Abstract
Iron overload disorders represent an important class of human diseases. Of the primary iron overload conditions, by far the most common and best studied is HFE-related hemochromatosis, which results from homozygosity for a mutation leading to the C282Y substitution in the HFE protein. This disease is characterized by reduced expression of the iron-regulatory hormone hepcidin, leading to increased dietary iron absorption and iron deposition in multiple tissues including the liver, pancreas, joints, heart and pituitary. The phenotype of HFE-related hemochromatosis is quite variable, with some individuals showing little or no evidence of increased body iron, yet others showing severe iron loading, tissue damage and clinical sequelae. The majority of genetically predisposed individuals show at least some evidence of iron loading (increased transferrin saturation and serum ferritin), but a minority show clinical symptoms and severe consequences are rare. Thus, the disorder has a high biochemical penetrance, but a low clinical prevalence. Nevertheless, it is such a common condition in Caucasian populations (1:100–200) that it remains an important clinical entity. The phenotypic variability can largely be explained by a range of environmental, genetic and physiological factors. Men are far more likely to manifest significant disease than women, with the latter losing iron through menstrual blood loss and childbirth. Other forms of blood loss, immune system influences, the amount of bioavailable iron in the diet and lifestyle factors such as high alcohol intake can also contribute to iron loading and disease expression. Polymorphisms in a range of genes have been linked to variations in body iron levels, both in the general population and in hemochromatosis. Some of the genes identified play well known roles in iron homeostasis, yet others are novel. Other factors, including both co-morbidities and genetic polymorphisms, do not affect iron levels per se, but determine the propensity for tissue pathology.
Keywords: Hemochromatosis, HFE, iron overload, environmental modifiers, genetic modifiers
Introduction
Haemochromatosis can be defined in a number of ways, but, in its broadest sense, it refers to a disease associated with excess iron in the body (1,2). Where that iron comes from is an important consideration. Iron loading syndromes that result from defects in proteins directly involved with body iron acquisition (dietary iron absorption) or its regulation are known as primary iron loading disorders. Secondary iron loading usually results from blood transfusions, which are frequently used to treat haemoglobinopathies or hemolytic syndromes, but excess iron can also be associated with various chronic liver diseases, including alcoholic liver disease. Conditions that require transfusion also frequently exhibit ineffective erythropoiesis, and this also drives an increase in dietary iron absorption. Because humans have a very limited capacity to excrete iron, excess iron can remain in the body for a considerable time.
This review will concern itself largely with primary iron overload and particularly hemochromatosis resulting from mutations in the homeostatic iron regulator (HFE) gene (3,4). HFE-related hemochromatosis (HH) is one of a family of mechanistically related diseases characterized by reduced expression of the iron regulatory hormone hepcidin (see below), but it is by far the most prevalent of these disorders and the one encountered most frequently clinically. HH is almost exclusively a Caucasian disease of northwestern European origin, and a single mutation in the HFE gene is responsible for most disease (5). Homozygosity for this mutation is found at highest frequency in Ireland [1:83], but is also prevalent (usually around 1:200) in other parts of Europe and globally where there are northern European-derived populations (6-9).
In hemochromatosis, intestinal iron absorption is inappropriately high for a given body iron load (10), and this characteristic, combined with the limited capacity of humans to excrete iron (11), leads to iron accumulation and its deposition in many body tissues. Although iron is essential for normal cellular function, it is also toxic when present in excess (12), and it is iron-related free radical-mediated tissue damage that underlies the clinical consequences of the condition.
The hemochromatosis phenotype
A typical person genetically predisposed to HH will accumulate extra iron from their diet from birth (3,4). The extra iron amounts to only several milligrams each day, so it can take decades for the body to accumulate sufficient iron to lead to clinical manifestations. Indeed, many individuals do not progress to clinical disease at all. Iron demands are particularly high during the growing years, so young people with HFE mutations rarely show overt symptoms. Nevertheless, even in childhood, evidence of iron overload may be apparent in the form of increased transferrin saturation (TSAT) in genetically predisposed individuals, and this correlates with the development of iron overload in young adult life (13). With the onset of menstruation, females also regularly lose iron through menstrual blood loss which slows their rate of iron accretion. Symptomatic HH normally appears between 40 and 60 years in men, and not until after menopause in women (3,4). Some forms of hemochromatosis lead to early iron loading and tissue damage, and such juvenile hemochromatosis will be considered briefly below.
The most useful markers of body iron status are TSAT, which reflects the amount of recently absorbed iron and iron being trafficked around the body, and serum ferritin (SF), which represents body iron stores (14). A raised TSAT is the first indication of excess iron, and SF rises later after iron has accumulated in the tissues (15). Newly absorbed iron first binds to transferrin (TF) in the blood and is transported to sites of iron utilization and storage (16). Under normal circumstances, TF is approximately 30% saturated with iron, giving it a considerable buffering capacity to deal with excess iron. In HH, the TSAT is typically greater than 60% in men or 50% in women, but can become fully saturated with high iron loading (17). In this situation, the level of non-transferrin-bound iron (NTBI) in the blood increases. NTBI is normally only present in the circulation at extremely low concentrations, but in HH it can reach much higher levels (18,19). This form of iron is very readily taken up by tissues and can be particularly toxic to cells (17,20). It seems likely that NTBI is the major form in which iron is delivered to the tissues in HH (17).
Once within cells, any iron not immediately used for metabolic purposes is stored in ferritin, a large, multi-subunit protein that acts as a nanocage to sequester iron in a non-damaging form (21). It is synthesized on demand as the cellular iron content increases, and intracellular ferritin levels can become extremely high, particularly in specialized iron storage cells, such as hepatocytes. Ferritin-bound iron can be mobilized if the systemic demand for iron rises, and this is why phlebotomy is such as effective treatment for HH. Small amounts of ferritin can also be released into the circulation, and the levels of this SF are proportional to body iron stores. This makes SF an extremely useful clinical marker for diagnosing and monitoring HH. The upper limit of the SF normal range is approximately 300 µg/L in men and 200 µg/L in women (14). In HH, SF usually, but not always, exceeds the normal range and can even reach several thousand. Levels of SF above 1,000 µg/L are strongly correlated with increased risk of tissue damage, and even death (22-24), but lower iron loads, giving SF values between the upper limit of normal and 1,000 µg/L, have also been shown to be clinically significant (25,26). Since SF can also be increased by inflammation (21), or released from damaged tissues, an elevated value is not specific for iron loading, although in the absence of inflammation or significant infection (e.g., metabolic syndrome, hepatitis), SF provides a reliable index of the body iron load.
Many tissues can become iron loaded in HH, with the liver, heart, pancreas, anterior pituitary, and joints being particularly severely affected (3). Consequently, clinical signs can include liver dysfunction, arthropathy, increased skin pigmentation, cardiomyopathy and diabetes mellitus. Iron loading is usually progressive, as is the development of symptoms (15), but the clinical picture can be quite variable, with different individuals manifesting different symptoms and at different times. Early symptoms can include lethargy, weakness, abdominal pain and weight loss. Since the liver is the first major organ that is exposed to newly absorbed iron, it is not surprising that liver iron levels can rise early in HH and become quite high (27). In a typical adult male, the amount of storage iron, i.e., iron in excess of immediate metabolic needs, is approximately 1 g, and in women it is approximately 300 mg (28). Much of this storage iron is in the liver. In HH, the amount of iron in the liver can reach 40 g or more (3). It is a testimony to the storage capacity of intracellular ferritin that it can sequester a large amount of iron for a long time before tissue damage becomes apparent. Advanced HH is associated with fibrosis and cirrhosis of the liver, and affected individuals are at a much higher risk of developing hepatocellular carcinoma (HCC), portal hypertension and end-stage liver disease (29). A large North American HH screening study (HEIRS) showed that men who were homozygous for the major HFE mutation had an odds ratio of 3.3 for liver disease (9). While in the past, a liver biopsy followed by the chemical measurement of its iron content was required to accurately assess the body iron load, the contemporary approach is to use magnetic resonance imaging (MRI) following by post-scan analysis of the data to generate an iron concentration (30). This method is also useful to assess body iron levels when the SF may be unreliable due to concomitant disease.
Arthropathy is another common feature of HH, and affects 25–50% of symptomatic patients (31,32). It particularly involves the second and third metacarpophalangeal joints of the hands initially, but can progress to involve other joints including the hips, wrists, ankles and knees. HH can also be associated with various bone abnormalities including reduced bone mass, altered bone microarchitecture, osteoporosis, osteopenia and fractures (33,34).
The pancreas, heart and anterior pituitary are also important sites of iron accumulation (3,4). Increased iron in the pancreatic Islets can reduce insulin production, and diabetes mellitus is a frequent complication of HH. In the heart, iron accumulates in the cardiomyocytes and this can lead to congestive heart failure and arrhythmias when iron loading is severe (35,36). Interestingly, despite these negative effects on cardiac function, HH is associated with a lower risk of coronary artery disease (37). Using a combination of human data and murine models to investigate the mechanisms underlying this phenomenon, it was demonstrated that HFE represses hepatocyte low-density lipoprotein (LDL) receptor expression, and consequently dysfunctional HFE is associated with increased receptor levels and enhanced plasma LDL-cholesterol clearance (38). Iron accumulation in the anterior pituitary can affect the production of a range of hormones and this characteristic underlies the hypogonadism that often accompanies the disease (39).
Treatment of most cases of hemochromatosis involves therapeutic phlebotomy (40). The removal of each 100 mL of blood removes approximately 40–50 mg of iron, so aggressive phlebotomy can rapidly deplete body iron levels. Most of the adverse consequences of HH resolve following iron removal, with the extent of resolution being dependent on the level of tissue damage prior to the commencement of phlebotomy (41-43). If the disease is treated early and before severe tissue pathology, an essentially complete return to normality can be achieved. However, if tissue damage is severe, e.g., advanced cirrhosis of the liver, recovery may be more limited. Individuals diagnosed and treated early have a normal life expectancy (44,45), while those diagnosed after the development of cirrhosis have a decreased life expectancy, even if they are de-ironed by phlebotomy (9). Some clinical features of HH may not resolve following phlebotomy, including the arthropathy and increased risk of liver cancer, suggesting that early and irreversible changes can occur in some tissues (46,47).
Hemochromatosis genetics
Although hemochromatosis was named as a disease over 130 years ago (48), for most of that time its precise origin was unclear. Marcel Simon in 1976 recognized that HH was an inherited disease that was linked to the human leukocyte antigen (HLA) region on human chromosome 6 (49). However, it was not until 1996 that the affected gene was identified as HFE (5), and thereafter the genetics of HH became well established. HH is the most common recessive, autosomally inherited genetic disease in humans. Almost all clinical HH is associated with homozygosity for a mutation which leads to the substitution of a tyrosine residue for a cysteine at position 282 of the HFE protein. This mutation is usually simply referred to as C282Y (or pCys282Tyr). The high prevalence of this mutation indicates a very strong founder effect, and potentially there is some selective advantage for the mutation as it has been retained in the gene pool for at least 4,000 years (50). The only other prevalent mutation in HFE leads to the H63D substitution. This mutation on its own is rarely associated with significant iron loading (9,51,52), but when present in compound heterozygosity with C282Y, moderate iron loading has been observed in some individuals. However, they are at no or only slightly increased risk of clinical sequelae (9,37,53), and often in these cases there are comorbidities or contributing environmental factors. Multiple other mutations in HFE have been described, but these are rare (54-56). Interestingly, deletion of the HFE gene is the most common cause of HH in Sardinia (57). The identification of the C282Y variant as the cause of the great majority of HH greatly simplified diagnosis, and today, testing for this mutation is routine when patients present with consistently raised SF and TSAT. In some cases, a typical clinical picture of HH may be seen, but the C282Y variant may not be present. In such cases more comprehensive testing of the genome may be warranted to search for other variants in HFE or other iron-related genes (46,58).
Within a few years of the identification of HFE, studies into the basic regulation of iron homeostasis, mainly in mice initially, led to the identification of the liver-derived peptide hormone hepcidin as the “master” regulator of body iron homeostasis (59,60). Hepcidin is secreted predominantly by the liver and regulates iron entry into the plasma by binding to the iron export protein ferroportin (FPN) and causing its internalisation and subsequent degradation (61). The expression of hepcidin itself is inversely related to body iron requirements (59,60). When requirements are high, hepcidin levels are low, more iron is released from stores, and more iron is absorbed from the diet. When iron requirements are low, such as in an iron loading situation, hepcidin levels are high and iron entry into the plasma is reduced. One might expect that in HH, hepcidin levels would be high because of increased body iron, but in fact, hepcidin expression is considerably reduced in HH (62). This explains why people with HH continue to absorb iron even though they are already carrying a considerable iron load, as they are not able to increase their hepcidin levels to restrict iron intake. These studies demonstrate that HFE acts as an upstream regulator of hepcidin (63). Interestingly, hepcidin expression can be increased by inflammation by a pathway that is independent of HFE (64), and thus chronic inflammation has the potential to suppress the iron loading phenotype (65).
Not all forms of inherited iron loading can be explained by mutations in the HFE gene (type 1 HH), and genetic analyses of these disorders have led to the identification of several other key players in body iron homeostasis (Table 1). Patients with mutations in the gene which encodes transferrin receptor 2 (TFR2) develop hemochromatosis (type 3) that is phenotypically very similar to HFE-related disease, although perhaps a little more severe (66). There are also very severe forms of iron loading known as juvenile hemochromatosis (type 2) where iron accumulates rapidly in the body in early postnatal life, with consequent early clinical presentation. Mutations in two genes have been found to lead to juvenile hemochromatosis, HFE2, the gene encoding hemojuvelin (HJV) (type 2A) and HAMP, which encodes hepcidin itself (type 2B) (67,68). Types 1–3 hemochromatosis share the common feature of reduced hepcidin expression, which provides the mechanistic basis for the iron loading (1,2,63). In the case of HFE- and TFR2-related HH, the reduction in hepcidin is relatively small and the iron loading is gradual. In the case of HJV- and HAMP-related disease, negligible hepcidin is produced and iron loading is rapid and severe. Since the hepcidin regulatory pathway plays such an important role in determining body iron levels, it might be expected that mutations in FPN, the molecular target of hepcidin, could also result in iron loading. This is indeed the case and many mutations in FPN have been described (69,70). Some of these affect the binding of hepcidin to FPN, meaning that hepcidin is unable to effectively reduce FPN levels (61,71) and thus regulate iron absorption. These patients (HH type 4B) have an elevated SF and TSAT, like patients with HFE-associated HH. Other mutations restrict the ability of FPN to transport iron (69,70) (HH type 4A or FPN disease). Affected individuals accumulate iron in various organs, but they do not absorb dietary iron efficiently, so SF is increased, but not TSAT. Interestingly, such patients develop relatively few clinical symptoms despite high tissue iron (70).
Table 1. Forms of hemochromatosis and some other iron loading disorders.
| Disorder | Gene symbol | OMIM | Type of disorder | Hepcidin level | Iron phenotype and clinical features | |
|---|---|---|---|---|---|---|
| HFE-related hemochromatosis (type 1) | HFE | 235200 | Primary iron overload | Low | Late onset (4th–5th decade of life) parenchymal iron overload; liver disease; arthropathy; hypogonadism; diabetes; cardiomyopathy | |
| Juvenile hemochromatosis (HJV-related) (type 2A) | HFE2 (HJV) | 602390 | Primary iron overload | Very low | Severe early onset (childhood) parenchymal iron overload; cardiac disease; liver cirrhosis; hypogonadism; diabetes; arthropathy | |
| Juvenile hemochromatosis (hepcidin-related) (type 2B) | HAMP | 613313 | Primary iron overload | Very low to absent | ||
| TFR2-related hemochromatosis (type 3) | TFR2 | 604250 | Primary iron overload | Low | Parenchymal iron overload; liver disease; arthropathy; hypogonadism; diabetes; cardiomyopathy; “intermediate” age of onset | |
| FPN-related hemochromatosis (transport defective) (type 4A) (ferroportin disease) | SLC40A1 (FPN1) | 606069 | Primary iron overload | Low to normal† | Reticuloendothelial iron overload (spleen and liver); raised SF but normal or reduced TSAT; fatigue, arthralgia | |
| FPN-related hemochromatosis (hepcidin resistance) (type 4B) | SLC40A1 (FPN1) | 606069 | Primary iron overload | High | Parenchymal iron overload; increased TSAT and SF; similar clinical features to HFE-related hemochromatosis | |
| β-thalassemia | HBB | 613985 | Hemoglobinopathy | Low to normal | Parenchymal and reticuloendothelial iron overload; anemia; reticulocytosis; cardiac diseases; liver disease; transfusion worsens the phenotype | |
| Sickle cell anemia | HBB | 603903 | Hemoglobinopathy | Low to normal | ||
| X-linked sideroblastic anemia | ALAS2 | 300751 | Hemoglobinopathy | Low† | ||
| Pyruvate kinase deficiency | PKLR | 266200 | Hemolytic anemia | Low | Parenchymal iron overload; anemia | |
| Hereditary spherocytosis | Heterogenous | 182900 | Hemolytic anemia | Low | Parenchymal iron loading; anemia; jaundice; splenomegaly | |
| Some other iron-loading disorders | ||||||
| Friedreich ataxia | FXN | 229300 | Mitochondrial iron overload | Unknown† | Mitochondrial iron overload; neurological and heart disease | |
| Hereditary atransferrinemia | TF | 209300 | Plasma protein deficiency | Low | Severe hemosiderosis of the heart and liver; microcytic anemia | |
| Hereditary aceruloplasminemia | CP | 604290 | Plasma protein deficiency | Low | Iron loading in the brain, retina, liver and pancreas; increased SF but low TSAT; movement disorders and cognitive impairment; retinal degeneration; cirrhosis; diabetes mellitus; microcytic anemia | |
†, predicted levels based on current knowledge of hepcidin regulation. ALAS2, delta aminolevulinate synthase 2; CP, ceruloplasmin; FPN, ferroportin; FXN, frataxin; HAMP, hepcidin anti-microbial peptide; HBB, hemoglobin beta; HJV, hemojuvelin; OMIM, Online Mendelian Inheritance in Man; PKLR, pyruvate kinase, liver and red blood cell; SLC40A1, solute carrier family 40 (iron-regulated transporter), member 1; TF, transferrin; TFR2, transferrin receptor 2; TSAT, transferrin saturation.
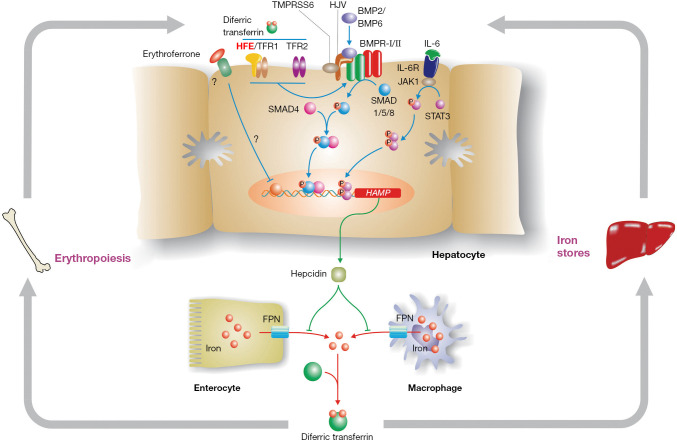
Some knowledge of the hepcidin regulatory pathway (Figure 1) is helpful in understanding disease penetrance in hemochromatosis (59,60). To modulate body iron homeostasis, hepcidin must respond to multiple signals, including body iron load, changes in erythropoiesis, inflammation, hypoxia and other stimuli. The bone morphogenetic protein (BMP)/SMAD signalling pathway is the central pathway modulating hepcidin expression. BMPs act through their cell surface receptors to phosphorylate SMAD1/5/8 which binds to SMAD4. The resulting complex enters the nucleus where it binds to and activates the HAMP promoter. Several BMPs have been shown to stimulate this pathway, including BMP2 and BMP6, and at least BMP6 is responsive to cellular iron levels. For hepcidin to be regulated by inflammation, an intact BMP/SMAD signalling pathway is necessary, but not sufficient. Pro-inflammatory cytokines, notably IL-6, will stimulate HAMP expression via the JAK/STAT3 signalling pathway. Several signals appear to be involved in communicating the iron requirements of the erythroid marrow to hepcidin, with erythroferrone being the best studied (72). This remains an area of active investigation.
Figure 1.
The hepcidin pathway and the regulation of body iron homeostasis. Hepcidin is the master regulator of body iron homeostasis. It is synthesized predominantly by hepatocytes and, after secretion, binds to the iron export protein ferroportin (FPN) and removes it from the surface of target cells. This in turn decreases iron export from these cells. Macrophages and intestinal enterocytes are prime targets, but most cells express FPN on their surface. The HAMP gene, which encodes hepcidin, is regulated by a complex series of upstream signalling pathways. The bone morphogenetic protein (BMP)/SMAD pathway is the core regulatory pathway that responds to body iron requirements. Mutations in various proteins that modulate signalling through this pathway lead to hemochromatosis by reducing hepcidin expression. These include hemojuvelin (HJV) which acts as a BMP co-receptor, and homeostatic iron regulator (HFE) and transferrin receptor 2 (TFR2), which modulate signalling through the BMP/SMAD pathway via mechanisms that are not yet fully understood. Increased body iron levels normally stimulate hepcidin expression via the BMP/SMAD pathway, while proinflammatory cytokines increase hepcidin by signalling through the JAK/STAT pathway. The suppression of hepcidin by enhanced erythropoiesis is, at least in part, mediated by erythroferrone.
While the roles of HJV, hepcidin and FPN are now well-defined, precisely how HFE and TFR2 influence HAMP expression is not fully understood. However, there is compelling evidence that both proteins are required for optimal signalling through the BMP/SMAD pathway (73). The proteins may form part of a large multi-protein complex with BMP receptors and co-receptors that enhances HJV-mediated BMP-SMAD signalling (74), but this has yet to be proven in vivo. Interestingly, mice lacking both HFE and TFR2 have a more severe phenotype than mice lacking either gene alone (75), suggesting that the roles of the two proteins are not fully interchangeable. A similar situation has been observed in humans (76).
Penetrance of HFE-associated hemochromatosis
The phenotypic presentation of individuals who are homozygous for the C282Y variant can be quite variable. Some individuals never accumulate significant amounts of iron nor show clinical manifestations, others show biochemical evidence of iron loading (increased SF and TSAT) but no clinical consequences, whereas others show iron loading in multiple tissues and clinical sequelae (3,4). The majority of C282Y homozygotes will not develop significant disease, even if they have raised iron indices, so HH is a disorder with low clinical penetrance. In a study published by Beutler and colleagues in 2002 that stimulated discussion in this area, only 1 of 152 C282Y homozygotes met their definition of “frank clinical hemochromatosis” (77). However, many considered this figure to be unrealistically low. The reconciliation of these two views comes with the understanding of how penetrance is defined. If a biochemical definition is used, the penetrance of C282Y homozygosity is quite high (in the Beutler study, SF was raised in 76% of men and 54% of women), but if a clinical definition of phenotype is used, penetrance is much lower. The clinical phenotype itself can vary from very subtle changes (e.g., fatigue, reduced quality of life) to the more overt consequences of pathological damage to organs. If mortality is used as an end point, then penetrance is very low indeed. Most clinicians would agree that severe iron-related disease is very infrequent in HH, but many C282Y homozygotes will develop some clinical signs, even if subtle. There is certainly evidence that iron loading does not need to be at a level that precipitates severe disease and organ damage to lead to adverse health outcomes (25,26).
Biochemical penetrance in HH is simple to ascertain as it is based on objective measurements such as SF, TSAT or body iron load. In most HH patients, the TSAT is often well above the normal range, and the SF concentration is also usually increased. The latter can range from mildly (300–1,000 µg/L) to grossly (>1,000 µg/L) elevated. In two large population-based studies of HH, up to 84% of male and 73% of female C282Y homozygotes had a raised TSAT, and 88% of male and 57% of female homozygotes had a raised SF (9,52). As would be expected from their limited physiological loss of iron, biochemical penetrance of HH is much higher in men than it is in women (9,52). Even though HH is a progressive iron loading disorder, the rate of body iron acquisition can vary. It is usually greatest in the first few decades of life, but thereafter diminishes and the SF level may remain steady or even decline (9,52,78). This is not unduly surprising. In HFE-related HH, hepcidin levels are reduced, but not absent, so the body retains some capacity to regulate its iron intake, and it will decline as the iron load increases (10).
Only a relatively small number of studies have objectively looked at clinical penetrance in HH. Allen et al. (52) genotyped 31,192 Australians and identified 203 C282Y homozygotes. All homozygotes, and a selection of other genotypes were examined by clinicians who were unaware of the genotype. This study showed that 28.4% of male, but only 1.2% of female, C282Y homozygotes developed iron overload-related disease, with disease being defined as biochemical evidence of iron overload in the presence of at least one clinical symptom (e.g., biopsy-proven fibrosis, HCC, arthritis) (52). Nevertheless, in the same study 82% of men and 54% of women had a raised SF (and 73% and 69% respectively had a raised TSAT), consistent with the observation that a predisposing genotype or raised body iron does not lead to overt disease in most individuals. An earlier study by Whitlock et al. (79) analyzed several longitudinal, population-based studies and found that 38–50% of C282Y homozygotes had a raised SF, but only 10–33% developed HH-related symptoms. These analyses appear representative of the true clinical picture. Consistent with their higher iron load, men are by far the most likely to develop end-organ damage in HH (9,52,80). In a more recent study, Pilling et al. examined the large UK Biobank population (451,243 participants; 2,890 C282Y homozygotes) and found that liver disease, diabetes mellitus and rheumatoid arthritis and osteoarthritis were over-represented (with odds ratios ranging from 1.5 to 4.3) in participants homozygous for the C282Y variant (37).
A SF of greater than 1,000 µg/L confers a clearly increased risk of tissue damage and clinical sequelae (23), but are individuals with more modest iron accumulation at increased risk of developing the complications of iron overload? Recent evidence suggests this is the case, with C282Y homozygotes with a SF value between the upper limit of normal and 1,000 µg/L having a lower mortality than the general population following phlebotomy therapy (25). Another study showed that there was a beneficial effect of phlebotomy in C282Y homozygotes with SF vales less than 1,000 µg/L, and in this case the effect was observable after only a few months of iron depletion (26). These studies demonstrate that there is a benefit in treating HH patients with mildly elevated iron indices as it will reduce their risk of subsequent clinical problems.
C282Y homozygosity is responsible for the vast majority of HH, but do other genotypes (particularly C282Y/+, C282Y/H63D, H63D/H63D and H63D/+) confer an increased risk of iron loading? In general, these other genotypes have only a very limited effect. Some C282Y or H63D heterozygotes have increased TSAT and SF, but they do not develop the complications of iron overload (9,51,81). For example, Pedersen and Milman (82) demonstrated that TSAT was elevated in 9% of C282Y heterozygotes, and 8% of H63D heterozygotes, with corresponding SF values of 9% and 12%. Very few in either group had both indices raised. Similarly, H63D homozygotes show very low biochemical penetrance. A small number may have raised iron indices, but clinical consequences are rare (83). The situation is a little different for C282Y/H63D compound heterozygotes. These individuals are more likely to have raised iron indices than simple heterozygotes (9,52), and in one study, 0.5–2.0% of compound heterozygotes developed clinical evidence of iron overload (53). In the HEIRS study, male compound heterozygotes were also more likely to report a history of liver disease (9), but a more recent meta-analysis did not show a link between compound heterozygosity and liver cirrhosis (84). Many compound heterozygotes who do show clinical sequelae will present with a co-morbidity (e.g., fatty liver, viral hepatitis), so it is often difficult to attribute their disease symptoms solely to increased iron (15).
Environmental, physiological and genetic modifiers of the HFE-hemochromatosis phenotype
A range of genetic, physiological and environmental factors can contribute to the variable penetrance in HH, and these can also operate at different levels. There may be variation in components of the iron homeostatic machinery such that different individuals take up and/or store different amounts of iron. Alternatively, the amount of iron accumulated by two individuals may be similar, but the pathological consequences of that iron may differ. For example, one individual may be particularly susceptible to the development of hepatic fibrosis. It is the net effect of these influences that determines whether someone genetically predisposed to HH will develop disease and how severe that will be.
Environmental and physiological factors play an important role in determining the severity of HH. This is perhaps best illustrated by the sex difference in the prevalence of iron loading and HH-related disease (9,37,52). Women and men do not fundamentally differ in their iron absorption mechanism, but women lose much more iron than men during their lifetime through both menstrual bleeding and childbirth (85,86), and testosterone suppresses hepcidin, favouring higher iron intake in men (87). Thus, it takes women much longer to accumulate iron to clinically significant levels, and this never happens at all in many women. After the menopause, women and men accumulate iron at similar rates (88). Other forms of blood loss, usually pathological, can also ameliorate the HH phenotype, and regular blood donation can limit body iron accumulation and hence disease expression, although this is not always the case (89-91).
Another physiological modulator to iron loading is the immune system. Interest in this area was piqued soon after the cloning of the HFE gene with the recognition that HFE was a non-classical major histocompatibility complex (MHC) class I-like protein (5), and is consistent with the earlier association of HH with the HLA system (49). However, even before HFE was identified, it was recognized that HH patients with more severe iron loading had abnormally high CD4/CD8 lymphocyte ratios (92) and that this reflected constitutively low CD8+ lymphocyte numbers (93-96). Although the mechanisms are not fully understood, HFE may act as a suppressor of CD8+ T cell activation and differentiation (97,98). In addition, primary defects in the immune system per se can lead to iron loading, and this could influence the hemochromatosis phenotype. Not only do mice lacking both HFE and β2-microglobulin have more severe iron overload than mice lacking only HFE (99), but mice lacking classical MHC class I proteins also accumulate excess iron (100,101). Indeed, extended HLA haplotypes have been associated with variations in iron loading in HH (95,96,102). The mechanisms linking both the adaptive and innate immune systems to iron homeostasis are only partly understood and this represents a fruitful area for further investigation.
In HFE-related HH, age is an important consideration as iron levels progressively increase over time in many, but not all, individuals. The older a person is, the more likely they are to have accumulated a sufficiently large amount of iron to show the clinical manifestations of the disease (103). Symptoms are rarely observed in younger patients and it is usually the 4th or 5th decade of life before significant health problems arise (3,4).
In most cases, the iron content of the diet is unlikely to exert a major influence on iron loading in HH, but it can be a contributing factor (104,105). Vegetarians and vegans will be relatively protected from iron loading and may take longer to manifest signs of HH, whereas C282Y homozygotes who eat a large amount of red meat may load relatively quickly (91). In addition, factors which influence the efficiency of iron absorption, either positively or negatively, could contribute in a small way to body iron load. For example, proton pump inhibitors may reduce dietary iron absorption, and individuals taking iron supplements or substances that increase iron absorption (such as large doses of vitamin C) may take up relatively more iron (106,107). C282Y homozygotes do not generally need to be too restrictive about their diet, but they should limit their intake of foods containing large amounts of iron or substances that may stimulate its absorption (89).
Other clinical conditions can also influence disease expression in HH. For example, excess alcohol consumption is frequently associated with an increased body iron load and this reflects a reduction in hepcidin expression (108,109). Also, C282Y homozygotes who consume excess alcohol have more severe liver disease and are more likely to progress to cirrhosis (110,111). The incidence of non-alcoholic fatty liver disease (NAFLD) is rising globally and it too can contribute to disease expression in HH (112,113). Even in the absence of HFE mutations, many patients with NAFLD develop mild iron overload (dysmetabolic iron overload syndrome) (114). Hepcidin levels are inappropriately high in NAFLD, likely reflecting increased inflammation, but why this does not limit iron absorption is unclear, and suggests the possibility of hepcidin resistance (115). Increased iron indices have also been demonstrated in autoimmune and viral hepatitis, and these correlate with inappropriately low hepcidin expression and potentially increased iron absorption, but hepcidin regulation in these conditions appears to be complex (113). Experimental studies have shown that the combination of iron and other hepatic toxins increases the severity of liver pathology (116-118).
Although environmental factors or co-morbidities appear to account for the majority of variation in the penetrance of HH, genetic modifiers also play a role. Perhaps the clearest demonstration that genetics contributes to variations in body iron status comes from studies with inbred mouse strains which vary widely in their capacity to accumulate and store iron (119,120). Any mutations/polymorphisms that affect the activity of proteins involved in iron homeostasis, and notably the hepcidin regulatory pathway, have the potential to alter body iron levels in HH.
A number of studies have specifically looked for genetic modifiers of HFE-related HH, while others have sought genetic explanations for variations in iron status in the general population that could also influence the HH phenotype. Some of these are summarized in Table 2. Some of the modifying variants are in genes encoding proteins of iron metabolism, so their involvement is not surprising. These include the hepcidin regulators BMP2 (134) and TMPRSS6 (133), and the iron reductase CYBRD1 (122,123), which is predicted to be involved in iron absorption, and the iron transport protein TF (132), but others are novel (121). One of these is GNPAT which encodes an enzyme involved in peroxisomal lipid metabolism (124,125). Precisely how GNPAT contributes to iron homeostasis is unclear, but in vitro studies have shown it to be a potential regulator of hepcidin expression (124). The examination of GNPAT variants in other populations has shown mixed results, some supporting its involvement in modulating HH risk, but others not (126-128). Polymorphisms in yet other genes (such as PCSK7 and PNPLA3) have been associated with liver disease risk (129-131), and these also may modulate the HH phenotype. The number of genetic modifiers of HH identified will undoubtedly increase in time, but it is most likely we are dealing with multiple polymorphisms having small effects rather than a few modifiers with large effects.
Table 2. Some genes containing polymorphisms/mutations that may potentially modify the HFE-related hemochromatosis phenotype.
| Gene symbol | Gene product | Function | Reference |
|---|---|---|---|
| ARNTL | Aryl hydrocarbon receptor nuclear translocator-like | Linked to TF expression; involved in circadian rhythm generation | (121) |
| BMP2 | Bone morphogenetic protein 2 | Upstream positive regulator of hepcidin | (121) |
| CYBRD1 | Duodenal cytochrome B | Iron reductase that may be involved in dietary iron absorption | (122,123) |
| FADS2 | Fatty acid desaturase 2 | Linked to TF expression; involved in lipid metabolism; changes in lipid and iron homeostasis are frequently associated | (121) |
| GNPAT | Glyceronephosphate O-acyltransferase | Peroxisomal protein involved in the production of plasmalogens, a type of lipid | (124-128) |
| NAT2 | N-acetyltransferase 2 | Linked to TF expression; involved in xenobiotic metabolism; link to iron homeostasis unclear. | (121) |
| PCSK7 | Proprotein convertase subtilisin/kexin type 7 | Serine protease involved in processing proproteins in the constitutive secretory pathway | (129,130) |
| PNPLA3 | Patatin like phospholipase domain-containing protein 3 (or 1-acylglycerol-3-phosphate O-acyltransferase or adiponutrin) | A multifunctional enzyme with both triacylglycerol lipase and acylglycerol O-acyltransferase activity; involved in lipid metabolism in adipocytes | (131) |
| TF | Transferrin | The major plasma iron transport protein | (121,132) |
| TMPRSS6 | Transmembrane serine protease 6 | Upstream negative regulator of hepcidin | (133) |
Summary and conclusions
There is extensive clinical experience with HH and, in most cases, the disorder can be easily diagnosed, particularly since the advent of genetic testing, and readily treated by phlebotomy. Biochemical penetrance in C282Y homozygotes is relatively high, but clinical penetrance is relatively low, and clinical sequelae are much more likely to be seen in men than in women. Variations in penetrance between individuals reflect a combination of physiological, environmental and genetic factors, and co-morbidities. The physiological and environmental factors are broadly appreciated, but studies of genetic modifiers are in their relative infancy. In time we will learn more about polymorphisms that affect iron loading and/or disease outcome, but we are likely looking at a genetic landscape where multiple loci each contribute a small effect. In the great majority of cases, HH patients will continue to be monitored using conventional blood iron status parameters, with the use of non-invasive imaging to monitor organ iron load and tissue pathology, and phlebotomy to deplete accumulated iron. However, with an increasingly advanced understanding of iron homeostasis mechanisms and disease modifiers, and bespoke therapeutic options on our doorstep, we now have the tools available to diagnose and treat unusual iron overload cases when they present.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editors (Ralf Weiskirchen and Wolfgang Stremmel) for the series “Unresolved Basic Issues in Hepatology” published in Annals of Translational Medicine. The article has undergone external peer review.
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-5512
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5512). The series “Unresolved Basic Issues in Hepatology” was commissioned by the editorial office without any funding or sponsorship. Dr. GJA reports personal fees from Protagonist Therapeutics, other from PharmaNutra, outside the submitted work. EBJ has no other conflicts of interest to declare.
References
- 1.Brissot P, Troadec MB, Loréal O, Brissot E. Pathophysiology and classification of iron overload diseases; update 2018. Transfus Clin Biol 2019;26:80-8. 10.1016/j.tracli.2018.08.006 [DOI] [PubMed] [Google Scholar]
- 2.Piperno A, Pelucchi S, Mariani R. Inherited iron overload disorders. Transl Gastroenterol Hepatol 2020;5:25. 10.21037/tgh.2019.11.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brissot P, Pietrangelo A, Adams PC, et al. Haemochromatosis. Nat Rev Dis Primers 2018;4:18016. 10.1038/nrdp.2018.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kowdley KV, Brown KE, Ahn J, Sundaram V. ACG Clinical Guideline: Hereditary hemochromatosis. Am J Gastroenterol 2019;114:1202-18. 10.14309/ajg.0000000000000315 [DOI] [PubMed] [Google Scholar]
- 5.Feder JN, Gnirke A, Thomas W, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 1996;13:399-408. 10.1038/ng0896-399 [DOI] [PubMed] [Google Scholar]
- 6.Byrnes V, Ryan E, Mayne P, et al. Neonatal screening for the HFE mutations in the Irish population. Gut 1999;1:527A. [Google Scholar]
- 7.Milman N, Pedersen P, Steig T, et al. Frequencies of the hereditary hemochromatosis allele in different populations. Comparison of previous phenotypic methods and novel genotypic methods. Int J Hematol 2003;77:48-54. 10.1007/BF02982602 [DOI] [PubMed] [Google Scholar]
- 8.Olynyk JK, Cullen DJ, Aquilia S, et al. A population-based study of the clinical expression of the hemochromatosis gene. N Engl J Med 1999;341:718-24. 10.1056/NEJM199909023411002 [DOI] [PubMed] [Google Scholar]
- 9.Adams PC, Reboussin DM, Barton JC, et al. Hemochromatosis and iron-overload screening in a racially diverse population. N Engl J Med 2005;352:1769-78. 10.1056/NEJMoa041534 [DOI] [PubMed] [Google Scholar]
- 10.McLaren GD, Nathanson NH, Jacobs A, et al. Regulation of intestinal iron absorption and mucosal iron kinetics in hereditary hemochromatosis. J Lab Clin Med 1991;117:390-401. [PubMed] [Google Scholar]
- 11.Widdowson EM, McCance RA. The absorption and excretion of iron before, during and after a period of very high intake. Biochem J 1937;31:2029-34. 10.1042/bj0312029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galaris D, Barbouti A, Pantopoulos K. Iron homeostasis and oxidative stress: An intimate relationship. Biochim Biophys Acta Mol Cell Res 2019;1866:118535. 10.1016/j.bbamcr.2019.118535 [DOI] [PubMed] [Google Scholar]
- 13.Porto G, Cruz E, Teles MJ, et al. HFE Related Hemochromatosis: Uncovering the Inextricable Link between Iron Homeostasis and the Immunological System. Pharmaceuticals (Basel) 2019;12:122. 10.3390/ph12030122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch S, Pfeiffer CM, Georgieff MK, et al. Biomarkers of Nutrition for Development (BOND)-Iron Review. J Nutr 2018;148:1001S-1067S. 10.1093/jn/nxx036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powell LW, Seckington RC, Deugnier Y. Haemochromatosis. Lancet 2016;388:706-16. 10.1016/S0140-6736(15)01315-X [DOI] [PubMed] [Google Scholar]
- 16.Anderson GJ, Vulpe CD. Mammalian iron transport. Cell Mol Life Sci 2009;66:3241-61. 10.1007/s00018-009-0051-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brissot P, Ropert M, Le Lan C, et al. Non-transferrin bound iron: a key role in iron overload and iron toxicity. Biochim Biophys Acta 2012;1820:403-10. 10.1016/j.bbagen.2011.07.014 [DOI] [PubMed] [Google Scholar]
- 18.Loréal O, Gosriwatana I, Guyader D, et al. Determination of non-transferrin-bound iron in genetic hemochromatosis using a new HPLC-based method. J Hepatol 2000; 32:727-33. 10.1016/S0168-8278(00)80240-8 [DOI] [PubMed] [Google Scholar]
- 19.Le Lan C, Loréal O, Cohen T, et al. Redox active plasma iron in C282Y/C282Y hemochromatosis. Blood 2005;105:4527-31. 10.1182/blood-2004-09-3468 [DOI] [PubMed] [Google Scholar]
- 20.Cabantchik ZI, Breuer W, Zanninelli G, et al. LPI-labile plasma iron in iron overload. Best Pract Res Clin Haematol 2005;18:277-87. 10.1016/j.beha.2004.10.003 [DOI] [PubMed] [Google Scholar]
- 21.Theil EC. Ferritin: the protein nanocage and iron biomineral in health and in disease. Inorg Chem 2013;52:12223-33. 10.1021/ic400484n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guyader D, Jacquelinet C, Moirand R, et al. Noninvasive prediction of fibrosis in C282Y homozygous hemochromatosis. Gastroenterology 1998;115:929-36. 10.1016/S0016-5085(98)70265-3 [DOI] [PubMed] [Google Scholar]
- 23.Allen KJ, Bertalli NA, Osborne NJ, et al. HFE Cys282Tyr homozygotes with serum ferritin concentrations below 1000 µg/L are at low risk of hemochromatosis. Hepatology 2010;52:925-33. 10.1002/hep.23786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barton JC, Barton JC, Acton RT, et al. Increased risk of death from iron overload among 422 treated probands with HFE hemochromatosis and serum levels of ferritin greater than 1000 µg/L at diagnosis. Clin Gastroenterol Hepatol 2012;10:412-6. 10.1016/j.cgh.2011.11.032 [DOI] [PubMed] [Google Scholar]
- 25.Bardou-Jacquet E, Morcet J, Manet G, et al. Decreased cardiovascular and extrahepatic cancer-related mortality in treated patients with mild HFE hemochromatosis. J Hepatol 2015;62:682-9. 10.1016/j.jhep.2014.10.025 [DOI] [PubMed] [Google Scholar]
- 26.Ong SY, Gurrin LC, Dolling L, et al. Reduction of body iron in HFE-related haemochromatosis and moderate iron overload (Mi-Iron): a multicentre, participant-blinded, randomised controlled trial. Lancet Haematol 2017;4:e607-14. 10.1016/S2352-3026(17)30214-4 [DOI] [PubMed] [Google Scholar]
- 27.Mehta KJ, Farnaud SJ, Sharp PA. Iron and liver fibrosis: Mechanistic and clinical aspects. World J Gastroenterol 2019;25:521-38. 10.3748/wjg.v25.i5.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook JD, Skikne BS, Lynch SR, et al. Estimates of iron sufficiency in the US population. Blood 1986;68:726-31. 10.1182/blood.V68.3.726.726 [DOI] [PubMed] [Google Scholar]
- 29.Kowdley KV, Brandhagen DJ, Gish RG, et al. Survival after liver transplantation in patients with hepatic iron overload: the national hemochromatosis transplant registry. Gastroenterology 2005;129:494-503. 10.1016/j.gastro.2005.05.004 [DOI] [PubMed] [Google Scholar]
- 30.Paisant A, d'Assignies G, Bannier E, et al. MRI for the measurement of liver iron content, and for the diagnosis and follow-up of iron overload disorders. Presse Med 2017;46:e279-87. 10.1016/j.lpm.2017.10.008 [DOI] [PubMed] [Google Scholar]
- 31.Carroll GJ, Breidahl WH, Olynyk JK. Characteristics of the arthropathy described in hereditary hemochromatosis. Arthritis Care Res (Hoboken) 2012;64:9-14. 10.1002/acr.20501 [DOI] [PubMed] [Google Scholar]
- 32.Kiely PD. Haemochromatosis arthropathy - a conundrum of the Celtic curse. J R Coll Physicians Edinb 2018; 48:233-8. 10.4997/JRCPE.2018.307 [DOI] [PubMed] [Google Scholar]
- 33.Guggenbuhl P, Deugnier Y, Boisdet JF, et al. Bone mineral density in men with genetic hemochromatosis and HFE gene mutation. Osteoporos Int 2005;16:1809-14. 10.1007/s00198-005-1934-0 [DOI] [PubMed] [Google Scholar]
- 34.Jeney V. Clinical impact and cellular mechanisms of iron overload-associated bone loss. Front Pharmacol 2017;8:77. 10.3389/fphar.2017.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Díez-López C, Comín-Colet J, González-Costello J. Iron overload cardiomyopathy: from diagnosis to management. Curr Opin Cardiol 2018;33:334-40. 10.1097/HCO.0000000000000511 [DOI] [PubMed] [Google Scholar]
- 36.Gulati V, Harikrishnan P, Palaniswamy C, et al. Cardiac involvement in hemochromatosis. Cardiol Rev 2014; 22:56-68. 10.1097/CRD.0b013e3182a67805 [DOI] [PubMed] [Google Scholar]
- 37.Pilling LC, Tamosauskaite J, Jones G, et al. Common conditions associated with hereditary haemochromatosis genetic variants: cohort study in UK Biobank. BMJ 2019;364:k5222. Erratum in: BMJ. 2019 Oct 23;367:l6157. doi: 10.1136/bmj.l6157. 10.1136/bmj.k5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demetz E, Tymoszuk P, Hilbe R, et al. The haemochromatosis gene Hfe and Kupffer cells control LDL cholesterol homeostasis and impact on atherosclerosis development. Eur Heart J 2020; 41:3949-59. 10.1093/eurheartj/ehaa140 [DOI] [PubMed] [Google Scholar]
- 39.McDermott JH, Walsh CH. Hypogonadism in hereditary hemochromatosis. J Clin Endocrinol Metab 2005; 90:2451-5. 10.1210/jc.2004-0980 [DOI] [PubMed] [Google Scholar]
- 40.Loréal O, Cavey T, Robin F, et al. Iron as a Therapeutic Target in HFE-Related Hemochromatosis: Usual and Novel Aspects. Pharmaceuticals (Basel) 2018;11:131. 10.3390/ph11040131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falize L, Guillygomarc'h A, Perrin M, et al. Reversibility of hepatic fibrosis in treated genetic hemochromatosis: a study of 36 cases. Hepatology 2006;44:472-7. 10.1002/hep.21260 [DOI] [PubMed] [Google Scholar]
- 42.Powell LW, Dixon JL, Ramm GA, et al. Screening for hemochromatosis in asymptomatic subjects with or without a family history. Arch Intern Med 2006; 166:294-301. 10.1001/archinte.166.3.294 [DOI] [PubMed] [Google Scholar]
- 43.Bardou-Jacquet E, Morandeau E, Anderson GJ, et al. Regression of fibrosis stage with treatment reduces long-term risk of liver cancer in patients with hemochromatosis caused by mutation in HFE. Clin Gastroenterol Hepatol 2020;18:1851-7. 10.1016/j.cgh.2019.10.010 [DOI] [PubMed] [Google Scholar]
- 44.Adams PC, Speechley M, Kertesz AE. Long-term survival analysis in hereditary hemochromatosis. Gastroenterology 1991;101:368. 10.1016/0016-5085(91)90013-B [DOI] [PubMed] [Google Scholar]
- 45.Niederau C, Fischer R, Pürschel A, et al. Long-term survival in patients with hereditary hemochromatosis. Gastroenterology 1996;110:1107-19. 10.1053/gast.1996.v110.pm8613000 [DOI] [PubMed] [Google Scholar]
- 46.Barton JC, Edwards CQ. HFE Hemochromatosis. 2000 Apr 3 [updated 2018 Dec 6]. In: Adam MP, Ardinger HH, Pagon RA, et al. editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle, 1993–2020. [Google Scholar]
- 47.Prabhu A, Cargill T, Roberts N, et al. Systematic review of the clinical outcomes of iron reduction in hereditary hemochromatosis. Hepatology 2020;72:1469-82. 10.1002/hep.31405 [DOI] [PubMed] [Google Scholar]
- 48.Ulvik RJ. Hereditary haemochromatosis through 150 years. Tidsskr Nor Laegeforen 2016;136:2017-21. 10.4045/tidsskr.15.1003 [DOI] [PubMed] [Google Scholar]
- 49.Simon M, Bourel M, Fauchet R, et al. Association of HLA-A3 and HLA-B14 antigens with idiopathic haemochromatosis. Gut 1976;17:332-4. 10.1136/gut.17.5.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cassidy LM, Martiniano R, Murphy EM, et al. Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc Natl Acad Sci USA 2016;113:368-73. 10.1073/pnas.1518445113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaloumis SG, Allen KJ, Bertalli NA, et al. Natural history of HFE simple heterozygosity for C282Y and H63D: a prospective 12-year study. J Gastroenterol Hepatol 2015;30:719-25. 10.1111/jgh.12804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allen KJ, Gurrin LC, Constantine CC, et al. Iron-overload-related disease in HFE hereditary hemochromatosis. N Engl J Med 2008;358:221-30. 10.1056/NEJMoa073286 [DOI] [PubMed] [Google Scholar]
- 53.Gurrin LC, Bertalli NA, Dalton GW, et al. HFE C282Y/H63D compound heterozygotes are at low risk of hemochromatosis-related morbidity. Hepatology 2009;50:94-101. 10.1002/hep.22972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barton JC, Edwards CQ, Acton RT. HFE gene: Structure, function, mutations, and associated iron abnormalities. Gene 2015;574:179-92. 10.1016/j.gene.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piperno A, Arosio C, Fossati L, et al. Two novel nonsense mutations of HFE gene in five unrelated Italian patients with hemochromatosis. Gastroenterology 2000;119:441-5. 10.1053/gast.2000.9369 [DOI] [PubMed] [Google Scholar]
- 56.Hamdi-Rozé H, Beaumont-Epinette MP, Ben Ali Z, et al. Rare HFE variants are the most frequent cause of hemochromatosis in non-c282y homozygous patients with hemochromatosis. Am J Hematol 2016;91:1202-5. 10.1002/ajh.24535 [DOI] [PubMed] [Google Scholar]
- 57.Le Gac G, Congiu R, Gourlaouen I, et al. Homozygous deletion of HFE is the common cause of hemochromatosis in Sardinia. Haematologica 2010;95:685-7. 10.3324/haematol.2009.016816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McDonald CJ, Ostini L, Wallace DF, et al. Next-generation sequencing: Application of a novel platform to analyze atypical iron disorders. J Hepatol 2015; 63:1288-93. 10.1016/j.jhep.2015.06.027 [DOI] [PubMed] [Google Scholar]
- 59.Ganz T, Nemeth E. Hepcidin and iron homeostasis. Biochim Biophys Acta 2012;1823:1434-43. 10.1016/j.bbamcr.2012.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silvestri L, Nai A, Dulja A, et al. Hepcidin and the BMP-SMAD pathway: An unexpected liaison. Vitam Horm 2019;110:71-99. 10.1016/bs.vh.2019.01.004 [DOI] [PubMed] [Google Scholar]
- 61.Nemeth E, Tuttle MS, Powelson J, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004;306:2090-3. 10.1126/science.1104742 [DOI] [PubMed] [Google Scholar]
- 62.Bridle KR, Frazer DM, Wilkins SJ, et al. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homeostasis. Lancet 2003;361:669-73. 10.1016/S0140-6736(03)12602-5 [DOI] [PubMed] [Google Scholar]
- 63.Darshan D, Frazer DM, Anderson GJ. Molecular basis of iron-loading disorders. Expert Rev Mol Med 2010;12:e36. 10.1017/S1462399410001687 [DOI] [PubMed] [Google Scholar]
- 64.Frazer DM, Wilkins SJ, Millard KN, et al. Increased hepcidin expression and hypoferraemia associated with an acute phase response are not affected by inactivation of HFE. Br J Haematol 2004;126:434-6. 10.1111/j.1365-2141.2004.05044.x [DOI] [PubMed] [Google Scholar]
- 65.van Deuren M, Kroot JJ, Swinkels DW. Time-course analysis of serum hepcidin, iron and cytokines in a C282Y homozygous patient with Schnitzler's syndrome treated with IL-1 receptor antagonist. Haematologica 2009;94:1297-300. 10.3324/haematol.2009.005975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Camaschella C, Roetto A, Calì A, et al. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet 2000;25:14-5. 10.1038/75534 [DOI] [PubMed] [Google Scholar]
- 67.Papanikolaou G, Samuels ME, Ludwig EH, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet 2004;36:77-82. 10.1038/ng1274 [DOI] [PubMed] [Google Scholar]
- 68.Roetto A, Papanikolaou G, Politou M, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet 2003;33:21-2. 10.1038/ng1053 [DOI] [PubMed] [Google Scholar]
- 69.Détivaud L, Island ML, Jouanolle AM, et al. Ferroportin diseases: functional studies, a link between genetic and clinical phenotype. Hum Mutat 2013;34:1529-36. 10.1002/humu.22396 [DOI] [PubMed] [Google Scholar]
- 70.Vlasveld LT, Janssen R, Bardou-Jacquet E, et al. Twenty years of ferroportin disease: a review or an update of published clinical, biochemical, molecular, and functional features. Pharmaceuticals (Basel) 2019;12:132. 10.3390/ph12030132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aschemeyer S, Qiao B, Stefanova D, et al. Structure function analysis of ferroportin defines the binding site and an alternative mechanism of action of hepcidin. Blood 2018;131:899-910. 10.1182/blood-2017-05-786590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kautz L, Jung G, Valore EV, et al. Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat Genet 2014;46:678-84. 10.1038/ng.2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiao X, Alfaro-Magallanes VM, Babitt JL. Bone morphogenic proteins in iron homeostasis. Bone 2020;138:115495. 10.1016/j.bone.2020.115495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.D’Alessio F, Hentze MW, Muckenthaler MU. The hemochromatosis proteins HFE, TfR2, and HJV form a membrane-associated protein complex for hepcidin regulation. J Hepatol 2012;57:1052-60. 10.1016/j.jhep.2012.06.015 [DOI] [PubMed] [Google Scholar]
- 75.Wallace DF, Summerville L, Crampton EM, et al. Combined deletion of Hfe and transferrin receptor 2 in mice leads to marked dysregulation of hepcidin and iron overload, Hepatology 2009;50:1992-2000. 10.1002/hep.23198 [DOI] [PubMed] [Google Scholar]
- 76.Pietrangelo A, Caleffi A, Henrion J, et al. Juvenile hemochromatosis associated with pathogenic mutations of adult hemochromatosis genes. Gastroenterology 2005;128:470-9. 10.1053/j.gastro.2004.11.057 [DOI] [PubMed] [Google Scholar]
- 77.Beutler E, Felitti VJ, Koziol JA, et al. Penetrance of 845G-->A (C282Y) HFE hereditary haemochromatosis mutation in the USA. Lancet 2002;359:211-8. 10.1016/S0140-6736(02)07447-0 [DOI] [PubMed] [Google Scholar]
- 78.Gurrin LC, Osborne NJ, Constantine CC, et al. The natural history of serum iron indices for HFE C282Y homozygosity associated with hereditary hemochromatosis. Gastroenterology 2008;135:1945-52. 10.1053/j.gastro.2008.08.056 [DOI] [PubMed] [Google Scholar]
- 79.Whitlock EP, Garlitz BA, Harris EL, et al. Screening for hereditary hemochromatosis: a systematic review for the U.S. Preventive Services Task Force. Ann Intern Med 2006;145:209-23. 10.7326/0003-4819-145-3-200608010-00009 [DOI] [PubMed] [Google Scholar]
- 80.Gan EK, Powell LW, Olynyk JK. Natural history and management of HFE-hemochromatosis. Semin Liver Dis 2011;31:293-301. 10.1055/s-0031-1286060 [DOI] [PubMed] [Google Scholar]
- 81.Bulaj ZJ, Griffen LM, Jorde LB, et al. Clinical and biochemical abnormalities in people heterozygous for hemochromatosis. N Engl J Med 1996;335:1799-805. 10.1056/NEJM199612123352403 [DOI] [PubMed] [Google Scholar]
- 82.Pedersen P, Milman N. Genetic screening for HFE hemochromatosis in 6,020 Danish men: penetrance of C282Y, H63D, and S65C variants. Ann Hematol 2009;88:775-84. 10.1007/s00277-008-0679-1 [DOI] [PubMed] [Google Scholar]
- 83.Gochee PA, Powell LW, Cullen DJ, et al. A population-based study of the biochemical and clinical expression of the H63D hemochromatosis mutation. Gastroenterology 2002;122:646-51. 10.1016/S0016-5085(02)80116-0 [DOI] [PubMed] [Google Scholar]
- 84.Ye Q, Qian BX, Yin WL, et al. Association between the HFE C282Y, H63D polymorphisms and the risks of non-alcoholic fatty liver disease, liver cirrhosis and hepatocellular carcinoma: An updated systematic review and meta-analysis of 5,758 cases and 14,741 controls. PLoS One 2016;11:e0163423. 10.1371/journal.pone.0163423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rushton DH, Barth JH. What is the evidence for gender differences in ferritin and haemoglobin? Crit Rev Oncol Hematol 2010;73:1-9. 10.1016/j.critrevonc.2009.03.010 [DOI] [PubMed] [Google Scholar]
- 86.Coad J, Conlon C. Iron deficiency in women: assessment, causes and consequences. Curr Opin Clin Nutr Metab Care 2011;14:625-34. 10.1097/MCO.0b013e32834be6fd [DOI] [PubMed] [Google Scholar]
- 87.Latour C, Kautz L, Besson-Fournier C, et al. Testosterone perturbs systemic iron balance through activation of epidermal growth factor receptor signaling in the liver and repression of hepcidin. Hepatology 2014;59:683-94. 10.1002/hep.26648 [DOI] [PubMed] [Google Scholar]
- 88.Warne CD, Zaloumis SG, Bertalli NA, et al. HFE p.C282Y homozygosity predisposes to rapid serum ferritin rise after menopause: a genotype-stratified cohort study of hemochromatosis in Australian women. J Gastroenterol Hepatol 2017. 32:797-802. 10.1111/jgh.13621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Adams PC, Barton JC. How I treat hemochromatosis. Blood 2010;116:317-25. 10.1182/blood-2010-01-261875 [DOI] [PubMed] [Google Scholar]
- 90.Barton JC, Preston BL, McDonnell SM, et al. Severity of iron overload in hemochromatosis: effect of volunteer blood donation before diagnosis. Transfusion 2001; 41:123-9. 10.1046/j.1537-2995.2001.41010123.x [DOI] [PubMed] [Google Scholar]
- 91.Cade JE, Moreton JA, O’Hara B, et al. Diet and genetic factors associated with iron status in middle-aged women. Am J Clin Nutr 2005;82:813-20. 10.1093/ajcn/82.4.813 [DOI] [PubMed] [Google Scholar]
- 92.Reimão R, Porto G, de Sousa M. Stability of CD4/CD8 ratios in man: new correlation between CD4/CD8 profiles and iron overload in idiopathic haemochromatosis patients. C R Acad Sci III 1991;313:481-7. [PubMed] [Google Scholar]
- 93.Porto G, Reimao R, Goncalves C, et al. Haemochromatosis as a window into the study of the immunological system: A novel correlation between CD8+ lymphocytes and iron overload. Eur J Haematol 1994;52:283-90. 10.1111/j.1600-0609.1994.tb00097.x [DOI] [PubMed] [Google Scholar]
- 94.Cruz E, Melo G, Lacerda R, et al. The CD8+ T-lymphocyte profile as a modifier of iron overload in HFE hemochromatosis: An update of clinical and immunological data from 70 C282Y homozygous subjects. Blood Cells Mol Dis 2006;37:33-9. 10.1016/j.bcmd.2006.04.004 [DOI] [PubMed] [Google Scholar]
- 95.Cruz E, Whittington C, Krikler SH, et al. A new 500 kb haplotype associated with high CD8+ T-lymphocyte numbers predicts a less severe expression of hereditary hemochromatosis. BMC Med Genet 2008;9:97. 10.1186/1471-2350-9-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thorstensen K, Kvitland MA, Irgens WO, et al. Iron loading in HFE p.C282Y homozygotes found by population screening: relationships to HLA-type and T-lymphocyte subsets. Scand J Clin Lab Invest 2017;77:477-85. 10.1080/00365513.2017.1342136 [DOI] [PubMed] [Google Scholar]
- 97.Macedo MF, Porto G, Costa M, et al. Low numbers of CD8+ T lymphocytes in hereditary haemochromatosis are explained by a decrease of the most mature CD8+ effector memory T cells. Clin Exp Immunol 2010;159:363-71. 10.1111/j.1365-2249.2009.04066.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Costa M, Cruz E, Oliveira S, et al. Lymphocyte gene expression signatures from patients and mouse models of hereditary hemochromatosis reveal a function of HFE as a negative regulator of CD8+ T-lymphocyte activation and differentiation in vivo. PLoS One 2015;10:e0124246. 10.1371/journal.pone.0124246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Levy JE, Montross LK, Andrews NC. Genes that modify the hemochromatosis phenotype in mice. J Clin Invest 2000;105:1209-16. 10.1172/JCI9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Santos M, Schilham MW, Rademakers LH, et al. Defective iron homeostasis in 2-microglobulin knockout mice recapitulates hereditary hemochromatosis in man. J Exp Med 1996;184:1975-85. 10.1084/jem.184.5.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cardoso EM, Macedo MG, Rohrlich P, et al. Increased hepatic iron in mice lacking classical MHC class I molecules. Blood 2002;100:4239-41. 10.1182/blood-2002-05-1565 [DOI] [PubMed] [Google Scholar]
- 102.Costa M, Cruz E, Barton JC, et al. Effects of highly conserved major histocompatibility complex (MHC) extended haplotypes on iron and low CD8+ T lymphocyte phenotypes in HFE C282Y homozygous hemochromatosis patients from three geographically distant areas. PLoS One 2013;8:e79990. 10.1371/journal.pone.0079990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Olynyk JK, St Pierre TG, Britton RS, et al. Duration of hepatic iron exposure increases the risk of significant fibrosis in hereditary hemochromatosis: a new role for magnetic resonance imaging. Am J Gastroenterol 2005;100:837-41. 10.1111/j.1572-0241.2005.41287.x [DOI] [PubMed] [Google Scholar]
- 104.Moretti D, van Doorn GM, Swinkels DW, et al. Relevance of dietary iron intake and bioavailability in the management of HFE hemochromatosis: a systematic review. Am J Clin Nutr 2013;98:468-79. 10.3945/ajcn.112.048264 [DOI] [PubMed] [Google Scholar]
- 105.Weiss G. Genetic mechanisms and modifying factors in hereditary hemochromatosis. Nat Rev Gastroenterol Hepatol 2010;7:50-8. 10.1038/nrgastro.2009.201 [DOI] [PubMed] [Google Scholar]
- 106.Vanclooster A, van Deursen C, Jaspers R, et al. Proton pump inhibitors decrease phlebotomy need in HFE hemochromatosis: Double-blind randomized placebo-controlled trial. Gastroenterology 2017;153:678-680.e2. 10.1053/j.gastro.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 107.Lane DJ, Richardson DR. The active role of vitamin C in mammalian iron metabolism: much more than just enhanced iron absorption! Free Radic Biol Med 2014;75:69-83. 10.1016/j.freeradbiomed.2014.07.007 [DOI] [PubMed] [Google Scholar]
- 108.Bridle K, Cheung TK, Murphy T, et al. Hepcidin is down-regulated in alcoholic liver injury: implications for the pathogenesis of alcoholic liver disease. Alcohol Clin Exp Res 2006;30:106-12. 10.1111/j.1530-0277.2006.00002.x [DOI] [PubMed] [Google Scholar]
- 109.Hsu CC, Kowdley KV. The effects of alcohol on other chronic liver diseases. Clin Liver Dis 2016;20:581-94. 10.1016/j.cld.2016.02.013 [DOI] [PubMed] [Google Scholar]
- 110.Fletcher LM, Dixon JL, Purdie DM, et al. Excess alcohol greatly increases the prevalence of cirrhosis in hereditary hemochromatosis. Gastroenterology 2002;122:281-9. 10.1053/gast.2002.30992 [DOI] [PubMed] [Google Scholar]
- 111.Scotet V, Merour MC, Mercier AY, et al. Hereditary hemochromatosis: effect of excessive alcohol consumption on disease expression in patients homozygous for the C282Y mutation. Am J Epidemiol 2003;158:129-34. 10.1093/aje/kwg123 [DOI] [PubMed] [Google Scholar]
- 112.Powell EE, Jonsson JR, Clouston AD. Metabolic factors and non-alcoholic fatty liver disease as co-factors in other liver diseases. Dig Dis 2010;28:186-91. 10.1159/000282084 [DOI] [PubMed] [Google Scholar]
- 113.Czaja AJ. Review article: iron disturbances in chronic liver diseases other than haemochromatosis - pathogenic, prognostic, and therapeutic implications. Aliment Pharmacol Ther 2019;49:681-701. 10.1111/apt.15173 [DOI] [PubMed] [Google Scholar]
- 114.Deugnier Y, Bardou-Jacquet É, Lainé F. Dysmetabolic iron overload syndrome (DIOS). Presse Med 2017;46:e306-11. 10.1016/j.lpm.2017.05.036 [DOI] [PubMed] [Google Scholar]
- 115.Rametta R, Dongiovanni P, Pelusi S, et al. Hepcidin resistance in dysmetabolic iron overload. Liver Int 2016;36:1540-8. 10.1111/liv.13124 [DOI] [PubMed] [Google Scholar]
- 116.Tsukamoto H, Horne W, Kamimura S, et al. Experimental liver cirrhosis induced by alcohol and iron. J Clin Invest 1995;96:620-30. 10.1172/JCI118077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tan TC, Crawford DH, Jaskowski LA, et al. Excess iron modulates endoplasmic reticulum stress-associated pathways in a mouse model of alcohol and high-fat diet-induced liver injury. Lab Invest 2013;93:1295-312. 10.1038/labinvest.2013.121 [DOI] [PubMed] [Google Scholar]
- 118.Tan TC, Crawford DH, Jaskowski LA, et al. A corn oil-based diet protects against combined ethanol and iron-induced liver injury in a mouse model of hemochromatosis. Alcohol Clin Exp Res 2013;37:1619-31. 10.1111/acer.12155 [DOI] [PubMed] [Google Scholar]
- 119.McLachlan S, Lee SM, Steele TM, et al. In silico QTL mapping of basal liver iron levels in inbred mouse strains. Physiol Genomics 2011;43:136-47. 10.1152/physiolgenomics.00025.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cavey T, Ropert M, de Tayrac M, et al. Mouse genetic background impacts both on iron and non-iron metals parameters and on their relationships. Biometals 2015;28:733-43. 10.1007/s10534-015-9862-8 [DOI] [PubMed] [Google Scholar]
- 121.Benyamin B, Esko T, Ried JS, et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat Commun 2014;5:4926. Erratum in: Nat Commun. 2015 Mar 30;6:6542. doi: 10.1038/ncomms7542. 10.1038/ncomms5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Constantine CC, Anderson GJ, Vulpe CD, et al. A novel association between a SNP in CYBRD1 and serum ferritin levels in a cohort study of HFE hereditary haemochromatosis. Br J Haematol 2009;147:140-9. 10.1111/j.1365-2141.2009.07843.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pelucchi S, Mariani R, Calza S, et al. CYBRD1 as a modifier gene that modulates iron phenotype in HFE p.C282Y homozygous patients. Haematologica 2012;97:1818-25. 10.3324/haematol.2012.062661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McLaren CE, Emond MJ, Subramaniam VN, et al. Exome sequencing in HFE C282Y homozygous men with extreme phenotypes identifies a GNPAT variant associated with severe iron overload. Hepatology 2015;62:429-39. 10.1002/hep.27711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barton JC, Chen WP, Emond MJ, et al. GNPAT p.D519G is independently associated with markedly increased iron stores in HFE p.C282Y homozygotes. Blood Cells Mol Dis 2017;63:15-20. 10.1016/j.bcmd.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bardou-Jacquet E, de Tayrac M, Mosser J, et al. GNPAT variant associated with severe iron overload in HFE hemochromatosis. Hepatology 2015;62:1917-8. 10.1002/hep.27854 [DOI] [PubMed] [Google Scholar]
- 127.Levstik A, Stuart A, Adams PC. GNPAT variant (D519G) is not associated with an elevated serum ferritin or iron removed by phlebotomy in patients referred for C282Y-linked hemochromatosis. Ann Hepatol 2016;15:907-10. [DOI] [PubMed] [Google Scholar]
- 128.Greni F, Valenti L, Mariani R, et al. GNPAT rs11558492 is not a major modifier of iron status: Study of Italian hemochromatosis patients and blood donors. Ann Hepatol 2017;16:451-6. 10.5604/01.3001.0009.8601 [DOI] [PubMed] [Google Scholar]
- 129.Stickel F, Buch S, Zoller H, et al. Evaluation of genome-wide loci of iron metabolism in hereditary hemochromatosis identifies PCSK7 as a host risk factor of liver cirrhosis. Hum Mol Genet 2014;23:3883-90. 10.1093/hmg/ddu076 [DOI] [PubMed] [Google Scholar]
- 130.Pelucchi S, Galimberti S, Greni F, et al. Proprotein convertase 7 rs236918 associated with liver fibrosis in Italian patients with HFE-related hemochromatosis. J Gastroenterol Hepatol 2016;31:1342-8. 10.1111/jgh.13315 [DOI] [PubMed] [Google Scholar]
- 131.Valenti L, Maggioni P, Piperno A, et al. Patatin like phospholipase domain containing-3 gene I148M polymorphism, steatosis, and liver damage in hereditary hemochromatosis. World J Gastroenterol 2012; 18:2813-20. 10.3748/wjg.v18.i22.2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.de Tayrac M, Roth MP, Jouanolle AM, et al. Genome-wide association study identifies TF as a significant modifier gene of iron metabolism in HFE hemochromatosis. J Hepatol 2015;62:664-72. 10.1016/j.jhep.2014.10.017 [DOI] [PubMed] [Google Scholar]
- 133.Benyamin B, Ferreira MA, Willemsen G, et al. Common variants in TMPRSS6 are associated with iron status and erythrocyte volume. Nat Genet 2009;41:1173-5. 10.1038/ng.456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Milet J, Le Gac G, Scotet V, et al. A common SNP near BMP2 is associated with severity of the iron burden in HFE p.C282Y homozygous patients: a follow-up study. Blood Cells Mol Dis 2010;44:34-7. 10.1016/j.bcmd.2009.10.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as