Abstract
Background
Activated epidermal growth factor receptor (EGFR) mutation is the main pathogenic cause of non-small cell lung cancer (NSCLC) in Asia. However, the impact of plasma EGFR mutation abundance, especially of the ultra-low abundance of EGFR mutation detected by highly sensitive techniques on clinical outcomes of first-line EGFR tyrosine kinase inhibitors (TKIs) for advanced NSCLC patients remains unclear.
Methods
We qualitatively detected baseline EGFR status of NSCLC tissues using amplification-refractory mutation system and quantified the plasma abundance of EGFR mutations through next-generation sequencing (NGS). Every 8–12 weeks, we performed dynamic detection of plasma mutation abundance and imaging evaluation. We analyzed the association between plasma abundance of EGFR sensitizing mutations, tumor size, tumor shrinkage percentage, concomitant TP53 mutations, and clinical response to TKIs.
Results
This prospective study enrolled 135 patients with advanced NSCLC. The objective response rate (ORR) and disease control rate (DCR) for EGFR mutation–positive patients were 50.0% and 87.0%, respectively. When the cutoff value of plasma EGFR mutation abundance was 0.1%, the ORRs of TKI-treated patients were significantly different (60.0% for the >0.1% group vs. 21.4% for the ≤0.1% group, P=0.028). Median progression-free survival (PFS) was significantly longer for participants with a mutation abundance above 0.1% compared to those with a 0.01–0.1% abundance (log rank, P=0.0115). There was no significant association between plasma abundance of EGFR sensitizing mutations and tumor size, tumor shrinkage percentage, or concomitant TP53 mutations. Cox multivariate analysis demonstrated that plasma mutation abundance was an independent predictive factor for PFS [hazard ratio (HR) 2.41, 95% confidence interval (CI): 1.12–5.20; P=0.025]. We identified 11 participants with the acquired T790M resistance mutation according to serial dynamic plasma samples.
Conclusions
Liquid biopsy screening based on highly sensitive NGS is reliable for detecting drug resistance and actionable somatic mutations. The plasma abundance of the EGFR driver mutation affected clinical response to EGFR-TKIs in advanced NSCLC patients; prolongation of PFS was also observed in patients with an ultra-low abundance of EGFR sensitizing mutations.
Keywords: Epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs), mutation abundance, circulating tumor DNA, non-small cell lung cancer (NSCLC), acquired T790M resistance
Introduction
Lung cancer is a leading cause of cancer-related incidence and mortality worldwide (1). Activated epidermal growth factor receptor (EGFR) mutation is the main cause of non-small cell lung cancer (NSCLC) among those of Asian ethnicity (2). It has been shown that EGFR tyrosine kinase inhibitors (TKIs) significantly prolong the progression-free survival (PFS) of NSCLC patients with TKI-sensitive EGFR mutations compared with traditional platinum-based doublet chemotherapy (3-6). However, approximately 20–30% of NSCLC patients with sensitive EGFR mutations do not respond to EGFR-TKI therapy (primary resistance), or they develop early progressive disease (PD) after treatment initiation (3,4,7).
Previous studies have found that gene polymorphism, concurrent genomic mutations, or tumor size can affect EGFR-TKI efficacy (8-10). Zhou et al. first demonstrated that the abundance of EGFR mutations could predict the extent of benefit from EGFR-TKI treatment for advanced NSCLC by using qualitative approaches with different sensitivities (11). Their results revealed that patients with a low abundance of EGFR mutations benefit more from EGFR-TKI therapy compared to those with wild-type EGFR, while patients with a high abundance of EGFR mutations benefit more than those with a low abundance of EGFR mutations. Later, Li et al. quantitatively analyzed the abundance of EGFR mutations using an amplification-refractory mutation system (ARMS+) method and corroborated the effect of EGFR abundance (12). Meanwhile, Li et al. revealed that the PFS of patients with a low abundance of EGFR mutations was similar to that of patients with wild-type EGFR. Thus, the impact of EGFR mutation abundance on clinical outcomes of advanced NSCLC patients, especially those with a very low abundance of EGFR mutations (13,14), remains unclear.
Highly sensitive techniques like droplet digital polymerase chain reaction (ddPCR) (15,16), quantitative polymerase chain reaction (qPCR)-Invader (17), and allele-targeted next-generation sequencing (NGS) (18-22) offer alternative methods for plasma EGFR mutation analysis, which can lower the detection limit to below 0.1%. In particular, NGS-based circulating tumor DNA (ctDNA) sequencing can detect multiple mutations in parallel fashion and has been increasingly used in clinical practice (23). However, neoplastic tissue biopsy remains the gold standard for tumor genotyping in NSCLC patients. Whether or not naïve NSCLC patients with a low abundance EGFR mutation in plasma as detected by highly sensitive techniques should be treated with EGFR-TKIs still requires elucidation.
In this study, we used the circulating single-molecule amplification and resequencing technology (cSMART) method to prospectively evaluate plasma EGFR mutation status at baseline and track the dynamic EGFR change during TKI therapy. The cSMART assay, a highly-sensitive detection platform based on NGS, is suitable for the quantitation of the EGFR mutations (24-26). The primary aim of this study was to further explore the relationship between low abundance plasma EGFR mutations as determined by high-sensitivity methods and the therapeutic efficacy of first-generation EGFR-TKIs. The secondary objective was to explore the dynamic changes in mutation status during therapy in relation to clinical outcomes. This study is registered at ClinicalTrials.gov (NCT02980536) in 2016.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7155).
Methods
Participants and clinical samples
Newly diagnosed patients with advanced stage lung adenocarcinoma at 9 participating hospitals in Inner Mongolia, China from November 2016 to December 2018 were recruited into this observational study. All participants were diagnosed pathologically with lung adenocarcinoma according to World Health Organization (WHO) criteria (2015). Clinical disease staging was determined using the tumor, node, metastasis (TNM) staging system of the International Association for the Study of Lung Cancer (version 7). The key inclusion criteria were as follows: initial, nonoperative NSCLC adenocarcinoma patients; stage III and IV; over 18 years old; Eastern Cooperative Oncology Group (ECOG) performance status score (PS) less than 2; clear measurable tumor lesion in the lung according to the Response Evaluation Criteria in Solid Tumors (RECIST, version 1.1) (27). Meanwhile, the exclusion criteria were as follows: major organ dysfunction and severe cardiopathy; brain metastasis–related syndrome; bone metastasis–related complications; allergy to TKIs; already received radiotherapy at the site of curative effect observation; receiving allogenic blood transfusion within 14 days of recruitment.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by institutional ethics committee of Baotou Cancer Hospital, Inner Mongolia, China. Written informed consent was taken from all individual participants included in the study. Participant information on sex, age, smoking history, EGFR mutational sites and mutation abundance, and treatment with first-generation first-line EGFR-TKIs or chemotherapy was collected.
Tissue and plasma mutation assays
Mutation detections in tissue and plasma were determined by the gold standard ARMS-PCR assay and cSMART assay, respectively. Tumor tissue specimens at baseline were prepared as formalin-fixed, paraffin-embedded (FFPE) sections and DNA was extracted with QIAamp DNA FFPE kit (Qiagen, Hilden, Germany). The DNA from FFPE tissue was analyzed for EGFR mutations in exon 18 to 21 by ARMS method using the commercially available AmoyDx kit (Amoy Diagnostics Co., Ltd., Xiamen, Fujian, China). The plasma DNA was extracted according to the manufacturer’s protocol formulated for MagMAX™ Cell-free DNA Isolation Kit (Applied Biosystems, Foster City, CA, USA). The cSMART assay, designed specifically for detection of hot spot oncogenic mutations in plasma, was performed as previously described (24-26). The known mutations specifically targeted in this study were EGFR mutations, including G719X (3 variants), exon 19 deletions (16 variants), exon 20 insertions (3 variants), S768I, T790M, C797S, and exon 21 point mutations, L858R and L861Q; Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations, including G12X (6 variants), G13D, Q61X (6 variants), and A146X (3 variants); ERBB2 mutations, including exon 20 insertions (5 variants); BRAF mutations, including V600X (4 variants); PIK3CA mutations, including R88Q, E542K, E545K/D, and A1047R/L; TP53 mutations, including R175C/H, R248W/Q, and R273C/H; and MET exon 14 skipping. Apart from EGFR mutations, mutations of KRAS, ERBB2, BRAF, TP53, PIK3CA, ROS1, cMET, and ALK genes were specifically targeted in the panel of the present study (28). The cSMART assay was also designed for detection of novel mutations within the nearby region picked up by the primers. The mutation abundance was defined as mutant allele read numbers over total allele read numbers.
Treatment and evaluation of objective response rate (ORR) and PFS
At baseline, all participants underwent physical examination, laboratory tests, and computed tomography (CT) scans of the chest and abdomen 1 week before treatment. Brain magnetic resonance imaging (MRI) was required only in participants with known or suspected brain metastasis.
Participants harboring sensitizing mutations detected by ARMS or cSMART at baseline received gefitinib (250 mg QD), icotinib (125 mg TID) or afatinib (40 mg BID) as first-line therapy throughout the course of the disease until disease progression, intolerable toxicity, participant refusal, or death. Dynamic changes of mutations in peripheral blood were detected every 8–12 weeks by cSMART assay. The participants without EGFR and other targeted gene mutations received a platinum-based first-line combined chemotherapy regimen, monotherapy S-1 (tegafur, gimeracil, or oteracil potassium) (40 mg BID), or combined therapy with bevacizumab monoclonal antibody according to guidelines. Those with ALK mutations were treated with crizotinib (250 mg BID).
The objective tumor response was evaluated every 8–12 weeks during treatment by investigators according to the stipulations of RECIST (version 1.1) (27). Tumor shrinkage was expressed as a relative change of the sum of the longest diameters of the target lesions based on computed tomography (CT) examination. Nontarget lesions and newly occurring lesions were not considered in the measurement of change in tumor size. The therapeutic effect of EGFR-TKI was evaluated as complete response (CR), disappearance of all target lesions; partial response (PR), with at least a 30% decrease in the sum of the longest diameter of target lesions with the baseline sum of the longest diameter as the reference; PD, with an increase of at least 20% from the baseline sum of the longest diameter or the presence of 1 or more new lesions; stable disease (SD), with less than 20% increase or less than 30% decrease in the size of target tumor lesions. The ORR was defined as achieving either CR or PR. The disease control rate (DCR) included CR, PR, and SD. The primary endpoint was PFS, which was defined as the time from initiation of treatment to disease progression according to RECIST criteria or death resulting from any cause, whichever occurred first.
Statistical analyses
The PFS was analyzed by Kaplan-Meier plots and the log-rank test was used to calculate the significance of the difference between groups. Participants with no disease progression or those who were lost to follow-up were censored at the last date of disease assessment for PFS or the last follow-up time. Multivariate Cox proportional hazards regression models were used to evaluate the association between independent prognosis factors (age, sex, EGFR mutation abundance etc.) and PFS. A receiver operating characteristic (ROC) curve analysis was used to determine the optimal cutoff value of mutation abundance, and the area under the curve (AUC) was calculated.
The ORR, DCR, and relationship between EGFR mutation abundance and clinical characteristics were calculated by Pearson’s chi-square test or Fisher’s exact test. Continuous variables were compared using the Mann-Whitney U test.
Data were analyzed using SPSS version 17.0 (IBM Corp., Armonk, NY, USA) or GraphPad Prism 8.1 (GraphPad Software, San Diego, CA, USA). The survival curve was drawn using GraphPad Prism version 8.1. A two-sided P value <0.05 was considered statistically significant. The last follow-up date was January 6, 2019.
Results
Patients characteristics and EGFR mutation status
The outline of the study is diagrammed in Figure 1. A total of 135 newly diagnosed advanced NSCLC patients were recruited as the study cohort. Tumor specimens of all but 1 participant were collected at baseline via tissue biopsy and detected by ARMS method. Meanwhile, 135 plasma samples were collected and analyzed with cSMART array. Overall, there were 134 patients who provided both tumor tissue and matched blood samples at baseline. The 1 participant without a tissue specimen collected had the EGFR exon 19 deletion (19Del) mutation in plasma. Of the 135 participants, 58 (43%) were female, and 66 (49%) were over 65 years of age. Totals of 56 (41.8%) and 66 (49.3%) participants were confirmed to harbor EGFR mutation in tumor tissues and plasma, respectively. A total of 6 participants were EGFR mutation positive in tissue but negative in their matched plasma (ARMS+/cSMART−), whereas 16 cases showed the opposite (ARMS−/cSMART+). In total, 50 participants were EGFR mutation positive in both tissue and plasma (ARMS+/cSMART+), 22 were EGFR mutation positive in either tissue or plasma (ARMS+/cSMART−, ARMS−/cSMART+), and 62 were wild-type EGFR (ARMS−/cSMART−). The overall, positive, and negative concordance rate of EGFR mutation status between tissue and plasma at baseline was 83.6% (112/134), 89.3% (50/56), and 79.5% (62/78), respectively. Among the 73 participants with positive EGFR mutations, 12 did not receive first-generation EGFR-TKI therapy, and another 7 either harbored a resistance mutation or were EGFR negative in plasma. Eventually, 54 participants who had EGFR sensitizing mutations and received first-generation EGFR-TKI therapy as first-line treatment were further assessed for prognosis and clinical outcomes. The demographics and clinical characteristics of the 54 EGFR mutation-positive participants are summarized in Table 1. Briefly, 35.2% of these participants were over 65 years old, 53.7% were female, 77.8% had an ECOG PS 0 or 1, 63.0% were T1–T2 stage, 53.7% were N0–N1, 20.4% were former or current smokers, and 37.0% carried an 19Del mutation.
Figure 1.
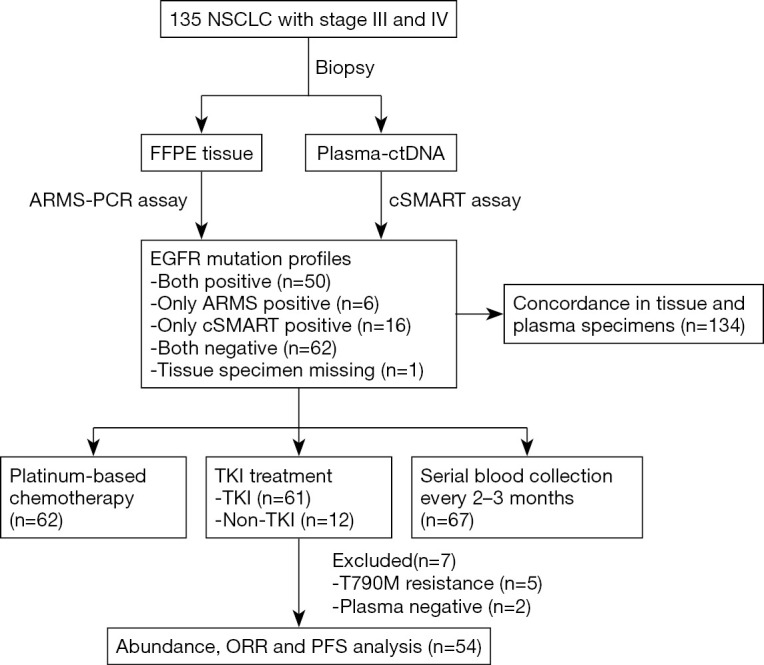
Patient enrolment and study overview. ORR, objective response rate; PFS, progression-free survival.
Table 1. Baseline demographic and clinical characteristics.
| Characteristics | Patients (n=54) | Percentage |
|---|---|---|
| Age in years | ||
| ≤65 | 35 | 64.8 |
| >65 | 19 | 35.2 |
| Gender | ||
| Male | 25 | 46.3 |
| Female | 29 | 53.7 |
| T stage | ||
| T1-2 | 34 | 63.0 |
| T3-4 | 20 | 37.0 |
| N stage | ||
| N0-1 | 29 | 53.7 |
| N2-3 | 25 | 46.3 |
| ECOG PS | ||
| 0 or 1 | 42 | 77.8 |
| >1 | 12 | 22.2 |
| EGFR mutations | ||
| Exon 19 deletion | 20 | 37.0 |
| Exon 21 L858R | 18 | 33.4 |
| Uncommon mutations | 16 | 29.6 |
| Smoking status | ||
| Never-smoker | 43 | 79.6 |
| Smoker | 11 | 20.4 |
The most prevalent EGFR mutations were exon19 non-frameshift deletion (24/72, 33.3% in tissue; 22/73, 31.5% in plasma) and EGFR L858R mutation (23/72, 31.9% in tissue; 20/73, 27.4% in plasma). The less prevalent mutations were EGFR G719X variants (3 tissue and 16 blood samples), S768I mutation [6, 6], L861Q mutation [2, 3], exon 20 insertion [1, 4], and resistance mutation T790M [1, 6]. Complex EGFR mutations were found in 4 tissue and 13 blood samples. The overall concordance rate between tissue and plasma was very high for 19Dels (97.0%, 130/134), L858R (96.3%,129/134), and S768I (100.0%, 6/6), but quite low for uncommon mutations, including G719X (18.8%, 3/16), T790M (16.7%, 1/6), and exon 20 insertions (0/5).
Among the 9 genes assayed in plasma by cSMART panel, the most common mutated genes in the EGFR mutation–positive cohort were TP53 (47.9%, 35/73), followed by KRAS (11.0%, 8/73), ALK (11.0%, 8/73), and PIK3CA (9.6%, 7/73). The comutation of ERBB2 or BRAF with EGFR was each found only once (1.4%).
Impact of plasma abundance of EGFR sensitizing mutations and response to EGFR-TKIs
Of the participants with EGFR sensitizing mutations in plasma, 27 (50%) had PR to TKIs, 20 (37.4%) had SD and 7 (12.6%) exhibited PD. The overall ORR and DCR were 50.0% and 87.0%, respectively (Table 2).
Table 2. EGFR mutation abundance and tumor response.
| EGFR abundance | PR (%) | SD (%) | PD (%) | ORR (%) | P value | DCR (%) | P value |
|---|---|---|---|---|---|---|---|
| ≤0.0622% | 3 | 5 | 2 | 30.00 | 0.293 | 80.00 | 0.601 |
| >0.0622% | 24 | 15 | 5 | 54.55 | 88.64 | ||
| ≤0.1% | 3 | 9 | 2 | 21.43 | 0.028 | 85.71 | >0.999 |
| >0.1% | 24 | 11 | 5 | 60.00 | 87.50 | ||
| ≤1.14% | 10 | 13 | 3 | 38.46 | 0.173 | 88.46 | >0.999 |
| >1.14% | 17 | 7 | 4 | 60.71 | 85.71 | ||
| ≤3.36% | 18 | 17 | 4 | 48.72 | 0.544 | 89.74 | 0.382 |
| >3.36% | 9 | 3 | 3 | 58.33 | 80.00 | ||
| Overall | 27 | 20 | 7 | 50.0 | 87.0 |
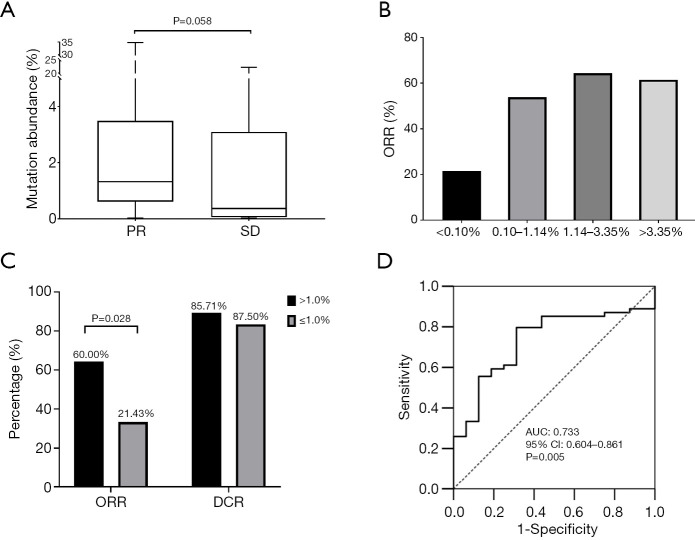
The abundance of EGFR sensitizing mutations in plasma varied from 0.02% to 47.76% with a median of 1.14%, as revealed by cSMART assay at baseline. The median abundance for participants with PR was higher than those with SD after EGRF-TKI treatment (1.32% vs. 0.36%, P=0.058) (Figure 2A), suggesting that the abundance of EGFR activating mutations might be associated with the objective response to EGFR-TKIs. Therefore, we used 25th percentile (0.10%), the median (1.14%), and 75th percentile (3.36%) of mutation abundance as the cutoff values to compare the response rates of the high and low abundance groups (Figure 2B). The difference in DCR between the high and low abundance groups was not significant with any of the cutoff values, but the difference in ORR between the high and low abundance groups was significant when the cutoff value was at 0.1%, (60.00% vs. 21.43%, P=0.028) (Figure 2C; Table 2). Furthermore, we used ROC analysis to estimate the cutoff values of high and low abundance EGFR mutations according to objective response, yielding an AUC of 0.733 [95% confidence interval (CI): 0.604–0.861, P=0.005] (Figure 2D). The responding cutoff value of mutation abundance was 0.062%. However, no significant difference in ORR or DCR was found at this cutoff (Table 2). Finally, 0.10% was used as the lowest threshold of EGFR-mutant abundance in peripheral blood for efficacy evaluation.
Figure 2.
EGFR mutation abundance and clinical response. (A) Box plot analysis for mutation abundance in patients with PR or SD. (B) ORR stratified by quantiles of EGFR sensitizing mutation abundance in plasma. (C) ORR and DCR of low and high mutation abundance groups. (D) ROC curve and AUC. EGFR, epidermal growth factor receptor; PR, partial response; SD, stable disease; DCR, disease control rate; ORR, objective response rate; ROC, receiver operating characteristic; AUC, area under ROC curve.
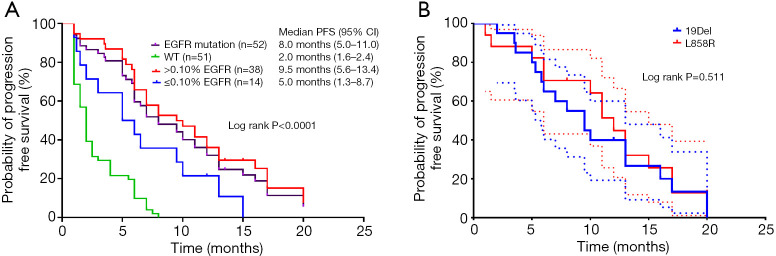
The participants were subdivided into two groups based on the 0.10% abundance value in TKI-naïve plasma samples (high: >0.10%; low: ≤0.10%). There was a significant difference in the PFS duration across the high abundance mutation, low abundance mutation, and wild-type EGFR groups (log rank, P<0.0001) (Figure 3A). The median PFS after EGFR-TKI treatment in the EGFR sensitizing mutation group (8.0 months, 95% CI: 5.0–11.0) was superior to that in the wild-type EGFR (2.0 months, 95% CI: 1.6–2.4; log rank P<0.0001) (Figure 3A). The median PFS for participants with an EGFR mutation abundance above the cutoff (9.5 months, 95% CI: 5.6–13.4) was markedly longer than for those with a low abundance of EGFR mutation (5.0 months, 95% CI: 1.3–8.7) (log rank P=0.0115). The PFS of participants with a low abundance of EGFR sensitizing mutations was significantly longer than that of those in the EGFR wild-type group (log rank P=0.0054). These results showed that participants with a high abundance of EGFR mutations in plasma demonstrated a better response to TKIs and longer PFS than those with wild-type EGFR or a low abundance of EGFR mutations. However, participants with a low abundance of EGFR mutations could also gain some benefits from TKI treatment compared to those with wild-type EGFR in terms of PFS. No significant difference in PFS was found for participants with 19Del or L858R mutations (Figure 3B).
Figure 3.
Kaplan-Meier analysis of PFS in patients with (A) high and low mutation abundance or (B) 19Del or L858R mutation (95% CI). The dotted line indicates 95% CI. 19Del, exon 19 deletion; PFS, progression-free survival; CI, confidence interval.
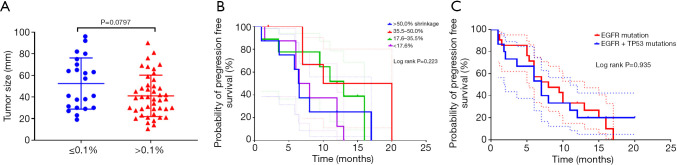
Tumor size change and concomitant mutations were also considered to be related to responsiveness. In order to assess the impact of tumor change and concomitant mutations, we further investigated the relationship between the abundance of EGFR sensitizing mutations with baseline tumor size, maximal tumor shrinkage after treatment, and concomitant TP53 gene mutations. The participants with lower EGFR mutation abundance had larger tumor sizes than those with higher EGFR mutation abundance [mean ± standard deviation (SD) 52.4±23.7 vs. 41.0±19.2 mm)]. However, the difference was not significant (P=0.0797) (Figure 4A). The median, 25th, and 75th percentile of the maximal tumor shrinkage (nadir size compared to baseline) was 35.5%, 17.6%, and 50.0%, respectively. Responders were further divided into four groups according to the degree of maximal tumor shrinkage in response: >50%, 35.5–50%, 17.6–35.5%, and <17.6%. No significant association was observed between tumor shrinkage and median PFS (log rank P=0.223) (Figure 4B), suggesting that tumor size change was not predictive of PFS in responders. In addition, there was no significant difference in PFS for participants with or without a concomitant TP53 mutation (log rank P=0.935) (Figure 4C).
Figure 4.
Association between EGFR mutation abundance, tumor size (A), tumor shrinkage (B), and TP53 concomitant mutation (C). The dotted line indicates 95% CI. CI, confidence interval; EGFR, epidermal growth factor receptor.
Univariate and multivariate analysis of PFS
The results in univariate analysis showed that EGFR mutation abundance >0.1% and ECOG PS 0 or 1 were significantly associated with better PFS. In the multivariate Cox proportional hazards regression model, mutation abundance, sex, smoking status, TNM stage, and ECOG PS were used as covariates. Multivariate analysis results demonstrated that mutation abundance [hazard ratio (HR) 2.41, 95% CI: 1.12–7.375.20; P=0.025] and ECOG PS (HR 1.83, 95% CI: 1.19–2.82; P=0.006) were independent predictive factors for PFS (Table 3).
Table 3. Univariate and multivariate Cox regression analyses of PFS for patients.
| Variable | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Abundance: ≤0.1% vs. >0.1% | 2.21 (0.96–4.96) | 0.011 | 2.41 (1.12–5.20) | 0.025 | |
| ECOG PS: >1 vs. 0 or 1 | 2.29 (0.86–6.12) | 0.029 | 1.83 (1.19–2.82) | 0.006 | |
| Age: ≤65 vs. >65 years | 0.83 (0.42–1.53) | 0.476 | |||
| Female vs. male | 0.84 (0.45–1.56) | 0.561 | |||
| T3+4 vs. T1+2 stage | 1.12 (0.57–2.16) | 0.728 | |||
| N2+3 vs. N0+1 stage | 0.97 (0.51–1.88) | 0.725 | |||
| Smokers vs. non-smokers | 1.31 (0.59–2.94) | 0.446 |
Dynamic monitoring for resistance mutation
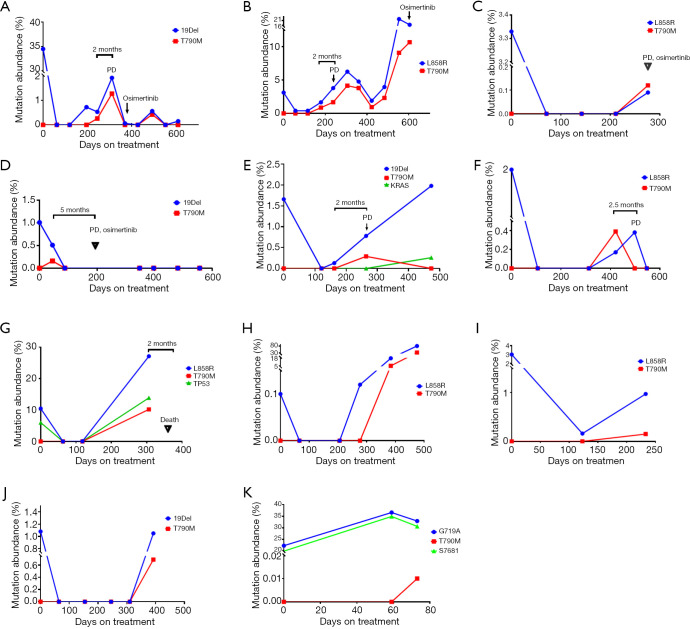
To identify the development of resistance mutation during TKI treatment, serial blood specimens (222 samples) in 67 patients with mutant EGFR were assayed every 8–12 weeks (Figure 5). At baseline, T790M resistance mutation was found in 6 cases (Table S1). Of them, only 1 participant (no. NM01ZMD) was identified as harboring concurrent 19Del and T790M resistance mutation in both tissue and plasma. A single participant had a L858R mutation in tissue but a T790M mutation in plasma (abundance: 0.06%). Two participants with EGFR mutation–negative tissue were found to carry T790M in plasma (abundance: 0.02%). Both T790M mutation and EGFR sensitizing mutation in plasma were found in 2 participants but the mutation abundance of T790M was very low (0.04%, 0.07%). Three participants with de novo T790M mutation received EGFR-TKI therapy, and T790M mutation was cleared from plasma during treatment. However, recurrent T790M was observed in 1 participant (no. NM01ZSH) for a duration of 12 months.
Figure 5.
Dynamic monitoring of the T790M resistance mutation during treatment. (A-K) The abundance changes of EGFR mutations for 11 patients. EGFR, epidermal growth factor receptor.
Acquired T790M resistance mutations were detected in 11 participants. The median T790M mutation abundance was 0.34% in the range of 0.01–10.2%. Of these 11 participants, 5 achieved PR and 6 achieved SD in response to TKIs (Table S2). In 2 of these participants with acquired T790M resistance mutation, T790M mutation was unexpectedly detected at the first molecular testing after initiating therapy (abundance 0.16% and 0.01%, respectively; Figure 5D,K). The 2 participants were considered as early resistant (1.5 and 2.0 months, respectively). The period of the presence of T790M mutation in the remaining 9 participants was 4–14 months with a mean of 10 months.
With the exception of 1 participant (Figure 5K), EGFR sensitizing mutation abundance in plasma decreased in response to first-generation EGFR-TKIs; 8 participants exhibited a complete plasmatic response (Figure 5A,C,D,E,F,G,H,J). Subsequently, plasmatic EGFR sensitizing mutations reemerged and mutation abundance increased. Plasmatic T790M was identified at a somewhat lower abundance than the EGFR sensitizing mutation. The T790M in plasma was detectable 2–5 months before imaging progression by RECIST criteria. A total of 4 participants were treated with the third-generation EGFR-TKI, osimertinib, after PD (Figure 5 A,B,C,D).
Discussion
In the present study, we investigated the association of EGFR-activating mutation abundance in plasma at baseline and the therapeutic outcomes of first-generation EGFR-TKI treatment in advanced NSCLC. In addition, dynamic changes of EGFR mutations from ctDNA during treatment were analyzed to monitor the emergence of drug resistance mutations and predict the clinical efficacy of EGFR-TKIs. We found that not only those participants with highly abundant EGFR mutations in pretreatment blood samples have longer PFS on EGFR-TKIs, but that even those with a very low abundance of EGFR sensitizing mutations could benefit from EGFR-TKI targeted therapy.
Liquid biopsy based on ctDNA is considered complementary to detection of EGFR mutations in tissues, as it has the potential to survey the whole tissue (5,28-31). In this study, we used cSMART assay with high sensitivity to detect EGFR mutations and abundance change in plasma. Our results showed that EGFR sensitizing mutation abundance in TKI-naïve plasma at baseline was associated with response to EGFR-TKIs regardless of mutation pattern, which was in accordance with prior conclusions (11,12). Importantly, we found that the participants with even a very low abundance (≤0.1%) in plasma had longer median PFS than those with wild-type EGFR (5.0 vs. 2.0 months). Moreover, the extremely low abundance of EGFR mutations was mostly detected in participants who had EGFR mutations only in ctDNA. A total of 16 cases in this investigation were found to harbor positive EGFR mutations in blood samples but were negative in the matched tissues. The EGFR mutation abundance was less than 0.1% in 11 cases (68.8%). Previous studies have suggested that the status or abundance of EGFR mutations in the tissues and/or ctDNA may have different predictive values with regard to response to EGFR-TKIs in different therapeutic backgrounds (29,31,32). Therefore, we also assessed EGFR-TKI efficacy in various EGFR mutation statuses in tissue and blood samples. The results showed that the median PFS for ARMS+/cSMART+ participants were significantly longer than that of those with ARMS-/cSMART+ (data not shown), which was consistent with another study that used a highly sensitive ddPCR method to assess EGFR sensitizing mutation abundance in plasma (31). Undoubtedly, as shown in these studies, participants with EGFR sensitizing mutations detected in both tissue and ctDNA (ARMS+/cSMART+) are indicated as bearing a high load of EGFR mutations, resulting in a better clinical outcome. Our results demonstrated that NSCLC patients with a low abundance EGFR sensitizing mutation, particularly those who were initially mutation negative by tumor biopsy, could also receive clinical benefit from EGFR-TKIs. Recently, Yan et al. demonstrated that, for patients with a low abundance of EGFR mutations, the median PFS was significantly longer in a combination group of EGFR-TKI plus chemotherapy than in an EGFR-TKI monotherapy group (median PFS: 7.9 vs. 5.9 months) (33). Thus, patients with a low abundance of EGFR mutations might achieve better clinical outcomes from EGFR-TKI treatment combined with chemotherapy.
It has been speculated based on comparative studies that the difference in abundance of EGFR mutations may be attributed to technical sensitivity (31) or tumor heterogeneity and clonal evolution (34). In terms of technology, cSMART assay has been shown to be highly sensitive and reliable for measuring levels of EGFR variants across a wide dynamic range, with the lowest mutation abundance reaching 0.01% (24-26,28). Due to the heterogeneity of tumor tissue, the collected biopsy specimen is not always representative of the whole tumor tissue. Heterogeneity may result in missed detection in some tumor specimens, which can be supplemented by ctDNA from a different region of the tumor (31,34). It is likely that ctDNA is derived from apoptotic or necrotic tumor cells. Therefore, the mutation abundance reflects the proportion of mutated cells in tumor tissue. Although large tumors might theoretically be more heterogeneous than smaller ones because of poor blood supply and hypoxia, we did not find a significant difference in tumor size for participants with high or low mutation abundance. Also, no association between the extent of target lesion shrinking to EGFR-TKI and PFS was found, which further supports previous results (35-37).
In order to exclude the influence of comutated genes, we further confirmed the impact of concomitant TP53 mutations on EGFR-TKI efficacy. No significant differences were observed in terms of response and PFS in participants with or without concomitant TP53 mutations, regardless of mutation status of KRAS or other actionable genes. The results were not in agreement with a recent study in which TP53 mutations reduced responsiveness to TKIs and worsened prognosis in EGFR-mutated NSCLC patients (10).
Acquired resistance is an inevitable process during therapy with EGFR-TKIs, usually developing after a median treatment period of 10–12 months (38). Mutation of T790M is the major resistance mechanism to EGFR-TKIs with a prevalence of 49–63% in the most frequent resistance-associated molecular alterations (38,39). Reports on the frequency of T790M in FFPE specimens for pretreatment patients have varied from 2–40% depending on the type and sensitivity of the detection method employed (40-42). In a study using a highly sensitive assay with a validated sensitivity of 0.1%, T790M mutation was detected in frozen tumor tissue at a rate of 2.8% in TKI-naïve NSCLC patients with EGFR sensitizing mutations (43). In our study, the frequency of the primary T790M mutation was found to be 1.8% for tissue (1/56) and 9.1% for blood samples (6/66), which was consistent with previously published results (40-44). Of note, the four T790M-positive samples missed by ARMS-PCR in tissue were found by cSMART assay to have very low T790M levels of 0.02–0.07%.
Wang et al. found that patients with pre-TKI plasma samples positive for T790M had significantly inferior PFS compared with pre-TKI–negative patients (45). We did not observe this phenomenon due to the limited samples with de novo T790M mutation. However, we observed that the abundance of EGFR sensitizing mutations decreased (or reached mutation clearance) in plasma after the initiation of EGFR-TKIs and subsequently started to increase along with the appearance of T790M. Meanwhile, T790M in plasma was detectable before clinical PD and a third-generation EGFR-TKI, osimertinib, was subsequently used to treat participants with acquired T790M mutation. Our findings and previous reports (16,22) were consistent with the hypothesis of the selection of resistant neoplastic clones operated by EGFR-TKI, that grow until becoming clinically relevant. In clinical practice, the abundance of both EGFR sensitizing mutations and T790M could vary under the selection pressure of treatment. Therefore, noninvasive dynamic monitoring for the abundance of EGFR sensitizing mutations and T790M is necessary, and potentially can be used to guide subsequent treatment by allowing early cessation of ineffective TKIs and commencement of a different TKI drug that counters the resistance mechanism.
There were several limitations in this study. First, the number of EGFR mutation–positive participants was limited. The different mutation types, including uncommon EGFR mutations, were analyzed together as EGFR mutation positive due to the small sample size. For these reasons, the comparison between subgroups should be interpreted with caution. Second, repeat biopsies were not performed to obtain matched tissue samples with blood after disease progression to confirm the presence of T790M. Third, the follow-up time of some participants should be extended. Finally, the cutoff value to distinguish between participants with a high and low abundance of EGFR sensitizing mutations was based on the detection technique in this study. Therefore, larger cohorts are needed to validate the impact of plasma mutation abundance in predicting treatment responsiveness.
Collectively, highly abundant EGFR mutations in pretreatment blood samples predicted longer PFS on EGFR-TKIs. The participants with a low abundance of EGFR sensitizing mutations quantified by a highly sensitive detection method could benefit from EGFR-TKI targeted therapy. Furthermore, noninvasive, dynamic assessment of plasmatic EGFR mutation using a highly sensitive detection method could monitor changes of EGFR sensitizing mutation abundance and T790M resistance mutation before clinical resistance, and may potentially be used to guide subsequent treatment.
Supplementary
The article’s supplementary files as
Acknowledgments
We are grateful to all the patients who participated in the study. The abstract of this study was presented as a poster at the European Society for Medical Oncology (ESMO) Annual Meeting 2019 in Barcelona, Spain.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by institutional ethics committee of Baotou Cancer Hospital, Inner Mongolia, China. Written informed consent was taken from all individual participants included in the study.
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7155
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7155
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7155). Dr. HZ and Dr. X Su reports other from Berry Oncology Corporation, outside the submitted work. The other authors have no conflicts of interest to declare.
(English Language Editors: J. Jones and J. Gray)
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Midha A, Dearden S, McCormack R. EGFR mutation incidence in nonsmall-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res 2015;5:2892-911. [PMC free article] [PubMed] [Google Scholar]
- 3.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 4.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 5.Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non–small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. 10.1200/JCO.2010.33.4235 [DOI] [PubMed] [Google Scholar]
- 6.Zhou C, Wu YL, Chen G, et al. Final overall survival results from a randomised, phase III study of erlotinib versus chemotherapy as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer (OPTIMAL, CTONG-0802). Ann Oncol 2015;26:1877-83. 10.1093/annonc/mdv276 [DOI] [PubMed] [Google Scholar]
- 7.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 8.Morgensztern D, Waqar S, Subramanian J, et al. Prognostic significance of tumor size in patients with stage III non-small-cell lung cancer A Surveillance, Epidemiology, and End Results (SEER) survey from 1998 to 2003. J Thorac Oncol 2012;7:1479-84. 10.1097/JTO.0b013e318267d032 [DOI] [PubMed] [Google Scholar]
- 9.Zhao M, Zhang Y, Cai W, et al. The Bim deletion polymorphism clinical profile and its relation with tyrosine kinase inhibitor resistance in Chinese patients with non-small cell lung cancer. Cancer 2014;120:2299-307. 10.1002/cncr.28725 [DOI] [PubMed] [Google Scholar]
- 10.Canale M, Petracci E, Delmonte A, et al. Impact of TP53 mutations on outcome in EGFR-mutated patients treated with first-line tyrosine kinase inhibitors. Clin Cancer Res 2017;23:2195-202. 10.1158/1078-0432.CCR-16-0966 [DOI] [PubMed] [Google Scholar]
- 11.Zhou Q, Zhang XC, Chen ZH, et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J Clin Oncol 2011;29:3316-21. 10.1200/JCO.2010.33.3757 [DOI] [PubMed] [Google Scholar]
- 12.Li X, Cai W, Yang G, et al. Comprehensive analysis of EGFR-mutant abundance and its effect on efficacy of EGFR TKIs in advanced NSCLC with EGFR mutations. J Thorac Oncol 2017;12:1388-97. 10.1016/j.jtho.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 13.Zhao ZR, Wang JF, Lin YB, et al. Mutation abundance affects the efficacy of EGFR tyrosine kinase inhibitor readministration in non-small-cell lung cancer with acquired resistance. Med Oncol 2014;31:810. 10.1007/s12032-013-0810-6 [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Zhang M, Tang WY, et al. Mutation abundance affects the therapeutic efficacy of EGFR-TKI in patients with advanced lung adenocarcinoma: A retrospective analysis. Cancer Biol Ther 2018;19:687-94. 10.1080/15384047.2018.1450115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X, Zhuo ML, Ye X, et al. Quantification of mutant alleles in circulating tumor DNA can predict survival in lung cancer. Oncotarget 2016;7:20810-24. 10.18632/oncotarget.8021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014;20:1698-705. 10.1158/1078-0432.CCR-13-2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang Z, Wan RX, Zou BJ, et al. Highly sensitive and specific real-time PCR by employing serial invasive reaction as a sequence identifier for quantifying EGFR mutation abundance in cfDNA. Anal Bioanal Chem 2018;410:6751-9. 10.1007/s00216-018-1316-z [DOI] [PubMed] [Google Scholar]
- 18.Bordi P, Del Re M, Danesi R, et al. Circulating DNA in diagnosis and monitoring EGFR gene mutations in advanced non-small cell lung cancer. Transl Lung Cancer Res 2015;4:584-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couraud S, Vaca-Paniagua F, Villar S, et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res 2014;20:4613-24. 10.1158/1078-0432.CCR-13-3063 [DOI] [PubMed] [Google Scholar]
- 20.Cabanero M, Tsao MS. Circulating tumour DNA in EGFR-mutant non-small-cell lung cancer. Curr Oncol 2018;25:S38-S44. 10.3747/co.25.3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman AM, Bratman S, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchetti A, Palma JF, Felicioni L, et al. Early prediction of response to tyrosine kinase inhibitors by quantification of EGFR mutations in plasma of NSCLC patients. J Thorac Oncol 2015;10:1437-43. 10.1097/JTO.0000000000000643 [DOI] [PubMed] [Google Scholar]
- 23.Murtaza M, Dawson SJ, Tsui DWY, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. 10.1038/nature12065 [DOI] [PubMed] [Google Scholar]
- 24.Lv W, Wei X, Guo R, et al. Noninvasive prenatal testing for Wilson disease by use of circulating single-molecule amplification and resequencing technology (cSMART). Clin Chem 2015;61:172-81. 10.1373/clinchem.2014.229328 [DOI] [PubMed] [Google Scholar]
- 25.Chai X, Ren P, Wei B, et al. A comparative study of EGFR oncogenic mutations in matching tissue and plasma samples from patients with advanced non-small cell lung carcinoma. Clin Chim Acta 2016;457:106-11. 10.1016/j.cca.2016.04.003 [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Cheng G, Han XH, et al. Application of single-molecule amplification and resequencing technology for broad surveillance of plasma mutations in patients with advanced lung adenocarcinoma. J Mol Diagn 2017;19:169-81. 10.1016/j.jmoldx.2016.09.008 [DOI] [PubMed] [Google Scholar]
- 27.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 28.Chen K, Zhao H, Shi Y, et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (DYNAMIC). Clin Cancer Res 2019;25:7058-67. 10.1158/1078-0432.CCR-19-1213 [DOI] [PubMed] [Google Scholar]
- 29.Bai H, Mao L, Wang H, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol 2009;27:2653-9. 10.1200/JCO.2008.17.3930 [DOI] [PubMed] [Google Scholar]
- 30.Mok T, Wu YL, Lee J, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res 2015;21:3196-203. 10.1158/1078-0432.CCR-14-2594 [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Duan J, Chen H, et al. Analysis of EGFR mutation status in tissue and plasma for predicting response to EGFR-TKIs in advanced non-small-cell lung cancer. Oncol Lett 2017;13:2425-31. 10.3892/ol.2017.5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 2007;97:778-84. 10.1038/sj.bjc.6603949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan X, Wang H, Li P, et al. Efficacy of first-line treatment with epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) alone or in combination with chemotherapy for advanced non-small cell lung cancer (NSCLC) with low-abundance mutation. Lung Cancer 2019;128:6-12. 10.1016/j.lungcan.2018.12.007 [DOI] [PubMed] [Google Scholar]
- 34.Bai H, Wang ZJ, Wang YY, et al. Detection and clinical significance of intratumoral EGFR mutational heterogeneity in Chinese patients with advanced non-small cell lung cancer. PLoS One 2013;8:e54170. 10.1371/journal.pone.0054170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu TH, Hsiue EHC, Lee JH, et al. Best response according to RECIST during first-line EGFR-TKI treatment predicts survival in EGFR mutation-positive non-small-cell lung cancer patients. Clin Lung Cancer 2018;19:e361-72. 10.1016/j.cllc.2018.01.005 [DOI] [PubMed] [Google Scholar]
- 36.Pan Y, Gao G, Chen X, et al. Larger tumors are associated with inferior progression-free survival of first-line EGFR-tyrosine kinase inhibitors and a lower abundance of EGFR mutation in patients with advanced non-small cell lung cancer. Thorac Cancer 2019;10:686-94. 10.1111/1759-7714.12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee CK, Lord S, Marschner I, et al. The value of early depth of response in predicting long-term outcome in EGFR-mutant lung cancer. J Thorac Oncol 2018;13:792-800. 10.1016/j.jtho.2018.03.010 [DOI] [PubMed] [Google Scholar]
- 38.Sequist LV, Waltman A, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. 10.1126/scitranslmed.3002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosaka T, Yatabe Y, Endoh H, et al. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 2004;64:8919-23. 10.1158/0008-5472.CAN-04-2818 [DOI] [PubMed] [Google Scholar]
- 41.Rosell R, Molina MA, Costa C, et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res 2011;17:1160-8. 10.1158/1078-0432.CCR-10-2158 [DOI] [PubMed] [Google Scholar]
- 42.Su KY, Chen HY, Li KC, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol 2012;30:433-40. 10.1200/JCO.2011.38.3224 [DOI] [PubMed] [Google Scholar]
- 43.Ye X, Zhu ZZ, Zhong L, et al. High T790M detection rate in TKI-naive NSCLC with EGFR sensitive mutation: truth or artifact? J Thorac Oncol 2013;8:1118-20. 10.1097/JTO.0b013e31829f691f [DOI] [PubMed] [Google Scholar]
- 44.Zhang S, Xia B, Jiang H, et al. Comprehensive profiling and quantitation of oncogenic mutations in non small-cell lung carcinoma using single molecule amplification and re-sequencing technology. Oncotarget 2016;7:50477-89. 10.18632/oncotarget.10464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Chen R, Wang SH, et al. Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS One 2014;9:e110780. 10.1371/journal.pone.0110780 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as