Abstract
Background
Cancer stem cells (CSCs) give rise to a diverse variety of differentiated cells, which comprise the bulk of the tumor microenvironment (TME). However, the exact multi-directional differentiation potential of CSCs has not been fully clarified. This study was designed to explore whether CSCs differentiate into cellular components of the TME to promote the growth of lung carcinoma.
Methods
The present of CD133+, CD2+, and CD133+CD2+ cells in both clinical lung adenocarcinoma tissue and non-small cell lung carcinoma (NSCLC) cell lines were monitored using polymerase chain reaction (PCR) Array, flow cytometry (FCM), quantitative real-time PCR (qRT-PCR) and immunohistofluorescence (IF). Stem-like properties of CD133+ cells and CD2+ cells were detected by sphere formation assay, IF, and western blot. Colony formation and xenograft tumors experiments were performed to assess the malignant behaviors of CD2+ cells. The differentiation of CD133+ cells to CD2+ Th17-like cells was observed by FCM. The interleukin (IL)-2/phosphorylated signal transducer and activator of transcription protein 5 (pSTAT5)/retinoic acid receptor-related orphan receptor gamma t (RORγt) signaling pathway was evaluated by western blot and FCM.
Results
We found that CD133+ cells within both clinical lung adenocarcinoma tissue and NSCLC cell lines included a subset of CD2-expressing cells, which were correlated with the grade of malignancy (r=0.7835, P<0.01) and exhibited stem-like properties. Then, we determined the tumorigenic effects of CD2 on the growth of transplanted Lewis lung carcinoma cells (LLC1) in C57/BL6 mice. The results indicated that CD2+ cells were effective in promoting tumor growth in vivo (P<0.01). Furthermore, we obtained direct evidence of an ability of CD133+ cells to transform to T-helper 17-like cells via an intermediate CD133+CD2+ progenitor cell that is able to secrete IL-17A and IL-23. Furthermore, we found that IL-2 can inhibit the production of T-helper 17-like cells (P<0.001) by modulating the activation of STAT5 signaling pathways to downregulate the expression of RORγt (P<0.001).
Conclusions
Our data demonstrates that Th17-like cells generated from CSCs support cancer progression. These findings enrich the definition of multidirectional differentiation potential of CSCs and improve the understanding of the role of CSCs in cancer progression, which aids the improvement and creation of therapies.
Keywords: Non-small cell lung carcinoma (NSCLC), cancer stem cells (CSCs), differentiation, tumor microenvironment (TME), Th17
Introduction
Non-small cell lung carcinoma (NSCLC) is the most commonly diagnosed cancer and the leading cause of cancer-associated death (1). The prognosis of NSCLC is poor, and the survival rate is only 15% after 5 years because many of these patients ultimately do not respond to chemo-radiotherapy, which is associated with unimaginable degree of cellular heterogeneity within a single tumor (2). The heterogeneity of tumor may correlates with cancer stem cells (CSCs) or tumor-initiating cells (TICs) which may be responsible for NSCLC treatment failure which includes local recurrence, distant metastasis, and the limitations of therapeutic agents (3). CSCs, which have been identified in most types of cancer, possess the capabilities of self-renewal, infinite proliferation, and multi-directional differentiation (4). Therefore, further efforts dedicated to uncovering the biological characteristics of these cells are urgently needed.
Tumor microenvironment (TME) is the cellular environment in which the CSCs exists (5). In addition to CSCs, the TME is composed of immune cells, blood vessels, fibroblasts, smooth muscle cells, some epithelial cells, and extracellular matrix (6). CSCs maintain intricate interactions with other malignant cells within the TME largely influencing the outcome of cancer growth and metastasis. They have been certified that TME related features, especially immune and stromal cell promote the development of diagnostic and prognostic assessment process of NSCLC (7). Moreover, the metabolic state and activities of the immune cells comprising the TME influences the proliferative and invasive capacity of tumor cells.
The immune cells in the TME are related to poor elimination of tumor cells, including a lack of strong cancer antigens or epitopes recognized by T cells, minimal activation of cancer-specific T cells, poor infiltration of T cells into tumors, downregulation of the major histocompatibility complex on cancer cells, as well as immunosuppressive factors and cells in the TME (8). In addition to cytokines and infiltrating immune cells, other cells in the TME also suppress the immune system (9). These cells may act as intermediaries, triggering a cascade of events that ultimately culminates in immunosuppression; however, the actual causes remain poorly defined.
It has been shown that a positive response of the body in eliminating tumor cells usually depends on the interaction of tumor cells with immune cells in the TME (10). When these interactions occur, the relationship between tumor cells and immune cells in the TME plays an important role in inhibiting or enhancing the immune response (11). The occurrence, development, and outcome of the tumor depends on the function of the immune cells in the TME and is determined by T-cell immunity (8). T cells provide signals for the identification and killing of cancer cells. These signals are essential to the protective immune response against tumors. The CSCs in the CSC niche, as well as in the TME, add complexity to tumor immunity (12,13). Owing to this complexity, further investigation is required to uncover the relationship between CSCs and T cells in the TME.
Recent studies on CSCs have shown that glioblastoma stem-like cells and colorectal CSCs are multipotent and have differentiation potential along tumor and endothelial lineages (14,15). Also, murine mammary gland tumor cells demonstrating CSC characteristics can become functional adipocytes (16), and neurons generated from CSCs support cancer progression (17). These observations indicate that the cells in the TME could enter through infiltration or result from CSC differentiation. Consistent with this hypothesis, the presence of some immune cells within tumors is ascribable to infiltration and trafficking, whereas others may actually originate from the tumor itself. The biomarkers and effects on the tumorigenesis and development of these immune cells with different origin need to be further clarified.
This study was designed to test the potential of CSCs to differentiate into neoplastic immune cells and evaluate the capacity of immune-like cells to participate in the process of tumorigenesis. The significance of neoplastic immune cells to the prognosis of NSCLC patients was also analyzed.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-980).
Methods
Cell culture and reagents
Human non-small cell lung cancer A549 cells and Lewis lung carcinoma cells (LLC1) obtained from the Shanghai Institutes for Biological Sciences were validated by short tandem repeat (STR) DNA profiling and tested for mycoplasma. Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 or Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1X penicillin/streptomycin (Gibco) at 37 °C in 5% CO2.
Cell isolation and culture
CD133+ cells were isolated by magnetic-activated cell sorting (MACS) with a CD133 MicroBead Tumor Tissue Kit (Miltenyi Biotec, 130-100-857) using methods described previously (18). The cells were cultured in serum-free media for sphere formation assays. When we explored whether the cytokines had a role in the differentiation, 10 ng/mL of interleukin (IL)-2, IL-3, IL-6, or transforming growth factor beta (TGF-β) was added to the culture for 24 hours. CD133+ cells from clinical tumor specimens were isolated by flow cytometry (FCM). CD2+ and CD2− cells were isolated by MACS with CD2 MicroBeads (Miltenyi Biotec, 130-091-114).
FCM
A single cell suspension was obtained by grinding and filtering tissue, and the A549 cell suspension was digested with trypsin. All cells were resuspended in wash buffer (PBS containing 0.1% BSA) and stained for surface markers using FITC Mouse Anti-Human CD2 (BD Biosciences, 555326), FITC Mouse Anti-Human CD4 (BD Biosciences, 550628), FITC Mouse IgG1, κ Isotype Control (BD Biosciences, 555748), CD133/1 Antibody, anti-human, APC (Miltenyi Biotec, 130-113-668), and isotype control antibodies, mouse IgG1, APC (Miltenyi Biotec, 130-113-196). FCM data were acquired with a FACSCalibur (BD Biosciences). All used antibodies were list in Table S1.
Cell immunofluorescence staining
Immunofluorescence staining was performed as described previously (18). Briefly, primary antibodies were added to cells and incubated for approximately 90 minutes at 4 °C. Nuclei were stained with 4'6-diamidino-2-phenylindole (DAPI). All immunofluorescence staining results of in vitro cell culture were replicated more than 5 times. The primary antibodies used were FITC Mouse Anti-Human CD2 (BD Biosciences, 555326) and CD133/1 Antibody, anti-human, APC (Miltenyi Biotec, 130-113-668). All used antibodies were list in Table S1.
Real-time quantitative polymerase chain reaction
The levels of messenger RNA (mRNA) expression were determined by quantitative polymerase chain reaction (qPCR) performed using an ABI PRISM 7500 Cycler (Applied Biosystems). Primers that resulted in amplification were designed from human structural genes and are listed in Table S2. Following isolation of RNA from cell lines and its conversion to cDNA, qPCR quantitation was performed as described previously (18). RNA isolation and reverse transcription of CD133+ cells from human lung tissue specimens were performed using the REPLI-g WTA Single Cell Kit (QIAGEN,150063).
PCR array
Total RNA was obtained from cells using a RNeasy Mini Kit (QIAGEN, 74104). During RNA purification, an RNase-Free DNase Set (QIAGEN, 79254) was used for efficient on-column digestion of tiny amounts of DNA. RNA concentration quality control was performed using 1% agarose electrophoresis and the NanoQ Micro-volume Spectrophotometer (Capital Bio). Template cDNA was made using an RT2 First Strand Kit (QIAGEN, 330401). RNA isolation and reverse transcription of CD133+ cells from human lung tissue specimens were performed following the same steps as those described for real-time quantitative PCR.
Next, the cDNA was mixed with an appropriate amount of RT2 SYBR Green ROX qPCR Master mix (QIAGEN, 330522). The mixture was then aliquoted into wells of the Human Terminal Differentiation Markers RT2 Profiler PCR Array (QIAGEN, 330231 PAHS-048ZA) or the Human Cell Surface Markers RT2 Profiler PCR Array (QIAGEN, 330231 PAHS-055ZA). PCR was performed, and the relative gene expression was determined with data from a real-time cycler (Applied Biosystems 7500), using the ΔΔCt method. All experiments were completed according to the manufacturers’ guidelines.
Enzyme-linked immunosorbent assay (ELISA)
After 12, 24, 36, and 48 hours of culture of isolated cells, supernatants were harvested. ELISAs were performed using the Human IL-17A ELISA kit (Invitrogen, KAC1591) and the Human IL-22 Sunny ELISA kit (Invitrogen, BMS2047). For each assay conducted, we followed the specific experimental steps set out in the manufacturers’ instructions.
Western blot
Cells were collected and lysed in RIPA lysis buffer with added phenylmethylsulfonyl fluoride (PMSF). We detected the sample protein concentration using the BCA method (MULTI SCIENCES, BCA Protein Quantitation Kit, LK-PQ0012). Equal amounts of protein were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes. The membranes were blocked using 5% nonfat milk and incubated overnight with the following primary antibodies: octamer-binding transcription factor-4 (Oct4) Antibody (Cell Signaling, 2750, 1:1,000), SRY-related high-mobility-group (HMG)-box protein-2 (Sox2) (D6D9) XP® Rabbit mAb (Cell Signaling, 3579, 1:1,000), Nanog (D73G4) XP® Rabbit mAb (Cell Signaling, 4903, 1:2,000), Signal transducer and activator of transcription protein 5 (Stat5) (D3N2B) Rabbit mAb (Cell Signaling, 25656, 1:1,000), Phospho-Stat5 (Tyr694) (D47E7) XP® Rabbit mAb (Cell Signaling, 4322, 1:1,000), retinoic acid receptor-related orphan receptor gamma t (RORγt) Monoclonal Antibody (Invitrogen, 14-6988-82, 1:800) and β-Actin (Cell Signaling, 4970, 1:1,000). After incubation, the membranes were washed and then incubated a second time with horseradish peroxidase-conjugated secondary antibodies. Finally, the blots on the membranes were visualized with enhanced chemiluminescence plus reagents (Life Technologies, Novex® Chemiluminescent Substrates, WP20005). All used antibodies were list in Table S1.
Animal experiment
For the animal study, 4- to 6-week-old male C57/BL6 mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The mice were housed in a conventional animal house under standard laboratory conditions, with a 12-hour light/12-hour dark cycle and a constant temperature (22±1 °C) and humidity (55%±5%). Standard rodent chow and tap water were freely available. All animal experiments were performed according to the Guidelines for the Care and Use of Laboratory Animals. The experimental protocols were approved by the Local Committee on Animal Care, Use and Protection of Hebei Medical University. The approval file for study related to animal experiment was No. 20190051. They were randomly assigned to 3 groups. After at least 5 days of acclimatization, the C57/BL6 mice were subcutaneously injected with 1×106 LLC1 cells in 100 µL PBS via the right flank. In the experiments, each group comprised 5 mice. The size of each subcutaneously transplanted tumor was measured with the Vevo 2100 Imaging Platform (Visual Sonics, Canada). The tumor volume was calculated with the following formula: tumor volume = 0.5 × long diameter × short diameter2.
Human lung tissue specimen experiment
For the experiment, we used fresh lung cancer tissue samples that were collected between 2018 and 2020 from surgically resected specimens under approved research protocols. The pathological diagnoses were made by the pathology department of a local hospital according to the TNM staging system (tumor size, node involvement, metastasis presence). Stage I, II, and IIIA NSCLC samples were identified as adenocarcinoma by pathological histology. None of the patients had a direct blood relationship, or had received radiotherapy or chemotherapy preoperatively. Informed written consent was obtained from all the patients involved in the study. This study was approved by the Institutional Review Board of Hebei Medical University (No. 20190066) and conformed to the provisions of the Declaration of Helsinki (as revised in 2013). Hematoxylin and eosin (H&E) staining of the adenocarcinoma tissues was performed as described previously (19). Some sections were then stained for tissue immunofluorescence staining following the manufacturers’ protocols. The primary antibodies used were Rabbit Anti-CD133 antibody (Abcam, ab19898), Mouse Anti-CD2 antibody (Abcam, ab193344), and Mouse Anti-CD4 antibody (Abcam, ab25804). The secondary antibodies were Alexa Fluor 488 Goat anti-Mouse IgG antibody (Invitrogen, A-21121) and Alexa Fluor 594 Goat anti-Rabbit IgG antibody (Invitrogen, R37117). All used antibodies were list in Table S1.
Cell Counting Kit-8 (CCK-8) assay
The viability of cells after the indicated treatment was measured by CCK-8 assay (Dojindo, Japan) according to the manufacturer’s instructions. Briefly, 5×103 cells/well were seeded into 96-well plates. When we explored whether the cytokines had a role in the survival of cells, 10 ng/mL of IL-2, IL-3, IL-6, or TGF-β was added to the culture for 24 hours. The absorbance of the converted dye was measured at 450 nm using a microplate reader (Thermo Fisher, Finland).
Colony formation assay
Cells were seeded into a 6-well plate and cultured for 10–14 days. Then, the colonies were fixed with 20% methanol and stained with 0.1% crystal violet dye. Representative images were obtained, and the colonies were counted.
Silencing of STAT5 by small interfering RNA (siRNA)
siRNA transfection was performed according to the manufacturer’s instructions. Briefly, 4×105 A549/DDP cells were seeded into 6-well plates for 12 hours in the culture medium. Then, STAT5 siRNA (Cell Signaling Technology, 6275) and control siRNA (Cell Signaling Technology, 6275) were transfected with HiPerFect Transfection Reagent (QIAGEN, 301704). Following 48 hours of incubation, STAT5 expression was measured by western blot and qPCR.
Statistical analysis
Data were analyzed using GraphPad Prism (GraphPad 8.0). Unless otherwise noted, data were presented as the mean and standard deviation (SD). The number of experimental and control groups was at least 5. One-way analysis of variance (ANOVA) and unpaired t-tests were used as appropriate. Spearman’s correlation tests were also performed. P values were considered significant at P<0.05.
Results
CD133+CD2+ cells existed in NSCLC and were correlated with a poor prognosis
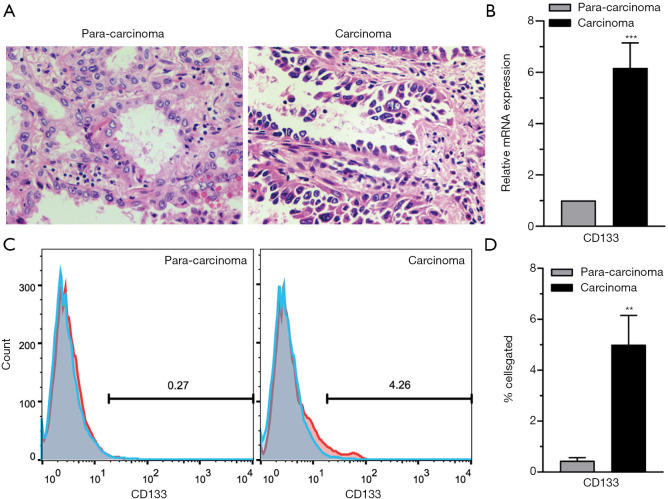
CD133 (also known as AC133 or prominin-1), a cell-surface glycoprotein, marks self-renewing cancer stem cells (CSCs) in a variety of solid tumors, including those in the brain, colon, pancreas, prostate, liver, and lung (20). CD133+ CSCs are known markers of chemo- and radio-resistance in multiple aggressive cancers, that may drive intra-tumoral heterogeneity (21). In the present research, tumor tissues were collected from NSCLC patients undergoing surgery and analyzed by H&E staining. The results showed that the carcinoma samples showed significant characteristics of NSCLC. Cancer cells in lung adenocarcinoma tissues were irregularly arranged and cytological atypia was present (Figure 1A). Then, we performed a series analysis of the presence of CD133+ cells in NSCLC and para-carcinoma tissues using real-time PCR and FCM. It was confirmed that the CD133 expression levels of both mRNA (Figure 1B) and protein (Figure 1C,D) were significantly higher in tumor tissues than in adjacent non-tumor tissues. All of these results suggested the presence of CD133+ cells in NSCLC.
Figure 1.
CD133+ cells exist in NSCLC. (A) Representative images of H&E staining of carcinoma and para-carcinoma tissues. Magnifications 200×. (B) The expression level of CD133 in 44 tumor tissues examined by real-time PCR. (C) Representative flow cytometry results for CD133 in carcinoma and para-carcinoma tissues from a patient. (D) The expression level of CD133 in 44 tumor tissues examined by flow cytometry. n=44. Data are shown as mean ± SD. **, P<0.01, ***, P<0.001. NSCLC, non-small cell lung carcinoma; H&E, hematoxylin and eosin; PCR, polymerase chain reaction.
Next, we isolated CD133+ cells from human lung adenocarcinoma tissue by fluorescence-activated cell sorting (FACS) to study the phenotypes and characteristics of CD133+ cells. After sorting, the CD133+ cells were enriched to a purity of 97% (Figure S1). Then, the gene expression profiles of these cells were assessed using the Human Terminal Differentiation Markers RT2 Profiler PCR Array. The result of the PCR array showed that CD133+ cells expressed specific genes in 13 tissues (Figure S2A and Table S3), including 7 bone marrow cell-related genes (Figure 2A). Bone marrow is known to be an important source of immune cells.
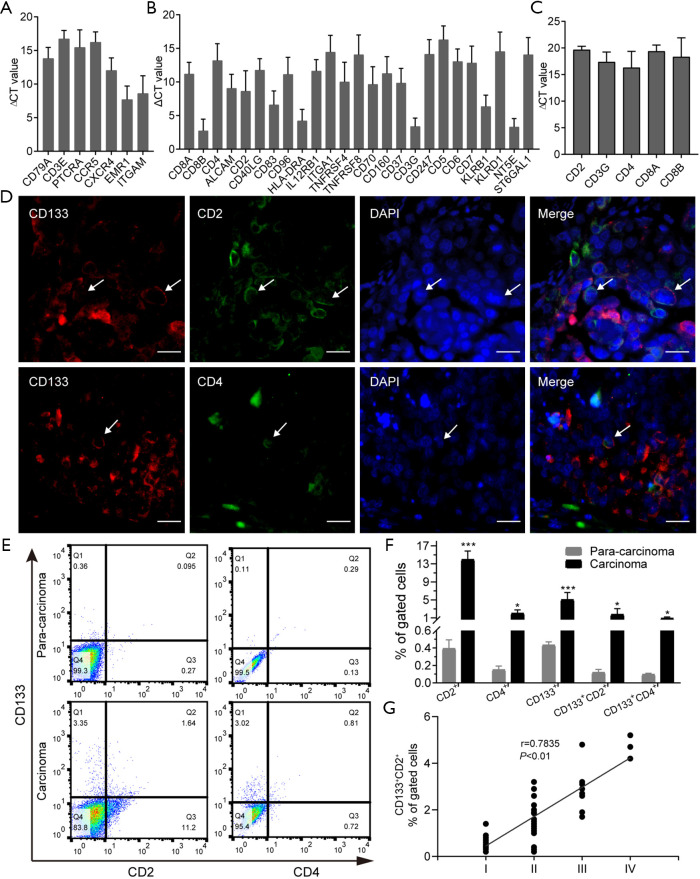
Figure 2.
CD133+ CD2+ cells exist in NSCLC and are correlated with poor prognosis. (A) Gene expression profiles of CD133+ cells from tumor tissue by the Human Terminal Differentiation Markers RT2 Profiler PCR Array. (B) Gene expression profiles of CD133+ cells from tumor tissue by the Human Cell Surface Markers RT2 Profiler PCR Array. (C) 5 classical markers of T-cell types detected by real-time PCR. (D) Immunofluorescence staining for CD133, CD2, and CD4 in tumor tissues. Red fluorescence: CD133 molecule. Green fluorescence: CD2 or CD4 molecules. Blue fluorescence: Nucleus. The white arrows refer to the cells co-expressing CD133 with CD2 or CD4. Scale bar: 100 µm. (E,F) Detection of CD133+, CD2+, and CD4+ cells in tumor tissues by flow cytometry. Data are shown as mean ± SD. *, P<0.05, ***, P<0.001. (G) Correlation of CD133 CD2 expression and TNM stage of NSCLC. NSCLC, non-small cell lung carcinoma; PCR, polymerase chain reaction; TNM, tumor node metastasis.
To confirm the specificity of the differentiation program, markers of other cell types were further examined using another PCR array (Human Cell Surface Markers RT2 Profiler PCR Array). The analysis showed that CD133+ cells expressed specific gene markers of 11 types of cells (Figure S2B and Table S4). Among them, T-cell marker genes accounted for the highest number of cell-specific genes expressed, with 25 key genes of different T-cell subtypes being detected (Figure 2B). To confirm the results of the PCR array, 5 classical markers (CD2, CD3G, CD4, CD8A, and CD8B) of 2 typical T-cell types were selected for detection by real-time PCR. CD133+ cells were indeed found to express these T-cell markers (Figure 2C). Next, we detected the expression of CD133, CD2, and CD4 in lung adenocarcinoma tissue. Immunohistofluorescence (IF) showed that CD133, CD2, and CD4 were expressed on the surface of the cell membrane, and that CD133 was co-expressed with CD2 and CD4 (Figure 2D). The results of FCM showed that the levels of CD2+, CD4+, CD133+, CD133+CD2+, and CD133+CD4+ cells in cancer tissues were significantly higher than those in para-carcinoma tissues (Figure 2E,F). These data indicated the presence of CD133+CD2+ cells in human lung adenocarcinoma. Furthermore, we analyzed the clinical features of 41 NSCLC patients with available clinical data (Table S5) and found a correlation between the presence of CD133+CD2+ cells and clinicopathological stage (Figure 2G). Our results indicated that CD133+CD2+ cells are present in NSCLC tumors and may serve as novel malignant markers for patients with NSCLC.
CD133+CD2+ cells were detected in the A549 cell line
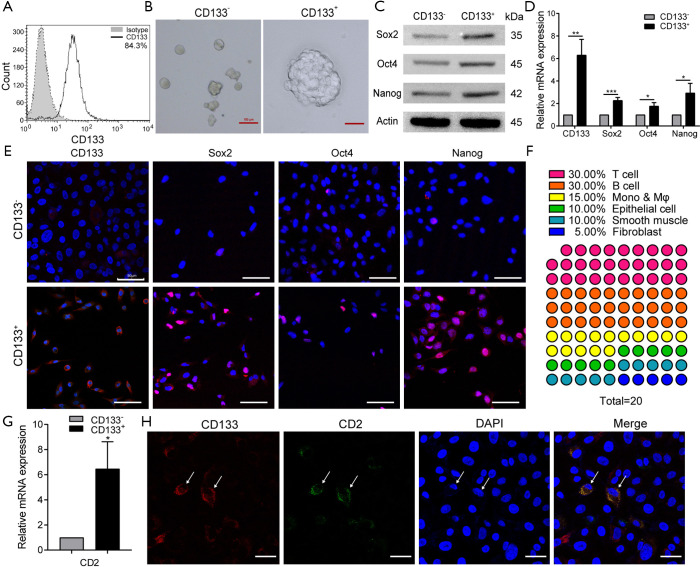
To further confirm the presence of CD133+CD2+ cancer cells in NSCLC, we detected the levels of these cells in the lung adenocarcinoma cell line A549. FCM analysis performed on A549 cells indicated that the 1.52% of A549 cells were CD133+ cells (Figure S3). Then, we enriched CD133+ A549 cells by performing MACS (Figure 3A), and performed a sphere formation assay. The results showed differences in the growth and proliferation of CD133+ and CD133− A549 subpopulations (Figure 3B). In addition, the pluripotency factors Sox2, Oct4, and Nanog (22) were highly expressed in CD133+ A549 cells compared to CD133− cells (Figure 3C,D,E).
Figure 3.
CD133+CD2+ cells also exist in the A549 cell line. (A) Representative detection of CD133+ cells purity after tumor tissues were sorted by MACS. (B) The sphere-forming abilities of CD133+ cells and CD133− cells. The pluripotency factors (CD133, Sox2, Oct4, and Nanog) of 2 types of cells were evaluated by western blot (C), real-time PCR (D), and cell immunofluorescence (E). Representative images of western blot and immunofluorescence. Data are presented as the mean ± SD of 3 replications. Scale bar: 100 µm. (F) The top 20 genes with the highest expression in CD133+ cells in the real-time RT2 Profiler PCR Array (Human Cell Surface Markers). (G) Real-time PCR to verify the CD2 expression in both types of cells. (H) Immunofluorescence staining of CD2 (green) and CD133 (red) of CD133+ cells. Blue: nucleus. Scale bar: 50 µm. All data are expressed as the mean ± SD. *, P<0.05, **, P<0.01, ***, P<0.001. All experiments were repeated at least 3 times. Sox2, SRY-related high-mobility-group (HMG)-box protein-2; Oct4, octamer-binding transcription factor-4; IL-2, interleukin-2; MACS, magnetic-activated cell sorting; PCR, polymerase chain reaction.
The gene expression profiles of 2 subpopulations were verified with the Human Cell Surface Markers PCR Array. The top 20 genes in the PCR chip of CD133+ cells were analyzed, and 75% of them were found to be immune system genes. Of them, T-cell-related genes accounted for the largest proportion (30%) (Figure 3F). This finding also extended our understanding of the differentiation capability of stem-like cells. CD2 is an excellent pan-T-cell marker and 1 of the earliest antigens, appearing on T lymphocytes before CD1 and after CD7. The level of CD2 mRNA in both kinds of cells was found to be decreased as tumor cells progressed from CD133+ cells to CD133− cells (Figure 3G). The melting curve (Figure S4A), agarose gel electrophoresis (Figure S4B), and sanger sequencing (Figure S4C) indicated it to be a specific amplification. According to all of these results, CD2 was expressed in A549 cells, and its expression in the CD133+ subpopulation was greater than that in CD133− cells. The results of immunofluorescence analyses demonstrated that CD2 could be co-expressed with CD133 on the cytomembrane (Figure 3H), Moreover, CD133− cells also expressed CD2 (Figure S5). To sum up, these results suggest that CD133+CD2+ and CD2+ subpopulations exist in the A549 cell line.
CD2+ cells differentiated from CD133+CD2+ cells and exhibited stem-like properties in NSCLC cell line
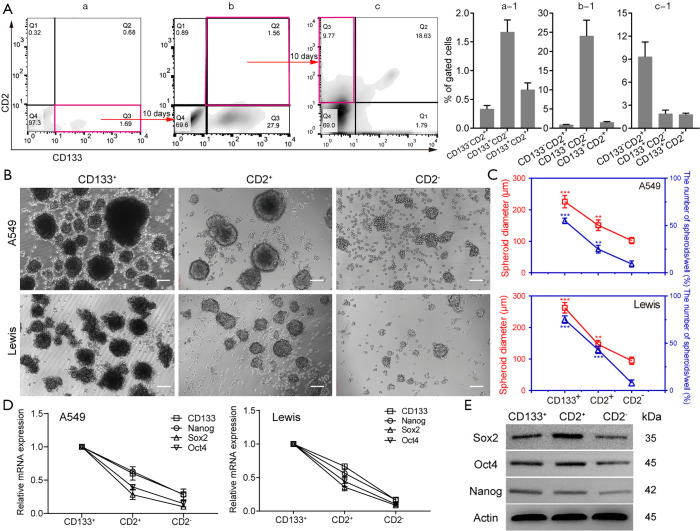
CD2 is a specific marker for T cells and can therefore be used to identify their presence. Therefore, the identification of the origin and features of CD2+ cells are of considerable interest. Dissociated A549 cells were divided into 4 groups: (I) CD133+/CD2−, (II) CD133+/CD2+ (double positive, DP), (III) CD133−/CD2+, and (IV) CD133−/CD2− (double negative, DN) (Figure 4Aa, a-1). To define the lineage potential further, CD133+CD2− (Figure 4Aa, Q3) cells were enriched and cultured for 10 days, which resulted in the downregulation of CD133, the upregulation of CD2, and notably, the reappearance of CD133+CD2+ A549 cells (Figure 4Ab, b-1). Then, we sorted CD133+CD2+ cells (Figure 4Ab, Q2) by FACS, and the same procedure was performed. After another 10 days, FCM analysis demonstrated the emergence of a CD133−CD2+ population (Figure 4Ac, Q1 and Figure 4Ac-1). The data confirmed that CD133+CD2+ progenitors in A549 cells could arise from the CD133+ cell population and were capable of differentiating into CD133−CD2+ cells of tumorous origin. To determine the stem-like properties of CD133−CD2+ cells, we performed a sphere formation assay. CD2+, CD2−, and CD133+ cells were sorted from A549 and Lewis cell lines by MACS. As shown in Figure 4B, the spheres formed by the 3 kinds of cells displayed different morphological characteristics. Measurement of the diameters and the number of spheroids in each well showed that CD2+ cells formed more and larger spheres than CD2− cells but fewer and smaller spheres than CD133+ cells (Figure 4C). The expression levels of stemness markers, including CD133, Nanog, Sox2, and Oct4, in CD2+ cells were higher than those in CD2− cells but lower than those in CD133+ cells (Figure 4D,E). Taken together, our observations indicated that CD2+ NSCLC cells exhibited stem-like properties.
Figure 4.
CD2+ cells differentiate from CD133+ cells and exhibit stronger stem-like properties. (A) The process of CD133+ cells differentiation into CD2+ cells observed by flow cytometry. The representative result is shown. (B,C) Sphere formation of different types of A549 and Lewis cells, with the sphere-forming capability evaluated based on the spheroid diameter and the number of spheroids. Scale bar: 100 µm. n=5 wells per group. Real-time PCR (D) and western blot (E) detection of pluripotency factors in 3 types of cells. Gene expression was normalized to the geometric mean of the housekeeping gene GAPDH. Data are shown as mean ± SD. All experiments were repeated at least 3 times. **, P<0.01; ***, P<0.001. Sox2, SRY-related high-mobility-group (HMG)-box protein-2; Oct4, octamer-binding transcription factor-4; PCR, polymerase chain reaction.
CD2+ NSCLC cells showed significantly higher proliferation and tumorigenic ability
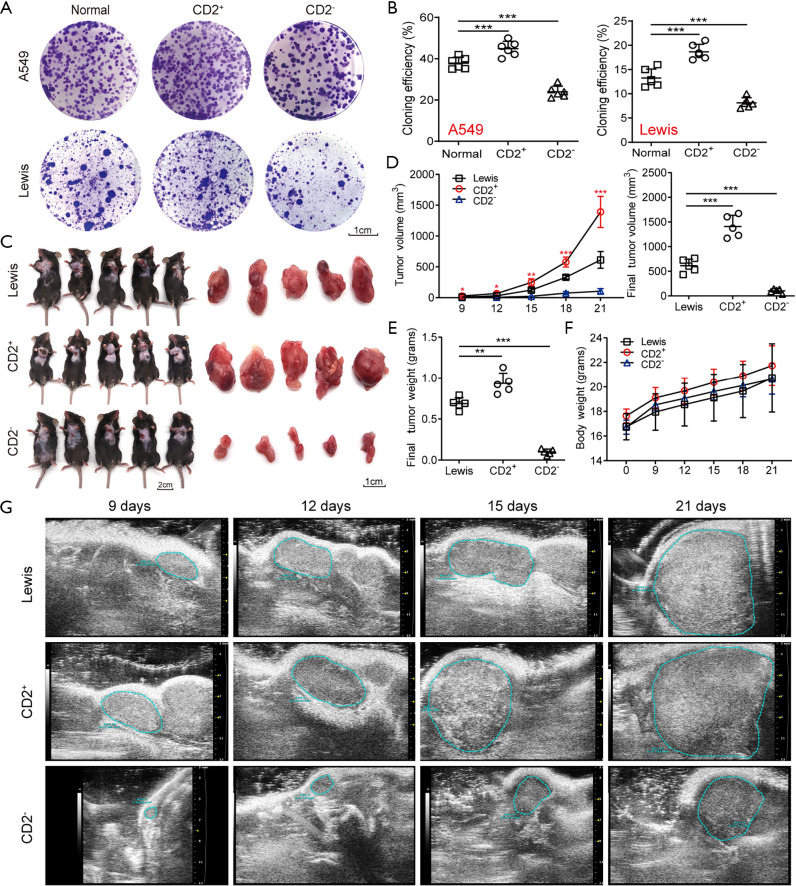
To evaluate the self-renewal and proliferation of CD2+ NSCLC cells in vitro, a colony formation assay was conducted. The results showed that the cloning efficiency of CD2+ cells was much higher than that of the parent and CD2− cells (Figure 5A,B). We also examined the tumorigenicity of CD2+ Lewis cells by injecting C57/BL6 mice with lung carcinoma cells. The effects were evaluated in 3 different groups of mice: the Lewis, CD2+ Lewis, and CD2− Lewis. The tumor growth curve and final tumor size data revealed that the average tumor volume and weight in the CD2+ group were significantly greater than those in the Lewis and CD2− groups (Figure 5C,D,E). No notable differences in body weight were observed among the mice in the 3 groups (Figure 5F). Ultrasound examination showed that the tumor size of each group started to show a significant difference from 9 days after modeling (Figure 5G). Together, these results indicated that CD2+ Lewis cells had strong proliferation ability and were effective in promoting tumor growth.
Figure 5.
CD2+ cells showed strong proliferation ability in vitro and in vivo. (A,B) Colony formation efficiency of different types of A549 and Lewis cells assessed by colony-forming assay. The colonies were stained with crystal violet dye. Representative images of colonies are shown on panel A. Cloning efficiency of these cells are shown on panel B. Scale bar: 1 cm. n=5 wells per group. (C) Representative images of animal model and tumor samples excised from different groups. (D,E) The average tumor volume and weight of different groups. (F) The average body weight of mice in different groups. (G) Representative ultrasound images of subcutaneously transplanted tumors at different times. n=5 for each group. Data are shown as mean ± SD. *, P<0.05, **, P<0.01, ***, P<0.001.
CD2+ NSCLC cell may be a Th17-like cell subpopulation
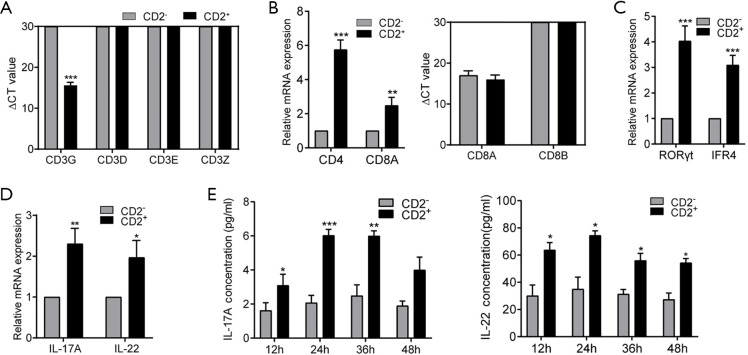
To further investigate whether CD2+ NSCLC cell was T cell-like, we performed real-time PCR for CD3, a T-cell co-receptor. The 4 chains of CD3 molecules (CD3G, CD3D, CD3E, and CD3Z) were not expressed in CD2− cells. Meanwhile, CD2+ cells expressed CD3G at a low level but did not express the other CD3 molecules (Figure 6A). We also analyzed the mRNA levels of CD4 and CD8. CD4 and CD8A were upregulated in CD2+ cells compared to CD2− cells (Figure 6B, left). Using the delta CT method after standardization to compare CD8A and CD8B with internal genes, we found that CD8A was expressed in both groups. However, CD8B had an infinite delta CT value, which meant that it was not expressed in the CD2+ cells and CD2- cells (Figure 6B, right). Therefore, subsequent experiments focused on CD4+ T-cell subsets. Next, we observed the expression of transcription factors and cytokines involved in T-cell polarization. The levels of retinoic acid RORγt and interferon regulatory factor 4 (IFR4) genes, which are specific transcription factors of Th17 cells, were significantly higher in CD2+ cells (Figure 6C). The same was true for the gene expression of the cytokines IL-17A and IL-22 (Figure 6D), the production of which by Th17 cells triggers inflammatory signaling cascades and increases NSCLC cell proliferation, migration, and invasion (23-25). Subsequently, the secretions of IL-17A and IL-22 in CD2+ and CD2− cell culture supernatants were detected by ELISA. After 24 hours of culture, the secretions of IL-17A and IL-22 reached a peak. At 12, 24, and 36 hours after culture, there were significant differences in the IL-17A and IL-22 secretion levels between the CD2+ and CD2− groups (Figure 6E). In short, the results of ELISA demonstrated that CD2+ cells secreted IL-17A and IL-22. These findings demonstrated that this CD2+ subpopulation had a similar phenotype to Th17 lymphocytes (26) and thus, could be referred to as CD2+ Th17-like NSCLC cells.
Figure 6.
CD2+ NSCLC cells may be Th17-like cells. (A,B,C,D) Gene expression levels of the indicated immune markers in CD2+ and CD2− cells by real-time PCR. (E) The levels of IL-17 and IL-22 in cell culture supernatants at different times as detected by ELISA. Data are shown as mean ± SD (n=5; *, P<0.05, **, P<0.01, ***, P<0.001). NSCLC, non-small cell lung carcinoma; PCR, polymerase chain reaction; IL, interleukin; ELISA, enzyme-linked immunosorbent assay.
IL-2-mediated phosphorylation of STAT5 suppressed the induction of CD2+ Th17-like cells
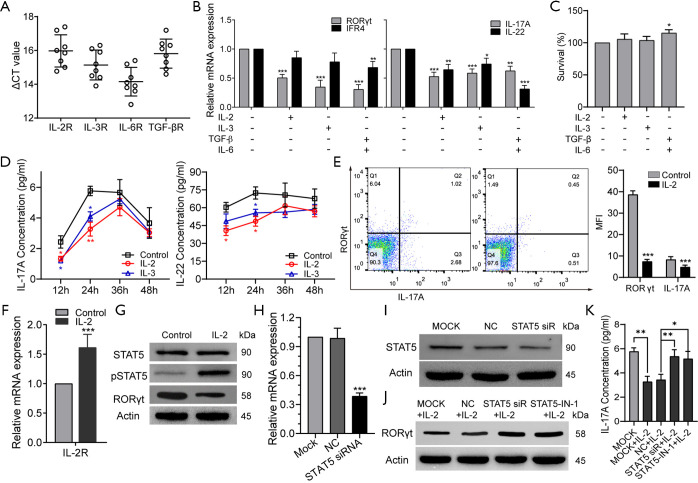
In the healthy body, IL-2 (27), IL-3 (28), TGFβ, and IL-6 are involved in the induction of normal Th17 cell development (29). Therefore, we explored whether these cytokines had a role in CD2+ Th17-like cell differentiation. To address this possibility, the expression levels of IL-2, IL-3, IL-6, and TGFβ receptors were determined in CD133+ subpopulations sorted from A549 cells. IL2R, IL-3R, IL-6R, and TGFβR were observed to be constitutively expressed (Figure 7A). Compared to cells cultured in medium alone, the addition of human IL-2, IL-3, IL-6, and TGFβ resulted in a marked reduction in the expression of RORγt, IFR4, IL-17A, and IL-22 (Figure 7B). Meanwhile, the results of CCK8 assays showed that cell survival was not affected by IL-2 or IL-3; however, the addition of exogenous IL-6 with TGFβ to the culture promoted the growth of CD133+ cells (Figure 7C). Treatment with IL-2 or IL-3 decreased the concentrations of IL-17A and IL-22 in culture supernatant, as determined by ELISA. The inhibitive effect of IL-2 was found to be more significant than that of IL-3, and the inhibition was most obvious after 24 hours of IL-2 treatment (Figure 7D). Therefore, IL-2 was selected as a priority for further study. The results of FCM showed that the addition of IL-2 decreased the proportion of cells expressing IL-17A and RORγt (Figure 7E). These findings revealed that the addition of human IL-2 resulted in a marked reduction in the production of Th17-like cells.
Figure 7.
IL-2 can inhibit the production of Th17-like cells by modulating the activation of pSTAT5. (A) The expression of cytokine receptors in CD133+ cells by real-time PCR. (B) RORγt, IFR4, IL-17A, and IL-22 gene expression of CD133+ cells in different groups. (C) The survival of CD133+ cells were examined by CCK-8 assay. (D) The levels of IL-17A and IL-22 in cell culture supernatants after the indicated treatments at different times as detected by ELISA. (E) Proportions of cells expressing IL-17A and RORγt with the addition of IL-2 as measured by FCM. (F) The levels of IL-2R in CD133+ cells after treatment with IL-2 as detected by real-time PCR. (G) Representative western blotting for STAT5, p-STAT5, and RORγt in CD133+ cells. (H,I) The significant inhibition of gene and protein expression of STAT5 by STAT5 siRNA. MOCK, solvent control. NC, control siRNA. siRNA, small interfering RNA. (J) Representative western blotting for RORγt. STAT5-IN-1, STAT5 inhibitor. (K) IL-17A expression examined by ELISA. Data represent 3 independent experiments. Data are shown as mean ± SD. *, P<0.05, **, P<0.01, ***, P<0.001. IL, interleukin; TGF-β, transforming growth factor β; RORγt, retinoid-related orphan nuclear receptor γt; STAT5, signal transducer and activator of transcription 5; p-STAT5, Phospho-signal transducer and activator of transcription 5; Th17, T helper cell 17; PCR, polymerase chain reaction; CCK-8, Cell Counting Kit-8; ELISA, enzyme-linked immunosorbent assay; FCM, flow cytometry; IL-2R, IL-2 receptor; siRNA, small interfering RNA.
The expression of the orphan nuclear receptor RORγt is widely known to be critical for Th17 differentiation. IL-2 is a potent activator of STAT5 transcription factors that have critical in vivo functions in lymphoid development (30). To test whether IL-2 could suppress CD2+ Th17-like cell differentiation through an intrinsic cell mechanism requiring STAT5, the effect of exogenous IL-2 on the expression of IL-2R was explored. Data showed that IL-2 acted to promote IL-2R expression (Figure 7F). The level of pSTAT5 was increased and that of RORγt was decreased after IL-2 treatment, whereas the expression of STAT5 was not affected (Figure 7G). This finding supported the hypothesis that IL-2 may suppress CD2+ Th17-like cell differentiation via modulation of pSTAT5. To clarify this process, we designed a STAT5 siRNA. As shown in Figure 7H and Figure 7I, the expression of STAT5 was decreased by STAT5 siRNA. The expression levels of RORγt in the STAT5 siRNA and pSTAT5 inhibitor groups were significantly higher than those in the non-transfected and negative control groups (Figure 7J). These observations demonstrated that STAT5 siRNA and pSTAT5 inhibitor reversed the inhibition of RORγt induced by IL-2. In other words, IL-2 inhibited the expression of RORγt via STAT5 phosphorylation. Furthermore, the IL-17A secreted by CD2+ Th17-like cells was significantly reduced by the addition of IL-2, and this response was reversed after transfection with STAT5 siRNA or the addition of pSTAT5 inhibitor (Figure 7K). Collectively, these results indicated that IL-2 could inhibit the production of Th17-like cells, possibly via the IL-2R by modulating the phosphorylation level of STAT5. This study demonstrates the multipotency of CD133+ cells and their ability to differentiate to CD2+ Th17-like cells that promote cancer progression. IL-2-mediated phosphorylation of STAT5 opposes the process of CD133+ cells differentiation to CD2+ Th17-like cells. Some other results also suggested that CSCs are multipotent and capable of differentiation along tumor and endothelial lineages, adipocytes, and neurons (Figure 8).
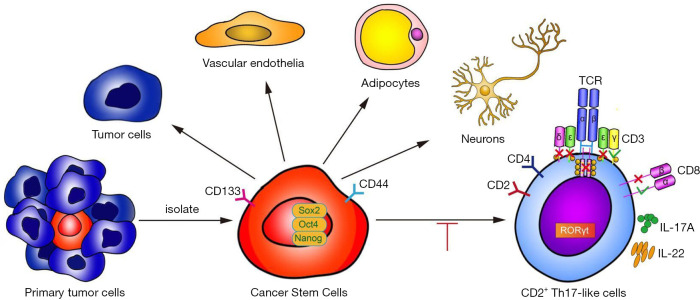
Figure 8.
Graphical Abstract. Previous studies have suggested that CSCs are multipotent and capable of differentiation along tumor and endothelial lineages, adipocytes, and neurons. This study demonstrates that CD133+ CSCs originating from lung carcinoma differentiate to CD2+ Th17-like cells. IL, interleukin; CSCs, cancer stem cells; Th17, T helper cell 17.
Discussion
The cells in the TME, such as cancer-associated vascular endothelial cells, fibroblasts, neuroendocrine cells, adipose cells, and immunocytes, are indispensable in all stages of human cancer development, including tumorigenesis, growth, progression, and metastasis (14,16,17,31). Several studies have suggested that CSCs possess multilineage differentiation potential, and they are thought to be the driving factor of intratumoral heterogeneity, cancer metastasis, and therapeutic resistance (4,32). Previous studies have shown that CD133+ cells in tumor tissues represent CSCs, while CD133− cells include a complex variety of cells, such as solid tumor cells, infiltrating immune cells, and neovascular endothelial cells (33). To better investigate molecules on the surface and the multi-directional differentiation potential of CSCs, as well as to understand the molecular basis through which CSCs promote tumor progression, we identified and isolated CSCs from NSCLC tissue, indicated by the surface expression of CD133. Our PCR array results showed that CSCs expressed specific genes in 13 tissues, including bone marrow cell-related genes, which suggested that CSCs might create a cellular environment for the tumor that includes tumor-related immune-like cells and is suitable for the survival of tumor cells.
Human T cells can differentiate from bone marrow and mature in the thymus (34), and the relationship between immune cells and tumor cells is extremely important for the development of cancer. We found that a portion of CD133+ cells expressed serial markers of immune cells, especially CD2 and CD4. CD133 is a CSCs marker, and CD2 is a surface antigen of the human T-lymphocyte lineage. The subsets of cells expressing both CD133 and CD2 molecules greatly aroused our interest. We found that CD133+CD2+ cells were present during each stage of NSCLC, and their expression increased along with the tumor grade, suggesting that CD2 together with CD133 may serve as a novel prognostic predictor for NSCLC patients.
CSCs make up an important portion of the TME; however, CSC differentiation is strictly regulated by the TME and its diverse types of stromal cells and extracellular matrix components (32). We found that CSCs expressed many markers of different types of cells, which comprised the bulk of the TME. Interestingly, the most expressed marker was associated with T cells. T cells play an important role in the antitumor immune response. The earliest marker for T cell differentiation is CD2, which is found on the surface of thymocytes and T cells (35). We found that CD2+ cells exhibit stem-like properties in NSCLC cells in vitro and promoted the growth of transplanted tumors in vivo. Thus, further study of the T-cell subtypes of these CD2+ cells and the regulation of their differentiation is of great significance. Many studies have arrived at the conclusion that immune cells are an important component of the TME and play a significant role in the progression of cancers (36). We found that CSCs isolated from an NSCLC cell line could differentiate into CD2+ Th17-like immune cells, which indicated that a portion of the immune cells in a TME may originate from CSCs. CD2+ Th17-like immune cells secreted the cytokines IL-17A and IL-22, which regulate the proliferation and metastasis of lung cancer cells (24,25). Improved characterization of the immunological properties of CSCs will contribute to the rational design of immunotherapeutic interventions to target these cells and may lead to the eradication of malignant diseases (37).
We demonstrated that CD133+ cells can give rise to CD2+ Th17-like cells. However, the Th17-like cells showed similar but not identical characteristics to Th17 lymphocytes in the immune system. Specifically, they lacked functional surface markers such as CD3. These CD2+ Th17-like cells were found to promote the growth of lung carcinoma cells. Further investigation is needed to fully demonstrate the specific functions of CD133+-derived CD2+ Th17-like cells. These CD2+ Th17-like cells may be one of the components of the blood–tumor barrier and act as a tumor immune guard. We speculate that the Th17-like cells can recognize and tolerate the tumor cells because they share the same origin, which may help tumor cells to complete the immune escape process. All of these possibilities are worthy of further study.
Inhibiting the production of Th17-like cells may be an effective approach for NSCLC therapy. We found that IL-2 inhibited the production of Th17-like cells, acting through the IL-2R by modulating the activation of STAT5 signaling.
Our research provides direct evidence that the CD2+ Th17-like tumor-related immune-like cells differentiate from CD133+ CSCs through an intermediate CD133+CD2+ progenitor cell and support cancer progression. Not only do these findings enrich the definition of the multidirectional differentiation potential of CSCs but from them, we can also infer that the inhibitory TME forms a blood-tumor barrier, which protects the tumor cells as the blood-brain barrier protects nerve cells (38) and as the placental barrier protects the fetus (39). This barrier is not only difficult for immune cells to overcome, but it is also hard for some macromolecule-targeted drugs to penetrate. However, how CSCs participate in the construction of the TME remains complex, and additional experiments and clinical studies are needed in the future.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank Huaxing Zhang for providing assistance with this study, and LetPub (www.letpub.com) for providing linguistic assistance during the preparation of this manuscript.
Funding: This work was supported by the Colleges and Universities in Hebei Province Scientific Research Project High Level Talents Fund (No. GCC2014004), the CAMS Innovation Fund for Medical Sciences (2019-12M-5-055), Natural Science Foundation of Hebei Province (H2020206579), and the Key Research and Development Program of Hebei Province (20327125D).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study conformed to the provisions of the Declaration of Helsinki (as revised in 2013). Informed written consent was obtained from all the patients involved in the study and the protocol was approved by the medical ethics committee of Hebei Medical University (No. 20190066). All animal experiments were performed according to the Guidelines for the Care and Use of Laboratory Animals. The experimental protocols were approved by the Local Committee on Animal Care, Use and Protection of Hebei Medical University. The approval file for study related to animal experiment was No. 20190051.
Footnotes
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-980
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-980
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-980). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Guilbaud E, Gautier EL, Yvan-Charvet L. Macrophage Origin, Metabolic Reprogramming and IL-1 Signaling: Promises and Pitfalls in Lung Cancer. Cancers (Basel) 2019;11:298. 10.3390/cancers11030298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schegoleva AA, Khozyainova AA, Fedorov AA, et al. Prognosis of Different Types of Non-Small Cell Lung Cancer Progression: Current State and Perspectives. Cell Physiol Biochem 2021;55:29-48. 10.33594/000000340 [DOI] [PubMed] [Google Scholar]
- 4.Makena MR, Ranjan A, Thirumala V, et al. Cancer stem cells: Road to therapeutic resistance and strategies to overcome resistance. Biochim Biophys Acta Mol Basis Dis 2020;1866:165339. 10.1016/j.bbadis.2018.11.015 [DOI] [PubMed] [Google Scholar]
- 5.Ghasemi F, Sarabi PZ, Athari SS, et al. Therapeutics strategies against cancer stem cell in breast cancer. Int J Biochem Cell Biol 2019;109:76-81. 10.1016/j.biocel.2019.01.015 [DOI] [PubMed] [Google Scholar]
- 6.Giraldo NA, Sanchez-Salas R, Peske JD, et al. The clinical role of the TME in solid cancer. Br J Cancer 2019;120:45-53. 10.1038/s41416-018-0327-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Zhou R. Tumor microenvironment related novel signature predict lung adenocarcinoma survival. PeerJ 2021;9:e10628. 10.7717/peerj.10628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JM, Chen DS. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann Oncol 2016;27:1492-504. 10.1093/annonc/mdw217 [DOI] [PubMed] [Google Scholar]
- 9.Relation T DM, Horwitz EM. Concise Review An (Im)Penetrable Shield How the Tumor Microenvironment Protects Cancer Stem Cells. Stem Cells 2017;35:1123-30. 10.1002/stem.2596 [DOI] [PubMed] [Google Scholar]
- 10.Murad JP, Tilakawardane D, Park AK, et al. Pre-conditioning modifies the TME to enhance solid tumor CAR T cell efficacy and endogenous protective immunity. Mol Ther 2021. [Epub ahead of print]. doi: . 10.1016/j.ymthe.2021.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang H, Fang J, Fan X, et al. Advances in Molecular Mechanisms for Traditional Chinese Medicine Actions in Regulating Tumor Immune Responses. Front Pharmacol 2020;11:1009. 10.3389/fphar.2020.01009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu HJ, Chu PY. Role of Cancer Stem Cells in Cholangiocarcinoma and Therapeutic Implications. Int J Mol Sci 2019;20:4154. 10.3390/ijms20174154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garg M. Epithelial Plasticity, Autophagy and Metastasis: Potential Modifiers of the Crosstalk to Overcome Therapeutic Resistance. Stem Cell Rev Rep 2020;16:503-10. 10.1007/s12015-019-09945-9 [DOI] [PubMed] [Google Scholar]
- 14.Wang R, Chadalavada K, Wilshire J, et al. Glioblastoma stem-like cells give rise to tumour endothelium. Nature 2010;468:829-33. 10.1038/nature09624 [DOI] [PubMed] [Google Scholar]
- 15.Shangguan W, Fan C, Chen X, et al. Endothelium originated from colorectal cancer stem cells constitute cancer blood vessels. Cancer Sci 2017;108:1357-67. 10.1111/cas.13262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishay-Ronen D, Diepenbruck M, Kalathur RKR, et al. Gain Fat-Lose Metastasis: Converting Invasive Breast Cancer Cells into Adipocytes Inhibits Cancer Metastasis. Cancer Cell 2019;35:17-32.e6. 10.1016/j.ccell.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 17.Lu R, Fan C, Shangguan W, et al. Neurons generated from carcinoma stem cells support cancer progression. Signal Transduct Target Ther 2017;2:16036. 10.1038/sigtrans.2016.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia X, Cong B, Zhang J, et al. CCK8 negatively regulates the TLR9-induced activation of human peripheral blood pDCs by targeting TRAF6 signaling. Eur J Immunol 2014;44:489-99. 10.1002/eji.201343725 [DOI] [PubMed] [Google Scholar]
- 19.Zhou W, Chen X, Hu Q, et al. Galectin-3 activates TLR4/NF-kappaB signaling to promote lung adenocarcinoma cell proliferation through activating lncRNA-NEAT1 expression. BMC Cancer 2018;18:580. 10.1186/s12885-018-4461-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim WT, Ryu CJ. Cancer stem cell surface markers on normal stem cells. BMB Rep 2017;50:285-98. 10.5483/BMBRep.2017.50.6.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vora P, Venugopal C, Salim SK, et al. The Rational Development of CD133-Targeting Immunotherapies for Glioblastoma. Cell Stem Cell 2020;26:832-44.e6. 10.1016/j.stem.2020.04.008 [DOI] [PubMed] [Google Scholar]
- 22.Balaji S, Santhi R, Kim U, et al. Cancer Stem Cells with Overexpression of Neuronal Markers Enhance Chemoresistance and Invasion in Retinoblastoma. Curr Cancer Drug Targets 2020;20:710-9. 10.2174/1568009620666200504112711 [DOI] [PubMed] [Google Scholar]
- 23.Wang D, Yu W, Lian J, et al. Th17 cells inhibit CD8(+) T cell migration by systematically downregulating CXCR3 expression via IL-17A/STAT3 in advanced-stage colorectal cancer patients. J Hematol Oncol 2020;13:68. 10.1186/s13045-020-00897-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Zhang Q, Wu Q, et al. Interleukin-22 secreted by cancer-associated fibroblasts regulates the proliferation and metastasis of lung cancer cells via the PI3K-Akt-mTOR signaling pathway. Am J Transl Res 2019;11:4077-88. [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Z, He D, Zhao S, et al. IL-17A/IL-17RA promotes invasion and activates MMP-2 and MMP-9 expression via p38 MAPK signaling pathway in non-small cell lung cancer. Mol Cell Biochem 2019;455:195-206. 10.1007/s11010-018-3483-9 [DOI] [PubMed] [Google Scholar]
- 26.Liu W, Li H, Zhang X, et al. Prostaglandin I2-IP signalling regulates human Th17 and Treg cell differentiation. Prostaglandins Leukot Essent Fatty Acids 2013;89:335-44. 10.1016/j.plefa.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 27.Alam F, Singh A, Flores-Malavet V, et al. CD25-Targeted IL-2 Signals Promote Improved Outcomes of Influenza Infection and Boost Memory CD4 T Cell Formation. J Immunol 2020;204:3307-14. 10.4049/jimmunol.2000205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahandideh B, Derakhshani M, Abbaszadeh H, et al. The pro-Inflammatory cytokines effects on mobilization, self-renewal and differentiation of hematopoietic stem cells. Hum Immunol 2020;81:206-17. 10.1016/j.humimm.2020.01.004 [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Sanchez ME, Mendoza L, Villarreal C, et al. A Minimal Regulatory Network of Extrinsic and Intrinsic Factors Recovers Observed Patterns of CD4+ T Cell Differentiation and Plasticity. PLoS Comput Biol 2015;11:e1004324. 10.1371/journal.pcbi.1004324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauché D, Joyce-Shaikh B, Fong J, et al. IL-23 and IL-2 activation of STAT5 is required for optimal IL-22 production in ILC3s during colitis. Sci Immunol 2020;5:eaav1080. 10.1126/sciimmunol.aav1080 [DOI] [PubMed] [Google Scholar]
- 31.Begum A, McMillan RH, Chang YT, et al. Direct Interactions With Cancer-Associated Fibroblasts Lead to Enhanced Pancreatic Cancer Stem Cell Function. Pancreas 2019;48:329-34. 10.1097/MPA.0000000000001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saijo A, Goto H, Nakano M, et al. Bone marrow-derived fibrocytes promote stem cell-like properties of lung cancer cells. Cancer Lett 2018;421:17-27. 10.1016/j.canlet.2018.02.016 [DOI] [PubMed] [Google Scholar]
- 33.Chen E, Zeng Z, Bai B, et al. The prognostic value of CSCs biomarker CD133 in NSCLC: a meta-analysis. Oncotarget 2016;7:56526-39. 10.18632/oncotarget.10964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terstappen LW, Huang S, Picker LJ. Flow cytometric assessment of human T-cell differentiation in thymus and bone marrow. Blood 1992;79:666-77. 10.1182/blood.V79.3.666.666 [DOI] [PubMed] [Google Scholar]
- 35.Cho DH, Kim MC, Park CI. Expression analysis data of T-cell surface antigen CD2 gene from rock bream (Oplegnathus fasciatus). Data Brief 2019;24:103832. 10.1016/j.dib.2019.103832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varn FS, Wang Y, Mullins DW, et al. Systematic Pan-Cancer Analysis Reveals Immune Cell Interactions in the Tumor Microenvironment. Cancer Res 2017;77:1271-82. 10.1158/0008-5472.CAN-16-2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harjes U. Tumour immunology: Tumours copy to escape. Nat Rev Cancer 2017;17:453. [DOI] [PubMed] [Google Scholar]
- 38.Bar-Klein G, Lublinsky S, Kamintsky L, et al. Imaging blood–brain barrier dysfunction as a biomarker for epileptogenesis. Brain 2017;140:1692-705. 10.1093/brain/awx073 [DOI] [PubMed] [Google Scholar]
- 39.Muoth C, Aengenheister L, Kucki M, et al. Nanoparticle transport across the placental barrier: pushing the field forward! Nanomedicine (Lond) 2016;11:941-57. 10.2217/nnm-2015-0012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as