Abstract
Background
Chryseobacterium indologenes (C. indologenes) has recently emerged as a cause of life-threatening nosocomial infections in humans. This study aims to investigate the clinical characteristics, homology, and antimicrobial patterns of C. indologenes clinical isolates at a teaching hospital in Shanghai, China.
Methods
A total of 135 consecutive non-replicate clinical C. indologenes isolates from January 2010 to December 2018 were collected at a tertiary care university hospital in Shanghai, China. Genetic relatedness of the isolates was performed by pulsed-field gel electrophoresis (PFGE). The antimicrobial susceptibility of these isolates was measured by the microdilution broth method. The prevalence of β-lactamase genes was investigated by polymerase chain reaction (PCR), while the quinolone resistance-determining regions (QRDRs) were sequenced.
Results
All 135 C. indologenes isolates were collected from hospitalized patients with an average age of 55 years. Most of these clinical isolates were derived from ascites (59.3%) or urine (23.7%) specimens. Eighty (80/135) of the strains were classified as clone D by PFGE. In vitro drug susceptibility tests showed that minocycline and trimethoprim-sulfamethoxazole had sound antibacterial effects. However, more than 86% of the tested strains were resistant to cephalosporins (ceftazidime, cefotaxime), β-lactamase/β-lactamase inhibitors (cefoperazone-sulbactam), and carbapenems (meropenem, imipenem). Metallo-β-lactamase blaIND and type A broad-spectrum β-lactamase genes blaCIA were present in 135 and 103 isolates, respectively. The clinical strains in our hospital mainly carried blaIND-2 (89.6%, 121/135). Compared with previous studies, these strains had a high rate of resistance to quinolones. The resistance rates to levofloxacin, ciprofloxacin, norfloxacin, gatifloxacin, and nemonoxacin were as high as 83.7–94.8%. The mutations at Ser83Val, Ser83Tyr, and Asp87Gly in the QRDRs of GyrA were significantly related to the resistance of C. indologenes to levofloxacin. All but one quinolone-resistant strain contained at least one significant mutation.
Conclusions
This study showed a clonal dissemination of C. indologenes isolates in infections at a tertiary care university hospital in Shanghai, China. Minocycline and trimethoprim-sulfamethoxazole had favorable in vitro antibacterial effects. However, the high resistance rate to β-lactams and quinolones was due to carrying β-lactamase (blaIND, blaCIA), and mutations in the QRDRs of GyrA.
Keywords: Chryseobacterium indologenes (C. indologenes), nosocomial infection, drug resistance, quinolone-resistance determining region
Introduction
Chryseobacterium indologenes (C. indologenes), is a non-lactose fermenting gram-negative bacillus formally classified under the Flavobacterium CDC group IIb (1). Since its first identification in 1993 from the tracheal aspirate of a patient with ventilator-associated pneumonia (VAP) as an opportunistic pathogen (2), C. indologenes has been notorious for causing nosocomial infections due to its presence in fluid-associated devices which serve as a potential reservoir of infection (3). C. indologenes causes many kinds of infections such as catheter-related bacteremia, urinary tract infection, biliary tract infection, peritonitis, surgical wound infection, and hospital-associated pneumonia (HAP), especially in immunocompromised patients (4-7). Nosocomial infections attributable to C. indologenes have been increasingly reported in numerous countries and have caused significant morbidity and mortality. A study conducted in Taiwan found that 98% (212/215) of C. indologenes infections were nosocomial, and the mortality rates of patients with bacteremia or pneumonia was 35.4% (40/113) (8). More recently, Cantero et al. reported an outbreak of C. indologenes infections in an intensive care unit of a Spanish hospital where mortality reached 25% (3).
C. indologenes exhibits resistance to the majority of antimicrobial agents such as carbapenems, cephalosporins, aminoglycosides, and chloramphenicol which are used to empirically treat infections caused by other gram-negative bacteria (1,9). Therefore, the infection caused by C. indologenes is challenging to manage and often results in unfavorable outcomes (10). In addition, according to the SENTRY Antimicrobial Surveillance Program, clinical C. indologenes isolates from Asia generally have higher resistance rates to cephalosporins and carbapenems than other continents (11). The resistance rate of the Chryseobacterium species to ceftazidime and imipenem from 1997–2001 in Latin America and North America was 40–42.9% and 73.3–85.7%. In contrast, the resistance rates to ceftazidime and imipenem in Asia manifested as 87.5% and 100% (11). It has been speculated that C. indologenes is intrinsically resistant to cephalosporins and carbapenems due to its production of molecular class A β-lactamase blaCIA and class B carbapenem-hydrolyzing β-lactamase blaIND (12,13).
Minocycline, trimethoprim-sulfamethoxazole, and quinolones have demonstrated favorable in vitro susceptibility test results. Among them, quinolones were the most active anti-infective agents with susceptibility rates of >95% in the SENTRY program (11). Together with their broad antibacterial spectrum and high tissue concentration, quinolones were recommended for the treatment of C. indologenes infections (14). However, the emergence of a quinolones-resistant C. indologenes strain has raised concern. It has been recently reported that the susceptibility rates to quinolones of C. indologenes isolates obtained in Taiwan during 2005–2017 have dropped to 16.7–19% (15). The quinolone resistance in C. indologenes has been attributed to alterations in DNA gyrase.
However, clinical reports and the molecular characterization of C. indologenes collected in mainland China are limited, resulting in relatively insufficient clinical evidence for the treatment of infectious diseases caused by the bacteria. This study aimed to explore the clinical characteristics, homology, and antimicrobial patterns of C. indologenes clinical isolates from January 2010 to December 2018 at a teaching hospital in Shanghai, China. Moreover, the distribution of blaCIA, blaIND, and the mutation of quinolone resistance-determining regions (QRDRs) were also investigated. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-933).
Methods
Bacterial isolates
A total of 135 consecutive non-replicate clinical C. indologenes isolates from January 2010 to December 2018 were collected at a tertiary care university hospital in Shanghai, China. A 16s-rRNA sequence analysis was performed for bacterial identification (type strain, DSM 16777T158; GenBank accession no. LN681561). All the clinical isolates were stored at −80 °C prior to use. These isolates were collected from different specimens including ascites, sputum, and urine. Medical records were anonymously collected to review clinical information, including age, sex, hospitalization department, and type of infection acquisition. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Huashan Hospital, Fudan University, China (approval number: KY2017-274). Individual consent for this retrospective analysis was waived. Nosocomial infection was defined as previously reported (16).
Homology analysis
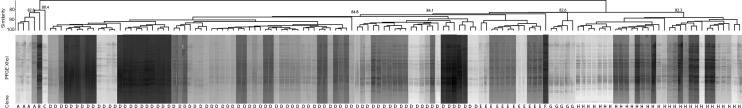
The homology analysis of clinical isolates was performed by pulsed-field gel electrophoresis (PFGE). Chromosomal DNA was prepared in agarose blocks and digested with XhoI. DNA fragments were separated by PFGE on a CHEF Mapper XA (Bio-Rad, Hercules, CA, USA) for 19 h at 14 °C, at 6 V/cm, a pulse angle of 120, and pulse times ranging from 0.5 to 13.6 s. We constructed dendrograms using the Dice coefficient and unweighted pair–group method using average linkage clustering. Isolates were regarded as a joint pulsed-field group if the Dice similarity coefficient >85% in this study. The result was analyzed by BioNumerisc software version 7.6 (Applied-Maths, Belgium)
Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) of 16 antimicrobial agents were measured by the microdilution broth method according to the Clinical and Laboratory Standards Institute 29th edition (CLSI 29th ed.), including cefotaxime, ceftazidime, piperacillin-tazobactam, cefoperazone-sulbactam, imipenem, meropenem, norfloxacin, levofloxacin, gatifloxacin, ciprofloxacin, moxifloxacin, nemonoxacin, amikacin, minocycline, trimethoprim-sulfamethoxazole (TMP-SMZ), and rifampicin (17). Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC27853, and Enterococcus faecalis ATCC29212 were used as quality control reference strains. The results were interpreted according to the criteria of “other non-Enterobacteriaceae” in CLSI 29th ed., except that the MICs of rifampin were interpreted according to the Enterococcus susceptibility breakpoints.
Detection and sequencing of β-lactamase genes and QRDRs
The presence of crucial β-lactamase [blaCIA, blaTEM, blaSHV, blaCTX-M (group1, group2,group9), blaKPC, blaOXA48, blaIMP, blaVIM, blaNDM] (12,18,19) and QRDRs (GyrA, GyrB, ParC, and ParE) (15) were amplified and sequenced by polymerase chain reaction (PCR) with primers and PCR conditions as previously described. The primers and PCR conditions for the amplification of blaIND are listed in Table S1.
Statistical analysis
Statistical analysis was conducted by a two-tailed Student’s t-test or chi-square test with SPSS Statistics 25 software (IBM, https://www.ibm.com), and a P value less than 0.05 was considered statistically significant.
Results
Bacterial isolates and site of isolation
Among 135 clinical C. indologenes isolates, 39 strains were isolated from 2010 to 2016, accounting for 28.9%. Since then, there has been a continual growth in the number of isolates. In 2017 and 2018, 66 strains and 30 strains were collected, accounting for 48.9% and 22.2%, respectively (Figure 1). The isolates were mainly collected from ascites (77/135, 57.0%) and urine (32/135, 23.7%). Other sites of isolation included sputum (18/135, 13.3%), bile (3/135, 2.2%), blood (2/135, 1.5%), wound secretions (1/135, 0.7%), and two isolates were missing information.
Figure 1.
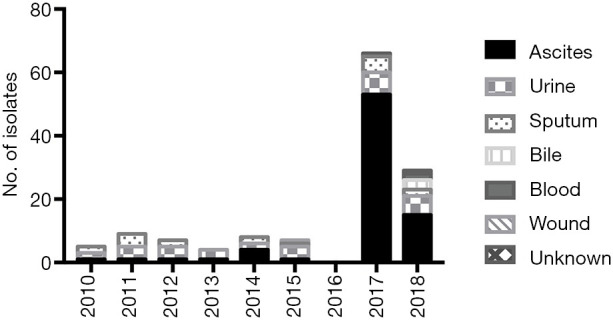
The distribution of 135 C. indologenes isolates according to the year and site of isolation. Unknown: missing information.
Demographic data and clinical characteristics
The demographic data and clinical characteristics of the 135 C. indologenes infections are summarized in Table 1. Male patients predominated (97/135, 71.9%), and the mean age was 55 years (range, 5–98 years), with 36 patients over 65 years. Most of these patients came from the general surgery department (79/135, 58.5%), followed by the intensive care unit (ICU) (8/135, 5.9%), the neurosurgery department (8/135, 5.9%), and the gerontology department (7/135, 5.2%). Remaining patients came from other departments including the infectious disease, nephrology, hematology, neurology and pneumology departments. Nosocomial infection accounted for the vast majority (84.4%, 114/135), three infections were identified as community-acquired, while others remained difficult to ascertain due to lack of information.
Table 1. Demographic data and clinical characteristics of patients with Chryseobacterium indologenes infection.
| Variables | Total (n=135) |
|---|---|
| Sex, n (%) | |
| Male | 97 (71.9) |
| Female | 38 (28.1) |
| Age | |
| Range, years | 5–98 |
| Median, years | 55 |
| Age ≥65, n (%) | 36 (26.7) |
| Department, n (%) | |
| General surgery | 79 (58.5) |
| Intensive care unit | 8 (5.9) |
| Neurosurgery | 8 (5.9) |
| Gerontology | 7 (5.2) |
| Infectious disease | 5 (3.7) |
| Nephrology | 4 (3.0) |
| Hematology | 3 (2.2) |
| Neurology | 2 (1.5) |
| Pneumology | 1 (0.7) |
| Others* | 18 (13.3) |
| Type of infection acquisition, n (%) | |
| Community-acquired | 3 (2.2) |
| Healthcare-associated | 114 (84.4) |
| Unclear* | 18 (13.3) |
*, information was hard to ascertain due to missing data.
Homology analysis
The homology analysis of 135 C. indologenes isolates was determined by PFGE. With a similarity coefficient of 85% as a cutoff value, eight significant clusters were found (Figure 2). Among them, the D type was the most prevalent, accounting for 59.3% (80/135), followed by the H and E types which accounted for 23.0% (31/135) and 8.9% (12/135), respectively.
Figure 2.
Dendrogram generated from the analysis of 135 C. indologenes isolates.
Antimicrobial susceptibility testing
The MICs results of 16 antimicrobial agents are shown in Table 2. Minocycline and trimethoprim/sulfamethoxazole appeared to be the most potent antimicrobial agents against C. indologenes, with a low resistance rate of 0.7% (1/135) and 2.2% (3/135), respectively. Rifampicin also showed a good antibacterial effect (resistance rate 23.7%). More than 86% of the tested strains were resistant to cephalosporins (cefotaxime and ceftazidime), β-lactamase/β-lactamase inhibitors (cefoperazone-sulbactam), carbapenems (meropenem and imipenem), and aminoglycosides (amikacin). Furthermore, most C. indologenes clinical isolates were resistant to quinolones, and the resistance rates to norfloxacin, levofloxacin, gatifloxacin, and ciprofloxacin were all over 83%, followed by moxifloxacin (resistance rate 56.3%). We also tested the new fluorine-free quinolone nemonoxacin. However, the result was unsatisfactory with a drug resistance rate of 94.8%. At the same time when the high level of resistance to quinolones was detected, the MIC50 values of norfloxacin, levofloxacin, gatifloxacin, ciprofloxacin, and nemonoxacin were >32 µg/mL.
Table 2. Antimicrobial susceptibility of 135 Chryseobacterium indologenes isolates (µg/mL).
| Antimicrobial agent | S (%) | I (%) | R (%) | MIC50 | MIC90 | Susceptibility breakpoint | MIC range |
|---|---|---|---|---|---|---|---|
| Cefotaxime | 0.0 | 5.9 | 94.1 | >128 | >128 | ≤8 | 1–128 |
| Ceftazidime | 6.7 | 5.9 | 87.4 | >128 | >128 | ≤8 | 1–128 |
| Piperacillin-tazobactam | 37.0 | 9.6 | 53.3 | >128/4 | >128/4 | ≤16/4 | 1/4–128/4 |
| Cefoperazone-sulbactam | 10.4 | 3.0 | 86.7 | >128/64 | >128/64 | ≤8/4 | 1/0.5–128/64 |
| Imipenem | 0.7 | 1.5 | 97.8 | >64 | >64 | ≤4 | 0.5–64 |
| Meropenem | 0.0 | 2.2 | 97.8 | >64 | >64 | ≤4 | 0.5–64 |
| Norfloxacin | 13.3 | 3.0 | 83.7 | >32 | >32 | ≤2 | 0.25–32 |
| Levofloxacin | 14.8 | 0.7 | 84.4 | 32 | >32 | ≤2 | 0.25–32 |
| Gatifloxacin | 14.1 | 1.5 | 84.4 | >32 | >32 | ≤2 | 0.25–32 |
| Ciprofloxacin | 12.6 | 0.7 | 86.7 | >32 | >32 | ≤1 | 0.25–32 |
| Moxifloxacin | 23.7 | 20.0 | 56.3 | 8 | >32 | ≤2 | 0.25–32 |
| Nemonoxacin | 5.2 | 0.0 | 94.8 | >32 | >32 | ≤2 | 0.25–32 |
| Amikacin | 0.7 | 1.5 | 97.8 | >128 | >128 | ≤16 | 1–128 |
| Minocycline | 98.5 | 0.7 | 0.7 | 2 | 4 | ≤4 | 0.5–64 |
| TMP-SMZ | 97.8 | 0.0 | 2.2 | 0.5/9.5 | 1/19 | ≤2/38 | 0.125/2.375–16/304 |
| Rifampicin | 72.6 | 3.7 | 23.7 | 0.5 | 16 | ≤1 | 0.25–32 |
S, susceptible; I, intermediate; R, resistant; MIC, minimal inhibitory concentration; MIC50, minimal inhibitory concentration for 50% isolates; MIC90, minimal inhibitory concentration for 50% isolates; TMP-SMZ, trimethoprim-sulfamethoxazole.
Detection of β-lactamase genes
All C. indologenes isolates carried the metallo-β-lactamase blaIND, and the type A broad-spectrum β-lactamase blaCIA was present in 103 isolates. According to the sequence results, blaIND-2 was found in 121 (89.6%) isolates, which accounted for the majority, followed by blaIND-7 (n=4), blaIND-3 (n=3), blaIND-14 (n=3), blaIND-5 (n=2), blaIND-8 (n=1), and blaIND-11 (n=1). None of these isolates had a combination of more than two types of blaIND. No blaTEM, blaSHV, blaCTX-M (group 1, group 2, group 9), blaKPC, blaOXA48, blaIMP, blaVIM, blaNDM were detected.
Mutations in the QRDRs
The mutations of amino acids in the QRDRs of C. indologenes are shown in Table 3. QRDR mutations of GyrA, GyrB, and ParE were detected. In the QRDR of GyrA, among the 115 levofloxacin non-susceptible isolates, 60 isolates carried mutations at Ser83Tyr, 50 isolates carried mutations at Ser83Val, 3 isolates carried mutations at Ser83Phe, and 62 isolates carried the mutation at Asp87Gly in GyrA. Compared with susceptible isolates, mutations at Ser83Tyr, Ser83Val, and Asp87Gly were significantly correlated with levofloxacin non-susceptibility (P<0.001). Amino acid mutations were detected in GyrB at codons 425 and 473, and in ParE at codon 539 but these mutations were not significantly related to the susceptibility of levofloxacin (P=0.081, P=0.081, P=0.362). No amino acid mutation was found in ParC. It is noteworthy that all but one resistant strain contained at least one significant mutation.
Table 3. Amino acid alterations in the quinolone resistance-determining regions of Chryseobacterium indologenes isolates.
| Amino acid | Susceptibility of levofloxacin, n (%) | P value | |
|---|---|---|---|
| Susceptible (n=20) | Non-susceptible (n=115) | ||
| Position 83 of GyrA | |||
| Serine | 20 (100.0) | 2 (1.7) | <0.001 |
| Valine | 0 (0.0) | 50 (43.5) | <0.001 |
| Tyrosine | 0 (0.0) | 60 (52.2) | <0.001 |
| Phenylalanine | 0 (0.0) | 3 (2.6) | >0.999 |
| Position 87 of GyrA | |||
| Aspartic acid | 19 (95.0) | 53 (46.1) | <0.001 |
| Glycine | 1 (5.0) | 62 (53.9) | |
| Position 425 of GyrB | |||
| Leucine | 2 (10.0) | 2 (1.7) | 0.1044 |
| Isoleucine | 18 (90.0) | 113 (98.3) | |
| Position 473 of GyrB | |||
| Lysine | 2 (10.0) | 2 (1.7) | 0.1044 |
| Arginine | 18 (90.0) | 113 (98.3) | |
| Position 539 of ParE | |||
| Valine | 20 (100.0) | 105 (91.3) | 0.3575 |
| Isoleucine | 0 (0.0) | 10 (8.7) | |
Analysis of quinolones-resistant level
The connection between the level of quinolones resistance and significant amino acid mutations at codons 83 and 87 of GyrA were analyzed. As shown in Table 4, the strain with a single amino acid substitution in GyrA at Ser83Val (n=50) and Ser83Tyr (n=1) was moderately resistant to levofloxacin (MIC50 =8 µg/mL). The isolates had two mutations in GyrA at Ser83Tyr and Asp87Gly (n=59) and showed a high level of levofloxacin resistance (MIC50 >32 µg/mL). We also found three strains carrying substitutions in GyrA at Phe83Ser and Asp87Gly. Although Phe83Ser was not related to levofloxacin resistance, the strains containing these two mutations also showed high levels of resistance (MIC50 >32 µg/mL). Strains carrying the GyrA subunit double-site mutations showed a 4–32 times higher MIC value for quinolones than the single-point mutation strains. The same trend was observed in gatifloxacin, ciprofloxacin, and moxifloxacin.
Table 4. The correlation between amino acid mutations at different sites of GyrA and MIC50 (µg/mL) of quinolones.
| No. amino acid mutation | No. isolates (n) | Amino acid of GyrA | MIC range (MIC50) | |||||
|---|---|---|---|---|---|---|---|---|
| 83 | 87 | Levofloxacin | Ciprofloxacin | Gatifloxacin | Moxifloxacin | |||
| Wild | 21 | – | – | ≤0.25–32 (0.5) | ≤0.25–>32 (0.5) | ≤0.25–16 [1] | ≤0.25–8 (≤0.25) | |
| 1 | 1 | – | Asp87Gly | 1 | 1 | 1 | 0.5 | |
| 1 | 50 | Ser83Val | – | 8–16 [8] | 16–>32 [32] | 4->32 [8] | 2–32 [4] | |
| 1 | 1 | Ser83Tyr | - | 8 | 32 | 32 | 4 | |
| 2 | 3 | Ser83Phe | Asp87Gly | 32–>32 (>32) | >32 | 16–>32 (>32) | 8–>32 (>32) | |
| 2 | 59 | Ser83Tyr | Asp87Gly | 8–>32 (>32) | >32 | 8–>32 [32] | 4–>32 [16] | |
MIC50, minimal inhibitory concentration for 50% isolates.
Discussion
C. indologenes is widely distributed in plants, soil and water, and in the past was rarely considered a human pathogen (9). Based on a PUBMED literature search using the keywords ‘Chryseobacterium indologenes infections’, we identified 44 studies published in the period 2010–2019 which are summarized in Table S2. Analysis of the scientific literature suggests that the highest frequency of C. indologenes infections occurs in Asia, several clinical cases were occurred in North America, South America, and Europe. C. indologenes infections were mainly sporadic, sometimes outbreaks could also be a threat in hospital. Susceptible populations are mainly infants (especially premature babies), the elderly, long-term hospitalized patients, patients with serious underlying diseases or immunocompromised. C. indologene can cause multiple types of infection, the most frequently reported infections were pneumonia and bacteraemia. The long-term indwelling devices especially tracheal tubes and intravascular catheters increased the risk of infection. It is worth noting that C. indologenes infections have also been gradually appearing in patients with normal immunity and no catheter implantation. In this study, we conducted a 9-year retrospective study and described a clonal dissemination of hospital-acquired infection caused by C. indologenes. Our findings are in accord with previous literature indicating that most cases of C. indologenes are due to nosocomial infection. It should be stressed that C. indologenes can be colonized and spread in the hospital environment through contaminated and humid medical equipment (20). Furthermore, with the development of medical technology and the increased use of various invasive procedures and broad-spectrum antibacterial drugs, there has been a phenomenal growth of nosocomial infection caused by C. indologenes (9). The clonal dissemination of multidrug-resistant bacteria in hospitals has become a public health problem that jeopardizes patients and increases medical burden. This is the first report to describe a clonal dissemination of C. indologenes in mainland China. According to our results, this clonal dissemination was related to an intra-abdominal infection after liver transplantation in the general surgery department, which highlights the need for active surveillance and implementation of infection control measures.
The susceptibilities of C. indologenes vary dramatically from one region to another, making it difficult to choose an effective drug for empirical treatment (11). Previous studies have demonstrated that Chryseobacterium sepsis is nearly uniformly resistant to extended-spectrum penicillin, aztreonam, cephalosporins, imipenem, meropenem, chloramphenicol, and aminoglycosides. Quinolones, minocycline, rifampicin, and trimethoprim-sulfamethoxazole are, however, usually effective (7,8,10,21). Quinolones used to be considered an appropriate antimicrobial agent to treat C. indologenes infections (22). However, according to a recent study, the resistance rate to quinolones is rising, especially for those strains isolated in Asia. In our study, in vitro drug susceptibility tests showed that minocycline and trimethoprim-sulfamethoxazole had sound antibacterial effects on C. indologenes, with resistance rates of 0.7% and 2.2%, respectively. Cephalosporins, β-lactam/β-lactamase inhibitors, carbapenems, and aminoglycosides showed insufficient activity against these pathogens, and our study showed a much higher resistance rate to these antimicrobial agents than previous reports. Moreover, the high resistance rate to quinolones suggested that they were no longer an appropriate choice. It is noteworthy that our study showed the lowest susceptibility to ciprofloxacin compared with other Asian studies (12.6% vs. 14.7–68%) (10,15,22,23).
Our study revealed that C. indologenes displayed an increased rate of resistance to previously potent antibiotics. Hence, we investigated the resistant mechanism of β-lactams and fluoroquinolones. Usually, C. indologenes shows intrinsic resistance to carbapenems and cephalosporins as a result of molecular class A β-lactamase blaCIA and class B carbapenem-hydrolyzing β-lactamase blaIND. These two β-lactamases are peculiar to the Chryseobacterium species. Lin et al. detected blaIND1-4 in clinical isolates of C. indologenes during 2004 to 2006 in Hefei, China, and reported that blaIND1 (32.1%, 9/28) and blaIND2 (35.7%, 10 /28) were the most prevalent (23). To date, twelve new blaIND alleles have been registered in GenBank, but as yet no investigation of the prevalence of all 16 blaIND alleles and blaCIA in clinical isolates has been conducted. In our study, we found that all isolates carried blaIND, and 76.2% (103/135) of isolates carried blaCIA. The dominant genotype was blaIND-2, accounting for 89.6% (121/135).
The mechanism of resistance to quinolones is mainly caused by mutations in QRDRs (GyrA, GyrB, ParC, and ParE subunits), efflux pumps (QepA and OqxAB), DNA topoisomerase protection protein Qnr, and quinolone acetyltransferase Aac (6’)-Ib-cr (24-27). The gene mutations in QRDRs resulting in changes of the enzyme’s affinity for quinolone is the primary resistant mechanism of quinolones (28). Quinolone resistance in C. indologenes was previously identified to be associated with the alterations in DNA gyrase. A study reported that mutations of Ser83Tyr and Asp87Tyr in the GyrA subunit lead to the non-susceptibility of C. indologenes (P<0.001, P=0.018) (15). In this study, we found that both the mutations at codons 83 and 87 of the GyrA were associated with quinolones resistance (P<0.001), and the mutation types were Ser83Val, Ser83Tyr, and Asp87Gly. When the hydroxyl-containing serine at position 83 of the GyrA mutates into valine or tyrosine, it will affect the formation of hydrogen bonds and the replacement of aliphatic chains. After replacing aspartic acid at position 87 with glycine, the negatively charged amino acids are replaced by uncharged amino acids, and the charge of the peptide chain changes accordingly. These changes in amino acids verify that the formation of hydrogen bonds and the presence of negatively charged amino acids play a vital role in the interaction of quinolones with gyrase-DNA complexes (26). This is the first study to report that Ser83Val and Asp87Gly are associated with quinolones resistance of C. indologenes. Among the Enterobacteriaceae, the most frequently identified mutations in GyrA were at codons 83 and 87 (29). Notably, double mutations in GyrA are related to high levels of resistance to fluoroquinolones in Escherichia coli isolates (30). We detected a similar phenomenon in C. indologenes insofar as double amino acid alterations in the QRDRs of GyrA were significantly associated with high levels of quinolones resistance.
The present results were limited by small sample size and retrospective single-center study design. A prospective multi-center study should be conducted to monitor antimicrobial agent resistance pattern and mechanisms of C. indologenes. In conclusion, this study reveals the clinical characteristics and antimicrobial susceptibility patterns of C. indologenes. A clonal dissemination of C. indologenes in Huashan Hospital has been observed. Minocycline and TMP-SMZ were the most suitable antimicrobial agents to treat infections caused by this pathogen. An increased resistance rate to β-lactams and quinolones was exhibited compared with previous studies. The resistance mechanism of β-lactams in C. indologenes is mediated by blaCIA and blaIND. The resistance mechanism of quinolones is related to amino acid mutations in the quinolone resistance-determining region of the GyrA subunit.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (grant number 81603163).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Huashan Hospital, Fudan University, China (approval number: KY2017-274). Individual consent for this retrospective analysis was waived.
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-933
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-933
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-933). The authors have no conflicts of interest to declare.
(English Language Editor: D. Fitzgerald)
References
- 1.Fraser SL, Jorgensen JH. Reappraisal of the antimicrobial susceptibilities of Chryseobacterium and Flavobacterium species and methods for reliable susceptibility testing. Antimicrob Agents Chemother 1997;41:2738-41. 10.1128/AAC.41.12.2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonten MJ, van Tiel FH, van der Geest S, et al. Topical antimicrobial prophylaxis of nosocomial pneumonia in mechanically ventilated patients. Microbiological observations. Infection 1993;21:137-9. 10.1007/BF01710529 [DOI] [PubMed] [Google Scholar]
- 3.Cantero M, Parra LM, Munez E, et al. A cluster of Chryseobacterium indologenes cases related to drainage water in intensive care units. Infect Control Hosp Epidemiol 2018;39:997-9. 10.1017/ice.2018.126 [DOI] [PubMed] [Google Scholar]
- 4.Lambiase A, Del Pezzo M, Raia V, et al. Chryseobacterium respiratory tract infections in patients with cystic fibrosis. J Infect 2007;55:518-23. 10.1016/j.jinf.2007.08.002 [DOI] [PubMed] [Google Scholar]
- 5.Antonello VS, Daht P, Polli J, et al. Ventilator-associated pneumonia in neonatal intensive care unit due to Chryseobacterium indologenes. Pediatr Infect Dis J 2017;36:e353-e355. 10.1097/INF.0000000000001730 [DOI] [PubMed] [Google Scholar]
- 6.González-Castro A, Alsasua A, Peñasco Y, et al. Tracheo-bronchitis and pneumonia associated with mechanical ventilation by Chryseobacterium indologenes. Rev Esp Anestesiol Reanim 2017;64:294-8. [DOI] [PubMed] [Google Scholar]
- 7.Esposito S, Russo E, De Simone G, et al. Transient bacteraemia due to Chryseobacterium indologenes in an immunocompetent patient: a case report and literature review. J Chemother 2015;27:324-9. 10.1179/1973947814Y.0000000206 [DOI] [PubMed] [Google Scholar]
- 8.Chen FL, Wang GC, Teng SO, et al. Clinical and epidemiological features of Chryseobacterium indologenes infections: analysis of 215 cases. J Microbiol Immunol Infect 2013;46:425-32. 10.1016/j.jmii.2012.08.007 [DOI] [PubMed] [Google Scholar]
- 9.Hsueh PR, Teng LJ, Yang PC, et al. Increasing incidence of nosocomial Chryseobacterium indologenes infections in Taiwan. Eur J Clin Microbiol Infect Dis 1997;16:568-74. 10.1007/BF02447918 [DOI] [PubMed] [Google Scholar]
- 10.Chang YC, Lo HH, Hsieh HY, et al. Identification, epidemiological relatedness, and biofilm formation of clinical Chryseobacterium indologenes isolates from central Taiwan. J Microbiol Immunol Infect 2015;48:559-64. 10.1016/j.jmii.2014.04.004 [DOI] [PubMed] [Google Scholar]
- 11.Kirby JT, Sader HS, Walsh TR, et al. Antimicrobial susceptibility and epidemiology of a worldwide collection of Chryseobacterium spp: report from the SENTRY Antimicrobial Surveillance Program (1997-2001). J Clin Microbiol 2004;42:445-8. 10.1128/JCM.42.1.445-448.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto T, Nagata M, Ishimine N, et al. Characterization of CIA-1, an Ambler class A extended-spectrum beta-lactamase from Chryseobacterium indologenes. Antimicrob Agents Chemother 2012;56:588-90. 10.1128/AAC.05165-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellais S, Léotard S, Poirel L, et al. Molecular characterization of a carbapenem-hydrolyzing beta-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol Lett 1999;171:127-32. [DOI] [PubMed] [Google Scholar]
- 14.Lin YT, Jeng YY, Lin ML, et al. Clinical and microbiological characteristics of Chryseobacterium indologenes bacteremia. J Microbiol Immunol Infect 2010;43:498-505. 10.1016/S1684-1182(10)60077-1 [DOI] [PubMed] [Google Scholar]
- 15.Lin JN, Lai CH, Yang CH, et al. Differences in clinical manifestations, antimicrobial susceptibility patterns, and mutations of fluoroquinolone target genes between Chryseobacterium gleum and Chryseobacterium indologenes. Antimicrob Agents Chemother 2019;63:e02256-18. 10.1128/AAC.02256-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control 2008;36:309-32. 10.1016/j.ajic.2008.03.002 [DOI] [PubMed] [Google Scholar]
- 17.Wayne P; Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-ninth Informational Supplement M100-S29. CLSI, 2019, USA.
- 18.Poirel L, Walsh TR, Cuvillier V, et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 2011;70:119-23. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 19.Dallenne C, Da Costa A, Decre D, et al. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 2010;65:490-5. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- 20.Beattie RE, Skwor T, Hristova KR. Survivor microbial populations in post-chlorinated wastewater are strongly associated with untreated hospital sewage and include ceftazidime and meropenem resistant populations. Sci Total Environ 2020;740:140186. 10.1016/j.scitotenv.2020.140186 [DOI] [PubMed] [Google Scholar]
- 21.Mirza HC, Tuncer O, Olmez S, et al. Clinical strains of Chryseobacterium and Elizabethkingia spp. isolated from pediatric patients in a university hospital: performance of MALDI-TOF MS-based identification, antimicrobial susceptibilities, and baseline patient characteristics. Microb Drug Resist 2018;24:816-21. 10.1089/mdr.2017.0206 [DOI] [PubMed] [Google Scholar]
- 22.Liang CY, Yang CH, Lai CH, et al. Genomic features, comparative genomic analysis, and antimicrobial susceptibility patterns of Chryseobacterium arthrosphaerae strain ED882-96 isolated in Taiwan. Genes 2019;10:309. 10.3390/genes10040309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin XH, Xu YH, Cheng J, et al. Heterogeneity of bla(IND) metallo-beta-lactamase-producing Chryseobacterium indologenes isolates detected in Hefei, China. Int J Antimicrob Agents 2008;32:398-400. 10.1016/j.ijantimicag.2008.04.028 [DOI] [PubMed] [Google Scholar]
- 24.Yamane K, Wachino J, Suzuki S, et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother 2007;51:3354-60. 10.1128/AAC.00339-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HB, Wang M, Park CH, et al. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother 2009;53:3582-4. 10.1128/AAC.01574-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hooper DC, Jacoby GA. Topoisomerase inhibitors: fluoroquinolone mechanisms of action and resistance. Cold Spring Harb Perspect Med 2016;6:a025320. 10.1101/cshperspect.a025320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother 2005;56:463-9. 10.1093/jac/dki245 [DOI] [PubMed] [Google Scholar]
- 28.Hooper DC. Mechanisms of action of antimicrobials: focus on fluoroquinolones. Clin Infect Dis 2001;32 Suppl 1:S9-S15. 10.1086/319370 [DOI] [PubMed] [Google Scholar]
- 29.Ostrer L, Khodursky RF, Johnson JR, et al. Analysis of mutational patterns in quinolone resistance-determining regions of GyrA and ParC of clinical isolates. Int J Antimicrob Agents 2019;53:318-24. 10.1016/j.ijantimicag.2018.12.004 [DOI] [PubMed] [Google Scholar]
- 30.Vila J, Ruiz J, Marco F, et al. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob Agents Chemother 1994;38:2477-9. 10.1128/AAC.38.10.2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as