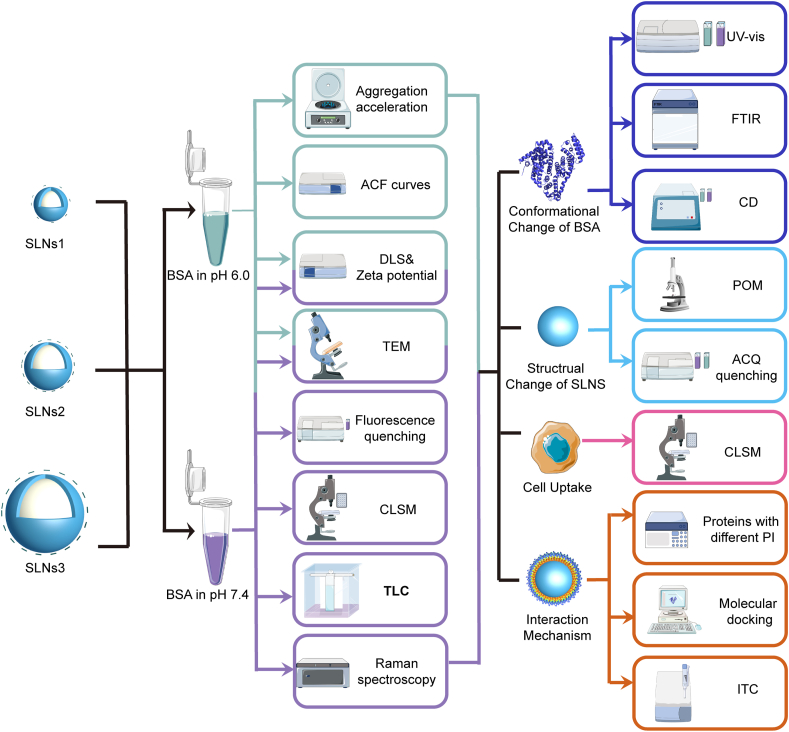
Abstract
When nanoparticles were introduced into the biological media, the protein corona would be formed, which endowed the nanoparticles with new bio-identities. Thus, controlling protein corona formation is critical to in vivo therapeutic effect. Controlling the particle size is the most feasible method during design, and the influence of media pH which varies with disease condition is quite important. The impact of particle size and pH on bovine serum albumin (BSA) corona formation of solid lipid nanoparticles (SLNs) was studied here. The BSA corona formation of SLNs with increasing particle size (120–480 nm) in pH 6.0 and 7.4 was investigated. Multiple techniques were employed for visualization study, conformational structure study and mechanism study, etc. “BSA corona-caused aggregation” of SLN2‒3 was revealed in pH 6.0 while the dispersed state of SLNs was maintained in pH 7.4, which significantly affected the secondary structure of BSA and cell uptake of SLNs. The main interaction was driven by van der Waals force plus hydrogen bonding in pH 7.4, while by electrostatic attraction in pH 6.0, and size-dependent adsorption was confirmed. This study provides a systematic insight to the understanding of protein corona formation of SLNs.
KEY WORDS: Protein corona, Solid lipid nanoparticles, BSA corona-Caused aggregation, Nanoparticle-protein interaction, Size effect, Cell uptake, Medium pH, Conformational structure
Graphical abstract
Different protein corona formation phenomena were shown in solid lipid nanoparticles (SLNS) with the various size and medium pH. The biological effect and the underlying interaction mechanisms were studied to provide a systematic perspective.
1. Introduction
The development of nanotechnology is opening a new avenue of research on drug-loading nanocarriers. By virtue of their unique physicochemical properties and biocompatibility, a wide variety of nanoparticles (NPs) like liposomes1, metal–organic framework2,3, polymeric nanoparticles4,5 and magnetic nanoparticles6 have been reported to improve the therapeutic effect7,8. Multiple functions were designed and confirmed in these innovative NPs and great experimental outcomes were shown as well.
However, only few of the NPs under development have processed into clinical or market up to now9,10. It is believed that the complicated in vivo environment may be the one to blame. When administrated in vivo, the NPs are exposed to complicated and diversified biological fluids with different pH, abundant biomolecules and immune cells, which dramatically impacts the biodistribution, metabolism, cellular internalization, and toxicity of administrated NPs11. Furthermore, the administration routes of NPs were diversified, and the biological environment the NPs exposed to varied from one disease to another. Hence, the specific physiological conditions must be taken into account when studying the NPs fate.
Among them, tremendous attentions have been paid to the formation of protein corona, a phenomenon where biomolecules in biological fluids like proteins inevitably bind to the surface of NPs and change the biological identity of the pristine NPs12,13. The surface identities of NPs will be more similar to that of proteins after the formation of protein corona. The in vivo fate of NPs with new identities endowed by protein corona may be less predictable14. It was reported that the NPs with protein corona may be eliminated by the mononuclear phagocytic system more quickly15, while other studies suggested that the protein adsorption may decrease the NPs–cell membrane adhesion and reduces the cell uptake16. Moreover, the biodistribution, biodegradation, pharmacokinetics and drug release profile were also be proved to be affected by the protein corona13,17,18. Thus, the formation of protein corona and its impacts are considered one of the keystones to the gap between in vitro design and in vivo therapeutic effect.
Thus, there is a pressing need for the understanding of protein‒NPs interactions to control the formation of protein corona. Several articles have suggested that the protein‒NPs interactions are associated with the physicochemical properties of the NPs and the media19,20. To control the formation of protein corona, the directional modification of the physicochemical properties of the NPs is an effective strategy. The modifications of surface charge, hydrophobicity and morphology of NPs are complicated and costly, which makes the NPs even harder for industrialization21. Compared to other properties of NPs, the control of NPs size is the most mature and commonly used methods during NPs designing. Besides, the medium pH of the lesion varies with the disease type and condition. Hence, the medium pH is considered one of the key factors during the protein corona formation of NPs.
A series of articles have been reported to illustrate the effect of particle size or media pH on protein corona formation in gold NPs22, 23, 24, silica NPs25,26 and polymeric NPs27. It was suggested that the amount and types of adsorption protein were significantly affected by these factors. However, the systemic studies about the interaction between solid lipid nanoparticles (SLNs) and protein are still missing. The conclusions or experiences obtained from the previous studies are not sufficient for comprehensive understanding the protein corona formation of SLNs. Unlike inorganic NPs, SLNs is a lipid-based nanocarrier which bears a core‒shell structure: a hydrophilic surfactant shell and a hydrophobic lipid core. In the past decades, SLNs was widely employed in the treatment of colon cancer28, cystic fibrosis29, bacterial infection30, sepsis31, etc., and great therapeutic effects were shown in these studies. Compared to metal or polymeric NPs, the clinical transformation of SLNs is also more promising for its simple and low-cost production process, high biocompatibility and excellent drug loading capacity. When the administrated SLNs absorbed into plasma and most human organs, they are exposed to near-neutral pH environment. Whilst, the environmental pH is significantly lower when SLNs are exposed to some specific acidic disease lesions including tumor32, inflammation sites33 or bacterial infection sites34. Thus, the understanding of protein corona formation of SLNs of those two pH environments is of utmost importance. The systematic study should be performed to fully understand the detailed mechanism and potential impact of protein corona formation of SLNs.
To this end, the protein corona formation of SLNs with different particle sizes in different pH was exploited in this proof-of-concept study. Bovine serum albumin (BSA) was chosen as the model protein for its abundant in vivo content, high stability and excellent intrinsic spectroscopic properties. Herein, the SLNs with three nanoparticle sizes were prepared, including 120 nm, 240 nm and 480 nm. The prepared SLNs were subjected to interaction with BSA in different pH, i.e., pH 6.0 (acidic disease lesions) and 7.4 (normal human organ), to characterize and compare the protein corona of different SLNs in different environmental conditions. The protein corona formation was confirmed and analyzed by morphology observation using transmission electron microscopy (TEM), confocal laser scanning microscope (CLSM), dynamic laser scattering (DLS) measurements, bicinchoninic acid (BCA) assay and fluorescence spectroscopy, etc. In addition, the impact of different pH on protein corona formation was investigated utilizing ultraviolet–Visible spectroscopy (UV–Vis), Fourier transform infrared spectroscopy (FTIR), circular dichroism (CD), and CLSM. Isothermal titration calorimetry (ITC), polarizing optical microscopy (POM), molecule docking technique and other techniques were carried out to claim the underlying mechanism. Besides, cell uptake of SLNs with BSA corona in different pH was studied. All experimental designs of the current study were shown in Scheme 1. We believe that our findings will provide new insight into the development and application of SLNs.
Scheme 1.
The experimental framework of the current study.
2. Materials and methods
2.1. Materials and cells
BSA and HEPES powders were purchased from neoFroxx GmbH (Einhausen, Germany). Sodium hydroxide (NaOH) and methanol were purchased from ZHIYUAN Chemical Reagent Factory (Tianjin, China). Cetyl palmitate (CP) was obtained from Aladdin Industrial Corporation (Shanghai, China). Tween 80 was purchased from Fuyu Fine Chemical Co., Ltd. (Tianjin, China). Acetonitrile was purchased from Saen Chemical Technology Co., Ltd. (Shanghai, China). FITC-BSA was obtained from Solarbio Science & Technology Co., Ltd. (Beijing, China). Aggregation caused quenching (ACQ) probes (P2 and P4) were generously gifted by Prof. Wei Wu’ group (Fudan University, Shanghai, China). Trypsin and ovalbumin were purchased from Coolaber Science & Technology Co., Ltd. (Beijing, China). Lysozyme was purchased from YINGMAO Analysis and Testing Co., Ltd. (Guangzhou, China). BCA protein assay kit was purchased from Cwbiotech Co., Ltd. (Beijing, China). Murine melanoma cells B16 and macrophages cells RAW 264.7 were purchased from Procell Life Science & Technology Co., Ltd. (Wuhan, China). Dulbecco's modified Eagle's medium (DMEM), 4′,6-diamidino-2-phenylindole (DAPI), 4% paraformaldehyde, Roswell Park Memorial Institute (RPMI) 1640 medium, fetal bovine serum (FBS) and phosphate buffer solution (PBS) were obtained from Gibco Life Technologies (New York, NY, USA). All water utilized in this study was purified by VEOLIA-ELGA system (Veolia group, Paris, France).
2.2. Preparation and characterization of SLNs
SLNs with three different hydrodynamic diameters (DH) including 120, 240 and 480 nm at pH 6.0 and 7.4 were prepared using a high-pressure homogenization method. CP and Tween 80 were chosen as the lipid and surfactant, respectively, and they were weighed according to Supporting Information Table S1. Tween 80 was introduced into 50 ml HEPES buffer solution with pH 6.0 or 7.4 as the aqueous phase. The lipids and aqueous phase were heated in a water bath to 70 or 80 °C for 20 min, and then the molten lipids were quickly poured into the aqueous phase. After 1 min of 10,000 rpm shearing with a batch high shear (FA25, FLUKO Equipment Co., Ltd., Shanghai, China), high pressure homogenization was carried out according to the conditions in Table S1 by a high-pressure homogenizer (EmulsiFlex-C3, Avestin, Inc., Ottawa, Canada). The prepared SLNs were stored at room temperature. The DH, size distributions, polydispersity index (PDI) and zeta potential of the prepared SLNs were determined using a Malvern Mastersizer 2000® (Malvern Instruments Ltd., UK). Thereto, after gently shaking for 15 s, the size distribution and zeta potential of SLNs after storing 0, 24 and 48 h at room temperature were determined to detect the storage stability of SLNs. The data were measured in a cuvette equilibrated at 25 °C prior to analysis.
2.3. DLS, zeta potential measurements and TEM in both pH
BSA was dissolved in the HEPES buffer solution in pH 6.0 or 7.4–12 μmol/L and incubated with SLNs. The DH, size distribution and zeta potential were determined by the DLS method in 0, 24 and 48 h, respectively. The DH, zeta potential and normalized scattered field autocorrelation functions (ACFs) curves for SLNs incubated with BSA at a series of concentrations were recorded using the Malvern Mastersizer 2000®. The TEM images of SLNs were taken using an electron microscope (JEM-1400, JEOL Ltd., Tokyo, Japan). The accelerating voltage was set at 120 kV.
2.4. Aggregation characterization in pH 6.0
The aggregation acceleration experiment was taken with a high-speed centrifugation method to simulate the aggregation state of long-term incubation of BSA and SLNs at pH 6.0. The SLNs were centrifuged for 30 min at 16,000 rpm with or without BSA using a high-speed centrifuge (GL-20C, Shanghai Anting Scientific Instruments Factory, China) at 4 °C. In most scenarios, the suspension was separated to two layers, and the BSA‒SLNs complexes were in the supernatant. The UV–Vis absorption (Abs) value of the subnatant was measured at 280 nm after 22 times dilution using a 1-cm path length quartz cell by an ultraviolet spectrometer (TU-1901, Purkinje General Instrument Co., Ltd., Beijing, China).
2.5. Relative amount of BSA adsorption
The relative amount of BSA adsorption was determined by BCA assay. Briefly, the SLNs was incubated with BSA by a predetermined concentration for 24 h. Then, the unadsorbed BSA was separated by ultrafiltration centrifugation (16,000 rpm, 30 min) and washed three times by HEPES buffer solution with different pH. The amount of unadsorbed BSA was measured according to the instruction of BCA protein assay kit and the relative BSA adsorption amount was calculated.
2.6. The protein corona formation characterization in pH 7.4
The ACQ probe P4 was involved in the CLSM tests to monitor the SLNs. The P4-labeled SLN were incubated with FITC-labeled BSA for 24 h. Then, the mixtures were imaged by FV3000 CLSM (Olympus, Tokyo, Japan) with excitation wavelength set at 488 nm and 640 nm and magnification set at 100. The image display and analysis were performed using the software Olympus FV31S.
With the intrinsic fluorescent property of BSA, the fluorescent quenching of BSA when incubated with SLNs at different concentrations for 0, 24 and 48 h were measured by Fluoromax-4 (HORIBA, USA) using a 3-cm quartz cuvette. The BSA concentration was set at 12 μmol/L, and the concentration range of SLNs was 0–2.5 mmol/L. Excitation was performed at 280 nm with a slit width of 3 nm and the emission was performed from 300 nm to 450 nm with a slit width of 3 nm. The measurement was conducted at pH 7.4.
2.7. The conformational change of BSA
To study the conformational change of BSA upon interaction with SLNs, the UV–Vis, FTIR and CD spectra at 0, 24 and 48 h were recorded. Specifically, the UV–Vis spectra were recorded from 200 nm to 800 nm using a 1.0 cm quartz cells by ultraviolet spectrophotometer (UV-2600, Shimadzu Co., Ltd., Japan) with sampling points every 1 nm. The CD spectra were acquired using a Chirascan spectropolarimeter (Chirascan, Applied Photophysics Ltd., UK) with a 1-mm path length quartz cell at room temperature, which were recorded from 200 to 300 nm with bandwidth set at 1 nm under a constant flow of nitrogen gas. All UV–Vis and CD spectra were measured three times and the data of HEPES buffer were subtracted as the blank. The FTIR spectra were recorded by the FTIR spectrometer (UATR Two, PerkinElmer, Waltham, MD, USA) in two pH values. The measurements were performed from 4000 to 400 cm−1 with a 4 cm−1 resolution in transmittance mode. All measurements were carried in triplicate and the spectra of air were subtracted as the blank.
2.8. Cells culture and cellular uptake
B16 cells were cultured in RPMI-1640 medium while RAW 264.7 were cultured in DMEM, supplemented with 10% FBS at 37 °C with 5% CO2. For cellular uptake study, the cells were seeded in 24‒well plates at a density of 8 × 104 cells per well and further cultured overnight. Then, the culture medium was replaced by the serum-free medium and the P4-loaded SLNs before or after 24 h incubation with BSA in different pH were added. After 3 h of incubation, the SLNs were washed twice by PBS and fixed by 4% paraformaldehyde for 10 min. Then, the redundant paraformaldehyde was washed by PBS and the cell nuclei were stained with DAPI for 10 min. Then, the cells were photographed by the CLSM with excitation wavelength set at 405 and 640 nm.
2.9. The interaction mechanism
The content of Tween 80 in SLNs was determined by a UV–Vis method reported before35. In brief, the SLNs were suspended by 100 mL of water and then subjected to ultrasonication (50 W, 40 kHz) for 10 min. The pretreated suspensions were filtered through a 0.1 μm microporous membrane (Membrana GmbH, Germany) and the UV–Vis adsorption at 233 nm was measured using an ultraviolet spectrometer (Purkinje General Instrument Co., Ltd.). The data was subjected to the calibration curves of Tween 80 and the content of Tween 80 in SLNs was determined.
ITC experiment was carried out using VP-ITC (Malvern Instruments Ltd., Malvern, UK). All samples were degassed before the analysis. The 12 μmol/L BSA solution contained in a motorized syringe (titrant) was introduced into a cell containing 1.2 μmol/L SLNs (substrate). The total injection was 25 after the primary equilibration, with 300 s intervals between the injections. The initial injection time lag was 60 s, and the stirring speed was set at 394 rpm under 37 °C. The blank experiment was conducted for baseline subtraction. The data were analyzed using the VP Viewer 2000 software (MICROCAL, Malvern Instruments Ltd., Malvern, UK) and the thermodynamic parameters were obtained.
2.10. Statistical analysis
Statistical analysis was performed using OriginPro 8.5 software. The significant differences between the data were determined based on P values < 0.05 by utilizing ANOVA or t-tests. All above experiments were measured three times except for special instructions.
3. Results and discussion
3.1. Preparation and characterization of SLNs
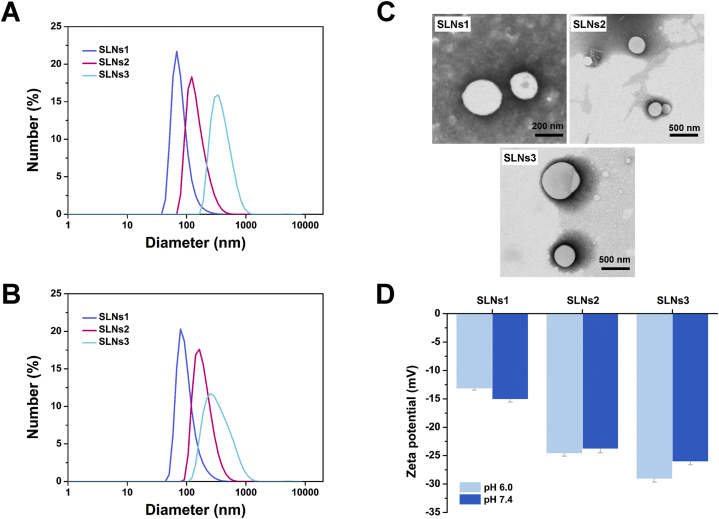
SLNs1 (120 nm), SLNs2 (240 nm) and SLNs3 (480 nm) were successfully prepared by a high-pressure homogenization method in pH 6.0 and 7.4 by CP and Tween 80. The DH, size distribution, PDI and zeta potential of them were shown in Fig. 1 and Table 1. The prepared SLNs have a narrow size distribution and a negative zeta potential in both pH values. The particle size of SLNs1–SLNs3 was significantly different in both pH (P < 0.05, Table 1). The TEM images (Fig. 1C) further revealed that the SLNs have a spherical shape and smooth surface. By measuring the size distribution and zeta potential evolution of SLNs in 48 h at room temperature, the storage stability was examined, and the results were shown in Supporting Information Fig. S1. The change of DH and zeta potential of SLNs were negligible in both pH, indicating that the SLNs were sufficiently stable for subsequent experiments.
Figure 1.
The particle size distribution of prepared SLNS in pH 6.0 (A) and 7.4 (B) (C) The TEM images of SLNS. (D) The Zeta-potential of prepared SLNS in different pH. (Data are expressed as mean ± SD, n = 3.
Table 1.
The DH and PDI of prepared SLNS.
| Formulation |
DH (nm) |
PDI |
||
|---|---|---|---|---|
| pH 6.0 | pH 7.4 | pH 6.0 | pH 7.4 | |
| SLNS1 | 134.3 ± 2.1 | 131.7 ± 0.6 | 0.205 ± 0.007 | 0.196 ± 0.007 |
| SLNS2 | 200.9 ± 4.7 | 217.8 ± 7.6 | 0.141 ± 0.017 | 0.131 ± 0.005 |
| SLNS3 | 449.2 ± 3.7 | 448.6 ± 6.0 | 0.213 ± 0.003 | 0.208 ± 0.026 |
Data are expressed as mean ± SD, n = 3.
3.2. DLS and zeta potential measurements in pH 6.0 and 7.4
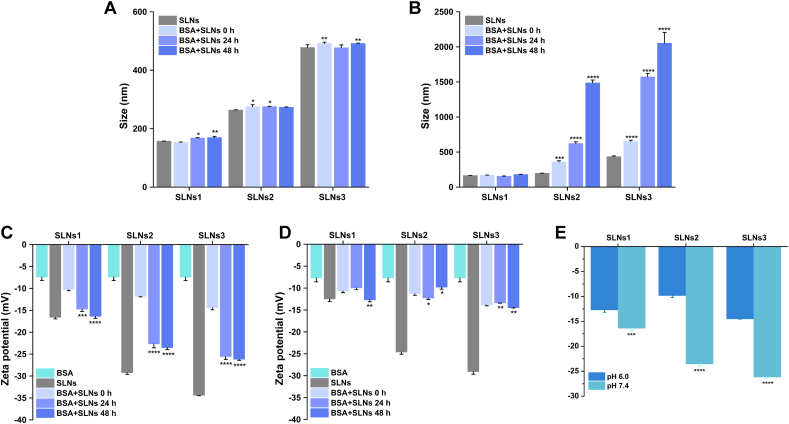
The BSA was dissolved in HEPES buffer solution at pH 6.0 or pH 7.4 for further experiment, and the maximum concentration of BSA was set at 12 μmol/L to maintain conformational stability (Supporting Information Fig. S2). The size and zeta potential of SLNs before and after incubation with BSA in 0, 24 and 48 h were measured, and the results were shown in Fig. 2 and Supporting Information Fig. S3. In pH 7.4 (Fig. 2A), the size of SLNs after incubation with BSA was only slightly larger than pristine SLNs in 48 h, presumably due to BSA adsorption. However, a significant increment of particle size was shown in pH 6.0 as soon as the BSA was added into SLNs except SLNs1 (Fig. 2B), and it became more obvious over time, which indicated that an aggregation of SLNs occurred in pH 6.0 immediately and the colloidal stability decreased. In addition, the zeta potential (absolute value) of SLNs after incubation with BSA were decreased compared to that of pristine SLNs in all samples, which was believed to be caused by the adsorption of low surface charge BSA on the high surface charge SLNs.
Figure 2.
The particle size of BSA incubated SLNS in 0, 24 and 48 h in pH 7.4 (A) and 6.0 (B). The zeta-potential of BSA incubated SLNS in 0, 24 and 48 h in pH 7.4 (C) and 6.0 (D). The zeta-potential of BSA incubated SLNS in different pH (E).∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001, Data are expressed as mean ± SD, n = 3, 0 h vs. other groups.
The increment of zeta potential of BSA incubated SLNs from 0 to 24 h in pH 7.4 (Fig. 2C) was extremely significant. This suggested that the adsorption of BSA took a longer time to reach the equilibrium. While in pH 6.0 (Fig. 2D), the zeta potential showed little difference among time points, suggesting that the adsorption was instant and stable. Furthermore, the zeta potential of SLNs in pH 6.0 was significantly lower than that in pH 7.4 after protein corona formation (Fig. 2E). The lower zeta potential meant the lower electrostatic repulsion between SLNs in pH 6.0, which might contribute to the process of aggregation and colloidal instability. This difference can be explained by the zwitterionic structure of BSA. When pH was higher than the PI of BSA (4.7), the higher the pH, the more negative charge was ionized. Thus, the electrostatic attraction between BSA and SLNs in pH 6.0 was much stronger than that in pH 7.4, which might lead to the more intense and rapid interaction. After adsorption, the lower pH (6.0) also lead to the more positive charged BSA, which endow the BSA coated SLNs with a lower zeta potential, inducing the aggregation of particles.
3.3. Colloidal stability of SLNs in pH 6.0
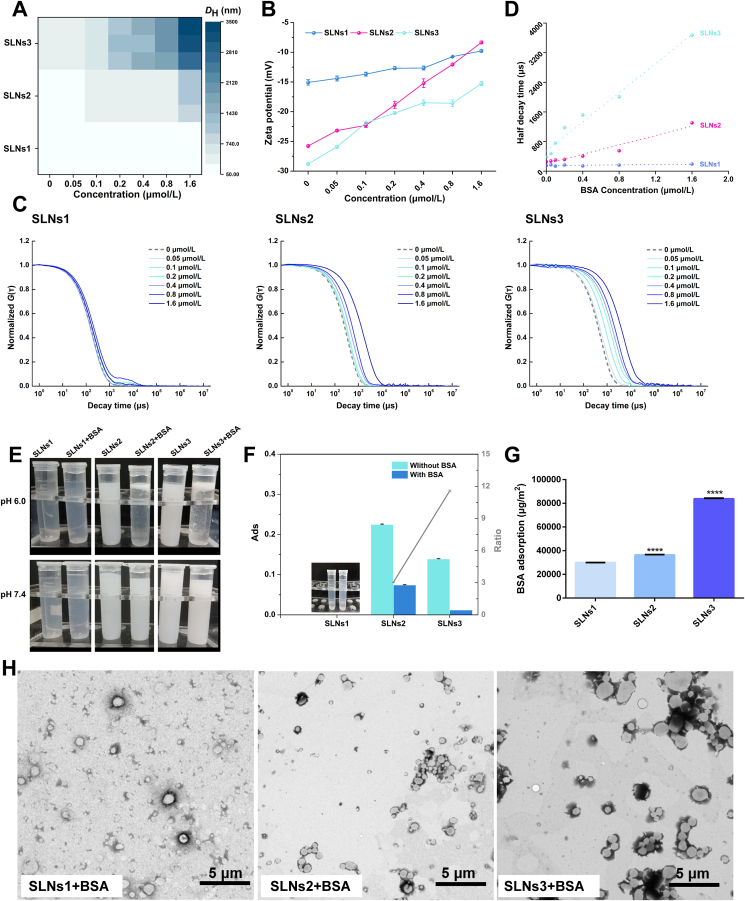
According to the DLS results, the aggregation of SLNs2 and SLNs3 was caused by the addition of BSA in pH 6.0. The TEM images of BSA incubated SLNs in pH 6.0 also show that the SLNs precipitated to micron-scale aggregates in pH 6.0 (Fig. 3H). To further confirm and analyze the colloidal stability of BSA‒SLNs complexes in this condition, a series of experiments were carried out.
Figure 3.
The colloidal stability of SLNS in pH 6.0: The heat map of DH–BSA concentration (A) and zeta-potential of BSA incubated SLNS in different BSA concentrations (B). The normalized ACF curves of BSA incubated SLNS in different BSA concentrations (C) and the half decay time-BSA concentration plots of SLNS–BSA (D). The images of SLNS with or without BSA in tubes (E) and the 280 nm Abs values of subnatant of BSA incubated SLNS after centrifugal and its ratio (with BSA vs. without BSA) (F). The relative BSA adsorption amount of SLNS determined by BCA assay (SLNS1 vs. other groups) (G). The TEM images of BSA incubated SLNS (H). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001, Data are expressed as mean ± SD, n = 3.
The effect of BSA concentration on the degree of SLNs aggregation was described by the particle size of SLNs with the addition of BSA at different concentrations. The heat map of DH vs. BSA concentration was shown in Fig. 3A. The darker the color, the larger the particle size. The particle size of BSA incubated SLNs became larger as the increment of BSA concentration apart from SLNs1, indicating that the amount of BSA was favorable to the aggregation of SLNs2 and SLNs3. In addition, the zeta potential of the samples was determined at the same time (Fig. 3B). With the incubation with the higher BSA concentration, the zeta potential was getting lower, corroborating the smaller repulsion force between SLNs and a less colloidal stability. Based on these results, we speculated that the aggregation of SLNs could be defined as “BSA corona-caused aggregation”. The BSA corona offered a new surface identity of SLNs: a more neutral charge, which might be one of the triggers of the aggregation.
To further explore the effect of particle size on the “BSA corona-caused aggregation” phenomenon, the normalized scattered field ACFs of SLNs in the presence of different concentrations of BSA were measured and the results are shown in Fig. 3C. The ACFs curves were fitted into the intensity autocorrelation function in Eq. (1):
| (1) |
where the G(τ) is the scattering intensity, B is the baseline, A is the intercept of ACFs curves, Dt is the diffusion coefficient, q is the scattering vector and τ is the decay time. According to Eq. (1), the larger the Dt value, the faster the scattering intensity decay, and the steeper the ACFs curve is. The correlation between Dt and DH can be explained by the Stokes–Einstein relation (Eq. (2)).
| (2) |
where KB is the Boltzmann's constant, T is the temperature and η is the viscosity of solvent. Hence, the negative correlation between Dt and DH was revealed, indicating that the decay time is proportional to the particle size. The ACFs curves of BSA incubated SLNs became flatter with the increase of BSA concentration, indicating the larger DH of the aggregates formed except for SLNs1 in pH 6.0. The half decay time was employed to compare the difference of ACFs curves of SLNs1‒SLNs3, and the half decay time‒BSA concentration plots were shown in Fig. 3D. The faster the plots raised, the greater the influence of BSA on colloidal stability. In accordance with the previous results, the boost rate of half decay time with the increment of BSA concentration was positively correlated with particle size of SLNs.
Then, the images of SLNs with or without BSA at pH 6.0 were taken after 48 h and shown in Fig. 3E. Compared to pristine SLNs, a significant layering behavior, viz. upper white layer and lower translucent layer, was recorded in the BSA incubated SLNs except SLNs1. This phenomenon demonstrated that the homogeneous suspension of SLNs was destroyed, and deterioration of colloidal stability took place. Hence, the aggregation acceleration experiment was carried out to simulate a much longer storage time and analyze the aggregation quantitatively. During the centrifugation process, the collisions and interactions between SLNs are strengthened and accelerated, accelerating the colloid unstable process of SLNs induced by BSA adsorption36,37. The ratio of 280 nm Abs of the subnatant between the pristine SLNs and BSA-incubated SLNs after centrifugation was employed to exam the degree of colloidal instability of SLNs. The higher the value, the less stable the SLNs was. As shown in Fig. 3F, the Abs of all BSA incubated SLNs was much lower than that of pristine SLNs, except that there was no obvious layering in SLNs1. This phenomenon demonstrated the colloidal instability of SLNs2 and SLNs3 after incubation with BSA. The plot of Abs ratio value increased with the increase of SLNs particle size, which suggested that the larger SLNs was more susceptible to aggregation upon interaction with BSA. The BCA assay was carried out to determine the relative amount of BSA adsorption. It was shown that the adsorption BSA amount was increased as the increment of particle size (Fig. 3G), which further proved the size-dependent of BSA adsorption.
Based on the above results, it could be concluded that the aggregation of SLNs except for SLNs1 occurred upon the addition of BSA in pH 6.0, which might be driven by the reduced electrostatic repulsion, suggesting the weakening of colloidal stability of SLNs under this circumstance. Furthermore, the effect of BSA on colloidal stability of SLNs positively depended on the particle size, time and BSA concentration within the experimental conditions.
3.4. The BSA corona formation in pH 7.4
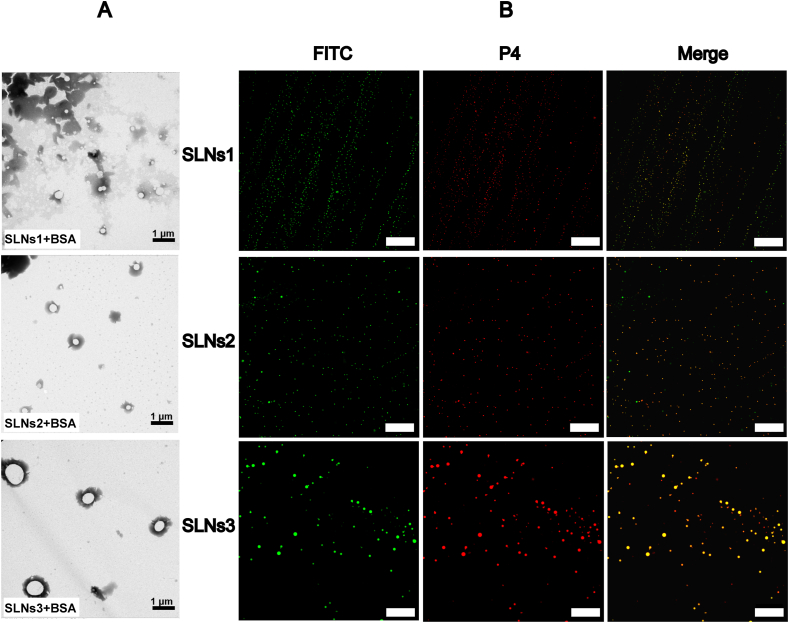
The formation of BSA corona in pH 7.4 was observed by the TEM images and CLSM images. As shown in Fig. 4A, the TEM images of SLNs after incubation with BSA maintained dispersed and uniform state, indicating no aggregation caused by the addition of BSA. As the protein corona was a thin layer, the morphology of BSA incubated SLNs was not significantly changed compared to the pristine SLNs. The confocal images of FITC-BSA incubated P4-SLNs in pH 7.4 were shown in Fig. 4B. FITC-BSA and P4-SLNs show green and red signals, respectively. The uniform distribution of the nanoparticles with four particle sizes was clearly shown in the fluorescence image. The majority of the green fluorescence of FITC-BSA was colocalized with the red fluorescence of P4-SLNs, suggesting that most of the BSA were adsorbed on the surface of SLNs and gave rise to the formation of protein corona.
Figure 4.
The TEM (A) and CLSM (B) images of BSA incubated SLNS in pH 7.4. Scale bar = 20 μm.
In addition, the formation of protein corona could also be confirmed by the Raman spectrum and thin layer chromatography (TLC) image. The disappearance of the characteristic band at 2880 cm−1 after incubation with BSA in Raman spectrum indicates the SLNs was coated by the protein corona (Supporting Information Fig. S4A). As for the TLC experiment, all SLNs was moved to the upper part of plate but part of BSA-incubated SLNs stayed at the origin, which might demonstrate that the change of polarity with protein corona formation (Fig. S4B).
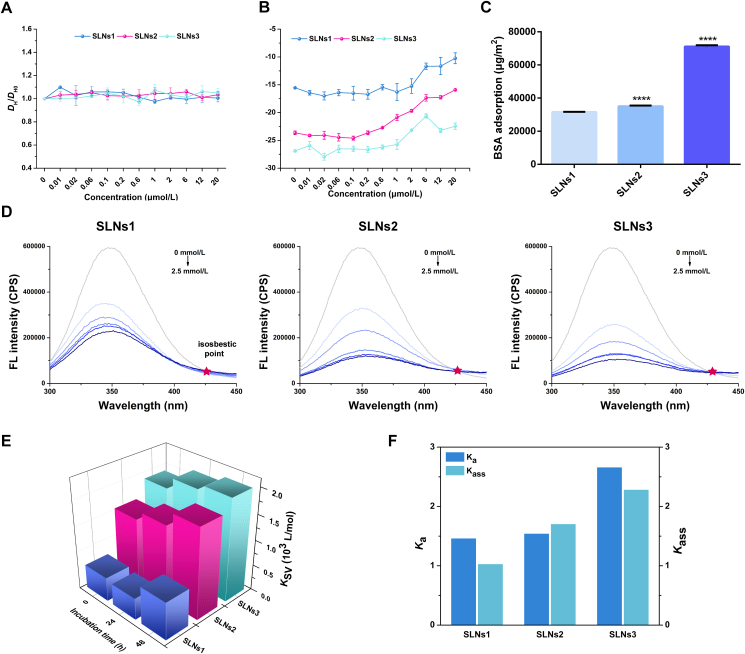
The particle size and surface charge measurement of SLNs after incubation with different concentration BSA solution were performed to further analyze the protein corona formation. As shown in Fig. 5A, there was no significant change in DH of SLNs before and after incubation with different concentrations of BSA, indicating the BSA adsorption occurred rather than aggregation. Also, the normalized ACF curves illustrated the maintaining of colloidal stability (Supporting Information Fig. S5). Similar to that in pH 6.0, the zeta potential of SLNs incubated with different BSA concentrations also getting lower with the higher BSA concentration in pH 7.4 (Fig. 5B). However, the zeta potential in pH 7.4 was not as low as that in pH 6.0 and thus the repulsion force was strong enough to maintain the SLNs dispersion. Then, the relative adsorption amount of BSA was measured by the BCA assay. The size dependency of BSA adsorption was also confirmed in pH 7.4 (Fig. 5C). These results indicated that the BSA was adsorbed on the surface of SLNs in particle size order.
Figure 5.
The protein corona formation of SLNS in pH 7.4. The particle size (A) and zeta-potential (B) curves of BSA incubated SLNS in different BSA concentrations. The relative BSA adsorption amount of SLNS determined by BCA assay (SLNS1 vs. other groups) (C). The fluorescence quenching of BSA after incubation with different concentrations of SLNS in 48 h and the red stars represent the isosbestic point (D). The KSV values of SLNS-BSA in 0, 24 and 48 h (E). The Ka values and the Kass values of SLNS-BSA in 48 h (F). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001, Data are expressed as mean ± SD, n = 3.
Then, to analyze the binding mode of BSA and SLNs in pH 7.4, the fluorescence quenching method was employed. With excitation at 280 nm, BSA exhibited strong and stable intrinsic fluorescence due to its two tryptophan (Trp) residues (Trp-212 and Trp-134), whereas SLNs and HEPES buffer did not show a considerable emission (Fig. S4C)38,39. The fluorescence quenching of BSA in the presence of SLNs in 0, 24 and 48 h was measured and the results were shown in Supporting Information Fig. S6 (0 and 24 h) and Fig. 5D (48 h). The fluorescence emission intensity of BSA was progressively decreased upon the addition of SLNs in all time points, indicating the interaction between BSA and SLNs. The isosbestic point (the red star) at about 430 nm indicated equilibrium was attained between BSA and SLNs and signified the formation of the ground state complex40. To further elucidate the fluorescence quenching mechanism caused by the SLNs, the Stern–Volmer equation was utilized to process the quenching data as shown in Eq. (3)41:
| (3) |
where F0 and F are the fluorescent intensities of BSA with or without SLNs, KSV is the quenching constant, Kq is the biomolecular-quenching-rate constant, τ0 is the average fluorescence lifetime of biomolecular which is about 10−8 s, and [Q] is the concentration of SLN incubated with BSA. According to the equation, the fluorescence quenching plots of BSA‒SLNs in pH 7.4 in all time points (Supporting Information Fig. S7) exhibited a good linear fitting, suggesting a single dynamic quenching mechanism or static quenching mechanism. The KSV values, which were used to evaluate the binding affinity of SLNs to BSA, were calculated based on the slope in different time points and shown in Fig. 5E. At all time points, the increased KSV values with the elevated SLNs size demonstrated a positive correlation between BSA‒SLNs interaction and SLNs size. Time evolution was also shown by that the KSV values increased over time, indicating a continuous process of interaction between BSA and SLNs. Furthermore, all Kq values of BSA‒SLNs complexes were higher than 2 × 1010 L/mol/s, the maximum scatter collision quenching constant, which meant that a static BSA quenching mechanism caused by the formation of a complex (like protein corona) is operative42.
As the interaction reaches equilibrium in 48 h, the binding molar ratio association constant (Kass) and number of binding sites (n) of BSA was evaluated by the Scatchard plot analysis43 as shown in Eq. (4):
| (4) |
where the linear regression with lg [Q] as the abscissa and as the ordinate was obtained and shown in Supporting Information Fig. S8A. The Kass was calculated by the intercept and the n was determined by the slope. Like the trend of KSV value, the Kass also showed a positive correlation with particle size (Fig. 5F), indicating a higher proportion of nonfluorescent fluorophore‒quencher complex in the BSA incubated SLNs with larger size.
Furthermore, the binding affinity constant (Ka) for BSA‒SLNs interaction was also calculated by the modified Stern–Volmer equation as in Eq. (5)44:
| (5) |
where fa is the fraction of accessible fluorescence. The plots of F0/(F0–F) vs. 1/[Q] result in 1/faKa as the slope and 1/fa as the intercept. The plots and the calculated results of Ka was shown in Fig. 5G. It was shown that the Ka of BSA‒SLNs increased with the increasing of DH, which was consistent with the patterns of Ksv and Kass values (Fig. 5F). Taken together, it could be concluded that the protein corona was formed in pH 7.4, which was positively correlated with particle size and evolved with time.
3.5. Conformational change of BSA
After confirming the integrity of SLNs after protein corona formation in both pH by employing the “on–off” ability of ACQ probes45, 46, 47 (Supporting Information Section 6). The conformational change of BSA was also detected. The conformation of proteins may change after they are adsorbed on NPs. Several studies suggested that the protein secondary structure might be altered significantly after interaction with model NPs22,48,49 while no significant conformational changes was found in other studies39,50. The UV–Vis, CD and FTIR spectra of BSA before and after incubated with SLNs were measured to clarify the conformational change of BSA interacted with SLNs in pH 6.0 and 7.4.
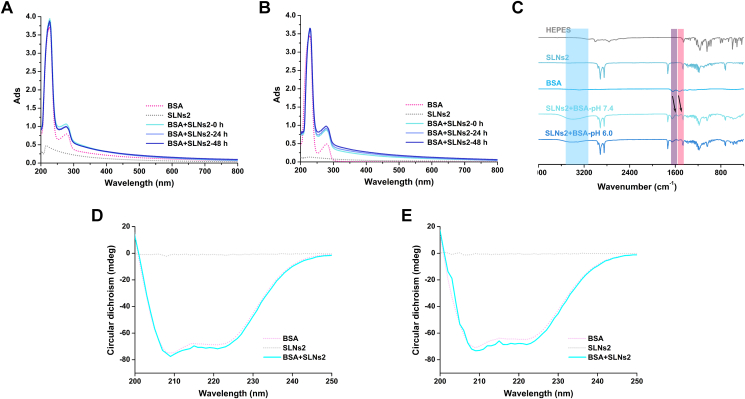
Analysis of UV–Vis spectra of BSA‒SLNs complexes was considered an effective approach to exam the interaction between protein and NPs. The adsorption peaks of BSA at 220 nm were due to the typical peptide-bond backbone structure while the adsorption peaks at 277 nm were ascribed to the aromatic amino acids (Trp, Tyr, and Phe)51. The UV–Vis adsorption changes of BSA at 0, 24 and 48 h with or without SLNs in pH 6.0 and 7.4 were shown in Fig. 6A and B and Supporting Information Fig. S9. Evolving with time, the adsorption peak of BSA at 277 nm after incubation with SLNs increased in pH 7.4 but decreased in pH 6.0, suggesting a different interaction mode. The hyperchromic shift for BSA with incubation with SLNs in pH 7.4 might contribute to the partial exposure of photon adsorbing amino acid residues, indicating the conformational change of BSA upon interaction with SLNs. In contrast, the aggregation of SLNs in pH 6.0 might result in the wrapping and hiding of these amino acid residues, which in turn led to the reduced Abs values.
Figure 6.
The conformational change of BSA upon interaction with SLNS. The UV–Vis spectra of BSA with or without SLNS2 in pH 6.0 (A) and pH 7.4 (B). The FTIR spectra of BSA with or without SLNS2 in different pH of SLNS2 (C). The CD spectra of BSA with or without SLNS2 in pH 6.0 (D) and pH 7.4 (E).
The FTIR spectra were investigated to further identify the structural changes of BSA upon binding with SLNs. Both protein amide I band 1700‒1600 cm−1 (mainly C O stretching vibrations) and amide II band 1600 ‒1500 cm−1 (C–N stretching vibrations coupled with a N–H in plane bending mode) correlated with the secondary structures of proteins52,53. The amide I bands were contributed from different secondary structures of protein, including 1620‒1645 cm−1 from β-sheet content, 1645‒1652 cm−1 from random coil content and 1652‒1662 cm−1 from α-helix content51. As shown in Fig. 6C and Supporting Information Fig. S10, the FTIR spectra of pure BSA displayed two characteristic bands at 1648 and 1535 cm−1, corresponding to protein amide I and amide II, respectively. However, with the binding with SLNs, the amide I band of BSA shifted from 1648 to 1656 cm−1 while the amide II band shifted from 1535 to 1543 cm−1 in both pH 6.0 and 7.4. The results suggest that the addition of SLNs caused a transformation of the secondary structure of BSA, like the increase of the proportion of α-helix. Furthermore, the broad band of BSA after incubation with SLNs at about 3400 cm−1 was assigned to the stretching vibrations of O–H and N–H, implying that the hydrogen bond might play a part in the binding between BSA and SLNs.
Furthermore, the conformational change of BSA upon binding with SLNs was analyzed by CD spectroscopy. The CD spectroscopy was considered a powerful tool to analyze the secondary structural change of protein. The CD spectra of BSA with or without SLNs in different time points were shown in Fig. 6D and E, and Supporting Information Fig. S11. Two negative bands at 208 and 220 nm in the UV region were exhibited and the values of which were correlated with the α-helix content of protein19. Similar to the FTIR results, the ellipticity values at 208 and 220 nm of BSA were significantly increased upon the binding with SLNs, indicating the increase of the proportion of α-helix.
In summary, it was suggested that the secondary structure of BSA was changed upon the interaction with BSA, viz. the exposure or hidden of photon adsorbing amino acid residues and the increment of the content of α-helix. However, further differences might be found in the tertiary or quaternary structure of BSA, and related studies need to be conducted. These findings implied that when SLNs was administrated in vivo, the secondary structure of proteins adsorbed on its surface might be changed and the biological functions of them might be affected in consequence. This process was unpredictable due to the complicated biological media. From this viewpoint, the protein corona formation needs to be controlled to prevent the potential influence on the fate of NPs or relevant normal physiological processes.
3.6. Cell uptake
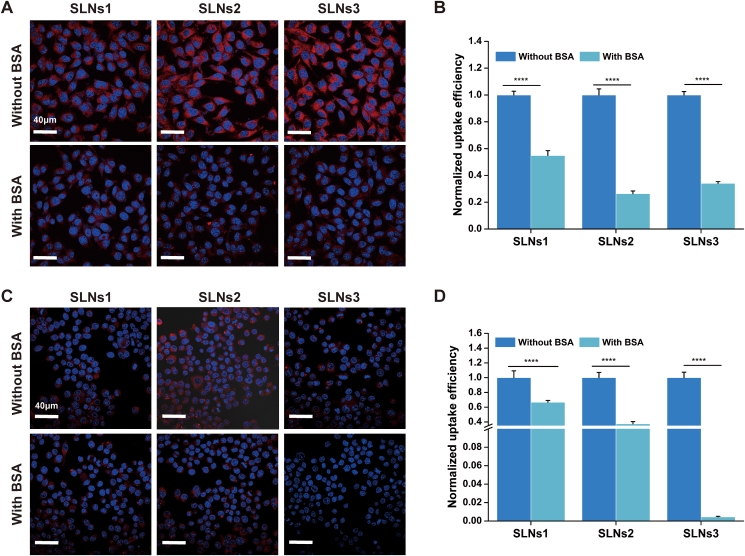
In order to investigate the effect of protein corona at the cellular level, the cellular uptake of SLNs was explored by CLSM. The B16 (melanoma cell, representing acidic environmental cells in tumor) and RAW 264.7 (macrophages cell, representing neutral environmental cells in plasma) were chosen as the model cells for pH 6.0 and 7.4, respectively. The cell nuclei were stained with DAPI (blue) and the SLNs was loaded with P4 (red). As shown in Fig. 7A, the red fluorescence of SLNs was significantly decreased after incubation with BSA in pH 6.0, indicating the B16 uptake of SLNs was decreased when BSA corona-caused aggregation occurs in pH 6.0. The mean fluorescence intensity was calculated and at least 60% reductions were shown (Fig. 7B). Given the NPs–cell interaction was strongly affected by the particle size of NPs, the cell uptake reduction might be caused by the increment of SLNs size. This meant the less efficient cell internalization and compromised therapeutic effect. Unexpectedly, SLNs1 which did not aggregate also showed reduced B16 uptake. It was speculated that the protein corona on the SLNs1 surface might enhance the hydrophilicity and affected the uptake of B16 and detailed mechanism would be investigated in the future.
Figure 7.
The cell uptake of SLNS before and after incubation with BSA in different pH: The CLSM images (A) and normalized uptake efficiency (B) of B16 uptake of SLNS before and after incubation with BSA in pH 6.0. The CLSM images (C) and normalized uptake efficiency (D) of RAW 264.7 uptake of SLNS before and after incubation with BSA in pH 7.4. ∗∗∗∗P < 0.0001, Data are expressed as mean ± SD, n = 3.
In pH 7.4, the reduction of macrophages uptake of SLNs was shown as well (Fig. 7C and D). According to previous studies, the BSA adsorption reduced the macrophages uptake in reticuloendothelial system because the enhanced surface hydrophilicity suppressed NPs–cell interaction16. It was indicated that the blood circulation time would be prolonged, and the therapeutic effect would be increased after the protein corona formation. Thus, the protein corona formation could have a favorable effect on the pharmacokinetics and pharmacodynamics of SLNs when exposed to normal organs.
These findings implied the necessity of environment-responsive NPs to smartly adjust the protein adsorption behavior. For instance, NPs that formed protein corona in neutral environment (prolonged circulation) and avoid protein absorption in acidic environment (enhanced uptake) would facilitate the therapy of specific diseases. It was important to figure out the underlying protein corona formation mechanism to for the design of ideal NPs.
3.7. Mechanism studies
3.7.1. The effect of surface hydrophilicity on the SLNs–BSA interaction
The Tween 80 content in SLNs was quantified by UV–Vis method and the results were shown in Table 2. As shown, the smaller the SLNs size, the higher the Tween 80 content was. Furthermore, the Tween 80 content of SLNs1 was much higher than that of the others, indicating that the surface of SLNs1 was most hydrophilic. According to the previous report, the surface hydrophobicity of NPs played an important role in the interaction with proteins, and the high hydrophilicity was a basic characteristic of materials with good anti-protein-adsorption ability54. It was hypothesized that the anti-protein-adsorption ability was highly associated with the hydration layer near the surface which formed a physical and energetic barrier to prevent protein adsorption on the surface55. From this standpoint, the high hydrophilic surface of SLNs1 might be a determinant of the weaker interaction level with BSA compared with SLNs2 and SLNs3.
Table 2.
The determined Tween 80 content in SLNS formulations.
| Formulation | Tween 80 content in SLNS (%, w/w) |
|---|---|
| SLNS1 | 31.742 ± 0.984 |
| SLNS2 | 7.775 ± 0.014 |
| SLNS3 | 2.927 ± 0.005 |
Data are expressed as mean ± SD, n = 3.
3.7.2. ITC assay
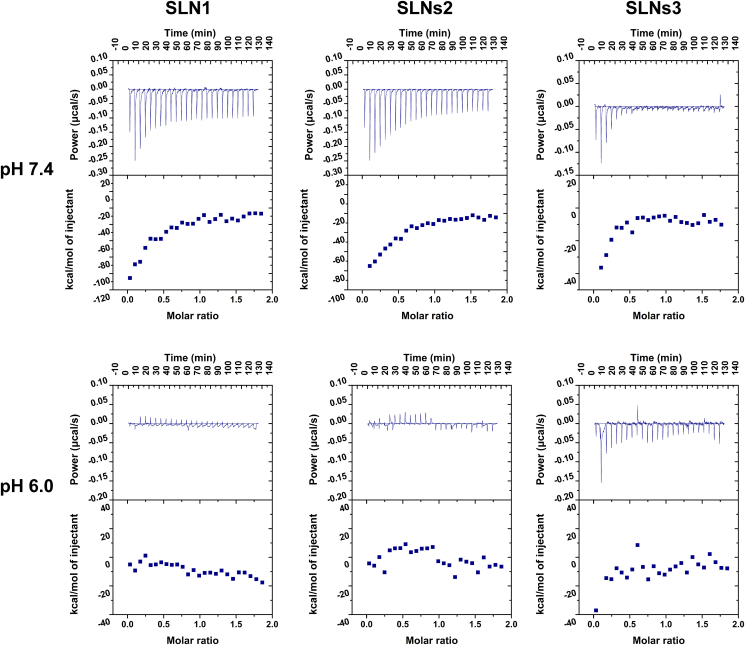
ITC gave more insight of the protein‒NPs binding mode from the thermodynamics approach, and it was employed to measure the enthalpy changes occurred through protein–nanoparticles interactions in many literatures54,56. The thermodynamic parameters including binding enthalpy (ΔH) and entropy (ΔS) could be determined through a single site binding model. According to the previous study, ΔH>0 and ΔS>0 corresponded to hydrophobic force, ΔH<0 and ΔS<0 corresponded to van der Waals force and hydrogen bond, and ΔH<0 and ΔS>0 corresponded to electrostatic interactions54. The ITC power–time data (upper panel) and the subsequent binding isotherm of BSA to SLNs (lower panel) were illustrated in Fig. 8. In pH 7.4, the negative injection signals implied an exothermic process of BSA‒SLNs interaction and the binding isotherm were fitted into single site binding model. The parameters obtained are shown in Supporting Information Table S2. The negative ΔH and ΔS values demonstrated that the van der Waals force and hydrogen bond, which were strong enough to negate the other interaction force like electrostatic attraction, played the main role in the interaction between BSA and SLNs.
Figure 8.
The ITC power-time data (upper panel) and the subsequent binding isotherm of BSA to SLNS (lower panel).
However, in pH 6.0, when BSA was injected to SLNs, weak and irregular enthalpy changes were recorded and the data could not be fitted by any binding models. According to the binding isotherm, endothermic and exothermic reactions are mixed in the interaction process, indicating the presence of multiple interaction forces. This phenomenon might be induced by the violent interaction amongst BSA, SLNs and protein corona-coated SLNs. The irregular binding isotherm might show the sum of these interaction forces like electrostatic attraction, hydrophobic effect, van der Waals force and hydrogen bonding. These results were strong proofs to the previous hypothesis that the aggregation of SLNs in pH 6.0 was BSA corona-caused aggregation.
3.7.3. Mechanism discussion
The interaction between Tween 80 and BSA was investigated, and the hydrophobic effect, van der Waals force and hydrogen bonding were proved to be the main interaction force between Tween 80 and BSA. The Tween 80–BSA interaction may attribute to the protein corona formation. In addition, several proteins with different PI were introduced to SLNs in different pH to monitor the aggregation state. The results show that the proteins with high PI would lead to SLNs aggregation more easily, which implied that the electrostatic attraction was a vital part of the interaction among NPs, proteins and protein corona coated NPs in low pH (Supporting Information Section 6).
In this study, multiple interactions existed between the prepared pristine SLNs, including electrostatic repulsion, hydrophobic effect, van der Waals force and hydrogen bonding. They could be divided into repulsion force (electrostatic repulsion) and attraction forces (other forces). The electrostatic repulsion force induced by the negative surface charge of pristine SLNs was larger than the attraction forces and consequently the dispersion state during storage was maintained. However, once the BSA solution was introduced into the SLNs, the adsorption of BSA on the SLNs surface occurred. The driven force varied under different circumstances. The main part was played by van der Waals force and hydrogen bond in pH 7.4, but by electrostatic attraction in pH 6.0. The larger SLNs showed greater interaction with BSA, which were in concordant with other reports25,57. In addition, the reaction rate of SLNs and BSA in different pH was also significantly different. The immediate BSA adsorption and protein corona-caused aggregation occurred as soon as the addition of BSA solution in pH 6.0, while the formation of BSA corona took a much longer process (up to 24 h) to reach equilibrium in pH 7.4.
It is worth mentioning that the BSA molecule arrangement on SLNs surface may also be impacted by the environment pH. It was reported that the BSA may be more compact near the PI, which in turn favored the BSA adsorption on SLNs surface58. Thus, in pH 6.0, the stronger BSA adsorption may be ascribed to the more compact arrangement of BSA molecule. In addition, due to the stronger repulsion between BSA on SLNs surface in pH 7.4, the coverage proportion of BSA on SLNs surface may be lower59. The different repulsion and BSA molecule arrangement may have an impact on the surface properties like charge intensity, and the detailed mechanism should be taken into further study. Thus, the surface property of SLNs was changed but not simply equivalent to that of BSA after corona formation.
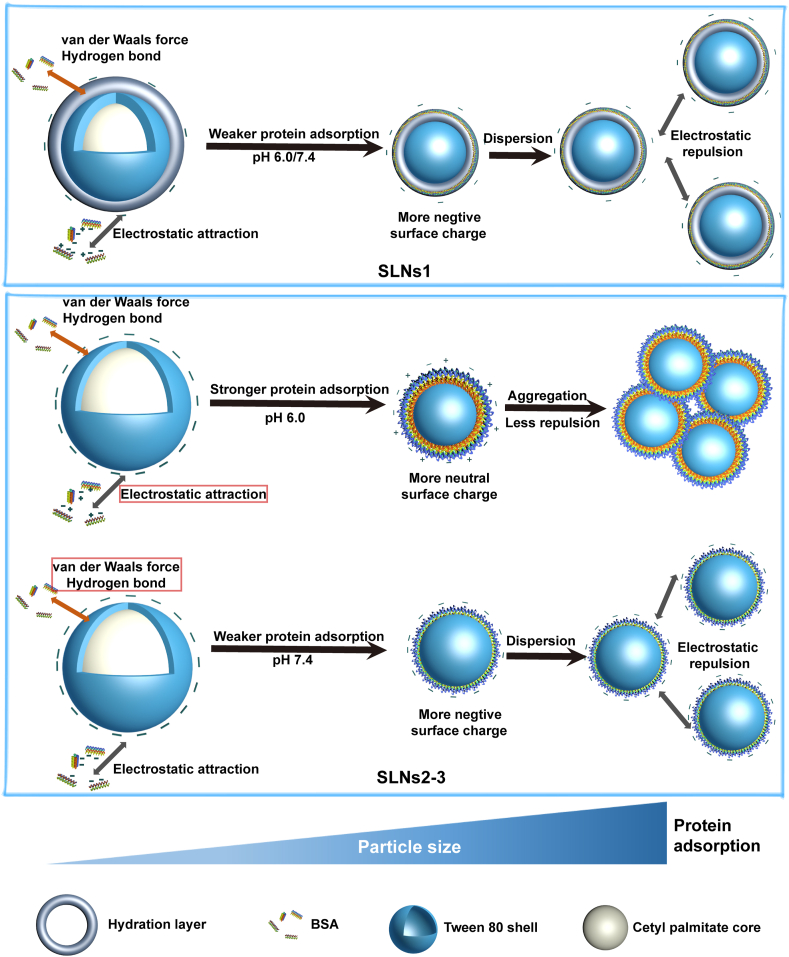
The surface properties of SLNs were changed with the formation of BSA corona. In pH 6.0, the electrostatic repulsion between SLNs was weakened by the more neutral surface charge and the equilibrium of repulsion and attraction forces was broken. The protein corona-caused aggregation phenomenon happened in SLNs2 and SLNs3. But for SLNs1, the smallest size and the most hydrophilic surfaces protect it from excessive protein adsorption and in turn show great colloidal stability. This conclusion could be proved by the introduction of Try and Lys with higher PI value than BSA, in which the larger electrostatic attraction between protein corona-coated SLNs1 also led to the aggregation. However, the adsorption of BSA did not bring such drastic changes of surface properties in pH 7.4 and the balance between repulsion and attraction forces of SLNs still existed. Thus, there was no aggregation found. These scenarios were summarized in Scheme 2.
Scheme 2.
The mechanism of BSA corona formation and the impact of particle size and pH. The main driving force was marked by the red box.
4. Conclusions
Herein, we reported the interaction between SLNs and BSA to explore the impact of particle size and pH on BSA corona formation and the consequent effects. Upon incubation with BSA, the BSA corona was formed. The BSA adsorption was driven by multiple forces. The main interaction was van der Waals force and hydrogen bonding in pH 7.4, while electrostatic attraction in pH 6.0. The positive correlation between particle size and protein adsorption were shown as well. Besides, the protein corona-caused aggregation occurred in pH 6.0 (except for SLNs1) due to the weakening of electrostatic repulsion, but no signs of aggregation were exhibited in pH 7.4. The small particle size and the hydrophilic surfaces of SLNs1 protected it from large amount BSA adsorption.
Upon protein corona formation, the conformational change of BSA including the exposure or hidden of photon-adsorbing amino acid residues and the increment of the content of α-helix happened, but the integrity of SLNs was maintained60, 61, 62. The B16 cell uptake (representing acidic environmental cells) was decreased by the size increment in pH 6.0, and RAW 264.7 macrophages phagocytosis (representing neutral environmental cells) was reduced by the surface hydrophilicity increment in pH 7.4.
Based on these findings, the effect of protein corona formation is highly variable in different conditions, but the fundamental rules of protein adsorption can be revealed. The small particle size and hydrophilic surface can reduce protein adsorption, and the protein PI should be taken into consideration in different pH values. Further, the approaches to regulate the adsorption behavior of proteins in complex biological media can also be implemented according to this proof-of-concept study. Our research would provide a systematic perspective of how medium factors and NPs properties affect the BSA binding and its biological effects.
Acknowledgments
The authors would like to acknowledge the project grants from National Natural Science Foundation of China (81703431 and 81673375) and from the Natural Science Fund Project of Guangdong Province (2016A030312013, China). The authors would like to thank Haijiao Xie from Shiyanjia Lab (www.shiyanjia.com) for the AutoDock molecular docking technique.
Author contributions
Wenhao Wang and Zhengwei Huang designed the research. Yanbei Li, Wenhua Wang and Jiayu Shi carried out the experiments. Fangqin Fu participated part of the experiments. Wenhao Wang performed data analysis and wrote the manuscript. Zhengwei Huang and Ying Huang revised the manuscript. Xin Pan and Chuanbin Wu supervised the project. All of the authors have read and approved the final manuscript.
Conflict of interest
The authors have no conflicts of interest to declare.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Appendix A Supporting information
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2020.10.023.
Contributor Information
Ying Huang, Email: huangy2007@jnu.edu.cn.
Chuanbin Wu, Email: wuchuanb@mail.sysu.edu.cn.
Appendix ASupplementary data
The following is the Supplementary data to this article:
Multimedia component 1
References
- 1.Stefanick J.F., Omstead D.T., Kiziltepe T., Bilgicer B. Dual-receptor targeted strategy in nanoparticle design achieves tumor cell selectivity through cooperativity. Nanoscale. 2019;11:4414–4427. doi: 10.1039/c8nr09431d. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y.B., Wu W.B., Liu J.J., Manghnani P.N., Hu F., Ma D., et al. Cancer-cell-activated photodynamic therapy assisted by Cu(II)-based metal-organic framework. ACS Nano. 2019;13:6879–6890. doi: 10.1021/acsnano.9b01665. [DOI] [PubMed] [Google Scholar]
- 3.Wan S.S., Cheng Q., Zeng X., Zhang X.Z. A Mn(III)-sealed metal-organic framework nanosystem for redox-unlocked tumor theranostics. ACS Nano. 2019;13:6561–6571. doi: 10.1021/acsnano.9b00300. [DOI] [PubMed] [Google Scholar]
- 4.Rezaei G., Mojtaba Daghighi S., Raoufi M., Esfandyari-Manesh M., Rahimifard M., Iranpur Mobarakeh V., et al. Synthetic and biological identities of polymeric nanoparticles influencing the cellular delivery: an immunological link. J Colloid Interface Sci. 2019;556:476–491. doi: 10.1016/j.jcis.2019.08.060. [DOI] [PubMed] [Google Scholar]
- 5.Sousa F., Dhaliwal H.K., Gattacceca F., Sarmento B., Amiji M.M. Enhanced anti-angiogenic effects of bevacizumab in glioblastoma treatment upon intranasal administration in polymeric nanoparticles. J Control Release. 2019;309:37–47. doi: 10.1016/j.jconrel.2019.07.033. [DOI] [PubMed] [Google Scholar]
- 6.Fedorenko S.V., Stepanov A., Sibgatullina G., Samigullin D., Mukhitov A.R., Petrov K.A., et al. Fluorescent magnetic nanoparticles for modulating the level of intracellular Ca2+ in motoneurons. Nanoscale. 2019;11:16103–16113. doi: 10.1039/c9nr05071j. [DOI] [PubMed] [Google Scholar]
- 7.Han Y., Gao Z.G., Chen L.Q., Kang L., Huang W., Jin M.J., et al. Multifunctional oral delivery systems for enhanced bioavailability of therapeutic peptides/proteins. Acta Pharm Sin B. 2019;9:902–922. doi: 10.1016/j.apsb.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C., Wang J.C., Wang Y.G., Gao H.L., Wei G., Huang Y.Z., et al. Recent progress in drug delivery. Acta Pharm Sin B. 2019;9:1145–1162. doi: 10.1016/j.apsb.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi J., Kantoff P.W., Wooster R., Farokhzad O.C. Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer. 2017;17:20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wicki A., Witzigmann D., Balasubramanian V., Huwyler J. Nanomedicine in cancer therapy: challenges, opportunities, and clinical applications. J Control Release. 2015;200:138–157. doi: 10.1016/j.jconrel.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 11.Jain P., Pawar R.S., Pandey R.S., Madan J., Pawar S., Lakshmi P.K., et al. In-vitro in-vivo correlation (IVIVC) in nanomedicine: is protein corona the missing link?. Biotechnol Adv. 2017;35:889–904. doi: 10.1016/j.biotechadv.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Charbgoo F., Nejabat M., Abnous K., Soltani F., Taghdisi S.M., Alibolandi M., et al. Gold nanoparticle should understand protein corona for being a clinical nanomaterial. J Control Release. 2018;272:39–53. doi: 10.1016/j.jconrel.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Bertrand N., Grenier P., Mahmoudi M., Lima E.M., Appel E.A., Dormont F., et al. Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics Nicolas. Nat Commun. 2017;8:1–8. doi: 10.1038/s41467-017-00600-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francia V., Yang K., Deville S., Reker-smit C., Nelissen I., Salvati A. Corona composition can affect the mechanisms cells use to internalize nanoparticles. ACS Nano. 2019;13:11107–11121. doi: 10.1021/acsnano.9b03824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch I., Salvati A., Dawson K.A. Protein-nanoparticle interactions: what does the cell see?. Nat Nanotechnol. 2009;4:546–547. doi: 10.1038/nnano.2009.248. [DOI] [PubMed] [Google Scholar]
- 16.Yan Y., Gause K.T., Kamphuis M.M.J., Ang C.S., O'Brien-Simpson N.M., Lenzo J.C., et al. Differential roles of the protein corona in the cellular uptake of nanoporous polymer particles by monocyte and macrophage cell lines. ACS Nano. 2013;7:10960–10970. doi: 10.1021/nn404481f. [DOI] [PubMed] [Google Scholar]
- 17.Schäffler M., Sousa F., Wenk A., Sitia L., Hirn S., Schleh C., et al. Blood protein coating of gold nanoparticles as potential tool for organ targeting. Biomaterials. 2014;35:3455–3466. doi: 10.1016/j.biomaterials.2013.12.100. [DOI] [PubMed] [Google Scholar]
- 18.MiclǍuş T., Beer C., Chevallier J., Scavenius C., Bochenkov V.E., Enghild J.J., et al. Dynamic protein coronas revealed as a modulator of silver nanoparticle sulphidation in vitro. Nat Commun. 2016;7:1–10. doi: 10.1038/ncomms11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Paoli Lacerda S.H., Park J.J., Meuse C., Pristinski D., Beker M.L., Karim A., et al. Interaction of gold nanoparticles with common human blood proteins. ACS Nano. 2009;26:365–379. doi: 10.1021/nn9011187. [DOI] [PubMed] [Google Scholar]
- 20.Givens B.E., Wilson E., Fiegel J. The effect of salts in aqueous media on the formation of the BSA corona on SiO2 nanoparticles. Colloids Surf B Biointerfaces. 2019;179:374–381. doi: 10.1016/j.colsurfb.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Tran S., DeGiovanni P.-J., Piel B., Rai P. Cancer nanomedicine: a review of recent success in drug delivery. Clin Transl Med. 2017;6:1–21. doi: 10.1186/s40169-017-0175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.del Caño R., Mateus L., Sánchez-Obrero G., Sevilla J.M., Madueño R., Blázquez M., et al. Hemoglobin bioconjugates with surface-protected gold nanoparticles in aqueous media: the stability depends on solution pH and protein properties. J Colloid Interface Sci. 2017;505:1165–1171. doi: 10.1016/j.jcis.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Dewald I., Isakin O., Schubert J., Kraus T., Chanana M. Protein identity and environmental parameters determine the final physicochemical properties of protein-coated metal nanoparticles. J Phys Chem C. 2015;119:25482–25492. [Google Scholar]
- 24.Piella J., Bastús N.G., Puntes V. Size-dependent protein-nanoparticle interactions in citrate-stabilized gold nanoparticles: the emergence of the protein corona. Bioconjugate Chem. 2017;28:88–97. doi: 10.1021/acs.bioconjchem.6b00575. [DOI] [PubMed] [Google Scholar]
- 25.Shang W., Nuffer J.H., Muñiz-Papandrea V.A., Colón W., Siegel R.W., Dordick J.S. Cytochrome c on silica nanoparticles: influence of nanoparticle size on protein structure, stability, and activity. Small. 2009;5:470–476. doi: 10.1002/smll.200800995. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S., Aswal V.K., Callow P. PH-dependent interaction and resultant structures of silica nanoparticles and lysozyme protein. Langmuir. 2014;30:1588–1598. doi: 10.1021/la403896h. [DOI] [PubMed] [Google Scholar]
- 27.Partikel K., Korte R., Stein N.C., Mulac D., Herrmann F.C., Humpf H.U., et al. Effect of nanoparticle size and PEGylation on the protein corona of PLGA nanoparticles. Eur J Pharm Biopharm. 2019;141:70–80. doi: 10.1016/j.ejpb.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 28.Shen M.Y., Liu T.I., Yu T.W., Kv R., Chiang W.H., Tsai Y.C., et al. Hierarchically targetable polysaccharide-coated solid lipid nanoparticles as an oral chemo/thermotherapy delivery system for local treatment of colon cancer. Biomaterials. 2019;197:86–100. doi: 10.1016/j.biomaterials.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 29.Nafee N., Husari A., Maurer C.K., Lu C., De Rossi C., Steinbach A., et al. Antibiotic-free nanotherapeutics: ultra-small, mucus-penetrating solid lipid nanoparticles enhance the pulmonary delivery and anti-virulence efficacy of novel quorum sensing inhibitors. J Control Release. 2014;192:131–140. doi: 10.1016/j.jconrel.2014.06.055. [DOI] [PubMed] [Google Scholar]
- 30.Ghanbar S., Fumakia M., Ho E.A., Liu S. A new strategy for battling bacterial resistance: turning potent, non-selective and potentially non-resistance-inducing biocides into selective ones. Nanomed Nanotechnol Biol Med. 2018;14:471–481. doi: 10.1016/j.nano.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 31.Wang J., Wang H., Zhu R., Liu Q., Fei J., Wang S. Anti-inflammatory activity of curcumin-loaded solid lipid nanoparticles in IL-1β transgenic mice subjected to the lipopolysaccharide-induced sepsis. Biomaterials. 2015;53:475–483. doi: 10.1016/j.biomaterials.2015.02.116. [DOI] [PubMed] [Google Scholar]
- 32.Wang T., Wang D., Yu H., Wang M., Liu J., Feng B., et al. Intracellularly acid-switchable multifunctional micelles for combinational photo/chemotherapy of the drug-resistant tumor. ACS Nano. 2016;10:3496–3508. doi: 10.1021/acsnano.5b07706. [DOI] [PubMed] [Google Scholar]
- 33.Torchilin V.P. Multifunctional stimuli-sensitive nanoparticulate systems for drug delivery. Nat Rev Drug Discov. 2014;13:813–827. doi: 10.1038/nrd4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto S., Yamazaki S., Shimizu T., Takeshima T., Fukuma S., Yamamoto Y., et al. Body temperature at the emergency department as a predictor of mortality in patients with bacterial infection. Via Medici. 2016;95:e3628. doi: 10.1097/MD.0000000000003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Z.W., Wu M.J., Ma C., Bai X.Q., Zhang X.J., Zhao Z.Y., et al. Spectroscopic quantification of surfactants in solid lipid nanoparticles. J Pharm Innov. 2019;15:155–162. [Google Scholar]
- 36.Wahgiman N.A., Salim N., Rahman M.B.A., Ashari S.E. Optimization of nanoemulsion containing gemcitabine and evaluation of its cytotoxicity towards human fetal lung fibroblast (MRC5) and human lung carcinoma (A549) cells. Int J Nanomed. 2019;14:7323–7338. doi: 10.2147/IJN.S212635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ballav N. High-conducting polyaniline via oxidative polymerization of aniline by MnO2, PbO2 and NH4VO3. Mater Lett. 2004;58:3257–3260. [Google Scholar]
- 38.Tayeh N., Rungassamy T., Albani J.R. Fluorescence spectral resolution of tryptophan residues in bovine and human serum albumins. J Pharmaceut Biomed Anal. 2009;50:107–116. doi: 10.1016/j.jpba.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 39.Yin M.M., Dong P., Chen W.Q., Xu S.P., Yang L.Y., Jiang F.L., et al. Thermodynamics and mechanisms of the interactions between ultrasmall fluorescent gold nanoclusters and human serum albumin, γ-globulins, and transferrin: a spectroscopic approach. Langmuir. 2017;33:5108–5116. doi: 10.1021/acs.langmuir.7b00196. [DOI] [PubMed] [Google Scholar]
- 40.Manjubaashini N., Kesavan M.P., Rajesh J., Daniel Thangadurai T. Multispectroscopic and bioimaging approach for the interaction of rhodamine 6G capped gold nanoparticles with bovine serum albumin. J Photochem Photobiol B Biol. 2018;183:374–384. doi: 10.1016/j.jphotobiol.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Yang H.Y., Wang M., Zhang Y.M., Li F., Yu S.N., Zhu L., et al. Conformational-transited protein corona regulated cell-membrane penetration and induced cytotoxicity of ultrasmall Au nanoparticles. RSC Adv. 2019;9:4435–4444. doi: 10.1039/c8ra10049g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaikh S.M.T., Seetharamappa J., Kandagal P.B., Manjunatha D.H. In vitro study on the binding of anti-coagulant vitamin to bovine serum albumin and the influence of toxic ions and common ions on binding. Int J Biol Macromol. 2007;41:81–86. doi: 10.1016/j.ijbiomac.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Jiang M., Xie M.X., Zheng D., Liu Y., Li X.Y., Chen X. Spectroscopic studies on the interaction of cinnamic acid and its hydroxyl derivatives with human serum albumin. J Mol Struct. 2004;692:71–80. [Google Scholar]
- 44.Lehrer S.S. Solute perturbation of protein fluorescence. the quenching of the tryptophyl fluorescence of model compounds and of lysozyme by iodide ion. Biochemistry. 1971;10:3254–3263. doi: 10.1021/bi00793a015. [DOI] [PubMed] [Google Scholar]
- 45.He H.S., Zhang J., Xie Y.C., Lu Y., Qi J.P., Ahmad E., et al. Bioimaging of intravenous polymeric micelles based on discrimination of integral particles using an environment-responsive probe. Mol Pharm. 2016;13:4013–4019. doi: 10.1021/acs.molpharmaceut.6b00705. [DOI] [PubMed] [Google Scholar]
- 46.Ahmad E., Feng Y.H., Qi J.P., Fan W.F., Ma Y.H., He H.S., et al. Evidence of nose-to-brain delivery of nanoemulsions: cargoes but not vehicles. Nanoscale. 2017;9:1174–1183. doi: 10.1039/c6nr07581a. [DOI] [PubMed] [Google Scholar]
- 47.Feng Y.H., He H.S., Li F.Q., Lu Y., Qi J.P., Wu W. An update on the role of nanovehicles in nose-to-brain drug delivery. Drug Discov Today. 2018;23:1079–1088. doi: 10.1016/j.drudis.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Dominguez-Medina S., Kisley L., Tauzin L.J., Hoggard A., Shuang B.D.S., Indrasekara A.S., et al. Adsorption and unfolding of a single protein triggers nanoparticle aggregation. ACS Nano. 2016;10:2103–2112. doi: 10.1021/acsnano.5b06439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang H.Y., Wang M., Zhang Y.M., Liu X.Y., Yu S.N., Guo Y.M., et al. Detailed insight into the formation of protein corona: conformational change, stability and aggregation. Int J Biol Macromol. 2019;135:1114–1122. doi: 10.1016/j.ijbiomac.2019.06.014. [DOI] [PubMed] [Google Scholar]
- 50.Grosjean R., Delacroix S., Gouget G., Beaunier P., Ersen O., Ihiawakrim D., et al. High pressures pathway toward boron-based nanostructured solids. Dalton Trans. 2017;47:7634–7639. doi: 10.1039/c8dt00932e. [DOI] [PubMed] [Google Scholar]
- 51.Wang G.K., Yan C.L., Gao S.Y., Liu Y.F. Surface chemistry of gold nanoparticles determines interactions with bovine serum albumin. Mater Sci Eng C. 2019;103:109856. doi: 10.1016/j.msec.2019.109856. [DOI] [PubMed] [Google Scholar]
- 52.Roach P., Farrar D., Perry C.C. Interpretation of protein adsorption: surface-induced conformational changes. J Am Chem Soc. 2005;127:8168–8173. doi: 10.1021/ja042898o. [DOI] [PubMed] [Google Scholar]
- 53.Chittur K.K. FTIR/ATR for protein adsorption to biomaterial surfaces. Biomaterials. 1998;19:357–369. doi: 10.1016/s0142-9612(97)00223-8. [DOI] [PubMed] [Google Scholar]
- 54.Bouchemal K. New challenges for pharmaceutical formulations and drug delivery systems characterization using isothermal titration calorimetry. Drug Discov Today. 2008;13:960–972. doi: 10.1016/j.drudis.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 55.Chen S.F., Li L.Y., Zhao C., Zheng J. Surface hydration: principles and applications toward low-fouling/nonfouling biomaterials. Polymer. 2010;51:5283–5293. [Google Scholar]
- 56.Nayak P.S., Borah S.M., Gogoi H., Asthana S., Bhatnagar R., Jha A.N., et al. Lactoferrin adsorption onto silver nanoparticle interface: Implications of corona on protein conformation, nanoparticle cytotoxicity and the formulation adjuvanticity. Chem Eng J. 2019;361:470–484. [Google Scholar]
- 57.Ajdari N., Vyas C., Bogan S.L., Lwaleed B.A., Cousins B.G. Gold nanoparticle interactions in human blood: A model evaluation. Nanomed Nanotechnol Biol Med. 2017;13:1531–1542. doi: 10.1016/j.nano.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 58.Revilla J., Elaïssari A., Carriere P., Pichot C. Adsorption of bovine serum albumin onto polystyrene latex particles bearing saccharidic moieties. J Colloid Interface Sci. 1996;180:405–412. [Google Scholar]
- 59.Baier G., Costa C., Zeller A., Baumann D., Sayer C., Araujo P.H.H., et al. BSA adsorption on differently charged polystyrene nanoparticles using isothermal titration calorimetry and the influence on cellular uptake. Macromol Biosci. 2011;11:628–638. doi: 10.1002/mabi.201000395. [DOI] [PubMed] [Google Scholar]
- 60.He H.S., Xie Y.C., Lv Y., Qi J.P., Dong X.C., Zhao W.L., et al. Bioimaging of intact polycaprolactone nanoparticles using aggregation-caused quenching probes: size-dependent translocation via oral delivery. Adv Healthc Mater. 2018;1800711:1–11. doi: 10.1002/adhm.201800711. [DOI] [PubMed] [Google Scholar]
- 61.Wang T., Qi J.P., Ding N., Dong X.C., Zhao W.L., Lu Y., et al. Tracking translocation of self-discriminating curcumin hybrid nanocrystals following intravenous delivery. Int J Pharm. 2018;546:10–19. doi: 10.1016/j.ijpharm.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 62.Chen Z.J., Lv Y.J., Qi J.P., Zhu Q.G., Lu Y., Wu W. Overcoming or circumventing the stratum corneum barrier for efficient transcutaneous immunization. Drug Discov Today. 2018;23:181–186. doi: 10.1016/j.drudis.2017.09.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1