Introduction
Several lines of evidence now attest that a considerable number of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants are circulating in many countries around the globe, thus reinforcing the need of continuous genomic surveillance to identify introduction events, monitor the diffusion of these new strains and assess their clinical and healthcare impact (1). New viral variants can be defined as “of concern” when their infectivity and/or pathogenicity and/or capacity to escape immune system recognition are significantly higher than the original “wild-type” strain, thus consequently boosting virus spread within the population, enhancing the risk of developing more severe illness and/or even causing epidemic rebounds. As specifically concerns SARS-CoV-2, at least three such variants have been identified in several different territories by both the European Centre for Disease Control and Prevention (ECDC) (2) and the US Centers for Disease Control and Prevention (CDC) (3) to-date.
SARS-CoV-2 variants of concern
The B.1.1.7 variant, which was originally identified in the UK, from where it soon spread replacing almost completely the former strains and becoming rapidly predominant, has already been detected in as many as 83 different countries (2). This SARS-CoV-2 strain is characterized by six polymorphisms in the sequence encoding for the spike protein and a dozen in other genomic regions, the most important of which is perhaps the amino acid change N501Y (Asparagine to Tyrosine substitution in position 501) of the spike protein (Table 1) (4). The main consequence is that viral particles bearing this polymorphism display clearly enhanced binding affinity to their natural host receptor angiotensin converting enzyme 2 (ACE2), which appears to result in higher viral load and enhanced transmissibility compared to the wild type virus (5,6). According to information garnered from viral spread in three English regions, this variant seems characterized by 56% increased transmission potential [95% credible interval (CrI), 50–74%] compared to the wild-type strain first reported in Wuhan, thus translating into a basic reproduction number (R0) of approximately 4.90 (Table 1) (2). Moreover, while it remains unclear and further investigation including adjustment for confounding factors (such as increased hospital patient burden) is required, some early data has suggested increased mortality for the B.1.1.7 strain (7).
Table 1. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concerns, as for February 23, 2021.
| Variant | Emergence | New mutations | R0 |
|---|---|---|---|
| Wild Type | China | – | 3.14 |
| B.1.1.7 | UK | T1001I, A1708D, I2230T, SGF 3675–3677 (deletion), HV 69-70 (deletion), Y144 (deletion), N501Y, A570D, P681H, T716I, S982A, D1118H, Q27stop, R52I, Y73C, D3L, S235F | 4.90 |
| B.1.351 | South Africa | T265I, K1655N, H2799Y, S2900L, K3353R, D4527Y, P4715L, T5912I, L18F, D80A, D215G, R246I, K417N, E484K, N501Y, D614G, A701V, Q57H, S171L, P71L, T205I | 4.71 |
| P1 (B.1.1.28.1) | Brazil | T733C, C2749T, S1188L, K1795Q, 11288-11296 (deletion), C12778T, C13860T, E5665D, L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, H 655Y, T1027I, E92K, 28269-28273 (insertion), P80R | 4.70–4.90 |
Biologically significant mutations are evidenced in bold. R0, basic reproduction number.
The B.1.351 variant, initially detected in South Africa, has also rapidly spread around the world. Thus SARS-CoV-2 strain is characterized by nine polymorphisms in the sequence encoding for the spike protein and a dozen in other genomic regions, the most important of which are again the amino acid change N501Y (Asparagine to Tyrosine substitution in position 501), but also the E484K polymorphism (Glutamic acid to Lysine substitution in position 484), and the amino acid change D614G (Aspartic acid to Glycine substitution in position 614) all in the spike protein (Table 1) (4). Beside the previously mentioned enhanced transmissibility of viral variants containing the N501Y mutations, additional concern has been raised that the E484K mutation within the receptor biding domain (RBD) of the spike protein may enable the virus to partially escape recognition by the host immune system, as attested be repeated evidence of decreased neutralization potency of convalescent plasma and sera from vaccinated individuals (8). The concomitant presence of the G614 polymorphism would then magnify virus infectivity. Therefore, patients infected with this strain may display higher viral load and shedding than those with the D614 polymorphism, thus being at higher risk of developing more severe illness and also being more infective (9). Preliminary evidence suggests that this B.1.351 variant may be characterized by 50% higher transmission potential (95% CrI, 20–113%) compared to the wild-type strain first reported in Wuhan, which would hence translate into a R0 of approximately 4.71 (Table 1) (2).
The third variant of concern, provisionally called P.1 (formerly known as B.1.1.28.1), was originally identified in Japan among returning travelers from Brazil (10). Compared to the ancestral strain B.1.1.28, this variant presents several mutations in the spike protein, encompassing also E484K and N501Y, which would make its biological properties rather similar to the B.1.351 strain. In particular, the presence of the N501Y mutation would suggest that this variant may also be characterized by ~50% increased transmissibility, and thereby by a predicted R0 comprised between 4.70–4.90.
There are little doubts that SARS-COV-2 has undergone a considerable number of mutations since it first reported in Wuhan, and it seems that will continue to do so as evidenced by the fact that up to 45 intra-host single-nucleotide SARS-CoV-2 variants can be identified in the vast majority of people with COVID-19 (11). This is not totally surprising since viruses which directly encode their genome in RNA, such as HIV and influenza along with SARS-CoV-2, insert mutations quite rapidly due to the fact that they reproduce inside the host, where enzymes copying RNA are vulnerable to errors. (12). However, unlike influenza, the replication machinery of SARS-CoV-2 (i.e., RNA-dependent-RNA polymerase) contains exonuclease activity with proofreading function, which should in theory result in a lower rate of new mutations (13). Thus, it has been postulated that otherwise silent mutations in (or in proteins interacting with) the RNA-dependent-RNA polymerase of SARS-CoV-2, which were first reported as early as February 2020 in the UK and Italy, may result in an increased mutation rate and, in part, explain the rapid emergence of intra-host variants and new strains (14).
It is thus conceivable that a new critical period may appear in the comings weeks and months, in which as more people are vaccinated, a higher pressure on the virus to escape our immune system will be made, ultimately increasing the likelihood of appearance of new mutations and persistent generation of new SARS-CoV-2 variants of concern (15). The B.1.427 and B.1.419 strains, which recently appeared in California, are paradigmatic examples, as they contain a L452R mutation which seems to reinforce the interplay between ACE2 and SARS-CoV-2 spike protein, thus enhancing by up to 25% its transmissibility and associating with 2- to 7-fold reduction in anti-SASR-CoV-2 neutralizing activity (16).
SARS-CoV-2 variants and herd immunity
The threshold of herd immunity is conventionally calculated as the rate of subjects within the general population who have acquired such an efficient immunity against a specific infection so that they will no longer contribute to the chain of transmission. Once the herd immunity threshold is overcome, the circulation of the pathogen is remarkably lowered, or even totally inhibited, thus contributing to gradually extinguishing the outbreak (17). The threshold of herd immunity is substantially dependent on the R0 (i.e., the estimated number of subjects who could be infected by a single carrier in a vulnerable population), and could be estimated as: [1]-[1]/[R0] (17). Notably, a highly frequent pathogen such as the influenza virus is characterized by a relatively low R0 (i.e., approximately 1.3) (18), thus meaning that immunization of a modest population rate (i.e., between 22–23%) would be sufficient to mitigate influenza circulation (Figure 1). Although several calculations have been attempted since the emergence of the first reported outbreak in Wuhan, a general consensus has been reached that the R0 of SARS-CoV-2 shall be comprised in a range between 2.69-3.59, with a mean value of approximately 3.14 (19). Even when considering the estimated effective reproduction number (Rt), which reflects the number of infections that can be potentially generated in a currently uninfected population (i.e., 3.18 for SARS-CoV-2; range 2.89–3.47), the herd immunity threshold would not change significantly (i.e., 69% vs. 68%) (19). This would actually translate into the concept that around 70% of the general population would need to be immunized before the circulation of SARS-CoV-2 could be interrupted (Figure 1). Owing to the fact that the R0 of the emerging SARS-CoV-2 variants B.1.1.7, B.1.351 and P1 (B.1.1.28.1) has been calculated at 50–56% higher than that of the wild type strain, this increased infective potential translates into the need for the herd immunity threshold to be further increased by around 10–12% without mitigating interventions, to achieve a new value approximating 80% (Figure 1). This cutoff is very close to that of highly communicable infectious pathologies such as smallpox and poliomyelitis (both around 75–85%).
Figure 1.
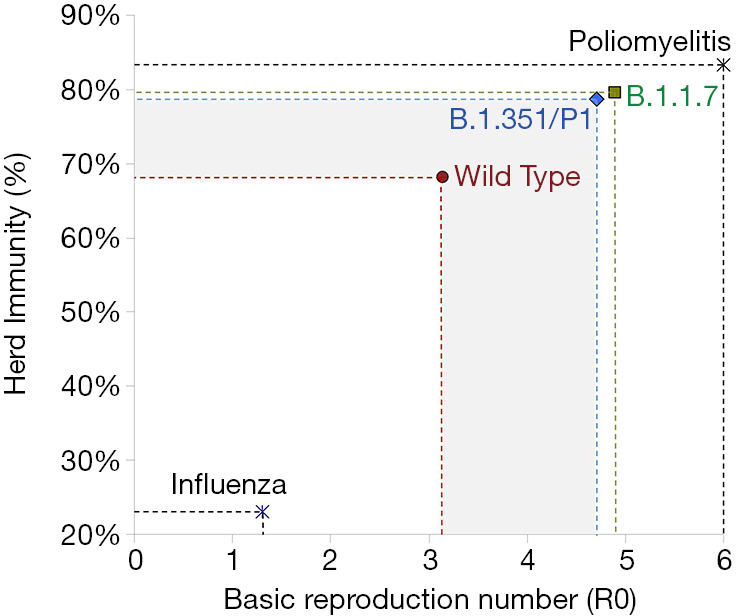
Herd immunity thresholds for influenza, poliomyelitis, wild type severe acute respiratory syndrome coronavirus (SARS-CoV-2) and its B.1.1.7, B.1.351 and P1 (B.1.1.28.1) variants.
What’s next?
The next and almost predictable question is then: will such a high herd immunity threshold be ever reachable by natural immunity, vaccination, or both? First, achievement of widespread natural immunity is unquestionably unethical due to the dramatic number of deaths that would be expected or the unsustainable burden that will be placed on healthcare systems worldwide (20). The current death rate of COVID-19 is 2.2% based on data daily updated by the John Hopkins University (21). With a current worldwide population of around 7848 million people (22), universal SARS-CoV-2 infection could cause over 173 million deaths, a number that is 3-fold higher than the casualties attributed to the dramatic Spanish Influenza pandemic outbreak in 1918–1919 (23), and even more than double the number of deaths that occurred during the second world war. With respect to the healthcare burden of universal natural infection, Italy has recorded a hospitalization rate of 5.1% in February 2021 (24), which would then translate into as many as 400 million hospital admissions if the entire worldwide population would be infected (or even more, considering countries with lower efficiency of the national healthcare system). Moreover, the long-term complications and sequelae of COVID-19 have yet to be fully understood and will likely create a further burden on the healthcare system for many years to come. Finally, due to a combination of waning of natural immunity and the risk for re-infection, especially with the new strains, appears significant, natural herd immunity may indeed be unachievable.
Thus given that natural herd immunity is unethical and perhaps even unreachable (25), we would need to concentrate most of our efforts on vaccination. The dramatic spread of the COVID-19 pandemic outbreak, causing paramount clinical, societal and economical consequences, has triggered an unprecedented hurdle race for obtaining a sufficient number of SARS-CoV-2 vaccines that could be administrated (probably even more than once) to the entire worldwide population (26). Preliminary evidence, garnered from a series of in vitro studies using some of these new SARS-CoV-2 variants/pseudoviral models, is however raising some doubts as to whether not only natural immunity, but also vaccination, may be fully efficient at preventing infections/transmission and/or development of severe diseases.
Growing evidence from the real world confirms now that some vaccines may be effective to reduce the risk of SARS-CoV-2 infection by 50–95% and, even more importantly, reduce the risk of developing symptomatic disease by 85–100% (27). Nonetheless, questions emerge to if these figures will be the same as newly emergent SARS-CoV-2 variants, especially those capable to escape immune recognition (e.g., B.1.351 and P1, among others), replace the former strains? There is no univocal answer to this question, and continuously referring to in vitro studies is, at least for the time being, not so straightforward. A preprint study submitted by Diamond and colleagues showed that the B.1.351 variant may be associated with nearly 10-fold loss-of-neutralization potency in vitro of human sera from recipients of BNT162b2 mRNA Covid-19 vaccine (28). Similar evidence has been provided in another preprint study submitted by Wu et al. (29), who also showed that serum collected from subjects immunized with the mRNA-1273 Covid-19 vaccine displayed an over 6-fold reduction of neutralizing potential when challenged with the same B.1.351 variant. Overall, this preliminary evidence would hence lead us to conclude that some of these emerging SARS-CoV-2 variants may generate a “perfect storm”, worse than the disease itself (30), whereby a higher herd immunity threshold would be needed for arresting their circulation, whilst this in turn could be more difficult to achieve due to their stronger “resistance” to the current generation of vaccines (Figure 2). Indeed, research is urgently needed to elucidate if less vulnerability to immunity is due only to changes in the RBD causing lower antibody affinity and thus reduced neutralizing activity, or if the new mutations induce a true mechanism of immune escape, which may be key to answering this question, tailoring a response, and providing updated vaccines/boosters.
Figure 2.
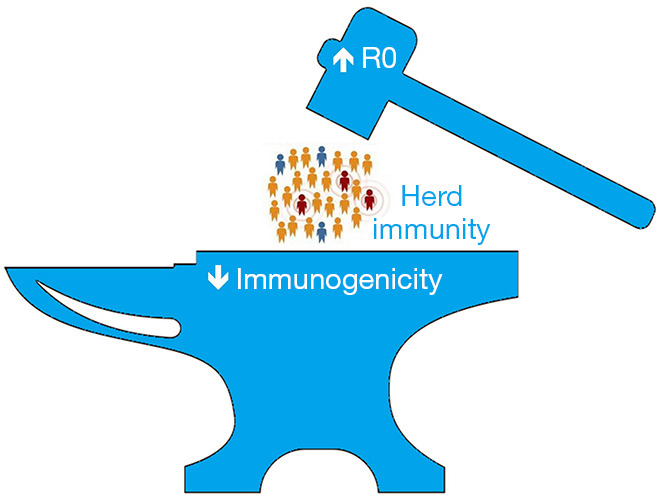
Herd immunity to severe acute respiratory syndrome coronavirus (SARS-CoV-2) seems to be squeezed between the higher basic reproduction number (R0) of emerging variants and their lower immunogenicity, which would ultimately make herd immunity threshold more challenging to be achieved.
Is the prospective totally dark?
Nonetheless, such pessimism shall be mitigated by at least three favorable and converging aspects. First, although the B.1.351, as well as other variants bearing the so-called “escape mutations”, may partially attenuate the neutralizing potential of antibodies developed after vaccination, the levels of immunoglobulin G (IgG) targeting the spike protein and/or the RBD is of at least 3-4 orders of magnitude (i.e., typically from 102 to 106) (31,32). Therefore, an up to 1-fold magnitude reduction of the neutralization potential would only partially reduce the efficiency of vaccine-generated antibodies to neutralize SARS-CoV-2 infection and/or pathogenicity. In the coming months, it will be essential to study the relationship between antibodies level/neutralizing activity and vaccine efficacy, in order to facilitate rapid understanding of the potential impact of polymorphisms as new mutant strains emerge.
Cellular immunity is the second essential aspect in this fight. Although this arm of the immune response is often overlooked due to inherent complexities beyond its routine assessment, several lines of evidence now attest that T cells are capable to kill virus-infected cells before they would spread from the upper respiratory tract, attenuate infectivity by containing the amount of virus circulating in an infected person, and even turn to be more effective against new SARS-CoV-2 variants since they target a vast array of different coronavirus antigens (33). Nonetheless, conflicting information has arisen with respect to the role of T cells in SARS-CoV-2, and further research would be urgently required.
Last but not least, the evidence provided so far that some SARS-CoV-2 variants could escape vaccine-induced neutralizing antibodies is only limited to in vitro studies, where mutants have been challenged with patients’ sera. No proof has been made available so far that our extraordinary immune system, post-vaccination, would not be able to adequately challenge ex vivo the newly emerging SARS-CoV-2 variants.
Conclusions
Although dark shadows are surrounding us, and the road ahead will present new challenges, we are still confident that armed with 21st century science and modern medicine, we will be able to overcome the latest struggle faced by mankind (34), just as we have in the past overcome the kaleidoscope of infectious diseases that have plagued humanity since its existence.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the editorial office, Annals of Translational Medicine for the series “Column in Laboratory Medicine”. The article did not undergo external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-893). The series “Column in Laboratory Medicine” was commissioned by the editorial office without any funding or sponsorship. GL serves as the unpaid Guest Editor of the series and serves as an unpaid Honorary Editors-in-Chief of Annals of Translational Medicine from June 2019 to May 2024. The authors have no other conflicts of interest to declare.
References
- 1.Fontanet A, Autran B, Lina B, et al. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet 2021;397:952-4. 10.1016/S0140-6736(21)00370-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Centre for Disease Control and Prevention. SARS-CoV-2 - increased circulation of variants of concern and vaccine rollout in the EU/EEA, 14th update. 15 February 2021. Last accessed, February 24, 2021. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/RRA-covid-19-14th-update-15-feb-2021.pdf
- 3.US Centers for Disease Control and Prevention. Emerging SARS-CoV-2 Variants. Last accessed, February 24, 2021.Available online: https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html
- 4.Tang JW, Toovey OTR, Harvey KN, et al. Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J Infect 2021;82:e8-e10. 10.1016/j.jinf.2021.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wise J. Covid-19: New coronavirus variant is identified in UK. BMJ 2020;371:m4857. 10.1136/bmj.m4857 [DOI] [PubMed] [Google Scholar]
- 6.Leung K, Shum MH, Leung GM, et al. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill 2021;26:2002106. 10.2807/1560-7917.ES.2020.26.1.2002106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iacobucci G. Covid-19: New UK variant may be linked to increased death rate, early data indicate. BMJ 2021;372:n230. [DOI] [PubMed] [Google Scholar]
- 8.Greaney AJ, Loes AN, Crawford KHD, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 2021;29:463-76.e6. 10.1016/j.chom.2021.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korber B, Fischer WM, Gnanakaran S, et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020;182:812-7.e19. 10.1016/j.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voloch CM, da Silva R, de Almeida LGP, et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. MedRxiv 2020.12.23.20248598; doi: https://doi.org/ 10.1101/2020.12.23.20248598. [DOI] [PMC free article] [PubMed]
- 11.Armero A, Berthet N, Avarre JC. Intra-Host Diversity of SARS-Cov-2 Should Not Be Neglected: Case of the State of Victoria, Australia. Viruses 2021;13:133. 10.3390/v13010133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callaway E. The coronavirus is mutating - does it matter? Nature 2020;585:174-7. 10.1038/d41586-020-02544-6 [DOI] [PubMed] [Google Scholar]
- 13.Manzanares-Meza LD, Medina-Contreras O. SARS-CoV-2 and influenza: a comparative overview and treatment implications. Bol Med Hosp Infant Mex 2020;77:262-73. 10.24875/BMHIM.20000183 [DOI] [PubMed] [Google Scholar]
- 14.Pachetti M, Marini B, Benedetti F, et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med 2020;18:179. 10.1186/s12967-020-02344-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahase E. Covid-19: What new variants are emerging and how are they being investigated? BMJ 2021;372:n158. [DOI] [PubMed] [Google Scholar]
- 16.Deng X, Garcia-Knight MA, Khalid MM, et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv [Preprint] 2021. doi: 10.1101/2021.03.07.21252647. [DOI]
- 17.Omer SB, Yildirim I, Forman HP. Herd Immunity and Implications for SARS-CoV-2 Control. JAMA 2020;324:2095-6. 10.1001/jama.2020.20892 [DOI] [PubMed] [Google Scholar]
- 18.Biggerstaff M, Cauchemez S, Reed C, et al. Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: a systematic review of the literature. BMC Infect Dis 2014;14:480. 10.1186/1471-2334-14-480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussein M, Toraih E, Elshazli R, et al. Meta-analysis on Serial Intervals and Reproductive Rates for SARS-CoV-2. Ann Surg 2021;273:416-23. 10.1097/SLA.0000000000004400 [DOI] [PubMed] [Google Scholar]
- 20.Neagu M. The bumpy road to achieve herd immunity in COVID-19. J Immunoassay Immunochem 2020;41:928-45. 10.1080/15321819.2020.1833919 [DOI] [PubMed] [Google Scholar]
- 21.John Hopkins University. Coronavirus Resource Center. Last accessed, February 24, 2021. Available online: https://coronavirus.jhu.edu/map.html
- 22.Worldometer. Current World Population. Last accessed, February 24, 2021. Available online: https://www.worldometers.info/world-population/
- 23.Martini M, Gazzaniga V, Bragazzi NL, et al. The Spanish Influenza Pandemic: a lesson from history 100 years after 1918. J Prev Med Hyg 2019;60:E64-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lab24. The daily curve in Italy. Last accessed, February 24, 2021. Available online: https://lab24.ilsole24ore.com/coronavirus/en/
- 25.Griffin S. Covid-19: Herd immunity is "unethical and unachievable," say experts after report of 5% seroprevalence in Spain. BMJ 2020;370:m2728. 10.1136/bmj.m2728 [DOI] [PubMed] [Google Scholar]
- 26.Gaebler C, Nussenzweig MC. All eyes on a hurdle race for a SARS-CoV-2 vaccine. Nature 2020;586:501-2. 10.1038/d41586-020-02926-w [DOI] [PubMed] [Google Scholar]
- 27.Abdool Karim SS, de Oliveira T. New SARS-CoV-2 Variants - Clinical, Public Health, and Vaccine Implications. N Engl J Med 2021. doi: . [Epub ahead of print]. 10.1056/NEJMc2100362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diamond M, Chen R, Xie X, et al. SARS-CoV-2 variants show resistance to neutralization by many monoclonal and serum-derived polyclonal antibodies. Preprint. Res Sq 2021. [preprint]. doi: . 10.21203/rs.3.rs-228079/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu K, Werner AP, Moliva JI, et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv [Preprint] 2021. doi: 10.1101/2021.01.25.427948. [DOI]
- 30.Lippi G, Sanchis-Gomar F, Henry BM. Coronavirus disease 2019 (COVID-19): the portrait of a perfect storm. Ann Transl Med 2020;8:497. 10.21037/atm.2020.03.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Widge AT, Rouphael NG, Jackson LA, et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N Engl J Med 2021;384:80-2. 10.1056/NEJMc2032195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahin U, Muik A, Vogler I, et al. BNT162b2 induces SARS-CoV-2-neutralising antibodies and T cells in humans. MedRxiv 2020.12.09.20245175; doi: https://doi.org/ 10.1101/2020.12.09.20245175. [DOI]
- 33.Ledford H. How 'killer' T cells could boost COVID immunity in face of new variants. Nature 2021;590:374-5. 10.1038/d41586-021-00367-7 [DOI] [PubMed] [Google Scholar]
- 34.Lippi G, Sanchis-Gomar F, Henry BM. COVID-19: unravelling the clinical progression of nature's virtually perfect biological weapon. Ann Transl Med 2020;8:693. 10.21037/atm-20-3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as


