Abstract
Background:
Population-level estimates of disease prevalence and control are needed to assess prevention and treatment strategies. However, available data often suffer from differential missingness. For example, population-level HIV viral suppression is the proportion of all HIV-positive persons with suppressed viral replication. Individuals with measured HIV status, and among HIV-positive individuals those with measured viral suppression, likely differ from those without such measurements.
Methods:
We discuss three sets of assumptions to identify population-level suppression in the intervention arm of the SEARCH Study (NCT01864603), a community randomized trial in rural Kenya and Uganda (2013-2017). Using data on nearly 100,000 participants, we compare estimates from i) an unadjusted approach assuming data are missing-completely-at-random (MCAR); ii) stratification on age-group, sex, and community; and, iii) targeted maximum likelihood estimation to adjust for a larger set of baseline and time-updated variables.
Results:
Despite high measurement coverage, estimates of population-level viral suppression varied by identification assumption. Unadjusted estimates were most optimistic: 50% (95%CI:46-54%) of HIV-positive persons suppressed at baseline, 80% (95%CI:78-82%) at Year 1, 85% (95%CI:83-86%) at Year 2, and 85% (95%CI:83-87%) at Year 3. Stratifying on baseline predictors yielded slightly lower estimates, and full adjustment reduced estimates meaningfully: 42% (95%CI:37-46%) of HIV-positive persons suppressed at baseline, 71% (95%CI:69-73%) at Year 1, 76% (95%CI:74-78%) at Year 2, and 79% (95%CI:77-81%) at Year 3.
Conclusions:
Estimation of population-level disease burden and control requires appropriate adjustment for missing data. Even in large studies with limited missingness, estimates relying on the MCAR assumption or baseline stratification should be interpreted cautiously.
Keywords: causal inference, HIV viral suppression, machine learning, missing data, Super Learner, TMLE
INTRODUCTION:
Accurate population-level estimates of disease prevalence and treatment coverage are needed to quantify disease burden and evaluate the success of programs for epidemic control. The data available to inform such estimates, however, are often susceptible to differential missingness. In other words, the missing-completely-at-random (MCAR) assumption rarely, if ever, holds.1-4 The field of HIV prevention and treatment provides an illustrative example. Consider the UNAIDS 90-90-90 target for the year 2020: 90% of all HIV-positive persons should know their status; 90% of those who know their status should be receiving antiretroviral therapy (ART); and 90% of those receiving ART should have suppressed HIV viral replication.5 Multiplying these proportions together yields an overall target, referred to here as “population-level suppression” - 73% of all HIV-positive persons should have suppressed HIV viral replication (Appendix). This target reflects the HIV care “cascade” from diagnosis, through treatment initiation and retention, to viral suppression.
While population-level suppression is widely used in assessing HIV care strategies, two recent systematic reviews noted the variability in both data quality and statistical approaches used for assessment.6,7 In particular, Granich et al.6 remarked on the challenges posed by incomplete data and inconsistent methodology, while Sabapathy et al.7 proposed a template to standardize data collection and evaluation. In this manuscript, we provide an in-depth demonstration of the methods used to estimate population-level suppression in the SEARCH Study, a cluster randomized trial in rural Kenya and Uganda (NCT01864603).8,9 We approach the missing data problem with a causal framework to define target parameters with counterfactuals, state identifiability assumptions sufficient to translate these targets into statistical quantities, and estimate the resulting statistical parameters.1-4,10-14 We refer the reader to companion papers for details on the trial.8,9
METHODS:
In general, the total number of HIV-positive persons in a population is unknown, and individuals with known HIV status are not necessarily representative of the general population. If, for example, persons with higher health-seeking behavior are more likely to test and less likely to be infected, then an unadjusted estimate of HIV prevalence (i.e. the proportion with HIV among those tested) is likely to underestimate the true prevalence of HIV in the population, even in the context of community-wide testing, as was implemented in recent Universal-Test-and-Treat trials.9,15-18
Likewise, measurement of plasma HIV RNA levels (viral loads) among HIV-positive individuals is generally incomplete and often depends on factors associated with viral suppression. For example, if viral loads are only measured at HIV clinic visits, then viral suppression among measured individuals will overestimate suppression among all HIV-positive persons, including newly diagnosed individuals who are not yet in-care and previously diagnosed individuals who are out-of-care. These familiar missing data challenges can be illustrated with a directed acyclic graph or another causal modeling approach (Figure 1).14,19-24
Figure 1:
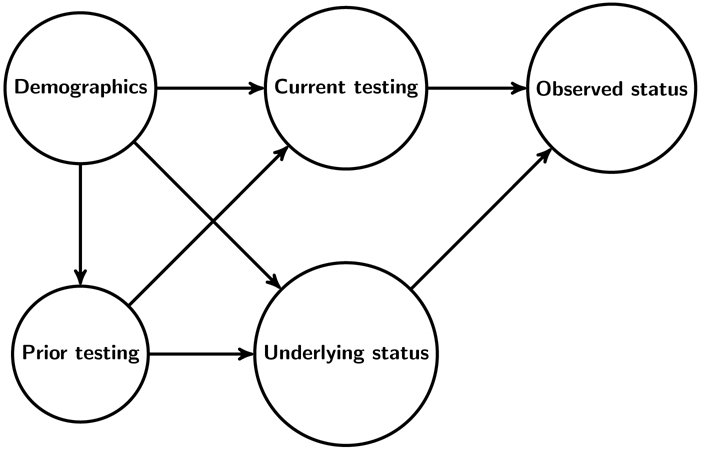
Simplified directed acyclic graph to represent the challenges posed by incomplete HIV testing. Demographics and prior testing are common causes of current testing (the hypothetical intervention node) and underlying HIV status (possible unobserved), both of which impact observed HIV status. Analogous challenges arise due to incomplete measurement of suppression among HIV-positive persons.
Overcoming these challenges requires knowledge of the data generating process. Consider the 16 communities in the intervention arm of the SEARCH Study. After a door-to-door census, we conducted community-wide testing annually through multidisease health fairs, followed by out-of-facility testing for residents who did not attend the fair.25,26 We linked participants over successive years with a fingerprint biometric. Prior diagnosis of HIV and ART use were ascertained through linkage to clinic records.8,27 A re-census was conducted 3 years after follow-up to determine interim deaths, out-migrations, and in-migrations.9
With this measurement scheme in mind, we describe the methods used in Petersen et al.8 and Havlir et al.9 to characterize viral suppression in the intervention arm at study baseline t=0, and annually thereafter t={1,2,3}. These cross-sectional analyses provide snapshots of population-level suppression among an open cohort of adult (≥15years) residents (allowing for entry due to age and in-migration, and exit due to death or outmigration). We note estimating viral suppression among a closed cohort of known baseline HIV-positive residents is a distinct goal, resulting in a different causal parameter, identifiability assumptions, and estimation approach.8,9
Causal parameters
Let be an indicator that an individual is HIV-positive at time t, irrespective of whether serostatus is measured. Likewise, let be a possibly unmeasured indicator of HIV viral suppression (<500cps/mL) at time t. Population-level suppression is the conditional probability of viral suppression given HIV-positive status: , or equivalently, the joint probability of being HIV-positive with suppression, divided by the probability of being HIV-positive (i.e. HIV prevalence): .
Ideally, anyone not already known to be HIV-positive (i.e. previously HIV-negative or HIV-unknown) would be tested at time t. Of course, this is never the case; further, missingness inherently depends on underlying HIV status - the status of an HIV-negative individual who does not test at time t is unknown, whereas the status of an HIV-positive individual not seen at time t might be known from prior testing. The problem is intensified after multiple rounds of community-wide testing, which provide multiple opportunities for prevalent HIV-positive persons to be diagnosed. To avoid this inherent dependence, we define as an indicator that an individual was seen at community-wide testing and had “known” HIV status at time t - due to a negative test result at time t, or a positive result at or before time t. We define observed HIV status as .
As with HIV testing, viral load measurement is incomplete; HIV-positive persons on whom viral load is measured may differ systematically from HIV-positive persons who are missing this measure. Define as an indicator of viral load measurement at time t, and define observed viral suppression as .
Three sets of identifiability assumptions
In the above, population-level suppression was expressed in terms of underlying indicators of HIV seropositivity and viral suppression: . We now present three sets of identifiability assumptions to write the numerator and denominator in this expression as parameters of the observed data distribution (see eAppendix 1 for proofs).
Unadjusted:
Suppose we are willing to assume that HIV prevalence among those seen at time t is representative of HIV prevalence among those not seen, and that viral suppression among HIV-positive persons with viral loads measured at time t is representative of suppression among HIV-positive persons without measured viral loads. More formally, suppose we make the following randomization assumptions as applied to missing data:1-4,10-14 and . If these assumptions hold, the numerator of population-level suppression is identified as
and denominator as .27 Taking the ratio of these yields the unadjusted statistical parameter:
| (Eq1) |
Baseline adjustment:
We can weaken the above assumptions on the missingness process by conditioning on baseline covariates. Specifically, let B denote mutually exclusive and exhaustive strata defined by age group, sex, and community of residence. Now suppose within each strata b, HIV prevalence among those seen at time t is representative of prevalence among those not seen, and within each strata b, suppression among HIV-positive persons with viral loads measured at time t is representative of suppression among HIV-positive persons without measured viral loads. More formally, we assume and .
Under these assumptions on missingness, we obtain the following identifiability result, corresponding to a hypothetical, dynamic intervention to first ensure knowledge of HIV status and then to ensure measurement of viral loads among HIV-positive persons:28-31
| (Eq2) |
In words, this is the strata-specific probability of viral suppression, given measurement and HIV-positive status; multiplied by the strata-specific probability of being HIV-positive, given measurement; and then standardized with respect to the distribution of strata. Identification of the denominator, population-level prevalence, follows from above:
| (Eq3) |
By taking the ratio of the numerator (Eq2) to the denominator (Eq3), we obtain a baseline-adjusted statistical parameter corresponding to population-level suppression under the above assumptions.
For the conditioning sets to be well defined, we also require the positivity assumption.11,32 Irrespective of age, sex, and community, there must be a positive probability of being seen with known HIV status , and for every strata in which some proportion of HIV-positive persons are seen, there must be a positive probability of viral load measurement: .
Time-varying adjustment:
While stratifying on certain baseline characteristics weakens our assumptions on missingness, there may be many other variables potentially impacting testing, underlying HIV status, and viral suppression among HIV-positive persons. In particular, ART use is a key determinant of viral suppression and may also be predictive of viral load measurement.
Define ARTt as an indicator of ART initiation prior to time t, and let Xt denote the remaining observed variables that are potentially predictive of both viral suppression and its measurement: the full set of baseline demographics (e.g. age, sex, marital status, education, occupation, alcohol use, mobility, wealth index, and community) together with prior HIV testing and suppression. While viral suppression without ART is possible, the UNAIDS target is focused on ART-induced suppression (Appendix).5 Therefore, we set to zero for persons not on ART. We further assume complete measurement of ART use, acknowledging that if ART use is incompletely captured, this assumption will lead to underestimation of suppression. Finally, for HIV-positive persons who have initiated ART, we assume that conditional on baseline and time-updated covariates Xt, suppression among those with a measured viral load at time t is representative of suppression among those with a missing viral load. More formally, we assume .
We also require the positivity assumption; all HIV-positive individuals who have initiated ART have a positive probability of having their viral load measured, regardless of their baseline and time-updated covariates: a.e.. Under these assumptions, we have the following identifiability result corresponding to a hypothetical intervention to ensure viral load measurement among ART initiators:28
| (Eq4) |
where the summation generalizes to an integral for continuous covariates. In words, this is the proportion of individuals who have started ART (and are, by implication, HIV-positive) in the total population (including both HIV-positive and HIV-negative persons) multiplied by the adjusted probability of being suppressed, given prior ART initiation.
For the denominator of HIV prevalence, we also consider an expanded adjustment set , consisting of all baseline demographics and prior HIV testing. For the subgroup without a prior HIV diagnosis, we assume that conditional on Lt, HIV prevalence among those tested at time t is representative of HIV prevalence among those not tested, or more formally, . We further assume positivity; previously undiagnosed persons have some chance of being tested regardless of their Lt values: a.e.. Under these assumptions, we have the following identifiability result corresponding to a hypothetical intervention to ensure HIV status is known:28
| (Eq5) |
where the summation generalizes to an integral for continuous covariates. In words, this is the proportion of the population previously known to be HIV-positive plus the adjusted proportion of the population newly known to be HIV-positive.
Taking the ratio of the numerator (Eq4) to the denominator (Eq5) yields a fully adjusted statistical parameter for population-level suppression under the above assumptions.
Estimation approaches
The unadjusted parameter (Eq1) can be estimated with the empirical proportion of the population with measured viral suppression. The baseline-adjusted parameter (Eq2÷Eq3) can also be estimated with empirical proportions. Specifically, we would generate covariate strata-specific estimates by taking empirical means, and then combine by standardizing across strata. A similar approach was used in the PopART (HPTN 071) Universal-Test-and-Treat trial with stratification factors including sex, age group and community.17,33,34 This approach corresponds to G-computation when fully-saturated regressions are used to estimate the conditional probability of the outcome, given measurement and the adjustment set (i.e. the “outcome regression”).28,35,36 It is further equivalent to inverse-weighting when fully-saturated regressions are used to estimate the conditional probability of measurement, given the adjustment set (i.e. the “propensity score”).37-39
When the adjustment set is higher dimensional, such as for our fully-adjusted parameter (Eq4÷Eq5), alternative approaches are needed to smooth over values of the covariates with weak support.32 We could, for example, use logistic regression with two-way interactions to estimate the propensity scores for inverse-weighting. This approach was used in a sensitivity analysis in the Ya Tsie Universal-Test-and-Treat trial (also called the Botswana Combination Prevention Project).18,40
Another approach is targeted maximum likelihood estimation (TMLE), which offers efficiency gains over inverse-weighting and allows for flexible adjustment for a large set of covariates through machine learning.24,41. We refer the reader to 42,43 for an introduction to TMLE and to 44 for a detailed comparison with other methods, such as parametric G-computation and augmented inverse-weighting. Briefly, TMLE combines estimates of outcome regression with estimates of the propensity score to achieve a number of desirable properties. TMLE is double robust - it is consistent if either the outcome regression is consistently estimated or the propensity score is consistently estimated. TMLE is also a substitution estimator, potentially improving robustness under strong confounding or rare outcomes.44-47
In eAppendix 2, we provide step-by-step descriptions of the unadjusted estimator, parametric G-computation, inverse-weighting estimator, and TMLE to adjust for missing outcomes.
Implementation
In the SEARCH Study, the primary approach used TMLE to estimate the fully adjusted parameter (Eq4÷Eq5). Within TMLE, Super Learner was implemented to estimate the outcome regressions and propensity scores (i.e. the conditional probability of measurement, given the adjustment set).48 Super Learner is an ensemble, machine learning method that uses cross-validation to build the optimal combination of predictions from a library of candidate algorithms. For ease of interpretation, reduced computational burden, and to avoid over-fitting, our Super Learner library was limited to logistic regressions (with various screening algorithms), generalized additive models (with various screening algorithms), and the mean (corresponding to the unadjusted estimator). We implemented TMLE fully stratified on community, allowing the outcome regressions and propensity scores to vary by community. Full implementation details, including computing code, are available in eAppendix 3.
For comparison, we also present the use of empirical proportions to estimate the unadjusted parameter (Eq1), and the baseline-adjusted parameters (Eq2÷Eq3) controlling for sex, age group (15-19 years, 20-29 years, 30-39 years, 40-49 years, 50-59 years, and 60+ years), and community.
We obtained statistical inference with influence curve standard errors, treating the community as the unit of independence. We conducted analyses in R v.3.6.1 (Vienna, Austria) with the ltmle_v1.1-0 and SuperLearner_v2.0-25 packages.49-51
Ethical considerations
The SEARCH Study was approved by the ethics committees at the University of California, San Francisco; the Kenya Medical Research Institute; and Makerere University School of Medicine in Uganda. As previously described,9 oral informed consent was provided participation in the census and health fairs; written informed consent was provided to receive ART outside of country guidelines.
RESULTS
The baseline characteristics of the study participants have been described elsewhere.8,9,25 In brief, approximately one-third of the 79,818 residents enumerated in the baseline census were from each study region, and nearly half of participants were aged 15-30 years; men constituted 45% (eTable 1). HIV status was determined on 89% (71,402) of residents at baseline (Table 1). After baseline, knowledge of HIV serostatus remained high with 77% of residents (69,175/90,047) seen at Year 1, 75% (71,577/95,599) at Year 2, and 81% (80,390/99,186) at Year 3. There were no obvious demographic differences between the enumerated population and those with known HIV serostatus (eTable 1).
Table 1:
Number and coverage of residents contributing to unadjusted estimates of population-level HIV viral suppression at the time of annual testing. Each column is a subset of the former. Changes in annual population size are due to additions from in-migrants and aging-in, and due to subtractions from death and outmigration. Years refer to time since study baseline, which varied by community (Year 0 ranging from June 2013 to June 2014).
| Resident (≥15 years) |
HIV serostatus known |
HIV-positive serostatus |
Viral load measured |
Viral replication suppressed |
|
|---|---|---|---|---|---|
| Year 0 | 79818 | 71402 | 7009 | 5332 | 2659 |
| Year 1 | 90047 | 69175 | 6526 | 6137 | 4906 |
| Year 2 | 95599 | 71577 | 6687 | 6276 | 5316 |
| Year 3 | 99186 | 80390 | 6991 | 6738 | 5737 |
Viral loads were measured for 76% of baseline HIV-positive residents (Table 1). Missing viral loads were more common at baseline due to early assay failures.26 Despite ~95% coverage of viral load measurement for the remaining years, baseline and time-varying characteristics differed for HIV-positive persons with measured versus missed HIV RNA levels (Table 2). In particular, HIV-positive women were more likely to have their viral load measured than HIV-positive men. After baseline, adolescents (15-24years) were more likely to be missed than older adults (25+years). Viral load measurement also differed notably by the time-varying characteristics; HIV-positive persons who were previously aware of their status, had evidence of starting ART, or had a history of suppressing viral replication were more likely to have their viral load measured than their counterparts.
Table 2:
Select baseline and time-varying characteristics of HIV-positive residents by year and by viral load measurement. Metrics in N (%).
| Total | Female | Male | 15-24 years |
25+ years |
Prior diagnosisa |
Prior ART useb |
Prior Supp.c |
|
|---|---|---|---|---|---|---|---|---|
| Year 0 | ||||||||
| Tested | 5332 | 3599 (67) | 1733 (33) | 687 (13) | 4645 (87) | 3856 (72) | 3149 (59) | |
| Missed | 1677 | 1060 (63) | 617 (37) | 226 (13) | 1451 (87) | 1081 (64) | 847 (51) | |
| Year 1 | ||||||||
| Tested | 6137 | 4100 (67) | 2037 (33) | 729 (12) | 5408 (88) | 5917 (96) | 5591 (91) | 2276 (37) |
| Missed | 389 | 238 (61) | 151 (39) | 54 (14) | 335 (86) | 310 (80) | 225 (58) | 92 (24) |
| Year 2 | ||||||||
| Tested | 6276 | 4168 (66) | 2108 (34) | 782 (12) | 5494 (88) | 6153 (98) | 5970 (95) | 4637 (74) |
| Missed | 411 | 247 (60) | 164 (40) | 89 (22) | 322 (78) | 333 (81) | 230 (56) | 141 (34) |
| Year 3 | ||||||||
| Tested | 6738 | 4603 (68) | 2135 (32) | 1023 (15) | 5715 (85) | 6480 (96) | 6376 (95) | 5108 (76) |
| Missed | 253 | 135 (53) | 118 (47) | 54 (21) | 199 (79) | 222 (88) | 173 (68) | 143 (57) |
Abbreviations: “ART”=antiretroviral therapy; “Supp”=Suppression.
Positive HIV test or Ministry of Health record of HIV care before the start of the community-specific health fair at year t.
ART use, as determined through Ministry of Health records or suppressed HIV RNA, before the start of the community-specific health fair at year t.
Suppressed HIV RNA before the start of the community-specific health fair at year t.
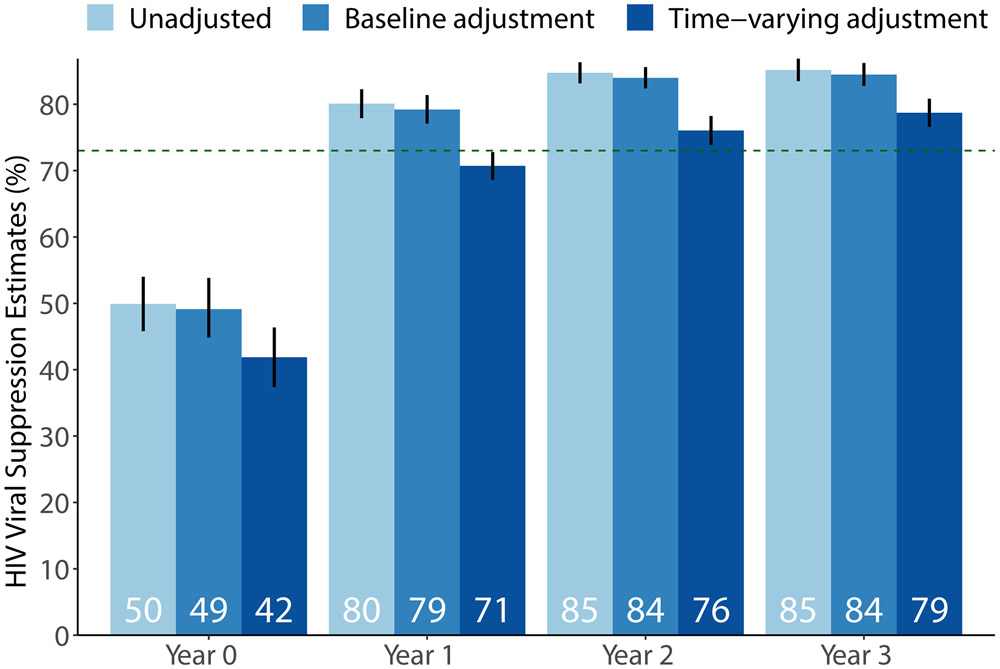
Estimates of population-level suppression varied meaningfully depending on the identifiability assumptions employed (Figure 2). At baseline, the unadjusted approach suggested that half of all HIV-positive residents had suppressed viral replication (50%; 95%CI: 46-54%). Stratifying on age group, sex, and community slightly reduced the estimate to 49% (95%CI: 45-54%). The lowest estimate of 42% (95%CI: 37-46%) was obtained after adjusting for the full set of baseline and time-varying characteristics.
Figure 2:
Estimates of population-level HIV viral suppression at the time of annual testing in the intervention arm of the SEARCH trial. Estimates were obtained with the empirical mean among those measured (“Unadjusted”), stratifying on sex, age group and community (“Baseline adjustment”), and using targeted maximum likelihood estimation (TMLE) with Super Learner to adjust for both baseline and time-varying characteristics (“Time-varying adjustment”). Horizontal dashed line at 73% suppression indicates UNAIDS target; vertical lines indicate 95% confidence intervals.
Deviations were pronounced in subsequent years (Figure 2). The unadjusted approach suggested that 80% (95%CI: 78-82%) of all HIV-positive residents were suppressed at Year 1, 85% (95%CI: 83-86%) at Year 2, and 85% (95%CI: 83-87%) at Year 3. Estimates adjusted for baseline covariate-strata were similar: 79% (95%CI: 77-81%) at Year 1, 84% (95%CI: 82-86%) at Year 2, and 84% (95%CI: 83-86%) at Year 3. Fully adjusted estimates were meaningfully lower: 71% (95%CI: 69-73%) at Year 1, 76% (95%CI: 74-78%) at Year 2, and 79% (95%CI: 77-81%) at Year 3.
DISCUSSION:
In an open cohort of nearly 100,000 residents in rural Kenya and Uganda, we compared three approaches for missing outcomes: (i) an unadjusted approach, the empirical proportion among those measured; (ii) stratification on age group, sex, and community; and (iii) TMLE with Super Learner to adjust for the full set of baseline and time-varying covariates. Despite low levels of missingness, estimates diverged by identifiability assumptions. The unadjusted approach consistently yielded the highest estimates; the fully adjusted approach consistently yielded the lowest estimates.
In the SEARCH Study, HIV serostatus and HIV RNA viral levels were obtained through multidisease testing at health fairs with follow-up for non-participants.25 Unlike clinic-based ascertainment, this approach reaches HIV-positive persons who are in-care or who are out-of-care.9,16-18 As a result, the MCAR assumption may seem reasonable.1-4 However, deviations between the unadjusted estimates and adjusted ones suggest there were meaningful differences in the population measured and population missed with respect to, among other factors, prior diagnosis, ART use, and viral suppression.
Both adjusted approaches were built on causal assumptions that (distinct subsets of) observed participant characteristics were sufficient to control for the common causes of measurement and health outcomes.1-4 When assuming baseline covariates were sufficient, we fully stratified on sex, age group, and community; as a result, this was equivalent to a fully non-parametric approach for the outcome regression in G-computation and to a fully non-parametric approach for the propensity score in inverse-weighting. Beyond age, sex, and community, there were, however, additional differences between those with measured versus missing viral loads, including differences in post-baseline variables; specifically, persons without prior diagnosis, ART initiation, or viral suppression were less likely to have their viral load measured. Therefore, the primary approach in the SEARCH Study was to weaken the assumptions on the missingness process by conditioning on a larger set of baseline and time-updated covariates.8,9,27 We estimated the fully adjusted statistical parameter using TMLE with Super Learner.
It is important to note, however, that the primary approach still relied on non-testable assumptions. If there is an unmeasured cause of measurement and health outcomes, none of the identifiability assumptions will hold and the wished-for causal parameters will diverge from the statistical estimands. In the SEARCH Study, we aimed to minimize this divergence by conditioning on a large set of baseline and time-varying variables and assessing the plausibility of assumptions with experts in HIV prevention. During estimation, we further aimed to avoid unsubstantiated assumptions and minimize bias due to model misspecification by using TMLE with Super Learner. This approach will provide a consistent point estimate if either the outcome regression or the propensity score is consistently estimated and will provide consistent inference (i.e. valid confidence intervals) when both are consistently estimated at sufficiently fast rates and satisfy the relevant empirical process conditions.52,53 Recent extensions to obtain double robust inference, while of interest, were not implemented in the current analyses.
While it is well recognized that estimates can only be interpreted causally if the identifiability assumptions hold, similar challenges apply when interpreting estimates adjusting for missing data.14 Specifically, our estimates are only truly representative of population-level metrics if the missingness assumptions hold13 and there is sufficient data support. These challenges, however, should not inspire inaction; instead, we often need to use available data to obtain timely estimates of disease burden and control under minimal assumptions, which are transparently stated and carefully evaluated.
In the running example, the identification choice had implications for policymaking and targeting resources. Both the unadjusted and baseline-adjusted approaches suggested the UNAIDS 90-90-90 target (73%-suppression) was surpassed within one year of the intervention and the UNAIDS 95-95-95 target (86%-suppression) was nearly achieved by the trial’s close.5 In contrast, the estimates controlling for time-updated covariates indicated the 90-90-90 target was achieved after two years, but there still was a substantial gap to the 95-95-95 target.
In summary, estimates of population-level HIV viral suppression continue to be the benchmark in assessing programmatic success in epidemic control. In four cross-sectional analyses of nearly 100,000 participants in the intervention arm of the SEARCH Study, we demonstrated the impact of missing data assumptions that can occur even in large studies with high measurement. While we focused on a specific example – estimating population-level HIV viral suppression - our results have broad applicability to missing data problems, commonly encountered in public health and medicine.3,4,10-13 We recommend adjustment for a large set of baseline and time-varying covariates that potentially influence both measurement and underlying status; TMLE with Super Learner is one approach to performing such adjustments.
Supplementary Material
Acknowledgments:
We thank the Ministry of Health of Uganda and of Kenya; our research teams and administrative teams in San Francisco, Uganda, and Kenya; collaborators and advisory boards; and especially all the communities and participants involved in the study.
Sources of Funding: This study was supported by grant numbers U01AI099959, UM1AI068636, and R01AI074345 from National Institute of Allergy and Infectious Diseases at the National Institutes of Health; by the President’s Emergency Plan for AIDS Relief; and by Gilead Sciences, which provided Truvada.
Appendix: UNAIDS 90-90-90 target and population-level suppression
For the moment assume complete measurement, and let HIVt be an indicator of HIV-positive serostatus at time t; Dxt be an indicator of having an HIV diagnosis by time t; ARTt be an indicator of antiretroviral therapy (ART) use at time t, and Suppt be an indicator of suppressed viral replication at time t. The UNAIDS 90-90-90 targets are a series of proportions or conditional probabilities:5
% of all HIV-positives who are diagnosed (first-90):
% of diagnosed who are on ART (second-90):
% on ART who are currently suppressed (third-90):
Multiplying together the three “90s” yields the proportion of all HIV-positive persons who are currently suppressed (i.e. population-level suppression):
Since each numerator and denominator is a population-level proportion, we can equivalently express the targets as follows: first-90=(number previously diagnosed)/(number HIV-positive), second-90=(number on ART)/(number previously diagnosed), third-90=(number virally suppressed)/(number on ART), and population-level suppression=(number virally suppressed)/(number HIV-positive).
Therefore, one could directly estimate population-level suppression, as we demonstrated here, or instead estimate each 90-90-90 target and multiply. These two approaches should yield identical results, as demonstrated in our previous work.8,9,27 However, deviations between the direct estimate and the multiplied-one can occur when making the missing-completely-at-random (MCAR) assumption.1-4 Specifically, under MCAR, the denominators of the third-90 and population-level suppression become conditional on having a viral load measured, which is almost always a subset of the population on ART and a subset of the population who is HIV-positive.
Footnotes
Conflicts of Interest: There is no conflict of interest
Computing code: Available at https://github.com/LauraBalzer/Far-From-MCAR
Data availability:
Data sufficient to reproduce the study findings will be made available approximately one year after completion of the ongoing trial (NCT01864603). Further inquiries can be directed to the SEARCH Scientific Committee at douglas.black@uscf.edu.
References:
- 1.Rubin DB. Inference and missing data. Biometrika. 1976;63(3):581–592. [Google Scholar]
- 2.Little RJA, Rubin DB. Statistical Analysis with Missing Data. 2nd ed. Wiley; 2002. [Google Scholar]
- 3.Perkins NJ, Cole SR, Harel O, et al. Principled Approaches to Missing Data in Epidemiologic Studies. Am J Epidemiol. 2018;187(3):568–575. doi: 10.1093/aje/kwx348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes RA, Heron J, Sterne JAC, Tilling K. Accounting for missing data in statistical analyses: multiple imputation is not always the answer. Int J Epidemiol. 2019;48(4):1294–1304. doi: 10.1093/ije/dyz032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joint United Nations Programme on HIV/AIDS (UNAIDS). 90-90-90 An Ambitious Treatment Target to Help End the AIDS Epidemic.; 2014. http://www.unaids.org/en/resources/documents/2014/90-90-90
- 6.Granich R, Gupta S, Hall I, Aberle-Grasse J, Hader S, Mermin J. Status and methodology of publicly available national HIV care continue and 90-90-90 targets: A systematic review. PLoS Med. 2017;14(4):e1002253. doi:doi: 10.1371/journal.pmed.1002253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabapathy K, Hensen B, Varsaneux O, Floyd S, Fidler S, Hayes R. The cascade of care following community-based detection of HIV in sub-Saharan Africa - A systematic review with 90-90-90 targets in sight. PloS One. 2018;13(7):e0200737. doi: 10.1371/journal.pone.0200737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen M, Balzer L, Kwarsiima D, Sang N, others. Association of implementation of a universal testing and treatment intervention with HIV diagnosis, receipt of antiretroviral therapy, and viral suppression among adults in East Africa. JAMA. 2017;317(21):2196–2206. doi: 10.1001/jama.2017.5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Havlir DV, Balzer LB, Charlebois ED, et al. HIV Testing and Treatment with the Use of a Community Health Approach in Rural Africa. N Engl J Med. 2019;381(3):219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robins JM. Robust estimation in sequentially ignorable missing data and causal inference models. In: 1999 Proceedings of the American Statistical Association. American Statistical Association; 2000:6–10. [Google Scholar]
- 11.Robins JM, Rotnitzky A, Zhao LP. Estimation of regression coefficients when some regressors are not always observed. J Am Stat Assoc. 1994;89(427):846–66. [Google Scholar]
- 12.Bang H, Robins JM. Doubly robust estimation in missing data and causal inference models. Biometrics. 2005;61:962–972. [DOI] [PubMed] [Google Scholar]
- 13.Mohan K, Pearl J, Tian J. Graphical models for inference with missing data. In: Advances in Neural Information Processing Systems 26. ; 2013. http://papers.nips.cc/paper/4899-graphical-models-for-inference-with-missing-data.pdf [Google Scholar]
- 14.Petersen ML, van der Laan MJ. Causal Models and Learning from Data: Integrating Causal Modeling and Statistical Estimation. Epidemiology. 2014;25(3):418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perriat D, Balzer LB, Hayes R, Lockman S, Walsh F, et al. Comparative Assessment of Five Large-Scale Studies of Universal HIV Testing and Treatment in Sub-Saharan Africa. J Int AIDS Soc. 2018;21(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwuji CC, Orne-Gliemann J, Larmarange J, et al. Universal test and treat and the HIV epidemic in rural South Africa: a phase 4, open-label, community cluster randomised trial. Lancet HIV. 2018;5(3):e116–e125. doi: 10.1016/S2352-3018(17)30205-9 [DOI] [PubMed] [Google Scholar]
- 17.Hayes RJ, Donnell D, Floyd S, et al. Effect of Universal Testing and Treatment on HIV Incidence — HPTN 071 (PopART). N Engl J Med. 2019;381(3):207–218. doi: 10.1056/NEJMoa1814556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makhema J, Wirth KE, Pretorius Holme M, et al. Universal Testing, Expanded Treatment, and Incidence of HIV Infection in Botswana. N Engl J Med. 2019;381(3):230–242. doi: 10.1056/NEJMoa1812281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neyman J Sur les applications de la theorie des probabilites aux experiences agricoles: Essai des principes (In Polish). English translation by D.M. Dabrowska and T.P. Speed (1990). Stat Sci. 1923;5:465–480. [Google Scholar]
- 20.Rubin DB. Estimating causal effects of treatments in randomized and nonrandomized studies. J Educ Psychol. 1974;66(5):688–701. doi: 10.1037/h0037350 [DOI] [Google Scholar]
- 21.Holland PW. Statistics and Causal Inference. J Am Stat Assoc. 1986;81(396):945–960. [Google Scholar]
- 22.Rubin DB. Comment: Neyman (1923) and Causal Inference in Experiments and Observational Studies. Stat Sci. 1990;5(4):472–480. [Google Scholar]
- 23.Pearl J Causality: Models, Reasoning and Inference. 2nd ed. Cambridge University Press; 2009. [Google Scholar]
- 24.van der Laan M, Rose S. Targeted Learning: Causal Inference for Observational and Experimental Data. Springer; 2011. [Google Scholar]
- 25.Chamie G, Clark TD, Kabami J, Kadede K, Ssemmondo E, others. A hybrid mobile HIV testing approach for population-wide HIV testing in rural East Africa. Lancet HIV. 2016;3(3):e111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain V, Liegler T, Kabami J, et al. Assessment of Population-Based HIV RNA Levels in a Rural East African Setting Using a Fingerprick-Based Blood Collection Method. Clin Infect Dis. 2013;56(4):598–605. doi: 10.1093/cid/cis881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balzer LB, Schwab J, Laan MJ van der Petersen ML. Evaluation of Progress Towards the UNAIDS 90-90-90 HIV Care Cascade: A Description of Statistical Methods Used in an Interim Analysis of the Intervention Communities in the SEARCH Study. University of California at Berkeley; 2017. http://biostats.bepress.com/ucbbiostat/paper357/ [Google Scholar]
- 28.Robins JM. A new approach to causal inference in mortality studies with sustained exposure periods–application to control of the healthy worker survivor effect. Math Model. 1986;7:1393–1512. doi: 10.1016/0270-0255(86)90088-6 [DOI] [Google Scholar]
- 29.Hernán MA, Lanoy E, Costagliola D, Robins JM. Comparison of dynamic treatment regimes via inverse probability weighting. Basic Clin Pharmacol Toxicol. 2006;98(3):237–242. [DOI] [PubMed] [Google Scholar]
- 30.van der Laan MJ, Petersen ML. Causal Effect Models for Realistic Individualized Treatment and Intention to Treat Rules. Int J Biostat. 2007;3(1):Article 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robins JM, Orellana L, Rotnitzky A. Estimation and extrapolation of optimal treatment and testing strategies. Stat Med. 2008;27(23):4678–4721. [DOI] [PubMed] [Google Scholar]
- 32.Petersen ML, Porter KE, Gruber S, Wang Y, van der Laan MJ. Diagnosing and responding to violations in the positivity assumption. Stat Methods Med Res. 2012;21(1):31–54. doi: 10.1177/0962280210386207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayes R, Floyd S, Schaap A, et al. A universal testing and treatment intervention to improve HIV control: One-year results from intervention communities in Zambia in the HPTN 071 (PopART) cluster-randomised trial. PLOS Med. 2017;14(5):e1002292. doi: 10.1371/journal.pmed.1002292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Floyd S, Ayles H, Schaap A, et al. Towards 90-90: Findings after two years of the HPTN 071 (PopART) cluster-randomized trial of a universal testing-and-treatment intervention in Zambia. PLOS ONE. 2018;13(8):e0197904. doi: 10.1371/journal.pone.0197904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taubman SL, Robins JM, Mittleman MA, Hernán MA. Intervening on risk factors for coronary heart disease: an application of the parametric G-formula. Int J Epidemiol. 2009;38(6):1599–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snowden JM, Rose S, Mortimer KM. Implementation of G-Computation on a Simulated Data set: demonstration of a Causal Inference Technique. Am J Epidemiol. 2011;173(7):731–738. doi: 10.1093/aje/kwq472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horvitz DG, Thompson DJ. A generalization of sampling without replacement from a finite universe. J Am Stat Assoc. 1952;47:663–685. doi: 10.2307/2280784 [DOI] [Google Scholar]
- 38.Rosenbaum PR, Rubin DB. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:41–55. doi: 10.2307/2335942 [DOI] [Google Scholar]
- 39.Hernán MA, Brumback B, Robins JM. Marginal structural models to estimate the causal effect of zidovudine on the survival of HIV-positive men. Epidemiology. 2000;11(5):561–570. [DOI] [PubMed] [Google Scholar]
- 40.Gaolathe T, Wirth KE, Holme MP, et al. Botswana’s progress toward achieving the 2020 UNAIDS 90-90-90 antiretroviral therapy and virological suppression goals: a population-based survey. Lancet HIV. 2016;3(5):e221–230. doi: 10.1016/S2352-3018(16)00037-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Laan MJ, Rubin DB. Targeted Maximum Likelihood Learning. Int J Biostat. 2006;2(1):Article 11. doi: 10.2202/1557-4679.1043 [DOI] [Google Scholar]
- 42.Schuler MS, Rose S. Targeted Maximum Likelihood Estimation for Causal Inference in Observational Studies. Am J Epidemiol. 2017;185(1):65–73. doi: 10.1093/aje/kww165 [DOI] [PubMed] [Google Scholar]
- 43.Luque-Fernandez MA, Schomaker M, Rachet B, Schnitzer ME. Targeted maximum likelihood estimation for a binary treatment: A tutorial. Stat Med. 2018;37(16):2530–2546. doi: 10.1002/sim.7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose S, van der Laan MJ. Why TMLE? In: van der Laan MJ, Rose S, eds. Targeted Learning: Causal Inference for Observational and Experimental Data. Springer; 2011. [Google Scholar]
- 45.Sekhon JS, Gruber S, Porter KE, van der Laan MJ. Propensity-Score-Based Estimators and C-TMLE. In: van der Laan MJ, Rose S, eds. Targeted Learning: Causal Inference for Observational and Experimental Data. Springer; 2011:343–364. [Google Scholar]
- 46.Gruber S, van der Laan MJ. Targeted minimum loss based estimator that outperforms a given estimator. Int J Biostat. 2012;8(1):Article 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balzer L, Ahern J, Galea S, van der Laan MJ. Estimating Effects with Rare Outcomes and High Dimensional Covariates: Knowledge is Power. Epidemiol Methods. 2016;5(1):1–18. doi: 10.1515/em-2014-0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Laan MJ, Polley EC, Hubbard AE. Super Learner. Stat Appl Genet Mol Biol. 2007;6(1):Article 25. doi: 10.2202/1544-6115.1309 [DOI] [PubMed] [Google Scholar]
- 49.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2018. http://www.R-project.org [Google Scholar]
- 50.Schwab J, Lendle S, Petersen M, van der Laan M. Ltmle: Longitudinal Targeted Maximum Likelihood Estimation.; 2017. http://CRAN.R-project.org/package=ltmle [Google Scholar]
- 51.Polley E, LeDell E, Kennedy C, van der Laan M. SuperLearner: Super Learner Prediction.; 2018. Accessed July 15, 2019. http://CRAN.R-project.org/package=SuperLearner [Google Scholar]
- 52.van der Laan MJ. Targeted estimation of nuisance parameters to obtain valid statistical inference. Int J Biostat. 2014;10(1):29–57. doi: 10.1515/ijb-2012-0038 [DOI] [PubMed] [Google Scholar]
- 53.Benkeser D, Carone M, Laan MJVD, Gilbert PB. Doubly robust nonparametric inference on the average treatment effect. Biometrika. 2017;104(4):863–880. doi: 10.1093/biomet/asx053 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sufficient to reproduce the study findings will be made available approximately one year after completion of the ongoing trial (NCT01864603). Further inquiries can be directed to the SEARCH Scientific Committee at douglas.black@uscf.edu.