the fraction of endothelial cells. The data were log transformed and normalized by the total expression. We then performed principal component (PC) analysis using the top 2,000 variable genes to determine significant PCs. 20 statistically significant PCs were selected based on the JackStraw plot and provided as input for constructing a shared-nearest-neighbors graph based on the Euclidean distance. Cells were clustered by the Louvain method with a resolution parameter 0.4. Uniform Manifold Approximation and Projection was used to visualize clustering results. Marker genes that define each cluster were identified by comparing cells in a specific cluster with ones in all other clusters using the Seurat package likelihood ratio test. Statistically significant differentially expressed genes between samples were identified in a pairwise manner using DESeq2, which is based on the negative binomial distribution model. Four subsets of liver endothelial cells were defined based on gene expression profiles as follows (Kalucka et al., 2020): (1) LSECs expressing Clec4g and Dnase1l3; (2) lymphatic endothelial cells expressing Ccl21a, Prss23, Ifi27l2a, and Timp2a; (3) macrovascular venous endothelial cells expressing Rspo3; and (4) macrovascular arterial endothelial cells expressing high levels of Sdc1, Clu, and Ehd4. CECs and macrovascular-like endothelial cells were seen almost exclusively in the Gimap5 mutant mouse. CECs were defined by high expression levels of Cd34, Col4a2, and Sparc (Aizarani et al., 2019). Macrovascular-like cells were annotated based on the absence of LSEC (Clec4g and Dnase1l3) and lymphatic (Ccl21a, Prss23, Ifi27l2a, and Timp2a) gene markers and the expression of liver macrovascular endothelial markers (Plac8, Rbp1, and low expression of Rspo3; Kalucka et al., 2020). scRNA-seq data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (accession no. GSE158988). The mouse Gimap5-dependent liver endothelial cell atlas (scRNA-seq data) can be interactively explored at https://cellbrowser.yalespace.org/gimap5_res0-3/?ds=gimap5.
GSEA
GSEA software version 4.0.3 (https://www.gsea-msigdb.org/gsea/index.jsp) was used to determine whether a predefined set of genes is enriched in the observed gene expression profile. Genes were ranked according to the fold change of gene expression. Given a predefined gene set, an enrichment score (ES) is calculated to measure the overrepresentation of members of that gene set appearing at the extremes (top or bottom) of the ranked gene list. The ES is then evaluated for significance using gene-based permutation tests (1,000). The ES, in addition to the permutation P value, indicates the degree to which the defined gene set is enriched in the gene expression data. The gene sets included the preranked list of genes differentially expressed among Gimap5-deficient and sufficient liver endothelial cells as compared with a background list of mouse Gata4-dependent liver endothelial regulated genes (Géraud et al., 2017).
Pseudotime trajectory analysis
Monocle3 in R was used to conduct the analysis (Trapnell et al., 2014). LSECs, CECs, and macrovascular-like cells were subset from the larger dataset in order to perform pseudotime analysis. A dimensionality value of 20 was used for dimension reduction.
Protein localization and transmembrane domain prediction
Interpro 83.0 (Blum et al., 2021) was used to predict transmembrane domains as well as protein localization using signal sequences. Gimap5 protein sequences for both mice and humans were uploaded and analyzed using SignalP for signal sequences and THMM for transmembrane domains.
Clinical features and liver biopsy findings in patients with recessive mutations in GIMAP5
Kindred 1
Subject P1-1 is a 25-yr-old male who is the offspring of a first cousin union. He was initially evaluated at 13 yr of age for ecchymosis. His exam was remarkable for short stature (below the third percentile), low weight (∼10th percentile), and splenomegaly. His laboratory tests revealed pancytopenia and mild elevation of transaminases. Abdominal ultrasound showed heterogeneous liver parenchyma, portosystemic collateral vasculature, and splenomegaly. Upper endoscopy showed large esophageal varices, and the patient underwent shunt surgery at 17 yr of age with improvement in esophageal varices and other collaterals. His most recent blood work revealed direct hyperbilirubinemia and worsening coagulopathy consistent with progression of underlying liver disease. His sister is subject P1-2, who is currently 23 yr old. She had no significant past medical history until 11 yr of age, when she presented with hemoptysis in the setting of presumed pneumonia. Like her brother, she had splenomegaly on exam. Her laboratory test results were remarkable for leukopenia, thrombocytopenia, and mildly elevated transaminases. Esophageal and gastric fundic varices were seen on endoscopy. Liver biopsies of both siblings were remarkable for portal venous abnormalities consistent with idiopathic portal hypertension (Fig. 2 A and Fig. S1). Specifically, the liver biopsy from P1-1 showed heterogeneity in the architecture and scarring. One aspect of the biopsy, which could be the subcapsular tissue, showed more scarring with few fibrous septa leading to vague nodularity, while the other aspect of the biopsy showed minimal scarring and no well-formed fibrous septa. The portal venules in these fibrous septa either were not identified or showed multiple dilated channels at the periphery of the portal tracts, with some of them extending into the sinusoids. The hepatic cord pattern was largely preserved, and there were no features to suggest cirrhosis. The fibrous septa and lobules showed very minimal and focal lymphocytic infiltrates (Fig. S1). CD34 immunostaining showed increased staining in the sinusoidal endothelium suggesting capillarization of the sinusoids, which extended up to zone 2 in many of the lobules (Fig. 2 A). The lobular parenchyma was devoid of any steatosis, hepatocytic ballooning, or acidophil necrosis. Focal and mild bile ductular proliferation was noted. Taken together, the findings are suggestive of incomplete septal cirrhosis and consistent with noncirrhotic portal hypertension. The biopsy of subject P1-2 consisted of a scant amount of liver tissue containing only few portal areas and was fragmented, making the interpretation difficult. However, there was no evidence of cirrhosis, and no well-formed fibrous septa were noted. The portal venules were not identifiable in the portal areas. There was no significant portal or lobular inflammation, and findings in the hepatic lobules were similar to those of subject P1-1.
Kindred 2
Subject P2-1, who died at 17 yr of age, is the offspring of a second cousin union and had recurrent episodes of fever of unknown etiology in early childhood.. He subsequently developed splenomegaly and pancytopenia. Imaging studies revealed heterogeneous hepatic parenchyma and regenerative nodules, splenomegaly, and multiple para-aortocaval lymph nodes. Upper endoscopy showed esophageal varices consistent with portal hypertension. This patient’s liver biopsy showed hepatic parenchyma with subtle nodularity. There is no portal inflammation, but only a few small portal areas are seen. Occasional foci of lobular inflammation are seen. The reticulin stain highlights the nodularity with zones of widened two-cell-thick plates bounded by a narrow, compressed cell plate consistent with nodular regenerative hyperplasia (Fig. S1). CD34 stain is positive in ∼50% of sinusoids, consistent with abnormal portal venous blood flow (Fig. 2 A). The trichrome stain suggests periportal fibrosis, but there is no evidence of cirrhosis (Fig. S1). The patient ultimately died from hypoxemic respiratory failure in the setting of Acinetobacter sepsis, Aspergillus infection, and a pulmonary embolus. His brother, subject P2-2, presented with fatigue and epistaxis at age 15 yr and died at 22 yr of age from liver failure and portal hypertension complications, including hyperbilirubinemia, ascites, and encephalopathy. Initially, he was found to have anemia, thrombocytopenia, and splenomegaly. He experienced multiple episodes of autoimmune hemolytic anemia with a positive Coombs test result. Autoantibodies associated with liver-related autoimmunity (antinuclear antibodies, antimitochondrial antibodies, anti–liver kidney microsome and anti–smooth muscle antibodies) were absent. Over time, he showed elevated transaminases, hyperbilirubinemia, and large esophageal varices and portal hypertensive gastropathy. His sister, subject P2-3, is a 33-yr-old female with no significant medical history until 17 yr of age, when she presented with fever, chills, and abdominal/back pain and was found to have splenomegaly. At 22 yr of age, she had an episode of bleeding in the setting of severe thrombocytopenia, which improved after i.v. Ig and corticosteroid treatment. She is currently asymptomatic. His other sister, subject P2-4, is a 31-yr-old female with splenomegaly and diffusely coarse liver parenchyma with a lobulated contour seen on imaging studies. She is currently asymptomatic. The maternal uncle of this sibship, subject P2-5, died at 44 yr of age from infection and liver complications. He was asymptomatic until 35 yr of age, when he presented with fatigue. He was found to have easy bruising, epistaxis, gingival bleeding, splenomegaly, thrombocytopenia, and elevated transaminases of unknown etiology. He subsequently developed jaundice, pancytopenia, and dyspnea. Contrast-enhanced abdominal imaging revealed splenomegaly and collateral circulation consistent with portal hypertension, coarse granular liver parenchyma, and one 2-cm hepatic lesion with arterial enhancement and portal venous washout highly suspicious for hepatocellular carcinoma.
Kindred 3
Subject P3-1 is a 7-yr-old male who is the offspring of a first cousin union. He was found to have hepatosplenomegaly at 2 yr of age. His laboratory tests were remarkable for pancytopenia and elevation of liver transaminases and γ-glutamyl transferase. His liver biopsy was small (barely 1 cm) with only three portal areas. The liver biopsy showed largely preserved architecture. There was one area of hepatocellular collapse that was devoid of any hemorrhage, inflammation, or scarring. Portal tracts appeared normal on a low-magnification image, except one enlarged portal area with fibrosis. On closer examination, the portal tract showed a sclerotic, narrow-caliber portal venule and multiple small venules. The reticulin stain showed collapse of reticulin framework in areas of hepatocyte loss, and the cord architecture was preserved in other areas (Fig. S1). The CD34 stain showed increased sinusoidal endothelial staining, particularly near the portal tracts, with a heterogeneous staining pattern in the hepatic lobules (Fig. 2). Abdominal ultrasound revealed ascites. Upper endoscopy showed esophageal varices.
Kindred 4
Subject P4-1 is a 21-yr-old male who presented at 14 yr of age with petechia. Per an initial report (Patterson et al., 2018), this patient suffered from recurrent thrombocytopenia, mild hemolytic anemia, neutropenia, and lymphopenia. The patient was given a working diagnosis of immunodeficiency with features of autoimmunity. Ultimately, he was diagnosed with GIMAP5 deficiency at 16 yr of age (Patterson et al., 2018). Recently, at the age of 21, he was admitted with worsening jaundice, pancytopenia, and splenomegaly and was found to have noncirrhotic portal hypertension. His liver biopsy was remarkable for incomplete fibrous septa and features of nodular regenerative hyperplasia (Fig. S1). Like patients P1-1, P2-1, and P3-1, this subject’s liver tissue revealed abnormal sinusoidal CD34 expression (Fig. 2 A). Computed tomographic angiography of the abdomen and magnetic resonance imaging per hepatoma protocol confirmed portal hypertension with splenorenal shunt and esophageal varices and showed no evidence of portal or hepatic vein thrombosis. Upper endoscopy confirmed esophageal varices. Subject P4-1 was the only individual in this cohort who underwent a direct portal venogram. He was found to have an increased portal pressure of 14 mm Hg (normal portal venous pressure ranges between 5 and 10 mm Hg). The patient underwent recent elective splenectomy for worsening splenomegaly and thrombocytopenia and is clinically stable with close outpatient follow-up.
Online supplemental material
Fig. S1 shows representative images of histological findings seen in liver biopsies from patients with rare biallelic mutations in GIMAP5. Fig. S2 demonstrates persistent liver abnormalities in Gimap5sph/sphRag1−/− mice. Fig. S3 depicts representative images of subcellular localization of Gimap5 in sorted liver endothelial cells (DAPI−CD45−CD31+). Table S1 lists rare recessive mutations in GIMAP5 identified in affected individuals in four unrelated kindreds.
Supplementary Material
lists rare recessive mutations in GIMAP5 identified in affected individuals in four unrelated kindreds.
Acknowledgments
We thank the patients and their families for their invaluable contributions to this work; Geoffrey W. Butcher (The Babraham Institute, Cambridge, U.K.) for kindly providing the MAC421 anti-Gimap5 antibody; the staff of the Yale Center for Genome Analysis, the Yale Flow Cytometry Facility, the Yale Comparative Research Core, Scott Roberts, and Sandra Zehentmeier for technical assistance; and Ralf Adams for permission to access Cdh5(PAC)-CreERT2 mice.
This work was in part supported by the National Institutes of Health Centers for Mendelian Genomics (U54 HG006504); the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under award K08 DK113109 (to S. Vilarinho); the Yale Liver Center (P30 DK034989); and the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. J. Brancale is supported by the National Institute of General Medical Sciences of the National Institutes of Health under award 1T32GM136651-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions: S. Vilarinho and R.P. Lifton participated in designing the study. S. Vilarinho, K. Drzewiecki, J. Brancale, M.A. Leney-Greene, J.P. Pereira, J. Zhang, and R.P. Lifton performed research and/or participated in data analysis. S. Vilarinho, S. Sari, B. Dalgiç, A.Ü. Aksu, G.E. Şahin, A. Ozen, S. Baris, E. Karakoc-Aydiner, M. Schmalz, K. Radhakrishnan, H.C. Su, and M.J. Lenardo contributed to recruitment and/or ascertainment of participants. J. Choi and J. Brancale analyzed single-cell RNA-sequencing data with input from J.P. Pereira and S. Vilarinho. D. Jain and D. Kleiner contributed with histological analysis. K. Hoebe contributed Gimap5flx/flx mice. K. Drzewiecki and S. Vilarinho wrote the first draft; J.P. Pereira, M.J. Lenardo, and R.P. Lifton helped in writing the paper; and all authors critically read and contributed to the final version of the manuscript.
References
- Adzhubei, I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., and Sunyaev S.R.. 2010. A method and server for predicting damaging missense mutations. Nat. Methods. 7:248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizarani, N., Saviano A., Sagar L., Mailly L., Durand S., Herman J.S., Pessaux P., Baumert T.F., and Grün D.. 2019. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 572:199–204. 10.1038/s41586-019-1373-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzu, V., Fonseca M., Duckworth A., Kennard L., Moini N., Qurashi M., Brais R., Davies S., Manson A., Staples E., et al. 2019. Liver disease is common in patients with common variable immunodeficiency and predicts mortality in the presence of cirrhosis or portal hypertension. J. Allergy Clin. Immunol. Pract. 7:2484–2486.e3. 10.1016/j.jaip.2019.04.016 [DOI] [PubMed] [Google Scholar]
- Barnes, M.J., Aksoylar H., Krebs P., Bourdeau T., Arnold C.N., Xia Y., Khovananth K., Engel I., Sovath S., Lampe K., et al. 2010. Loss of T cell and B cell quiescence precedes the onset of microbial flora-dependent wasting disease and intestinal inflammation in Gimap5-deficient mice. J. Immunol. 184:3743–3754. 10.4049/jimmunol.0903164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yakov, G., Kapuria D., Marko J., Cho M.H., Pittaluga S., Kleiner D.E., Koh C., Holland S., Uzel G., and Heller T.. 2018. Liver disturbances in activated phosphoinositide 3-kinase δ syndrome. J. Allergy Clin. Immunol. Pract. 6:1763–1765. 10.1016/j.jaip.2018.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, M., Chang H.Y., Chuguransky S., Grego T., Kandasaamy S., Mitchell A., Nuka G., Paysan-Lafosse T., Qureshi M., Raj S., et al. 2021. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49:D344–D354. 10.1093/nar/gkaa977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Yu M., Dai X., Zogg M., Wen R., Weiler H., and Wang D.. 2011. Critical role for Gimap5 in the survival of mouse hematopoietic stem and progenitor cells. J. Exp. Med. 208:923–935. 10.1084/jem.20101192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter, T.I., Chandra A., Bacon C.M., Babar J., Curtis J., Screaton N., Goodlad J.R., Farmer G., Steele C.L., Leahy T.R., et al. 2017. Clinical spectrum and features of activated phosphoinositide 3-kinase δ syndrome: a large patient cohort study. J. Allergy Clin. Immunol. 139:597–606.e4. 10.1016/j.jaci.2016.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeve, L.D. 2015. Liver sinusoidal endothelial cells in hepatic fibrosis. Hepatology. 61:1740–1746. 10.1002/hep.27376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado, I., Carrasco M., Cano E., Carmona R., García-Carbonero R., Marín-Gómez L.M., Soria B., Martín F., Cano D.A., Muñoz-Chápuli R., et al. 2014. GATA4 loss in the septum transversum mesenchyme promotes liver fibrosis in mice. Hepatology. 59:2358–2370. 10.1002/hep.27005 [DOI] [PubMed] [Google Scholar]
- GBD 2017 Disease and Injury Incidence and Prevalence Collaborators . 2018. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 392:1789–1858. 10.1016/S0140-6736(18)32279-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géraud, C., Koch P.S., Zierow J., Klapproth K., Busch K., Olsavszky V., Leibing T., Demory A., Ulbrich F., Diett M., et al. 2017. GATA4-dependent organ-specific endothelial differentiation controls liver development and embryonic hematopoiesis. J. Clin. Invest. 127:1099–1114. 10.1172/JCI90086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri, Y. 2012. Endothelial dysfunction in the regulation of cirrhosis and portal hypertension. Liver Int. 32:199–213. 10.1111/j.1478-3231.2011.02579.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalucka, J., de Rooij L.P.M.H., Goveia J., Rohlenova K., Dumas S.J., Meta E., Conchinha N.V., Taverna F., Teuwen L.A., Veys K., et al. 2020. Single-cell transcriptome atlas of murine endothelial cells. Cell. 180:764–779.e20. 10.1016/j.cell.2020.01.015 [DOI] [PubMed] [Google Scholar]
- Kircher, M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., and Shendure J.. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna, G., Paez J., and Cardier J.E.. 2004. Expression of the hematopoietic stem cell antigen Sca-1 (LY-6A/E) in liver sinusoidal endothelial cells: possible function of Sca-1 in endothelial cells. Stem Cells Dev. 13:528–535. 10.1089/scd.2004.13.528 [DOI] [PubMed] [Google Scholar]
- Malamut, G., Ziol M., Suarez F., Beaugrand M., Viallard J.F., Lascaux A.S., Verkarre V., Bechade D., Poynard T., Hermine O., et al. 2008. Nodular regenerative hyperplasia: the main liver disease in patients with primary hypogammaglobulinemia and hepatic abnormalities. J. Hepatol. 48:74–82. 10.1016/j.jhep.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Moon, A.M., Singal A.G., and Tapper E.B.. 2020. Contemporary epidemiology of chronic liver disease and cirrhosis. Clin. Gastroenterol. Hepatol. 18:2650–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta, T., Nasreen M., Seike T., Goji A., Ohigashi I., Miyazaki T., Ohta T., Kanno M., and Takahama Y.. 2006. IAN family critically regulates survival and development of T lymphocytes. PLoS Biol. 4:e103. 10.1371/journal.pbio.0040103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, A.Y., Leney-Greene M., Lynberg M., Xu X., Zheng L., Zhang Y., Matthews H., Chao B., Morawski A., Jiang P., et al. 2021. Human immunodeficiency reveals GIMAP5 as lymphocyte-specific regulator of senescence. bioRxiv. (Preprint posted February 23, 2021) 10.1101/2021.02.22.432146 [DOI]
- Patterson, A.R., Endale M., Lampe K., Aksoylar H.I., Flagg A., Woodgett J.R., Hildeman D., Jordan M.B., Singh H., Kucuk Z., et al. 2018. Gimap5-dependent inactivation of GSK3β is required for CD4+ T cell homeostasis and prevention of immune pathology. Nat. Commun. 9:430. 10.1038/s41467-018-02897-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, A.R., Bolcas P., Lampe K., Cantrell R., Ruff B., Lewkowich I., Hogan S.P., Janssen E.M., Bleesing J., Khurana Hershey G.K., et al. 2019. Loss of GTPase of immunity-associated protein 5 (Gimap5) promotes pathogenic CD4+ T-cell development and allergic airway disease. J. Allergy Clin. Immunol. 143:245–257.e6. 10.1016/j.jaci.2018.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poisson, J., Lemoinne S., Boulanger C., Durand F., Moreau R., Valla D., and Rautou P.E.. 2017. Liver sinusoidal endothelial cells: physiology and role in liver diseases. J. Hepatol. 66:212–227. 10.1016/j.jhep.2016.07.009 [DOI] [PubMed] [Google Scholar]
- Rentzsch, P., Witten D., Cooper G.M., Shendure J., and Kircher M.. 2019. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47:D886–D894. 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothweiler, S., Terracciano L., Tornillo L., Dill M.T., Heim M.H., and Semela D.. 2014. Downregulation of the endothelial genes Notch1 and ephrinB2 in patients with nodular regenerative hyperplasia. Liver Int. 34:594–603. 10.1111/liv.12261 [DOI] [PubMed] [Google Scholar]
- Schaffner, F., and Poper H.. 1963. Capillarization of hepatic sinusoids in man. Gastroenterology. 44:239–242. 10.1016/S0016-5085(63)80130-4 [DOI] [PubMed] [Google Scholar]
- Schulteis, R.D., Chu H., Dai X., Chen Y., Edwards B., Haribhai D., Williams C.B., Malarkannan S., Hessner M.J., Glisic-Milosavljevic S., et al. 2008. Impaired survival of peripheral T cells, disrupted NK/NKT cell development, and liver failure in mice lacking Gimap5. Blood. 112:4905–4914. 10.1182/blood-2008-03-146555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, D., Ghobadi F., Boulay G., Ilangumaran S., Lavoie C., and Ramanathan S.. 2017. GTPase of the immune-associated nucleotide protein 5 regulates the lysosomal calcium compartment in T lymphocytes. Front. Immunol. 8:94. 10.3389/fimmu.2017.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris Consortium . 2018. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 562:367–372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., and Rinn J.L.. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32:381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilarinho, S., and Lifton R.P.. 2012. Liver transplantation: from inception to clinical practice. Cell. 150:1096–1099. 10.1016/j.cell.2012.08.030 [DOI] [PubMed] [Google Scholar]
- Vilarinho, S., and Lifton R.P.. 2016. Pioneering a global cure for chronic hepatitis C virus infection. Cell. 167:12–15. 10.1016/j.cell.2016.08.038 [DOI] [PubMed] [Google Scholar]
- Vilarinho, S., Sari S., Yilmaz G., Stiegler A.L., Boggon T.J., Jain D., Akyol G., Dalgic B., Günel M., and Lifton R.P.. 2016. Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension. Hepatology. 63:1977–1986. 10.1002/hep.28499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Nakayama M., Pitulescu M.E., Schmidt T.S., Bochenek M.L., Sakakibara A., Adams S., Davy A., Deutsch U., Lüthi U., et al. 2010. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 465:483–486. 10.1038/nature09002 [DOI] [PubMed] [Google Scholar]
- Ward, C., Lucas M., Piris J., Collier J., and Chapel H.. 2008. Abnormal liver function in common variable immunodeficiency disorders due to nodular regenerative hyperplasia. Clin. Exp. Immunol. 153:331–337. 10.1111/j.1365-2249.2008.03711.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, M., Staniczek T., Kürschner S.W., Schmid C.D., Schönhaber H., Cordero J., Kessler L., Mathes A., Sticht C., Neßling M., et al. 2021. Endothelial GATA4 controls liver fibrosis and regeneration by preventing a pathogenic switch in angiocrine signaling. J. Hepatol. 74:380–393. 10.1016/j.jhep.2020.08.033 [DOI] [PubMed] [Google Scholar]
- Wong, V.W., Saunders A.E., Hutchings A., Pascall J.C., Carter C., Bright N.A., Walker S.A., Ktistakis N.T., and Butcher G.W.. 2010. The autoimmunity-related GIMAP5 GTPase is a lysosome-associated protein. Self Nonself. 1:259–268. 10.4161/self.1.3.12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C., Gao X., Wei Q., Nakahara F., Zimmerman S.E., Mar J., and Frenette P.S.. 2018. Stem cell factor is selectively secreted by arterial endothelial cells in bone marrow. Nat. Commun. 9:2449. 10.1038/s41467-018-04726-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists rare recessive mutations in GIMAP5 identified in affected individuals in four unrelated kindreds.